Summary
Signals in many biological processes can be amplified by recruiting multiple copies of regulatory proteins to a site of action. Harnessing this principle, we have developed a novel protein scaffold, a repeating peptide array termed SunTag, which can recruit multiple copies of an antibody-fusion protein. We show that the SunTag can recruit up to 24 copies of GFP, thereby enabling long-term imaging of single protein molecules in living cells. We also use the SunTag to create a potent synthetic transcription factor by recruiting multiple copies of a transcriptional activation domain to a nuclease-deficient CRISPR/Cas9 protein and demonstrate strong activation of endogenous gene expression and re-engineered cell behavior with this system. Thus, the SunTag provides a versatile platform for multimerizing proteins on a target protein scaffold and is likely to have many applications in imaging and in controlling biological outputs.
Introduction
Recruitment of multiple copies of a protein to a target substrate (e.g. DNA, RNA, or protein) presents a general principle for signal amplification in biological systems. For example, binding of multiple copies of a transcription factor to a single promoter dramatically enhances transcriptional activation of the target gene (Anderson and Freytag, 1991; Chen et al., 1992; Pettersson and Schaffner, 1990). Similarly, the recruitment of multiple copies of an RNA binding protein to an mRNA can result in potent regulation of translation (Pillai et al., 2004; Pique et al., 2008). Protein localization and interactions also can be modulated by the copy number of interaction sites within a polypeptide sequence. For example, many nuclear proteins contain multiple nuclear localization signal (NLS) sequences, which control robustness of nuclear import (Luo et al., 2004).
The principle of signal amplification via protein multimerization has also been widely used in imaging and engineering of biological systems. A commonly used method to study RNA localization, even at the single molecule level, is to insert multiple copies (as many as 24) of the MS2 binding RNA hairpin into a target RNA molecule, which then recruit many MS2-GFP fusion proteins, fluorescently labeling the RNA molecule with many GFP molecules (Bertrand et al., 1998; Fusco et al., 2003). The activity of a RNA-binding protein can also be studied by artificially tethering it to an RNA in multiple copies using the MS2 system (Coller and Wickens, 2007). Similar multimerization approaches have also been used to fluorescently label a specific region of a chromosome. For example, the LacO operon can be inserted into a chromosomal locus in many tandem repeats and then visualized by the recruitment of many copies of GFP-LacI (Gordon et al., 1997). GFP-tagged DNA-binding proteins, such as the CRISPR-associated protein Cas9, can also be used to fluorescently label a native repetitive DNA sequence, as such repetitive sequences recruit many copies of the GFP-tagged DNA binding proteins (Chen et al., 2013). Furthermore, as with native transcriptional regulation, a gene can be artificially activated when a binding site for a synthetic transcription factor is placed upstream of a gene in multiple copies; this principle is employed in the “Tet-On” system for inducible transgene expression (Huang et al., 1999; Sadowski et al., 1988). Taken together, these studies demonstrate the power of introducing multiple copies of protein binding sites within RNA or DNA for the purpose of signal amplification.
Despite the success of multimerizing nucleic acid based motifs within RNA and DNA, for protein recruitment no comparable and generic system exists for controlling copy number of protein-protein interactions. For fluorescence imaging, fusion of 3 copies of GFP to a protein of interest has been used to increase signal intensity, but a further increase in the copy number of fluorescent proteins is challenging due to their size (~25 kDa) and bacterial recombination when constructing DNA plasmids encoding such proteins. Here, we describe a new synthetic system for recruiting as many as 24 copies of a protein to a target polypeptide chain. We demonstrate that this approach can be used to create bright fluorescent signals for single molecule protein imaging in living cells, through the recruitment of 24 copies of GFP to a target protein. We also demonstrate that the system can be used to modulate gene expression through the recruitment of multiple copies of gene regulatory effector domains to a nuclease-deficient CRISPR/Cas9 protein targeted to specific sequences in the genome. The ability to amplify biological signals through controlled protein multimerization will likely have many additional uses in biological research and biotechnology.
Results
Development of the SunTag, a system for recruiting multiple protein copies to a polypeptide scaffold
Protein multimerization on a single RNA or DNA template is made possible by identifying protein domains that bind with high affinity to a relatively short nucleic acid motif. We therefore sought a protein-based system with similar properties, specifically a protein that can bind tightly to a short peptide sequence (Figure 1A–B). Antibodies are capable of binding to short, unstructured peptide sequences with high affinity and specificity, and, importantly, peptide epitopes can be designed that differ from naturally occurring sequences in the genome. Furthermore, while antibodies generally do not fold properly in the cytoplasm, single chain variable fragment (scFv) antibodies, in which the epitope binding regions of the light and heavy chains of the antibody are fused to form a single polypeptide, have been successfully expressed in soluble form in cells (Colby et al., 2004; Lecerf et al., 2001; Worn et al., 2000).
Figure 1. Identification of an antibody-peptide pair that binds tightly in vivo.
(A–B) Schematic of the antibody-peptide labeling strategy. (B) schematic of the experiment described in (C) in which the mitochondrial targeting domain of mitoNEET (yellow box, mito) fused to mCherry and 4 tandem copies of a peptide recruits a GFP-tagged intracellular antibody to mitochondria (C) ScFv-GCN4-GFP was co-expressed with either mito-mCherry-4xGCN4peptide (lower panel) or mito-mCherry-FKBP as a control (upper panel) in U2OS cells, and cells were imaged using spinning disk confocal microscopy. Scale bars, 10 µm. See also figure S1.
We expressed three previously developed single-chain antibodies (Colby et al., 2004; Lecerf et al., 2001; Worn et al., 2000) fused to EGFP in U2OS cells, and co-expressed their cognate peptides (multimerized in 4 tandem copies) fused to the cytoplasmic side of the mitochondrial protein mitoNEET (Colca et al., 2004) (referred to here as Mito, figure S1A). We then assayed whether the antibody-GFP fusion proteins would be recruited to the mitochondria by fluorescence microscopy, which would indicate binding between antibody and peptide (Figure 1B). Of the three antibody-peptide pairs tested, only the GCN4 antibody-peptide pair showed robust and specific binding, while not disrupting normal mitochondrial morphology (Figure 1C and figure S1B). Thus, we focused our further efforts on the GCN4 antibody-peptide pair.
The GCN4 antibody was optimized to allow intracellular expression in yeast (Worn et al., 2000). In human cells, however, we still observed some protein aggregates of scFv-GCN4-GFP at high expression levels (Figure S2A). To improve scFv-GCN4 stability, we added a variety of N- and C-terminal fusion proteins known to enhance protein solubility, and found that fusion of superfolder-GFP (sfGFP) alone (Pedelacq et al., 2006), or along with the small solubility tag GB1 (Gronenborn et al., 1991) to the C-terminus of the GCN4 antibody almost completely eliminated protein aggregation, even at high expression levels (Figure S2A). Thus, we performed all further experiments with scFv-GCN4-sfGFP-GB1 (hereafter referred to as scFv-GCN4-GFP).
