Abstract
Serine protease inhibitors (serpins) are a diverse family of proteins that is conserved across taxa. The diversity of Amblyomma americanum serpins (AAS) is far more complex than previously thought as revealed by discovery of 57 and 33 AAS transcripts that are respectively expressed in male and female A. americanum ticks, with 30 found in both. While distinct reproductively, both male and female metastriate ticks, such as A. americanum, require a blood meal. Thus, 30 AAS sequences found in both male and female ticks could play important role(s) in regulating tick feeding and thus represent attractive candidates for anti-tick vaccine development. Of significant interest, 19 AAS sequences expressed in male and female ticks are also part of the 48 AAS sequences expressed in fed female tick salivary glands or midguts; two organs through which the tick interacts with host blood and immune response factors. Considered the most important domain for serpin function, the reactive center loop (RCL) is further characterized by a single ‘P1’ site amino acid residue, which is central to determining the protease regulated by the serpin. In this study, a diversity of 17 different P1 site amino acid residues were predicted, suggesting that A. americanum serpins potentially regulate a large number of proteolytic pathways. Our data also indicate that some serpins in this study could regulate target protease common to all tick species, in that more than 40% of AAS show 58–97% inter-species amino acid conservation. Of significance, 24% of AAS showed 62–100% inter-species conservation within the functional RCL domain, with 10 RCLs showing ≥90–100% conservation. In vertebrates, serpins with basic residues at the P1 site regulate key host defense pathways, which the tick must evade to feed successfully. Interestingly, we found that AAS sequences with basic or polar uncharged residues at the putative P1 site are more likely to be conserved across tick species. Another notable observation from our data is that AAS sequences found only in female ticks and those found in both males and females, but not those found only in male ticks, were highly conserved in other tick species. While descriptive, this study provides the basis for more in-depth studies exploring the roles of serpins in tick feeding physiology.
Keywords: Amblyomma americanum, serine protease inhibitors (serpins), tick feeding physiology, orthologous serpins
INTRODUCTION
Ticks are among the most successful ectoparasites of humans and animals. In livestock production, an estimated 80% of the world’s cattle are affected by tick-borne diseases (TBD), of which the most important include theileriosis, babesiosis, anaplasmosis, and heartwater (Marcelino et al., 2012). In addition to pathogen transmission, tick feeding has been documented to cause severe direct effects including paralysis, exsanguination, reduced livestock productivity, and damage to skin impacting the economic value of hides (Klompen et al., 1996; Jongejan and Uilenberg, 2004). In public health, ticks have been implicated in the transmission of 17 diseases affecting humans, and the list continues to grow (Day, 2011; Dantas-Torres et al., 2012; Savage et al., 2013).
Amblyomma americanum has emerged among the most important tick species in public health in the United States. A. americanum transmits multiple TBD agents, including Ehrlichia chaffeensis, E. ewingii, Rickettsia amblyommii, Francisella tularensis, the as yet undescribed causative agent of southern tick associated rash illness (STARI), Cytauxzoon felis, Theileria cervi, and the emerging human Heartland virus (Waldrup et al., 1992; James et al., 2001; Childs and Paddock, 2003; Telford and Goethert, 2004; Telford III et al., 2008; de la Fuente et al., 2008; Goddard, 2009; Schulze et al., 2011; Savage et al., 2013). For many years, this tick was mainly distributed in the southeastern United States, but has now been reported as established in 32 states throughout the Southeast, South Central, and Midwest regions as well as along the eastern seaboard as far north as Maine (Springer et al., 2014). Its emerging geographic expansion and role as a vector of many important human disease agents makes A. americanum an important consideration in strategies to improve public health.
With a lack of effective vaccines against TBD agents, the prevention of these infections in humans and animals depends on the control of ticks, which is currently acaricide based. However, acaricide use comes with many disadvantages, including the threat of food and environmental contamination and resistance development to these chemicals by ticks. A promising alternative strategy to the chemical control of ticks is to vaccinate hosts against the ticks themselves. The prerequisite to the development of an effective vaccine however, is the identification of effective molecular targets against tick feeding success and/or pathogen transmission. Among the emerging candidates for vaccine target antigens are members of the serine protease inhibitor (serpin) family. In animals, pathways critical to life, such as blood coagulation, complement activation, and inflammation are tightly regulated by serpins (Moore et al., 1993; Gettins, 2002; Tekin et al., 2005; Huntington, 2006; Huntington, 2011; Gatto et al., 2013). Furthermore, dysfunctional serpin activity in humans has been cited to cause numerous diseases including cirrhosis, emphysema, blood coagulation disorders, and dementia (Stein and Carrell, 1995; Davis et al., 1999; Gooptu and Lomas, 2009; Mocchegiani et al., 2011; Benson and Wilkes, 2012; Bosche et al., 2012; Gatto et al., 2013). The tick feeding style of lacerating host tissue and imbibing host blood which bleeds into the feeding site, is expected to provoke tissue repair and immune response mechanisms such as platelet aggregation, inflammation, blood clotting, and complement pathways (Ribeiro, 1989; Wikel et al., 1994), all of which are serpin-regulated. Thus, to complete feeding, ticks have to overcome serpin-regulated host defense pathways. From this perspective, it is conceivable that ticks may utilize serpins to block these host defenses against tick feeding (Muleng et al., 2001; Mulenga et al., 2002).
Serpin-encoding cDNAs have now been cloned from several tick species (Nene et al., 2002; Sugino et al., 2003; Mulenga et al., 2003a; Mulenga et al., 2003b; Imamura et al., 2005; Ribeiro et al., 2006; Imamura et al., 2006; Prevot et al., 2007; Chalaire et al., 2011; Yu et al., 2013). On the basis of unique putative functional domain reactive center loops (RCLs), at least 45 serpins are expressed in I. scapularis (Mulenga et al., 2009). Similarly, Mulenga et al., (Mulenga et al., 2007) and Karim et al., (Karim et al., 2011) have reported at least 17 and 32 different serpin transcripts expressed in A. americanum and A. maculatum, respectively. Data are now emerging which support the idea that some tick-encoded serpins are functional inhibitors associated with counter defense against anti-tick responses in the host, such as inflammation, complement activation, platelet aggregation, and blood clotting (Imamura et al., 2005; Prevot et al., 2009; Chmelar et al., 2011; Chmelar et al., 2012; Mulenga et al., 2013). In other studies, a significant reduction in feeding efficiency has been observed in ticks which fed on animals immunized with recombinant tick serpins (Imamura et al., 2005; Imamura et al., 2006; Prevot et al., 2007; Imamura et al., 2008; Kaewhom et al., 2009; Jittapalapong et al., 2010) suggesting a prime importance of serpins in tick feeding physiology.
An observation in our lab is that while overall amino acid conservation levels for serpins are around 35–45%, there are some serpins which show much higher conservation across all tick species investigated. We believe that highly conserved serpins could play crucial role(s) in tick physiology. The goal of this study was two-fold: first, to identify serpin transcripts expressed in unfed and fed A. americanum ticks, and second, to conduct a global intra- and inter-tick species bioinformatic analysis of A. americanum serpins (AAS) and other tick serpins. This study has described 57 and 33 AAS sequences that were respectively found only in male and female ticks, and a further 30 that were found in both. Nearly half of the serpin sequences expressed in A. americanum are predicted to regulate pathways important to all tick species, as they show 58–97% amino acid conservation in both metastriate and prostriate ticks. Although this study is descriptive, data presented here provide a foundation for further in depth studies on the roles of serpins in tick physiology.
MATERIALS AND METHODS
Identification and sequence analysis of A. americanum serpin (AAS) transcripts
AAS sequences used in this study were obtained by data mining of de novo assembled A. americanum transcriptomes (unpublished data), serendipitously while attempting to clone other targets, and from GenBank (Mulenga et al., 2007). A. americanum transcriptomes were assembled from Illumina sequence reads (BioProject accession number PRJNA226980) of 24 and 96h fed female phage display cDNA expression libraries, unfed and fed male and unfed and 24h fed female whole ticks, as well as 48, 96 and 120h fed tick dissected salivary gland (SG) and midgut (MG) tissues, using two approaches. In the first approach libraries were individually assembled with source library information for each contig retained, and in the second approach reads from all sources were combined and assembled (unpublished).
Mining and identification of putative AAS sequences was accomplished in two steps. In the first step, assembled contigs were subjected to batch blastx screening against tick sequences in GenBank. In the second step, contig sequences with matches to serpin sequences were manually inspected to confirm the presence of two consensus amino acid motifs: the reactive center loop (RCL) “"p17 [E]-p16 [E/K/R]-p15 [G]-p14 [T/S]-p13 [X]-p12-9 [AGS]-p8-1 [X]-p1' -4'” in the C-terminus, and the ‘NAVYFKG’ motif in the N-terminus (Carrell et al., 1987; Miura et al., 1995; Gettins, 2002). Sequences with a unique RCL sequence were identified as new and assigned an AAS number. Sequences without an RCL region were declared partial in the C-terminus region. For these sequences, two comparisons were made using Bl2seq-blastp (NCBI). First, these sequences were compared to each other to cluster contigs representing the same serpin. Next, we compared these clusters to AAS containing an RCL to eliminate redundancy between these groups. In addition to consensus amino acid motifs and secondary structure, a typical serpin ranges from 350–450 amino acids long (Gettins, 2002). Thus, sequences that had a starting methionine and were at least 350 amino acid residues long were considered putatively full-length. Full-length sequences were subjected to SignalP Version 4 web server to detect signal peptides (Petersen et al., 2011).
Relative AAS transcript abundance
To get insight into relative abundance, Illumina reads were mapped back to assembled contigs using the map reads to reference option in CLC genomics workbench vers. 6.4.2. Relative abundance values were adjusted to account for contig size and total library reads using the following equation: ey = (nyNxL1x/nxNyLy) × ex, where ex and ey represent normalized relative abundance levels of AAS transcript in libraries X and Y, Nx and Ny represent the total number of reads in libraries X and Y, nx and ny represent the number of reads related to the specific AAS transcript in libraries X and Y, and Lx and Ly represent the length of the contig related to the specific AAS transcript in libraries X and Y.
Phylogeny and comparative sequence analysis among AAS sequences
To determine relationships between AAS sequences, a guide phylogeny tree was constructed using the neighbor-joining method in MacVector vers 12 DNA analysis software (MacVector Inc., Cary, North Carolina). Sequences were first aligned using T-coffee, then a phylogeny tree out-rooted from human antithrombin (CAA48690), was constructed using the neighbor-joining method set to the default bootstrap setting of 1000 replications and differences adjusted using the absolute # differences setting. Subsequently, AAS sequences that clustered together on the phylogeny tree were subjected to pairwise sequence alignment analyses using MacVector. For partial sequences, amino acid identity levels were determined based on available sequences.
Comparative analyses of AAS to other tick serpin sequences
To investigate relationships among all available tick serpins, AAS and other tick serpins from publically available databases were subjected to phylogeny analysis and multiple sequence alignment analyses using alignment tools at NCBI and MacVector vers 12. This analysis was done at the whole amino acid sequence and RCL levels. At the whole amino acid sequence level, amino acid sequences were subjected to batch pairwise comparisons using Bl2seq-blastp (NCBI) to identify AAS orthologs in other tick species. The serpin RCL is an important functional domain, which determines what protease is regulated by a candidate serpin. Thus, to investigate the relationship of AAS sequences with other tick serpins at the functional level, the neighbor-joining method in MacVector (MacVector Inc.), was used to construct guide phylogeny trees using putative RCLs. To manage the huge dataset, RCLs were first divided into four groups based on charge and polarity characteristics of the amino acid residue at the putative P1 site: polar basic, polar acidic, polar uncharged, and hydrophobic. A separate tree was constructed for each group. Next intra-clade pairwise alignments of RCLs were performed using MacVector to determine identity levels.
RESULTS
Amblyomma americanum male and female ticks express large numbers of serpin transcripts
Data mining of transcriptomes from fed and unfed male and female whole ticks, as well as dissected 48, 96 and 120h female SG and MG transcriptomes, identified 28 and 57 AAS sequences respectively found only in females and males, respectively, and an additional 30 found in both (Table 1, Supplemental Tables 1 and 2). Mulenga et al., (2007) described 17 AAS (here after identified as AAS1–17) sequences that were expressed in 120h fed ticks. Of these 17 AAS sequences, this study found 7 sequences in both males and females, and five AAS sequences in females, while the remaining five were not found at all. Taken together, these studies show the total number of AAS sequences found only in female ticks to be 33 (Table 1, Supplemental Table 1). Please note that AAS 77 and 78 were found only in the combined A. americanum transcriptome were source library information for assembled contigs was not retained, and thus source information is unknown (Supplemental Table 1). Overall, 87 and 63 AAS sequences were found in male and female ticks, respectively (Supplemental Table 1 and 2).
Table 1.
