Abstract
Many forms of antibody-based targeted therapeutics, including antibody drug conjugates, utilize the internalizing function of the targeting antibody to gain intracellular entry into tumor cells. Ideal antibodies for developing such therapeutics should be capable of both tumor-selective binding and efficient endocytosis. The macropinocytosis pathway is capable of both rapid and bulk endocytosis, and recent studies have demonstrated that it is selectively up-regulated by cancer cells. We hypothesize that receptor-dependent macropinocytosis can be achieved using tumor-targeting antibodies that internalize via the macropinocytosis pathway, improving potency and selectivity of the antibody-based targeted therapeutic. Although phage antibody display libraries have been utilized to find antibodies that bind and internalize to target cells, no methods have been described to screen for antibodies that internalize specifically via macropinocytosis. We hereby describe a novel screening strategy to identify phage antibodies that bind and rapidly enter tumor cells via macropinocytosis. We utilized an automated microscopic imaging-based, High Content Analysis platform to identify novel internalizing phage antibodies that colocalize with macropinocytic markers from antibody libraries that we have generated previously by laser capture microdissection-based selection, which are enriched for internalizing antibodies binding to tumor cells in situ residing in their tissue microenvironment (Ruan, W., Sassoon, A., An, F., Simko, J. P., and Liu, B. (2006) Identification of clinically significant tumor antigens by selecting phage antibody library on tumor cells in situ using laser capture microdissection. Mol. Cell. Proteomics. 5, 2364–2373). Full-length human IgG molecules derived from macropinocytosing phage antibodies retained the ability to internalize via macropinocytosis, validating our screening strategy. The target antigen for a cross-species binding antibody with a highly active macropinocytosis activity was identified as ephrin type-A receptor 2. Antibody-toxin conjugates created using this macropinocytosing IgG were capable of potent and receptor-dependent killing of a panel of EphA2-positive tumor cell lines in vitro. These studies identify novel methods to screen for and validate antibodies capable of receptor-dependent macropinocytosis, allowing further exploration of this highly efficient and tumor-selective internalization pathway for targeted therapy development.
There is significant interest in the development of targeted therapeutics such as antibody drug conjugates that have the potential to improve the therapeutic window of cytotoxic drugs by delivering them specifically and intracellularly to cancer cells (2–6). The pathway by which the targeted agent enters tumor cells can influence both the uptake efficiency and the intracellular fate of the internalized agent, both of which contribute to the cytotoxic potency (6, 7). Among known endocytic routes, macropinocytosis provides an intriguing pathway for cellular entry as it is a form of bulk uptake and can therefore efficiently and rapidly internalize targeting agents. Macropinosomes comprise large, endocytic vesicles that range from 0.2 μm to 3 μm in size, which are up to 30-fold larger than the 0.1 μm average size of protein-coated, endocytic vesicles (8). Additionally, studies have shown that macropinocytosis is selectively up-regulated in Ras-transformed cancers (a common oncogenic mutation in human cancers) and plays an important role in tumor cell homeostasis by serving as an amino acid supply route (9), suggesting that targeted therapeutics based on antibodies that internalize via the macropinocytosis pathway may provide additional tumor-specificity against a wide variety of human cancers.
To therapeutically explore the utility of antibodies that gain entry into tumor cells via receptor-dependent macropinocytosis, a generally applicable method needs to be developed that readily identifies such antibodies. Although phage antibody display libraries have been extensively used to select for antibodies that internalize into tumor cells, no methods have been developed to uncover antibodies capable of cellular entry through the macropinocytosis pathway. To address this need, we developed a high content analysis (HCA)1-based screening strategy that employs automated image-based analysis to identify phage antibodies that colocalize with Texas Red-conjugated 70 kDa neutral dextran (ND70-TR), a macropinocytosis marker. We used the HCA protocol to screen single chain variable fragment (scFv) phage antibody display library selection outputs that we have generated previously by laser capture microdissection (LCM)-based selection on live tumor cells and tissues (1), and identified antibodies that are capable of efficient internalization via macropinocytosis. We performed kinetics and subcellular colocalization studies for phage antibodies as well as full-length immunoglobulin G (IgG) molecules derived from the parental scFvs and identified a highly active, macropinocytosing antibody that rapidly internalizes and colocalizes with early endosomal and lysosomal markers. We further identified the target antigen as EphA2 by immunoprecipitation and mass spectrometry. To confirm internalization by an independent functional assay and to demonstrate therapeutic potential, we created an antibody-toxin conjugate and showed potent and specific cytotoxic activity against a panel of EphA2-positive tumor cell lines. To our knowledge, this is the first description of a generally applicable screening strategy to uncover macropinocytosing antibodies, enabling further exploration of this class of antibody–antigen pairs for the development of effective antibody-targeted therapeutics.
