Abstract
Motivation: DNA methylation is an epigenetic change occurring in genomic CpG sequences that contribute to the regulation of gene transcription both in normal and malignant cells. Next-generation sequencing has been used to characterize DNA methylation status at the genome scale, but suffers from high sequencing cost in the case of whole-genome bisulfite sequencing, or from reduced resolution (inability to precisely define which of the CpGs are methylated) with capture-based techniques.
Results: Here we present a computational method that computes nucleotide-resolution methylation values from capture-based data by incorporating fragment length profiles into a model of methylation analysis. We demonstrate that it compares favorably with nucleotide-resolution bisulfite sequencing and has better predictive power with respect to a reference than window-based methods, often used for enrichment data. The described method was used to produce the methylation data used in tandem with gene expression to produce a novel and clinically significant gene signature in acute myeloid leukemia. In addition, we introduce a complementary statistical method that uses this nucleotide-resolution methylation data for detection of differentially methylated features.
Availability: Software in the form of Python and R scripts is available at http://bioserv.mps.ohio-state.edu/premer and is free for non-commercial use.
Contact: pearlly.yan@osumc.edu; bundschuh@mps.ohio-state.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
DNA methylation, the addition of a methyl group to cytosine residues in a CpG dinucleotide pair, is one of the best characterized epigenetic changes in eukaryotes and plays a pivotal role in silencing gene transcription in diverse biologic settings, including embryonic development, lyonization, autoimmunity and cancer (Feinberg and Tycko, 2004; Richardson, 2007; Smith and Meissner, 2013). In the context of cancer, with the discovery that DNA methylation status may carry prognostic information and with the advent of novel DNA hypomethylating agents (i.e. aza-nucleosides), the study of the biologic, diagnostic, prognostic and pharmacodynamic role of DNA methylation has immediate relevance in the design of novel treatment approaches.
A variety of methods to determine DNA methylation have been developed and can be broadly categorized on the basis of methylation readout (bisulfite converted versus capture-based techniques) and of sample or data throughput. Throughput ranges from low-throughput analysis of a small number of loci [e.g. methylation profiling of 24 tumor-suppressor gene promoter CpG islands (Seeber et al., 2010)] to array-based methods with on the order of 480 000 probes in the case of the Infinium HumanMethylation450 BeadChip, to next-generation sequencing (NGS)-based approaches that offer high-resolution broad genomic coverage at extremely high throughput. Supplementary Text and Supplementary Table S1 further describe these techniques and categorize methylation analysis techniques in terms of readout and throughput.
The gold standard for the characterization of genome-wide DNA methylation status is whole-genome bisulfite sequencing (WGBS). With the current NGS platforms, data yields in excess of 300 GB per flow cell, the costs associated with data generation and analysis for this approach still remain out of reach, especially if a relatively large number of patient samples need to be evaluated at diagnosis and at sequential time points after treatment. Genome complexity-reduction methods such as reduced representation bisulfite sequencing (RRBS) (Gu et al., 2011; Meissner et al., 2005), double-enzyme RRBS (Wang et al., 2013), Infinium arrays (Dedeurwaerder et al., 2011) and GoldenGate Methylation assay (Bibikova and Fan, 2009) have been developed to make bisulfite-based approaches more suitable for large-scale studies. However, one notable drawback of these methods is the limited proportion of the genome and types of regions these assays interrogate (Bock et al., 2010). Additionally, researchers should be aware that the bisulfite conversion process may occur with limited efficiency and cannot distinguish between the two alternative bases, 5-methylcytosine and 5-hydroxy-methylcytosine (Jin et al., 2010), which are likely to have different biologic significance: in contrast to 5-methylcytosine’s transcriptional repression, the hydroxymethylated bases may have a transcriptionally activating effect (Ficz et al., 2011).
Capture-based methylated DNA enrichment techniques (Supplementary Table S1) avoid the majority of the problems associated with WGBS (cost; complexity), RRBS and arrays (lengthy procedure that is difficult to automate; limited genomic scope) and of bisulfite chemistry in general [potential for incomplete conversion; DNA degradation; the inability to distinguish methylation from hydroxymethylation (Jin et al., 2010)]. Despite the cost advantage of the capture-based approaches, there has been some reluctance to adopt these methods owing to a lack of a nucleotide-level methylation signal. In fact, because the base unit of information is a DNA fragment captured by a methylcytosine-binding protein or antibody and sequenced, it is presupposed that at least some CpGs within each sequenced fragment are methylated because of the capture/enrichment step, but it is uncertain precisely which CpG dinucleotides within the fragment are methylated.
Current algorithms associated with these approaches focus on the methylation signal within genomic ‘windows’, i.e. genomic intervals of a predetermined nucleotide length. While in the past we have also used 500 bp windows along the entire genome to bin enrichment-based sequence ‘reads’ in the analysis of the global methylation effect of the hypomethylating agent decitabine in acute myeloid leukemia (AML) patients (Yan et al., 2012), we more recently recognized the untapped potential of transforming read-based data into nucleotide-level data, a solution that we present here. Nucleotide scale methylation values computationally derived from captured fragment data are inherently an approximation and therefore cannot completely replace bisulfite-based data. However, this novel approach of transforming read-based data into nucleotide-based signals not only removes artifacts associated with window-based algorithms (e.g. window-based methods may erroneously assign sequencing reads to windows devoid of CG dinucleotides; Fig. 1) but also produces flexible intermediate data, unconstrained by genomic region boundaries, for subsequent analyses. Compared with other current methods, Probabilistic Extension of Methylated Reads at CpG resolution (PrEMeR-CG) has the advantage of not requiring an extra sequencing experiment such as an artificially methylated sample (Riebler et al., 2014) or a paired restriction enzyme sample (Stevens et al., 2013) to arrive at CpG resolution signal.
