Abstract
Some viruses and most eukaryotic cells have microRNAs that regulate the expression of many genes. Although many viral miRNAs have been identified, only a few have been included in in vivo functional studies. Here we show that a Py-encoded miRNA downregulates the expression of the pro-apoptotic factor Smad2, resulting in the suppression of the apoptosis pathway. To study the Py miRNA in an in vivo context, a miRNA-deficient mutant virus was created on the background of the LID virus strain which establishes a rapid and lethal infection in newborn mice. Apoptosis analysis on kidney tissues indicates that the pro-apoptotic pathway is targeted in the infected host as well. Suppression of apoptosis through targeting of Smad2 by the Py miRNA is expected to synergize with anti-apoptotic effects previously attributed to the polyoma tumor antigens in support of virus replication in the natural host.
Keywords: Mouse polyomavirus, microRNA, apoptosis, Smad2 a cellular target, animal infection
Introduction
microRNAs (miRNAs) are non-coding small RNAs that regulate the expression of many genes and have been proposed as useful biomarkers for diverse diseases with possibly therapeutic implications. Most eukaryotic cells have been confirmed to utilize miRNAs for posttranscriptional gene regulation (Bartel, 2009; Garzon, Calin, and Croce, 2009; Kumar et al., 2007). Some viruses including retrovirus, herpesvirus and polyomavirus have been shown to encode miRNAs that are involved in the regulation of viral replication, immune evasion and cellular apoptotic pathways (Cullen, 2010; Dahlke et al., 2012; Grundhoff and Sullivan, 2011; Imperiale, 2014; Kincaid and Sullivan, 2012; Pfeffer et al., 2004; Sullivan et al., 2005).
Studies of the highly oncogenic mouse polyomavirus (Py) have identified viral replicative and transforming functions as well as genetic determinants of the host that underlie susceptibility or resistance to tumor induction by the virus (Benjamin, 2001; Benjamin, 2007). Decades of investigations have revealed the virus’s structural, molecular biological, and genetic characteristics and how they affect virus replication, cell transformation and tumorigenesis (Imperiale, 2007). The protein networks altered in Py-induced oncogenesis overlap with those affected in many human cancers (Cheng et al., 2009; Fluck and Schaffhausen, 2009; Freund et al., 1994; Rozenblatt-Rosen et al., 2012). The first identification of a miRNA encoded by a polyomavirus was in SV40 where it was shown to target and downregulate the overlapping large T antigen. This study raised the intriguing possibility that this miRNA may serve as part of an immune evasion mechanism (Sullivan et al., 2005). We subsequently identified an analogous Py miRNA with an autoregulatory role downregulating early viral gene expression (Sullivan et al., 2009). Results of attempts to demonstrate an effect of the Py miRNA on immune evasion or viral clearance in vivo were negative (Sullivan et al., 2009). The results of this initial study were inconclusive with respect to demonstrating or ruling out a physiological role of the Py miRNA. Here we turn our attention to host genes as possible targets of Py miRNA and show that the pro-apoptotic factor Smad2 is downregulated by Py miRNA resulting in suppression of apoptosis in vivo.
Results
The Py microRNA targets Smad2 and reduces the apoptotic rate in infected cells
We have previously identified Py-encoded miRNAs (Sullivan et al., 2009) and showed that they inhibit Py large and middle T (tumor) antigen expression. To investigate their possible roles in the regulation of host gene expression, we first turned to bioinformatic target prediction programs. Interestingly, one of the putative targets predicted by TargetScanMouse Custom Version 5.1 (Lewis, Burge, and Bartel, 2005) was TgfβRI, while another web-based program RNAhybrid (Kruger and Rehmsmeier, 2006) predicted Smad2 as a potential target of Py-miRNA. The putative binding site in the Smad2 3′UTR region is shown in Figure 1A and Supplemental Figure 1. The ability to block apoptotic responses of the host is critical particularly for viruses such as polyoma with long replication cycles (36–48hrs). Since TgfβRI and Smad2 are important regulators of the programed cell death machinery (Heldin, Landstrom, and Moustakas, 2009; Hough, Radu, and Dore, 2012; Moustakas and Heldin, 2009), we undertook further investigations of TgfβRI and Smad2 as possible targets of the Py miRNA.
