Abstract
Inactivation of cell survival factors is a crucial step in apoptosis. The phosphoinositide 3 kinase (PI3K) and AKT signaling pathway promotes cell growth, proliferation and survival and its deregulation causes cancer. How this pathway is suppressed to promote apoptosis is poorly understood. Here we report the identification of a CED-3 caspase substrate in C. elegans, CNT-1, that upon cleavage by CED-3 during apoptosis activates an N-terminal phosphoinositide-binding fragment (tCNT-1), which translocates from cytoplasm to plasma membrane to block AKT binding to phosphatidylinositol (3,4,5)-triphosphate (PIP3), thereby disabling AKT activation and its pro-survival activity. Our findings reveal a new mechanism that negatively regulates AKT cell signaling to promote apoptosis and that may restrict cell growth and proliferation in normal cells.
One major unanswered question in apoptosis, an essential process in animal development and homeostasis, is how the cell-suicide program is executed in a highly coordinated fashion so that the dying cell is disassembled and removed swiftly without causing any deteriorating outcomes1. The cell death execution process is activated by caspases, which are Aspartate-specific cysteine proteases that cleave multiple substrates to orchestrate cellular disassembly, including chromosome fragmentation, nuclear membrane breakdown, and phagocytosis of apoptotic cells1,2. Numerous caspase substrates have been identified in mammals and some are involved in cell death execution in vivo2. For example, the PAK2 kinase is activated by caspase-3 cleavage, which is required for fragmentation of dying cells into apoptotic bodies3. In C. elegans, many apoptosis regulators and executors are conserved4, including the CED-3 caspase and CED-3 activator CED-4 (homologous to the human caspase activator Apaf-1), both of which are essential for apoptosis in C. elegans. How CED-3 kills the cells by proteolytic cleavage is poorly understood. Recently, the dicer ribonuclease (DCR-1) was found to be converted into a deoxyribonuclease upon CED-3 cleavage during apoptosis, which initiates apoptotic chromosome fragmentation5. Similarly, CED-3 cleavage of the mitochondrial fission protein DRP-1 and the multipass transmembrane protein CED-8 generates C-terminal cleavage products that promote mitochondria elimination and phosphatidylserine externalization in apoptotic cells, respectively6,7. Thus, CED-3 cleavage of DCR-1, DRP-1 and CED-8 activates three different cell disassembly events that facilitate cell killing. In this study, we report a new CED-3 substrate, CNT-1, whose cleavage by CED-3 activates an N-terminal cleavage product, truncated CNT-1 (tCNT-1), that promotes apoptosis by suppressing the AKT pro-survival activity.
In mammals, the PI3K and AKT pathway is critical for cell growth, proliferation, survival, and metabolism8,9. Hyperactivation of this pathway results in cancers, revealing the oncogenic potential of PI3K and AKT signaling components10–12, whereas impaired signaling in this pathway causes diabetes and cardiovascular disease13,14. In C. elegans, the AKT homologues, AKT-1 and AKT-2, act in the insulin and insulin-like growth factor signaling (IIS) pathway to regulate lifespan, development, metabolism, and stress resistance15–17. In the IIS pathway, the insulin receptor-like protein DAF-2 activates the PI3K complex AGE-1/AAP-118–20, leading to the generation of PIP3 on the inner leaflet of the plasma membrane. PIP3 may then recruit serine–threonine kinases PDK-1, AKT-1, AKT-2, and SGK-1 to the plasma membrane by engaging their pleckstrin homology (PH) domains21–23, as has been shown with mammalian AKT9. PDK-1 likely phosphorylates and activates AKT-1, AKT-2, and SGK-1, which negatively regulate the activity of the forkhead transcription factor DAF-1624,25, preventing its translocation from the cytosol to the nucleus26,27. The PI3K and AKT pathway is negatively regulated by the lipid phosphatase DAF-18 (homologous to human Phosphatase and Tensin Homolog, PTEN), which dephosphorylates and converts PIP3 to PIP2 and blocks recruitment of AKT kinases to the plasma membrane28–31. Mutations in components of this pathway lead to changes in lifespan, development, metabolism, and stress responses in C. elegans32,33. In addition, loss-of-function (lf) mutations in the akt-1 and akt-2 genes cause hypersensitivity to ionizing radiation-induced germ cell apoptosis34, suggesting that AKT kinases also play a pro-survival role in C. elegans.
In this study, we set out to understand how caspases inactivate cell survival factors to promote apoptosis and demonstrate a new mechanism that inactivates AKT kinases. CNT-1, upon cleavage by CED-3, generates tCNT-1 that acquires a potent phosphoinositide-binding activity and translocates from the cytoplasm to the plasma membrane, where it outcompetes AKT kinases for binding to PIP3, suppresses the PI3K and AKT cell survival pathway, and promotes apoptosis.
RESULTS
cnt-1 promotes apoptosis downstream of ced-3
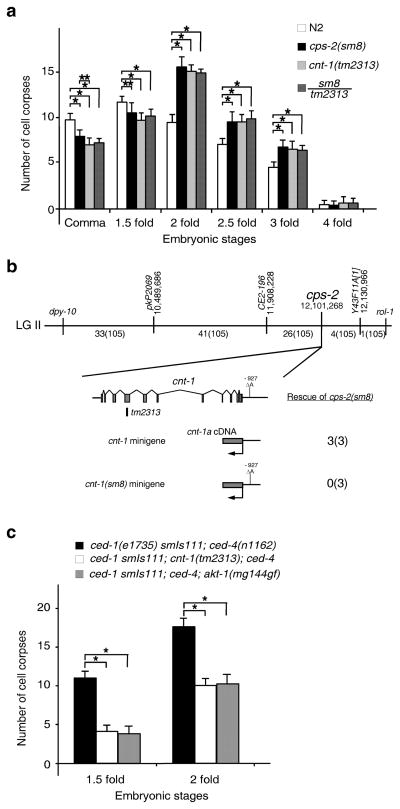
To identify genes acting downstream of CED-3 to kill cells, we performed a genetic screen to isolate suppressors of ectopic neuronal death induced by activated CED-3 (acCED-3)35 and isolated a recessive mutation, sm8, which defines a new gene cps-2 (CED-3 Protease Suppressor) on linkage group (LG) II (METHODS).
We investigated whether cps-2 affects apoptosis by examining embryonic cell death in cps-2(sm8) animals. Compared with wild type N2 embryos, cps-2(sm8) embryos had fewer apoptotic cell corpses in early stages of embryogenesis (comma and 1.5-fold stages) and more cell corpses in later stages (Fig. 1a), displaying a characteristic delay-of-cell-death phenotype observed in mutants defective in genes acting downstream of ced-37,35–37. Three-point mapping and single-nucleotide polymorphism (SNP) mapping placed cps-2(sm8) at a position of 12,103,628 base pair (bp) on LGII (Fig. 1b). Since there are no available fosmid or cosmid clones in this region for transformation rescue experiments, we performed an RNA interference (RNAi) screen on nine candidate genes in this region and found that RNAi treatment of cnt-1 caused a similar delay-of-cell-death defect (Supplementary Table 1). We then introduced into cps-2(sm8) animals a cnt-1 minigene, which contains a full-length cnt-1 cDNA fused to 1944 bp of the cnt-1 promoter, and found that it rescued the cps-2(sm8) defect (Fig. 1b and Supplementary Fig. 1a). Moreover, an existing cnt-1 deletion mutation (tm2313) caused a similar delay-of-cell-death defect and failed to complement cps-2(sm8) (Fig. 1a), indicating that tm2313 and sm8 are allelic. Sequencing analysis of cps-2(sm8) animals revealed a nucleotide deletion at 927 bp upstream from the cnt-1 translational start, but no mutation in the coding region of cnt-1 and 1031 bp 3′ untranslated region. Since a cnt-1 minigene carrying this nucleotide deletion failed to rescue the cell death defect in cps-2(sm8) animals (Fig. 1b and Supplementary Fig. 1a) and since neither cnt-1 mRNA nor CNT-1 protein was detected in cps-2(sm8) animals (Supplementary Fig. 1b, c), sm8 likely disrupts a cis-element important for cnt-1 expression. These results indicate that cps-2 is cnt-1. cnt-1(tm2313) was thus used in all subsequent experiments.
Figure 1.
Cloning and characterization of cps-2. (a) Embryonic cell corpses were counted in animals with the indicated genotype. (b) Mapping and cloning of cps-2. The top bar represents the genetic map with two genetic markers (dpy-10 and rol-1) and three SNPs (pkP2069, CE2-196, and Y43F11A[1]) used for mapping cps-2(sm8). Numbers below are the fractions of the 105 recombinant events that occur between two genetic markers and three SNPs, respectively. The bottom panel shows the cnt-1 coding region, the sm8 mutation, the tm2313 deletion, and rescuing results of the cps-2(sm8) mutant by the Pcnt-1CNT-1a::3xFlag and the Pcnt-1(sm8)CNT-1a::3xFlag constructs. Three independent transgenic arrays were examined for each rescue experiment. (c) Embryonic cell corpses were counted in animals with the indicated genotype. In a and c, the y axis represents average number of cell corpses and error bars represent standard deviation (s.d.). Statistical significance values were determined by two-way ANOVA, followed by Bonferroni comparison (n = 15 embryos at each stage). * P < 0.001; ** P < 0.05.
We examined whether delay of cell death caused by cnt-1(tm2313) results in survival of cells that normally die, yielding extra “undead” cells in the anterior pharynx of mutant animals35. Like wild-type animals, cnt-1(tm2313) animals had few extra undead cells (Supplementary Table 2). However, in sensitized backgrounds such as animals weakly defective in ced-3 and ced-4, cnt-1(tm2313) significantly increased the number of extra cells observed in these weak cell death mutants, indicating that cnt-1 promotes apoptosis.
