Abstract
With the continuing interest in deciphering the interplay between protein function and conformational changes, small fluorescence probes will be especially useful for tracking changes in the crowded protein interior space. Presently, we describe the potential utility of six unnatural amino acid fluorescence donors structurally related to tryptophan and show how they can be efficiently incorporated into a protein as fluorescence probes. We also examine the various photophysical properties of the new Trp analogues, which are significantly redshifted in their fluorescence spectra relative to tryptophan. In general, the Trp analogues were well tolerated when inserted into E. coli DHFR, and did not perturb enzyme activity, although substitution for Trp22 did result in a diminution in DHFR activity. Further, it was demonstrated that D and E at position 37 formed efficient FRET pairs with acridon-2-ylalanine (Acd) at position 17. The same was also true for a DHFR construct containing E at position 79 and Acd at position 17. Together, these findings demonstrate that these tryptophan analogues can be introduced into DHFR with minimal disruption of function, and that they can be employed for the selective study of targeted conformational or electrostatic changes in proteins, even in the presence of unmodified tryptophans.
Keywords: fluorescence, amino acid, tryptophan analogues, FRET, protein biosynthesis
1. Introduction
Following the changes in the fluorescence spectrum of a protein is a common technique for monitoring protein conformational and electrostatics transitions due to ligand binding, protein-protein associations, or chemical transformations. The fluorescence signal can result from naturally occurring amino acids (e.g. tryptophan and tyrosine) or can be introduced chemically (e.g. via thio linked Alexa dyes). The sensitivity of fluorescence techniques allows the monitoring of subtle biophysical changes under equilibrium or non-equilibrium conditions. Therefore, protein conformation and function changes are widely studied through the use of Förster resonance energy transfer (FRET) by measuring changes in the efficiency of energy transfer between a donor and acceptor.1,2 The acceptor may also be a fluorescent molecule, leading to another longer wavelength emission, or a quencher.2-6 With the appropriate pairs, FRET can be used to quantitatively determine distance changes that are related to protein processes.
Typically, a FRET system in a single protein may include two different variants of green fluorescent protein (GFP),1,7-9 or two large polycyclic aromatic molecules.10,11 Additionally, the large polycyclic aromatic fluorophores/quenchers are often chemically tethered to the proteins via flexible linkers.10,11 However, the large size of a fluorophore can significantly perturb protein structure and function. This is particularly of concern when trying to monitor conformational changes in a crowded protein interior space, such as an enzyme active site. Moreover, the flexible tethers used for attaching large fluorophores provide the reporter molecules with still more conformational freedom, independent of actual protein conformational changes. In fact, this is a vastly under-appreciated and problematic issue; a recent study has pointed out that many observed reaction-coupled protein motions might be completely disconnected from the actual chemical transformation.12 Thus, to measure subtle conformational changes in proteins, it is important to develop smaller fluorophores/quenchers that can be efficiently incorporated into the normal peptide backbone, thus limiting their degrees of freedom unrelated to changes in protein structure.
A biosynthetic method has been employed to site-specifically incorporate smaller size fluorescent amino acids into proteins.13-18 A few studies have incorporated a donor and an acceptor amino acids into a single protein, through decoding a four-base codon CGGG and a nonsense codon UAG in the presence of an aminoacyl-tRNACCCG and an aminoacyl-tRNACUA.19-22 Such small amino acid FRET pairs have been shown to be useful for studying the reorganization of protein structure20-22 or for monitoring protein backbone cleavage.19 In previous studies, we have incorporated a series of biphenyl-L-phenylalanine derivatives into E. coli dihydrofolate reductase (DHFR) as a fluorescence donor and L-(7-hydroxycoumarin-4-yl)ethylglycine as an acceptor, to study DHFR conformational changes.21,22 In these reports, all of the amino acids studied were well tolerated at position 17 of DHFR, which is sterically accessible. However, DHFR displayed a range of sensitivities to the individual amino acids at position 115, which points into the active site of DHFR. This finding demonstrated the importance of identifying fluorescent amino acids that can minimally perturb protein structures to permit study of their subtle conformational changes.
Among the most thoroughly investigated Trp analogues are the azatryptophans, which have proved to be almost ideal isosteric substitutes for natural tryptophan in cellular proteins.23,24 Among the azaindoles under study, 4- and 7-azaindoles have exhibited the largest Stokes shifts in steady-state fluorescence measurements.23,24 They are highly biocompatible and as azatryptophans they can be introduced into target protein sequences by ribosomal translation. Recently, N-methylated 4- and 7-azaindoles have been found to have fluorescence properties better than the parent 4- and 7-azaindoles because they retain the pronounced red shift characteristic of the parent 4- and 7-azaindoles, but with more intense fluorescence.23
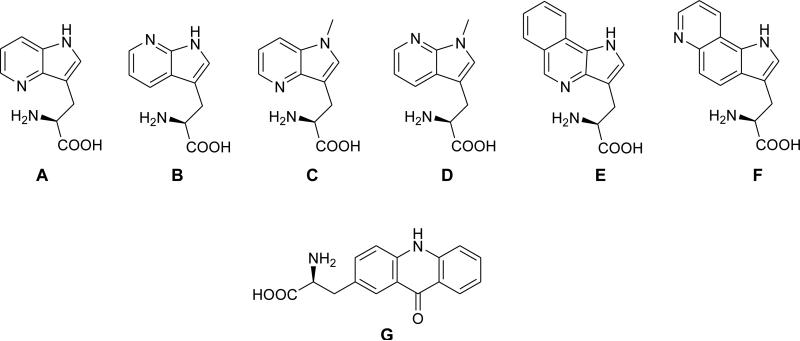
Prompted by the need to develop smaller and sensitive fluorophores that can be readily accommodated within a protein without perturbation of the conformation and function of that protein, we have synthesized more hydrophobic N-methylated 4- and 7-azatryptophans and also two tricyclic tryptophan derivatives (Figure 1).25 The asymmetric syntheses of the Trp analogues were accomplished by a stereoselective strategy utilizing the Schöllkopf chiral reagent,25 and the photophysical properties of these Trp analogues were characterized. While size is one important factor of FRET donor/acceptor pairs, intrinsic qualities of the molecules such as their rotational degrees of freedom and propensity to interact with water can also be important. In our previous studies, we have reported a series of hydrophobic amino acids as the fluorescence donor.21,22 We have now explored a series of derivatives of tryptophan, which acts as an important probe for fluorescence analysis of protein structure and function. Spectral isolation of individual residues is typically difficult in proteins containing more than one tryptophan residue. Thus, it is desirable to substitute tryptophan by a fluorophore having different spectroscopic properties (quantum yield, excitation and emission wavelengths) while at the same time maintaining native protein conformation and function.
Figure 1.
Tryptophan analogues (A-F) and L-acridon-2-ylalanine (Acd, G) which were incorporated into dihydrofolate reductase.
These Trp analogues possess many photophysical properties that are unique from normal Trp, suggesting that they can be used to selectively monitor a targeted conformational change even in the presence of multiple native Trp residues (Tables 1 and 2). In general, the Trp analogues induce minimal effects on the enzyme activity when incorporated into DHFR. Two analogues (D and E) having favorable fluorescence properties were used to demonstrate that these Trp derivatives can function as donors to form efficient FRET pairs with acridon-2-ylalanine (Acd, G)26,27 as the acceptor. Thus, these compounds will have considerable value for a variety of fluorescence studies.
Table 1.
Fluorescence and Absorption Energy Maxima and Fluorescence Lifetimes of N-Acetylated Methyl Esters of Trp Analogues
| tryptophan derivatives | neutral species () | cationic species () | ||||
|---|---|---|---|---|---|---|
| λ abs, max | λ em, max | fluorescence lifetime, τ | λ abs, max | λ em, max | fluorescence lifetime, τ | |
| A | 290 nm | 390 nm | τ1 = 0.94 ± 0.02 ns | 286, 340 nm | 449 nm | τ1 = 3.51 ± 0.08 ns |
| τ2 = 2.16 ± 0.09 ns | τ2 = 4.84 ± 0.05 ns | |||||
| B | 289 nm | 371 nm | τ1 = 0.17 ± 0.01 ns | 292 nm | 437 nm | τ1 = 1.11 ± 0.00 ns |
| τ2 = 4.84 ± 1.00 ns | τ2 = 2.45 ± 0.22 ns | |||||
| C | 292 nm | 404 nm | τ1 = 1.49 ± 0.05 ns | 291 nm | 456 nm | τ1 = 3.79 ± 0.10 ns |
| τ2 = 2.98 ± 0.20 ns | τ2 = 4.74 ± 0.01 ns | |||||
| D | 288 nm | 391 nm | τ1 = 3.67 ± 1.00 ns | 295 nm | 382 nm | τ1 = 2.05 ± 0.11 ns |
| τ2 = 14.6 ± 0.2 ns | τ2 = 7.63 ± 0.26 ns | |||||
| E | 262 nm | 413 nm | τ1 = 3.02 ± 0.02 ns | 259 nm | 493 nm | τ1 = 1.13 ± 0.02 ns |
| τ2 = 5.79 ± 0.02 ns | τ2 = 3.84 ± 0.06 ns | |||||
| F | 272 nm | 421 nm | τ1 = 1.76 ± 0.20 ns | 287 nm | 526 nm | τ1 = 2.32 ± 0.09 ns |
| τ2 = 3.53 ± 0.09 ns | τ2 = 3.69 ± 0.06 ns | |||||
| Tryptophana | 280 nm | 350 nm | τ1 = 3.16 ± 0.1 ns | 277 nm | 362 nm | τ1 = 3.2 ± 0.1 ns |
| τ2 = 0.50 ± 0.20 ns | τ2 = 0.52 ± 0.15 ns | |||||
| τ3 = 9.2 ± 0.1 ns | ||||||
Tryptophan fluorescence lifetimes were measured in sodium tetraborate buffer, pH 8.0, and HCl–water solution at pH 3.5.44
Table 2.
