Abstract
Little is known about the population of eukaryotic viruses in the human gut (“virome”) or the potential role it may play in disease. We used a metagenomic approach to define and compare the eukaryotic viromes in pediatric diarrhea cohorts from two locations (Melbourne and Northern Territory, Australia). We detected viruses known to cause diarrhea, non-pathogenic enteric viruses, viruses not associated with an enteric reservoir, viruses of plants, and novel viruses. Viromes from Northern Territory children contained more viral families per sample than viromes from Melbourne, which could be attributed largely to an increased number of sequences from the families Adenoviridae and Picornaviridae (genus enterovirus). qRT-PCR/PCR confirmed the increased prevalence of adenoviruses and enteroviruses. Testing of additional diarrhea cohorts by qRT-PCR/PCR demonstrated statistically different prevalences in different geographic sites. These findings raise the question of whether the virome plays a role in enteric diseases and conditions that vary with geography.
Keywords: Diarrhea, Virome, Metagenomics
Highlights
-
•
22 different viral families detected in pediatric diarrhea.
-
•
More viral families in diarrhea from Northern Territory than diarrhea from Melbourne.
-
•
Adenoviridae and Picornaviridae more common in Northern Territory than in Melbourne.
-
•
qRT-PCR/PCR confirmed the increased prevalence of adenoviruses and enteroviruses.
-
•
Viromes in patients with diarrhea differ between two different geographic sites.
Background
It is well established that disease prevalence varies in different geographical regions of the world. Improvements in hygiene and decreased microbial exposure in childhood have been hypothesized to be responsible for the increased occurrence of allergies, autoimmune disorders, and inflammatory bowel disease in the westernized world (Strachan, 1989). On the other hand, conditions such as environmental enteropathy and decreased oral vaccine efficacy are seen in the developing world. Environmental enteropathy is a diffuse villous atrophy of the small bowel, which is ubiquitous in children in the developing world (Campbell et al., 2003, Menzies et al., 1999). It has been observed that environmental enteropathy reverses on transfer to an environment with improved hygiene and sanitation (Lindenbaum, 1968, Lindenbaum et al., 1971). Clearly, the environment is an important factor in the development of human disease.
The human gut contains a diverse microbial community, and it is postulated that many disorders of digestion and growth including diarrhea, inflammatory bowel disease, environmental enteropathy, and malnutrition could be related to perturbations in this biomass. Significant effort has been made to understand the bacterial component of the human stool microbiome and the role it may play in human disease. For example, a dysbiosis or shift in the relative abundances of the bacterial taxa has been associated with obesity (Turnbaugh et al., 2009), inflammatory bowel disease (Frank et al., 2007, Sartor, 2008), diabetes (Larsen et al., 2010), and necrotizing enterocolitis (Mai et al., 2011). The bacterial microbiome can be influenced by several factors including diet, geography, and phage populations. Studies comparing the bacterial gut communities from healthy populations in different parts of the world show that the bacterial microbiome varies with geography (De Filippo et al., 2010, Yatsunenko et al., 2012).
A few studies have examined the human bacteriophage virome in stool. All of the studies reported to date have been done in healthy humans. These findings have shown that the phage virome changes rapidly in the first week of life (Breitbart et al., 2008), interpersonal variation is high while intrapersonal diversity is low (Reyes et al., 2010), diet plays an important role (Minot et al., 2011), and that the phage virome evolves quickly (Minot et al., 2013).
By contrast, little is known about the enteric eukaryotic virome or the potential role it, and variation within it, may play in human disease. In monkeys, pathogenic infection with simian immunodeficiency virus is associated with expansion of the enteric virome, including both RNA viruses and DNA viruses (Handley et al., 2012). A limited number of studies have begun to define the eukaryotic stool viromes of people with diarrhea (Finkbeiner et al., 2008, Nakamura et al., 2009, Phan et al., 2012, Smits et al., 2014, van Leeuwen et al., 2010), non-polio acute flaccid paralysis (Victoria et al., 2009), and healthy adults (Zhang et al., 2006). However, a limitation of the previous human studies is that none have explicitly compared eukaryotic viromes between geographic sites, disease states or age groups, and thus it is not known what factors might influence the composition of the human virome.
Acute diarrhea is one of the leading causes of mortality worldwide, and viruses are known to play a major etiologic role. As a significant fraction of cases are of unexplained etiology, many metagenomic studies have focused upon identifying novel candidate microbial agents. Because previous metagenomic studies demonstrated significant virus diversity in patients with diarrhea (Finkbeiner et al., 2008) we focused this study on comparing the eukaryotic virus populations in stools of children with diarrhea collected from two different locations, Melbourne, Australia and the Northern Territory, Australia.
Results
Sequencing statistics and cohort demographics
Metagenomic sequencing was performed on 43 stool samples from Melbourne and 44 stool samples from Northern Territory. The Melbourne and Northern Territory cohorts were 53% and 50% female, respectively (p=non-significant). The average age of the Melbourne cohort was 25.4 months, while the average age of the Northern Territory cohort was 15.7 months (p=0.002 (t-test)). 454 sequencing of the Melbourne and Northern Territory samples yielded 646,630 and 406,252 total reads and 76,347 and 118,420 unique reads respectively. The average sequence length of the Melbourne and the Northern Territory samples was 339 and 329 respectively. 83.6% of the unique reads possessed detectable similarity to sequences in GenBank while the remaining 16.4% of reads had no significant similarity to sequences in GenBank. Of the 162,784 classifiable sequences 2.5% were viral, 75.8% bacteria, 4.4% phage, 8.9% fungi, 7.0% human, and 1.4% other (plant, fish, etc.). Further breakdown of these statistics by cohort is shown in Table 1.
Table 1.
Sequencing statistics by cohort.