Since sm8 inhibits acCED-3-induced neuronal death, we examined whether cnt-1 functions downstream of ced-3, using an integrated transgene (smIs111) expressing acCED-3 from the egl-1 promoter38. The cell death initiating gene egl-1 functions upstream of ced-3 to induce apoptosis and is mainly expressed in dying cells39. When we placed smIs111 in the ced-4(n1162) mutant, in which almost all physiological cell deaths are blocked, acCED-3 still induced ectopic cell death (Fig. 1c). cnt-1(tm2313) significantly reduced the number of ectopic deaths in smIs111; ced-4(n1162) embryos, indicating that cnt-1 likely acts downstream of ced-3 to promote apoptosis.
CNT-1 is cleaved sequentially at two sites by CED-3
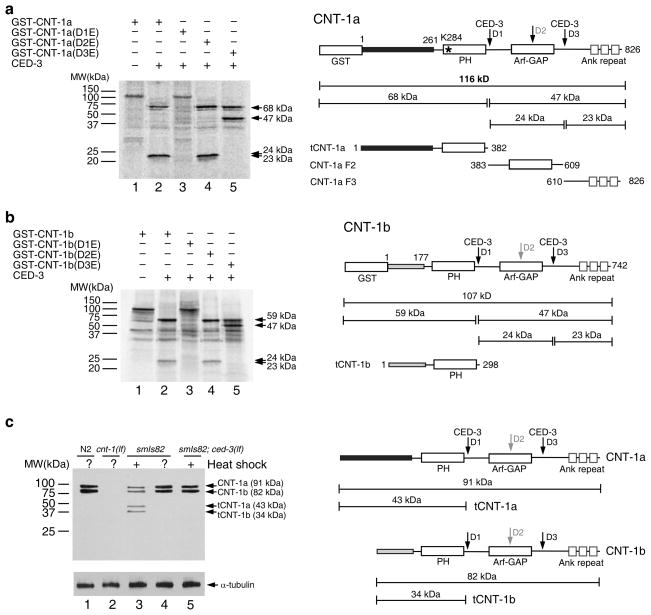
Given that cnt-1 acts downstream of ced-3, we tested whether CNT-1 is a substrate of CED-3 protease. We incubated 35S-Methionine labeled glutathione S-transferase (GST) fusion with the longer CNT-1 isoform, GST-CNT-1a, with CED-3 and found that it was cleaved by CED-3, yielding two species of approximately 68 and 24 kilodalton (kDa) (Fig. 2a). Since GST-CNT-1a is 116 kDa, this cleavage pattern suggests that the 24 kDa species might contain two cleavage products of similar sizes and that CNT-1a is cleaved at two different sites. Based on the consensus CED-3 cleavage sites (DXXD; X can be any amino acid)40, we identified three potential CED-3 cleavage sites, Asp382 (DSVD), Asp508 (DSID), and Asp609 (DVQD). We then generated Aspartic acid to Glutamic acid substitutions in each potential cleavage site, which is named D1, D2 and D3, respectively, and tested CED-3 cleavage of the mutant proteins. The D1E mutation completely abolished cleavage of CNT-1a by CED-3 (Fig. 2a), suggesting that D1 is required for CED-3 cleavage. The D2E mutation did not alter the cleavage pattern (Fig. 2a), indicating that D2 is not a CED-3 cleavage site in vitro. The D3E mutation abolished the 24 kDa species, and instead, produced a new 47 kDa band along with the 68 kDa band (Fig. 2a), confirming that CED-3 cleavage at D3 generates two cleavage products of similar sizes (23 and 24 kDa). Because D1E also blocked cleavage at the D3 site, CED-3 cleavage at the D1 site is required for cleavage at the D3 site. In CNT-1b, a shorter isoform of CNT-1 that has the same 564 amino acid C-terminus, including a pleckstrin homology (PH) domain, but has a different 177-residue N-terminal sequence (Fig. 2b), three identical CED-3 cleavage sites were found (Asp298, Asp424, and Asp525). We observed similar CED-3 cleavage patterns with GST-CNT-1b and GST-CNT-1b mutants carrying D1E, D2E and D3E, respectively (Fig. 2b). These data indicate that D1 is the primary CED-3 cleavage site in CNT-1 and is required for subsequent CED-3 cleavage at D3.
Figure 2.
Cleavage of CNT-1 by CED-3 in vitro and in vivo. (a) CED-3 cleavage assay of GST-CNT-1a. Left, 35S-Methionine labeled GST-CNT-1a and its mutant derivatives were incubated with or without CED-3 and resolved by 15% SDS polyacrylamide gel (PAGE). CED-3 cleavage products are indicated by arrows. Right, a schematic diagram of GST-CNT-1a with the sizes of the CED-3 cleavage products indicated (tCNT-1a, CNT-1a F2, and CNT-1a F3). Three predicted CED-3 cleavage sites are indicated with arrows. The black box at the N-terminus represents the region that is different from CNT-1b. The Lysine residue in the PH domain (K284) critical for lipid binding is indicated with an asterisk. (b) CNT-1b cleavage by CED-3. Left, GST-CNT-1b and its mutant derivatives were synthesized, labeled, digested with CED-3, and resolved on SDS-PAGE as described in a. Right, a schematic diagram of GST-CNT-1b with the sizes of the CED-3 cleavage products indicated. The gray box at the N-terminus represents the region that is different from CNT-1a. (c) Immunoblotting analysis of C. elegans embryos to show CED-3-mediated cleavage of CNT-1 in vivo. Left, embryos with the indicated genotype were treated with or without heat-shock, lysed and resolved on SDS-PAGE (see METHODS). Antibodies used were anti-CNT-1 antibody (upper panel) or an anti-alpha-tubulin antibody (lower panel)(Uncropped image in Supplementary Fig. 1e). Right, a schematic diagram of CNT-1a, tCNT-1a, CNT-1b, and tCNT-1b and their sizes.
To examine if CNT-1 is a CED-3 substrate in vivo, we probed for CNT-1 cleavage products in worm lysates using an antibody raised against the N-terminal cleavage product of CNT-1b (tCNT-1b), which recognizes recombinant tCNT-1b and tCNT-1a (Supplementary Fig. 1d). In wild-type animals, we detected two bands consistent with the sizes of full-length CNT-1a and CNT-1b, which were absent in the cnt-1(tm2313) mutant (Fig. 2c). The absence of cleaved CNT-1 products is due to the fact that few apoptosis occurs in wild-type embryos. In animals carrying an integrated transgene (smIs82) that induces global expression of death initiator EGL-1 and widespread apoptosis under the control of heat-shock promoters (PhspEGL-1)6, the same two bands were observed prior to heat-shock treatment (Fig. 2c). After heat shock treatment, the intensity of these two bands was reduced and two smaller bands consistent with the sizes of tCNT-1a and tCNT-1b appeared. The appearance of tCNT-1a and tCNT-1b bands was CED-3-dependent, because a ced-3(n717) lf mutation abolished these two bands in smIs82 animals. Therefore, CNT-1 is cleaved by CED-3 in C. elegans.
Cleavage of CNT-1 activates tCNT-1 proapoptotic activity
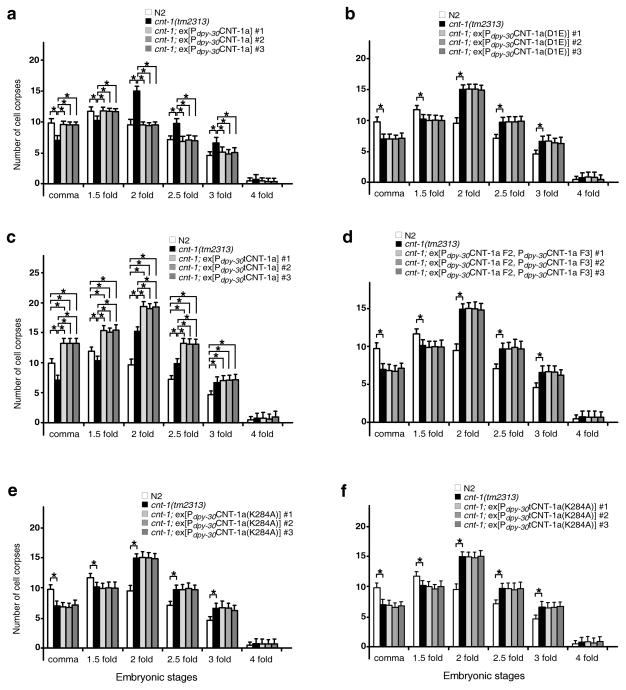
We determined whether CNT-1 cleavage by CED-3 is important for apoptosis. Expression of CNT-1a or CNT-1b from a globally active dpy-30 promoter fully rescued the delay-of-cell-death defect in cnt-1(tm2313) animals (Fig. 3a and Supplementary Fig. 2a). However, Pdpy-30CNT-1a(D1E) and Pdpy-30CNT-1b(D1E), expressing CNT-1 mutants resistant to CED-3 cleavage, did not rescue the cnt-1(tm2313) mutant (Fig. 3b and Supplementary Fig. 2b), indicating that cleavage of CNT-1 by CED-3 is critical for its proapoptotic function. Expression of tCNT-1a or tCNT-1b, at 20% of the endogenous CNT-1 level (Supplementary Fig. 2c, d), led to increased cell death in most embryonic stages compared to wild type and cnt-1(tm2313) embryos (Fig. 3c and Supplementary Fig. 2e). The increased cell corpses observed in animals carrying Pdpy-30tCNT-1a or Pdpy-30tCNT-1b were due to ectopic cell death, because some pharyngeal cells that normally live were randomly lost in transgenic larvae (Supplementary Table 3). In comparison, co-expression of the other two CNT-1a CED-3 cleavage fragments, CNT-1a F2 and CNT-1a F3 (Fig. 2a), did not induce ectopic cell death or rescue the cnt-1(tm2313) mutant (Fig. 3d), whereas co-expression of all three CED-3 cleavage products did (Supplementary Fig. 2f). These results indicate that tCNT-1, activated by CED-3 cleavage, is responsible for CNT-1’s proapoptotic activity.
