Background: Factors that influence the cytoadherence property of P. falciparum are not completely understood.
Results: Targeted disruption of the gene that encodes a RESA-like protein in P. falciparum results in stable and increased chondroitin 4-sulfate-dependent cytoadherence capacity.
Conclusion: A RESA-like protein gene regulates PfEMP1 expression and cytoadherence of P. falciparum.
Significance: Our findings provide a significant insight into the regulation of P. falciparum cytoadherence.
Keywords: Cell Adhesion, Cell Surface Protein, Chondroitin Sulfate, Glycosaminoglycan, Malaria, Parasite, Plasmodium
Abstract
The Plasmodium falciparum erythrocyte membrane protein 1 (PfEMP1) family proteins mediate the adherence of infected erythrocytes to microvascular endothelia of various organs, including the placenta, thereby contributing to cerebral, placental, and other severe malaria pathogenesis. Several parasite proteins, including KAHRP and PfEMP3, play important roles in the cytoadherence by mediating the clustering of PfEMP1 in rigid knoblike structures on the infected erythrocyte surface. The lack of a subtelomeric region of chromosome 2 that contains kahrp and pfemp3 causes reduced cytoadherence. In this study, microarray transcriptome analysis showed that the absence of a gene cluster, comprising kahrp, pfemp3, and four other genes, results in the loss of parasitized erythrocytes adhering to chondroitin 4-sulfate (C4S). The role of one of these genes, PF3D7_0201600/PFB0080c, which encodes PHISTb (Plasmodium helical interspersed subtelomeric b) domain-containing RESA-like protein 1 expressed on the infected erythrocyte surface, was investigated. Disruption of PFB0080c resulted in increased var2csa transcription and VAR2CSA surface expression, leading to higher C4S-binding capacity of infected erythrocytes. Further, PFB0080c-knock-out parasites stably maintained the C4S adherence through many generations of growth. Although the majority of PFB0080c-knock-out parasites bound to C4S even after culturing for 6 months, a minor population bound to both C4S and CD36. These results strongly suggest that the loss of PFB0080c markedly compromises the var gene switching process, leading to a marked reduction in the switching rate and additional PfEMP1 expression by a minor population of parasites. PFB0080c interacts with VAR2CSA and modulates knob-associated Hsp40 expression. Thus, PFB0080c may regulate VAR2CSA expression through these processes. Overall, we conclude that PFB0080c regulates PfEMP1 expression and the parasite's cytoadherence.
Introduction
Plasmodium falciparum, the most virulent among several species of parasites that infect humans, is responsible for >90% of all deaths due to malaria (1–3). The severity of malaria caused by P. falciparum depends on the ability of parasites to sequester in the microvasculature of various organs through the adherence of infected red blood cells (IRBCs)4 to endothelial cell surface molecules, including CD36, CD31, and ICAM-1, and in the intervillous space of placenta via chondroitin 4-sulfate (C4S) (1–8). Thus, falciparum malaria manifests in multiple systemic clinical conditions and organ-related fatal pathologies, including cerebral and pregnancy-associated malaria (9, 10). The proteins that mediate parasite cytoadherence are a family of antigenically variant proteins called P. falciparum erythrocyte membrane protein 1 (PfEMP1) expressed on the IRBC surface. Different members of the PfEMP1 family of proteins bind distinct host adhesion molecules, contributing to organ-specific sequestration of P. falciparum (11–15).
The erythrocyte membrane-tethered PfEMP1s are clustered in relatively rigid, knobby protrusions formed by the accumulation of a conical shaped assembly of many molecules of the knob-associated histidine-rich protein (KAHRP) beneath the erythrocyte membrane (16–19). The acidic cytoplasmic ∼45-kDa polypeptide portion of PfEMP1 binds positively charged KAHRP, which interacts with erythrocyte membrane skeletal proteins and stabilizes knob assembly, whereas the extracellular portion mediates IRBC binding to host cell adhesion molecules (20, 21). The knob structure is further stabilized through the interaction of several other parasite proteins, including P. falciparum erythrocyte membrane protein 3 (PfEMP3), ring-infected erythrocyte surface antigen (RESA) encoded by the PF3D7_0102200 gene (PFA00110w; gene ID according to an earlier version of PlasmoDB), mature parasite-infected erythrocyte surface antigen (MESA) encoded by PF3D7_050800 (PFE0040c), and possibly other exported proteins, with erythrocyte skeletal proteins (16, 19, 22, 23, 24). The clustering of PfEMP1 in the knob appears to allow for the multivalent interactions with host receptors, thereby enhancing the strength of IRBC binding to endothelial surface molecules in the vascular bed or to the C4S chains of chondroitin sulfate proteoglycan (CSPG) in the placenta. The lack of knob structures due to the deficiency in KAHRP expression results in reduced IRBC adherence to host receptors, such as CD36 (17). Deficiency in PfEMP3 also significantly reduces IRBC adherence to CD36 (25). However, the absence of RESA enhances avidity of IRBC binding to CD36, whereas a lack of MESA is without effect on cytoadherence capacity (25).
Previous studies have shown that subtelomeric chromosomal deletions in P. falciparum lead to the loss of cytoadherence (17, 26). Although this phenomenon is commonly seen in cultured parasites, it has also been reported to occur in field isolates (27). It is likely that a loss of cytoadherence is prevalent in vivo, and the resulting non-adherent parasites are likely to be cleared from the circulation by the spleen, whereas adherent parasites are selected for and propagated. Earlier work has shown that the deletion of a ∼55-kb subtelomeric region in chromosome 9 that contains the cytoadherence-linked asexual gene (clag9), critical for the cell surface expression of PfEMP1, results in a loss of cytoadherence (28, 29). Similarly, a subtelomeric deletion of ∼100 kb in chromosome 2 results in significantly reduced cytoadherence because this region contains the kahrp and pfemp3 genes, which are crucial for the assembly of a rigid knob and a clustering of PfEMP1 in the knob (17).
A PfEMP1 named VAR2CSA that is exclusively expressed on the surface of C4S-binding P. falciparum-infected IRBCs specifically mediates the sequestration of IRBCs in the placenta by binding to the C4S chains of placental CSPG (13, 30, 31). The IRBC adherence in the placenta contributes to pregnancy-associated malaria, characterized by several clinical conditions, including low birth weight, fetal loss, and maternal deaths (10). In efforts to gain insight into parasite proteins that might play role in PfEMP1-mediated adherence of IRBCs to C4S, we initially performed a comparative analysis of parasite transcripts in C4S-binding parasites and parasites not selected for binding (predominantly non-binding parasites with ∼1–2% binding parasites), using a genome-wide oligonucleotide microarray. The results showed that transcripts of contiguous genes on chromosome 2, including PF3D7_0201500 (PFB0075c), PF3D7_0201600 (PFB0080c, which encodes RESA-like protein 1 (RLP1)), PF3D7_0201700 (PFB0085c), PF3D7_0201600 (PFB0090c, KAHsp40), PF3D7_0201900 (PFB0095c, PfEMP3), and PF3D7_0202000 (PFB0100c, KAHRP), were transcribed at relatively high levels in the C4S-binding parasites but were down-regulated or missing in the C4S-nonbinding parasites. The roles of KAHRP and PfEMP3 in rigid knob formation and strengthening of the adhesive interactions between PfEMP1s and their receptors have been well established (16, 17, 19, 22). Recently, PF3D7_0201800 (PFB0090c) has been shown to be an Hsp40, named KAHsp40, that interacts with KAHRP, PfEMP1, PfEMP3, Hsp70, and Hsp101 (32, 33). KAHsp40 functions as a chaperone for proteins involved in knob assembly formation (32, 33). A targeted disruption of PFB0085c showed no obvious change in PfEMP1 expression and C4S adherence ability of parasites, whereas that of PFB0090c resulted in var gene switching to express different PfEMP1 (34). In this study, we show that PFB0080c is expressed on the IRBC surface and analyzed its role in VAR2CSA-mediated binding of IRBCs to the C4S chains of placental CSPG. Further, we found that the disruption of PFB0080c results in parasites stably maintaining the C4S-adherent phenotype through many cell cycles. We also found that a minor population of C4S-binding parasites lacking PFB0080c acquired CD36 binding characteristics, resulting in IRBCs adhering to both C4S and CD36. Thus, our results demonstrate that PFB0080c regulates VAR2CSA expression and var gene switching processes. The regulation of VAR2CSA expression by PFB0080c appears to involve more than one mechanism, including their interactions with one another and regulation of the expression of other genes, such as PFB0090c; each mechanism may partially contribute.
EXPERIMENTAL PROCEDURES
Parasites
P. falciparum parasites were cultured using O-positive human RBCs in RPMI 1640 medium containing 10–20% human O-positive plasma and 50 μg/ml gentamycin under 90% nitrogen, 5% oxygen, and 5% carbon dioxide atmosphere. The parasite cultures were synchronized as described previously (35, 36).
Selection of C4S- and CD36-adherent P. falciparum Parasites
The C4S-adherent P. falciparum populations from 3D7 and NF54 strains (3D7-CSA and NF54-CSA parasites, respectively) were selected by panning on plastic dishes coated with CSPG purified from human placenta as described previously (36, 37). The CD36-adherent parasites from PFB0080c knock-out 3D7-CSA parasites (see below) were similarly selected by panning on CD36-coated plastic dishes. The adherent parasites were cultured at 3% hematocrit as outlined above. Unless otherwise stated, prior to binding analysis, the adherent parasites were reselected by two or three rounds of panning on plates coated with purified placental CSPG (36) or CD36 (38). The IRBCs were harvested at the early to mid-trophozoite stages.
IRBC-CSPG Adhesion and Adhesion Inhibition Assays
The binding of IRBCs to C4S and the inhibition of IRBC binding to C4S were performed using CSPG purified from human placenta as reported previously (36, 37, 39). Briefly, the CSPG dissolved (400 ng/ml) in PBS, pH 7.2, was coated onto plastic Petri dishes as circular spots, blocked with 1% BSA in PBS. The spots were overlaid with a 2% suspension of washed parasite culture (∼20% parasitemia) in PBS. After 30 min, the unbound cells were washed off, and the bound cells fixed with 2% glutaraldehyde, stained with Giemsa, and counted using light microscopy (35, 36).
For analyzing the inhibition of IRBC binding, solutions of glycosaminoglycans in PBS at twice the desired final concentrations were mixed with equal volumes of 4% suspension of IRBCs in PBS. The suspensions were incubated at room temperature for 30 min and then layered on CSPG-coated spots on Petri dishes. After 30 min at room temperature, the unbound cells were washed off, and the bound cells were fixed with 2% glutaraldehyde, stained, and analyzed as above.
Isolation of Parasite RNA
Approximately 100 μl of cell pellets obtained from the synchronous cultures of 3D7-CSA and PFB0080c-knock-out 3D7-CSA parasites (∼20% parasitemia) at 18–20 h postinvasion were suspended in 1 ml of TRIzol (Invitrogen) and extracted with 200 μl of chloroform. In each case, total RNA in the aqueous phase was precipitated with 0.5 ml of isopropyl alcohol, and washed with 75% ethanol. The RNA was air-dried, dissolved in 30 μl of diethylpyrocarbonate-treated water, treated with DNase for 15 min at room temperature, and purified using RNA purification columns (Invitrogen). The RNA (500 ng) in each case was reverse-transcribed using SuperScript II reverse transcriptase (Invitrogen). The PCR products (cDNA) were analyzed by electrophoresis on 2% agarose gels, and the cDNA was used for PCR amplification.
