ABSTRACT
Neutrophils engulf and kill bacteria using oxidative and nonoxidative mechanisms. Despite robust antimicrobial activity, neutrophils are impaired in directing Salmonella clearance and harbor viable intracellular bacteria during early stages of infection that can subsequently escape to more-permissive cell types. The mechanisms accounting for this immune impairment are not understood. We report that Salmonella limits exposure to oxidative damage elicited by d-amino acid oxidase (DAO) in neutrophils by expressing an ABC importer specific for d-alanine, a DAO substrate found in peptidoglycan stem peptides. A Salmonella dalS mutant defective for d-alanine import was more susceptible to killing by DAO through exposure to greater oxidative stress during infection. This fitness defect was reversed by selective depletion of neutrophils or by inhibition of DAO in vivo with a small-molecule inhibitor. DalS-mediated subversion of neutrophil DAO is a novel host-pathogen interaction that enhances Salmonella survival during systemic infection.
IMPORTANCE
Neutrophils engulf Salmonella during early stages of infection, but bacterial killing is incomplete. Very little is known about how Salmonella survives in neutrophils to gain access to other cell types during infection. In this study, we show that d-amino acid oxidase (DAO) in neutrophils consumes d-alanine and that importing this substrate protects Salmonella from oxidative killing by neutrophil DAO. Loss of this importer results in increased bacterial killing in vitro, in neutrophils, and in a mouse model of infection, all phenotypes that are lost upon inhibition of DAO. These findings add mechanistic insight into a novel host-pathogen interaction that has consequences on infection outcome.
INTRODUCTION
The d-amino acid oxidase (DAO) catalyzes the flavin-dependent deamination of certain d-amino acids to yield an α-ketoacid, ammonium ion, and hydrogen peroxide (1, 2). For example, DAO regulates d-serine levels in the brain, where this amino acid coactivates glutamate-dependent N-methyl-d-aspartate receptors on postsynaptic neurons (1). Owing to its ability to generate hydrogen peroxide and its widespread conservation among vertebrates and invertebrates (3), historically, DAO was considered a potential component of the innate host defense system. Work from the 1960s identified DAO activity in the purified granule fraction of human neutrophils (4). This activity was later localized by electron microscopy to the neutrophil phagosome following engulfment of latex beads in the presence of d-alanine (5). Studies of gnotobiotic mice showed DAO in the kidney in response to d-alanine liberated from the microbiota (6), and mice with a spontaneous mutation in DAO were shown to be more susceptible to infection with Staphylococcus aureus (7), further linking DAO to an understudied host defense system that is responsive to microbial input. Mammals do not synthesize d-alanine. However, in bacteria, it constitutes the terminal amino acid in peptidoglycan stem peptides, making it a potential discriminator between self and nonself in the context of immunity.
In neutrophil phagosomes, hydrogen peroxide liberated from DAO-catalyzed dehydrogenation of d-alanine would be accessible to myeloperoxidase (MPO), which catalyzes the formation of hypochlorous acid (HOCl) from chloride and hydrogen peroxide. Although debated (8), the latter reaction has for some time been considered the clinically relevant and major microbicidal pathway in neutrophils, with reaction products being directly toxic to bacteria or going on to form secondary chloramines (9–11). Experimental work indicates that hydrogen peroxide is rate limiting for MPO-catalyzed halogenation in neutrophils (12), suggesting that an auxiliary source of peroxide, in addition to dismutation of superoxide formed from the more widely studied NADPH oxidase, would be beneficial toward neutrophil killing activity.
Previously, we identified an ATP-binding cassette transporter in Salmonella enterica that imports d-alanine (13). This transporter underwent regulatory evolution for expression in the intracellular environment following host cell invasion (14), providing a clue that its function was related to intracellular survival. Structural work confirmed the specificity and chiral selectivity for d-alanine, showing that DalS, the periplasmic binding component, restricts the beta-carbon of alanine to a d-configuration due to steric hindrance of the l-isomer (13). Interestingly, DalS was dispensable for Salmonella enterica serovar Typhimurium growth in vitro and for peptidoglycan composition. However, d-alanine import was required for competitive fitness during infection of mice, and mice infected with DalS-deficient Salmonella had a significantly extended survival time than mice infected with wild-type Salmonella (13). Our previous results suggested a possible interaction between DalS and the host immune system; however, its function remained enigmatic. Using a murine model of systemic infection with the pathogen Salmonella enterica serovar Typhimurium, which is widely used as a model for the host-restricted Salmonella serovar Typhi, we show that DalS helps protect Salmonella from DAO-dependent killing in neutrophils. Salmonella mutants with a deletion of dalS were exposed to greater DAO-dependent oxidative stress during host infection, leading to a loss of competitive fitness and increased killing by neutrophils. However, this defect could be repaired upon inhibition of DAO activity or by host neutropenia. Thus, DalS-mediated subversion of neutrophil DAO is an important host-pathogen interaction that enhances bacterial survival during early stages of infection. These data help explain, in part, the incomplete killing of Salmonella by neutrophils, allowing secondary dissemination to more-permissive cell types (15).
RESULTS
DalS protects S. Typhimurium from the antimicrobial activity of DAO.
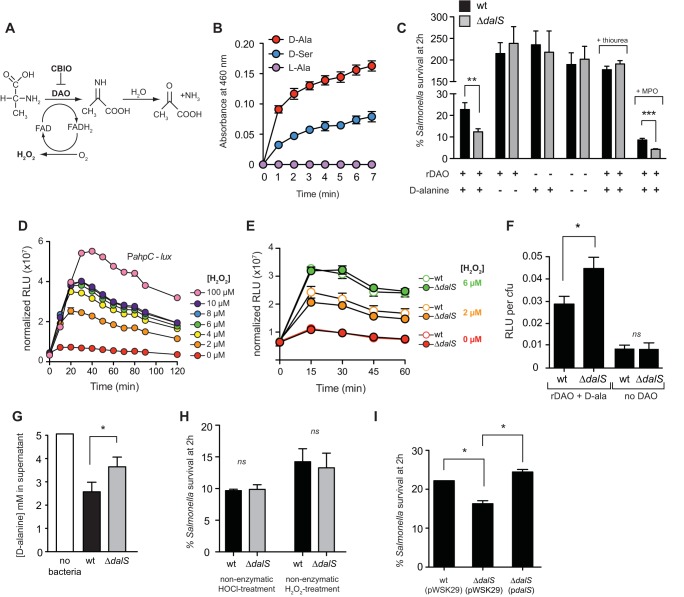
The flavin-dependent deamination of d-alanine by DAO yields the α-ketoacid, ammonium ion, and hydrogen peroxide (Fig. 1A). To study the susceptibility of S. enterica serovar Typhimurium to the activity of DAO, we purified recombinant DAO and confirmed its hydrogen peroxide-generating activity in the presence of 5 mM d-alanine or 50 mM d-serine (Fig. 1B; see Fig. S1 in the supplemental material). Increasing the concentration of d-serine was necessary, as this substrate produces only 10% of the Vmax activity that d-alanine produces (4). DalS-deficient bacteria were more sensitive to killing by purified DAO in the presence of d-alanine (Fig. 1C). This killing was blocked by the addition of thiourea, a potent hydroxyl radical scavenger that mitigates the toxic effects of hydrogen peroxide on bacterial cells by reducing the formation of hydroxyl radicals from hydrogen peroxide (16, 17). As expected, the addition of myeloperoxidase to the in vitro reaction increased the magnitude of killing of both wild-type and dalS mutants, yet the enhanced susceptibility of ΔdalS bacteria persisted (Fig. 1C). These data established that hydrogen peroxide was the toxic DAO reaction product in these in vitro reactions and that Salmonella mutants lacking DalS are more susceptible to DAO-dependent killing.
