Abstract
The rodent incisor is one of a number of organs that grow continuously throughout the life of an animal. Continuous growth of the incisor arose as an evolutionary adaptation to compensate for abrasion at the distal end of the tooth. The sustained turnover of cells that deposit the mineralized dental tissues is made possible by epithelial and mesenchymal stem cells residing at the proximal end of the incisor. A complex network of signaling pathways and transcription factors regulates the formation, maintenance, and differentiation of these stem cells during development and throughout adulthood. Research over the past 15 years has led to significant progress in our understanding of this network, which includes FGF, BMP, Notch, and Hh signaling, as well as cell adhesion molecules and microRNAs. This review surveys key historical experiments that laid the foundation of the field and discusses more recent findings that definitively identified the stem cell population, elucidated the regulatory network, and demonstrated possible genetic mechanisms for the evolution of continuously growing teeth.
Keywords: hypselodont, tissue regeneration, tooth, dental, renewal
In 1768, the year James Cook boarded the HMS Endeavour on his first voyage of discovery, a French naturalist named Auguste Fougeroux documented a finding of his own. He noted in Observations Anatomiques that the teeth of a rabbit, unlike those of humans, grow continuously (Fougeroux de Bondaroy, 1768). This intriguing phenomenon was experimentally confirmed some 40 years later by Oudet, who cut off rabbit incisors at the gingival (or gum) level and found that these teeth indeed regenerated (Oudet, 1823). These first steps by Fougeroux and Oudet laid the foundation for the discovery two centuries later that the continuous growth of incisors in rabbits and rodents is fueled by adult stem cells that reside in the proximal end of the tooth and generate all necessary cell types throughout the animal’s life.
Over the past several years, the adult mouse incisor has emerged as an attractive model system for the study of adult stem cells. Such cells are present in many different organs and are required for homeostasis as well as injury repair. Studies using mouse genetics, as well as other experimental approaches such as explant cultures, have deepened our understanding of the signaling pathways and genetic networks that are involved in the formation and the renewal of the rodent incisor. Here, we review the current state of the field of incisor stem cells.
The mouse incisor as a model system for stem cell biology
Teeth consist of three parts – crowns, roots, and supporting structures – and they are anchored in maxillary and mandibular bones by periodontal ligaments. These ligaments extend from the bone and insert into the outermost layer of the tooth root, called cementum. The crown of the tooth is exposed to the oral cavity and provides masticatory function. It is covered by the hardest substance in the body, enamel, which is produced by the epithelially-derived ameloblasts. Underneath enamel is dentin, which is laid down by the odontoblasts of mesenchymal origin. Dentin encloses the dental pulp, which contains the neurovascular bundle of the tooth. In the root portion of the tooth, dentin is covered by cementum.
There is a great diversity among mammals in terms of the number and shape of teeth. Humans possess 20 primary teeth and 32 adult teeth; the adult teeth are comprised of 8 incisors, 4 canines, 8 premolars, and 12 molars. The primary teeth appear at around 6 months of age and are fully shed by the early teen years. Once the tooth erupts into the oral cavity, the dental epithelial tissue is lost, such that adult human teeth lose the potential to regenerate enamel, and the remaining mesenchymal tissues have only a limited capacity to regenerate dentin, cementum, and pulp. In contrast, mice, which are an important and commonly used model for investigation of tooth development, exhibit a highly specialized dentition. They possess 4 incisors and 12 molars, which are separated by a toothless area called the diastema.
All rodents, including mice, have incisors that grow throughout their lifetime, and this growth is counterbalanced by continuous wear. The continuous formation of enamel and dentin is made possible by the presence of active adult epithelial and mesenchymal stem cells. The epithelial stem cells, which are the principal focus of this review, reside in a niche called the cervical loop; the mesenchymal stem cells in the dental pulp are not yet as well characterized as their epithelial counterparts.
Identification of incisor epithelial stem cells
With the emergence of comparative anatomy in the late 1800s, it was concluded that continuous incisor growth is common to all extant species of glires (rodents and lagomorphs) (Cope, 1888), and the advent of histological and microscopic techniques in the early 20th century allowed for closer scrutiny of the incisors of these species (Addison, 1915). These early studies suggested that the constant supply of enamel was provided by cells residing in the proximal soft tissue, which was called the “enamel organ”.
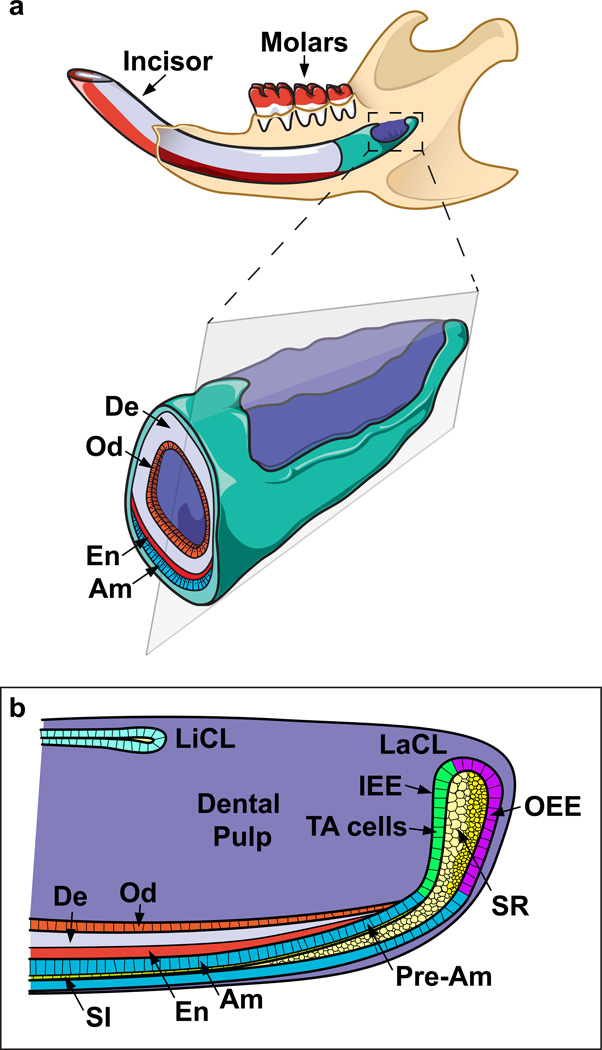
The initial studies of incisor growth utilized mechanical demarcations via cuts along the erupted enamel. These enabled observation of tooth renewal as well as rough measurements of the growth rate (Addison, 1915). Later investigations using tritiated thymidine autoradiography showed that the mouse incisor grows at the rate of ~365 microns per day (Smith and Warshawsky, 1975a). These were followed by more extensive histological studies (Warshawsky and Smith, 1974; Smith and Warshawsky, 1975b; Moe et al., 1979) that provided detailed descriptions of the morphological structures of the enamel organ and identified the labial and lingual cervical loops (liCL and laCL). The laCL was postulated to house cells that would give rise to enamel-secreting ameloblasts. These pioneering studies made important contributions to the modern understanding of ameloblast development and maturation and led to the later discovery that dental epithelial stem cells reside in the stellate reticulum (SR) and outer enamel epithelium (OEE) of the laCL. These stem cells give rise to the rapidly dividing transit-amplifying (TA) cells, which further differentiate into pre-ameloblasts (Figure 1). In turn, those cells give rise to mature ameloblasts as they progress distally through the proximal presecretory, secretory, and maturation zones (Warshawsky and Smith, 1974). On the other hand, when compared to the laCL, the liCL is relatively small in size, has less proliferating cells, and does not contain cells that normally form ameloblasts. Thus, the enamel is distributed asymmetrically, as it is only present on the labial surface of the tooth, in contrast to the softer dentin, which wraps around the entire incisor. This creates a difference in hardness between the outer and inner surfaces of the tooth, and it enables abrasion through mastication to primarily file down the inner (dentin) side of incisor, generating a sharp tip.
Figure 1. Schematics of the rodent jaw and incisor cervical loops.
(a) The incisor is mostly embedded in the jawbone, and the stem cell compartment is located at the proximal end of the tooth. The stem cells continuously supply ameloblasts (Am) and odontoblasts (Od), cells that are responsible for replacing the worn enamel (En) and dentin (De), respectively. (b) The incisor epithelial stem cells are located in the outer enamel epithelium (OEE) and stellate reticulum (SR) of the labial cervical loop (laCL), as shown in this diagram of a sagittal section of the proximal incisor. These cells are quiescent but give rise to transit-amplifying (TA) cells in the inner enamel epithelium (IEE) that undergo massive proliferation. The TA cells in turn give rise to pre-ameloblasts (Pre-Am) and subsequently the fully differentiated ameloblasts, as well as cells in the stratum intermedium (Si). Two morphologically distinct cell types can be identified in the SR, one adjacent to the OEE and the other adjacent to the TA cells. Compared to the laCL, the lingual cervical loop (liCL) is smaller and does not normally give rise to ameloblasts. Finally, mesenchymal stem cells that generate odontoblasts are believed to reside in the proximal dental pulp.
Analysis of the mitotic activity of epithelial cells in different regions of the incisor further solidified the evidence for an active population of progenitor cells. Tritiated thymidine labeling revealed that cells within the adult rat enamel organ first proliferated and subsequently became post-mitotic as they exited the presecretory zone and moved distally (Smith and Warshawsky, 1975a). These early studies also suggested the possibility of several stem cell types within the cervical loop, as discussed later. Smith (1980) postulated that distinct compartments may exist within the OEE, inner enamel epithelium (IEE), and SR. Treatment of animals with mitotic arrest agents, such as vinblastine, resulted in decreased velocity of ameloblast migration along the incisor, hinting that proper incisor growth and cell movement require proliferation (Samperiz et al., 1985). The next major advance came in the 1990s, when molecular techniques provided clues about the signaling cascades regulating the stem cell niche and identified potential markers for dental stem cells (Harada et al., 1999). When BrdU (5-bromo-2'-deoxyuridine) was pulsed in cultured explants and followed by a prolonged chase, label-retaining cells (LRCs) were found. This result was intriguing, as in a number of tissues the stem cells divide infrequently, and thus nuclear labeling is maintained for extended periods without dilution (Fuchs, 2009). BrdU LRCs were subsequently identified in vivo in the SR and OEE 60 days after BrdU was injected into postnatal day 0 (P0) pups (Seidel et al., 2010), strengthening the idea that stem cells reside in these compartments. Similar results were obtained using a genetic system, in which the expression of a tetracycline operator regulated transgene that encodes a GFP tagged nucleosomal Histone 2B protein (H2B-GFP) is controlled in the dental epithelium with a keratin 5 promoter driven tetracycline activator (K5-tTA). These studies found LRCs in the same region 2 months after the cessation of H2B-GFP induction (Li et al., 2012; Seidel et al., 2010), pointing to a group of slow cycling cells in the rodent incisor that were likely to be stem cells.