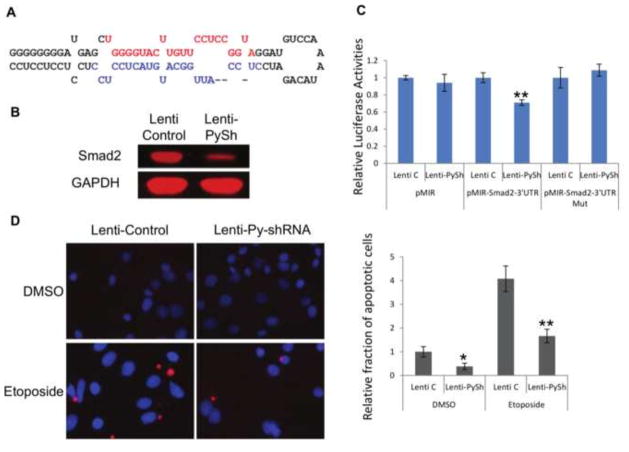
Fig. 1. The Py-miRNA targets Smad2 and results in reduced apoptosis.
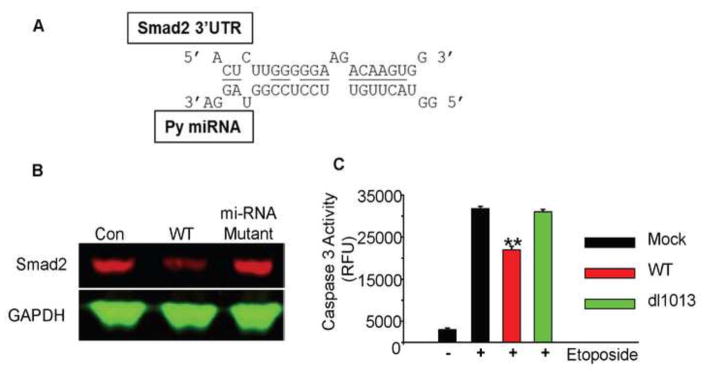
A. Sequence comparison between Py-miRNA and murine Smad2 3′UTR. B. Western blot of Smad2 expression between uninfected, WT-infected, and miRNA-deficient mutant Py-infected baby mouse kidney cells. Smad2 expression was lowered with WT Py virus, but not with miRNA-deficient mutant Py. C. Caspase 3 activity assay of WT-infected and miRNA-deficient mutant Py-infected IMCD cells treated with etoposide. WT but not mutant virus inhibits etoposide induced apoptosis (** p-value < 0.01).
We first infected baby mouse kidney cells with wild type virus (PTA) or miRNA-deficient mutant virus (PTA-dl1013) (Magnusson and Berg, 1979; Sullivan et al., 2009) to determine whether the expression levels of TgfβRI and Smad2 are altered by Py-miRNA. Cell lysates were prepared at 40 hours post infection and subjected to western blot analysis. The protein level of TgfβRI was not altered by viral infection (data not shown). The Smad2 protein, however, was downregulated by the wild type but not by the mutant virus. The level of Smad2 in mutant infected cells remained similar to that of the uninfected control (Figure 1B). These results suggest that Py-miRNA may be involved in the repression of Smad2. As targeting the host’s programmed cell death pathway would be advantageous if not essential for viral infections, an apoptosis analysis with the wild type and mutant strains was performed. Mouse kidney epithelial cells (IMCD) were infected with PTA or mutant PTA-dl1013 and treated with etoposide (25 μM for 16 hours) to induce apoptosis. Cell lysates were mixed with fluorogenic peptides as substrates for proapoptotic caspase-3. The caspase-3 enzyme activity was significantly reduced in the wild type PTA-infected cells compared to that in miRNA-deficient mutant–infected cells or the uninfected control. These results indicate that Py miRNA targets the host’s apoptotic pathway by downregulating Smad2 (Figure 1C).
Lentiviruses harboring Py-shRNA targets Smad2 and the apoptotic pathway
To independently test the role of Py short hairpin (Py-shRNA, precursor to Py miRNA) in suppressing programed cell death, we generated a lentiviral vector harboring the Py-shRNA (Figure 2A). Lentiviral particles with Py-shRNA were prepared along with empty vector control lentivirus (pLKO) and used to infect IMCD cells. First its ability to downregulate the expression of Smad2 was reexamined by western blot. As seen in Figure 2B, Smad2 protein levels were reduced with Py-shRNA-harboring lentivirus compared to control virus-infected cells, indicating that the introduction of Py-shRNA-harboring lentiviral particles into mouse cells leads to decreased expression of Smad2. To determine if the 3′UTR region of Smad2 is important for the target recognition and inhibition by Py-shRNA, pMIR-luciferase vectors with or without the 3′UTR region of Smad2 were constructed and used in dual luciferase assays. IMCD cells were transfected with pRenilla and pMIR control, pMIR-Smad2-3′UTR or pMIR-Smad2-3′UTR deleted vector, and then infected with control lenti or lenti with Py-shRNA. At 48 hours post infection, cells were lysed and subjected to a luciferase analysis (Figure 2C). The enzyme activity of vector including 3′UTR of Smad2 was significantly reduced with lenti-Py-shRNA, but the vector lacking the 3′UTR of Smad2 did not show any difference with lenti-Py-shRNA, demonstrating that Py-shRNA represses the expression of Smad2 by targeting its 3′UTR region.
