Abstract
Phosphoinositides are a class of phospholipids generated by the action of phosphoinositide kinases with key regulatory functions in eukaryotic cells. Here, we present the atomic structure of phosphatidylinositol 4-kinase type IIα (PI4K IIα), in complex with ATP solved by X-ray crystallography at 2.8 Å resolution. The structure revealed a non-typical kinase fold that could be divided into N- and C-lobes with the ATP binding groove located in between. Surprisingly, a second ATP was found in a lateral hydrophobic pocket of the C-lobe. Molecular simulations and mutagenesis analysis revealed the membrane binding mode and the putative function of the hydrophobic pocket. Taken together, our results suggest a mechanism of PI4K IIα recruitment, regulation, and function at the membrane.
Keywords: crystal structure, kinase, membrane, Monte Carlo simulations, phosphatidyl inositol
Introduction
The importance of inositol lipids in many aspects of eukaryotic biology has now been widely recognized [1]. As phosphorylation products of phosphatidylinositol (PI), they are generated by a plethora of enzymes that phosphorylate specific positions of the inositol ring, yielding the seven known inositol lipid isomers [1]. These lipids control membrane dynamics lipid distribution, ion channels, transporters, and intracellular signaling among emerging roles in viral entry and replication [1]. Phosphorylation of PI at the 4-position by PI 4-kinases (PI4Ks) and subsequently by phosphatidylinositol 4-phosphate (PI4P) 5-kinases has been recognized as the major route of PI(4,5)P2 synthesis [2]. Although PI(4,5)P2 is one of the best studied phosphoinositides with connections to a number of plasma membrane-linked processes [1], it is increasingly evident that its precursor, PI4P plays independent and critical regulatory roles in various membrane compartments, most notably in the plasma membrane [3], Golgi [4], and late endosomes/lysosomes [5–7]. The recent identification of PI4P as a key phosphoinositide required for the replication of a number of RNA viruses [8] has drawn attention to the significant gaps in our understanding of the biogenesis of this lipid by PI4K enzymes at the molecular level.
PI4P is made by four distinct PI4K enzymes in mammals [9], all of which have homologues in lower eukaryotes [10]. The highest PI4K activity associated with isolated membranes is attributed to the so-called type II enzymes that are tightly membrane-bound and require detergents to be extracted from membranes [11]. In contrast, type III PI4Ks are peripherally membrane associated soluble proteins whose recruitment to membranes involves interaction with other protein [9]. Bioinformatics analysis revealed that the catalytic domains of type III PI4Ks show high homology to the PI3Ks and PI-kinase-related protein kinases, whereas type II PI4Ks form a distinct family [12,13]. Additionally, the biology of type III PI4Ks has been explored primarily by elegant yeast studies where the larger PI4K IIIα enzyme (and the yeast orthologue, Stt4p) is mostly responsible for the synthesis of the PM pool of PI4P, whereas PI4K IIIβ (or its yeast orthologue, Pik1p) mostly functions in the Golgi and post-Golgi compartments [10]. Mammalian type II PI4Ks (alpha and beta) are proteins whose membrane association and activity is highly dependent on palmitoylation [14,15], and they are present in the PM and in a number of endomembrane compartments including the TGN and early and late endosomes [5,16,17]. Type II PI4K enzymes are tightly linked to vesicular trafficking by contributing to the recruitment of the clathrin adaptors AP1, AP3, some GGAs, and VAMP3 [6,17–19] and are particularly important for the lysosomal targeting of EGF receptors [5] or enzymes of sphingolipid metabolism [7]. The activity of the PI4K IIα enzyme is regulated by cholesterol [20], which also affects the palmitoylation and localization of the protein [21].
The dissimilarity of the type II PI4Ks to any other kinase families has presented difficulties in gaining any reliable insight into the structure of these molecules. Here, we present the X-ray structure of the PI4K IIα at 2.8 Å resolution. The structure provides a critical template for this unique family of type II PI4K enzymes. The structure, which revealed non-typical kinase fold and an unexpected hydrophobic pocket with a second ATP bound, will greatly facilitate further study of these proteins and allow targeted manipulation of their interactions with regulatory components.
