Abstract
Genome-wide characterization of the in vivo cellular response to perturbation is fundamental to understanding how cells survive stress. Identifying the proteins and pathways perturbed by small molecules affects biology and medicine by revealing the mechanisms of drug action. We used a yeast chemogenomics platform that quantifies the requirement for each gene for resistance to a compound in vivo to profile 3250 small molecules in a systematic and unbiased manner. We identified 317 compounds that specifically perturb the function of 121 genes and characterized the mechanism of specific compounds. Global analysis revealed that the cellular response to small molecules is limited and described by a network of 45 major chemogenomic signatures. Our results provide a resource for the discovery of functional interactions among genes, chemicals, and biological processes.
Chemical genomics is a powerful approach for understanding in vivo mechanisms of drug action. The ability to interpret molecular-level responses in a cellular context has led to therapies for intractable diseases (1). “Guilt-byassociation” approaches allow mechanisms of untested compounds to be inferred on the basis of profile similarity to established drugs (2, 3). Loss-of-function genetic screens provide direct mechanistic insight as they report genes that when deleted, confer drug sensitivity. Here, we used yeast genomic tools (4) in loss-of-function assays to systematically characterize the cellular response to small-molecule perturbation by screening 3250 compounds using a haploinsufficiency profiling (HIP) and homozygous profiling (HOP) chemogenomic platform (5–7). HIP exploits drug-induced haploinsufficiency (8), as measured by a growth or fitness defect (FD) observed in a heterozygous strain deleted for one copy of the drug’s target gene. HIP identifies candidate protein targets by measuring the drug-induced FDs of ~1100 heterozygous strains representing the yeast essential genome (5, 6). In the complementary HOP assay, drug-induced FDs are reported for ~4800 homozygous deletion strains, identifying the nonessential genes required to buffer the targeted pathways (7, 9). Each combined HIPHOP profile provides a genome-wide view of the cellular response to a specific compound.
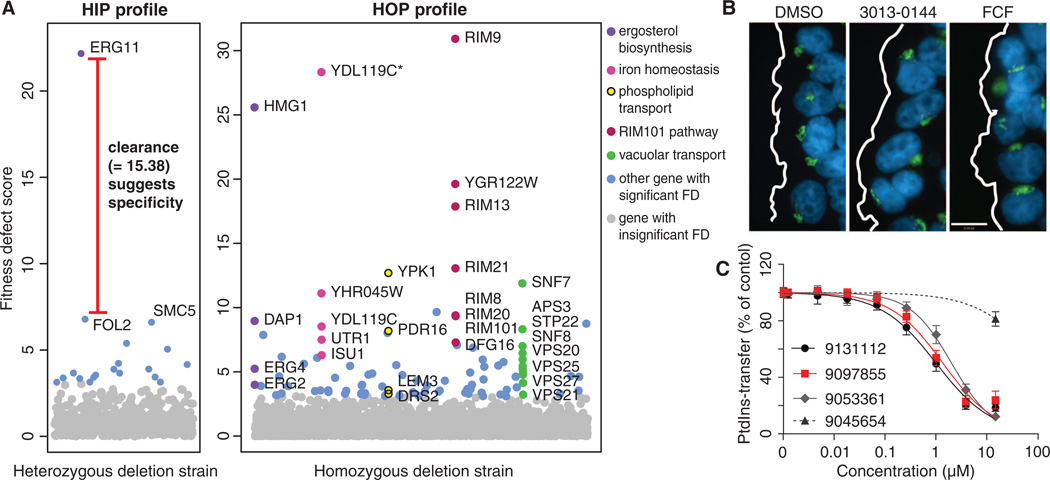
By prescreening 50,000 diverse druglike small molecules, we identified 3250 compounds that inhibited wild-type yeast growth (~95% of unknown mechanism; table S1 and fig. S1). Each compound was profiled genome-wide, and FDs were measured for each strain; larger scores representing a greater requirement for the deleted gene to resist chemical treatment (10). For example, the Erg11Δ/ERG11 strain represents a “hit” as it had the largest FD in the fluconazole HIP profile and passed significance and specificity thresholds (10). Fluconazole inhibits the protein Erg11, thus demonstrating the ability of HIP to identify targets in vivo (Fig. 1A). Fluconazole HOP identified mechanisms that buffer the ergosterol pathway, including the requirement for iron (Fig. 1A). Gene Ontology (GO) enrichments are provided for each profile (fig. S2) (10). Additional relationships among genes, profiles, pathways, and compounds can be explored with the interactive online HIPHOP chemogenomic database http://chemogenomics.pharmacy.ubc.ca/HIPHOP/ (10).
