SUMMARY
The development of targeted therapeutics for neuroblastoma, the third most common tumor in children, has been limited by a poor understanding of growth signaling mechanisms unique to the peripheral nerve precursors from which tumors arise. In this study, we combined genetics with gene expression analysis in the peripheral sympathetic nervous system to implicate arginase 1 and GABA signaling in tumor formation in vivo. In human neuroblastoma cells, either blockade of ARG1 or benzodiazepine-mediated activation of GABA-A receptors induced apoptosis and inhibited mitogenic signaling through AKT and MAPK. These results suggest that ARG1 and GABA influence both neural development and neuroblastoma, and that benzodiazepines in clinical use may have potential for neuroblastoma therapy.
INTRODUCTION
Neuroblastoma, a common and deadly pediatric tumor of the peripheral nervous system, is derived from sympathetic neural progenitors that normally differentiate into post-mitotic neurons. This tumor likely arises from disruption of growth pathways specific to the neural lineage, as mutations in canonical growth pathways seen in epithelial, glial, and hematopoietic tumors are rarely observed. Amplification of the MYCN proto-oncogene is a hallmark of high-risk neuroblastoma (Brodeur et al., 1984; Schwab et al., 1984), and a transgenic mouse model driven by MYCN develops tumors with histological and genomic characteristics of human neuroblastoma (Hackett et al., 2003; Weiss et al., 1997). In this model, tumor incidence varies among mouse strains, with the strain FVB/N showing complete resistance to tumor formation.
In this study, we exploited this genetic variance to identify novel molecular pathways driving neuroblastoma development. We first identified tumor susceptibility loci through classical genetic linkage mapping in genetically heterogeneous backcross mice. We next analyzed gene expression as a function of genetic variation to identify quantitative trait loci (eQTL) in sympathetic superior cervical ganglia (SCG) in these mice. We then used eQTL localizing to tumor susceptibility loci to identify candidate tumor susceptibility genes. Liver arginase (Arg1), a component of both the urea cycle and the gamma-amino butyric acid (GABA) biosynthetic pathway, was the strongest eQTL at the most prominent tumor susceptibility locus. Multiple genes and eQTL involved in GABA neurotransmitter signaling were located at secondary susceptibility loci, providing an intriguing genetic link between this pathway and neuroblastoma biology. We validated this link using compounds targeting the ARG1-GABA pathway in human neuroblastoma cell lines. Nor-NOHA, an ARG1 inhibitor, inhibited growth and viability of cells. More strikingly, activation of GABA-A signaling using a benzodiazepine derivative induced apoptosis and was associated with decreased activity of PI3K/AKT and MAPK signaling, central pathways in cell growth and survival. These data, implicating arginine metabolism and GABA signaling in the pathogenesis of neuroblastoma, present a combination of novel therapeutic targets. Additionally, demonstration that the neurotransmitter GABA inhibits growth of tumor cells derived from a neural lineage highlights an emerging role for neurotransmitters in regulating development of the peripheral nervous system.
RESULTS
Penetrance of neuroblastoma in TH-MYCN mice is strain-dependent
Transgenic mice were generated on a Balb/c x C57B6/J background with ~10% incidence of tumors. After two back-crosses into strain FVB/NJ, incidence decreased to zero (Figure S1A). Conversely, incidence increased steadily with each successive backcross into strain 129/SvJ, to ~60% (Chesler et al., 2007). Levels of transgene expression were similar between strains (Figure S1B).
We crossed resistant transgenic FVB/NJ mice to susceptible wild-type 129/SvJ. Four percent of F1 mice developed tumors, suggesting that resistance was dominant. To generate a genetically diverse population for linkage mapping, we backcrossed transgenic F1 animals to wild-type 129/SvJ mice. Incidence of tumors in the resulting N1 backcross generation was 38% (N=203), with an average survival (109 days) identical to that of mice carrying the transgene in a pure 129/SvJ background (Figure S1C, (Chesler et al., 2007)).
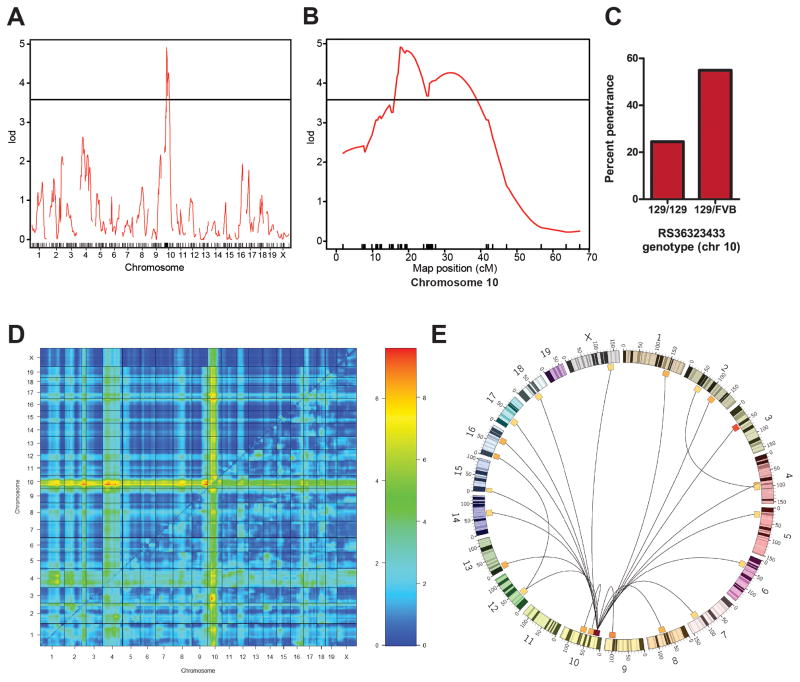
Linkage analysis identifies a tumor susceptibility modifier on chromosome 10
To identify genomic loci associated with tumor susceptibility, we genotyped 203 mice using a combination of microsatellite and SNP markers (Table S1). Interval mapping for linkage to tumor susceptibility produced a maximum LOD (log of odds) score on chromosome 10 at marker RS36323433 (LOD=4.1, significant at a 5% genome-wide error rate). Interestingly, tumor susceptibility was influenced strongly by gender. Sex as an interacting covariate increased the LOD score to 4.9 (Figure 1A, B). This locus was not significant in female mice but was in males (LOD=4.3, N=82). It may be relevant in this regard that neuroblastoma is slightly more prevalent in boys (1.3:1) than in girls (Hale et al., 1994). Contrary to the expected allele segregation at this locus, heterozygous mice were tumor prone (55%), while mice homozygous for the 129/SvJ alleles were resistant (25% incidence) (Figures 1C and S1D). The absence of a single locus at which an FVB/N allele conferred complete resistance to tumor formation (a trait of FVB/N mice) suggested that the genetics underlying patterns of resistance and susceptibility in pure inbred strains is complex.
Figure 1. A locus on chromosome 10 and multiple secondary loci are linked to tumor susceptibility.
(A) LOD plot for tumor susceptibility shows a single significant locus on chromosome 10. Horizontal line indicates 5% genome-wide significance threshold (LOD=3.58, 1000 permutations). (B) LOD plot of chromosome 10 only. Hash marks on the horizontal axis indicate marker positions. Horizontal line indicates 5% genome-wide significance threshold (LOD=3.58, 1000 permutations). (C) Effect plot for the marker closest to the maximum LOD score (RS36323433) showing incidence of tumors as a function of genotype. (D) Two-QTL analysis using sex as an interacting covariate reveals that multiple secondary loci interact with the chromosome 10 locus. The top left half shows the results of the additive model, and the bottom right half shows results of the “full” model. The bar on the right displays the correspondence between color on the chart and LOD scores (the left side of the bar corresponds to the additive analysis on the top left half of the plot, while the right side of the bar, with a different scale, corresponds to the full model analysis on the bottom right). Locations of the maximum LOD scores are shown in Table S2. (E) Circos plot illustrating the two-way interactions showin in (D). Pairs of loci with a LOD greater than 6 are connected.