Fig. 2. Lentiviruses harboring Py-shRNA also target Smad2 and the pro-apoptotic pathway.
A. Polyomavirus short hairpin inserted into pLKO generating a Py-shRNA lentiviral clone that was used in immunoblotting and apoptosis assays (red nucleotides, estimated position of the 5p miRNA; blue nucleotides, estimated position of the 3p miRNA). B. Smad2 protein expression was reduced with Py-shRNA lentivirus in infected IMCD cells. C. pMIR-Luciferase-Smad2-3′UTR showed reduced luciferase activities with Lenti-Py-shRNA while control pMIR and pMIR-Smad2-3′UTR mutant vectors showed no changes with Lenti-Py-shRNA (** p-value < 0.01). D. Mouse IMCD cells were infected with control or Py-shRNA lentivirus, then treated with either DMSO or etoposide. Infected cells were fixed on coverslips, and TUNEL assays were performed to compare the rates of apoptosis. Left – Representative images of the TUNEL-assayed coverslips. Red: apoptotic; Blue: DAPI stain. Right – Apoptotic cells were counted in each sample and normalized to control lentivirus infected, DMSO treated samples (* p-value < 0.05; ** p-value < 0.01).
TUNEL assays were also performed to test whether the Py-shRNA is involved in blocking apoptosis. IMCD cells on coverslips were infected with lenti-pLKO or lenti-Py-shRNA, treated with DMSO or etoposide, then fixed and TUNEL assayed for apoptosis. The number of apoptotic cells (red) were counted in each sample and compared (Figure 2D). More apoptotic cells were seen with control lenti-pLKO than with lenti-Py-shRNA. Moreover, under conditions of etoposide treatment to induce apoptosis, the lenti-Py-shRNA strongly inhibited the response confirming that the Py-shRNA functions to block apoptosis.
The Py miRNA inhibits apoptosis in kidneys of infected mice
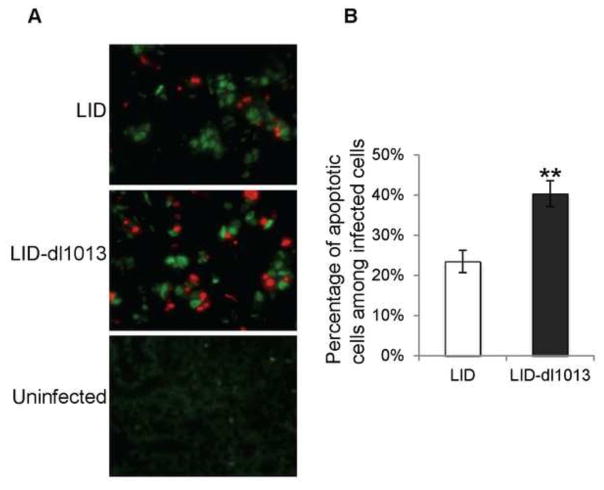
A previous study using wild type and microRNA-deficient mutant viruses on a standard wild type (PTA) virus background showed similar levels of infection in kidneys of neonatally infected mice (Sullivan et al., 2009). The overall infection rate was low, however, possibly obscuring differences. Here we investigate the role of the Py miRNA by comparing wild type and mutant viruses on the background of the LID virus strain. LID is a highly virulent strain of Py that induces a lethal infection of newborn mice (Bauer et al., 1995; Bolen et al., 1985). Mice succumb within a few weeks due to a widely disseminated infection accompanied by extensive tissue destruction most prominently in the kidney. LID owes its virulence to a single amino acid substitution in the major capsid protein VP1 (V296A) which reduces its van der Waals interaction with sialic acid receptors. The lower avidity of receptor binding by LID facilitates release from cell debris and promotes virus spread. We reasoned that by virtue of LID’s ability to induce an acute and rapidly spreading infection, an effect of the Py miRNAs could be more readily assessed using this viral genetic background. Newborn mice were injected with LID or LID-dl1013 and sacrificed at day 8 post injection. Kidney sections from age-matched uninfected mice were prepared as controls. The fixed slides were incubated with anti-digoxigenin (red) and anti-polyoma large T antigen (green) antibodies to identify and quantify apoptotic cells among infected cells. Roughly 3,000 infected cells from seven LID and ten LID-dl1013-injected mice were examined by fluorescence microscopy. As seen in Figure 3A and 3B, the percentages of apoptotic cells in kidneys of LID-dl1013-infected mice were significantly higher than in kidneys of LID-infected mice, confirming that Py miRNA blocks apoptosis in the infected host.