Very tight binding of the antibody-peptide pair in vivo is critical for the formation of multimers on a protein scaffold backbone. To determine the dissociation rate of the GCN4 antibody-peptide interaction, we performed fluorescence recovery after photobleaching (FRAP) experiments on scFv-GCN4-GFP bound to the mitochondrial-localized mito-mCherry-4xGCN4pep. After photobleaching, very slow GFP recovery was observed (half-life of ~5–10 min (Figure 2A–B)), indicating that the antibody bound very tightly to the peptide. It is also important to optimize the spacing of the scFv-GCN4 binding sites within the protein scaffold so that they could be saturated by scFv-GCN4, since steric hindrance of neighboring peptide binding sites is a concern. We varied the spacing between neighboring GCN4 peptides and quantified the antibody occupancy on the peptide array using the mitochondrial localization assay described above combined with quantitative fluorescence microscopy (Figure 2C–E and figure S2B). The ratio of GFP fluorescence (from the scFv-GCN4-GFP antibody) to mCherry fluorescence (present in one copy on the mito-4xGCN4pep scaffold) on the mitochondria provided a measure of the number of antibodies recruited to the protein scaffold. To correct for different intensities of the two fluorescent proteins, this ratio was normalized to the GFP-mCherry fluorescence intensity ratio of a control protein in which GFP and mCherry were directly fused (Figure S2B). We compared a short (GGSGG) and long (GGSGGSGGSGGSGG) linker separating the 22 a.a. GCN4 peptide and found an average GFP:mCherry corrected intensity ratio of 3.4 ± 1.5 and 4.1 ± 1.6, respectively (n=40 cells) (Figure 2C–E). Complete saturation of all binding sites would result in a GFP:mCherry molar ratio of 4, while a maximal molar ratio of 2 is expected if two antibodies could not bind to adjacent peptides due to steric hindrance. Thus, the results from this experiment indicate that a spacer as short as five amino acids sufficiently separates peptides to allow binding of antibodies to neighboring peptides. Importantly, in a peptide array containing 24 tandem copies of the peptide, separated by 5 a.a. linkers, we found an average GFP:mCherry corrected fluorescence intensity ratio of 24 (Figure 2C,E). These results show that full antibody occupancy can be achieved with as many as 24 copies of a peptide binding site. Taken together, these results show that this optimized GCN4 antibody-peptide pair meets all the requirements for an effective system for recruiting many copies of a protein to a polypeptide scaffold in a controlled fashion. As the GCN4 antibody-peptide pair allows ultra-bright fluorescent labeling of molecules, we named the tagging system SUperNova (SunTag) after the very bright stellar explosion.
Figure 2. Characterization of the off-rate and stoichiometry of the binding interaction between the scFv-GCN4 antibody and the GCN4 peptide array in vivo.
(A) Mito-mCherry-24xGCN4pep was co-transfected with scFv-GCN4-GFP in HEK293 cells and their co-localization on mitochondria in a single cell is shown (−10 sec). At 0 sec, the mitochondria-localized GFP signal was photobleached in a single Z-plane using a 472nm laser, and fluorescence recovery was followed by time-lapse microscopy. Scale bar, 5 µm. (B) The fluorescence recovery after photobleaching (FRAP) was quantified for 20 cells. (C–E) Indicated constructs were transfected in HEK293 cells and images were acquired 24 hr after transfection with identical image acquisition settings. Representative images are shown in (C). Note that the GFP signal intensity in the mito-mCherry-24xGCN4pep + scFv-GCN4-GFP is highly saturated when the same scaling is used as in the other panels. Bottom row shows a zoom of a region of interest: dynamic scaling was different for the GFP and mCherry signals, so that both could be observed. Scale bars, 10 mm. (D–E) Quantifications of the GFP:mCherry fluorescence intensity ratio on mitochondria after normalization. Each dot represents a single cell and dashed lines indicates the average value. See also figure S2.
Single molecule imaging in living cells using SunTag
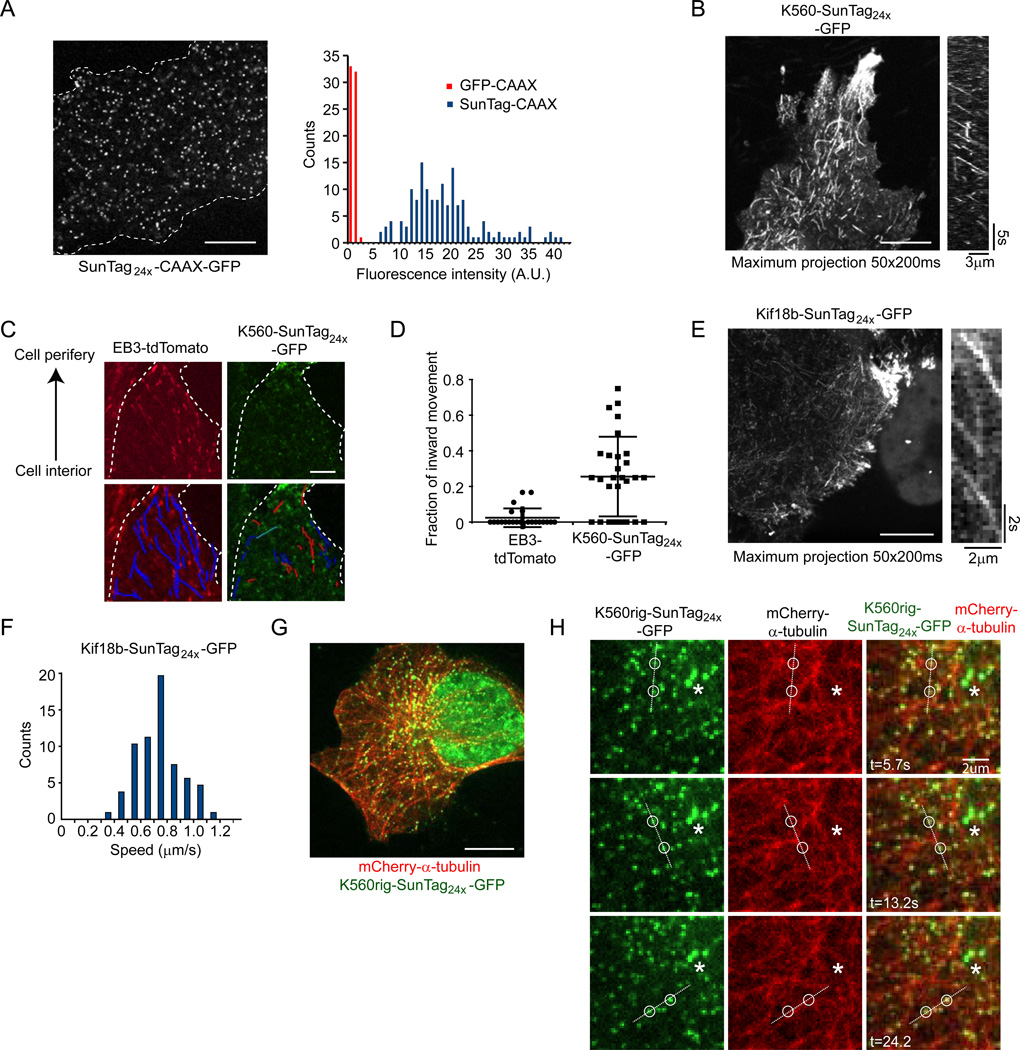
Single molecule imaging is a powerful emerging tool in cell biology; in our first application of the SunTag, we tested whether SunTag24x (24 copies of the peptide binding site) could be used for single molecule imaging in living cells (all imaging in this study was performed at 37° C). We first fused a plasma membrane targeting domain (CAAX) to the C-terminus of the SunTag24x (SunTag24x-CAAX) (See Supplemental Experimental procedures for details on nomenclature). SunTag24x-CAAX was co-expressed with the scFv-GCN4-GFP antibody fused to a nuclear localization sequence (NLS) to sequester unbound antibody into the nucleus (together referred to as SunTag24x-CAAX-GFP), which resulted in the appearance of bright punctae on the plasma membrane (Figure 3A). Individual fluorescent punctae could be visualized diffusing in the plane of the membrane, either by spinning disk confocal microscopy (Figure 3A and Movie S1) or total internal fluorescence microscopy (TIRF; not shown). The fluorescence intensity of single SunTag-CAAX foci was on average 18-fold brighter than that of single sfGFP-CAAX molecules when using identical imaging conditions (Figure 3A), largely consistent with one antibody-GFP fusion bound to each peptide in the 24x peptide array. This result also supports the conclusion that individual fluorescence foci are indeed single SunTag24x-CAAX molecules. As the SunTag foci were much brighter than single GFP, it was possible to use lower excitation laser power to detect single foci; when using 10mW laser power when imaging SunTag24x-CAAX compared with 55mW for sfGFP-CAAX, we observed that the signal-to-noise (single molecule brightness compared to neighboring background) was still 5.5-fold greater and the rate of photobleaching was more than 10-fold lower for the SunTag compared to single GFP (half-life was 6.8 s; n=150 molecules in 5 cells for SunTag24x-CAAX vs 0.57 s; n=60 molecules in 4 cells for sfGFP-CAAX). Thus, due to the brighter signal, SunTag24x-CAAX could be imaged at lower laser intensities than sfGFP-CAAX and single foci could be imaged substantially longer.
