Abstract
The Achromobacter is a genus in the family Alcaligenaceae, comprising fifteen species isolated from different sources, including clinical samples. The ability to detect and correctly identify Achromobacter species, particularly A. xylosoxidans, and differentiate them from other phenotypically similar and genotypically related Gram-negative, aerobic, non-fermenting species is important for patients with cystic fibrosis (CF), as well as for nosocomial and other opportunistic infections. Traditional phenotypic profile-based analyses have been demonstrated to be inadequate for reliable identifications of isolates of Achromobacter species and genotypic-based assays, relying upon comparative 16S rRNA gene sequence analyses are not able to insure definitive identifications of Achromobacter species, due to the inherently conserved nature of the gene. The uses of alternative methodologies to enable high-resolution differentiation between the species in the genus are needed. A comparative multi-locus sequence analysis (MLSA) of four selected ‘house-keeping’ genes (atpD, gyrB, recA, and rpoB) assessed the individual gene sequences for their potential in developing a reliable, rapid and cost-effective diagnostic protocol for Achromobacter species identifications. The analysis of the type strains of the species of the genus and 46 strains of Achromobacter species showed congruence between the cluster analyses derived from the individual genes. The MLSA gene sequences exhibited different levels of resolution in delineating the validly published Achromobacter species and elucidated strains that represent new genotypes and probable new species of the genus. Our results also suggested that the recently described A. spritinus is a later heterotypic synonym of A. marplatensis. Strains were analyzed, using whole-cell Matrix-Assisted Laser Desorption/Ionization Time-Of-Flight mass spectrometry (MALDI-TOF MS), as an alternative phenotypic profile-based method with the potential to support the identifications determined by the genotypic DNA sequence-based MLSA. The MALDI-TOF MS data showed good accordance in strain groupings and identifications by the MLSA data.
Introduction
The Gram-negative, aerobic, non-fermenting bacteria are ubiquitously present in various ecosystems, important for environmental and biotechnological applications and many of these microorganisms have become problematic in hospital settings. Species of Pseudomonas, Burkholderia, Acinetobacter and Stenotrophomonas are the leading nosocomial pathogens in this expanding group [1], [2] and genera of the family Alcaligenaceae, i.e., Alcaligenes, Ralstonia, Achromobacter, etc., are emerging, as well, as significant pathogens in notable patient populations [3], particularly those suffering from cystic fibrosis (CF). Recent studies have reported as many as 5 to 10% of colonizing bacteria in respiratory tract samples from CF patients are Achromobacter species [1]; earlier studies also reported approximately 5% of CF patients examined were colonized with A. xylosoxidans [4]; 3 to 4% of CF patients exhibit chronic colonizations and approximately 2% sporadic colonizations [5], [6]. Infections by A. xylosoxidans in CF patients have been observed to lead to decline in lung function [7], [8]. The ability to detect and correctly identify Achromobacter species, particularly A. xylosoxidans, and differentiate them from other phenotypically similar and genotypically related Gram-negative, aerobic, non-fermenting species is increasingly important. Misidentifications compromise infection control measures and confound efforts to recognise the epidemiology of infections. The growing number of species and increasing complexity of bacterial taxonomy and the expansion of virulence and antibiotic resistance present significant challenges, requiring new development and periodic optimisation of identification protocols for new, as well as already described taxa.
Achromobacter is one of 19 genera belonging to the family Alcaligenaceae, in the class Betaproteobacteria [9]–[12] and the taxonomy of Achromobacter has been closely intertwined with that of the genus Alcaligenes [10]; several species of Alcaligenes have been reclassified as Achromobacter. Achromobacter comprises 15 species: A. xylosoxidans (ex Yabuuchi and Ohyama 1971) Yabuuchi and Yano 1981, sp. nov., nom. rev. emend. (Type species of the genus) [10]; A. ruhlandii (Packer and Vishniac 1955) (Yabuuchi et al. 1998, comb. nov.) [10]; A. piechaudii (Kiredjian et al. 1986) Yabuuchi et al. 1998, comb. nov. [10]; A. denitrificans (Ruger and Tan 1983) Coenye et al. 2003, comb. nov. [13]; A. insolitus Coenye et al. 2003, sp. nov. [14]; A. spanius Coenye et al. 2003, sp. nov. [14]; A. marplatensis Gomila et al. 2011, sp. nov. [15]; A. animicus; A. mucicolens; A. pulmonis; and A. spiritinus Vandamme et al., 2013a [16]. Recently, four new species have been described: A. insuavis sp. nov.; A. aegrifaciens sp. nov.; A. anxifer sp. nov.; and A. dolens Vandamme et al., 2013b [17]. These species were isolated from different sources, including clinical samples. A. xylosoxidans is widely distributed in the environment and is also recognized to be an opportunistic human pathogen, associated with a variety of infections, including bacteraemia, meningitis, pneumonia, and peritonitis [18]–[20]. Nosocomial outbreaks attributed to disinfectant solutions, dialysis fluids, saline solutions and deionised water contaminated with this species have been described [21], [22]. A. xylosoxidans presents significant problems for persistent infection of the respiratory tract in persons with CF [4], although the precise role in contributing to pulmonary decline in this patient population is not clear. However, due to the well-known difficulties in differentiating the species of Achromobacter, it must be appreciated that isolates identified as ‘A. xylosoxidans’ may, in fact, comprise different Achromobacter species. A. ruhlandii is considered to be a soil inhabitant, although it has been associated also with human clinical conditions [23]. A. piechaudii has been isolated from human clinical samples, including blood, as well as from soil [24]. A. insolitus and A. spanius were isolated initially from leg wound and blood samples, respectively [14]. A. denitrificans strains are found typically in soil and water but can occasionally be found in human clinical samples [13], [23]. A. animicus, A. mucicolens, A. pulmonis, A. spiritinus [16], [25], A. aegrifaciens, A. anxifer, A. dolens and A. insuavis [17] were isolated from the sputa of patients with and without CF, as well as from water and sludge.
Traditional phenotypic-based analyses have been demonstrated to be inadequate for reliable, definitive identifications of Achromobacter species [12], [15], [25]. Recognizing the limitations of phenotypic-based identifications of bacteria, genotypic-based phylogeny has been recommended as the basis for the taxonomy of microorganisms [26]. While whole genome sequences ultimately will constitute the paramount reference standard for microbial phylogeny, sequence determinations of ‘biomarker’ genes, such as those for 16S rRNA, will continue to provide a basis for determining microbial phylogenetic relationships, taxonomy and identification. The comparative analyses of 16S rRNA gene sequences are used as part of the standard protocol by clinical laboratories, particularly for reliable, initial, estimates of the identifications of isolates. However, comparative 16S rRNA gene sequence analyses do not engender the resolution required to ensure definitive delineation of closely related bacterial species, due to the conserved nature of the gene; all known Achromobacter species exhibit sequence dissimilarities less than 1% to each other. Protein-coding ‘house-keeping’ gene sequence data provide higher-resolution differentiation for reliable identification of closely related species [27] and compilations of multiple gene sequences complement their differentiation [28]. During the course of this study, two research groups developed independent multiple-locus approaches to facilitate the analyses of Achromobacter species [25], [29]. While multi-locus sequence analysis (MLSA) and multi-locus sequence typing (MLST) approaches enhance the insight into the systematic relationships and population dynamics of bacterial taxa, such approaches are complex and not always practical for rapid and cost-effective microbial identifications for clinical diagnoses. The ideal DNA sequence-based protocol for clinical diagnostics would be one in which a single target gene would afford the resolution necessary for reliable species-level differentiation and identifications, elucidated by one or two sequencing reactions. The focus of this study was to assess selected house-keeping gene sequences to identify marker genes that can be used for reliable differentiation and identification of clinical isolates of Achromobacter species.
Analyses, using Matrix-Assisted Laser Desorption/Ionization Time-Of-Flight mass spectrometry (MALDI-TOF MS) have not been evaluated previously for the potential for identification of the individual species in the Achromobacter genus. The Achromobacter species reference data included in the VITEK MS IVD (BioMérieux, Inc.) identification system are limited to A. xylosoxidans and A. denitrificans, although the SARAMIS software (Anagnostec GmbH/bioMérieux, Inc.) [30] included the seven species of Achromobacter that were validly published until 2010; the MALDI Biotyper (Bruker Corp.) identification system includes A. xylosoxidans, A. ruhlandii, A. piechaudii, A. denitrificans, A. insolitus, and A. spanius [31]. A comprehensive assessment of MALDI-TOF MS for identifying all species of Achromobacter offers a complementary methodology for comparison with DNA sequence-based approaches. The MALDI-TOF MS identifications of the Achromobacter species, analysed in this study are compared and correlated with the MLSA identifications.