Updated list of Amblyomma americanumserpin (AAS) transcripts
| Table 1A: AAS sequences found in male ticks | ||||||||
|---|---|---|---|---|---|---|---|---|
| AAS# ID | Accession# | Comment | AAS# ID | Accession# | Comment | AAS# ID | Accession# | Comment |
| 44 | GAYW01000312 | Partial | 73 | GAYW01000241 | Partial | 95 | GAYW01000268 | Partial |
| 46 | GAYW01000194° | Partial | 74 | GAYW01000254 | Partial | 96 | GAYW01000269 | Partial |
| 48 | GAYW01000205° | Full, NSP | 75 | GAYW01000255 | Partial | 97 | GAYW01000270 | Partial |
| 49 | GAYW01000207 | Partial | 79 | GAYW01000231 | Partial | 98 | GAYW01000271 | Partial |
| 50 | GAYW01000208° | Partial | 80 | GAYW01000243 | Partial | 99 | GAYW01000272 | Partial |
| 52 | GAYW01000285 | Partial | 81 | GAYW01000232 | Partial | 100 | GAYW01000273 | Partial |
| 53 | GAYW01000287 | Full, NSP | 82 | GAYW01000247 | Partial | 101 | GAYW01000275 | Partial |
| 55 | GAYW01000292° | Partial | 83 | GAYW01000249 | Partial | 102 | GAYW01000277 | Partial |
| 56 | GAYW01000294 | Full, NSP | 84 | GAYW01000251 | Partial | 103 | GAYW01000279 | Partial |
| 57 | GAYW01000295 | Full, NSP | 85 | GAYW01000235 | Partial | 104 | GAYW01000280 | Partial |
| 58 | GAYW01000298 | Full, NSP | 86 | GAYW01000237 | Partial | 105 | GAYW01000281 | Partial |
| 59 | GAYW01000302 | Full, NSP | 87 | GAYW01000256 | Partial | 106 | GAYW01000282 | Partial |
| 60 | GAYW01000304 | Full, NSP | 88 | GAYW01000258 | Partial | 107 | GAYW01000283 | Partial |
| 61 | GAYW01000310 | Partial | 89 | GAYW01000259 | Partial | 111 | GAYW01000225 | Partial |
| 62 | GAYW01000311 | Partial | 90 | GAYW01000262 | Partial | 113 | GAYW01000240 | Partial |
| 64 | GAYW01000246 | Partial | 91 | GAYW01000264 | Partial | 114 | GAYW01000230 | Partial |
| 68 | GAYW01000308° | Partial | 92 | GAYW01000265 | Partial | 120 | GAYW01000253° | Partial |
| 70 | GAYW01000197 | Partial | 93 | GAYW01000266 | Partial | 121 | GAYW01000257° | Partial |
| 71 | GAYW01000313 | Partial | 94 | GAYW01000267 | Partial | 122 | GAYW01000239 | Partial |
| Table 1B: AAS sequences found in female ticks | ||||||||
|---|---|---|---|---|---|---|---|---|
| AAS# ID | Accession# | Comment | AAS# ID | Accession# | Comment | AAS# ID | Accession# | Comment |
| 2 | ABS87354 | Full, SP | 20 | GAYW01000324° | Full, SP | 43 | GAYW01000156 | Partial |
| 5 | ABS87357 | Full, SP | 22 | GAYW01000149° | Partial | 45 | GAYW01000131 | Partial |
| 9 | ABS87361 | Full, SP | 26 | GAYW01000039° | Partial | 65 | GAYW01000368 | Partial |
| 11 | ABS87363 | Full, NSP | 29 | GAYW01000365° | Partial | 69 | GAYW01000330 | Partial |
| 12 | ABS87364 | Full, SP | 32 | GAYW01000130 | Partial | 76 | ^ | Full, NSP |
| 13 | ABS87365 | Full, SP | 36 | GAYW01000364° | Full, NSP | 109 | GAYW01000172° | Partial |
| 14 | ABS87366 | Full, SP | 37 | GAYW01000322° | Partial | 112 | GAYW01000397 | Partial |
| 15 | ABS87367 | Full, SP | 38 | GAYW01000044 | Partial | 115 | GAYW01000125 | Partial |
| 16 | ABS87368 | Full, SP | 40 | GAYW01000047 | Partial | 116 | GAYW01000070 | Partial |
| 17 | ABS87369 | Full, SP | 41 | GAYW01000021 | Partial | 117 | GAYW01000067 | Partial |
| 19 | GAYW01000076° | Full, SP | 42 | GAYW01000022 | Partial | 118 | GAYW01000073 | Partial |
| Table 1C: AAS sequences found in both male and female ticks | ||||||||
|---|---|---|---|---|---|---|---|---|
| AAS# ID | Accession# | Comment | AAS# ID | Accession# | Comment | AAS# ID | Accession# | Comment |
| 1 | ABS87353 | Full, SP | 24 | GAYW01000037° | Partial | 47 | GAYW01000316° | Full, NSP |
| 3 | ABS87355 | Full, SP | 25 | GAYW01000015° | Full, NSP | 51 | GAYW01000367° | Partial |
| 4 | ABS87356 | Full, SP | 27 | GAYW01000017° | Full, SP | 54 | GAYW01000363° | Partial |
| 6 | ABS87358 | Full, SP | 28 | GAYW01000019° | Full, NSP | 63 | GAYW01000286° | Partial |
| 7 | ABS87359 | Full, NSP | 30 | GAYW01000321° | Full, SP | 66 | GAYW01000309° | Partial |
| 8 | ABS87360 | Full, SP | 31 | GAYW01000315° | Full, SP | 67 | GAYW01000317° | Full, NSP |
| 10 | ABS87362 | Full, SP | 33 | GAYW01000184 | Partial | 72 | GAYW01000344° | Partial |
| 18 | GAYW01000325° | Full, NSP | 34 | GAYW01000293° | Full, NSP | 108 | GAYW01000260° | Partial |
| 21 | GAYW01000077° | Full, SP | 35 | GAYW01000288° | Partial | 110 | GAYW01000276° | Partial |
| 23 | GAYW01000078° | Full, SP | 39 | GAYW01000018° | Full, SP | 119 | GAYW01000221° | Partial |
| Table 1D: AAS sequences with no source information | |||||
|---|---|---|---|---|---|
| AAS# ID | Accession# | Comment | AAS# ID | Accession# | Comment |
| 77 | ^ | Partial | 78 | Clust1-1-153 | Partial |
Full = full length open reading frame, SP = signal peptide is present, NSP = no signal peptide;
indicates more than one accession number associated with AAS (see Supplemental Table 1);
contig sequence was too short for GenBank submission (<200bp), hyperlinked in Supplemental Table 1.
Assignment of AAS identification numbers was done arbitrarily in three steps. In the first step, assembled A. americanum transcriptomes were subjected to batch blastx scanning against tick serpin sequence entries at NCBI. This analysis identified 388 contigs that encoded putative serpins. A typical serpin is characterized by a unique RCL region (Gettins, 2002). Thus in the second step, we conducted a manual inspection of all 388 contigs and found 61 previously unknown AAS RCL sequences that did not show identity to previously described AAS1–17 (Mulenga et al., 2007). AAS sequences encoding the 61 new RCLs were assigned the identifications of AAS18–78, according to the order in which they were discovered. 233 of the 388 contigs did not have RCL regions. These sequences were subjected to intra-contig comparisons using the bl2seq-blastn function at NCBI. This analysis identified 44 contig sequences that encoded previously unreported AAS sequences and were designated as AAS79–122 (Table 1). This brought the total number of unique transcripts identified in this study to 105. A typical serpin molecule is 350–450 amino acids long (Gettins, 2002). On this basis, 40 of the 122 AAS sequences were determined to have complete open reading frames (ORF), as well as the consensus serpin amino-terminus motif (NAVYFKG), and the start methionine. Of the 40 AAS ORFs, 23 are predicted to have signal peptides (Table 1).
Majority of female AAS transcripts expressed in fed salivary glands and midgut tissues
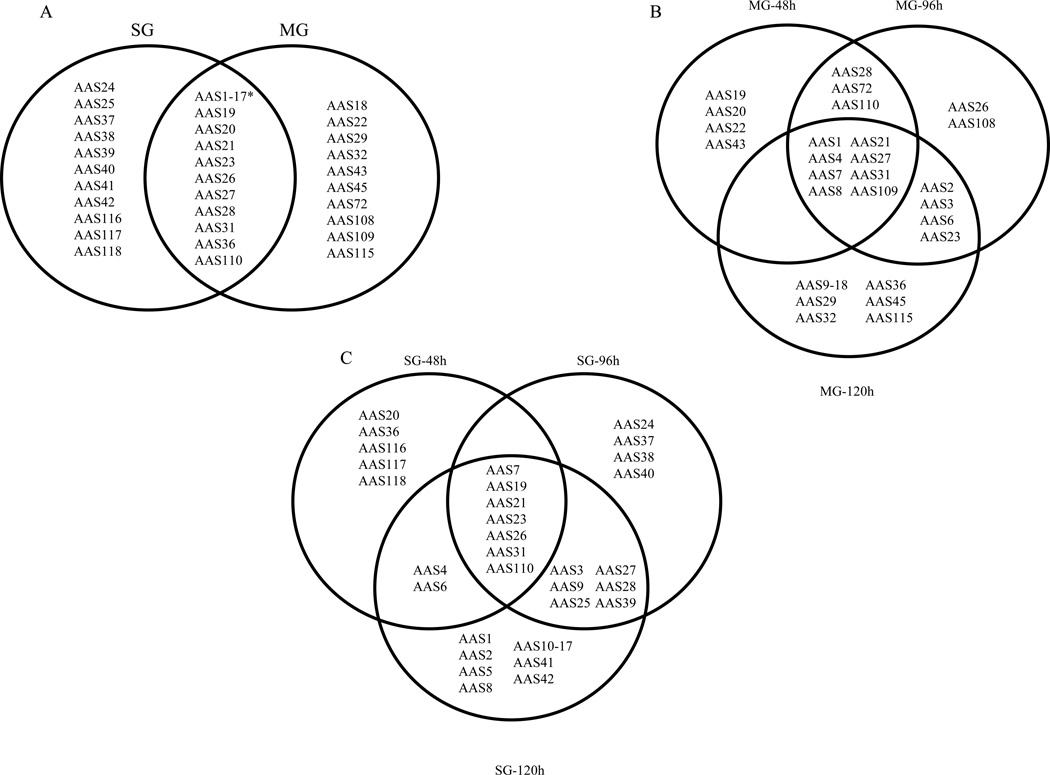
Figures 1A–C summarizes the different AAS sequences found in MG and SG of 48, 96 and 120h fed female ticks. Data previously reported for AAS1–17 are indicated with an asterisk in Figure1A, and data for MG and SG at 120h is taken from Mulenga et. al., (2007). Of the 63 AAS sequences found in female tick transcriptomes to date, 48 have been found in 48, 96, and 120h MG and/or SG. Of these 48, 10 and 12 AAS sequences were respectively found only in MG or in SG, while the remaining 26 were found in both (Figure 1A). This translates to a total of 36 and 38 different AAS sequences found in MG and in SG, respectively. Of the 36 AAS found in MG, seven (AAS1, 4, 7, 8, 21, 31, and 109) were found at all-time points, four (AAS27, 28, 72, and 110) were found at both 48 and 96h time points, and four (AAS19, 20, 22 and 43), one (AAS108), and 16 (AAS9–18, 23, 29, 32, 36, 45, and 115) were found at the 48, 96, or 120h time points, respectively (Figure 1B). Likewise, of the 38 AAS found in SG (Figure 1C), five (AAS7, 19, 21, 23, and 110) were found at all-time points, seven (AAS3, 9, 25, 27, 28, 31, and 39) were found at the 96 and 120h time points, and five (AAS20, 36, 116, 117, and 118), five (AAS24, 26, 37, 38 and 40), and 12 (AAS1, 2, 5, 8, 10–17, 41, and 42) were found in 48, 96, or 120h SG, respectively.
Figure 1. Adult female A. americanum salivary gland (SG) and midgut (MG) expressed serpin (AAS) transcripts.
(A) Total AAS sequences found in MG and SG and both at all tested time points, (B) Apparent temporal and spatial distribution of AAS transcripts found in MG (B), and SG (C). Please note that 17 previously characterized SG and MG expressed AAS transcripts [42] are not included here.