Figure 3.
Cleavage of CNT-1 by CED-3 activates its proapoptotic activity. Cell corpses were scored in embryos with the indicated genotypes and at the indicated embryonic stages. The y axis represents the average number of cell corpses and error bars represent s.d. Statistical significance values were determined by two-way ANOVA, followed by Bonferroni comparison (n = 15 embryos at each stage). * P < 0.001.
cnt-1 promotes cell death by inhibiting the AKT pathway
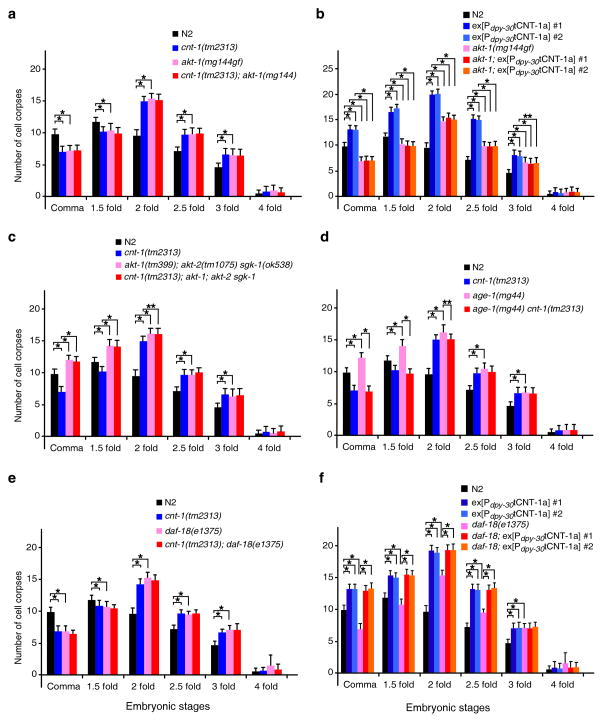
Because the only recognizable motif in tCNT-1 is the PH domain (Fig. 2), a potential phosphoinositide (PI) binding domain41, and because one of the PI species, PIP3, activates AKT kinases (also containing a PH domain) to promote cell survival8,9, we asked whether C. elegans AKT kinases participate in regulating apoptosis and genetically interact with cnt-1. An activating mutation in akt-1, akt-1(mg144gf)22, caused a delay of cell death defect similar to that of the cnt-1(tm2313) mutant (Fig. 4a). Combination of cnt-1(tm2313) and akt-1(mg144gf) did not enhance this phenotype, suggesting that akt-1 and cnt-1 function in the same pathway. Since the ced-3(n2438); akt-1(mg144gf) double mutant had more extra undead cells than ced-3(n2438) or akt-1(mg144gf) single mutants (Supplementary Table 4), this suggests that akt-1 inhibits cell death. Epistasis analysis using animals carrying a Pdpy-30tCNT-1a transgene showed that akt-1(mg144gf) completely blocked ectopic cell death induced by Pdpy-30tCNT-1a (Fig. 4b), indicating that akt-1 likely acts downstream of cnt-1 to inhibit cell death. Consistently, akt-1(mg144gf) suppressed ectopic cell death induced by smIs111, a transgene expressing acCED-3, to the same extent as cnt-1(tm2313)(Fig. 1c), indicating that akt-1 can act downstream of CED-3 to promote cell survival.
Figure 4.
cnt-1 acts downstream of age-1 but upstream of akt-1, akt-2, and sgk-1 to promote apoptosis. Cell corpses were scored in the indicated strains as in Fig. 3. The y axis represents average number of cell corpses and error bars represent s.d. Statistical significance values were determined by two-way ANOVA, followed by Bonferroni comparison (n = 15 embryos at each stage). * P < 0.001; ** P < 0.05.
We also examined animals carrying a lf mutation in akt-1, akt-2 or sgk-1, three AKT kinases acting in parallel in the IIS pathway22,23. akt-1(tm399), akt-2(tm1075), or sgk-1(ok538) each did not cause a detectable cell death defect, whereas all three combinations of double mutations and the triple mutation combination caused increased cell death throughout embryogenesis (Supplementary Fig. 3a), indicating that the presence of at least two AKT and SGK kinases is important for cell survival. Since cnt-1(tm2313) failed to suppress ectopic cell death in the triple mutant (Fig. 4c and Supplementary Table 5), these results suggest that cnt-1 functions upstream of akt-1, akt-2, and sgk-1 to promote cell death.
AKT kinases phosphorylate proteins to transduce signals in various pathways22,42,43. To examine if the kinase activity of AKT is important for its anti-apoptotic activity, we expressed the kinase-defective mutants of AKT-1 and AKT-2, AKT-1(K222M) and AKT-2(K209M), in the akt-1(tm399); akt-2(tm1075) mutant22. Expression of wild type AKT-1 or AKT-2 rescued the increased cell death phenotype of the akt-1(tm399); akt-2(tm1075) mutant, whereas expression of AKT-1(K222M) or AKT-2(K209M) did not (Supplementary Fig. 4a, b), indicating that the kinase activity of AKT is required for its pro-survival function.
A key substrate of AKT kinases in the IIS pathway is DAF-16, a homologue of the Foxo transcriptional factor24,25,42. A daf-16 lf mutation, mu86, caused a delay-of-cell-death defect similar to that of cnt-1(tm2313) or akt-1(mg144gf) animals, but did not enhance their cell death defects (Supplementary Fig. 3b, c), indicating that daf-16, akt-1 and cnt-1 function in the same pathway to affect apoptosis. Moreover, daf-16(mu86) fully suppressed ectopic cell death induced by tCNT-1a (Supplementary Fig. 3d) and increased the number of extra undead cells in ced-3(n2438) animals (Supplementary Table 4), suggesting that daf-16 acts downstream of cnt-1 to promote cell death.
cnt-1 acts downstream of PI3K to promote cell death
C. elegans AKT kinases are likely activated by PIP3 and thus regulated by AGE-1, a PI3K that phosphorylates PIP2 to generate PIP319,20. An age-1(mg44) lf mutation caused an increased cell death phenotype similar to that of akt-1(tm399); akt-2(tm1075) sgk-1(ok538) animals (Fig. 4c, d), but did not enhance the increased cell death phenotype caused by Pdpy-30tCNT-1a (Supplementary Fig. 3e), suggesting that age-1 inhibits and tCNT-1 promotes apoptosis in the same pathway. Opposite to what was observed with the akt-1(lf); akt-2(lf) sgk-1(lf) triple mutant (Fig. 4c), cnt-1(tm2313) blocked both the increased cell death and the missing cell phenotypes caused by age-1(mg44) (Fig. 4d and Supplementary Table 5), indicating that cnt-1 likely acts downstream of age-1 to promote apoptosis.
PIP3 is dephosphorylated and converted to PIP2 by the PTEN phosphatase8,9, which is encoded by daf-18 in C. elegans28–31. The daf-18(e1375) lf mutation caused a delay-of-cell-death phenotype similar to that of cnt-1(tm2313) animals (Fig. 4e) and did not exacerbate cnt-1(tm2313) cell death defect, suggesting that daf-18 and cnt-1 act in the same pathway. daf-18(e1375) also increased the number of extra cells in ced-3(n2438) animals (Supplementary Table 4). daf-18(e1375), however, did not block ectopic cell death induced by tCNT-1a (Fig. 4f), indicating that daf-18 acts upstream of, or in parallel to, cnt-1 to promote cell death, possibly at the level of PIP3 regulation. Taken together, our results suggest that cnt-1 and daf-18 act in the same pathway to promote cell death and that cnt-1 operates downstream of age-1 and upstream of akt-1, akt-2, sgk-1 and daf-16.
Cleavage of CNT-1 activates its phosphoinositide binding
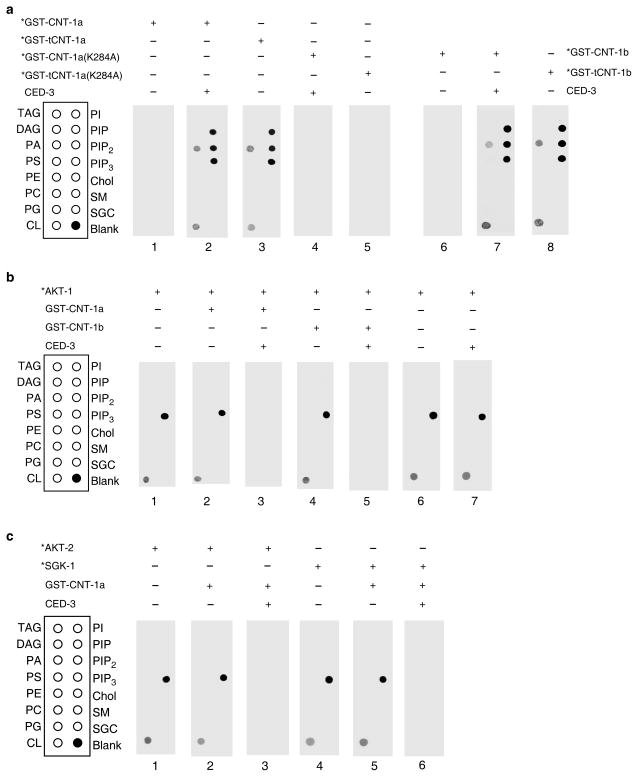
Given that age-1 activates AKT kinases through PIP3 and daf-18 inactivates this pathway through dephosphorylation of PIP3, we hypothesized that tCNT-1, with a PH domain, promotes cell death by interfering with the binding of PIP3 by AKT kinases. We examined whether tCNT-1 binds PIP3 using an in vitro lipid-binding assay. 35S-Met labeled GST-CNT-1a on its own did not bind any lipid, but showed strong binding to PIP, PIP2 and PIP3 and weak binding to phosphatidic acid (PA) and cardiolipin (CL) after pre-treated with CED-3 (Fig. 5a). GST-tCNT-1a displayed an identical lipid-binding pattern to that of CED-3-treated GST-CNT-1a (Fig. 5a). So did GST-CNT-1b pretreated with CED-3 and GST-tCNT-1b (Fig. 5a). When we altered one of the highly conserved, phosphoinositide-binding lysine residues in the PH domain of CNT-1a (K284A)44, neither CNT-1a(K284A) pretreated with CED-3 nor tCNT-1a(K284A) showed any lipid-binding activity (Fig. 5a). Moreover, expression of CNT-1a(K284A) or tCNT-1a(K284A) in cnt-1(tm2313) animals did not rescue the cell death defect or cause ectopic cell death (Fig. 3e, f). Since the K284A mutation did not alter the expression levels of CNT-1a proteins in vitro or in vivo (Supplementary Fig. 5), these results indicate that the PH domain is required for tCNT-1 to acquire phosphoinositide-binding after CED-3 cleavage and to promote apoptosis.