| Northern Territory | Melbourne | |
|---|---|---|
| Average read length | 329 nt | 339 nt |
| Total reads | 406,252 | 646,630 |
| Unique reads | 118,420 | 76,347 |
| Unique viral reads | 2754 (2.3%) | 1266 (1.7%) |
| Unique bacterial reads | 73,164 (61.8%) | 50,196 (65.7%) |
| Unique phage reads | 3417 (2.9%) | 3680 (4.8%) |
| Unique fungi reads | 12,643 (10.7%) | 1897 (2.5%) |
| Unique human reads | 8183 (6.9%) | 3212 (4.2%) |
| Unique other reads | 1055 (0.9%) | 1317 (1.7%) |
| No similarity to sequences in GenBank | 17,204 (14.5%) | 14,779 (19.4%) |
Viruses detected by metagenomic analysis
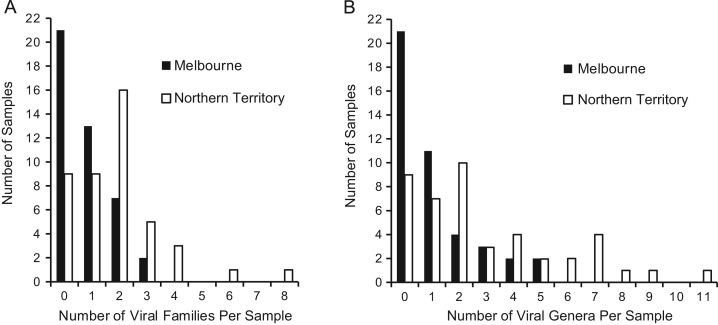
Since all of the stool samples sequenced were from children with acute diarrhea, it was anticipated that a known diarrhea virus would be detected in many of the samples. Twenty-four (28%) of the 87 samples contained sequences derived from the families Reoviridae, Caliciviridae, Adenoviridae or Astroviridae. Strikingly, sequences from many additional virus families were detected in these samples as well as in samples that did not contain sequences from the four canonical diarrheagenic virus families. The most commonly detected virus family was the Anelloviridae, which was present in 37 samples. In total, viral sequences were identified in 57 (66%) of the 87 Australian samples while 35/87 (40%) of the samples had sequences from 2 or more viral families (Supplementary Table 1). Thirty-five of the 44 (80%) samples from the Northern Territory had one or more viral families detected which was more than the 22 of the 43 (51%) samples from Melbourne that had virus detected. One sample from Northern Territory contained sequences from 8 different viral families ( Fig. 1a). Overall, sequences from 22 different viral families were detected in these 87 samples. These included viruses from families known to reside in the gastrointestinal tract such as Picornaviridae (Pallansch and Roos, 2007), Anelloviridae (Okamoto et al., 1998), Circoviridae (Li et al., 2010), and Orthomyxoviridae (Wootton et al., 2006) and known plant viruses, including members of the families Betaflexiviridae, Bromoviridae, Endornaviridae, and Virgaviridae, consistent with previous studies that reported detection of plant viruses (Finkbeiner et al., 2008, Victoria et al., 2009, Zhang et al., 2006). In addition, viruses not previously thought to have an enteric reservoir were detected. For example, human parainfluenza 3 was detected in one sample. The most common viral families detected were Anelloviridae (37 samples), Picornaviridae (26 samples) and Adenoviridae (10 samples). As with all sequencing based studies, our findings are limited by the depth of sequencing achieved. It is possible that deeper sequencing may detect additional viruses present at low abundance.
Fig. 1.
Number of viral families (A) and genera (B) in Northern Territory and Melbourne. Diarrhea samples from the Northern Territory contained more viral families and genera per sample than diarrhea samples from Melbourne.
A number of samples contained what are likely to be novel viruses that shared only very limited sequence identity with known viruses. For example, one sample contained 110 unique sequences with limited similarity to viruses in the order of Picornavirales. Assembly of these reads and sequences that did not share detectable sequence similarity with anything in the database yielded 9 contigs that shared highest sequence similarity with viruses in the order Picornavirales. The longest contig of 9898 nt (601 reads, 18× coverage) shared only 28% amino acid identity to the Israel acute paralysis virus of bees and is likely to be almost the complete genome. A second contig of 7748×nt (322 reads, 12× coverage) shared 28% amino acid identity to the Kashmir bee virus. The two novel viruses shared 82% nt identity with each other. These contigs have been deposited in GenBank [accession numbers KJ420969–KJ420970]. In other samples, sequences were also identified from divergent DNA and RNA viruses, including small DNA viruses from the family Anelloviridae and RNA virus families including Endornaviridae, Betaflexiviridae, Partitiviridae, and Virgaviridae. It is unknown if these newly described viruses are capable of infecting humans or if they are dietary passengers that infect food ingested by the individual.
Comparison of viromes
For each stool specimen, it would be ideal to define the number of distinct virus species present and the relative abundance of each species. However, a number of technical challenges and limitations precluded this form of analysis. First, highly variable coverage levels of the viruses ranging from a single read to hundreds of reads of a virus were obtained, presumably due to variations in viral load as well as the relative abundance of the virus compared to other non-viral nucleic acid molecules in the sample. For samples with low coverage levels, it is impossible to determine whether multiple reads that align to different regions of the same reference virus are derived from the same or related but distinct viruses; therefore, it is challenging to accurately define the number of virus species present. Furthermore estimation of the viral load or copy number from the number of reads obtained is complicated by the sequence independent PCR step, which may introduce amplification biases.
Due to these considerations, we conservatively chose to define viral diversity based on the presence or absence of sequences derived from each of the 95 virus families defined by the ICTV. Therefore, the virome of an individual sample was defined as the number and type of viral taxa with at least one representative sequence present in the sample. These conservative criteria will likely underestimate the true viral diversity of the sample. Similar analyses were also performed by quantifying the presence or absence of sequences from each virus genus.
Virome comparison by gender
To assess whether gender influences the virome, we compared the number of viral families detected per sample by gender in the Northern Territory and Melbourne cohorts and as an aggregate cohort. There was no statistically significant difference (Wilcoxon) in the number of viral families per sample by gender (p=0.912 (regardless of location), p=0.444 (Melbourne), p=0.952 (Northern Territory)). We then assessed if there were gender differences at the individual virus family level. No individual virus family had a statistically significant difference.
Virome comparison by geographic location
To determine if the virome differs by geographic location we compared the number of viral families and genera per sample in each location (Fig. 1). Diarrhea samples from the Northern Territory contained more viral families per sample than diarrhea from Melbourne (p=0.0002 (Wilcoxon)). This was also true at the genus level (p=0.0003 (Wilcoxon)). To control for differences for the larger number of unique reads generated in the Northern Territory samples compared to the Melbourne samples (Table 1), we randomly selected unique reads from the Northern Territory samples to achieve the same sampling depth of the Melbourne samples 100 times. All 100 iterations showed that samples from the Northern Territory contained more viral families (and viral genera) per sample than the samples from Melbourne (p<0.05 (Wilcoxon)). This result demonstrates that the significant difference between Northern Territory and Melbourne is not due to the higher number of unique reads obtained for Northern Territory samples. To determine if a particular viral family drove this difference we compared the prevalence of each individual virus family in the two cohorts. The highly abundant anelloviruses were detected with similar frequencies in the two cohorts (Northern Territory, 52.3%, Melbourne, 32.6%; (p=0.083 (Fisher׳s exact))). Two families, the Picornaviridae (p=0.0003 (Fisher׳s exact)) and the Adenoviridae (p=0.015 (Fisher׳s exact)) were statistically different between the two locations. As there are many picornaviruses known to infect humans, we then determined which picornavirus genera were present. Sequences from the following genera in the family Picornaviridae were detected: Cardiovirus, Enterovirus, Klassevirus, Kobuvirus, and Parechovirus ( Table 2). The genus Enterovirus was the most frequently detected and was the only one that was statistically different between the two locations (p<0.001 (Fisher׳s exact)).
Table 2.
Number of samples with picornavirus genera detected by sequencing.
| Northern Territory | Melbourne | |
|---|---|---|
| Cardiovirus | 1 | 1 |
| Enterovirus | 17 | 2 |
| Klassevirus | 1 | 0 |
| Kobuvirus | 1 | 0 |
| Parechovirus | 6 | 2 |
As the average age of the two cohorts was statistically significantly different (25.4 months (Melbourne) vs. 15.7 months (Northern Territory)), we evaluated if the difference in age was confounding the difference seen by location. We assessed if there was a correlation between age and the total number of viral families present. No correlation was detected in either Northern Territory (Spearman, rho −0.271 (p=0.075)) or Melbourne (Spearman, rho −0.113 (p=0.472)). Furthermore, to assess if the relationship between location and total number of viral families per sample was stable regardless of age, we performed a semiparametric analysis of covariance in which the number of total viral families per sample was rank transformed. After adjusting for age, both the number of viral families (p=0.001) and the number of viral genera (p=0.001) between the two sites were still statistically different. Therefore, age differences between the Northern Territory and Melbourne cohorts do not contribute significantly to the virome differences between the two regions.
