Abstract
AIM: To characterize the CagA variable region of Helicobacter pylori isolates from Chinese patients.
METHODS: DNA fragments in CagA variable region were amplified and sequenced respectively from genomic DNA of 19 isolates from patients with gastric cancer and 20 isolates from patients with chronic gastritis. The tendency of phosphorylation in tyrosine(s) of CagA proteins was evaluated subsequently by phosphorylation assay in vivo and in vitro respectively.
RESULTS: About 97.44% (38/39) H pylori isolates possessed CagA gene. CagA+ strains contained 2-4 tandem five-amino-acid motifs EPIYA but only one EPIYA had repeated sequence in CagA variable region in different isolates. There was no significant difference between the number of EPIYA motifs in H pylori from patients with different diseases. However, only tyrosine site in EPIYA within repeated sequence could be phosphorylated by AGS cells in vivo although all tyrosine sites in EPIYA could be phosphorylated in vitro.
CONCLUSION: CagA in Chinese has no functional difference in perturbing cellular signal pathway among different H pylori isolates.
Keywords: Helicobacter pylori, cag pathogencity island, CagA, Tyrosine phosphorylation
INTRODUCTION
Helicobacter pylori (H pylori) has been recognized as the causative agent of chronic gastric inflammation, which can progress further to a variety of diseases such as peptic ulcer and mucosa-associated lymphoid tissue (MALT) lymphoma or adenocarcinoma[1-3]. Type I isolates of H pylori possess a major disease-associated genetic component, the cag pathogenicity island (PAI) which encodes a type IV secretion apparatus and virulence factors such as the immunodominant CagA protein[4,5]. Recent studies have demonstrated that the CagA protein is translocated from the bacterium into the host cell and induces reorganization of the host cell actin cytoskeleton, activation of Rho GTPases Rac1 and Cdc42, recruitment of transcription factors NF-KB and AP-1, activation of proto-oncogenes c-fos and c-jun and release of cytokines and chemokines[6-10]. Stein et al[11] have confirmed that the C-terminal half of CagA (namely CagA variable region) is phosphorylated in tyrosine sites by c- src/lyn protein kinase family once translocated into eukaryotic cells. Higashi and coworkers[12] found a src homology 2 (SH2) containing tyrosine phosphatase 2 (SHP-2) was an intracellular target of CagA protein. SHP-2 is an important mediator molecule in several cell signal transductions and rearrangement of the actin cytoskeleton which plays a crucial role in inducing the scattering (‘hummingbird’) phenotype in gastric epithelial cells (AGS)[12,13]. Phosphorylated CagA disturbs signal transductions by binding to SHP-2 in host cells[10]. However, CagA protein varies in size in different strains, this size variation raises an intriguing possibility that the biological activity of CagA can vary from one strain to the next, which may influence the pathogenicities of different strains[14-16]. China is one of the nations where the infection incidence of H pylori is among the highest in the world and more than 90% isolated strains possess PAI gene, the biological activity of H pylori virulence factor CagA isolated from Chinese remains unclear so far[17,18]. In the present report, we cloned and sequenced the variable region of CagA DNA fragments in 19 strains from patients with gastric cancer and 20 strains from patients with chronic gastritis. Further, we evaluated the ability of CagA protein to phosphorylate in tyrosine(s).
MATERIALS AND METHODS
A total of 39 H pylori clinical strains including 19 strains from patients with gastric cancer and 20 strains from patients with chronic gastritis were isolated from the biopsy specimens in the Second Affiliated Hospital of Zhejiang University College of Medicine between October 2000 and April 2002. The diagnosis obtained by endoscopy and histology was recorded for all patients from whom the strains were isolated. There were 22 males and 17 females, with their age ranging 36-68 years (median age 48.6 years).
Isolation and culture of H pylori strains
Gastric mucosa supernatant was directly cultured in the PYL plate (Biomerux Company, France) and incubated in microaerophilic atmosphere for 4 d at 37 °C. Bacterial clones were identified by both biochemical (including rapid urease reaction, peroxidase test and Gram’s staining) and serological reactions. Following primary isolation, the bacteria were inoculated into Columbia agar (Biomerux Company, France) with 50 mL/L frozen-melting sheep blood, 100 mL/L fetal bovine serum, and Skirrow’s antibiotic supplement (Biomerux Company, France) in a microaerophilic atmosphere for 5 d at 37 °C, washed 3 times with 100 mmol/L PBS, pH 7.4. H pylori genomic DNA was extracted according to the instructions of bacterial genome extraction kit (Sangon Engineering Biotechnology Company, Shanghai, China).