Relative AAS transcript abundance
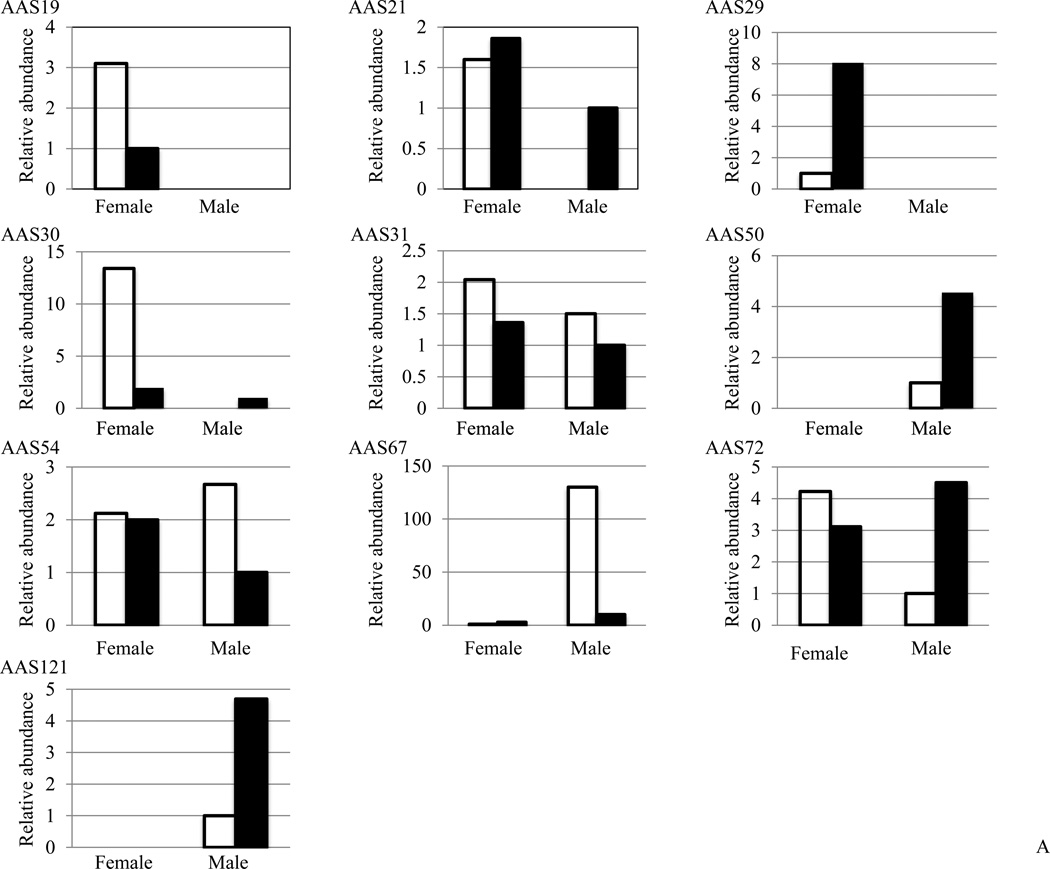
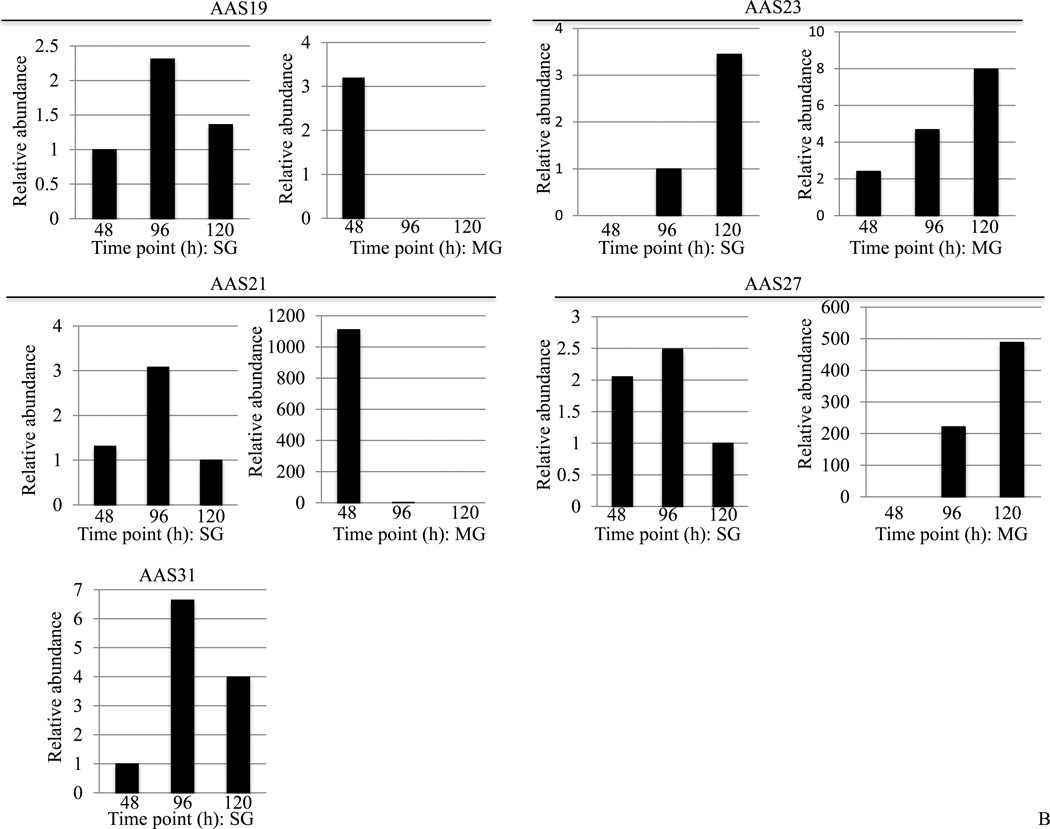
We successfully mapped reads back to de novo assembled AAS contigs. This analysis identified 20 of 122 AAS transcripts with variable sequence reads in different libraries (Figure 2A and 2B, Supplemental Table3). For 8 (AAS25, 28, 39, 51, 66, 108, 110, and 119) of the 20 AAS sequences, differences between sequence reads were minimal (Supplemental Table 3) and we concluded that these apparently occurred in equivalent abundance in all libraries. Figure 2A summarizes differential abundance for ten (AAS19, 21, 23, 27, 29, 30, 31, 50, 54, 67, 72, 121) in unfed and fed, male and female whole tick libraries. Of ten AAS detected at unfed and fed time points, five are more abundant in unfed ticks (AAS19, 30, 31, 54, and 67), four are more abundant in fed ticks (AAS21, 29, 50, and 121), and one (AAS72) showed a mixed pattern with abundance in unfed females, but not in unfed males, and a transcript increase in fed male ticks (Figure 2A). It is also notable that AAS19, 21, 29, and 30 are abundant in female ticks, while AAS50, 67, and 121 are predominant in male ticks. Of the 31 AAS sequences summarized in Figure 1, we determined differential abundance in SG and MG for five transcripts, AAS19, 21, 23, 27, and 31 (Figure 2B), with differences for the remaining sequences being minimal to negligible (Supplemental Table 3). While AAS23 transcript abundance increases with feeding in both SG and MG, the remaining show an apparent dichotomous pattern. Transcripts for AAS19, 21, 27, and 31 are highest at the 96h time point, and lower at the 120h time point in SG. In MG, AAS19 and 21 are abundant at 48h, and absent or low at 96 and 120h time points, while AAS27 and 31 increase with feeding (Figure 2B).
Figure 2. Relative abundance of A. americanum serpin (AAS) transcripts: Sequence reads were mapped back to de novo assembled contigs in different libraries.
Relative abundance values in: (2A) unfed (empty bars) and fed (black filled bars) male and female ticks, and (2B) dissected 48–120h salivary glands (SG) and midguts (MG) were calculated using the formula described in materials and methods..
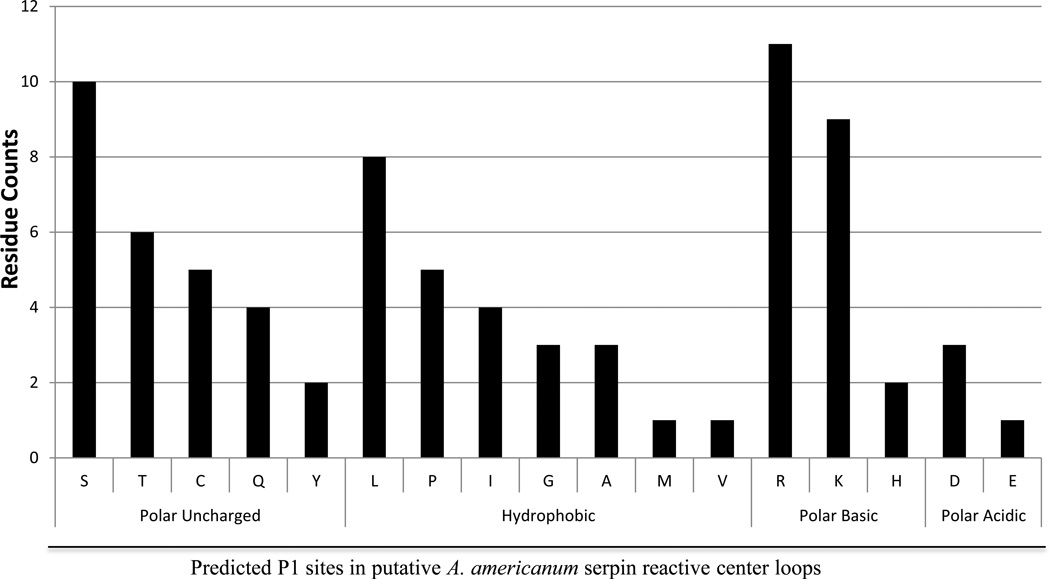
A diversity of seventeen amino acid residues is predicted at P1 sites of AAS putative RCLs
Table 2 lists 78 predicted reactive center loops (RCLs) from 27 (Table 2A) and 22 (Table 2B) AAS sequences found in female and male ticks, the 27 found in both (Table 2C), and 2 of an undetermined source (Table 2D). Please note that Table 2A includes previously characterized AAS1–17 (Mulenga et al., 2007). We would like to note that the RCL regions for AAS68 and 71 (Table 2B), and AAS77 (Table 2D) are partial (marked with an asterisk in Table 2), however based on the available sequence we were still able to conclude these RCLs to be unique. Additionally, while the predicted RCL for AAS8 and 9, 4 and 12, and 13 and 15 are identical, these sequences differ in the N-terminus region by 13–25 amino acids (Mulenga et al., 2007). Numbering of amino acid residues in the RCL is based on the standard nomenclature developed by Schechter and Berger (Schechter and Berger, 1967), in which amino acid residues at the N-terminal end of the scissile bond (P1-P1) are not primed and those on the C-terminal end are primed: “"p17 [E]-p16 [E/K/R]-p15 [G]-p14 [T/S]-p13 [X]-p12-9 [AGS]-p8-1 [X]-p1' -4'” (Miura et al., 1995; Gettins, 2002). Molecular analysis predictions of the P1 site assume that there are 17 amino acid residues between the beginning of the RCL hinge region (P17), and the scissile bond (P1-P1’), (Hopkins and Stone, 1995). Based on these conventions, a diversity of 17 different amino acid residues is predicted at the P1 sites of AAS1–78 (except for AAS71 which has a partial RCL excluding the P1 site, Table 2B). The P1 residues for AAS1–78 have the following charge and polarity properties: 27 (~ 35%) are polar uncharged [S (10/26), C (5/26), T (6/26), Q (4/26), Y (2/26)], 25 (~32%) are hydrophobic [L (8/25), I (4/25), P (5/25), G (3/25), M (1/25), A (3/25), V (1/25)], 21 (27%) are polar basic [R (11/21), K (9/21), H (1/21)], and four residues (~ 5%) are polar acidic [D (3/4), E (1/4)] (Figure 3). It is noteworthy that 11 of the 22 AAS sequences found in MG and SG have basic residues at the predicted P1 site.
Table 2.
Reactive Center Loops (RCLs) of A. americanumserpins
| Table 2A: Predicted RCLs found in female ticks | |||
|---|---|---|---|
| AAS ID | RCL amino acid sequence | AAS ID | RCL amino acid sequence |
| 2 | EEGTVAAGVTSVRVKPKSFAR | 29 | EEGTEAAAATAVTVVDGCMPR |
| 5 | EEGTVAAAVTGLSVTPLVVPP | 32 | EDGTEPEVAPTNAVFQPAVRT |
| 9 | EEGSPATAVTGVIMYTQSAFV | 36 | EEGTEAAAATGMRIQLKTRVK |
| 11 | EEGTVPTAVPGILLVGLVARH | 37 | EEGTEAAAATAVVMMCRSAAM |
| 12 | EEGTIAAAVTGLSFVPISALH | 38 | EEGTIATAVTGLSFAPISALH |
| 13 | EEGTVAAAVTGLSFPHLVVPP | 40 | EKGRAAAGVAARAFYTRAGDH |
| 14 | EEGTVATAVTGISLVALSALH | 41 | EEGSEAAGATGVVFVELIAVR |
| 15 | EEGTVAAAVTGLSFPHLVVPP | 42 | EKGTEAAAATAVMMMACCMSA |
| 16 | EEGTAAEAVTGLSITPLAVPP | 43 | EDGTEPEAAATNAVFQPAVRT |
| 17 | EEGTVAAAVTGLSSIALSSVG | 45 | EEGTEAAAATGVVMMCDSLPM |
| 19 | EEGSEAAAVTGFVIQLRTAAF | 65 | EEGSETDSATLMRISGKAXCE |
| 20 | EVGTRAVAATEAQFVSKSLVH | 69 | EEGTIAAAVTGLSFVATASFN |
| 22 | EEGSEAAGATGVIFYTKSAIV | 76 | EEGTIAAAVTGSLFRAHLGSP |
| 26 | EEGTEAAAATATVAMFGSAPS | ||
| Table 2B: Predicted RCLs Found in male ticks | |||
|---|---|---|---|
| AAS ID | RCL amino acid sequence | AAS ID | RCL amino acid sequence |
| 44 | EEGTEAAAATAIITTECCIMP | 59 | EEGSWPVTYTEHVLSTGDPVT |
| 46 | EEGSEAAGATGVIFVETIAVR | 60 | ENGSSAAAVTGTTLHKSVHVP |
| 48 | EEGTEAAAGSASILTRRDAVE | 61 | EGGTDASSATAMTSLACSATM |
| 49 | EDGTDSEAARANEVPEPAFSI | 62 | EEGTEPEAATANALIESAGSI |
| 50 | EEGSETDSATLMRISGKAAEE | 64 | ARGGRAVSNEVQSTTSATTAA |
| 52 | ENGTVAAAASAAIGVGSAGPS | 68 | EDGTVAASTAALAFHADRPF* |
| 53 | EGGTEPGPATAGEASAPAGPE | 70 | EEGTIATAVTGLSFVATASFN |
| 55 | ENGTVAAAATAAEGGSSSGIM | 71 | EVGTKA* |
| 56 | EEGLGEACRPPNPLPATMIAF | 73 | GAGGRPPSSNDSREAGTSPAK |
| 57 | ENGTGAPAAEGSIYSPAFRRR | 74 | ERSTSRMPKYTGAQGAPGTSS |
| 58 | EEGTEAAAATGMTLMMCGAMV | 75 | RRGPKTVAAQAVAKEAARTAK |
| Table 2C: Predicted RCLs found in both male and female ticks | |||
|---|---|---|---|
| AAS ID | RCL amino acid sequence | AAS ID | RCL amino acid sequence |
| 1 | EEGTVAAGVTSVRVKQKHSAR | 30 | EEGTVATAVTGLSNTRILDDS |
| 3 | EEGTGAAGVPSVGGKPKSFAR | 31 | EEGSEAAAVTGVVINTRTIGG |
| 4 | EEGTIAAAVTGLSFVPISALH | 33 | ENGTVAAAASAALLVGSAGPN |
| 6 | EEGTVATAVTGISLALSALHT | 34 | ENGTKAAAATTALGSNSFYVP |
| 7 | EEGTEAAAATGIAMMLMCARF | 35 | EKGTVAAASAAAAGGSSLYNP |
| 8 | EEGSQAAAVTGVIIYTQSAFV | 39 | EKGTVASASTVAIIVSRIGTP |
| 10 | EEGSQAAAVTGVIIYTQSAFV | 47 | EEGTEAAAATAMPAANSCEMF |
| 18 | EEGSEAAGATAVIFFTRGGSS | 51 | EEGTEAAAATAIIATECCIMP |
| 21 | EKGTEAVALSSGIIRHSKTPG | 54 | EDGVEGLFLTPLIMMCYAGVS |
| 23 | EEGTVAAAVTSIRMRMKSSRR | 63 | EDGNEIARTSALVTEVVSKVA |
| 24 | EEGTVATAVTGLSFVATASFN | 66 | EKGRAAAGVAARTFYTRAGDH |
| 25 | EEGTEAAAATAVVMMCYSLPM | 67 | EEGSEAAAATAVLIETRSDVP |
| 27 | EEGTEAAAASGVVGVNRIGID | 72 | EEPVRLDLHVSTLRLAERTDL |
| 28 | EEGTEAAAATGMVAMARCAII | ||
| Table 2D: Predicted RCLs, source undetermined | |||
|---|---|---|---|
| AAS ID | RCL amino acid sequence | AAS ID | RCL amino acid sequence |
| 77 | EEGSQATAVTGVIIYTQS* | 78 | EEGTEAAAATSVVMMCDSLPM |
Amino acid residues at putative P1 site are bolded;
indicates a partial RCL sequence.