Since the segregation of genotypes with tumor susceptibility at the chromosome 10 locus did not match the patterns of tumor susceptibility in the parent strains, we next considered a more complex genetic model to explain susceptibility. A 2-QTL test identified several loci that interacted with the chromosome 10 locus, with similar LOD scores (Figure 1D and 1E, Table S2). Interestingly, while all of these loci interacted with the chromosome 10 locus, they did not interact with each other.
Expression QTL analysis identifies Arg1 as a candidate modifier
The 95% confidence interval of the peak on chromosome 10 spanned 47Mb and contained over 281 genes, complicating identification of candidates. Hypothesizing that susceptibility to tumors may be governed by differential expression of genes within this locus, we compared mRNA expression levels in neural crest-derived sympathetic SCG from transgenic 129/SvJ and FVB/NJ male mice. We identified 9,820 genes that were differentially expressed genome-wide between the two strains (Figure S1C, Table S3), including 137 within the 95% confidence interval for the chromosome 10 susceptibility locus (representing almost half of all genes within the locus). Surprisingly, when male and female 129/SvJ and FVB/NJ mice were compared, only 8 genes were differentially expressed between sexes. With the exception of a RIKEN clone on chromosome 2, all mapped to either the X or Y chromosomes (Figure S1D, Table S3), excluding gender-specific gene expression as a mechanism for the gender effect at the chromosome 10 locus.
Gene expression levels can be influenced by complex interactions among cis- and trans- acting factors. One method for distinguishing these factors involves generating a genetically heterogeneous population (such as our backcross population), measuring gene expression levels, and treating the expression level of each gene as a quantitative trait for subsequent linkage mapping (expression QTL or eQTL (Brem et al., 2002; Schadt et al., 2003)). Through this method, cis- and trans- acting alleles influencing gene expression are decoupled from each other, and genes whose differential expression is due to cis-acting factors at a locus can be distinguished from genes under the control of trans-acting factors at other loci. Genes with eQTL overlapping with QTL for physiological phenotypes have been shown to control these phenotypes (Schadt et al., 2003; Yang et al., 2009). We thus utilized this technique to identify eQTL within our chromosome 10 locus, as well as at the numerous other secondary loci identified in the 2-QTL tumor susceptibility analysis.
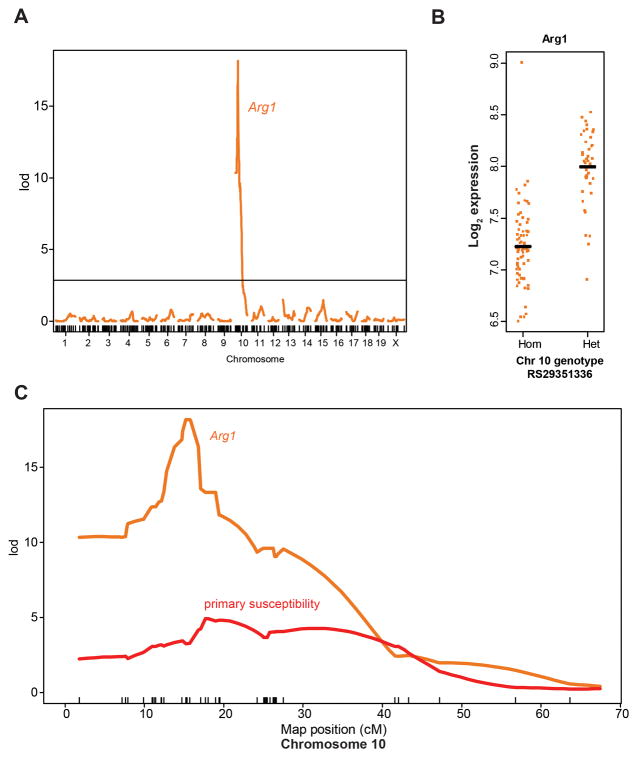
We measured mRNA expression levels in 116 SCG from the backcross population and tested for linkage between gene expression levels and germline genetic variation. We identified 342 eQTL acting both locally and on other chromosomes (Table S4). Four eQTL mapped to the susceptibility locus on chromosome 10, with Arg1 (liver arginase) showing the strongest eQTL. When this measurement was refined by interval mapping, Arg1 expression was linked to a QTL at chromosome 10 with a LOD score of 18.2 (Figure 2A). Mice heterozygous at that locus had almost 2-fold higher expression of Arg1 compared to mice homozygous for the 129/SvJ allele (Figures 2B and S2A). Of the four eQTL mapping to the locus, the Arg1 eQTL overlapped most directly and with the highest LOD score (Figures S2B and S2C), making Arg1 our top candidate gene (Figure 2C, Table 1). Notably, among 137 genes differentially expressed at the chromosome 10 locus in parental strains, only four, including Arg1, showed significant eQTL mapping to the locus (Table S4). These data suggest that differential expression of the other genes in the parents was due in part to trans-acting factors, illustrating the power of eQTL analysis in dissecting control of gene expression at a genetic locus and filtering candidate genes.
Figure 2. An eQTL for Arg1 co-localizes with the tumor susceptibility locus on chromosome 10.
(A) Interval mapping for Arg1 expression, the most significant eQTL in the chromosome 10 region, showing a LOD score of 18.2 on chromosome 10, centered at the physical location of the Arg1 gene. Horizontal line indicates 5% genome-wide significance threshold. (B) Log2 Arg1 expression level as a function of genotype at SNP RS29351336 on chromosome 10. (C) A plot of LOD scores on chromosome 10 for susceptibility (red line) and the Arg1 eQTL (orange line) showing co-localization of the peaks.
Table 1.
Candidate genes at susceptibility loci
| A. Candidate eQTL at susceptibility loci | ||||||
|---|---|---|---|---|---|---|
| chromosome | susceptibility locus (Mb) | LOD | eQTL gene | eQTL locus (Mb) | eQTL p-val | perm. p-val |
| 10 | 28 | 5.0 | Arg1 | 25 | 2.92E-20 | <0.001 |
| 2 | 166 | 7.0 | Gabra3 | 178 | 5.57E-34 | <0.001 |
| 4 | 115 | 7.4 | Slc6a1 | 129 | 2.81E-05 | 0.001 |
| B. non-eQTL GABA genes at susceptibility loci | ||||