FIG 1 .
DalS protects S. Typhimurium from the antimicrobial activity of DAO. (A) DAO catalyzes the flavin-dependent deamination of d-alanine to yield the α-ketoacid, NH4+, and hydrogen peroxide. (B) Recombinant DAO (rDAO) produces hydrogen peroxide upon the addition of d-alanine and d-serine as measured by peroxidase-coupled oxidation of o-dianisidine. Data are the means ± standard errors (error bars) from three experiments. (C) S. Typhimurium dalS mutants are more susceptible to the reaction products of DAO than the wild type (wt) is. Bacteria were incubated with purified DAO in the presence (+) or absence (−) of exogenous d-alanine. Survival data are the means plus standard errors (error bars) from three independent experiments. (D) The OxyR-dependent PahpC-lux reporter strain is sensitive to hydrogen peroxide. Data are mean relative light units (RLU) normalized to the optical density of the culture. (E) Wild-type (wt) Salmonella and dalS mutant sense nonenzymatic hydrogen peroxide equally. Data are the means ± standard errors from three experiments. RLU data are normalized to the optical density of the culture. Reporter activities for the wild type and dalS mutants for each of the treatment groups are not significantly different. (F) A dalS mutant is exposed to greater hydrogen peroxide stress compared to the wild type following exposure to DAO and d-alanine. Data are from strains carrying the PahpC-lux reporter and are the means plus standard errors from three separate experiments. (G) S. Typhimurium lacking DalS is defective for d-alanine import. The ability of Salmonella to remove d-alanine (5 mM) from the culture medium was estimated by DAO-catalyzed peroxidase-coupled oxidation of o-dianisidine. (H) ΔdalS mutants have wild-type sensitivity to exogenous hydrogen peroxide (0.1%) and HOCl (0.01%) generated nonenzymatically. (I) Sensitivity of dalS mutants to DAO-dependent killing is normalized to wild-type levels upon complementation with a dalS-encoding plasmid (pdalS). All experiments were performed independently three times. Values that are statistically significantly different by the Mann-Whitney test are indicated by bars and asterisks as follows: *, P < 0.05; **, P < 0.01; ***, P < 0.001. Values that are not statistically significantly different (ns) are indicated.
To determine whether DalS-deficient bacteria allowed a higher concentration of enzymatic hydrogen peroxide product to form from DAO, we constructed an S. Typhimurium reporter strain, previously validated to report hydrogen peroxide stress, by fusing the OxyR-dependent ahpC promoter (18, 19) to luxCDABE from Photorhabdus luminescens. We confirmed the activity of this strain in response to hydrogen peroxide as low as 2 µM and showed dose-dependent luminescence in response to peroxide up to 100 µM (Fig. 1D), well below the steady-state peroxide concentration in neutrophil phagosomes estimated to be ~5 µM by kinetic models (10). Importantly, we confirmed that this reporter responded equally in the wild type and dalS mutants exposed to nonenzymatically generated hydrogen peroxide at relevant concentrations (Fig. 1E). However, upon exposure to hydrogen peroxide generated enzymatically by DAO in the presence of d-alanine, reporter activity was significantly greater in dalS mutants than in the wild type (Fig. 1F). This was consistent with an impaired ability of the dalS mutant to reduce an initial 5 mM extracellular concentration of d-alanine to the same extent as the wild type (Fig. 1G), in agreement with our previous data (13). For a control, both wild-type and ΔdalS bacteria were killed to equivalent levels following exposure to nonenzymatic sources of hydrogen peroxide or hypochlorous acid (HOCl) (Fig. 1H), indicating that increased killing of the dalS mutant was DAO dependent and not due to an inherent sensitivity to hydrogen peroxide or HOCl. Complementing ΔdalS with the dalS gene under the control of its native promoter restored bacterial survival to wild-type levels in the presence of DAO and d-alanine (Fig. 1I). Together these data indicated that DalS-deficient bacteria allowed a higher concentration of DAO-dependent hydrogen peroxide product to form, resulting in increased killing.
Salmonella lacking DalS is more susceptible to DAO killing in neutrophils.
Neutrophils express DAO (4, 5) and are an early host cell target for Salmonella (20, 21). To test whether the S. Typhimurium dalS mutant was more susceptible to the microbicidal activity of neutrophils, we infected purified neutrophils with wild-type Salmonella or dalS mutants and monitored bacterial survival. After 2 h, 40% of wild-type Salmonella had survived, whereas only ~25% of the dalS mutants remained viable (Fig. 2A). To confirm that this killing phenotype was dependent on DAO, we used the chemical inhibitor 6-chloro-1,2-benzisoxazol-3(2H)-one (CBIO) that blocks the dehydrogenation reaction catalyzed by DAO (22) (Fig. 2B). We also confirmed that CBIO had no effect on S. Typhimurium growth and did not bind to the S. Typhimurium DalS protein as measured by fluorescence thermal shift of purified DalS (see Fig. S2 in the supplemental material) (13). In the presence of CBIO, the ability of neutrophils to kill Salmonella was impaired, and there was no longer a survival defect of the dalS mutant (Fig. 2A). To confirm that this killing phenotype was linked to DAO-dependent hydrogen peroxide stress, we infected neutrophils with the wild type and dalS mutants carrying the OxyR-dependent ahpC reporter in the presence or absence of CBIO. Consistent with the killing phenotypes observed, peroxide reporter activity in the dalS mutant was double that seen in wild-type Salmonella during neutrophil infections (Fig. 2C). However, when DAO activity was inhibited with CBIO, the reporter activities from the wild type and dalS mutants were reduced by ~35% and ~65%, respectively, and were no longer different (Fig. 2C). Together these data established that DalS enhances S. Typhimurium survival in neutrophils in a manner that depends on functional DAO.
FIG 2 .
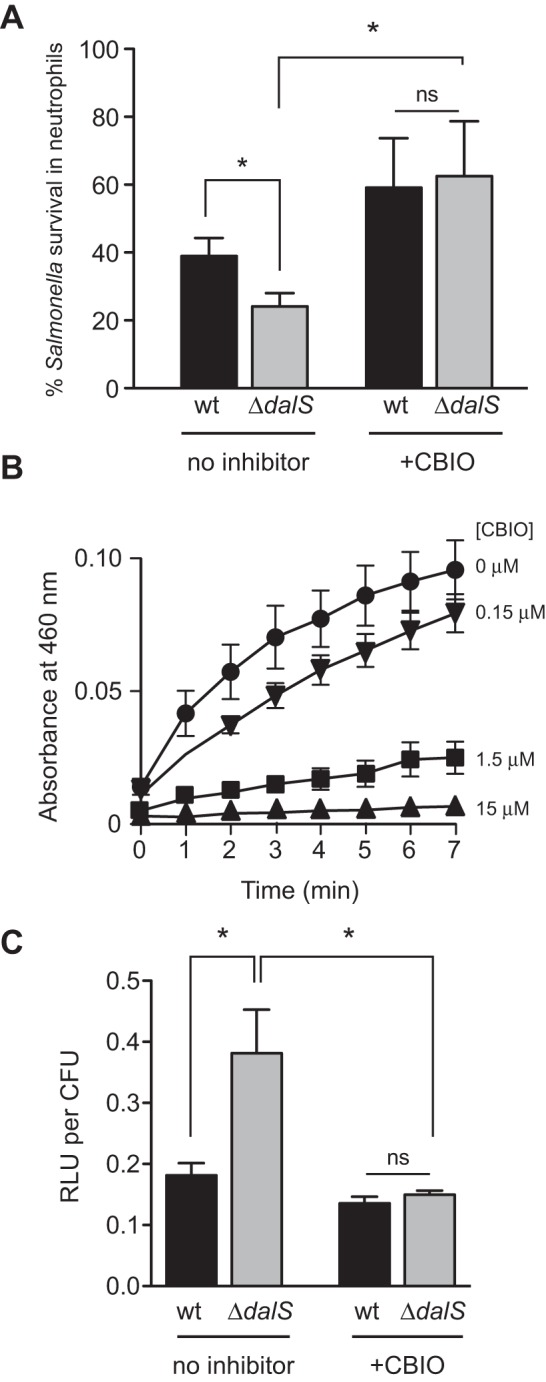
DalS-deficient Salmonella bacteria are killed more efficiently in neutrophils in a DAO-dependent manner. (A) S. Typhimurium ΔdalS bacteria are sensitized to neutrophil killing. Bacterial killing by purified neutrophils was measured after 2 h. Survival was measured as the ratio of the number of viable bacteria at 2 h compared to the initial number of internalized bacteria. The sensitivity of ΔdalS mutants to neutrophil killing was inhibited by DAO inhibition with CBIO. Data are from three experiments. (B) CBIO inhibits DAO enzyme activity in vitro. Different concentrations of CBIO (0, 0.15, 1.5, and 15 µM) were tested. Data are the means ± standard errors (error bars) from three experiments. (C) Salmonella bacteria lacking DalS are exposed to greater hydrogen peroxide stress in neutrophils. Hydrogen peroxide stress levels are restored when DAO is inhibited with CBIO. Data are from strains carrying the PahpC-lux reporter 30 min following infection of purified neutrophils and are the means plus standard error from three separate experiments. Statistical significance is indicated as follows: *, P < 0.05 by the Mann-Whitney test; ns, not significantly different.