Amplification of DNA fragments of CagA variable region in clinical isolates
A total of 39 DNA samples were amplified by high fidelity PCR master (Roche Company, Germany) with upstream primer: 5’-GACGAGCCTATTTATGCT-3’ and downstream primer: 5’-GCCTCATCAAAATCAATTGT-3’. The reaction mixture was incubated in a thermal cycler. PCR conditions were as follows: denaturation at 94 °C for 2 min followed by 30 cycles of denaturation at 94 °C for 15 s, annealing at 48 °C for 30 s, and extension at 72 °C for 30 s, with a final extension reaction for 7 min[14]. Samples were analyzed on an 1% agarose gel in 1×Tris base-EDTA buffer. DNA fragments were excised and purified from gel with kit for DNA sequence respectively.
Construction of pcDNA3.1(+) and pET15b expression vectors containing CagA DNA fragments with 2, 3 and 4 EPIYAs as well as their mutants respectively
Genomic DNA containing CagA fragments with 2, 3 and 4 EPIYAs from the above clinical isolates were used as templates, respectively. Two subfragments (about 800 bp) from CagA variable region in each strain could be amplified by high fidelity overlapped PCR with primers containing BamH I or Xho I[19]. The upstream primer was 5’-TCGGATCCAATTGGAGAGCAGAA-3’ and the downstream primer: 5’-GCTAGGCTCCAAAGCGGCCGCTCTTGCTTCCTTACTAG-3’ for construction 1. The upstream primer was 5’-ATGCCTGGACCAAGAGCGGCCGCTTTCGATC-3’ and the downstream primer was 5’-GACTCGAGTTAAGATTTTTGGAAACCAC-3’ for construction 2. After ligation at Not I site, the PCR products were digested with double restriction endonuclease BamHI/Xho I, and an 1509 bp CagA fragment was cloned into pcDNA3.1(+) mammalian expression vector (Invitrogen Company, USA). The mutants (referred to as EPIFA) of CagA variable region in which tyrosine in the repeated sequence was mutated to phenylalanine were produced by using a Chameleon site-directed mutagenesis kit (Stratagene Company, USA, data not shown), including EPIFA with 1, 2 and 3 EPIYAs, respectively. The procedure was strictly performed under the manufacturer’s instructions. pcDNA3.1(+) containing CagA or mutants was constructed as template, wild and mutated CagA fragments with EPIYA(s) were subsequently subcloned into Nde I/BamH I sites of pET15b expression vectors (Novagen Company, USA) and BL21 DE3 plysS E.coli component cells (Novagen Company, USA) were transformed. The recombinant proteins were induced by IPTG at the concentration of 0.4 mmol/L for 2 h, and purified by the His.Bind purification kit (Novagen Company, USA) for evaluation of the phosphorylation at tyrosine site(s) of CagA protein in vitro.
Phosphorylation assay
Phosphorylation assay in vivo Human AGS gastric epithelial cells were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum (Invitrogen Company, USA) before transfection. Cells were transfected with pcDNA3.1(+) expression vectors containing CagA or mutants with 2-4 EPIYA motifs respectively by Lipofectamine 2000 reagent (Invitrogen Company, USA) according to the manufacturer’s protocols. Fifty milligrams of plasmid was transfected into AGS cells by 40 μL of Lipofectamine 2000 reagent. Cells were harvested 72 h after transfection and lysed in lysis buffer containing 1 mmol/L EDTA, 1 mmol/L orthovanadate, 1 mmol/L phenylmethylsulfonyl fluoride (PMSF), 1 μmol/L leupeptin, 1 μmol/L pepstatin. The cell lysate was mixed with anti-CagA-antibody-sepharose 4B gel (a generous gift from Dr. Yi-Xiong Wang in Right Biotechnology, China) at 4 °C for 60 min, gently mixed, washed 3 times with 100 mmol/L PBS, pH7.4, denatured with Tris base-choride buffer containing 100 mmol/L dithiothreitol, centrifuged at 13000 g for 20 min. The supernatant collected was subjected to electrophoresis and transferred to a nitrocellulose membrane filter (Hybond Company,USA), blocked with 5% milk powder, incubated in at 1:250 diluted monoclonal antibodies against phosphorylated tyrosine (Y99, Santa Cruz Company, USA) at 37 °C for 2 h, washed 5 times. Then 1:100 diluted biotinylated sheep antibodies against mouse (Maxim Biotechnology Company, USA) were added and washed 5 times, and incubated with streptoavidin-peroxidase conjugate (Maxim Biotechnology Company, USA) for 60 min. Finally, the color was developed by ECL substrate.