Materials and Methods
Bacterial strains and growth conditions
All strains used in this study were obtained from the Culture Collection University of Gothenburg (CCUG; www.ccug.se), including the type strains of eleven validly published species of the genus Achromobacter: A. xylosoxidans CCUG 56438T (the type species of the genus); A. ruhlandii CCUG 57103T; A. piechaudii CCUG 724T; A. denitrificans CCUG 407T; A. insolitus CCUG 47057T; A. spanius CCUG 47062T; A. marplatensis CCUG 56371T; A. animicus CCUG 61966T; A. mucicolens CCUG 61961T; A. pulmonis CCUG 61972T; A. spiritinus CCUG 61968T; and 46 well-characterized strains of Achromobacter species of clinical and environmental origin. The sources of these isolates are diverse, including clinical samples (human sputum, respiratory tract from CF patients, synovial fluid samples, eye secretions, mucous samples of human cheek, choledochal cyst secretions, human tracheal secretions, pleural fluids, human wounds, bronchoalveolar lavage, otitis media, and environmental samples (soil, chicken tracheal samples, laboratory wash system) (Table S1). The type strain of the type species of the genus Bordetella, B. pertussis CCUG 30873T, was included as an out-group. Isolates were cultured on 5% Blood Agar and on Nutrient Agar media, at 30°C, 24–48 hours.
DNA extraction, PCR amplification and DNA sequencing
Bacterial genomic DNA for PCR amplifications was extracted as previously described [32]. Five genes were selected for the MLSA: 16S rRNA; atpD (encoding the β subunit of ATP synthase), gyrB (encoding the β-subunit of DNA gyrase); recA (encoding the α-subunit of recombinase); and rpoB (encoding the β subunit of the RNA polymerase). Primers used for PCR amplifications and sequencing are listed in Table 1. PCR amplification primers for 16S rRNA genes, gyrB and rpoB were described previously [33]–[35]. Primers for the amplification of gyrB and rpoB were modified, according to the sequences of analyzed type strains. Primers used for amplification of atpD and recA were derived through alignment of the gene sequences from available whole genome sequence data of bacteria in the family Alcaligenaceae: Bordetella avium 197N; Bordetella pertussis Tohama I; Bordetella petrii DSM 12804; Burkholderia cenocepacia AU1054; Burkholderia mallei ATCC 23344; Burkholderia ambifaria MC40-6; Ralstonia solanacearum GMI1000; Ralstonia pickettii 12J; Cupriavidus metallidurans CH34; Cupriavidus necator H16; Cupriavidus taiwanensis LMG 19424; Janthinobacterium sp.; and Herminiimonas arsenicoxydans ULPAs1. Primers were designed for amplification of the sequence regions with the highest incidence of polymorphic sites in these genes. Internal sequencing primers were designed according to the alignments of the sequences of the type strains. PCR amplifications were carried out as previously described [15]. The amplification reactions were performed in an Eppendorf thermocycler, with an initial denaturation step of 5 min at 94°C, followed by 35 cycles of 1 min at 94°C, 1 min at the annealing temperature for each gene (55°C for 16S rRNA gene and 58°C for atpD, gyrB and recA genes) and 1 min 30 sec at 72°C. For the atpD gene, the initial denaturation step was 95°C for 2 min, followed by 35 cycles of 1 min at 95°C, 1 min at 60°C and 1 min 30 s at 72°C. Following the amplification cycles, samples were incubated at 72°C for 10 min and then cooled to 4°C. PCR amplicons of the targeted genes were purified, using the MultiScreen HTS PCR 96-well Filter Plates (Millipore) and sequenced directly, using the ABI PRISM BigDye Terminator Cycle Sequencing Kit version 3.1, according to the instructions of the manufacturer (Applied Biosystems, Inc.). Sequences were determined, using an ABI PRISM 3100 Avant-Genetic Analyzer and a 3130 Genetic Analyzer (Applied Biosystems).
Table 1. Primers used for PCR-amplification and sequencing in this study.
| Gene | Primer | Sequence (5′→ 3′) | Product Size (bp) | Reference | |
| 16S rRNA | 16F27 | PCR | AGAGTTTGATCMTGGCTCAG | 1400 | Lane, 1991 |
| 16R1492 | PCR | TACGGYTACCTTGTTACGACTT | Lane, 1991 | ||
| 16F357 | Sequencing | ACTCCTACGGGAGGCAGCAG | Lane, 1991 | ||
| 16R518 | Sequencing | CGTATTACCGCGGCTGCTGG | Lane, 1991 | ||
| gyrB | gyrB1F | PCR | ACAACGGCCGCGGSATTCC | 1020 | Tayeb et al., 2008* |
| UgyrBR | PCR | GCNGGRTCYTTYTCYTGRCA | Yamamoto et al., 2000 | ||
| gyrBF433 | Sequencing | ACAATGGCGTSAAGATCCGC | This study | ||
| gyrBR599 | Sequencing | AGCTGTCGTTCCACTGCATCG | This study | ||
| rpoB | rpoB-F | PCR | NGGCGAAATGGCDGARAACC | 1040 | Tayeb et al., 2008* |
| rpoB-R | PCR | NNGARTCYTCGAAGTGGTAACC | Tayeb et al., 2008* | ||
| rpoBF404 | Sequencing | GTACGGCTTCCTGGAAACGC | This study | ||
| rpoBR607 | Sequencing | GCAMGGCACGGCCTGGCG | This study | ||
| recA # | recAF126 | PCR | NCAGATYGARAAGCAGTTTYGG | 770 | This study |
| recAF196 | Sequencing | AGGTNGTNTCSACSGGNTCGC | This study | ||
| recAR928 | PCR | RCCGYYRTAGSYRTACCASGC | This study | ||
| recAR1015 | PCR | CGCGNAYNYKRTTYTCGATCTC | This study | ||
| atpD | atpDF30 | PCR | YTTCTTGGCCTTYTCGAAGGC | 900 | This study |
| atpDF63 | Sequencing | CCGACCATGTAGAASGCCTG | This study | ||
| atpDR1130 | Sequencing | GCATCATGGACGTGCTSGG | This study | ||
| atpDR1172 | PCR | GGCRMNCCGATYTCGGTGCC | This study |
*This primer has been modified from the original primer.
For recA gene, the primer combination recAF126 and recAR928 was used. When the PCR failed, recAF126 and recAR1015 combination was used.
Sequence analysis
Sequences obtained for each of the genes analyzed were assembled, manually corrected and compared to publically available sequences in Genbank, using the BLAST (Basic Local Alignment Search Tool) algorithm of the NCBI (National Center for Biotechnology Information) [36]. Alignments were performed by a hierarchical multiple alignment method implemented in the program Clustal X [37]. Sequences aligned automatically were checked manually. The evolutionary distances derived from the pair-wise differences between sequences (Jukes-Cantor correction, [38]) were calculated, using the program DNADIST, included in the phylogenetic inference package (PHYLIP 3.69) [39]. Cluster analyses and phylogenetic trees were constructed, using the Neighbor-joining distance method. Bootstrap analyses, with 1,000 repeats, were performed, using the PHYLIP program. Bootstrap values greater than 500 are indicated in the respective trees. The topologies of phylogenetic trees were visualized with the program TreeView [40]. In addition to individual cluster analyses determined for each gene, a tree derived from the concatenated sequences of the protein-coding genes was also constructed to compare the robustness of the Achromobacter intra-generic branching order. Similarity matrices were calculated, using the Bionumerics, version 7 software (Applied Maths NV, Sint-Martens-Latem, Belgium), and evolutionary reconstruction analyses were conducted, using the MEGA 5 program [41].
Allele diversity and polymorphic sites
Allele diversity and polymorphic sites were calculated with the DnaSP package, version 3.51 [42].
Nucleotide sequence accession numbers
The nucleotide sequences determined in this study have been deposited in the EMBL database under the following accession numbers: 16S rRNA gene, HG423398 to HG423445; atpD, HG454790 to HG454847; gyrB, HG454848 to HG454905; recA, HG454906 to HG454963 and rpoB, HG454964 to HG455021. GeneBank accession numbers for the sequences of the strains used in the study are available in Table S1.