Microarray Analysis
Highly synchronous cultures of C4S-selected (NF54-CSA) and non-selected (NF54-NS) P. falciparum NF54 strain (∼30% parasitemia) were harvested at 18 h postinvasion. The cell pellet (∼1 ml) was suspended in 6 ml of ice-cold PBS, layered on successive 80, 60, and 40% stepwise Percoll cushions, and centrifuged at 1860 × g at 4 °C. Only the pellets containing IRBCs (the late ring stage parasites) and RBCs at the bottom of the tubes underneath the Percoll cushions were collected to obtain a nearly homogenous population of late ring parasites; cell debris and IRBCs on the top of the Percoll cushions were discarded. Cell pellets were washed with incomplete RPMI 1640 medium and lysed with 0.05% saponin in ice-cold PBS, pH 7.2. The released parasites were collected by centrifugation at 4 °C, and total RNA was extracted as described previously (40). Briefly, the parasite culture pellets were dissolved in TRIzol (Invitrogen), and 0.2 volume of chloroform was added, mixed thoroughly, and centrifuged. Total RNA in the aqueous phase was precipitated with isopropyl alcohol at room temperature for 10 min, and the precipitate was collected by centrifugation at 4 °C and dissolved in diethylpyrocarbonate-treated water. Total RNA was purified on RNeasy mini spin columns (Qiagen, Valencia, CA).
Eight micrograms of total RNA were used for cDNA synthesis. An oligo(dT) primer containing a phage T7 promoter at its 5′ end was used to prime the cDNA synthesis reaction. A second strand of cDNA was then synthesized and used as a template for in vitro transcription in the presence of biotinylated ribonucleotide (Enzo Life Sciences, Farmingdale, NY). The labeled cRNA was then fragmented, hybridized to the array, and stained with a phycoerythrin-streptavidin conjugate.
Hybridizations to a custom malaria parasite array (scrMalaria) that contains probes for 5159 P. falciparum genes were carried out with 15 μg of fragmented cRNA at 45 °C for 16 h, and then the hybridization solution was removed, and the arrays were stained and washed according to Affymetrix protocols (40). Arrays were scanned at an emission wavelength of 560 nm at 3-μm resolution using a confocal scanner (Affymetrix), and the signal intensity for each sequence feature on the array was determined using the 70th percentile method in Microarray Suite 5 (Affymetrix). All array measurements were processed using a match-only integral distribution algorithm for high density oligonucleotide array analysis, as described previously (41, 42).
Quantitative RT-PCR
Approximately 500 ng of parasite RNA was separately reverse-transcribed using SuperScript II reverse transcriptase (Invitrogen), and the cDNA was used for quantitative PCR analysis. The PCR was performed using 200 μm gene-specific primers (Table 1), 2× SYBR Green PCR mix and cDNA in an ABI Prism model 7900HT instrument (ABI Prism quantitative PCR kit from Applied Biosystems). The cDNA levels for each gene in the PCR mixture were normalized to that of fructose bisphosphate aldolase, and data analysis was performed by the ΔΔCt method.
TABLE 1.
List of P. falciparum genes specific primers used for quantitative RT-PCR analysis
| Gene ID | Primer sequence |
|---|---|
| PFL0030c (VAR2CSA) | |
| Forward | 5′-ACGATTGGTGGGAAACAAAT-3′ |
| Reverse | 5′-CCCCATTCTTTTATCCATCG-3′ |
| PFB0100c (KAHRP) | |
| Forward | 5′-GTGGACCTGCCGCTATAGAT-3′ |
| Reverse | 5′-TACCGTGAGAACCATCGTGT-3′ |
| PFB0095c (PfEMP3) | |
| Forward | 5′-GCTGAAGAACCAGACGATGA-3′ |
| Reverse | 5′-TCTGGAGCTCTAAATTTATCAGGA-3′ |
| PFB0090c | |
| Forward | 5′-TTGGAGGATCATCTCCGTTT-3′ |
| Reverse | 5′-TTCTTCGACTTTGTCCATGC-3′ |
| PFB0080c | |
| Forward | 5′-TGGCCTGTAGATTGTTATGGAG-3′ |
| Reverse | 5′-TTAAATTTCAAAGCACGTCCA-3′ |
| PFB0075c | |
| Forward | 5′-GGAATTGCTAAATGAATGGGA-3′ |
| Reverse | 5′-TGTCTGTTTCATCATTTGTTTCA-3′ |
| PFI1730w (CLAG9) | |
| Forward | 5′-CATGTTGCGTTTATCTGTACACGAT-3′ |
| Reverse | 5′-TGAGTTGGATGGTTGGTTTTTTT-3′ |
| PF14_0425 (fructose bisphosphate aldolase) | |
| Forward | 5′-TGTACCACCAGCCTTACCAG-3′ |
| Reverse | 5′-TTCCTTGCCATGTGTTCAAT-3′ |
| PF07_0073 (seryl t-RNA synthetase) | |
| Forward | 5′-TGAATGCCCCAAAAAAATTACC-3′ |
| Reverse | 5′-TTGGGCGGTGGTTGCTA-3′ |
Knock-out of the PF3D7_0201600/PFB0080c Gene
A 971-nucleotide fragment at the 5′ region of PFB0080c covering a 625-nucleotide portion of the promoter region and a 950-nucleotide fragment at the 3′ region covering 714 nucleotides of the UTR were PCR-amplified from genomic DNA of 3D7-CSA using the primers shown in Table 1. The products were cloned into pCC1 vector at NcoI and EcoRI, and SpeI and SacII restriction sites, respectively. The pCC1 vector contains a human dihydrofolate reductase cassette driven by the calmodulin promoter flanked by multiple cloning sites for inserting targeting sequences to act as targets for homologous recombination (43). The recombinant plasmid, pCC1-ΔPFB0080c, was transformed into Escherichia coli XL10 competent cells, plated on agar plates, and the ampicillin-resistant colonies were selected and grown. Plasmids were prepared using a maxiprep kit (Qiagen). The purified plasmid was transfected to 3D7-CSA parasites, which were freshly panned on placental CSPG-coated plates, as described by Dietsch et al. (44). Briefly, ∼100 μg of purified pCC1-ΔPFB0080c plasmid was resuspended in 100 μl of cytomix solution (25 mm HEPES and 10 mm potassium phosphate containing 120 mm KCl, 0.15 mm CaCl2, 5 mm MgCl2, and 2 mm EGTA, pH 7.6) and electroporated into washed, fresh RBCs in incomplete RPMI 1640 medium at 310 V and a capacitance of 950 microfarads. The transfected cells were washed twice with RPMI medium, and the pCC1-ΔPFB0080c plasmid-loaded RBCs were added to purified schizont stage IRBCs and cultured. After two or three cycles of growth, the plasmid-loaded RBCs were added, and the parasites were further cultured. The transfected parasites were selected by culturing in RPMI 1640 medium containing 5 nm WR99210 (positive selection), and the drug-resistant parasites were obtained after 3 weeks of culturing. After culturing for 2 months in the presence of WR99210 (Jacobus Pharmaceuticals, Princeton, NJ), the resistant parasites were subjected to negative selection against the cdup (cytosine deaminase/uracil phosphoribosyltransferase) gene product by growth in the presence of 0.4 μm 5′-fluorocytosine (ICN Biomedicals) to obtain PFB0080c gene-disrupted (ΔPFB0080c) parasites by double homologous recombination with the integrated human dihydrofolate reductase cassette. The parasites were cultured under drug pressure, and genomic DNA was isolated as outlined below and analyzed for the disruption of the PFB0080c gene by PCR using primers corresponding to the coding region, bp 884–954 (see Table 1).
Isolation of Parasite Genomic DNA
Wild type 3D7-CSA and ΔPFB0080c parasites obtained under WR99210 pressure and those obtained under both WR99210 and 5-fluorocytosine pressure were cultured to 20–30% parasitemia. At the late trophozoite stage, cultures were harvested, and parasites from IRBCs were released by treatment with 0.05% saponin in ice-cold PBS, pH 7.2. The released parasites were pelleted at 1860 × g at 4 °C for 20 min and washed with ice-cold PBS. The parasite pellets were suspended in 10 mm Tris-HCl, 20 mm EDTA, pH 8.0, containing 0.5% SDS and proteinase K (25 μg/ml) and incubated at 37 °C for 3 h. The solution was extracted three times with phenol and chloroform and then treated with RNase. The DNA was precipitated by adding 3 m sodium acetate to a final concentration of 0.3 m, followed by 2.5 volumes of ethanol. Precipitated DNA was recovered by centrifugation and washed with 70% ethanol, its concentration was estimated by measuring the OD at 260 nm, and it was used for the analysis of PFB0080c by PCR.
Expression and Purification of PFB0080c
A 939-nucleotide fragment (nucleotides 361–1260) in the open reading frame of the PFB0080c gene that encodes amino acid sequence 121–420 of PFB0080c was amplified by PCR of P. falciparum genomic DNA and cloned into pGEX4T3 vector at SalI and NotI sites in frame with the GST gene sequence using the following primers: 5′-CGTTGTGTCGACAATATGTTAACAAAGAAAATGAAAATTG-3′ (forward) and 5′-CGTTTAGCGGCCGCTGATGGATCCTCATTTGCTCC-3′ (reverse). The clones containing PFB0080c inserts were screened by restriction digestion analysis and confirmed by nucleotide sequence analysis. The recombinant plasmid was transformed into BL21(De3)RIPL E. coli for protein expression. Bacteria were grown to an A600 of 0.8, and protein expression was induced with 1 mm isopropyl β-d-1-thiogalactopyranoside at 37 °C. After 3 h, the bacteria were lysed by sonication and centrifuged in a Sorvall RC5B centrifuge, and supernatants were loaded onto glutathione-agarose columns. The columns were washed with 20 mm Tris-HCl, pH 7.5, containing 150 mm NaCl and 0.1% Triton X-100. The bound GST-PFB0080c fusion protein was eluted with 50 mm Tris-HCl, pH 8.0, containing 15 mm glutathione.
Production of Anti-PFB0080c Antibodies
Four BALB/c mice were immunized by intramuscular administration of the purified recombinant PFB0080c protein (25 μg/mice) mixed with Titermax adjuvant. After 21 days, mice were given a booster injection of 25 μg of the protein in Titermax adjuvant. An additional booster dose of 25 μg of protein was administered 21 days after the first boost. Two weeks later, blood was collected from individual mice serum prepared. The antibody titer as assessed by ELISA was ranged from 1:80,000 to 1:160,000.