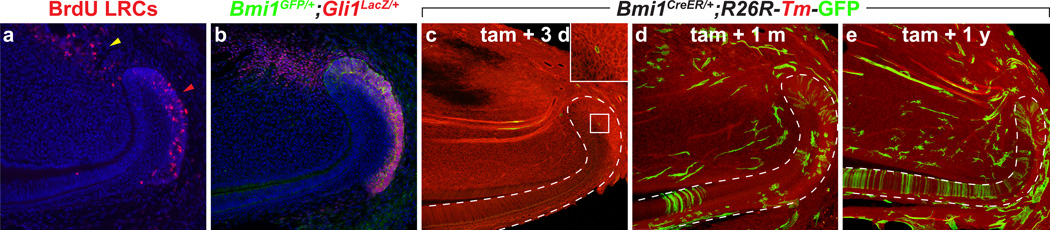
Definitive identification of stem cells requires either lineage-tracing or transplantation experiments. Historically, candidate stem cell markers were chosen based on a number of criteria, such as being targets of major signaling pathways that are active within the putative stem cell niche (Joyner and Zervas, 2006). Recently, several stem cell markers in the cervical loop were identified using in vivo lineage tracing. The first such marker found, the sonic hedgehog (Shh) responsive gene Gli1, is expressed in LRCs, and lineage tracing showed that Gli1-expressing cells give rise to differentiated ameloblasts and cells in the stratum intermedium (SI). This study used the Gli1CreER transgene, which can be conditionally induced by tamoxifen to activate the expression of Cre-responsive reporters (Seidel et al., 2010). The next two markers, Bmi1 and Sox2, were identified based on their expression in other stem cell systems, including the brain, blood, intestine, and several other organs (Molofsky et al., 2003; Park et al., 2003; Sangiorgi and Capecchi, 2008; Suh et al., 2007; Arnold et al., 2011). Bmi1 expression in the adult incisor coincides with the BrdU label-retention domain and with Gli1 expression (Figure 2). Bmi1-expressing cells also generate progeny that populate the differentiated ameloblasts (Biehs et al., 2013). Functionally, Bmi1 plays a critical role in promoting proliferation and self-renewal by suppressing the expression of Ink4a/Arf, a locus that encodes the cell cycle inhibitors p16 and p19. Importantly, Bmi1 also actively prevents inappropriate differentiation by inhibiting the expression of Hox genes. As a result, in the absence of Bmi1, stem cell self-renewal decreases, resulting in the loss of enamel deposition over time. Sox2 also marks incisor epithelial stem cells, although its expression in the P2 mouse incisor differs from that of Gli1 and Bmi1 in the adults, as the domain of expression is broader, extending anteriorly towards the TA region and into the SR (Juuri et al., 2012). These expression differences may be due to discrepancies in the stages when the samples were examined, or they may reflect the presence of distinct stem cell subpopulations in the cervical loop, similar to what has been reported in the stem cell niche of the gut (Tian et al., 2011). Nonetheless, Sox2-expressing cells contribute to all epithelial lineages in the incisor and may do so by migrating from the SR directly to the IEE and TA regions, as opposed to Gli1 and Bmi1 expressing cells, which appear to populate the OEE. Finally, Sox2 expression is induced by FGF8 signaling and further fine-tuned by microRNAs (miRNAs). This finding, as discussed in the next section, is consistent with a number of studies showing that incisor stem cells are regulated by an intricate network of signaling pathways, transcription factors, and miRNAs.
Figure 2. Bmi1 and Gli1 mark incisor stem cells.
(a and b) BrdU label retaining cells (LRCs) reside in the OEE and SR of the laCL (red arrow head) and the proximal mesenchyme (yellow arrow head). The same population also expresses Bmi1 (green) and Gli1 (red). DAPI is blue. (c–e) Bmi1CreER;R26R-Tm-GFP mice were induced at 6 weeks with tamoxifen and chased for the indicated time period to lineage trace Bmi1 expressing stem cells, which give rise to ameloblasts. Cells that have undergone recombination are green and non-Bmi1 lineage is red. Mesenchymal Bmi1 expressing cells also give rise to progeny.
Development and regulation of incisor epithelial stem cells
The formation of the incisor progresses through comparable stages as the well-studied molar, and many of the molecules that govern molar development similarly regulate incisor formation (Tummers and Thesleff, 2009). This process is characterized by reciprocal epithelial-mesenchymal interactions, which are widely used in the development of many organs, such as the limb and the skin appendages (Fuchs and Horsley, 2008; Zeller et al., 2009). The mammalian dental epithelium originates from oral ectoderm, while the mesenchyme derives from cranial neural crest, which arises from the margins of the neuroepithelium and undergoes a ventral-lateral migration. Neural crest-derived mesenchyme forms many dental tissues, such as dentin, pulp, and periodontal ligaments, whereas the ectoderm-derived epithelium mainly produces the enamel-secreting ameloblasts and their supporting cells (Tucker and Sharpe, 2004; Chai et al., 2000; Rothova et al., 2012).
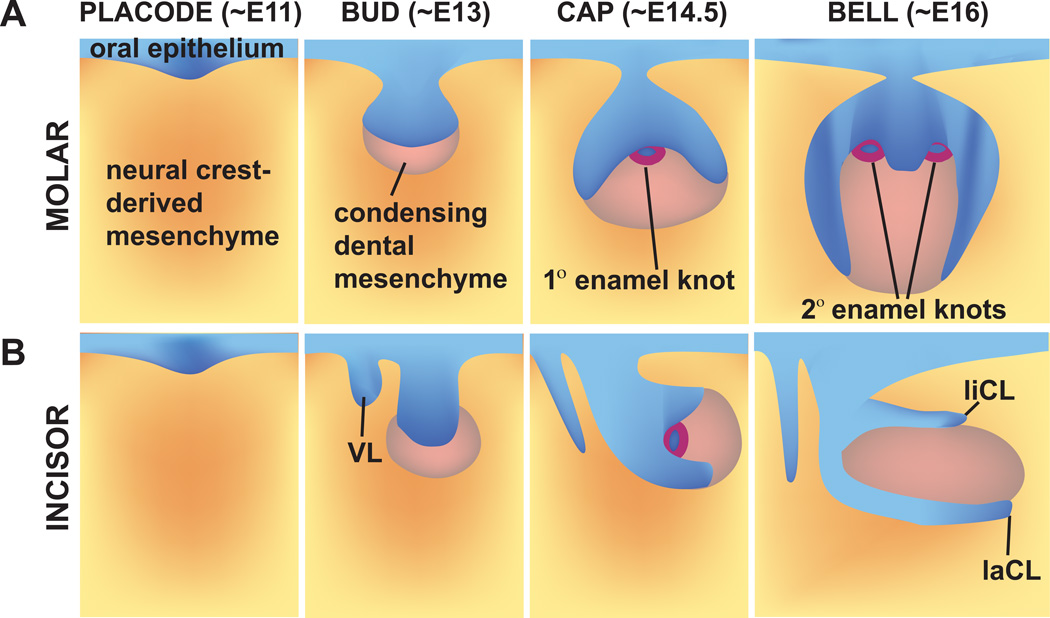
In mice and mouse-like rodents (e.g., rats and voles), tooth development begins between embryonic day (E) 8.5–10. At that time, the first signaling molecules involved in development are expressed, followed by the thickening of the epithelium into a placode; in mice, this occurs at E11. Subsequently, a series of developmental stages called bud, cap, and bell occur (Figure 3). During the bud stage (E12.5–13.5 in mice), the epithelium begins to invaginate into the underlying mesenchyme, which responds by condensing to form the dental papilla. During the transition from bud to cap stage, a transient signaling center called the enamel knot (EK), characterized by expression of several secreted factors, forms.
Figure 3. Schematic depiction of mouse molar (a) and incisor (b) tooth development.
Tooth development begins with thickening and invagination of the oral epithelium into the underlying mesenchyme at ~E11. At the bud stage (~E13), the mesenchyme condenses. At the cap stage (~E14.5), the enamel knot, a central signaling center, appears. At the bell stage (~E16), the secondary enamel knots, corresponding to the future location of cusps, form. In addition, the extracellular matrices of enamel and dentin are excreted by the differentiating ameloblasts and odontoblasts, respectively. Tooth development is similar in the incisor and molar, with a few key differences being the proximal-distal “rotation” of the incisor at the bell stage, as well as the absence of secondary enamel knots in the incisor. VL, vestibular lamina.
The cap stage (E14–15.5 in mice) is characterized by further epithelial and mesenchymal proliferation. The proliferating dental epithelium surrounds the dental papilla, and the mesenchyme proliferates and forms the dental follicle. During the late cap stage, significant morphologic differences between the developing incisor and molar emerge, as the developing incisor begins to grow parallel to the long axis of the jaw. During the bell stage (E16–E18 in mice), the final tooth shape and the tooth-specific cell types become apparent. The primary EKs are replaced by secondary EKs in the molars, which correspond to locations and numbers of future tooth cusps (Jernvall et al., 1994). Thus, monocuspid teeth like incisors and canines exhibit only one primary EK, whereas the multicuspid premolars and molars exhibit several both a primary EK as well as secondary EKs.