Figure 3. The SunTag allows long-term single molecule fluorescence imaging in the cytoplasm.
(A–H) U2OS cells were transfected with indicated SunTag24x constructs together with the scFv-GCN4-GFP-NLS, and were imaged by spinning disk confocal microscopy 24 hr after transfection. (A) A representative image of SunTag24x-CAAX-GFP is shown (left), as well as the fluorescence intensities quantification of the foci (right, blue bars). As a control, U2OS were transfected with sfGFP-CAAX and of fluorescence intensities of single sfGFP-CAAX molecules were also quantified (red bars). The average fluorescence intensity of the single sfGFP-CAAX was set to 1. Dotted line marks the outline of the cell (left). Scale bar, 10 mm. (B) Cells expressing K560-SunTag24x–GFP were imaged by spinning disk confocal microscopy (image acquisition every 200 ms). Movement is revealed by a maximum intensity projection of 50 time-points (left) and a kymograph (right). Scale bar, 10 µm. (C–D) Cells expressing both EB3-tdTomato and K560-SunTag24x–GFP were imaged and moving particles were tracked manually. Red and blue tracks (lower panel) indicate movement towards the cell interior and periphery, respectively (C). The duration of the movie was 20 s. Scale bar, 5 µm. Dots in (D) represent individual cells with between 5–20 moving particles scored per cell. The mean and standard deviation is indicated. (E–F) Cells expressing Kif18b–SunTag24x–GFP were imaged with a 250 ms time interval. Images in (E) show a maximum intensity projection (50 time-points, left) and a kymograph (right). Speeds of moving molecules was quantified from 10 different cells (F). Scale bar, 10 µm. (G–H) Cells expressing both mCherry-α-tubulin and K560rig-SunTag24x–GFP were imaged with a 600 ms time interval. The entire cell is shown in (G), while H shows zoomed-in stills of a time series from the same cell. Open circles track two foci on the same microtubule, which is indicated by the dashed line. Asterisks indicate stationary foci. Scale bars, 10 and 2 µm (G and H), respectively. See also figure S3 and Movies S1–S6.
Single molecule imaging in the interior of the cell is more difficult than at the plasma membrane due to lower signal to background and the inability to use TIRF microscopy. We therefore tested whether the SunTag could be used to image single molecules deep inside the cell. When the SunTag24x was expressed as a cytosolic protein (SunTag24x-GFP) or a nuclear protein by fusing it to a nuclear import sequence (NLS-SunTag24x-GFP), bright foci were observed rapidly diffusing in the cytoplasm or nucleus, respectively, using spinning disk confocal microscopy (Figure S3A and Movie S1). Thus, SunTag provides a very visible fluorescent marker for single molecules within the cell interior.
Since the SunTag with 24 bound GFPs would be ~1.4 mDa, much larger than a single GFP tag, we wished to determine the effect on diffusion. Using single molecule tracking (See Experimental Procedures section), we found that SunTag24x-GFP-CAAX molecules diffused more slowly in the plasma membrane than GFP-CAAX (diffusion coefficients of 1.3 ± 0.28 µm2/s, n = 924 molecules in 3 cells vs 0.12 ± 0.02 µm2/s, n = 3767 molecules in 4 cells (mean ± SEM), respectively) (Figure S3B–C).
Similarly, single molecule tracking revealed a diffusion coefficient of single cytoplasmic SunTag24x-GFP foci of 0.76 ± 0.06 µm2/s (mean ± SEM; n = 1369 tracks in 6 cells) (Figure S3D). This value is reduced compared to an average sized protein tagged with a single GFP (~20 µm2/s) (Baum et al., 2014), but well within the expected range of a multimer of 24 GFP molecules (Baum et al., 2014). Together, these results show, as expected, that the relatively large SunTag reduces protein diffusion compared to a single GFP.
We next imaged SunTag in other intracellular compartments. The mitoNEET mitochondrial targeting domain fused to the SunTag24x was expressed in U2OS cells together with scFv-GCN4-GFP and imaged using spinning disk confocal microscopy. Bright punctae could be observed very rapid diffusing in the mitochondrial membrane (Movie S1). We also examined a histone H2B-SunTag24x– GFP fusion and found that punctae exhibited slow, highly confined diffusion in the nucleus (D = 2.1 ± 0.6 × 103 nm2/s) (Figure S3E–F and Movie S2), consistent with an interaction with DNA. Together, these results show that the SunTag24x can be targeted to different regions of the cell and used as a single molecule fluorescent reporter.
We next tested whether the SunTag could be used to make single molecule measurements of cytoskeletal motors moving in vivo. Previous studies have imaged single motor proteins fused to three copies of GFP using TIRF microscopy (Cai et al., 2009), but the signal is relatively weak and imaging by TIRF microscopy is limited to molecules that are very close to the glass surface (<200 nm). We first fused SunTag24x to a truncated version of kinesin-1 (termed K560), which is a processive motor that lacks its cargo binding domain. Spinning disk confocal imaging (10 frames/sec) of K560-SunTag24x-GFP revealed bright foci moving unidirectionally throughout the cell with an average speed of 1.29 ± 0.24 µm/s (Figure 3B, Figure S3G and Movie S3). Due to the very low photobleaching, we were able to measure run lengths of single K560-SunTag24x-GFP molecules, revealing an average run length of 1.28 ± 0.63 µm (Figure S3H), which is consistent with previous measurements (0.91 µm) (Cai et al., 2009). These results show that the SunTag allows long-term single molecule imaging of functional cytoskeletal motor proteins in vivo.
Interestingly, when we imaged motility of K560-SunTag24x-GFP (which moves exclusively towards plus-ends of microtubules), we found that a substantial fraction of K560-SunTag24x-GFP motors moved towards the cell interior, indicating that the microtubule tracks for these motors have their plus-ends directed inwards (Figure 3C–D). This was surprising, as microtubules are generally thought to be oriented with their plus-ends outwards. Indeed, in these same cells, when microtubule polarity was assessed using a conventional method of visualizing EB3-tdTomato, a protein that selectively marks the growing plus ends of microtubule (Akhmanova and Steinmetz, 2008), then microtubules plus-ends were found to be oriented almost exclusively towards the cell periphery (Figure 3C–D). By fusing the microtubule minus-end binding protein Camsap2 to SunTag, we indeed could confirm that a subset of microtubules had their minus-ends near the cell periphery in these cells (Figure S3I and Movie S4). Thus, live-cell imaging using K560-SunTag24x-GFP as a reporter for microtubule polarity suggests that cells contain a subpopulation of microtubules that are not growing (and hence not interacting with EB3) and have inverted polarity with respect to the cell periphery.
We next sought to test whether the SunTag could be used to study cytoskeletal motors whose motility have not been characterized. Kif18b is a member of the kinesin superfamily which has been shown to track with growing microtubule plus-ends and regulate their dynamics (Stout et al., 2011; Tanenbaum et al., 2011). However, it is currently unclear whether individual Kif18b molecules track the growing plus-ends by rapidly binding and releasing the plus-end, as is the case for other plus-end tracking proteins (Akhmanova and Steinmetz, 2008), or whether it walks along the microtubule, remaining at the tip as it elongates. While Kif18b motility has not been directly measured, homologs of Kif18b were found to move at rates in vitro that are far too slow to keep up with microtubule growth (<100 nm/s) , arguing against the latter model. To analyze Kif18b’s motility in vivo, we expressed full length Kif18b with a C-terminal SunTag24x in U2OS cells. Surprisingly, and unlike what was reported for its homologs, single KIF18b-SunTag24x-GFP molecules translocated along microtubules at high speeds (635 ± 163 nm/s; mean ± s.d.) (Figure 3E–F and Movie S5), which is substantially faster than the speed of microtubule growth in these cells (Tanenbaum et al., 2006). These results reveal that the SunTag can be used to determine the physical properties of molecular motors in vivo.
