Abstract
BRPF1 is part of the MOZ HAT complex and contains a unique combination of domains typically found in chromatin-associated factors, which include PHD fingers, a bromodomain and a PWWP domain. Bromodomains are conserved structural motifs generally known to recognize acetylated histones, and the BRPF1 bromodomain preferentially selects for H2AK5ac, H4K12ac and H3K14ac. We solved the X-ray crystal structures of the BRPF1 bromodomain in complex with the H2AK5ac and H4K12ac histone peptides. Site-directed mutagenesis on residues in the BRPF1 bromodomain-binding pocket was carried out to investigate the contribution of specific amino acids on ligand binding. Our results provide critical insights into the molecular mechanism of ligand binding by the BRPF1 bromodomain, and reveal that ordered water molecules are an essential component driving ligand recognition.
Keywords: Epigenetics, bromodomain-PHD finger protein 1, X-ray crystallography, site-directed mutagenesis, isothermal titration calorimetry, circular dichroism
Introduction
In eukaryotic cells, in order to facilitate proper gene regulation and nuclear organization, most of the genome is packaged into a polymeric structure consisting of repetitive units of nucleosomes [1, 2]. Each nucleosome consists of approximately 146 base pairs of super-helical nuclear DNA folded around an octameric structure of four core histone proteins: two H2A/H2B dimers and an H3/H4 tetramer [3]. These histone proteins contain an N-terminal tail region and a C-terminal globular domain. The N-terminal histone tails are presumably flexible and protrude from the nucleosome core [4]. These tail regions are also rich in lysine and, to a lesser extent, arginine and serine [2], and are targets of numerous post-translational modifications (PTMs) such as acetylation, methylation, phosphorylation, and ubiquitination, among others [4]. An emerging model suggests that the collective combination of these covalent PTMs regulate gene expression and has been termed the “histone code” [1, 4–6].
Modulators of the “histone code” can be separated into three groups which include the “writers”, “erasers”, and “readers” of histone PTMs [7]. The “writers” and “erasers’ are enzymes such as histone acetyltransferases (HATs), and histone deacetylases (HDACs), respectively, that can modify chromatin by adding or removing different PTMs. The epigenetic “readers” are proteins that do not alter the histones, but rather discriminatively detect PTMs located at specific positions on each histone. The regulation of chromatin dynamics by these modulators dictates the outcome of numerous nuclear processes including transcription, DNA repair, recombination, and replication [8–11]. These processes are central to cell homeostasis, as alterations in chromatin structure contribute to the development of cancer and other human diseases [8, 9, 12].
In humans, the t(8;16)(p11;p13) translocation of the MOZ (monocytic leukemia zinc-finger protein, also known as KAT6A or MYST3) HAT is associated with a subtype of acute myeloid leukemia (AML) with a particularly poor outcome [13, 14]. In this abnormality, the N-terminal region of MOZ is fused to the C-terminal part of the transcription co-activator CREB binding protein (CBP) [14]. MOZ has also been found translocated to the CBP homolog p300 [15], to the transcriptional intermediary binding factor 2 (TIF2) [16], and to the nuclear receptor co-activator 3 (NcoA3) transcription factor [17]. MOZ functions as a multi-subunit HAT complex and acetylates free histones H3, H4, H2A and H2B in vitro [18, 19]. Acetylation of histones located near gene promoters is associated with up-regulation of gene transcription, and the acetylation activity of MOZ has been shown to control expression of homeobox (HOX) genes [20]. The MOZ HAT also plays a direct role in hematopoiesis and is essential for the development and maintenance of hematopoietic stem cells (HSCs) [21].
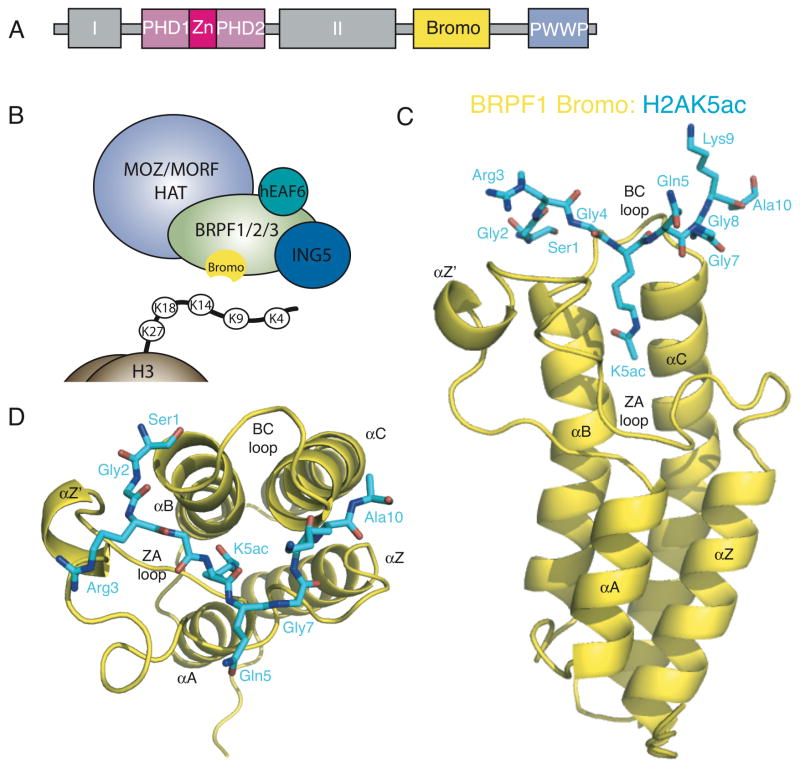
MOZ forms a tetrameric complex with the ING5 (inhibitor of growth 5), hEAF6 (homolog of Esa1-associated factor 6), and the bromodomain-PHD finger proteins (BRPF1, -2, or -3) (Figure 1B) [8, 18]. BRPF proteins have been shown to bridge the association of MOZ with ING5 and hEAF6, thereby promoting its acetyltransferase activity [18]. Deletion mapping studies also revealed that the acetyltransferase domain of MOZ is sufficient for BRPF1 interaction [18]. BRPF proteins therefore play a key role in assembling and activating MOZ HAT complexes. BRPF1 contains a unique combination of domains typically found in chromatin-associated factors, including a double plant homeodomain (PHD) and zinc finger (ZnF) assembly (PZP), a bromodomain, and a chromo/Tudor-related Pro-Trp-Trp-Pro (PWWP) domain (Figure 1A). PHD fingers are a conserved C3HC4 zinc finger motif commonly found in nuclear proteins that regulate transcription and chromatin remodeling [22]. The first PHD finger (PHD1) of BRPF2 has been shown to recognize the unmodified histone H3 tail, while the second PHD finger (PHD2) interacts non-specifically with DNA [23, 24]. The PWWP domain is necessary for the association of BRPF1 with condensed chromatin and is able to recognize H3K36me3 [25, 26]. These domains within BRPF1, in concert with additional chromatin reader domains located in other subunits of the complex, help recruit MOZ to distinct sites of active chromatin [23, 26].
Figure 1.
The BRPF1 bromodomain recognizes acetylated histones. (A) Schematic representation of domain modules within the human BRPF1 protein. (B) The MOZ/MORF HAT complex. (C) A ribbon diagram of the human BRPF1 bromodomain (yellow) in complex with the histone H2AK5ac peptide (blue sticks). (D) Top view of the H2AK5ac peptide in the binding pocket of the BRPF1 bromodomain. The structures were generated with Pymol [65].
Bromodomains are evolutionary conserved structural motifs composed of about 110 amino acids that were first identified in the Drosophila protein brahma (hence the name “bromo”), and are generally known to recognize acetylated histone residues [27]. All bromodomains share a conserved fold that comprises a left-handed bundle of four α-helices (αZ, αA, αB, αC), linked by loop regions of variable length (ZA and BC loops), which line the acetyllysine (Kac) binding site and determine ligand-binding specificity [28, 29]. The BRPF1 bromodomain is a member of subfamily IV of the human bromodomains (as defined by the phylogenetic tree generated by Filippakopoulos et al., 2012 [29]), which also includes the bromodomains of the BRD1, BRD9, BRPF1/3, ATAD2 and ATAD2b proteins. Although the three-dimensional structure of each of these bromodomains is known [29, 30], the specific histone ligands recognized by these bromodomains, and the molecular basis directing their substrate specificity is largely undiscovered.