Phosphorylation assay in vitro Nonidet P40-soluble fraction of AGS cells was performed as described by Stein et al[11]. Ten milligrams of CagA or mutant recombinant proteins with EPIYAs was incubated with 10 μL of cell lysate in a total reaction volume of 40 μL 250 mmol/L Tris-Cl buffer (pH7.4) containing 100 Ci gamma-32P ATP (Perkin Elmer company, USA), 62.5 mmol/L MnCl2, 312.5 mmol/L MgCl2, 5 mmol/L EGTA, 1 mmol/L sodium-o-vanadate. Phosphorylation was carried out at 30 °C for 10 min and the mixed solution was subjected to SDS-polyacrylamide gel electrophoresis. After electrophoresis, the gel was exposed at 4 °C for 24 h, respectively.
RESULTS
Amino acid sequence of CagA variable region
Thirty-eight of 39 isolates possessed CagA gene and the positive rate of CagA+ was 97.44% (38/39). The nucleotide sequences of CagA were quite similar among the 38 strains with CagA+. However, the variation in the number of repetitions of the 5-amino acid EPIYA sequence in the 3’ region of CagA was observed. Amino acid sequences were found to be 2-4 tandem five-amino-acid motifs EPIYA between each of which, there was an interval of 13-44 amino acids in CagA variable region. The case of CagA protein with 3 tandem EPIYAs was most common in all clinical isolates. There were 3 types of CagA variable region among the 38 isolates, namely CagA with 2 EPIYAs, CagA with 3 EPIYAs and CagA with 4 EPIYAs (Figure 1). To explore the relationship between the number of EPIYAs in isolates and disease outcomes, we adopted χ2 test to evaluate the correlation and found that there was no significant difference in the number of tandem five-amino-acid motif EPIYAs in isolates from gastric cancer and chronic gastritis (Table 1), P>0.05. It implied that the variation of the number of EPIYAs might not relate to the disease outcome (gastric cancer vs chronic gastritis). Subsequently the analysis of sequences showed that each CagA+ strain contained only one repeated sequence in the C-terminal half of CagA protein, namely genetic structure of EPIYA-TIDDL-FPLKRHDKVEDLSKV (this EPIYA was usually referred to as D1 region, TIDDL as D2 region, FPLKRHDKVEDLSKV as D3 region[11]). There were two obviously differences in this structure compared with Western standard strain NCTC11637. One difference was found in the number of repeated sequences. There was only one repeated sequence in each Chinese isolate compared with three repeated sequences in NCTC11637. The other was found in the key amino acid sequences in D2 region of repeated sequences. The amino acid sequence in D2 region was DFD in each H pylori from Chinese patients, and was DDL in NCTC11637 and other Western isolates[11,14]. The variation of two amino acids in D2 region conferred Chinese CagA protein to more perfectly match the SHP-2 consensus motif (PY-S/T/A/V/I-X-V/I/L-X-W/F) than the Western strains[20]. Hence, we speculated that Chinese CagA protein might be more able to activate SHP-2 molecules.
Figure 1.

Amino acid sequence characteristics of CagA variable region. CZU-1,2,3: Isolates from Chinese patients stand for the three types of CagA variable region respectively; CZU-1: CagA with 2 EPIYAs; CZU-2: CagA with 3 EPIYAs and CZU-3: CagA with 4 EPIYAs. The red letters stand for the deletion of amino acids in CZU-1 and the letters underline the mean extra amino acids in CZU-3 when compared to CZU-2. NCTC11637 was a Western standard strain.