Molar Absorptivities and Quantum Yields of N-Acetylated Methyl Esters of Trp Analogues in MeOH
| Trp analogue | ε (M−1cm−1) | Φ F |
|---|---|---|
| tryptophan | 6900 | 0.18 |
| A | 7870 | 0.04 |
| B | 8920 | 0.02 |
| C | 7740 | 0.06 |
| D | 8070 | 0.30 |
| E | 18100 | 0.10 |
| F | 15500 | 0.03 |
2. Results
2.1. Synthesis of Fluorescent Aminoacyl pdCpAs Esters
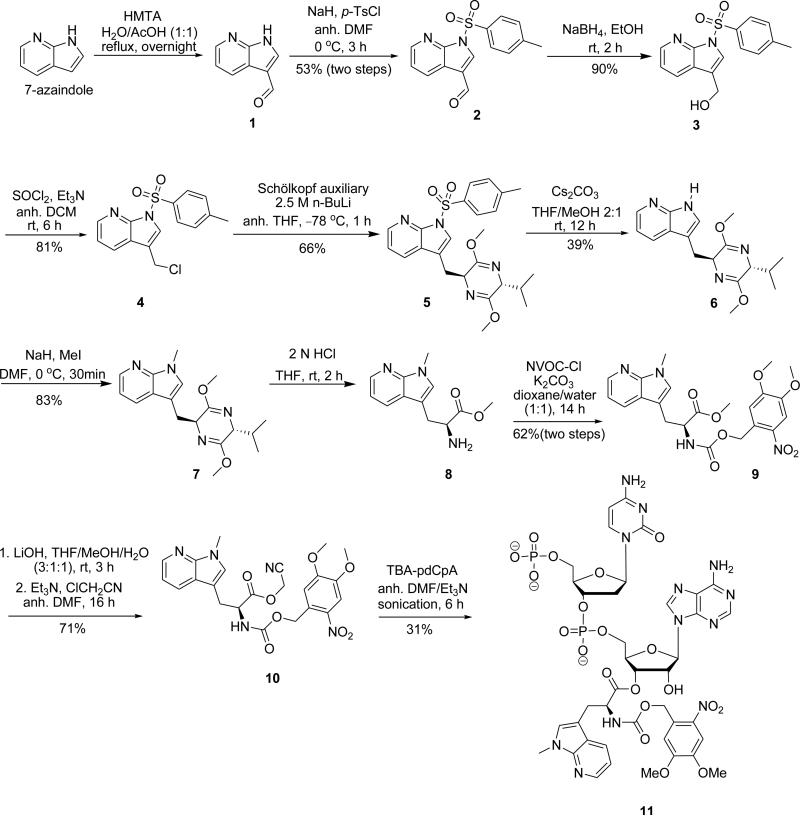
The synthesis of the aminoacylated pdCpA derivative of amino acid D (Figure 1) was accomplished starting from commercially available 7-azaindole which was first formylated under Duff reaction conditions28 to yield aldehyde 1, the latter of which was subjected to N-tosylation with p-toluenesulfonyl chloride (p-TsCl) and sodium hydride to afford 2 in 53% overall yield (Scheme 1).29 Reduction of aldehyde2 to alcohol 3 using NaBH4 in EtOH proceeded almost quantitatively.28 Chlorination of alcohol 3 with thionyl chloride afforded 4 in 81% yield.30 Asymmetric synthesis of the amino acid precursor was carried out using the Schöllkopf reagent ((R)-2,5-dihydro-3,6-dimethoxy-2-isopropylpyrazine).31 Regioselective lithiation (n-BuLi, THF, −78 °C) of the chiral auxiliary produced the lithium enolate, which afforded the adduct 5 from 4 with high diastereoselectivity (only one diastereomer was detectable in the 1H NMR and 13C NMR spectra). N-Detosylation of 5 was performed using cesium carbonate in 2:1 THF/MeOH to yield 6 in 39% yield.32 N-methylation of 6 with methyl iodide and sodium hydride afforded 7 in 83% yield.29 Mild hydrolysis (2 N HCl) afforded the α-substituted amino acid methyl ester 831 which was protected as the NVOC carbamate to yield 9 in 62% overall yield.21 Methyl ester 9 was then hydrolyzed to afford the free acid which was subsequently treated with chloroacetonitrile to afford the requisite cyanomethyl ester 10 in 31% yield.21
Scheme 1.
Synthetic Route Employed for the Preparation of Amino Acid D and its Aminoacyl-pdCpA.
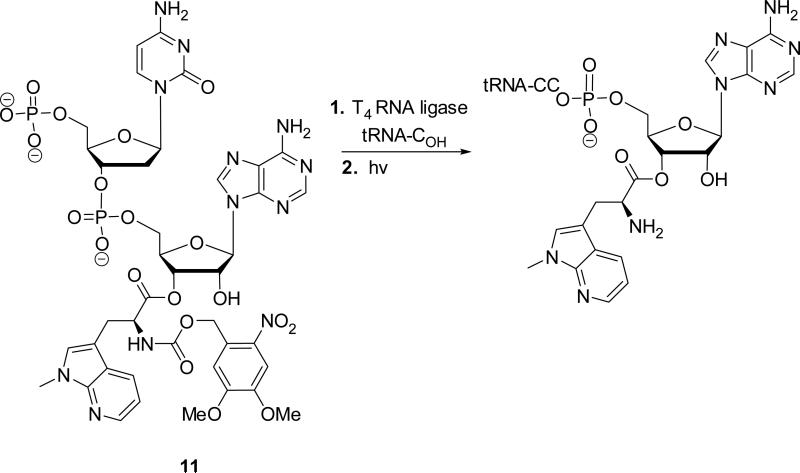
In order to evaluate the properties of D as a constituent of the protein DHFR, this amino acid was used to activate a suppressor tRNA (Scheme 2). Treatment of cyanomethyl ester 10 with the tris(tetrabutylammonium) salt of pdCpA33 in anhydrous DMF afforded pdCpA ester 11 in 52% yield. The aminoacylated dinucleotide was ligated to an abbreviated tRNACUA-COH transcript in the presence of T4 RNA ligase and ATP to afford NVOC-aminoacyl-tRNA. The activated tRNA was deprotected by UV irradiation to afford free aminoacyl-tRNA.21
Scheme 2.
Preparation of Suppressor tRNACUA Activated with Amino Acid D.
The synthesis of the aminoacylated pdCpA derivative of amino acid A (Figure 1) employed a similar route (Scheme 1 of the Supplementary data). Initially, commercially available 4-azaindole was formylated to yield 12 which was N-tosylated with p-TsCl and NaH, affording 13 in 23% overall yield.29 Reduction of aldehyde 13 with NaBH4 in EtOH afforded alcohol 14 in 87% yield.28 Chlorination of 14 with thionyl chloride then provided 15. Regioselective lithiation (n-butyllithium, THF, −78°C) of the Schöllkopf reagent produced the lithium enolate which afforded adduct 16 in 62% yield with high diastereoselectivity.31 Mild hydrolysis (2 N HCl) provided α-substituted amino acid methyl ester 1729 which was protected as the NVOC carbamate to yield 18 in 33% overall yield.21 N-Detosylation of 18 using cesium carbonate in 2:1 THF/MeOH afforded ester 19.32 Hydrolysis of 19 then afforded the free acid, which was activated as cyanomethyl ester 20 in 52% overall yield.21 Treatment of the cyanomethyl ester with a solution of the tris (tetrabutylammonium) salt of pdCpA33 in dry DMF gave the corresponding aminoacylated pdCpA containing amino acid A in 42% yield.
The syntheses of the aminoacylated pdCpA derivatives of amino acid B and C (Figure 1) followed the same procedures as for amino acids A and D, respectively (Schemes 2 and 3 of the Supplementary data). The synthesis of the aminoacylated pdCpA derivative of amino acid G (Figure 1) was accomplished starting from commercially available Boc-4-aminophenylalanine which was first methylated with thionyl chloride in methanol to yield the methyl ester 33 (Scheme 4 of the Supplementary data). Treatment of 33 with 2-chlorobenzoic acid then gave 34 in 47% yield.26 Cyclodehydration of 34 to provide acridinone derivative 35 using hot polyphosphoric acid (PPA) caused loss of the Boc protecting group.26 Accordingly, free amine 35 was protected as the NVOC carbamate, yielding 36 in 10% overall yield from 34.21 Methyl ester 36 was then treated with LiOH to afford the free acid, which was subsequently treated with chloroacetonitrile to afford the requisite cyanomethyl ester 37 in 40% yield.21 Treatment of the cyanomethyl ester with a solution of tris (tetrabutylammonium) salt of pdCpA33 in dry DMF gave the corresponding aminoacylated pdCpA containing amino acid G in 34% yield.
The synthesis of the N-acetylated methyl esters of Trp analogues is illustrated in Scheme 5 of the Supplementary data. Compounds 40 and 42 were prepared from intermediates 17 and 22, respectively, which were acetylated using Ac2O;34 tosyl group removal was effected using cesium carbonate.32 Compounds 43 and 44 were synthesized by acetylating intermediates 29 and8, respectively (Scheme 5 of the Supplementary data).
2.2 Activation of Suppressor tRNACUA and Synthesis of Modified DHFRs
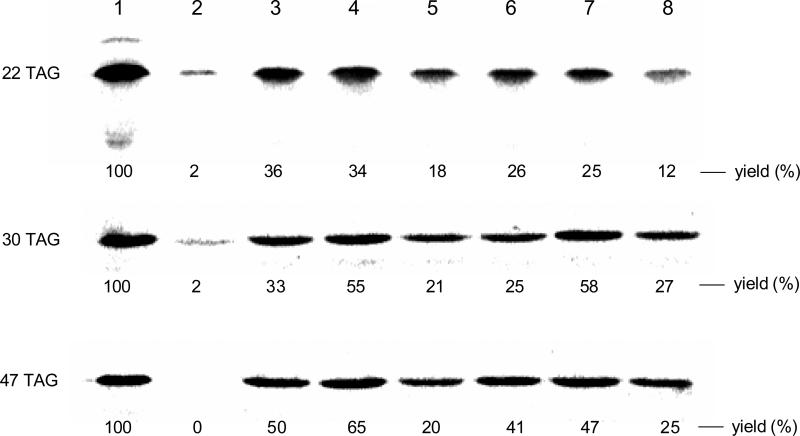
The individual N-NVOC protected aminoacylated pdCpA derivatives were ligated to a suppressor tRNACUA lacking its 3′-terminal cytidine and adenosine residues (tRNACUA-COH35) via the agency of T4 RNA ligase. As shown in Figure 1 of the Supplementary data, this assay afforded full length tRNA transcripts activated with amino acids A – F. The NVOC protecting groups were then removed by exposure to high intensity UV light at 4 °C. The aminoacyl-tRNAs so obtained were employed in an in vitro cell free transcription-translation system, which was programmed with DHFR DNA plasmids containing TAG codons at the positions corresponding to residues Trp22, Trp30 or Trp47 of DHFR. Modified DHFR synthesis was carried out in the presence of tryptophanyl-tRNACUA derivatives. As shown in Figure 2, each of the six tryptophanyl-tRNAs afforded good suppression of the UAG codons at positions 22, 30 and 47 of DHFR mRNAs, with suppression yields ranging from 12 to 65% compared to wild-type DHFR. Each of the modified DHFRs contained a hexahistidine fusion peptide at the N-terminus of DHFR,35 providing a convenient means to purify the proteins on a Ni-NTA column.36 Final purification of each was then accomplished on a DEAE-Sepharose CL-6B column. The purification is illustrated in Figure 2 of the Supplementary data for the DHFR containing amino acid D at position 22.