Fig. 3. The apoptosis mechanism is targeted by the Py-miRNA in infected mice.
Kidneys were harvested from C3H/BiDa mice neonatally-infected with LID Py or mutant, non-miRNA expressing LID-dl1013 Py at day 8 post injection. Kidney sections were subjected to apoptosis analyses (TUNEL) with the apoptosis marker anti-digoxigenin (red) and anti-polyoma large T antigen (green) antibodies. A. Representative images of kidney stains from each group. An uninfected kidney sample is shown as a control. B. Quantification of apoptosis rates in the LID and LID-dl1013 infected groups. The percentage of cells that double-stained for apoptosis and Py-infection were quantified between the two groups. Roughly 3,000 infected cells from each group were counted and are plotted (** p-value < 0.01).
Discussion
Viruses have to prevent rapid cell death responses by their host cells for efficient infection and spread. This is particularly critical for viruses such as those of the polyoma group whose replication cycles are long (36–48 hrs) compared to typical host apoptotic responses (4–8 hrs) (Spencer and Sorger, 2011). In this report, we have used target prediction tools and biochemical approaches to identify the proapoptotic factor Smad2 as a target of downregulation by the Py miRNA. The miRNA-mediated pathway of apoptosis inhibition by polyoma is expected to synergize with other anti-apoptotic effects previously attributed to the polyoma viral T antigens. The polyoma middle T antigen inhibits apoptosis by activating the PI3-kinase-Akt pathway (Dahl et al., 1998) and the large T antigen by interaction with p150, a transcription factor with pro-apoptotic and growth arrest functions (Gu et al., 2011; Li et al., 2001; Li et al., 2004; Liu et al., 2007). Polyoma thus employs overlapping and potentially redundant mechanisms to inhibit apoptotic responses of the host, utilizing both early (T antigens) and late (PymiRNA) gene expression.
Several viral miRNAs have been shown to target proapoptotic genes of their hosts. Epstein-Barr virus-encoded miRNA targets the pro-apoptotic gene PUMA as well as another pro-apoptotic gene Bim (Choy et al., 2008; Marquitz et al., 2011), and Marek’s disease virus type 1 (MDV1)-encoded miRNA suppresses apoptosis in cell culture by targeting Smad2 (Xu et al., 2011). The functional importance of viral miRNAs during infection of the natural host has not always been possible to establish. To explore the functions of the Py miRNA in the mouse, we generated a miRNA-deficient mutant virus on the background of the virulent LID strain. The mutant virus was significantly less effective than the wild type in blocking apoptosis. This was examined in kidneys of neonatally infected mice which serves as the major site of virus amplification. Inhibition of apoptosis is expected to result in enhanced infection not only in the kidney but also in other vital tissues. Further studies will be needed to evaluate the role of Py miRNA in the establishment of acute as well as long term persistent infection, induction of tumors, as well as interactions with the host immune system.
The Py miRNA may target cellular genes in addition to Smad2. miRNAs of other members of the polyomavirus family have been shown or predicted to target a variety of cellular genes that serve the virus in terms of promoting virus replication or evading recognition by the host immune system. Human polyomaviruses BK and JC possess an identical miRNA which targets the stress-induced ligand ULBP3, and this inhibition contributes to immune evasion (Bauman et al., 2011). Human genes that are involved in B cell proliferation or cell cycle modulation have been predicted as potential targets of the Merkel cell polyomavirus-encoded miRNA although they await further validation (Lee et al., 2011). DUSP8 (M3/8), a negative regulator of c-Jun N-terminal kinases (JNK), is targeted by SV40-encoded miRNA, indicating that SV40 miRNA may be involved in the regulation of MAPK pathways (Chen et al., 2013; Muda et al., 1996). The miRNAs encoded by these small DNA viruses provides an efficient way to regulate the expression of multiple genes of their hosts.