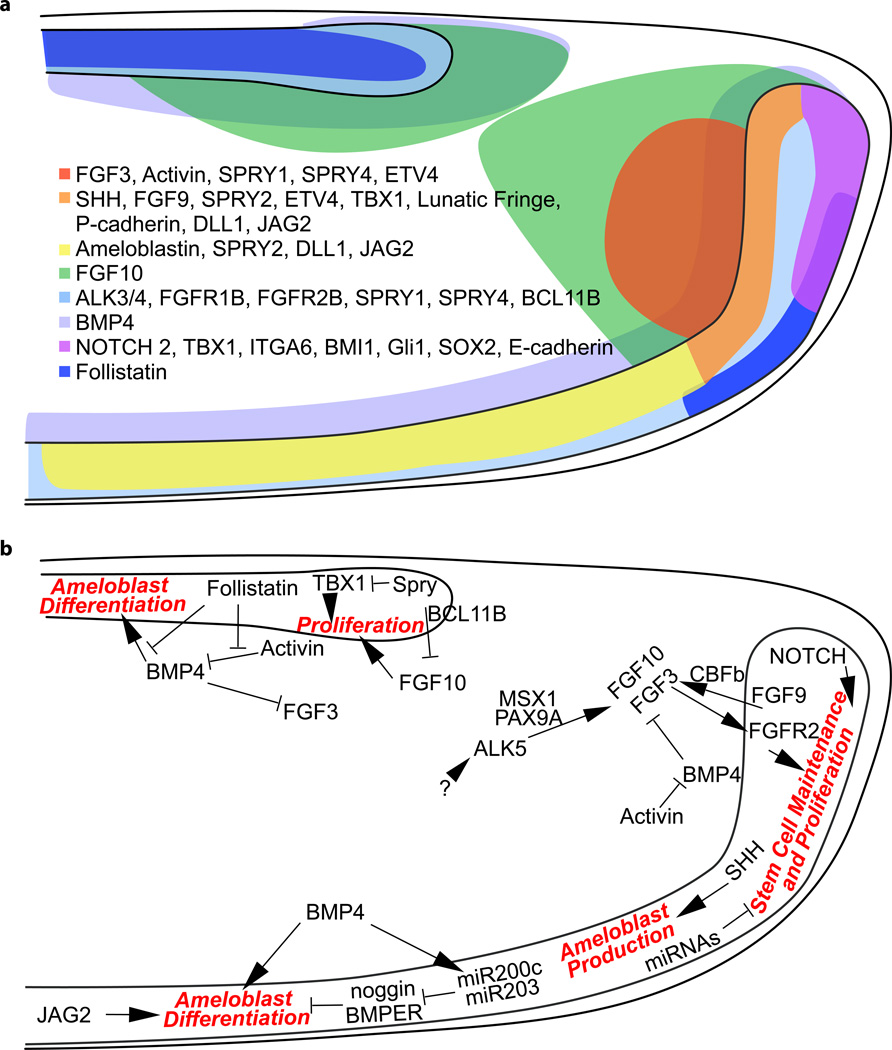
Several signaling pathways are critical for the formation and maintenance of the incisor epithelial stem cells both during development and in adults (Figure 4). Among the best studied of these is FGF signaling. During incisor organogenesis, Fgf3 and Fgf10 initially have overlapping expression in the dental papilla (the condensing mesenchyme) through E14 in the cap stage incisor bud (Harada et al., 2002). From E16 (early bell stage) onwards to adulthood, Fgf10 is expressed in the mesenchyme neighboring both the lingual and labial IEE of the forming cervical loops, which express the FGF receptor genes Fgfr1b and Fgfr2b. Fgf3 expression, in contrast, is only present in the mesenchyme adjacent to the labial IEE (Harada et al., 1999, 2002; Kettunen et al., 2000; Wang et al., 2007). Functionally, these mesenchymally-expressed FGFs, although not required for the early differentiation of ameloblasts during development, are crucial for the proliferation and survival of epithelial stem cells in the developing cervical loops (Harada et al., 2002; Wang et al., 2007). This is evident in the Fgf10−/− knockout embryos, where the cervical loop forms initially but eventually regresses due to reduced growth and increased apoptosis (Harada et al., 2002). Teeth in Fgf3−/− mice, on the other hand, are largely normal, probably due to the redundant function of Fgf10. However, Fgf3−/−;Fgf10+/− compound mutants exhibit a severely hypoplastic laCL and reduced enamel formation, indicating that the levels of FGF signaling are important for the maintenance of the incisor epithelial stem cell pool (Wang et al., 2007). Consistent with this result, FGF receptor 2(IIIb) null mice have no discernible incisors at birth (De Moerlooze et al., 2000). Conditional removal of Fgfr2 or misexpression of a dominant negative FGFR2b in the incisor epithelium demonstrated that FGF signaling acts tissue-specifically to maintain the epithelial stem cell niche both during development and adulthood (Lin et al., 2009; Parsa et al., 2010). Additional FGFs, such as Fgf9, are expressed in the incisor epithelium during development (Porntaveetus et al., 2011; Kettunen and Thesleff, 1998) and may play key roles in initially activating the mesenchymal expression of FGFs (Wang et al., 2007; Bei and Maas, 1998). Supporting this view, genetic ablation of the core binding factor β, which binds to Runx transcription factors and is required for the epithelial expression of Fgf9, abrogated the expression of Fgf3 and Fgf10 in the developing dental mesenchyme (Kurosaka et al., 2011).
Figure 4.
(a) Expression pattern of signaling molecules and transcription factors that regulate the formation and maintenance of the cervical loops. (b) The regulatory network for incisor stem cells. Key cellular processes (red), including stem cell maintenance, proliferation and differentiation, are regulated by an intricate network of signaling molecules and transcription factors, which either promote (as indicated by→) or inhibit (as indicated by —|) the downstream events, and ultimately maintain the homeostasis of the stem cell niche.
Several studies also highlighted the importance of spatially and quantitatively balanced FGF signaling in the incisor for maintenance of the asymmetry of the organ, in which ameloblasts and enamel are restricted to the labial side. One important class of FGF regulators is the intracellular antagonists encoded by the Sprouty genes (Spry1, 2, and 4), which are expressed in both the lingual and labial epithelia as well as the adjacent mesenchyme (Klein et al., 2008). When Sprouty genes are deleted and the inhibitory signal is removed, such as in Spry4−/−;Spry2+/− mutants, cells in both lingual and labial epithelia and mesenchyme exhibit increased sensitivity to FGF signaling. This leads to ectopic mesenchymal Fgf3 and Fgf10 expression and the formation of lingual ameloblasts (Klein et al., 2008). The Sprouty proteins may act in part through indirect regulation of the transcription factors, TBX1 and BCL11B, which are up- and downregulated respectively in the Spry4−/−;Spry2+/− liCL at E16.5 (Caton et al., 2009; Kyrylkova et al., 2012). Functional studies demonstrated that TBX1 promotes incisor epithelial proliferation by inhibiting PITX2 transcriptional activity, which is in turn responsible for the expression of p21, an inhibitor of cell cycling (Cao et al., 2010a). Thus, Tbx1 null incisors cultured in kidney capsules were hypoplastic and devoid of enamel (Caton et al., 2009). On the other hand, removal of Bcl11b resulted in an inverted Fgf3/10 expression pattern in the lingual and labial mesenchyme at E16.5, leading to expansion of the liCL and formation of lingual ameloblasts, accompanied by a reduction in size of the laCL and abnormal labial ameloblast development (Kyrylkova et al., 2012). Lastly, a hypomorphic allele of Bcl11b promoted adult TA cell proliferation and maintained epithelial stem cell number, although whether this occurred in an Fgf3-dependent or independent mechanism is not clear (Katsuragi et al., 2013).
FGF activity and the regulation of incisor asymmetry are modulated by two TGFβ family molecules, BMP4 and Activin, during incisor development. BMP4 is expressed symmetrically throughout the dental mesenchyme and indirectly represses Fgf3 (but not Fgf10) expression in the lingual mesenchyme. Activin is expressed more robustly in the labial mesenchyme, and bead implantation studies in E16 incisor explants demonstrated that Activin counteracts the effect of BMP4 (Wang et al., 2007) This ensures the maintenance of Fgf3 expression in the labial mesenchyme and thus promotes stem cell proliferation (Wang et al., 2007). Furthermore, the residual Activin activity on the lingual side is antagonized by Follistatin (encoded by Fst), which is expressed in the lingual dental epithelium and preserves the inhibitory effect of BMP4 on lingual Fgf3 expression. Consequently, Fst null embryos develop ectopic Fgf3 expression in the lingual mesenchyme, which leads to the expansion of the liCL and the formation of lingual ameloblasts and enamel (Wang et al., 2007). Conversely, misexpression of Fst throughout the dental epithelium caused reduced Fgf3 expression and a subsequent reduction in proliferation and laCL size (Wang et al., 2007). BMP4 also promotes ameloblast differentiation in the more distal labial epithelium, and this is inhibited in the lingual epithelium by locally expressed Follistatin to maintain incisor asymmetry (Wang et al., 2004). Consistent with the idea that BMP4 functions in two regions of the developing incisor, misexpression of the BMP inhibitor Noggin resulted in overgrowth of incisors due to increased proliferation in the progenitor cell population in the cervical loop. However, the mutant incisors did not form enamel, as ameloblast differentiation that is normally induced by BMP signals was prevented (Plikus et al., 2005). Finally, TGFβ receptor type I (Alk5/Tgfbr1) in the dental mesenchyme regulates proper tooth initiation and development of the incisor epithelium (Zhao et al., 2008, 2011). When Alk5 was deleted specifically in the dental mesenchyme, the expression of Fgf3 and Fgf10 was downregulated in the mesenchyme, leading to reduced proliferation and fewer LRCs in the cervical loop, which could be rescued by exogenous FGF10 in incisor explant culture (Zhao et al., 2011). The activation of mesenchymal Fgf expression occurs at least in part through the transcription factors MSX1 and PAX9, which interact to initiate the expression of Fgf3 and Fgf10 at E12.5, and thus the subsequent formation of the incisors (Zhao et al., 2008; Bei and Maas, 1998; Nakatomi et al., 2010).
In addition to FGF and TGF-β/BMP signaling, signaling through the Hedgehog (Hh) pathway is an important regulator of incisor epithelial stem cells in adult mice. As mentioned earlier, Gli1 positive Hh-responsive cells were found in both the epithelial stem cell compartment and the dental pulp. These cells respond to Sonic hedgehog (SHH) that is expressed in the TA cells and maturing ameloblasts (Bitgood and McMahon, 1995; Seidel et al., 2010). Blocking of the pathway in vivo using a Smoothened inhibitor demonstrated that Hh signaling is required for the continuous generation of ameloblasts in the adult (Seidel et al., 2010). The expression of Shh itself appears to be partially regulated by epithelial Fgf9, at least during development. In this context, ectopic FGF9 suppressed Shh expression, while Fgf9 deficiency resulted in a proximal shift of the Shh expressing domain (Kurosaka et al., 2011). Interestingly, this parallels some aspects of the developing limb, where FGF dependent Etv4/5 is required to limit the expression of Shh in the posterior limb bud mesenchyme and suppress Shh activity anteriorly (Mao et al., 2009; Zhang et al., 2009). It is not yet known whether Etv family proteins have similar functions in the developing mouse incisor.