|---|---|---|---|---|
| chromosome | susceptibility locus (Mb) | LOD | gene | gene location (Mb) |
| 1 | 131 | 7.8 | Dbi | 122 |
| 9 | 117 | 7.8 | Trak1 | 121 |
| 17 | 35 | 6.9 | Gabbr1 | 37 |
| 7 | 144 | 6.3 | Oat | 140 |
| C. Gene descriptions | ||
|---|---|---|
| gene | location (chromosome) | description |
| Arg1 | 10 | liver arginase |
| Gabra3 | X* | GABA-A receptor, subunit alpha 3 |
| Slc6a1 | 6* | GABA transporter, removes GABA from synaptic cleft |
| Dbi | 1 | diazepam binding inhibitor; modulates the action of the GABA receptor |
| Trak1 | 9 | trafficking protein, kinesin binding 1, regulates GABA receptor |
| Gabbr1 | 17 | GABA-B receptor, 1 |
| Oat | 7 | ornithine aminotransferase (produces glutamate, a GABA precursor) |
controlled by trans eQTL
eQTL for GABA-related genes map to secondary susceptibility loci
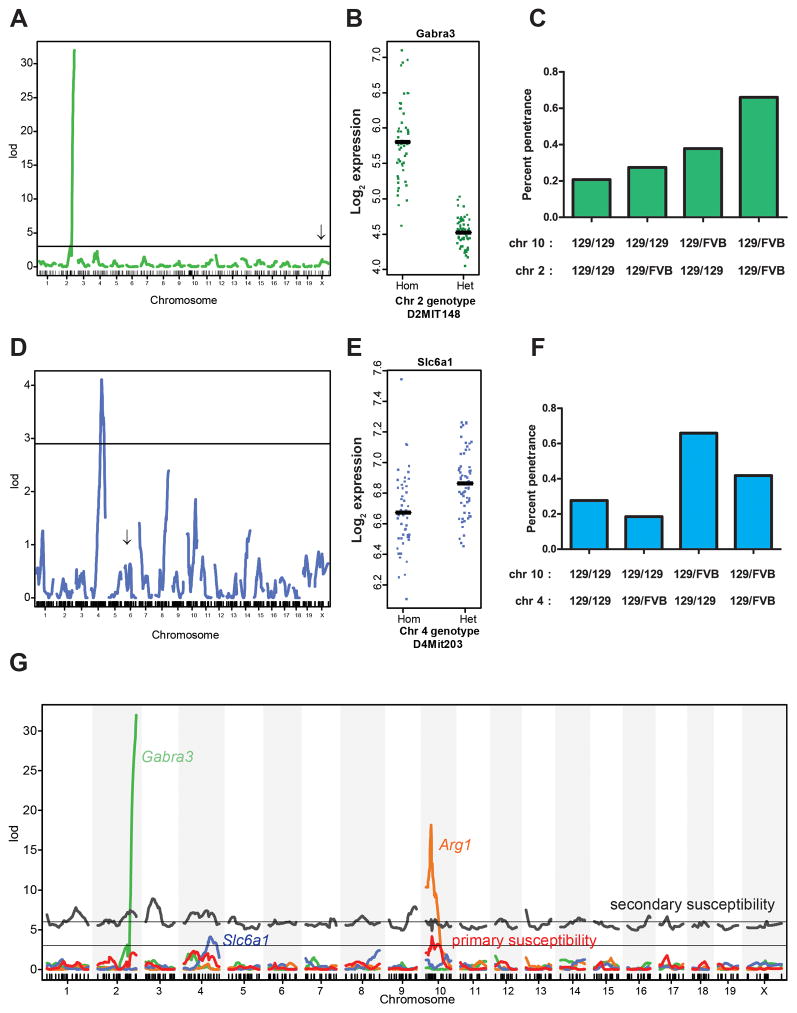
We next looked for eQTL that mapped to secondary tumor susceptibility loci that interacted with the chromosome 10 locus. We noted eQTL for genes related to GABA neurotransmitter signaling at two of the loci (Table 1). Notably, a trans-eQTL on chromosome 2 controlled expression of the Gabra3 receptor subunit on the X chromosome (Figure 3A–C). This eQTL overlapped directly with the secondary susceptibility locus on chromosome 2 (Figure S3A, Table 1). Mice harboring alleles resulting in high Arg1 expression and low Gabra3 expression showed the strongest susceptibility to tumors (Figure 3C). Similarly, an eQTL for the GABA transporter Slc6a1 (Figure 3D–F) mapped to a secondary susceptibility locus on chromosome 4 (Figure S3B, Table 1), though the gene was on chromosome 6. We noted that the differences in gene expression for Gabra3 and Slc6a1 were also noted in the parental strains (Figure S3C and S3D). The overlap of Gabra3, Slc6a1, and Arg1 eQTL with primary and secondary susceptibility loci is illustrated genome-wide in Figure 3G.
Figure 3. Secondary susceptibility loci co-localize with eQTL for GABA-related genes.
(A) A trans-eQTL on chromosome 2 controls expression of the GABA-A receptor subunit 3 (Gabra3) on the X chromosome (arrow) (LOD=30.9). (B) Gabra3 expression as a function of D2MIT148 genotype. (C) Plot of tumor incidence as a function of genotypes at the chromosome 2 and 10 loci. (D) Expression QTL for the Slc6a1 GABA transporter on chromosome 4 (LOD=4.1). Arrow indicates the physical location of Slc6a1 on chromosome 6. (E) Slc6a1 expression as a function of D4MIT203 genotype. (F) Tumor incidence as a function of genotypes at loci on chromosomes 4 and 10. (G) Genome-wide plot of LOD scores for the Gabra3 (green), Slc6a1 (blue), Arg1 (orange) eQTL and tumor susceptibility for a single-locus model (red) and the combined effect of the peak of the chromosome 10 locus plus points across the rest of the genome (gray), showing correspondence between eQTL peaks and primary and secondary susceptibility loci. Two-dimensional plots for Gabra3 and Slc6a1 are shown in Figure S3.
We next investigated secondary susceptibility loci that lacked GABA-related eQTL candidates and found several GABA-related genes mapping within 10Mb of susceptibility peaks. The locus on chromosome 1 (LOD 7.8) flanked by markers RS5056599 (116MB) and D1MIT1001 (131Mb), LOD 7.8) lies in close proximity to Dbi (diazepam binding inhibitor, 122Mb), a gene that modulates GABA receptor activity (Gray et al., 1986). Similarly, the locus on chromosome 9 centered near 117Mb (D9MIT201, LOD 7.9) is 4 Mb from the Trak1 gene, which encodes a trafficking factor that modulates GABA receptor homeostasis (Gilbert et al., 2006). The locus on chromosome 7 centered at 144Mb (RS13479509, LOD 6.4) is 4 MB from ornithine aminotransferase (Oat), which converts ornithine to glutamate (the substrate for GABA synthesis). Finally, the locus on chromosome 17 centered near 35Mb (D17MIT231, LOD 6.7) is 2 Mb from the GABA-B receptor 1. Together, at least 6 secondary susceptibility loci co-localized with genes in the GABA pathway, and/or eQTL controlling these genes (Table 1).
Expression of GABA pathway genes identified by linkage and eQTL analysis predict survival in neuroblastoma
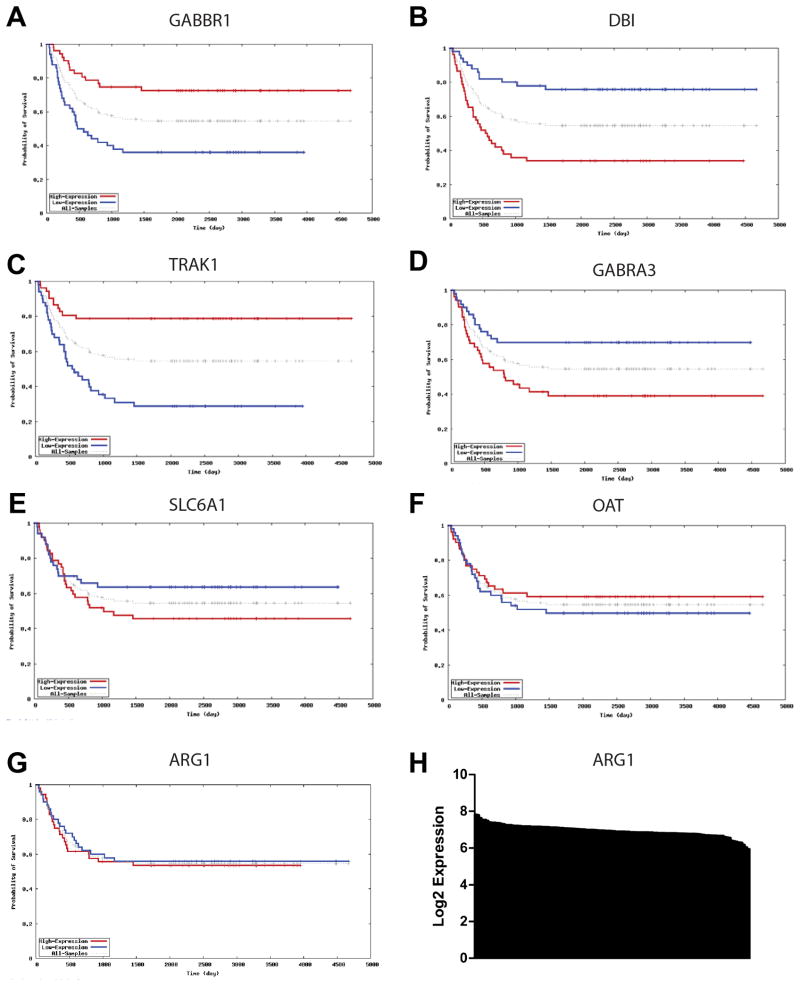
We next investigated whether expression of Arg1 and the GABA pathway genes correlated to outcomes in human neuroblastoma. We analyzed a database of gene expression profiles of human neuroblastoma samples (Asgharzadeh et al., 2006) available at http://home.ccr.cancer.gov/oncology/oncogenomics. Strikingly, all of the genes in the GABA pathway either mapping to tumor susceptibility loci or controlled by eQTL at these loci showed a significant correlation with survival (figure 4A–F). ARG1 did not show a strong correlation with survival in human neuroblastomas (Figure 4G). However, the population was relatively homogeneous with respect to ARG1 expression (Figure 4H), hindering our ability to segregate tumors based on expression levels.