Materials and methods
Cells
Baby mouse kidney cells were grown in DMEM with 10% newborn calf serum, and 293 FT, NIH 3T3 and IMCD mouse kidney epithelial cells (were grown in DMEM with 10% fetal bovine serum.
Plasmids and viruses
The pMIR luciferase reporter (Life Technologies, Carlsbad, CA, USA) was used to clone mouse Smad2 3′UTR. PCR fragments were prepared with a primer set 5′ TTACTAGTCTAGGAGTAAAGGGAGCGGGTTGG and 5′ TTAAGCTTCAATGGGGTACAATGCTGTAA for WT and a primer set 5′ TTACTAGTCTAGGAGTAAAGGGAGCGGGTTGG and 5′ TTAAGCTTTGGCTGGCTAAGGAGTGACAAGAAC for the deletion mutant. Each insert was cloned into the HindIII/SpeI site (downstream of the luciferase reporter gene) of the pMIR vector, and their structures were confirmed by sequencing. The pRL-TK vector was from Promega (Madison, WI, USA).
The lenti viral vector pLKO (Sigma Aldrich, St Louis, MO, USA) was used to clone the Py short hairpin with these synthetic nucleotides: PyshF 5′CCGGGGGGGGGGATGAGCTGGGGTACTTGTTCCTCCGGTAGGATGTCCAAATACAGATCC TCCATTGGCATGTACTCCTCCTCCTCCTCCTCCTTTTTG and PyshR 5′AATTCAAAAAGGAGGAGGAGGAGGAGGAGTACATGCCAATGGAGGATCTGTATTTGGAC ATCCTACCGGAGGAACAAGTACCCCAGCTCATCCCCCCCC
The newly constructed pLKO-Py-shRNA was sequenced to confirm its structure and used to make lenti viral particles in 293 FT cells with ViraPower Packaging Mix (Life Technologies). The viral supernatants were harvested after 48–72 hours, spun at 3,000 rpm for 10 min and filtered with 0.45 μm Millex syringe filters (EMD Millipore Corporation, Billerica, MA, USA). Virus was then concentrated by mixing with 5X PEG precipitation solution (40% Polyethylene Glycol, 0.4M NaCl and 2mM EDTA), stored overnight at 4°C and centrifuged at 3,000 rpm for 30 min. Pelleted virus was resuspended in PBS buffer and stored at −80° C.
A LID version of dl1013 (a derivative of the strain A2) was constructed with QuikChange site-directed mutagenesis kit (Agilent Technologies, La Jolla, CA, USA), a primer set (5′-GGAGTACATGCCAATGGAACCGGAGGAACAAGTACCCC / 5′-GGGGTACTTGTTCCTCCGGTTCCATTGGCATGTACTCC) and LID-pBluescript plasmid DNA as a template. After confirming the structure, the virus portion of the plasmid was transfected into NIH 3T3 cells, and viruses were harvested at 9 days post transfection.
Py-miRNA target predictions
The seed sequences (5′ GGGGUAC and 5′ CCAUUGG) of the Py-miRNAs were analyzed with TargetScanMouse Custom Version 5.1 (Lewis, Burge, and Bartel, 2005) for prediction of targets. Another web-based program RNAhybrid (Kruger and Rehmsmeier, 2006) was employed to predict the Py-miRNA binding sites in 3′UTR regions of mouse genes.
Immunoblot analysis
Cells were lysed in PBS containing 1% NP40 and protease inhibitors (Roche, South San Francisco, CA, USA). Immunoblotting was carried out on cell extracts using an Odyssey infrared imaging system (LI-COR Biosciences, Lincoln, NE).
Fluorometric caspase-3 activity assay
Fifty micrograms of whole-cell lysates was incubated with 200 nM Ac-DEVD-AMC (Ac is N-acetyl, DEVD is Asp-Glu-Val-Asp, and AMC is 7-amino-4-methylcoumarin) (BD Biosciences; Franklin Lakes, NJ) in reaction buffer (20 mM HEPES [pH 7.4], 2 mM dithiothreitol [DTT], 10% glycerol) at 37°C for 1 h. The reaction was monitored by fluorescence emission at 465 nm (excitation at 360 nm) and measured with a fluorescence plate reader.