Our recent studies revealed the interaction of the BRPF1 bromodomain with multiple acetyllysine residues on the N-terminus of histones, and demonstrated that it preferentially binds to the H2AK5ac, H4K12ac and H3K14ac histone ligands [31]. However, the molecular mechanism driving recognition of the acetylated histone tails by the BRPF1 bromodomain has not been elucidated. In this study, we present the X-ray crystal structures of the BRPF1 bromodomain in complex with two of its histone peptide ligands, H2AK5ac and H4K12ac. Importantly this is the first bromodomain structure solved in complex with an H2AK5ac ligand. These structures were used to evaluate the relative contribution of specific bromodomain binding site residues to ligand recognition via site-directed mutagenesis coupled with isothermal titration calorimetry (ITC). Through these studies we are able to describe the molecular basis of histone acetyllysine recognition by the BRPF1 bromodomain for the first time. A better understanding of this process will ultimately be useful in the rational design of pharmaceutical agents against acute myeloid leukemia and other cancers.
Materials and Methods
Cloning, expression, and purification of the BRPF1 bromodomain
Isolated cDNA from human BRPF1 was kindly provided by Dr. Xiang-Jiao Yang. The BRPF1 bromodomain region (residues 629–742) was amplified and subcloned into the pDEST15 vector (Invitrogen) as described previously [31]. BRPF1 bromodomain mutants E36A, I88A, F89A, N83A, A84S, C79A, and N78A, were generated using the QuikChange® mutagenesis procedure (Stratagene) as described in Champagne et al., 2006 [32]. All mutants were produced by conventional PCR using the pDEST15 plasmid containing the BRPF1 bromodomain as a template. The DNA sequence for each mutant was verified before being expressed in E. coli Rosetta™ 2(DE3)pLysS competent cells (Novagen).
The wild-type or mutant BRPF1 bromodomain proteins were expressed in E. coli Rosetta™ 2 and purified essentially as described in [31]. For ITC or circular dichroism (CD) experiments the 117 residue BRPF1 bromodomain proteins (114 from the bromodomain and -GPL from the N-terminal GST tag) were concentrated and dialyzed into buffers consisting of 20 mM NaNH4 pH 7.0 and 150 mM NaCl, or 50 mM NaNH4 pH 7.0 and 50 mM NaCl, respectively. For X-ray crystallography the protein was further purified using gel filtration chromatography (HiPrep 16/60 Sephacryl S-100 High Resolution, GE Healthcare) and equilibrated with wash buffer (20 mM Tris pH 8.0, 150 mM NaCl and 1 mM DTT). The purified BRPF1 bromodomain was then concentrated to 69.9 mg/mL at 4°C for crystallization experiments.
Crystallization
Prior to crystallization 2.3 mM of the purified BRPF1 bromodomain protein was mixed with 5 mM of either the H2A(res 1–12)K5ac or H4(res 4–17)K12ac (SGRG Kac QGGKARA or GKGGKGLG Kac GGAKR, respectively) modified histone peptide in a 1.5 mL microcentrifuge tube. Crystallization screens using the sitting drop method were performed by mixing 1 μL of the protein-ligand solution with 1 μL of the reservoir solution in 96-well VDX plates with a reservoir volume of 100 μL.
Crystals of the BRPF1 bromodomain bound to H2AK5ac grew at 4°C from Hampton Research Crystal Screen I condition number 26 (0.2 M NH4SO4, 0.1 M MES monohydrate pH 6.5, and 30% w/v polyethylene glycol monomethyl ether (PEG) 5,000). Initial crystals were further optimized using the hanging drop method in 24 well VDX plates (Hampton Research). The pH of all buffers was adjusted with HCl or NaOH. The H2AK5ac crystals were improved through seeding techniques and the addition of 1% propylene glycol to the crystallization condition. The crystals grew to ~ 0.30, 0.10, 0.045 mm3 in a P21 space group with the unit cell parameters a= 60.9 Å, b= 55.6 Å, c= 82.1 Å and β = 93.6°. All crystals were flash cooled in liquid nitrogen at 100 K.
The H4K12ac bromodomain-ligand complex grew at 4°C from Hampton Research Crystal Screen II, condition 30 (0.1 M HEPES pH 7.5, 10% w/v polyethylene glycol (PEG) 6,000, 5% v/v (+/−)-2-methyl-2,4-pentanediol (MPD)). Crystals were reproduced using the hanging drop method in a 24 wells VDX plates (Hampton Research) and grew to ~ 0.35, 0.08, 0.03 mm3 in space group P43212 with the unit cell parameters a=, b = 75.1 Å, c= 86.3 Å. The H4K12ac-bromodomain crystals were cryoprotected by sequentially transferring them into reservoir solution supplemented with 10% and 15% MPD before flash freezing in liquid nitrogen at 100 K. Additional details on the crystallization methods and initial data collection can be found in Lubula et al., 2014 [33].
Data Collection, Structure Determination and Refinement
The crystal diffraction data for both crystal forms was collected on beamline X29 at the Brookhaven National Laboratory (BNL) synchrotron at a wavelength of 1.075 Å. Data for the BRPF1 bromo-H2AK5ac crystal were processed with the HKL-2000 software package [34]. Data collection statistics are reported in Table 1. The structure was solved using molecular replacement, and BRD1 (PDB ID 3RCW) was used as the starting model. Molecular replacement was performed with HKL-3000 [35] integrated with MOLREP [36] and selected programs from the CCP4 package [37]. ARP/wARP [38] and BUCCANEER [39] were used to rebuild the initial model. The model was later manually updated using COOT to add the H2AKac histone ligand [40] and refined with REFMAC [41]. MOLPROBITY [42] and ADIT [43] were used for structure validation. The final R-factor and R-free values of the model are 20.39 % and 25.26 %, respectively, for data from 41.02 Å to 1.80 Å. There was clear electron density for residues 1–117 of the BRPF1 bromodomain in chains A, B, C and D, as well as for histone H2AK5ac residues 1–10 in chains E, F, G and H (peptide residues 11–12 had no density observed).
Table 1.
Crystallographic data collection and refinement statistics of the BRPF1 bromodomain in complex with H2AK5ac and H4K12ac.
| Bromodomain + H4K12ac | Bromodomain + H2AK5ac | |
|---|---|---|
| No. of crystals | 1 | 1 |
| Beamline | BNL X29 | BNL X29 |
| Wavelength (Å) | 1.075 | 1.075 |
| Detector | ADSC Quantum-315r CCD | ADSC Quantum-315r CCD |
| Crystal-to-detector distance (mm) | 250 | 225 |
| Rotation range per image (°) | 1 | 0.5 |
| Total rotation range (°) | 360 | 284 |
| Exposure time per image (sec) | 12 | 12 |
| Resolution range (Å) | 34.43–1.94 | 41.02–1.80 |
| Space group | P43212 | P21 |
| Unit cell parameters | ||
| a, b, c (Å) | 75.1, 75.1, 86.3 | 60.9, 55.6, 82.1 |
| α, β, γ (°) | 90.0, 90.0, 90.0 | 90.0, 93.6, 90.0 |
| Unique reflections | 18780 | 48421 |
| Rwork/Rfree (%) | 17.55/21.29 | 20.39/25.26 |
| Redundancy | 27.7 (27.2) | 4.0 (3.9) |
| Mean I/σ(I) | 25.84 (14.66) | 21.04 (3.0) |
| Completeness (%) | 99.6 (96.9) | 99.9 (100) |
| Rmerge (%)* | 13.9 (27.9) | 8.1 (38.6) |
| Wilson B factor (Å2) | 26.5 | 21.0 |
| Number of non-hydrogen atoms | ||
| Protein | 997 | 4267 |
| Water | 143 | 722 |
| RMSD | ||
| Bond lengths (Å) | 0.007 | 0.019 |
| Bond angles (°) | 0.95 | 1.78 |
| Ramachandran plot | ||
| Residues in most favored regions | 93.5% | 94.0% |
| Residues in additional allowed regions | 5.6% | 6.0% |
| Residues in generously allowed regions | 0.9% | 0.0% |
| Residues in disallowed regions | 0.0% | 0.0% |
Rmerge = SUM (ABS(I − <I>)) / SUM (I)
Data for the BRPF1 Bromo-H4K12ac crystal were processed with the HKL2000 software package, and the statistics are shown in Table 1. The structure was solved by molecular replacement using the ligand-free BRPF1 bromodomain structure (PDB code: 2D9E) as the starting model. Molecular replacement was performed with PHASER [44] from the CCP4 package [37]. The PHENIX [45] program was used for automatic model building and density modification. The H4K12ac peptide bound to the BRPF1 bromodomain molecule was built into the density map using the COOT program [40] and the structure of the complex was refined against 1.94 Å diffraction data with simulated annealing and restraint minimization methods using the PHENIX [45] program. PROCHECK [46], MOLPROBITY [42] and ADIT [43] were used for structure validation. The final R-factor and R-free values of the model are 17.55 % and 21.29 %, respectively, for data from 34.43 Å to 1.94 Å. There was clear electron density for residues 4–117 of the BRPF1 bromodomain (chain A), and for residues 6–13 (chain B) in the histone H4K12ac ligand. H4K12ac histone peptide residues 11, 12 and 14–17 had no observable density.