Figure 4. Expression of GABA genes correlates with survival in human neuroblastomas.
Survival curves based on the dataset from (Asgharzadeh et al., 2006) available at http://home.ccr.cancer.gov/oncology/oncogenomics are shown for (A) the GABA-B receptor, (B) DBI, (C) Trak1, (D) GABRA-3, (E) SLC6A1, (F) OAT, and (G) ARG1. (H) A plot of ARG1 expression levels for all tumors arranged from highest to lowest along the X-axis, showing relatively homogeneous expression levels.
Inhibition of ARG1 decreases viability of human neuroblastoma cells
The higher-expressing Arginase allele conferring tumor susceptibility is nested within an overall resistant genetic background in purebred mice (either FVB/NJ or FVB/NJ x 129/SvJ F1). The allelic variation in secondary loci (harboring components of the GABA pathway) only showed an effect in combination with alleles at other loci. This genetic complexity precluded validation in vivo. We therefore took an in vivo approach to test whether these biochemical pathways had relevance to human tumor biology. As there is little precedent for these pathways in regulation of cell growth and thus little basis for the specific downstream pathways involved, we assessed the effect of compounds targeting this pathway on overall growth and viability.
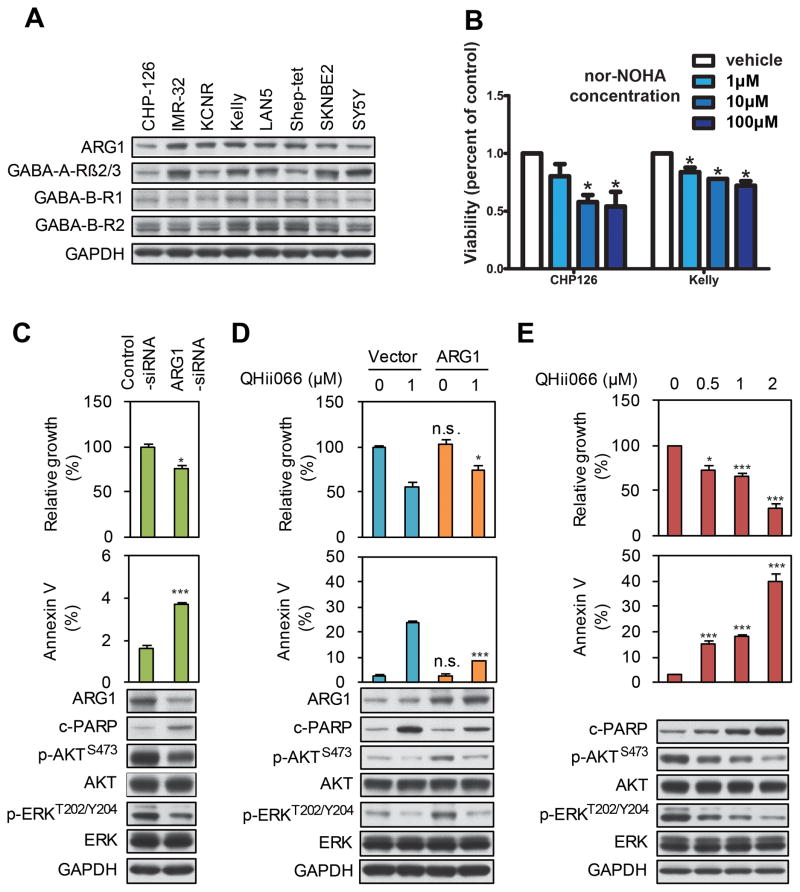
We detected ARG1 by western blot across a panel of human neuroblastoma cell lines (Figure 5A). We treated two lines, Kelly and CHP-126, with the reversible arginase inhibitor nor-NOHA (N-Omega-hydroxy-nor-arginine) (Tenu et al., 1999) to test whether inhibition of arginase could impair tumor growth. Viability was decreased in CHP-126 and Kelly cells across a range of doses from 1μM to 100μM (Figure 5B). As nor-NOHA is not a particularly potent inhibitor in neuroblastoma cell lines, we next tested siRNA against ARG1. ARG1 siRNA induced a significant decrease in growth, an increase in the apoptotic markers Annexin V cleaved PARP, and reduced levels of phosphorylated AKT and ERK, both central components of mitogenic signaling pathways (Figures 5C and S4B). We next tested whether overexpression of ARG1 had the opposite effect. While overexpression alone did not produce a notable effect on growth, it did increase levels of phosphorylated AKT and ERK, and partially rescued the inhibitory effect of the benzodiazepine QHii066 (described below) (Figures 5D and S4C). We conclude that ARG1 expression modulates viability and proliferation in human neuroblastoma cells.
Figure 5. ARG1 inhibition and GABA activation suppress neuroblastoma cell growth and survival.
(A) Expression of ARG1, GABA-A, and GABA-B receptors in neuroblastoma cell lines. Cell lines as indicated were harvested, lysed and analyzed by Western blot using the antisera indicated. (B) WST-1 assay showing a dose-dependent decrease in viability following 72h treatment of neuroblastoma cells with varying doses of the ARG1 inhibitor nor-NOHA. (C) LAN5 cells were transfected with either control or ARG1 siRNA. Cell viability was measured by WST-1. Apoptosis was measured by flow cytometry for the apoptotic marker annexin V. An aliquot of cells was analyzed by immunoblot using antisera indicated. (D) LAN-5-pBABE-vector and LAN-5-pBABE-ARG1 cells were treated with DMSO or with the GABA-A agonist QHii066. Cell viability, apoptosis, and protein markers were measured as in panel (C). (E) LAN-5 cells were treated with the GABA-A agonist QHii066 for 3 days. Cell viability, apoptosis, and protein markers were measured as in panel (C). For panels B–E, data shown are mean ± SD of triplicate measurements. NS, not significant vs control/vehicle using student’s t-test; * indicates p<0.05; *** indicates p<0.001.
GABA-A activation induces apoptosis in neuroblastoma
We next investigated the role of GABA signaling in the control of cell growth and survival. GABA-A and GABA-B receptors were detected in all cell lines tested (Figures 5A and S4A). To assess whether modulation of this pathway influenced cell growth and survival, we treated cells with a potent and selective GABA-A receptor agonist QHii066, a benzodiazepine derivative (He et al., 2000; Huang et al., 2000). QHii066 slowed cell growth and induced apoptosis in a dose-dependent fashion in both LAN-5 (Figure 5E) and Kelly (Figure S4D). Immunoblotting revealed increased PARP cleavage and reduced abundance of phosphorylated AKT and ERK in cells treated with QHii066. Flow cytometry demonstrated G0/G1 arrest (Figure S4E). These data suggest that specific activation of the GABA-A receptor decreases cell viability, induces apoptosis, and suppresses growth and survival signaling pathways in neuroblastoma cell lines.
DISCUSSION
The pathways regulating development of the peripheral nervous system are distinct from those controlling other cell types. As a consequence, common genomic aberrations driving tumorigenesis in epithelial, hematopoietic, and glial tumors rarely show abnormalities in neuroblastoma, a tumor of the sympathetic peripheral nervous system. Genetic analysis in humans and model systems has been key to identifying mechanisms driving the disease. Here, we used a genetically engineered mouse model for neuroblastoma to uncover a novel signaling pathway relevant to human disease.
Though the influence of strain background on tumor penetrance is frequently observed in mouse models of cancer, few genes underlying this susceptibility have been identified (Crawford et al., 2008a; Ewart-Toland et al., 2003; MacPhee et al., 1995; Mao et al., 2006; Park et al., 2005; Wakabayashi et al., 2007), mostly due to the limited resolution of quantitative trait linkage mapping. Analysis of gene expression as a function of genotype (i.e., treating gene expression levels as heritable traits and performing linkage analysis) has facilitated identification of genes underlying physiological variation (Brem et al., 2002; Schadt et al., 2003; Yang et al., 2009). eQTL analysis has identified several genes modifying tumor susceptibility in mouse models for cancer (Crawford et al., 2008b; La Merrill et al., 2010; Quigley et al., 2009). In the current study, eQTL analysis of the peripheral nervous system identified Arg1 as a candidate neuroblastoma modifier gene at chromosome 10, as well as a particularly prominent eQTL on chromosome 2 governing expression of a GABA-A neurotransmitter receptor subunit in trans on the X chromosome (a phenomenon that would have been missed by conventional analysis of candidate genes at the susceptibility loci). Numerous other susceptibility loci overlapped with other GABA-related genes, suggesting a mechanism for the pattern of genetic linkage to tumor susceptibility, and implicating an interaction between up-regulation of arginase activity and down-regulation of GABA receptors as cooperating mechanisms promoting susceptibility to tumors.