Preparation of Parasite Protein Extract
About 100 μl of parasite culture (∼15% parasitemia) pellets were treated with 0.05% saponin in incomplete RPMI 1640 medium and centrifuged at 1860 × g. The pellets were washed with PBS, pH 7.2, and then extracted with 200 μl of PBS, containing 0.5% Triton X-100 and a mixture of protease inhibitors. The supernatants were collected, and the insoluble materials were then extracted with 15 μl of PBS, 2% SDS as reported previously (29). The protein contents in the 2% SDS extracts were estimated by the Bradford assay, as reported (29).
Western Blotting Analysis
The 2% SDS extract of 3D7-CSA parasites (20 μg of total parasite proteins/lane) were electrophoresed on 10% SDS-polyacrylamide mini gels under reducing conditions. The protein bands in gels were transferred onto nitrocellulose membranes. The membranes were blocked with 2% BSA in PBS and incubated with 1:1000 diluted mouse anti-PFB0080c antiserum or 1:1000 diluted mouse anti-Pf39 monoclonal antibody (MRA-87 obtained from MR4). Bound antibodies were detected with horseradish peroxidase-conjugated goat anti-mouse IgG and chemiluminescence detection reagent.
Immunofluorescence Analysis of PFB0080c and VAR2CSA
Thin smears of enriched IRBCs from wild type 3D7-CSA and ΔPFB0080c parasite cultures on glass slides were fixed for 10 min with methanol/acetone (1:1, v/v), cooled at −20 °C, and blocked with 5% BSA in PBS, pH 7.2, for 2 h. The slides were then incubated with 1:100 diluted mouse anti-PFB0080c antiserum for 1 h at room temperature. The slides were washed and then incubated with 1:100 diluted FITC-conjugated goat anti-mouse IgG (BD Biosciences) and 0.4 μm DAPI for 30 min. The stained cells were imaged using an inverted Nikon Eclipse TE2000-U fluorescent microscope equipped with a ×60 oil immersion objective lens. Similar areas of the cells were also photomicrographed under bright field with the same resolution.
For immunofluorescence analysis of surface-displayed PFB0080c, enriched, live IRBCs from 3D7-CSA parasite cultures were blocked with 5% BSA and immunostained. The IRBCs were incubated with 1:50 diluted mouse anti-PFB0080c antiserum at room temperature for 30 min. The cells were washed twice with PBS and then incubated with either 1:100 diluted FITC-conjugated goat anti-mouse IgG (BD Biosciences) and 0.4 μm DAPI or 1:100 diluted phycoerythrin-conjugated goat anti-mouse IgG (Invitrogen) and 0.4 μm DAPI. The immunostained IRBCs were washed with PBS. Fluorescent and light micrographs were viewed and recorded as above.
Immunofluorescence analysis of VAR2CSA in parental 3D7-CSA and ΔPFB0080c parasites was performed as above. The non-permeabilized, enriched IRBCs were blocked with 5% BSA and treated with 1:50 diluted rabbit anti-VAR2CSA serum followed by 1:100 diluted biotinylated sheep anti-rabbit IgG. After washing, cells were incubated with 1:300 diluted phycoerythrin-conjugated streptavidin (BD Biosciences) and 0.4 μm DAPI. The anti-VAR2CSA serum was raised in rabbit against the recombinant protein corresponding to Duffy binding-like domain 5ϵ of VAR2CSA, expressed in a baculovirus-insect cell system (45).
Analysis of PFB0080c and VAR2CSA on the IRBC Surface by Flow Cytometry
3D7-CSA and ΔPFB0080c parasite IRBCs (10–15% parasitemia) harvested at 20–24 h postinvasion were blocked with 5% BSA in PBS for 1 h at room temperature. The IRBCs were then incubated with 1:10 diluted mouse anti-PFB0080c antiserum or rat anti-VAR2CSA antiserum raised against the recombinant full-length extracellular portion of VAR2CSA expressed in a baculovirus-insect cell system (31) in PBS containing 2% BSA for 1 h. After washing three times with PBS, cells were incubated for 30 min with 1:100 diluted Alexa Fluor 488-conjugated goat anti-mouse IgG or donkey anti-rat IgG (Invitrogen) in PBS containing 2% BSA in PBS. Further, the IRBCs were stained with ethidium bromide (2.5 μg/ml) for 30 min and analyzed by using a FACSCalibur flow cytometer (BD Biosciences). IRBCs similarly immunostained with mouse or rat preimmune serum were analyzed as controls. Data analysis was performed using FlowJo software (Treestar, Ashland, OR).
PFB0080c and VAR2CSA Binding Analysis by ELISA
Ninety-six-well microtiter plates were coated with 5 μg/ml purified GST-tagged recombinant PFB0080c protein, control GST protein, and BSA in sodium bicarbonate buffer, pH 9.5, at 4 °C overnight. After washing with PBS, pH 7.2, the plates were blocked with 5% fat-free milk in PBS at room temperature for 2 h and then incubated for 1 h with His-tagged full-length recombinant VAR2CSA protein (31) at concentrations ranging from 0.125 to 3 μg/ml. The plates were washed four times with PBS and incubated with 1:1000 diluted anti anti-His5 antibody (Qiagen, Valencia, CA) in PBS containing 2% fat-free milk for 1 h, followed by 1:5000 HRP-conjugated goat anti-mouse IgG (GE Healthcare) for 1 h. The plates were washed with PBS containing 0.05% Tween 20 and treated with 3,3′,5,5′-tetramethylbenzidine substrate (Sigma-Aldrich). The color development was stopped with 0.2 n HCl, and absorbance at 450 nm was measured using a plate reader.
Statistical Analysis
Results are expressed as either mean ± S.E. One-way analysis of variance (Prism GraphPad version 3.0. Software, San Diego, CA) was used to calculate the statistical difference among experimental groups. p values of <0.05 were considered as statistically significant.
RESULTS
The C4S-adherent P. falciparum Parasites Express PFB0080c and Other Genes of PFB0075c-PFB0100c Cluster
Several P. falciparum proteins encoded by genes in the subtelomeric regions of chromosomes, including CLAG9, KAHRP, PfEMP3, and RESA, are known to play important roles in the PfEMP1-mediated adherence of IRBCs to host receptors. These proteins are essential for PfEMP1 transport, formation of rigid knob assemblies, and dense clustering of PfEMP1 in knobs or expression of PfEMP1 on the surface of infected erythrocytes (16, 17, 19, 22, 25). To determine whether additional genes were involved in parasite cytoadherence, we initially assessed the levels of transcripts in C4S-binding parasites and non-binding parasites by whole genome microarray analysis (supplemental Table S1). Synchronous cultures of parasites were harvested at 18 h postinvasion and fractionated on Percoll density centrifugation to obtain nearly homogenous late stage parasites. The mRNA from each culture was isolated, labeled, and hybridized to a high density microarray containing probes to 5146 P. falciparum gene coding regions, of which 4816 were probed by six or more independent and unique 25-mer sequences (47). A relative expression level for each gene on the microarray was computed using a match-only integral distribution algorithm, and these levels were compared. Altogether, 136 genes showed more than 3-fold higher expression by the CSA-binding parasites, but given that it is difficult to perfectly synchronize two independent P. falciparum samples, changes in the transcripts of some genes may not be significant. However, much larger changes in transcript level were observed for var2csa (PF3D7_1200600/PFL0030c) and the genes from a subtelomeric segment of chromosome 2 that encodes PF3D7_0201500 (PFB0075c), PF3D7_0201600 (PFB0080c, RLP1), PF3D7_0201700 (PFB0085c), PF3D7_0201600 (PFB0090c, KAHsp40), PF3D7_0201900 (PFB0095c, PfEMP3), and PF3D7_0202000 (PFB0100c, KAHRP). Transcript levels for these were between 3.7- and 163-fold higher in the C4S-binding parasites compared with non-binding parasites (Table 2). Examination of the relative expression levels for the non-C4S-selected parasites showed that in most cases, genes in this region were expressed at nearly background levels (∼25 units; see supplemental Table S1). To confirm these results, RT-PCR analysis was performed, and this showed that the transcripts of all genes in the PFB0075c-PFB0100c gene cluster were present at high levels in C4S-binding parasites, whereas these transcripts were either absent or present only at negligibly low levels in non-selected parasites (data not shown). Quantitative RT-PCR using primers shown in Table 1 confirmed the high abundance of the transcripts of these genes in C4S-adherent parasites (Table 2). Given that the subtelomeric end of chromosome 2 containing the PFB0075c-PFB0100c cluster is prone to deletion during genetic recombination in parasites (27) and is missing in P. falciparum strains, such as Dd2 (48), the data suggest that the high levels of transcripts observed for these genes in C4S-selected parasites compared with those in non-selected parasites was due to the deletion of a subtelomeric section of chromosome 2 in the majority of non-selected parasites. This deletion potentially includes the entire distal region of chromosome 2, whose genes were expressed in neither binding nor nonbinding parasites at 18–20 h postinvasion.
TABLE 2.
List of highly up-regulated P. falciparum genes in C4S-selected parasites compared to non-selected parasites as assessed by microarray and quantitative RT-PCR analyses
| Gene ID | Characteristics of the expressed proteins | Up-regulation of transcriptsa |
|
|---|---|---|---|
| Microarray analysisb | Quantitative RT-PCR analysis | ||
| -fold | |||
| PFL0030c | VAR2CSA PfEMP1 | 9 | 354 |
| (C4S binding) | |||
| PFB0100c | KAHRP | 83 | 576 |
| (knob formation) | |||
| PFB0095c | PfEMP3 | 164 | 298 |
| (knob stabilization) | |||
| PFB0090c | KAHsp40, DnaJ domain | 18 | 75 |
| (chaperone) | |||
| PFB0085c | PHISTb and DnaJ domains | 3.7 | NDc |
| (uncharacterized protein) | |||
| PFB0080c | PHISTb domain | 48 | 112 |
| (RLP1) | |||
| PFB0075c | HYP9 domain | 20 | 244 |
| (integral membrane protein)b | |||
a Initial microarray analysis gave a qualitative assessment of gene transcripts present at high levels in C4S-binding parasites compared with non-binding parasites (supplemental Table S1). Subsequent real-time RT-PCR analysis quantified the levels of highly abundant gene transcripts in C4S-binding parasites. The upper and lower confidence levels for gene expression, measured by microarray analysis, are given in supplemental Table S1. Because microarray measurements are not linear at the upper ends, the data are not expected to be in agreement with those determined by quantitative RT-PCR analysis.
b Ref. 47.
c Not determined.
A search of P. falciparum gene expression profiles in the PlasmoDB database showed that genes of the PFB0075c-PFB0100c cluster are expressed early during the ring stage, coinciding with the timing of the expression of the var2csa gene PF3D7_1200600 (PFL0030c) (35). PFB0100c (KAHRP) and PFB0095c (PfEMP3) have been extensively characterized, and their functional roles in cytoadherence have been elucidated (16, 17, 19, 22). The proteins encoded by the remaining four genes of the cluster contain PHIST, DnaJ, or hyp domains (see Table 2), and such proteins may be exported to infected erythrocytes (34). These proteins have been predicted to play important roles in parasite survival, virulence, or cytoadherence (34). Although some functional information is available for the proteins encoded by PFB0085c and PFB0090c (32–34), the products of PFB0075c and PFB0080c have not been experimentally characterized
PF3D7_020600/PFB0080c Gene-encoded Protein Is Expressed on the Surface of C4S-binding IRBCs
Although it is known that PF3D7_020700 (PFB0085c) and PF3D7_020900 (PFB0090c) are not essential for the cytoadherence property of P. falciparum (34), the contributions of PF3D7_020500 (PFB0075c) and PF3D7_020600 (PFB0080c) have not been studied. To determine whether the protein encoded by PFB0080c is expressed by C4S-binding parasites and to examine its subcellular localization, we expressed and purified a recombinant protein corresponding to the 107–420-amino acid portion of PFB0080c/RLP1 in E. coli and raised antibodies in mice. Western blotting using the antiserum showed that PFB0080c was robustly expressed by C4S-selected parasites and was not present in non-selected parasites (Fig. 1). The observed ∼55 kDa band agrees with the calculated molecular mass of 58.6 kDa for PFB0080c/RLP1.