Figure 5.
CED-3-activated phosphoinositide binding of CNT-1 blocks PIP3 binding by AKT and SGK kinases. (a) Lipid binding assays of CNT-1 in the presence of CED-3. The indicated proteins labeled with 35S-Methionine (*) were incubated with or without CED-3 and then incubated with membrane strips (see METHODS). (b) AKT-1 lipid binding assay in the presence of CNT-1 and CED-3. Unlabeled GST-CNT-1a and GST-CNT-1b were incubated with CED-3 and then added to membrane strips with radiolabeled AKT-1(*) as described in METHODS. (c) AKT-2 and SGK-1 lipid binding assay in the presence of CNT-1 and CED-3. Radiolabeled AKT-2(*) and SGK-1(*) were incubated with membrane strip as described in (b). In all panels, lipids spotted on the strips are: triacylglyceride (TAG), diacylglyceride (DAG), phosphatidic acid (PA), phosphatidylserine (PS), phosphatidylethanolamine (PE), phosphatidylcholine (PC), phosphatidylglycerol (PG), cardiolipin (CL), phosphatidylinositol (PI), Phosphatidylinositol (4)-phosphate (PIP), Phosphatidylinositol (4,5)-bisphosphate (PIP2), Phosphatidylinositol (3,4,5)-trisphosphate (PIP3), cholesterol (Chol), sphingomyelin (SM), and 3-sulfogalactosylceramide (SGC).
tCNT-1 blocks PIP3 binding by AKT and SGK kinases
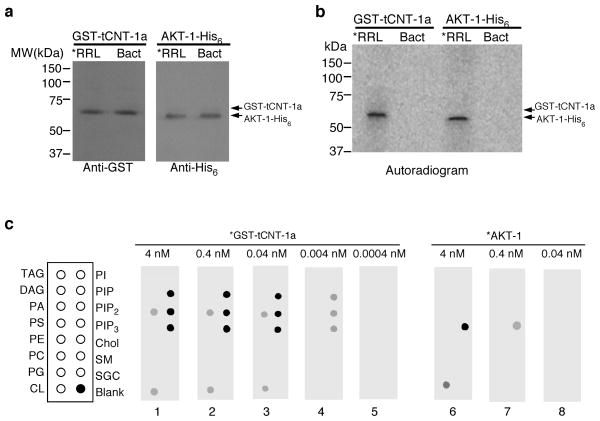
We tested whether tCNT-1 interferes with binding of AKT and SGK kinases to PIP321–23. 35S-Met-labeled AKT-1 alone bound strongly to PIP3 and weakly to cardiolipin and this binding was not affected by addition of unlabeled GST-CNT-1a or GST-CNT-1b (Fig. 5b). Addition of GST-CNT-1a or GST-CNT-1b pre-treated with CED-3 completely blocked PIP3 binding by AKT-1 (Fig. 5b), whereas incubation of AKT-1 with CED-3 did not alter AKT-1 lipid binding (Fig. 5b). Likewise, both AKT-2 and SGK-1 bound strongly to PIP3 and this binding was blocked by GST-CNT-1a pre-treated with CED-3 (Fig. 5c). To probe how tCNT-1 inhibits PIP3 binding by AKT-1, we compared their PIP3 binding affinity. We first determined the concentrations of 35S-Met-labeled GST-tCNT-1a and AKT-1 synthesized in rabbit reticulocyte lysate (RRL) by comparing them with recombinant GST-tCNT-1a and AKT-1 proteins with known concentrations, which were purified from bacteria (Bact) but showed no lipid binding activity (Fig. 6a). Labeled GST-tCNT-1a and AKT-1-His6 displayed comparable radioactive signal intensity at the same concentrations (Fig. 6b). GST-tCNT-1a displayed strong PIP3 binding at 0.04 nM, weak binding at 0.004 nM, and no binding at 0.0004 nM (Fig. 6c). By contrast, AKT-1 showed strong PIP3 binding at 4 nM, weak binding at 0.4 nM, and no binding at 0.04 nM (Fig. 6c), indicating that tCNT-1a binds PIP3 with two order of magnitude higher affinity than AKT-1. Because the expression level of CNT-1 in C. elegans is approximately 70% higher than that of AKT-1 (Supplementary Fig. 6), the combination of higher CNT-1 concentrations and much higher binding affinity to PIP3 than those of AKT-1 allows tCNT-1 to block PIP3 binding by AKT kinases and thus their activation.
Figure 6.
PIP3 binding activity of tCNT-1a is stronger than that of AKT-1 by two orders of magnitude. (a) Immunoblotting of 5.1 μl of GST-tCNT-1a and 2.9 μl of AKT-1-His6 proteins synthesized and radiolabeled in rabbit reticulocyte lysate (*RRL) and 1 pmol of GST-tCNT-1a or AKT-1-His6 proteins purified from bacteria (Bact) using an anti-GST or an anti-His6 antibody (see METHODS). (b) Autoradiogram of 1 pmol of 35S-Met-labeled GST-tCNT-1a and AKT-1-His6 proteins made in RRL shows comparable radioactive signal intensity. (c) Lipid binding assays of GST-tCNT-1a or AKT-1-His6. Radiolabeled (*) GST-tCNT-1a or AKT-1-His6 with the indicated concentrations were incubated with lipid membrane strips as described in Fig. 5.
tCNT-1 translocates to plasma membrane upon CED-3 cleavage
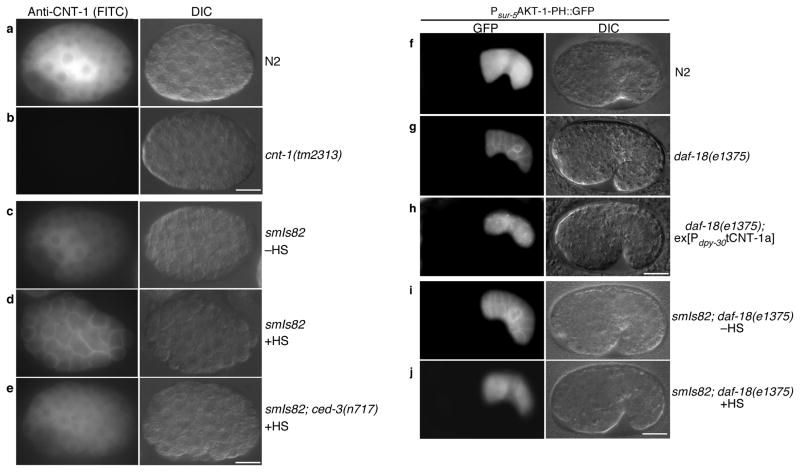
Several proteins containing the PH domain translocate from cytosol to plasma membrane via a PIP3-mediated mechanism45–47. We analyzed subcellular localization of CNT-1 using an antibody raised against tCNT-1b (Supplementary Fig. 1d). CNT-1 localized in the cytoplasm of all cells in wild type embryos (Fig. 7a) but was not detected in cnt-1(tm2313) embryos (Fig. 7b), demonstrating the specificity of the antibody. In smIs82 (PhspEGL-1) embryos that were induced to undergo global apoptosis and widespread CED-3 activation through heat-shock treatment, CNT-1 staining was observed on the plasma membrane of all cells, in addition to staining in the cytoplasm (Fig. 7d and Supplementary Fig. 7c, d), indicating that a portion of CNT-1 translocated from cytosol to plasma membrane upon apoptosis activation. By contrast, in smIs82 embryos without heat-shock treatment or in smIs82; ced-3(n717) embryos with heat-shock treatment, CNT-1 remained in the cytoplasm (Fig. 7c, e and Supplementary Fig. 7a, b), indicating that CNT-1 translocation from cytosol to plasma membrane is apoptosis- and CED-3-dependent. In cnt-1(tm2313); smIs82 embryos expressing CNT-1a(D1E), the CED-3 uncleavable form of CNT-1a, CNT-1a(D1E) did not translocate to the plasma membrane after heat-shock treatment (Supplementary Fig. 7g–i). By contrast, in cnt-1(tm2313); smIs82 embryos expressing CNT-1a, CNT-1a was observed on plasma membrane after heat-shock treatment (Supplementary Fig. 7j–l). These results confirm that CED-3 cleavage is required for CNT-1 translocation to plasma membrane during apoptosis. Consistently, loss of cnt-1 partially suppressed and expression of tCNT-1a enhanced ectopic cell death induced by smIs82 (Supplementary text and Supplementary Fig. 4c d), providing further support to the finding that cleavage of CNT-1 and activation of tCNT-1 is an important downstream event of CED-3 activation and apoptosis.