While Arg1 is associated with the urea cycle in liver, expression of Arg1 in sympathetic nerve ganglia (Yu et al., 2003) that do not carry out the urea cycle suggested a role in other biochemical pathways. In neurons, Arg1 is part of the GABA synthesis pathway, producing ornithine, a precursor of glutamate and of GABA. The biochemical link between Arg1 and the GABA pathway potentially explains our unique genetic pattern of several secondary loci interacting with a single gene/locus. Only one cytoplasmic arginase gene exists (Arg1). However, the GABA signaling pathway may be perturbed at numerous genomic loci, including several components of the GABA-A receptor, genes for which are dispersed throughout the genome. Thus, in a genetic model involving perturbation of arginase and GABA signaling, many GABA-related genes could interact with the single Arg1 gene on chromosome 10 with similar effects (disrupting two connected pathways), while GABA genes may not interact with each other (as multiple disruptions would have redundant effects on the same pathway). This model is consistent with the pattern of genetic interactions we observed in our system.
Several downstream outputs of the arginase pathway could account for increased expression of Arg1 predisposing mice to tumors. Arginase may act as an immunosuppressant (Bak et al., 2008; Yachimovich-Cohen et al., 2010), alter levels of nitric oxide synthases (NOSs)(Ciani et al., 2004; Jenkins et al., 1995), or alter levels of ornithine, the substrate for polyamine synthesis linked to tumorigenesis (Gerner and Meyskens, 2004) and neural proliferation (Huang et al., 2007). The rate-limiting polyamine synthetic enzyme, ornithine decarboxylase (ODC), is a well-established target for MYCN (Bello-Fernandez et al., 1993). ODC inhibitors inhibit neuroblastoma development in vitro and in TH-MYCN mice (Hogarty et al., 2008; Koomoa et al., 2008), and are currently in clinical trials for neuroblastoma (http://clinicaltrials.gov/show/NCT01586260). Ornithine is also a substrate for synthesis of glutamate and GABA. Notably, ornithine aminotransferase (Oat), which converts ornithine to glutamate, maps to a susceptibility locus on chromosome 7 (Table 1). Since GABA inhibits neuronal growth and promotes differentiation (Andang et al., 2008), at least one ultimate output of Arg1 activity (GABA) could inhibit both induction and further growth of tumors, driving selection for secondary genetic lesions that disable this pathway.
While Arg1 activity has not been studied extensively in cancer, inhibition of arginase has been shown to disrupt growth of breast cancer cells (Singh et al., 2000). Arginase expression has also been linked to neuronal viability. Clinically, ARG1 mutations cause arginemia, characterized by neurodegeneration (OMIM 207800). While hepatic production of neurotoxic metabolites are speculated to cause this neurodegeneration (De Deyn et al., 1991; Deignan et al., 2008), our data, coupled with detection of Arg1 expression in neural tissues (Yu et al., 2001; Yu et al., 2002; Yu et al., 2003), suggest that Arg1 may be involved in growth and survival signaling in neuroblasts, and inhibition of Arg1 may have an intrinsic cytotoxic effect in neurons and neuroblastoma cells.
The observation that mice with lower expression of GABA-A receptor subunits are tumor prone is consistent with GABA’s known role in neuronal cell growth and differentiation (Andang et al., 2008; Le-Corronc et al., 2011), and suggests that the association of down-regulation of GABA-A receptors with aggressive human neuroblastomas has biological significance (Roberts et al., 2004). However, since the receptor is formed as a pentameric combination of 19 possible subunits, with changing expression patterns and biological roles (Le-Corronc et al., 2011), testing this hypothesis is not straightforward. GABA signaling negatively regulates growth of neural crest stem cells (Andang et al., 2008). In our hands, neuroblastoma cells expressing the GABA-A receptor showed decreased viability, increased apoptosis, and diminished activity of mitogenic signaling pathways in response to GABA-A activation, supporting a role of GABA signaling in tumor cell growth and survival.
Neuroblastoma is a common pediatric tumor with a unique biology, making the development of novel targeted therapeutics problematic. As a result, improvements in clinical outcomes have been modest over the last several decades. Familial studies have identified genes such as ALK (Janoueix-Lerosey et al., 2008; Mosse et al., 2008) and PHOX2B (Mosse et al., 2004; Trochet et al., 2004) that drive neuroblastoma in a subset of human cases. Conversely, high-powered genome-wide association studies (Capasso et al., 2009; Diskin et al., 2012; Nguyen le et al., 2011; Wang et al., 2011) have succeeded in identifying numerous genes that confer slightly increased risk for the disease. However, the complexity of the disease has confounded the search for druggable pathways. This complexity is illustrated by the current study. We observed a stark variation in tumor susceptibility between two closely-related laboratory mouse strains in a relatively tightly-controlled model system. Though seemingly straightforward, genetic analysis of this variation revealed a strikingly complex system of genetic interactions that required an intensive genomic analysis to decipher. While aggregated genetic evidence implicated the GABA pathway, none of the components were strong enough to be detected alone without an additional genetic interaction, in a well-controlled model system with limited genetic variability compared to human studies. While the current study was successful in identifying a pharmacologically-tractable pathway for therapeutic intervention, it illustrates the difficulty in identifying and characterizing the complex biochemical pathways that influence tumor development.
Arginase inhibitors are under investigation for highly prevalent diseases such as hypertension (Durante et al., 2007). Even more encouragingly, QHii066, the GABA agonist used in this study, is closely related to the benzodiazepine diazepam (Valium), with a similar mechanism of action. The implication of GABA and other neurotransmitters in neuroblastoma growth, and the demonstration that a benzodiazepine derivative can induce apoptosis, opens up the potential to apply numerous clinically-approved drugs used in neurology and psychology as chemotherapeutic agents. These results also highlight a possible connection between the role of neurotransmitters in nervous system development and the regulation of neuroblastoma growth.
EXPERIMENTAL PROCEDURES
Mice
All mice were obtained from the Jackson Labs (Bar Harbor, ME), and were housed and treated following UCSF IACUC guidelines. Backcross mice were generated by crossing TH-MYCN transgenic FVB/N mice to wild-type 129/SvJ, and subsequently crossing F1 offspring to wild-type 129/SvJ. Mice were observed for at least one year to be considered tumor-free. Tumor-negative backcross mice were followed until one year of age (the latest tumor was detected at 342 days). Superior cervical ganglia (SCG) were surgically isolated and snap-frozen in liquid nitrogen. SCG were isolated from the parental control groups at 21 days. DNA was isolated from spleen and genotyped using a combination of microsatellite and SNP markers. RNA from SCG was analyzed using Affymetrix Mouse Exon arrays. Susceptibility linkage was analyzed using R-QTL. Differential gene expression was assessed using Significance Analysis of Microarrays, and eQTL were calculated using custom software as described previously (Quigley et al., 2009). Cell viability and cell-cycle status were assessed using a WST-1 assay and flow cytometry.
Taqman analysis of transgene expression
Taqman expression analysis was performed on 6 mice (3 female, 3 male) from each strain. Proprietary assays for human MYCN and controls L18 and mGUS were obtained from Applied Biosystems (Carlsbad, CA). MYCN relative to mGUS is shown in Figure S1B.
Genotyping
DNA was isolated from spleen tissue using a proteinase K lysis followed by phenol chloroform extraction. Microsatellite marker genotyping was carried out by the Marshfield Clinic (Marshfield, WI), and CIDR (Baltimore, MD). SNP genotyping was performed using template-directed primer extension with fluorescence polarization detection (FP-TDI (Hsu et al., 2001), Acycloprime II, Perkin Elmer, Waltham, MA) and SNPStream (Bell et al., 2002) 48-plex (Beckman Coulter, Brea, CA). Markers and map positions are shown in Table S1. The marker set had an average spacing of 8 MB genome-wide (excluding the high density of markers on chromosome 10).