Isothermal Titration Calorimetry
ITC experiments were carried out at 5°C with a MicroCal ITC200 instrument (GE Healthcare). Each of the BRPF1 bromodomain mutant proteins and histone peptide samples were dialyzed into a 20 mM NaH2PO4-Na2HPO4 (pH 7.0) and 150 mM NaCl buffer. Calorimetric titration was performed by titrating each histone tail ligand (5 mM) into 0.2 mM of the mutant BRPF1 bromodomain protein in the sample cell in a series of 19 individual injections of 2 μL, at time intervals of 150 seconds. These were preceded by a preliminary injection of 0.5 μL of the 5 mM peptide sample. To determine the heat of dilution of the titrant peptides in the experimental buffer, control experiments were conducted under identical conditions. As part of data analysis, this was deducted from the experimental data. The obtained change-in-heat peaks were then analyzed by the Origin 7.0 program (OriginLab Corporation) and used to calculate binding affinities. Experiments in which binding occurred were performed in triplicate, while non-binding experiments were performed in duplicate.
Circular Dichroism Spectroscopy
Circular Dichroism (CD) spectra were recorded on a JASCO J-815 CD Spectrometer (JASCO, Japan) at 25°C in a 1.6 cm cell. BRPF1 bromodomain wild type (WT) or mutant proteins were dialyzed in 50 mM NaH2PO4-Na2HPO4 pH 7.0, 50 mM NaCl and diluted to between 0.1 μM and 0.5 μM in concentration. CD spectra were measured from 199–260 nm. Two spectra were measured and averaged for each mutant bromodomain protein sample and the wild-type protein. Spectra were analyzed using the K2D3 [47, 48] structure prediction software to determine the percent α-helical and β-sheet content.
Results and Discussion
Overall structure of the BRPF1 bromodomain
As discussed above, the BRPF1 subunit is part of the MOZ HAT complex, and it contains multiple chromatin reader domains known to direct the MOZ acetyltransferase activity to chromatin substrates [18, 49]. To further understand the role of the bromodomain in recruiting BRPF1 to histones it is imperative to characterize the molecular mechanism of BRPF1 bromodomain-mediated acetyllysine recognition. To this end we determined the crystal structures of the BRPF1 bromodomain in complex with its H2A(1-12)K5ac and H4(4-17)K12ac histone peptide ligands (PDB IDs 4QYL and 4QYD, respectively). The crystallographic data collection and refinement statistics are listed in Table 1. The bromodomain of BRPF1 in complex with the H2AK5ac peptide crystallized in a monoclinic space group P21, with four protein-ligand complexes in the crystallographic asymmetric unit, while the BRPF1 bromo-H4K12ac structure was in a tetragonal space group P43212 with only one protein-ligand complex in the asymmetric unit. Both of the crystal structures were solved by molecular replacement using the unpublished bromodomains of Peregrin for the H4K12ac structure (PDB ID 2D9E) and BRD1 for the H2AK5ac structure (PDB ID 3RCW) as the initial models, respectively. In the crystal containing the H2AK5ac ligand, the four protein monomers are in identical conformations and are superimposable on each other with RMSD values of 0.15–0.18 Å over 117 pairs of identical Cα atoms [50]. The BRPF1 bromodomain bound to H4K12ac (Chain A) is also in the same conformation as the H2AK5ac-bound structure (Chain D) with an RMSD of 0.94 Å over 113 identical Cα atoms [50]. The overall architecture of the BRPF1 bromodomains exhibit structural features characteristic of other bromodomain modules and contain the typical left-handed four-helix bundle topology (Figure 1C). These four helices, sequentially named αZ, αA, αB, αC, respectively (from the N-terminus), are connected by two loops: a long loop connecting helices αZ and αA (the ZA loop) and a short loop connecting helices αB and αC (the BC loop). The ZA loop is packed against the BC loop to form a hydrophobic pouch that constitutes the binding pocket. Figure 1C shows the side view of the 1.8 Å BRPF1 bromo-H2AK5ac complex and depicts the overall bromodomain fold. Figure 1D shows the top view where the H2AK5ac ligand is oriented between the ZA and BC loops with the acetylated lysine protruding down into the deep binding pocket formed between the four alpha-helical bundle. Structural alignment of the H2AK5ac- and H4K12ac-bound BRPF1 bromodomain structures to the apo BPRF1 bromodomain structure (PDB ID 2D9E) produces an RMSD of 0.652, which indicates that no major conformational changes in the overall fold of the bromodomain occurs upon ligand binding.
Acetyllysine coordination of H2AK5ac and H4K12ac
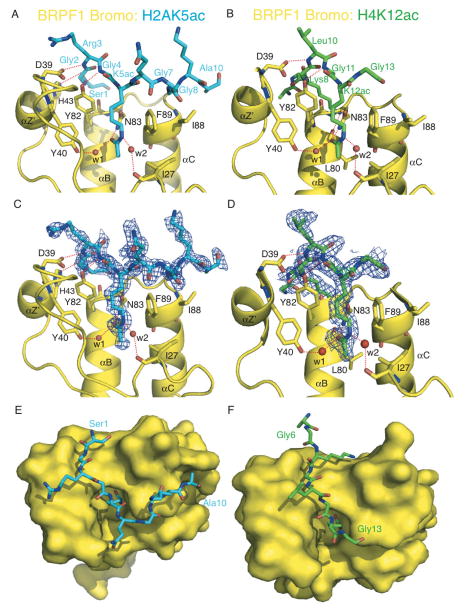
The crystal structures of the BRPF1 bromodomain in complex with the H2AK5ac and H4K12ac histone peptide ligands reveal that these two protein-peptide complexes are maintained by a network of H-bond interactions (Figure 2A and 2B). These As observed in other bromodomains, and our previous molecular dynamic models of the BRPF1 bromo-ligand interaction, the acetyllysine residue of the histone peptides is deeply inserted into the hydrophobic binding pocket of the BRPF1 bromodomain (Figure 2E and 2F) [27, 31, 51]. In this pocket, the carbonyl oxygen of the N-acetyl group on both the H2AK5ac and H4K12ac histone peptide ligands forms a H-bond interaction with the amide nitrogen of the N83 residue in the BRPF1 bromodomain (Figure 2A and 2B). This particular interaction is highly conserved among bromodomains and is the major mode of recognition of the acetylated histone ligand [28]. The importance of the direct hydrogen bond between bromodomain residue N83 and the acetylated lysine moiety is highlighted by the complete loss of ligand binding observed when N83 is mutated to alanine (Figure 3A and 3B, red trace, Supplemental Figs 1–3). In both structures, acetyllysine coordination is also stabilized by H-bond contacts through ordered water molecules in the binding pocket. The carbonyl oxygen of the acetyllysine moiety is connected to residue Y40 through water (w1), while the ε-amino group forms a hydrogen bond to the backbone carbonyl of I27 mediated by another water molecule (w2).
Figure 2.