MALDI-TOF MS analyses
Matrix-Assisted Laser Desorption/Ionization-Time-Of-Flight mass spectrometry (MALDI-TOF MS) profiles for the strains studied were performed at Anagnostec, GmbH, Germany [30] and at the CCUG and the Department of Clinical Microbiology, Sahlgrenska University Hospital. Strain biomass were analyzed on a Flexi Mass stainless steel target, using a whole-cell protocol with 1 µL matrix solution of saturated α-cyano-4 hydroxy-cinnamic acid in a mixture of acetonitrile:ethanol:water (1∶1∶1) acidified with 3% (v/v) trifluoroacetic acid. For each strain, mass spectra were prepared in duplicate and analyzed on an AXIMA Confidence instrument (Shimadzu/Kratos, Manchester, UK), in the linear positive ion extraction mode. Mass spectra were accumulated from 100 profiles, each from five nitrogen laser pulse cycles, by scanning the entire sample spot. Ions were accelerated with pulsed extraction at a voltage of 20 kV. Raw mass spectra were processed automatically for baseline correction and peak recognition. Resulting mass fingerprints were exported to the SARAMIS (Spectral Archiving and Microbial Identification System, Release 3.36, Anagnostec GmbH, Germany) analysis program and compared to reference spectra. The percentage similarities of identical mass peaks were calculated and used to generate dendrograms, applying single-linkage agglomerate calculations. The spectra strains were also analyzed using the VITEK MS IVD (bioMérieux, Inc.) version 2.0.
Phenotypic profiling
Phenotypic characterizations were performed on all strains, using customized protocols of the CCUG Typing Laboratory (http://www.ccug.se/default.cfm?navID=160) for Gram-negative, non-fermentative bacteria. The various tests included the API commercial strips for physiological profiling, API 20NE and API ZYM (bioMérieux, Inc.), and other metabolic and morphological features (Table S2).
DNA-DNA hybridization
Total genomic DNA was isolated using the Wizard Genomic DNA Purification Kit (Promega), following the manufacturer’s instructions, and DNA-DNA hybridizations were performed, in duplicate, using a non-radioactive method described previously [43]. Reference DNA of A. marplatensis CCUG 56371T and A. spiritinus CCUG 61988T were double-labeled with DIG-11-dUTP and Biotin-16-dUTP, using a Nick Translation Kit (Boehringer, Mannheim, Germany). Labeled DNA was hybridized with the DNA of A. marplatensis CCUG 56371T, A. marplatensis CCUG 61988T, A. ruhlandii CCUG 57103T, A. xylosoxidans CCUG 56438T and A. spiritinus strains CCUG 61969 and CCUG 61970.
Results
Multi-locus sequence analysis
An MLSA assessment of five selected house-keeping genes has been carried out for the Achromobacter species and applied to 46 strains of clinical and environmental origin. The sequences of the 16S rRNA gene, atpD, gyrB, recA, and rpoB were analyzed for the type strains of 11 validly published species of the genus. Partial sequences of the 16S rRNA genes, including nucleotide positions 28 to 500 (Escherichia coli 16S rRNA gene sequence numbering) were extracted from the nearly-complete sequences and the partial-sequence similarities determined and compared with those of the nearly-complete gene sequences. The 16S rRNA gene sequence similarities between the type strains of the species ranged from 99.1% to 100% for single-primer partial sequences (472 nucleotide positions) and from 99.0% to 100% for the nearly complete gene sequences (Figure S1a in File S1). The differences between the pair-wise similarities of the partial 16S rRNA gene sequences and the nearly complete gene sequences of the type strains of the Achromobacter species differed by an average of only 0.24%. Thus, the partial 16S rRNA gene sequences, using a single primer reaction for determining the sequences of the 5′-region of the gene, provided pair-wise similarities that are comparable with the similarities determined from nearly complete sequence comparisons. However, the problem with using 16S rRNA gene sequences is the inherently low level of resolution that exists between the sequences of the different species. Consequently, cluster analysis of the Achromobacter species, based upon their 16S rRNA gene sequences, exhibited little or no delineation between the species and was not further used on the MLSA study.
In the cases of protein-coding gene comparisons, the respective similarity tables were generated showing the ranges of similarities between the nucleotide sequences and also for the translated amino acid sequences of the species (Figures S2–S5 in File S1). Alignment and comparative sequence analyses of the selected house-keeping genes, atpD, gyrB, recA, and rpoB, exhibited lower levels of similarity and the respective gene trees showed greater discrimination than the 16S rRNA gene sequences between the species of Achromobacter (Figures S2–S5 in File S1). The sequence similarities for the house-keeping genes between the type strains of the Achromobacter species ranged from 94.6% to 99.3% for atpD (Figure S2a in File S1), 91.7% to 98.5% for gyrB (Figure S3a in File S1), 89.9% to 97.9% for recA (Figure S4a in File S1), and 92.1% to 99.8% for rpoB (Figure S5a in File S1). The branching orders of the sequences of the different house-keeping genes showed a high degree of consistency, i.e., the most closely related species were observed to be the same in all the genes studied (Figures S2b–S5b in File S1). The type strains of two pairs of species: A. xylosoxidans CCUG 56438T and A. ruhlandii CCUG 57103T; and A. marplatensis CCUG 56371T and A. spiritinus CCUG 61968T exhibited the highest pair-wise similarities in all comparative house-keeping gene sequence analyses.
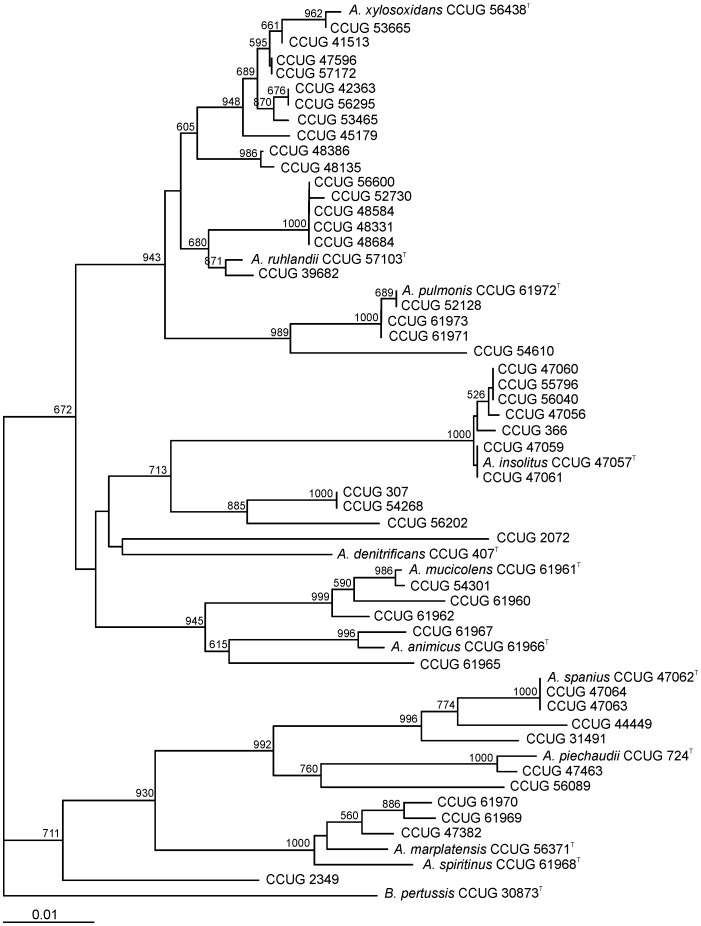
Forty-six strains (Table S1) that had been isolated from clinical and environmental samples and identified prior to this study as Achromobacter species were analyzed by 16S rRNA gene and by house-keeping gene sequence comparisons. Using the sequencing primer, 16R518 (Table 1), 472 nucleotide positions of the 16S rRNA gene were used for initial estimations of taxonomic identities. For the house-keeping gene sequences, the sequencing primers (Table 1) used determined: 621 nucleotide positions for recA; 528 nucleotide positions for gyrB; 527 nucleotide positions for rpoB; and 513 nucleotide positions for atpD. The pair-wise sequence similarities determined between the clinical strains and the type strains of the Achromobacter species ranged from 97.6% to 100% for the 16S rRNA gene, 92.4% to 100% for atpD, 89.7% to 100% for gyrB, 87.4% to 100% for recA, and 91.1% to 100% for rpoB. Individual sequence similarities and evolutionary distances were calculated and dendrograms from the cluster analyses were generated for recA (Figure 1) and for each other house-keeping gene (Figures S6–S8 in File S1). The recA, as well as atpD, gyrB, and rpoB cluster analyses demonstrated greater discrimination than the 16S rRNA gene tree (Figure S9 in File S1) for the species in the Achromobacter genus. A concatenated analysis of atpD, gyrB, recA and rpoB genes was also performed (Figure S10 in File S1).
Figure 1. Phylogenetic tree of the strains of Achromobacter used in this study based on the phylogenetic analysis of recA gene.