Figure 3. Diversity and counts for amino acid residues at the putative P1 position in reactive center loops of the first 78 A. americanum serpin sequences.
Amino acid residues at putative P1 sites were determined based on molecular analysis predictions of the P1 site that assume that there are 17 amino acid residues between the beginning of the RCL hinge region (P17), and the scissile bond (P1-P1’) [54].
Some AAS sequences are highly identical
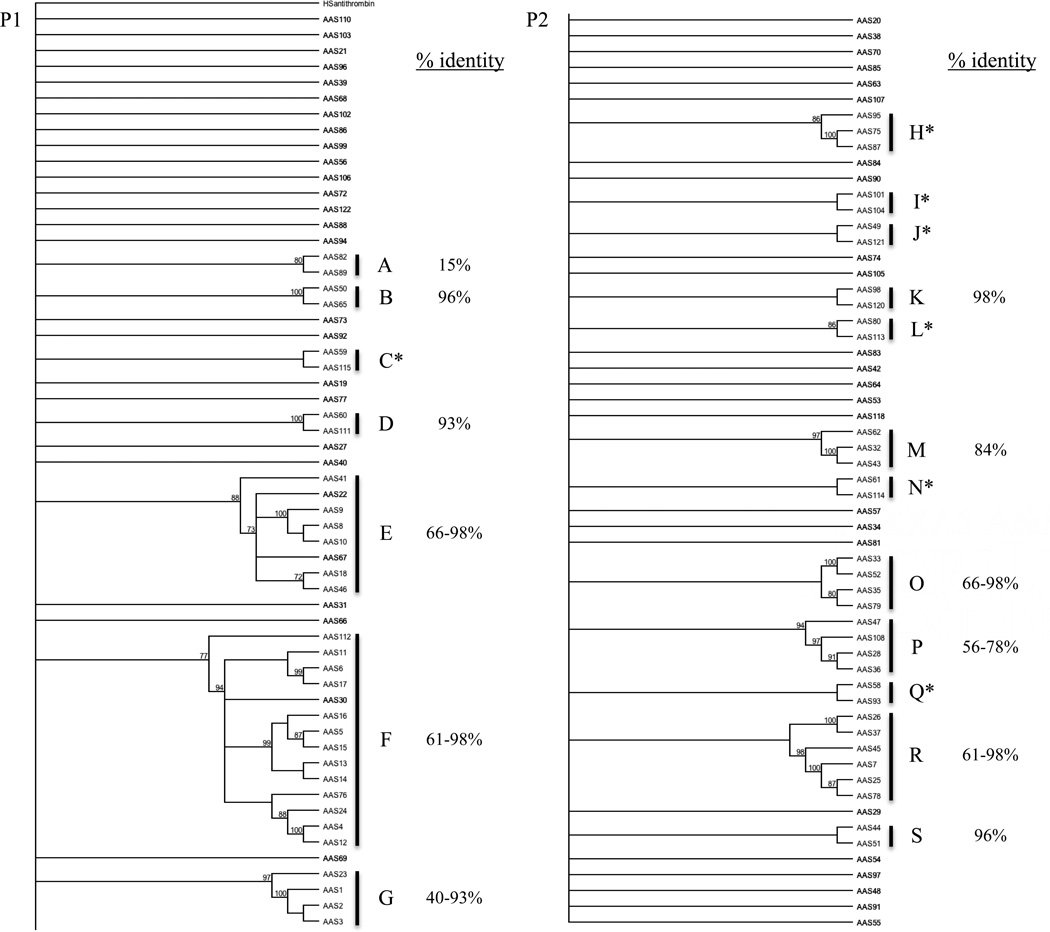
To gauge the relationships between AAS sequences, amino acid sequences were subjected to phylogeny analysis, except for AAS87 and 109, which were too short for an informative alignment (Figure 4). We would like to note that due to the large number of sequences, we split the tree into two parts, Figures 4P1 and 4P2. As shown in Figure 4, 68 of the 122 AAS sequences segregated into 19 clusters labeled A-S, and the remaining sequences did not cluster. Of the 19 clusters, eight have more than two sequences: E (AAS8–10, 18, 22, 41, 46, and 67), F (AAS4–6, 11–17, 24, 30, and 112), G (AAS1–3, 23), H (AAS75, 87, and 95), M (AAS32, 43, and 62), O (AAS33, 35, 52 and 79), P (AAS28, 36, 47, and 108), and R (AAS7, 25, 26, 37, 45, and 78), and the remaining 11 clusters have single pairs. When subjected to pairwise sequence alignment analysis, sequences in clusters A, B, D, E, F, G, K, M, O, P, R, and S showed variable amino acid identity levels of 15, 96, 93, 66–98, 61–98, 40–93, 98, 84 (excluding AAS62), 66–98, 56–78, 61–98, and 96%, respectively (Figure 4P1 and 4P2). In the remaining clusters (marked with asterisks), amino acid identity levels were below 15%. We would like to caution here that some sequences being compared are partial, and therefore the picture of these relationships might be incomplete. From pairwise alignment analyses, two general patterns emerged. In the first pattern, differences between two sequences were scattered throughout the alignment (not shown). In the second pattern, found between AAS25 and AAS45, and AAS33 and AAS52, differences were restricted to the C-terminus region within the RCL (not shown).
Figure 4. Phylogeny relationship of A. americanum serpin (AAS) sequences.
Translated AAS amino acid sequences and human antithrombin were aligned using T-coffee in in MacVector version 12. A bootstrap supported phylogeny tree was then constructed with human antithrombin (CAA48690) as the out-group using neighbor-joining method. Clades containing more than 2 AAS sequences are labeled “A” to “T”. Amino acid identity levels within each cluster are indicated. Clusters where amino acid identities were below 15% are marked with an asterisk (*) sign.
Close to half of AAS sequences have orthologs in other tick species
To determine if any AAS sequences in this study had orthologs in other tick species, inter-species comparisons were performed. A search for other tick serpin sequences in publically available databases retrieved 165 serpin sequences across nine tick species (Table 3). Pairwise comparisons of these serpins to AAS1–122 identified 50 AAS sequences that were conserved in other tick species. To manage the high number of sequences, the data are presented in separate tables: A. americanum versus A. maculatum (Karim et al., 2011) and A. variegatum (Ribeiro et al., 2011) (Table 4A), and versus R. pulchellus (direct submission), R. appendiculatus (Mulenga et al. 2003b), R. microplus (Tirloni et al., 2014), The Gene Index Project, http://compbio.dfci.harvard.edu/tgi/], R. haemaphysaloides (direct submission), H. longicornis (Imamura et al., 2005), I. scapularis (GenBank direct submissions, The Gene Index Project), and I. ricinus (Leboulle et al., 2002b) (Table 4B). As shown in Table 4A, AAS4–6, 8–19, 22–27, 29, 30, 41, 46, and 47 show ≥75–96% amino acid identity to A. maculatum serpin sequences AEO35533, AEO35520, AEO34312, AEO34447, AEO34313, AEO34314, AEO32541, AEO34349, AEO34279, AEO34218, AEO34217, AEO33019, and AEO32217, while the remaining sequences showed <75% amino acid conservation. Likewise, in Table 4B, AAS19–22, 41, 42, and 65 show amino acid identities of ≥75–91% to 7 R. pulchellus serpins: JAA54307, JAA54309, JAA543410, JAA54167, JAA54314, JAA54313, respectively, while the rest showed amino acid identities of <75%. Additionally, AAS7 shows 75 and 77% identity to R. microplus EST89704 and R. pulchellus JAA54312, and AAS19 and 21 show 82–96% identity to R. microplus TC17409, TC22658, and EST767976, while AAS41, 42, and 54 show 65, 79 and 90% identity to partially characterized R. appendiculatus AAK61378, and AAK61376, and R. microplus TC16456, respectively. Four AAS (24, 38, 69, and 70) showed 70% or greater identity to the same partially characterized R. appendiculatus AAK61377. Of the 50 AAS sequences conserved in other tick species, only 11 sequences (AAS19–21, 25, 37, 42, 44, 45, 54, 66 and 78) appear to be conserved in prostriate ticks (Table 4B). Except for AAS19, which showed 81 and 82% amino acid identity to I. ricinus ABI94058 and I. scapularis XP_00245308, respectively, all other AAS sequences showed 58–70% conservation with prostriate tick sequences (Table 4B).
Table 3.
Other tick serpins downloaded from publically available databases
Table 4.
|
A. americanum serpins (AAS) that have orthologs in other Amblyomma spp ticks | |||||
|---|---|---|---|---|---|
| AAS ID |
Other tick best match | % ID |
AAS ID |
Other tick best match | % ID |
| 1 | Amac-AEO32759 | 71 | 25 | Amac-AEO35533 | 81 |
| 2 | Amac-AEO32759 | 71 | Amac-AEO35520 | 76 | |
| 3 | Amac-AEO32759 | 70 | Amac-AEO32774 | 70 | |
| 4 | Amac-AEO34312 | 79 | Amac-AEO32541 | 63 | |
| Amac-AEO34279 | 89 | Amac-AEO32217 | 64 | ||
| 5 | Amac-AEO34312 | 75 | Amac-AEO32160 | 67 | |
| Amac-AEO34279 | 77 | Amac-AEO32154 | 70 | ||
| 6 | Amac-AEO34312 | 75 | 26 | Amac-AEO35533 | 64 |
| Amac-AEO34279 | 82 | Amac-AEO35520 | 64 | ||
| 7 | Amac-AEO35533 | 87 | Amac-AEO32774 | 62 | |
| Amac-AEO35520 | 86 | Amac-AEO32541 | 77 | ||
| Amac-AEO32774 | 72 | Amac-AEO32217 | 79 | ||
| Amac-AEO32541 | 65 | Amac-AEO32160 | 62 | ||
| Amac-AEO32217 | 66 | Amac-AEO32154 | 61 | ||
| Amac-AEO32160 | 70 | 27 | Amac-AEO34349 | 81 | |
| Amac-AEO32154 | 71 | 28 | Amac-AEO34314 | 62 | |
| 8 | Amac-AEO34447 | 82 | 29 | Amac-AEO35533 | 62 |
| Amac-AEO34313 | 64 | Amac-AEO35520 | 59 | ||
| Amac-AEO33019 | 71 | Amac-AEO32774 | 67 | ||
| 9 | Amac-AEO34447 | 79 | Amac-AEO32160 | 61 | |
| Amac-AEO34313 | 62 | Amac-AEO32154 | 65 | ||
| Amac-AEO33019 | 69 | 30 | Amac-AEO34312 | 76 | |
| 10 | Amac-AEO34447 | 82 | Amac-AEO34279 | 76 | |
| Amac-AEO34313 | 64 | 36 | Amac-AEO34314 | 59 | |
| Amac-AEO33019 | 72 | Amac-AEO35533 | 67 | ||
| 11 | Amac-AEO34447 | 63 | 37 | Amac-AEO35520 | 65 |
| Amac-AEO34313 | 69 | Amac-AEO32774 | 71 | ||
| 12 | Amac-AEO34447 | 78 | Amac-AEO32541 | 70 | |
| Amac-AEO34313 | 89 | Amac-AEO32217 | 73 | ||
| 13 | Amac-AEO34447 | 74 | Amac-AEO32160 | 68 | |
| Amac-AEO34313 | 78 | Amac-AEO32154 | 71 | ||
| 14 | Amac-AEO34447 | 74 | 41 | Avar-DAA34478 | 73 |
| Amac-AEO34313 | 80 | ||||