Figure 2.
Autoradiogram of a 15% SDS-polyacrylamide gel (100 V, 2 h) illustrating the incorporation of tryptophan analogues into positions 22 (upper panel), 30 (middle panel) and 49 (lower panel) of DHFR. Lane 1, wild-type DHFR expression; lane 2, modified DHFR DNA in the presence of abbreviated suppressor tRNACUA-COH; lane 3, incorporation of amino acid A; lane 4, incorporation of amino acid B; lane 5, incorporation of amino acid C; lane 6, incorporation of amino acid D; lane 7, incorporation of amino acid E; lane 8, incorporation of amino acid F. Phosphorimager analysis was performed using an Amersham Biosciences Storm 820 equipped with ImageQuant version 5.2 software from Molecular Dynamics.
The enzymatic activities of the modified DHFRs were judged by their ability to consume NADPH (Table 3) under steady-state conditions. Since the substrates (dihydrofolate and NADPH) are in excess of the enzyme, the rate constants measured represent the enzyme turnover efficiency. Replacement of Trp22, which is in the catalytically relevant Met20 loop subdomain of DHFR,37 with tryptophan analogues A – D resulted in reduction in enzyme activity (the turnover rate constants were found to be ~22 – 51% of that obtained for wild-type DHFR38 under the same assay conditions). Substitution of the bulkier tricyclic amino acids E and F at position 22 resulted in a more significant reduction of DHFR activity. The differences in enzyme activity between the six modified DHFRs likely reflects the location of the modified Trps near the substrate binding site (Met20 loop).37,39 Replacement of the two tryptophan residues (Trp30 and Trp47), which are not located on the catalytically important Met20 loop,40 did not affect the activity of the enzyme. The results demonstrated that these tryptophan derivatives have properties as fluorescence donors suitable for minimal perturbation of protein structures, thus potentially allowing the study of conformational changes in DHFR.
Table 3.
Enzymatic Activities of DHFRs Singly Modified at Positions 22, 30 or 47
| DHFR | position 22 (Trp) (kD, s−1) | position 30 (Trp) (kD, s−1) | position 47 (Trp) (kd, s−1) |
|---|---|---|---|
| wild-type DHFR38 | 12 | 12 | 12 |
| A | 6.1 ± 0.5a | 13.2 ± 0.4 | 11.5 ± 0.4 |
| B | 5.4 ± 0.4 | 12.6 ± 0.4 | 12.4 ± 0.4 |
| C | 2.6 ± 0.2 | 12.0 ± 0.4 | 13.8 ± 0.4 |
| D | 3.7 ± 0.2 | 13.9 ± 0.5 | 13.2 ± 0.5 |
| E | 0.7 ± 0.1 | 12.5 ± 0.4 | 11.9 ± 0.4 |
| F | 0.8 ± 0.1 | 13.4 ± 0.5 | 12.8 ± 0.4 |
Standard deviation is based on data from three experiments.
2.3 Tryptophan-based Fluorophores
We characterized the various photophysical properties of the six tryptophan-based fluorophores by measuring their molar absorptivities, quantum yields, emission maxima, absorption maxima, and fluorescence lifetimes of the N-acetylated methyl esters of the Trp analogues in methanol (Tables 1 and 2). At pH 6.5, L-tryptophan has maximum absorption and fluorescence emission centered at 280 and ~350 nm, respectively.41,42 N-Acetylation and esterification have been shown not to affect the spectrophotometric properties of aromatic amino acids like tryptophan and tyrosine. Also, it is known that the fluorescence decay of tryptophan is biphasic, which can be best described by two fluorescence lifetimes of ~3.15 and ~0.5 ns. This has been attributed to different rotamers or conformers of the alkyl side chain of tryptophan.42,43
Table 1 shows that these Trp derivatives possess spectrophotometric properties that differ from tryptophan itself, and in some cases the differences are quite significant. Moreover, by adjusting the pH of the medium, we found that the neutral and protonated species can have very different photophysical properties. This allows us to attribute the measured data strictly to one dominant species in solution and avoids the complexity of contributions from a mixture of two species. In general, the absorption maxima of the Trp analogues do not differ greatly (~ ±10 nm) from native tryptophan, with derivative E showing the largest difference. In contrast, the fluorescence emission spectra of the Trp derivatives are shifted (bathochromic shift) from normal tryptophan to a greater degree. This is especially true for the tricyclic compounds E and F. Further, for all six Trp derivatives (both neutral and protonated species), we observed biphasic fluorescence decay. The measured fluorescence lifetimes of the six Trp derivatives are also different from that of normal tryptophan. Interestingly, the biggest difference was found in D where the two fluorescence lifetimes are significantly longer than tryptophan.
To explore the capability of these tryptophan derivatives as FRET donors, we selected two analogues (D and E) and incorporated them into DHFR. As shown in Table 2, analogue D has a higher molar absorbance coefficient (8070) and a higher quantum yield (0.30) than tryptophan (6900 and 0.18, respectively); and analogue E has a 2.6-fold greater molar absorbance coefficient than tryptophan, albeit at a lower (0.10) quantum yield. Both of the analogues exhibit stronger fluorescence emission and should thus minimize the background from the tryptophans in DHFR. The emission wavelength maxima of tryptophan analogues D and E are at 391 nm and 413 nm, respectively (Table 1). To utilize the emissions of these two fluorescence donors in FRET, we selected acridon-2-ylalanine (G) as the fluorescence acceptor; this fluorophore has absorption peaks at 388 and 407 nm in H2O and emission peaks at ~ 420 and 450 nm.26 Using these pairs, one can selectively excite G through the emission from D/E while keeping the five native tryptophans of DHFR intact.
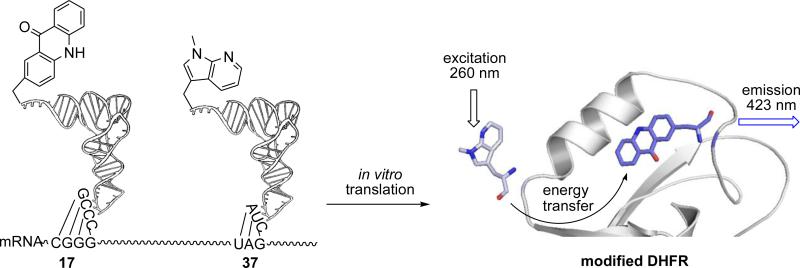
Acridon-2-ylalanine has been used as a fluorescence acceptor for FRET studies previously.27 It has been reported to have a small size (222 Å3), long lifetime (τ ~ 15 ns), high quantum yield (Φ = 0.95 in water) and a blue-wavelength fluorescence emission.27 Once the FRET pairs were selected, we calculated the Förster distance (R0) of these donors and acceptor, following the standard protocol.27 The R0 between D and G is 27.4 Å; and the R0 between E and G is 29.5 Å. The Glu17 residue of E. coli DHFRs is located in the Met 20 loop, which exhibits conformational changes upon substrate and cofactor binding and product release.39,45-47 In our previous studies, we demonstrated that position 17 of DHFR tolerated small fluorescent amino acids.21,22 In the present study, we replaced Glu17 with the acceptor G. For the donor positions, we selected Asn37 and Asp79 to fit the calculated R0 values, as their distances to E17 are 33 Å and 31 Å, respectively (PBD 1RA1). As shown in Scheme 3 and Figure 3, by decoding a nonsense codon UAG and a four-base codon CGGG in the presence of the aminoacyl-tRNACUA and aminoacyl-tRNACCCG, the donors (D or E) and the acceptor (G) amino acids were incorporated into a single protein of DHFR at positions 37 and 17, respectively (see Figures 1 and 3 of the Supplementary data for the tRNA ligations). 21,22 The yields of incorporation were 8.3 and 8.8 %, relative to wild type. We also incorporated E into position 79 and G into position 17 of DHFR with a relative yield of 11% compared to wild type (see Figure 4 of the Supplementary data). As a control, the singly modified DHFRs containing a donor or an acceptor at the corresponding sites were also prepared in yields ranging from 27 to 63% (see Figure 5 of the Supplementary data). As shown in Table 4, even though the acceptor G at position 17 decreased the rate of NADPH consumption to 7.1 s−1, donor D did not further perturb the activity of DHFR when introduced into position 37. Only a small incremental decrease was observed when E was introduced into positions 37 or 79. These findings provide further evidence that these Trp analogues are suitable candidates as small fluorophores that possess less potential for disturbing the native protein structure and enzyme activity.
Scheme 3.
Strategy Employed for Incorporation of a Pair of Fluorophores into DHFR at Positions 17 and 37 for FRET Study.
Figure 3.
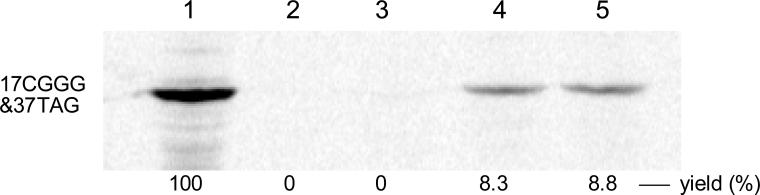
Autoradiogram of a 15% SDS-polyacrylamide gel (100 V, 2 h) illustrating the incorporation of tryptophan analogues into positions 17 and 37 of DHFR. Lane 1, wild-type DHFR expression; lane 2, modified DHFR DNA (17CGGG; 37TAG) in the presence of abbreviated suppressor tRNACCCG-COH; lane 3, modified DHFR DNA in the presence of L-acridon-2-ylalanyl-tRNACCCG; lane 4, incorporation of amino acids D and G; lane 5, incorporation of amino acids E and G.
Table 4.
Enzymatic Activities of Modified DHFRs Containing FRET Pairs
| DHFR | kD, s−1 |
|---|---|
| wild type DHFR38 | 12 |
| 37D | 11.8 ± 0.4a |
| 37E | 12.1 ± 0.4 |
| 79E | 11.2 ± 0.5 |
| 17G | 7.1 ± 0.5 |
| 17G37D | 7.1 ± 0.4 |
| 17G37E | 6.1 ± 0.4 |
| 17G79E | 5.2 ± 0.2 |
Standard deviation is based on data from three experiments.