Table 1.
Comparison of the number of tandem five-amino-acid motif EPIYAs in strains from gastric cancer and gastritis.
| Case |
EPIYA (n) |
|||
| 2 | 3 | 4 | ||
| Gastritis | 20 | 4 | 13 | 3 |
| Gastric cancer | 18 | 3 | 12 | 3 |
χ2 test, χ2 = 0.301, P>0.05.
Tyrosine phosphorylation sites in EPIYA
We used the phosphorylation assay in vivo and in vitro to identify whether and which tyrosine site(s) in EPIYA could be phosphorylated. In phosphorylation assay in vitro, CagA recombinant proteins with 2, 3 and 4 EPIYAs as well as their mutants EPIFA with 1, 2 and 3 EPIYAs could be labeled with 32P respectively. Moreover, CagA protein with 4 EPIYAs became more marked with 32P than that with 2 or 3 EPIYAs and their mutants with 1, 2 and 3 EPIYAs (Figure 2: A-F). It implied that the tyrosine phosphorylation site in EPIYA both within and outside the repeated sequences of CagA recombinant proteins could be phosphorylated by AGS cell lysate in vitro, and CagA protein with 4 EPIYAs became more phosphorylated than that with 2 or 3 EPIYAs and their mutants. However, only one tyrosine site in EPIYA within the repeated sequences could be phosphorylated and no tyrosine in EPIYA outside the repeated sequences could be phosphorylated in vivo by AGS cells when transfected pcDNA3.1(+) mammalian expression vectors containing CagA with 2, 3 and 4 EPIYAs as well as EPIFA with 1, 2 and 3 EPIYAs, since the mutants of EPIFA with 1, 2 and 3 EPIYAs could not react with antibody to phosphorylated tyrosine in transfected AGS cells (Figure 2: G-L). Moreover, CagA protein with 4 EPIYAs did not become more phosphorylated than that with 2 or 3 EPIYAs. The outcomes were similar to that of infection of AGS cells with H pylori bacteria containing CagA with 2, 3 and 4 EPIYAs, respectively (data not shown). Hence, we speculated that the number of EPIYA motifs might not relate to the ability of tyrosine to phosphorylate on CagA protein in vivo, which could only be determined by the number of EPIYA repeated sequences. As a result, this outcome raised an intriguing possibility that the ability of Chinese CagA to phosphorylate in tyrosine was equal to one another in different isolated strains of type I because each CagA protein contained only one repeated sequence in clinical isolates from Chinese patients with either gastric adenocarcinoma or gastritis.
Figure 2.
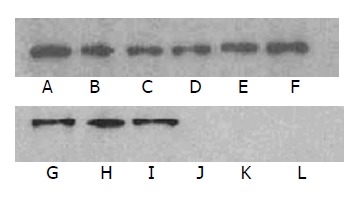
Tyrosine phosphorylation sites in EPIYA in vitro and in vivo. A-F: tyrosine phosphorylation in vitro. Lane A: CagA with 4 EPIYAs; lane B: CagA with 3 EPIYAs; lane C: CagA with 2 EPIYAs; lane D: EPIFA with 1 EPIYA; lane E: EPIFA with 2 EPIYAs; lane F: EPIFA with 3 EPIYAs. lane G-L: tyrosine phosphorylation in vivo. G: CagA with 4 EPIYAs; H: CagA with 3 EPIYAs; I: CagA with 2 EPIYAs; lane J: EPIFA with 3 EPIYAs; lane K: EPIFA with 2 EPIYAs; lane L: EPIFA with 1 EPIYA.