Coordination of the H2AK5ac and H4K12ac histone ligands by the BRPF1 bromodomain. (A) The BRPF1 bromodomain (yellow) in complex with H2AK5ac (blue) ligand. Hydrogen bonds are indicated by a red dotted line. (B) The BRPF1 bromodomain (yellow) in complex with the H4K12ac (green) ligand. (C) The Fo-Fc omit map representing the bound histone H2AK5ac peptide (blue) in complex with the BRPF1 bromodomain (yellow) contoured at 1σ. (D) The Fo-Fc omit map representing the bound histone H4K12ac peptide (green) in complex with the BRPF1 bromodomain (yellow) contoured at 1σ. (E) Surface representation of the H2AK5ac binding groove. (F) Surface representation of the H4K12ac ligand binding pocket.
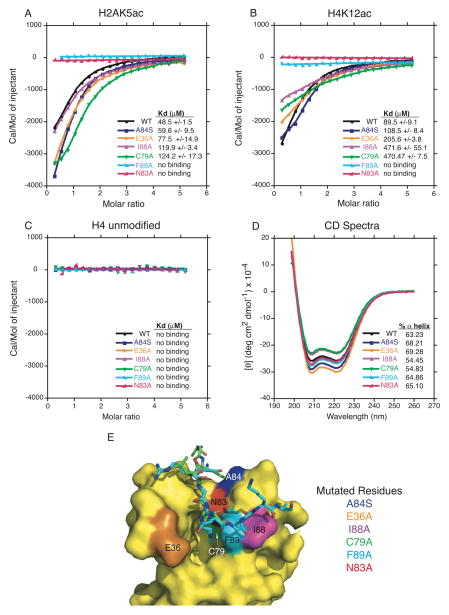
Figure 3.
ITC measurements of interactions between mutant BRPF1 bromodomain proteins and histone tail ligands. (A) Superposed exothermic ITC enthalpy plots for the binding of the BPRF1 bromodomain wild type and mutant proteins to the H2A(1-12)K5ac peptide. The insert lists the measured binding constants. (B) Superposed exothermic ITC enthalpy plots for binding of the BRPF1 bromodomain wild type and mutant proteins to the H4(4-17)K12ac peptide. The insert lists the measured binding constants. (C) Superposed exothermic ITC enthalpy plots for the binding of the BRPF1 bromodomain wild type and mutant proteins to the H4(4-17) unmodified peptide. The insert lists the measured binding constants. (D) Circular dichroism spectra in the far-UV region of the BRPF1 bromodomain wild type and mutant proteins. The percent alpha-helical content of each protein is listed in the insert. (E) Surface representation of the BRPF1 bromodomain showing specific point mutations introduced into the binding pocket by site-directed mutagenesis and the superimposed structures of the histone ligands in the binding pocket. The two BRPF1 bromodomain structures were superimposed using Pymol [65].
Molecular mechanism of histone ligand recognition
In the H2AK5ac-bound structure, in addition to the acetyllysine contacts described above, there was clear electron density for the first ten residues in the histone peptide (Figure 2C), with only Arg11 and Ala12 unordered. The median B factor for the H2AK5ac ligand (10 residues) is 25 Å2 for chains E, F, G and H, respectively, and all modeled residues have an average occupancy more than 90%. Other bromodomain-ligand contacts observed in this complex include hydrogen bonds between D39, H43 and Y82 of the BRPF1 bromodomain to the backbone amino group of Arg3, the backbone carbonyl of Gly2, and the backbone amino of Gly4 in the histone peptide, respectively (Figure 2A). There are also two important hydrophobic interactions between F89 and I88 of the bromodomain and Gly7 and Gly8 in the histone ligand.
In the H4K12ac-bound structure there was clear electron density for eight of the fourteen histone peptide residues (Figure 2D). In addition to the acetyllysine contacts, hydrogen bonds are observed between D39 and Y82 in the BRPF1 bromodomain to the backbone amino groups of Leu10 and Gly11, respectively. There are also two hydrogen bonds between the ε-amino group of Lys8 in the H4K12ac peptide and the backbone carbonyls of L80 and N83 of the bromodomain. The hydrophobic residues F89 and I88 are positioned in the bromodomain binding pocket to contact peptide residues Gly14 and Ala15, but these two residues in the H4K12ac ligand were unordered, with no observed density (also missing from the density map were residues Gly4, Lys5, Lys16 and Arg17). The median B factor for the H4K12ac ligand (8 residues) is 31 Å2, and all modeled residues have an average occupancy more than 90%.
The extensive network of intermolecular contacts observed between the BRPF1 bromodomain and histone peptide residues adjacent to the acetyllysine in both the H2AK5ac and H4K12ac ligand-bound structures indicates that these interactions play a role in the binding specificity. The hydrophobic contact between F89 and the Kac +2 position in the histone peptide (Gly7 in H2AK5ac, and Gly14 in H4K12ac) also appears to be a critical component of histone recognition as demonstrated by the complete loss of ligand binding in the F89A mutant (Figure 3A and 3B, cyan trace, Supplemental Figs 1–3). The F89 residue is located at the entrance of the acetyllysine binding pocket and is positioned to make additional hydrophobic contacts with the methylene groups in the acetyllysine side chain (Figure 2A and 2B). To further elucidate the molecular mechanism driving recognition of the H2AK5ac and H4K12ac acetylated histone peptides by the BRPF1 bromodomain, we designed four additional mutant proteins with point mutations at A84S, E36A, I88A and C79A (Figure 3E). Each of these residues are located proximal to the acetyllysine binding pocket, and were selected based on an observed direct contact with the histone ligand in the crystal structure or on the presences of significant chemical shift changes displayed in our previously published NMR titration studies [31]. Once expressed, these mutant BRPF1 bromodomain proteins were analyzed using ITC to measure their effect on ligand binding affinities as compared to the wild type (WT) protein. Of note, the mutations introduced into the BRPF1 bromodomain did not cause any significant change in the overall structure, or unfolding of the mutant proteins. This is demonstrated by the circular dichroism (CD) spectroscopy studies we performed on each of the wild-type and mutant proteins. As depicted in Figure 3D and Table 2, there is a less than 6% change in α-helical content observed between the WT and mutant BRPF1 bromodomain proteins.
Table 2.
Percentages of α-helix and β-strand composition in wild type (WT) and mutant BRPF1 proteins calculated from circular dichroism experiments.
| Protein | % α-helix* | % β-strand* |
|---|---|---|
| BRPF1 WT | 63.23 | 2.54 |
| BRPF1 A84S | 68.21 | 1.49 |
| BRPF1 E36A | 69.28 | 1.27 |
| BRPF1 I88A | 54.45 | 6.99 |
| BRPF1 C79A | 54.83 | 7.36 |
| BRPF1 F89A | 64.86 | 1.48 |
| BRPF1 N83A | 65.10 | 1.61 |
The % α-helix and β-strand were calculated using the program K2D3 [47].
Residue C79 is located at the bottom of the hydrophobic binding pocket of the BRPF1 bromodomain, and although it does not make a direct contact to the acetylated lysine, the C79A mutation results in a 2.5–5 fold loss of binding affinity with the H2AK5ac and H4K12ac ligands, likely due to changes in the binding pocket composition or electrostatics. The I88A mutation has a similar effect on the ligand binding affinity as the C79A mutation, but this residue plays an important role in making hydrophobic contacts to the backbone residues of the histone ligands as observed in the crystal structures. As might be expected based on the structures, the E36A and A84A mutations located more distal to the acetyllysine binding pocket had relatively moderate affects on histone binding affinity. Interestingly, E36A showed a strong chemical shift change upon binding of the histone binding in our previous NMR experiments suggesting this residue might be critical for ligand coordination [31]. However, the examination of the crystal structures reveals that E36 does not directly contact the histone peptides. Instead, the E36 side chain changes conformation in the ZA loop to accommodate histone ligand binding (Figure 4A). These observations concur with previous studies that have shown the ligand binding selectivity of bromodomains is driven by a network of interactions that coordinate both the acetyllysine and surrounding histone peptide residues [30, 51–53].