| 15 | Amac-AEO34447 | 73 | 46 | Avar-DAA34478 | 73 |
| Amac-AEO34313 | 77 | Amac-AEO34447 | 69 | ||
| Amac-AEO34313 | 76 | ||||
| Amac-AEO33019 | 75 | Amac-AEO33019 | 75 | ||
| 16 | Amac-AEO34447 | 72 | |||
| Amac-AEO34313 | 75 | 47 | Amac-AEO34314 | 79 | |
| 17 | Amac-AEO34447 | 75 | 67 | Amac-AEO34313 | 70 |
| Amac-AEO34313 | 79 | Amac-AEO33019 | 74 | ||
| 18 | Amac-AEO34447 | 68 | |||
| Amac-AEO34313 | 76 | ||||
| 19 | Amac-AEO32217 | 91 | |||
| Amac-AEO34218 | 91 | ||||
| 22 | Avar-DAA34478 | 71 | |||
| Amac-AEO34447 | 69 | ||||
| Amac-AEO34313 | 74 | ||||
| Amac-AEO33019 | 86 | ||||
| 23 | Amac-AEO32759 | 66 | |||
| 24 | Amac-AEO34312 | 77 | |||
| Amac-AEO34279 | 86 | ||||
| B: AAS amino acid identity to serpins in metastriata and prostriata ticks | ||||||||
|---|---|---|---|---|---|---|---|---|
| AAS ID | Other tick best match | % ID | AAS ID | Other tick best match | % ID | AAS ID | Other tick best match | %ID |
| 1 | Rhaem-AFX65224 | 61 | 25 | 45 | Rpulc-JAA54306 | 66 | ||
| Rapp-AAK61377 | 69 | Rapp-AAK61377 | 66 | Rpulc-JAA54314 | 71 | |||
| Rmic-AAP75707 | 68 | Rapp-AAK61375 | 62 | Rpulc-JAA54313 | 71 | |||
| Rmic-TC16466 | 73 | Rmic-TC24850 | 66 | Rpulc-JAA54312 | 68 | |||
| Rmic-EST89704 | 69 | Ir-XP_02399745 | 61 | |||||
| 4 | Rhaem-AFX65224 | 59 | Rpulc-JAA54306 | 70 | Ir-XP_002408111 | 60 | ||
| Rapp-AAK61377 | 66 | Rpulc-JAA54314 | 63 | |||||
| Rmic-AAP75707 | 65 | Rpulc-JAA54313 | 66 | 46 | Rapp-AAK61378 | 64 | ||
| Rmic-TC16466 | 67 | Rpulc-JAA54312 | 71 | Rpulc-JAA54312 | 73 | |||
| Hlong-BAD11156 | 73 | Rpulc-JAA54311 | 64 | |||||
| Ir-CAB55818.2 | 61 | 47 | Rpulc-JAA63611 | 59 | ||||
| 6 | Rapp-AAK61377 | 68 | Is-XP_002434444 | 59 | ||||
| Rmic-AAP75707 | 66 | Ir-00245308 | 70 | 50 | Rpulc-JAA54309 | 76 | ||
| Rmic-TC16466 | 69 | Ir-EW874987 | 70 | |||||
| Hlong-BAD11156 | 69 | 54 | Rmic-TC16466 Is-XP_002401187 |
90 58 |
||||
| 65 | Rpulc-JAA54309 | 80 | ||||||
| Rhaem-AFX65224 | 74 | Rmic-TC24850 | 61 | |||||
| Rmic-TC24850 | 69 | Rmic-EST89704 | 61 | 66 | Rpulc-JAA54310 Is-XP_002415891 |
70 59 |
||
| Rmic-EST89704 | 75 | Rpulc-JAA54306 | 61 | |||||
| 7 | Rpulc-JAA54306 | 72 | 26 | Rpulc-JAA54314 | 66 | 67 | Rpulc-JAA54310 | 68 |
| Rpulc-JAA54314 | 69 | Rpulc-JAA54313 | 63 | |||||
| Rpulc-JAA54313 | 69 | Rpulc-JAA54312 | 62 | Rapp-AAK61377 | 70 | |||
| Rpulc-JAA54312 | 77 | Rpulc-JAA54311 | 66 | 69 | Rmic-AAP75707 | 70 | ||
| Hlong-BAD11156 | 74 | Hlong-BAD11156 | 68 | |||||
| Rmic-TC24850 | 60 | Rapp-AAK61377 | 71 | |||||
| 8 | Rpulc-JAA54310 | 68 | 29 | Rmic-EST89704 | 62 | 70 | Rmic-AAP75707 | 71 |
| 9 | Rpulc-JAA54310 | 66 | Rpulc-JAA54310 | 74 | Hlong-BAD11156 | 73 | ||
| 10 | Rpulc-JAA54310 | 66 | 30 | Rapp-AAK61377 | 66 | 78 | Rpulc-JAA54306 | 65 |
| 11 | Rmic-TC16466 Hlong-BAD11156 |
62 62 |
Rmic-AAP75707 Rmic-TC16466 |
66 68 |
Rpulc-JAA54313 Rpulc-JAA54312 |
61 70 |
||
| Hlong-BAD11156 | 70 | Rpulc-JAA54314 | 63 | |||||
| 12 | Rmic-TC16466 Hlong-BAD11156 |
72 74 |
Rmic-AHC98652 | 62 | ||||
| 36 | Rpulc-JAA63611 | 69 | Rmic-AHC98653 | 68 | ||||
| 13 | Rmic-TC16466 | 67 | Rpuc-JAA62387 | 68 | Rmic-AHC98662 | 61 | ||
| 14 | Rmic-TC16466 | 70 | Rapp-AAK61376 | 61 | ||||
| 15 | Rmic-TC16466 | 66 | Rmic-EST89704 | 65 | Rhaem-AFX65225 | 68 | ||
| Rpulc-JAA54306 | 63 | Is-XP_00240811 | 61 | |||||
| 16 | Rmic-TC16466 | 66 | ||||||
| Rpulc-JAA54306 | 60 | Is-XP_002399745 | 61 | |||||
| 17 | Rmic-TC16466 | 69 | Rpulc-JAA54314 | 67 | Ir-JAA66228 | 61 | ||
| 18 | Rpulc-JAA54310 | 72 | 37 | Rpulc-JAA54313 | 64 | Ir-JAA72595 | 61 | |
| Rpulc-JAA54312 | 63 | Ir-JAA66227 | 61 | |||||
| Rmic-TC22658 | 95 | Rpulc-JAA54311 | 67 | |||||
| Rmic-EST67697 | 82 | Ir-CAB55818.2 | 60 | |||||
| 19 | Rpulc-JAA54307 | 87 | Is-ACI46630 | 61 | ||||
| Ir-ABI94058 | 81 | 114 | Rpulc-JAA54310 | 75 | ||||
| Is-XP_00245308 | 82 | |||||||
| Rapp-AAK61377 | 71 | Rpulc-JAA63611 | 67 | |||||
| 38 | Rmic-AAP75707 | 70 | 121 | Rpulc-JAA62387 | 67 | |||
| Rpulc-JAA54310 | 79 | |||||||
| 20 | Is-XP_002401986 | 58 | 39 | Ir-XP_002401986 | 68 | |||
| Rmic-TC17409 | 96 | 40 | Rpulc-JAA54310 | 67 | ||||
| Rpulc-JAA63258 | 91 | Rapp-AAK61378 | 65 | |||||
| 21 | Ir-JAB72483 | 67 | 41 | Rpulc-JAA54310 | 76 | |||
| Is-XP_002401986 | 70 | |||||||
| Rapp-AAK61376 | 79 | |||||||
| 22 | Rapp-AAK61378 | 61 | Rpulc-JAA54314 | 79 | ||||
| 23 | Rhaem-AFX65224 | 71 | 42 | Rpulc-JAA54313 | 77 | |||
| Rpulc-JAA54310 | 75 | Is-XP_002403236 | 61 | |||||
| Is-XP_002407493 | 61 | |||||||
| Rapp-AAK61377 | 71 | |||||||
| 24 | Rmic-AAP75707 | 71 | 44 | Rpulc-JAA54314 | 59 | |||
| Rmic-TC16466 | 73 | Ir-XP_02399745 | 58 | |||||
Amac = A. maculatum, Avar = A. variegatum, % amino acid identities are bed
Rapp = R. appendiculatus, Rmic = R. microplus, Rhaem = R. haemaphysaloides, Rpulc = R. pulchellus, Ir = I. ricinus, Is = I. scapularis, and Hlong = H. longicornis. % amino acid identities are bolded
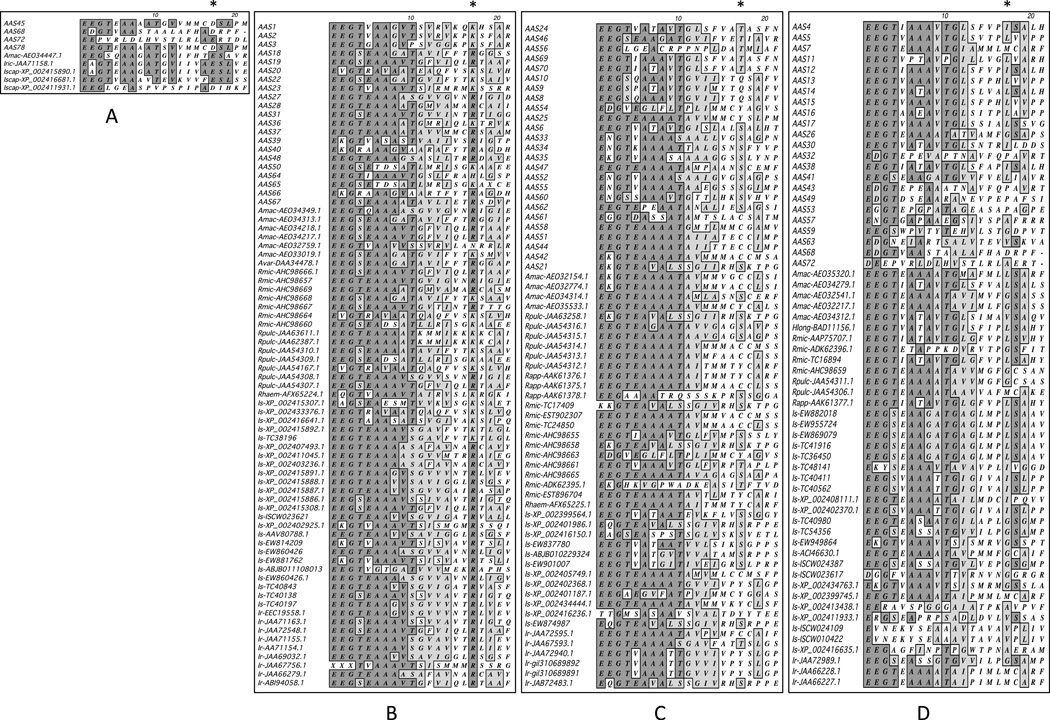
Table 5 summarizes the 29 AAS RCL sequences which show at least 62% sequence conservation in other tick species. Preliminary manual inspection of RCLs showed high identities between sequences where the predicted P1 site is of the same charge and polarity, therefore all 212 tick serpin sequences for which an RCL could be determined were first divided into one of four groups: polar uncharged, polar basic, polar acidic, and hydrophobic. RCL sequences (Figure 5A–D) were then subjected to phylogeny analysis (not shown). Lastly, RCLs from all tick species were subjected to pairwise sequence alignments. Identity levels for the 29 highly conserved AAS RCLs, (AAS4, 7, 12, 14, 18–23, 25–29, 31, 37, 38, 42, 44, 45, 47, 50, 52, 54, 58, 65, 69, and 70) ranged from approximately 62–100% (Table 5). Of these 29 RCLs, seven (AAS7, 19–21, 23, 27, and 42) are conserved in both metastriate and prostriate ticks, with the remaining being conserved only in metastriate ticks. Seven AAS RCLs (AAS18, 20–22, 27, 42, and 50) showed 95–100% conservation with at least one RCL from another species. For AAS25, RCL identity to its partially characterized ortholog in R. appendiculatus (AAK61375), was higher, at 81% identity, than for whole sequence identity, at 62%. Three AAS RCLs (AAS7, 19, and 20) showed >80% identity to at least one RCL from a prostriate species. The identities between remaining RCL sequences ranged between ~62–76% (Table 5). It is notable that the RCL for AAS19 is 100% identical to serpin RCLs across several tick species in multiple genera, and including both metastriate and prostriate ticks. Of the 29 conserved AAS RCLs, 38% (11/29) (AAS18–20, 22, 23, 27, 28, 31, 37, 50, 65) have basic amino acid residues, 35% (10/29) (AAS21, 25, 42, 44, 47, 52, 54, 58, 69, and 70) have polar uncharged residues, and 24% (7/29) (AAS4, 7, 12, 14, 26, 29, 38, and 42) have hydrophobic residues, while only one sequence, AAS45, has a polar acidic residue at the putative P1 site. It is interesting to note that for the 10 most highly inter-species conserved RCL sequences (those which are ≥90%), the majority (7/10) have basic P1 residues, while the remaining three have polar uncharged P1 residues.
Table 5.
