SUMMARY
While the catalog of mammalian transcripts and their expression levels in different cell types and disease states is rapidly expanding, our understanding of transcript function lags behind. We present a robust technology enabling systematic investigation of the cellular consequences of repressing or inducing individual transcripts. We identify rules for specific targeting of transcriptional repressors (CRISPRi), typically achieving 90–99% knockdown with minimal off-target effects, and activators (CRISPRa) to endogenous genes via endonuclease-deficient Cas9. Together they enable modulation of gene expression over a ~1000-fold range. Using these rules, we construct genome-scale CRISPRi and CRISPRa libraries, each of which we validate with two pooled screens. Growth-based screens identify essential genes, tumor suppressors and regulators of differentiation. Screens for sensitivity to a cholera-diphtheria toxin provide broad insights into the mechanisms of pathogen entry, retro-translocation and toxicity. Our results establish CRISPRi and CRISPRa as powerful tools that provide rich and complementary information for mapping complex pathways.
INTRODUCTION
Dramatic advances in sequencing technology have catalogued a universe of transcribed loci—greatly exceeding the number of canonical protein-coding open reading frames (ORFs)—which collectively are responsible for carrying out the instructions encoded by the genome (Djebali et al., 2012). A central challenge now is to understand the biological role of these transcripts and how quantitative differences in their expression define cellular states in normal development and in disease. Despite intense efforts, the function of many protein-coding genes remains poorly defined. Even less is known about the biological roles of most non-canonical transcripts such as enhancer RNAs, upstream antisense RNAs, lncRNAs, or other intergenic RNAs (Cech and Steitz, 2014). Efforts to address this deficiency in our knowledge would be greatly aided by techniques that are capable of dynamically and precisely controlling the expression of individual transcripts.
One way to explore the function of genes is to disrupt their expression through repression. The dominant tool for programmed knockdown of mRNAs is RNA interference (RNAi) (Chang et al., 2006). However, RNAi has pervasive problems with off-target effects, which can be especially confounding in the context of large-scale screens (Adamson et al., 2012; Jackson et al., 2003; Sigoillot et al., 2012). Additionally, because RNAi is mediated by cytoplasmic argounaute proteins, gene silencing through this approach is best suited to depletion of cytosolic mRNA targets.
An alternative emerging strategy is the use of programmable genome editing methods that permanently delete or modify DNA using designable, sequence-specific endonucleases such as zinc finger, transcription activator-like effector (TALE) nucleases, or CRISPR (clustered regularly interspaced short palindromic repeats)/Cas9 (CRISPR-associated protein 9) proteins (Gaj et al., 2013; Sander and Joung, 2014). A series of elegant studies recently exploited the readily programmable nature of Cas9, in which the specificity is determined by a short guide (sg)RNA, to enable genome-scale loss-of-function screens (Koike-Yusa et al., 2014; Shalem et al., 2014; Wang et al., 2014). These studies established CRISPR-mediated cutting as a powerful screening technology complementary to RNAi and haploid mutagenesis screens (Carette et al., 2009). Nonetheless, screening approaches based on genome editing are currently focused on loss-of-function studies involving irreversible frameshift disruptions, limiting their utility for the study of essential genes and long noncoding RNAs. Additionally, double-stranded DNA breaks can be cytotoxic (Huang et al., 1996; Jackson, 2002). Finally, indels formed from error-prone DNA repair are often short and in-frame, which could limit the ability to disable all of the alleles of a gene.
A programmable DNA binding protein that can recruit an effector domain to turn transcription on and off in a dynamic and quantitative manner offers, in principle, a more flexible tool for interrogating the many transcripts in complex genomes. Pioneering experiments with designed chimeric zinc finger and TALE proteins fused to transcription effector domains demonstrate that such an approach can modulate transcription of endogenous genes (Beerli et al., 1998, 2000; Zhang et al., 2011). However, as each transcript target requires a unique fusion protein, expanding these methods to genome-scale is arduous.
Recently, we and others have used catalytically inactive Cas9 (dCas9) fusion proteins guided by gene-specific sgRNAs to localize effector domains to specific DNA sequences to either repress (CRISPRi) or activate (CRISPRa) transcription of target genes (Gilbert et al., 2013; Sander and Joung, 2014). To date, a small number of sgRNAs have been tested, leaving unanswered whether CRISPRi/a is a feasible strategy for globally interrogating gene function and, if so, how best to target a gene to activate or repress transcription while minimizing off-target effects.
Here, we describe the development and application of a method for high-specificity, genome-scale modulation of transcription of endogenous genes in human cells using CRISPRi/a. To accomplish this, we first performed a saturating screen in which we tested the activity of every unique sgRNA broadly tiling around the transcription start sites (TSSs) of 49 genes known to modulate cellular susceptibility to ricin (Bassik et al., 2013). From this, we extracted distinct rules for regions where either CRISPRi or CRISPRa maximally changes the expression of endogenous genes, as well as rules for predicting off-target effects, providing an algorithm to design two genome-scale libraries targeting each gene with 10 sgRNAs. We validated these libraries by screening for genes that control cell growth and response to a chimeric cholera/diphtheria fusion toxin (CTx-DTA) (Guimaraes et al., 2011). These experiments demonstrate that our CRISPRi/a screening platform is robust, showing high reproducibility and activity with undetectable intrinsic toxicity. More generally, we establish that transcriptional repression is inducible, reversible, and can target essential genes. We demonstrate that we can use CRISPRi and CRISPRa to control transcript levels for endogenous genes across a high dynamic range. We also provide extensive evidence that properly designed CRISPRi reagents are highly specific. As such, these methods represent transformative tools for defining transcript function across the breadth of transcripts encoded by the human genome.
RESULTS
A High-Throughput Tiling Screen Defines Rules for CRISPRi Activity at Endogenous Genes
CRISPRi can repress transcription by directly blocking RNA polymerase activity (dCas9) or through effector domain-mediated transcriptional silencing (dCas9-KRAB) (Gilbert et al., 2013; Qi et al., 2013). In order to better understand and optimize CRISPRi activity, we used a pooled high-throughput screen to define rules that determine CRISPRi repression of endogenous genes. We targeted 49 genes that we had previously shown to modulate cellular susceptibility to the AB toxin ricin (Bassik et al., 2013). The extent of gene repression for these genes typically has a monotonic relationship with the ricin resistance phenotype, allowing us to use a ricin resistance score calculated by monitoring sgRNA frequencies in a pooled screen to indirectly measure transcriptional repression.
Using massively parallel oligonucleotide synthesis, we generated a library of sgRNAs that tile the DNA in a 10-kilobase window around the TSS of these 49 genes (54,810 total sgRNAs) (Bassik et al., 2009) (Figure 1A). We also included 1,000 negative control sgRNAs derived from scrambled sequences corresponding to the same windows.
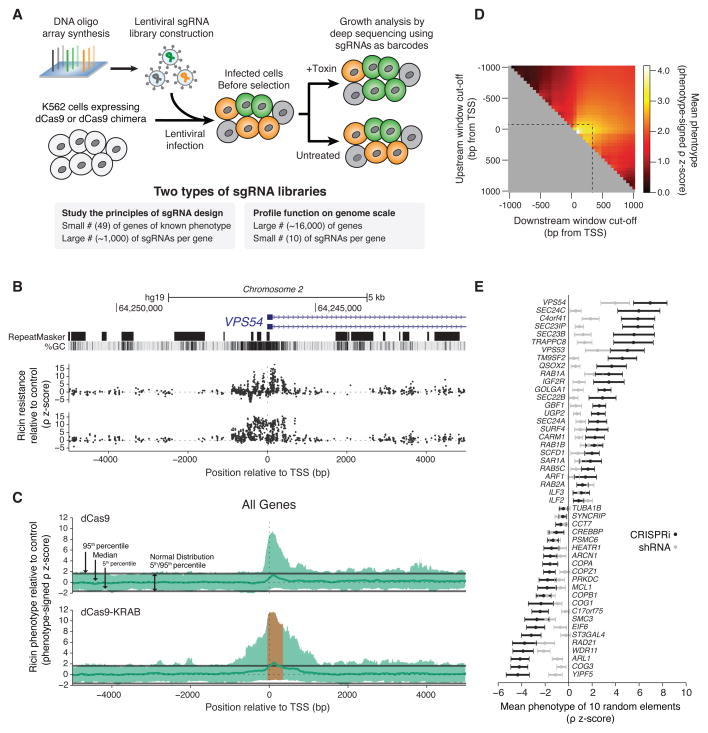
Figure 1. A Tiling sgRNA Screen Defines Rules for CRISPRi Activity at Endogenous Genes in Human Cells.
(A) Massively parallel determination of growth or toxin resistance phenotypes caused by sgRNAs in mammalian cells expressing dCas9 or dCas9 fusion constructs. (B) UCSC tracks showing the genomic organization, GC content, and repetitive elements around the TSS of a representative gene, VPS54, across a 10kb window targeted by the tiling sgRNA library. sgRNA ricin resistance phenotypes (as z-scores, see Figure S1 and Experimental Procedures) in dCas9 and dCas9-KRAB expressing K562 cells are depicted in black on the top and bottom, respectively. See also Figure S2A for more examples. (C) Sliding-window analysis of all 49 genes targeted in a tiling sgRNA library. Green line: median sgRNA activity in a defined window for all genes. Orange region: observed average window of maximum CRISPRi activity. Data displayed as a phenotype signed z-score, excluding all guides longer than 22bp. (D) CRISPRi activity for all 49 genes in defined windows relative to the TSS of each gene. (E) Ricin resistance phenotypes, comparing CRISPRi sgRNAs selected by our rules to RNAi, for genes previously established to cause ricin resistance phenotypes when knocked down by RNAi. Mean phenotype-signed z-score for 10 subsampled sgRNAs or shRNAs. See also Figure S2F.
We packaged this tiling library of sgRNAs into lentiviral particles and transduced K562 human myeloid leukemia cells stably expressing dCas9 or a dCas9-KRAB fusion protein, which we have previously described (Gilbert et al., 2013). We harvested populations of cells expressing the library either at the outset of the experiment, after growth under standard conditions, or following ricin treatment. We then counted the frequency of each sgRNA in the library in each sample using deep sequencing to determine how each sgRNA in the library modulates cell growth and cellular susceptibility to ricin phenotypes. We defined these phenotypes quantitatively as gamma (γ) and rho (ρ), respectively (See Figure S1A and (Kampmann et al., 2013)).