FIGURE 1.
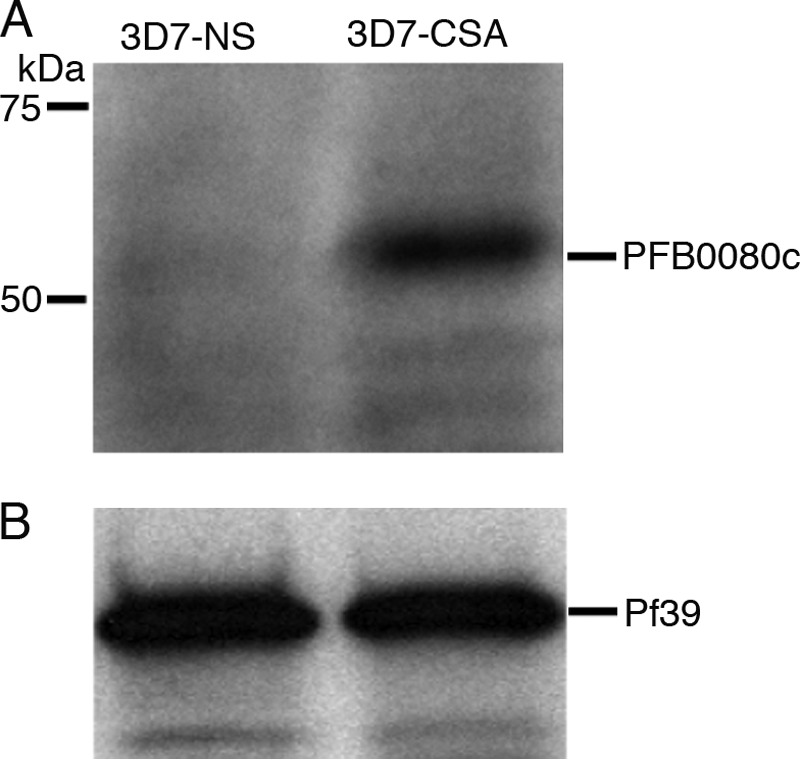
C4S-adherent, but not non-adherent, P. falciparum parasites express PFB0080c protein. Western blot analysis of proteins from 3D7 C4S-selected (3D7-CSA) and non-selected (3D7-NS) parasites was performed using mouse anti-PFB0080c antiserum. A, the antiserum of PFB0080c recognized a band of PFB0080c at the expected size of ∼55 kDa only in C4S-selected parasites. B, the parasite protein Pf39 served as a loading control. The molecular mass and the electrophoretic mobility of standard proteins are shown.
Immunofluorescence analysis of C4S-binding IRBCs using anti-PFB0080c/RLP1 antiserum showed a strong expression of PFB0080c on the surface of C4S-selected, permeabilized IRBCs, exhibiting a characteristic punctate “beads on a string”-like staining pattern similar to that observed for PFEMP1 on the IRBC surface (Fig. 2A). A weak immunostaining of PFB0080c/RLP1 was also seen in the cytoplasm. No immunostaining of PFB0080c/RLP1 was observed on the surface of the non-adherent IRBCs (Fig. 2B). Immunostaining of live, non-permeabilized IRBCs using anti-PFB0080c/RLP1 antiserum followed by FITC-conjugated goat anti-mouse or phycoerythrin-conjugated goat anti-mouse secondary antibodies also showed a punctate beads on a string-like staining pattern on the surface of erythrocytes; no cytoplasmic staining was observed (Fig. 2, C and D). Together, these results clearly demonstrated that PFB0080c/RLP1 is expressed on the IRBC surface.
FIGURE 2.
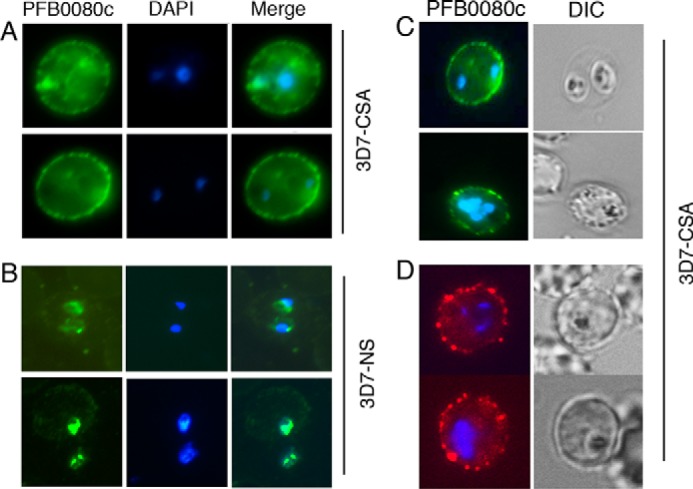
PFB0080c is expressed on the IRBC surface of C4S-selected P. falciparum. A–D, immunofluorescence analysis of IRBCs was performed using anti-PFB0080c antiserum, followed by either FITC-conjugated goat anti-mouse IgG (A–C) or phycoerythrin-conjugated goat anti-mouse IgG (D) secondary antibody. A and B, permeabilized C4S-adherent (3D7-CSA) IRBCs (A) and non-selected (3D7-NS) IRBCs (B). C and D, live 3D7-CSA IRBCs. PFB0080c is strongly expressed on the surface of C4S-adherent IRBCs but not in 3D7-NS IRBCs.
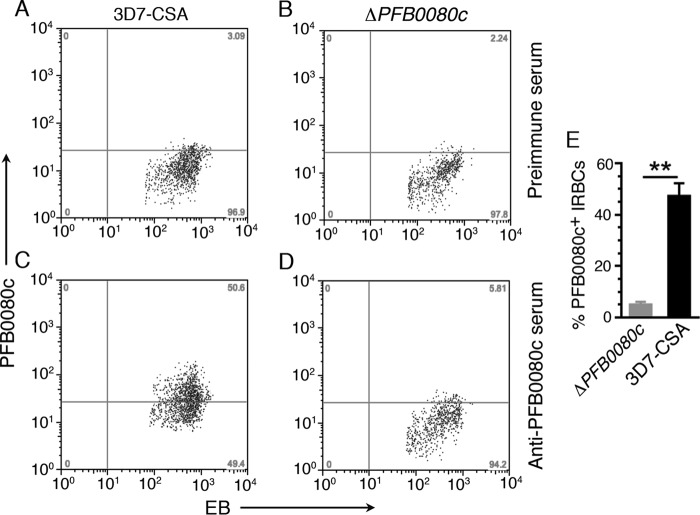
To confirm and quantify the surface expression of PFB0080c/RLP1, we stained live RBCs infected with 3D7-CSA or PFB0080c-knock-out (ΔPFB0080c) parasites (see below) using either preimmune serum or an anti-PFB0080c antiserum and analyzed the stained cells by flow cytometry (Fig. 3, A–D). The 3D7-CSA parasites strongly expressed PFB0080c on the IRBC surface (Fig. 3C). In contrast, few of the ΔPFB0080c IRBCs were bound by anti-PFB0080c antiserum, similar to the number staining positive with control preimmune serum (see Fig. 3, A and B). Quantification of the combined results of two independent experiments showed that ∼48% of 3D7-CSA IRBCs are PFB0080c-positive, whereas only ∼5% of ΔPFB0080c IRBCs were PFB0080c-positive (Fig. 3E). These results confirm that PFB0080c/RLP1 is expressed on the IRBC surface.
FIGURE 3.
Live IRBC staining and flow cytometry confirm that PFB0080c is expressed on the IRBC surface. IRBCs of 3D7-CSA and ΔPFB0080c parasites were treated with mouse anti-PFB0080c antiserum or preimmune mouse serum followed by Alexa Fluor 488-conjugated goat anti-mouse IgG and ethidium bromide. The ethidium bromide (EB)-gated IRBCs were analyzed for PFB0080c by flow cytometry. A–D, dot plots of PFB0080c surface expression in gated IRBCs in one of two independent experiments each performed in duplicate. E, quantification of PFB0080c-positive IRBCs of 3D7-CSA and ΔPFB0080c parasites. Data are expressed as mean ± S.E. (error bars). **, p < 0.01.
Because PFB0080c/RLP1 is a PHIST domain-containing protein and such proteins constitute a part of export machinery of parasites, some of which are localized to the knobs in the IRBC surface (34), it is possible that this protein is involved in either protein transport or membrane function of the infected erythrocytes, potentially by mediating interactions with other proteins, such as VAR2CSA. PFB0080c/RLP1 transcripts peak during the ring stage between 10 and 15 h postinvasion of erythrocytes (PlasmoDB). Thus, the timing of the expression of PFB0080c/RLP1 coincides with that of VAR2CSA (protein) and strong binding of IRBCs to C4S even at the late ring stage (17, 35).
PFB0080c Plays a Role in the Expression of PfEMP1 and Cytoadherence
To determine whether PFB0080c has a role in P. falciparum cytoadherence, the gene was disrupted by double crossover homologous recombination as described previously (43). To accomplish this, we constructed a plasmid, pCC1-ΔPFB0080c, by using the strategy depicted in Fig. 4 to knock-out the gene and transfected it into 3D7-CSA parasites freshly selected by panning on plates coated with purified placental CSPG. The transfected parasites were selected by WR99210 and 5-fluorocytosine (ancoban) treatments and tested for the disruption of PFB0080c. PCR analysis using the primers outlined in Table 1 showed the presence of a band corresponding to the expected region of PFB0080c sequence in 3D7-CSA parental parasites; however, this band was completely absent in the ΔPFB0080c parasites selected under the pressure of both WR99210 and 5-fluorocytosine drugs (Fig. 5A). The absence of PFB0080c in the IRBCs of ΔPFB0080c parasites is also evident from flow cytometry analysis (see Fig. 3; compare A, B, and D).
FIGURE 4.

Schematic diagram of PFB0080c gene disruption strategy. The 5′ end 971-bp fragment (shown in red) and a 3′ end 950-bp fragment (blue) covering the promoter, coding, and UTR regions of PFB0080c were cloned as shown into the pCC1 vector. The C4S-adherent 3D7 parasites were transfected with the plasmid. Integration of the human dhfr cassette into the coding region of PFB0080c by double homologous recombination resulted in PFB0080c disruption. The gene-disrupted (ΔPFB0080c) parasites were selected by positive and negative selections under drug pressure using WR99210 and 5-fluorocytosine, respectively. ΔPFB0080c parasites were cultured under WR99210 and 5-fluorocytosine drug pressure for further analysis.