MATERIALS AND METHODS
Tissue Culture
Prostate cancer cell lines DU145 and LNCaP, breast cancer cell line MDA-MB-231, lung cancer cell line A549, cervical cancer cell line HeLa, epidermoid carcinorma cell line A431, and human embryonic kidney (HEK) 293A cell line were purchased from the American Type Culture Collection (ATCC). Human foreskin normal fibroblast line Hs27 was purchased from UCSF Cell Culture Core Facility. Benign prostatic hyperplasia (BPH-1) cells were originally obtained from Dr. Gerald Cunha's lab at UCSF (10) and maintained in the lab. All cells were grown in high-glucose, l-glutamine, and sodium pyruvate-supplemented complete Dulbecco's modified Eagle's medium (DMEM) (Caisson Labs, North Logan, UT) with the addition of 10% fetal bovine serum (Fisher Scientific) and penicillin-streptomycin solution (Axenia BioLogix, Dixon, CA). Cells were grown in 5% CO2 at 37 °C on tissue culture-treated flasks (BD Biosciences). Cells were passaged utilizing 0.25% trypsin-EDTA (Invitrogen, Carlsbad, CA).
Preparation of Phage Antibody Display Library Selection Output for Screening
Phage antibody library selection outputs generated previously by LCM-based selection on prostate tumor tissues (1) were streaked onto 2X YT agar plates containing 12.5 μg/ml tetracycline to yield monoclonal phage antibodies. In this phage display system, the scFv was fused to pIII in the fd phage display vector that carries genes necessary for phage assembly (11, 12). Individual colonies were inoculated in 2X YT containing 12.5 μg/ml tetracycline and grown in deep 96-well plates (Fisher Scientific) at 37 °C with 225 RPM shaking for 18 h. The plates were centrifuged to pellet the bacteria and supernatants containing phage particles were transferred into a new 96-well plate for HCA experiments (see below). Positive clones from initial HCA screenings were re-tested using purified phage using polyethylene glycol (PEG8000) as previously described (1, 13–15). Antibody sequences were determined using 96-well plate-based DNA sequencing (Functional Biosciences, Madison, WI).
Recombinant Antibody Cloning, Expression, and Purification
For IgG production, heavy and light chain variable fragments were subcloned into IgG-AbVec (kindly provided by Dr. Patrick Wilson at University of Chicago) γ and λ mammalian expression vectors, as previously described, to produce secretable IgG1 antibodies (16). For scFv-Fc fusion production, scFv was subcloned from phage into pFUSE-hIgG1 Fc2 (InvivoGen, San Diego, CA). Mammalian transfection complexes containing antibody expression DNA and polyethylenimine (Sigma-Aldrich) in Opti-MEM (Invitrogen) were added to HEK 293A cells in the presence of serum-free DMEM containing Nutridoma-SP (Roche) and penicillin-streptomycin. Antibody-containing media were harvested after 4 days and affinity-purified using protein A agarose (Pierce/Fisher). Antibody concentrations were determined using the Blitz® Bio-Layer interferometry System (ForteBio, Menlo Park, CA).
HCA Screening
Supernatants from 96-well bacterial culture plates (see above) were used for initial HCA screening. DU145 cells were seeded in 96-well plates (BD Biosciences) overnight, and incubated with phage-containing supernatants and 50 μg/ml TexasRed-conjugated 70-kDa neutral dextran (ND70-TR, Invitrogen) in DMEM/10% FBS at 37 °C with 5% CO2 overnight. Cells were washed 3× with PBS, fixed with 4% paraformaldehyde (Santa Cruz Biotechnology, Santa Cruz, CA) in PBS for 10 min, washed 3x in PBS, and then permeabilized in PBS containing 1% fraction V bovine serum albumin (Fisher Scientific) and 0.1% TritonX-100 (Sigma). Phage were detected with 3.5 μg/ml biotin-conjugated, rabbit anti-fd bacteriophage (Sigma-Aldrich) for 1h at RT followed by 1 μg/ml Alexa Fluor® 488-conjugated streptavidin (Jackson ImmunoResearch, West Grove, PA) for 15 min at RT. Hoechst 33342 (Thermo Scientific) at 1 μg/ml for 30 min at RT was used to detect nuclei. The 96-well plates were imaged on a CellInsight™ NXT HCS platform (Thermo Scientific) with a semi-aprochromat 20× LUCPLFLN objective (Olympus) utilizing >6 fields per well with a minimum of 300 cells per well. Pearson's correlation coefficient analysis between ND70-TR and phage particles were conducted using Thermo Scientific HCS Studio software suite on all imaged fields and averaged per well.