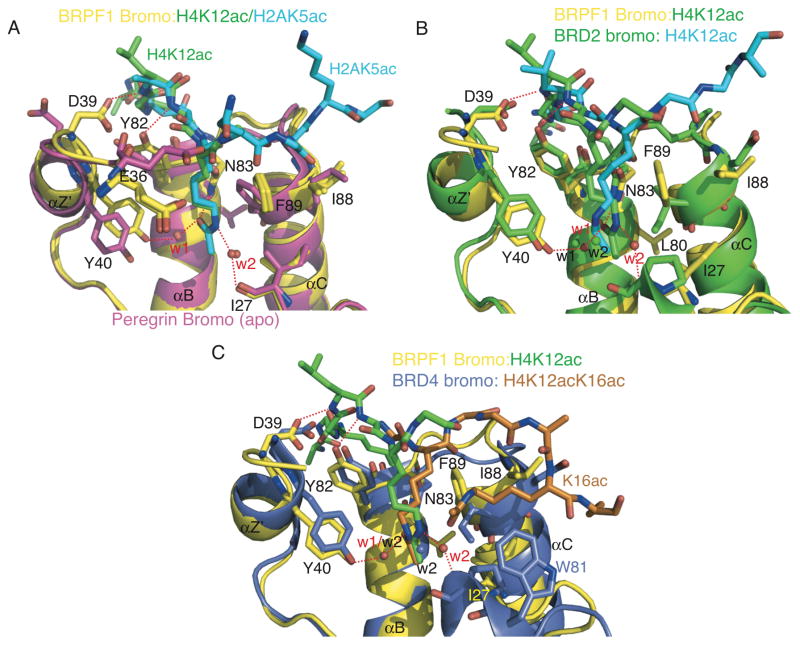
Figure 4.
Structural comparison of bromodomain ligand coordination. (A) The BRPF1 bromodomain (yellow) in complex with H2AK5ac (blue) ligand or in complex with the H4K12ac (green) ligand. The BRPF1 bromodomain structure is superimposed onto the ‘apo’ BRPF1 bromodomain structure (magenta) using Pymol [65]. Hydrogen bonds are indicated by a red dotted line, and water molecules in the ligand-bound structures are indicated by red spheres and labels. (B) The BRPF1 bromodomain (yellow) in complex with the H4K12ac (green) ligand superimposed onto the BRD2 bromodomain crystal structure (green, PDB ID 2DVQ) in complex with the H4K12ac ligand (blue). Water molecules from the BRPF1 structure are indicated by red spheres and labels, and green spheres with black labels indicate water molecules from the BRD2 structure. (C) The BRPF1 bromodomain (yellow) in complex with the H4K12ac (green) ligand superimposed onto the BRD4 bromodomain crystal structure (blue, PDB ID 3UVX) in complex with the diacetylated histone H4K12acK16ac ligand (orange). Water molecules from the BRPF1 structure are indicated by red spheres and labels, and water molecules from the BRD4 structure are indicated by blue spheres with black labels. Residue numbering is for the BRPF1 bromodomain, except where indicated by corresponding colors.
As seen with other chromatin reader domains, histone ligand binding does not induce major conformational changes in the BRPF1 bromodomain. Structural alignment of the apo BRPF1 bromodomain structure solved by NMR (PDB ID 2D9E), with the H2AK5ac- and H4K12ac-bound structures, reveals only minor adjustments in the side chains of amino acid residues in the binding pocket (Figure 4A). Residues I88 and F89 are re-oriented to make hydrophobic contacts to the peptide backbone and the methylene groups of K5ac and K12ac, respectively. The side chain of D39 flips into the binding pocket to form a hydrogen bond to the histone peptide. Residues Y40 and Y82 also make slight conformational changes to accommodate hydrogen bond formation to the ligands (Figure 4A).
Comparison with other BRPF1 bromodomain-ligand models
An unpublished NMR structure of the BRPF1 bromodomain in complex with the H4K5ac ligand is deposited in the PDB (PDB ID 2RS9). In this structure the acetyllysine is coordinated by an H-bond contact between the carbonyl oxygen of the N-acetyl group of the histone H4K5ac ligands and the amide nitrogen of the N83 residue in the BRPF1 bromodomain. However, when compared to our BRPF1 bromo-H4K12ac structure, the H4K5ac ligand is oriented in the opposite direction in the binding pocket, with F89 contacting the peptide at the Kac −2 position (Gly4) rather than at the Kac +2 position (Gly14). There are also no additional hydrogen bonds observed between adjacent residues in the H4K5ac histone peptide and the BRPF1 bromodomain structure, which could be attributed to the relatively weak 1.2 mM binding affinity the bromodomain has for this ligand [31].
Previously, we published the results of molecular dynamic (MD) simulation experiments to predict the ligand binding contacts between the BRPF1 bromodomain and its histone ligands [31]. The hydrogen bond contacts between the BRPF1 bromodomain N83 residue and acetyllysine in the H2AK5ac and H4K12ac histone ligands are conserved between the crystal structures and the MD models of ligand coordination. However, the prediction of hydrogen bond contacts between the histone peptides and the BRPF1 bromodomain were limited because ordered waters in the binding pocket were not depicted in the MD simulations. We were unable to model these interactions since the unpublished NMR structures available for the BRPF1 bromodomain (PDB ID 2RS9 and 2D9E) did not contain information about water molecules.
Recently, Vidler et al., 2012 discussed the importance of five ordered water molecules, which appear to be a conserved feature of the bromodomain binding pocket [54]. They observed that these ordered waters are found in deposited bromodomain crystal structures regardless of the presence or absence of a bound ligand, suggesting that water plays a significant role in the structural composition of the binding pocket, and affects the selectivity and modality of ligand binding [54]. We carried out structural alignments of the ligand-free (“apo”) structures of the human PCAF bromodomain (PDB ID 3GG3) and the BRD1 bromodomain (PDB ID 3RCW), to compare them with the ligand-bound (“holo”) structure of the first bromodomain of BRD4 (PDB ID 3UVX) and our crystal structures the BRPF1 bromodomain (in complex with the H2AK5ac and H4K12ac ligands). Analysis of these structures reveals that the water mediating hydrogen-bonding between the hydroxyl group of Y40 and the carbonyl oxygen of the acetyllysine moiety (w1 in Figure 2A and 2B) is present in the binding pocket regardless of the absence or presence of the ligand. Conversely, the second water in our crystal structures (w2 in Figure 2A and 2B) is absent in the apo structures, and this water is only seen in the holo structures where it bridges between a backbone carbonyl of the bromodomains to the epsilon nitrogen of the acetylated lysine in the histone ligands. This suggests that the ordered waters in the bromodomain binding pocket contribute to the molecular mechanism driving binding of the histone H2AK5ac and H4K12ac ligands in the BRPF1 bromodomain, and may be a conserved mode of acetyllysine recognition for other bromodomains as well.
Comparison with other bromodomain subfamilies
Despite the low degree of overall sequence homology, bromodomains have a highly conserved overall structural fold comprising four α-helices (αZ, αA, αB, and αC) linked by highly variable loop regions (ZA and BC loops) that form a hydrophobic binding pocket [27]. However, the surface properties of bromodomains have been shown to be highly diverse [29]. The electrostatic potential of the surface area around the acetylated lysine binding pocket ranges from highly positively to strongly negatively charged, suggesting that individual bromodomains recognize largely different ligands [29]. Furthermore, it has been suggested that bromodomain peptide recognition is dependent on the pattern of multiple PTMs rather than on a single acetylation site [29]. Bromodomains in different subfamilies are therefore expected to use a variety of recognition modes to select for their particular acetylated histone ligand(s).
Currently there are no other bromodomain structures in complex with a histone H2AK5ac ligand deposited into the PDB, but the structure of the second bromodomain of the BRD2 in complex with H4K12ac has been solved. BRD2 belongs to the human BET family of bromodomains in subfamily II, which also includes BRD3, BRD4 and BRDT, and these proteins have a unique architecture with two tandem bromodomains and a conserved extra-terminal domain [55]. In the intact nuclei, the BET protein BRD2 activates transcription by targeting histone H4K12ac, one of the active marks of chromatin [55]. BRD2 has also been shown to bind to the histone H4K5ac peptide, and more weakly recognizes the histone H4K16ac and H3K9ac modifications [55]. Although both BRD2 and the BRPF1 bromodomains belong to two separate sub-families, the modality of histone H4K12ac recognition is similar. Figure 4B shows the structural alignment of the BRPF1 and BRD2 (PDB ID 2DVQ) bromodomains bound to the H4K12ac peptide. The RMSD of these two structures is 1.4, mainly due to slight conformational changes in the alpha-helical regions. In the bromodomain binding pocket, the histone H4K12ac ligand backbone is oriented in the same direction in both structures, and the amino acid side chains coordinating the histone peptide are in nearly identical conformations. In both structures, the acetyllysine side chain of H4K12ac sits in the deep hydrophobic binding pocket. In BRD2, the N-acetyl group of K12ac is positioned so the carbonyl forms a hydrogen bond with the amide group of N156. The hydrogen bond network coordinating the acetyllysine through two ordered waters (w1 and w2) is also conserved between the BRPF1 and BRD2 bromodomain structures, with only slight shifts in location of the waters to accommodate the orientation of the acetyllysine side chain in the binding pocket. The acetylated lysine of H4K12 as is also surrounded by the hydrophobic residues P98 (I27), I162 (F89) in BRD2 [55], which contribute to the binding pocket formation and modality of ligand selection in the same way we observe for the BRPF1 bromodomain.