Linkage analysis
Interval mapping was performed using the R-QTL(Broman et al., 2003) package in the R statistical language. Genotypes flagged as probable errors by R-QTL were discarded. The genetic map positions were determined using the physical map positions (NCBI 37/mm9), followed by re-estimation of the map using R-QTL, and likely mis-mapped markers were discarded. Linkage analysis was performed on a 1-cM grid. Genome-wide significance thresholds were determined by running 1000 permutations for each dataset. Interval analysis was performed using the binary mode of the “EM” model. All results reported as significant were significant at a 5% genome-wide error rate. 95% confidence intervals (CI) were determined using the lodint function in R-QTL. Genes within the confidence intervals were determined by counting all genes in the UCSC genome assembly mapping between markers flanking the 95% CI.
Expression Arrays
RNA from superior cervical ganglia was isolated using the RNEasy kit (QIAGEN, Valencia, CA), as we found these buffers were more effective at disrupting the ganglia than Trizol. 1μg of RNA was used as a starting template for the RiboMinus rRNA subtraction (Invitrogen, Carlsbad, CA) followed by the ST labeling protocol (Affymetrix, Santa Clara, CA). Labeled samples were hybridized to Affymetrix Mouse Exon 1.0 arrays. Array quality control was performed using the Affymetrix Expression Console. 2-way comparisons between homogenous groups (e.g. male vs. females or 129/SvJ vs FVB/N males) were performed using the Significance Analysis of Microarrays (SAM) package (Tusher et al., 2001) using a 5% false discovery rate. Results are presented in Table S3 and plotted in Figures S1C and S1D. Array data are available at http://www.ncbi.nlm.nih.gov/geo/, accession number GSE59675
eQTL analysis
Arrays were normalized using RMA in the XPS package (http://www.bioconductor.org/packages/2.6/bioc/html/xps.html). eQTL were calculated as described in (Quigley et al., 2009); briefly, linkage between gene expression and loci was assessed by linear regression with genome-wide significance assessed using a FDR-based method.
Cell Lines and Reagents
Neuroblastoma cell lines were obtained from ATCC. Cells were grown in RPMI media with 10% FBS and antibiotics with the exception of SK-N-BE(2) (DMEM/F12, 10% serum) and IMR-32 (DMEM, 10% serum plus non-essential amino acids). Nor-NOHA was obtained from Bachem (Torrance, CA) and dissolved in DMSO. QHii066 was synthesized at the University of Wisconsin-Milwaukee as described previously (United States Patent US 7,119,196B2).
Western blotting
Equal amounts of total protein were loaded for 4%–12% SDS-polyacrylamide gel electrophoresis and transferred to nitrocellulose membranes. After blocking, membranes were blotted with ARG1 (R&D Systems, Minneapolis MN), GABA B R1, Erk (Santa Cruz Biotech, Santa Cruz, CA), GABA A Rβ2/3, GABA B R2, β-tubulin (Millipore, Billerica, MA), cleaved-PARP (Asp214), p-Akt (Ser473), Akt, pErk (Thr202/Tyr204) (Cell Signaling Technology, Danvers, MA), and GAPDH (Upstate Biotechnology, Lake Placid NY). Antibodies were detected with HRP-linked anti-mouse, anti-rabbit (Amersham/GE, Piscataway, NJ), or anti-sheep IgG (Calbiochem/EMD, Gibbstown, NJ), followed by enhanced chemiluminescence (Amersham/GE, Piscataway, NJ).
siRNA Transfection
Control siRNA (Santa Cruz Biotechnology) and Arg1 siRNA (Dharmacon) were transfected using lipofectamine 2000 (Invitrogen, Carlsbad, CA). Cells were harvested for analysis after 72h
Retroviral Transduction
Human ARG1 cDNA was obtained from the IMAGE consortium, cloned into pENTR4 vector (Invitrogen, Carlsbad, CA), then subcloned into a GATEWAY compatible pBABE vector. To generate retrovirus, the packaging cell line HEK 293T was co-transfected with pBABE vector or pBABE-ARG1, along with plasmids expressing gag/pol and VSVg (vesicular stomatitis virus glycoprotein), using Effectene transfection reagent (Qiagen). At 48 hours posttransfection, the viral supernatants were filtered and used to infect cells. LAN-5 and Kelly cells were transduced and selected for two weeks with 0.5μg/ml puromycin. Cells were harvested for analysis after 72h of subsequent treatment.
Viability
Cells were seeded in 12-well plates and treated as indicated for 48 hours. Viability was determined using the WST-1 assay (Roche) and analyzed using spectrophotometric analysis (absorbance at 450nm) after 60 minutes.
Flow cytometry
Cells were plated on 6-well dishes and treated as indicated for 24 hours. They were then harvested, fixed in 70% ethanol for 30 minutes, then stained with 5 μg/ml propidium iodide (PI) containing 125 unit/ml RNase. All samples were analyzed on a FACSCaliber flow cytometer (Becton Dickenson, Franklin Lakes, NJ) and DNA histograms were modeled offline using ModFit-LT (Verity Software, Topsham ME). Apoptosis was detected by flow cytometry for annexin V-FITC per the manufacturer’s protocol (Annexin V-FITC detection kit, BioVision, Milpitas, CA) using FlowJo software (Tree Star, Inc, Ashland, OR).
Supplementary Material
Acknowledgments
The authors are grateful to the Marshfield Clinic and CIDR for genotyping, and to Fernando Pardo Manuel de Villena, Gary Churchill, Saunak Sen, Denise Lind, and Roger Nicoll for experimental advice and comments on the manuscript. This work was supported by the March of Dimes, the Concern Foundation, the UCSF Academic Senate, NIH R01CA102321, R01NS055750, R01MH046851, P30CA82103, U01CA176287, and ARRA supplement for R01NS055750 (WAW), as well as MH096463 and NS076517 (JMC).