We also tested whether the SunTag could be used to image single cytoskeletal filament dynamics in dense networks using a novel type of fluorescence speckle microscopy (FSM). Traditional FSM visualizes and tracks identifiable fluorescent “speckles” that arise from the stochastic variations in the incorporation of fluorescently-labeled actin or tubulin monomers into complex cytoskeletal networks (Waterman-Storer et al., 1998). However, due to the stochastic number of fluorophores contained within a diffraction-limited spot in traditional FSM, the brightness of fluorescent speckles varies considerably. Furthermore, since multiple fluorophores give rise to an individual speckle, a speckle can contain fluorescently labeled monomers that are present in different filaments which is problematic for following single filament dynamics. Therefore, we examined whether single SunTag molecules could be used to create positional marks to follow the movements of individual microtubules in living cells. For this purpose, we fused SunTag24x to an ATP hydrolysis defective K560 mutant that forms a tight, rigor-bond with microtubules, but does not translocate along them (termed K560rig). When K560rig-SunTag24x-GFP was expressed in cells at low levels, individual GFP foci colocalized with microtubules (visualized by α-tubulin-mCherry), thus providing a sparse labeling pattern of the microtubule network (Figure 3G–H and Movie S6). While the microtubule network appeared largely static when the microtubules were imaged directly with mCherry-tubulin (Movie S6), imaging of K560rig-SunTag24x-GFP revealed many microtubules undergoing translocation events in cells (Figure 3H and Movie S6).
Microtubule translocation occurred with an average speed of 1.1 ± 0.3 µm/s (mean ± SD; n =21 events in 4 cells), a rate suggesting that they were propelled by microtubule-based motor proteins. The frequency of microtubule translocation was 0.17 min−1 per cell and these movements had a duration of 1.9 ± 0.7 sec (mean ± SD, n=21 events in 4 cells). Angular movements of the microtubule axis also could be observed in cases where a single microtubules had two or more K560rig-SunTag24x-GFP molecules bound as fiducial markers (Figure 3H and Movie S6). These results reveal that the SunTag provides a powerful tool to study movements of individual microtubule filaments in dense microtubule networks in living cells.
Optimizing the SunTag for higher protein expression
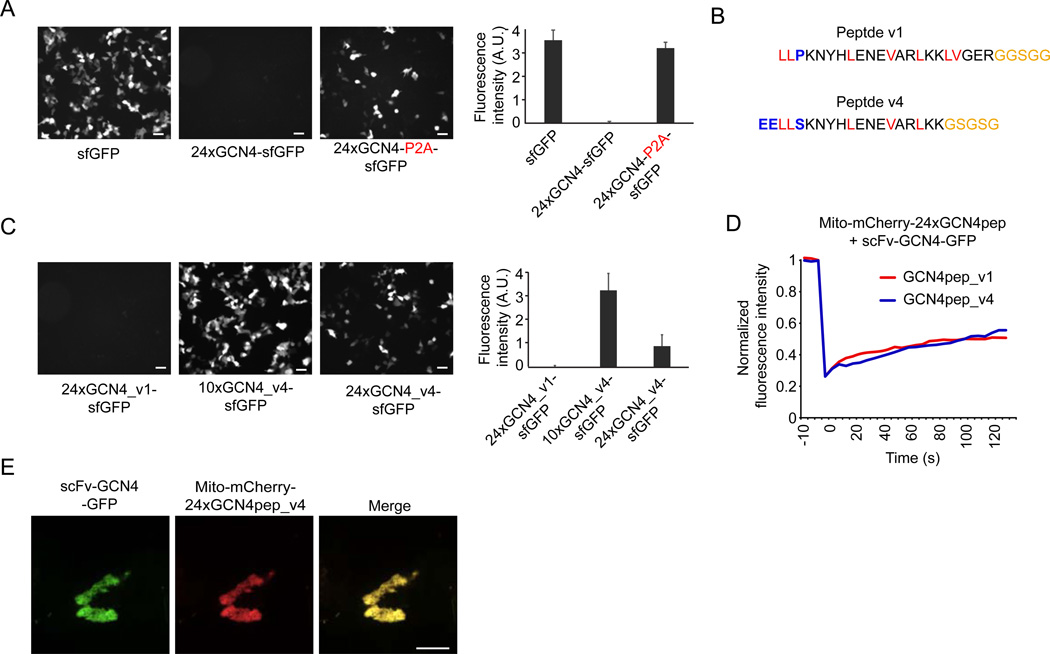
The first generation construct of SunTag24x (termed v1) described in the previous sections was expressed at extremely low levels, which, while useful for single molecule imaging, may be suboptimal for other applications that require higher or physiological concentrations of the protein. Based on the number of foci observed when the SunTag24x was co-expressed with scFv-GCN4-GFP, we estimate that only a few hundred protein copies of v1 SunTag24x are expressed per cell. Consistent with this, when GFP was directly fused to the v1 SunTag24x a very low overall GFP signal was observed by epifluorescence compared to the expression of sfGFP alone on the same promoter and plasmid (Figure 4A). The very low expression level of the SunTag24x may be due to either a problem with the mRNA (poor synthesis, stability or translation) or an instability of the peptide array after its translation. To distinguish between these possibilities, we inserted a viral P2A ribosome skipping sequence in between the 24xGCN4 peptide array and GFP, which allows synthesis of two distinct proteins (i.e. 24xGCN4 peptide array and GFP) from the same mRNA. Insertion of the P2A site in between 24xGCN4 peptide and GFP dramatically increased GFP expression (Figure 4A), indicating that the mRNA is present and efficiently translated, strongly suggesting that poor protein stability explains the low expression of the 24xGCN4 peptide array.
Figure 4. An optimized peptide array for high expression.
(A) Indicated constructs were transfected in HEK293 cells and imaged 24 hr after transfection using wide-field microscopy. All images were acquired using identical acquisition parameters. Representative images are shown (left) and fluorescence intensities were quantified (n=3) (right). (B) Sequence of the first and second generation GCN4 peptide (modified or added residues are colored blue, hydrophobic residues are red and linker residues are yellow). (C–E) Indicated constructs were transfected in HEK293 cells and imaged 24 hr after transfection using widefield (C) or spinning disk confocal (D–E) microscopy. (C) Representative images are shown (left) and fluorescence intensities were quantified (n=3) (right). (D–E) GFP signal on mitochondria was photobleached and fluorescence recovery was determined over time. The graph (E) represents an average of 6 cells per condition. (E) shows an image of a representative cell before photobleaching. Scale bars in (A,C), 50 µm, scale bars in (D,E), 10 µm. See also Movie S7.
The GCN4 peptide contains many hydrophobic residues (Figure 4B) and is largely unstructured in solution (Berger et al., 1999); thus, the poor expression of the peptide array could be due to its unstructured and hydrophobic nature. To test this idea, we designed several modified peptide sequence that were predicted to increase α-helical propensity and reduce hydrophobicity. One of these optimized peptides (v4, Figure 4B) was expressed moderately well as a 24x peptide array and even higher expression was achieved with a 10x peptide array (Figure 4C). Importantly, fluorescence imaging revealed that the scFv-GCN4 antibody robustly bound to the GCN4 v4 peptide array in vivo and FRAP analysis suggests that the scFv-GCN4 antibody dissociates with a similar slow off-rate from the GCN4 v4 peptide array as the original peptide (Figure 4D–E). Furthermore, K560 motility could be observed when it was tagged with the optimized v4 24x peptide array, indicating that the optimized v4 peptide array did not interfere with protein function (Movie S7). Together, these results identify a new version of the peptide array that can be used for applications requiring higher expression.