Luciferase dual assays
The vectors pRL-TK and pMIR, pMIR-mSmad2 3′URT WT or pMIR-mSmad2 3′URT del mutant were introduced into IMCD cells with Lipofectamine 2000 (Life Technologies), and then those cells were infected with control lenti or lenti-Py-shRNA. After 48 hours of incubation at 37 °C, cells were subjected to luciferase activity assays (Dual-Luciferase Reporter System). Firefly luciferase values were normalized to those of Renilla luciferase.
TUNEL assays
All mice were bred and maintained in a specific-pathogen-free animal facility at Harvard Medical School. Protocols for animal studies have been reviewed and approved by the Harvard Medical Area Standing Committee on Animals (“HMA IACUC”), and are in accordance with PHS policy on Care and Use of Laboratory Animals under the guidance of the Office of Laboratory Animal Welfare (OLAW) within the NIH.
New born C3H/BiDa mice were injected intraperitoneally with ~50μl of virus suspension containing 106 pfu of the LID or LID-dl1013 strain of Py. Infected mice were then held in a dedicated infected animal facility. Infected mice and uninfected control mice (age-matched) were sacrificed 8 days post-inoculation, and the kidney tissues were immediately removed and frozen. Tissue section slides were prepared at the Harvard Rodent Histopathology Core.
TUNEL assays were performed using ApopTag Red In Situ Apoptosis Detection Kit (EMD Millipore Corporation) as described in the manufacturer’s protocols with slight modifications. Briefly, tissue sections were fixed in neutral buffered formalin, washed and incubated for 5 min in antigen retrieval solution (HistoReveal; Abcam, Cambridge, MA, USA). After washing, they were treated with Proteinase K and then incubated with TdT enzyme for 1 hr at 37° C. After washing, they were incubated with 1:50 anti-Py tumor antigen (generated in rat; Benjamin Lab) for 16 hrs at 4° C. Then Anti-digoxigenin conjugate (rhodamine) and anti-rat IgG (Oregon Green) were used for 1 hr at room temperature followed by washing. Samples were mounted with a mounting medium containing DAPI and viewed by fluorescence microscopy.
For quantification, stained slides were read by a researcher blinded to the tissue source type using a Nikon Fluorescent Microscope and accompanying software. Briefly, for each tissue, 8–10 fields were chosen and imaged for red and green fluorescence, with the merged images used to count the fraction of infected (green-fluorescing) cells that were concurrently undergoing apoptosis (red-fluorescing). Results were averaged across all tissues and the groups compared using t-tests.
Supplementary Material
Supplementary Materials Figure S1. Py-miRNA may target Smad2 3′UTR (predicted by RNAhybrid).
AUTHOR HIGHLIGHTS.
A mouse polyomavirus-encoded microRNA represses Smad2 expression.
Programmed cell death machinery is targeted by the polyoma microRNA.
The polyoma microRNA blocks apoptosis in infected mouse kidney tissues.
Acknowledgments
This study was supported by Texas A&M University-Kingsville Research Award (to CKS; 160330-00016) and National Cancer Institute R01 (to TLB; CA-092320).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer PH, Bronson RT, Fung SC, Freund R, Stehle T, Harrison SC, Benjamin TL. Genetic and structural analysis of a virulence determinant in polyomavirus VP1. J Virol. 1995;69 (12):7925–31. doi: 10.1128/jvi.69.12.7925-7931.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauman Y, Nachmani D, Vitenshtein A, Tsukerman P, Drayman N, Stern-Ginossar N, Lankry D, Gruda R, Mandelboim O. An identical miRNA of the human JC and BK polyoma viruses targets the stress-induced ligand ULBP3 to escape immune elimination. Cell Host Microbe. 2011;9(2):93–102. doi: 10.1016/j.chom.2011.01.008. [DOI] [PubMed] [Google Scholar]
- Benjamin TL. Polyoma virus: old findings and new challenges. Virology. 2001;289(2):167–73. doi: 10.1006/viro.2001.1124. [DOI] [PubMed] [Google Scholar]
- Benjamin TL. Polyoma Viruses. In: Fox MTDJJ, Quimby FW, Barthold AW, Newcomer CE, Smith AL, editors. In The Mouse in Biomedical Research. 2. Academic Press; 2007. pp. 105–139. [Google Scholar]