Footnotes
AUTHOR CONTRIBUTIONS:
CSH performed the majority of the experiments and data analysis. DQ performed the eQTL and statistical analysis and helped prepare the manuscript. RW, CC, JWC WCG, and YB performed biochemical assays. DDG, WCG and KN assisted with mouse work. LP, JC, JM, and PK provided technical support for genotyping data generation and analysis. YKS, JW, JC, SK, TVD, and JK provided technical support for expression array data generation and analysis. SR synthesized QHii066. JMC and YJC helped design biochemical experiments. AB, JK, YJC, JMC and TVD helped design experiments and prepare the manuscript. WAW supervised the project. QWF supervised and performed the majority of the in vitro analysis. CSH and WAW designed experiments and wrote the paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andang M, Hjerling-Leffler J, Moliner A, Lundgren TK, Castelo-Branco G, Nanou E, Pozas E, Bryja V, Halliez S, Nishimaru H, et al. Histone H2AX-dependent GABA(A) receptor regulation of stem cell proliferation. Nature. 2008;451:460–464. doi: 10.1038/nature06488. [DOI] [PubMed] [Google Scholar]
- Asgharzadeh S, Pique-Regi R, Sposto R, Wang H, Yang Y, Shimada H, Matthay K, Buckley J, Ortega A, Seeger RC. Prognostic significance of gene expression profiles of metastatic neuroblastomas lacking MYCN gene amplification. J Natl Cancer Inst. 2006;98:1193–1203. doi: 10.1093/jnci/djj330. [DOI] [PubMed] [Google Scholar]
- Bak SP, Alonso A, Turk MJ, Berwin B. Murine ovarian cancer vascular leukocytes require arginase-1 activity for T cell suppression. Mol Immunol. 2008;46:258–268. doi: 10.1016/j.molimm.2008.08.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell PA, Chaturvedi S, Gelfand CA, Huang CY, Kochersperger M, Kopla R, Modica F, Pohl M, Varde S, Zhao R, et al. SNPstream UHT: ultra-high throughput SNP genotyping for pharmacogenomics and drug discovery. Biotechniques. 2002;(Suppl):70–72. 74, 76–77. [PubMed] [Google Scholar]
- Bello-Fernandez C, Packham G, Cleveland JL. The ornithine decarboxylase gene is a transcriptional target of c-Myc. Proc Natl Acad Sci U S A. 1993;90:7804–7808. doi: 10.1073/pnas.90.16.7804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brem RB, Yvert G, Clinton R, Kruglyak L. Genetic dissection of transcriptional regulation in budding yeast. Science. 2002;296:752–755. doi: 10.1126/science.1069516. [DOI] [PubMed] [Google Scholar]
- Brodeur GM, Seeger RC, Schwab M, Varmus HE, Bishop JM. Amplification of N-myc in untreated human neuroblastomas correlates with advanced disease stage. Science. 1984;224:1121–1124. doi: 10.1126/science.6719137. [DOI] [PubMed] [Google Scholar]
- Broman KW, Wu H, Sen S, Churchill GA. R/qtl: QTL mapping in experimental crosses. Bioinformatics. 2003;19:889–890. doi: 10.1093/bioinformatics/btg112. [DOI] [PubMed] [Google Scholar]
- Capasso M, Devoto M, Hou C, Asgharzadeh S, Glessner JT, Attiyeh EF, Mosse YP, Kim C, Diskin SJ, Cole KA, et al. Common variations in BARD1 influence susceptibility to high-risk neuroblastoma. Nat Genet. 2009;41:718–723. doi: 10.1038/ng.374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesler L, Goldenberg DD, Seales IT, Satchi-Fainaro R, Grimmer M, Collins R, Struett C, Nguyen KN, Kim G, Tihan T, et al. Malignant progression and blockade of angiogenesis in a murine transgenic model of neuroblastoma. Cancer Res. 2007;67:9435–9442. doi: 10.1158/0008-5472.CAN-07-1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciani E, Severi S, Contestabile A, Bartesaghi R. Nitric oxide negatively regulates proliferation and promotes neuronal differentiation through N-Myc downregulation. J Cell Sci. 2004;117:4727–4737. doi: 10.1242/jcs.01348. [DOI] [PubMed] [Google Scholar]
- Crawford NP, Alsarraj J, Lukes L, Walker RC, Officewala JS, Yang HH, Lee MP, Ozato K, Hunter KW. Bromodomain 4 activation predicts breast cancer survival. Proc Natl Acad Sci U S A. 2008a;105:6380–6385. doi: 10.1073/pnas.0710331105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford NP, Walker RC, Lukes L, Officewala JS, Williams RW, Hunter KW. The Diasporin Pathway: a tumor progression-related transcriptional network that predicts breast cancer survival. Clin Exp Metastasis. 2008b;25:357–369. doi: 10.1007/s10585-008-9146-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Deyn PP, Marescau B, Macdonald RL. Guanidino compounds that are increased in hyperargininemia inhibit GABA and glycine responses on mouse neurons in cell culture. Epilepsy Res. 1991;8:134–141. doi: 10.1016/0920-1211(91)90081-p. [DOI] [PubMed] [Google Scholar]
- Deignan JL, Marescau B, Livesay JC, Iyer RK, De Deyn PP, Cederbaum SD, Grody WW. Increased plasma and tissue guanidino compounds in a mouse model of hyperargininemia. Mol Genet Metab. 2008;93:172–178. doi: 10.1016/j.ymgme.2007.09.016. [DOI] [PubMed] [Google Scholar]
- Diskin SJ, Capasso M, Schnepp RW, Cole KA, Attiyeh EF, Hou C, Diamond M, Carpenter EL, Winter C, Lee H, et al. Common variation at 6q16 within HACE1 and LIN28B influences susceptibility to neuroblastoma. Nat Genet. 2012;44:1126–1130. doi: 10.1038/ng.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durante W, Johnson FK, Johnson RA. Arginase: a critical regulator of nitric oxide synthesis and vascular function. Clin Exp Pharmacol Physiol. 2007;34:906–911. doi: 10.1111/j.1440-1681.2007.04638.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewart-Toland A, Briassouli P, de Koning JP, Mao JH, Yuan J, Chan F, MacCarthy-Morrogh L, Ponder BA, Nagase H, Burn J, et al. Identification of Stk6/STK15 as a candidate low-penetrance tumor-susceptibility gene in mouse and human. Nat Genet. 2003;34:403–412. doi: 10.1038/ng1220. [DOI] [PubMed] [Google Scholar]
- Gerner EW, Meyskens FL., Jr Polyamines and cancer: old molecules, new understanding. Nat Rev Cancer. 2004;4:781–792. doi: 10.1038/nrc1454. [DOI] [PubMed] [Google Scholar]
- Gilbert SL, Zhang L, Forster ML, Anderson JR, Iwase T, Soliven B, Donahue LR, Sweet HO, Bronson RT, Davisson MT, et al. Trak1 mutation disrupts GABA(A) receptor homeostasis in hypertonic mice. Nat Genet. 2006;38:245–250. doi: 10.1038/ng1715. [DOI] [PubMed] [Google Scholar]
- Gray PW, Glaister D, Seeburg PH, Guidotti A, Costa E. Cloning and expression of cDNA for human diazepam binding inhibitor, a natural ligand of an allosteric regulatory site of the gamma-aminobutyric acid type A receptor. Proc Natl Acad Sci U S A. 1986;83:7547–7551. doi: 10.1073/pnas.83.19.7547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett CS, Hodgson JG, Law ME, Fridlyand J, Osoegawa K, de Jong PJ, Nowak NJ, Pinkel D, Albertson DG, Jain A, et al. Genome-wide array CGH analysis of murine neuroblastoma reveals distinct genomic aberrations which parallel those in human tumors. Cancer Res. 2003;63:5266–5273. [PubMed] [Google Scholar]
- Hale G, Gula MJ, Blatt J. Impact of gender on the natural history of neuroblastoma. Pediatr Hematol Oncol. 1994;11:91–97. doi: 10.3109/08880019409141905. [DOI] [PubMed] [Google Scholar]
- He X, Huang Q, Ma C, Yu S, McKernan R, Cook JM. Pharmacophore/receptor models for GABA(A)/BzR alpha2beta3gamma2, alpha3beta3gamma2 and alpha4beta3gamma2 recombinant subtypes. Included volume analysis and comparison to alpha1beta3gamma2, alpha5beta3gamma2, and alpha6beta3gamma2 subtypes. Drug Des Discov. 2000;17:131–171. [PubMed] [Google Scholar]
- Hogarty MD, Norris MD, Davis K, Liu X, Evageliou NF, Hayes CS, Pawel B, Guo R, Zhao H, Sekyere E, et al. ODC1 is a critical determinant of MYCN oncogenesis and a therapeutic target in neuroblastoma. Cancer Res. 2008;68:9735–9745. doi: 10.1158/0008-5472.CAN-07-6866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu TM, Chen X, Duan S, Miller RD, Kwok PY. Universal SNP genotyping assay with fluorescence polarization detection. Biotechniques. 2001;31:560, 562, 564–568. doi: 10.2144/01313rr01. passim. [DOI] [PubMed] [Google Scholar]