Results and Discussion
PI4K IIα crystal structure
The proline-rich N-termini of the enzyme contains physiologically important binding sites for ubiquitin ligase Itch [22] and clathrin adaptor complex 3 [6] but is predicted to be disordered, and its deletion does not affect the kinase activity [23]. Since this N-terminus lowers solubility and expression yields of the recombinant enzyme, it was omitted in the constructs used for biochemical analysis (called here pseudo-wt). For crystallographic analysis, additional 12 residues at the C-terminus were deleted and the palmitoylated LCCPCCF motif was replaced with T4 lysozyme to increase solubility and to improve crystallization properties (Fig 1A). We obtained crystals that diffracted to 2.8 Å and belonged to the orthorhombic P21212 space group with one molecule per asymmetric unit. The structure was subsequently solved by MR-SAD and refined to Rwork = 21.15% and Rfree = 25.11%. (Table1). The first ordered residue in the structure was R101, and the last one was A454. We were able to trace the entire chain from R101 to A454 except for two short disordered loops between residues S231 – I251 and Y318 – D331. The structure revealed a non-typical kinase fold that could be divided into the N-lobe and C-lobe with the ATP binding groove between (Fig 1B and C, detailed Fig 2A). The N-lobe consists of four helices (H1-H4) packed around a core of four small antiparallel β-sheets (S1-S4). The C-lobe has two β-sheets closely surrounded by helices H5, H6, and H7. Helices H8, H9, and H10 are packed in a ‘roof’ arrangement above both lobes and represent the most distant part of the kinase from the membrane. Surprisingly, the structure revealed another ATP molecule bound in a lateral hydrophobic pocket partly supported by the H11 helix (Fig 1B, Supplementary Fig S2) (detailed Fig 2B). While this study was under review, a structural study of PI4K IIIβ that has a more typical PI-kinase fold was published [24]. Superimposition of the whole kinase domains of PI4K IIα and PI4K IIIβ (Fig 1D) showed that only helix H3 of the N-lobe can be superimposed on the corresponding regions of the PI4K IIIβ typical kinase domain. However, helix H6 and sheets S5, S6 of the C-lobe can be superimposed on the corresponding regions of the PI4K IIIβ typical kinase domain. The dissimilarity of the N-lobes and the similarity of the C-lobes become apparent when the superimpositions are done separately for each lobe (Supplementary Fig S1 C and D). Comparison of the structures of PI4K IIα and of its closest structural relative found in a search of Protein Data Bank (Supplementary Fig S1A)—the cell translocating kinase A (ctkA), a Ser/Thr kinase from Helicobacter pylori [25]—shows a possible evolutionary connection between the type II PI4Ks and Ser/Thr protein kinases.
Figure 1. The overall fold of PI4K IIα.
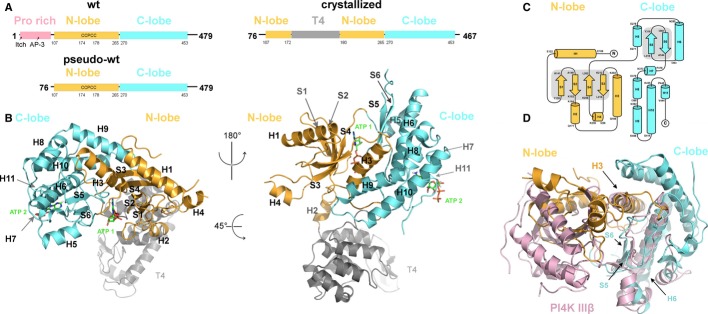
- Wt, pseudo-wt, and crystallized PI4K IIα. Binding sites for ubiquitin ligase Itch and AP3 are shown within the proline rich of the wt enzyme (left). The N-terminus is deleted in the pseudo-wt construct (lower left) used for biochemical assays. In the crystallized construct (right), the last 12 residues are deleted as well and the palmitoylated CCPCC motif is replaced with T4 lysozyme.
- Overall fold of PI4K IIα. On the left, view oriented to show the ATP binding pocket localized between the N- and C-lobes. On the right, view rotated 180° and tilted to place T4 lysozyme in the back. The kinase has 11 α-helices (numbered H1–H11), H1 to H4 are located in the N-lobe, where they surround two pairs of antiparallel β-sheets (S1–S4). A pair of β-sheets (S5, S6) in the C-lobe is closely surrounded by helices H5, H6, and H7, while the helices H8, H9, and H10 form a ‘roof’. A second ATP is bound in a lateral hydrophobic pocket of the C-lobe that is partially supported by helix H11. The palmitic acid residues would occupy the loop that is replaced by the T4 lysozyme. The N-lobe is depicted in orange, the C-lobe in cyan, the T4 lysozyme in gray.
- Topology plot of PI4K IIα. The enzyme's N-lobe consists of four α-helices and four β-sheets, and the C-lobe is composed of seven α-helices and two β-sheets. The color code is as in (A).
- Superimposition of PI4K IIα and PI4K IIIβ. Only helix H3 of the N-lobe can be superimposed on the corresponding regions of the PI4K IIIβ typical kinase domain. On the other hand, from the C-lobe helix H6 and sheets S5, S6 superimpose well. The N-lobe and C-lobe of PI4K IIα are in orange and cyan, respectively. The PI4K IIIβ kinase domain is shown in pink.
Table 1.