Figure 7.
CED-3-dependent translocation of CNT-1 to the plasma membrane blocks AKT-1 membrane recruitment. (a–e) Immunostaining of CNT-1 in C. elegans embryos. FITC (left) and Differential Interference Contrast (DIC, right) images of the stained embryos with the indicated genotype were shown. –HS, no heat-shock treatment. +HS, with heat-shock treatment. (f–j) Microscopy analysis of AKT-1-PH::GFP localization in C. elegans embryos. All strains contained the same ex[Psur-5AKT-1-PH::GFP] transgenic array. Embryos with the indicated genotype were examined with or without heat-shock treatment. GFP (left) and DIC (right) images of transgenic embryos were shown. In all panels (a–j), scale bar represents 10 μm.
tCNT-1 inhibits association of AKT-1 to the plasma membrane
AKT-1 also contains a PH domain and its human homolog translocates to the plasma membrane through binding to PIP348. Given the limited amount of PIP3 on the plasma membrane, the higher concentration of CNT-1 in vivo than AKT-1, and the much stronger PIP3 binding affinity of tCNT-1 than that of AKT-1, we examined if tCNT-1 blocks AKT-1 translocation to the plasma membrane. The first 180 residues of AKT-1, including its PH domain (residues 15–118), were fused to GFP and expressed from the sur-5 gene promoter (Psur-5AKT-1-PH::GFP). In wild type embryos carrying Psur-5AKT-1-PH::GFP, GFP was diffuse within the cells (Fig. 7f). When this transgene was crossed into daf-18(e1375) animals that are deficient in converting PIP3 to PIP2 and thus have elevated levels of PIP3, in the plasma membrane, bright GFP signals were seen on the plasma membrane, suggesting that AKT-1-PH::GFP translocated to the plasma membrane through binding to PIP3 (Fig. 7g). Expression of tCNT-1a abolished AKT-1-PH::GFP localization to the plasma membrane in daf-18(e1375) embryos (Fig. 7h), indicating that tCNT-1 inhibited association of AKT-1 to the plasma membrane. Similarly, in smIs82; daf-18(e1375) animals carrying Psur-5AKT-1-PH::GFP, some AKT-1-PH::GFP localized to the plasma membrane when without heat-shock treatment (Fig. 7i). After heat shock treatment, AKT-1-PH::GFP disappeared from the plasma membrane (Fig. 7j), indicating that endogenous tCNT-1 induced by smIs82 is sufficient to block AKT-1 association with plasma membrane and thus AKT-1 activation and function.
DISCUSSION
The physiological relevance of caspase substrates is an understudied area that limits our understanding of apoptosis. We have identified multiple components acting downstream of CED-3 to promote apoptosis using a sensitized CED-3 protease suppressor (cps) screen35. The cell death defects of each cps mutant are mild or undetectable, due to the fact that multiple CED-3 downstream pathways act in parallel to kill the cell5–7,35. In this study, we characterized one of the cps genes, cps-2 (cnt-1), whose inactivation delays cell death and suppresses apoptosis in sensitized genetic backgrounds. Importantly, CNT-1 is a substrate of CED-3 both in vitro and in vivo. A mutation blocking CNT-1 cleavage by CED-3 abolishes its pro-apoptotic activity, whereas tCNT-1 alone is sufficient to induce apoptosis. These results establish that CNT-1 is an in vivo CED-3 target.
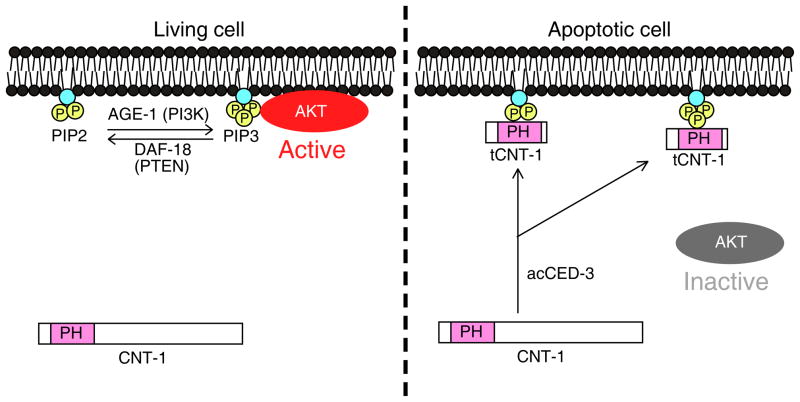
Despite containing a PH domain, a potential phosphoinositide-binding motif, CNT-1 does not bind any phospholipids in vitro or associate with plasma membrane in vivo. Upon activated by CED-3 cleavage during apoptosis, tCNT-1 acquires strong binding to phosphoinositides and translocates to the plasma membrane where it outcompetes AKT kinases for PIP3 binding to block recruitment and activation of AKT kinases at the plasma membrane, thereby inactivating the AKT survival pathway to accelerate cell killing (Fig. 8). Therefore, CED-3 activates a downstream death execution event by cleaving CNT-1, which then inactivates the AKT cell survival pathway. Our study elucidates a previously unknown, regulatory link between cell death execution and cell survival signaling.
Figure 8.
A model of CED-3-activated suppression of AKT signaling by CNT-1 in C. elegans. In non-apoptotic cells, some AKT kinases are recruited to the plasma membrane and activated by PIP3 to transduce the survival signal. In apoptotic cells, CNT-1 is cleaved by activated CED-3 to generate potent phosphoinositide-binding tCNT-1, which translocates to the plasma membrane from the cytoplasm. tCNT-1 outcompetes AKT kinases for binding to PIP3 and thus displaces and inactivates AKT kinases, leading to loss of the survival signal and apoptosis.
Given the critical roles of the AKT kinases in cell growth, survival and metabolism, the activities of AKT kinases are tightly regulated at several different levels9,14. Association of AKT with PIP3 generated by PI3K at the plasma membrane is the critical first step, followed by sequential phosphorylation of AKT by PDK1 and mTORC2, leading to full activation of AKT. At each step, negative regulators are in place to fine-tune the level of AKT activation. The PTEN phosphatase converts PIP3 back to PIP2, reducing or blocking AKT activation49,50. Dephosphorylation of AKT by phosphatases, including protein phosphatase 2A and phosphatase PHLPP, also reduces or blocks AKT activation51–54. In this study, we report a new mechanism that could rapidly shut down AKT at the level of PIP3 binding and plasma membrane recruitment, through proteolytic activation of a PH-containing protein to generate a competing phosphoinositide-binding activity. Given the presence of hundreds of PH-domain proteins in eukaryotes with unknown functions41, this could be a general and conserved mechanism for regulating membrane-associated cell signaling or activity. There are several potential human CNT-1 homologs, ACAP1, ACAP2 and ACAP3, but none has been reported to have a role in apoptosis. It will be interesting to investigate if these ACAP proteins might act like CNT-1 to inhibit AKT survival signaling in humans.
AKT kinases and PI3K are required for the IIS pathway in C. elegans. Inactivation of these genes increases life span and stress resistance32,33. Interestingly, loss of cnt-1 does not affect these two processes (Supplementary Fig. 8), indicating that CNT-1 is an apoptosis-specific inhibitor of the PI3K and AKT pathway. AKT is a major drug target in treating cancer, however, clinical development of AKT inhibitors has been restricted by the fact that AKT is involved in multiple important cellular events, which inevitably will lead to numerous on-target or off-target side effects9,14,55. Targeted inactivation of AKT in cancer cells to induce apoptosis through a CNT-1-like mechanism could present a new therapeutic strategy when combined with other anti-cancer therapies.
ONLINE METHODS
Strains and culture conditions
C. elegans strains were maintained using standard procedures56. The following alleles were used in the genetic analyses: LGI, daf-16(mu86), ced-1(e1735), smIs13, smIs111; LGII, dpy-10(e128), age-1(mg44), cnt-1(tm2313), cps-2(sm8), rol-1(e91); LGIII, daf-2(e1370), ced-4(n1162, n2273), smIs82; LGIV, daf-18(e1375), ced-3(n717, n2438), dpy-4(e1166); LGV, akt-1(mg144gf, tm399), him-5(e1490); LGX, akt-2(tm1075), sgk-1(ok538).
Isolation of the cps-2(sm8) mutation
Ethyl methane sulfonate (EMS) mutagenesis was performed on animals carrying an integrated transgene (smIs1) containing both Pmec-7acCED-3 and Pmec-3GFP35. Pmec-7acCED-3 drives acCED-3 expression in six mechanosensory neurons and the ectopic death of these non-essential neurons, which are also labeled by GFP (Pmec-3GFP). Progeny of the mutagenized animals were screened for mutants with increased survival of the fluorescent mechanosensory neurons. From a screen of 3,000 mutagenized haploid genomes, we isolated several recessive mutations, including sm8, which defines a new gene cps-2 (CED-3 Protease Suppressor) on linkage group (LG) II. For the complementation test result shown in Fig. 1a, the complete genotype of cps-2(sm8)/cnt-1(tm2313) is smIs13/+; cps-2(sm8)/cnt-1(tm2313); him-5(e1490)/+. smIs13 is an integrated line of Psur-5SUR-5::GFP.
Quantification of cell corpses and extra cells
The number of cell corpses in living embryos and the number of extra cells in the anterior pharynx of L4 larvae were scored using Nomarski optics as described previously35.
CNT-1 antibody
Amino acids 1–298 of CNT-1b (tCNT-1b) fused with Glutathione S-transferase (GST) were expressed in Escherichia coli strain BL21(DE3) and affinity purified from the soluble fraction of the bacterial lysate using Glutathione Sepharose 4B beads (GE Helthcare). Rats were immunized with purified GST-tCNT-1b proteins (Spring Valley Laboratories, Inc.). CNT-1 antibodies were affinity purified from terminal bleeds using purified GST-tCNT-1a proteins as described previously57.