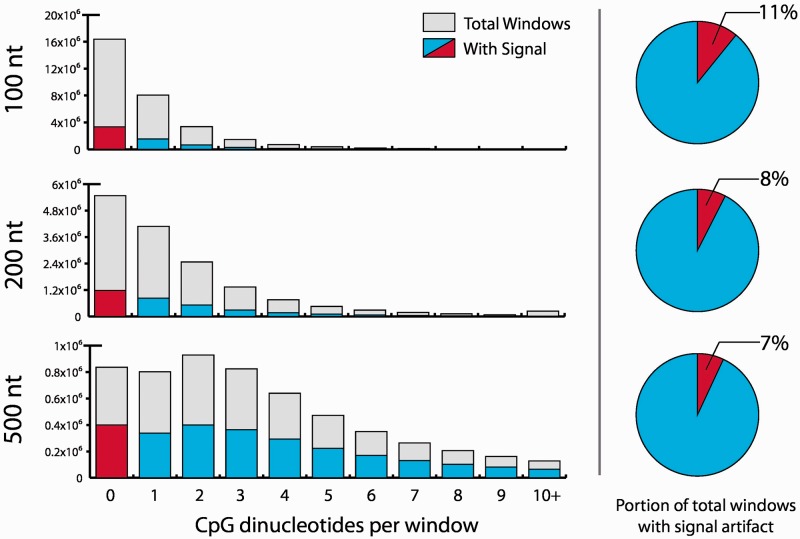
Fig. 1.
When methylation signal is determined by dividing the genome into windows, many windows with no CpGs contained within them may have methylation values ascribed to them. Here, window sizes of 100, 200 and 500 nt are all shown to exhibit this binning artifact. Data shown are from the present study
PrEMeR-CG is a computational approach that harnesses the implicit information associated with library fragment profiles to infer nucleotide-resolution methylation values in addition to read counts data, and Methylation Modeling Analysis using GEEs (MethMAGE), a complementary statistical method that uses the nucleotide-resolution methylation data in the detection of differentially methylated features (DiMeFs), previously annotated genomic regions in which the methylation profile differs between different groups (e.g. those treated with a hypomethylating agent versus those that are not). Because enrichment-based methylome analysis is easily automated and suitable for large sample size studies, our method provides a critical bridge between the practicality of capture-based studies and the resolution of WGBS. This fact is demonstrated by the large AML cohort in which PrEMeR-CG derived methylation was able to stratify patients based on outcome and was combined with gene expression to create a novel prognostic gene signature, which was validated in other patient cohorts (Marcucci et al., 2014).
2 METHODS
2.1 MethylCap-seq library generation and sequencing
Under an institutional review board-approved protocol and with the informed consent of patients, bone marrow (BM) aspirates were procured from 10 patients with AML. Genomic DNA was extracted from BM mononuclear cells and sonicated. Methylated DNA fragments were enriched with a biotinylated methyl-binding protein pulldown technique. Libraries were constructed and the methylated fragments were sequenced on an Illumina GAIIx yielding ∼ reads of 36 nt per sample. Sample quality control was evaluated using criteria described in our quality control module (Trimarchi et al., 2012). For complete details of laboratory methods, see the Supplementary Text.
2.2 MethylCap-seq read alignment
Passed filter (using default Illumina pass filter settings) sequencing reads were processed to collapse duplicates (i.e. all reads with the same sequence information) to a single read to control for polymerase chain reaction artifacts. Non-duplicate reads were then aligned to the human reference genome NCBI 36.1/hg18 using Bowtie (Langmead et al., 2009). Alignment parameters allowed for two mismatches in a 32 bp seed and suppressed all reads that mapped to multiple locations in the genome.
2.3 Assignment of methylation values from MethylCap-seq reads
For MethylCap-seq reads, methylation values were assigned by one of two methods. For the 500 bp window method, we used our previously published algorithm (Rodriguez et al., 2012; Yan et al., 2012), which bins the genome into 500 bp windows and assigns a normalized ‘reads per million’ metric to each window. The method takes each read and extends it to the average fragment size for the sample. These fragments are then assigned to the 500 bp window that contains more than half of the extended fragment, and the total count of these reads in a 500 bp genomic window represents its methylation value. For the PrEMeR-CG method, we used the algorithm described in Section 3.
2.4 Reduced representation bisulfite sequencing
RRBS sequencing libraries were generated from the same 10 AML patients described above using a published protocol (Gu et al., 2011). They were sequenced on the Illumina HiSeq 2000 to generate 50 bp single-end reads. Sequenced reads were aligned using the Bismark aligner (Krueger and Andrews, 2011).
2.5 Comparison of methylation calling accuracy
To quantify the accuracy of methylation determination, we compared the methylation signals of two MethylCap-seq–based analysis methods with that obtained with the RRBS method in matching samples. In order not to give an advantage to either window-based or CpG-resolution MethylCap-seq analysis methods, we analyzed the performance of each method at both CpG and 500 bp window resolution. When evaluating performance at CpG resolution, the methylation values of the CpG resolution method could be used directly (i.e. each CpG could be assigned a distinct value according to the method described below), whereas for the window-based method, the methylation value of a window calculated according to the method described in detail elsewhere (Rodriguez et al., 2012; Yan et al., 2012) was assigned to all CpGs in that window. Similarly, when evaluating performance at window resolution, the methylation values of the window-based method could be used directly, whereas for the CpG resolution method, each window was assigned the mean methylation value of all CpGs in that window.