S. Typhimurium lacking DalS is sensitized to neutrophil DAO in vivo.
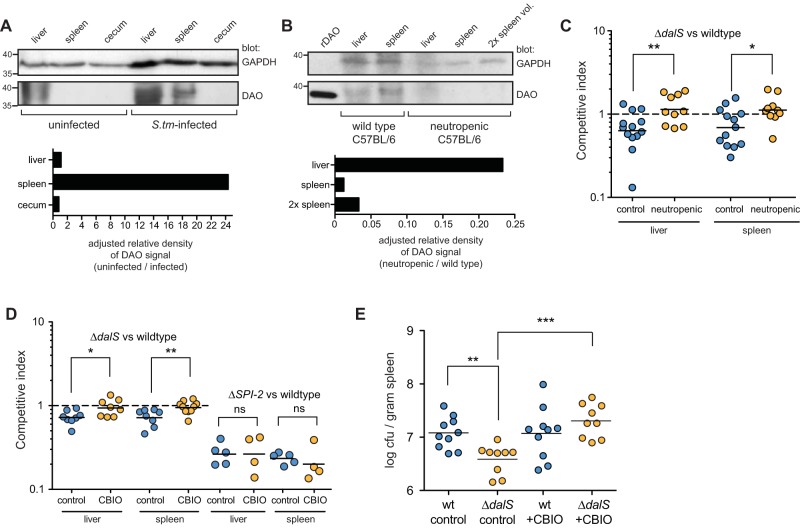
Neutrophils are the primary source of DAO among polymorphonuclear leukocytes (4, 5), and their numbers increase rapidly in the spleen in response to S. Typhimurium infection (19, 21). Given this, we reasoned that the levels of splenic DAO would increase following S. Typhimurium infection due to neutrophil influx. We collected spleens from infected and uninfected mice and probed them for DAO using a DAO antibody. Uninfected mice had no detectable DAO in the spleen or cecum by this method (Fig. 3A). In contrast, splenic DAO increased dramatically upon S. Typhimurium infection (Fig. 3A). Hepatic DAO has been detected in some studies but not others (23). We detected DAO in livers isolated from uninfected mice, but this did not increase dramatically upon infection when normalized to a control host protein (Fig. 3A). To verify that neutrophils were the source of splenic DAO, we rendered mice neutropenic by antibody-based neutrophil depletion and verified this by flow cytometry (see Fig. S3 in the supplemental material). In infected neutropenic mice, splenic DAO became undetectable, and liver DAO was reduced to ~25% of that seen in nonneutropenic controls (Fig. 3B). The residual DAO signal in neutropenic mouse liver is likely from hepatocytes, which are a source of DAO activity in a variety of animals (24).
FIG 3 .
S. Typhimurium bacteria lacking DalS are more susceptible to neutrophil DAO killing in vivo. (A) Western blot of DAO in liver, spleen, and cecum from uninfected mice or mice infected with S. Typhimurium (S.tm). The positions of molecular weight markers (in kilodaltons) are shown to the left of the blots. The relative density of the DAO signal adjusted to the GAPDH signal is shown below the Western blot. (B) Neutropenic mice fail to accumulate splenic DAO following S. Typhimurium infection. Neutropenic mice were infected with S. Typhimurium, and organs were probed for DAO (2× spleen vol., twice the amount of sample loaded). (C) Neutropenia normalizes the fitness defect of the S. Typhimurium ΔdalS mutant. Control mice and neutropenic mice were infected with an equal mixture of wild-type S. Typhimurium and S. Typhimurium ΔdalS mutant for 2 days. The competitive index data from two experiments are shown. Each symbol represents the value for an individual mouse. The mean value for the group of mice is shown by the short horizontal black line. The broken line shows a competitive index of 1. (D) Inhibition of DAO in vivo restores virulence to ΔdalS mutants. Mice were infected with an equal mixture of wild-type S. Typhimurium and S. Typhimurium ΔdalS (first set) or an equal mixture of wild-type S. Typhimurium and S. Typhimurium ΔSPI-2 (second set) and treated with either carboxymethyl cellulose as a control or the DAO inhibitor CBIO. The competitive index data from two experiments are shown. (E) CBIO restores dalS mutant defect in singly infected mice. Mice were infected with either wild-type (wt) S. Typhimurium or S. Typhimurium dalS mutant and treated with CBIO or CMC control for 24 h. The number of CFU was normalized to spleen mass. Values that are statistically significantly different by the Mann-Whitney test are indicated by bars and asterisks as follows: *, P < 0.05; **, P < 0.01; ***, P < 0.001. Values that are not statistically significantly different (ns) are indicated.
Given that DalS protected S. Typhimurium from neutrophil DAO, we hypothesized that depleting neutrophils would restore virulence to the dalS mutant in vivo. Indeed, in neutropenic mice, the S. Typhimurium dalS mutant competed equally with wild-type bacteria, whereas they remained significantly attenuated in control treated animals containing neutrophils (Fig. 3C). To verify that this restoration of virulence in the mutant was linked to the attendant loss of DAO following neutrophil depletion, we alternatively inhibited DAO activity in infected mice by in vivo administration of CBIO (25). These data showed that the effect of neutropenia on the dalS mutant was phenocopied by DAO inhibition (Fig. 3D). The restoration of virulence to dalS mutants following DAO inhibition in vivo was specific to DalS and not a generalized immune dysfunction, because a Salmonella mutant (ΔSPI2) containing a lesion in the type III secretion system involved in intracellular replication (26) remained attenuated in mice in the presence of CBIO (Fig. 3D). To confirm the importance of DalS for Salmonella during infection and to further connect this virulence factor to host DAO, single infections were performed with the wild type or the dalS mutant in mice in which DAO was inhibited or not inhibited. Mice infected with wild-type S. Typhimurium carried a significantly higher splenic bacterial burden than mice infected with a dalS mutant (Fig. 3E), confirming that DalS contributes to bacterial fitness in the host. Treatment of mice with the DAO inhibitor CBIO significantly increased the bacterial load associated with ΔdalS infection (Fig. 3E). Together, these experiments clearly linked the phenotype of the dalS mutant to the expression and activity of neutrophil DAO.