FIGURE 5.

PFB0075c-PFB0100c cluster and clag9 are intact in PFB0080c knock-out P. falciparum. A, a 100-bp fragment corresponding to the deleted region of PFB0080c was PCR-amplified from the genomic DNA isolated from the parental C4S-adherent and ΔPFB0080c parasites selected under WR99210 and 5-fluorocytosine (FC) pressure. Although the PFB0080c fragment is present in parental C4S-adherent parasites, it is completely absent in ΔPFB0080c parasites selected under the pressure of both WR99210 and 5-fluorocytosine. B, fragments corresponding to the coding regions of the indicated genes (clag9 present in chromosome 9 and genes of the PFB0075c-PFB0100c cluster in chromosome 2) that are located in the subtelomeric regions were amplified from the genomic DNA of ΔPFB0080c parasites using the primers listed in Table 1. Seryl tRNA was analyzed as a control. All genes except for PFB0080c were amplified, indicating that subtelomeric regions of chromosomes 2 and 9 that contribute to cytoadherence are intact in ΔPFB0080c parasites.
Because telomeric end deletion in several chromosomes of parasites occurs spontaneously during culturing, we wondered whether subtelomeric regions of chromosomes 2 and 9, which contribute to cytoadherence, had remained intact during the disruption of PFB0080c and subsequent drug selection for disrupted parasites. PCR analysis of genomic DNA showed that the telomeric end of chromosome 2 was intact in the ΔPFB0080c parasites, as indicated by the presence of pfemp3, kahrp, PFB0075c, and PFB0090c (Fig. 5B). Also, the observed PCR amplification of clag9 indicated that the subtelomeric region of chromosome 9 that contains clag9 and is prone to deletion during culturing, was intact in ΔPFB0080c parasites. Thus, it appeared that the integrity of chromosomes 2 and 9 was maintained in PFB0080c-knock-out parasites.
To test whether deletion of PFB0080c affected VAR2CSA expression on the IRBC surface, we performed immunofluorescence analysis of non-permeabilized parental 3D7-CSA and ΔPFB0080c parasites by immunolabeling with rabbit anti-VAR2CSA antibody. The results showed that both parental and PFB0080c-knock-out parasites efficiently expressed VAR2CSA on the surface of IRBCs (Fig. 6, A and B).
FIGURE 6.

PFB0080c-knock-out P. falciparum express VAR2CSA on the surface of infected erythrocytes. A and B, immunofluorescence analysis of VAR2CSA in live ΔPFB0080c IRBCs was performed using rabbit anti-VAR2CSA antiserum followed by biotin-conjugated sheep anti-rabbit IgG secondary antibody and phycoerythrin-conjugated streptavidin. Shown are the immunofluorescent and light micrographs of parental 3D7-CSA parasites (A) and ΔPFB0080c parasites (B); both parasite types expressed VAR2CSA protein. DIC, differential interference contrast.
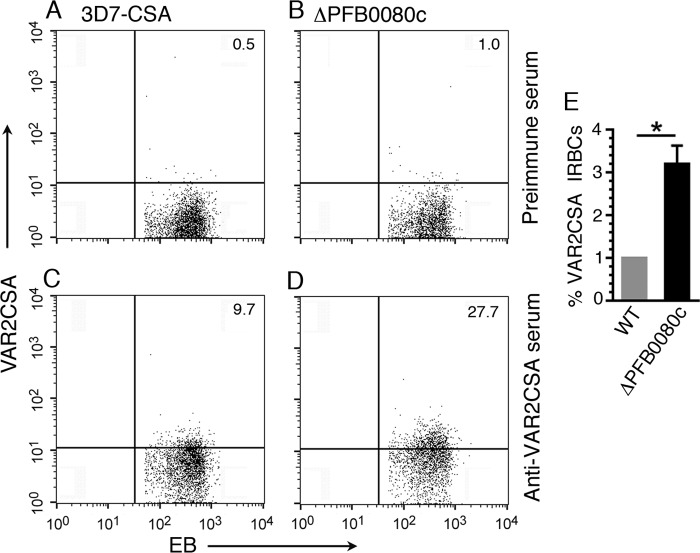
To quantify the amount of VAR2CSA protein on the IRBC surface, we immunostained live RBCs infected with 3D7-CSA or ΔPFB0080c parasites using either an anti-VAR2CSA antiserum or the preimmune serum and analyzed them by flow cytometry (Fig. 7, A–D). Compared with 3D7-CSA IRBCs, more IRBCs of ΔPFB0080c parasites expressed VAR2CSA (Fig. 7, A–D). Quantification revealed that VAR2CSA expression on the surface of ΔPFB0080c parasites was ∼3.2-fold higher than that on the surface of 3D7-CSA IRBCs (Fig. 7E). These results indicate that PFB0080c may play a role in the regulation of VAR2CSA expression or its presentation on the IRBC surface.
FIGURE 7.
PFB0080c-knock-out parasites express higher levels of VAR2CSA on the surface of parasite-infected erythrocytes. 3D7-CSA and ΔPFB0080c IRBCs were treated with either rat VAR2CSA antiserum or rat preimmune serum followed by Alexa-Fluor 488-conjugated goat anti-rat IgG and ethidium bromide (EB). The ethidium bromide-gated IRBCs were analyzed for VAR2CSA by flow cytometry. A–D, dot plots of VAR2CSA surface expression in gated IRBCs in a representative of two independent experiments each performed in duplicate. E, quantification of VAR2CSA-positive IRBCs of 3D7-CSA and ΔPFB0080c parasites. Data are expressed as mean ± S.E. (error bars). *, p < 0.05.
To further elucidate whether the regulation of VAR2CSA expression by PFB0080c is transcriptional or translational, we analyzed var2csa mRNA levels in 3D7-CSA and ΔPFB0080c parasites. The var2csa mRNA level in 3D7-CSA parasites was 2.6-fold higher than in ΔPFB0080c parasites (Fig. 8). These results are consistent with higher levels of VAR2CSA protein on the surface of ΔPFB0080c parasites compared with 3D7-CSA parasites (see Fig. 7). The data also suggest that PFB0080c regulates VAR2CSA expression at the transcriptional level.
FIGURE 8.

PFB0080c knock-out P. falciparum express genes of PFB0075c-PFB00100c cluster. RNA preparations from 3D7-CSA and ΔPFB0080c parasites were reverse-transcribed using SuperScript II reverse transcriptase, and the cDNA thus obtained was analyzed for the indicated genes by quantitative PCR using SYBR Green PCR mix and nucleotide primers listed in Table 1. The analysis was performed in triplicate. The relative levels of gene expression were normalized to fructose bisphosphate aldolase. Data are expressed as mean values ± S.E. (error bars). *, p < 0.05; **, p < 0.01.
Because it is possible that the higher levels of VAR2CSA expression in ΔPFB0080c parasites are due to changes in the expression of one or more genes in the surrounding region of chromosome 2 (i.e. the PFB0075c-PFB0100c cluster), we assessed transcription of these nearby genes. The mRNA level of PFB0090c was significantly higher in ΔPFB0080c parasites than in 3D7-CSA (Fig. 8). This is interesting, given that PFB0090c is a KAHsp40 chaperone, which interacts with knob-associated proteins, such as PfEMP1, KAHRP, and PfEPMP3, in addition to the chaperones, Hsp70 and Hsp101 (32, 33). Thus, increased expression of PFB0090c could contribute at least partly to higher levels of VAR2CSA expression. In contrast to PFB0090c, mRNA levels of PFB0085c, PFB0095c, and PFB00100c in ΔPFB0080c parasites were similar to those in 3D7-CSA parasites (Fig. 8). Although the transcription of PFB0075c in ΔPFB0080c parasites was slightly higher than in 3D7-CSA parasites, the increase was not statistically significant. Finally, because previous studies have shown that clag9 is required for P. falciparum cytoadherence (28, 29), we also assessed the expression of clag9. There was no observable difference in its expression in 3D7-CSA and PFB0080c parasites (see Fig. 8), ruling out the contribution of CLAG9 to higher expression of VAR2CSA by PFB0080c parasites.
It is also possible that PFB0080c interacts directly with VAR2CSA and that this interaction may modulate the cytoadherence capacity of parasites or function as a cue for feedback regulation of VAR2CSA expression. Therefore, we performed an ELISA to determine whether VAR2CSA binds PFB0080c. We found that VAR2CSA bound to PFB0080c in a dose-dependent manner (Fig. 9). This interaction had no effect on IRBC adherence because PFB0080c protein was unable to inhibit IRBC binding to placental CSPG (not shown). Further, anti-PFB0080c antiserum also had no effect on IRBC binding to CSPG (not shown). Thus, PFB0080c may control VAR2CSA expression by interacting with it during protein transport and/or at the IRBC surface. Further, because deficiency in PFB0080c increases the IRBC binding capacity, the interaction of PFB0080c with VAR2CSA is not essential for the stability of functional VAR2CSA protein.
FIGURE 9.

PFB0080c binds to VAR2CSA in a dose-dependent manner. ELISA analysis was performed to assess the binding of the purified GST-tagged recombinant PFB0080c to the His-tagged recombinant full-length extracellular portion of VAR2CSA. Ninety-six-well microtiter plates were coated with 5 μg/ml PFB0080c and control proteins GST and BSA. After blocking the wells, VAR2CSA at the indicated concentrations was added to each well. The bound VAR2CSA was detected using mouse anti-His5 antibody and HRP-conjugated goat anti-mouse secondary antibody as described under “Experimental Procedures.” The analysis was performed three times, each in duplicate. The data are expressed as mean values ± S.E. (error bars). *, p < 0.01.
Given that PFB0080c regulates VAR2CSA expression, it was also of interest to determine whether the increased surface expression of VAR2CSA contributes to its functional property (i.e. the capacity of IRBC to bind to the C4S chains of placental CSPG). To accomplish this objective, we measured the binding capacity of ΔPFB0080c parasite IRBCs by incubating separately with bovine tracheal CSA, C4S containing 33% 4-sulfate, chondroitin 6-sulfate, and hyaluronic acid and allowing the treated cells to bind to the placental CSPG coated on plastic Petri dishes (36, 49, 50). The IRBCs of ΔPFB0080c parasites strongly bound to C4S, as evident by the concentrations of C4S (80 μg/ml) required for the complete inhibition of IRBC binding to placental CSPG (Fig. 10A); this C4S-inhibitory concentration is even higher than that observed for parental 3D7-CSA parasites (35, 36, 49, 50). As seen previously for parental C4S-adherent IRBCs (35, 36, 49, 50), both bovine tracheal CSA and C4S containing 33% 4-sulfate inhibited the binding of ΔPFB0080c IRBCs to the placental CSPG in a dose-dependent manner, whereas chondroitin 6-sulfate and hyaluronic acid showed only marginal inhibition of IRBC binding to placental CSPG (Fig. 10A). These results demonstrated that the ΔPFB0080c parasites specifically and strongly bind to the C4S chains of placental CSPG. Notably, the observed high binding capacity of ΔPFB0080c IRBCs (see Fig. 10A) is consistent with the high levels of VAR2CSA expression by the ΔPFB0080c parasites (see Fig. 7).