Interestingly, BRD4 bromodomain has been shown to coordinate the H4K12acK16ac double modification (PDB ID 3UVX, Figure 4C) [29]. Coordination of the K12ac is consistent with what is observed in the BRPF1 and BRD2 bromodomain structures (Figure 4A and 4B). However, the H4K16ac is bound in an adjacent hydrophobic pocket formed by P82, W81 and I146 in the BRD4 structure. In the BRPF1 structure W81 is a charged asparagine (N26), while a more flexible isoleucine side chain in BRD4 (I146) replaces F89 in the BRPF1 bromo structure. These two amino acid substitutions create critical hydrophobic contacts necessary for coordination of the second H4K16ac group, which would likely not be possible in the BRPF1 bromodomain. Importantly, the two ordered waters are also observed in the H4K12ac binding pocket of the BRD4 bromodomain structure, and coordinate the carbonyl oxygen and epsilon nitrogen of the K12ac moiety as described above. Thus, these two ordered waters appear to be a general feature of the H4K12ac ligand binding mechanism.
The PCAF and CBP/p300 bromodomains, which belong to the first and third bromodomain subfamilies, are able to recognize histone H3K36ac and H4K20ac peptide ligands, respectively. The NMR solution structures of these bromodomains in complex with their ligands indicate that they have unique modes of recognition for their acetylated histone ligands [53]. PCAF has been shown to preferably interact with acetylation sites with a hydrophobic residue at (Kac +1) position and a positively charged or aromatic residue at (Kac +3) [53]. On the other hand the CBP bromodomain has been shown to favor bulky hydrophobic residues at (Kac +1) and (Kac +2), a positively charged residue at (Kac −1), and an aromatic residue at (Kac −2) [53]. Despite the variability in the mode of acetylated histone recognition, all bromodomains have been shown to coordinate the acetylated lysine moiety via H-bond interactions to the highly conserved asparagine in the binding pocket. However, the ordered water molecules are not observed in these structures since they were solved using NMR techniques.
BRPF1 bromodomain as a drug target
As mentioned previously, the t(8;16)(p11;p13) translocation of the MOZ HAT complex contributes to the progression of AML [14, 56, 57]. In this abnormality, the MOZ N-terminal domain fuses to the C-terminal part of the CBP transcriptional co-activator resulting in highly active complex [18]. The BRPF proteins have been shown to bridge the association of MOZ with ING5 and hEAF6, thereby promoting MOZ acetyltransferase activity [18]. The BRPF1 subunit continues to associate with the MOZ HAT complex even in the translocated proteins and thereby plays an important role in MOZ-related regulation of transcription, hematopoiesis, leukemogenesis, and other developmental processes [18, 58]. Our results indicate that the BRPF1 bromodomain contributes to recruiting the MOZ complex to distinct sites of active chromatin. Thus, preventing this process may be a useful tool in helping patients suffering from MOZ-associated AML. Inhibitors of human bromodomains are currently under intense investigation as a potential therapeutic strategy in multiple disease types [59, 60]. In one study, a short hairpin RNA screen suggested that inhibition of the BRD4 bromodomain might be an effective treatment for AML [61]. Furthermore, an inhibitor of the BET family of bromodomains, JQ1, is currently under study as a potential drug for HIV reactivation, heart disease, and as a non-hormonal male contraceptive [62–64]. In a computational analysis of bromodomains by Vidler et al., 2012, the BRPF1 bromodomain was classified as being “druggable” based on signature residues in its binding pocket [54]. The structural data and mutational analysis presented here are an important step in outlining the molecular basis of histone acetyllysine recognition by the BRPF1 bromodomain. A better understanding of this process will ultimately be useful in the rational design of pharmaceutical agents to treat AML and other cancers.
In conclusion, the results presented offer important insights into the role of the bromodomain in recruiting BRPF1, and subsequently the MOZ HAT complex, to histones. Specifically, to better understand the modality by which the BRPF1 bromodomain interacts with its ligands, we solved the crystal structures of the BRPF1 bromodomain in complex with its H2AK5ac and H4K12ac histone ligands. These structures reveal that a network of H-bond and hydrophobic contacts coordinate the acetyllysine moiety and make specific contacts to flanking residues in the histone tail. We also identified two ordered water molecules in the binding pocket, which mediate coordination of the acetyllysine moiety. To gain a better understanding of the molecular mechanism driving specificity, we also carried out site-directed mutagenesis in combination with ITC studies. Our results indicate that the two most important contacts for ligand coordination include the highly conserved N83 residue, which coordinates the acetyllysine directly, as well as F89, which creates a critical hydrophobic contact with the histone tail and shapes the bromodomain binding pocket wall. Our results reinforce previous studies showing that ligand selectivity of bromodomains is a composite of both the acetylated lysine residue coordination, and contacts to peptide residues flanking the acetyllysine in the histone tail. The BRPF1 bromodomain interacts with a region of MOZ that remains intact after leukemic translocations. Thus, the structural and functional characterization of the BRPF1 bromodomain-histone interaction is important for unraveling its role in modulating the genomic binding targets of the MOZ HAT, and for the development of future therapeutics for MOZ-related leukemias.
Supplementary Material
Highlights.
Solved the crystal structure of the BRPF1 bromodomain bound to the H2AK5ac ligand.
Solved the crystal structure of the BRPF1 bromodomain bound to the H4K12ac ligand.
Site-directed mutagenesis identifies critical binding pocket residues.
ITC experiments quantified the effect mutations have on ligand binding affinity.
We outline the molecular mechanism of acetyllysine binding by the BRPF1 bromodomain.
Ordered water molecules are an essential component driving ligand recognition.
Acknowledgments
We are grateful to Dr. Robert Sweet, Dr. Rebecca Page, Dr. William Grath, and all the staff members providing support during the 2013 RapiData course at the Brookhaven National Laboratory for their help with crystal data collection and processing. We are equally thankful to Dr. XJ Yang for providing us with the BRPF1 cDNA. We also thank Dr. Matthew Liptak and Amanda Roffman in the Department of Chemistry at the University of Vermont for their advice and assistance on setting up circular dichroism spectroscopy experiments. This study was supported by a National Institutes of Health grant, NIGMS 1R15GM104865, and an ACPHS internal research award to KCG. DNA sequencing was performed in the Vermont Cancer Center DNA Analysis Facility. Crystal growth, screening and initial data collection was carried out at the Center for X-ray Crystallography at the University of Vermont, which is supported by the National Institutes of Health grants P01CA098993 and R01CA52040, awarded by the National Cancer Center.