Many sgRNAs potently repress gene expression, as evidenced by their impact on ricin sensitivity (Figure 1B and Figure S2A). Plotting this data for all 49 genes showed that active sgRNAs cluster around or just downstream from the TSS of each gene for dCas9-KRAB and dCas9, respectively (Figure 1C). We saw that strong CRISPRi activity is obtained by targeting dCas9-KRAB to a window of DNA from −50 to +300 bp relative to the TSS of a gene, with a maximum in the ~50–100 bp region just downstream of the TSS (Figure 1C–D). This suggested that optimal activity leverages the combined activity of dCas9 interference along with repression from the KRAB domain. We also observed that sgRNAs with protospacer lengths of 18–21 base pairs were significantly more active than sgRNAs containing longer protospacers (Figure S2B). Nucleotide homopolymers had a strongly negative effect on sgRNA activity (Figure S2D). However, neither the DNA strand that was targeted nor the sgRNA GC content across a broad range strongly correlated with sgRNA activity (Figure S2C and S2E).
To evaluate the feasibility of genome-scale genetic screens based on CRISPRi, we compared the strength of phenotypes obtained with CRISPRi to our previously published shRNA data. We applied the rules described above to randomly subsampled data from our sgRNA tiling library, picking sets of 10 or 24 sgRNAs. We calculated a normalized phenotype z-score by dividing mean phenotypes for each gene by the standard deviation of sgRNA phenotypes from the non-targeting control set (Figure S1B). We see significant ricin phenotypes for each of the 49 genes. Moreover, in virtually every case the normalized ricin phenotype z-score or p-value is stronger (in many cases far stronger) than seen with a comparably-sized shRNA library (generated by sub-sampling our published data) (Figure 1E and Figure S2F).
CRISPRi Transcriptional Silencing is Highly Sensitive to Mismatches between the Target DNA Site and the sgRNA
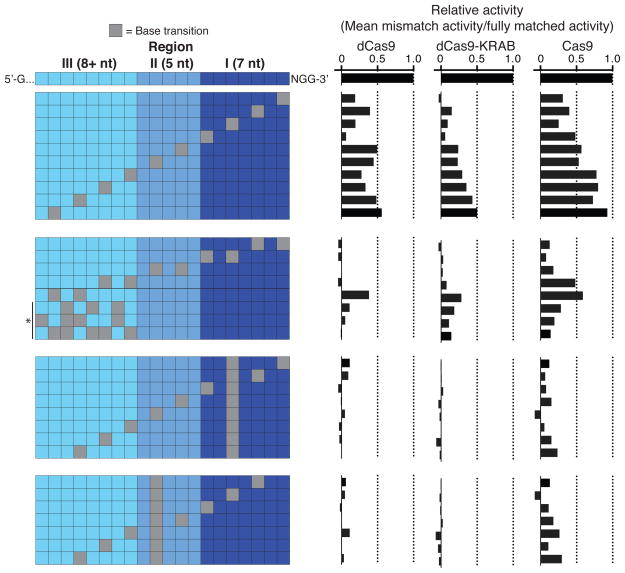
To assess CRISPRi off-target activity at endogenous genes, we selected a set of 30 sgRNAs from our tiling library (6 sgRNAs/gene targeting 5 genes). For each of these sgRNAs, we tested the activity of a series of derivative sgRNAs with a variable number and position of mismatches (Figure 2). This experiment allowed us to measure the relative amount of gene repression for sgRNAs with or without mismatch base pairing targeting the same DNA locus. We found that even a single mismatch at the 3′ end of the protospacer decreased CRISPRi activity on average, while combinations of mismatches that pass our off-target filter abolished activity (Figure 2, Figure S3, and Extended Experimental Procedures). From this analysis, we concluded that properly designed CRISPRi sgRNAs have minimal off-target transcriptional repression activity.
Figure 2. CRISPRi Activity is Highly Sensitive to Mismatches Between the sgRNA and DNA sequence.
On- and off-target activity of dCas9, dCas9-KRAB and Cas9 for sgRNAs with a varying number and position of mismatches. Off-target activity of sgRNAs with mismatches is displayed as percent of the on-target activity for the corresponding sgRNA without mismatches. Asterisk indicates sgRNAs with 3, 4, or 5 mismatches randomly distributed across region 3 of the sgRNA sequence. Data is displayed for each mismatch position as the mean of all sgRNAs with that mismatch; see Figure S3 for individual sgRNA activities. sgRNAs were included in the analysis only if the fully matched guide was highly active (phenotype-signed z-score ≥ 4); N=5 for dCas9, N=11 for dCas9-KRAB, and N=10 for Cas9.
A High-Throughput Tiling Screen Defines Rules for CRISPRa Activity at Endogenous Genes
We recently developed an improved CRISPRa method, termed sunCas9, in which expression of a single sgRNA with one binding site is sufficient to robustly activate transcription (Tanenbaum et al.) In the sunCas9 system, a single dCas9 fusion protein bound to DNA recruits multiple copies of the activating effector domain, thus amplifying our ability to induce transcription (Figure 3A).
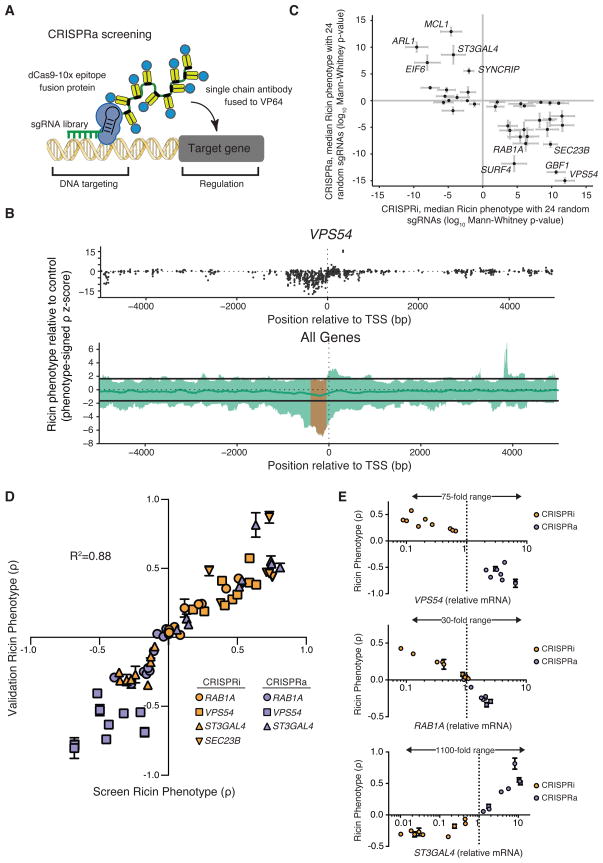
Figure 3. A Tiling sgRNA Screen Defines Rules for CRISPRa Activity at Endogenous Genes in Human Cells.
(A) A schematic of the dCas9-SunTag + scFV−VP64 + sgRNA system for CRISPRa. (B) Activity of sgRNAs in K562 cells stably expressing each component of CRISPRa, as a function of the distance of the sgRNA site to the TSS of the targeted gene (Phenotype-signed z-scores; therefore, negative values represent opposite results than from knockdown). Top, sgRNAs targeting VPS54; Bottom, sliding-window analysis of all 49 genes targeted by our tiling library in green. Green line, median activity; orange, window of maximal activity. Guides longer than 22bp were excluded. See also Figure S4. (C) CRISPRa phenotypes and CRISPRi (dCas9-KRAB) phenotypes are anti-correlated for select genes. For each gene, a p-value is calculated using CRISPRi/a sgRNA activity relative to a negative control distribution for 24 sub-sampled sgRNAs. (D) CRISPRi knockdown and CRISPRa activation of the same gene can have opposing effects on ricin resistance in both primary screens and single sgRNA validation experiments (mean ± standard deviation of 3 replicates). (E) Modulation of expression levels for 3 genes by CRISPRi and CRISPRa as quantified by qPCR plotted against the ricin resistance phenotype (mean ± standard deviation of 3 replicates) measured for each sgRNA.
To define rules for optimal CRISPRa sgRNA design, we used our tiling library, which targets genes capable of modulating cellular sensitivity to ricin. We previously showed for several of the genes in this tiling library that knockdown and plasmid overexpression resulted in opposite ricin phenotypes (Bassik et al., 2013). For example, knockdown of SEC23B sensitized cells to ricin, whereas SEC23B overexpression desensitized cells to ricin. These observations suggested that we should be able to observe reversed phenotypes in this tiling screen arising from CRISPRa activity.
We transduced K562 cells stably expressing the sunCas9 system (Figure 3A) with the sgRNA tiling library and screened for ricin phenotypes as described for CRISPRi above. Analysis of data for individual genes or averaged data for all 49 genes demonstrated that many sgRNAs for each gene affected ricin resistance (Figure 3B and Figure S4A–B). Our negative control sgRNAs showed very little activity and were not correlated between biological replicate screens, suggesting that CRISPRa activity is specific. We observed a peak of active sgRNAs for CRISPRa at −400 to −50 bp upstream from the TSS (Figure 3B). This activity pattern fits with a model in which each VP16 domain can bind the mediator complex and recruit basal transcription machinery, activating transcription when spaced appropriately from a TSS (Mittler et al., 2003). With this system, we have shown we can turn on genes that are poorly expressed and increase the expression of well-expressed genes (Figure 3E). Overall, our CRISPRi/a tiling screens provide rules for how CRISPRi/a controls expression of endogenous genes.
An Allelic CRISPRi/a Series of Transcript Activation and Repression Shows that Protein Abundance Dynamically Modulates the Cellular Response to Ricin
For many genes, we do not know how the relative abundance of the encoded protein relates to its function. We observed a marked anti-correlation in our ricin screens between CRISPRa phenotypes and CRISPRi phenotypes for individual genes (Figure 3C). As the genes targeted by our tiling library were selected based on a knockdown phenotype, all genes showed phenotypes in the CRISPRi screen, but only a subset showed phenotypes in the CRISPRa screen.