Fig. 1. Validation of chemical-genetic probes.
(A) Fluconazole HIPHOP profile. Fitness defect (FD) scores plotted for each deletion strain. HIP (left) identifies the established drug target Erg11. HOP (right) identifies processes directly (e.g., sterol biosynthesis) and indirectly (e.g., iron ion homeostasis) related to ERG11 function. Significant FDs (standard normal distribution P ≤ 0.001) are labeled except those (blue) not covered by the highlighted processes; *, dubious gene overlapping labeled gene. (B) Cdc12 inhibitor. In a wound-healing assay, HeLa cells with dimethyl sulfoxide (DMSO), 1 µM 3013-0144, and 5 µM forchlorfenuron (FCF) were fixed and stained as described, with DNA stained blue and antibodies against the Golgi visualized via green fluorescence (10). DMSO-treated cells show the Golgi reoriented toward the wound edge (white line); in contrast, 3013-0144 inhibited Golgi reorientation as effectively as FCF (scale bar, 10 µm). (C) Dose-dependent inhibition of the phosphatidylinositol (PtdIns) transfer activity of purified recombinant Sec14 (10). Transfer of radiolabeled PtdIns as a percentage of the untreated control (y axis), measured in the presence of 9131112, 9097855, 9053361*, and 9045654 (an inactive derivative) at the indicated concentrations (x axis). Data are mean ± SD (N = 3). *9053361 did not qualify as a HIP hit, but was nonetheless validated.
In total, HIP identified 317 compounds that specifically perturb the function of 121 essential genes. To distinguish these compounds from drugs or credentialed chemical probes, we refer to them as “chemical-genetic probes,” and to their interacting gene partners as “HIP hits” (10). Consistent with the ability of HIP to identify protein targets, these specific interactions were significantly enriched for established compound-target pairs (hypergeometric test P < 10−4) including drugs approved by the U.S. Food and Drug Administration (e.g., rapamycin) and chemical probes (e.g., cerulenin) (table S2 and fig. S3). These drugs and probes target homologous proteins in yeast and mammalian cells, suggesting that some of our uncharacterized compounds may function similarly in mammalian cells, even though yeast required about fives times as much compound to inhibit growth by 20% [minimum 20% inhibitory concentration (IC20) = ~244 nM, median = ~100 µM; fig. S4 and table S3]. This observation is consistent with published data (11, 12) and reflective of yeast’s robust xenobiotic defenses. Using quantitative growth assays (fig. S5 and table S2), we confirmed dose-dependent drug-induced haploinsufficiency for 63 compound-gene pairs, 54 of them novel (figs. S6 to S9). Specific chemical-genetic probes were tested for inhibitory activities in cell-free assays (IC50 range 1 to 500 µM, median = ~23 µM) and/or cell-based assays (IC50 range 30 nM to 100 mM, median = 60 µM). For example, we validated inhibitors of actin (0136-0228) and tubulin (1327-0036) in yeast and mammalian cell-based assays (IC50 range 30 nM to 100 µM) and in in vitro polymerization assays (IC50 range20 to25µM; fig.S10).An in vitro assay suggested that compound 1327-0036 binds to the colchicine-binding site on the tubulin dimer (fig. S10). Another compound (3013-0144) perturbed the septin Cdc12 (IC50 = 1 µM), exhibiting about five times more activity than forchlorfenuron, a known septin inhibitor (13) (Fig. 1B). Three compounds that perturb Sec14, a conserved phosphatidylinositol transfer protein, were also biochemically validated (IC50= ~1 µM; Fig. 1C), and we have recently shown that two additional inhibitors are effective in vivo and in vitro (14). The specificity of these compounds for Sec14 was further validated by demonstrating that suppressed double mutants cki1Δ sec14Δ (15) and kes1Δ sec14Δ (16) were resistant to these inhibitors (fig. S7). This experimental support provides encouraging proof-of-concept data and underscores the need for further characterization of putative protein inhibitors (see fig. S5 for the structures of all validated inhibitors).