DAO contributes to bacterial peroxide stress independent of NOX2.
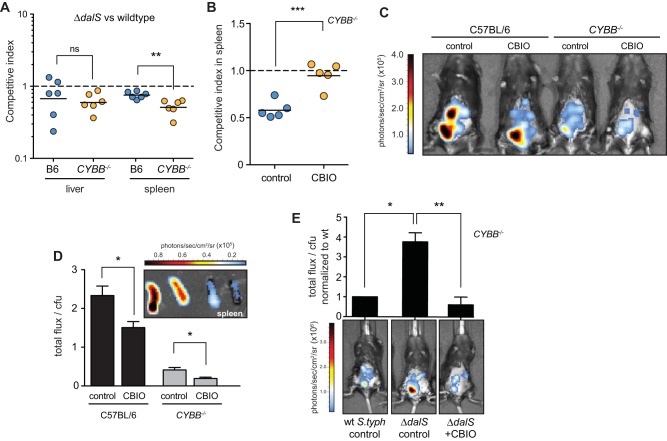
The NADPH oxidase system in neutrophils is a source of superoxide that can spawn an array of reactive oxygen species. Prominent among these is hydrogen peroxide that is consumed by myeloperoxidase to generate hypochlorous acid (HOCl) within the neutrophil phagosome (27). However, our experiments indicated that neutrophils killed S. Typhimurium ΔdalS mutant bacteria more effectively than wild-type bacteria even in the presence of functional NADPH oxidase, suggesting that DAO is a relevant source of innate immune activity. If this were the case, then the fitness defect in the ΔdalS mutant would persist in a host lacking NADPH oxidase. To quantify the contribution of DAO to host protection independent of NADPH oxidase, we infected CYBB/gp91/NOX2 mice lacking the flavocytochrome b-245 heavy chain, an essential component of NADPH oxidase. In the liver, dalS mutant bacteria remained attenuated and at levels similar to those in wild-type mice. Interestingly, in the spleen where neutrophils accumulate after infection (15, 21), the defect of dalS mutants was significantly amplified in CYBB−/− mice compared to their defect in wild-type mice (Fig. 4A). The fitness defect of the dalS mutant was repaired upon treatment of CYBB−/− mice with CBIO (Fig. 4B), implicating DAO as a relevant source of innate immune protection during S. Typhimurium infections. These data indicated that DalS increases early Salmonella survival, particularly in the spleen, even in the absence of NADPH oxidase.
FIG 4 .
DAO contributes to bacterial peroxide stress independent of NOX2. (A) DalS mutants remain attenuated in mice lacking functional NOX2/NADPH oxidase. Wild-type mice (C57BL/6 [B6]) or CYBB−/− mice were infected with an equal mixture of wild-type S. Typhimurium and S. Typhimurium ΔdalS mutant, and the competitive index was calculated after 2 days. (B) DalS mutant defect is restored with CBIO treatment in NADPH oxidase-deficient mice. CYBB−/− mice were infected with an equal mixture of wild-type S. Typhimurium and S. Typhimurium ΔdalS mutant and treated with CBIO or carboxymethyl cellulose as a control. The competitive index was calculated after 2 days. (C) DAO elicits peroxide stress in mice. Wild-type and CYBB−/− mice with functional (control) or inhibited DAO (CBIO) were infected with the peroxide reporter strain and imaged after 2 h. Images represent 5-s integrations and are representative of the images from three separate experiments. (D) Ex vivo imaging of excised spleens from mice in panel B. (Inset) Images were integrated over 5 s and plotted as total photon flux normalized to viable colony counts. (E) DalS mutants are exposed to greater peroxide stress than the wild type in vivo. CYBB−/− mice were infected with wild-type S. Typhimurium (wt S.typh) or ΔdalS mutants containing the OxyR-dependent luminescent reporter and then treated with the DAO inhibitor CBIO or carboxymethyl cellulose (control). Representative whole-body images from infected mice are shown with normalized luminescence flux plotted above each image. The values plotted in the graph represent the values for three mice per group. Values that are statistically significantly different by the Mann-Whitney test are indicated by bars and asterisks as follows: *, P < 0.05; **, P < 0.01; ***, P < 0.001.
To confirm that this source of bactericidal peroxide was DAO, we used the OxyR-dependent ahpC-lux transcriptional reporter constructed earlier and performed whole-animal in vivo imaging of infected mice to quantify hydrogen peroxide stress in S. Typhimurium following DAO inhibition. S. Typhimurium produced OxyR-dependent luminescence in both wild-type mice and in CYBB−/− mice lacking NADPH oxidase, which was reduced in both cases when mice were treated with the DAO inhibitor CBIO (Fig. 4C). Importantly, at these early time points after infection, the bacterial loads in the groups of mice were found to be similar at necropsy (data not shown). OxyR-dependent luminescence in wild-type mice was greater than that from CYBB−/− mice, a result that was expected because NADPH oxidase activity is broadly conserved among immune cells that interact with Salmonella (21, 28), whereas DAO is restricted to neutrophils (4, 5). Luminescence normalized to the bacterial load in the spleen was quantified in both wild-type and CYBB−/− mice by ex vivo imaging of freshly excised organs (Fig. 4D). This analysis showed that inhibition of DAO activity in both wild-type and CYBB−/− mice significantly reduced OxyR-dependent luminescence from splenic S. Typhimurium by ~60%, confirming that DAO was a relevant source of hydrogen peroxide during infection in the presence or absence of NOX2.
Our previous results showed that S. Typhimurium lacking DalS is exposed to greater DAO-dependent oxidative stress during infection. We quantified this in mice lacking NADPH oxidase by infecting CYBB−/− mice with DalS-deficient Salmonella carrying the OxyR-dependent luminescence reporter. OxyR-dependent luminescence in mice infected with the ΔdalS mutant was significantly greater than from mice infected with wild-type Salmonella. Treatment of mice with CBIO to inhibit DAO activity significantly reduced this luminescence signal (Fig. 4E). Together, these experiments revealed that splenic S. Typhimurium is exposed to DAO-dependent peroxide stress in mice and that DalS reduces the magnitude of this stress to promote infection.
DISCUSSION
The majority of work on Salmonella infection of immune cells has focused on macrophages. However, neutrophils are the major cell type infected by Salmonella during the first 2 days of infection. In the gut, 70% of luminal S. Typhimurium bacteria are inside neutrophils at day 1 (29), and in the spleen, a neutrophil-enriched population harbored 100% of the viable intracellular S. Typhimurium population in the first 24 h (20) and 70% of this bacterial population on day 2 with a concomitant increase in the number of infected macrophages (21). Despite robust antimicrobial activity of neutrophils, which indeed kills many Salmonella bacteria during infection (15, 30), neutrophils are unable to direct full S. Typhimurium clearance (31). This incomplete killing is thought to allow Salmonella to spread to a more permissive macrophage population, which has been shown to generate sublethal oxidative bursts (15). The mechanisms that account for this immune subversion of neutrophils are not known but likely involve resistance to the microbicidal effector functions of these cells (32). Interestingly, despite the importance of this host-pathogen interaction, very little is known mechanistically about how Salmonella survives in neutrophils. Our data are consistent with neutrophil DAO functioning to exert bactericidal activity on S. Typhimurium at early stages of infection. These data provide insight into a host-pathogen interaction in systemic neutrophils that has bearing on the outcome of Salmonella infection. It is likely important that this immune subversion mechanism is also conserved in S. Typhi, as the DalS transport system is 98% identical in this human pathogen.