Abbreviations
- AML
acute myeloid leukemia
- BRPF1
bromodomain-PHD finger protein 1
- CBP
CREB binding protein
- CD
circular dichroism
- HAT
histone acetyltransferase
- HDAC
histone deacetylase
- hEAF6
homolog of Esa1-associated factor 6
- HOX
homeobox
- HSCs
hematopoietic stem cells
- ING5
inhibitor of growth 5
- ITC
isothermal titration calorimetry
- MD
molecular dynamic
- MOZ
monocytic leukemic zinc-finger
- NcoA3
nuclear receptor co-activator 3
- NMR
nuclear magnetic resonance
- PHD
plant homeodomain
- PTM
post-translational modification
- PWWP
proline-tryptophan-tryptophan-proline
- TIF2
transcriptional intermediary binding factor 2
- ZnF
zinc finger
Footnotes
Structured summary of protein interactions:
BRPF1 and H4K12ac bind by x-ray crystallography (View interaction)
BRPF1 and H2AK5ac bind by x-ray crystallography (View interaction)
H4K12ac and BRPF1 bind by isothermal titration
calorimetry (1, 2, 3, 4, 5, 6, 7)
H2AK5ac and BRPF1 bind by isothermal titration
calorimetry (1, 2, 3, 4, 5, 6, 7)
Database: structural data are available in RCSB Protein Data Bank under the accession number(s) 4QYD and 4QYL.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–5. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 2.Sterner DE, Berger SL. Acetylation of histones and transcription-related factors. Microbiol Mol Biol Rev. 2000;64:435–59. doi: 10.1128/mmbr.64.2.435-459.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–60. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 4.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 5.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–80. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 6.Zaidi SK, Young DW, Montecino M, Lian JB, Stein JL, van Wijnen AJ, Stein GS. Architectural epigenetics: mitotic retention of mammalian transcriptional regulatory information. Mol Cell Biol. 2010;30:4758–66. doi: 10.1128/MCB.00646-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baker LA, Allis CD, Wang GG. PHD fingers in human diseases: disorders arising from misinterpreting epigenetic marks. Mutation research. 2008;647:3–12. doi: 10.1016/j.mrfmmm.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doyon Y, Cayrou C, Ullah M, Landry AJ, Cote V, Selleck W, Lane WS, Tan S, Yang XJ, Cote J. ING tumor suppressor proteins are critical regulators of chromatin acetylation required for genome expression and perpetuation. Moll Cell. 2006;21:51–64. doi: 10.1016/j.molcel.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 9.Gellert M. V(D)J recombination: RAG proteins, repair factors, and regulation. Annu Rev Biochem. 2002;71:101–32. doi: 10.1146/annurev.biochem.71.090501.150203. [DOI] [PubMed] [Google Scholar]
- 10.Santoro R, Li J, Grummt I. The nucleolar remodeling complex NoRC mediates heterochromatin formation and silencing of ribosomal gene transcription. Nat Genet. 2002;32:393–6. doi: 10.1038/ng1010. [DOI] [PubMed] [Google Scholar]
- 11.Iizuka M, Matsui T, Takisawa H, Smith MM. Regulation of replication licensing by acetyltransferase Hbo1. Mol Cell Biol. 2006;26:1098–108. doi: 10.1128/MCB.26.3.1098-1108.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boussouar F, Jamshidikia M, Morozumi Y, Rousseaux S, Khochbin S. Malignant genome reprogramming by ATAD2. Biochimica et biophysica acta. 2013;1829:1010–4. doi: 10.1016/j.bbagrm.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 13.Borrow J, Stanton VP, Jr, Andresen JM, Becher R, Behm FG, Chaganti RS, Civin CI, Disteche C, Dube I, Frischauf AM, Horsman D, Mitelman F, Volinia S, Watmore AE, Housman DE. The translocation t(8;16)(p11;p13) of acute myeloid leukaemia fuses a putative acetyltransferase to the CREB-binding protein. Nat Genet. 1996;14:33–41. doi: 10.1038/ng0996-33. [DOI] [PubMed] [Google Scholar]
- 14.Borrow J, Shearman AM, Stanton VP, Jr, Becher R, Collins T, Williams AJ, Dube I, Katz F, Kwong YL, Morris C, Ohyashiki K, Toyama K, Rowley J, Housman DE. The t(7;11)(p15;p15) translocation in acute myeloid leukaemia fuses the genes for nucleoporin NUP98 and class I homeoprotein HOXA9. Nat Genet. 1996;12:159–67. doi: 10.1038/ng0296-159. [DOI] [PubMed] [Google Scholar]
- 15.Kitabayashi I, Aikawa Y, Yokoyama A, Hosoda F, Nagai M, Kakazu N, Abe T, Ohki M. Fusion of MOZ and p300 histone acetyltransferases in acute monocytic leukemia with a t(8;22)(p11;q13) chromosome translocation. Leukemia. 2001;15:89–94. doi: 10.1038/sj.leu.2401983. [DOI] [PubMed] [Google Scholar]
- 16.Carapeti M, Aguiar RC, Goldman JM, Cross NC. A novel fusion between MOZ and the nuclear receptor coactivator TIF2 in acute myeloid leukemia. Blood. 1998;91:3127–33. [PubMed] [Google Scholar]
- 17.Esteyries S, Perot C, Adelaide J, Imbert M, Lagarde A, Pautas C, Olschwang S, Birnbaum D, Chaffanet M, Mozziconacci MJ. NCOA3, a new fusion partner for MOZ/MYST3 in M5 acute myeloid leukemia. Leukemia. 2008;22:663–5. doi: 10.1038/sj.leu.2404930. [DOI] [PubMed] [Google Scholar]
- 18.Ullah M, Pelletier N, Xiao L, Zhao SP, Wang K, Degerny C, Tahmasebi S, Cayrou C, Doyon Y, Goh SL, Champagne N, Cote J, Yang XJ. Molecular architecture of quartet MOZ/MORF histone acetyltransferase complexes. Mol Cell Biol. 2008;28:6828–43. doi: 10.1128/MCB.01297-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Champagne N, Pelletier N, Yang XJ. The monocytic leukemia zinc finger protein MOZ is a histone acetyltransferase. Oncogene. 2001;20:404–9. doi: 10.1038/sj.onc.1204114. [DOI] [PubMed] [Google Scholar]
- 20.Camos M, Esteve J, Jares P, Colomer D, Rozman M, Villamor N, Costa D, Carrio A, Nomdedeu J, Montserrat E, Campo E. Gene expression profiling of acute myeloid leukemia with translocation t(8;16)(p11;p13) and MYST3-CREBBP rearrangement reveals a distinctive signature with a specific pattern of HOX gene expression. Cancer Res. 2006;66:6947–54. doi: 10.1158/0008-5472.CAN-05-4601. [DOI] [PubMed] [Google Scholar]
- 21.Katsumoto T, Aikawa Y, Iwama A, Ueda S, Ichikawa H, Ochiya T, Kitabayashi I. MOZ is essential for maintenance of hematopoietic stem cells. Genes Dev. 2006;20:1321–30. doi: 10.1101/gad.1393106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pena PV, Davrazou F, Shi X, Walter KL, Verkhusha VV, Gozani O, Zhao R, Kutateladze TG. Molecular mechanism of histone H3K4me3 recognition by plant homeodomain of ING2. Nature. 2006;442:100–3. doi: 10.1038/nature04814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qin S, Jin L, Zhang J, Liu L, Ji P, Wu M, Wu J, Shi Y. Recognition of Unmodified Histone H3 by the First PHD Finger of Bromodomain-PHD Finger Protein 2 Provides Insights into the Regulation of Histone Acetyltransferases Monocytic Leukemic Zinc-finger Protein (MOZ) and MOZ-related factor (MORF) J Biol Chem. 2011;286:36944–55. doi: 10.1074/jbc.M111.244400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu L, Qin S, Zhang J, Ji P, Shi Y, Wu J. Solution structure of an atypical PHD finger in BRPF2 and its interaction with DNA. J Struct Biol. 2012;180:165–73. doi: 10.1016/j.jsb.2012.06.014. [DOI] [PubMed] [Google Scholar]
- 25.Laue K, Daujat S, Crump JG, Plaster N, Roehl HH, Kimmel CB, Schneider R, Hammerschmidt M. The multidomain protein Brpf1 binds histones and is required for Hox gene expression and segmental identity. Development. 2008;135:1935–46. doi: 10.1242/dev.017160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vezzoli A, Bonadies N, Allen MD, Freund SM, Santiveri CM, Kvinlaug BT, Huntly BJ, Gottgens B, Bycroft M. Molecular basis of histone H3K36me3 recognition by the PWWP domain of Brpf1. Nat Struct Mol Biol. 2010;17:617–9. doi: 10.1038/nsmb.1797. [DOI] [PubMed] [Google Scholar]
- 27.Dhalluin C, Carlson JE, Zeng L, He C, Aggarwal AK, Zhou MM. Structure and ligand of a histone acetyltransferase bromodomain. Nature. 1999;399:491–496. doi: 10.1038/20974. [DOI] [PubMed] [Google Scholar]
- 28.Mujtaba S, Zeng L, Zhou MM. Structure and acetyl-lysine recognition of the bromodomain. Oncogene. 2007;26:5521–7. doi: 10.1038/sj.onc.1210618. [DOI] [PubMed] [Google Scholar]