Statistics of crystallographic data collection and refinement
| Data collection | ||
|---|---|---|
| Crystal | Native PI4K IIα | SeMet PI4K IIα |
| Space group | P21212 | P21212 |
| Cell dimension | a = 104.8 Å, b = 79.5 Å, c = 78.7 Å, | a = 102.3 Å, b = 79.5 Å, c = 79.1 Å |
| X-ray source | BESSY ID 14-2 | BESSY ID 14-1 |
| Wavelength, Å | 0.91841 | 0.97984 |
| Resolution, Å | 36.84–2.77 (2.87–2.77) | 49.16–3.00 (3.10–3.00) |
| No. of unique reflections | 16256 (1631) | 13505 (1325) |
| I/σ (I) | 12.45 (2.02) | 10.99 (3.07) |
| Rmerge | 15.9 | 20.9 |
| Data completeness, % | 93.87 (95.77) | 99.92 (98.95) |
| Multiplicity | 5.8 | 12.9 |
| Refinement | |
|---|---|
| Rwork, % | 21.15 (27.10) |
| Rfree, % | 25.11 (36.45) |
| Number of non-hydrogen atoms | 3975 |
| Protein residues | 484 |
| rms bond angle deviation, ° | 0.013 |
| rms bond angle deviation, Å | 1.46 |
| Average B-factor | 58.30 |
| Ramachandran (outliers/favored) | 0.21%/97% |
Numbers in parentheses refer to the highest resolution shell of the respective dataset.
Figure 2. Detailed view and mutagenesis analysis of ATP binding sites.
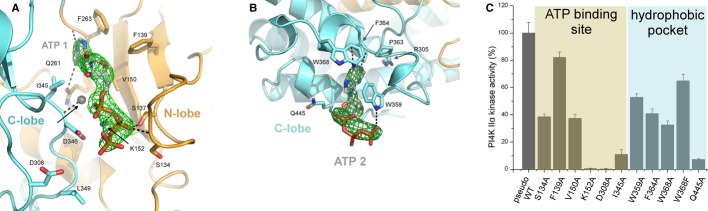
- Detailed view of the ATP binding pocket. Hydrogen bonds between ATP and residues S137, K152, Q261, D346 as well as hydrogen bonds with a coordinated water molecule and protein backbone shown as dotted black lines. The catalytically important residue D308 surprisingly does not form a direct contact with ATP as it is spaced 4.55 Å away from the ATP's closest γ-phosphate oxygen. The unbiased Fo-Fc map contoured at 2 sigma is shown around the ATP molecule. The protein backbone is shown in cartoon representation, N-lobe colored in orange, C-lobe in cyan. ATP and side chains of selected residues are shown in stick representation and colored according to elements. Oxygen is shown in red, nitrogen in blue, phosphor in orange, kinase carbon atoms are colored according to domain assignment, and ATP's carbons are in silver. Water molecule is shown as a sphere. Hydrogen atoms are not shown.
- Detailed view of the lateral hydrophobic binding pocket of the C-lobe. The pocket is formed by the hydrophobic residues W359, F364, W368 with some help of P363. The adenine ring forms only two hydrogen bonds with the protein backbone, a third hydrogen bond is formed between the W359 indole ring and the γ-phosphate of ATP. The unbiased Fo-Fc map contoured at 2 sigma is shown around the ATP molecule. The figure is colored as in (A).
- Mutagenesis analysis of the ATP binding site and the hydrophobic pocket. Selected residues of the ATP binding site were mutated to alanines, and the kinase activity was measured in vitro using recombinant proteins and the luminescent ADP-Glo kinase assay [35]. The luminescence signal was normalized to the pseudo-wt activity. The mutation of all the tested residues of the ATP binding site had profound effect on the kinase activity with the exception of F139 that is not in close vicinity of the ATP. The most profound effect was the mutations of K152 and D308. Similarly, all mutations tested in the hydrophobic pocket had profound effect. Noteworthy, the mutations W368A had a two times stronger phenotype than mutation W368F presumably because phenylalanine is also a hydrophobic residue albeit not as much as a tryptophan residue.
In summary, the structure of the PI4K IIα serves as a template for the type II PI4K enzymes. It reveals a non-typical kinase fold and an unexpected hydrophobic pocket with a second ATP bound. Its flattened fold brings the catalytic center close to the membrane-anchored substrate [26] with the γ-phosphate of the ATP pointing directly to the lipid membrane (Fig 3A).
Figure 3. Membrane binding mode of PI4K IIα.
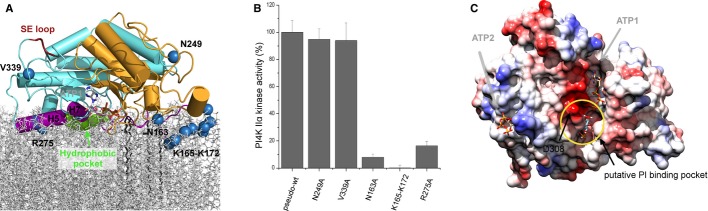
- Simulation-based model of PI4K IIα on the lipid bilayer. The kinase is bound to the membrane by several regions of the kinase that include helices H5 and H7 (highlighted in violet), the lateral hydrophobic pocket and the entire flexible segment between Pro164 and Asp182, which contains the four palmitoylated Cys residues. These regions are in direct contact with the lipid membrane, and the γ-phosphate of the ATP points toward the membrane. Residues selected for mutagenesis analysis are highlighted as blue spheres. The hydrophobic pocket is highlighted in green and the surface exposed (SE) loop in red.