- Bolen JB, Fisher SE, Chowdhury K, Shan TC, Williams JE, Dawe CJ, Israel MA. A determinant of polyomavirus virulence enhances virus growth in cells of renal origin. J Virol. 1985;53(1):335–9. doi: 10.1128/jvi.53.1.335-339.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CJ, Cox JE, Kincaid RP, Martinez A, Sullivan CS. Divergent MicroRNA targetomes of closely related circulating strains of a polyomavirus. J Virol. 2013;87(20):11135–47. doi: 10.1128/JVI.01711-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, DeCaprio JA, Fluck MM, Schaffhausen BS. Cellular transformation by Simian Virus 40 and Murine Polyoma Virus T antigens. Semin Cancer Biol. 2009;19(4):218–28. doi: 10.1016/j.semcancer.2009.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy EY, Siu KL, Kok KH, Lung RW, Tsang CM, To KF, Kwong DL, Tsao SW, Jin DY. An Epstein-Barr virus-encoded microRNA targets PUMA to promote host cell survival. J Exp Med. 2008;205(11):2551–60. doi: 10.1084/jem.20072581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen BR. Five questions about viruses and microRNAs. PLoS Pathog. 2010;6(2):e1000787. doi: 10.1371/journal.ppat.1000787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl J, Jurczak A, Cheng LA, Baker DC, Benjamin TL. Evidence of a role for phosphatidylinositol 3-kinase activation in the blocking of apoptosis by polyomavirus middle T antigen. J Virol. 1998;72(4):3221–6. doi: 10.1128/jvi.72.4.3221-3226.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlke C, Maul K, Christalla T, Walz N, Schult P, Stocking C, Grundhoff A. A microRNA encoded by Kaposi sarcoma-associated herpesvirus promotes B-cell expansion in vivo. PLoS One. 2012;7(11):e49435. doi: 10.1371/journal.pone.0049435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fluck MM, Schaffhausen BS. Lessons in signaling and tumorigenesis from polyomavirus middle T antigen. Microbiol Mol Biol Rev. 2009;73(3):542–63. doi: 10.1128/MMBR.00009-09. Table of Contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freund R, Bauer PH, Crissman HA, Bradbury EM, Benjamin TL. Host range and cell cycle activation properties of polyomavirus large T-antigen mutants defective in pRB binding. J Virol. 1994;68(11):7227–34. doi: 10.1128/jvi.68.11.7227-7234.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garzon R, Calin GA, Croce CM. MicroRNAs in Cancer. Annu Rev Med. 2009;60:167–79. doi: 10.1146/annurev.med.59.053006.104707. [DOI] [PubMed] [Google Scholar]
- Grundhoff A, Sullivan CS. Virus-encoded microRNAs. Virology. 2011;411(2):325–43. doi: 10.1016/j.virol.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu H, Li D, Sung CK, Yim H, Troke P, Benjamin T. DNA-binding and regulatory properties of the transcription factor and putative tumor suppressor p150(Sal2) Biochim Biophys Acta. 2011;1809(4–6):276–83. doi: 10.1016/j.bbagrm.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heldin CH, Landstrom M, Moustakas A. Mechanism of TGF-beta signaling to growth arrest, apoptosis, and epithelial-mesenchymal transition. Curr Opin Cell Biol. 2009;21(2):166–76. doi: 10.1016/j.ceb.2009.01.021. [DOI] [PubMed] [Google Scholar]
- Hough C, Radu M, Dore JJ. Tgf-beta induced Erk phosphorylation of smad linker region regulates smad signaling. PLoS One. 2012;7(8):e42513. doi: 10.1371/journal.pone.0042513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imperiale MJ. Polyomavirus miRNAs: the beginning. Curr Opin Virol. 2014;7C:29–32. doi: 10.1016/j.coviro.2014.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imperiale MJ, Major EO. Polyomaviruses. In: Knipe DM, Howley PM, editors. Fields Virology. 5. Vol. 2. Lippincott Williams & Wilkins; Philadelphia, PA: 2007. pp. 2263–2298. [Google Scholar]
- Kincaid RP, Sullivan CS. Virus-encoded microRNAs: an overview and a look to the future. PLoS Pathog. 2012;8(12):e1003018. doi: 10.1371/journal.ppat.1003018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruger J, Rehmsmeier M. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res. 2006;34(Web Server issue):W451–4. doi: 10.1093/nar/gkl243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet. 2007;39(5):673–7. doi: 10.1038/ng2003. [DOI] [PubMed] [Google Scholar]