PCR of adenovirus, astrovirus, coronovirus, enterovirus, norovirus, and rotavirus
In order to independently confirm the sequencing results, we used PCR to define the prevalence of the most frequently detected viruses for which pan-family or pan-genus primers could be used including: adenovirus, astrovirus, enterovirus, norovirus, and rotavirus. We additionally screened the samples for a family not detected by sequencing, the Coronaviridae.
There was good concordance between the sequencing results and the qPCR for adenovirus. All ten of the positive samples by sequencing were confirmed by our qPCR. Additionally, there were two samples that were negative by sequencing, but positive by qPCR. The adenovirus qPCR was capable of detecting five copies per reaction (Supplementary Fig. 1a). The two samples that sequencing failed to identify as adenovirus positive were the two with the lowest copy number by qPCR (6.9×102 and 1.2×103 copies/g stool).
There were three samples sequencing positive for astrovirus. One of these samples was PCR positive. No samples were PCR positive and sequencing negative. One of the samples positive by sequencing and negative by PCR had 165 reads whose top blast hits were astroviruses. Close inspection of the consensus sequence of these reads showed 3 and 4 mismatches in the forward and reverse primer regions, respectively, which is a likely explanation for why it was not detected by PCR. The other sample positive by sequencing had only a single read, which was not in the region targeted by the primers, so it is unknown if sequence variation in the primer binding sites could be an explanation for this sample as well.
Nineteen samples were positive by sequencing for the enterovirus genus, including 3 that contained rhinovirus species and 16 that contained enterovirus species. We utilized a pan-enterovirus assay (Verstrepen et al., 2001) that does not detect rhinovirus species to evaluate concordance. Twelve of the 16 sequencing positive samples were qRT-PCR positive. The sensitivity limit of the enterovirus qRT-PCR assay was 500 copies as defined by its standard curve (Supplementary Fig. 1b), so it is possible that these four discordant samples were below the limit of detection. In addition, there was one sample that was qRT-PCR positive, but sequencing negative.
Six samples were positive by sequencing for norovirus GII. All of these samples were detected by qRT-PCR. There was one additional sample that was qRT-PCR positive but sequencing negative. No samples were sequencing or qRT-PCR positive for norovirus GI.
There were eight samples sequencing positive for rotavirus. All eight of these samples were RT-PCR positive as well. Additionally, there were four samples from the Northern Territory that were positive for rotavirus by RT-PCR, but negative by sequencing. The sequencing based analysis of rotaviruses was not statistically different between the two sites (p=0.2658). However, because the RT-PCR assay detected four additional positive samples, rotavirus prevalence as measured by RT-PCR was statistically significant (p=0.0264). One factor that complicates interpretation of this difference is that fact that different vaccines were used in the two different regions. In Melbourne RotaTeq (Merck), a live human-bovine pentavalent reassortant vaccine is used, while Rotarix (GlaxoSmithKline), a live attenuated human vaccine strain, is used in the Northern Territory. The stool viral load of Rotarix has been found to be 100 fold higher than children receiving RotaTeq (Hsieh et al., 2014) which may have impacted the differences seen.
No samples were positive by sequencing for coronaviruses. One sample was positive by RT-PCR. This amplicon was cloned and Sanger sequenced and confirmed to be coronavirus 229E, which has been reported in diarrhea samples previously (Risku et al., 2010). Overall there was good concordance between the sequencing data and the PCR screening. The level of concordance observed was similar to that observed in other studies comparing next generation sequencing to PCR (Wylie et al., 2012).
PCR of adenovirus and enterovirus in additional samples
To explore whether the differences seen in adenovirus and enterovirus, could be seen when examining a larger number of samples, we tested additional samples from Melbourne and Northern Territory. Further testing for rotavirus was not pursued as the vaccination status of each subject was unknown and could be a potential confounder. We tested samples from Melbourne (n=159) and Northern Territory (n=165), including all of the samples that were analyzed by metagenomic sequencing. Enteroviruses were more prevalent in the Northern Territory (18.0%) compared to Melbourne (0.6%) by qRT-PCR and adenoviruses were more abundant in the Northern Territory (18.8%) than in Melbourne (6.3%) by PCR confirming the metagenomic results.
To explore the generalizability of these results, we assessed the prevalence of enteroviruses and adenoviruses in additional diarrhea cohorts from Seattle (n=80) and The Gambia (n=160) ( Table 3a–c), which yielded enterovirus positivity rates of 0% and 28.8%, respectively. Adenovirus positivity was even higher in The Gambia (41.9%), while comparable adenovirus frequencies were detected in Seattle (21.3%), and Northern Territory (18.8%).
Table 3.
qPCR of adenovirus and enterovirus in additional samples.
| Adenovirus qPCR positive | Enterovirus qRT-PCR positive | |
|---|---|---|
| (a) qPCR results. | ||
| Melbourne (n=159) | 10 (6.3%) | 1 (0.6%) |
| Northern Territory (n=165) | 31 (18.8%) | 29 (18.0%) |
| Seattle (n=80) | 17 (21.3%) | 0 (0%) |
| The Gambia (n=160) | 67 (41.9%) | 46 (28.8%) |
| Northern Territory | Seattle | The Gambia | |
|---|---|---|---|
| (b) Adenovirus qPCR site comparison p values | |||
| Melbourne | 0.0042 | 0.0036 | 0.0006 |
| Northern Territory | 0.9981 | 0.0006 | |
| Seattle | 0.0096 | ||
| The Gambia | |||
| Northern Territory | Seattle | The Gambia | |
|---|---|---|---|
| (c) Enterovirus qRT-PCR site comparison p values | |||
| Melbourne | 0.0006 | 0.9796 | 0.0006 |
| Northern Territory | 0.0006 | 0.0967 | |
| Seattle | 0.0006 | ||
| The Gambia | |||
Discussion
We examined the diversity of viral communities in stools from 87 children with diarrhea from two different geographic locations collected in the same time period using standardized inclusion criteria at both sites. This study design enabled us to minimize variables while defining and comparing the viromes of patients collected from the two sites.
Many virus families were equally prevalent in the two cohorts, including common diarrhea causing viruses such as norovirus (Caliciviridae) and astrovirus (Astroviridae). In addition, some viruses that have no known pathogenic properties, such as anelloviruses, were frequently detected in both cohorts. Viruses with limited identity to non-human viruses (ex. Israeli acute paralysis virus) were detected. It is unknown whether these represent novel human pathogens or are dietary passengers. Other studies of reclaimed water (Rosario et al., 2009), raw sewage (Cantalupo et al., 2011, Ng et al., 2012), and stool (Victoria et al., 2009) have also found invertebrate viruses. Moreover, it is striking that many samples had multiple viral families detected beyond the viruses typically associated with diarrhea. Most notable was the sample with 8 different viral families detected which raises the question of whether there may be some synergy between viruses (i.e. do some infections make the gut more permissive to infection by additional viruses?). Furthermore, these observations suggest the possibility that viruses are present in the human gut in a fashion analogous to the well-established bacterial microbiome. Longitudinal studies are needed to shed further light on the nature of the eukaryotic virome.