FIGURE 10.
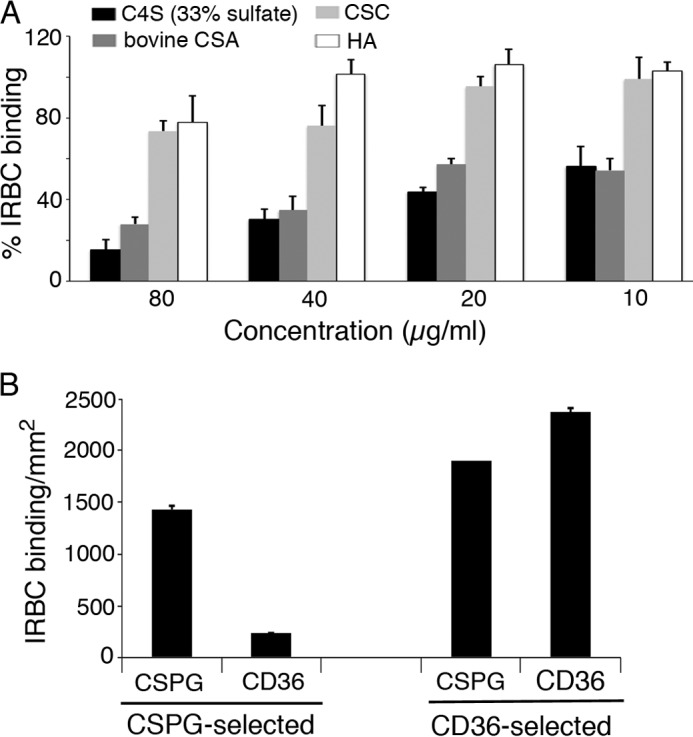
Disruption of PFB0080c confers stable maintenance of P. falciparum-infected erythrocytes binding to C4S, and a minor population bind to both C4S and CD36. The C4S binding specificity and binding capacity were determined by analyzing the inhibition of IRBCs binding to the purified placental CSPG by the indicated soluble glycosaminoglycans at different concentrations. Assays were done in duplicate. The number of IRBCs bound per mm2 in an untreated control sample ranged from 1500 to 3000, depending upon the parasitemia (10–20%). A, plotted values are presented as the percentage of glycosaminoglycan-pretreated IRBCs bound per mm2 relative to the binding of untreated control IRBCs, which was considered as 100% binding. B, after culturing for 5–6 months, ΔPFB0080c parasites were selected for CD36 and C4S binding by panning on plastic plates coated with CD36 and placental CSPG, respectively. In each case, the IRBCs were assessed for binding to CD36 and placental CSPG. Assays were performed in duplicate, and the number of IRBCs bound per mm2 is shown. IRBCs selected by panning on CD36 bound to both CD36 and CSPG, whereas IRBCs selected by panning on CSPG bound predominantly to CSPG and only at low levels to CD36. Data are expressed as mean values ± S.E. (error bars).
Because ΔPFB0080c parasites efficiently express VAR2CSA, we wondered whether PFB0080c/RLP1 might play a role in the stable expression of VAR2CSA when parasites are continuously cultured though many cell cycles. To test this, we analyzed the C4S-binding capacity of ΔPFB0080c parasites in continuous culture for ∼6 months without panning on CSPG. The parasites bound to C4S in a manner similar to that of freshly selected parasites. These results are in contrast to a ∼75% decrease in the binding capacity of 3D7-CSA IRBCs after continuously culturing for 2 months (35). Together, these data indicate that deletion of PFB0080c results in the efficient expression of VAR2CSA through many generations of parasite growth.
Further, we analyzed the ability of ΔPFB0080c parasites, which were continuously cultured for 5–6 months, for binding to CD36. A small population of IRBCs could bind to CD36 (Fig. 10B). We selected this CD36-binding parasite population and tested for binding to placental CSPG and CD36. Interestingly, these IRBCs, although selected for binding to CD36, exhibited binding specificity to both C4S and CD36 (Fig. 10B). In contrast, IRBCs selected for placental CSPG binding bound predominantly to CSPG and only at low levels to CD36 (Fig. 10B). The results indicate that the loss of PFB0080c, in addition to conferring stable expression of C4S-binding VAR2CSA through many generations of parasite growth, allows for the expression of CD36-binding PfEMP1 at a slow rate by a small population of parasites, which also express VAR2CSA. These results further indicate that PFB0080c/RLP1 is involved in the regulation of PfEMP1 expression. In future studies, it would be interesting to know whether C4S-adherent ΔPFB0080c parasites express, in addition to VAR2CSA, ICAM-1 and other adhesive receptor-specific PfEMP1s, thereby binding to multiple receptors.
DISCUSSION
The results of this study show that C4S-adherent P. falciparum parasites express, in addition to kahrp and pfemp3, four other genes that together constitute a gene cluster, PF3D7_0201500-PF3D7_0202000 (PFB0075-PFB0100c), located in the subtelomeric left arm of chromosome 2. However, non-adherent parasites did not express these genes, probably due to a spontaneous subtelomeric deletion that occurred during continuous culturing of these parasites. Together, the results strongly suggest that these genes play significant roles in parasite cytoadherence. Although it is well known that kahrp and pfemp3 play important roles in cytoadherence (16–18, 22), little or nothing is known about the four other parasite genes, all of which encode proteins exported to the infected erythrocytes.
A previous study assessed the roles of PFB0085c and PFB0090c in cytoadherence by targeted gene disruption in CS2 parasites, which stably express VAR2CSA and thus maintain the C4S-adherent ability through many generations of parasite growth (34). Disruption of PFB0085c has been shown to have no effect on IRBC adherence to C4S (34). These results together with our observation that PFB0085c expression in ΔPFB0080c parasites is unchanged compared with C4S-CSA parasites suggest that PFB0085c is not critical for PfEMP1 expression. However, because PFB0085c is expressed by C4S-adherent parasites (see Table 2) and given that it is an exported protein possessing the DnaJ domain, it is possible that this protein functions as a chaperone in exporting cytoadherence machinery or proteins unrelated to cytoadherence to infected erythrocytes. In parasites lacking PFB0085c, other protein(s) having a redundant function may perform its role.
In contrast to PFB0085c, disruption of PFB0090c, which has recently been shown to encode DnaJ domain-containing KAHsp40 (32, 33), resulted in a marked reduction in expression of VAR2CSA and a negligible level of C4S-binding capacity in CS2 parasites (34). However, upon panning of PFB0090c-knock-out parasites on CSA-coated plates, C4S-adherent parasites were selected. Therefore, it appears that deletion of this gene leads to a switching by CS2 parasite strain to express different PfEMP1s, including VAR2CSA (34). Considering that CS2 parasites maintain stable C4S adherence characteristics and that a lack of PFB0090c resulted in parasite switching to express different PfEMP1s, KAHsp40 appears to play an important role in the stable expression of VAR2CSA over a long period of parasite culturing.
Based on the results presented here, we conclude that PFB0080c also modulates PfEMP1 expression, but differently than PFB0090c, by contributing to the mechanism involved in the regulation of var gene switching. The following observations support this conclusion. We reported earlier that, after about 2 months of continuous culturing, only ∼30% of C4S-adherent 3D7 parasites retain C4S-binding ability, and the remainder lost their ability to bind to C4S (35). In fact, it was necessary to reselect C4S-adherent 3D7 parasites every 6–8 weeks of culturing by panning on CSPG-coated plates to obtain IRBCs that efficiently bound to C4S (35, 36, 49, 50). Further, the binding strength of freshly selected 3D7-CSA parasites, as determined by the ability of bovine CSA to inhibit the IRBC binding to the purified placental CSPG (IC50 of CSA ∼10 μg/ml), was substantially decreased after culturing for ∼2 months (IC50 of CSA ∼3 μg/ml) (35). In contrast to the parental 3D7-CSA parasites, the ΔPFB0080c 3D7 parasites efficiently expressed VAR2CSA and stably maintained C4S-binding ability even after continuous culturing for ∼6 months, and only a minor population switched to express additionally CD36-adherent PfEMP1. Surprisingly, unlike parental 3D7-CSA parasites, the ΔPFB0080c parasites strongly bound to the placental CSPG, exhibiting an IC50 of >10 μg/ml with CSA even after ∼6 months of continuous culturing. Additionally, a minor population of ΔPFB0080c parasites also bound to CD36, exhibiting dual binding characteristics (i.e. IRBCs bound to both CSPG and CD36). These results suggest that this population of parasites express both CD36-binding PfEMP1 and VAR2CSA.
Although the expression of PFB0090c protein by ΔPFB0080c parasites remains to be determined, based on the observed increase in transcription, it is tempting to suggest that PFB0080c modulates the stable expression of VAR2CSA partly by controlling PFB0090c expression. As shown in Fig. 8, deletion of PFB0080c resulted in the up-regulation of both PFB0090c and var2csa transcription. PFB0090c/KAHsp40 is a knob-associated chaperone protein that interacts with PfEMP1 and several other knob-associated proteins (32, 33). Thus, PFB0090c may contribute to one or more processes, such as (i) folding of VAR2CSA into functional protein, (ii) transporting it efficiently through erythrocyte cytosol, and (iii) efficient surface display of VAR2CSA. Further, it has been shown that the deletion of PFB0090c results in the loss of the C4S-adherent property of CS2 parasites, which otherwise stably maintain the C4S-adherent property (34). However, after further culturing and selection of ΔPFB0090c parasites, the IRBCs could bind to CSA; thus, it appears that the lack of PFB0090c resulted in var gene switching. Therefore, increased expression of PFB0090c is expected to markedly slow down var gene switching to stably express PfEMP1. The stable expression of VAR2CSA over a long period of continuous culturing of parasites may at least in part be due to the increased expression of PFB0090c upon PFB0080c deletion.
Several studies have shown that PfEMP1 expression is mutually exclusive (51, 52), meaning that a parasite only expresses one type of PfEMP1 at given time, thus binding specifically to a single receptor. Although this is the case in most parasite strains, some can exhibit a dual/multiple binding property. For example, similar to our observations here with ΔPFB0080c parasites, it has been reported that IRBCs expressing type 41 PfEMP1 can bind to both ICAM-1 and CD36 (17). It has also been shown that individual parasites of the HB3 stain can simultaneously express both copies of duplicated var2csa genes (53). Furthermore, a recent study has shown that deletion of PfSet2 resulted in the dysregulation of var gene expression, leading to simultaneous expression of different PfEMP1s on the surface (54). Thus, it appears that an exception to the monoallelic transcription rule is possible whenever genes affecting the regulation of var genes are altered. Together, the above observations suggest that deletion of PFB0080c allows for the efficient and stable expression of VAR2CSA and additionally expression of two or more PfEMP1 having different adhesive specificity by a population of C4S-adherent parasites to exhibit two or possibly more binding characteristics.