- Lee S, Paulson KG, Murchison EP, Afanasiev OK, Alkan C, Leonard JH, Byrd DR, Hannon GJ, Nghiem P. Identification and validation of a novel mature microRNA encoded by the Merkel cell polyomavirus in human Merkel cell carcinomas. J Clin Virol. 2011;52(3):272–5. doi: 10.1016/j.jcv.2011.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120(1):15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Li D, Dower K, Ma Y, Tian Y, Benjamin TL. A tumor host range selection procedure identifies p150(sal2) as a target of polyoma virus large T antigen. Proc Natl Acad Sci U S A. 2001;98 (25):14619–24. doi: 10.1073/pnas.251447198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Tian Y, Ma Y, Benjamin T. p150(Sal2) is a p53-independent regulator of p21(WAF1/CIP) Mol Cell Biol. 2004;24(9):3885–93. doi: 10.1128/MCB.24.9.3885-3893.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Adler AS, Segal E, Chang HY. A transcriptional program mediating entry into cellular quiescence. PLoS Genet. 2007;3(6):e91. doi: 10.1371/journal.pgen.0030091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnusson G, Berg P. Construction and analysis of viable deletion mutants of polyoma virus. J Virol. 1979;32(2):523–9. doi: 10.1128/jvi.32.2.523-529.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquitz AR, Mathur A, Nam CS, Raab-Traub N. The Epstein-Barr Virus BART microRNAs target the pro-apoptotic protein Bim. Virology. 2011;412(2):392–400. doi: 10.1016/j.virol.2011.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moustakas A, Heldin CH. The regulation of TGFbeta signal transduction. Development. 2009;136(22):3699–714. doi: 10.1242/dev.030338. [DOI] [PubMed] [Google Scholar]
- Muda M, Theodosiou A, Rodrigues N, Boschert U, Camps M, Gillieron C, Davies K, Ashworth A, Arkinstall S. The dual specificity phosphatases M3/6 and MKP-3 are highly selective for inactivation of distinct mitogen-activated protein kinases. J Biol Chem. 1996;271(44):27205–8. doi: 10.1074/jbc.271.44.27205. [DOI] [PubMed] [Google Scholar]
- Pfeffer S, Zavolan M, Grasser FA, Chien M, Russo JJ, Ju J, John B, Enright AJ, Marks D, Sander C, Tuschl T. Identification of virus-encoded microRNAs. Science. 2004;304(5671):734–6. doi: 10.1126/science.1096781. [DOI] [PubMed] [Google Scholar]
- Rozenblatt-Rosen O, Deo RC, Padi M, Adelmant G, Calderwood MA, Rolland T, Grace M, Dricot A, Askenazi M, Tavares M, Pevzner SJ, Abderazzaq F, Byrdsong D, Carvunis AR, Chen AA, Cheng J, Correll M, Duarte M, Fan C, Feltkamp MC, Ficarro SB, Franchi R, Garg BK, Gulbahce N, Hao T, Holthaus AM, James R, Korkhin A, Litovchick L, Mar JC, Pak TR, Rabello S, Rubio R, Shen Y, Singh S, Spangle JM, Tasan M, Wanamaker S, Webber JT, Roecklein-Canfield J, Johannsen E, Barabasi AL, Beroukhim R, Kieff E, Cusick ME, Hill DE, Munger K, Marto JA, Quackenbush J, Roth FP, DeCaprio JA, Vidal M. Interpreting cancer genomes using systematic host network perturbations by tumour virus proteins. Nature. 2012;487(7408):491–5. doi: 10.1038/nature11288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer SL, Sorger PK. Measuring and modeling apoptosis in single cells. Cell. 2011;144(6):926–39. doi: 10.1016/j.cell.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan CS, Grundhoff AT, Tevethia S, Pipas JM, Ganem D. SV40-encoded microRNAs regulate viral gene expression and reduce susceptibility to cytotoxic T cells. Nature. 2005;435(7042):682–6. doi: 10.1038/nature03576. [DOI] [PubMed] [Google Scholar]
- Sullivan CS, Sung CK, Pack CD, Grundhoff A, Lukacher AE, Benjamin TL, Ganem D. Murine Polyomavirus encodes a microRNA that cleaves early RNA transcripts but is not essential for experimental infection. Virology. 2009;387(1):157–67. doi: 10.1016/j.virol.2009.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S, Xue C, Li J, Bi Y, Cao Y. Marek’s disease virus type 1 microRNA miR-M3 suppresses cisplatin-induced apoptosis by targeting Smad2 of the transforming growth factor beta signal pathway. J Virol. 2011;85(1):276–85. doi: 10.1128/JVI.01392-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Materials Figure S1. Py-miRNA may target Smad2 3′UTR (predicted by RNAhybrid).