Cross-tick species conserved A. americanum serpin reactive center loops
| Serpin ID | Conserved RCL amino acid sequence |
*% ID |
Serpin ID | Conserved RCL amino acid sequence |
*% ID |
|---|---|---|---|---|---|
| AAS4* | EEGTIAAAVTGLSFVPISALH | - | AAS26 | EEGTEAAAATATVAMFGSAPS | - |
| Amac-AE034279 | EEGTIATAVTGLSFVALSALS | 81 | Amac-AE032541 | EEGTEAAAATAVIMVFGSASS | 76 |
| Rapp-AAK61377 | EEGTIATAVTGLGFVPLSAHY | 76 | Amac-AE032217 | EEGTEAAAATAVIMLFGSASS | 76 |
| Rmic-AAP75707 | EEGTIATAVTGLGFVPLSVHY | 71 | AAS27 | EEGTEAAAASGVVGVNRIGID | - |
| Rmic-AHC98654 | EEGTIATAVTGLGFVPLSAHY | 71 | Is-EW860426 | EEGTEAAAASGVVAVNRLIGV | 66 |
| Rmic-AHC98661 | EEGTVAAAVTGLFVRPTAPLP | 71 | Is-XP_002411045 | EEGTEAAAASGVVMTRRAIEG | 75 |
| Rmic-AHC98655 | EEGTIAAAVTGLFVMPSSSLY | 81 | Amac-AE033019 | EEGTQAAAASGVVGVNRIGIE | 90 |
| Hlong-BAD11156 | EEGTVATAVTGISFIPLSAHY | 67 | Rpulc-JAA54308 | EEGTEAAAVSGVVSVNRIGIE | 86 |
| AAS7 | EEGTEAAAATGIAMMLMCARF | - | Rmic-AHC98657 | EEGTEAAAVTGVIGVNRIGIE | 81 |
| Amac-AE035320 | EEGTEAAAATGMAFMLLSARF | 81 | AAS28 | EEGTEAAAATGMVAMARCAII | - |
| Ir-JAA66227 | EEGTEAAAATAIPIMLMCARF | 86 | Rmic-AHC98669 | EEGTEAAAATGMVAMARCASM | 90 |
| Ir-JAA72595 | EEGTEAAAATAVPVMFCCAIF | 66 | AAS29 | EEGTEAAAATAVTVVDGCMPR | - |
| Rhaem-AFX65225 | EEGTEAAAATAITMMTYCARF | 81 | Rmic-AHC98662 | EEGTEAAAATAVMMVACCMSS | 71 |
| Rapp-AAK61376 | EEGTEAAAATAITMMTYCARF | 81 | AAS31 | EEGSEAAAVTGVVINTRTIGG | - |
| Rpulc-JAA54312 | EEGTEAAAATAITMMTYCARF | 81 | Rmic-AHC98667 | EEGSEAAAVTGVTINTRTTTG | 81 |
| AAS12 | EEGTIAAAVTGLSFVPISALH | - | Rmic-AHC98657 | EEGTEAAAVTGVIGVNRIGIE | 71 |
| Amac-AE034279 | EEGTIATAVTGLSFVALSALS | 81 | AAS37 | EEGTEAAAATAVVMMCRSAAM | - |
| Rapp-AAK61377 | EEGTIATAVTGLGFVPLSAHY | 71 | Rpul-JAA54314 | EEGTEAAAATAVMMMACCMSS | 66 |
| Rmic-AAP75707 | EEGTIATAVTGLGFVPLSVHY | 71 | Rmic-AHC98662 | EEGTEAAAATAVMMVACCMSS | 66 |
| Rmic-AHC98654 | EEGTIATAVTGLGFVPLSAHY | 71 | Rmic-AHC98669 | EEGTEAAAATGMVAMARCASM | 71 |
| Rmic-AHC98661 | EEGTVAAAVTGLFVRPTAPLP | 71 | AAS38 | EEGTIATAVTGLSFAPISALH | - |
| Rmic-AHC98655 | EEGTIAAAVTGLFVMPSSSLY | 81 | Amac-AE034279 | EEGTIATAVTGLSFVALSALS | 86 |
| Hlong-BAD11156 | EEGTVATAVTGISFIPLSAHY | 67 | Rapp-AAK61377 | EEGTIATAVTGLGFVPLSAHY | 81 |
| AAS14 | EEGTVATAVTGISLVALSALH | - | Rmic-AHC98654 | EEGTIATAVTGLGFVPLSAHY | 71 |
| Amac-AE034279 | EEGTIATAVTGLSFVALSALS | 81 | AAS42 | EKGTEAAAATAVMMMACCMSA | - |
| Rapp-AAK61377 | EEGTIATAVTGLGFVPLSAHY | 67 | Rpulc-JAA54314 | EEGTEAAAATAVMMMACCMSS | 90 |
| Rmic-AAP75707 | EEGTIATAVTGLGFVPLSVHY | 62 | Amac-AE032774.1 | EKGTEAAAATAVMMVACCLSI | 86 |
| Hlong-BAD11156 | EEGTVATAVTGISFIPLSAHY | Amac-AE032154.1 | EKGTEAAAATAVMMVGCCLSI | 81 | |
| AAS18 | EEGSEAAGATAVIFFTRGGSS | - | Rmic-AHC98662 | EEGTEAAAATAVMMVACCMSS | 86 |
| Amac-AE034313 | EEGSEAAGATAVIFFTRGGIP | 90 | Rmic-AHC98652 | EEGTEAAAATAVMMAACCLSS | 81 |
| Avar-DAA34478 | EEGSEAAGATAVIFFTRGGAP | 90 | Rapp-AAK61375 | EEGTEAAAATAVMMAACCLSS | 81 |
| Rmic-AHC98668 | EEGTIAAAVTGLFVMPSSSLY | 71 | Is-XP_002405749 | EEGTEAAAATAMVMLCCMSFP | 76 |
| AAS19 | EEGSEAAAVTGFVIQLRTAAF | - | AAS44 | EEGTEAAAATAIITTECCIMP | - |
| Rpulc-JAA54307 | EEGSEAAAVTGFVIQLRTAAF | 100 | Rmic-AHC98662 | EEGTEAAAATAVMMVACCMSS | 66 |
| Amac-AE034218 | EEGSEAAAVTGFVIQLRTAAF | 100 | AAS45 | EEGTEAAAATGVVMMCDSLPM | - |
| Amac-AE034217 | EEGSEAAAVTGFVIQLRTAAF | 100 | Rmic-AHC98669 | EEGTEAAAATGMVAMARCASM | 71 |
| Ir-JAA72548 | EEGSEAAAVTGFVIQLRTAAF | 100 | AAS47 | EEGTEAAAATAMPAANSCEMF | - |
| Ir-ABI94058 | EEGSEAAAVTGFVIQLRTAAF | 100 | Amac-AE034314 | EEGTEAAAATAMLASNSCERF | 86 |
| Is-XP_002415308 | EEGSEAAAVTGFVIQLRTAAF | 100 | AAS50 | EEGSETDSATLMRISGKAAEE | - |
| Rmic-TC22658 | EEGSEAAAVTGFVIQLRTAAF | 100 | Rpulc-JAA54309 | EEGSEADSATLLRISGKAAEE | 90 |
| AAS20 | EVGTRAVAATEAQFVSKSLVH | - | Rmic-AHC98660 | EEGSEADSATLLRISGKAAEE | 90 |
| Rpulc-JAA54167 | EVGTRAVAATQAQFVSKSLVH | 95 | AAS52 | ENGTVAAAASAAIGVGSAGPS | - |
| Is-ISCW016489 | EEGTRAVAATQAQFVSKSLVQ | 90 | Rpulc-JAA54315 | EEGTEAAAATAAVGAGSAGPS | 76 |
| Rmic-AHC98664 | EVGTRAVAATQAQFVSKSLVH | 95 | AAS54 | EDGVEGLFLTPLIMMCYAGVS | - |
| AAS21 | EKGTEAVALSSGIIRHSKTPG | - | Rmic-AHC98663 | EDGVEGLFLTPLIMMCYAGVS | 100 |
| Rpulc-JAA63258 | EKGTEAVALSSGIIRHSKTPG | 100 | AAS58 | EEGTEAAAATGMTLMMCGAMV | - |
| Rmic-AHC98658 | EKGTEAVALSSGIVRHSKTPG | 95 | Rmic-AHC98669 | EEGTEAAAATGMVAMARCASM | 71 |
| Is-XP_002401986 | EQGTEAVALSSGIVRHSRPPE | 95 | AAS65 | EEGSETDSATLMRISGKA-CE | - |
| Ir-JAB72483 | EQGTEAVALSSGIVRHSRPPE | 90 | Rpulc-JAA54309 | EEGSEADSATLLRISGKAAEE | 81 |
| Rmic-AHC98658 | EKGTEAVALSSGIVRHSKTPG | 95 | Rmic-AHC98660 | EEGSEADSATLLRISGKAAEE | 88 |
| AAS22 | EEGSEAAGATGVIFYTKSAIV | - | AAS69 | EEGTIAAAVTGLSFVATASFN | - |
| Amac-AE034349 | EEGSEAAGATGVIFYTKSMVV | 90 | Rmic-AHC98661 | EEGTVAAAVTGLFVRPTAPLP | 71 |
| Rpulc-JAA54315 | EEGSEAAAATAVIFYTKSAAV | 86 | AAS70 | EEGTIATAVTGLSFVATASFN | - |
| Rmic-AHC98668 | EEGTIAAAVTGLFVMPSSSLY | 90 | Rmic-AHC98654 | EEGTIATAVTGLGFVPLSAHY | 66 |
| AAS23 | EEGTVAAAVTSIRMRMKSSRR | - | |||
| Is-XP_002434763 | EKGTVAAAVTSISMRMGSSLA | 76 | |||
| AAS25 | EEGTEAAAATAVVMMCYSLPM | - | |||
| Rmic-AHC98653 | EEGTEAAAATAVTLMTYCARI | 66 | |||
| Rmic-AHC98662 | EEGTEAAAATAVMMVACCMSS | 66 | |||
Amac = A. maculatum, Is = I. scapularis, Ir = I. ricinus, Rapp= R. appendiculatus, Rmic = R. microplus, Rpulc = R. pulchellus, Rhaem = R. haemaphyssaloides, Avar = A. variegatum, Hlong= H. longicornis. Amino acid residues at P1 sites are bed.
Figure 5. Multiple sequence alignment of tick serpin reactive center loops (RCL).
Predicted RCLs in serpins found in this study and other tick serpins downloaded from GenBank were subjected to multiple sequence alignment using T-coffee in MacVector vrsion 12. Asterisk (*) sign denotes predicted amino acid residues at P1 sites: (A) polar acidic, (B) polar basic, (C) hydrophobic, and (D) polar uncharged. Identical amino acid residues are shaded gray. AAS = Amblyomma americanum serpins, Amac = A. maculatum, Avar = A. variegatum, Rmic = Rhipicephalus microplus, Rpulc = R. pulchellus, Rhaem= R. haemaphysalis, Rapp = R. appendiculatus, Hlong = Haemaphysalis longicornis, Is = Ixodes scapularis, Ir = I. ricinus.
DISCUSSION
This study provides an update of unique serpin-coding sequences expressed in unfed and fed A. americanum male and female ticks. While this is the first report of a very large number of serpin transcripts from male ticks, data presented here are not unusual. High numbers of serpin sequences were reported in ticks I. scapularis (Mulenga et al., 2009), R. pulchellus (direct submission), and A. maculatum (Karim et al., 2011), in mosquitoes Anopheles gambiae, Culex quinquefasciatus, and Aedes aegypti (Rawlings et al., 2012), in Bombyx mori (Zou et al., 2009), Tribolium (Zou et al., 2007), Drosophila (Reichhart, 2005), in mouse and human genomes (Puente and López-Otín, 2004; Gatto et al., 2013; Heit et al., 2013), and in Arabidopsis and Oryza sativa (Fluhr et al., 2012). Such large serpin counts across a great diversity of taxa indicate the importance of this protein family in regulating homeostasis in most branches of life. While serpin counts in other tick species do not approach the number for A. americanum as reported here, the discrepancy is likely due to the fact that this is the first analysis of tick transcriptomes of its scope with both male and female tissues at various time points having been explored.
Although based on the design of this study we are unable to conclude expression patterns, the observation of a greater diversity of AAS transcripts in male than female ticks is interesting. We speculate that the high diversity of AAS transcripts found in male ticks could be explained by the timing of reproductive physiological changes. Male metastriate tick species such as A. americanum feed for only a short time prior to completion of spermatogenesis and mating (Kiszewski et al., 2001). On the other hand, many female reproductive activities including vitellogenin synthesis and deposition into oocytes, ovulation, fertilization, and oviposition do not occur until after several days of feeding (Kiszewski et al., 2001; Sonenshine and Roe, 2013). The majority of AAS which were found exclusively in males in this study, were from male ticks fed for at least three days and were potentially ready to mate, while female ticks in this study were fed up to 120h with reproductive activities just beginning. As a result, it is possible that there are additional serpins important for female reproductive physiology, that are expressed at later feeding time points than those identified in this study. Although empirical data will be needed, it’s interesting to note that AAS50 and 121 both found in male ticks were abundant after feeding when the male tick has entered its physiologically reproductive state. It will be interesting to investigate expression of serpins in the immature stages, where there is a lack of sexual dimorphism. This will inform on which serpins are indeed involved in sex-specific physiology, and which are involved in other feeding-related physiology such as host-defense modulation. Evidence in insects, Drosophila and Aedes aegypti, indicated that serpins are among the male reproductive gland proteins transferred to females during mating (Coleman et al., 1995; Sirot et al., 2008). Although there are distinct differences between biology of ticks and insects, it is plausible that some male A. americanum serpins found in this study could serve as reproductive proteins.
The SG and MG represent two major organs through which the tick interacts with its host and with pathogens. Thus, the identification of 31 previously unknown AAS transcripts in SG and MG is interesting. Although relative expression analysis done in this study is limited, the expression patterns of AAS21 and AAS27 are notable. Based on relative abundance determined in this study AAS21 abundance is 1,000-fold higher at 48h MG compared to SG, while AAS27 increases 500 fold at 120h in MG more than SG. It will be interesting to investigate role(s) of AAS21 and AAS27 in tick feeding. Another important goal of this study was to identify A. americanum tick serpins that are expressed in both male and female metastriata ticks. We believe that these could represent those that are important to tick feeding regulation. Despite obvious differences in their biology, both male and female ticks must interact with host defense mechanisms before mating. Indeed, there is evidence that like females, male ticks express anti-inflammatory molecules such as histamine-binding proteins (Paesen et al., 1999; Bior et al., 2002), which are potentially involved in facilitating male tick feeding. Thus, the 16 AAS transcripts found in both male and female ticks, and in 48–120h SG and MG could represent those that are important to tick feeding success.
The high number of AAS sequences in this study could be explained by gene duplication and exon shuffling in the RCL region as suggested by high amino acid identity among some AAS sequences that clustered together on the phylogeny tree and those that showed differences restricted to the RCL region. Serpin diversity by gene duplication and subsequent divergence has been reported in a number of organisms including humans (Heit et al., 2013), mice (Borriello and Krauter, 1990; Hancock, 2005) and B. mori (Zou et al., 2009). In An. gambiae serpin genes clustering phylogenetically were found in clusters on the same chromosome, indicating that they could be duplicated genes (Suwanchaichinda and Kanost, 2009). Similarly, in I. scapularis 11 highly identical serpins were found on the same supercontig (Mulenga et al, 2009). The observation of differences restricted to the RCL has been observed in transcripts that are products of alternatively spliced exons. This phenomenon was reported in Manduca sexta (Jiang and Kanost, 1997), in B. mori (Zou et al., 2009), and in Ctenocephalides felis (Brandt et al., 2004). We also observed a very curious pattern between AAS4 and 12, and AAS13 and 15, in which RCLs were identical with differences restricted to outside of the RCL (Mulenga et al., 2007). Whether or not these observations are consistent with events in vivo requires further investigation. However, it is interesting to note that in this study we observed a similar pattern between serpin sequences in other tick species: A. maculatum AEO34217 and AEO34218, and R. pulchellus JAA62387 and JAA63611, where the RCL is the same and differences in sequences are outside of the RCL region. We speculate that if consistent with events in vivo, AAS transcripts that share the same RCL sequence could function as redundant proteins, or could regulate the same protease under different spatio-temporal conditions.