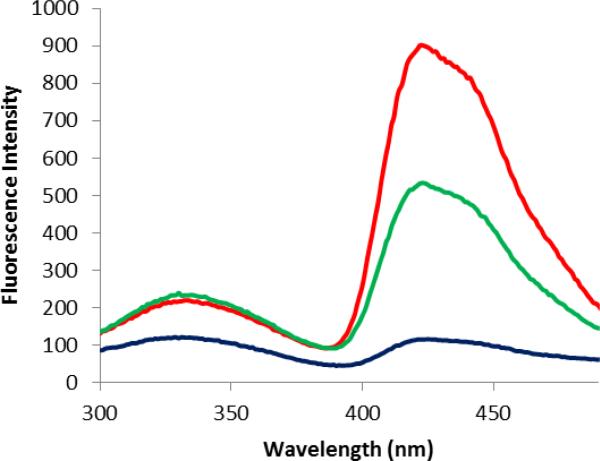
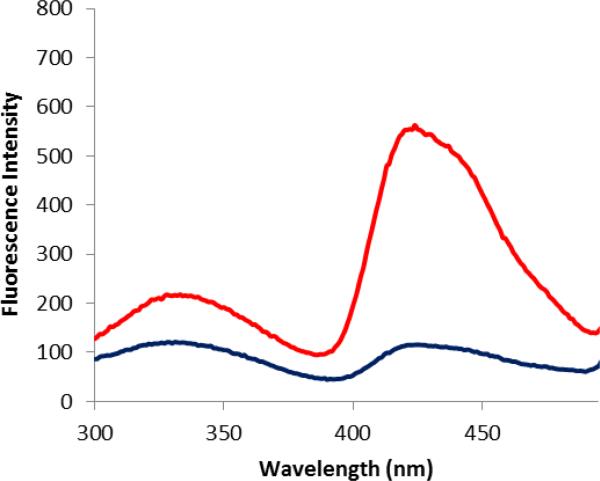
After purification on a Ni-NTA column and a DEAE-Sepharose CL-6B column, the doubly modified DHFRs were excited at 260 nm, which was chosen in order to decrease the interference of the tryptophans in DHFR (having λmax 280 nm). This is especially advantageous for the E-G donor—acceptor system because the absorption maximum of E is blueshifted (262 nm; Table 1) from native Trp. As shown (Figure 4), the modified DHFR containing the fluorescent acceptor G at position 17 and the donor E at position 37 exhibited efficient FRET emission peaks at 420 nm. Similarly, the modified DHFR containing the fluorescent acceptor G at position 17 and the donor D at position 37 also exhibited a stronger FRET emission at 420 nm. Comparatively, the four remaining tryptophans in DHFR displayed only very weak FRET emissions at the same wavelength. As shown in Figure 5, the modified DHFR containing the fluorescence acceptor G at position 17 and the donor E at position 79 also exhibited efficient FRET. These results demonstrated that donors D or E can form a FRET pair with the acceptor G in DHFR at different positions. While acridon-2-ylalanine (G) functioned entirely acceptably in its role as a FRET acceptor in these experiments, it would obviously be to good advantage to develop smaller acceptors for the new Trp donors described here, such that both the donor and acceptor could be embedded entirely within the folded protein structure.
Figure 4.
Fluorescence emission spectra of DHFRs containing amino acid G at position 17 (black trace), G at position 17 and D at position 37 (red trace), and G at position 17 and E at position 37 (green trace). The spectra were recorded at pH 8.0 following excitation at 260 nm.
Figure 5.
Fluorescence emission spectra of DHFRs containing amino acid G at position 17 (black trace), G at position 17 and E at position 79 (red trace). The spectra were recorded at pH 8.0 following excitation at 260 nm.
3. Discussion
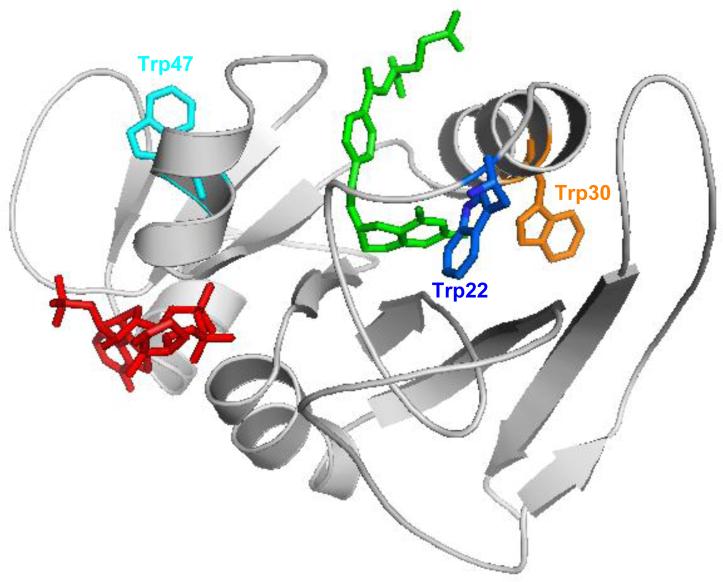
In this study, we demonstrated the incorporation of six tryptophan derivatives (A – F) into DHFR at three different positions to explore their effect on DHFR function. We showed that these Trp-based fluorophores exhibit spectrophotometric properties that are distinct from tryptophan itself. In the X-ray crystallographic structure of DHFR, residue Trp22 is involved in substrate binding through H-bonds between the residue and an ordered water molecule.39 Replacement of this residue may disorder substrate binding and affect the function of DHFR. This is consistent with the finding that incorporation of tricyclic amino acids E and F into position 22 reduced the activity of the modified DHFRs to 6 and 7% of the wild-type enzyme. As shown in Figure 6, Trp30 and Trp47 are located in two α-helices. Neither of them involves substrate or cofactor binding. In these cases, none of the six tryptophan derivatives significantly affected DHFR activity when they replaced the tryptophan residue. These results demonstrated that these tryptophan analogues have intrinsic properties that can minimize the perturbation of protein structure, thus permitting their use for the study of protein conformational changes.
Figure 6.
Structure of wild-type E. coli DHFR (PBD entry 1RX6), with Trp22 shown in blue, Trp30 in orange and Trp47 in cyan. The substrate tetrahydrofolate is shown in green and the cofactor NADPH is shown in red.
After comparing their structures and photophysical properties, we selected one bicyclic Trp analogue (D) and one tricyclic Trp derivative (E) as fluorescence donors to study their FRET properties in DHFR with the highly efficient acceptor G. We incorporated these two donors into position 37 of DHFR, and also incorporated E at position 79. Positions 37 and 79 have similar distances to E17 (33 Å and 31 Å, respectively). All three constructs retained reasonable enzymatic activity (Table 4). The DHFRs modified at positions of 37 and 79 with D or E had essentially the same NADPH consuming activities as wild type. In comparison, the DHFR modified at position 17 with the acceptor G exhibited decreased activity (to 59% of wild type). For the doubly modified DHFRs containing G at position of 17 and D or E at positions of 37 or 79, the NADPH consuming activities ranged from 43 – 59% of wild type. When excited at 260 nm, all three doubly modified DHFRs exhibited efficient FRET (Figures 4 and 5). Among them, the modified DHFR containing G at position 17 and D at position 37 exhibited the best FRET. These Trp analogues can either be used alone or in a FRET pair in the presence of other native Trp residues, because of their spectral differences. Also, it is known that the spectral properties of tryptophan are sensitive to changes in the electrostatic environment in/around a protein. 48,49 Thus, these small Trp-based fluorophores might be employed to study subtle changes in the conformational and electrostatic environment in the crowded protein interior space without drastically interfering with normal protein folding or enzyme activity. E. coli DHFR has five native Trp residues three of which (22, 30, 47) are in or near the crowded enzyme active site, which would have very little tolerance for larger fluorescence probes. The results here demonstrate that the six Trp-based fluorophores generally induce minimal catalytic disturbance when incorporated at such positions.
4. Conclusions
Several tryptophan analogues have been prepared, characterized with respect to their photophysical properties, and incorporated into three different positions of dihydrofolate reductase. Incorporation at positions 30 and 47 in lieu of tryptophan afforded proteins having essentially full activity, while substitution for Trp22 reduced activity somewhat in every case, especially for tricyclic analogues E and F. Two of the tryptophan analogues (D and E) were employed in FRET experiments at position 37, using acridon-2-ylalanine as the acceptor at position 17, and were found to produce FRET efficiently. The results demonstrate the utility of selected tryptophan analogues as participants in energy transfer experiments in DHFR, even in the presence of other unmodified tryptophans.
5. Experimental section
5.1. General Experimental Procedures
All experiments requiring anhydrous conditions were conducted in flame-dried glassware fitted with rubber septa under a positive pressure of dry nitrogen, unless otherwise noted. Reactions were performed at room temperature unless otherwise indicated. Analytical thin layer chromatography was performed using glass plates pre-coated with silica gel (0.25 mm, 60 Å pore size, 230-400 mesh, Silicycle) impregnated with a fluorescent indicator (254 nm). TLC plates were visualized by exposure to ultraviolet light (UV). Flash-column chromatography was performed employing silica gel (60 Å pore size, 40-63 μm, standard grade, Silicycle). An acetone cooling bath was cooled to the appropriate temperature by the addition of small portions of dry ice.
1H NMR and 13C NMR spectra were recorded on Varian INOVA 400 (400 MHz) and Varian INOVA 500 (500 MHz) spectrometers at 25 °C. Proton chemical shifts are expressed in parts per million (ppm, δ scale) and are referenced to residual protium in the NMR solvent (CDCl3, DMSO-d6 or CD3OD). Splitting patterns are designated as follows: s, singlet; br s, broad singlet; d, doublet; dd, doublet of doublets; t, triplet; q, quartet; m, multiplet. High resolution mass spectra were obtained at the Arizona State University CLAS High Resolution Mass Spectrometry Facility or the Michigan State University Mass Spectrometry Facility. HPLC purification was performed with a Waters 600 pump coupled with a Varian ProStar 340 detector and a Grace Econosil C18 column (250 × 10 mm, 5 μm). The tetra-n-butylammonium (TBA) salt of pdCpA was prepared using Dowex 50W×8, 200-400 mesh activated in its TBA form.