Hydrogen peroxide production is rate limiting for MPO-catalyzed halogenation and killing of bacteria in neutrophils (12). These data suggest that an auxiliary source of peroxide, in addition to dismutation of superoxide formed from the more widely studied NADPH oxidase, would be beneficial toward neutrophil killing activity. In mice lacking NADPH oxidase, we showed that Salmonella indeed senses an auxiliary source of peroxide stress that can be inhibited by CBIO, a specific small-molecule inhibitor of DAO. This DAO-catalyzed source of oxidative stress was biologically relevant because it elicited a larger response in DalS mutants and was able to better control growth of Salmonella dalS mutants that lack the d-alanine import system. In our in vitro experiments with purified DAO, exogenous d-alanine was added to elicit Salmonella killing by DAO, but d-alanine derived from bacteria was sufficient for killing by DAO in neutrophils. Neutrophils are a rich source of membrane-perturbing antimicrobial peptides, which might sensitize Salmonella to the effects of neutrophil DAO due to liberation of d-alanine during membrane damage. This is supported by the fact that neutrophil antimicrobial peptides act synergistically with peptidoglycan recognition proteins to kill bacteria (33). In addition, bacterial killing by purified DAO is likely mediated by the terminal hydrogen peroxide product that forms, requiring higher concentrations to reach toxic levels. However, DAO-derived hydrogen peroxide in neutrophils would be converted by MPO to more-toxic secondary products. Indeed, steady-state levels of hydrogen peroxide in the low micromolar range appear sufficient to support killing in neutrophil phagosomes (10).
Salmonella has been reported to actively disrupt the trafficking of NADPH oxidase to the Salmonella-containing vacuole (34), although the molecular basis for this observation has not been uncovered. The subversion by Salmonella of this additional oxidative stress might provide a more complete inhibition of oxidative killing in neutrophils, allowing Salmonella time to access more-permissive splenic macrophages at later stages of infection (15). As predicted by our model, the presence of DalS confers a selective advantage to Salmonella in wild-type mice, and it confers a higher selective advantage in NADPH oxidase-deficient mice, but it is dispensable when DAO is inhibited or if neutrophils are depleted. These data highlight a novel interaction between host and microbe that adds to the growing complexity of bacterial pathogenesis. In this context, evasion of neutrophil DAO is a virulence mechanism operating in conjunction with other mechanisms of immune subversion that each confer different but important fitness gains on the infecting pathogen.
Patients with chronic granulomatous disease (CGD) that lack NADPH oxidase activity are highly susceptible to a variety of bacterial infections. However, these patients are rarely infected with catalase-negative organisms (35), suggesting that a source of hydrogen peroxide stress independent of NADPH oxidase may be selective for certain infections in CGD patients (8). A proposed explanation for this was that the bacteria themselves generate enough hydrogen peroxide to mediate MPO-catalyzed halogenation. However, this idea has been challenged because catalase-negative bacteria are equally virulent in a CGD model system and produce less peroxide (approximately 2 orders of magnitude less) than CGD phagocytes with nonprotective levels of residual NADPH oxidase activity (36). Instead, others have argued that the ability of CGD neutrophils to kill even catalase-positive organisms suggests an incomplete loss of hydrogen peroxide and/or an alternate system of intracellular killing (37). Our results are consistent with both of these suggestions. These findings provide rationale for the targeted augmentation of DAO activity in certain immune deficiencies. For example, polyethylene glycol-conjugated DAO can restore bactericidal activity to congenitally defective CGD neutrophils in vitro (38). In this disease, even modest residual oxidative activity offers significant protection from severe illness and greater likelihood of long-term survival (39). Thus, augmenting oxidative killing mechanisms in neutrophils may have clinical benefit in some cases.
MATERIALS AND METHODS
Ethics statement.
All animal experiments were conducted according to guidelines set by the Canadian Council on Animal Care using protocols approved by the Animal Review Ethics Board at McMaster University.
Bacteria, cloning, and reagents.
All bacterial strains are isogenic derivatives of Salmonella enterica serovar Typhimurium strain SL1344 and are listed in Table S1 in the supplemental material. The mRNA encoding porcine d-amino acid oxidase (DAO) was amplified by PCR and cloned with a 6-histidine tag as previously described (7) using primers BRT139 (GGA ATT CCA TAT GCG TGT GGT GGT GAT TGG) and BRT140 (GGA AGA TCT TCA GTG GTG GTG GTG GTG GTG GAG GTG GGA TGG TGG CAT T). DNA was digested with BglII and NdeI, and pET3a was digested with BamHI and NdeI and then ligated. All cloning steps were confirmed by sequencing, and plasmids were transformed into Escherichia coli BL21(DE3). The OxyR-dependent ahpC-lux transcriptional fusion was generated as described previously with slight modifications (19). Three hundred base pairs upstream of the S. Typhimurium ahpC gene was amplified using primers BRT169 (CGG GAT CCG TAA TGT AGA GCG CAA CAC TT) and BRT171 (CGT ACG TAT ACT TCC TCC GTG TTT TCG TT) and cloned as a SnaBI/BamHI fragment in pGEN-luxCDABE.
Protein purification.
Purification of the six-histidine-tagged DalS (DalS-6HIS) was performed as described previously (13). Briefly, E. coli BL21(DE3) carrying pDalS-6HIS was grown in LB at 37°C and then induced with 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at 22°C for an additional 3 h. The cells were lysed by sonication, and soluble supernatant was loaded onto a nickel-nitrilotriacetic acid (Ni-NTA) bead column (Qiagen) and eluted with 80 mM imidazole. Recombinant DAO was purified by growing E. coli BL21(DE3) carrying pDAO-6HIS in LB at 37°C to an A600 of 0.55, and then the bacteria were induced with 0.1 mM IPTG and grown at 37°C for an additional 20 h. Cultures were centrifuged at 4,000 × g for 13 min at 12°C, resuspended in lysis buffer (20 mM Tris [pH 7.5], 0.5 M NaCl, protease inhibitors), and lysed via sonication (Misonix sonicator, Ultrasonic processor S-400, at 40% amplitude with 6 pulses of 30 s at 1-min intervals). Lysates were pelleted at 10,000 × g for 30 min at 4°C. Supernatant was loaded onto a Ni-NTA bead column and washed with an imidazole gradient (10, 20, and 40 mM) in Tris-buffered saline (TBS) (40 mM Tris [pH 7.5], 0.5 M NaCl). DAO-6HIS was eluted in TBS containing 80 to 320 mM imidazole, and its purity was determined by SDS-PAGE. Purified DAO was dialyzed in TBS, concentrated using 3K Amicon Ultra Centrifugal filters (catalog no. UFC800324; Millipore) and stored at −80°C.
Kinetic assays. (i) DAO production of hydrogen peroxide.
Enzymatic activity of DAO was measured by colorimetric assay using peroxidase-coupled oxidation of o-dianisidine as previously described (1). Briefly, DAO (5 µg/ml), 5 mM amino acid (d-alanine, d-serine, d-valine, and l-alanine; Sigma), horseradish peroxidase (6.6 µg/ml) (Sigma), and 6-chloro-1,2-benzisoxazol-3(2H)-one (CBIO) (0, 0.15, 1.5, and 15 µM) (Sigma) were incubated at room temperature in phosphate-buffered saline (PBS) containing o-dianisidine (75 µg/ml), and A460 was measured.
(ii) Estimation of d-alanine concentration.
S. Typhimurium was grown to mid-log phase in LPM medium (pH 5.8) (40) at 37°C. Bacteria were washed twice in phosphate-buffered saline, and 1 × 107 S. Typhimurium bacteria were incubated with 5 mM d-alanine for 3 h at 37°C in LPM medium. Cultures were pelleted at 10,000 g for 2 min. The supernatant was filter sterilized and incubated with DAO (5 µg/ml) and horseradish peroxidase (6.6 µg/ml) (Sigma) at room temperature in PBS containing 75 µg/ml o-dianisidine. A460 was measured after 1 h. The d-alanine concentration was determined by comparison to a standard curve.