Activation of gene transcription using Cas9-SunTag
Since the SunTag system could be used for amplification of a fluorescence signal, we tested whether it also could be used to amplify regulatory signals involved in gene expression. Transcription of a gene is strongly enhanced by recruiting multiple copies of transcriptional activators to endogenous or artificial gene promoters (Anderson and Freytag, 1991; Chen et al., 1992; Pettersson and Schaffner, 1990). Thus, we thought that robust, artificial activation of gene transcription might also be achieved by recruiting multiple copies of a synthetic transcriptional activator to a gene using the SunTag. Recently, a highly versatile, synthetic transcriptional activator was developed by fusing the herpes virus transcriptional activation domain VP16 (or 4 copies of VP16, termed VP64) to a nuclease-deficient mutant of the CRISPR effector protein Cas9 (dCas9), which can be targeted to any sequence in the mammalian genome using sequence-specific small guide RNAs (sgRNAs) (Cheng et al., 2013; Farzadfard et al., 2013; Gilbert et al., 2013; Hu et al., 2014; Kearns et al., 2014; Maeder et al., 2013; Mali et al., 2013; Perez-Pinera et al., 2013). While targeting of dCas9-VP64 was able to increase transcription of the targeted gene, the level of gene activation using dCas9-VP64 was generally very low, most often less than 2-fold (Cheng et al., 2013; Hu et al., 2014; Mali et al., 2013; Perez-Pinera et al., 2013), thus severely limiting the potential use of this system. However, consistent with multiple transcriptional activators being required to stimulate robust transcription, a GFP reporter containing multiple binding sites for a single sgRNA was potently activated by dCas9-VP64 (Gilbert et al., 2013). In addition, recruitment of multiple copies of dCas9-VP64 to a native promoter using multiple non-overlapping sgRNAs also greatly enhanced transcriptional activation (Cheng et al., 2013; Hu et al., 2014; Maeder et al., 2013; Mali et al., 2013; Perez-Pinera et al., 2013). We therefore wondered whether recruitment of multiple VP64 domains to a single molecule of dCas9 using the SunTag would enhance the ability of dCas9 to activate endogenous transcription (See Figure 5A).
Figure 5. dCas9-SunTag allows genetic rewiring of cells through activation of endogenous genes.
(A) Schematic of gene activation by dCas9-VP64 and dCas9-SunTag-VP64. dCas9 binds to a gene promoter through its sequence specific sgRNA (red line). Direct fusion of VP64 to dCas9 (top) results in a single VP64 domain at the promoter which poorly activates transcription of the downstream gene. In contrast, recruitment of many VP64 domains using the SunTag potently activates transcription of the gene (bottom). (B–D) K562 cells stably expressing dCas9-VP64 or dCas9-SunTag10x-VP64 were infected with lentiviral particles encoding indicated sgRNAs, as well as BFP and a puromycin resistance gene and selected with 0.7 µg/ml puromycin for 3 days to kill uninfected cells. (B–C) Cells were stained for CXCR4 using a directly labeled α-CXCR4 antibody and fluorescence was analyzed by FACS. (D) Trans-well migration assays (see experimental procedures) were performed with indicated sgRNAs. Results are displayed as the fold change in directional migrating cells over control cell migration. (E) dCas9-VP64 or dCas9-SunTag10x-VP64 induced transcription of CDKN1B with several sgRNAs. mRNA levels were quantified by qPCR. (F) Doubling time of control cells or cells expressing indicated sgRNAs was determined (See Experimental Procedures section). Graphs in (C, D and F) are averages of three independent experiments. Graph in (E) is average of two biological replicates, each with two or three technical replicates. All error bars indicated standard error of the mean (SEM). See also figure S4.
To test whether dCas9 could be tagged with the SunTag, we used an imaging-based approach in which dCas9-SunTag24x_v4 was co-expressed with scFv-GCN4-GFP and targeted to telomeres using a telomere-specific sgRNA (dCas9-SunTag24x_v4-GFP) When examined by fluorescence microscopy, very bright dots were observed in the nucleus, similar to previous work with dCas9 directly labeled with GFP (dCas9-GFP) (Chen et al., 2013) (Figure S4A). Analysis of dCas9-SunTag24x_v4-GFP data showed that telomere labeling was 19.7 ± 6.2 -fold (mean ± SD) (n=100 telomeres in 5 cells (dCas-GFP) or n=120 telomeres in 6 cells (dCas9-SunTag24x_v4-GFP)) brighter compared to those labeled by the dCas9 directly fused to GFP, consistent with the recruitment of ~24 copies of GFP to a single dCas9 molecule (Figure S4A–B). As a control, in the absence of the sgRNA targeting the telomere, nuclear GFP fluorescence was diffuse (Figure S4A). Analysis of telomere mobility revealed a very similar diffusion coefficient for telomeres labeled with dCas9-GFP and dCas9-SunTag24x_v4-GFP (3.1 ± 0.6 × 103 nm2/s (n=511 telomeres in 8 cells) vs 2.8 ± 0.8 × 103 nm2/s (255 telomeres in 5 cells), respectively) (Figure S4C), indicating that SunTag labeling of telomeres did not alter their mobility. Thus, dCas9-SunTag can efficiently recruit multiple proteins to a single genomic locus and can be used for very bright labeling of telomeres.
Next, scFv-GCN4-GFP was fused to VP64 to test whether recruitment of multiple VP64 domains to a promoter would enhance transcription of the downstream gene. K562 cell lines were generated expressing either dCas9-VP64 (Gilbert et al., 2013) alone or co-expressing dCas910x_v4 with GCN4-sfGFP-NLS-VP64 (hereafter referred to as dCas9-SunTag-VP64). dCas9-SunTag10x_v4 was used for these experiments, as we found similar maximal activation and less cell-to-cell variation in gene expression compared to the dCas9-SunTag24x_v4 (data not shown). As a target gene, we selected CXCR4, a transmembrane receptor known to stimulate cell migration, which is normally poorly expressed in K562 cells. dCas9-VP64 and dCas9-SunTag10x_v4-VP64 expressing cells were infected with a lentivirus that encoded either a control sgRNA or an sgRNA targeting CXCR4 (sgCXCR4; three different sgRNA were tested). Five days after lentivirus infection, the levels of CXCR4 protein were determined. We found minimal activation of CXCR4 expression using dCas9-VP64 with the three sgRNAs tested (Figure 5C). In contrast, strong activation (10–50-fold) was observed with all three CXCR4 sgRNAs using dCas9-SunTag10x_v4-VP64 (Figure 5B–C). These results show that robust transcriptional activation can be achieved by SunTag-dependent multimerization of transcriptional activation domains at an endogenous gene promoter using a single sgRNA.
We next wished to test whether transcriptional regulation using dCas9-SunTag10x_v4-VP64 could induce a biological response. CXCR4 is a chemokine receptor that stimulates cell migration in response to activation by SDF1a (Brenner et al., 2004). We tested whether activation of CXCR4 in K562 could induce migration in response to SDF1a using a transwell migration assay. We found that activating CXCR4 expression using dCas9-SunTag10x_v4-VP64 dramatically stimulated cell migration by an order of magnitude (Figure 5D). In contrast, very weak (<2-fold) enhancement of cell migration was observed using CXCR4 activation by dCas9-VP64 (Figure 5D). This result indicates that dCas9-SunTag10x_v4-VP64-dependent gene activation is sufficiently potent to affect the behavior of these cells. Surprisingly, cells expressing the highest level of CXCR4 showed less cell migration, suggesting there may be an optimum level of CXCR4 expression for stimulation of cell migration (compare Figure 5C with 5D).
CXCR4 is normally expressed at very low levels in K562 cells, so we tested whether the expression of a well-expressed gene could also be increased using SunTag-dependent transcriptional activation. For this purpose, we chose the cell cycle inhibitor CDKN1B (also known as p27kip1). CDKN1B mRNA expression level was determined in both dCas9-VP64 and dCas9-SunTag-VP64 cells with four different sgRNAs. Very little activation of CDKN1B transcription was observed using dCas9-VP64 (28% increase in mRNA at best) (Figure 5E), while 3/4 sgRNAs robustly activated CDKN1B in dCas9-SunTag10x_v4-VP64 cells (330% for the best sgRNA) (Figure 5E). Furthermore, as expected for increased levels of the cell cycle inhibitor CDKN1B, activation of CDKN1B with dCas9-SunTag10x_v4-VP64 significantly reduced cell growth (Figure 5F). In contrast, activation of CDKN1B with dCas9-VP64 had little impact on cell growth (Figure 5F). Taken together, these results show that the SunTag-dependent signal amplification robustly enhances transcriptional activation by dCas9-VP64 and allows functional re-engineering of cell behavior through precise control of gene expression.
Discussion
Amplification of biological signals is important for many biological processes as well as for imaging and bioengineering. Here, we have developed a versatile protein tagging system, the SunTag, which can be used to increase fluorescence of genetically-encoded proteins as well as amplify gene expression. The SunTag system provides a proof-of-concept of the power of controlled protein multimerization and could form the basis for developing other protein multimerization strategies.