Immunoblotting analysis
C. elegans embryos were harvested with M9, centrifuged at 500 × g to remove bacteria, and then sonicated in the PBS buffer. After centrifugation at 25,400 × g to remove debris, 2 x SDS loading buffer was added and heated at 95° for 3 minutes. The samples were resolved with 15% SDS-PAGE and transferred to the PVDF membrane (Millipore). The membrane was first blocked with 5% nonfat dry milk in PBST (PBS + 0.1% Tween 20) and washed 2 times with PBST for 5 minutes each. The membrane was then incubated with affinity-purified anti-CNT-1 antibody in 1:1,000 dilution for 1 hour at room temperature and washed three times with PBST for 5 minutes each. It was then incubated with HRP-conjugated goat anti-rat IgG antibody (Jackson Immunoresearch #112-035-003; validation is provided on the manufacturer’s website) with 1:2,000 dilution for 1 hour at room temperature, washed three times with PBST, and detected using enhanced chemiluminescent substrate (Pierce). For alpha-tubulin immunoblotting, anti-alpha-tubulin antibody (Developmental Studies Hybridoma Bank #AA4.3; validation is provided on the manufacturer’s website) was used at 1:5,000 dilution and HRP-conjugated goat anti-mouse IgG antibody (Jackson Immunoresearch #115-035-003; validation is provided on the manufacturer’s website) was used at 1:3,000 dilution. For anti-GST and anti-His6 immunoblotting, a mouse anti-GST antibody (Santa Cruz Laboratory #sc-138; validation is provided on the manufacturer’s website) or a rabbit anti-His6 antibody (Santa Cruz Laboratory #sc-803; validation is provided on the manufacturer’s website) was used at 1:2,000 dilution with a HRP-conjugated goat anti-mouse IgG antibody or a HRP-conjugated goat anti-rabbit IgG antibody (Jackson Immunoresearch #111-035-003; validation is provided on the manufacturer’s website) at 1:3,000 dilution. For anti-AKT-1 immunoblotting, an affinity purified goat anti-AKT-1 antibody (Bethyl Laboratories)60 was used at 1:10,000 dilution with a HRP-conjugated donkey anti-goat IgG antibody (Jackson Immunoresearch #705-035-003; validation is provided on the manufacturer’s website) at 1:10,000 dilution.
Heat shock experiments
The heat shock experiments were carried out as described previously5.
CED-3 cleavage assay
CED-3 protease assays were done as described previously40. Briefly, proteins of interest were synthesized and labeled with 35S-Methionine using TNT Transcription and Translation coupled system (Promega) and incubated with or without 5 ng of purified CED-3 at 30°C for 1 hour. The reactions were then resolved by 15% SDS-PAGE gel and subjected to autoradiography.
Lipid binding assay
Various CNT-1 proteins were synthesized and labeled with 35S-Methionine using the TNT system and incubated with or without purified CED-3 as described above. The membrane strip containing various lipids (Echelon Biosciences) was blocked in 3% fatty-acid free bovine serum albumin (Sigma) in PBST (PBS + 0.01% Tween 20) for 1 hour and washed 3 times with PBST. The membrane strip was then incubated with 2 μl of the reticulocyte lysate containing the protein of interest in PBST at 1:1,000 dilution for 1 hour at room temperature. After washing 3 times with PBST, the membrane strip was subjected to autoradiography. For the PIP3 competition assay, AKT-1, AKT-2 and SGK-1 were synthesized and labeled with 35S-Methionine, while GST-CNT-1 was synthesized with unlabeled Methionine. After GST-CNT-1 was pretreated with CED-3, the lysate containing 35S-Methionine-labeled AKT-1 (20 μl) and the lysate containing unlabeled GST-CNT-1 (2 μl) were added to the membrane strip in 2 ml of PBST at the same time.
Determination of in vitro synthesized protein concentrations
The concentrations of GST-tCNT-1a and AKT-1-His6 synthesized in rabbit reticulocyte lysate (also labeled by 35S-Methionine*) were determined by the immunoblotting analysis, comparing the signal intensity of these proteins with that of recombinant GST-tCNT-1a (12 μg/ml) or AKT-1-His6 (20 μg/ml) purified from bacteria. Recombinant GST-tCNT-1a or AKT-1-His6 purified from bacteria did not show any lipid binding activity, probably due to misfolding of the proteins. Briefly, 5.1 μl of GST-tCNT-1a or 2.9 μl of AKT-1-His6 synthesized in rabbit reticulocyte lysate (*RRL) or 1 pmol of GST-tCNT-1a or AKT-1-His6 purified from bacteria (Bact) were resolved by 15% SDS-PAGE and subjected to immunoblotting analysis using an anti-GST or an anti-His6 antibody as described in Immunoblotting analysis. Since the amounts of two GST-tCNT-1a proteins are comparable, the concentration of GST-tCNT-1a from RRL is approximately 196 nM. Since the amounts of two AKT-1-His6 proteins are also comparable, the concentration of AKT-1-His6 from RRL is approximately 345 nM.
Transgenic animals
Transgenic animals were generated as described previously58. Pcnt-1CNT-1a::3x flag or Pcnt-1(sm8)CNT-1a::3xFlag was injected into cps-2(sm8) animals at 20 μg/ml along with the pTG96 plasmid (at 20 μg/ml) as a co-injection marker. The pTG96 plasmid contains a sur-5::gfp translational fusion that is expressed in all somatic cells at most developmental stages59. Pdpy-30CNT-1a, Pdpy-30CNT-1b, Pdpy-30CNT-1a(D1E), Pdpy-30CNT-1b(D1E), Pdpy-30tCNT-1a, Pdpy-30tCNT-1b, Pdpy-30CNT-1 F2, Pdpy-30CNT-1 F3, Pdpy-30CNT-1a(K284A), and Pdpy-30tCNT-1a(K284A) were injected into cnt-1(tm2313) animals at 20 μg/ml along with the pTG96 plasmid (at 20 μg/ml), respectively. Transgenic arrays containing Pdpy-30tCNT-1a were then crossed into the akt-1(mg144gf), age-1(mg44) or daf-16(mu86) mutant for further analyses. Psur-5AKT-1-PH::GFP was injected into daf-18(e1375) animals at 20 μg/ml along with the pRF4 (at 20 μg/ml), a dominant rol-6 construct. One of the transgenic arrays was then crossed into N2 animals and smIs82; daf-18(e1375) animals or injected with Pdpy-30tCNT-1a at 20 μg/ml along with Psur-5mCherry (at 20 μg/ml) as the second transgenic marker to generate transgenic animals containing both Pdpy-30tCNT-1a and Psur-5AKT-1-PH::GFP. Pdpy-30AKT-1, Pdpy-30AKT-1(K222M), Pdpy-30AKT-2, or Pdpy-30AKT-2(K209M) was injected into akt-1(tm399)/nT1; akt-2(tm1075) animals at 20 μg/ml along with the pTG96 plasmid (at 20 μg/ml), respectively.
Molecular biology
Full-length cnt-1a cDNA and cnt-1b cDNA were amplified using reverse transcription polymerase chain reaction (RT-PCR) and then subcloned into the pET-41b vector via its Spe I and Not I sites to generate the pET-41b-CNT-1a and pET-41b-CNT-1b expression vector. To construct various pET-41b-CNT-1 mutant expression vectors [pET-41b-CNT-1a(D1E), pET-41b-CNT-1a(D2E), pET-41b-CNT-1a(D3E), pET-41b-CNT-1b(D1E), pET-41b-CNT-1b(D2E), pET-41b-CNT-1b(D3E), and pET-41b-CNT-1a(K284A)], pET-41b-CNT-1a or pET-41b-CNT-1b was used as a DNA template to make the indicated amino acid substitutions using a Quick Change mutagenesis kit (Stratagene). pET-41b-tCNT-1a and pET-41b-tCNT-1b were constructed by subcloning PCR fragment coding for amino acids 1–382 of CNT-1a and 1–298 of CNT-1b into pET-41b via Spe I and EcoR V sites, respectively. To construct Pcnt-1CNT-1a::3x flag, a 1944 bp Sph I–Nhe I PCR fragment containing the cnt-1 promoter was fused with a 2640 bp Nhe I–Sma I PCR fragment containing the full-length cnt-1a cDNA, which was then subcloned into a modified pPD95.79 vector, in which the gfp coding region was replaced with a 3x flag sequence. To construct Pdpy-30CNT-1a and Pdpy-30CNT-1b, full-length cnt-1a and cnt-1b cDNAs were PCR amplified and subcloned into pS235 via its Sph I and Xma I sites, which contains the promoter of the C. elegans dpy-30 gene. Quick Change mutagenesis was then performed to obtain Pdpy-30CNT-1a(D1E) and Pdpy-30CNT-1b(D1E). Pdpy-30tCNT-1a and Pdpy-30tCNT-1b were constructed by subcloning PCR fragment coding for amino acids 1–382 of CNT-1a and 1–298 of CNT-1b into pS235 via its Nhe I and Xma I sites. To construct Pdpy-30CNT-1a F2 and Pdpy-30CNT-1a F3, PCR fragments coding for amino acids 383–609 and 610–826 of CNT-1a were subcloned into pS235 via Nhe I and Xma I sites. Pdpy-30CNT-1a(K284A) and Pdpy-30tCNT-1a(K284A) were generated by Quick Change mutagenesis. To construct pET-21b-AKT-1, pET-21b-AKT-2, and pET-21b-SGK-1 expression vectors, akt-1, akt-2, and sgk-1 full-length cDNAs were PCR amplified and subcloned into pET-21b through its Nhe I and Xho I sites. To construct Psur-5AKT-1-PH::GFP expression vector, a PCR fragment coding for amino acids 1–180 of AKT-1 was subcloned into a modified pPD95.77 vector, which contains the promoter of the sur-5 gene. Pdpy-30AKT-1 or Pdpy-30AKT-2 was constructed by subcloning AKT-1 or AKT-2 cDNA fragment into the pS235 vector via its Nhe I and Sma I sites. Quick-change mutagenesis was then performed to generate kinase-defective mutants of AKT-1 and AKT-2 that contain K222M and K209M substitutions, respectively.