We demonstrated that the diarrhea samples from the Northern Territory had more viral families per sample than the samples from Melbourne. Further analysis demonstrated that the families Adenoviridae, Picornaviridae, and Reoviridae were more common in the Northern Territory than in Melbourne. Furthermore, we found that within the picornaviruses the genus Enterovirus was the most commonly detected and was preferentially found in the Northern Territory. These results clearly demonstrate that enteric viromes in patients with diarrhea can differ between two different geographic sites.
There are many possible explanations for the observed population-specific differences in the human diarrhea stool viromes. It is plausible that environmental factors such as diet, living conditions, water quality, hygiene and/or socioeconomic status could dictate the composition of the stool virome. The samples from Melbourne mainly represent patients living in a westernized and urban setting, while the samples from the Northern Territory represent patients largely from remote communities scattered across a large area. Health indicators among children in the Northern Territory, including for a range of infectious diseases, are worse than for children elsewhere in Australia (Gracey and King, 2009, Ruben and Walker, 1995). The paucity of enteroviruses in the Seattle cohort and their comparative abundance in children from The Gambia is consistent with the model that these viruses are associated with children living in impoverished conditions. Culture-based studies from several decades ago of healthy children also demonstrated a greater frequency of excretion of enteroviruses among persons of lower socioeconomic status (Honig et al., 1956, Otatume and Addy, 1975). Additionally, a recent study in Seattle showed no enterovirus detection in stools from children with diarrhea (Braun et al., 2012).
From the sequencing data there was not a statistically significant difference in the prevalence of rotavirus between Melbourne and the Northern Territory. RT-PCR confirmed the presence of rotavirus in these samples and identified four additional positive samples in the Northern Territory. These additional positive samples then made rotavirus statically different between Melbourne and the Northern Territory. It is possible that these differences are related to the different rotavirus vaccinations given, the proportion of children that were vaccinated, or differences in vaccine efficacy.
While we observed differences in the prevalence of adenoviruses between Melbourne and the Northern Territory, broader analysis of additional diarrhea cohorts (Seattle and The Gambia) yielded a more complex picture. As with the enteroviruses, the highest rate of adenovirus positivity was in The Gambia (42%) and the lowest was in Melbourne (6%). The 42% adenovirus prevalence in The Gambia is similar to previously described reports from some other locations (France and Kenya) (Berciaud et al., 2012, Magwalivha et al., 2010). Unlike the situation with enteroviruses, adenoviruses were detected in approximately equal frequencies in Northern Territory and Seattle. Further studies are needed in larger populations, over multiple years, in these sites to ascertain whether these observations truly reflect geographic differences, or whether additional factors such as seasonality, year-to-year variation, or community outbreaks contribute to these findings.
Conclusions
This study demonstrates major inter-population differences in the human stool eukaryotic virome. The potential etiologic role of the human stool virome in disorders with profound geographic differences remains to be seen. In one hypothesis, these disorders, which include environmental enteropathy, oral vaccine failures, autoimmune diseases, inflammatory bowel disease, and allergic conditions, could be consequences of perturbations in the stool virome. To evaluate this and other hypotheses, it will be important to define the childhood stool virome in other geographic locations as well in health and other states of disease.
Methods
All stool samples were obtained with parental/guardian consent under protocols approved by Human Studies Committees from Seattle Children׳s Hospital, University of Maryland Baltimore, the Central Australian Human Research Ethics Committee, the Human Research Ethics Committee of the Menzies School of Health Research, the Northern Territory Department of Health and Families, the Royal Children׳s Hospital, and the Joint Ethics Committee of the Gambia Government/Medical Research Council Unit. The use of deidentified samples from the previous studies was approved by the Human Studies Committee of Washington University in St. Louis.
Australian stool samples
Stool samples were collected from children less than 5 years old admitted to The Royal Children׳s Hospital, Melbourne, Victoria, Australia and Alice Springs Hospital or Royal Darwin Hospital Northern Territory, Australia, with acute diarrhea between December 2009 and September 2010. These samples were collected under the same inclusion criteria and were archived at −70 oC in Melbourne. Rotavirus vaccine was part of the routine childhood vaccinations throughout the collection period. In Melbourne RotaTeq (Merck), a live human-bovine pentavalent reassortant vaccine is used, while Rotarix (GlaxoSmithKline), a live attenuated human vaccine strain is used in the Northern Territory. 165 samples from the Northern Territory and 159 samples from Melbourne were randomly selected for this study. Of these, 44 samples from Northern Territory and 43 samples from Melbourne were sequenced.
Seattle stool samples
Stool samples were collected from children less than 5 years old who were evaluated for self-defined diarrhea at the Seattle Children׳s Hospital Emergency Department from 2003 to 2005 as part of a prospective study (Denno et al., 2012). Eighty samples were randomly selected from this cohort for the adenovirus and enterovirus qPCR prevalence phase of this study.
Gambian stool samples
Stool samples were collected from children with moderate-to-severe diarrhea less than 5 years old from 2008 to 2009 as part of the Global Enteric Multi-center Study (GEMS) (Kotloff et al., 2012). Diarrhea was defined as new (onset after >7 diarrhea free days), acute (onset within the previous 7 days) and having one of the following criteria: sunken eyes, loss of skin turgor, intravenous hydration administered or prescribed, visible blood in loose stools, or admission to hospital with diarrhea (Kotloff et al., 2013). Samples were collected from children located in the Upper River Division (URD), a rural region in The Gambia. 160 diarrhea stools were randomly selected from this cohort for the adenovirus and enterovirus qPCR prevalence phase of this study.
Nucleic acid preparation and sequencing
Frozen stool was resuspended in 6 volumes of sterile PBS and centrifuged. Resulting supernatants were passed through a 0.45 um filter. Total nucleic acids (RNA and DNA) were isolated from the filtrate using the Ampliprep DNA extraction machine (Roche). In order to evaluate samples for both RNA and DNA viruses the total nucleic acids were randomly amplified. Specifically, RNA present in the sample was converted to cDNA by reverse transcription. Following second strand synthesis, the double-stranded cDNA and any single-stranded or double-standed DNA were then PCR amplified using unique sequence primers which were used as a barcode to assign sequences to their corresponding sample (Wang et al., 2003). The products were then pooled, adapter ligated and sequenced using the 454 GS FLX Titanium platform (454 Life Science).