The chemicals used for synthesis were purchased from Aldrich Chemical Co., Sigma Chemical Co. or Combi Blocks. THF was distilled under argon from sodium-benzophenone ketyl and CH2Cl2 was distilled under argon from calcium hydride.Ni-NTA agarose was obtained from Qiagen Inc. DNA oligonucleotides were purchased from Integrated DNA Technologies. DEAE-Sepharose, ammonium persulfate, acrylamide, N, N′-methylene-bis-acrylamide, acetic acid, potassium glutamate, ammonium acetate, dithiothreitol, magnesium acetate, phospho(enol)pyruvate, Escherichia coli tRNA, isopropyl β-D-thiogalactopyranoside (IPTG), ATP, GTP, CTP, UTP, cAMP, amino acids, rifampicin, and formamide were obtained from Sigma-Aldrich. Tris and SDS were obtained from Bio-Rad Laboratories (Hercules, CA). [35S]-methionine (1000 Ci/mmol, 10 μCi/μL) was purchased from PerkinElmer Inc. Protease inhibitor (complete, EDTA-free) was obtained from Boehringer Mannheim Corp. T4 RNA ligase and T4 polynucleotide kinase were purchased from New England Biolabs Inc.
Phosphorimager analysis was performed using an Amersham Biosciences Storm 820 equipped with ImageQuant version 5.2 software from Molecular Dynamics. UV spectral measurements were made using a Perkin-Elmer Lamdba 20 UV/vis spectrometer. Fluorescence was monitored using a Varian Cary Eclipse Fluorescence Spectrophotometer.
5.2. Synthesis of pdCpA Derivatives
The synthesis of the pdCpA derivative of amino acid D is outlined in Scheme 1. The syntheses of pdCpA derivatives of amino acids A – C and G26,27 are outlined in Schemes 1, 2, 3 and 4 of the Supplementary data, respectively, and the detailed experimental procedures and compound characterizations are provided in the Supplementary data. The syntheses of the aminoacylated pdCpA derivatives of E and F have been reported previously.25
5.2.1. 1-Tosyl-1H-pyrrolo[2,3-b]pyridine-3-carbaldehyde (2)
To a stirred solution containing 1.00 g (8.47 mmol) of 1H-pyrrolo[2,3-b]pyridine in 10 mL of 1:1 AcOH/H2O was added 1.78 g (12.7 mmol) of hexamethylenetetramine (HMTA). The reaction mixture was heated at reflux under argon overnight. The reaction mixture was cooled to 0 °C and the formed precipitate was filtered and dried under vacuum. 1H-pyrrolo[2,3-b]pyridine-3-carbaldehyde (1) was obtained as a colorless solid and was used directly in the next step without further purification. To a stirred solution containing 757 mg (5.17 mmol) of 1 in 20 mL of anhydrous DMF at 0 °C was added 414 mg (10.4 mmol) of NaH. The reaction mixture was stirred at 0 °C for 10 min under argon and then 1.48 g (7.76 mmol) p-TsCl was added. The reaction mixture was stirred at 0 °C under argon for 3 h, diluted with 100 mL of NaHCO3 and extracted with two 50-mL portions of EtOAc. The combined organic phase was dried (MgSO4) and concentrated under diminished pressure. The residue was purified by chromatography on a silica gel column (10 × 4 cm). Elution with 4:1 hexanes/ethyl acetate gave 1-tosyl-1H-pyrrolo[2,3-b]pyridine-3-carbaldehyde (2) as a yellow solid: yield 1.34 g (53% for two steps); silica gel TLC Rf 0.35 (1:1 hexanes/ethyl acetate); 1H NMR (CDCl3, 400 MHz) δ 2.40 (s, 3H), 7.27-7.34 (m, 3H), 8.16 (d, 2H, J = 8.4 Hz), 8.39 (s, 1H), 8.50 (d, 1H, J = 2.4 Hz), 8.51 (d, 1H, J = 5.6 Hz) and 10.04 (s, 1H); 13C NMR (CDCl3, 400 MHz) δ21.8, 119.1, 119.5, 120.7, 120.8, 128.8, 130.1, 131.5, 134.4, 135.9, 146.5, 146.8 and 185.3; mass spectrum (APCI), m/z 301.0658 (M+H)+ (C15H13N2O3S requires m/z 301.0647).
5.2.2. 3-Hydroxymethyl-1-tosyl-1H-pyrrolo[2,3-b]pyridine (3)
To a suspension of 1.34 g (4.46 mmol) of 1-tosyl-1H-pyrrolo[2,3-b]pyridine-3-carbaldehyde (2) in 20 mL of EtOH was added 222 mg (8.92 mmol) of NaBH4 and the reaction mixture was stirred at room temperature for 2 h, then diluted with 100 mL of NaHCO3. The formed precipitate was filtered and dried under vacuum. 3-Hydroxymethyl-1-tosyl-1H-pyrrolo[2,3-b]pyridine (3) was obtained as an off-white solid: yield 1.20 g (90%); silica gel TLC Rf 0.1 (1:1 hexanes/ethyl acetate); 1H NMR (CDCl3, 400 MHz) δ 2.29 (s, 3H), 4.75 (s, 2H), 7.10-7.20 (m, 3H), 7.62 (s, 1H), 7.88 (dd, 1H, J = 8.0 and 1.6 Hz), 7.99 (d, 2H, J = 8.0 Hz) and 8.37 (dd, 1H, J = 4.4 and 1.6 Hz); 13C NMR (CDCl3, 400 MHz) δ 21.7, 57.4, 118.8, 118.9, 122.0, 123.9 128.2, 128.7, 129.8, 135.5, 145.3, 145.4 and 147.8; mass spectrum (APCI), m/z 303.0805 (M+H)+ (C15H15N2O3S requires m/z 303.0803).
5.2.3. 3-(Chloromethyl)-1-tosyl-1H-pyrrolo[2,3-b]pyridine (4)
To a cooled (−10 °C) solution containing 1.20 g (3.9 mmol) of 3-hydroxymethyl-1-tosyl-1H-pyrrolo[2,3-b]pyridine (3) in 20 mL of anhydrous CH2Cl2 and 2.18 mL (1.58 g, 15.6 mmol) of Et3N was added dropwise 0.58 mL (947 mg, 7.93 mmol) of SOCl2 . The reaction mixture was allowed to warm slowly to room temperature and was then stirred for 6 h, diluted with 50 mL of NaHCO3, and extracted with two 50-mL portions of EtOAc. The combined organic phase was dried (MgSO4) and concentrated under diminished pressure. The residue was purified by chromatography on a silica gel column (10 × 2 cm). Elution with 4:1 hexanes/ethyl acetate gave 3-(chloromethyl)-1-tosyl-1H-pyrrolo[2,3-b]pyridine (4) as a brown solid: yield 1.01 g (81%); silica gel TLC Rf 0.42 (4:1 hexanes/ethyl acetate); 1H NMR (CDCl3, 400 MHz) δ 2.33 (s, 3H), 4.69 (s, 2H), 7.18-7.25 (m, 3H), 7.74 (s, 1H), 7.92 (d, 1H, J = 1.2 Hz), 7.94 (d, 1H, J = 1.6 Hz), 8.06 (d, 1H, J = 8 Hz) and 8.43 (dd, 1H, J = 4.8 and 1.2 Hz); 13C NMR (CDCl3, 400 MHz) δ 21.7, 37.5, 15.4, 119.0, 121.5, 124.9, 128.2, 128.5, 129.8, 135.2, 145.5, 145.6 and 147.5; mass spectrum (APCI), m/z 321.0469 (M+H)+ (C15H14N2O2SCl requires m/z 321.0465).
5.2.4. 3-(((2S,5R)-5-Isopropyl-3,6-dimethoxy-2,5-dihydropyrazin-2-yl)methyl)-1-tosyl-1H-pyrrolo[2,3-b]pyridine (5)
To a stirred solution containing 0.62 mL (650 mg, 3.53 mmol) of Schöllkopf's reagent in 10 mL of anhydrous THF at −78 °C was added 0.45 mL (308 mg, 4.82 mmol) of 2.5M BuLi. The reaction mixture was stirred at −78 °C for 30 min under argon and then a solution containing 1.03 g (3.21 mmol) of 3-(chloromethyl)-1-tosyl-1H-pyrrolo[2,3-b]pyridine (4) in 10 mL of anhydrous THF was added. The reaction mixture was stirred for 1 h under argon at −78 °C, then diluted with 50 mL of satd aq NH4Cl and extracted with two 50-mL portions of EtOAc. The combined organic phase was dried (MgSO4) and concentrated under diminished pressure. The residue was purified by chromatography on a silica gel column (10 × 2 cm). Elution with 2:1 hexanes/ethyl acetate gave 3-(((2S,5R)-5-isopropyl-3,6-dimethoxy-2,5-dihydropyrazin-2-yl)methyl)-1-tosyl-1H-pyrrolo[2,3-b]pyridine (5) as a yellow oil: yield 783 mg (52%); silica gel TLC Rf 0.15 (4:1 hexanes/ethyl acetate); 1H NMR (CDCl3, 400 MHz) δ 0.51(d, 3H, J = 6.8 Hz), 0.78 (d, 3H, J = 8 Hz), 1.99-2.03 (m, 1H), 2.22 (s, 3H), 3.07-3.12 (m, 3H), 3.57 (s, 3H), 3.60 (s, 3H), 4.22-4.25 (m, 1H), 7.02-7.05 (m, 1H), 7.13 (d, 1H, J = 8 Hz), 7.36 (s, 1H), 7.74 (dd, 1H, J = 8.0 and 1.6 Hz), 7.87 (d, 2H, J = 8.4 Hz) and 8.29 (dd, 1H, J =4.8 and 1.6 Hz); 13C NMR (CDCl3, 400 MHz) δ 16.4, 18.8, 21.5, 29.1, 31.3, 52.1, 52.3, 55.5, 60.4, 114.9, 118.3, 123.7, 124.6, 127.5, 128.2, 129.5, 135.5, 144.6, 144.8, 147.1, 161.8 and 164.0; mass spectrum (APCI), m/z 469.1921 (M+H)+ (C24H29N4O4S requires m/z 469.1909).