(iii) FTS assay.
DalS binding was determined by fluorescent thermal shift (FTS) assay as described previously (13). Briefly, the assay was performed with a LightCycler 480 system (Roche) (498-nm excitation, 610-nm emission). Each reaction mixture contained DalS-6HIS (10 mM), SyPro Orange (5 µM) (Invitrogen), and the indicated amino acid or CBIO in a mixture of 100 mM HEPES and 150 mM NaCl (pH 7.5) in a 96-well plate (Roche). A change in the melting temperature (ΔTm) of 2°C upon the addition of ligand was considered positive binding.
Bactericidal activity assays with purified DAO.
S. Typhimurium was grown to mid-log phase in LPM medium at 37°C and washed twice in PBS. A total of 1 × 107 S. Typhimurium were incubated with 5 µg/ml DAO and 5 mM d-alanine in the presence or absence of 150 mM thiourea, 0.1% hydrogen peroxide, 0.01% HOCl, or 2.5 U/ml myeloperoxidase (MPO) in LPM medium at 37°C with shaking for 2 h. Cultures were serially diluted in PBS and plated on LB to determine killing activity. The number of CFU were determined and normalized to 0-h growth.
Isolation of mouse peritoneal neutrophils and bactericidal activity.
Female 6- to 10-week-old C57BL/6 mice (Charles River) were injected with 1 ml of 2% Biogel (Bio-Rad) in PBS. Neutrophils were harvested via peritoneal lavage 12 to 16 h later with 6 ml RPMI medium (10% fetal bovine serum [FBS], 1× HEPES, 1× sodium pyruvate, 1× β-mercaptoethanol, 1× essential amino acids). Cells were passed through a 40-µm cell strainer to remove Biogel, and purity was determined by Giemsa staining (Sigma). Approximately 2 × 107 neutrophils were obtained per mouse. Neutrophils were exposed to 3 µM CBIO in PBS or PBS alone at 37°C and 5% CO2 in RPMI medium for 60 min. Overnight cultures of wild-type S. Typhimurium and S. Typhimurium ΔdalS mutant with or without ahpC-luxCDABE were diluted in RPMI medium and added to neutrophils at a multiplicity of infection of 1:100 in tissue culture wells. Infected cells were incubated at 37°C and 5% CO2 in RPMI medium for 30 min to allow bacterial uptake. Infected cells were washed five times in PBS and incubated for an additional 90 min in RPMI medium or for 30 min for ahpC-luxCDABE assays. Luminescence was measured in bacteria containing the ahpC-lux reporter. Bacteria were washed five times with PBS and lysed with 250 µl of lysis buffer (1% Triton X-100, 0.1% SDS). Lysates were serially diluted and plated for CFU determination. Bacterial killing was determined as the ratio of CFU at 2 h to the CFU at 0.5 h and normalized to survival of wild-type bacteria.
Animal experiments.
For in vivo experiments, C57BL/6 (Charles River) or B6.129S6-CYBBtm1Din/J (Jackson Laboratory) mice were infected via the peritoneum with 2 × 105 Salmonella for competitive experiments as described previously (41). At 24 h (single infections) or 48 to 60 h (competitive infections) after infection, mice were sacrificed by cervical dislocation, and the livers and spleens were harvested. Organs were homogenized in PBS with a mixer mill (10 min, 30 Hz) (Retsch), serially diluted in PBS, and plated on LB containing streptomycin. Colonies were replica plated onto LB agar containing either chloramphenicol or streptomycin to determine the ratio of the wild type to mutant colonies. To induce neutropenia, mice were injected intraperitoneally (i.p.) with 0.15 mg anti-Ly6G clone 1A8 antibody (BioXcell) daily for two consecutive days prior to infection. To confirm neutropenia by flow cytometry, blood was mixed with acid-citrate-dextrose (ACD) anticoagulant and placed on ice. Cells were harvested by centrifugation at 5,000 × g for 10 min at 4°C. Red blood cells were eliminated with ACK lysis buffer (150 mM ammoniumchloride, 10 mM potassium hydrogen carbonate, 1 mM EDTA). The cells were first Fc blocked with anti-mouse CD16/CD32 (1:100; eBioscience) antibody and then stained with anti-mouse CD3e-PE-CF594 (anti-mouse CD3e labeled with phycoerythrin and CF594) (1:200; BD Biosciences), anti-mouse CD11b-PE (1:200; eBioscience), and anti-mouse Gr-1 APC-eFluor 780 (anti-mouse Gr-1 labeled with allophycocyanin and eFluor 780) (1:200; eBioscience) antibodies. All samples were run using an LSRII flow cytometer (BD Biosciences) and analyzed using FlowJo software (Tree Star, Inc.). For inhibition of DAO in vivo, mice were injected i.p. with 25 mg of CBIO/kg of body weight in 0.5% carboxymethyl cellulose (CMC) (Sigma) or 0.5% CMC only every 6 h during the course of infection.
Immunoblotting.
Neutropenic or immune replete C57BL/6 mice were infected with 2 × 105 S. Typhimurium in 0.1 M HEPES (pH 8.0) and 0.9% NaCl. The spleens, livers, and ceca were removed 48 h after infection and homogenized as described above in a solution of 50 µM Tris, 150 µM NaCl, and protease inhibitor cocktail (Roche), pH 7.5. Samples were centrifuged at 12,000 × g for 20 min at 4°C. Supernatants were diluted in an equal volume of SDS-PAGE sample buffer (100 mM Tris-HCl [pH 6.8], 20% [vol/vol] glycerol, 4% [wt/vol] SDS, 0.002% [wt/vol] bromophenol blue, and 100 mM dithiothreitol [DTT]) and boiled for 5 min. Five-microliter portions from the samples, including 5 µg/ml recombinant DAO labeled with histidine (rDAO-HIS) as a control, were separated on 12% polyacrylamide gels and blotted with goat anti-DAO (1:2,000; Sigma) and sheep anti-goat IgG-HRP (sheep anti-goat IgG labeled with horseradish peroxidase) (1:5,000; Abcam). Blots were stripped and reprobed using mouse anti-GAPDH (antibody against glyceraldehyde-3-phosphate dehydrogenase) (1:1,000; Novus Biologicals) and goat anti-mouse IgG-HRP (1:10,000; Cedarlane). Conjugated HRP was detected using chemiluminescence (Western Lightning Plus; PerkinElmer).
Bioluminescent reporter assay.
S. Typhimurium containing the PahpC-luxCDABE reporter was grown to mid-log phase in 96-well plates. Bacteria were treated with hydrogen peroxide (0, 2, 4, 6, 8, 10, and 100 µM), and A600 and luminescence were measured every 10 min. Luminescence was normalized to the A600.
In vivo bioluminescent imaging.
C57BL/6 or B6.129S6-CYBBtm1Din/J mice were treated with 25 mg/kg CBIO in 0.5% CMC or 0.5% CMC only and infected i.p. with 1 × 107 S. Typhimurium containing the ahpC-luxCDABE reporter 1 h after treatment. Mice were anesthetized (2% isoflurane carried in 2% oxygen) 2 h postinfection and imaged for 5 s in a Spectrum in vivo Imaging System (IVIS) (Caliper Life Sciences). The spleens were isolated and imaged ex vivo. Tissues were homogenized as described above, serially diluted, and plated on LB agar to obtain total CFU per organ. Total flux was normalized to tissue CFU.
Statistical analysis.