Analysis of sequencing reads
Sequences were analyzed using a custom bioinformatics pipeline (VirusHunter) (Zhao et al., 2013). Briefly, sequences were deconvoluted based on their barcode (Supplemental Table 2). To reduce the risk of cross contamination between samples barcodes were required to be present at both ends of the read with a perfect match and was trimmed before any further analysis. To reduce the number of near identical sequences introduced from PCR bias, sequences were clustered using CD-HIT (Li and Godzik, 2006) requiring 95% identity over 95% of the sequence. The longest sequence from each cluster was chosen as the representative unique sequence. Sequences were then masked by RepeatMasker (http://www.repeatmasker.org). If a sequence did not contain a stretch of at least 50 consecutive non-“N” nucleotides or if greater than 40% of the total length of the sequence was masked, it was removed from further analysis. The remaining high quality unique sequences were then sequentially compared to (1) human genome using BLASTn, (2) nt database by BLASTn and (3) nr database using BLASTx. Minimum E-value cutoffs of 1×10−10 for BLASTn and 1×10−5 for BLASTx were used. Sequences were then binned based on the top BLAST hit to human, mouse, fungal, bacterial, phage, viral, and other. Sequences aligning to both a virus and another kingdom with the same E-value were classified as ambiguous and not subjected to further analysis. Viral sequences were binned into families based on the taxonomical identity of the best BLAST hit as defined by the NCBI Taxonomy database. Results from this approach have been previously compared directly to other similar BLAST-based taxonomic classifiers such as MEGAN (Handley et al., 2012). Samples were considered positive for a family (or genus) if at least one unique read was present. From previous experience with stool virome analysis, alignments to particular viral families in the NCBI Taxonomy database (herpesviridae, iridoviridae, mimiviridae, phycodnaviridae, poxviridae, unclassified dsDNA viruses, and environmental samples) have a high frequency of false positives, usually due to low complexity and/or repetitive sequence motifs yielding artefactual alignments. Thus, all sequences aligning to these families were manually evaluated and those deemed to be false positives were removed from further analysis.
Assembly of viral genome
Assembly was performed on one sample, which was noted to contain 110 unique sequences with limited similarity to viruses in the order of Picornavirales.
Sequences identified as viral as well as sequences that had no significant hit to any sequence in the NR and NT databases were assembled using Newbler (454 Life Sciences, Branford, CT) with default parameters.
Real-time RT-PCR (qRT-PCR) screening for enterovirus
A previously described pan-enterovirus TaqMan assay targeting conserved sequences in the 5′UTR region of the genome (Verstrepen et al., 2001) was used to assess enterovirus prevalence in all cohorts. The qRT-PCR was performed using the One-Step RT-PCR TaqMan kit (Applied Biosystems). The 25 μL reaction included 5 μL of extracted sample, 12.5 pmol of each primer, and 6.25 pmol of probe. The following cycling conditions were used: 48 °C for 30 min, 95 °C for 10 min, 40 cycles of 95 °C for 15 s and 60 °C for 1 min. To generate a standard curve for this assay, in vitro transcribed RNA was generated from a plasmid containing the region of interest using MAXIscript (Ambion) per the manufacturer׳s protocol. Serial dilutions of this in vitro transcribed RNA from 5×106 to 5 copies were used to generate a standard curve and a limit of detection of 500 copies was defined (Supplemental Fig. 1b). Samples were tested in a 96-well plate format with 8 negative controls and 1 positive control per plate. The threshold of all plates was set at a standard value, and samples were counted as positive if their cycle threshold was <38.00.
qPCR screening for adenovirus
A pan-adenovirus TaqMan assay that detects all adenovirus species was used to assess adenovirus prevalence in all cohorts (Jothikumar et al., 2005). qPCR was performed with the TaqMan Universal PCR Master Mix (Applied Biosystems). The 25 μL reaction included 5 μL of extracted sample, 12.5 pmol of each primer, and 4 pmol of probe. The following cycling conditions were used: 50 °C for 2 min, 95 °C for 10 min, 45 cycles of 95 °C for 15 s and 60 °C for 1 min. To establish a standard curve for this assay, a plasmid containing the region of interest was used in serial dilutions of 5×106 to 5 copies and a limit of detection of 5 copies was defined (Supplementary Fig. 1a). Samples were tested in a 96-well plate format with 8 negative controls and 1 positive control per plate. The threshold of all plates was set at a standard value, and samples were counted as positive if their cycle threshold was <38.00.
Real-time RT-PCR (qRT-PCR) screening for norovirus
We modified a previously described assay to detect norovirus GI and norovirus GII separately (Rolfe et al., 2007). This was used to independently confirm our sequencing results. The qRT-PCR was performed using the One-Step RT-PCR TaqMan kit (Applied Biosystems). The primers and probes were as previously described (Rolfe et al., 2007) except RING2P was labeled with VIC instead of JOE. The 25 μL reaction included 5 μL of extracted sample, 12.5 pmol of each primer, and 6.25 pmol of probe. The following cycling conditions were used: 50 °C for 30 min, 95 °C for 10 min, 50 cycles of 95 °C for 15 s and 56 °C for 1 min. To generate a standard curve for this assay, in vitro transcribed RNA was generated from plasmids containing the regions of interest using MEGAscript (Ambion) per the manufacturer׳s protocol. Serial dilutions of these in vitro transcribed RNA from 5×106 to 5 copies were used to generate a standard curve and a limit of detection of 5 copies was defined (Supplementary Fig. 1c and d) for both assays. Samples were tested in a 96-well plate format with 8 negative controls and 1 positive control per plate. The threshold of all plates was set at a standard value, and samples were counted as positive if their cycle threshold was <38.00.
Reverse transcription PCR (RT-PCR) for astrovirus
A previously described astrovirus consensus RT-PCR was used to independently confirm our sequencing results (Finkbeiner et al., 2009). Qiagen OneStep RT-PCR kit was used to screen 3 μL of extracted material from each sample using the consensus primers SF0073 (5′-GATTGGACTCGATTTGATGG-3′) and SF0076 (5′-CTGGCTTAACCCACATTCC-3′′) which target the ORF 1b (RNA polymerase) of astroviruses under the following cycling conditions: 30 min RT step, 94 °C hold for 10 min, followed by 40 cycles of 94 °C for 30 s, 52 °C for 30 s, and 72 °C for 50 s.
RT-PCR for rotavirus
To independently confirm our sequencing results a previously described RT-PCR to detect group A rotavirus was used (Rao et al., 1995). RT-PCR screening was done using the Qiagen OneStep RT-PCR kit. Each 25 μL reaction used 3 μL of extracted material from each sample and forward (5′-ATGCTCAAGATGGAGTCT-3′) and reverse (5′-GGTCACATAACGCCCCTAT-3′) primers which target nonstructural protein (NSP) 3 under the following cycling conditions: 30 min RT step, 94 °C hold for 10 min, followed by 40 cycles of 94 °C for 30 s, 56 °C for 30 s, and 72 °C for 1 min.
RT-PCR for coronavirus
A previously described pan-coronavirus RT-PCR was used to independently confirm our sequencing results (Vijgen et al., 2008). Qiagen OneStep RT-PCR kit was used to screen 3μL of extracted material from each sample using the primers Cor-FW (5′–ACWCARHTVAAYYTNAARTAYGC-3′) and Cor-RV (5′-TCRCAYTTDGGRTARTCCCA-3′) which target a 251 bp region of the RNA polymerase of coronaviruses under the following cycling conditions: 30 min RT step, 94 °C hold for 10 min, followed by 50 cycles of 94 °C for 30 s, 48 °C for 30 s, and 72 °C for 1 min. PCR amplicons were cloned into pCR4 (Invitrogen) and sequenced using standard Sanger sequencing technology.