5.2.5. 3-(((2S, 5R-5-Isopropyl-3,6-dimethoxy-2,5-dihydropyrazin-2-yl)methyl)-1H-pyrrolo[2,3-b]pyridine (6)
To a stirred solution containing 303 mg (0.65 mmol) of 3-(((2S,5R)-5-isopropyl-3,6-dimethoxy-2,5-dihydropyrazin-2-yl)methyl)-1-tosyl-1H-pyrrolo[2,3-b]pyridine (5) in 5 mL of 2:1 THF/methanol was added 685 mg (1.94 mmol) of Cs2CO3. The reaction mixture was stirred at room temperature for 12 h under argon, then diluted with 20 mL of brine and extracted with two 50-mL portions of EtOAc. The combined organic phase was dried (MgSO4) and concentrated under diminished pressure. The residue was purified by chromatography on a silica gel column (10 × 2 cm). Elution with ethyl acetate gave 3-(((2S,5R)-5-isopropyl-3,6-dimethoxy-2,5-dihydropyrazin-2-yl)methyl)-1H-pyrrolo[2,3-b]pyridine (6) as a yellow oil: yield 77 mg (39%); silica gel TLC Rf 0.3 (1:1 hexanes/ethyl acetate); 1H NMR (CDCl3, 400 MHz) δ 0.60 (d, 3H, J = 6.8 Hz), 0.91 (d, 3H, J = 6.8 Hz), 2.10-2.13 (m, 1H), 3.26-3.29 (m, 2H), 3.36-3.38 (m, 1H), 3.65 (s, 3H), 3.68 (s, 3H), 4.34-4.35 (m, 1H), 7.02 (dd, 1H, J = 4.4 and 4.4 Hz), 7.08 (s, 1H), 7.95 (d, 1H, J = 6.8 Hz), ), 8.26 (d, 1H, J = 4.8 Hz) and 11.52 (s, 1H); 13C NMR (CDCl3, 400 MHz) δ 16.5, 19.0, 29.7, 31.4, 52.3, 52.4, 56.7, 60.5, 109.8, 114.5, 121.3 123.9, 127.8, 142.0, 148.8, 162.8 and 163.9; mass spectrum (APCI), m/z 315.1829 (M+H)+ (C17H23N4O2 requires m/z 315.1821).
5.2.6. 3-(((2S, 5R)-5-Isopropyl-3,6-dimethoxy-2,5-dihydropyrazin-2-yl)methyl)-1-methyl-1H-pyrrolo[2,3-b]pyridine (7)
To a stirred solution containing 76 mg (0.24 mmol) of 3-(((2S,5R)-5-isopropyl-3,6-dimethoxy-2,5-dihydropyrazin-2-yl)methyl)-1H-pyrrolo[2,3-b]pyridine (6) in 3 mL of anhydrous DMF at 0 °C was added 11.5 mg (0.48 mmol) of NaH followed by 0.03 mL (69 mg, 0.48 mmol) of MeI. The reaction mixture was stirred at 0 °C under argon for 30 min, then diluted with 100 mL of NaHCO3 and extracted with two 50-mL portions of EtOAc. The combined organic phase was dried (MgSO4) and concentrated under diminished pressure. The residue was purified by chromatography on a silica gel column (10 × 2 cm). Elution with ethyl acetate gave 3-(((2S,5R)-5-isopropyl-3,6-dimethoxy-2,5-dihydropyrazin-2-yl)methyl)-1-methyl-1H-pyrrolo[2,3-b]pyridine (7) as yellow oil: yield 68 mg (83%); silica gel TLC Rf 0.4 (1:1 hexanes/ethyl acetate); 1H NMR (CDCl3, 400 MHz) δ 0.59 (d, 3H, J = 6.8 Hz), 0.91 (d, 3H, J = 7.2 Hz), 2.09-2.13 (m, 1H), 3.22 (d, 2H, J = 4.4 Hz), 3.40-3.41 (m, 1H), 3.62 (s, 3H), 3.66 (s, 3H), 3.79 (s, 3H), 4.28-4.29 (m, 1H), 6.86 (s, 1H), 6.95-6.98 (m, 1H), 7.87 (dd, 1H, J = 8.0 and 1.6 Hz), and 8.25 (dd, 1H, J =8.4 and 1.6 Hz); 13C NMR (CDCl3, 400 MHz) δ 16.6, 19.1, 29.6, 31.1, 31.5, 52.2, 52.3, 56.7, 60.6, 108.9, 114.8, 121.3 127.6, 127.7, 142.6, 147.7, 162.8 and 163.9; mass spectrum (APCI), m/z 329.1977 (M+H)+ (C18H25N4O2 requires m/z 329.1977).
5.2.7. Methyl (S)-2-((4,5-dimethoxy-2-nitrobenzyloxy)carbonyl)-3-(1-methyl-1H-pyrrolo[2,3-b]pyridin-3-yl)propionate (9)
To a stirred solution containing 67.0 mg (0.21 mmol) of 3-(((2S,5R)-5-isopropyl-3,6-dimethoxy-2,5-dihydropyrazin-2-yl)methyl)-1-methyl-1H-pyrrolo[2,3-b]pyridine (7) in 4 mL of THF at 0 °C was added 4 mL of 2 N aq HCl. The reaction mixture was stirred at room temperature for 2 h. The reaction mixture was then poured slowly into 30 mL of NaHCO3 and then extracted with two 30-mL portions of EtOAc. The combined organic phase was dried (MgSO4) and concentrated under diminished pressure. Methyl (S)-2-amino-3-(1-methyl-1H-pyrrolo[2,3-b]pyridin-3-yl)propionate was obtained as a yellow oil and was used directly in the next step without further purification. To a stirred solution containing 36 mg (0.15 mmol) of methyl (S)- 2-amino-3-(1-methyl-1H-pyrrolo[2,3-b]pyridin-3-yl)propionate (8) in 2 mL of 1:1 dioxane/water was added 75 mg (0.54 mmol) of K2CO3 followed by 52 mg (0.24 mmol) of NVOC-Cl. The reaction mixture was stirred at room temperature for 14 h under argon, then diluted with 50 mL of brine and extracted with two 50-mL portions of EtOAc. The combined organic phase was dried (MgSO4) and concentrated under diminished pressure. The residue was purified by chromatography on a silica gel column (10 × 2 cm). Elution with 1:3 hexanes/ethyl acetate gave methyl (S)-2-((4,5-dimethoxy-2-nitrobenzyloxy)carbonylamino)-3-(1-methyl-1H-pyrrolo[2,3-b]pyridin-3-yl)propionate (9) as a yellow oil: yield 58 mg (62% for two steps); silica gel TLC Rf 0.3 (1:1 hexanes/ethyl acetate); 1H NMR (CDCl3, 400 MHz) δ 3.24-3.26 (m, 2H), 3.66 (s, 3H), 3.78 (s, 3H), 3.83 (s, 3H), 3.88 (s, 3H), 4.65-4.67 (m, 1H), 5.46 (d, 1H, J = 8 Hz), 5.57 (d, 1H, J = 8.4 Hz), 6.88 (s, 1H), 6.96-6.99 (m, 2H), 7.63 (s, 1H), 7.76 (d, 1H, J = 7.2 Hz), 8.27 (dd, 1H, J = 4.8 and 1.2 Hz) and 8.60 (d, 1H, J = 7.2 Hz); 13C NMR (CDCl3, 400 MHz) δ 27.9, 31.1, 52.5, 54.5, 56.4, 56.4, 63.9, 106.8, 108.1, 110.0, 115.3, 120.3, 126.8, 127.7, 127.8, 139.6, 143.1, 147.8, 148.0, 153.6, 155.3 and 172.2; mass spectrum (APCI), m/z 473.1673 (M+H)+ (C22H25N4O8 requires m/z 473.1672).
5.2.8. Cyanomethyl (S)-2-((4,5-dimethoxy-2-nitrobenzyloxy)carbonylamino)-3-(1-methyl-1H-pyrrolo[2,3-b]pyridin-3-yl)propionate (10)
To a stirred solution containing 31.0 mg (0.07 mmol) of methyl (S)-2-((4,5-dimethoxy-2-nitrobenzyloxy)carbonyl)-3-(1-methyl-1H-pyrrolo[2,3-b]pyridin-3-yl)propionate (9) in 1 mL of 1:3:1 water/THF/methanol was added 214 μL (0.21 mmol) of 1 N LiOH. The reaction mixture was stirred at room temperature for 3 h, and then concentrated under diminished pressure. The residue was dissolved in 1 mL of anh DMF and 30.0 μL (22.0 mg, 0.21 mmol) of Et3N was added followed by 13.0 μL (16.0 mg, 0.21 mmol) of chloroacetonitrile. The reaction mixture was stirred at 23 °C for 16 h, then diluted with 20 mL of NaHCO3 and extracted with two 50-mL portions of EtOAc. The combined organic phase was dried (MgSO4) and concentrated under diminished pressure. The residue was purified by chromatography on a silica gel column (10 × 2 cm). Elution with ethyl acetate gave cyanomethyl (S)-2-((4,5-dimethoxy-2-nitrobenzyloxy)carbonylamino)-3-(1-methyl-1H-pyrrolo[2,3-b]pyridin-3-yl)propionate (10) as a light yellow solid: yield 22 mg (71%); silica gel TLC Rf 0.4 (1:1 hexanes/ethyl acetate); 1H NMR (CDCl3, 400 MHz) δ 3.32 (d, 2H, J = 5.6 Hz), 3.84 (s, 3H), 3.89 (s, 3H), 3.93 (s, 3H), 4.62-4.81 (m, 3H), 5.43-5.55 (m, 2H), 6.90 (s, 1H), 7.04-7.07 (m, 2H), 7.69 (s, 1H), 7.83 (d, 1H, J = 7.6 Hz), 8.33 (dd, 1H, J = 4.8 and 1.6 Hz); 13C NMR (CDCl3, 400 MHz) δ 27.9, 31.3, 49.1, 54.6, 56.5, 56.6, 64.3, 105.9, 108.3, 110.3, 113.9, 115.7, 120.2, 126.8, 127.5, 128.1, 139.9, 143.5, 147.9, 148.4, 153.7, 155.4 and 170.7; mass spectrum (APCI), m/z 498.1638 (M+H)+ (C23H24N5O8 requires m/z 498.1625).