We selected loci for comparison as follows. For the nucleotide-resolution comparison, we selected all CpGs that had a minimum coverage of 10 RRBS reads and for which the pulldown-based methylation call was non-zero. Similarly, for the window-resolution comparison, we selected genomic windows of fixed width, which had at least two CpGs with at least 10 covering RRBS reads each and for which the window pulldown-based methylation signal was non-zero.
To compare the two different methods, we constructed receiver-operator characteristic (ROC) curves using a threshold on each method’s methylation call as a high/low binary discriminator and the corresponding methylation call from paired RRBS analysis as benchmark, or measure of truth. Curves were then constructed by plotting the true-positive rate versus the false-positive rate in ROC space for 100 values of the respective discriminator. Noting that RRBS methylation signal is highly concentrated about 0 and 1, consistent with the binary nature of true methylation, we defined an RRBS signal of <50% as ‘low’ and 50% as ‘high’.
2.6 Determination of DiMeFs
Methylation status in a region can be compared between two groups (generically ‘A’ and ‘B’, but in practice perhaps ‘drug treated’ and ‘control’, or ‘mutated’ and ‘unmutated’), and if different, the region is designated a DiMeF. DiMeFs are genomic regions that have defined boundaries usually based on previous annotation as opposed to the dynamically determined boundaries typically associated with differentially methylated regions. DiMeFs were determined using a generalized estimating equation (GEE) set up as follows. Suppose we have n subjects and is a vector of nucleotide-resolution methylation signals of subject i observed at CpG sites within a region. Then, we model a mean structure with identity link by
| (1) |
with Xi being a vector of indicator function taking the value 1 when subject i belongs to group A and 0 otherwise. Then the estimator of is obtained by solving β such that
| (2) |
where in which Ai is a diagonal matrix with the empirical marginal variances on the diagonal, is a working correlation matrix and is an over-dispersion parameter. For the working correlation, an autoregressive AR(1) model is used:
| (3) |
Such a model is appropriate because the correlation of methylation signals in CpGs is related to proximity of the CpGs within the region. For further details on setting up the model, see Aerts et al. (2002). Finally, we tested on versus for each region and selected significant regions based on a false discovery rate (FDR) < 0.05.
2.7 Calculations
All calculations were performed in Python 2.7 and in R version 2.15 (R Development Core Team, 2012) with the geepack package (Højsgaard et al., 2006), which is available from the CRAN repository.
3 RESULTS
3.1 Sequencing
Genomic DNA from BM samples from 10 AML patients was extracted and divided into two aliquots; one aliquot was enriched for methylated DNA (see Section 2). Libraries were prepared and sequenced on an Illumina GAIIx sequencer resulting in an average yield of 21 629 060 reads per sample (range 17 541 841–26 311 159). For validation purposes, the other aliquot was subjected to RRBS (see Section 2) yielding on average 14 651 361 reads per sample (range 6 289 882–25 039 287).
3.2 Methylation calling with window-based method
One of the major shortcomings of window-based methods in the analysis of MethylCap-seq data is the fact that windows impose artificial boundaries along the genome, which can result in analysis artifacts. To demonstrate this, we used our previously published window-based analysis tool (Rodriguez et al., 2012; Yan et al., 2012) to determine methylation values from our MethylCap-seq data. Figure 1 depicts the number of windows with attributed methylation signal compared with all windows containing the indicated number of CpGs for various window sizes. Using this approach, we demonstrated that the window-based method assigns methylation even in windows that contain no CpGs; this artifact is a result of methylated CpGs in neighboring windows pulling down fragments that span the window boundary and thus are counted even in CpG-less windows. Despite the disadvantage of containing artifacts, window methods provide global methylation information for a large portion of the genome.
3.3 Methylation calling from pulldown data at CpG resolution
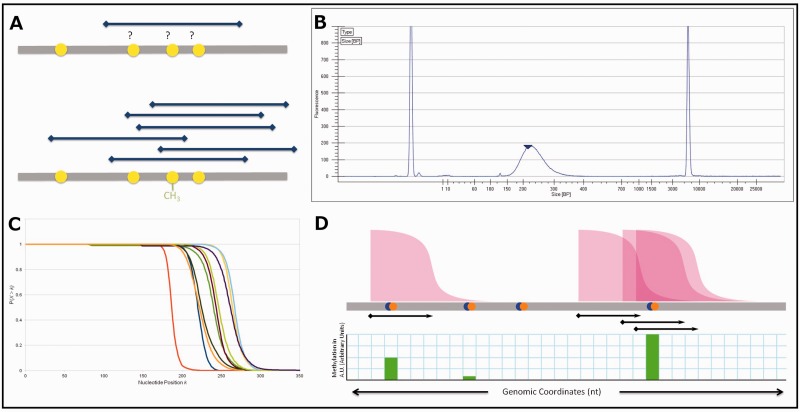
Motivated by the recognition of artifacts in window-based methods for the analysis of MethylCap-seq data, we strove to develop a method that could use these data to infer methylation values at CpG resolution; such a method is inherently free from artifacts associated with window boundaries. An existing boundary-free tool, BALM (Lan et al., 2011), uses a tag-shifting method with a bi-asymmetric-Laplace model to identify methylated loci. When implemented on our data, we noted that BALM covers a significantly reduced portion of the genome than, e.g. the 500 bp window method (Supplementary Table S2), and we wanted to develop a method that preserved this global coverage. The idea behind our new method, which we call PrEMeR-CG, is outlined in Figure 2A. Although each fragment can localize one or more methylated CpGs necessarily only to within the fragment length (Fig. 2A top), the overall distribution of fragments still allows inference of methylation at higher resolution by integrating information about the overlap of several sequenced fragments (Fig. 2A bottom). In particular, our method proceeds as follows:
Fig. 2.