Statistical analysis
Two-tailed p values were derived using Wilcoxon rank-sum, chi-square, Fisher׳s exact, or t-tests, as indicated. Wilcoxon rank-sum test, a non-parametric test, was used to compare ordinal data. Fisher׳s exact test was used for categorical data (ex. presence or absence). Assessment of correlation was done using Spearman Rank. For the site comparisons of qPCR results, chi-square with Sidak adjustment for multiple comparisons was used. Statistical analyses were performed using SAS (Cary, NC) software version 9.2.
Availability of supporting data
Sequences (human reads removed) have been uploaded to the MG-RAST (http://metagenomics.anl.gov/) server under the project name Australian Pediatric Diarrhea [MG-Rast ID 4529846.3–4529932.3].
Acknowledgments
This work was funded in part by National Institutes of Health [Grant U54 AI057160] to the Midwest Regional Center of Excellence for Biodefense and Emerging Infectious Disease Research, US Department of Agriculture [Grant 0202238], Bill & Melinda Gates Foundation [OPP1016839], Children׳s Discovery Institute (MD-FR-2013-292), the Morphology and Biobank Cores of the Washington University School of Medicine Digestive Diseases Research Core Center [P30DK052574], and National Institutes of Health-National Center for Advancing Translational Sciences [UL1 TR000448 sub award KL2 TR000450 to L.R.H.]. D.W. holds an Investigator in the Pathogenesis of Infectious Disease Award from the Burroughs Wellcome Fund. C.D.K. is supported by a Career Development Award fellowship from the National Health and Medical Research Council, Australia (607347).
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.virol.2014.09.012.
Contributor Information
Lori R. Holtz, Email: Holtz_l@kids.wustl.edu.
Song Cao, Email: scao@path.wustl.edu.
Guoyan Zhao, Email: GZhao@pathology.wustl.edu.
Irma K. Bauer, Email: Bauer_I@kids.wustl.edu.
Donna M. Denno, Email: ddenno@u.washington.edu.
Eileen J. Klein, Email: eileen.klein@seattlechildrens.org.
Martin Antonio, Email: mantonio@mrc.gm.
O. Colin Stine, Email: ostin001@umaryland.edu.
Thomas L. Snelling, Email: tsnelling@ichr.uwa.edu.au.
Carl D. Kirkwood, Email: carl.kirkwood@mcri.edu.au.
David Wang, Email: davewang@wustl.edu.
Appendix A. Supplementary materials
Supplementary Data Supplemental Fig. 1. Real time standard curves. (A) Real time PCR screening assay for adenovirus standard curve. A plasmid containing the region of interest was used in serial dilutions of 5×106 to 5 copies to generate a standard curve. Real time RT-PCR screening assay for enterovirus (B), norovirus GI (C) and norovirus GII (D) standard curves. In vitro transcribed RNA from 5×106 to 5 copies were used to generate a standard curve.
Supplementary data Supplemental Table 1. Viral families detected by sample. Number of unique reads shown.
Supplementary data Supplemental Table 2. Barcoding sequence for each sample.
References
- Berciaud S., Rayne F., Kassab S., Jubert C., Faure-Della Corte M., Salin F., Wodrich H., Lafon M.E. Adenovirus infections in Bordeaux University Hospital 2008–2010: clinical and virological features. J. Clin. Virol. 2012;54:302–307. doi: 10.1016/j.jcv.2012.04.009. [DOI] [PubMed] [Google Scholar]
- Braun L., Renaud C., Fairchok M., Kuypers J., Englund J., Martin E. Human parechovirus and other enteric viruses in childcare attendees in the era of rotavirus vaccines. J. Pediatr. Infect. Dis. Soc. 2012;1:136–143. doi: 10.1093/jpids/pis005. [DOI] [PubMed] [Google Scholar]
- Breitbart M., Haynes M., Kelley S., Angly F., Edwards R.A., Felts B., Mahaffy J.M., Mueller J., Nulton J., Rayhawk S., Rodriguez-Brito B., Salamon P., Rohwer F. Viral diversity and dynamics in an infant gut. Res. Microbiol. 2008;159:367–373. doi: 10.1016/j.resmic.2008.04.006. [DOI] [PubMed] [Google Scholar]
- Campbell D.I., Murch S.H., Elia M., Sullivan P.B., Sanyang M.S., Jobarteh B., Lunn P.G. Chronic T cell-mediated enteropathy in rural west African children: relationship with nutritional status and small bowel function. Pediatr. Res. 2003;54:306–311. doi: 10.1203/01.PDR.0000076666.16021.5E. [DOI] [PubMed] [Google Scholar]
- Cantalupo P.G., Calgua B., Zhao G., Hundesa A., Wier A.D., Katz J.P., Grabe M., Hendrix R.W., Girones R., Wang D., Pipas J.M. Raw sewage harbors diverse viral populations. MBio. 2011;2(5) doi: 10.1128/mBio.00180-11. pii: e00180-11. doi: 10.1128/mBio.00180-11. Print 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Filippo C., Cavalieri D., Di Paola M., Ramazzotti M., Poullet J.B., Massart S., Collini S., Pieraccini G., Lionetti P. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc. Natl. Acad. Sci. USA. 2010;107:14691–14696. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denno D.M., Shaikh N., Stapp J.R., Qin X., Hutter C.M., Hoffman V., Mooney J.C., Wood K.M., Stevens H.J., Jones R., Tarr P.I., Klein E.J. Diarrhea etiology in a pediatric emergency department: a case control study. Clin. Infect. Dis.: Off. Publ. Infect. Dis. Soc. Am. 2012;55:897–904. doi: 10.1093/cid/cis553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkbeiner S.R., Allred A.F., Tarr P.I., Klein E.J., Kirkwood C.D., Wang D. Metagenomic analysis of human diarrhea: viral detection and discovery. PLoS Pathog. 2008;4:e1000011. doi: 10.1371/journal.ppat.1000011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkbeiner S.R., Le B.M., Holtz L.R., Storch G.A., Wang D. Detection of newly described astrovirus MLB1 in stool samples from children. Emerg. Infect. Dis. 2009;15:441–444. doi: 10.3201/1503.081213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank D.N., St, Amand A.L., Feldman R.A., Boedeker E.C., Harpaz N., Pace N.R. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl. Acad. Sci. USA. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gracey M., King M. Indigenous health. Part 1: determinants and disease patterns. Lancet. 2009;374:65–75. doi: 10.1016/S0140-6736(09)60914-4. [DOI] [PubMed] [Google Scholar]