Imaging applications of the SunTag
The SunTag represents the brightest genetically-encoded fluorescent tagging system available and has several major advantages over existing imaging methods. First, due to its extremely high signal, a low expression level of SunTag-proteins is sufficient for imaging and thus avoids potential problems associated with protein overexpression. For example, we have found that overexpression of GFP-mitoNEET is detrimental to mitochondrial function (data not shown). However, we have achieved very bright images of mitochondria with much lower expression of mitoNEET-SunTag24x-GFP than can be achieved by single copy GFP tagging. Second, bright labeling of both organelles and single molecules allows imaging with much lower light illumination, which could minimize phototoxicity during long-term imaging. Third, bright labeling using the SunTag will likely be beneficial for automated tracking, especially single molecule tracking in vivo, as such algorithms work best with a high signal-to-noise ratio. Fourth, the SunTag allows single molecule imaging deep inside the cytoplasm and nucleus, in contrast to TIRF microscopy, which is mainly applicable to molecules close to the cell membrane. Finally, SunTag, which can be used to produce bright, single molecule labels on cytoskeletal filaments, has advantages over traditional FSM, which relies on stochastic fluctuations in the distribution of many fluorophores (Waterman-Storer et al., 1998).
We also show that SunTag is a powerful single molecule reporter of intracellular processes. For example, K560-SunTag movements revealed a stable subset of microtubules with reversed polarity in U2OS cells, which was not evident from tracking growing microtubules with EB3-tdTomato. We demonstrate that K560rig-SunTag can be used to observe microtubule sliding in the cytoplasm; this method also could be powerful for studying microtubule movements in the mitotic spindle or axon, where very high microtubule densities preclude visualization of single microtubules by traditional imaging with GFP-tubulin. dCas9-SunTag allows much brighter labeling of genomic loci than dCas9 directly fused to GFP. Previous work revealed that dCas9-GFP could also be used to image a non-repetitive DNA sequence if 36 non-overlapping sgRNAs were used (Chen et al., 2013). It will be interesting to test whether a non-repetitive DNA sequence can be imaged using dCas9-SunTag with a smaller number of sgRNAs. Overall, these results show that the SunTag is a versatile tool for single molecule imaging and very bright labeling of intracellular structures and organelles.
In this study, we have developed two different versions of the SunTag; v1 and v4. The major advantage of the v1 scaffold is that it is expressed at very low levels, simplifying single molecule imaging. However, using weak promoters to drive expression of the v4 scaffold, we have found that is also possible to perform single molecule imaging. We also found that the v1 scaffold shows some aggregation at higher protein concentrations, while the v4 scaffold did not. Thus, for many imaging applications, the v4 scaffold expressed from weak promoters (see Supplemental Experimental Procedures) may be superior.
While imaging using the SunTag has many advantages, the large size of the tag itself represents a potential drawback. The SunTag has the potential to interfere with the activity or half-life when fused to a protein. These caveats are true of single GFP tagging as well, but are more a concern with the SunTag due to its much larger size (1,400 kDa, with 24 bound antibody-GFPs). As is true of any tagging strategy, the suitability of SunTag will need to be determined on a protein-by-protein basis and taking into account the goals of a particular experiment. Nonetheless, in this study, we provide examples of where the SunTag does not interfere with protein activity (kinesin motility) or localization (MitoNeet, Camsap2, Histone H2B, dCas9). In this study, we mostly used SunTag as a single molecule reporter (e.g. an in vivo motor activity assay, microtubule polarity and movement, chromatin dynamics and telomere localization and motility), and in many of those cases we used truncated or inactivated proteins rather than native proteins. For studies that require examining functions of particular proteins in their native state, it will be important to test protein functionality, for example by genetic complementation experiments.
Using SunTag to Control Gene Transcription and Engineer Cell Behavior
The second application of the SunTag, for which we provide a proof-of-concept, is the amplification of biological signaling pathways. Transcriptional regulation is a powerful example, as transcriptional output is strongly dependent on the number of transcriptional activators recruited to the gene promoter (Anderson and Freytag, 1991; Chen et al., 1992; Pettersson and Schaffner, 1990). Indeed, previous attempts to activate transcription of endogenous genes using a single dCas9 or TALE fused to the transcriptional activation domain VP64 generally resulted in weak transcriptional activation. However, several studies showed that robust gene activation was possible using multiple sgRNAs targeting the same promoter or when multiple sgRNA binding sites were present in an artificial promoter (Cheng et al., 2013; Gilbert et al., 2013; Hu et al., 2014; Maeder et al., 2013; Mali et al., 2013; Perez-Pinera et al., 2013). In contrast, our results demonstrate that the dCas9-SunTag transcriptional system can robustly activate the expression of an endogenous gene using a single sgRNA. This method not only simplifies single gene activation, but also opens possibilities for activating multiple genes simultaneously, potentially allowing complex genetic re-wiring of cells or organisms. For example, generation of induced pluripotent stem cells (iPS) requires expression of four transcription factors (Takahashi and Yamanaka, 2006) and induced human cardiomyocytes requires expression of five factors (Fu et al., 2013). It will be very interesting to test whether such cell fate decisions can be directed by activation of endogenous genes using the SunTag, rather than through gene overexpression with transfected plasmids.
The ability to upregulate gene expression using dCas9-SunTag with a single sgRNA also opens the door to large scale genetic screens to uncover phenotypes that result from increased gene expression. This application will be especially important for understanding the effects of gene upregulation in cancer. In addition, large scale activation screens could be used to identify proteins that promote induced pluripotency (Takahashi and Yamanaka, 2006) or, conversely, promote differentiation to a specific cell lineage.
SunTag might also have applications in promoting transcriptional silencing. Previous work has found that Cas9 fused to transcriptional silencing domain fuse can inhibit gene-specific transcription (Gilbert et al., 2013), but in most cases some residual transcription was observed. Recruitment of many transcriptional silencing domains to a single promoter via SunTag-dCas9 could potentially enhance gene silencing for loss-of-function studies. This approach could complement wildtype Cas9 nuclease-mediated gene knockout and might be especially useful for the analysis of essential genes and non-coding RNAs. In addition, multiple types of transcriptional activators or repressors could be recruited to a single scaffold, which may be required for maximal transcriptional activation or repression.
Advantage of the SunTag over direct fusion of protein multimers
Here, we tag single molecules with up to 24 copies of GFP or 10 copies of VP64 using the SunTag system. Indirect labeling of proteins using the SunTag has several major advantages over direct fusion of many copies of the fluorescent proteins or regulatory domains to the protein of interest. First, it is likely very difficult to generate plasmids containing 24 copies of a protein due to the large size and bacterial plasmid recombination. Second, the SunTag allows a high level of flexibility, as only a single genetic element needs to be modified to generate an entirely new protein multimer. Third, previous attempts to increase transcriptional output by increasing the copy number of VP64 in a direct protein fusion were not effective (Cheng et al., 2013), indicating that the molecular configuration of the SunTag is more efficient for transcriptional activation than a single chain fusion.
Future Developments of the SunTag System
The SunTag system is based on a scFv antibody and its epitope. Antibody-based systems have several major advantages, including their very high affinity and their ability to recognize short peptides. However, a major drawback of antibodies is that they are generally poorly expressed in the cytoplasm and are relatively large in size. Several alternatives to antibodies have been developed, including DARPins, which are small, highly stable proteins that can be evolved in vitro to bind their epitope with high affinity (Binz et al., 2004). Such proteins could form the basis of future work on a second generation SunTag system. Furthermore, development of a second, orthogonal system could allow two color single molecule imaging, as well as dual transcriptional regulation that could allow activation and silencing of different genes in the same cell. Finally, another important future tool will be creating an inducible recruitment system, which could, for example, enable temporal gene regulation. In summary, SunTag, and its potential future derivatives that operate using the same principles, represents a powerful system that allows single molecule imaging and transcriptional activation of endogenous genes, and which could find much broader uses in biological research and bioengineering.
Experimental procedures
Cell culture and plasmids
HEK293 and U2OS cells were grown in DMEM/10% FCS/Pen/Strep. K562 cells were grown in RPMI, containing 25 mM HEPES, with 10% FCS/Pen/Strep. HEK293 and U2OS cells were transfected with PEI (Sigma) and Fugene 6 (Roche), respectively. Sequences and discussion of constructs are provided in the Supplemental Experimental Procedures and Supplemental table 1.