- Mutagenesis analysis of the membrane binding model. Two residues predicted not to contact the membrane (V339 and N249), two residues predicted to be important for membrane binding (N163 and R275) and the entire K165–K172 membrane binding segment were chosen for in vitro mutagenesis analysis using recombinant and the luminescent ADP-Glo kinase assay. The luminescence signal was normalized to the pseudo-wt activity. While the mutations V339A or N249A had no effect on the kinase activity, both mutations N163A and R275A had significant effect and when the entire membrane binding segment K165–K172 was mutated to alanines, the kinase was inactive.
- The inositol binding pocket. Docking studies with inositol-1-P identified a putative PI binding pocket. The catalytically important aspartate residue D308 does not directly contact the ATP, instead it is a part of the putative PI binding pocket. The surface of the kinase is colored accordingly to electrostatic potential. ATPs and inositol-1-P are shown in stick representation.
The ATP binding groove
The catalytic ATP binding groove lies between the N- and C-lobes (Fig 1B). ATP is held in position by hydrogen bonds with side chains of residues K152, S137, D346, and with the protein backbone (Fig 2A). Mutagenesis analysis (Fig 2C) showed that residues defining the ATP binding site (e.g. S134, V150, I345) are important for catalytic activity, whereas mutation of a residue further away (F139A) has no significant impact on activity. We also show that the residue K152 making direct hydrogen bond with the α-phosphate is indispensable for catalytic activity. Since the D308 is too far away from the ATP, the known D308A kinase dead mutation can be only explained by D308 being part of the inositol binding pocket and/or being important for positioning a water molecule for catalysis. Unfortunately, we were unable to get crystal structure with inositol or inositol-1-P bound. However, modeling suggests that inositol can be fitted without structural or energetic constraints in a pocket in close vicinity to the ATP's γ-phosphate (Fig 3C).
Membrane binding mode of PI4K IIα
Molecular simulations show that when the kinase is tightly bound to the membrane, the ATP molecule, located in the catalytic site between the N- and C-lobes, points directly toward the lipid bilayer. Notably, the lateral hydrophobic pocket is then exposed toward the lipid bilayer (Fig 3A). This suggests that it could bind a hydrophobic ligand present within the lipid membrane in the physiological environment. In a second group of solutions obtained in our simulations, the kinase is in a ‘loose’ state where it is tethered to the membrane only by the four palmitoyl groups, the segment between K165 and K172, and the lateral hydrophobic pocket (Supplementary Fig S3C). This, probably non-physiological, second configuration is described and discussed in more detail in the Supplementary Information.
To test our model, we chose two residues at the predicted membrane interface (N163, R275) and two control residues (N249, V339) predicted not to interact with the membrane for mutational analysis. As predicted, the mutations N163A or R275A significantly lowered the kinase activity, whereas the mutations N249A or V339A had no effect (Fig 3B). In addition, when the segment 165-KWTKWLQK-172 that is predicted most important for membrane binding was mutated to alanines, the kinase became inactive (Fig 3B). However, even though that the mutants expressed at the same level as wild-type enzyme both in mammalian and in bacterial cells and that the recombinant mutant proteins behaved as wild-type during size exclusion chromatography, we cannot completely rule out that the mutants had somewhat lower kinase activity due to folding problems.
Based on these data, we propose that the enzyme is kept at the membrane by multiple interactions: First, its palmitoylation plays a very important role. Second, the amphipathic segment 165-KWTKWLQK-172 adjacent to the palmitoylation sites also makes a significant contribution, probably through immersion of hydrophobic residues into the membrane with the basic residues interacting with the phosphorylated lipid head group—a common theme among membrane binding proteins [27,28]. Third, a hydrophobic pocket with W359 and W368 also helps aligning the enzyme with the membrane. The last contributing regions identified are helices H5 and H7. The impact of the latter interactions on enzymatic activity is exaggerated in assays using recombinant enzymes that are not palmitoylated when expressed in bacteria (Fig 3). This explains why the combined mutations of W359 and W368 only partially reduced the activity of the enzyme purified from mammalian cells or assayed in the cells in situ (Fig 4).
Figure 4. Kinase activity of human PI4KIIα mutants assayed in situ and in solution.
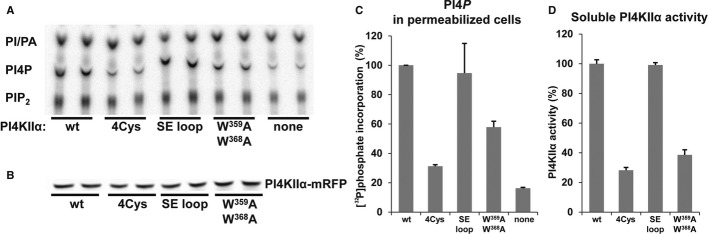
- COS-7 cells transfected with the indicated PI4KIIα-RFP plasmids were pretreated with 10 μM wortmannin (Wm) to inhibit the endogenous type III PI 4-kinases, then permeabilized with digitonin and incubated with [γ-32P]ATP and Wm for 15 min prior to phospholipid extraction. Shown is a representative TLC autoradiogram.
- Western blot showing the expression levels of PI4KIIα-RFP constructs used in (A).
- Quantification of phosphoinositide levels from two in situ PI4KIIα experiments shown in (A) performed in duplicate. Shown are grand averages ± range.