Treatment groups were compared using a nonparametric Mann-Whitney test. All analyses were performed using Graph Prism 4.0 (GraphPad Software Inc. San Diego, CA). A P value of 0.05 or less was considered significant.
SUPPLEMENTAL MATERIAL
Recombinant DAO produces hydrogen peroxide upon the addition of d-alanine. DAO-dependent oxidation of 0 to 20 mM d-alanine as measured by peroxidase-coupled oxidation of o-dianisidine. Data are the means with standard errors for three experiments. Download
The DAO inhibitor CBIO does not bind to DalS or impair bacterial growth. (A) Fluorescence thermal shift experiments were conducted with purified DalS without ligand and in the presence of d-alanine, d-valine, or CBIO as described in Materials and Methods. Data are the means with standard errors from three separate experiments. (B) Wild-type S. Typhimurium and dalS mutants were grown in the presence or absence of CBIO in the culture medium. Optical density was measured over time. Data are the means with standard errors from three separate experiments. Download
Flow cytometry verifying neutrophil depletion in neutropenic animals. The neutrophil gate was defined as CD3-CD11b+ Gr-1hi. Download
List of strains used in this study
ACKNOWLEDGMENTS
We are grateful to members of the Coombes lab for helpful discussions on this work, and we thank Gerry Wright, Eric Brown, and Tim Gilberger for comments on the manuscript.
This work was supported by the Canadian Institutes of Health Research (MOP 82704), the Canada Foundation for Innovation, and the Canada Research Chairs program (to B.K.C.). S.A.R.-Y. and B.R.T. are recipients of Ontario Graduate Scholarships. B.K.C. is the Canada Research Chair in Infectious Disease Pathogenesis.
B.R.T. designed and carried out the in vitro experiments and performed the in vivo experiments. S.A.R.-Y. assisted with neutrophil isolations, provided advice on depletion experiments, and did the fluorescence-activated cell sorting (FACS) analysis. B.K.C. designed experiments, analyzed data, wrote the paper, and acquired funding for the experiments.
We declare that we have no competing financial interests related to this work.
Footnotes
Citation Tuinema BR, Reid-Yu SA, Coombes BK. 2014. Salmonella evades d-amino acid oxidase to promote infection in neutrophils. mBio 5(6):e01886-14. doi:10.1128/mBio.01886-14.
REFERENCES
- 1. Sacchi S. 2013. d-Serine metabolism: new insights into the modulation of d-amino acid oxidase activity. Biochem. Soc. Trans. 41:1551–1556. 10.1042/BST20130184. [DOI] [PubMed] [Google Scholar]
- 2. Umhau S, Pollegioni L, Molla G, Diederichs K, Welte W, Pilone MS, Ghisla S. 2000. The X-ray structure of d-amino acid oxidase at very high resolution identifies the chemical mechanism of flavin-dependent substrate dehydrogenation. Proc. Natl. Acad. Sci. U. S. A. 97:12463–12468. 10.1073/pnas.97.23.12463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pollegioni L, Piubelli L, Sacchi S, Pilone MS, Molla G. 2007. Physiological functions of d-amino acid oxidases: from yeast to humans. Cell. Mol. Life Sci. 64:1373–1394. 10.1007/s00018-007-6558-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cline MJ, Lehrer RI. 1969. d-Amino acid oxidase in leukocytes: a possible d-amino-acid-linked antimicrobial system. Proc. Natl. Acad. Sci. U. S. A. 62:756–763. 10.1073/pnas.62.3.756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Robinson JM, Briggs RT, Karnovsky MJ. 1978. Localization of d-amino acid oxidase on the cell surface of human polymorphonuclear leukocytes. J. Cell Biol. 77:59–71. 10.1083/jcb.77.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lyle LR, Jutila JW. 1968. d-Amino acid oxidase induction in the kidneys of germ-free mice. J. Bacteriol. 96:606–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nakamura H, Fang J, Maeda H. 2012. Protective role of d-amino acid oxidase against Staphylococcus aureus infection. Infect. Immun. 80:1546–1553. 10.1128/IAI.06214-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Segal AW. 2005. How neutrophils kill microbes. Annu. Rev. Immunol. 23:197–223. 10.1146/annurev.immunol.23.021704.115653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lehrer RI, Hanifin J, Cline MJ. 1969. Defective bactericidal activity in myeloperoxidase-deficient human neutrophils. Nature 223:78–79. 10.1038/223078a0. [DOI] [PubMed] [Google Scholar]
- 10. Winterbourn CC, Hampton MB, Livesey JH, Kettle AJ. 2006. Modeling the reactions of superoxide and myeloperoxidase in the neutrophil phagosome: implications for microbial killing. J. Biol. Chem. 281:39860–39869. 10.1074/jbc.M605898200. [DOI] [PubMed] [Google Scholar]
- 11. Grisham MB, Jefferson MM, Melton DF, Thomas EL. 1984. Chlorination of endogenous amines by isolated neutrophils. Ammonia-dependent bactericidal, cytotoxic, and cytolytic activities of the chloramines. J. Biol. Chem. 259:10404–10413. [PubMed] [Google Scholar]
- 12. Rosen H, Crowley JR, Heinecke JW. 2002. Human neutrophils use the myeloperoxidase-hydrogen peroxide-chloride system to chlorinate but not nitrate bacterial proteins during phagocytosis. J. Biol. Chem. 277:30463–30468. 10.1074/jbc.M202331200. [DOI] [PubMed] [Google Scholar]
- 13. Osborne SE, Tuinema BR, Mok MC, Lau PS, Bui NK, Tomljenovic-Berube AM, Vollmer W, Zhang K, Junop M, Coombes BK. 2012. Characterization of DalS, an ATP-binding cassette transporter for d-alanine, and its role in pathogenesis in Salmonella enterica. J. Biol. Chem. 287:15242–15250. 10.1074/jbc.M112.348227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tomljenovic-Berube AM, Mulder DT, Whiteside MD, Brinkman FS, Coombes BK. 2010. Identification of the regulatory logic controlling Salmonella pathoadaptation by the SsrA-SsrB two-component system. PLoS Genet. 6:e1000875. 10.1371/journal.pgen.1000875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Burton NA, Schürmann N, Casse O, Steeb AK, Claudi B, Zankl J, Schmidt A, Bumann D. 2014. Disparate impact of oxidative host defenses determines the fate of Salmonella during systemic infection in mice. Cell Host Microbe 15:72–83. 10.1016/j.chom.2013.12.006. [DOI] [PubMed] [Google Scholar]
- 16. Imlay JA, Chin SM, Linn S. 1988. Toxic DNA damage by hydrogen peroxide through the Fenton reaction in vivo and in vitro. Science 240:640–642. 10.1126/science.2834821. [DOI] [PubMed] [Google Scholar]
- 17. Kohanski MA, Dwyer DJ, Hayete B, Lawrence CA, Collins JJ. 2007. A common mechanism of cellular death induced by bactericidal antibiotics. Cell 130:797–810. 10.1016/j.cell.2007.06.049. [DOI] [PubMed] [Google Scholar]
- 18. Christman MF, Morgan RW, Jacobson FS, Ames BN. 1985. Positive control of a regulon for defenses against oxidative stress and some heat-shock proteins in Salmonella typhimurium. Cell 41:753–762. 10.1016/S0092-8674(85)80056-8. [DOI] [PubMed] [Google Scholar]