Hierarchically clustering all HIPHOP profiles allowed classification of cellular response types into major (covering ~36% of profiles), minor (~40%), or unique (~24%) signatures. Each major response was defined by a characteristic gene signature in a cluster of more than four profiles, while most minor signatures were associated with three to four profiles (fig. S11 and table S4). Several minor signatures point to compelling biology, including a signature representing the response to three chemical-genetic probes that share a 2,5-dimethylpyrrole chemical moiety and putatively target the geranylgeranyltransferase complex RAM2/CDC43 (table S2). Unique signatures (one to two profiles each) include distinctive drugs (e.g., methotrexate) and chemical-genetic probes (e.g., 0kpi-0099, fig. S12).
We focused on the highest-confidence major cellular response signatures, which represent ~70% of our chemical-genetic probes (10). Of these 45 signatures, 33 were enriched for known gene function, 40 represent ≥1 HIP hits, and 11 represent ≥2 compounds of known mechanism (Fig. 2). Five of these 11 signatures are enriched for compounds with similar bioactivity [hypergeometric test false discovery rate (FDR) ≤ 0.1; table S5], supporting mechanism prediction for related compounds (10) (Fig. 2). For example, the exosome signature compounds included four chemotherapeutics known to target this complex (hypergeometric test P < 10−10; Fig. 3) (5, 6), allowing a similar mechanism to be inferred for two additional compounds. Similarly, we predict DNA damage as the underlying mechanism for 20 uncharacterized compounds with the same signature as established DNA-damaging agents (table S5). Response signatures also provide hypotheses applicable to mammalian cells. For example, trichlorophene induced mitochondrial stress. Trichlorophene-treated immortalized human leukemia cells confirmed a mitochondrial-specific mechanism; exhibiting increased generation of mitochondrial reactive oxygen species and a reduction in reserve oxygen capacity (fig. S13). Signatures also yielded new information about well-characterized compounds; e.g., the tubulin inhibitors nocodazole and benomyl induced a signature containing tubulin biogenesis and SWR1 complex genes, a biological link between cytoarchitecture and chromatin structure supported by genetic interaction data (17).
Fig. 2. The cellular response is defined by a network of chemogenomic response signatures.
Each circular node represents a major signature; size is proportional to confidence in the signature (10). Node color: dark blue if GO enriched (hypergeometric test FDR ≤ 0.1), pale blue otherwise. Node border color: green, signature represents two or more compounds of known mechanism (select compound names are shown, and are in bold if they drive bioactivity class enrichment); red, signature represents chemical-genetic probes (select HIP hits are shown, and are in bold if validation data are provided). Signatures are connected to chemical moiety nodes where signature compounds are enriched (hypergeometric test FDR ≤ 0.1). Boxes indicate signatures discussed in the text. ERAD, endoplasmic reticulum–associated degradation; RNA pol III, RNA polymerase III; ROS, reactive oxygen species; TOFA, 5-tetradecoxyfuran-2-carboxylic acid; TRP, tryptophan.