Distance matrix was calculated by the Jukes-Cantor method. Dendrogram was generated by neighbor-joining. Bordetella pertussis CCUG 30873T was used as an outgroup. The bar indicates sequence divergence. Bootstrap values of more than 500 (from 1000 replicates) are indicated at the nodes.
The individual protein-coding gene dendrograms showed high levels of congruence in the branching order of the species. Overall, the recA dissimilarities were greater than those observed for the other three genes assessed in this study, providing the best discrimination between species of Achromobacter and, in all cases, clear inter-species delineations, except for A. marplatensis and A. spiritinus. The recA pair-wise similarities between the type strains of the species ranged from 89.9% (between A. piechaudii and A. pulmonis and between A. spanius and A. pulmonis) to 97.6% and 97.9% (between A. ruhlandii and A. xylosoxidans and between A. marplatensis and A. spiritinus, respectively).
Some of the clinical and environmental strains tested were closely related to one of the species type strains (Table 2), but others (7 strains) did not cluster closely to the type strain of any described species, with similarities below the highest recA similarity noted for the pair-wise similarities between the type strains of the most closely related species, probably representing novel species. The same result was observed when cluster analyses derived from concatenated gene sequences were considered. High bootstrap values for the branching points confirmed the robustness of the gene sequence trees. Comparative analyses of translated nucleotide sequences to amino acid sequences for the genes studied showed that the topologies of the cluster analyses were maintained, although calculated dissimilarities were lower than those observed for nucleotide sequences.
Table 2. Genotypic analyses and individual recA- and concatenated MLSA-based identifications of clinical and environmental strains of Achromobacter species.
| Concatenated MLSA | recA analysis | |||
| Strain | Sequence similarity# (%) | Closest species match or speciesassignation | Sequence similarity (%) | Closest species match or speciesassignation* |
| CCUG 307* | 96.8 | A. denitrificans | 95.3 | A. denitrificans |
| CCUG 366 | 99.4 | A. insolitus | 99.7 | A. insolitus |
| CCUG 2072* | 95.6 | A. denitrificans | 93.6 | A. denitrificans |
| CCUG 2349* | 95.6 | A. ruhlandii | 94.5 | A. ruhlandii |
| CCUG 31491 | 98.7 | A. spanius | 97.9 | A. spanius |
| CCUG 39682 | 99.3 | A. ruhlandii | 99.5 | A. ruhlandii |
| CCUG 41513 | 99.5 | A. xylosoxidans | 99.4 | A. xylosoxidans |
| CCUG 42363 | 98.8 | A. xylosoxidans | 98.7 | A. xylosoxidans |
| CCUG 44449 | 98.7 | A. spanius | 97.9 | A. spanius |
| CCUG 45179 | 98.9 | A. xylosoxidans | 98.3 | A. xylosoxidans |
| CCUG 47056 | 99.6 | A. insolitus | 99.7 | A. insolitus |
| CCUG 47059 | 99.8 | A. insolitus | 100 | A. insolitus |
| CCUG 47060 | 99.5 | A. insolitus | 99.8 | A. insolitus |
| CCUG 47061 | 99.8 | A. insolitus | 100 | A. insolitus |
| CCUG 47063 | 100 | A. spanius | 100 | A. spanius |
| CCUG 47064 | 100 | A. spanius | 100 | A. spanius |
| CCUG 47382I | 99.2/99.0 | A. spiritinus/A. marplatensis | 98.2/98.0 | A. marplatensis/A. spiritinus |
| CCUG 47463 | 99.5 | A. piechaudii | 99.3 | A. piechaudii |
| CCUG 47596 | 99.3 | A. xylosoxidans | 99.2 | A. xylosoxidans |
| CCUG 48135 | 96.9 | A. ruhlandii | 98.2 | A. ruhlandii |
| CCUG 48331 | 98.6 | A. xylosoxidans/A. ruhlandii | 98.5 | A. ruhlandii |
| CCUG 48386 | 97.6 | A. ruhlandii | 98.4 | A. ruhlandii |
| CCUG 48584 | 98.6 | A. xylosoxidans/A. ruhlandii | 98.5 | A. ruhlandii |
| CCUG 48684 | 98.6 | A. xylosoxidans/A. ruhlandii | 98.5 | A. ruhlandii |
| CCUG 52128 | 99.9 | A. pulmonis | 100 | A. pulmonis |
| CCUG 52730 | 98.6 | A. ruhlandii | 98.4 | A. ruhlandii |
| CCUG 53465 | 98.6 | A. xylosoxidans | 98.7 | A. xylosoxidans |
| CCUG 53665 | 99.7 | A. xylosoxidans | 99.8 | A. xylosoxidans |
| CCUG 54268* | 96.7 | A. denitrificans | 95.3 | A. denitrificans |
| CCUG 54301 | 97.7 | A. mucicolens | 99.8 | A. mucicolens |
| CCUG 54610* | 97.2 | A. ruhlandii | 96.9 | A. pulmonis |
| CCUG 55796 | 99.7 | A. insolitus | 99.8 | A. insolitus |
| CCUG 56040 | 99.6 | A. insolitus | 99.8 | A. insolitus |
| CCUG 56089* | 96.7 | A. spanius | 95.5 | A. piechaudii |
| CCUG 56202* | 96.4 | A. denitrificans | 95.2 | A. denitrificans |
| CCUG 56295 | 98.7 | A. xylosoxidans | 98.7 | A. xylosoxidans |
| CCUG 56600 | 98.6 | A. xylosoxidans/A. ruhlandii | 98.5 | A. ruhlandii |
| CCUG 57172 | 99.6 | A. xylosoxidans | 99.2 | A. xylosoxidans |
| CCUG 61960 | 98.6 | A. mucicolens | 98.5 | A. mucicolens |
| CCUG 61962 | 99.0 | A. mucicolens | 98.5 | A. mucicolens |
| CCUG 61965* | 97.6 | A. animicus | 95.9 | A. animicus |
| CCUG 61967 | 98.9 | A. animicus | 99.2 | A. animicus |
| CCUG 61969I | 99.2/99.1 | A. spiritinus/A. marplatensis | 98.7/98.2 | A. marplatensis/A. spiritinus |
| CCUG 61970I | 99.1/98.8 | A. spiritinus/A. marplatensis | 98.4/97.9 | A. marplatensis/A. spiritinus |
| CCUG 61971 | 99.9 | A. pulmonis | 99.8 | A. pulmonis |
| CCUG 61973 | 99.9 | A. pulmonis | 99.8 | A. pulmonis |
*indicates novel species, i.e., with recA similarities <97.6% to a recognized species.
determined from the concatenated sequences of atpD, gyrB, recA and rpoB.
Vandamme et al., 2013a described A. spiritinus as a species, distinct from A. marplatensis.
Determination of gene sequence polymorphic sites
The number of polymorphic sites and the allele diversity for the house-keeping genes were determined for the 57 strains included in this study (Table S3). The number of polymorphic sites in the four protein-coding loci studied ranged from 70 for atpD gene to 151 for recA gene. The recA gene sequence was the most discriminating of the genes analyzed, with the highest average number of nucleotide differences.
In order to compare the results of this study with those obtained in two other studies that described DNA sequence-based methods for the analyses of Achromobacter species [25], [29], the number of polymorphic sites for the gene sequence regions analyzed in those studies, i.e., atpD, recA and rpoB, were compared for the type strains of eleven validly published species (Table S4). For atpD, recA and rpoB, different regions of the gene sequences were analysed and, in all three cases, the sequence regions analysed in this study contained more polymorphic sites, providing potential for higher degrees of discrimination between species. Results were also compared with nrdA gene sequences (Table S4), proposed as a single locus sequencing tool for Achromobacter speciation (Spilker et al., 2013). The short and long nrdA sequences exhibited 60 and 115 polymorphic sites, respectively; and 27.6 and 50.4 average number of nucleotide differences, respectively.
MALDI-TOF MS analyses
The type strains of the species of Achromobacter and the strains of Achromobacter species analyzed in this study were all identified as ‘A. denitrificans/A. xylosoxidans’, using the VITEK MS IVD Database (bioMérieux, Inc.) as reference. However, using the SARAMIS Database (bioMérieux, Inc.) [30], the MALDI-TOF MS mass signal profiles of the type strains of the eleven species were distinct (Figure 2). The MALDI-TOF MS branching order of the species in Achromobacter was observed to be different from that derived from the recA gene and the concatenated sequences of the house-keeping gene MLSA (Figure 1 and Figure S10 in File S1). However, in the cases of all Achromobacter species, the most closely related species determined by MALDI-TOF MS was observed to be the closest related species also by MLSA. The strains clustering with the type strains of a given species, by MALDI-TOF MS profiling, i.e., at 70% similarity delineation, were observed to cluster, in most cases, with the type strain of the respective species by concatenated house-keeping gene MLSA, as well as by single-gene recA cluster analysis. The closely related species, A. xylosoxidans and A. ruhlandii showed a close relationship by MALDI-TOF MS analysis, with A. ruhlandii branching outside of the cluster of the A. xylosoxidans type strain and other strains clustering with A. xylosoxidans by MLSA, albeit with similarities lower than 70%. A. marplatensis and A. spiritinus grouped closely together by MALDI-TOF MS analysis, in two delineated branches, with a similarity at approximately 70%.