DISCUSSION
The correlation between H pylori as a class I carcinogen and gastric adenocarcinoma is not completely clarified and has attracted much attention so far. CagA gene as a marker for cag pathogenicity island associated with more virulent H pylori strains may be one of the most important contributors in the process of gastric carcinogenesis, because the antibody titer of CagA is much higher in gastric carcinoma than in non- gastric carcinoma and the detectable rate of CagA antibody in human beings is significantly higher in high occurrence region of gastric cancer than in low occurrence area. CagA+ strains can significantly increase the risk for developing severe gastritis and gastric carcinoma compared with CagA- H pylori strains[21-23]. A research suggests that the development of more prominent gastritis and severe atrophy in CagA+ patients is an indicator for the importance of CagA rather than H pylori load[24]. indicating that CagA protein plays an important role in pathogenesis of H pylori. In general, CagA from different H pylori has a significant difference because of the number of repeated sequences of EPIYA in C-terminal half of CagA although N-terminal of CagA protein is quite conserved. Hence, the highlight in study of CagA is whether this diversity of CagA is correlated with disease outcomes. Azuma et al[15] found that there were more EPIYAs in H pylori from atrophic gastritis and gastric cancer than in H pylori from gastritis. The frequencies of genotypes of CagA with more than 4 EPIYAs were significantly higher in atrophic gastritis than in duodenal ulcer. One-third of strains with more than 4 EPIYAs were in gastric cancer. They concluded that H pylori infection with the CagA genotype with more than 4 EPIYAs might correlate with the pathogenesis of atrophic gastritis and gastric cancer[15]. In contrast, CagA protein varied in the number (from 1 to 3) of repeated sequences of EPIYA in different strains isolated from Western countries[11,16]. We demonstrated in this work that each Chinese CagA variable region contained 2-4 tandem EPIYA motifs and only one EPIYA had repeated sequences. There was no significant difference between the number of EPIYA motifs in CagA variable regions in H pylori isolated from patients with either gastritis or gastric carcinoma. There were three types of CagA variable region among the 38 isolates, namely CagA with 2 EPIYAs, CagA with 3 EPIYAs and CagA with 4 EPIYAs. But no CagA with more than 4 EPIYAs or deletion of cagE was observed in these Chinese H pylori isolates. It appears that there is a difference in the 3 -variable region of CagA of H pylori isolates from Chinese and Japanese[15].
The ability of CagA protein to phosphorylate in tyrosine(s) is the premise of CagA protein perturb cellular signal pathway and induction of cellular response. Higashi and coworkers[12] have found that a tyrosine phosphatase -2 containing c-src homology 2 is an intracellular target of tyrosine-phosphorylated CagA protein. The phosphorylase activity of SHP-2 by binding to tyrosine-phosphorylated CagA protein has a close relationship with the “hummingbird” phenotype of AGS cells. Besides SHP-2 as an intracellular target of tyrosine-phosphorylated CagA protein, CagA protein could interact with Grb2 and phosphorylate CagA protein which is indispensable for the Grb2 binding[25], resulting in the activation of the Ras/MEK/ERK pathway and cell proliferation. Hence, the ability of phosphorylated CagA protein could reflect the level of biological function of CagA and might play a key role in the pathogenicity of H pylori. Except that one tyrosine site in repeated sequences is phosphorylated after CagA DNA is transfected into gastric epithelial cells, no other tyrosine sites in EPIYA outside the repeated sequences could be phosphorylated in vivo although all the tyrosine sites in EPIYA could be phosphorylated in vitro, indicating that sequence diversity of tyrosine phosphorylation motifs on CagA is not related to the presence of tyrosine phosphorylation. The reason is that the membrane tethering of CagA protein is necessary for the tyrosine- phosphorylated EPIYA in host cells. The other obvious difference that might influence the phosphorylation ability of CagA is the amino acid sequence in D2 region in repeated sequences[16]. Each Chinese isolate is DFD which could more perfectly match the SHP-2 consensus motifs than DDL in NCTC11637 and other Western isolated strains. It implies that Chinese CagA has a much stronger ability to perturb cell signal pathway. Above all, the fact raises an intriguing possibility that Chinese CagA has no functional difference in perturbing cellular signal pathway among different isolates of type I although there is a diversity in C-terminal half of CagA.
ACKNOWLEDGEMENTS
The excellent technical assistance of Jie Lin and Wen-Zi Jiang is gratefully acknowledged. We also thank Professor Xin-You Xie of the Sir Run Run Shaw’s Hospital for reading the manuscript.