(A) Conceptual underpinning of nucleotide-resolution methylation inference from pulldown data. A single DNA fragment does not yield information on which of the CpGs covered by it are methylated (top sequence), but an ensemble of such fragments contains methylation information (bottom sequence). (B) Example Bioanalyzer output. The distribution shown describes the length in nucleotides of fragments, which comprise the DNA library. At the extremes are calibration markers. (C) CCDFs for the 10 samples in this study derived from Bioanalyzer data. This is the probability (y axis) that any given fragment X is longer than the indicated nucleotide position k along the x axis. (D) Schematic diagram of probabilistic read extension. The cumulative distribution function shown in (C) is attached to every sequencing read. Then, each CpG receives contributions to its methylation signal from all cumulative distribution functions overlapping the CpG
Each aligned read corresponds to one DNA fragment in the sequencing library captured owing to the presence of methylated cytosine(s) within the fragment. The length of the DNA fragment from which any given sequencing read originated is unknown, but the Bioanalyzer high sensitivity DNA chip used during library preparation (see Section 2) produces a histogram describing fragment length frequencies in the library (Fig. 2B). We used the Bioanalyzer fragment profile information to construct a probability function for each sample. This probability function for a sample is defined as the probability that any given fragment from that sample has a length greater than a particular value k; this is also called the complementary cumulative distribution function (CCDF; equation 4; Fig. 2C).
| (4) |
Next, each sequencing read is extended along its hypothetical fragment, and any CpG included in the extension is assigned a weighted portion of that read’s signal. The methylation call mi imparted by a read to the CpG at the ith position within a given DNA fragment is calculated first by determining the probability that a fragment containing that CpG existed in the sample, then dividing by the expected number of CpGs (using and summing) that likely lie within the fragment giving rise to the read, yielding:
| (5) |
for , where C is the set of nucleotide indices in the extended read that are CpG locations. That is, mi and are calculated only at CpGs within the fragment. In this step, the methylation value at each CpG is thus its fractional share of all CpGs, weighted according to the probability that any and all CpGs in the calculation existed in the fragment that generated the read. For example, if a CpG is within range (described by the fragment distribution profile) of N reads (even if the read does not extend over the CpG), the signal imparted to that CpG is the sum of N probabilities that the fragment was that long. This is depicted graphically in Figure 2D. The two left most CpGs in the figure lie within the CCDF graph of one fragment: first with near 100% probability, and the second CpG with near zero, but non-zero, probability. If those probabilities were 0.95 and 0.05, each CpG would receive that signal. Finally, the so-computed methylation values at individual CpGs within the fragment are summed over all overlapping fragments (i.e. all reads within a predefined maximal fragment size about the CpG’s position) to determine an overall probable methylation signal at that CpG dinucleotide (Fig. 2D). Because each sample has a differing number of total passed-filter reads that successfully align to the genome, the methylation values must also be normalized for total read yield. We used reads per million aligned reads as a normalization factor to compare across samples.
3.4 Comparison with window-based method
While our CpG resolution method is by definition free from window boundary artifacts, we needed to quantitatively compare its accuracy in determining methylation with that of a window-based method. To eliminate the requirement for extrinsic information [such as an artificially methylated control sample (Riebler et al., 2014) or a paired restriction enzyme sample (Stevens et al., 2013)] and to focus specifically on the effect of resolution, we chose our own previously published method (Rodriguez et al., 2012; Yan et al., 2012) as comparison.
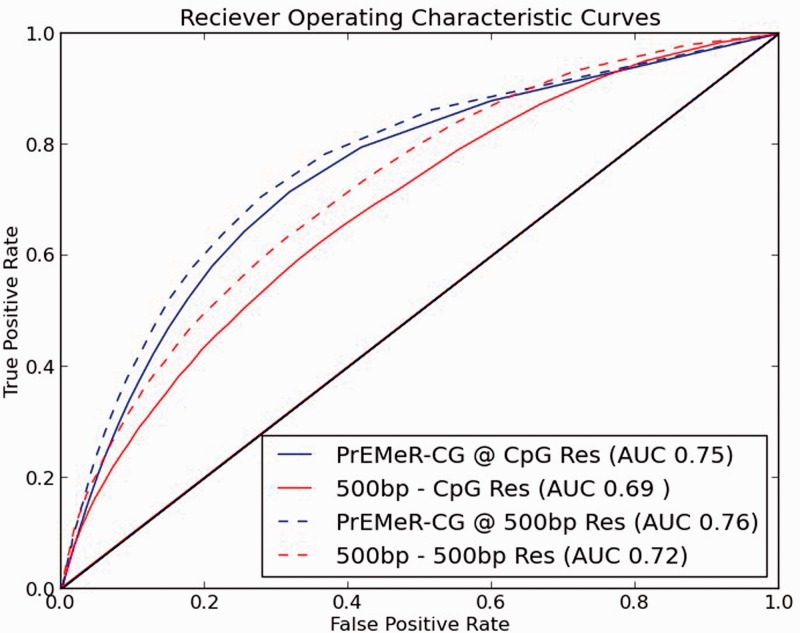
We used RRBS-derived methylation values from the same 10 samples as our MethylCap-seq data as a measure of true methylation status for purpose of comparison. We then used PrEMeR-CG as well as our prior window-based method to assign relative methylation values and applied various levels of discrimination to call ‘high methylation’ or ‘low methylation’ from the capture-based data. By comparing these classifications with the RRBS single-base resolution standard, we constructed two sets of ROC curves as described in Section 2. Figure 3 shows genome-wide ROC curves in four cases: curves were generated for PrEMeR-CG and the prior 500 bp window calling method, each at nucleotide resolution and at 500 bp resolution (Supplementary Fig. S1 shows the same results for each of the 10 samples separately). When evaluating methylation calling according to the RRBS standard, PrEMeR-CG has a higher area under the curve (AUC: 0.75 and 0.76), representing higher accuracy in methylation determination and outperforming the window-based method (AUC: 0.69 and 0.72) irrespective of the resolution at which the performance is evaluated. Although these AUCs are somewhat low, high concordance between MethylCap-seq and RRBS is not anticipated because of the differences in these methodolgies discussed in Section 1. The same analysis was repeated at the 100 bp window resolution with similar outcomes (Supplementary Fig. S2).