To validate results from both the CRISPRi and CRISPRa tiling screens, we selected an allelic series of sgRNAs by phenotype from the screen and re-tested each sgRNA individually (38 sgRNAs targeting 4 genes). Our results show that our CRISPRi/a screens produced reliable phenotype scores, robustly reproduced upon retesting, and that CRISPRi/a can activate and repress the transcription of endogenous genes over a wide dynamic range (up to ~1000-fold) (Figure 3D–E), enabling systematic interrogation of how gene dosage controls cellular functions of interest.
A Robust and Highly Specific Genome-Scale CRISPRi Screening Platform
The results of our tiling CRISPRi screen established our ability to pick active sgRNAs with low off-target activity and provided a set of rules enabling us to design a robust genome-scale sgRNA library. We chose a library size of 10 sgRNAs/gene for the following reasons. Over half of the sgRNAs conforming to these rules gave clear ricin phenotypes. For a library with 10 sgRNAs/gene, 94% of the genes would thus have 2 or more highly active sgRNAs. Finally, computational sub-sampling of the phenotypic data from our tiling library data to 10 sgRNAs/gene and calculation of z-scores for hit genes indicated that a library with 10 sgRNAs/gene would reliably detect hit genes (Figure 1E).
We synthesized and cloned a genome-scale CRISPRi sgRNA library targeting 15,977, human protein-coding genes (10 sgRNAs/TSS, targeting 20,898 TSS) with 11,219 non-targeting control sgRNAs for a total of 206,421 sgRNAs (Table S2). To evaluate this library, we first screened for genes essential for cell growth in K562 cells. Briefly, K562 cells stably expressing dCas9-KRAB were transduced in replicate with the entire genome-scale library, and each replicate was grown for 10 days at a minimum library coverage of 3,750 cells/sgRNA in a single spinner flask.
To characterize our screening methodology and library design, we examined the correlation between screen replicates and our negative control distributions. Individual sgRNAs reproducibly showed dramatic depletion (up to 256-fold) over a 10-day screen, demonstrating that individual sgRNAs can have profound effects on cell growth (Table S2) (Figure 4A). The distribution of our negative-control sgRNAs was very narrow with little correlation between replicates (Spearman R = 0.036), suggesting that the off-target activity of these controls is very low (Figure 4A). Indeed, 99.7% of our negative controls had no detectable activity. The observed specificity is consistent with our previously published RNA-seq data (Gilbert et al., 2013).
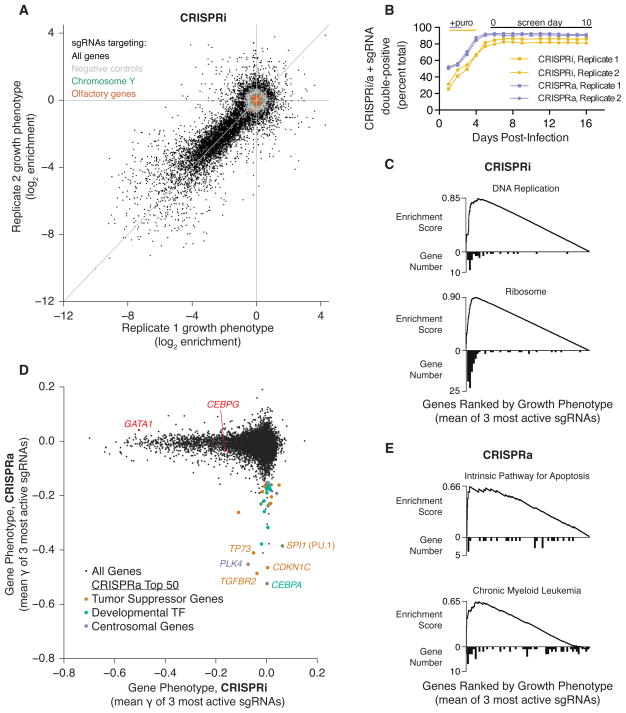
Figure 4. Genome-Scale CRISPRi and CRISPRa Screens Reveal Genes Controlling Cell Growth.
(A) sgRNA phenotypes from a genome-scale CRISPRi screen for growth in human K562 cells (black). Three classes of negative control sgRNAs are color-coded: non-targeting sgRNAs (grey), sgRNAs targeting Y-chromosomal genes (green) and sgRNAs targeting olfactory genes (orange). (B) Co-expression of sgRNAs and dCas9-KRAB or dCas9-SunTag + scFV−VP64 is not toxic in K562 cell lines over 16 days. (C) Gene set enrichment analysis (GSEA) for hits from the CRISPRi screen. A histogram of gene distribution is shown under the GSEA curve. (D) CRISPRi versus CRISPRa gene phenotypes for genome-scale growth screens (black). For the 50 genes in the CRISPRa screen with the most negative growth phenotype, each gene was annotated and labeled based on evidence of activity as a tumor suppressor (orange), developmental transcription factor (green), or in regulation of the centrosome (purple). Two additional CRISPRi hit genes that are discussed in the text are labeled in red. See Table S4 for annotations and references. (E) GSEA for hits from the CRISPRa growth screen. A histogram of gene distribution is shown under the GSEA curve.
To further explore the prevalence of off-target effects, we examined two classes of genes not expected to show any on-target activity in our screen: olfactory receptors and genes on the Y chromosome. The sgRNAs that target these genes were designed and picked in the same manner as the rest of library; however, olfactory receptors should not be expressed in this cell type and, as K562 cells are derived from a female donor, sgRNAs that target genes on the Y chromosome lack a DNA target. As with the negative controls, these genes show no phenotype on average and exhibit very little correlation between replicates (Spearman R = 0.057 for olfactory genes and −0.052 for Y-targeting) (Figure 4A). We also observed no evidence of non-specific toxicity due to expression of dCas9-KRAB and our sgRNA library in K562 cells suggesting that dCas9 bound to the genome is not toxic under these conditions (Figure 4B). Thus, CRISPRi is highly specific and non-toxic.
To identify hit genes in this screen, we used a metric of average growth phenotype (γ) for the top three sgRNAs for each gene (see Experimental Procedures and Table S3). Among the top hits were genes involved in essential cellular functions, including translation, transcription and DNA replication (Figure 4C and Figure S5A) (Huang et al., 2009a, 2009b), thus validating our approach as a screening platform.
A Genome-Scale CRISPRa Screening Platform
The results of our CRISPRa tiling screen established our ability to confidently measure gene phenotypes resulting from inducing expression with single sgRNAs. As with the CRISPRi tiling screens, our data enabled the development of a set of rules that allowed construction of a genome-scale CRISPRa library. Many of these rules overlapped with those of CRISPRi (e.g., sgRNA length and sequence preferences). A key difference is that the optimal window for targeting sgRNAs for CRISPRa lies upstream of the TSS (Figure 3B). We therefore constructed an independent CRISPRa library, designing 10 sgRNAs between 400 to 50 base pairs upstream of each TSS for 15,977 human genes, along with 5,968 non-targeting control sgRNAs, for a total of 198,810 sgRNAs.
We evaluated our CRISPRa platform in a screen for genes that affect cell growth when induced in K562 cells constitutively expressing the sunCas9 system. Replicate screens were conducted as described above. The magnitude of growth defects seen in our CRISPRa screen was comparable to that of the above CRISPRi screen, although fewer sgRNAs caused a growth phenotype (Figure S5B and Table S2). We analyzed control sgRNAs with no genomic target or Y chromosome targets and found minimal phenotypes, which lacked substantial correlation between experimental replicates (Spearman R = 0.155 and R = 0.010, respectively), indicating that the phenotype distribution observed in non-targeting controls was primarily a result of stochastic noise rather than off-target effects. Furthermore, the fraction of cells expressing sgRNAs and the sunCas9 system was stable over the course of the experiment, indicating that there was no general toxicity associated with the CRISPRa platform (Figure 4B). These data suggest that CRISPRa is also specific and non-toxic.
Defining Regulators of Survival and Differentiation in Human Cells by CRISPRa
We then investigated the genes whose induction caused cells to deplete over the course of our CRISPRa screen. We scored genes by the average γ of the three most active sgRNAs as above, and compared these phenotypes to those observed in the CRISPRi screen (Figure 4D and Table S3). The results from the two screens had little overlap, suggesting that few genes are both essential and toxic upon induction, and that wild-type expression levels of genes are generally optimal for K562 growth. Whereas CRISPRi hits are naturally limited to expressed genes, CRISPRa hits included genes across a broad range of endogenous expression levels (Figure S5D). We observed that the majority of genes that inhibited growth in the CRISPRa screen fell into three overlapping classes.
The first class was tumor suppressor genes: 18 of the top 50 genes, including six of the top seven, are known to have potent tumor suppressor activity (Vogelstein et al., 2013; Zhao et al., 2013). These genes include p53-related protein TP73, cell cycle inhibitors CDKN1C (p57) and CDKN1A (p21), apoptotic factors BAK1 and BCL2L11 (BIM), and chromatin remodeling factor ARID1A (Figure 4D and Table S4). Gene set enrichment analysis (GSEA) confirmed this observation, highlighting several genes important in the intrinsic pathway of apoptosis or in chronic myeloid leukemia (CML) homeostasis consistent with the origin of K562 cells as a clonal isolate from a CML blast crisis (ATCC) (Figure 4E). Similarly, top gene ontology annotations included “positive regulation of apoptosis” and “regulation of cell cycle” (Figure S5C). While tumor suppressors are classically considered to be mutated early in cancer progression (Vogelstein et al., 2013), these results demonstrate than many potential tumor suppressor genes remain functional but down-regulated, and suggest that CRISPRa can be used to pinpoint intact pathways and vulnerabilities in tumor cells.
Transcription factor families with well-established roles in tissue development and differentiation represent another class of growth hits, accounting for 16 of the top 50 genes (K562 cells have known potential to undergo erythroid differentiation). These genes include CCAAT/Enhancer-binding proteins (CEBP), Homeobox genes, Forkhead box genes, Ikaros family zinc finger proteins, and hematopoietic differentiation factor SPI1 (PU.1) (Figure 4D and Table S4) (Spitz and Furlong, 2012). This observation is reflected in enriched annotations relating to multicellularity, cell differentiation, and development (Figure S5C).