Confocal Analysis
DU145 cells were seeded in 8-well Lab-Tek II chambered coverglass (Thermo Scientific) overnight for confocal microscopy studies. Cells were incubated with antibodies (IgGs at 10 μg/ml or purified phage at 109 cfu/ml) and 50 μg/ml ND70-TR in DMEM/10% FBS at 37 °C with 5% CO2 for indicated periods (see text), washed, fixed, and permeabilized as described above. To label subcellular structures, rabbit antibodies against early endosomes, lysosomes, caveolin-2, and clathrin heavy chain (Cell Signaling) were added to permeabilized cells at 1:100 dilutions for 3 h at RT. Cell-associated human IgGs were detected with 1 μg/ml Alexa Fluor® 647-conjugated goat anti-human IgG (Jackson ImmunoResearch) for 30 min at RT. Cell-associated phage were detected with 3.5 μg/ml biotin-conjugated, rabbit anti-fd bacteriophage for 1 h at RT followed by 1 μg/ml Alexa Fluor® 488-conjugated streptavidin (Jackson ImmunoResearch) for 15 min at RT. Antibodies against organelles were detected with Alexa Fluor® 488- or phycoerythrin-conjugated goat anti-rabbit for 30 min at RT. Hoechst 33342 at 1 μg/ml for 30 min at RT was used to detect nuclei. Cells in eight-well glass chambered coverglass were then imaged on the FluoView® FV10i laser confocal microscope (Olympus) equipped with two galvanometer scanning mirrors. Confocal images were taken with an Olympus 60× phase contrast water-immersion objective with NA 1.2. Image analyses including Pearson's and Mander's correlation coefficients, Z-projection, Z-projection dissection, and 3D renderings were performed with the included Olympus confocal software suite.
Internalization Kinetics Assay
DU145 cells seeded in eight-well chambered coverglass were pulsed with antibodies at 10 μg/ml in complete DMEM/FBS for 30 min at 4 °C, followed by a chase in 37 °C warmed, complete DMEM/FBS, and incubated at 37 °C with 5% CO2. Individual wells at varying time points were then washed in PBS and fixed in 4% paraformaldehyde before undergoing immunofluorescence as described above. For flow cytometry-based internalization kinetics assay, DU145 cells were seeded in six-well plate, then treated with antibodies at 10 μg/ml for varying amounts of time. Then cells were trypsinized, probed with anti-human secondary antibody, and analyzed on a flow cytometer. Cytochalasin D (Sigma) was resuspended in DMSO and cells were pulsed with 50 μg/ml of the drug in serum-free DMEM at 37 °C, followed by a chase in complete DMEM/FBS containing the drug and antibodies.
Immunoprecipitation of the Target Antigen
Purified HCA-F1 scFv-Fc fusions were first chemically cross-linked to protein A agarose beads. Briefly, antibodies were affinity-bound onto protein A agarose (Invitrogen) in a tube. Beads were then spun down and washed with 0.2 m sodium borate, pH 9.0. Dry dimethyl pimelimidate (DMP, Sigma) was added to the beads in the presence of sodium borate to yield a final concentration of 13 mg/ml and incubated at RT for 30 min. Beads were washed with sodium borate and DMP crosslinking was repeated a second time. Chemical crosslinking was terminated through washes with 0.2 m ethanolamine, pH 8.0, for 2 h at RT. Finally, unconjugated antibodies were eluted from beads using 0.1 m glycine, pH 2.8, followed by washes with PBS. Exposed surface proteins on DU145 cells were biotinylated using EZ-Link Sulfo-NHS-LC-Biotin (Thermo Pierce) according to manufacturer's recommendations and then lysed using standard RIPA buffer (50 mm Tris, pH 7 - 8, 150 mm NaCl, 0.1% SDS, 0.5% sodium deoxycholate, and 1% Nonidet P-40). Immunoprecipitation was performed as described previously (17, 18). Briefly, 5 mg of biotinylated lysates were first precleared against protein A agarose for 1 h at RT and then incubated with scFv-Fc-conjugated protein A beads overnight at 4 °C. Beads were then washed with 500 mm NaCl in PBS, spun down, and boiled in SDS sample buffer to be run on two 4–12% Tris-glycine SDS-PAGE gels (Invitrogen). One gel was GelCode-stained (Thermo) and the other gel was used for Western blotting using standard procedures. Horseradish peroxidase-conjugated streptavidin was used in the Western blot to assess which protein band to extract from the GelCode-stained gel.