Fig. 3.
ROC curves comparing the sensitivity and specificity of PrEMeR-CG to the 500 bp window method demonstrate PrEMeR-CG’s superior match to the reference bisulfite sequenced samples at both CpG resolution as well as 500 bp window resolution
3.5 Utilization of CpG-resolution data in the detection of DiMeFs
One of the end goals of methylation analysis is to determine changes between conditions in different genomic regions of interest. To use the inferred single-base resolution data derived from PrEMeR-CG, a new statistical approach is needed that does not require averaging methylation signals across regions while identifying DiMeFs. For this expressed purpose, we developed a method called MethMAGE, a domain-specific use of the GEE.
A primary feature of the nucleotide-resolution data generated by PrEMeR-CG is that the signals at neighboring CpG sites are correlated (Supplementary Fig. S3), both because they are derived from fragments of finite size and because of biological correlations between methylation levels at neighboring CpGs. In fact, we believe that one of the main advantages of PrEMeR-CG is the possibility to explicitly incorporate the correlation structure of neighboring CpGs in DiMeF calling. In addition, the CpG methylation signals are not normally distributed and have a relatively high proportion of non-detectable signal (zero values). The GEE approach is non-parametric and flexible with respect to these signals and explicitly takes into account correlations, and therefore, is well suited to model the nucleotide-resolution data generated by PrEMeR-CG. More specifically, for each genomic feature, GEE builds a linear model of methylation signals at CpG sites with working covariance structure (see Section 2). Because there is a decreasing correlation between CpG sites with increasing distance along the genome, we chose an autoregressive AR(1) model to describe the covariance structure. We note that this autoregressive AR(1) covariance structure is a major difference compared with the previous GEE-based differential methylation calling package A-clust (Sofer et al., 2013) (in addition to the difference that MethMAGE tests methylation in features of fixed bounds to call DiMeFs that accommodate annotation whereas A-clust determines regions to test by dynamically clustering correlated CpGs). We believe that the exchangeable working correlation used by A-clust is appropriate for the small clusters and short distances that typically separate the CpGs in the clusters detected by A-clust. In contrast, the AR(1) correlation used by MethMAGE is autoregressive and more suitable for the expected correlation of methylation in larger annotated features. The mean difference between case and control groups is estimated by solving generalized estimation equations using the GEE function from the R package geepack (see Section 2). From the fitted data, a P-value is generated for each feature, and DiMeFs were then called with the FDR controlled at 0.05 according to the Benjamini–Hochberg procedure (1995).
To demonstrate the utility of MethMAGE, DiMeFs were called between groups that were defined by mutation status. The choice of mutation status as a classifier was made based on our previously published work, which showed that the methylation of DiMeFs specific to known mutations are prognostic (Marcucci et al., 2014). We selected RefSeq gene promoters, defined as 1000 bp up- and downstream of the annotated transcription start sites of RefSeq genes, as well as CpG islands, defined via their UCSC genome browser track (Gardiner-Garden and Frommer, 1987; Meyer et al., 2013), as features because their methylation status is associated with gene regulation. To define two groups, we chose among the many different known AML mutations shown to be associated with DiMeFs in (Marcucci et al., 2014) the FLT3-ITD (internal tandem duplication of the FLT3 gene) status as the grouping criterion because it provided the most balanced separation of our group of 10 patients (seven FLT3 wild type and three FLT3-ITD patients). MethMAGE is also able to eliminate features if they do not have sufficient coverage among the sample groups for a statistical test of the model to be meaningful. For the sample groups used here, 94% of the promoters and 91% of the CpG islands contained sufficient coverage to be included in MethMAGE (Table 1). MethMAGE identified 181 DiMeFs in promoters and 584 DiMeFs in CpG islands, whereas MEDIPS identified four DiMeFs in promoters and four DiMeFs in CpG islands, while zero DiMeFs were detected by the t-test, and the negative binomial-based edgeR and DEseq methods (Supplementary Table S2). Analysis details can be found in the Supplementary Material.
Table 1.