The complementary nature of the CRISPRi and CRISPRa screens is nicely illustrated by results from two gene pairs (SPI1/GATA1 and CEBPA/CEBPG) in which one member of each pair inhibits the function of the other. GATA1 and CEBPG were strong hits in the CRISPRi screen, consistent with their roles as inhibitors of myeloid differentiation. By contrast, both SPI1 and CEBPA were robust hits in our CRISPRa activation screen. These observations are consistent with the inhibitory functions of SPI1 and CEBPA: silencing of CEBPA leads to de-repression of CEBPG (Alberich-Jordà et al., 2012) and the protein encoded by SPI1 (PU.1) is a direct binding partner of GATA-1 and inhibits its transcriptional activity (Zhang et al., 2000).
Finally, several hit genes have key roles in mitosis. PLK4 controls centrosome duplication, and overexpression of the gene in U2OS cells leads to increased centriole number (Habedanck et al., 2005). The proteins encoded by KIF18B and KIF2C form a complex that destabilizes microtubules during mitosis (Tanenbaum et al., 2011).
Overall, the results from our paired CRISPRi/a growth screens demonstrate that complementary information can be obtained by loss- and gain-of-function genetic screens, and highlight the utility of the platform for future studies into tumor biology and cell differentiation.
Dynamically Controlling Gene Expression with CRISPRi
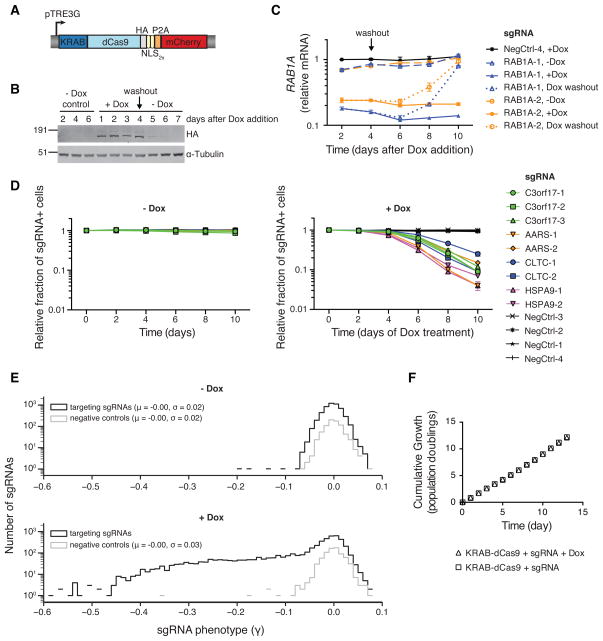
The ability to reversibly tune the expression of select transcripts would be a powerful tool for evaluating transcript function. To evaluate the applicability of CRISPRi to this purpose, we cloned a lentiviral expression construct that places an optimized KRAB-dCas9 fusion protein under the control of a doxycycline-inducible promoter (Figure 5A–B). Induced expression of KRAB-dCas9 robustly depletes transcript levels from sgRNA-targeted genes (Figure 5C and S5E). To further assess dynamic control of CRISPRi, we inducibly repressed several genes identified in our genome-scale CRISPRi growth screen (Figure S5G). Cells that express sgRNAs targeting these essential genes showed almost no growth phenotype in the absence of doxycycline, but rapidly and robustly disappeared from the population upon addition of doxycycline (Figure 5D). Additionally, gene repression and resulting phenotypes were reversible (Figure 5C and S5E–F), indicating that KRAB-dCas9 does not create a permanently repressive chromatin state at targeted promoters.
Figure 5. CRISPRi Gene Silencing is Inducible, Reversible, and Non-Toxic.
(A) Expression construct encoding an inducible KRAB-dCas9 fusion protein. (B) Western blot analysis of inducible KRAB-dCas9 in the absence, presence, and after washout of doxycycline. (C) Relative RAB1A expression levels (as quantified by qPCR) in inducible CRISPRi K562 cells transduced with RAB1A-targeting sgRNAs in the absence, presence, and after washout of doxycycline. Mean ± standard error of technical replicates (N=2) normalized to control cells (assayed in the presence of doxycycline) from the day 2 time point. (D) Competitive growth assays performed with inducible CRISPRi K562 cells transduced with the indicated sgRNAs in the presence and absence of doxycycline. Data is represented as the mean ± standard deviation of replicates (N=3). See also Figure S5G. (E) A CRISPRi sublibrary screen for effects on cell growth was performed with inducible CRISPRi K562 cells in the presence and absence of doxycycline. (F) Cumulative growth curves from the sublibrary screen represented in (E) show no bulk changes to growth caused by induction of KRAB-dCas9. Mean ± standard deviation of replicate infections each screened in duplicate.
To test our ability to dynamically control expression of essential genes on a larger scale, we cloned a sublibrary targeting 426 manually curated genes (10 sgRNAs/TSS or 4,923 targeting sgRNAs plus 750 non-targeting controls). This library was transduced into K562 cells stably expressing our inducible KRAB-dCas9 fusion protein, and cell growth effects were then evaluated in the presence and absence of doxycycline. Only 4 sgRNAs were depleted strongly in the absence of doxycycline; however, with induction of KRAB-dCas9, many sgRNAs were strongly depleted (Figure 5E). Negative control sgRNAs again produced a narrow distribution of phenotypes with little correlation between biological replicates with or without doxycycline. Additionally, we found no evidence that targeted KRAB-dCas9 generally impedes cell growth (Figure 5F). Taken together, these results demonstrate CRISPRi is non-toxic, inducible and reversible.
A Genome-Scale CRISPRi Screen Reveals Pathways and Complexes that Govern Response to Cholera and Diphtheria Toxin
To test the performance of our CRISPRi approach for detecting genes controlling a more complex cellular phenotype, we performed a genome-scale CRISPRi screen for genes that modulate sensitivity to a chimeric toxin composed of the diphtheria toxin catalytic A subunit covalently linked to cholera toxin (CTx-DTA, Figure 6A). This chimera had been previously developed to provide a growth readout for cholera intoxication (Guimaraes et al., 2011). Some aspects of both cholera and diphtheria toxin entry and toxicity are well characterized but open questions remain. The cell surface receptor for cholera toxin is the GM1a ganglioside (Van Ness et al., 1980). After endocytosis, the toxin traffics via the Golgi to the endoplasmic reticulum (ER), from which it retro-translocates into the cytosol, possibly through the ER-associated degradation (ERAD) machinery. Once in the cytosol, the DTA moiety ADP-ribosylates the diphthamide residue in Elongation Factor 2, halting translation and killing the cell (Figure 6A).
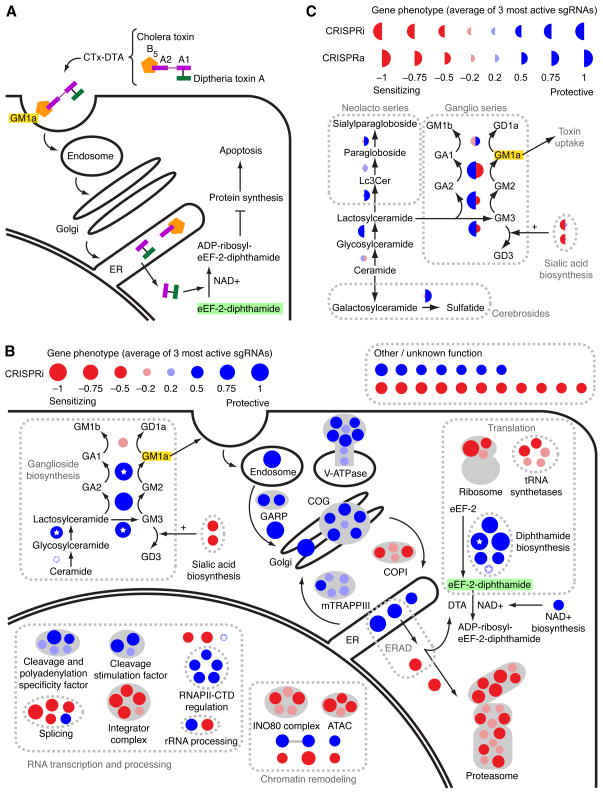
Figure 6. Genome-Scale CRISPRi and CRISPRa Screens Reveal Known and New Pathways and Complexes Governing the Response to a Cholera-Diphtheria Fusion Toxin (CTx-DTA).
(A) Model for CTx-DTA binding, retrograde trafficking, retro-translocation and cellular toxicity. (B) Overview of top hit genes detected by the CTx-DTA screen. Dark red and blue circles: Top 50 sensitizing and protective hits, respectively. Light red and blue circles: further hits that fall into the same protein complexes or pathways as top 50 hits. Circle area is proportional to phenotype strength. White stars denote genes identified in a previous haploid mutagenesis screen (Guimaraes et al., 2011). See also Figure S6 for hit gene names. (C) CRISPRi and CRISPRa hits in sphingolipid metabolism. Display as in (B), except that the left and right sides of each circle represent the phenotypes in the CRISPRi and CRISPRa screens, respectively.
K562 cells expressing the CRISPRi sgRNA library and dCas9-KRAB were either grown under standard conditions or treated with several pulses of CTx-DTA over the course of 10 days. We observed highly correlated enrichment and depletion of many sgRNAs between replicates, indicating that CRISPRi can identify genes that modulate both resistance and sensitivity to a selective pressure (Table S2).
We ranked genes by the average phenotype of their three strongest sgRNAs (Table S3, Figure 6B, and Figure S6). GSEA revealed that KEGG pathways enriched for top protective hit genes were “Infection with Vibrio cholerae” and “Glycosphingolipid biosynthesis, ganglio-series” (Figure S7B), while gene sets for top sensitizing genes included “ribosome” and “proteasome” (Figure S7B). Since the diphtheria toxin catalytic subunit inhibits translation, depletion of the ribosome can be expected to sensitize cells to the toxin. Disruption of the proteasome also sensitizes cells to CTx-DTA, suggesting that the cytosolic toxin is a substrate for proteasomal degradation. Taken together, the unbiased GSEA analysis provides support for the high specificity in hit gene identification by our CRISPRi approach.