- Characterization of kinase activities of PI4KIIα-mRFP mutants affinity-purified from COS-7 cells. ADP-Glo luminescence signal was normalized to the wild-type activity after subtracting the background signal detected from untransfected cells. Shown are means ± SEM from triplicate measurements obtained in two experiments.
The hydrophobic pocket
An intriguing feature of the solved structure is the hydrophobic pocket located at the C-lobe that is occupied by a second ATP molecule. It is predominantly formed by W359, F464, and W368 residues with some help from P363 (Fig 2B). In this pocket, the ATP molecule is stabilized by only two hydrogen bonds to the backbone of residues F452 and Y453, a third hydrogen bond is formed between the γ-phosphate and the nitrogen of the W359 indole ring. It is likely that the second ATP is found in the structure only because of the lack of a more suitable hydrophobic ligand in the crystallization condition. In the real cellular environment, at the surface or within the lipid bilayer, other, more suitable hydrophobic ligands could be presented. It is tempting to speculate that the recruitment or activity of the kinase is regulated by a ligand presented on the membrane suitable for binding to the hydrophobic pocket. The hydrophobic pocket could sterically accommodate the head group of several phospholipids, including the inositol ring of PI and cholesterol that has been implied in the control of PI4K IIα palmitoylation and activity is cholesterol (Supplementary Fig S2) [20,21]. To address its functional relevance, we performed mutagenesis studies. We used permeabilized cells expressing the various mutant enzymes to assay the enzyme activity in its natural environment. We compared the double mutant within the hydrophobic pocket (mutation W359,368A) with the 4Cys palmitoylation-deficient mutant (the palmitoylated motif CCPCC is mutated to SSPSS) and with an additional mutant in which a surface exposed (SE) hydrophobic loop not predicted to be important for membrane binding in our simulations was mutated (332-WVVV-335 mutated to 332-SAAA-335) (Fig 4). We found that the palmitoylation-deficient 4Cys mutant showed the strongest impairment in kinase activity in both the permeabilized cell (Fig 4C) and in vitro assay (Fig 4D). No major effect was observed when the solvent exposed hydrophobic loop was mutated. Mutations within the hydrophobic pocket facing the membrane significantly reduced but did not eliminate the kinase activity. It is noteworthy that mutations W359A or W368A in the bacterially expressed and thus non palmitoylated pseudo-wt protein (Fig 2C) showed stronger impairment of kinase activity than the double mutant W359,368A expressed in the mammalian cells. Since the latter proteins is palmitoylated, these results suggest that the hydrophobic pocket is important for membrane binding or proper positioning and its importance becomes critical when the enzyme lacks palmitoylation. We also tested the localization of the mutant enzyme and found no obvious difference compared to wild-type except for the 4Cys mutation, which made a larger fraction of the enzyme cytoplasmic and enhanced the localization in the plasma membrane (Supplementary Fig S4). Further studies are in progress in cellular systems to address whether these mutations change the cholesterol requirement of the enzyme or if there is any specific lipid ligand that interacts with this site. However, the fact that these mutants show reduced activity even by the in vitro assays against lipid micelles suggests that this site may be promiscuous in its membrane interaction.
Concluding remarks
Taken together, our results provide structural information on PI4P synthesis at the Golgi or other membranes. They explain how the PI4K IIα interacts with the membrane. The existence of a hydrophobic pocket facing the membrane can form the structural basis for recruitment or regulation of the enzymatic activity of PI4K IIα by a membrane components. While this study was under review, another structural study on PI4K IIα was published [29]. Zhou and colleagues used a different approach and obtained crystals in which the two PI4KIIα molecules in the asymmetric unit pack one against each other at the putative membrane binding surface leaving the hydrophobic pocket empty. Nevertheless, the major findings of the two studies are in good agreement. While Zhou et al found that mutation of the W359 and W368 residues completely eliminated the enzymatic activity, our studies showed only a reduction. This difference may be due to the fact that they tested the effects of their mutations only with the recombinant protein expressed in bacteria in which all cysteines were already mutated. Our analysis using enzymes assayed in intact cells should be more informative in this respect. Regardless of these small differences, the structural information obtained here should help design further studies and aid rational drug design to better understand the roles of these enzymes in the control of trafficking and signaling in eukaryotic cells.
Materials and Methods
Plasmid construction, protein expression, and purification
PI4KIIα-mRFP was prepared from the previously described PI4KIIα-GFP construct [30] by replacing the entire GFP coding sequence with that coding the mRFP. Human PI4KIIα residues 76–479 was cloned into pRSFD vector (Novagen) with a N-terminal 6xHis tag followed by a GB1 solubility tag and TEV protease cleavage site using restriction cloning. Mutations were generated using the QuikChange kit (Stratagene) or the Phusion Site-Directed Mutagenesis Kit (Thermo Scientific). For crystallographic studies, the cysteine-rich motif LeuCysCycProCycCycPhe (residues 173–179) was replaced by a GlyThrGly linker followed by T4 lysozyme using the QuikChange kit (Stratagene) and the last 12 amino acids were deleted. The resulting protein had improved solubility and crystallization properties.