- 19. Aussel L, Zhao W, Hébrard M, Guilhon AA, Viala JP, Henri S, Chasson L, Gorvel JP, Barras F, Méresse S. 2011. Salmonella detoxifying enzymes are sufficient to cope with the host oxidative burst. Mol. Microbiol. 80:628–640. 10.1111/j.1365-2958.2011.07611.x. [DOI] [PubMed] [Google Scholar]
- 20. Dunlap NE, Benjamin WH, Jr, Berry AK, Eldridge JH, Briles DE. 1992. A “safe-site” for Salmonella typhimurium is within splenic polymorphonuclear cells. Microb. Pathog. 13:181–190. 10.1016/0882-4010(92)90019-K. [DOI] [PubMed] [Google Scholar]
- 21. Geddes K, Cruz F, Heffron F. 2007. Analysis of cells targeted by Salmonella type III secretion in vivo. PLoS Pathog. 3:e196. 10.1371/journal.ppat.0030196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ferraris D, Duvall B, Ko YS, Thomas AG, Rojas C, Majer P, Hashimoto K, Tsukamoto T. 2008. Synthesis and biological evaluation of d-amino acid oxidase inhibitors. J. Med. Chem. 51:3357–3359. 10.1021/jm800200u. [DOI] [PubMed] [Google Scholar]
- 23. Konno R, Sasaki M, Asakura S, Fukui K, Enami J, Niwa A. 1997. d-Amino-acid oxidase is not present in the mouse liver. Biochim. Biophys. Acta 1335:173–181. 10.1016/S0304-4165(96)00136-5. [DOI] [PubMed] [Google Scholar]
- 24. D’Aniello A, D’Onofrio G, Pischetola M, D’Aniello G, Vetere A, Petrucelli L, Fisher GH. 1993. Biological role of d-amino acid oxidase and d-aspartate oxidase. Effects of d-amino acids. J. Biol. Chem. 268:26941–26949. [PubMed] [Google Scholar]
- 25. Gong N, Gao ZY, Wang YC, Li XY, Huang JL, Hashimoto K, Wang YX. 2011. A series of d-amino acid oxidase inhibitors specifically prevents and reverses formalin-induced tonic pain in rats. J. Pharmacol. Exp. Ther. 336:282–293. 10.1124/jpet.110.172353. [DOI] [PubMed] [Google Scholar]
- 26. Knodler LA, Vallance BA, Hensel M, Jäckel D, Finlay BB, Steele-Mortimer O. 2003. Salmonella type III effectors PipB and PipB2 are targeted to detergent-resistant microdomains on internal host cell membranes. Mol. Microbiol. 49:685–704. [DOI] [PubMed] [Google Scholar]
- 27. Winterbourn CC, Kettle AJ. 2013. Redox reactions and microbial killing in the neutrophil phagosome. Antioxid. Redox Signal. 18:642–660. 10.1089/ars.2012.4827. [DOI] [PubMed] [Google Scholar]
- 28. Nauseef WM. 2008. Nox enzymes in immune cells. Semin. Immunopathol. 30:195–208. 10.1007/s00281-008-0117-4. [DOI] [PubMed] [Google Scholar]
- 29. Loetscher Y, Wieser A, Lengefeld J, Kaiser P, Schubert S, Heikenwalder M, Hardt WD, Stecher B. 2012. Salmonella transiently reside in luminal neutrophils in the inflamed gut. PLoS One 7:e34812. 10.1371/journal.pone.0034812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mayadas TN, Cullere X, Lowell CA. 2014. The multifaceted functions of neutrophils. Annu. Rev. Pathol. 9:181–218. 10.1146/annurev-pathol-020712-164023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cheminay C, Chakravortty D, Hensel M. 2004. Role of neutrophils in murine salmonellosis. Infect. Immun. 72:468–477. 10.1128/IAI.72.1.468-477.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Amulic B, Cazalet C, Hayes GL, Metzler KD, Zychlinsky A. 2012. Neutrophil function: from mechanisms to disease. Annu. Rev. Immunol. 30:459–489. 10.1146/annurev-immunol-020711-074942. [DOI] [PubMed] [Google Scholar]
- 33. Royet J, Dziarski R. 2007. Peptidoglycan recognition proteins: pleiotropic sensors and effectors of antimicrobial defences. Nat. Rev. Microbiol. 5:264–277. 10.1038/nrmicro1620. [DOI] [PubMed] [Google Scholar]
- 34. Vazquez-Torres A, Xu Y, Jones-Carson J, Holden DW, Lucia SM, Dinauer MC, Mastroeni P, Fang FC. 2000. Salmonella pathogenicity island 2-dependent evasion of the phagocyte NADPH oxidase. Science 287:1655–1658. 10.1126/science.287.5458.1655. [DOI] [PubMed] [Google Scholar]
- 35. Winkelstein JA, Marino MC, Johnston RB, Jr, Boyle J, Curnutte J, Gallin JI, Malech HL, Holland SM, Ochs H, Quie P, Buckley RH, Foster CB, Chanock SJ, Dickler H. 2000. Chronic granulomatous disease. Report on a national registry of 368 patients. Medicine (Baltimore) 79:155–169. 10.1097/00005792-200005000-00003. [DOI] [PubMed] [Google Scholar]
- 36. Messina CG, Reeves EP, Roes J, Segal AW. 2002. Catalase negative Staphylococcus aureus retain virulence in mouse model of chronic granulomatous disease. FEBS Lett. 518:107–110. 10.1016/S0014-5793(02)02658-3. [DOI] [PubMed] [Google Scholar]
- 37. Pitt J, Bernheimer HP. 1974. Role of peroxide in phagocytic killing of pneumococci. Infect. Immun. 9:48–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nakamura H, Fang J, Mizukami T, Nunoi H, Maeda H. 2012. PEGylated d-amino acid oxidase restores bactericidal activity of neutrophils in chronic granulomatous disease via hypochlorite. Exp. Biol. Med. (Maywood) 237:703–708. 10.1258/ebm.2012.011360. [DOI] [PubMed] [Google Scholar]
- 39. Kuhns DB, Alvord WG, Heller T, Feld JJ, Pike KM, Marciano BE, Uzel G, DeRavin SS, Priel DA, Soule BP, Zarember KA, Malech HL, Holland SM, Gallin JI. 2010. Residual NADPH oxidase and survival in chronic granulomatous disease. N. Engl. J. Med. 363:2600–2610. 10.1056/NEJMoa1007097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Coombes BK, Brown NF, Valdez Y, Brumell JH, Finlay BB. 2004. Expression and secretion of Salmonella pathogenicity island-2 virulence genes in response to acidification exhibit differential requirements of a functional type III secretion apparatus and SsaL. J. Biol. Chem. 279:49804–49815. 10.1074/jbc.M404299200. [DOI] [PubMed] [Google Scholar]
- 41. Coombes BK, Wickham ME, Lowden MJ, Brown NF, Finlay BB. 2005. Negative regulation of Salmonella pathogenicity island 2 is required for contextual control of virulence during typhoid. Proc. Natl. Acad. Sci. U. S. A. 102:17460–17465. 10.1073/pnas.0505401102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Recombinant DAO produces hydrogen peroxide upon the addition of d-alanine. DAO-dependent oxidation of 0 to 20 mM d-alanine as measured by peroxidase-coupled oxidation of o-dianisidine. Data are the means with standard errors for three experiments. Download
The DAO inhibitor CBIO does not bind to DalS or impair bacterial growth. (A) Fluorescence thermal shift experiments were conducted with purified DalS without ligand and in the presence of d-alanine, d-valine, or CBIO as described in Materials and Methods. Data are the means with standard errors from three separate experiments. (B) Wild-type S. Typhimurium and dalS mutants were grown in the presence or absence of CBIO in the culture medium. Optical density was measured over time. Data are the means with standard errors from three separate experiments. Download
Flow cytometry verifying neutrophil depletion in neutropenic animals. The neutrophil gate was defined as CD3-CD11b+ Gr-1hi. Download
List of strains used in this study