Members of the Notch signaling pathway demonstrate intriguing expression patterns in the incisor, and several studies have pointed to a role for these genes in regulation of the stem cells. In mammals, three Notch receptors (Notch 1–3) are expressed in the newborn mouse incisor. Whereas Notch 1 expression is restricted to the SR, Notch 2 is expressed in the outer OEE and the underlying SR (Harada et al., 1999). Notch 3, on the other hand, was reported to be expressed only in the dental mesenchyme (Harada et al., 1999), although cervical loop expression has also been reported (Mitsiadis et al., 1998). The genes encoding the Notch ligands, Delta-like 1 (Dll1) and Jagged 2 (Jag2), as well as Lunatic fringe (Lfng), encoding the β1–3 N-acetylglucosamine transferase that modifies Notch receptors, are also expressed in the cervical loop dental epithelium (Harada et al., 1999; Mitsiadis et al., 1998, 2010). The expression of Jag2 and Lfng is regulated by FGF and BMP signaling (Harada et al., 1999; Mitsiadis et al., 2010). The Notch responsive gene, Hes1, is expressed in the SR, and when cervical loops dissected from newborn mice were cultured with the Notch signaling inhibitor, DAPT, they had reduced proliferation and increased apoptosis, leading to an overall reduction in size (Felszeghy et al., 2010). Furthermore, deletion of Jag2 in mice resulted in abnormal odontoblast and ameloblast differentiation, as well as the loss of matrix deposition due to downregulated Tbx1 expression (Mitsiadis et al., 2010). Finally, experiments using the rat cervical loop derived dental epithelial cell line, HAT-7, showed that Notch signaling may play a role in regulating the differentiation of SI from IEE (Harada et al., 2006; Kawano et al., 2002). In this context, both the inclusion of Jagged1 protein in culture and overexpression of the Notch1 internal domain resulted in an increase of SI cells, whereas a neutralizing Jagged1 antibody inhibited their differentiation (Harada et al., 2006).
The aforementioned signaling events positively regulate the incisor stem cells. In contrast, initial studies of the Wnt pathway have proposed that signaling inhibition, rather than activation, takes place during incisor renewal. Despite the presence of several Wnt ligands in both the epithelium and mesenchyme of the cervical loop at E16.5 and E18.5, canonical Wnt reporter (BATgal and TOPgal) signals are absent, and the Wnt responsive gene, Axin2, is not expressed (Suomalainen and Thesleff, 2010). It is somewhat surprising that Wnt signaling is not involved in the process of incisor renewal, considering that it plays critical roles in other epithelial stem cell niches, such as the skin and the intestine (Huelsken et al., 2001; He et al., 2004; Haegebarth and Clevers, 2009). Additionally, the intestinal stem cell marker Lgr5, which is a Wnt target gene, is expressed in the SR both during development and adulthood (Suomalainen and Thesleff, 2010; Chang et al., 2013), suggesting that Lgr5 is either regulated by other signaling pathways in the incisor or by low levels of Wnt activity that were not detected in previous experiments. Consistent with this, Axin2 is weakly expressed in the differentiating ameloblasts, indicative of some level of Wnt activity. Overexpression of Wnt3, a canonical Wnt, in the dental epithelium using the Keratin 14 promoter caused progressive loss of ameloblasts in postnatal mice (Millar et al., 2003), and upregulation of canonical Wnt signaling through expression of constitutively active β-catenin in the adult dental epithelium resulted in increased proliferation and expansion of the cervical loop (Liu et al., 2010). These studies indicated that uncontrolled Wnt signaling is undesirable in the incisor stem cell niche, but they did not directly address the requirement for Wnt signaling, either canonical or non-canonical, in the regulation of incisor stem cells. Therefore, the functional role of LGR5 and a closely related protein, LGR6, in the adult incisor stem cells still remain to be addressed.
In addition to signaling pathways, cell-cell and cell-extracellular matrix interactions, which are indispensable for the maintenance of stem cells in several systems (Chen et al., 2013), are likewise important in the incisor stem cells. In one example, the deletion of integrin β3 (CD61) resulted in a smaller cervical loop and reduced proliferation in the TA region in adult mice (Yoshida et al., 2013). In another study, the importance of cell-cell adhesion was highlighted by studying the role of E-cadherin in adult incisor stem cells. Conditional removal of E-cadherin in the cervical loop caused reduction in LRCs, increased proliferation in the TA cells, and decreased cell migration in the differentiating ameloblasts (Li et al., 2012). Deletion of two other cell adhesion molecules, PERP and Nectin, disrupted epithelial integrity, causing detachment of ameloblasts from the SI in postnatal and adult mice, respectively (Barron et al., 2008; Jheon et al., 2011a); this phenotype is comparable to the E-cadherin mutants. Finally, incisor epithelial stem cells are regulated by other factors, such as the heparin binding molecules midkine (MK) and heparin binding growth associated molecule (HB-GAM), which regulate the expression of each other and synergize with FGF signaling to promote cell proliferation in the developing cervical loop (Mitsiadis et al., 2008).
An additional level of regulation is provided by miRNAs, which modulate and fine-tune transcriptional networks in several stem cell systems (Yi and Fuchs, 2011). For example, miR-203 is upregulated during epidermal stratification, repressing the expression of the transcription factor p63 to promote differentiation and stratification (Lena et al., 2008; Yi et al., 2008). In the incisor, the complete loss of miRNA genesis through conditional deletion of Dicer1, which processes pre-miRNAs into mature miRNAs, resulted in phenotypes ranging from the formation of supernumerary incisors to ectopic budding of the cervical loops, depending on the time point at which deletion occurred (Cao et al., 2010b; Michon et al., 2010; Oommen et al., 2012). These phenotypes were attributed to increased proliferation in the cervical loop progenitor cells that was accompanied by impaired ameloblast differentiation. Microarray analysis identified differentially expressed miRNAs in the liCL, the laCL, and the ameloblasts (Jheon et al., 2011b), indicating that specific miRNAs function in different compartments of the incisor. In particular, miR200c maintains cell adhesion and promotes ameloblast differentiation by antagonizing the BMP inhibitor, noggin (Cao et al., 2013). MiR203 also facilitates BMP signaling by repressing BMPER (Bmp-binding endothelial cell precursor-derived regulator) expression. Both miR200c and miR203 are targets of the transcription factor PITX2, and their expression can be induced by BMP signaling (Cao et al., 2013), highlighting the complex interactions between miRNAs, signaling pathways and transcriptional networks that fine tune the regulatory machinery in order to ensure proper cell differentiation during both tooth development and renewal.
Incisor mesenchymal stem cells
As with the epithelially-derived ameloblasts, the mesenchymally-derived odontoblasts require a constant supply of new cells in order to accommodate the continuous growth of the incisor. Thus, a group of mesenchymal stem cells must exist and perform a function similar to their epithelial counterparts. It is generally thought that these cells are located at the apical end of the incisor between the laCL and liCL. This notion was bolstered by the findings that these cells retain BrdU labeling (Seidel et al., 2010) and respond to odontoblast damage in explant cultures (Feng et al., 2011). However, our understanding of the biology of the incisor stem cells in the mesenchyme has lagged behind the epithelium. Interestingly, Bmi1 and Gli1, markers for the cervical loop epithelial stem cells, are also expressed in the BrdU retaining mesenchymal cells (Biehs et al., 2013; Seidel et al., 2010), reinforcing the idea that these cells are stem cells. Nonetheless, further experiments are required to address the lineage and biology of Bmi1 and Gli1 expressing cells in the mesenchyme, as well as the functional roles of Bmi1 and Gli1 in this population. Ring1a/b, which are components of the core Polycomb Repressive Complex 1 (PRC1) that contains BMI1, are expressed in both the incisor epithelium and apical mesenchyme and are required for proper cell proliferation in both epithelium and mesenchyme, as well as for the correct formation of enamel and dentin (Lapthanasupkul et al., 2012). Thus, Polycomb group proteins play crucial roles in both the epithelial and mesenchymal stem cell compartments to ensure adequate cell proliferation and differentiation.
Evolution of the continuously growing incisor
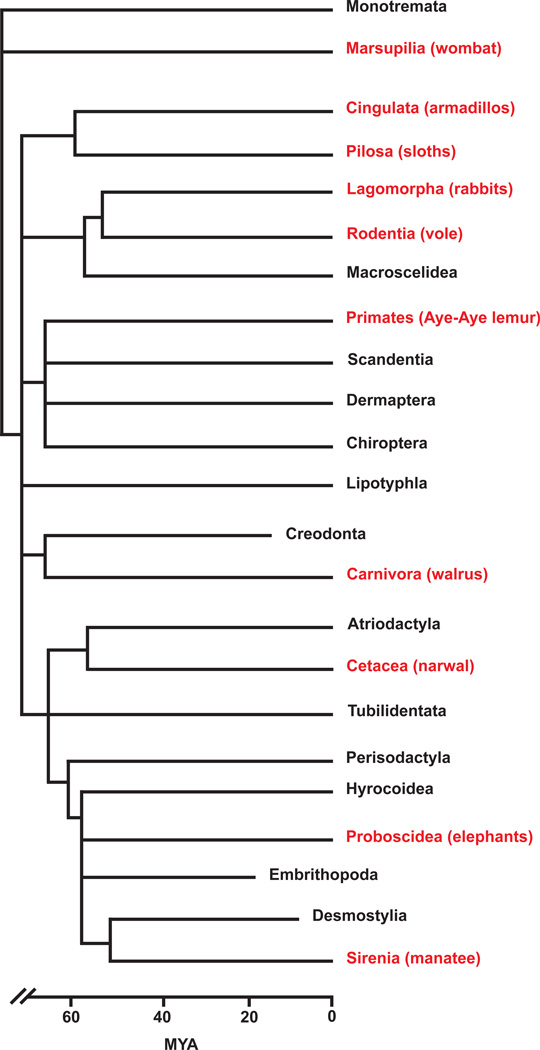
The ability to grow teeth continuously is not restricted to rodents. This property can be found in many other species, including the incisors of a primate, the Aye-aye lemur (Mittermeier et al., 1994); the molars of several other rodents, such as Sibling voles, Meadow voles, and guinea pigs (Hunt, 1959; Phillips and Oxberry, 1972; Tummers and Thesleff, 2003); and the molars of the order Xenarthra, represented by the sloths and armadillos (Naples, 1982; Tummers and Thesleff, 2008). The fossil record demonstrates that hypselodont, or continuously growing, teeth are a derived feature that has evolved many times in mammals (Figure 5).