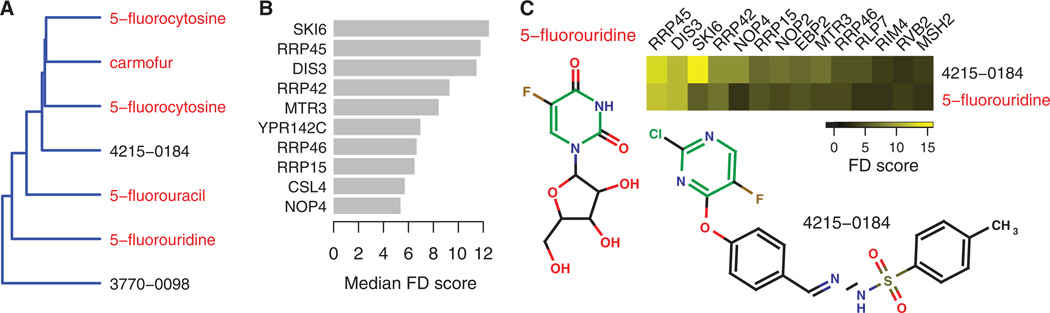
Fig. 3. Mechanism inference with the exosome signature.
(A) Dendrogram of the exosome profiles extracted from the dendrogram of all HIPHOP profiles. Profiled compounds with established mechanisms are shown in red. (B) The exosome signature. For each gene in the signature, the bar plot indicates the median FD score across the exosome profiles. (C) Mechanism inferred by signature similarity. Scores of genes exhibiting significant fitness defects in the profiles of 5-fluorouridine and an uncharacterized compound (4215-0184) associated with the exosome signature. Both compounds contain a 5-fluoropyrimidine substructure (green). Guilt-by-association infers that 4215-0184 inhibits the exosome.
Our signatures are recognizable in other genome-wide data sets, supporting their biological relevance. Yeast large-scale genetic interaction data (18) revealed that our response signatures were observed in 12% of 380 genetic profiles that lacked GO enrichment (table S6). In some cases, the signatures provided annotation for uncharacterized genes. For example, genes known to genetically interact with YPL109C (18) are not enriched for any GO-based function, yet were significantly enriched for our ubiquinone biosynthesis and proteasome signature (hypergeometric test FDR ≤ 0.1), suggesting a related function. Our signatures also identify links between biological processes (fig. S14). For example, the ubiquinone biosynthesis and proteasome signature links these two processes by 38 gene pairs exhibiting correlated fitness, or “cofitness.” Independent support for this observation is provided by nine genetic interactions (18) and one physical interaction (17) (table S7). Cofitness also supported a functional relationship between diphthamide biosynthesis and histone exchange in the NEO1-PIK1 signature (table S7).
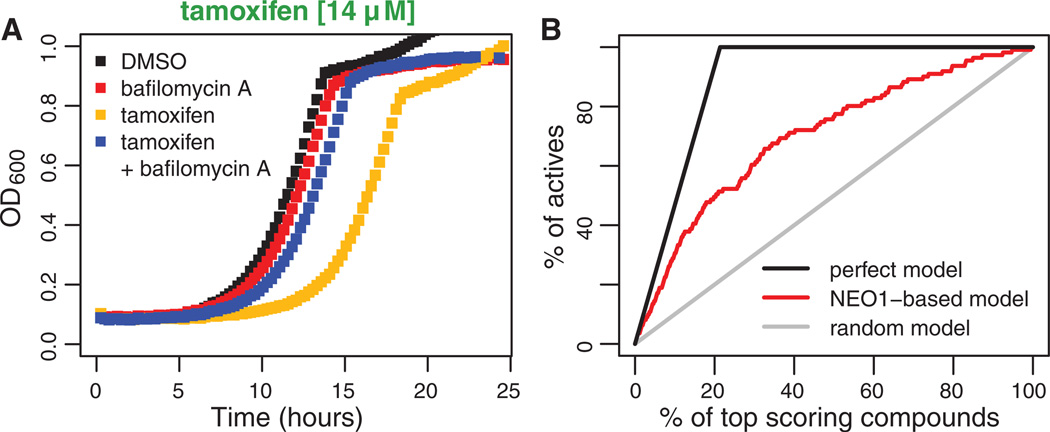
Approximately half (n = 28) of the major response signatures are associated with compounds significantly enriched for chemical moieties (hypergeometric test FDR ≤ 0.1; table S8 and Fig. 2), suggesting that specific molecular structural properties can drive a cellular response. For example, the NEO1 and NEO1-PIK1 signatures (Fig. 2) are characterized by NEO1 haploinsufficiency induced by cationic amphiphilic drugs (CADs; fig. S15). CADs are associated with drug-induced phospholipidosis (DIPL), a human phospholipid storage disorder (19, 20) caused by diverse therapeutics. At a cellular level, DIPL arises from the selective accumulation of CADs in the acidic vacuole and lysosome, in yeast and mammalian cells, respectively. Consistent with published yeast genetic studies, we confirmed that inhibition of yeast vacuolar adenosine triphosphatase by bafilomycin A alleviates the FD induced by CADs (21). Furthermore, we found that bafilomycin A rescued CAD-induced NEO1 haploinsuffiency (Fig. 4A and fig. S8). Structural features of