Determination of protein expression levels in C. elegans
200 wild-type N2 C. elegans embryos at 1.5-fold stage were collected in the PBS buffer and sonicated before adding 2 x SDS buffer. The N2 embryonic lysate and 1 pmol of GST-CNT-1a or AKT-1-His6 proteins purified from bacteria were resolved by SDS-PAGE and subsequently subjected to immunoblotting using an affinity-purified anti-CNT-1 antibody or anti-AKT-1 antibody as described in Immunoblotting analysis, respectively. To determine the expression level from the immunoblotting images, the signal intensity of a protein band was measured using the ImageJ software and subtracted by the background intensity, the value of which was used to determine the average expression level of the protein.
Immunostaining on C. elegans embryos
Mix-stage C. elegans animals were harvested with M9, bleached with 12% NaOCl and 1.5 M NaOH, and centrifuged at 500 × g to remove bacteria and debris. Embryos were washed 3 times with ddH2O, mounted on poly-L-lysine coated slides, and frozen at −80° for 10 minutes. They were fixed with Methanol for 15 minutes on ice and washed with PBS before being stained with anti-CNT-1 antibody at 1:500 dilution in PBS for 1 hour. After the embryos were washed three times with PBS, they were stained with Alexa Fluor 488–conjugated anti-rat IgG antibody (Molecular Probes #A-11006; validation is provided on the manufacturer’s website) at 1:1,000 dilution in PBS for 1 hour. They were then washed three times with PBS and visualized using an Axioplan 2 Nomarski Microscope (Carl Zeiss MicroImaging Inc.).
RT-PCR
The primer sequences for cnt-1 is: 5′ CTCTGTGCCAAGACTGGATGC-GGGC 3′ and 5′ CGTTGCCTGGGACGCGGCGGAC 3′, and the primer sequences for rpl-26 is: 5′ ATGAAGGTCAATCCGTTCGT 3′ and 5′ AGGACACGTCCAGTG-TTTCC 3′.
Lifespan analysis and thermo-tolerance assay
For lifespan analysis, animals were grown at 20° until the L4 larval stage and then transferred to new plates (10 animals per plates) at 25°. Animals were scored every day subsequently and moved periodically to keep growth conditions mold free. Animals were scored as dead if they failed to respond to a gentle tap on the head and tail with a platinum wire21. For thermo-tolerance assay, animals were grown at 20° until the L4 larval stage and then transferred to new plates (10 animals per plates) at 33°. Animals were scored every 2 hours for the first 24 hours, subsequently scored every 12 hours until 60 hours. Animals were scored as dead if they failed to respond to a gentle tap on the head and tail with a platinum wire.
RNAi experiments
RNAi experiments were carried out using a bacteria-feeding protocol described previously61.
Supplementary Material
Acknowledgments
We thank B. Derry (Hospital for Sick Children) for anti-AKT-1 antibodies, M. Han (University of Colorado) for some RNAi clones, Y. Kohara (Japan National Institute of Genetics) for akt-1 cDNA, M. Valencia and Y. Shi for making some of the constructs, S. Mitani (Tokyo Women’s Medical University) and G. Ruvkun (Massachusetts General Hospital) for strains, R.R. Skeen-Gaar for assistance in generating transgenic strains, and B.L. Harry, T. Blumenthal, B. Olwin, and J.M. Espinosa for comments on the manuscript. Some of the worm strains used in this study were kindly provided by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health. This work was supported by US National Institutes of Health (grants R01 GM59083, R01 GM79097 and R01 GM088241 to D.X.).
Footnotes
AUTHOR CONTRIBUTIONS
A.N. and D.X. conceived the research and designed experiments. A.N. carried out and analyzed experiments. K.D.S. assisted in some experiments. A.N. and D.X. wrote the paper.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Steller H. Mechanisms and genes of cellular suicide. Science. 1995;267:1445–9. doi: 10.1126/science.7878463. [DOI] [PubMed] [Google Scholar]
- 2.Crawford ED, Wells JA. Caspase substrates and cellular remodeling. Annu Rev Biochem. 2011;80:1055–87. doi: 10.1146/annurev-biochem-061809-121639. [DOI] [PubMed] [Google Scholar]
- 3.Rudel T, Bokoch GM. Membrane and morphological changes in apoptotic cells regulated by caspase-mediated activation of PAK2. Science. 1997;276:1571–4. doi: 10.1126/science.276.5318.1571. [DOI] [PubMed] [Google Scholar]
- 4.Metzstein MM, Stanfield GM, Horvitz HR. Genetics of programmed cell death in C. elegans: past, present and future. Trends Genet. 1998;14:410–6. doi: 10.1016/s0168-9525(98)01573-x. [DOI] [PubMed] [Google Scholar]
- 5.Nakagawa A, Shi Y, Kage-Nakadai E, Mitani S, Xue D. Caspase-dependent conversion of Dicer ribonuclease into a death-promoting deoxyribonuclease. Science. 2010;328:327–34. doi: 10.1126/science.1182374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Breckenridge DG, et al. Caenorhabditis elegans drp-1 and fis-2 regulate distinct cell-death execution pathways downstream of ced-3 and independent of ced-9. Mol Cell. 2008;31:586–97. doi: 10.1016/j.molcel.2008.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen YZ, Mapes J, Lee ES, Skeen-Gaar RR, Xue D. Caspase-mediated activation of Caenorhabditis elegans CED-8 promotes apoptosis and phosphatidylserine externalization. Nat Commun. 2013;4:2726. doi: 10.1038/ncomms3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cully M, You H, Levine AJ, Mak TW. Beyond PTEN mutations: the PI3K pathway as an integrator of multiple inputs during tumorigenesis. Nat Rev Cancer. 2006;6:184–92. doi: 10.1038/nrc1819. [DOI] [PubMed] [Google Scholar]
- 9.Luo J, Manning BD, Cantley LC. Targeting the PI3K–Akt pathway in human cancer: rationale and promise. Cancer Cell. 2003;4:257–62. doi: 10.1016/s1535-6108(03)00248-4. [DOI] [PubMed] [Google Scholar]
- 10.Staal SP, Hartley JW, Rowe WP. Isolation of transforming murine leukemia viruses from mice with a high incidence of spontaneous lymphoma. Proc Natl Acad Sci U S A. 1977;74:3065–7. doi: 10.1073/pnas.74.7.3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li J, et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 1997;275:1943–7. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- 12.Vivanco I, Sawyers CL. The phosphatidylinositol 3–Kinase AKT pathway in human cancer. Nat Rev Cancer. 2002;2:489–501. doi: 10.1038/nrc839. [DOI] [PubMed] [Google Scholar]
- 13.Cho H, et al. Insulin resistance and a diabetes mellitus-like syndrome in mice lacking the protein kinase Akt2 (PKB beta) Science. 2001;292:1728–31. doi: 10.1126/science.292.5522.1728. [DOI] [PubMed] [Google Scholar]
- 14.Hers I, Vincent EE, Tavare JM. Akt signalling in health and disease. Cell Signal. 2011;23:1515–27. doi: 10.1016/j.cellsig.2011.05.004. [DOI] [PubMed] [Google Scholar]
- 15.Finch CE, Ruvkun G. The genetics of aging. Annu Rev Genomics Hum Genet. 2001;2:435–62. doi: 10.1146/annurev.genom.2.1.435. [DOI] [PubMed] [Google Scholar]
- 16.Kenyon C. The plasticity of aging: insights from long-lived mutants. Cell. 2005;120:449–60. doi: 10.1016/j.cell.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 17.Wolff S, Dillin A. The trifecta of aging in Caenorhabditis elegans. Exp Gerontol. 2006;41:894–903. doi: 10.1016/j.exger.2006.06.054. [DOI] [PubMed] [Google Scholar]
- 18.Kimura KD, Tissenbaum HA, Liu Y, Ruvkun G. daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science. 1997;277:942–6. doi: 10.1126/science.277.5328.942. [DOI] [PubMed] [Google Scholar]
- 19.Morris JZ, Tissenbaum HA, Ruvkun G. A phosphatidylinositol-3-OH kinase family member regulating longevity and diapause in Caenorhabditis elegans. Nature. 1996;382:536–9. doi: 10.1038/382536a0. [DOI] [PubMed] [Google Scholar]
- 20.Wolkow CA, Munoz MJ, Riddle DL, Ruvkun G. Insulin receptor substrate and p55 orthologous adaptor proteins function in the Caenorhabditis elegans daf-2/insulin-like signaling pathway. J Biol Chem. 2002;277:49591–7. doi: 10.1074/jbc.M207866200. [DOI] [PubMed] [Google Scholar]
- 21.Paradis S, Ailion M, Toker A, Thomas JH, Ruvkun G. A PDK1 homolog is necessary and sufficient to transduce AGE-1 PI3 kinase signals that regulate diapause in Caenorhabditis elegans. Genes Dev. 1999;13:1438–52. doi: 10.1101/gad.13.11.1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paradis S, Ruvkun G. Caenorhabditis elegans Akt/PKB transduces insulin receptor-like signals from AGE-1 PI3 kinase to the DAF-16 transcription factor. Genes Dev. 1998;12:2488–98. doi: 10.1101/gad.12.16.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hertweck M, Gobel C, Baumeister R. C elegans SGK-1 is the critical component in the Akt/PKB kinase complex to control stress response and life span. Dev Cell. 2004;6:577–88. doi: 10.1016/s1534-5807(04)00095-4. [DOI] [PubMed] [Google Scholar]