Figure 5. Phylogenetic tree depicting estimated divergence times of mammalian clades based on the fossil record.
Clades shown in red include hypselodont species, whereas the clades shown in black do not. MYA – million years ago.
The ancestral state of the mammalian tooth is called brachydonty and is characterized by a relatively short, low crown and long root. Brachydont teeth, such as the mouse molar or all human teeth, do not grow continuously but do form a cervical loop during development and crown morphogenesis. Once crown formation is complete, the development of the root is initiated. This begins with the thinning out of the cervical loop and the loss of the SR. The remaining double layered epithelium is known as Hertwig’s epithelial root sheath (HERS), which eventually becomes a fenestrated network, facilitating the formation of cementum, the connection of the tooth to the jaw bones through the periodontal ligament, and thus the maturation of the root (Ten Cate, 1996; Thomas, 1995).
An increase in crown height became more prevalent in mammals living in the increasingly arid Miocene epoch as an adaptive response to the fibrous and abrasive diets that are common in dry environments (Jernvall and Fortelius, 2002). The formation of a high crowned, or hypsodont, tooth is thought to be the result of delayed root formation, leading to prolonged crown formation (Tummers and Thesleff, 2003). It is therefore postulated that hypsodonty is an intermediate state between brachydonty and hypselodonty during evolution, and hypselodonty is a condition in which the tooth continues its crown formation without undergoing root genesis (MacFadden, 2000; Tummers and Thesleff, 2003). This is clear in the rodent incisor, which, throughout the animal’s life, possesses a crown analogue at the labial side, where the laCL continues to produce ameloblasts and enamel, and a root analogue at the lingual side, which, despite being fragmented and reduced in size, appears to contain stem cells that contribute to incisor epithelium renewal (Seidel et al., 2010). Similarly, the continuously growing molars in sibling voles also maintain cervical loop structures that house the epithelial stem cells (Tummers and Thesleff, 2003, 2008). In contrast, the molars of sloths and armadillos lack enamel and form HERS, resembling the root analogue of the rodent incisors. The sharpening of the tooth in these cases is a result of the differences in dentin densities at each side of the tooth (Naples, 1982; Tummers and Thesleff, 2008). Consequently, all continuously growing teeth appear to have evolved to maintain a supply of stem cells from the cervical loop, but this may have occurred through modulation of different aspects of the regulatory network, which is reflected in the variations of the developmental design and the anatomy of these teeth.
Recent studies employing an evolutionary-developmental biology approach and functional analysis in mice have shed light on the potential mechanisms leading to the evolution of the continuously growing teeth. One important milestone for the derivation of hypselodonty is the maintenance of the cervical loop structure. By comparing gene expression in mouse incisors, mouse molars, and continuously growing vole molars, it was found that expression of Fgf10 in the mesenchyme and components of the Notch signaling pathway in the epithelium was maintained in the continuously growing teeth, while the disappearance of their expression was correlated with root formation and loss of the cervical loop stem cell niche in non-continuously growing teeth (Tummers and Thesleff, 2003). Interestingly, Fgf10 null mouse incisors underwent root formation when cultured under kidney capsules. Conversely, overexpression of Fg10 in the mouse molar dental papilla by means of electroporation resulted in the formation of cervical loop like structures, although it is unclear whether these were functionally equivalent to the incisor apical buds (Yokohama-Tamaki et al., 2006). Consequently, it is possible that during evolution, the preserved expression of Fgf10 and other genes prolonged the presence of the cervical loop during development, which first led to the formation of the high-crowned teeth and culminated in the transition to hypselodonty.
Another key step in the evolution of continuously growing teeth is the modification of the crown to root transition, a process that is currently poorly understood. Mouse genetic studies and in vitro culturing strategies have provided some basic knowledge of root formation in mouse molars. For example, molars from mice carrying a spontaneous mutant Ptch1 allele, which have altered Hh activity, develop smaller roots due to decreased cell proliferation in HERS (Nakatomi et al., 2006). Insulin-like growth factor-I and vasoactive intestinal peptide also exhibit proliferative effects that promote root sheath elongation (Fujiwara et al., 2005; Xu et al., 2012), and the initiation of HERS formation is aided by the termination of EGF expression, which otherwise maintains the cervical loop (Fujiwara et al., 2009). As HERS elongates apically, its maintenance depends on proper odontoblast differentiation, which is regulated by canonical Wnt signaling. This is evident in mutants that lack β-catenin, and thus Wnt signaling, in the forming odontoblasts (Kim et al., 2013; Zhang et al., 2013a). In these mutants, odontoblasts failed to differentiate and form dentin, and even though HERS elongated, its cellular integrity was perturbed and roots were not formed. Excessive β-catenin/Wnt signaling also perturbs normal root development, suggesting that correct spatiotemporal regulation of Wnt activity is required for correct odontogenesis and root formation (Bae et al., 2013). Several transcription factors have also been found to be critical for root formation. For example, the LIM homeodomain transcription factor LHX6 is required for molar root development, and Lhx6-null mice have severely affected root structures (Zhang et al., 2013b). The transcription factors nuclear factor I-C (NFI-C) and MSX2 also play important roles in root formation. Mice carrying mutations in NFI-C lack roots completely (Steele-Perkins et al., 2003), whereas Msx2 null mice have abnormal root morphogenesis (Aïoub et al., 2007). MSX2 function is required to maintain Bmp2 expression, indicative of the role of Bmp signaling in root formation. In agreement with this, Bmp2/4 are expressed in the mesenchyme of the root tip, and misexpression of noggin in the epithelium resulted in delayed root formation and patterning defects (Plikus et al., 2005; Yamashiro et al., 2003). Epithelium-specific deletion of SMAD4, which is critical in mediating BMP/TGFβ signaling, resulted in downregulation of Shh and Nfic expression and defects in HERS elongation (Huang et al., 2010). Intriguingly, removal of BMP receptor type1A (BMPR1A) in the epithelium did not overtly affect HERS formation but converted crown epithelia into the root lineage due to elevated Wnt signaling, suggesting that BMP and TGFβ signaling may regulate different aspects of crown to root transition (Yang et al., 2013). Ectodysplasin (Eda), which encodes a membrane bound signaling molecule and causes X-linked hypohidrotic ectodermal dysplasia when mutated, has also been shown to play an important role in the process of crown to root transition. Excess EDA in the epithelium by means of overexpression was sufficient to maintain cervical loops, despite HERS formation and a complete loss of enamel, a phenotype that is reminiscent of the sloth molars (Tummers and Thesleff, 2008). Finally, it is intriguing that differential cell proliferation and motility between IEE and OEE has been observed, suggesting that cell movement in the epithelium may be important for root formation (Sakano et al., 2012). Consequently, as knowledge of the underlying mechanisms that regulate crown to root transition increases, it will become possible to analyze how these pathways may be modulated during evolution to prevent root formation, and thus to enable the formation of a continuously growing tooth.
Conclusions and Perspectives
Research in the past 15 years has identified the presence of a group of epithelial stem cells in the mouse incisor and elucidated several important regulatory mechanisms that govern the formation and maintenance of these cells. However, many outstanding questions remain. For instance, it is unclear whether all epithelial cells in the cervical loop stem cell niche are equivalent. As discussed earlier, it is possible that Gli1 and Bmi1 mark different stem cells compared to Sox2. Indeed, when Shh activity is blocked by a smoothened inhibitor, a group of Shh non-responsive stem cells in the incisor epithelium can still give rise to SI cells (Seidel et al., 2010), suggesting that there are at least two types of stem cells. There also appear to be Lgr5-expressing SR cells adjacent to the OEE (Chang et al., 2013), and as Lgr5 is a stem cell marker in other systems (Schuijers and Clevers, 2012), it is possible that these SR cells represent yet another subgroup of stem cells. It will therefore be important to examine the molecular and functional differences between these populations. A powerful approach to address this question will be to perform expression profiling using gene expression microarray or RNA sequencing at the single cell level (Trimarchi et al., 2008; Tang et al., 2009). Results from such studies could also shed light on the transcriptional differences between quiescent stem cells and proliferative TA cells. Indeed, the rodent incisor, with its conveyor belt-like cell progression, represents a tractable system for studying mechanisms that trigger the transition from stem cells to the TA state. By uncovering signaling pathways or cellular factors that are crucial for stem cell proliferation and differentiation, we can learn about processes that may instruct future efforts toward the rational utilization of stem cells for regenerative medicine. To that end, several in vitro systems have been established that allow stem cells to be maintained for a prolonged period of time before induction of differentiation (Chavez et al., 2012; Chang et al., 2013). These systems could potentially be combined with tissue engineering and newly developed material fabrication techniques to make components of teeth.
Another interesting question is the developmental origin of the incisor stem cells. How are they formed initially? Are they set aside early or are they produced from dividing progenitor cells at later times in development? Will it be possible to generate incisor stem cells by expression of key transcription factors, similar to the induced pluripotent stem cell process? Finally, from an evolutionary point of view, the developmental mechanisms that result in hypselodonty remain largely unknown. While Fgf10 appears to be involved (Yokohama-Tamaki et al., 2006), the upstream and downstream events have not been discovered. By answering these questions, we will not only deepen our understanding of incisor stem cells, but also extend our current knowledge of other adult stem cell systems.
Acknowledgments
Grant support: R01-DE021420 and DP2-OD00719 to O.D.K., F32-DE023705 to J.K.H, F30-DE022482 to V.M.