Within the RCL region, the amino acid at the P1 site is considered most important in determining the target protease(s) that is/are regulated by a candidate serpin (Gettins, 2002; Huntington, 2006; Huntington, 2011). Accordingly, the observation that 17 different amino acid residues were predicted at P1 sites of putative RCLs identified in this study indicates the potential diversity of proteases that may be regulated by these serpins. Our analysis of AAS RCL P1 sites showed a near-even distribution of residues across charge/polarity types with the exception of polar acidic similar to M. sexta, B. mori (Zou et al., 2009), plants (Roberts et al., 2004; Roberts and Hejgaard, 2008), and humans (Cassar and Hunter, 2013) where polar acidic P1 residues in serpins appear to be rare. Prediction of the P1 site amino acid residue is not in itself sufficient to determine the protease that may be regulated by a candidate serpin; empirical evidence is required. However, there is ample evidence that serpins with basic residues at the P1 site regulate trypsin and tryspin-like proteases (Gettins, 2002, Li et al., 1999, Leboulle et al., 2002a; Leboulle et al., 2002b; Prevot et al., 2006). Thus it is interesting that the most highly conserved AAS RCLs in this study have basic P1 residues. From the perspective of tick feeding biology, serpins with basic P1 residues could represent those that ticks use to evade trypsin-like protease-mediated host defense pathways such as blood clotting. We would like to caution the reader here that in both plants and in non-hematophagous organisms such as Drosophila, the majority of serpins have basic P1 amino acid residues (Reichhart, 2005; Fluhr et al., 2012). It is also interesting to note that two tick anti-coagulant serpins, HLS2 (orthologous to AAS12), in H. longicornis (BAD11156) (Imamura et. a., 2005), and Iris in I. ricinus (AJ269658) (Prevot et. al., 2006), contain hydrophobic residues at their P1 sites. Further experiments are therefore required in order to determine proteases/pathways that may be regulated by the serpins described in this study.
In nature, different tick species may infest the same animal host and implicitly, these different tick species will face the same host defense mechanisms. It is conceivable that different tick species could utilize conserved proteins to interact with the same host, such as the 50 cross-tick species conserved AAS sequences identified in this study. It is notable that only ~22% (11/50) were conserved in both metastriate and prostriate tick species. These could be particularly interesting candidates for further functional studies given that in general, amino acid identities between metastriate and prostriate tick proteins sequences are low. In particular, AAS25, which was detected in fed males and females, and in SG, is orthologous to Iris, found in SG and saliva of I. ricinus. Iris was demonstrated to inhibit lymphocyte proliferation, as well as the immune system cytokines IFN-γ and IL-6 (Leboulle et. al., 2002a; Leboulle et. al., 2002b). Similarly, Highly conserved AAS proteins represent very interesting candidates for development of universal anti-tick vaccines as advocated (Maritz-Olivier et al., 2007).
The RCL is important to serpin function (Gettins, 2002), and thus it is interesting to note that some AAS RCLs are 90–100% conserved in other tick species. These serpins could be involved in regulating target proteases that are important to all ticks. Based on our inter-species comparative sequence analysis, it is interesting to note that AAS sequences with polar basic P1 sites, followed by polar uncharged P1 sites, were likely to be conserved in other tick species. It is noteworthy that hydrophobic P1 residues, though prevalent, tended to show little conservation in other tick species. This suggests that proteases that can interact with hydrophobic Pl residues may be involved in species-specific physiology, and might be diverging at a fast rate. Although the present study is descriptive, it contributes significantly to the picture of serpin transcript diversity expressed by male and female A. americanum ticks. Additionally, based on the data in this analysis, the picture of serpin transcript diversity in other tick species is likely to be far from complete.
Supplementary Material
ACKNOWLEDGEMENTS
This research was supported by National Institute Health grants (AI081093, AI093858, AI074789, AI074789-01A1S1) to AM. Authors would like to thank tick labs at Texas A&M and Oklahoma State universities for supplying ticks used in this research.
References
- Benson HL, Wilkes DS. Matrix metalloproteinases in T cell mediated pulmonary diseases. Front. Biosci. (Elite Ed) 2012;4:2162–2169. doi: 10.2741/533. [DOI] [PubMed] [Google Scholar]
- Bior AD, Essenberg RC, Sauer JR. Comparison of differentially expressed genes in the salivary glands of male ticks, Amblyomma americanum and Dermacentor andersoni. Insect Biochem. Mol. Biol. 2002;32:645–655. doi: 10.1016/s0965-1748(01)00143-6. [DOI] [PubMed] [Google Scholar]
- Borriello F, Krauter KS. Reactive site polymorphism in the murine protease inhibitor gene family is delineated using a modification of the PCR reaction (PCR + 1) Nucleic Acids Research. 1990;18:5481–5487. doi: 10.1093/nar/18.18.5481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosche B, Molcanyi M, Noll T, Kochanek M, Kraus B, Rieger B, El Majdoub F, Dohmen C, Löhr M, Goldbrunner R, Brinker G. Occurrence and recurrence of spontaneous chronic subdural haematoma is associated with a factor XIII deficiency. Clin. Neurol. Neurosurg. 2012;115:13–18. doi: 10.1016/j.clineuro.2012.03.045. [DOI] [PubMed] [Google Scholar]
- Brandt KS, Silver GM, Becher AM, Gaines PJ, Maddux JD, Jarvis EE, Wisnewski N. Isolation, characterization, and recombinant expression of multiple serpins from the cat flea, Ctenocephalides felis. Arch. Insect Biochem. Physiol. 2004;55:200–214. doi: 10.1002/arch.10139. [DOI] [PubMed] [Google Scholar]
- Carrell RW, Pemberton PA, Boswell DR. The serpins: Evolution and adaptation in a family of protease inhibitors. Cold Spring Harb. Symp. Quant. Biol. 1987;52:527–535. doi: 10.1101/sqb.1987.052.01.060. [DOI] [PubMed] [Google Scholar]
- Cassar NJ, Hunter GJ. Serpins: Form, function, and dysfunction. Xjenza-Online. 2013;501:51. [Google Scholar]
- Chalaire KC, Kim TK, Garcia-Rodriguez H, Mulenga A. Amblyomma americanum (L.) (Acari: Ixodidae) tick salivary gland serine protease inhibitor (serpin) 6 is secreted into tick saliva during tick feeding. J. Exp. Biol. 2011;214:665–673. doi: 10.1242/jeb.052076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childs JE, Paddock CD. The ascendancy of Amblyomma americanum as a vector of pathogens affecting humans in the United States. Annu. Rev. Entomol. 2003;48:307–337. doi: 10.1146/annurev.ento.48.091801.112728. [DOI] [PubMed] [Google Scholar]
- Chmelar J, Calvo E, Pedra JHF, Francischetti IMB, Kotsyfakis M. Tick salivary secretion as a source of antihemostatics. J. Proteomics. 2012;75:3842–3854. doi: 10.1016/j.jprot.2012.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chmelar J, Oliveira CJ, Rezacova P, Francischetti IM, Kovarova Z, Pejler G, Kopacek P, Ribeiro JM, Mares M, Kopecky J, Kotsyfakis M. A tick salivary protein targets cathepsin G and chymase and inhibits host inflammation and platelet aggregation. Blood. 2011;117:736–744. doi: 10.1182/blood-2010-06-293241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman S, DrÄhn B, Petersen G, Stolorov J, Kraus K. A Drosophila male accessory gland protein that is a member of the serpin superfamily of proteinase inhibitors is transferred to females during mating. Insect Biochem. Mol. Biol. 1995;25:203–207. doi: 10.1016/0965-1748(94)00055-m. [DOI] [PubMed] [Google Scholar]
- Dantas-Torres F, Chomel BB, Otranto D. Ticks and tick-borne diseases: A one health perspective. Trends Parasitol. 2012;28:437–446. doi: 10.1016/j.pt.2012.07.003. [DOI] [PubMed] [Google Scholar]
- Davis RL, Shrimpton AE, Holohan PD, Bradshaw C, Feiglin D, Collins GH, Sonderegger P, Kinter J, Becker LM, Lacbawan F, Krasnewich D, Muenke M, Lawrence DA, Yerby MS, Shaw CM, Gooptu B, Elliott PR, Finch JT, Carrell RW, Lomas DA. Familial dementia caused by polymerization of mutant neuroserpin. Nature. 1999;401:376–379. doi: 10.1038/43894. [DOI] [PubMed] [Google Scholar]
- Day MJ. One health: The importance of companion animal vector-borne diseases. Parasit. Vectors. 2011;4 doi: 10.1186/1756-3305-4-49. 49-3305-4-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Fuente J, Estrada-Pena A, Venzal JM, Kocan KM, Sonenshine DE. Overview: Ticks as vectors of pathogens that cause disease in humans and animals. Front. Biosci. 2008;13:6938–6946. doi: 10.2741/3200. [DOI] [PubMed] [Google Scholar]
- Fluhr R, Lampl N, Roberts TH. Serpin protease inhibitors in plant biology. Physiol. Plantarum. 2012;145:95–102. doi: 10.1111/j.1399-3054.2011.01540.x. [DOI] [PubMed] [Google Scholar]
- Gatto M, Iaccarino L, Ghirardello A, Bassi N, Pontisso P, Punzi L, Shoenfeld Y, Doria A. Serpins, immunity and autoimmunity: Old molecules, new functions. Clin. Rev. Allergy Immunol. 2013;45:267–280. doi: 10.1007/s12016-013-8353-3. [DOI] [PubMed] [Google Scholar]
- Gettins PG. Serpin structure, mechanism, and function. Chem. Rev. 2002;102:4751–4804. doi: 10.1021/cr010170+. [DOI] [PubMed] [Google Scholar]
- Goddard J. Infectious diseases and arthropods. Totowa, New Jersey: Springer; 2009. [Google Scholar]
- Gooptu B, Lomas DA. Conformational pathology of the serpins: Themes, variations, and therapeutic strategies. Annu. Rev. Biochem. 2009;78:147–176. doi: 10.1146/annurev.biochem.78.082107.133320. [DOI] [PubMed] [Google Scholar]
- Hancock JM. Gene factories, microfunctionalization and the evolution of gene families. Trends Genetics. 2005;21:591–595. doi: 10.1016/j.tig.2005.08.008. [DOI] [PubMed] [Google Scholar]
- Heit C, Jackson BC, McAndrews M, Wright MW, Thompson DC, Silverman GA, Nebert DW, Vasiliou V. Update of the human and mouse SERPIN gene superfamily. Hum. Genomics. 2013;7:22. doi: 10.1186/1479-7364-7-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins PC, Stone SR. The contribution of the conserved hinge region residues of alpha1-antitrypsin to its reaction with elastase. Biochemistry. 1995;34:15872–15879. doi: 10.1021/bi00048a033. [DOI] [PubMed] [Google Scholar]
- Huntington JA. Serpin structure, function and dysfunction. J. Thrombosis Haemostasis. 2011;9:26–34. doi: 10.1111/j.1538-7836.2011.04360.x. [DOI] [PubMed] [Google Scholar]
- Huntington JA. Shape-shifting serpins – advantages of a mobile mechanism. Trends Biochem. Sci. 2006;31:427–435. doi: 10.1016/j.tibs.2006.06.005. [DOI] [PubMed] [Google Scholar]
- Imamura S, Konnai S, Vaz Ida S, Yamada S, Nakajima C, Ito Y, Tajima T, Yasuda J, Simuunza M, Onuma M, Ohashi K. Effects of anti-tick cocktail vaccine against Rhipicephalus appendiculatus. Jpn. J. Vet. Res. 2008;56:85–98. [PubMed] [Google Scholar]
- Imamura S, Junior IdSV, Sugino M, Ohashi K, Onuma M. A serine protease inhibitor (serpin) from Haemaphysalis longicornis as an anti-tick vaccine. Vaccine. 2005;23:1301–1311. doi: 10.1016/j.vaccine.2004.08.041. [DOI] [PubMed] [Google Scholar]
- Imamura S, Namangala B, Tajima T, Tembo ME, Yasuda J, Ohashi K, Onuma M. Two serine protease inhibitors (serpins) that induce a bovine protective immune response against Rhipicephalus appendiculatus ticks. Vaccine. 2006;24:2230–2237. doi: 10.1016/j.vaccine.2005.10.055. [DOI] [PubMed] [Google Scholar]
- James AM, Liveris D, Wormser GP, Schwartz I, Montecalvo MA, Johnson BJB. Borrelia lonestari infection after a bite by an Amblyomma americanum tick. J. Infect. Dis. 2001;183:1810–1814. doi: 10.1086/320721. [DOI] [PubMed] [Google Scholar]
- Jiang H, Kanost MR. Characterization and functional analysis of 12 naturally occurring reactive site variants of serpin-1 from Manduca sexta. J. Biol. Chem. 1997;272:1082–1087. doi: 10.1074/jbc.272.2.1082. [DOI] [PubMed] [Google Scholar]
- Jittapalapong S, Kaewhom P, Pumhom P, Canales M, De La Fuente J, Stich RW. Immunization of rabbits with recombinant serine protease inhibitor reduces the performance of adult female Rhipicephalus microplus. Transboundary Emerg. Dis. 2010;57:103–106. doi: 10.1111/j.1865-1682.2010.01108.x. [DOI] [PubMed] [Google Scholar]
- Jongejan F, Uilenberg G. The global importance of ticks. Parasitology. 2004;129(Suppl):S3–S14. doi: 10.1017/s0031182004005967. [DOI] [PubMed] [Google Scholar]
- Kaewhom P, Sirinarumitr T, Chantakru S, Jittapalapong S. Humoral antibody responses of rabbits immunized with anti-tick vaccine using SERPIN recombinant protein. Kasetsart Journal, Natural Sciences. 2009;43:69–76. [Google Scholar]