- 29.Filippakopoulos P, Picaud S, Mangos M, Keates T, Lambert JP, Barsyte-Lovejoy D, Felletar I, Volkmer R, Muller S, Pawson T, Gingras AC, Arrowsmith CH, Knapp S. Histone recognition and large-scale structural analysis of the human bromodomain family. Cell. 2012;149:214–31. doi: 10.1016/j.cell.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun H, Liu J, Zhang J, Shen W, Huang H, Xu C, Dai H, Wu J, Shi Y. Solution structure of BRD7 bromodomain and its interaction with acetylated peptides from histone H3 and H4. Biochem Biophys Res Commun. 2007;358:435–41. doi: 10.1016/j.bbrc.2007.04.139. [DOI] [PubMed] [Google Scholar]
- 31.Poplawski A, Hu K, Lee W, Natesan S, Peng D, Carlson S, Shi X, Balaz S, Markley JL, Glass KC. Molecular Insights into the Recognition of N-Terminal Histone Modifications by the BRPF1 Bromodomain. J Mol Biol. 2014;426:1661–76. doi: 10.1016/j.jmb.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Champagne KS, Piscitelli E, Francklyn CS. Substrate recognition by the hetero-octameric ATP phosphoribosyltransferase from Lactococcus lactis. Biochemistry. 2006;45:14933–43. doi: 10.1021/bi061802v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lubula MY, Poplawaski A, Glass KC. Crystallization and preliminary X-ray diffraction analysis of the BRPF1 bromodomain in complex with its H2AK5ac and H4K12ac histone-peptide ligands. Acta Cryst F. 2014:F70. doi: 10.1107/S2053230X14018433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods Enzymol. 1997;276:307–26. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 35.Minor W, Cymborowski M, Otwinowski Z, Chruszcz M. HKL-3000: the integration of data reduction and structure solution--from diffraction images to an initial model in minutes. Acta Crystallogr D Biol Crystallogr. 2006;62:859–66. doi: 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- 36.Vagin AaTA. MOLREP: an automated program for molecular replacement. J Appl Cryst. 1997;30:1022–1025. [Google Scholar]
- 37.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235–42. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perrakis A, Morris R, Lamzin VS. Automated protein model building combined with iterative structure refinement. Nature structural biology. 1999;6:458–63. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- 39.Cowtan K. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr D Biol Crystallogr. 2006;62:1002–11. doi: 10.1107/S0907444906022116. [DOI] [PubMed] [Google Scholar]
- 40.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 41.Murshudov GN, Skubak P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, Vagin AA. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr. 2011;67:355–67. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, 3rd, Snoeyink J, Richardson JS, Richardson DC. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–83. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang H, Guranovic V, Dutta S, Feng Z, Berman HM, Westbrook JD. Automated and accurate deposition of structures solved by X-ray diffraction to the Protein Data Bank. Acta Crystallogr D Biol Crystallogr. 2004;60:1833–9. doi: 10.1107/S0907444904019419. [DOI] [PubMed] [Google Scholar]
- 44.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. Journal of applied crystallography. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–21. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Cryst. 1993;26:283–291. [Google Scholar]
- 47.Louis-Jeune C, Andrade-Navarro MA, Perez-Iratxeta C. Prediction of protein secondary structure from circular dichroism using theoretically derived spectra. Proteins. 2011 doi: 10.1002/prot.23188. [DOI] [PubMed] [Google Scholar]
- 48.Andrade MA, Chacon P, Merelo JJ, Moran F. Evaluation of secondary structure of proteins from UV circular dichroism spectra using an unsupervised learning neural network. Protein Eng. 1993;6:383–90. doi: 10.1093/protein/6.4.383. [DOI] [PubMed] [Google Scholar]
- 49.Carlson S, Glass KC. The MOZ Histone Acetyltransferase in Epigenetic Signaling and Disease. Journal of cellular physiology. 2014 doi: 10.1002/jcp.24617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang Y, Skolnick J. TM-align: a protein structure alignment algorithm based on the TM-score. Nucleic Acids Research. 2005;33:2302–9. doi: 10.1093/nar/gki524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mujtaba S, He Y, Zeng L, Yan S, Plotnikova O, Sachchidanand Sanchez R, Zeleznik-Le NJ, Ronai Z, Zhou MM. Structural mechanism of the bromodomain of the coactivator CBP in p53 transcriptional activation. Mol Cell. 2004;13:251–63. doi: 10.1016/s1097-2765(03)00528-8. [DOI] [PubMed] [Google Scholar]
- 52.Owen DJ, Ornaghi P, Yang JC, Lowe N, Evans PR, Ballario P, Neuhaus D, Filetici P, Travers AA. The structural basis for the recognition of acetylated histone H4 by the bromodomain of histone acetyltransferase gcn5p. EMBO J. 2000;19:6141–9. doi: 10.1093/emboj/19.22.6141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zeng L, Zhang Q, Gerona-Navarro G, Moshkina N, Zhou MM. Structural basis of site-specific histone recognition by the bromodomains of human coactivators PCAF and CBP/p300. Structure. 2008;16:643–52. doi: 10.1016/j.str.2008.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vidler LR, Brown N, Knapp S, Hoelder S. Druggability analysis and structural classification of bromodomain acetyl-lysine binding sites. J Med Chem. 2012;55:7346–59. doi: 10.1021/jm300346w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Umehara T, Nakamura Y, Jang MK, Nakano K, Tanaka A, Ozato K, Padmanabhan B, Yokoyama S. Structural basis for acetylated histone H4 recognition by the human BRD2 bromodomain. J Biol Chem. 2010;285:7610–8. doi: 10.1074/jbc.M109.062422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brown T, Swansbury J, Taj MM. Prognosis of patients with t(8;16)(p11;p13) acute myeloid leukemia. Leuk Lymphoma. 2012;53:338–41. doi: 10.3109/10428194.2011.614703. [DOI] [PubMed] [Google Scholar]
- 57.Panagopoulos I, Fioretos T, Isaksson M, Samuelsson U, Billstrom R, Strombeck B, Mitelman F, Johansson B. Fusion of the MORF and CBP genes in acute myeloid leukemia with the t(10;16)(q22;p13) Hum Mol Genet. 2001;10:395–404. doi: 10.1093/hmg/10.4.395. [DOI] [PubMed] [Google Scholar]
- 58.Perez-Campo FM, Borrow J, Kouskoff V, Lacaud G. The histone acetyl transferase activity of monocytic leukemia zinc finger is critical for the proliferation of hematopoietic precursors. Blood. 2009;113:4866–74. doi: 10.1182/blood-2008-04-152017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Papavassiliou KA, Papavassiliou AG. Bromodomains: pockets with therapeutic potential. Trends in molecular medicine. 2014 doi: 10.1016/j.molmed.2014.06.004. [DOI] [PubMed] [Google Scholar]
- 60.Filippakopoulos P, Knapp S. Targeting bromodomains: epigenetic readers of lysine acetylation. Nature reviews Drug discovery. 2014;13:337–56. doi: 10.1038/nrd4286. [DOI] [PubMed] [Google Scholar]
- 61.Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, Magoon D, Qi J, Blatt K, Wunderlich M, Taylor MJ, Johns C, Chicas A, Mulloy JC, Kogan SC, Brown P, Valent P, Bradner JE, Lowe SW, Vakoc CR. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478:524–8. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Spiltoir JI, Stratton MS, Cavasin MA, Demos-Davies K, Reid BG, Qi J, Bradner JE, McKinsey TA. BET acetyl-lysine binding proteins control pathological cardiac hypertrophy. Journal of molecular and cellular cardiology. 2013;63:175–9. doi: 10.1016/j.yjmcc.2013.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Banerjee C, Archin N, Michaels D, Belkina AC, Denis GV, Bradner J, Sebastiani P, Margolis DM, Montano M. BET bromodomain inhibition as a novel strategy for reactivation of HIV-1. Journal of leukocyte biology. 2012;92:1147–54. doi: 10.1189/jlb.0312165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Matzuk MM, McKeown MR, Filippakopoulos P, Li Q, Ma L, Agno JE, Lemieux ME, Picaud S, Yu RN, Qi J, Knapp S, Bradner JE. Small-molecule inhibition of BRDT for male contraception. Cell. 2012;150:673–84. doi: 10.1016/j.cell.2012.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific; Palo Alto, CA: 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.