References
- Addison AJ. The structure and growth of the incisor teeth of the albino rat. J. Morphol. 1915;26:43–96. [Google Scholar]
- Aïoub M, Lézot F, Molla M, Castaneda B, Robert B, Goubin G, Néfussi JR, Berdal A. Msx2 −/− transgenic mice develop compound amelogenesis imperfecta, dentinogenesis imperfecta and periodental osteopetrosis. Bone. 2007;41:851–859. doi: 10.1016/j.bone.2007.07.023. [DOI] [PubMed] [Google Scholar]
- Arnold K, Sarkar A, Yram MA, Polo JM, Bronson R, Sengupta S, Seandel M, Geijsen N, Hochedlinger K. Sox2(+) adult stem and progenitor cells are important for tissue regeneration and survival of mice. Cell Stem Cell. 2011;9:317–329. doi: 10.1016/j.stem.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae CH, Lee JY, Kim TH, Baek JA, Lee JC, Yang X, Taketo MM, Jiang R, Cho ES. Excessive Wnt/β-catenin signaling disturbs tooth-root formation. J. Periodontal Res. 2013;48:405–410. doi: 10.1111/jre.12018. [DOI] [PubMed] [Google Scholar]
- Barron MJ, Brookes SJ, Draper CE, Garrod D, Kirkham J, Shore RC, Dixon MJ. The cell adhesion molecule nectin-1 is critical for normal enamel formation in mice. Hum. Mol. Genet. 2008;17:3509–3520. doi: 10.1093/hmg/ddn243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bei M, Maas R. FGFs and BMP4 induce both Msx1-independent and Msx1-dependent signaling pathways in early tooth development. Development. 1998;125:4325–4333. doi: 10.1242/dev.125.21.4325. [DOI] [PubMed] [Google Scholar]
- Biehs B, Hu JK-H, Strauli NB, Sangiorgi E, Jung H, Heber R-P, Ho S, Goodwin AF, Dasen JS, Capecchi MR, et al. BMI1 represses Ink4a/Arf and Hox genes to regulate stem cells in the rodent incisor. Nat. Cell Biol. 2013;15:846–852. doi: 10.1038/ncb2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitgood MJ, McMahon AP. Hedgehog and Bmp genes are coexpressed at many diverse sites of cell-cell interaction in the mouse embryo. Dev. Biol. 1995;172:126–138. doi: 10.1006/dbio.1995.0010. [DOI] [PubMed] [Google Scholar]
- Cao H, Florez S, Amen M, Huynh T, Skobe Z, Baldini A, Amendt BA. Tbx1 regulates progenitor cell proliferation in the dental epithelium by modulating Pitx2 activation of p21. Dev. Biol. 2010a;347:289–300. doi: 10.1016/j.ydbio.2010.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H, Wang J, Li X, Florez S, Huang Z, Venugopalan SR, Elangovan S, Skobe Z, Margolis HC, Martin JF, et al. MicroRNAs play a critical role in tooth development. J. Dent. Res. 2010b;89:779–784. doi: 10.1177/0022034510369304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H, Jheon A, Li X, Sun Z, Wang J, Florez S, Zhang Z, McManus MT, Klein OD, Amendt BA. The Pitx2:miR-200c/141:noggin pathway regulates Bmp signaling and ameloblast differentiation. Development. 2013;140:3348–3359. doi: 10.1242/dev.089193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caton J, Luder H-U, Zoupa M, Bradman M, Bluteau G, Tucker AS, Klein O, Mitsiadis TA. Enamel-free teeth: Tbx1 deletion affects amelogenesis in rodent incisors. Dev. Biol. 2009;328:493–505. doi: 10.1016/j.ydbio.2009.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai Y, Jiang X, Ito Y, Bringas P, Jr, Han J, Rowitch DH, Soriano P, McMahon AP, Sucov HM. Fate of the mammalian cranial neural crest during tooth and mandibular morphogenesis. Development. 2000;127:1671–1679. doi: 10.1242/dev.127.8.1671. [DOI] [PubMed] [Google Scholar]
- Chang JYF, Wang C, Jin C, Yang C, Huang Y, Liu J, McKeehan WL, D’Souza RN, Wang F. Self-renewal and multilineage differentiation of mouse dental epithelial stem cells. Stem Cell Res. 2013;11:990–1002. doi: 10.1016/j.scr.2013.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez MG, Yu W, Biehs B, Harada H, Snead ML, Klein OD. Characterization of Dental Epithelial Stem Cells from the Mouse Incisor with 2D and 3D Platforms. Tissue Eng. Part C Methods. 2012 doi: 10.1089/ten.tec.2012.0232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Lewallen M, Xie L. Adhesion in the stem cell niche: biological roles and regulation. Development. 2013;140:255–265. doi: 10.1242/dev.083139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cope E. The Mechanical Causes of the Origin of the Dentition of the Rodentia. Am. Nat. 1888;22:3–23. [Google Scholar]
- Felszeghy S, Suomalainen M, Thesleff I. Notch signalling is required for the survival of epithelial stem cells in the continuously growing mouse incisor. Differ. Res. Biol. Divers. 2010;80:241–248. doi: 10.1016/j.diff.2010.06.004. [DOI] [PubMed] [Google Scholar]
- Feng J, Mantesso A, Bari CD, Nishiyama A, Sharpe PT. Dual origin of mesenchymal stem cells contributing to organ growth and repair. Proc. Natl. Acad. Sci. 2011;108:6503–6508. doi: 10.1073/pnas.1015449108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fougeroux de Bondaroy AD. Observations Anatomiques. Hist Acad Roy Sci Paris. 1768:47–48. [Google Scholar]
- Fuchs E. The Tortoise and the Hair: Slow-Cycling Cells in the Stem Cell Race. Cell. 2009;137:811–819. doi: 10.1016/j.cell.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs E, Horsley V. More than one way to skin. Genes Dev. 2008;22:976–985. doi: 10.1101/gad.1645908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara N, Tabata MJ, Endoh M, Ishizeki K, Nawa T. Insulin-like growth factor-I stimulates cell proliferation in the outer layer of Hertwig’s epithelial root sheath and elongation of the tooth root in mouse molars in vitro. Cell Tissue Res. 2005;320:69–75. doi: 10.1007/s00441-004-1065-5. [DOI] [PubMed] [Google Scholar]
- Fujiwara N, Akimoto T, Otsu K, Kagiya T, Ishizeki K, Harada H. Reduction of Egf signaling decides transition from crown to root in the development of mouse molars. J. Exp. Zoolog. B Mol. Dev. Evol. 2009;312B:486–494. doi: 10.1002/jez.b.21268. [DOI] [PubMed] [Google Scholar]
- Haegebarth A, Clevers H. Wnt signaling, lgr5, and stem cells in the intestine and skin. Am. J. Pathol. 2009;174:715–721. doi: 10.2353/ajpath.2009.080758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada H, Kettunen P, Jung HS, Mustonen T, Wang YA, Thesleff I. Localization of putative stem cells in dental epithelium and their association with Notch and FGF signaling. J. Cell Biol. 1999;147:105–120. doi: 10.1083/jcb.147.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada H, Toyono T, Toyoshima K, Yamasaki M, Itoh N, Kato S, Sekine K, Ohuchi H. FGF10 maintains stem cell compartment in developing mouse incisors. Development. 2002;129:1533–1541. doi: 10.1242/dev.129.6.1533. [DOI] [PubMed] [Google Scholar]
- Harada H, Ichimori Y, Yokohama-Tamaki T, Ohshima H, Kawano S, Katsube K, Wakisaka S. Stratum intermedium lineage diverges from ameloblast lineage via Notch signaling. Biochem. Biophys. Res. Commun. 2006;340:611–616. doi: 10.1016/j.bbrc.2005.12.053. [DOI] [PubMed] [Google Scholar]
- He XC, Zhang J, Tong W-G, Tawfik O, Ross J, Scoville DH, Tian Q, Zeng X, He X, Wiedemann LM, et al. BMP signaling inhibits intestinal stem cell self-renewal through suppression of Wnt-beta-catenin signaling. Nat. Genet. 2004;36:1117–1121. doi: 10.1038/ng1430. [DOI] [PubMed] [Google Scholar]
- Huang X, Xu X, Bringas P, Jr, Hung YP, Chai Y. Smad4-Shh-Nfic signaling cascade-mediated epithelial-mesenchymal interaction is crucial in regulating tooth root development. J. Bone Miner. Res. Off. J. Am. Soc. Bone Miner. Res. 2010;25:1167–1178. doi: 10.1359/jbmr.091103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsken J, Vogel R, Erdmann B, Cotsarelis G, Birchmeier W. beta-Catenin controls hair follicle morphogenesis and stem cell differentiation in the skin. Cell. 2001;105:533–545. doi: 10.1016/s0092-8674(01)00336-1. [DOI] [PubMed] [Google Scholar]
- Hunt A. A description of the molar teeth and investing tissues of normal guinea pigs. J. Dent. Res. 1959;38:216–231. doi: 10.1177/00220345590380020301. [DOI] [PubMed] [Google Scholar]
- Jernvall J, Fortelius M. Common mammals drive the evolutionary increase of hypsodonty in the Neogene. Nature. 2002;417:538–540. doi: 10.1038/417538a. [DOI] [PubMed] [Google Scholar]
- Jernvall J, Kettunen P, Karavanova I, Martin LB, Thesleff I. Evidence for the role of the enamel knot as a control center in mammalian tooth cusp formation: non-dividing cells express growth stimulating Fgf-4 gene. Int. J. Dev. Biol. 1994;38:463–469. [PubMed] [Google Scholar]
- Jheon AH, Mostowfi P, Snead ML, Ihrie RA, Sone E, Pramparo T, Attardi LD, Klein OD. PERP regulates enamel formation via effects on cell-cell adhesion and gene expression. J. Cell Sci. 2011a;124:745–754. doi: 10.1242/jcs.078071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jheon AH, Li C-Y, Wen T, Michon F, Klein OD. Expression of microRNAs in the stem cell niche of the adult mouse incisor. PloS One. 2011b;6:e24536. doi: 10.1371/journal.pone.0024536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyner AL, Zervas M. Genetic inducible fate mapping in mouse: establishing genetic lineages and defining genetic neuroanatomy in the nervous system. Dev. Dyn. 2006;235:2376–2385. doi: 10.1002/dvdy.20884. [DOI] [PubMed] [Google Scholar]