The proteins were expressed in E. coli BL21 Star and purified using affinity, anion exchange, and size exclusion chromatography as usual in our laboratory [31–33] and are detailed in the Supplementary Information.
Crystallization and data collection
Before setting up crystallization drops, PI4K IIα was supplemented with ATP and MgCl2. The crystals grew in 2 days, belonged to the P21212 space group and diffracted to 2.77 Å resolution. The structure was solved using MR-SAD with T4 lysozyme as a search model and the anomalous signal from SeMet containing crystals as detailed in the Supplementary Information. The structure was refined to Rwork = 21.15% and Rfree = 25.11%.
Replica Exchange Monte Carlo simulations
To efficiently sample physical configurations of the membrane-bound kinase, we used a coarse-grained model developed to simulate proteins both on membranes and in solution [34]. The detailed description of the coarse-grained model and simulation procedures are in the Supplementary Information. Briefly, we performed Replica Exchange Monte Carlo simulations with soft harmonic restrains imposed on the z-coordinates of the four palmitoylated Cys residues to mimic their attachment to the membrane. We analyzed the resulting 10,000 configurations as described in Supplementary Information and identified the residues that contribute to membrane interactions.
Docking studies
The docking runs were performed in AutoDock Vina using the default scoring function. The 3D structures of the docked molecules were built using ACD/ChemSketch 12.01 (http://www.acdlabs.com), and the geometry was optimized with MOPAC2012 (http://www.OpenMOPAC.net) using PM7 method. Further details are in the Supplementary Information.
Cell culture
COS-7 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100 U/ml of penicillin, and 100 μg/ml of streptomycin. COS-7 cells seeded on 12-well plates were transfected for 24 h with PI4KIIα-mRFP constructs using Lipofectamine 2000 (Life Technologies, Carlsbad, CA).
In situ PI4KIIα kinase assay
Transfected COS-7 cells were incubated for 10 min in complete medium containing 10 μM wortmannin, followed by incubation in permeabilization medium containing 110 mM KCl, 10 mM NaCl, 5 mm MgCl2, 20 mM Hepes at pH 7.4, 2 mM EGTA, 0.05% bovine serum albumin, 15 μg/ml digitonin, 0.3 mM ATP, 12.5 μCi/ml [γ-32P]ATP at 37°C, in the presence of 10 μM wortmannin. Reactions were terminated after 15 min using perchloric acid (5% final concentration). Inositol lipids were then extracted, separated by thin layer chromatography, and quantitated by phosphorimaging as described previously [30].
In vitro soluble PI4KIIα kinase assay
Lysates from COS-7 cells transiently transfected with PI4KIIα-mRFP constructs were prepared as previously described before [35]. Lysates were then incubated for 1 h with streptavidin-conjugated Dynabeads (Life Technologies, Carlsbad, CA), prebound with biotin-conjugated anti-RFP antibody (Rockland, Gilbertsville, PA). PI4KIIα-bound Dynabeads were washed and subjected to ADP-Glo kinase assay as described [35]. Recombinant proteins were used in the ADP-Glo kinase assay at 85 nM concentration. Details are stated in the Supplementary Information.
Data deposition
The crystallography, atomic coordinates, and structure factors have been deposited in the RCSB Protein Data Bank, http://www.pdb.org (accession code 4PLA).
Acknowledgments
Crystallographic data were collected at ESRF beamlines ID 14-4 and ID-23-1 and at beamlines 14-1 and 14-2 operated by the Helmholtz-Zentrum Berlin (HZB) at the BESSY II electron storage ring. This project was supported by MarieCurie FP7-PEOPLE-2012-CIG, project number 333916 (to E.B.), by Project InterBioMed LO1302 from Ministry of Education of the Czech Republic, and by the Academy of Sciences Czech Republic (RVO: 61388963). The work of B.R. was supported by the Polish National Science Centre Grant No. 2012/05/B/NZ1/00631. The work of M.J., E.W., and T.B. is supported by the intramural research program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development of the NIH, Bethesda, MD, USA.