Figure 2. Dendogram of relatedness between the Achromobacter species strains analyzed based on MALDI-TOF MS analysis.
The VITEK MS IVD database (bioMérieux, Inc.) for bacterial identifications does not yet include all species of the Achromobacter genus; the only identifications possible for most strains of Achromobacter species has been an identification of ‘A. denitrificans/A. xylosoxidans’.
Phenotypic characterization
The colonies of all strains grown on nutrient agar were whitish in color, small, i.e., 1 to 2 mm in diameter, mucoid, nonhemolytic, catalase positive and oxidase positive. All strains were analyzed by API ZYM and API 20NE metabolic profiling and by a series of customized tests that have been applied by the CCUG Typing Lab (www.ccug.se). The phenotypic analyses did not allow consistent, effective differentiation characterization of the strains of a given species from those of other species (Table S2).
DNA-DNA hybridization analyses
In all cases of the genes analyzed, the type strains of A. marplatensis (CCUG 56371T) and A. spiritinus (CCUG 61968T) possessed the highest pair-wise similarities among the recognized species (98.8% concatenated sequence similarities and 97.9% in the case of recA gene). Such high sequence similarities for the A. marplatensis-A. spiritinus type strains pairs, when all other type strains of Achromobacter, except A. ruhlandii and A. xylosoxidans, exhibited significantly lower pair-wise sequence similarities, suggested the possibility of misclassification of A. spiritinus. Further analyses, including DNA-DNA hybridization (DDH), were done to confirm and clarify the taxononomic affiliation of A. spiritinus. The DDH similarity between A. marplatensis and A. spiritinus showed values greater than 80%, confirming that A. spiritinus should be considered to be the same species as A. marplatensis. DDH similarity values between A. marplatensis and the types strains used as controls, A. ruhlandii and A. xylosoxidans, were lower than 44%; when A. spiritinus was labelled, the values were lower than 55%. These data, were not in accordance with the recent description and valid publication of A. spiritinus sp. nov. [16] and led us to contact the curators of the Achromobacter PubMLST database, where the MLST profiles of both species were re-examined. The results of re-examination of the MLST data concluded that the original MLST data for the A. marplatensis type strain was incorrect, such that the MLST data of A. spiritinus, determined later, was not recognised to be indistinct. The MLST data for the A. marplatensis type strain has been corrected in the PubMLST database and these sequence data have been made available to the public (http://pubmlst.org/achromobacter/).
Discussion
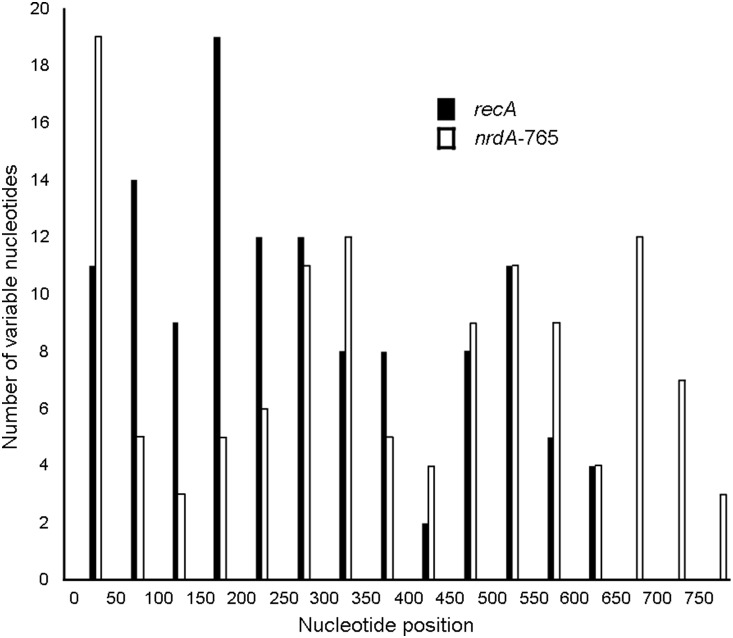
The MLSA in this study assessed selected house-keeping gene targets that could be applied as biomarkers for identification of the species of the genus Achromobacter. In the course of this study, two different research groups, in independent studies, described MLST and MLSA approaches for high-resolution discrimination of strains of individual species (i.e., by MLST) and the differentiation of Achromobacter species (i.e., by MLSA) [25], [29]. Spilker et al. (2013) assessed the ability of nrdA sequence analysis, one of the seven genes described in the previous MLSA scheme, to differentiate Achromobacter species [6], [25]. However, these studies did not focus on defining a protocol that could be applied for the reliable, rapid and cost-effective differentiation and identification of clinically-relevant strains of Achromobacter species for the diagnoses of infections. Our analyses have shown that recA sequence analyses provide a higher resolution tool for the identification of Achromobacter species. It is clear from studies reported in the literature and from the results of this study that traditional metabolic profile-based phenotypic testing does not allow for consistent differentiation of the species of Achromobacter [12], [15], [16], [25]. Furthermore, comparative sequence analysis of 16S rRNA genes, i.e., partial sequences or nearly complete gene sequences, possess limited value for differentiating and identifying Achromobacter species, due to the inherent high degrees of similarity between the sequences of the different species. Thus, additional biomarker targets of functionally-conserved, ‘house-keeping’ gene sequences, such as atpD, gyrB, recA and rpoB, offer potential, alternative species-level identification tools, since they are present in all species of the genus and they exhibit degrees of variation higher than what is observed for the 16S rRNA genes. The focus of this study was to elucidate a single house-keeping gene that would prove applicable for the delineation and identification of all species of the genus Achromobacter and develop a reliable, rapid and cost-effective protocol based upon comparative DNA sequence analyses. The results obtained from sequence alignments and similarity determinations demonstrated that the RecA gene has the most discriminatory sequence with the highest degree of inter-species variation. The branching order of the Achromobacter species, derived from cluster analyses, is maintained, for the most part, for all individual gene sequence analyses, as well as a concatenated analysis of the four protein-coding genes analyzed in this study (i.e., atpD, gyrB, recA, and rpoB). However, recA was observed to provide the greatest degree of discrimination between the most closely related species. The recA pair-wise similarities between the type strains of the Achromobacter species ranged from 89.9% to 97.6%. The species pair, A. marplatensis CCUG 56371T compared with A. spiritinus CCUG 61968T, possessed the highest pair-wise similarities among the recognized species (97.9%), suggesting the possibility of misclassifications between them. Further DDH analysis and MLSA sequencing confirmed and clarified the taxonomic affiliation of A. spiritinus as a later synonym of A. marplatensis. From these determinations, it could be concluded that isolates and strains can be identified to the species level if they exhibit recA similarities of 98.0%, or greater, to a recognized species. If recA similarities between isolates and strains are 97.6%, or lower, to all known species, the identification will be inconclusive. The sequence data were compared also with the gene sequence data for NrdA (nrdA), proposed as a single locus sequencing tool for Achromobacter speciation by Spilker et al., 2013 (Figure S11 in File S1 and Figure 3) [6]. Both genes allowed good discrimination between Achromobacter species with similar pair-wise similarities among them, although recA showed a higher number of polymorphic sites than nrdA.
Figure 3. The variation in recA and nrdA-765 sequences among Achromobacter species.
The y-axis shows the number of nucleotide positions, within 50-nucleotide position intervals (x-axis), in the respective sequences that exhibit variation between the type strains of the species of Achromobacter.
According to the cluster analyses derived from the individual, as well as the concatenated gene sequences, the type strains of the species formed stable lineages within the genus, providing the reference points for closely related strains, while other strains were seen to diverge from the known species, representing probable new species in Achromobacter.