- Handley S.A., Thackray L.B., Zhao G., Presti R., Miller A.D., Droit L., Abbink P., Maxfield L.F., Kambal A., Duan E., Stanley K., Kramer J., Macri S.C., Permar S.R., Schmitz J.E., Mansfield K., Brenchley J.M., Veazey R.S., Stappenbeck T.S., Wang D., Barouch D.H., Virgin H.W. Pathogenic simian immunodeficiency virus infection is associated with expansion of the enteric virome. Cell. 2012;151:253–266. doi: 10.1016/j.cell.2012.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honig E.I., Melnick J.L., Isacson P., Parr R., Myers I.L., Walton M. An endemiological study of enteric virus infections: poliomyelitis, coxsackie, and orphan (ECHO) viruses isolated from normal children in two socioeconomic groups. J. Exp. Med. 1956;103:247–262. doi: 10.1084/jem.103.2.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh Y.C., Wu F.T., Hsiung C.A., Wu H.S., Chang K.Y., Huang Y.C. Comparison of virus shedding after lived attenuated and pentavalent reassortant rotavirus vaccine. Vaccine. 2014;32:1199–1204. doi: 10.1016/j.vaccine.2013.08.041. [DOI] [PubMed] [Google Scholar]
- Jothikumar N., Cromeans T.L., Hill V.R., Lu X., Sobsey M.D., Erdman D.D. Quantitative real-time PCR assays for detection of human adenoviruses and identification of serotypes 40 and 41. Appl. Environ. Microbiol. 2005;71:3131–3136. doi: 10.1128/AEM.71.6.3131-3136.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotloff K.L., Blackwelder W.C., Nasrin D., Nataro J.P., Farag T.H., van Eijk A., Adegbola R.A., Alonso P.L., Breiman R.F., Faruque A.S., Saha D., Sow S.O., Sur D., Zaidi A.K., Biswas K., Panchalingam S., Clemens J.D., Cohen D., Glass R.I., Mintz E.D., Sommerfelt H., Levine M.M. The Global Enteric Multicenter Study (GEMS) of diarrheal disease in infants and young children in developing countries: epidemiologic and clinical methods of the case/control study. Clin. Infect. Dis.: Off. Publ. Infect. Dis. Soc. Am. 2012;55(Suppl. 4):S232–S245. doi: 10.1093/cid/cis753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotloff K.L., Nataro J.P., Blackwelder W.C., Nasrin D., Farag T.H., Panchalingam S., Wu Y., Sow S.O., Sur D., Breiman R.F., Faruque A.S., Zaidi A.K., Saha D., Alonso P.L., Tamboura B., Sanogo D., Onwuchekwa U., Manna B., Ramamurthy T., Kanungo S., Ochieng J.B., Omore R., Oundo J.O., Hossain A., Das S.K., Ahmed S., Qureshi S., Quadri F., Adegbola R.A., Antonio M., Hossain M.J., Akinsola A., Mandomando I., Nhampossa T., Acacio S., Biswas K., O׳Reilly C.E., Mintz E.D., Berkeley L.Y., Muhsen K., Sommerfelt H., Robins-Browne R.M. and Levine M.M., Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study, Lancet. 2013 Jul 20;382(9888):209-22. 10.1016/S0140-6736(13)60844-2. Epub 2013 May 14. [DOI] [PubMed]
- Larsen N., Vogensen F.K., van den Berg F.W., Nielsen D.S., Andreasen A.S., Pedersen B.K., Al-Soud W.A., Sorensen S.J., Hansen L.H., Jakobsen M. Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PLoS One. 2010;5:e9085. doi: 10.1371/journal.pone.0009085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Kapoor A., Slikas B., Bamidele O.S., Wang C., Shaukat S., Masroor M.A., Wilson M.L., Ndjango J.B., Peeters M., Gross-Camp N.D., Muller M.N., Hahn B.H., Wolfe N.D., Triki H., Bartkus J., Zaidi S.Z., Delwart E. Multiple diverse circoviruses infect farm animals and are commonly found in human and chimpanzee feces. J. Virol. 2010;84:1674–1682. doi: 10.1128/JVI.02109-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- Lindenbaum J. Small intestine dysfunction in Pakistanis and Americans resident in Pakistan. Am. J. Clin. Nutr. 1968;21:1023–1029. doi: 10.1093/ajcn/21.9.1023. [DOI] [PubMed] [Google Scholar]
- Lindenbaum J., Gerson C.D., Kent T.H. Recovery of small-intestinal structure and function after residence in the tropics. I. Studies in Peace Corps volunteers. Ann. Intern. Med. 1971;74:218–222. doi: 10.7326/0003-4819-74-2-218. [DOI] [PubMed] [Google Scholar]
- Magwalivha M., Wolfaardt M., Kiulia N.M., van Zyl W.B., Mwenda J.M., Taylor M.B. High prevalence of species D human adenoviruses in fecal specimens from Urban Kenyan children with diarrhea. J. Med. Virol. 2010;82:77–84. doi: 10.1002/jmv.21673. [DOI] [PubMed] [Google Scholar]
- Mai V., Young C.M., Ukhanova M., Wang X., Sun Y., Casella G., Theriaque D., Li N., Sharma R., Hudak M., Neu J. Fecal microbiota in premature infants prior to necrotizing enterocolitis. PLoS One. 2011;6:e20647. doi: 10.1371/journal.pone.0020647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menzies I.S., Zuckerman M.J., Nukajam W.S., Somasundaram S.G., Murphy B., Jenkins A.P., Crane R.S., Gregory G.G. Geography of intestinal permeability and absorption. Gut. 1999;44:483–489. doi: 10.1136/gut.44.4.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minot S., Bryson A., Chehoud C., Wu G.D., Lewis J.D., Bushman F.D. Rapid evolution of the human gut virome. Proc. Natl. Acad. Sci. USA. 2013;110:12450–12455. doi: 10.1073/pnas.1300833110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minot S., Sinha R., Chen J., Li H., Keilbaugh S.A., Wu G.D., Lewis J.D., Bushman F.D. The human gut virome: inter-individual variation and dynamic response to diet. Genome Res. 2011;21:1616–1625. doi: 10.1101/gr.122705.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura S., Yang C.S., Sakon N., Ueda M., Tougan T., Yamashita A., Goto N., Takahashi K., Yasunaga T., Ikuta K., Mizutani T., Okamoto Y., Tagami M., Morita R., Maeda N., Kawai J., Hayashizaki Y., Nagai Y., Horii T., Iida T., Nakaya T. Direct metagenomic detection of viral pathogens in nasal and fecal specimens using an unbiased high-throughput sequencing approach. PLoS One. 2009;4:e4219. doi: 10.1371/journal.pone.0004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng T.F., Marine R., Wang C., Simmonds P., Kapusinszky B., Bodhidatta L., Oderinde B.S., Wommack K.E., Delwart E. High variety of known and new RNA and DNA viruses of diverse origins in untreated sewage. J. Virol. 2012;86:12161–12175. doi: 10.1128/JVI.00869-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto H., Akahane Y., Ukita M., Fukuda M., Tsuda F., Miyakawa Y., Mayumi M. Fecal excretion of a nonenveloped DNA virus (TTV) associated with posttransfusion non-A-G hepatitis. J. Med. Virol. 1998;56:128–132. [PubMed] [Google Scholar]
- Otatume S., Addy P. Ecology of enteroviruses in tropics. I. Circulation of enteroviruses in healthy infants in tropical urban area. Jpn. J. Microbiol. 1975;19:201–209. doi: 10.1111/j.1348-0421.1975.tb00869.x. [DOI] [PubMed] [Google Scholar]
- Pallansch M., Roos R. Enteroviruses: polioviruses, coxsackieviruses, echoviruses, and newer enteroviruses. In: Knipe D., Howley P, editors. Fields Virology. 5th ed. Lippincott Williams and Wilkins; Philadelphia: 2007. pp. 839–894. [Google Scholar]