NEO1 and NEO1-PIK1 compounds proved predictive of response; a statistical structure-based model performed about seven times better than random in identifying compounds that induce NEO1 haploinsufficiency (fig. S16; percentage of correct predictions in cross-validation = 99%) (10). Our yeast-based model suggests that haploinsufficiency of NEO1, and by extension, its human homologs (ATP9A and B), may prove useful as a biomarker to identify DIPL-causing compounds (Fig. 4B).
Fig. 4. NEO1-based signatures and drug-induced phospholipidosis (DIPL).
(A) Rescue of CAD-induced NEO1 haploinsufficiency. Tamoxifen-induced NEO1 haploinsufficiency is rescued by bafilomycin A. Growth of the neo1Δ/NEO1 strain treated with these compounds was monitored by measuring the optical density at 600 nm (OD600) (y axis) for 24 hours (x axis). (B) Prediction of DIPL. The plot shows the percentage of DIPL-causing compounds (actives; y axis) identified among the top-scoring compounds (x axis).
Our systems-level view of the cellular response to small molecules provides a resource for the exploration of multifaceted relationships among genes, biological processes, chemical structures, and response signatures. Although not previously captured by any existing GO category, in retrospect, we detect signatures present in other large-scale genomic data sets suggesting that they may be used to address the challenges of incomplete gene annotation and integration of diverse genome-wide data sets. It is likely that we have identified all major signatures (within similar chemical space in yeast), as we observed saturation in our screen. Reanalysis of our prior chemogenomic data set (7) revealed that ~60% of the 45 signatures could be detected (fig. S17 and table S4), and simulation demonstrates that 80% of our 45 major clusters would be identified after screening <30% of the compounds (fig. S18). We expect that these signatures therefore represent fundamental small-molecule response systems that are present across eukaryotic cells. Accordingly, we expect that many of our 317 chemical-genetic probes will be directly applicable to mammalian cell biology and may support novel targets as opportunities to pursue for therapeutic intervention (5, 22, 23).
Supplementary Material
Acknowledgments
Supported by the National Human Genome Research Institute (RO1 003317-07) (C.N., G.G., and R.W.D.); Canadian Cancer Society Research Institute (#020380) (C.N. and G.G.); Canadian Institute for Health Research (MOP-81340) (G.G.), MOP-79368 (G.W.B.), and MOP-700724 (W.S.T.); Canadian Research Chair (G.G.); Charles H. Best Institute (A.Y.L.); Belgian National Fund for Scientific Research and the Interuniversity Poles of Attraction Program (M.B.); French National Research Agency (J.C.); Marie Curie Fellowship (I.M.W.); Leukemia and Lymphoma Society (A.D.S.); Robert A. Welch Foundation (V.A.B.); and NIH grants (GM103504) (G.D.B.) and GM44530 (V.A.B.).