5.2.9. (S)-2-((4,5-Dimethoxy-2-nitrobenzyloxy)carbonylamino)-3-(1-methyl-1H-pyrrolo[2,3-b]pyridin-3-yl)pdCpA (11)
To a stirred solution containing 5.20 mg (4.00 μmol) of pdCpA tetrabutylammonium salt in 100 μL of 9:1 anh DMF/anhydrous triethylamine was added 10.4 mg (21.0 μmol) of cyanomethyl (S)-2-((4,5-dimethoxy-2-nitrobenzyloxy)carbonylamino)-3-(1-methyl-1H-pyrrolo[2,3-b]pyridin-3-yl)propionate (10). The reaction mixture was sonicated for 2 h. The reaction mixture was purified by HPLC on a C18 reversed phase column (250 × 10 mm) using a linear gradient of 99:1 → 1:99 50 mM aq ammonium acetate (pH 4.5) / acetonitrile. The retention time of the desired product was 23.4 min. The fractions containing the product were lyophilized to afford 11 as a colorless solid: yield 1.3 mg (31%); mass spectrum (ESI), 1075.2356 (M-H)− (C40H45N12O20P2 requires m/z 1075.2348).
5.3. Measurement of Photophysical Properties of Tryptophan Derivatives A - F
The UV/vis absorption spectra (220-400 nm) were recorded using a Cary 100 Bio UV/vis spectrophotometer thermostated at 22 °C. The sample compounds were prepared as 5 × 10−5 M solutions in anhydrous methanol in 1-cm path length quartz cuvettes. To generate the neutral species, 1 mM 4-ethylmorpholine-HCl was added to buffer the system at 8.3 in methanol. To generate the cationic species, 5 mM HCl (from a stock of standardized 1 M HCl solution) was added to set the system at 2.3 in methanol. The CH3OH2+ concentrations were determined potentiometrically using a combination glass electrode calibrated with Fisher certified standard aqueous buffers (pH 4.00 and 10.00) as described previously.50 The values51 in methanol were obtained by subtracting a correction constant of -2.24 from the electrode readings with the autoprotolysis constant for methanol taken as 10−16.77 M2.
Fluorescence emission spectra (300-600 nm) were collected at room temperature using a FluoroMax-4 spectrofluorometer (5 nm bandpass) equipped with a Horiba Jobin Yvon NanoLED (peak = 281 nm).
The fluorescence lifetime (TCSPC) determinations were performed at room temperature with a NanoLED-281 excitation source (1 MHz refresh rate) while the emission wavelength (10 nm bandpass) was set to the specific λem indicated in Table 1. Data were collected with Data Station v. 2.5 and analyzed using DAS6 software. A typical experiment contained 1-5 × 10−5 M tryptophan derivative with either 1 mM 4-ethylmorpholine-HCl buffer ( = 8.3) or 5 mM HCl ( = 2.3) in methanol. Fluorescence decay measurements were performed in duplicate and the averaged values are listed in Table 1. In water, Ludox CL colloidal silica solutions (0.01%) were used as light scattering standards.
Fluorescence quantum yields (Table 2) of fluorescent compounds were determined using the gradient method.52 Tryptophan derivatives A – F were dissolved in methanol. Solutions of each compound were made such that the UV absorption at the maximum wavelength were 0.02, 0.04, 0.06, 0.08 and 0.1. Two standards, 2-aminopyridine (ΦF 0.60, λex 295 nm) and anthracene (ΦF 0.27, λex 340 nm), were used to calculate the fluorescent quantum yield of the tryptophan analogues according to the formula Φx = Φs × (Gradx × nx2)/(Grads × ns2), where Grad is gradient of the plot of integrated intensity versus absorbance, n is the refractive index of the solvent, s is the standard of known ΦF, and x is the tested sample.52
5.4. Ligation of Suppressor tRNACUA-COH with Tryptophan Analogues and Deprotection of the N-NVOC Group
The yeast suppressor tRNAPheCUA was prepared as reported previously.35 Activation of the suppressor tRNACUA was carried out in 200 μL (total volume) of 100 mM Hepes buffer, pH 7.5, containing 2.0 mM ATP, 15 mM MgCl2, 200 μg of suppressor tRNA-COH, 4.0 A260 units of N-NVOC protected aminoacyl-pdCpA (5-10 fold molar excess), 15% DMSO and 400 units of T4 RNA ligase. After incubation at 37 °C for 1 h, the reaction was quenched by the addition of 20 μL of 3 M NaOAc, pH 5.2, followed by 600 μL of ethanol. The reaction mixture was incubated at –20 °C for 30 min, then centrifuged at 15,000 × g at 4 °C for 30 min. The supernatant was decanted carefully and the tRNA pellet was washed with 200 μL of 70% ethanol, then dissolved in 60 μL of RNase free H2O. The efficiency of ligation was estimated by 8% denaturing PAGE (pH 5.2).53 The NVOC protecting group was removed by exposure to high intensity UV light at 4 °C for 3 min.54
5.5 In vitro Translation of DHFR Containing Tryptophan Analogues at Positions 17, 22, 30, 37 and 47
The modified DHFR plasmid was obtained by site-directed mutagenesis as described previously using the wild-type DHFR plasmid or modified DHFR (17CGGG) as the template.55 The DNA primer for mutagenesis at position 22 was 5'-GCATGGAAAACGCCATGCCGTAGAACCTGCCTGCCGATCTCGC-3'; the primer for mutagenesis at position 30 was 5'-CCTGCCTGCCGATCTCGCCTAGTTTAAACGCAACACCTTAAATAAACC-3'; the primer for mutagenesis at position 37 was 5'- CGCCTGGTTTAAACGCAACACCTTATAGAAACCCGTGATTATGGGCCGC-3'; the primer for mutagenesis at position 47 was 5'-GTGATTATGGGCCGCCATACCTAGGAATCAATCGGTCGTCCGTTG-3'.
The in vitro expression mixture (500 μL total volume) contained 50 μg of modified DHFR (TAG at position 22, 30, 37, 47, and/or CGGG at position 17) plasmid DNA, 200 μL of premix (35 mM Tris-acetate, pH 7.0, 190 mM potassium glutamate, 30 mM ammonium acetate, 2.0 mM dithiothreitol, 11 mM magnesium acetate, 20 mM phospho(enol)pyruvate, 0.8 mg/mL of E. coli tRNA, 0.8 mM IPTG, 20 mM ATP and GTP, 5 mM CTP and UTP and 4 mM cAMP), 100 μM of each of the 20 amino acids, 150 μCi of [35S]-L-methionine, 10 μg/μL rifampicin, 150 μL of S-30 extract from E. coli strain BL21(DE3), 150 μg of deprotected misacylated tRNACUA and/or 150 μg of deprotected acridon-2-ylalanyl-tRNACCCG.56 The reaction mixture was incubated at 30 °C for 45 min to maximize the yield.57 Plasmid DNA containing the gene for wild-type DHFR was used as the positive control, and an abbreviated tRNA (tRNA-COH) lacking any amino acid was used as the negative control. An aliquot containing 2 μL of reaction mixture was removed, treated with 2 μL of loading buffer and heated at 90 °C for 2 min. The sample was analyzed by 15% SDS-PAGE at 100 V for 2 h.
5.6. Purification of DHFR Analogues
The analogues of DHFR containing an N-terminal hexahistidine fusion peptide were purified by Ni-NTA chromatography.58 The in vitro translation reaction mixture (500 μL) was diluted with 1500 μL of 50 mM Tris-HCl, pH 8.0, containing 300 mM NaCl and 10 mM imidazole, and mixed gently with 150 μL of a 50% slurry of Ni-NTA resin at 4 °C for 1 h. Then the mixture was applied to a column and washed with 800 μL of 50 mM Tris-HCl, pH 8.0, containing 300 mM NaCl and 20 mM imidazole. Finally, the DHFR analogue was washed three times with 200 μL of 50 mM Tris-HCl, pH 8.0, containing 30 mM NaCl and 150 mM imidazole. The final three Ni-NTA column eluates were combined and applied to a 90-μL DEAE-Sepharose CL-6B column.59 The column was washed with 300 μL of 50 mM Tris-HCl, pH 8.0, containing 100 mM NaCl, 300 μL of 50 mM Tris-HCl, pH 8.0, containing 200 mM NaCl, and then three 100-μL portions of 50 mM Tris-HCl, pH 8.0, containing 300 mM NaCl. Aliquots of each fraction were analyzed by 15% SDS-PAGE.
5.7. Enzymatic Activities of DHFR Analogues
The enzymatic activities of wild-type and the modified DHFRs containing tryptophan derivatives were measured by the consumption of NADPH in 1 mL of MTEN buffer (containing 50 mM 2-(N-morpholino) ethanesulfonic acid (MES), 25 mM Tris, 25 mM ethanolamine, 100 mM NaCl, 0.1 mM EDTA and 10 mM β-mercaptoethanol, pH 7.0) as described.38 MTEN buffer (0.97 mL) was mixed with 10 μL of 10 mM β-nicotinamide adenine dinucleotide phosphate reduced form (NADPH) and 100 ng of protein.60 The reaction mixture was incubated at 37 °C for 3 min. Then 20 μL of 5 mM dihydrofolate in MTEN buffer, pH 7.0, was added to the above mixture, and the A340 value was monitored over a period of 10 min with a spectrophotometer thermostatted at 25 °C. The absorbance vs time traces were fitted to a standard single exponential decay to obtain the enzyme turnover rate constants. Under each condition, at least three independent kinetic runs were conducted, and the average rate constants were used for analysis. It should be pointed out that the enzyme activity described herein refers to the E. coli DHFR rate constants, which have been shown to involve the release of THF from the E:NADPH:THF product complex.38
5.8. Fluorescence Spectra of DHFR Analogues
The fluorescence spectra of modified DHFRs containing tryptophan derivatives were measured using a Varian Cary Eclipse Fluorescence Spetrophotometer with the excitation slit as 10 nm and emission slit as 10 nm. The protein samples (0.2 – 1.0 μM) were excited with ultraviolet light close to the excitation maxima, and the emission spectra were recorded.