The dendrogram derived from MALDI-TOF MS mass peak profiles (Figure 2) shows species clusters similar to the ones obtained when comparing individual or concatenated house-keeping gene sequences. Considering the genotypic-based phylogeny as the basis for microbial systematics and taxonomy, the phenotypic-based MALDI-TOF MS analyses were observed to be more accurate for identifications of strains than the traditional phenotypic characterization. The species identifications, using MALDI-TOF MS and the commercially available databases, to date, have not included all of the species of the Achromobacter genus. The VITEK MS IVD database (bioMérieux, Inc.) contains only the species A. xylosoxidans and A. denitrificans, while the MALDI Biotyper database (Bruker Corp.) includes A. insolitus, A. spanius, A. piechaudii and A. ruhlandii, as well. In previous studies comparing the performance of MALDI-TOF MS, comparative 16S rRNA gene sequence analysis was used as the reference method, which, as previously mentioned, does not exhibit resolution adequate for definitive species-level identifications [44], [45]. The protein mass peak profile similarities between bacterial species of a genus indicates that the spectra combined with species-specific protein peaks are able to delineate most species of Achromobacter from each other, using MALDI-TOF MS. In the case of this study, the MALDI-TOF MS analyses of the Achromobacter species provided an alternative method that could be correlated with the resolution of the Achromobacter species by selected house-keeping gene sequence analyses.
Complete genome sequence analyses are considered to be the ultimate method for deriving the phylogenetic relationships of bacterial taxa; phylogeny is recognised to be the reference backbone of bacterial taxonomy. However, until genome sequencing is established as routine in microbiology laboratories, complementary methods are needed that provide accurate, rapid and cost-effective identifications for clinical diagnostics, biotechnology applications and environmental studies. In conclusion, selected house-keeping gene sequence analyses, i.e., comparative recA analysis is a robust method for deriving reliable identifications of clinically-relevant isolates and strains of the species in the genus Achromobacter. Because an exact recA similarity ‘cut-off’ for species identifications is not absolutely defined from sequence analyses alone, the results from alternative methods, such as MALDI-TOF MS analyses, may be correlated with the sequence data to provide insight into definitive recA similarity values that can be used for species-level identification.
Supporting Information
Strains of Achromobacter species and GenBank accession numbers for the sequences used in this study. Accession numbers indicated in bold are for sequences determined in this study.
(PDF)
Biochemical characteristics of all Achromobacter species strains examined. +, positive; –, negative; w, weak.
(PDF)
Genetic diversity of the selected loci among the Achromobacter type strains and the clinical isolates analyzed in this study.
(PDF)
Genetic diversity values for the loci atpD, recA and rpoB obtained in our study compared with the results obtained for different authors, and genetic diversity for nrdA gene analysed for other authors. For those genetic diversity calculations only the seven type strains commons in all studies were considered.
(PDF)
Contains the following files: Figure S1. Gene sequence similarities and evolutionary relationships for the type strains of the Achromobacter species. (a) 16S rRNA gene sequence similarities for the type strains of the Achromobacter species. Nearly-complete 16S rRNA gene sequence similarities are in the lower diagonal; partial 16S rRNA gene sequence similarities are in the upper diagonal. (b) Evolutionary distances were computed using the Jukes-Cantor method and are in the units of the number of base substitutions per site. The analysis involved 11 nucleotide sequences. All positions containing gaps and missing data were eliminated. There were a total of 1346 positions in the final dataset. Figure S2. Gene sequence similarities and evolutionary relationships for the type strains of the Achromobacter species. (a) atpD gene sequence similarities for the type strains of the Achromobacter species gene sequence similarities are on the lower diagonal; amino acid sequence similarities are on the upper diagonal. (b) The sequence relationships were inferred, using the UPGMA method. The analysis involved 11 nucleotide sequences. All positions containing gaps and missing data were eliminated. There were a total of 727 positions in the final dataset. Figure S3. Gene sequence similarities and evolutionary relationships for the type strains of the Achromobacter species. (a) gyrB gene sequence similarities for the type strains of the Achromobacter species gene sequence similarities are on the lower diagonal; aminoacid sequence similarities are on the upper diagonal. (b) The sequence relationships were inferred, using the UPGMA method. The analysis involved 11 nucleotide sequences. All positions containing gaps and missing data were eliminated. There were a total of 593 positions in the final dataset. Figure S4. Gene sequence similarities and evolutionary relationships for the type strains of the Achromobacter species. (a) recA gene sequence similarities for the type strains of the Achromobacter species gene sequence similarities are on the lower diagonal; aminoacid sequence similarities are on the upper diagonal. (b) The sequence relationships were inferred, using the UPGMA method. The analysis involved 11 nucleotide sequences. All positions containing gaps and missing data were eliminated. There were a total of 621 positions in the final dataset. Figure S5. Gene sequence similarities and evolutionary relationships for the type strains of the Achromobacter species. (a) rpoB gene sequence similarities for the type strains of the Achromobacter species gene sequence similarities are on the lower diagonal; aminoacid sequence similarities are on the upper diagonal. (b) The sequence relationships were inferred, using the UPGMA method. The analysis involved 11 nucleotide sequences. All positions containing gaps and missing data were eliminated. There were a total of 598 positions in the final dataset. Figure S6. Phylogenetic tree of the 57 Achromobacter strains studied based on the analysis of 513 bp of the atpD gene. Distance matrix was calculated by the Jukes-Cantor method. Dendrogram was generated by neighbor-joining. Bordetella pertussis CCUG 30873T was used as an outgroup. The bar indicates sequence divergence. Bootstrap values of more than 500 (from 1000 replicates) are indicated at the nodes. Figure S7. Phylogenetic tree of the 57 Achromobacter strains studied based on the analysis of 528 bp of the gyrB gene. Distance matrix was calculated by the Jukes-Cantor method. Dendrogram was generated by neighbor-joining. Bordetella pertussis CCUG 30873T was used as an outgroup. The bar indicates sequence divergence. Bootstrap values of more than 500 (from 1000 replicates) are indicated at the nodes. Figure S8. Phylogenetic tree of the 57 Achromobacter strains studied based on the analysis of 527 bp of the rpoB gene. Distance matrix was calculated by the Jukes-Cantor method. Dendrogram was generated by neighbor-joining. Bordetella pertussis CCUG 30873T was used as an outgroup. The bar indicates sequence divergence. Bootstrap values of more than 500 (from 1000 replicates) are indicated at the nodes. Figure S9. Phylogenetic tree of the 57 Achromobacter strains studied based on the analysis of 398 bp of the 16S rRNA gene. Distance matrix was calculated by the Jukes-Cantor method. Dendrogram was generated by neighbor-joining. Bordetella pertussis CCUG 30873T was used as an outgroup. The bar indicates sequence divergence. Bootstrap values of more than 500 (from 1000 replicates) are indicated at the nodes. Figure S10. Phylogenetic tree of the strains of Achromobacter used in this study based on the phylogenetic analysis of four concatenated genes (atpD, gyrB, recA and rpoB). Distance matrices were calculated by the Jukes-Cantor method. Dendrograms were generated by neighbor-joining. Bordetella pertussis CCUG 30873T was used as an outgroup. The bar indicates sequence divergence. Bootstrap values of more than 500 (from 1000 replicates) are indicated at the nodes. Figure S11. Gene sequence similarities and evolutionary relationships for the type strains of the Achromobacter species. (a) nrdA gene sequence similarities of a 765 pb region for the type strains of the Achromobacter species gene sequence similarities are on the lower diagonal; nrdA gene sequence similarities of the short region are on the upper diagonal.
(PDF)
Acknowledgments
The authors acknowledge the technical expertise of the CCUG staff for the phenotypic profile and the assistance of the Achromobacter PubMLST database curators. The authors thank Dr. Peter Vandamme for his collaboration and discussion regarding the taxonomy of Achromobacter. Margarita Gomila is the recipient of a postdoctoral contract from the University of the Balearic Islands funded by the Spanish Ministry of Education, Culture and Sports through the International Excellence Campus Program. Margarita Gomila was supported for exchange to the CCUG through fellowships from the Spanish Ministry of Education and Science, by means of the José Castillejo Program (2008) and from a FEMS Research Fellowship 2009–1.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.