We further defined the 50 hits with the strongest protective effect and the 50 hits with the strongest sensitizing effect as “top hits” (all of these are far outside of the range seen with otherwise matched negative control sgRNAs). We characterized these genes by assigning them to cellular pathways and protein complexes according to their previously characterized roles (Figure 6B and Figure S6). Our CRISPRi screen identified a protective effect of knockdown for all top hits recovered in the previously published haploid mutagenesis screen (white stars in Figure 6B). The two top pathways identified by haploid mutagenesis as modulating cellular sensitivity to CTx-DTA are the diphthamide biosynthetic pathway (required to generate eEF-2-diphthamide, the target of diphtheria toxin) and the ganglioside biosynthetic pathway (required to produce GM1a, the cell-surface receptor for cholera toxin). Our screen also identified many additional core components of each pathway. While knockdown of all hits in the diphthamide biosynthesis pathway had a protective effect, the results for ganglioside biosynthesis genes showed a more complex pattern: knockdown of enzymes involved in the production of GM1a were protective, whereas knockdown of enzymes that catalyze the production of other gangliosides (including GM1b) was sensitizing. These results provide genetic confirmation that GM1a is the relevant cell-surface receptor for CTx-DTA and more broadly illustrate the value of being able to reliably detect both sensitizing and protective genes to dissect biological pathways.
Many of the top hits are components of cellular pathways and protein complexes previously identified in experiments to be important for retrograde trafficking and retro-translocation of other toxins such as ricin and Shiga toxin (Bassik et al., 2013; Smith et al., 2009). Retro-translocation of the catalytic chain of CTx has been proposed to be mediated by the ER-associated degradation (ERAD) pathway, although this pathway was not identified in previous genetic screens. Consistent with this proposed role for the ERAD machinery, knockdown of members of the ERAD E3 ubiquitin ligase complex, SYVN1 (encoding Hrd1) and SEL1L (the mammalian homolog of yeast Hrd3) rendered cells resistant to CTx-DTA. Factors that mediate cytosolic degradation of ERAD substrates (in particular UBXN4, also known as UBXD2 or erasin, and the proteasome) appeared as sensitizing hits, suggesting that they may reduce cytosolic levels of the toxin’s catalytic subunit in WT cells.
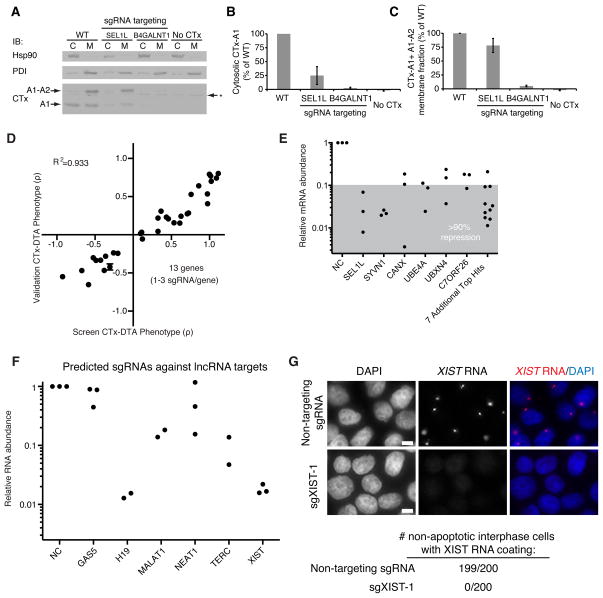
To validate the suggested role of the identified ERAD factors in toxin retro-translocation from the ER to the cytosol, we quantified the amount of CTx chains in the cytosol and membrane fractions. As expected, SEL1L knockdown resulted in a dramatic reduction of cytosolic CTx-A1, whereas levels in the membrane fraction were much less affected (Figure 7A–C). By contrast, knockdown of B4GALNT1, an enzyme required for the synthesis of the CTx receptor GM1a, resulted in a nearly complete absence of CTx chains from both the cytosolic and the membrane fraction (Figure 7A–C).
Figure 7. CRISPRi Strongly Represses Gene Expression of Both Protein-Coding and Non-Coding Genes, Resulting in Reproducible Phenotypes.
(A–C) Cells expressing a negative control sgRNA or an sgRNA targeting SEL1L or B4GALNT1 were incubated with cholera toxin and fractionated to quantify cholera toxin present in the cytosolic and membrane fractions by Western blot. B4GALNT1 repression blocks toxin uptake whereas SEL1L repression prevents toxin retro-translocation from the membrane fraction to the cytosol. (D) Validation of CTx-DTA screen phenotypes with single sgRNA re-test experiments. Data is represented as the mean ± standard deviation of replicates (N=3). (E) CRISPRi knockdown of 13 hit genes (28 sgRNAs; same sgRNAs as 7D) identified in the CTx-DTA screen was quantified by qPCR. The gray shaded region denotes sgRNAs showing at least 90% knockdown for each gene. (F) CRISPRi knockdown of 6 lncRNA genes was quantified by qPCR. 2–3 sgRNAs computationally predicted to target each gene were cloned and transduced into K562 cells expressing dCas9-KRAB. (G) K562 cells expressing dCas9-KRAB were transduced with either a non-targeting sgRNA or an sgRNA targeting the XIST locus (sgXIST-1). The cells were then stained with DAPI and an RNA FISH probe for the XIST transcript. 200 non-apoptotic interphase cells in each condition were scored for XIST RNA coating. XIST is undetectable in cells transduced with sgXIST-1. Scale bar represents 5μm.
An open question in CTx biology is how the toxin traverses the Golgi network (Wernick et al., 2010). Our screen revealed that COG and GARP complexes, which tether late endosomes to the trans-Golgi network or modulate intra-Golgi retrograde transport (Bonifacino and Rojas, 2006) are critical host factors for CTx-DTA. These and other complexes and pathways we identify here (Figure 6B), including several involved in RNA processing, had not previously been linked to cholera toxin biology, highlighting the potential of CRISPRi as a discovery platform. Importantly, many top hits—even those not previously implicated in cholera or diphtheria pathogenesis—were tightly clustered in well-defined protein complexes and pathways. For several of these, the vast majority of components were hits, suggesting that CRISPRi screens can approach saturation.
Potent Phenotypes and Knockdown Levels Achieved by the Genome-Scale CRISPRi Library
To validate the results from this screen, we re-tested sgRNAs that putatively modulate cellular response to CTx-DTA in mechanistically diverse ways. For each sgRNA, we quantified both the ricin phenotypes as well as the change in abundance of the targeted transcript by qPCR. Our re-test experiments were highly correlated with data from the primary screen (Figure 7D). In our validation experiments for the tiling ricin screen and the genome-scale CTx-DTA screen, the activities of 71 out of 72 sgRNAs were robustly confirmed and were highly correlated (R2=0.879) with the results obtained in the primary screen. Finally, analysis of mRNA levels by qPCR data showed robust repression, with ~80–99% knockdown for each sgRNA and at least 90% for every gene (Figure 7E).
A Genome-Scale CRISPRa Screen of Cholera-diphtheria Toxin Complements and Extends CRISPRi Results
To further explore the biological insights gained from CRISPRa screening, we performed a genome scale CRISPRa screen for genes that modulate sensitivity to CTx-DTA (Table S2–3). As with the CRISPRi screen, GSEA revealed the specificity of the detected hits (Figure S7B). For some of the top hits, CRISPR-mediated transcriptional activation and repression caused opposite phenotypes (e.g. enzymes in ganglioside biosynthesis, Figure 6C), similar to what we observed for genes controlling ricin sensitivity (Figure 3C).
CRISPRa also revealed additional and highly complementary information as illustrated by analysis of glycosphingolipid biosynthesis pathways. Induction of enzymes in the neolacto branch of sphingolipid biosynthesis protected cells from CTx-DTA (Figure 6C and Figure S7A,C). This pathway is a parallel branch to the ganglioside branch, which produces the CTx-DTA receptor GM1a. Our findings suggest that upregulation of the neolacto branch diverts the common precursor lactosylceramide away from the ganglioside branch. Similarly, upregulation of the sulfatide-generating enzyme GAL3ST1 has a protective effect, presumably by diverting ceramide from the sphingolipid to the cerebroside pathway (Figure 6C). These results highlight the capacity of CRISPRa to complement CRISPRi by querying the consequences of upregulating pathways that may otherwise be inactive.
Effective Knockdown of non-coding RNAs
Finally, we investigated whether CRISPRi was able to repress the transcription of long non-coding RNAs (lncRNAs), a class of transcripts that have been difficult to systematically perturb by other methods (Bassett et al., 2014). Using our CRISPRi library design algorithm, we selected and cloned up to three sgRNAs each targeting six characterized lncRNAs (GAS5, H19, MALAT1, NEAT1, TERC, XIST) (Geisler and Coller, 2013) with good evidence of expression in K562 cells. We transduced the sgRNAs into cells expressing dCas9-KRAB and quantified the amount of transcript knockdown by qPCR. We achieved >80% knockdown for all but one of the lncRNA genes tested (Figure 7F). Overall, more than 50% of the sgRNAs yielded >85% knockdown. We confirmed the strong repression of XIST by RNA fluorescence in situ hybridization (FISH) and observed no residual expression along the X chromosome (Figure 7G). These results demonstrate that CRISPRi can effectively repress lncRNA expression, enabling future systematic studies of non-coding gene function.
DISCUSSION
Here, we establish CRISPRi and CRISPRa as robust tools for systematically manipulating transcription of endogenous genes in human cells. We demonstrate that CRISPRi/a can be used to rapidly screen for both loss-of-function and gain-of-function phenotypes in a pooled format. We identify both known and unexpected genes that control growth of K562 cells or that modulate sensitivity to a toxin (CTx-DTA). We also show that we can use CRISPRi/a to create allelic series of gene expression, spanning a broad range from ~100-fold repression to ~10-fold induction, allowing us to define how the abundance of a protein or transcript relates to its function.
A key feature of CRISPRi is the low incidence of off-target effects, as evidenced by the near-absence of activity for three large and distinct classes of negative control sgRNAs in our genome-scale CRISPRi library. This feature simplifies validation and interpretation of screening results. The observed specificity likely stems from two distinct properties of our system. First, CRISPRi/a complexes bound outside a narrow window around the TSS largely fail to modulate transcription; this dramatically shrinks the sequence space across the genome where off-target binding could produce significant off-target activity. Second, CRISPRi activity is highly sensitive to mismatches between the sgRNA and target DNA, suggesting that off-target binding of dCas9 observed in ChIP-seq experiments is too transient to impact transcription (Duan et al., 2014; Kuscu et al., 2014; Wu et al., 2014).