Antigen Identification by Mass Spectrometry Analysis
Extracted gel bands were trypsin-digested and analyzed via tandem mass spectrometry (MS/MS, University of Minnesota). Charge state deconvolution and deisotoping were not performed. All MS/MS samples were analyzed using Sequest (Thermo Fisher Scientific; version 27, rev. 12). Sequest was set up to search the rs_human9606_031313_cRAP123 database (unknown version, 36,010 entries) assuming the digestion enzyme trypsin. Sequest was searched with a fragment ion mass tolerance of 0.80 Da and a parent ion tolerance of 0.079 Da and 0.32 Da. Iodoacetamide derivative of cysteine and oxidation of methionine were specified as fixed and variable modifications, respectively, in Sequest. Scaffold (version 4.0.5, Proteome Software Inc.) was used to validate protein identifications to create peak lists. Peptide identifications were accepted if they could be established at greater than 95.0% probability by the Peptide Prophet algorithm (19). Protein identifications were accepted if they could be established at greater than 90.0% probability and contained at least two identified peptides. Protein probabilities were assigned by the Protein Prophet algorithm (20). Peptide and protein false discovery rates, as determined by Protein Prophet algorithm, are 0.4% and 0.1%, respectively. Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony.
Antibody–Toxin Cytotoxicity Assay
The human IgG HCA-F1 was biotinylated with EZ-Link Sulfo-NHS-LC-Biotin (Thermo Pierce) according to manufacturer's recommendations. A panel of tumor and nontumorigenic cell lines (see text) were seeded in 96-well plates at a density of 1000–2000 cells per well and grown for 16 h at 37 °C in 5% CO2. Biotinylated IgG HCA-F1 was then incubated with streptavidin-ZAP (saporin conjugated with streptavidin, Advanced Targeting Systems) at a molar ratio of 1:1 and incubated on ice for 30 min to form the antibody-toxin (saporin) conjugate, which was then added to cells and incubated for 96 h at 37 °C in 5% CO2. Cell viability was then determined by CCK-8 assay (Dojindo, Rockville, Maryland) according to manufacturer's recommendations using the Synergy HT microtiter plate reader (Bio-Tek, Winooski, VT). The half maximal effective concentration (EC50) values were determined by curve fit using Prism (GraphPad Software, San Diego, CA).
RESULTS
HCA-based Screening Strategy
The HCA-based strategy that we used to identify antibodies capable of internalizing into tumor cells via macropinocytosis is outlined in Fig. 1A. The key feature is the development of an HCA platform that allows quantitative measurement of colocalization between phage antibodies and a macropinocytic marker, ND70-TR. To identify clinically relevant macropinocytosing antibodies, we screened phage antibody library selection outputs that we have generated previously by laser capture microdissection (LCM)-based selection, which are highly enriched for internalizing antibodies that bind to prostate tumor cells in situ residing in the tumor tissue microenvironment (1).
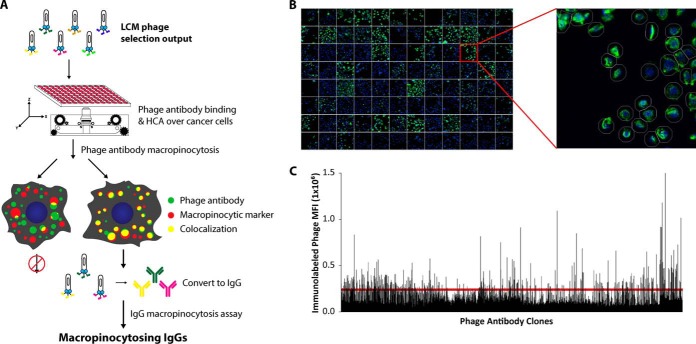
Fig. 1.