- Juuri E, Saito K, Ahtiainen L, Seidel K, Tummers M, Hochedlinger K, Klein OD, Thesleff I, Michon F. Sox2+ Stem Cells Contribute to All Epithelial Lineages of the Tooth via Sfrp5+ Progenitors. Dev. Cell. 2012;23:317–328. doi: 10.1016/j.devcel.2012.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsuragi Y, Anraku J, Nakatomi M, Ida-Yonemochi H, Obata M, Mishima Y, Sakuraba Y, Gondo Y, Kodama Y, Nishikawa A, et al. Bcl11b transcription factor plays a role in the maintenance of the ameloblast-progenitors in mouse adult maxillary incisors. Mech. Dev. 2013;130:482–492. doi: 10.1016/j.mod.2013.05.002. [DOI] [PubMed] [Google Scholar]
- Kawano S, Morotomi T, Toyono T, Nakamura N, Uchida T, Ohishi M, Toyoshima K, Harada H. Establishment of dental epithelial cell line (HAT-7) and the cell differentiation dependent on Notch signaling pathway. Connect. Tissue Res. 2002;43:409–412. doi: 10.1080/03008200290000637. [DOI] [PubMed] [Google Scholar]
- Kettunen P, Thesleff I. Expression and function of FGFs-4,-8, and -9 suggest functional redundancy and repetitive use as epithelial signals during tooth morphogenesis. Dev. Dyn. 1998;211:256–268. doi: 10.1002/(SICI)1097-0177(199803)211:3<256::AID-AJA7>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Kettunen P, Laurikkala J, Itäranta P, Vainio S, Itoh N, Thesleff I. Associations of FGF-3 and FGF-10 with signaling networks regulating tooth morphogenesis. Dev. Dyn. 2000;219:322–332. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1062>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Kim TH, Bae CH, Lee JC, Ko SO, Yang X, Jiang R, Cho ES. β-catenin is required in odontoblasts for tooth root formation. J. Dent. Res. 2013;92:215–221. doi: 10.1177/0022034512470137. [DOI] [PubMed] [Google Scholar]
- Klein OD, Lyons DB, Balooch G, Marshall GW, Basson MA, Peterka M, Boran T, Peterkova R, Martin GR. An FGF signaling loop sustains the generation of differentiated progeny from stem cells in mouse incisors. Development. 2008;135:377–385. doi: 10.1242/dev.015081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurosaka H, Islam MN, Kuremoto K-I, Hayano S, Nakamura M, Kawanabe N, Yanagita T, Rice DPC, Harada H, Taniuchi I, et al. Core binding factor beta functions in the maintenance of stem cells and orchestrates continuous proliferation and differentiation in mouse incisors. Stem Cells. 2011;29:1792–1803. doi: 10.1002/stem.722. [DOI] [PubMed] [Google Scholar]
- Kyrylkova K, Kyryachenko S, Biehs B, Klein O, Kioussi C, Leid M. BCL11B Regulates Epithelial Proliferation and Asymmetric Development of the Mouse Mandibular Incisor. PLoS ONE. 2012;7:e37670. doi: 10.1371/journal.pone.0037670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapthanasupkul P, Feng J, Mantesso A, Takada-Horisawa Y, Vidal M, Koseki H, Wang L, An Z, Miletich I, Sharpe PT. Ring1a/b polycomb proteins regulate the mesenchymal stem cell niche in continuously growing incisors. Dev. Biol. 2012;367:140–153. doi: 10.1016/j.ydbio.2012.04.029. [DOI] [PubMed] [Google Scholar]
- Lena AM, Shalom-Feuerstein R, Rivetti di Val Cervo P, Aberdam D, Knight RA, Melino G, Candi E. miR-203 represses “stemness” by repressing DeltaNp63. Cell Death Differ. 2008;15:1187–1195. doi: 10.1038/cdd.2008.69. [DOI] [PubMed] [Google Scholar]
- Li C-Y, Cha W, Luder H-U, Charles R-P, McMahon M, Mitsiadis TA, Klein OD. E-cadherin regulates the behavior and fate of epithelial stem cells and their progeny in the mouse incisor. Dev. Biol. 2012;366:357–366. doi: 10.1016/j.ydbio.2012.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Kwon H-J, Harada H, Ohshima H, Cho S-W, Jung H-S. Expression patterns of ABCG2, Bmi-1, Oct-3/4, and Yap in the developing mouse incisor. Gene Expr. Patterns GEP. 2011;11:163–170. doi: 10.1016/j.gep.2010.11.001. [DOI] [PubMed] [Google Scholar]
- Lin Y, Cheng Y-SL, Qin C, Lin C, D’Souza R, Wang F. FGFR2 in the dental epithelium is essential for development and maintenance of the maxillary cervical loop, a stem cell niche in mouse incisors. Dev. Dyn. 2009;238:324–330. doi: 10.1002/dvdy.21778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Dangaria S, Andl T, Zhang Y, Wright AC, Damek-Poprawa M, Piccolo S, Nagy A, Taketo MM, Diekwisch TGH, et al. beta-Catenin initiates tooth neogenesis in adult rodent incisors. J. Dent. Res. 2010;89:909–914. doi: 10.1177/0022034510370090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacFadden BJ. Cenozoic Mammalian Herbivores from the Americas: Reconstructing Ancient Diets and Terrestrial Communities. Annu. Rev. Ecol. Syst. 2000;31:33–59. [Google Scholar]
- Mao J, McGlinn E, Huang P, Tabin CJ, McMahon AP. Fgf-dependent Etv4/5 activity is required for posterior restriction of Sonic Hedgehog and promoting outgrowth of the vertebrate limb. Dev. Cell. 2009;16:600–606. doi: 10.1016/j.devcel.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michon F, Tummers M, Kyyrönen M, Frilander MJ, Thesleff I. Tooth morphogenesis and ameloblast differentiation are regulated by micro-RNAs. Dev. Biol. 2010;340:355–368. doi: 10.1016/j.ydbio.2010.01.019. [DOI] [PubMed] [Google Scholar]
- Millar SE, Koyama E, Reddy ST, Andl T, Gaddapara T, Piddington R, Gibson CW. Over- and ectopic expression of Wnt3 causes progressive loss of ameloblasts in postnatal mouse incisor teeth. Connect. Tissue Res. 2003;44(Suppl 1):124–129. [PubMed] [Google Scholar]
- Mitsiadis TA, Hirsinger E, Lendahl U, Goridis C. Delta-Notch Signaling in Odontogenesis: Correlation with Cytodifferentiation and Evidence for Feedback Regulation. Dev. Biol. 1998;204:420–431. doi: 10.1006/dbio.1998.9092. [DOI] [PubMed] [Google Scholar]
- Mitsiadis TA, Caton J, De Bari C, Bluteau G. The large functional spectrum of the heparin-binding cytokines MK and HB-GAM in continuously growing organs: the rodent incisor as a model. Dev. Biol. 2008;320:256–266. doi: 10.1016/j.ydbio.2008.05.530. [DOI] [PubMed] [Google Scholar]
- Mitsiadis TA, Graf D, Luder H, Gridley T, Bluteau G. BMPs and FGFs target Notch signalling via jagged 2 to regulate tooth morphogenesis and cytodifferentiation. Development. 2010;137:3025–3035. doi: 10.1242/dev.049528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittermeier RA, Tattersall I, Konstant WR, Meyers DM, Mast RB. Lemurs of Madagascar. Washington, D.C: Conservation International; 1994. [Google Scholar]
- Moe H, Thorball N, Nielsen HW. Structural alterations in proliferating, remodeling, and regressing tooth pulp arterioles. Cell Tissue Res. 1979;203:339–354. doi: 10.1007/BF00233263. [DOI] [PubMed] [Google Scholar]
- De Moerlooze L, Spencer-Dene B, Revest JM, Hajihosseini M, Rosewell I, Dickson C. An important role for the IIIb isoform of fibroblast growth factor receptor 2 (FGFR2) in mesenchymal-epithelial signalling during mouse organogenesis. Development. 2000;127:483–492. doi: 10.1242/dev.127.3.483. [DOI] [PubMed] [Google Scholar]
- Molofsky AV, Pardal R, Iwashita T, Park I-K, Clarke MF, Morrison SJ. Bmi-1 dependence distinguishes neural stem cell self-renewal from progenitor proliferation. Nature. 2003;425:962–967. doi: 10.1038/nature02060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatomi M, Morita I, Eto K, Ota MS. Sonic hedgehog signaling is important in tooth root development. J. Dent. Res. 2006;85:427–431. doi: 10.1177/154405910608500506. [DOI] [PubMed] [Google Scholar]
- Nakatomi M, Wang X-P, Key D, Lund JJ, Turbe-Doan A, Kist R, Aw A, Chen Y, Maas RL, Peters H. Genetic interactions between Pax9 and Msx1 regulate lip development and several stages of tooth morphogenesis. Dev. Biol. 2010;340:438–449. doi: 10.1016/j.ydbio.2010.01.031. [DOI] [PubMed] [Google Scholar]
- Naples VL. Cranial osteology and function in the tree sloths, Bradypus and Choloepus. American Museum novitates. Am Museum Novitiates. 1982;2739:1–41. [Google Scholar]
- Oommen S, Otsuka-Tanaka Y, Imam N, Kawasaki M, Kawasaki K, Jalani-Ghazani F, Anderegg A, Awatramani R, Hindges R, Sharpe PT, et al. Distinct roles of microRNAs in epithelium and mesenchyme during tooth development. Dev. Dyn. 2012;241:1465–1472. doi: 10.1002/dvdy.23828. [DOI] [PubMed] [Google Scholar]
- Oudet J. Expériences sur l’accroissement continué et la reproduction des dents chez les lapins. J Physiol Expér Pathol Tomes 3 and 4. 1823 [Google Scholar]
- Park I, Qian D, Kiel M, Becker MW, Pihalja M, Weissman IL, Morrison SJ, Clarke MF. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature. 2003;423:302–305. doi: 10.1038/nature01587. [DOI] [PubMed] [Google Scholar]
- Parsa S, Kuremoto K-I, Seidel K, Tabatabai R, Mackenzie B, Yamaza T, Akiyama K, Branch J, Koh CJ, Al Alam D, et al. Signaling by FGFR2b controls the regenerative capacity of adult mouse incisors. Development. 2010;137:3743–3752. doi: 10.1242/dev.051672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips CJ, Oxberry B. Comparative histology of molar dentitions of Microtus and Clethrionomys, with comments on dental evolution in microtine rodents. J. Mammal. 1972;53:1–20. [PubMed] [Google Scholar]
- Plikus MV, Zeichner-David M, Mayer J-A, Reyna J, Bringas P, Thewissen JGM, Snead ML, Chai Y, Chuong C-M. Morphoregulation of teeth: modulating the number, size, shape and differentiation by tuning Bmp activity. Evol. Dev. 2005;7:440–457. doi: 10.1111/j.1525-142X.2005.05048.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porntaveetus T, Otsuka-Tanaka Y, Basson MA, Moon AM, Sharpe PT, Ohazama A. Expression of fibroblast growth factors (Fgfs) in murine tooth development. J. Anat. 2011;218:534–543. doi: 10.1111/j.1469-7580.2011.01352.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothova M, Thompson H, Lickert H, Tucker AS. Lineage tracing of the endoderm during oral development. Dev. Dyn. Off. Publ. Am. Assoc. Anat. 2012;241:1183–1191. doi: 10.1002/dvdy.23804. [DOI] [PubMed] [Google Scholar]