Footnotes
The data reported in this paper are tabulated in the supplementary materials, and access to the entire data set is available for query and in a variety of downloadable formats at http://chemogenomics.pharmacy.ubc.ca/HIPHOP/. The raw microarray data are archived in the ArrayExpress database (www.ebi.ac.uk/arrayexpress) under accession no. E-MTAB-2391.
Supplementary Materials
www.sciencemag.org/content/344/6180/208/suppl/DC1
Materials and Methods
Figs. S1 to S24
Tables S1 to S10
References (24–65)
References and Notes
- 1.Barrett PM, Alagely A, Topol EJ. Hum. Mol. Genet. 2012;21(R1):R66–R71. doi: 10.1093/hmg/dds345. [DOI] [PubMed] [Google Scholar]
- 2.Lamb J, et al. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 3.Iorio F, et al. Proc. Natl. Acad. Sci. U.S.A. 2010;107:14621–14626. doi: 10.1073/pnas.1000138107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Giaever G, et al. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- 5.Giaever G, et al. Proc. Natl. Acad. Sci. U.S.A. 2004;101:793–798. doi: 10.1073/pnas.0307490100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lum PY, et al. Cell. 2004;116:121–137. doi: 10.1016/s0092-8674(03)01035-3. [DOI] [PubMed] [Google Scholar]
- 7.Hillenmeyer ME, et al. Science. 2008;320:362–365. doi: 10.1126/science.1150021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Giaever G, et al. Nat. Genet. 1999;21:278–283. doi: 10.1038/6791. [DOI] [PubMed] [Google Scholar]
- 9.Parsons AB, et al. Nat. Biotechnol. 2004;22:62–69. doi: 10.1038/nbt919. [DOI] [PubMed] [Google Scholar]
- 10.Materials and methods are available as supplementary material on Science Online.
- 11.Wallace IM, et al. Chem. Biol. 2011;18:1273–1283. doi: 10.1016/j.chembiol.2011.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Torres NP, Lee AY, Giaever G, Nislow C, Brown GW. Assay Drug Dev. Technol. 2013;11:299–307. doi: 10.1089/adt.2012.503. [DOI] [PubMed] [Google Scholar]
- 13.Hu Q, Nelson WJ, Spiliotis ET. J. Biol. Chem. 2008;283:29563–29571. doi: 10.1074/jbc.M804962200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nile AH, et al. Nat. Chem. Biol. 2014;10:76–84. doi: 10.1038/nchembio.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cleves AE, et al. Cell. 1991;64:789–800. doi: 10.1016/0092-8674(91)90508-v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fang M, et al. EMBO J. 1996;15:6447–6459. [PMC free article] [PubMed] [Google Scholar]
- 17.Chatr-aryamontri A, et al. Nucleic Acids Res. 2013;41:D816–D823. doi: 10.1093/nar/gks1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Costanzo M, et al. Science. 2010;327:425–431. doi: 10.1126/science.1180823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anderson N, Borlak J. FEBS Lett. 2006;580:5533–5540. doi: 10.1016/j.febslet.2006.08.061. [DOI] [PubMed] [Google Scholar]
- 20.Reasor MJ, Kacew S. Exp. Biol. Med. (Maywood) 2001;226:825–830. doi: 10.1177/153537020122600903. [DOI] [PubMed] [Google Scholar]
- 21.Rainey MM, Korostyshevsky D, Lee S, Perlstein EO. Genetics. 2010;185:1221–1233. doi: 10.1534/genetics.110.117846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Škrtić M, et al. Cancer Cell. 2011;20:674–688. doi: 10.1016/j.ccr.2011.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blackman RK, et al. PLOS ONE. 2012;7:e29798. doi: 10.1371/journal.pone.0029798. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.