As shown in Figs. 2 and 3, PFB0080c/RLP1 is expressed on the surface of parasite-infected erythrocytes. This observation is consistent with the features of PFB0080c/RLP1 that the protein contains an N-terminal peptide signal sequence for endoplasmic reticulum translocation and a canonical Plasmodium export element (PEXEL), a pentapeptide motif at the N-terminal end, for protein export to erythrocytes (55). Additionally, PFB0080c/RLP1 has a PHIST domain, a conserved domain consisting of ∼150 amino acids forming four consecutive α-helices (56). About 70 P. falciparum proteins contain PHIST domains, comprising a large family of exported proteins. A notable characteristic of PHIST family proteins is that they are expressed at the early ring stage, and some of these proteins appear to contribute to the rigidity of infected erythrocytes and cytoadherence (56). One of the prominent members of the PHIST family of proteins is RESA (19, 57, 58). A previous study has shown that a lack of RESA results in a significant increase in the capacity of CD36-binding parasites (25). Whereas parasites with disruptions in the genes encoding KAHRP or PfEMP3 showed, respectively, 14 and 70% CD36-adherent capacity, as compared with parental 3D7 parasites, those with the deletion of the subtelomeric end of chromosome 2 had a 19% adherent capacity. Based on these results, it was predicted that the loss of a RESA-like homolog, which is located in the subtelomeric end of chromosome 2 might increase the adherent capacity (25). The N-terminal end of PFB0080c resembles RESA. Our observations that targeted disruption of PFB0080c leads to a stable cytoadherence and that the adherent capacity of parasites is higher than that of the parental 3D7 parasites when measured after culturing for several months suggest that PFB0080c is similar to RESA in function. Thus, we have named this protein RESA-like protein 1 (RLP1).
Because RLP1 is expressed on the surface of infected erythrocytes, an important question is how exactly the lack of RLP1 results in the stable maintenance of C4S binding by parasites. Obviously, parasites must efficiently express VAR2CSA during culturing through many generations of parasite growth without significantly switching to express a different PfEMP1 and avoiding telomeric end deletions. As discussed above, because deletion of PFB0090c/KAHsp90 destabilizes stable expression of VAR2CSA in C4S-adherent parasites (34), one likely mechanism is that higher expression of KAHsp40 by ΔPFB0080c parasites contributes partly to the stable expression of VAR2CSA. Further, the deletion of PFB0080c may also result in other transcriptional alterations, leading to the changes in the regulatory mechanism that controls var gene switching/a single var gene expression. Several possible scenarios can be envisioned. (i) The genomic structural changes introduced by the integration of the deletion cassette prevent the frequent subtelomeric recombination events from occurring readily (46), thereby stabilizing VAR2CSA expression. (ii) The loss of RLP1 results in the up-regulated expression of several proteins including KAHsp40 may contribute to the steady and stable expression of VAR2CSA through many generations of parasite growth. (iii) The interaction of RLP1 with VAR2CSA shown here (and possibly its interactions with other proteins) is a cue for var gene switching. Thus, the absence of RLP1 results in the stable expression of PfEMP1 and the maintenance of chromosome integrity. In any event, overall, our data demonstrate that RLP1 modulates the expression of both PfEMP1 and KAHsp40; the latter in turn appears to contribute to the stable expression of VAR2CSA.
Supplementary Material
Acknowledgments
We thank Dr. Xianzhu Wu (Pennsylvania State University College of Medicine) for help in the preparation of flow cytometry figures and Dr. Irvin Sherman (University of California San Diego, School of Medicine) for valuable help in editing the manuscript.
This work was supported, in whole or in part, by National Institutes of Health, NIAID, Grants R01 AI45086 (to D. C. G.) and R01 AI10358 (to E. A. W.).

This article contains supplemental Table S1.
- IRBC
- infected red blood cell
- RBC
- red blood cell
- C4S
- chondroitin 4-sulfate
- CSA
- chondroitin sulfate A
- CSPG
- chondroitin sulfate proteoglycan
- ICAM-1
- intercellular cell adhesion molecule 1
- KAHRP
- knob-associated histidine-rich protein
- KAHsp40
- knob-associated Hsp40
- MESA
- mature parasite-infected erythrocyte surface antigen
- PHIST
- Plasmodium helical interspersed subtelomeric
- RESA
- ring-infected erythrocyte surface antigen
- var gene
- variant gene
- VAR2CSA
- PfEMP1 that mediates IRBC adherence to C4S/CSA
- NS
- non-selected.
REFERENCES
- 1. Snow R. W., Guerra C. A., Noor A. M., Myint H. Y., Hay S. I. (2005) The global distribution of clinical episodes of Plasmodium falciparum malaria. Nature 434, 214–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hay S. I., Guerra C. A., Gething P. W., Patil A. P., Tatem A. J., Noor A. M., Kabaria C. W., Manh B. H., Elyazar I. R., Brooker S., Smith D. L., Moyeed R.A., Snow R.W. (2009) A world malaria map: Plasmodium falciparum endemicity in 2007. PLoS Med. 6, e1000048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. World Heath Organization (2012) World malaria report 2013. World Heath Organization, Geneva, Switzerland [Google Scholar]
- 4. Ockenhouse C. F., Ho M., Tandon N. N., Van Seventer G. A., Shaw S., White N. J., Jamieson G. A., Chulay J. D., Webster H. K. (1991) Molecular basis of sequestration in severe and uncomplicated Plasmodium falciparum malaria: differential adhesion of infected erythrocytes to CD36 and ICAM-1. J. Infect. Dis. 164, 163–169 [DOI] [PubMed] [Google Scholar]
- 5. Baruch D. I., Gormely J. A., Ma C., Howard R. J., Pasloske B. L. (1996) Plasmodium falciparum erythrocyte membrane protein 1 is a parasitized erythrocyte receptor for adherence to CD36, thrombospondin, and intercellular adhesion molecule 1. Proc. Natl. Acad. Sci. U.S.A. 93, 3497–3502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fried M., Duffy P. E. (1996) Adherence of Plasmodium falciparum to chondroitin sulfate A in the human placenta. Science 272, 1502–1504 [DOI] [PubMed] [Google Scholar]
- 7. Weatherall D. J., Miller L. H., Baruch D. I., Marsh K., Doumbo O. K., Casals-Pascual C., Roberts D. J. (2002) Malaria and the red cell. Hematol. Am. Soc. Hematol. Edu. Program 35–57 [DOI] [PubMed] [Google Scholar]
- 8. Brabin B. J., Romagosa C., Abdelgalil S., Menéndez C., Verhoeff F. H., McGready R., Fletcher K. A., Owens S., D'Alessandro U., Nosten F., Fischer P. R., Ordi J. (2004) The sick placenta: the role of malaria. Placenta 25, 359–378 [DOI] [PubMed] [Google Scholar]
- 9. Miller L. H., Baruch D. I., Marsh K., Doumbo O. K. (2002) The pathogenic basis of malaria. Nature 415, 673–679 [DOI] [PubMed] [Google Scholar]
- 10. Desai M., ter Kuile F. O., Nosten F., McGready R., Asamoa K., Brabin B., Newman R. D. (2007) Epidemiology and burden of malaria in pregnancy. Lancet Infect. Dis. 7, 93–104 [DOI] [PubMed] [Google Scholar]
- 11. Su X. Z., Heatwole V. M., Wertheimer S. P., Guinet F., Herrfeldt J. A., Peterson D. S., Ravetch J. A., Wellems T. E. (1995) The large diverse gene family var encodes proteins involved in cytoadherence and antigenic variation of Plasmodium falciparum-infected erythrocytes. Cell 82, 89–100 [DOI] [PubMed] [Google Scholar]
- 12. Smith J. D., Chitnis C. E., Craig A. G., Roberts D. J., Hudson-Taylor D. E., Peterson D. S., Pinches R., Newbold C. I., Miller L. H. (1995) Switches in expression of Plasmodium falciparum var genes correlate with changes in antigenic and cytoadherent phenotypes of infected erythrocytes. Cell 82, 101–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Flick K., Chen Q. (2004) var genes, PfEMP1 and the human host. Mol. Biochem. Parasitol. 134, 3–9 [DOI] [PubMed] [Google Scholar]
- 14. Kraemer S. M., Smith J. D. (2006) A family affair: var genes, PfEMP1 binding, and malaria disease. Curr. Opin. Microbiol. 9, 374–380 [DOI] [PubMed] [Google Scholar]
- 15. Salanti A., Dahlbäck M., Turner L., Nielsen M. A., Barfod L., Magistrado P., Jensen A. T., Lavstsen T., Ofori M. F., Marsh K., Hviid L., Theander T. G. (2004) Evidence for the involvement of VAR2CSA in pregnancy-associated malaria. J. Exp. Med. 200, 1197–1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Waller K. L., Nunomura W., An X., Cooke B. M., Mohandas N., Coppel R. L. (2003) Mature parasite-infected erythrocyte surface antigen (MESA) of Plasmodium falciparum binds to the 30-kDa domain of protein 4.1 in malaria-infected red blood cells. Blood 102, 1911–1914 [DOI] [PubMed] [Google Scholar]
- 17. Horrocks P., Pinches R. A., Chakravorty S. J., Papakrivos J., Christodoulou Z., Kyes S. A., Urban B.C., Ferguson D. J., Newbold C. I. (2005) PfEMP1 expression is reduced on the surface of knobless Plasmodium falciparum infected erythrocytes. J. Cell Sci. 118, 2507–2518 [DOI] [PubMed] [Google Scholar]
- 18. Rug M., Prescott S. W., Fernandez K. M., Cooke B. M., Cowman A. F. (2006) The role of KAHRP domains in knob formation and cytoadherence of P. falciparum-infected human erythrocytes. Blood 108, 370–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pei X., Guo X., Coppel R., Bhattacharjee S., Haldar K., Gratzer W., Mohandas N., An X. (2007) The ring-infected erythrocyte surface antigen (RESA) of Plasmodium falciparum stabilizes spectrin tetramers and suppresses further invasion. Blood 110, 1036–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Waller K. L., Cooke B. M., Nunomura W., Mohandas N., Coppel R. L. (1999) Mapping the binding domains involved in the interaction between the Plasmodium falciparum knob-associated histidine-rich protein (KAHRP) and the cytoadherence ligand P. falciparum erythrocyte membrane protein 1 (PfEMP1). J. Biol. Chem. 274, 23808–23813 [DOI] [PubMed] [Google Scholar]
- 21. Mayer C., Slater L., Erat M. C., Konrat R., Vakonakis I. (2012) Structural analysis of the Plasmodium falciparum erythrocyte membrane protein 1 (PfEMP1) intracellular domain reveals a conserved interaction epitope. J. Biol. Chem. 287, 7182–7189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Maier A. G., Cooke B. M., Cowman A. F., Tilley L. (2009) Malaria parasite proteins that remodel the host erythrocyte. Nat. Rev. Microbiol. 7, 341–354 [DOI] [PubMed] [Google Scholar]