Outline of screening strategy and data from the first step of the screening, i.e. phage binding to DU145 cells. A, Schematic of HCA screening to identify macropinocytosis-dependent antibodies. HCA instruments allow automated high throughput detection of antibody colocalization with a macropinocytosis marker. The starting materials for the screening are sublibraries generated previously by us from LCM-based phage antibody library selection (1) that are enriched for internalizing phage antibodies binding to tumor cells in situ. B, DU145 cells were incubated in 96-well plates with phage-containing supernatants for 24 h at 37 °C in complete DMEM and10% FBS. Nuclei were stained with Hoechst 33342. Bound phages were immunolabeled with anti-fd antibodies (green). Zoomed insert portrays software-based, automated cell analysis, measuring mean fluorescence intensities (MFI) of immunolabeled phages. Over 300 cells were quantified for each phage clone. C, Plot of MFI values of immunolabeled phage binding to cell for 1439 phage clones. Red horizontal line represents MFI of ∼250,000, the threshold for prioritizing clones for further internalization analysis.
Analysis of Phage Antibody Binding Patterns by Automated Fluorescence Microscopy
Phage-infected bacteria were arrayed into 96-well plates and phage-containing supernatants were incubated with prostate cancer DU145 cells in 96-well plates in the presence of complete DMEM/10% FBS for 24 h at 37 °C. Phage antibody binding patterns were analyzed by automated fluorescent microscopy (Fig. 1B). A broad range of patterns of cell-associated phages was observed but internalization could not be clearly determined (supplemental Fig. S1). Image-based quantitation of phage binding was performed to generate a mean fluorescence intensity (MFI) value for each phage antibody (Fig. 1C). We selected the top 25% (MFI > 250,000) or 360 phage clones for more detailed analysis of internalizing properties (Fig. 1C). FACS analysis of a fraction of these phage clones on DU145 cells yielded MFI values consistent with the microscopic imaging-based analysis (supplemental Fig. S2).
HCA Identifies Phage Antibodies that Internalize via Macropinocytosis
Previous methods to select and screen for internalizing phage antibodies have utilized low pH, high salt washes in an attempt to strip surface-bound phage antibodies. Although this approach can be successful, strong binding, high-affinity phage antibodies may be resistant to even these harsh conditions. We tested several of the strongest binding phage antibody clones on fixed cells, which are incapable of internalization, and found that binding was resistant to low pH, high salt washes (supplemental Fig. S3). Thus, new methods are required to identify high-affinity internalizing phage antibodies.
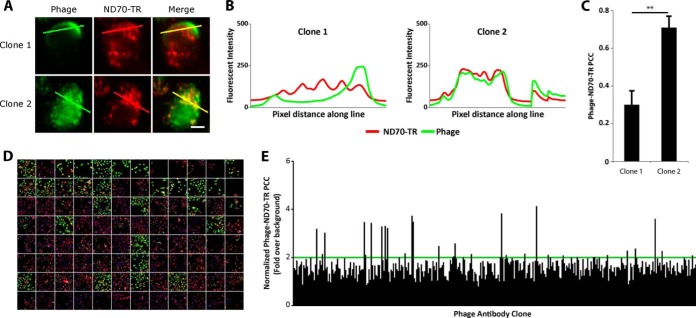
To screen for phage antibody clones that internalize into DU145 cells via macropinocytosis, we performed HCA on the strongest binding clones (top 25%, or 360) utilizing ND70-TR as a fluid-phase macropinocytic marker (21, 22). Previous studies have established that fluorescent high molecular weight dextrans can be used to label macropinosomes (21). Phage antibody-containing supernatants were co-incubated with ND70-TR over DU145 cells in culture media for 24h at 37 °C. Following washing, fixing, and permeabilization, cell-associated phage were detected by anti-phage antibody, and subjected to HCA to assess colocalization with ND70-TR (Fig. 2D). An initial image analysis revealed that some phage antibodies internalized into cells and colocalized with ND70-TR, primarily in juxtanuclear structures, whereas other clones exhibited poor colocalization with ND70-TR (Fig. 2A, B). Next, we performed a quantitative analysis by measuring the Pearson's correlation coefficient (PCC) between immunolabeled phage and ND70-TR fluorescence. High PCC values identified phage antibodies that exhibited strong colocalization with ND70-TR, whereas low PCC values identified phage antibodies that exhibited poor colocalization with ND70-TR (Fig. 2C). About 10%, or 36 clones, possessed greater than twofold PCC values when compared with controls (Fig. 2E). Following sequencing, 14 unique antibody sequences were identified from the 36 clones.