Imaging and quantitation
Cells were grown in 96-well glass bottom dishes and were imaged on an inverted Nikon TI spinning disk confocal microscope with the Nikon Perfect Focus system, 100× 1.45 NA objective, an Andor EM-CCD camera, and Micro-Manager software (Edelstein et al., 2010). Images in Figures 4A,C were acquired using widefield illumination, a 20x air objective, and a Hamamatsu CMOS Flash 4.0 camera.
To determine the number of antibodies bound to a single peptide array, a sfGFP-linker-mCherry fusion protein was imaged using image acquisition parameters for which GFP and mCherry fluorescence intensities were equal. Using the same acquisition parameters, fluorescence intensity ratio of the mito-mCherry-peptide array and GFP-tagged antibody was determined.
To measure fluorescence intensities, a circular region of interest (ROI) with a diameter of 0.5 µm was applied and an average background (same ROI measured in five different areas of the cell that did not contain a fluorescent foci) was subtracted. Kinesin run lengths and speeds were determined by kymographs analysis (Sirajuddin et al., 2014). Single molecule tracking was performed in 2D using Fiji Trackmate. MSD plots were generated in Matlab using the X,Y tracking coordinates generated by Trackmate. Diffusion coefficients were calculated based on MSD plots.
Quantification of protein and mRNA levels
To determine the levels of CXCR4 and CDKN1B transcriptional activation, K562 cells stably expressing either dCas9-VP64-BFP or dCas9-SunTag10x_v4 together with scFv-GCN4-GFP-VP64 were infected with lentivirus encoding individual sgRNAs (sequences provided in the Supplemental Experimental Procedures) targeting theCXCR4 and CDKN1B promoters, as well as BFP and a puromycin resistance gene. Cells were then selected with 0.7–1 µg/ml puromycin for 3 days. Measurements of CXCR4 protein levels was performed by FACS as described previously (Gilbert et al., 2013). For the measurement of CDKN1B mRNA levels, total RNA was isolated with Trizol (Ambion) and cDNA was synthesized using the Superscript cDNA synthesis kit VILO (Life Technologies). qPCR primers are listed in the Supplemental Experimental Procedures.
Transwell migration
Recombinant human SDF1a (Peprotech) was used as a chemoattractant for the migration assay. K562 cells were cultured in RPMI-1640 with 2% FCS for 16 hr. 75,000 cells were counted and resuspended in RPMI-1640/2% FCS and added to the upper chamber of 24-well Transwell inserts (8-micron pore size polyethylene terephthalate, Millipore), and 200 ng/mL SDF1a was added to the lower chamber. The number of K562 cells that migrated to the lower chamber was quantified after 5 hr by flow cytometry on a BD Bioscience LSR-II flow cytometer.
K562 growth rate measurements
K562 cells stably expressing either dCas9-VP64-BFP alone or dCas9-SunTag10x_v4 together with scFv-GCN4-GFP-VP64 were infected with lentivirus encoding indicated sgRNAs together with BFP at an MOI of approximately 0.3. Two days after infection, the fraction of BFP positive cells was determined by FACS. Cells were grown for two weeks, after which the fraction of BFP positive cells was re-measured. In cells infected with a control sgRNA, the fraction of BFP positive cells remained constant over time. In parallel, the cell doubling time of uninfected cells was determined. Using the cell doubling time and the fraction of BFP positive cells at day 3 and day 14, the growth rate of BFP positive cells was determined compared to uninfected control cells to determine the growth rate of sgRNA expressing cells.
Supplementary Material
Highlights.
-
-
SunTag allows controlled protein multimerization on a protein scaffold.
-
-
SunTag enables long-term single molecule imaging in living cells.
-
-
SunTag greatly improves CRISPR-based activation of gene expression.
Acknowledgments
We thank Evan Whitehead for making the dCas9–2xNLS-10xGCN4 and scFv-GCN4-sfGFP-2xNLS-VP64 plasmids, Vicki Allan for the EB3-tdTomato plasmid, Xiaokun Shu for the IFP2.0 protein and Max Horlbeck for designing the CDKN1B sgRNAs. We also thank Nico Stuurman for help with microscopy, Kate Carbone for help with single particle tracking, Andreas Plückthun for helpful discussions and the Vale lab members for helpful comments. M.E.T. was financially supported by a fellowship from the Dutch Cancer Society (KWF). L.S.Q. acknowledges support from the UCSF Center for Systems and Synthetic Biology, NIH Office of The Director (OD), and National Institute of Dental & Craniofacial Research (NIDCR). This work was in part supported by NIH P50 (grant GM081879, L.S.Q.), NIH Director’s Early Independence Award (grant OD017887, L.S.Q.), NIH R01 (grant DA036858, L.S.Q. and J.S.W.), NIH P50 (grant GM102706, J.S.W), NIH U01 (grant CA168370, J.S.W), the Leukemia and Lymphoma Society (L.A.G.) and the Howard Hughes Medical Institute (R.D.V. and J.S.W).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akhmanova A, Steinmetz MO. Tracking the ends: a dynamic protein network controls the fate of microtubule tips. Nature reviews Molecular cell biology. 2008;9:309–322. doi: 10.1038/nrm2369. [DOI] [PubMed] [Google Scholar]
- Anderson GM, Freytag SO. Synergistic activation of a human promoter in vivo by transcription factor Sp1. Molecular and cellular biology. 1991;11:1935–1943. doi: 10.1128/mcb.11.4.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum M, Erdel F, Wachsmuth M, Rippe K. Retrieving the intracellular topology from multi-scale protein mobility mapping in living cells. Nature communications. 2014;5:4494. doi: 10.1038/ncomms5494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger C, Weber-Bornhauser S, Eggenberger J, Hanes J, Pluckthun A, Bosshard HR. Antigen recognition by conformational selection. FEBS letters. 1999;450:149–153. doi: 10.1016/s0014-5793(99)00458-5. [DOI] [PubMed] [Google Scholar]
- Bertrand E, Chartrand P, Schaefer M, Shenoy SM, Singer RH, Long RM. Localization of ASH1 mRNA particles in living yeast. Molecular cell. 1998;2:437–445. doi: 10.1016/s1097-2765(00)80143-4. [DOI] [PubMed] [Google Scholar]
- Binz HK, Amstutz P, Kohl A, Stumpp MT, Briand C, Forrer P, Grutter MG, Pluckthun A. High-affinity binders selected from designed ankyrin repeat protein libraries. Nature biotechnology. 2004;22:575–582. doi: 10.1038/nbt962. [DOI] [PubMed] [Google Scholar]
- Brenner S, Whiting-Theobald N, Kawai T, Linton GF, Rudikoff AG, Choi U, Ryser MF, Murphy PM, Sechler JM, Malech HL. CXCR4-transgene expression significantly improves marrow engraftment of cultured hematopoietic stem cells. Stem Cells. 2004;22:1128–1133. doi: 10.1634/stemcells.2003-0196. [DOI] [PubMed] [Google Scholar]
- Cai D, McEwen DP, Martens JR, Meyhofer E, Verhey KJ. Single molecule imaging reveals differences in microtubule track selection between Kinesin motors. PLoS biology. 2009;7:e1000216. doi: 10.1371/journal.pbio.1000216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Gilbert LA, Cimini BA, Schnitzbauer J, Zhang W, Li GW, Park J, Blackburn EH, Weissman JS, Qi LS, et al. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell. 2013;155:1479–1491. doi: 10.1016/j.cell.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Azizkhan JC, Lee DC. The binding of transcription factor Sp1 to multiple sites is required for maximal expression from the rat transforming growth factor alpha promoter. Oncogene. 1992;7:1805–1815. [PubMed] [Google Scholar]