- 23. Kilili G. K., LaCount D. J. (2011) An erythrocyte cytoskeleton-binding motif in exported Plasmodium falciparum proteins. Eukaryot. Cell 10, 1439–1447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Parish L. A., Mai D. W., Jones M. L., Kitson E. L., Rayner J. C. (2013) A member of the Plasmodium falciparum PHIST family binds to the erythrocyte cytoskeleton component band 4.1. Malar. J. 12, 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cooke B. M., Glenister F. K., Mohandas N., Coppel R. L. (2002) Assignment of functional roles to parasite proteins in malaria-infected red blood cells by competitive flow-based adhesion assay. Br. J. Haematol. 117, 203–211 [DOI] [PubMed] [Google Scholar]
- 26. Lanzer M., de Bruin D., Wertheimer S. P., Ravetch J. V. (1994) Transcriptional and nucleosomal characterization of a subtelomeric gene cluster flanking a site of chromosomal rearrangements in Plasmodium falciparum. Nucleic Acids Res. 22, 4176–4182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Biggs B. A., Kemp D. J., Brown G. V. (1989) Subtelomeric chromosome deletions in field isolates of Plasmodium falciparum and their relationship to loss of cytoadherence in vitro. Proc. Natl. Acad. Sci. U.S.A. 86, 2428–2432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Trenholme K. R., Gardiner D. L., Holt D. C., Thomas E. A., Cowman A. F., Kemp D. J. (2000) clag9: A cytoadherence gene in Plasmodium falciparum is essential for binding of parasitized erythrocytes to CD36. Proc. Natl. Acad. Sci. U.S.A. 97, 4029–4033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Goel S., Valiyaveettil M., Achur R. N., Goyal A., Mattei D., Salanti A., Trenholme K. R., Gardiner D. L., Gowda D. C. (2010) Dual stage synthesis and crucial role of cytoadherence-linked asexual gene 9 in the surface expression of malaria parasite var proteins. Proc. Natl. Acad. Sci. U.S.A. 107, 16643–16648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Srivastava A., Gangnard S., Round A., Dechavanne S., Juillerat A., Raynal B., Faure G., Baron B., Ramboarina S., Singh S. K., Belrhali H., England P., Lewit-Bentley A., Scherf A., Bentley G.A., Gamain B. (2010) Full-length extracellular region of the var2CSA variant of PfEMP1 is required for specific, high-affinity binding to CSA. Proc. Natl. Acad. Sci. U.S.A. 107, 4884–4889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Khunrae P., Dahlbäck M., Nielsen M. A., Andersen G., Ditlev S. B., Resende M., Pinto V. V., Theander T. G., Higgins M. K., Salanti A. (2010) Full-length recombinant Plasmodium falciparum VAR2CSA binds specifically to CSPG and induces potent parasite adhesion-blocking antibodies. J. Mol. Biol. 397, 826–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Acharya P., Chaubey S., Grover M., Tatu U. (2012) An exported heat shock protein 40 associates with pathogenesis-related knobs in Plasmodium falciparum infected erythrocytes. PLoS One 7, e44605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Grover M., Chaubey S., Ranade S., Tatu U. (2013) Identification of an exported heat shock protein 70 in Plasmodium falciparum. Parasite 20, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Maier A. G., Rug M., O'Neill M.T., Brown M., Chakravorty S., Szestak T., Chesson J., Wu Y., Hughes K., Coppel R. L., Newbold C., Beeson J. G., Craig A., Crabb B. S., Cowman A. F. (2008) Exported proteins required for virulence and rigidity of Plasmodium falciparum-infected human erythrocytes. Cell 134, 48–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Madhunapantula S. V., Achur R. N., Gowda D. C. (2007) Developmental stage- and cell cycle number-dependent changes in characteristics of Plasmodium falciparum-infected erythrocyte adherence to placental chondroitin-4-sulfate proteoglycan. Infect. Immun. 75, 4409–4415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Alkhalil A., Achur R. N., Valiyaveettil M., Ockenhouse C. F., Gowda D. C. (2000) Structural requirements for the adherence of Plasmodium falciparum-infected erythrocytes to chondroitin sulfate proteoglycans of human placenta. J. Biol. Chem. 275, 40357–40364 [DOI] [PubMed] [Google Scholar]
- 37. Achur R. N., Valiyaveettil M., Alkhalil A., Ockenhouse C. F., Gowda D. C. (2000) Characterization of proteoglycans of human placenta and identification of unique chondroitin sulfate proteoglycans of the intervillous spaces that mediate the adherence of Plasmodium falciparum-infected erythrocytes to the placenta. J. Biol. Chem. 275, 40344–40356 [DOI] [PubMed] [Google Scholar]
- 38. Greenwalt D. E., Lipsky R. H., Ockenhouse C. F., Ikeda H., Tandon N. N., Jamieson G. A. (1992) Membrane glycoprotein CD36: a review of its roles in adherence, signal transduction, and transfusion medicine. Blood 80, 1105–1115 [PubMed] [Google Scholar]
- 39. Achur R. N., Valiyaveettil M., Gowda D. C. (2003) The low sulfated chondroitin sulfate proteoglycans of human placenta have sulfate group-clustered domains that can efficiently bind Plasmodium falciparum-infected erythrocytes. J. Biol. Chem. 278, 11705–11713 [DOI] [PubMed] [Google Scholar]
- 40. Le Roch K. G., Zhou Y., Blair P. L., Grainger M., Moch J. K., Haynes J. D., De La Vega P., Holder A. A., Batalov S., Carucci D. J., Winzeler E. A. (2003) Discovery of gene function by expression profiling of the malaria parasite life cycle. Science 301, 1503–1508 [DOI] [PubMed] [Google Scholar]
- 41. Young J. A., Fivelman Q. L., Blair P. L., de la Vega P., Le Roch K. G., Zhou Y., Carucci D. J., Baker D. A., Winzeler E. A. (2005) The Plasmodium falciparum sexual development transcriptome: A microarray analysis using ontology-based pattern identification. Mol. Biochem. Parasitol. 143, 67–79 [DOI] [PubMed] [Google Scholar]
- 42. Zhou Y., Abagyan R. (2002) Match-only integral distribution (MOID) algorithm for high-density oligonucleotide array analysis. BMC Bioinformatics 3, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Maier A. G., Braks J. A., Waters A. P., Cowman A. F. (2006) Negative selection using yeast cytosine deaminase/uracil phosphoribosyl transferase in Plasmodium falciparum for targeted gene deletion by double crossover recombination. Mol. Biochem. Parasitol. 150, 118–121 [DOI] [PubMed] [Google Scholar]
- 44. Deitsch K. W., Calderwood M. S., Wellems T. E. (2001) Malaria: cooperative silencing elements in var genes. Nature 412, 875–876 [DOI] [PubMed] [Google Scholar]
- 45. Barfod L., Nielsen M. A., Turner L., Dahlbäck M., Jensen A. T., Hviid L., Theander T. G., Salanti A. (2006) Baculovirus-expressed constructs induce immunoglobulin G that recognizes VAR2CSA on Plasmodium falciparum-infected erythrocytes. Infect. Immun. 74, 4357–4360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bopp S. E., Manary M. J., Bright A. T., Johnston G. L., Dharia N. V., Luna F. L., McCormack S., Plouffe D., McNamara C. W., Walker J. R., Fidock D. A., Denchi E. L., Winzeler E. A. (2013) Mitotic evolution of Plasmodium falciparum shows a stable core genome but recombination in antigen families. PLoS Genet. 9, e1003293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Le Roch K. G., Zhou Y., Batalov S., Winzeler E. A. (2002) Monitoring the chromosome 2 intraerythrocytic transcriptome of Plasmodium falciparum using oligonucleotide arrays. Am. J. Trop. Med. Hyg. 67, 233–243 [DOI] [PubMed] [Google Scholar]
- 48. Kidgell C., Volkman S. K., Daily J., Borevitz J. O., Plouffe D., Zhou Y., Johnson J. R., Le Roch K., Sarr O., Ndir O., Mboup S., Batalov S., Wirth D. F., Winzeler E. A. (2006) A systematic map of genetic variation in Plasmodium falciparum. PLoS Pathog. 2, e57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gowda A. S. P., Madhunapantula S. V., Achur R. N., Valiyaveettil M., Bhavanandan V. P., Gowda D. C. (2007) Structural basis for the adherence of Plasmodium falciparum-infected erythrocytes to chondroitin 4-sulfate and design of novel photoactivable reagents for the identification of parasite adhesive proteins. J. Biol. Chem. 282, 916–928 [DOI] [PubMed] [Google Scholar]
- 50. Achur R. N., Kakizaki I., Goel S., Kojima K., Madhunapantula S. V., Goyal A., Ohta M., Kumar S., Takagaki K., Gowda D. C. (2008) Structural interactions in chondroitin 4-sulfate mediated adherence of Plasmodium falciparum infected erythrocytes in human placenta during pregnancy-associated malaria. Biochemistry 47, 12635–12643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Dzikowski R., Li F., Amulic B., Eisberg A., Frank M., Patel S., Wellems T. E., Deitsch K. W. (2007) Mechanisms underlying mutually exclusive expression of virulence genes by malaria parasites. EMBO Rep. 8, 959–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhang Q., Huang Y., Zhang Y., Fang X., Claes A., Duchateau M., Namane A., Lopez-Rubio J. J., Pan W., Scherf A. (2011) Critical role of perinuclear filamentous actin in spatial repositioning and mutually exclusive expression of virulence genes in malaria parasites. Cell Host Microbe. 10, 451–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Brolin K. J., Ribacke U., Nilsson S., Ankarklev J., Moll K., Wahlgren M., Chen Q. (2009) Simultaneous transcription of duplicated var2csa gene copies in individual Plasmodium falciparum parasites. Genome Biol. 10, R117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Jiang L., Mu J., Zhang Q., Ni T., Srinivasan P., Rayavara K., Yang W., Turner L., Lavstsen T., Theander T. G., Peng W., Wei G., Jing Q., Wakabayashi Y., Bansal A., Luo Y., Ribeiro J. M. C., Scherf A., Aravind L., Zhu J., Zhao K., Miller L. H. (2013) PfSETvs methylation of histone H3K36 represses virulence genes in Plasmodium falciparum. Nature 499, 223–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Marti M., Baum J., Rug M., Tilley L., Cowman A. F. (2005) Signal mediated export of the proteins from the malaria parasite to the host erythrocyte. J. Cell Biol. 171, 587–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sargeant T. J., Marti M., Caler E., Carlton J. M., Simpson K., Speed T. P., Cowman A. F. (2006) Lineage-specific expansion of proteins exported to erythrocytes in malaria parasites. Genome Biol. 7, R12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Cowman A. F., Coppel R. L., Saint R. E., Favaloro J., Crewther P. E., Stahl H.-D., Bianco A. E., Brown G. V., Anders R. F., Kemp D. J. (1984) The ring-infected erythrocyte surface antigen (RESA) polypeptide of Plasmodium falciparum contains two separate blocks of tandem repeats encoding antigenic epitopes that are naturally immunogenic in man. Mol. Biol. Med. 2, 207–221 [PubMed] [Google Scholar]
- 58. Silva M. D., Cooke B. M., Guillotte M., Buckingham D. W., Sauzet J.-P., Le Scanf C., Contamin H., David P., Mercereau-Puijalon O., Bonnefoy S. (2005) A role for the Plasmodium falciparum RESA protein in resistance against heat shock demonstrated using gene disruption. Mol. Microbiol. 56, 990–1003 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.