Fig. 2.
Colocalization of phage antibodies with the macropinocytosis marker ND70-TR. A, Epifluorescent images of DU145 cells that were incubated with phage-containing supernatants and 50 μg/ml ND70-TR (red) for 24 h at 37 °C. Cell-associated phage were then detected by biotin-labeled anti-fd antibody followed by streptavidin-AlexaFluor 488 (green). Colocalization results in color overlap (yellow). Scale bar denotes 20 μm. B, To analyze colocalization, arbitrary lines were drawn across cells and fluorescent intensities along the drawn line were plotted for phages (green) and ND70-TR fluorescence (red). Co-variation of line intensity indicates colocalization. Representative images of two different phage antibodies with differing colocalization patterns are shown. C, Pearson's correlation coefficient (PCC) was quantified and averaged from >30 cells per phage conditions. Error bars denote S.E. for n = 3; * and ** indicate p values of <0.05 and <0.01, respectively, using two-tailed student's T-tests assuming unequal variance. D, Colocalization screening. DU145 cells were plated onto 96-well plates and incubated with phages and ND70-TR (red) for 24 h at 37 °C. Cells were immunolabeled against bacteriophages (green) and nuclei were stained with Hoechst 33342 (blue). E, Mean PCC between immunolabeled phage and ND70-TR of 360 phage clones, quantified from minimum of 300 cells per phage clone. PCC values were normalized to control phage clones that exhibited poor internalization. Green horizontal line represents 200% of control, a threshold for further analysis.
To rule out that the observed differences in internalization patterns are caused by different scFv display levels on phage, we performed Western blot analysis to determine the status of scFv-pIII fusion for six cell-binding phage antibodies with varying PCC values and found similar display levels (supplemental Fig. S4), which are consistent with the multivalent fd phage display format. It thus appears that the different internalization behavior exhibited by various phage antibodies reflects an intrinsic property of the scFv component.
Endocytosed Phages Macropinocytose en Route to Lysosomes in DU145 Cells
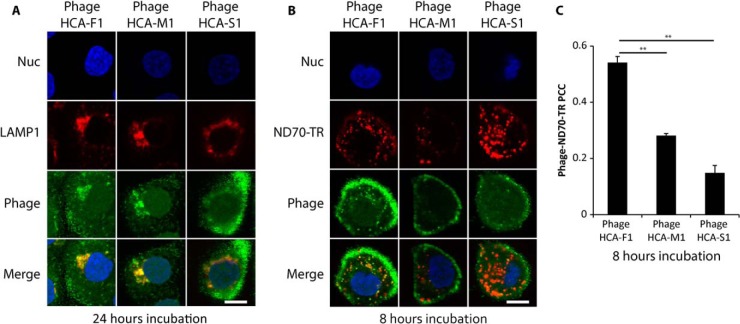
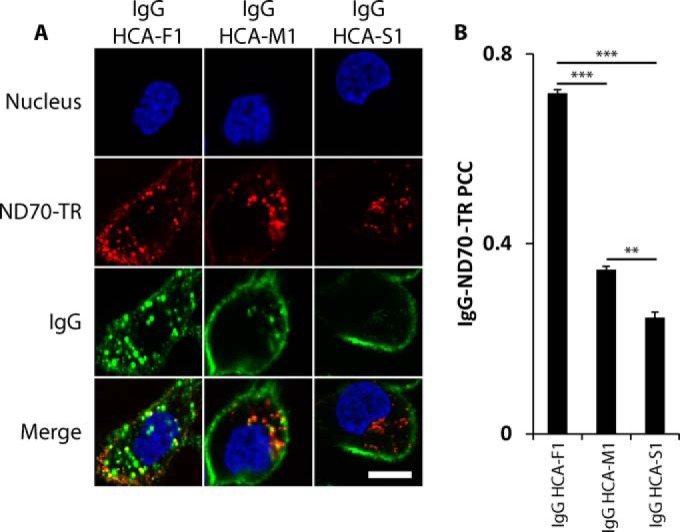
We further characterized three phage antibody clones, named HCA-F1, HCA-M1, and HCA-S1 (sequences shown in supplemental Table S1), two of which possessed high (HCA-F1 and HCA-M1, > twofold PCC values over control) and one with low (HCA-S1, < twofold PCC value over control) correlation between immunolabeled phages and ND70-TR. Using fluorescent confocal microscopy, we investigated whether these clones could internalize into juxtanuclear structures coinciding with lysosomal markers. After 24 h incubation with DU145 cells, phage antibodies colocalized with lysosomal-associated membrane protein 1 (LAMP1). Phages HCA-F1 and HCA-M1 were visible as compact, vesicular structures present in a juxtanuclear area, whereas phage HCA-S1 exhibited poor internalization (Fig. 3A). Computed 3D tomography also demonstrated that endocytosed phage HCA-F1 colocalized with internalized ND70-TR (supplemental Movie S1). We also examined whether phages could be visualized within early endosomes during early stages of endocytosis, however, phages did not colocalize with the endosomal marker, early endosomal antigen 1 (EEA1) (data not shown), suggesting that either the phages transited quickly through early endosomes or bypassed the early endosomes en route to lysosomes.