Author contributions
AB, DC, MJ, MK, AD, EW performed experiments. BR performed computer simulations. BR, DPK, RN, TB, and EB analyzed data. TB and EB wrote the manuscript. EB designed the study.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting Information
Supplementary information for this article is available online: http://embor.embopress.org
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Information
Review Process File
References
- 1.Balla T. Phosphoinositides: tiny lipids with giant impact on cell regulation. Physiol Rev. 2013;93:1019–1137. doi: 10.1152/physrev.00028.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berridge MJ, Irvine RF. Inositol trisphosphate, a novel second messenger in cellular signal transduction. Nature. 1984;312:315–321. doi: 10.1038/312315a0. [DOI] [PubMed] [Google Scholar]
- 3.Hammond GRV, Fischer MJ, Anderson KE, Holdich J, Koteci A, Balla T, Irvine RF. PI4P and PI(4,5)P-2 are essential but independent lipid determinants of membrane identity. Science. 2012;337:727–730. doi: 10.1126/science.1222483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.D'Angelo G, Vicinanza M, Wilson C, De Matteis MA. Phosphoinositides in Golgi complex function. Subcell Biochem. 2012;59:255–270. doi: 10.1007/978-94-007-3015-1_8. [DOI] [PubMed] [Google Scholar]
- 5.Minogue S, Waugh MG, De Matteis MA, Stephens DJ, Berditchevski F, Hsuan JJ. Phosphatidylinositol 4-kinase is required for endosomal trafficking and degradation of the EGF receptor. J Cell Sci. 2006;119:571–580. doi: 10.1242/jcs.02752. [DOI] [PubMed] [Google Scholar]
- 6.Craige B, Salazar G, Faundez V. Phosphatidylinositol-4-kinase type II alpha contains an AP-3-sorting motif and a kinase domain that are both required for endosome traffic. Mol Biol Cell. 2008;19:1415–1426. doi: 10.1091/mbc.E07-12-1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jovic M, Kean MJ, Szentpetery Z, Polevoy G, Gingras AC, Brill JA, Balla T. Two phosphatidylinositol 4-kinases control lysosomal delivery of the Gaucher disease enzyme, beta-glucocerebrosidase. Mol Biol Cell. 2012;23:1533–1545. doi: 10.1091/mbc.E11-06-0553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Altan-Bonnet N, Balla T. Phosphatidylinositol 4-kinases: hostages harnessed to build panviral replication platforms. Trends Biochem Sci. 2012;37:293–302. doi: 10.1016/j.tibs.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Minogue S, Waugh MG. The phosphatidylinositol 4-kinases: don't call it a comeback. Subcell Biochem. 2012;58:1–24. doi: 10.1007/978-94-007-3012-0_1. [DOI] [PubMed] [Google Scholar]
- 10.Strahl T, Thorner J. Synthesis and function of membrane phosphoinositides in budding yeast, Saccharomyces cerevisiae. Biochim Biophys Acta. 2007;1771:353–404. doi: 10.1016/j.bbalip.2007.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carpenter CL, Cantley LC. Phosphoinositide kinases. Curr Opin Cell Biol. 1996;8:153–158. doi: 10.1016/s0955-0674(96)80060-3. [DOI] [PubMed] [Google Scholar]
- 12.Barylko B, Gerber SH, Binns DD, Grichine N, Khvotchev M, Sudhof TC, Albanesi JP. A novel family of phosphatidylinositol 4-kinases conserved from yeast to humans. J Biol Chem. 2001;276:7705–7708. doi: 10.1074/jbc.C000861200. [DOI] [PubMed] [Google Scholar]
- 13.Minogue S, Anderson JS, Waugh MG, dos Santos M, Corless S, Cramer R, Hsuan JJ. Cloning of a human type II phosphatidylinositol 4-kinase reveals a novel lipid kinase family. J Biol Chem. 2001;276:16635–16640. doi: 10.1074/jbc.M100982200. [DOI] [PubMed] [Google Scholar]
- 14.Barylko B, Mao YS, Wlodarski P, Jung G, Binns DD, Sun HQ, Yin HL, Albanesi JP. Palmitoylation controls the catalytic activity and subcellular distribution of phosphatidylinositol 4-kinase II{alpha} J Biol Chem. 2009;284:9994–10003. doi: 10.1074/jbc.M900724200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jung G, Wang J, Wlodarski P, Barylko B, Binns DD, Shu H, Yin HL, Albanesi JP. Molecular determinants of activation and membrane targeting of phosphoinositol 4-kinase IIbeta. Biochem J. 2008;409:501–509. doi: 10.1042/BJ20070821. [DOI] [PubMed] [Google Scholar]
- 16.Balla A, Tuymetova G, Tsiomenko A, Varnai P, Balla T. A plasma membrane pool of phosphatidylinositol 4-phosphate is generated by phosphatidylinositol 4-kinase type-III alpha: studies with the PH domains of the oxysterol binding protein and FAPP1. Mol Biol Cell. 2005;16:1282–1295. doi: 10.1091/mbc.E04-07-0578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang YJ, Wang J, Sun HQ, Martinez M, Sun YX, Macia E, Kirchhausen T, Albanesi JP, Roth MG, Yin HL. Phosphatidylinositol 4 phosphate regulates targeting of clathrin adaptor AP-1 complexes to the Golgi. Cell. 2003;114:299–310. doi: 10.1016/s0092-8674(03)00603-2. [DOI] [PubMed] [Google Scholar]