- Cheng AW, Wang H, Yang H, Shi L, Katz Y, Theunissen TW, Rangarajan S, Shivalila CS, Dadon DB, Jaenisch R. Multiplexed activation of endogenous genes by CRISPR-on, an RNA-guided transcriptional activator system. Cell research. 2013;23:1163–1171. doi: 10.1038/cr.2013.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colby DW, Garg P, Holden T, Chao G, Webster JM, Messer A, Ingram VM, Wittrup KD. Development of a human light chain variable domain (V(L)) intracellular antibody specific for the amino terminus of huntingtin via yeast surface display. Journal of molecular biology. 2004;342:901–912. doi: 10.1016/j.jmb.2004.07.054. [DOI] [PubMed] [Google Scholar]
- Colca JR, McDonald WG, Waldon DJ, Leone JW, Lull JM, Bannow CA, Lund ET, Mathews WR. Identification of a novel mitochondrial protein ("mitoNEET") cross-linked specifically by a thiazolidinedione photoprobe. American journal of physiology Endocrinology and metabolism. 2004;286:E252–E260. doi: 10.1152/ajpendo.00424.2003. [DOI] [PubMed] [Google Scholar]
- Coller J, Wickens M. Tethered function assays: an adaptable approach to study RNA regulatory proteins. Methods in enzymology. 2007;429:299–321. doi: 10.1016/S0076-6879(07)29014-7. [DOI] [PubMed] [Google Scholar]
- Edelstein A, Amodaj N, Hoover K, Vale R, Stuurman N. Computer control of microscopes using microManager Current protocols in molecular biology / edited by Frederick M Ausubel [et al] Chapter. 2010;14 doi: 10.1002/0471142727.mb1420s92. Unit14 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farzadfard F, Perli SD, Lu TK. Tunable and Multifunctional Eukaryotic Transcription Factors Based on CRISPR/Cas. ACS synthetic biology. 2013;2:604–613. doi: 10.1021/sb400081r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu JD, Stone NR, Liu L, Spencer CI, Qian L, Hayashi Y, Delgado-Olguin P, Ding S, Bruneau BG, Srivastava D. Direct Reprogramming of Human Fibroblasts toward a Cardiomyocyte-like State. Stem cell reports. 2013;1:235–247. doi: 10.1016/j.stemcr.2013.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fusco D, Accornero N, Lavoie B, Shenoy SM, Blanchard JM, Singer RH, Bertrand E. Single mRNA molecules demonstrate probabilistic movement in living mammalian cells. Current biology : CB. 2003;13:161–167. doi: 10.1016/s0960-9822(02)01436-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, Stern-Ginossar N, Brandman O, Whitehead EH, Doudna JA, et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154:442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon GS, Sitnikov D, Webb CD, Teleman A, Straight A, Losick R, Murray AW, Wright A. Chromosome and low copy plasmid segregation in E. coli: visual evidence for distinct mechanisms. Cell. 1997;90:1113–1121. doi: 10.1016/s0092-8674(00)80377-3. [DOI] [PubMed] [Google Scholar]
- Gronenborn AM, Filpula DR, Essig NZ, Achari A, Whitlow M, Wingfield PT, Clore GM. A novel, highly stable fold of the immunoglobulin binding domain of streptococcal protein G. Science. 1991;253:657–661. doi: 10.1126/science.1871600. [DOI] [PubMed] [Google Scholar]
- Hu J, Lei Y, Wong WK, Liu S, Lee KC, He X, You W, Zhou R, Guo JT, Chen X, et al. Direct activation of human and mouse Oct4 genes using engineered TALE and Cas9 transcription factors. Nucleic acids research. 2014;42:4375–4390. doi: 10.1093/nar/gku109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CJ, Spinella F, Nazarian R, Lee MM, Dopp JM, de Vellis J. Expression of green fluorescent protein in oligodendrocytes in a time- and level-controllable fashion with a tetracycline-regulated system. Mol Med. 1999;5:129–137. [PMC free article] [PubMed] [Google Scholar]
- Kearns NA, Genga RM, Enuameh MS, Garber M, Wolfe SA, Maehr R. Cas9 effector-mediated regulation of transcription and differentiation in human pluripotent stem cells. Development. 2014;141:219–223. doi: 10.1242/dev.103341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecerf JM, Shirley TL, Zhu Q, Kazantsev A, Amersdorfer P, Housman DE, Messer A, Huston JS. Human single-chain Fv intrabodies counteract in situ huntingtin aggregation in cellular models of Huntington's disease. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:4764–4769. doi: 10.1073/pnas.071058398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M, Pang CW, Gerken AE, Brock TG. Multiple nuclear localization sequences allow modulation of 5-lipoxygenase nuclear import. Traffic. 2004;5:847–854. doi: 10.1111/j.1600-0854.2004.00227.x. [DOI] [PubMed] [Google Scholar]
- Maeder ML, Linder SJ, Cascio VM, Fu Y, Ho QH, Joung JK. CRISPR RNA-guided activation of endogenous human genes. Nature methods. 2013;10:977–979. doi: 10.1038/nmeth.2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Aach J, Stranges PB, Esvelt KM, Moosburner M, Kosuri S, Yang L, Church GM. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nature biotechnology. 2013;31:833–838. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedelacq JD, Cabantous S, Tran T, Terwilliger TC, Waldo GS. Engineering and characterization of a superfolder green fluorescent protein. Nature biotechnology. 2006;24:79–88. doi: 10.1038/nbt1172. [DOI] [PubMed] [Google Scholar]
- Perez-Pinera P, Kocak DD, Vockley CM, Adler AF, Kabadi AM, Polstein LR, Thakore PI, Glass KA, Ousterout DG, Leong KW, et al. RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nature methods. 2013;10:973–976. doi: 10.1038/nmeth.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersson M, Schaffner W. Synergistic activation of transcription by multiple binding sites for NF-kappa B even in absence of co-operative factor binding to DNA. Journal of molecular biology. 1990;214:373–380. doi: 10.1016/0022-2836(90)90187-q. [DOI] [PubMed] [Google Scholar]
- Pillai RS, Artus CG, Filipowicz W. Tethering of human Ago proteins to mRNA mimics the miRNA-mediated repression of protein synthesis. RNA. 2004;10:1518–1525. doi: 10.1261/rna.7131604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pique M, Lopez JM, Foissac S, Guigo R, Mendez R. A combinatorial code for CPE-mediated translational control. Cell. 2008;132:434–448. doi: 10.1016/j.cell.2007.12.038. [DOI] [PubMed] [Google Scholar]
- Sadowski I, Ma J, Triezenberg S, Ptashne M. GAL4-VP16 is an unusually potent transcriptional activator. Nature. 1988;335:563–564. doi: 10.1038/335563a0. [DOI] [PubMed] [Google Scholar]
- Sirajuddin M, Rice LM, Vale RD. Regulation of microtubule motors by tubulin isotypes and post-translational modifications. Nature cell biology. 2014;16:335–344. doi: 10.1038/ncb2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stout JR, Yount AL, Powers JA, Leblanc C, Ems-McClung SC, Walczak CE. Kif18B interacts with EB1 and controls astral microtubule length during mitosis. Molecular biology of the cell. 2011;22:3070–3080. doi: 10.1091/mbc.E11-04-0363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Tanenbaum ME, Galjart N, van Vugt MA, Medema RH. CLIP-170 facilitates the formation of kinetochore-microtubule attachments. The EMBO journal. 2006;25:45–57. doi: 10.1038/sj.emboj.7600916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanenbaum ME, Macurek L, van der Vaart B, Galli M, Akhmanova A, Medema RH. A complex of Kif18b and MCAK promotes microtubule depolymerization and is negatively regulated by Aurora kinases. Current biology : CB. 2011;21:1356–1365. doi: 10.1016/j.cub.2011.07.017. [DOI] [PubMed] [Google Scholar]
- Waterman-Storer CM, Desai A, Bulinski JC, Salmon ED. Fluorescent speckle microscopy, a method to visualize the dynamics of protein assemblies in living cells. Current biology : CB. 1998;8:1227–1230. doi: 10.1016/s0960-9822(07)00515-5. [DOI] [PubMed] [Google Scholar]
- Worn A, Auf der Maur A, Escher D, Honegger A, Barberis A, Pluckthun A. Correlation between in vitro stability and in vivo performance of anti-GCN4 intrabodies as cytoplasmic inhibitors. The Journal of biological chemistry. 2000;275:2795–2803. doi: 10.1074/jbc.275.4.2795. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.