DiMeFs generated by MethMAGE
| Feature | Number of features | Features evaluated | DiMeFs identified by MethMAGE |
|---|---|---|---|
| CpG islands | 27 640 | 22 562 | 589 |
| RefSeq gene promoters | 39 945 | 37 584 | 181 |
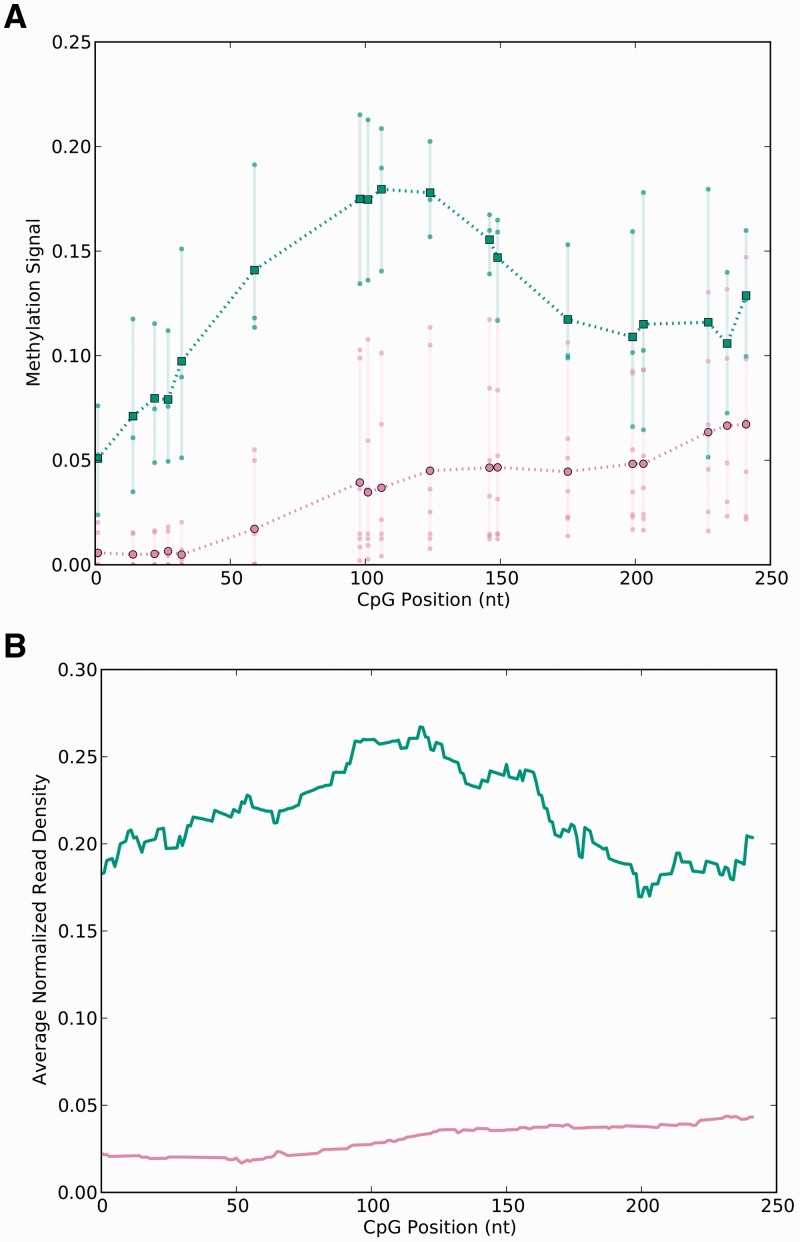
It is known that P-values generated by the GEE can become unreliable for small numbers of samples, and it is thus difficult to assess whether these DiMeFs are false-positive findings resulting from inaccurate estimation of P-values or are biologically meaningful DiMeFs. However, the methylation profiles of the examples shown in Figure 4 and Supplementary Figures S4 and S5 demonstrate that the regions MethMAGE identified appear to have different methylation profiles between the wild type and FLT3-ITD groups. In addition, the problem of unreliable P-value estimations, and thus possible false-positive findings, should become less of an issue when larger numbers of samples are compared, given the asymptotic property of GEE.
Fig. 4.
Representative features detected as differentially methylated by MethMAGE. (A) Methylation signal of each CpG is represented as a dot (lower curve [purple online] for FLT3 wild type and upper curve [green online] for FLT3-ITD samples) and the line segments connect the means of these values. (B) Normalized read density of the same region calculated by extending each read using the cumulative distribution functions for each sample, normalizing each sample to reads per million and averaging over the samples in each of the two groups
To assess the performance of MethMAGE, paired RRBS samples were used to determine which promoters and CpG Islands were differentially methylated (RRBS DiMeFs) and which were not differentially methylated (RRBS non-DiMeFs).
These RRBS DiMeFs and non-DiMeFs, called between the wild type and FLT3-ITD group, were compared with the DiMeFs reported by MethMAGE. Among the RRBS non-DiMeFs, MethMAGE demonstrated a low false-positive rate, detecting only a single DiMeF (Supplementary Table S4). MethMAGE also reported 60% of the RRBS DiMeFs demonstrating relatively good sensitivity considering the differences previously discussed between RRBS and MethylCap data (Supplementary Table S4). An example of some of the features identified as a DiMeF can be seen in Figure 4 and Supplementary Figures S4 and S5.
4 DISCUSSION
DNA methylation is well established as an epigenetic regulator in a wide variety of biological processes (Feinberg and Tycko, 2004; Richardson, 2007; Smith and Meissner, 2013), and recent years have seen tremendous progress in the laboratory and computational techniques of methylation analysis (Bock, 2012). Bock et al. analyzed two pairs of samples using two enrichment methods (MeDIP and MethylCap) coupled with NGS and two bisulfite-based methods (RRBS and Infinium assay) (Bock et al., 2010). Compared with MeDIP-seq, MethylCap-seq gave rise to peaks of methylated DNA with higher dynamic range (higher peaks and lower baseline signals), and the authors noted that bisulfite-based methods generally confirmed the enrichment-based methods. Overall, when compared with RRBS, the MethylCap-seq method had richer and more even coverage of the entire genome, including within-key features such as CpG islands and promoter regions, methylation of which is known to affect transcription.
Capture-based methods in combination with NGS offer the potential of cost-effective whole-genome methylation analysis, with the added benefit of the possibility to distinguish alternative cytosine modifications (Jin et al., 2010). Many capture-based sequencing methods have been implemented with success, assessing methylation in genomic windows, typically 50 bp and larger. We have previously published a technique for the analysis of MethylCap-seq data in which the genome is divided into 500 bp bins and genome-wide and per-feature differential methylation patterns can be described in terms of these bins (Rodriguez et al., 2012; Yan et al., 2012).