- 24.Lin K, Dorman JB, Rodan A, Kenyon C. daf-16: An HNF-3/forkhead family member that can function to double the life-span of Caenorhabditis elegans. Science. 1997;278:1319–22. doi: 10.1126/science.278.5341.1319. [DOI] [PubMed] [Google Scholar]
- 25.Ogg S, et al. The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature. 1997;389:994–9. doi: 10.1038/40194. [DOI] [PubMed] [Google Scholar]
- 26.Lee RY, Hench J, Ruvkun G. Regulation of C. elegans DAF-16 and its human ortholog FKHRL1 by the daf-2 insulin-like signaling pathway. Curr Biol. 2001;11:1950–7. doi: 10.1016/s0960-9822(01)00595-4. [DOI] [PubMed] [Google Scholar]
- 27.Lin K, Hsin H, Libina N, Kenyon C. Regulation of the Caenorhabditis elegans longevity protein DAF-16 by insulin/IGF-1 and germline signaling. Nat Genet. 2001;28:139–45. doi: 10.1038/88850. [DOI] [PubMed] [Google Scholar]
- 28.Gil EB, Malone Link E, Liu LX, Johnson CD, Lees JA. Regulation of the insulin-like developmental pathway of Caenorhabditis elegans by a homolog of the PTEN tumor suppressor gene. Proc Natl Acad Sci U S A. 1999;96:2925–30. doi: 10.1073/pnas.96.6.2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mihaylova VT, Borland CZ, Manjarrez L, Stern MJ, Sun H. The PTEN tumor suppressor homolog in Caenorhabditis elegans regulates longevity and dauer formation in an insulin receptor-like signaling pathway. Proc Natl Acad Sci U S A. 1999;96:7427–32. doi: 10.1073/pnas.96.13.7427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ogg S, Ruvkun G. The C. elegans PTEN homolog, DAF-18, acts in the insulin receptor-like metabolic signaling pathway. Mol Cell. 1998;2:887–93. doi: 10.1016/s1097-2765(00)80303-2. [DOI] [PubMed] [Google Scholar]
- 31.Rouault JP, et al. Regulation of dauer larva development in Caenorhabditis elegans by daf-18, a homologue of the tumour suppressor PTEN. Curr Biol. 1999;9:329–32. doi: 10.1016/s0960-9822(99)80143-2. [DOI] [PubMed] [Google Scholar]
- 32.Dorman JB, Albinder B, Shroyer T, Kenyon C. The age-1 and daf-2 genes function in a common pathway to control the lifespan of Caenorhabditis elegans. Genetics. 1995;141:1399–406. doi: 10.1093/genetics/141.4.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Larsen PL, Albert PS, Riddle DL. Genes that regulate both development and longevity in Caenorhabditis elegans. Genetics. 1995;139:1567–83. doi: 10.1093/genetics/139.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Quevedo C, Kaplan DR, Derry WB. AKT-1 regulates DNA-damage-induced germline apoptosis in C. elegans. Curr Biol. 2007;17:286–92. doi: 10.1016/j.cub.2006.12.038. [DOI] [PubMed] [Google Scholar]
- 35.Parrish J, et al. Mitochondrial endonuclease G is important for apoptosis in C. elegans. Nature. 2001;412:90–4. doi: 10.1038/35083608. [DOI] [PubMed] [Google Scholar]
- 36.Parrish JZ, Xue D. Functional genomic analysis of apoptotic DNA degradation in C. elegans. Mol Cell. 2003;11:987–96. doi: 10.1016/s1097-2765(03)00095-9. [DOI] [PubMed] [Google Scholar]
- 37.Stanfield GM, Horvitz HR. The ced-8 gene controls the timing of programmed cell deaths in C. elegans. Mol Cell. 2000;5:423–33. doi: 10.1016/s1097-2765(00)80437-2. [DOI] [PubMed] [Google Scholar]
- 38.Kokel D, Li Y, Qin J, Xue D. The nongenotoxic carcinogens naphthalene and para-dichlorobenzene suppress apoptosis in Caenorhabditis elegans. Nat Chem Biol. 2006;2:338–45. doi: 10.1038/nchembio791. [DOI] [PubMed] [Google Scholar]
- 39.Conradt B, Horvitz HR. The C. elegans protein EGL-1 is required for programmed cell death and interacts with the Bcl-2-like protein CED-9. Cell. 1998;93:519–29. doi: 10.1016/s0092-8674(00)81182-4. [DOI] [PubMed] [Google Scholar]
- 40.Xue D, Shaham S, Horvitz HR. The Caenorhabditis elegans cell-death protein CED-3 is a cysteine protease with substrate specificities similar to those of the human CPP32 protease. Genes Dev. 1996;10:1073–83. doi: 10.1101/gad.10.9.1073. [DOI] [PubMed] [Google Scholar]
- 41.Park WS, et al. Comprehensive identification of PIP3-regulated PH domains from C. elegans to H sapiens by model prediction and live imaging. Mol Cell. 2008;30:381–92. doi: 10.1016/j.molcel.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tullet JM, et al. Direct inhibition of the longevity-promoting factor SKN-1 by insulin-like signaling in C. elegans. Cell. 2008;132:1025–38. doi: 10.1016/j.cell.2008.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Datta SR, Brunet A, Greenberg ME. Cellular survival: a play in three Akts. Genes Dev. 1999;13:2905–27. doi: 10.1101/gad.13.22.2905. [DOI] [PubMed] [Google Scholar]
- 44.Isakoff SJ, et al. Identification and analysis of PH domain-containing targets of phosphatidylinositol 3-kinase using a novel in vivo assay in yeast. Embo J. 1998;17:5374–87. doi: 10.1093/emboj/17.18.5374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parent CA, Blacklock BJ, Froehlich WM, Murphy DB, Devreotes PN. G protein signaling events are activated at the leading edge of chemotactic cells. Cell. 1998;95:81–91. doi: 10.1016/s0092-8674(00)81784-5. [DOI] [PubMed] [Google Scholar]
- 46.Meili R, et al. Chemoattractant-mediated transient activation and membrane localization of Akt/PKB is required for efficient chemotaxis to cAMP in Dictyostelium. Embo J. 1999;18:2092–105. doi: 10.1093/emboj/18.8.2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Servant G, et al. Polarization of chemoattractant receptor signaling during neutrophil chemotaxis. Science. 2000;287:1037–40. doi: 10.1126/science.287.5455.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fayard E, Tintignac LA, Baudry A, Hemmings BA. Protein kinase B/Akt at a glance. J Cell Sci. 2005;118:5675–8. doi: 10.1242/jcs.02724. [DOI] [PubMed] [Google Scholar]
- 49.Stambolic V, et al. Negative regulation of PKB/Akt-dependent cell survival by the tumor suppressor PTEN. Cell. 1998;95:29–39. doi: 10.1016/s0092-8674(00)81780-8. [DOI] [PubMed] [Google Scholar]
- 50.Wu X, Senechal K, Neshat MS, Whang YE, Sawyers CL. The PTEN/MMAC1 tumor suppressor phosphatase functions as a negative regulator of the phosphoinositide 3-kinase/Akt pathway. Proc Natl Acad Sci U S A. 1998;95:15587–91. doi: 10.1073/pnas.95.26.15587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kuo YC, et al. Regulation of phosphorylation of Thr-308 of Akt, cell proliferation, and survival by the B55alpha regulatory subunit targeting of the protein phosphatase 2A holoenzyme to Akt. J Biol Chem. 2008;283:1882–92. doi: 10.1074/jbc.M709585200. [DOI] [PubMed] [Google Scholar]
- 52.Padmanabhan S, et al. A PP2A regulatory subunit regulates C. elegans insulin/IGF-1 signaling by modulating AKT-1 phosphorylation. Cell. 2009;136:939–51. doi: 10.1016/j.cell.2009.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gao T, Furnari F, Newton AC. PHLPP: a phosphatase that directly dephosphorylates Akt, promotes apoptosis, and suppresses tumor growth. Mol Cell. 2005;18:13–24. doi: 10.1016/j.molcel.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 54.Brognard J, Sierecki E, Gao T, Newton AC. PHLPP and a second isoform, PHLPP2, differentially attenuate the amplitude of Akt signaling by regulating distinct Akt isoforms. Mol Cell. 2007;25:917–31. doi: 10.1016/j.molcel.2007.02.017. [DOI] [PubMed] [Google Scholar]
- 55.Cheng GZ, et al. Advances of AKT pathway in human oncogenesis and as a target for anti-cancer drug discovery. Curr Cancer Drug Targets. 2008;8:2–6. [PubMed] [Google Scholar]
- 56.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang X, et al. C. elegans mitochondrial factor WAH-1 promotes phosphatidylserine externalization in apoptotic cells through phospholipid scramblase SCRM-1. Nat Cell Biol. 2007;9:541–9. doi: 10.1038/ncb1574. [DOI] [PubMed] [Google Scholar]
- 58.Mello CC, Kramer JM, Stinchcomb D, Ambros V. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. Embo J. 1991;10:3959–70. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gu T, Orita S, Han M. Caenorhabditis elegans SUR-5, a novel but conserved protein, negatively regulates LET-60 Ras activity during vulval induction. Mol Cell Biol. 1998;18:4556–64. doi: 10.1128/mcb.18.8.4556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Perrin AJ, et al. Noncanonical control of C. elegans germline apoptosis by the insulin/IGF-1 and Ras/MAPK signaling pathways. Cell Death Differ. 2013;20:97–107. doi: 10.1038/cdd.2012.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tabara H, et al. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell. 1999;99:123–32. doi: 10.1016/s0092-8674(00)81644-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.