- Karim S, Singh P, Ribeiro JMC. A deep insight into the sialotranscriptome of the gulf coast tick, Amblyomma maculatum. PLoS ONE. 2011;6:e28525. doi: 10.1371/journal.pone.0028525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman W, Phillips J. Ion and water balance in the ixodid tick Dermacentor andersoni I. routes of ion and water excretion. J. Exp. Biol. 1973;58:523–536. [Google Scholar]
- Kiszewski AE, Matuschka F, Spielman A. Mating strategies and spermiogenesis in ixodid ticks. Annu. Rev. Entomol. 2001;46:167–182. doi: 10.1146/annurev.ento.46.1.167. [DOI] [PubMed] [Google Scholar]
- Klompen JS, Black WC, 4th, Keirans JE, Oliver JH., Jr Evolution of ticks. Annu. Rev. Entomol. 1996;41:141–161. doi: 10.1146/annurev.en.41.010196.001041. [DOI] [PubMed] [Google Scholar]
- Leboulle G, Crippa M, Decrem Y, Mejri N, Brossard M, Bollen A, Godfroid E. Characterization of a novel salivary immunosuppressive protein from Ixodes ricinus ticks. J. Biol. Chem. 2002a;277:10083–10089. doi: 10.1074/jbc.M111391200. [DOI] [PubMed] [Google Scholar]
- Leboulle G, Rochez C, Louahed J, Ruti B, Brossard M, Bollen A, Godfroid E. Isolation of Ixodes ricinus salivary gland mRNA encoding factors induced during blood feeding. Am. J. Trop. Med. Hyg. 2002b;66:225–233. doi: 10.4269/ajtmh.2002.66.225. [DOI] [PubMed] [Google Scholar]
- Li J, Wang Z, Canagarajah B, Jiang H, Kanost M, Goldsmith EJ. The structure of active serpin 1K from Manduca sexta. Structure. 1999;7:103–109. doi: 10.1016/s0969-2126(99)80013-6. [DOI] [PubMed] [Google Scholar]
- Marcelino I, de Almeida AM, Ventosa M, Pruneau L, Meyer DF, Martinez D, Lefrançois T, Vachiéry N, Coelho AV. Tick-borne diseases in cattle: Applications of proteomics to develop new generation vaccines. J. Proteomics. 2012;75:4232–4250. doi: 10.1016/j.jprot.2012.03.026. [DOI] [PubMed] [Google Scholar]
- Maritz-Olivier C, Stutzer C, Jongejan F, Neitz AW, Gaspar AR. Tick anti-hemostatics: Targets for future vaccines and therapeutics. Trends Parasitol. 2007;23:397–407. doi: 10.1016/j.pt.2007.07.005. [DOI] [PubMed] [Google Scholar]
- Miura Y, Kawabata S, Wakamiya Y, Nakamura T, Iwanaga S. A limulus intracellular coagulation inhibitor type 2. purification, characterization, cDNA cloning, and tissue localization. J. Biol. Chem. 1995;270:558–565. doi: 10.1074/jbc.270.2.558. [DOI] [PubMed] [Google Scholar]
- Mocchegiani E, Giacconi R, Costarelli L. Metalloproteases/anti-metalloproteases imbalance in chronic obstructive pulmonary disease: Genetic factors and treatment implications. Curr. Opin. Pulm. Med. 2011;17(Suppl. 1):S11–S19. doi: 10.1097/01.mcp.0000410743.98087.12. [DOI] [PubMed] [Google Scholar]
- Moore A, Penfold LM, Johnson JL, Latchman DS, Moore HDM. Human sperm-egg binding is inhibited by peptides corresponding to core region of an acrosomal serine protease inhibitor. Mol. Reprod. Dev. 1993;34:280–291. doi: 10.1002/mrd.1080340308. [DOI] [PubMed] [Google Scholar]
- Mulenga A, Sugino M, Nakajim M, Sugimoto C, Onuma M. Tick-encoded serine proteinase inhibitors (serpins); potential target antigens for tick vaccine development. J. Vet. Med. Sci. 2001;63:1063–1069. doi: 10.1292/jvms.63.1063. [DOI] [PubMed] [Google Scholar]
- Mulenga A, Khumthong R, Chalaire KC. Ixodes scapularis tick serine proteinase inhibitor (serpin) gene family; annotation and transcriptional analysis. BMC Genomics. 2009;10:217. doi: 10.1186/1471-2164-10-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulenga A, Kim T, Ibelli AMG. Amblyomma americanum tick saliva serine protease inhibitor 6 is a cross-class inhibitor of serine proteases and papain-like cysteine proteases that delays plasma clotting and inhibits platelet aggregation. Insect Mol. Biol. 2013;22:306–319. doi: 10.1111/imb.12024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulenga A, Misao O, Sugimoto C. Three serine proteinases from midguts of the hard tick Rhipicephalus appendiculatus; cDNA cloning and preliminary characterization. Exp. Appl. Acarol. 2003a;29:151–164. doi: 10.1023/a:1024278402288. [DOI] [PubMed] [Google Scholar]
- Mulenga A, Tsuda A, Onuma M, Sugimoto C. Four serine proteinase inhibitors (serpin) from the brown ear tick, Rhiphicephalus appendiculatus; cDNA cloning and preliminary characterization. Insect Biochem. Mol. Biol. 2003b;33:267–276. doi: 10.1016/s0965-1748(02)00240-0. [DOI] [PubMed] [Google Scholar]
- Mulenga A, Tsuda A, Sugimoto C, Onuma M. Blood meal acquisition by ticks; molecular advances and implications for vaccine development. Jpn. J. Vet. Res. 2002;49:261–272. [PubMed] [Google Scholar]
- Mulenga A, Khumthong R, Blandon MA. Molecular and expression analysis of a family of the Amblyomma americanum tick lospins. J. Exp. Biol. 2007;210:3188–3198. doi: 10.1242/jeb.006494. [DOI] [PubMed] [Google Scholar]
- Nene V, Lee D, Quackenbush J, Skilton R, Mwaura S, Gardner MJ, Bishop R. AvGI, an index of genes transcribed in the salivary glands of the ixodid tick Amblyomma variegatum. Int. J. Parasitol. 2002;32:1447–1456. doi: 10.1016/s0020-7519(02)00159-5. [DOI] [PubMed] [Google Scholar]
- Paesen GC, Adams PL, Harlos K, Nuttall PA, Stuart DI. Tick histamine-binding proteins: Isolation, cloning, and three-dimensional structure. Mol. Cell. 1999;3:661–671. doi: 10.1016/s1097-2765(00)80359-7. [DOI] [PubMed] [Google Scholar]
- Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- Prevot PP, Adam B, Boudjeltia KZ, Brossard M, Lins L, Cauchie P, Brasseur R, Vanhaeverbeek M, Vanhamme L, Godfroid E. Anti-hemostatic effects of a serpin from the saliva of the tick Ixodes ricinus. J. Biol. Chem. 2006;281:26361–26369. doi: 10.1074/jbc.M604197200. [DOI] [PubMed] [Google Scholar]
- Prevot P-P, Couvreur B, Denis V, Brossard M, Vanhamme L, Godfroid E. Protective immunity against ixodes ricinus induced by a salivary serpin. Vaccine. 2007;25:3284–3292. doi: 10.1016/j.vaccine.2007.01.008. [DOI] [PubMed] [Google Scholar]
- Prevot P, Beschin A, Lins L, Beaufays J, Grosjean A, Bruys L, Adam B, Brossard M, Brasseur R, Zouaoui Boudjeltia K, Vanhamme L, Godfroid E. Exosites mediate the anti-inflammatory effects of a multifunctional serpin from the saliva of the tick Ixodes ricinus. FEBS Journal. 2009;276:3235–3246. doi: 10.1111/j.1742-4658.2009.07038.x. [DOI] [PubMed] [Google Scholar]
- Puente XS, López-Otín C. A genomic analysis of rat proteases and protease inhibitors. Genome Res. 2004;14:609–622. doi: 10.1101/gr.1946304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings ND, Barrett AJ, Bateman A. MEROPS: The database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 2012;40:D343–D350. doi: 10.1093/nar/gkr987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichhart J. Tip of another iceberg: Drosophila serpins. Trends Cell Biol. 2005;15:659–665. doi: 10.1016/j.tcb.2005.10.001. [DOI] [PubMed] [Google Scholar]
- Ribeiro JM, Anderson JM, Manoukis NC, Meng Z, Francischetti IM. A further insight into the sialome of the tropical bont tick, Amblyomma variegatum. BMC Genomics. 2011;12:136. doi: 10.1186/1471-2164-12-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro JC. Role of saliva in tick/host interactions. Exp. Appl. Acarol. 1989;7:15–20. doi: 10.1007/BF01200449. [DOI] [PubMed] [Google Scholar]
- Ribeiro JMC, Alarcon-Chaidez F, B. Francischetti IM, Mans BJ, Mather TN, Valenzuela JG, Wikel SK. An annotated catalog of salivary gland transcripts from Ixodes scapularis ticks. Insect Biochem. Mol. Biol. 2006;36:111–129. doi: 10.1016/j.ibmb.2005.11.005. [DOI] [PubMed] [Google Scholar]
- Roberts TH, Hejgaard J, Saunders NF, Cavicchioli R, Curmi PM. Serpins in unicellular eukarya, archaea, and bacteria: Sequence analysis and evolution. J. Mol. Evol. 2004;59:437–447. doi: 10.1007/s00239-004-2635-6. [DOI] [PubMed] [Google Scholar]
- Roberts TH, Hejgaard J. Serpins in plants and green algae. Funct. Integr. Genomics. 2008;8:1–27. doi: 10.1007/s10142-007-0059-2. [DOI] [PubMed] [Google Scholar]
- Sasaki T. Patchwork-structure serpins from silkworm (Bombyx mori) larval hemolymph. Eur. J. Biochem. 1991;202:255–261. doi: 10.1111/j.1432-1033.1991.tb16370.x. [DOI] [PubMed] [Google Scholar]
- Savage HM, Godsey MS, Jr, Lambert A, Panella NA, Burkhalter KL, Harmon JR, Lash RR, Ashley DC, Nicholson WL. First detection of heartland virus (Bunyaviridae: Phlebovirus) from field collected arthropods. Am. J. Trop. Med. Hyg. 2013;89:445–452. doi: 10.4269/ajtmh.13-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schechter I, Berger A. On the size of the active site in proteasesIpapain. Biochem. Biophys. Res. Commun. 1967;27:157–162. doi: 10.1016/s0006-291x(67)80055-x. [DOI] [PubMed] [Google Scholar]
- Schulze TL, Jordan RA, White JC, Roegner VE, Healy SP. Geographical distribution and prevalence of selected Borrelia, Ehrlichia and Rickettsia infections in Amblyomma americanum (Acari: Ixodidae) in New Jersey. J. Am. Mosq. Control Assoc. 2011;27:236–244. doi: 10.2987/11-6111.1. [DOI] [PubMed] [Google Scholar]
- Sirot LK, Poulson RL, Caitlin McKenna M, Girnary H, Wolfner MF, Harrington LC. Identity and transfer of male reproductive gland proteins of the dengue vector mosquito, Aedes aegypti: Potential tools for control of female feeding and reproduction. Insect Biochem. Mol. Biol. 2008;38:176–189. doi: 10.1016/j.ibmb.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenshine DE, Roe RM. Biology of ticks. Oxford University Press; 2013. [Google Scholar]
- Springer YP, Eisen L, Beati L, James AM, Eisen RJ. Spatial distribution of counties in the continental united states with records of occurrence of Amblyomma americanum (Ixodida: Ixodidae) J. Med. Entomol. 2014;51:342–351. doi: 10.1603/me13115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein PE, Carrell RW. What do dysfunctional serpins tell us about molecular mobility and disease? Nat. Struct. Biol. 1995;2:96–113. doi: 10.1038/nsb0295-96. [DOI] [PubMed] [Google Scholar]
- Sugino M, Imamura S, Mulenga A, Nakajima M, Tsuda A, Ohashi K, Onuma M. A serine proteinase inhibitor (serpin) from ixodid tick Haemaphysalis longicornis; cloning and preliminary assessment of its suitability as a candidate for a tick vaccine. Vaccine. 2003;21:2844–2851. doi: 10.1016/s0264-410x(03)00167-1. [DOI] [PubMed] [Google Scholar]
- Suwanchaichinda C, Kanost MR. The serpin gene family in Anopheles gambiae. Gene. 2009;442:47–54. doi: 10.1016/j.gene.2009.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tekin S, Padua MB, Brad AM, Hansen PJ. Antiproliferative actions of ovine uterine serpin. Am. J. Reproductive Immunol. 2005;53:136–143. doi: 10.1111/j.1600-0897.2005.00257.x. [DOI] [PubMed] [Google Scholar]
- Telford S, III, Goethert H, Bowman A, Nuttall P. Emerging and emergent tick-borne infections. Ticks: biology, disease and control. 2008:344–376. [Google Scholar]
- Telford S, Goethert H. Emerging tick-borne infections: Rediscovered and better characterized, or truly ‘new’? Parasitology. 2004;129:S301–S327. doi: 10.1017/s0031182003004669. [DOI] [PubMed] [Google Scholar]
- Tirloni L, Seixas A, Mulenga A, da Silva Vaz I, Jr, Termignoni C. A family of serine protease inhibitors (serpins) in the cattle tick Rhipicephalus (Boophilus) microplus. Exp. Parasitol. 2014;137:25–34. doi: 10.1016/j.exppara.2013.12.001. [DOI] [PubMed] [Google Scholar]
- Waldrup K, Moritz J, Baggett D, Magyar S, Wagner G. Monthly incidence of Theileria cervi and seroconversion to Babesia odocoilei in white-tailed deer (Odocoileus virginianus) in Texas. J. Wildlife Dis. 1992;28:457–459. doi: 10.7589/0090-3558-28.3.457. [DOI] [PubMed] [Google Scholar]
- Whisstock JC, Silverman GA, Bird PI, Bottomley SP, Kaiserman D, Luke CJ, Pak SC, Reichhart J, Huntington JA. Serpins flex their muscle: II. Structural insights into target peptidase recognition, polymerisation, and transport functions. J. Biol. Chem. 2010;285:24307–24312. doi: 10.1074/jbc.R110.141408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikel SK, Ramachandra RN, Bergman DK. Tick-induced modulation of the host immune response. Int. J. Parasitol. 1994;24:59–66. doi: 10.1016/0020-7519(94)90059-0. [DOI] [PubMed] [Google Scholar]
- Yu Y, Cao J, Zhou Y, Zhang H, Zhou J. Isolation and characterization of two novel serpins from the tick Rhipicephalus haemaphysaloides. Ticks Tick-borne Dis. 2013;4:297–303. doi: 10.1016/j.ttbdis.2013.02.001. [DOI] [PubMed] [Google Scholar]
- Zou Z, Evans JD, Lu Z, Zhao P, Williams M, Sumathipala N, Hetru C, Hultmark D, Jiang H. Comparative genomic analysis of the Tribolium immune system. Genome Biol. 2007;8:R177. doi: 10.1186/gb-2007-8-8-r177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Z, Picheng Z, Weng H, Mita K, Jiang H. A comparative analysis of serpin genes in the silkworm genome. Genomics. 2009;93:367–375. doi: 10.1016/j.ygeno.2008.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.