Still, it is inescapable that bisulfite-based methods provide inherently higher resolution than capture-based methods, and it is notable that methylation at an individual CpG dinucleotide has demonstrated biological importance (Claus et al., 2012). Because of the foregoing limitations in bisulfite as well as capture-based methods and to make more accurate methylation calls, we were motivated to devise a comprehensive method for analyzing capture-based data that could provide genome-wide nucleotide-resolution information as well as define DiMeFs between groups, but that would be free from external requirements (e.g. would not require a positive control sample treated with the prokaryotic methyltransferase M.SssI or paired normal samples).
In this article, we proposed a method to provide methylation values at individual CpG dinucleotides, thereby eliminating many of the issues associated with larger genomic windows. For example, once PrEMeR-CG calculates a methylation signal for each CpG, it is straightforward to assign methylation signal to any genomic region by averaging over all the CpGs contained in this region. This stands in contrast to window-based methods in which the raw data must be reanalyzed every time a new type of genomic region is introduced or one risks including DNA fragments that overlap a predefined window, which in turn overlaps the genomic region without the fragment itself actually overlapping the genomic region. Although the assignment of fractional methylation signal to CpGs within a read is straightforward, the key innovation in the PrEMeR-CG method is the recognition that CpGs in a pulled-down fragment may exist outside the read, and incorporating sequencing-library fragment length statistics to create a probabilistic model of methylation by virtually extending reads along their hypothetical fragments. Sequencing-library fragment statistics, although not typically available with public datasets, are almost certainly available to the laboratories and groups who generated the data: fragment-length profiling is an important preparatory step in sequencing to confirm the lack of adapter dimers as well as to ensure a specific average fragment length. To our knowledge, we are the first group to use this highly available yet underused information in DNA methylation analysis.
Using paired RRBS data from the same samples, we compared PrEMeR-CG with our prior 500 bp window-based method and demonstrated superior discriminatory power. In addition, we provide a non-parametric statistical method (MethMAGE) for detecting changes in methylation within a defined genomic region (feature) and demonstrated its utility in identifying regions with methylation differences.
We recognize there are limitations in the present method. First, these methylation calls are only inferences suitable for discovery and hypothesis generation; important findings must be validated by alternative techniques. Second, in any pulldown experiment there is an inherent bias vis-à-vis the actual fragments pulled down: density and spatial arrangement of methylated CpGs are known to affect pulldown (Fraga et al., 2003), and there may exist some other unknown factors that influence the affinity of the methyl-binding domain protein for the DNA. Finally, using RRBS as the standard for comparison in the generation of the ROC curves may be complicated by the potential differences in methylation affinities assessed by the two methods (i.e. RRBS may report both 5-methylcytosine and 5-hydroxymethylcytosine, while MethylCap may report only 5-methylcytosine). This confounding factor may explain the somewhat low reported AUC values.
As DNA methylation analysis is increasingly important in the clinical arena, sample preparation is moving away from manual preparation to automation. In manual sample preparation, a gel-based approach is used for fragment selection. It produces a tight distribution of fragment sizes although ranges can still vary significantly from sample to sample as depicted in Figure 2C. With automation, fragment selection is performed by paramagnetic bead selection, which produces a broader size distribution (data not shown) and a longer fall from one to zero in the CCDF. This heightens the importance of a probabilistic method such as PrEMeR-CG to take into account library fragment size.
An added incentive in using PrEMeR-CG is its accuracy in estimating the empirical distribution of fragment size in a sequencing library. We compared the PrEMeR-CG-derived fragment size to fragment size calculated from aligning paired-end reads to the genome and found excellent concordance of the distributions (Supplementary Figure S6). Paired-end data from MethylCap-Seq libraries will allow assignment of the true length for each fragment rather than using our probabilistic approach. However, paired-end experiments are more costly. To that end, our method is the most cost-effective means to infer nucleotide-level methylation values for a large clinical trial. The power of this approach is well illustrated by the derivation of a prognostically significant signature by combining gene expression and methylation data in a cohort of 134 AML patients (Marcucci et al., 2014).
In conclusion, we have presented an algorithm for inferring nucleotide-resolution methylation signal from a capture-based technique and a companion statistical test to detect DiMeFs using this nucleotide-level information. Discriminatory power was demonstrated using biological samples (acute leukemia BM blasts), and our approach warrants further study in future experiments.
Supplementary Material
ACKNOWLEDGEMENTS
The authors gratefully acknowledge Benjamin AT Rodriguez for his help in sample preparation.
Funding: This work was supported by the National Institutes of Health (CA016058, CA140158, CA101140 and CA102031); National Science Foundation (DMS-1042946 and agreement No. 0931642); and in part by an allocation of computing time from the Ohio Supercomputer Center.
Conflict of interest: none declared.