Footnotes
Supported by the National High Technology Research and Development Program of China, No. 2001AA227111
Edited by Wang XL Proofread by Chen WW
References
- 1.Uemura N, Okamoto S, Yamamoto S, Matsumura N, Yamaguchi S, Yamakido M, Taniyama K, Sasaki N, Schlemper RJ. Helicobacter pylori infection and the development of gastric cancer. N Engl J Med. 2001;345:784–789. doi: 10.1056/NEJMoa001999. [DOI] [PubMed] [Google Scholar]
- 2.Brenner H, Arndt V, Stürmer T, Stegmaier C, Ziegler H, Dhom G. Individual and joint contribution of family history and Helicobacter pylori infection to the risk of gastric carcinoma. Cancer. 2000;88:274–279. doi: 10.1002/(sici)1097-0142(20000115)88:2<274::aid-cncr5>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 3.Maeda S, Yoshida H, Ogura K, Yamaji Y, Ikenoue T, Mitsushima T, Tagawa H, Kawaguchi R, Mori K, Mafune Ki, et al. Assessment of gastric carcinoma risk associated with Helicobacter pylori may vary depending on the antigen used: CagA specific enzyme-linked immunoadsorbent assay (ELISA) versus commercially available H. pylori ELISAs. Cancer. 2000;88:1530–1535. [PubMed] [Google Scholar]
- 4.Censini S, Lange C, Xiang Z, Crabtree JE, Ghiara P, Borodovsky M, Rappuoli R, Covacci A. cag, a pathogenicity island of Helicobacter pylori, encodes type I-specific and disease-associated virulence factors. Proc Natl Acad Sci USA. 1996;93:14648–14653. doi: 10.1073/pnas.93.25.14648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Covacci A, Telford JL, Del Giudice G, Parsonnet J, Rappuoli R. Helicobacter pylori virulence and genetic geography. Science. 1999;284:1328–1333. doi: 10.1126/science.284.5418.1328. [DOI] [PubMed] [Google Scholar]
- 6.Backert S, Moese S, Selbach M, Brinkmann V, Meyer TF. Phosphorylation of tyrosine 972 of the Helicobacter pylori CagA protein is essential for induction of a scattering phenotype in gastric epithelial cells. Mol Microbiol. 2001;42:631–644. doi: 10.1046/j.1365-2958.2001.02649.x. [DOI] [PubMed] [Google Scholar]
- 7.Segal ED, Cha J, Lo J, Falkow S, Tompkins LS. Altered states: involvement of phosphorylated CagA in the induction of host cellular growth changes by Helicobacter pylori. Proc Natl Acad Sci USA. 1999;96:14559–14564. doi: 10.1073/pnas.96.25.14559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Churin Y, Kardalinou E, Meyer TF, Naumann M. Pathogenicity island-dependent activation of Rho GTPases Rac1 and Cdc42 in Helicobacter pylori infection. Mol Microbiol. 2001;40:815–823. doi: 10.1046/j.1365-2958.2001.02443.x. [DOI] [PubMed] [Google Scholar]
- 9.Keates S, Hitti YS, Upton M, Kelly CP. Helicobacter pylori infection activates NF-kappa B in gastric epithelial cells. Gastroenterology. 1997;113:1099–1109. doi: 10.1053/gast.1997.v113.pm9322504. [DOI] [PubMed] [Google Scholar]
- 10.Meyer-ter-Vehn T, Covacci A, Kist M, Pahl HL. Helicobacter pylori activates mitogen-activated protein kinase cascades and induces expression of the proto-oncogenes c-fos and c-jun. J Biol Chem. 2000;275:16064–16072. doi: 10.1074/jbc.M000959200. [DOI] [PubMed] [Google Scholar]
- 11.Stein M, Bagnoli F, Halenbeck R, Rappuoli R, Fantl WJ, Covacci A. c-Src/Lyn kinases activate Helicobacter pylori CagA through tyrosine phosphorylation of the EPIYA motifs. Mol Microbiol. 2002;43:971–980. doi: 10.1046/j.1365-2958.2002.02781.x. [DOI] [PubMed] [Google Scholar]