CRISPRa screening provides a new approach for exploring the diversity of transcripts across complex genomes. Gene activation has been used to dissect the limiting component of a biochemical process, identify the molecular target of a drug, or activate key rate-limiting steps in a pathway (Davis et al., 1987; Rine et al., 1983). Recently, a combinatorial cDNA overexpression screen identified genes that, when co-expressed, reprogram fibroblasts into pluripotent stem cells (Takahashi and Yamanaka, 2006). CRISPRa should greatly accelerate similar searches for combinations of factors with emergent properties. In addition, CRISPRa will likely provide insight into cellular pathways where redundancy hampers loss-of-function genetic approaches. CRISPRa will also enable the exploration of cellular states in which otherwise inactive pathways are induced, and thereby reveal functional coupling within complex cellular networks and suggest potential therapeutic strategies.
Our ability to control transcription with high specificity simplifies the analysis and validation of high-throughput screening data. The genome-scale CRISPRi and CRISPRa libraries described here contain 10 sgRNAs per TSS. The resulting library size allows each to be screened in a population of 200 million cells, which can be easily grown in a single spinner flask. Furthermore, the observed high specificity and an improved understanding of rules governing sgRNA activity should enable us to create even more compact sgRNA libraries. Additionally, an sgRNA library designed to activate or repress a broader range of transcripts in the human genome could reveal the function of many non-canonical RNAs encoded in the human genome. As most non-coding transcripts are nuclear and lack an open reading frame, methods that directly modulate transcription are optimally suited for interrogating the function of these RNAs (Derrien et al., 2012).
Systematic genetic interaction (GI) maps are powerful tools for revealing gene functions within pathways or complexes (Bassik et al., 2013; Boone et al., 2007; Costanzo et al., 2010). A CRISPRa GI map or a combined CRISPRi/a GI map could yield rich novel biology and help elucidate how networks of proteins dictate cellular function. More generally, quantitative methods of turning on and off one or multiple transcripts represent a critical tool for understanding how expression of the genes encoded in our genomes controls cell function and fate.
EXPERIMENTAL PROCEDURES
CRISPRi/a Libraries
Tiling libraries
sgRNAs were designed targeting 49 genes (see Figure 1E) previously identified in shRNA screens as having ricin resistance phenotype. All possible sgRNAs within a 10kb window around the gene TSS and meeting certain criteria were included (see Extended Experimental Procedures). Negative controls were designed based on scrambled sequences from these 10kb windows and filtered by the same criteria as targeting sgRNAs.
Genome-scale CRISPRi/a libraries
Genes were selected from the entire set of protein coding genes, although a subset of genes with a RPKM of 0 in a K562 cell RNA-seq expression data set were excluded. sgRNAs conforming to rules including low predicted off-targets and minimal length (see Figure S2 and Extended Experimental Procedures) were selected from a window of −50 to +300bp (CRISPRi) or −400 to −50bp (CRISPRa) with respect to the TSS. Negative controls we designed in the same way based on scrambled sequence derived from the same window of several hundred genes.
Library Cloning
Oligonucleotides encoding sgRNAs designed as described above were synthesized as pooled libraries. These were then cloned into lentiviral vectors for expression from a U6 promoter (see Extended Experimental Procedures).
Cell Line Construction
For constitutive and inducible CRISPRi screens, polyclonal cells expressing dCas9/KRAB fusion proteins driven from an SFFV or TRE3G promoter, respectively, were generated by viral transduction. For CRISPRa screens, a clonal cell line expressing dCas9-GCNx10 and a scFV-sfGFP-VP64 fusion was generated (See Extended Experimental Procedures).
Growth and Toxin Screens
Cells were grown at minimum library coverage of 1,000 for tiling screens and 3,750 for genome-scale screens. For growth screens cells were grown in spinner flasks and harvested at 0 and 10 days after puromycin selection. For toxin screens, cells were treated with pulses of ricin or CTx-DTA (Bassik et al., 2013; Guimaraes et al., 2011) and harvested when sufficient selective pressure relative to untreated cells had been applied. Briefly, DNA was isolated, the cassette encoding the sgRNA was amplified by PCR, and relative sgRNA abundance was determined by next generation sequencing as previously described (Bassik et al., 2013; Kampmann et al., 2014).
Supplementary Material
Sequences of the sgRNA protospacers (Tab 1) and qPCR primers (Tab 2) used for individual validation experiments in this study.
sgRNA IDs, transcripts targeted, protospacer sequences, and growth (γ) and CTx-DTA sensitivity (ρ) phenotypes for the genome-scale CRISPRi (Tab 1) and CRISPRa libraries (Tab 2). Phenotypes are average of two biological replicates.
Gene phenotypes for growth (γ) and CTx-DTA (ρ) phenotypes calculated from the 3 most active sgRNAs as described. Phenotypes are average of two biological replicates.
Categorization of the 50 genes with the most negative growth phenotype (γ). Genes were annotated as tumor suppressor genes, developmental transcription factors, and/or involved in centrosomal processes, as determined by published surveys of frequently mutated genes in cancer, individual reports, and gene family membership.
The formulas for deriving cellular phenotype from measurements of cell fraction in a population at discrete timepoints. Cells with a given genotype have an intrinsic growth rate g, often expressed as cell doublings per day. The log2 enrichment (log2e) of cells with a specific genotype in a population can be calculated from the fraction of cells in the population at the endpoint t versus that fraction at t0. In order to express this as the growth rate relative to wild-type (γ), log2e is normalized to the median log2e of the negative control set and then divided by t. Similarly, the phenotype of cells exposed to a selective pressure (e.g. toxin treatment) can calculated from the log2e of treated and untreated populations to obtain ρ, which is +1 for completely resistant cells and −1 for cells with 2-fold sensitivity to the pressure relative to wild-type. (B) In order to quantify the strength of individual sgRNA activity relative to noise in the experiment, sgRNA phenotype was divided by the standard deviation of negative control phenotypes to yield the z-score. For analyses of sgRNA strength in tiling screens, in which genes had known knockdown phenotypes for either sensitivity or resistance, z-scores were re-signed to give positive values where the phenotypes agreed with the expected phenotype and negative values otherwise.
(A) For several example genes, the phenotypes observed for sgRNAs expressed in dCas9 or dCas9-KRAB cells as a function of their position with respect to the TSS are depicted. Each point is an sgRNA. (B) Shorter sgRNAs have, on average, significantly higher activity. Each point represents an sgRNA, with lines connecting related sgRNAs that target the same PAM site but have increasing protospacer base pair length. Black lines represent the median activity for sgRNAs of specific lengths. sgRNAs are depicted if there are multiple sgRNAs targeting the same site and at least one sgRNA at that site is highly active (phenotype-signed z-score 5). (C) sgRNA sequences with very high or very low GC content are less active. (D) The presence of homopolymers (AAAA, GGGG, UUUU) within an sgRNA reduces activity on average. (E) The DNA strand targeted by an sgRNA has no effect on activity. (F) A comparison of a subset of sgRNAs selected based on CRISPRi activity rules versus our previously published shRNA library. For each gene, the Mann-Whitney p-value was calculated using sgRNA or shRNA activity relative to a negative control distribution using 24 shRNAs or 24 sgRNAs. The 24 shRNAs and sgRNAs were randomly selected 100 times from the shRNA library and sgRNAs meeting position and length rules, respectively, and the median and SD log10 p-value are displayed.
The on- and off- target activity of dCas9, dCas9-KRAB and Cas9 was measured for a series of sgRNAs with a varying number and position of mismatches. Each sgRNA is a point with the related mismatch series connected by lines. The measured off-target activity of each sgRNA with one or more mismatch is displayed as percent of the on-target activity for the corresponding sgRNA with 0 mismatches. The sgRNA series denoted with a star represents sgRNAs with 3, 4, or 5 mismatch base pairs randomly distributed across region 3 of the sgRNA sequence. sgRNAs were included in the analysis only if the fully matched guide was highly active (phenotype-signed z-score >= 4); N=5 for dCas9, 11 for dCas9-KRAB, and 10 for Cas9.
(A) The activity of sgRNAs in a CRISPRa cell line as a function of the distance of the sgRNA site to the TSS of the targeted gene for four example genes. Top, ARL1 and ST3GAL4 activation results in ricin resistance; bottom, SURF4 and RAB1A activation results in ricin sensitivity. (B) A sliding-window average only for genes with a significant CRISPRa ricin resistance phenotype targeted by our test library is shown in green. The median activity is shown with a green line while the window of maximal activity is shown in orange. The data is displayed as a phenotype-signed z-score.
(A) The top 10 DAVID annotation clusters identified in our CRISPRi growth screen are strongly enriched for known essential cellular processes. (B) Growth phenotypes from a genome-scale CRISPRa screen carried out in duplicate in K562 cells. Two classes of negative control sgRNAs are shown: non-targeting sgRNAs (grey) and sgRNAs targeting Y-chromosomal genes (green). (C) The top 10 DAVID annotation clusters identified in our CRISPRa growth screen. (D) RNA-sequencing RPKM (reads per kilo base per million) from K562 cells plotted for three gene sets: all genes and those genes identified to have activation or repression growth phenotypes by CRIPSRa or CRISPRi, respectively. (E) ST3GAL4 expression levels (relative to ACTB, as quantified by qPCR) in inducible CRISPRi K562 cells transduced with ST3GAL4-targeting sgRNAs in the absence, presence, and after washout of doxycycline. Data is normalized to control cells (assayed in the presence of doxycycline) at each time point and represented as the mean of technical replicates ± standard error. (F) Reversibility of ricin sensitivity (ST3GAL4-1) and resistance (RAB1A-2) phenotypes induced by CRISPRi. Inducible CRISRPi cells transduced with the indicated sgRNAs were challenged with 0.4 ng/mL ricin toxin in the presence and absence of doxycycline. Ricin challenge was performed before (Untreated) and after 2 days of doxycycline pre-treatment (+Dox), and following removal of doxycycline from tissue culture media and an 8 day recovery period (Dox washout). Rho values were calculated as described. Data represent the mean of ricin challenged replicates (N=3) ± standard deviation. (G) CRISPRi knockdown (mRNA levels relative to ACTB) of hit genes identified in the genome-scale growth screen (9 sgRNAs) was quantified by qPCR after two days of doxycycline treatment. The gray shaded region denotes sgRNAs showing at least 90% knockdown for each gene. See also Figure 5D.