Supplementary Material
Acknowledgement
This study was supported by Research Grant GM 092946 from the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplementary data
Supplementary data associated with this article can be found, in the online version, at http://
References and notes
- 1.Tsien RY. Annu. Rev. Biochem. 1998;67:509. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- 2.Selvin PR. Nat. Struct. Biol. 2000;7:730. doi: 10.1038/78948. [DOI] [PubMed] [Google Scholar]
- 3.Phillips SR, Wilson LJ, Borkman RF. Curr. Eye Res. 1986;5:611. doi: 10.3109/02713688609015126. [DOI] [PubMed] [Google Scholar]
- 4.Zhang P, Beck T, Tan W. Angew. Chem., Int. Ed. 2001;40:402. [PubMed] [Google Scholar]
- 5.Marti AA, Jockusch S, Li Z, Ju J, Turro NJ. Nucleic Acids Res. 2006;34:e50. doi: 10.1093/nar/gkl134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jockusch S, Marti AA, Turro NJ, Li Z, Li X, Ju J, et al. Photochem. Photobiol. Sci. 2006;5:493. doi: 10.1039/b600213g. [DOI] [PubMed] [Google Scholar]
- 7.Miyawaki A, Llopis J, Heim R, McCaffery JM, Adams JA, Ikura M, et al. Nature. 1997;388:882. doi: 10.1038/42264. [DOI] [PubMed] [Google Scholar]
- 8.Suzuki Y, Yasunaga T, Ohkura R, Wakabayashi T, Sutoh K. Nature. 1998;396:380. doi: 10.1038/24640. [DOI] [PubMed] [Google Scholar]
- 9.Tsien RY, Miyawaki A. Science. 1998;280:1954. doi: 10.1126/science.280.5371.1954. [DOI] [PubMed] [Google Scholar]
- 10.Zhang Z, Rajagopalan PT, Selzer T, Benkovic SJ, Hammes GG. Proc. Natl. Acad. Sci. U.S.A. 2004;101:2764. doi: 10.1073/pnas.0400091101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Antikainen NM, Smiley RD, Benkovic SJ, Hammes GG. Biochemistry. 2005;44:16835. doi: 10.1021/bi051378i. [DOI] [PubMed] [Google Scholar]
- 12.Liu CT, Wang L, Goodey NM, Hanoian P, Benkovic SJ. Biochemistry. 2013;52:5332. doi: 10.1021/bi400858m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cornish VW, Benson DR, Altenbach CA, Hideg K, Hubbell WL, Schultz PG. Proc. Natl. Acad. Sci. U.S.A. 1994;91:2910. doi: 10.1073/pnas.91.8.2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mendel D, Cornish VW, Schultz PG. Annu. Rev. Biophys. Biomol. Struct. 1995;24:435. doi: 10.1146/annurev.bb.24.060195.002251. [DOI] [PubMed] [Google Scholar]
- 15.Steward LE, Collins CS, Gilmore MA, Gilmore MA, Carlson JE, Ross JB, et al. J. Am. Chem. Soc. 1997;119:6. [Google Scholar]
- 16.Hohsaka T, Kajihara D, Ashizuka Y, Murakami H, Sisido MJ. Am. Chem. Soc. 1999;121:34. [Google Scholar]
- 17.Hohsaka T, Muranaka N, Komiyama C, Matsui K, Takaura S, Abe R, et al. FEBS Lett. 2004;560:173. doi: 10.1016/S0014-5793(04)00099-7. [DOI] [PubMed] [Google Scholar]
- 18.Hamada H, Kameshima N, Szymanska A, Wegner K, Łankiewicz L, Shinohara H, et al. Bioorg. Med. Chem. 2005;13:3379. doi: 10.1016/j.bmc.2005.03.014. [DOI] [PubMed] [Google Scholar]
- 19.Anderson RD, III, Zhou J, Hecht SM. J. Am. Chem. Soc. 2002;124:9674. doi: 10.1021/ja0205939. [DOI] [PubMed] [Google Scholar]
- 20.Kajihara D, Abe R, Iijima I, Komiyama C, Sisido M, Hohsaka T. Nat. Methods. 2006;3:923. doi: 10.1038/nmeth945. [DOI] [PubMed] [Google Scholar]
- 21.Chen S, Fahmi NE, Wang L, Bhattacharya C, Benkovic SJ, Hecht SM. J. Am. Chem. Soc. 2013;135:12924. doi: 10.1021/ja403007r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen S, Fahmi NE, Bhattacharya C, Wang L, Jin Y, Benkovic SJ, et al. Biochemistry. 2013;52:8580. doi: 10.1021/bi401275v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Merkel L, Hoesl MG, Albrecht M, Schmidt A, Budisa N. ChemBioChem. 2010;11:305. doi: 10.1002/cbic.200900651. [DOI] [PubMed] [Google Scholar]
- 24.Lepthien S, Hoesl MG, Merkel L, Budisa N. Proc. Natl. Acad. Sci. U.S.A. 2008;105:16095. doi: 10.1073/pnas.0802804105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Talukder P, Chen S, Arce PM, Hecht SM. Org. Lett. 2014;16:556. doi: 10.1021/ol403429e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Szymanska A, Wegner K, Lankiewicz L. Helv. Chim. Acta. 2003;86:3326. [Google Scholar]
- 27.Speight LC, Muthusamy AK, Goldberg JM, Warner JB, Wissner RF, Willi TS, et al. J. Am. Chem. Soc. 2013;135:18806. doi: 10.1021/ja403247j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Su W, Jia H, Dai G. Certain triazolopyridines and triazolopyrazines, compositions thereof and methods of use therefor. 2011 WO 2011079804 A1.
- 29.Yao CH, Song JS, Chen CT, Yeh TK, Hsieh TC, Wu SH, et al. Eur. J. Med. Chem. 2012;55:32. doi: 10.1016/j.ejmech.2012.06.053. [DOI] [PubMed] [Google Scholar]
- 30.Ramakrishna VSN, Shirsath VS, Kambhampati RS, Rao VSVV, Jasti V. N-arylsulfonyl-3-substituted indoles having serotonin receptor affinity, process for their preparation and pharmaceutical composition containing them. 2004 WO 2004048330 A1.
- 31.Schöllkopf U, Groth U, Westphalen K, Deng C. Synthesis. 1981;12:969. [Google Scholar]
- 32.Bajwa JS, Chen G, Prasad K, Repic O, Blacklock TJ. Tetrahedron Lett. 2006;47:6425. [Google Scholar]
- 33.Robertson SA, Noren CJ, Anthony-Cahill SJ, Griffith MC, Schultz PG. Nucleic Acids Res. 1989;17:9649. doi: 10.1093/nar/17.23.9649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johansson S, Redeby T, Altamore TM, Nilsson U, Börje A. Chem. Res. Toxicol. 2009;22:1774. doi: 10.1021/tx9001435. [DOI] [PubMed] [Google Scholar]
- 35.Karginov VA, Mamaev SV, An H, Van Cleve MD, Hecht SM, Komatsoulis GA, et al. J. Am. Chem. Soc. 1997;119:8166. [Google Scholar]
- 36.Janknecht R, de Martynoff G, Lou J, Hipskind RA, Nordheim A, Stunnenberg HG. Proc. Natl. Acad. U.S.A. 1991;88:8972. doi: 10.1073/pnas.88.20.8972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bolin JT, Filman DJ, Matthews DA, Hamlin RC, Kraut JJ. Biol. Chem. 1982;257:13650. [PubMed] [Google Scholar]
- 38.Fierke CA, Johnson KA, Benkovic SJ. Biochemistry. 1987;26:4085. doi: 10.1021/bi00387a052. [DOI] [PubMed] [Google Scholar]
- 39.Bystroff C, Oatley SJ, Kraut J. Biochemistry. 1990;29:3263. doi: 10.1021/bi00465a018. [DOI] [PubMed] [Google Scholar]
- 40.Bhabha G, Lee J, Ekiert DC, Gam J, Wilson IA, Dyson HJ, Benkovic SJ, Wright PE. Science. 2011;332:234. doi: 10.1126/science.1198542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Edelhoch H. Biochemistry. 1967;6:1948. doi: 10.1021/bi00859a010. [DOI] [PubMed] [Google Scholar]
- 42.Szabo AG, Rayner DM. J. Am. Chem. Soc. 1980;102:554. [Google Scholar]
- 43.Wignaendts van Resandt RW, Vogel RH, Provencher SW. Rev. Sci. Instrum. 1982;53:1392. [Google Scholar]
- 44.Gudgin E, Lopez-Delgado R, Ware WR. Can. J. Chem. 1981;59:1037. [Google Scholar]
- 45.Sawaya MR, Kraut J. Biochemistry. 1997;36:586. doi: 10.1021/bi962337c. [DOI] [PubMed] [Google Scholar]
- 46.Osborne MJ, Schnell J, Benkovic SJ, Dyson HJ, Wright PE. Biochemistry. 2001;40:9846. doi: 10.1021/bi010621k. [DOI] [PubMed] [Google Scholar]
- 47.McElheny D, Schnell JR, Lansing JC, Dyson HJ, Wright PE. Proc. Natl. Acad. Sci. U.S.A. 2005;102:5032. doi: 10.1073/pnas.0500699102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Burstein EA, Vedenkina NS, Ivkova MN. Photochem. Photobiol. 1973;18:263. doi: 10.1111/j.1751-1097.1973.tb06422.x. [DOI] [PubMed] [Google Scholar]
- 49.Vivian JT, Callis PR. Biophys. J. 2001;80:2093. doi: 10.1016/S0006-3495(01)76183-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gibson G, Neverov AA, Brown RS. Can. J. Chem. 2003;81:495. [Google Scholar]
- 51.For the designation of pH in non-aqueous solvents we use the forms recommended by the IUPAC, Compendium of Analytical Nomenclature. Definitive Rules 1997 3rd ed., Blackwell, Oxford, U. K. (1998). Thus refers to the measured pH in a non-aqueous solvent referenced to that solvent. Since the autoprotolysis constant of MeOH is 10−16.77, neutral is 8.4.
- 52.Lakowicz JR. Principles of Fluorescence Spectroscopy. 3rd ed. Springer; New York: 2006. [Google Scholar]
- 53.Varshney U, Lee CP, RajBhandary UL. J. Biol. Chem. 1991;266:24712. [PubMed] [Google Scholar]
- 54.Lodder M, Golovine S, Hecht SM. J. Org. Chem. 1997;62:778. doi: 10.1021/jo971692l. [DOI] [PubMed] [Google Scholar]
- 55.Sawano A, Miyawaki A. Nucleic Acids Res. 2000;28:E78–1. doi: 10.1093/nar/28.16.e78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pratt JM. Transcription and Translation: A Practical Approach. IRL Press; Oxford: 1984. [Google Scholar]
- 57.Lines JA, Yu Z, Dedkova LM, Chen S. Biochem. Biophys. Res. Commun. 2014;443:308. doi: 10.1016/j.bbrc.2013.11.095. [DOI] [PubMed] [Google Scholar]
- 58.Chen S, Zhang Y, Hecht SM. Biochemistry. 2011;50:9340. doi: 10.1021/bi201291p. [DOI] [PubMed] [Google Scholar]
- 59.Chen S, Wang L, Fahmi NE, Benkovic SJ, Hecht SM. J. Am. Chem. Soc. 2012;134:18883. doi: 10.1021/ja307179q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Maglia G, Javed MH, Allemann RK. Biochem. J. 2003;374:529. doi: 10.1042/BJ20030412. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.