- Sakano M, Otsu K, Fujiwara N, Fukumoto S, Yamada A, Harada H. Cell dynamics in cervical loop epithelium during transition from crown to root: implications for Hertwig’s epithelial root sheath formation. J. Periodontal Res. 2012;48:262–267. doi: 10.1111/jre.12003. [DOI] [PubMed] [Google Scholar]
- Samperiz MM, Blumen G, Merzel J. Effect of vinblastine on the cell cycle and migration of ameloblasts of mouse incisors as shown by autoradiography using 3H-thymidine. Cell Tissue Kinet. 1985;18:493–503. doi: 10.1111/j.1365-2184.1985.tb00691.x. [DOI] [PubMed] [Google Scholar]
- Sangiorgi E, Capecchi MR. Bmi1 is expressed in vivo in intestinal stem cells. Nat. Genet. 2008;40:915–920. doi: 10.1038/ng.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuijers J, Clevers H. Adult mammalian stem cells: the role of Wnt, Lgr5 and R-spondins. EMBO. J. 2012;31:2685–2696. doi: 10.1038/emboj.2012.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidel K, Ahn CP, Lyons D, Nee A, Ting K, Brownell I, Cao T, Carano RAD, Curran T, Schober M, et al. Hedgehog signaling regulates the generation of ameloblast progenitors in the continuously growing mouse incisor. Development. 2010;137:3753–3761. doi: 10.1242/dev.056358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CE. Cell turnover in the odontogenic organ of the rat incisor as visualized by graphic reconstructions following a single injection of 3H-thymidine. Am. J. Anat. 1980;158:321–343. doi: 10.1002/aja.1001580307. [DOI] [PubMed] [Google Scholar]
- Smith CE, Warshawsky H. Cellular renewal in the enamel organ and the odontoblast layer of the rat incisor as followed by radioautography using 3H-thymidine. Anat. Rec. 1975a;183:523–561. doi: 10.1002/ar.1091830405. [DOI] [PubMed] [Google Scholar]
- Smith CE, Warshawsky H. Histological and three dimensional organization of the odontogenic organ in the lower incisor of 100 gram rats. Am. J. Anat. 1975b;142:403–429. doi: 10.1002/aja.1001420402. [DOI] [PubMed] [Google Scholar]
- Steele-Perkins G, Butz KG, Lyons GE, Zeichner-David M, Kim H-J, Cho M-I, Gronostajski RM. Essential role for NFI-C/CTF transcription-replication factor in tooth root development. Mol. Cell. Biol. 2003;23:1075–1084. doi: 10.1128/MCB.23.3.1075-1084.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh H, Consiglio A, Ray J, Sawai T, D’Amour KA, Gage FH. In vivo fate analysis reveals the multipotent and self-renewal capacities of Sox2+ neural stem cells in the adult hippocampus. Cell Stem Cell. 2007;1:515–528. doi: 10.1016/j.stem.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suomalainen M, Thesleff I. Patterns of Wnt pathway activity in the mouse incisor indicate absence of Wnt/beta-catenin signaling in the epithelial stem cells. Dev. Dyn. 2010;239:364–372. doi: 10.1002/dvdy.22106. [DOI] [PubMed] [Google Scholar]
- Tang F, Barbacioru C, Wang Y, Nordman E, Lee C, Xu N, Wang X, Bodeau J, Tuch BB, Siddiqui A, et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat. Methods. 2009;6:377–382. doi: 10.1038/nmeth.1315. [DOI] [PubMed] [Google Scholar]
- Ten Cate AR. The role of epithelium in the development, structure and function of the tissues of tooth support. Oral Dis. 1996;2:55–62. doi: 10.1111/j.1601-0825.1996.tb00204.x. [DOI] [PubMed] [Google Scholar]
- Thomas HF. Root formation. Int. J. Dev. Biol. 1995;39:231–237. [PubMed] [Google Scholar]
- Tian H, Biehs B, Warming S, Leong KG, Rangell L, Klein OD, de Sauvage FJ. A reserve stem cell population in small intestine renders Lgr5-positive cells dispensable. Nature. 2011;478:255–259. doi: 10.1038/nature10408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimarchi JM, Stadler MB, Cepko CL. Individual retinal progenitor cells display extensive heterogeneity of gene expression. PloS One. 2008;3:e1588. doi: 10.1371/journal.pone.0001588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker A, Sharpe P. The cutting-edge of mammalian development; how the embryo makes teeth. Nat. Rev. Genet. 2004;5:499–508. doi: 10.1038/nrg1380. [DOI] [PubMed] [Google Scholar]
- Tummers M, Thesleff I. Root or crown: a developmental choice orchestrated by the differential regulation of the epithelial stem cell niche in the tooth of two rodent species. Development. 2003;130:1049–1057. doi: 10.1242/dev.00332. [DOI] [PubMed] [Google Scholar]
- Tummers M, Thesleff I. Observations on continuously growing roots of the sloth and the K14-Eda transgenic mice indicate that epithelial stem cells can give rise to both the ameloblast and root epithelium cell lineage creating distinct tooth patterns. Evol. Dev. 2008;10:187–195. doi: 10.1111/j.1525-142X.2008.00226.x. [DOI] [PubMed] [Google Scholar]
- Tummers M, Thesleff I. The importance of signal pathway modulation in all aspects of tooth development. J. Exp. Zoolog. B Mol. Dev. Evol. 2009;312B:309–319. doi: 10.1002/jez.b.21280. [DOI] [PubMed] [Google Scholar]
- Wang X-P, Suomalainen M, Jorgez CJ, Matzuk MM, Werner S, Thesleff I. Follistatin regulates enamel patterning in mouse incisors by asymmetrically inhibiting BMP signaling and ameloblast differentiation. Dev. Cell. 2004;7:719–730. doi: 10.1016/j.devcel.2004.09.012. [DOI] [PubMed] [Google Scholar]
- Wang X-P, Suomalainen M, Felszeghy S, Zelarayan LC, Alonso MT, Plikus MV, Maas RL, Chuong C-M, Schimmang T, Thesleff I. An integrated gene regulatory network controls stem cell proliferation in teeth. PLoS Biol. 2007;5:e159. doi: 10.1371/journal.pbio.0050159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warshawsky H, Smith CE. Morphological classification of rat incisor ameloblasts. Anat. Rec. 1974;179:423–446. doi: 10.1002/ar.1091790403. [DOI] [PubMed] [Google Scholar]
- Xu J, Kawashima N, Fujiwara N, Harada H, Ota MS, Suda H. Promotional effects of vasoactive intestinal peptide on the development of rodent Hertwig’s epithelial root sheath. Congenit. Anom. 2012;52:162–167. doi: 10.1111/j.1741-4520.2012.00371.x. [DOI] [PubMed] [Google Scholar]
- Yamashiro T, Tummers M, Thesleff I. Expression of bone morphogenetic proteins and Msx genes during root formation. J. Dent. Res. 2003;82:172–176. doi: 10.1177/154405910308200305. [DOI] [PubMed] [Google Scholar]
- Yang Z, Hai B, Qin L, Ti X, Shangguan L, Zhao Y, Wiggins L, Liu Y, Feng JQ, Chang JYF, et al. Cessation of Epithelial Bmp Signaling Switches the Differentiation of Crown Epithelia to the Root Lineage in a β-Catenin-Dependent Manner. Mol. Cell. Biol. 2013;33:4732–4744. doi: 10.1128/MCB.00456-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi R, Fuchs E. MicroRNAs and their roles in mammalian stem cells. J. Cell Sci. 2011;124:1775–1783. doi: 10.1242/jcs.069104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi R, Poy MN, Stoffel M, Fuchs E. A skin microRNA promotes differentiation by repressing "stemness". Nature. 2008;452:225–229. doi: 10.1038/nature06642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokohama-Tamaki T, Ohshima H, Fujiwara N, Takada Y, Ichimori Y, Wakisaka S, Ohuchi H, Harada H. Cessation of Fgf10 signaling, resulting in a defective dental epithelial stem cell compartment, leads to the transition from crown to root formation. Development. 2006;133:1359–1366. doi: 10.1242/dev.02307. [DOI] [PubMed] [Google Scholar]
- Yoshida T, Iwata T, Umemoto T, Shiratsuchi Y, Kawashima N, Sugiyama T, Yamato M, Okano T. Promotion of mouse ameloblast proliferation by Lgr5 mediated integrin signaling. J. Cell. Biochem. 2013;114:2138–2147. doi: 10.1002/jcb.24564. [DOI] [PubMed] [Google Scholar]
- Zeller R, López-Ríos J, Zuniga A. Vertebrate limb bud development: moving towards integrative analysis of organogenesis. Nat. Rev. Genet. 2009;10:845–858. doi: 10.1038/nrg2681. [DOI] [PubMed] [Google Scholar]
- Zhang R, Yang G, Wu X, Xie J, Yang X, Li T. Disruption of Wnt/β-catenin signaling in odontoblasts and cementoblasts arrests tooth root development in postnatal mouse teeth. Int. J. Biol. Sci. 2013a;9:228–236. doi: 10.7150/ijbs.5476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Verheyden JM, Hassell JA, Sun X. FGF-regulated Etv genes are essential for repressing Shh expression in mouse limb buds. Dev. Cell. 2009;16:607–613. doi: 10.1016/j.devcel.2009.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Gutierrez D, Li X, Bidlack F, Cao H, Wang J, Andrade K, Margolis HC, Amendt BA. The LIM homeodomain transcription factor LHX6: a transcriptional repressor that interacts with pituitary homeobox 2 (PITX2) to regulate odontogenesis. J. Biol. Chem. 2013b;288:2485–2500. doi: 10.1074/jbc.M112.402933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Oka K, Bringas P, Kaartinen V, Chai Y. TGF-β type I receptor Alk5 regulates tooth initiation and mandible patterning in a type II receptor-independent manner. Dev. Biol. 2008;320:19–29. doi: 10.1016/j.ydbio.2008.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Li S, Han D, Kaartinen V, Chai Y. Alk5-mediated transforming growth factor β signaling acts upstream of fibroblast growth factor 10 to regulate the proliferation and maintenance of dental epithelial stem cells. Mol. Cell. Biol. 2011;31:2079–2089. doi: 10.1128/MCB.01439-10. [DOI] [PMC free article] [PubMed] [Google Scholar]