Funding Statement
Margarita Gomila is the recipient of a postdoctoral contract from the University of the Balearic Islands funded by the Spanish Ministry of Education, Culture and Sports through the International Excellence Campus Program. Margarita Gomila was supported for exchange to the CCUG through fellowships from the Spanish Ministry of Education and Science, by means of the José Castillejo Program (2008) and from a FEMS Research Fellowship 2009-1. This work was supported also by funding from Västra Götaland Region (Sweden) projects ALFGBG-210591, VGFOUREG-157801 and VGFOUREG-232981, and from Spanish MINECO, through projects CGL2011-24318 and Consolider CSD2009-00006. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. LiPuma JJ (2010) The changing microbial epidemiology in cystic fibrosis. Clin Microbiol Rev 23:299–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Peleg AY, Hooper DC (2010) Hospital-acquired infections due to Gram-negative bacteria. N Engl J Med 362:1804–1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Amoureux L, Bador J, Fardeheb S, Mabille C, Couchot C, et al. (2013) Detection of Achromobacter xylosoxidans in hospital, domestic, and outdoor environmental samples and comparison with human clinical isolates. Appl Environ Microbiol 79:7142–7149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. De Baets F, Schelstraete P, Van Daele S, Haerynck F, Vaneechoutte M (2007) Achromobacter xylosoxidans in cystic fibrosis: prevalence and clinical relevance. J Cyst Fibros 6:75–78. [DOI] [PubMed] [Google Scholar]
- 5. Barrado L, Brañas P, Orellana MA, Martínez MT, García G, et al. (2013) Molecular characterization of Achromobacter isolates from cystic fibrosis and non-cystic fibrosis patients in Madrid, Spain. J Clin Microbiol 51:1927–1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Spilker T, Vandamme P, LiPuma JJ (2013) Identification and distribution of Achromobacter species in cystic fibrosis. J Cyst Fibros 12:298–301. [DOI] [PubMed] [Google Scholar]
- 7.De Baets F, Schelstraete P, Haerynck F, Van Bierviet S, DeBruyne R, et al. (2013) Achromobacter xylosoxidans induced bronchiolitis obliterans in cystic fibrosis. Pediatr Pulmonol; :doi:10.1002/ppul.22864.. [DOI] [PubMed]
- 8. Hansen CR, Pressler T, Ridderberg W, Johansen HK, Skov M (2013) Achromobacter spcies in cystic fibrosis: cross-infection caused by indirect patient-to-patient contact. J Cyst Fibros 6:609–615. [DOI] [PubMed] [Google Scholar]
- 9. De Ley J, Segers P, Kersters K, Mannheim W, Lievens A (1986) Intra- and intergeneric similarities of the Bordetella ribosomal ribonucleic acid cistrons: proposal for a new family, Alcaligenaceae . Int J Syst Bacteriol 36:405–414. [Google Scholar]
- 10. Yabuuchi E, Kawamura Y, Kosako Y, Ezaki T (1998) Emendation of genus Achromobacter and Achromobacter xylosoxidans (Yabuuchi and Yano) and proposal of Achromobacter ruhlandii (Packer and Vishniac) comb. nov., Achromobacter piechaudii (Kiredjian, et al.) comb. nov., and Achromobacter xylosoxidans subsp. denitrificans (Rüger and Tan) comb. nov. Microbiol Immunol 42:429–438. [DOI] [PubMed] [Google Scholar]
- 11. Blümel S, Mark B, Busse HJ, Kämpfer P, Stolz A (2001) Pigmentiphaga kullae gen. nov., sp. nov., a novel member of the family Alcaligenaceae with the ability to decolorize azo dyes aerobically. Int J Syst Evol Microbiol 51:1867–1871. [DOI] [PubMed] [Google Scholar]
- 12.Busse HJ, Auling G (2005) Genus II. Achromobacter Yabuuchi and Yano 1981, 477VP emend. Yabuuchi, Kawamura, Kosako and Ezaki 1998a, 1083. In:Brenner DJ, Krieg NR, Staley JTeditors. Bergey’s Manual of Systematic Bacteriology, vol. 2: The Proteobacteria, second edition. Springer. pp. 658–662.
- 13. Coenye T, Vancanneyt M, Cnockaert MC, Falsen E, Swings J, et al. (2003a) Kerstersia gyiorum gen. nov., sp. nov., a novel Alcaligenes faecalis-like organism isolated from human clinical samples, and reclassification of Alcaligenes denitrificans Rüger and Tan 1983 as Achromobacter denitrificans comb. nov. Int J Syst Evol Microbiol 53:1825–1831. [DOI] [PubMed] [Google Scholar]
- 14. Coenye T, Vancanneyt M, Falsen E, Swings J, Vandamme P (2003b) Achromobacter insolitus sp. nov. and Achromobacter spanius sp. nov., from human clinical samples. Int J Syst Evol Microbiol 53:1819–1824. [DOI] [PubMed] [Google Scholar]
- 15. Gomila M, Tvrzová L, Teshim A, Sedlácek I, González-Escalona N, et al. (2011) Achromobacter marplatensis sp. nov., isolated from a pentachlorophenol-contaminated soil. Int J Syst Evol Microbiol 61:2231–2237. [DOI] [PubMed] [Google Scholar]
- 16. Vandamme P, Moore ERB, Cnockaert M, De Brandt E, Svensson-Stadler L, et al. (2013a) Achromobacter animicus sp. nov., Achromobacter mucicolens sp. nov., Achromobacter pulmonis sp. nov. and Achromobacter spiritinus sp. nov., from human clinical samples. Syst Appl Microbiol 36:1–10. [DOI] [PubMed] [Google Scholar]
- 17. Vandamme P, Moore ERB, Cnockaert M, Peeters C, Svensson-Stadler L, et al. (2013b) Classification of Achromobacter genogroups 2, 5, 7 and 14 as Achromobacter insuavis sp. nov., Achromobacter aegrifaciens sp. nov., Achromobacter anxifer sp. nov. and Achromobacter dolens, sp. nov., respectively. Syst App Microbiol 36:474–482. [DOI] [PubMed] [Google Scholar]
- 18. D’Amato RF, Salemi M, Mathews A, Cleri DJ, Reddy G (1988) Achromobacter xylosoxidans (Alcaligenes xylosoxidans subsp. xylosoxidans) meningitis associated with a gunshot wound. J Clin Microbiol 26:2425–2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Duggan JM, Goldstein SJ, Chenoweth CE, Kauffman CA, Bradley SF (1996) Achromobacter xylosoxidans bacteremia: report of four cases and review of the literature. Clin Infect Dis 23:569–576. [DOI] [PubMed] [Google Scholar]
- 20. Liu L, Coenye T, Burns JL, Whitby PW, Stull TL, et al. (2002) Ribosomal DNA-Directed PCR for identification of Achromobacter (Alcaligenes) xylosoxidans recovered from sputum samples from cystic fibrosis patients. J Clin Microb 40:1210–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Spear JB, Fuhrer J, Kirby BD (1988) Achromobacter xylosoxidans (Alcaligenes xylosoxidans subsp. xylosoxidans) bacteremia associated with a well-water source; case report and review of the literature. J Clin Microbiol 26:598–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Vu-Thien H, Darbord JC, Moissenet D, Dulot C, Dufourcq JB, et al. (1998) Investigation of an outbreak of wound infections due to Alcaligenes xylosoxidans transmitted by chlorhexidine in a burn unit. Eur J Clin Microbiol Infect Dis 17:724–726. [DOI] [PubMed] [Google Scholar]
- 23.Kersters K, De Ley J (1984) Genus Alcaligenes Castellani and Chalmers 1919, 936AL. In:Krieg NR, Holt JGeditors. Bergey’s Manual of Systematic Bacteriology, vol. 1. Baltimore: Williams & Wilkins. pp. 361–373.
- 24. Kiredjian M, Holmes B, Kersters K, Guilvout I, De Ley J (1986) Alcaligenes piechaudii, a new species from human clinical specimens and the environment. Int J Syst Bacteriol 36:282–287. [Google Scholar]
- 25. Spilker T, Vandamme P, Lipuma JJ (2012) A multilocus sequence typing scheme implies population structure and reveals several putative novel Achromobacter species. J Clin Microbiol 50:3010–3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wayne LG, Brenner DJ, Colwell RR, Grimont PAD, Kandler O, et al. (1987) Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int J Syst Bacteriol 37:463–464. [Google Scholar]
- 27. Stackebrandt E, Frederiksen W, Garrity GM, Grimont PAD, Kämpfer P, et al. (2002) Report of the ad hoc committee for the re-evaluation of the species definition in bacteriology. Int J Syst Evol Microbiol 52:1043–1047. [DOI] [PubMed] [Google Scholar]
- 28. Hanage WP, Fraser C, Spratt BG (2006) Sequences, sequence clusters and bacterial species. Philos Trans R Soc Lond B Biol Sci 361:1917–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ridderberg W, Wang M, Nørskov-Lauritsen N (2012) Multilocus sequence analysis of isolates of Achromobacter from patients with cystic fibrosis reveals infecting species other than Achromobacter xylosoxidans . J Clin Microbiol 50:2688–2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kallow W, Erhard M, Shah HN, Raptakis E, Welker M (2010) MALDI TOF MS for microbial identification: years of experimental development to an established protocol. In:Shah HN, Gharbia SE, Encheva Veditors. Mass Spectrometry for Microbial Proteomics, John Wiley & Sons, London. pp. 255–276.