- 18.Wang J, Sun HQ, Macia E, Kirchhausen T, Watson H, Bonifacino JS, Yin HL. PI4P promotes the recruitment of the GGA adaptor proteins to the trans-Golgi network and regulates their recognition of the ubiquitin sorting signal. Mol Biol Cell. 2007;18:2646–2655. doi: 10.1091/mbc.E06-10-0897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jovic M, Kean MJ, Dubankova A, Boura E, Gingras AC, Brill JA, Balla T. Endosomal sorting of VAMP3 is regulated by PI4K2A. J Cell Sci. 2014;127:3745–3756. doi: 10.1242/jcs.148809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Waugh MG, Minogue S, Chotai D, Berditchevski F, Hsuan JJ. Lipid and peptide control of phosphatidylinositol 4-kinase IIalpha activity on Golgi-endosomal Rafts. J Biol Chem. 2006;281:3757–3763. doi: 10.1074/jbc.M506527200. [DOI] [PubMed] [Google Scholar]
- 21.Lu D, Sun HQ, Wang H, Barylko B, Fukata Y, Fukata M, Albanesi JP, Yin HL. Phosphatidylinositol 4-kinase IIalpha is palmitoylated by Golgi-localized palmitoyltransferases in cholesterol-dependent manner. J Biol Chem. 2012;287:21856–21865. doi: 10.1074/jbc.M112.348094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mossinger J, Wieffer M, Krause E, Freund C, Gerth F, Krauss M, Haucke V. Phosphatidylinositol 4-kinase IIalpha function at endosomes is regulated by the ubiquitin ligase Itch. EMBO Rep. 2012;13:1087–1094. doi: 10.1038/embor.2012.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barylko B, Wlodarski P, Binns DD, Gerber SH, Earnest S, Sudhof TC, Grichine N, Albanesi JP. Analysis of the catalytic domain of phosphatidylinositol 4-kinase type II. J Biol Chem. 2002;277:44366–44375. doi: 10.1074/jbc.M203241200. [DOI] [PubMed] [Google Scholar]
- 24.Burke JE, Inglis AJ, Perisic O, Masson GR, McLaughlin SH, Rutaganira F, Shokat KM, Williams RL. Structures of PI4KIIIbeta complexes show simultaneous recruitment of Rab11 and its effectors. Science. 2014;344:1035–1038. doi: 10.1126/science.1253397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim do J, Park KS, Kim JH, Yang SH, Yoon JY, Han BG, Kim HS, Lee SJ, Jang JY, Kim KH, et al. Helicobacter pylori proinflammatory protein up-regulates NF-kappaB as a cell-translocating Ser/Thr kinase. Proc Natl Acad Sci USA. 2010;107:21418–21423. doi: 10.1073/pnas.1010153107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rao VD, Misra S, Boronenkov IV, Anderson RA, Hurley JH. Structure of type IIbeta phosphatidylinositol phosphate kinase: a protein kinase fold flattened for interfacial phosphorylation. Cell. 1998;94:829–839. doi: 10.1016/s0092-8674(00)81741-9. [DOI] [PubMed] [Google Scholar]
- 27.Hurley JH, Boura E, Carlson LA, Rozycki B. Membrane budding. Cell. 2010;143:875–887. doi: 10.1016/j.cell.2010.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boura E, Hurley JH. Structural basis for membrane targeting by the MVB12-associated beta-prism domain of the human ESCRT-I MVB12 subunit. Proc Natl Acad Sci USA. 2012;109:1901–1906. doi: 10.1073/pnas.1117597109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou Q, Li J, Yu H, Zhai Y, Gao Z, Liu Y, Pang X, Zhang L, Schulten K, Sun F, et al. Molecular insights into the membrane-associated phosphatidylinositol 4-kinase IIalpha. Nat Commun. 2014;5:3552. doi: 10.1038/ncomms4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Balla A, Tuymetova G, Barshishat M, Geiszt M, Balla T. Characterization of type II phosphatidylinositol 4-kinase isoforms reveals association of the enzymes with endosomal vesicular compartments. J Biol Chem. 2002;277:20041–20050. doi: 10.1074/jbc.M111807200. [DOI] [PubMed] [Google Scholar]
- 31.Rezabkova L, Boura E, Herman P, Vecer J, Bourova L, Sulc M, Svoboda P, Obsilova V, Obsil T. 14-3-3 protein interacts with and affects the structure of RGS domain of regulator of G protein signaling 3 (RGS3) J Struct Biol. 2010;170:451–461. doi: 10.1016/j.jsb.2010.03.009. [DOI] [PubMed] [Google Scholar]
- 32.Boura E, Rezabkova L, Brynda J, Obsilova V, Obsil T. Structure of the human FOXO4-DBD-DNA complex at 1.9 A resolution reveals new details of FOXO binding to the DNA. Acta Crystallogr D Biol Crystallogr. 2010;66:1351–1357. doi: 10.1107/S0907444910042228. [DOI] [PubMed] [Google Scholar]
- 33.Nemecek D, Boura E, Wu W, Cheng N, Plevka P, Qiao J, Mindich L, Heymann JB, Hurley JH, Steven AC. Subunit folds and maturation pathway of a dsRNA virus capsid. Structure. 2013;21:1374–1383. doi: 10.1016/j.str.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim YC, Hummer G. Coarse-grained models for simulations of multiprotein complexes: application to ubiquitin binding. J Mol Biol. 2008;375:1416–1433. doi: 10.1016/j.jmb.2007.11.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tai AW, Bojjireddy N, Balla T. A homogeneous and nonisotopic assay for phosphatidylinositol 4-kinases. Anal Biochem. 2011;417:97–102. doi: 10.1016/j.ab.2011.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Information
Review Process File