- Phan T.G., Vo N.P., Bonkoungou I.J., Kapoor A., Barro N., O׳Ryan M., Kapusinszky B., Wang C., Delwart E. Acute diarrhea in West African children: diverse enteric viruses and a novel parvovirus genus. J. Virol. 2012;86:11024–11030. doi: 10.1128/JVI.01427-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao C.D., Das M., Ilango P., Lalwani R., Rao B.S., Gowda K. Comparative nucleotide and amino acid sequence analysis of the sequence-specific RNA-binding rotavirus nonstructural protein NSP3. Virology. 1995;207:327–333. doi: 10.1006/viro.1995.1087. [DOI] [PubMed] [Google Scholar]
- Reyes A., Haynes M., Hanson N., Angly F.E., Heath A.C., Rohwer F., Gordon J.I. Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature. 2010;466:334–338. doi: 10.1038/nature09199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risku M., Lappalainen S., Rasanen S., Vesikari T. Detection of human coronaviruses in children with acute gastroenteritis. J. Clin Virol. 2010;48:27–30. doi: 10.1016/j.jcv.2010.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolfe K.J., Parmar S., Mururi D., Wreghitt T.G., Jalal H., Zhang H., Curran M.D. An internally controlled, one-step, real-time RT-PCR assay for norovirus detection and genogrouping. J. Clin. Virol. 2007;39:318–321. doi: 10.1016/j.jcv.2007.05.005. [DOI] [PubMed] [Google Scholar]
- Rosario K., Nilsson C., Lim Y.W., Ruan Y., Breitbart M. Metagenomic analysis of viruses in reclaimed water. Environ. Microbiol. 2009;11:2806–2820. doi: 10.1111/j.1462-2920.2009.01964.x. [DOI] [PubMed] [Google Scholar]
- Ruben A.R., Walker A.C. Malnutrition among rural aboriginal children in the Top End of the Northern Territory. Med. J. Austr. 1995;162:400–403. doi: 10.5694/j.1326-5377.1995.tb124678.x. [DOI] [PubMed] [Google Scholar]
- Sartor R.B. Microbial influences in inflammatory bowel diseases. Gastroenterology. 2008;134:577–594. doi: 10.1053/j.gastro.2007.11.059. [DOI] [PubMed] [Google Scholar]
- Smits S.L., Schapendonk C.M., van Beek J., Vennema H., Schurch A.C., Schipper D., Bodewes R., Haagmans B.L., Osterhaus A.D., Koopmans M.P. New viruses in idiopathic human diarrhea cases, the Netherlands. Emerg. Infect. Dis. 2014;20:1218–1222. doi: 10.3201/eid2007.140190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strachan D.P. Hay fever, hygiene, and household size. Br. Med. J. 1989;299:1259–1260. doi: 10.1136/bmj.299.6710.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh P.J., Hamady M., Yatsunenko T., Cantarel B.L., Duncan A., Ley R.E., Sogin M.L., Jones W.J., Roe B.A., Affourtit J.P., Egholm M., Henrissat B., Heath A.C., Knight R., Gordon J.I. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Leeuwen M., Williams M.M., Koraka P., Simon J.H., Smits S.L., Osterhaus A.D. Human picobirnaviruses identified by molecular screening of diarrhea samples. J. Clin. Microbiol. 2010;48:1787–1794. doi: 10.1128/JCM.02452-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verstrepen W.A., Kuhn S., Kockx M.M., Van De Vyvere M.E., Mertens A.H. Rapid detection of enterovirus RNA in cerebrospinal fluid specimens with a novel single-tube real-time reverse transcription-PCR assay. J. Clin. Microbiol. 2001;39:4093–4096. doi: 10.1128/JCM.39.11.4093-4096.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victoria J.G., Kapoor A., Li L., Blinkova O., Slikas B., Wang C., Naeem A., Zaidi S., Delwart E. Metagenomic analyses of viruses in stool samples from children with acute flaccid paralysis. J. Virol. 2009;83:4642–4651. doi: 10.1128/JVI.02301-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijgen L., Moes E., Keyaerts E., Li S., Van Ranst M. A pancoronavirus RT-PCR assay for detection of all known coronaviruses. Methods Mol. Biol. 2008;454:3–12. doi: 10.1007/978-1-59745-181-9_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Urisman A., Liu Y.T., Springer M., Ksiazek T.G., Erdman D.D., Mardis E.R., Hickenbotham M., Magrini V., Eldred J., Latreille J.P., Wilson R.K., Ganem D., DeRisi J.L. Viral discovery and sequence recovery using DNA microarrays. PLoS Biol. 2003;1:E2. doi: 10.1371/journal.pbio.0000002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wootton S.H., Scheifele D.W., Mak A., Petric M., Skowronski D.M. Detection of human influenza virus in the stool of children. Pediatr. Infect. Dis. J. 2006;25:1194–1195. doi: 10.1097/01.inf.0000245097.95543.11. [DOI] [PubMed] [Google Scholar]
- Wylie K.M., Mihindukulasuriya K.A., Sodergren E., Weinstock G.M., Storch G.A. Sequence analysis of the human virome in febrile and afebrile children. PLoS One. 2012;7:e27735. doi: 10.1371/journal.pone.0027735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatsunenko T., Rey F.E., Manary M.J., Trehan I., Dominguez-Bello M.G., Contreras M., Magris M., Hidalgo G., Baldassano R.N., Anokhin A.P., Heath A.C., Warner B., Reeder J., Kuczynski J., Caporaso J.G., Lozupone C.A., Lauber C., Clemente J.C., Knights D., Knight R., Gordon J.I. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Breitbart M., Lee W.H., Run J.Q., Wei C.L., Soh S.W., Hibberd M.L., Liu E.T., Rohwer F., Ruan Y. RNA viral community in human feces: prevalence of plant pathogenic viruses. PLoS Biol. 2006;4:e3. doi: 10.1371/journal.pbio.0040003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao G., Krishnamurthy S., Cai Z., Popov V.L., Travassos da Rosa A.P., Guzman H., Cao S., Virgin H.W., Tesh R.B., Wang D. Identification of novel viruses using VirusHunter—an automated data analysis pipeline. PLoS One. 2013;8:e78470. doi: 10.1371/journal.pone.0078470. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data Supplemental Fig. 1. Real time standard curves. (A) Real time PCR screening assay for adenovirus standard curve. A plasmid containing the region of interest was used in serial dilutions of 5×106 to 5 copies to generate a standard curve. Real time RT-PCR screening assay for enterovirus (B), norovirus GI (C) and norovirus GII (D) standard curves. In vitro transcribed RNA from 5×106 to 5 copies were used to generate a standard curve.
Supplementary data Supplemental Table 1. Viral families detected by sample. Number of unique reads shown.
Supplementary data Supplemental Table 2. Barcoding sequence for each sample.
Data Availability Statement
Sequences (human reads removed) have been uploaded to the MG-RAST (http://metagenomics.anl.gov/) server under the project name Australian Pediatric Diarrhea [MG-Rast ID 4529846.3–4529932.3].