- 12.Higashi H, Tsutsumi R, Muto S, Sugiyama T, Azuma T, Asaka M, Hatakeyama M. SHP-2 tyrosine phosphatase as an intracellular target of Helicobacter pylori CagA protein. Science. 2002;295:683–686. doi: 10.1126/science.1067147. [DOI] [PubMed] [Google Scholar]
- 13.Maroun CR, Naujokas MA, Holgado-Madruga M, Wong AJ, Park M. The tyrosine phosphatase SHP-2 is required for sustained activation of extracellular signal-regulated kinase and epithelial morphogenesis downstream from the met receptor tyrosine kinase. Mol Cell Biol. 2000;20:8513–8525. doi: 10.1128/mcb.20.22.8513-8525.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hsu PI, Hwang IR, Cittelly D, Lai KH, El-Zimaity HM, Gutierrez O, Kim JG, Osato MS, Graham DY, Yamaoka Y. Clinical presentation in relation to diversity within the Helicobacter pylori cag pathogenicity island. Am J Gastroenterol. 2002;97:2231–2238. doi: 10.1111/j.1572-0241.2002.05977.x. [DOI] [PubMed] [Google Scholar]
- 15.Azuma T, Yamakawa A, Yamazaki S, Fukuta K, Ohtani M, Ito Y, Dojo M, Yamazaki Y, Kuriyama M. Correlation between variation of the 3' region of the cagA gene in Helicobacter pylori and disease outcome in Japan. J Infect Dis. 2002;186:1621–1630. doi: 10.1086/345374. [DOI] [PubMed] [Google Scholar]
- 16.Higashi H, Tsutsumi R, Fujita A, Yamazaki S, Asaka M, Azuma T, Hatakeyama M. Biological activity of the Helicobacter pylori virulence factor CagA is determined by variation in the tyrosine phosphorylation sites. Proc Natl Acad Sci USA. 2002;99:14428–14433. doi: 10.1073/pnas.222375399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang L, Jiang J, Pan KF, Liu WD, Ma JL, Zhou T. Infection of H pylori with CagA strain in a high-risk area of gastric cancer. Shijie Huaren Xiaohua Zazhi. 1998;6:40–41. [Google Scholar]
- 18.Zhang L, Yan XJ, Zhang LX, Han FC, Zhang NX, Hou Y, Liu YG. Seroepidemiological study of Hp and CagA Hp infection. Shijie Huaren Xiaohua Zazhi. 2000;8:389–392. [Google Scholar]
- 19.Tomb JF, White O, Kerlavage AR, Clayton RA, Sutton GG, Fleischmann RD, Ketchum KA, Klenk HP, Gill S, Dougherty BA, et al. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 20.De Souza D, Fabri LJ, Nash A, Hilton DJ, Nicola NA, Baca M. SH2 domains from suppressor of cytokine signaling-3 and protein tyrosine phosphatase SHP-2 have similar binding specificities. Biochemistry. 2002;41:9229–9236. doi: 10.1021/bi0259507. [DOI] [PubMed] [Google Scholar]
- 21.Enroth H, Kraaz W, Engstrand L, Nyrén O, Rohan T. Helicobacter pylori strain types and risk of gastric cancer: a case-control study. Cancer Epidemiol Biomarkers Prev. 2000;9:981–985. [PubMed] [Google Scholar]
- 22.Zhang L, Ma J, Pan K. Studies on infection of Helicobacter pylori and their subtypes cagA+ and hspA+ in population living in the areas with high and low incidence of gastric cancer. Zhonghua YuFang YiXue ZaZhi. 1998;32:67–69. [PubMed] [Google Scholar]
- 23.Miehlke S, Kirsch C, Agha-Amiri K, Günther T, Lehn N, Malfertheiner P, Stolte M, Ehninger G, Bayerdörffer E. The Helicobacter pylori vacA s1, m1 genotype and cagA is associated with gastric carcinoma in Germany. Int J Cancer. 2000;87:322–327. [PubMed] [Google Scholar]
- 24.Zhou JC, Xu CP, Zhang JZ. The research development of CagA/CagA of Helicobacter pylori mocular biology. Shijie Huaren Xiaohua Zazhi. 2001;9:560–562. [Google Scholar]
- 25.Mimuro H, Suzuki T, Tanaka J, Asahi M, Haas R, Sasakawa C. Grb2 is a key mediator of helicobacter pylori CagA protein activities. Mol Cell. 2002;10:745–755. doi: 10.1016/s1097-2765(02)00681-0. [DOI] [PubMed] [Google Scholar]