- 31. Seng P, Drancourt M, Gouriet F, La Scola B, Fournier PE, et al. (2009) Ongoing revolution in bacteriology: routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin Infect Dis 49:543–551. [DOI] [PubMed] [Google Scholar]
- 32. Gomila M, Ramirez A, Lalucat J (2007) Diversity of environmental Mycobacterium isolates from hemodialysis water as shown by a multigene sequencing approach. Appl Environ Microbiol 73:3787–3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lane DJ (1991) Nucleic acid techniques in bacterial systematics. In:Stackebrand E, Goodfellow Meditors. 16S/23S rDNA sequencing. Wiley, Chichester, United Kingdom. pp. 115–175.
- 34. Tayeb LA, Lefevre M, Passet V, Diancourt L, Brisse S, et al. (2008) Comparative phylogenies of Burkholderia, Ralstonia, Comamonas, Brevundimonas and related organisms derived from rpoB, gyrB and rrs gene sequences. Res Microbiol 159:169–177. [DOI] [PubMed] [Google Scholar]
- 35. Yamamoto S, Kasai H, Arnold DL, Jackson RW, Vivian A, et al. (2000) Phylogeny of the genus Pseudomonas: intrageneric structure reconstructed from the nucleotide sequences of gyrB and rpoD genes. Microbiology 146:2385–2394. [DOI] [PubMed] [Google Scholar]
- 36. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410. [DOI] [PubMed] [Google Scholar]
- 37. Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The Clustal X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucl Acids Res 25:4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jukes TH, Cantor CR (1969) Evolution of protein molecules. In:Munro HNeditors. Mammalian protein metabolism. Academic Press, Inc., New York, NY. pp. 21–132.
- 39. Felsenstein J (1989) PHYLIP – phylogeny inference package (version3.2). Cladistics 5:164–166. [Google Scholar]
- 40. Page RDM (1996) TREEVIEW: an application to display phylogenetic trees on personal computers. Comput Appl Biosci 12:357–358. [DOI] [PubMed] [Google Scholar]
- 41. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, et al. (2011) MEGA5: Molecular evolutionary genetics analysis using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony methods. Mol Biol Evol 28:2371–2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rozas J, Rozas R (1999) DnaSP version 3: an integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics 15:174–175. [DOI] [PubMed] [Google Scholar]
- 43. Ziemke F, Hofle MG, Lalucat J, Rossello-Mora R (1998) Reclassification of Shewanella putrefaciens Owen’s genomic group II as Shewanella baltica sp. nov. Int J Syst Bacteriol 48:179–186. [DOI] [PubMed] [Google Scholar]
- 44. Fernández-Olmos A, García-Castillo M, Morosini MI, Lamas A, Máiz L, et al. (2012) MALDI-TOF MS improves routine identification of non-fermenting Gram negative isolates from cystic fibrosis patients. J Cyst Fibros 11:59–62. [DOI] [PubMed] [Google Scholar]
- 45. Jacquier H, Carbonnelle E, Corvec S, Illiaquer M, Le Monnier A, et al. (2011) Revisited distribution of nonfermenting Gram-negative bacilli clinical isolates. Eur J Clin Microbiol Infect Dis 30:1579–1586. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Strains of Achromobacter species and GenBank accession numbers for the sequences used in this study. Accession numbers indicated in bold are for sequences determined in this study.
(PDF)
Biochemical characteristics of all Achromobacter species strains examined. +, positive; –, negative; w, weak.
(PDF)
Genetic diversity of the selected loci among the Achromobacter type strains and the clinical isolates analyzed in this study.
(PDF)
Genetic diversity values for the loci atpD, recA and rpoB obtained in our study compared with the results obtained for different authors, and genetic diversity for nrdA gene analysed for other authors. For those genetic diversity calculations only the seven type strains commons in all studies were considered.
(PDF)
Contains the following files: Figure S1. Gene sequence similarities and evolutionary relationships for the type strains of the Achromobacter species. (a) 16S rRNA gene sequence similarities for the type strains of the Achromobacter species. Nearly-complete 16S rRNA gene sequence similarities are in the lower diagonal; partial 16S rRNA gene sequence similarities are in the upper diagonal. (b) Evolutionary distances were computed using the Jukes-Cantor method and are in the units of the number of base substitutions per site. The analysis involved 11 nucleotide sequences. All positions containing gaps and missing data were eliminated. There were a total of 1346 positions in the final dataset. Figure S2. Gene sequence similarities and evolutionary relationships for the type strains of the Achromobacter species. (a) atpD gene sequence similarities for the type strains of the Achromobacter species gene sequence similarities are on the lower diagonal; amino acid sequence similarities are on the upper diagonal. (b) The sequence relationships were inferred, using the UPGMA method. The analysis involved 11 nucleotide sequences. All positions containing gaps and missing data were eliminated. There were a total of 727 positions in the final dataset. Figure S3. Gene sequence similarities and evolutionary relationships for the type strains of the Achromobacter species. (a) gyrB gene sequence similarities for the type strains of the Achromobacter species gene sequence similarities are on the lower diagonal; aminoacid sequence similarities are on the upper diagonal. (b) The sequence relationships were inferred, using the UPGMA method. The analysis involved 11 nucleotide sequences. All positions containing gaps and missing data were eliminated. There were a total of 593 positions in the final dataset. Figure S4. Gene sequence similarities and evolutionary relationships for the type strains of the Achromobacter species. (a) recA gene sequence similarities for the type strains of the Achromobacter species gene sequence similarities are on the lower diagonal; aminoacid sequence similarities are on the upper diagonal. (b) The sequence relationships were inferred, using the UPGMA method. The analysis involved 11 nucleotide sequences. All positions containing gaps and missing data were eliminated. There were a total of 621 positions in the final dataset. Figure S5. Gene sequence similarities and evolutionary relationships for the type strains of the Achromobacter species. (a) rpoB gene sequence similarities for the type strains of the Achromobacter species gene sequence similarities are on the lower diagonal; aminoacid sequence similarities are on the upper diagonal. (b) The sequence relationships were inferred, using the UPGMA method. The analysis involved 11 nucleotide sequences. All positions containing gaps and missing data were eliminated. There were a total of 598 positions in the final dataset. Figure S6. Phylogenetic tree of the 57 Achromobacter strains studied based on the analysis of 513 bp of the atpD gene. Distance matrix was calculated by the Jukes-Cantor method. Dendrogram was generated by neighbor-joining. Bordetella pertussis CCUG 30873T was used as an outgroup. The bar indicates sequence divergence. Bootstrap values of more than 500 (from 1000 replicates) are indicated at the nodes. Figure S7. Phylogenetic tree of the 57 Achromobacter strains studied based on the analysis of 528 bp of the gyrB gene. Distance matrix was calculated by the Jukes-Cantor method. Dendrogram was generated by neighbor-joining. Bordetella pertussis CCUG 30873T was used as an outgroup. The bar indicates sequence divergence. Bootstrap values of more than 500 (from 1000 replicates) are indicated at the nodes. Figure S8. Phylogenetic tree of the 57 Achromobacter strains studied based on the analysis of 527 bp of the rpoB gene. Distance matrix was calculated by the Jukes-Cantor method. Dendrogram was generated by neighbor-joining. Bordetella pertussis CCUG 30873T was used as an outgroup. The bar indicates sequence divergence. Bootstrap values of more than 500 (from 1000 replicates) are indicated at the nodes. Figure S9. Phylogenetic tree of the 57 Achromobacter strains studied based on the analysis of 398 bp of the 16S rRNA gene. Distance matrix was calculated by the Jukes-Cantor method. Dendrogram was generated by neighbor-joining. Bordetella pertussis CCUG 30873T was used as an outgroup. The bar indicates sequence divergence. Bootstrap values of more than 500 (from 1000 replicates) are indicated at the nodes. Figure S10. Phylogenetic tree of the strains of Achromobacter used in this study based on the phylogenetic analysis of four concatenated genes (atpD, gyrB, recA and rpoB). Distance matrices were calculated by the Jukes-Cantor method. Dendrograms were generated by neighbor-joining. Bordetella pertussis CCUG 30873T was used as an outgroup. The bar indicates sequence divergence. Bootstrap values of more than 500 (from 1000 replicates) are indicated at the nodes. Figure S11. Gene sequence similarities and evolutionary relationships for the type strains of the Achromobacter species. (a) nrdA gene sequence similarities of a 765 pb region for the type strains of the Achromobacter species gene sequence similarities are on the lower diagonal; nrdA gene sequence similarities of the short region are on the upper diagonal.
(PDF)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.