Same representation as Figure 6B, with added gene names.
(A) CRISPRi and CRISPRa hits in sphingolipid metabolism. Same representation as Figure 6C, with added gene names (B) Gene set enrichment analysis of top hits from genome-scale CRISPRi and CRISPRa screens for sensitivity to CTx-DTA. (C) CRISPRi versus CRISPRa gene CTx-DTA phenotypes. Phenotypes for genome-scale CTx-DTA CRISPRi and CRISPRa screens (black) were calculated based on the average resistance phenotype (ρ) of the three most active sgRNAs. Hit genes from either screen that were featured in Figure 6 as part of the glycosphingolipid biosynthesis pathway, proteasome, or diphthamide biosynthesis pathway are labele as indicated.
HIGHLIGHTS.
CRISPRi and CRISPRa provide complementary information for mapping complex pathways
CRISPRi/a expression series (up to ~1000 fold) reveal how gene dose controls function
CRISPRi provides strong (typically 90–99%) knockdown with minimal off-target effects
Genome-scale screens elucidate pathways controlling cholera/diphtheria toxicity
Acknowledgments
The authors thank M. Tanenbaum, J. Tsai, K. Kostova, J. Zalatan, W. Lim, S. Weissman, J. Doudna, and R. Vale for unpublished reagents, technical advice and helpful discussion. L.S.Q. acknowledges support from the UCSF Center for Systems and Synthetic Biology. This work was supported by NIH P50 GM102706 (J.S.W.), NIH P50 GM081879 (L.S.Q., E.H.W.), NIH U01 CA168370 and NIH R01 DA036858 (J.S.W.), as well as the Howard Hughes Medical Institute (J.E.V., Y.C., M.K., M.A.H., J.S.W.). B.A. is an HHMI Fellow of the Damon Runyon Cancer Research Foundation (DRG-[2182-14]). L.A.G. is a Fellow of the Leukemia and Lymphoma Society. M.A.H. is supported by the UCSF Medical Scientist Training Program. M.K. is supported by NCI/NIH Pathways to Independence Award K99CA181494.
Footnotes
AUTHOR CONTRIBUTIONS
BA, LAG, MAH, MK, JSW were primarily responsible for the conception, design, and interpretation of the experiments and wrote the manuscript. BA, YC, LAG, MAH, MK, JEV, and EHW conducted experiments. LAG cloned dCas9 chimeras and sgRNA expression constructs, constructed cell lines, carried out tiling screens, and conducted validation experiments. MAH designed libraries, carried out tiling and genome-scale screens, and analyzed screen data. BA constructed the inducible cell line and conducted all inducible experiments. CG and HLP contributed to CTx-DTA studies. BP contributed to XIST studies. MCB and LSQ contributed to the conception and interpretation of the experiments.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adamson B, Smogorzewska A, Sigoillot FD, King RW, Elledge SJ. A genome-wide homologous recombination screen identifies the RNA-binding protein RBMX as a component of the DNA-damage response. Nat Cell Biol. 2012;14:318–328. doi: 10.1038/ncb2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberich-Jordà M, Wouters B, Balastik M, Shapiro-Koss C, Zhang H, Di Ruscio A, DiRuscio A, Radomska HS, Ebralidze AK, Amabile G, et al. C/EBPγ deregulation results in differentiation arrest in acute myeloid leukemia. J Clin Invest. 2012;122:4490–4504. doi: 10.1172/JCI65102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett AR, Akhtar A, Barlow DP, Bird AP, Brockdorff N, Duboule D, Ephrussi A, Ferguson-Smith AC, Gingeras TR, Haerty W, et al. Considerations when investigating lncRNA function in vivo. eLife. 2014;3:e03058. doi: 10.7554/eLife.03058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassik MC, Lebbink RJ, Churchman LS, Ingolia NT, Patena W, LeProust EM, Schuldiner M, Weissman JS, McManus MT. Rapid creation and quantitative monitoring of high coverage shRNA libraries. Nat Methods. 2009;6:443–445. doi: 10.1038/nmeth.1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassik MC, Kampmann M, Lebbink RJ, Wang S, Hein MY, Poser I, Weibezahn J, Horlbeck MA, Chen S, Mann M, et al. A systematic mammalian genetic interaction map reveals pathways underlying ricin susceptibility. Cell. 2013;152:909–922. doi: 10.1016/j.cell.2013.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beerli RR, Segal DJ, Dreier B, Barbas CF. Toward controlling gene expression at will: specific regulation of the erbB-2/HER-2 promoter by using polydactyl zinc finger proteins constructed from modular building blocks. Proc Natl Acad Sci U S A. 1998;95:14628–14633. doi: 10.1073/pnas.95.25.14628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beerli RR, Dreier B, Barbas CF. Positive and negative regulation of endogenous genes by designed transcription factors. Proc Natl Acad Sci U S A. 2000;97:1495–1500. doi: 10.1073/pnas.040552697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifacino JS, Rojas R. Retrograde transport from endosomes to the trans-Golgi network. Nat Rev Mol Cell Biol. 2006;7:568–579. doi: 10.1038/nrm1985. [DOI] [PubMed] [Google Scholar]
- Boone C, Bussey H, Andrews BJ. Exploring genetic interactions and networks with yeast. Nat Rev Genet. 2007;8:437–449. doi: 10.1038/nrg2085. [DOI] [PubMed] [Google Scholar]
- Carette JE, Guimaraes CP, Varadarajan M, Park AS, Wuethrich I, Godarova A, Kotecki M, Cochran BH, Spooner E, Ploegh HL, et al. Haploid genetic screens in human cells identify host factors used by pathogens. Science. 2009;326:1231–1235. doi: 10.1126/science.1178955. [DOI] [PubMed] [Google Scholar]
- Cech TR, Steitz JA. The noncoding RNA revolution-trashing old rules to forge new ones. Cell. 2014;157:77–94. doi: 10.1016/j.cell.2014.03.008. [DOI] [PubMed] [Google Scholar]
- Chang K, Elledge SJ, Hannon GJ. Lessons from Nature: microRNA-based shRNA libraries. Nat Methods. 2006;3:707–714. doi: 10.1038/nmeth923. [DOI] [PubMed] [Google Scholar]
- Costanzo M, Baryshnikova A, Bellay J, Kim Y, Spear ED, Sevier CS, Ding H, Koh JLY, Toufighi K, Mostafavi S, et al. The genetic landscape of a cell. Science. 2010;327:425–431. doi: 10.1126/science.1180823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RL, Weintraub H, Lassar AB. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987;51:987–1000. doi: 10.1016/0092-8674(87)90585-x. [DOI] [PubMed] [Google Scholar]
- Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan J, Lu G, Xie Z, Lou M, Luo J, Guo L, Zhang Y. Genome-wide identification of CRISPR/Cas9 off-targets in human genome. Cell Res. 2014 doi: 10.1038/cr.2014.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaj T, Gersbach CA, Barbas CF. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013;31:397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler S, Coller J. RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts. Nat Rev Mol Cell Biol. 2013;14:699–712. doi: 10.1038/nrm3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, Stern-Ginossar N, Brandman O, Whitehead EH, Doudna JA, et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154:442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimaraes CP, Carette JE, Varadarajan M, Antos J, Popp MW, Spooner E, Brummelkamp TR, Ploegh HL. Identification of host cell factors required for intoxication through use of modified cholera toxin. J Cell Biol. 2011;195:751–764. doi: 10.1083/jcb.201108103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habedanck R, Stierhof YD, Wilkinson CJ, Nigg EA. The Polo kinase Plk4 functions in centriole duplication. Nat Cell Biol. 2005;7:1140–1146. doi: 10.1038/ncb1320. [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009a;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009b;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang LC, Clarkin KC, Wahl GM. Sensitivity and selectivity of the DNA damage sensor responsible for activating p53-dependent G1 arrest. Proc Natl Acad Sci U S A. 1996;93:4827–4832. doi: 10.1073/pnas.93.10.4827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson SP. Sensing and repairing DNA double-strand breaks. Carcinogenesis. 2002;23:687–696. doi: 10.1093/carcin/23.5.687. [DOI] [PubMed] [Google Scholar]
- Jackson AL, Bartz SR, Schelter J, Kobayashi SV, Burchard J, Mao M, Li B, Cavet G, Linsley PS. Expression profiling reveals off-target gene regulation by RNAi. Nat Biotechnol. 2003;21:635–637. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
- Kampmann M, Bassik MC, Weissman JS. Integrated platform for genome-wide screening and construction of high-density genetic interaction maps in mammalian cells. Proc Natl Acad Sci U S A. 2013;110:E2317–E2326. doi: 10.1073/pnas.1307002110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampmann M, Bassik MC, Weissman JS. Functional genomics platform for pooled screening and generation of mammalian genetic interaction maps. Nat Protoc. 2014;9:1825–1847. doi: 10.1038/nprot.2014.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike-Yusa H, Li Y, Tan EP, Velasco-Herrera MDC, Yusa K. Genome-wide recessive genetic screening in mammalian cells with a lentiviral CRISPR-guide RNA library. Nat Biotechnol. 2014;32:267–273. doi: 10.1038/nbt.2800. [DOI] [PubMed] [Google Scholar]
- Kuscu C, Arslan S, Singh R, Thorpe J, Adli M. Genome-wide analysis reveals characteristics of off-target sites bound by the Cas9 endonuclease. Nat Biotechnol. 2014;32:677–683. doi: 10.1038/nbt.2916. [DOI] [PubMed] [Google Scholar]
- Mittler G, Stühler T, Santolin L, Uhlmann T, Kremmer E, Lottspeich F, Berti L, Meisterernst M. A novel docking site on Mediator is critical for activation by VP16 in mammalian cells. EMBO J. 2003;22:6494–6504. doi: 10.1093/emboj/cdg619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Ness BG, Howard JB, Bodley JW. ADP-ribosylation of elongation factor 2 by diphtheria toxin. Isolation and properties of the novel ribosyl-amino acid and its hydrolysis products. J Biol Chem. 1980;255:10717–10720. [PubMed] [Google Scholar]
- Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rine J, Hansen W, Hardeman E, Davis RW. Targeted selection of recombinant clones through gene dosage effects. Proc Natl Acad Sci U S A. 1983;80:6750–6754. doi: 10.1073/pnas.80.22.6750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol. 2014;32:347–355. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelsen TS, Heckl D, Ebert BL, Root DE, Doench JG, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigoillot FD, Lyman S, Huckins JF, Adamson B, Chung E, Quattrochi B, King RW. A bioinformatics method identifies prominent off-targeted transcripts in RNAi screens. Nat Methods. 2012;9:363–366. doi: 10.1038/nmeth.1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RD, Willett R, Kudlyk T, Pokrovskaya I, Paton AW, Paton JC, Lupashin VV. The COG complex, Rab6 and COPI define a novel Golgi retrograde trafficking pathway that is exploited by SubAB toxin. Traffic Cph Den. 2009;10:1502–1517. doi: 10.1111/j.1600-0854.2009.00965.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitz F, Furlong EEM. Transcription factors: from enhancer binding to developmental control. Nat Rev Genet. 2012;13:613–626. doi: 10.1038/nrg3207. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Tanenbaum ME, Macurek L, van der Vaart B, Galli M, Akhmanova A, Medema RH. A complex of Kif18b and MCAK promotes microtubule depolymerization and is negatively regulated by Aurora kinases. Curr Biol CB. 2011;21:1356–1365. doi: 10.1016/j.cub.2011.07.017. [DOI] [PubMed] [Google Scholar]
- Tanenbaum ME, Gilbert LA, Qi LS, Weissman JS, Vale RD. A versatile protein tagging system for signal amplification in single molecule imaging and gene regulation. Revis. doi: 10.1016/j.cell.2014.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Kinzler KW. Cancer genome landscapes. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Wei JJ, Sabatini DM, Lander ES. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014;343:80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernick NLB, Chinnapen DJF, Cho JA, Lencer WI. Cholera toxin: an intracellular journey into the cytosol by way of the endoplasmic reticulum. Toxins. 2010;2:310–325. doi: 10.3390/toxins2030310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Scott DA, Kriz AJ, Chiu AC, Hsu PD, Dadon DB, Cheng AW, Trevino AE, Konermann S, Chen S, et al. Genome-wide binding of the CRISPR endonuclease Cas9 in mammalian cells. Nat Biotechnol. 2014;32:670–676. doi: 10.1038/nbt.2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Cong L, Lodato S, Kosuri S, Church GM, Arlotta P. Efficient construction of sequence-specific TAL effectors for modulating mammalian transcription. Nat Biotechnol. 2011;29:149–153. doi: 10.1038/nbt.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Zhang X, Iwama A, Yu C, Smith KA, Mueller BU, Narravula S, Torbett BE, Orkin SH, Tenen DG. PU. 1 inhibits GATA-1 function and erythroid differentiation by blocking GATA-1 DNA binding. Blood. 2000;96:2641–2648. [PubMed] [Google Scholar]
- Zhao M, Sun J, Zhao Z. TSGene: a web resource for tumor suppressor genes. Nucleic Acids Res. 2013;41:D970–D976. doi: 10.1093/nar/gks937. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sequences of the sgRNA protospacers (Tab 1) and qPCR primers (Tab 2) used for individual validation experiments in this study.
sgRNA IDs, transcripts targeted, protospacer sequences, and growth (γ) and CTx-DTA sensitivity (ρ) phenotypes for the genome-scale CRISPRi (Tab 1) and CRISPRa libraries (Tab 2). Phenotypes are average of two biological replicates.
Gene phenotypes for growth (γ) and CTx-DTA (ρ) phenotypes calculated from the 3 most active sgRNAs as described. Phenotypes are average of two biological replicates.
Categorization of the 50 genes with the most negative growth phenotype (γ). Genes were annotated as tumor suppressor genes, developmental transcription factors, and/or involved in centrosomal processes, as determined by published surveys of frequently mutated genes in cancer, individual reports, and gene family membership.
The formulas for deriving cellular phenotype from measurements of cell fraction in a population at discrete timepoints. Cells with a given genotype have an intrinsic growth rate g, often expressed as cell doublings per day. The log2 enrichment (log2e) of cells with a specific genotype in a population can be calculated from the fraction of cells in the population at the endpoint t versus that fraction at t0. In order to express this as the growth rate relative to wild-type (γ), log2e is normalized to the median log2e of the negative control set and then divided by t. Similarly, the phenotype of cells exposed to a selective pressure (e.g. toxin treatment) can calculated from the log2e of treated and untreated populations to obtain ρ, which is +1 for completely resistant cells and −1 for cells with 2-fold sensitivity to the pressure relative to wild-type. (B) In order to quantify the strength of individual sgRNA activity relative to noise in the experiment, sgRNA phenotype was divided by the standard deviation of negative control phenotypes to yield the z-score. For analyses of sgRNA strength in tiling screens, in which genes had known knockdown phenotypes for either sensitivity or resistance, z-scores were re-signed to give positive values where the phenotypes agreed with the expected phenotype and negative values otherwise.
(A) For several example genes, the phenotypes observed for sgRNAs expressed in dCas9 or dCas9-KRAB cells as a function of their position with respect to the TSS are depicted. Each point is an sgRNA. (B) Shorter sgRNAs have, on average, significantly higher activity. Each point represents an sgRNA, with lines connecting related sgRNAs that target the same PAM site but have increasing protospacer base pair length. Black lines represent the median activity for sgRNAs of specific lengths. sgRNAs are depicted if there are multiple sgRNAs targeting the same site and at least one sgRNA at that site is highly active (phenotype-signed z-score 5). (C) sgRNA sequences with very high or very low GC content are less active. (D) The presence of homopolymers (AAAA, GGGG, UUUU) within an sgRNA reduces activity on average. (E) The DNA strand targeted by an sgRNA has no effect on activity. (F) A comparison of a subset of sgRNAs selected based on CRISPRi activity rules versus our previously published shRNA library. For each gene, the Mann-Whitney p-value was calculated using sgRNA or shRNA activity relative to a negative control distribution using 24 shRNAs or 24 sgRNAs. The 24 shRNAs and sgRNAs were randomly selected 100 times from the shRNA library and sgRNAs meeting position and length rules, respectively, and the median and SD log10 p-value are displayed.
The on- and off- target activity of dCas9, dCas9-KRAB and Cas9 was measured for a series of sgRNAs with a varying number and position of mismatches. Each sgRNA is a point with the related mismatch series connected by lines. The measured off-target activity of each sgRNA with one or more mismatch is displayed as percent of the on-target activity for the corresponding sgRNA with 0 mismatches. The sgRNA series denoted with a star represents sgRNAs with 3, 4, or 5 mismatch base pairs randomly distributed across region 3 of the sgRNA sequence. sgRNAs were included in the analysis only if the fully matched guide was highly active (phenotype-signed z-score >= 4); N=5 for dCas9, 11 for dCas9-KRAB, and 10 for Cas9.
(A) The activity of sgRNAs in a CRISPRa cell line as a function of the distance of the sgRNA site to the TSS of the targeted gene for four example genes. Top, ARL1 and ST3GAL4 activation results in ricin resistance; bottom, SURF4 and RAB1A activation results in ricin sensitivity. (B) A sliding-window average only for genes with a significant CRISPRa ricin resistance phenotype targeted by our test library is shown in green. The median activity is shown with a green line while the window of maximal activity is shown in orange. The data is displayed as a phenotype-signed z-score.
(A) The top 10 DAVID annotation clusters identified in our CRISPRi growth screen are strongly enriched for known essential cellular processes. (B) Growth phenotypes from a genome-scale CRISPRa screen carried out in duplicate in K562 cells. Two classes of negative control sgRNAs are shown: non-targeting sgRNAs (grey) and sgRNAs targeting Y-chromosomal genes (green). (C) The top 10 DAVID annotation clusters identified in our CRISPRa growth screen. (D) RNA-sequencing RPKM (reads per kilo base per million) from K562 cells plotted for three gene sets: all genes and those genes identified to have activation or repression growth phenotypes by CRIPSRa or CRISPRi, respectively. (E) ST3GAL4 expression levels (relative to ACTB, as quantified by qPCR) in inducible CRISPRi K562 cells transduced with ST3GAL4-targeting sgRNAs in the absence, presence, and after washout of doxycycline. Data is normalized to control cells (assayed in the presence of doxycycline) at each time point and represented as the mean of technical replicates ± standard error. (F) Reversibility of ricin sensitivity (ST3GAL4-1) and resistance (RAB1A-2) phenotypes induced by CRISPRi. Inducible CRISRPi cells transduced with the indicated sgRNAs were challenged with 0.4 ng/mL ricin toxin in the presence and absence of doxycycline. Ricin challenge was performed before (Untreated) and after 2 days of doxycycline pre-treatment (+Dox), and following removal of doxycycline from tissue culture media and an 8 day recovery period (Dox washout). Rho values were calculated as described. Data represent the mean of ricin challenged replicates (N=3) ± standard deviation. (G) CRISPRi knockdown (mRNA levels relative to ACTB) of hit genes identified in the genome-scale growth screen (9 sgRNAs) was quantified by qPCR after two days of doxycycline treatment. The gray shaded region denotes sgRNAs showing at least 90% knockdown for each gene. See also Figure 5D.
Same representation as Figure 6B, with added gene names.
(A) CRISPRi and CRISPRa hits in sphingolipid metabolism. Same representation as Figure 6C, with added gene names (B) Gene set enrichment analysis of top hits from genome-scale CRISPRi and CRISPRa screens for sensitivity to CTx-DTA. (C) CRISPRi versus CRISPRa gene CTx-DTA phenotypes. Phenotypes for genome-scale CTx-DTA CRISPRi and CRISPRa screens (black) were calculated based on the average resistance phenotype (ρ) of the three most active sgRNAs. Hit genes from either screen that were featured in Figure 6 as part of the glycosphingolipid biosynthesis pathway, proteasome, or diphthamide biosynthesis pathway are labele as indicated.