- Huang Q, He X, Ma C, Liu R, Yu S, Dayer CA, Wenger GR, McKernan R, Cook JM. Pharmacophore/receptor models for GABA(A)/BzR subtypes (alpha1beta3gamma2, alpha5beta3gamma2, and alpha6beta3gamma2) via a comprehensive ligand-mapping approach. J Med Chem. 2000;43:71–95. doi: 10.1021/jm990341r. [DOI] [PubMed] [Google Scholar]
- Huang Y, Higginson DS, Hester L, Park MH, Snyder SH. Neuronal growth and survival mediated by eIF5A, a polyamine-modified translation initiation factor. Proc Natl Acad Sci U S A. 2007;104:4194–4199. doi: 10.1073/pnas.0611609104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janoueix-Lerosey I, Lequin D, Brugieres L, Ribeiro A, de Pontual L, Combaret V, Raynal V, Puisieux A, Schleiermacher G, Pierron G, et al. Somatic and germline activating mutations of the ALK kinase receptor in neuroblastoma. Nature. 2008;455:967–970. doi: 10.1038/nature07398. [DOI] [PubMed] [Google Scholar]
- Jenkins DC, Charles IG, Thomsen LL, Moss DW, Holmes LS, Baylis SA, Rhodes P, Westmore K, Emson PC, Moncada S. Roles of nitric oxide in tumor growth. Proc Natl Acad Sci U S A. 1995;92:4392–4396. doi: 10.1073/pnas.92.10.4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koomoa DL, Yco LP, Borsics T, Wallick CJ, Bachmann AS. Ornithine decarboxylase inhibition by alpha-difluoromethylornithine activates opposing signaling pathways via phosphorylation of both Akt/protein kinase B and p27Kip1 in neuroblastoma. Cancer Res. 2008;68:9825–9831. doi: 10.1158/0008-5472.CAN-08-1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Merrill M, Gordon RR, Hunter KW, Threadgill DW, Pomp D. Dietary fat alters pulmonary metastasis of mammary cancers through cancer autonomous and non-autonomous changes in gene expression. Clin Exp Metastasis. 2010;27:107–116. doi: 10.1007/s10585-009-9302-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le-Corronc H, Rigo JM, Branchereau P, Legendre P. GABA(A) Receptor and Glycine Receptor Activation by Paracrine/Autocrine Release of Endogenous Agonists: More Than a Simple Communication Pathway. Mol Neurobiol. 2011 doi: 10.1007/s12035-011-8185-1. [DOI] [PubMed] [Google Scholar]
- MacPhee M, Chepenik KP, Liddell RA, Nelson KK, Siracusa LD, Buchberg AM. The secretory phospholipase A2 gene is a candidate for the Mom1 locus, a major modifier of ApcMin-induced intestinal neoplasia. Cell. 1995;81:957–966. doi: 10.1016/0092-8674(95)90015-2. [DOI] [PubMed] [Google Scholar]
- Mao JH, Saunier EF, de Koning JP, McKinnon MM, Higgins MN, Nicklas K, Yang HT, Balmain A, Akhurst RJ. Genetic variants of Tgfb1 act as context-dependent modifiers of mouse skin tumor susceptibility. Proc Natl Acad Sci U S A. 2006;103:8125–8130. doi: 10.1073/pnas.0602581103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosse YP, Laudenslager M, Khazi D, Carlisle AJ, Winter CL, Rappaport E, Maris JM. Germline PHOX2B mutation in hereditary neuroblastoma. Am J Hum Genet. 2004;75:727–730. doi: 10.1086/424530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosse YP, Laudenslager M, Longo L, Cole KA, Wood A, Attiyeh EF, Laquaglia MJ, Sennett R, Lynch JE, Perri P, et al. Identification of ALK as a major familial neuroblastoma predisposition gene. Nature. 2008;455:930–935. doi: 10.1038/nature07261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen le B, Diskin SJ, Capasso M, Wang K, Diamond MA, Glessner J, Kim C, Attiyeh EF, Mosse YP, Cole K, et al. Phenotype restricted genome-wide association study using a gene-centric approach identifies three low-risk neuroblastoma susceptibility Loci. PLoS Genet. 2011;7:e1002026. doi: 10.1371/journal.pgen.1002026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park YG, Zhao X, Lesueur F, Lowy DR, Lancaster M, Pharoah P, Qian X, Hunter KW. Sipa1 is a candidate for underlying the metastasis efficiency modifier locus Mtes1. Nat Genet. 2005;37:1055–1062. doi: 10.1038/ng1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quigley DA, To MD, Perez-Losada J, Pelorosso FG, Mao JH, Nagase H, Ginzinger DG, Balmain A. Genetic architecture of mouse skin inflammation and tumour susceptibility. Nature. 2009;458:505–508. doi: 10.1038/nature07683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts SS, Mori M, Pattee P, Lapidus J, Mathews R, O’Malley JP, Hsieh YC, Turner MA, Wang Z, Tian Q, et al. GABAergic system gene expression predicts clinical outcome in patients with neuroblastoma. J Clin Oncol. 2004;22:4127–4134. doi: 10.1200/JCO.2004.02.032. [DOI] [PubMed] [Google Scholar]
- Schadt EE, Monks SA, Drake TA, Lusis AJ, Che N, Colinayo V, Ruff TG, Milligan SB, Lamb JR, Cavet G, et al. Genetics of gene expression surveyed in maize, mouse and man. Nature. 2003;422:297–302. doi: 10.1038/nature01434. [DOI] [PubMed] [Google Scholar]
- Schwab M, Varmus HE, Bishop JM, Grzeschik KH, Naylor SL, Sakaguchi AY, Brodeur G, Trent J. Chromosome localization in normal human cells and neuroblastomas of a gene related to c-myc. Nature. 1984;308:288–291. doi: 10.1038/308288a0. [DOI] [PubMed] [Google Scholar]
- Singh R, Pervin S, Karimi A, Cederbaum S, Chaudhuri G. Arginase activity in human breast cancer cell lines: N(omega)-hydroxy-L-arginine selectively inhibits cell proliferation and induces apoptosis in MDA-MB-468 cells. Cancer Res. 2000;60:3305–3312. [PubMed] [Google Scholar]
- Tenu JP, Lepoivre M, Moali C, Brollo M, Mansuy D, Boucher JL. Effects of the new arginase inhibitor N(omega)-hydroxy-nor-L-arginine on NO synthase activity in murine macrophages. Nitric Oxide. 1999;3:427–438. doi: 10.1006/niox.1999.0255. [DOI] [PubMed] [Google Scholar]
- Trochet D, Bourdeaut F, Janoueix-Lerosey I, Deville A, de Pontual L, Schleiermacher G, Coze C, Philip N, Frebourg T, Munnich A, et al. Germline mutations of the paired-like homeobox 2B (PHOX2B) gene in neuroblastoma. Am J Hum Genet. 2004;74:761–764. doi: 10.1086/383253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakabayashi Y, Mao JH, Brown K, Girardi M, Balmain A. Promotion of Hras-induced squamous carcinomas by a polymorphic variant of the Patched gene in FVB mice. Nature. 2007;445:761–765. doi: 10.1038/nature05489. [DOI] [PubMed] [Google Scholar]
- Wang K, Diskin SJ, Zhang H, Attiyeh EF, Winter C, Hou C, Schnepp RW, Diamond M, Bosse K, Mayes PA, et al. Integrative genomics identifies LMO1 as a neuroblastoma oncogene. Nature. 2011;469:216–220. doi: 10.1038/nature09609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss WA, Aldape K, Mohapatra G, Feuerstein BG, Bishop JM. Targeted expression of MYCN causes neuroblastoma in transgenic mice. EMBO J. 1997;16:2985–2995. doi: 10.1093/emboj/16.11.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yachimovich-Cohen N, Even-Ram S, Shufaro Y, Rachmilewitz J, Reubinoff B. Human embryonic stem cells suppress T cell responses via arginase I-dependent mechanism. J Immunol. 2010;184:1300–1308. doi: 10.4049/jimmunol.0804261. [DOI] [PubMed] [Google Scholar]
- Yang X, Deignan JL, Qi H, Zhu J, Qian S, Zhong J, Torosyan G, Majid S, Falkard B, Kleinhanz RR, et al. Validation of candidate causal genes for obesity that affect shared metabolic pathways and networks. Nat Genet. 2009;41:415–423. doi: 10.1038/ng.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Iyer RK, Kern RM, Rodriguez WI, Grody WW, Cederbaum SD. Expression of arginase isozymes in mouse brain. J Neurosci Res. 2001;66:406–422. doi: 10.1002/jnr.1233. [DOI] [PubMed] [Google Scholar]
- Yu H, Iyer RK, Yoo PK, Kern RM, Grody WW, Cederbaum SD. Arginase expression in mouse embryonic development. Mech Dev. 2002;115:151–155. doi: 10.1016/s0925-4773(02)00089-8. [DOI] [PubMed] [Google Scholar]
- Yu H, Yoo PK, Aguirre CC, Tsoa RW, Kern RM, Grody WW, Cederbaum SD, Iyer RK. Widespread expression of arginase I in mouse tissues. Biochemical and physiological implications. J Histochem Cytochem. 2003;51:1151–1160. doi: 10.1177/002215540305100905. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.