REFERENCES
- Aerts M, et al., editors. Topics in Modelling of Clustered Data. Number 96 in Monographs on Statistics and Applied Probability. Boca Raton: Chapman & Hall/CRC; 2002. [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol. 1995;57:289–300. [Google Scholar]
- Bibikova M, Fan J-B. GoldenGate assay for DNA methylation profiling. Methods Mol. Biol. 2009;507:149–163. doi: 10.1007/978-1-59745-522-0_12. PMID: 18987813. [DOI] [PubMed] [Google Scholar]
- Bock C. Analysing and interpreting DNA methylation data. Nat. Rev. Genet. 2012;13:705–719. doi: 10.1038/nrg3273. PMID: 22986265. [DOI] [PubMed] [Google Scholar]
- Bock C, et al. Quantitative comparison of genome-wide DNA methylation mapping technologies. Nat. Biotechnol. 2010;28:1106–1114. doi: 10.1038/nbt.1681. PMID: 20852634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claus R, et al. Quantitative DNA methylation analysis identifies a single CpG dinucleotide important for ZAP-70 expression and predictive of prognosis in chronic lymphocytic leukemia. J. Clin. Oncol. 2012;30:2483–2491. doi: 10.1200/JCO.2011.39.3090. PMID: 22564988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dedeurwaerder S, et al. Evaluation of the infinium methylation 450K technology. Epigenomics. 2011;3:771–784. doi: 10.2217/epi.11.105. PMID: 22126295. [DOI] [PubMed] [Google Scholar]
- Feinberg AP, Tycko B. The history of cancer epigenetics. Nat. Rev. Cancer. 2004;4:143–153. doi: 10.1038/nrc1279. PMID: 14732866. [DOI] [PubMed] [Google Scholar]
- Ficz G, et al. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature. 2011;473:398–402. doi: 10.1038/nature10008. PMID: 21460836. [DOI] [PubMed] [Google Scholar]
- Fraga MF, et al. The affinity of different MBD proteins for a specific methylated locus depends on their intrinsic binding properties. Nucleic Acids Res. 2003;31:1765–1774. doi: 10.1093/nar/gkg249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. J. Mol. Biol. 1987;196:261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- Gu H, et al. Preparation of reduced representation bisulfite sequencing libraries for genome-scale DNA methylation profiling. Nat. Protoc. 2011;6:468–481. doi: 10.1038/nprot.2010.190. PMID: 21412275. [DOI] [PubMed] [Google Scholar]
- Højsgaard S, et al. The R package geepack for generalized estimating equations. J. Stat. Softw. 2006;15:1–11. [Google Scholar]
- Jin S-G, et al. Examination of the specificity of DNA methylation profiling techniques towards 5-methylcytosine and 5-hydroxymethylcytosine. Nucleic Acids Res. 2010;38:e125. doi: 10.1093/nar/gkq223. PMID: 20371518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger F, Andrews SR. Bismark: a flexible aligner and methylation caller for bisulfite-seq applications. Bioinformatics. 2011;27:1571–1572. doi: 10.1093/bioinformatics/btr167. PMID: 21493656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan X, et al. High resolution detection and analysis of cpg dinucleotides methylation using MBD-seq technology. PLoS One. 2011;6:e22226. doi: 10.1371/journal.pone.0022226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, et al. Ultrafast and memory-efficient alignment of short dna sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcucci G, et al. Epigenetics meets genetics in acute myeloid leukemia: Clinical impact of a novel seven-gene score. J. Clin. Oncol. 2014;32:548–556. doi: 10.1200/JCO.2013.50.6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner A, et al. Reduced representation bisulfite sequencing for comparative high-resolution DNA methylation analysis. Nucleic Acids Res. 2005;33:5868–5877. doi: 10.1093/nar/gki901. PMID: 16224102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer LR, et al. The UCSC genome browser database: extensions and updates 2013. Nucleic Acids Res. 2013;41:D64–D69. doi: 10.1093/nar/gks1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. R: A Language and Environment for Statistical Computing. 2012. R Foundation for Statistical Computing, Vienna, Austria. ISBN 3-900051-07-0. [Google Scholar]
- Richardson B. Primer: epigenetics of autoimmunity. Nat. Clin. Pract. Rheumatol. 2007;3:521–527. doi: 10.1038/ncprheum0573. PMID: 17762851. [DOI] [PubMed] [Google Scholar]
- Riebler A, et al. Baymeth: improved DNA methylation quantification for affinity capture sequencing data using a flexible bayesian approach. Genome Biol. 2014;15:R35. doi: 10.1186/gb-2014-15-2-r35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez BAT, et al. Methods for high-throughput MethylCap-Seq data analysis. BMC Genomics. 2012;13(Suppl. 6):S14. doi: 10.1186/1471-2164-13-S6-S14. PMID: 23134780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeber LMS, et al. Methylation profiles of endometrioid and serous endometrial cancers. Endocr. Relat. Cancer. 2010;17:663–673. doi: 10.1677/ERC-10-0014. PMID: 20488783. [DOI] [PubMed] [Google Scholar]
- Smith ZD, Meissner A. DNA methylation: roles in mammalian development. Nat. Rev. Genet. 2013;14:204–220. doi: 10.1038/nrg3354. PMID: 23400093. [DOI] [PubMed] [Google Scholar]
- Sofer T, et al. A-clustering: a novel method for the detection of co-regulated methylation regions, and regions associated with exposure. Bioinformatics. 2013;29:2884–2891. doi: 10.1093/bioinformatics/btt498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens M, et al. Estimating absolute methylation levels at single-CpG resolution from methylation enrichment and restriction enzyme sequencing methods. Genome Res. 2013;23:1541–1553. doi: 10.1101/gr.152231.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimarchi M, et al. Enrichment-based DNA methylation analysis using next-generation sequencing: sample exclusion, estimating changes in global methylation, and the contribution of replicate lanes. BMC Genomics. 2012;13(Suppl. 8):S6. doi: 10.1186/1471-2164-13-S8-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, et al. Double restriction-enzyme digestion improves the coverage and accuracy of genome-wide CpG methylation profiling by reduced representation bisulfite sequencing. BMC Genomics. 2013;14:11. doi: 10.1186/1471-2164-14-11. PMID: 23324053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan P, et al. Genome-wide methylation profiling in decitabine-treated patients with acute myeloid leukemia. Blood. 2012;120:2466–2474. doi: 10.1182/blood-2012-05-429175. PMID: 22786882. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.