Fig. 3.
Confocal analysis of phage antibody internalization by DU145 cells. Confocal Z-slices of DU145 cells incubated with purified phage for A, 24 h at 37 °C or B, 8 h at 37 °C in the presence of ND70-TR. Cells were immunolabeled against phages (green), lysosomes (LAMP1, red), and nuclei (Hoechst 33342, blue). Scale bar: 20 μm. C, Mean PCC of internalized phages and ND70-TR. Over 30 cells were analyzed per phage antibody. ** denotes two-tailed t test p values of <0.01. Error bars represent S.E. for n = 3.
Phage Macropinocytose into DU145 Cells with Varying Kinetics
We next examined whether phage antibodies HCA-F1, HCA-M1, and HCA-S1 can exhibit distinguishable internalization kinetics. Whereas two phage antibodies HCA-F1 and HCA-M1 displayed a similar internalization pattern after a 24 h incubation, only phage HCA-F1 was capable of internalizing into DU145 cells after an 8 h incubation (Fig. 3B). PCC analysis between fluorescently immunolabeled, internalized phages and ND70-TR after an 8 h incubation showed significant differences between the three phage antibodies (Fig. 3C). Mander's correlation coefficient analysis, which is similar to PCC analysis but places weight on fluorescent intensity, also corroborated these differences (data not shown).
Internalization of IgGs Derived from scFvs
We cloned scFv from phages HCA-F1, HCA-M1, and HCA-S1 into full-length human IgG1 expression constructs and purified IgGs from transiently transfected human embryonic kidney (HEK) 293A cell supernatants. The purified IgGs HCA-F1, HCA-M1, and HCA-S1 demonstrated binding to DU145 cells via flow cytometry (supplemental Fig. S5) and colocalized with internalized ND70-TR in DU145 cells in a similar fashion to their parental phage antibodies (Fig. 4A). 3D computed tomography showed that IgG HCA-F1 possesses the most robust internalization properties, internalizing almost immediately upon incubation with cells and yielding very low amounts of detectable IgG on the surface of the cell after 90 min of incubation (supplemental Movie S2). Similar to the data from the phage experiments, the PCC value between immunolabeled IgG and ND70-TR was significantly higher for IgG HCA-F1 than either IgG HCA-M1 or IgG HCA-S1 (Fig. 4B).
Fig. 4.
Internalization and colocalization analysis of IgGs derived from scFvs. A, DU145 cells co-incubated with three IgGs with different internalization properties at 10 μg/ml and 50 μg/ml ND70-TR (red) for 90 min at 37 °C. Cells were immunolabeled against IgG using anti-human Fc (green). Nuclei were stained with Hoechst 33342 (blue). Single confocal Z-slice images are shown. Scale bar: 20 μm. B, PCC analysis of colocalization of IgGs HCA-F1, HCA-M1, and HCA-S1 with ND70-TR using Z-slices crossing the entire cell, quantitating a minimum of 10 cells. ** and *** denote two-tailed t test p values of <0.01 and <0.001, respectively. Error bars represent S.E. for n = 3.
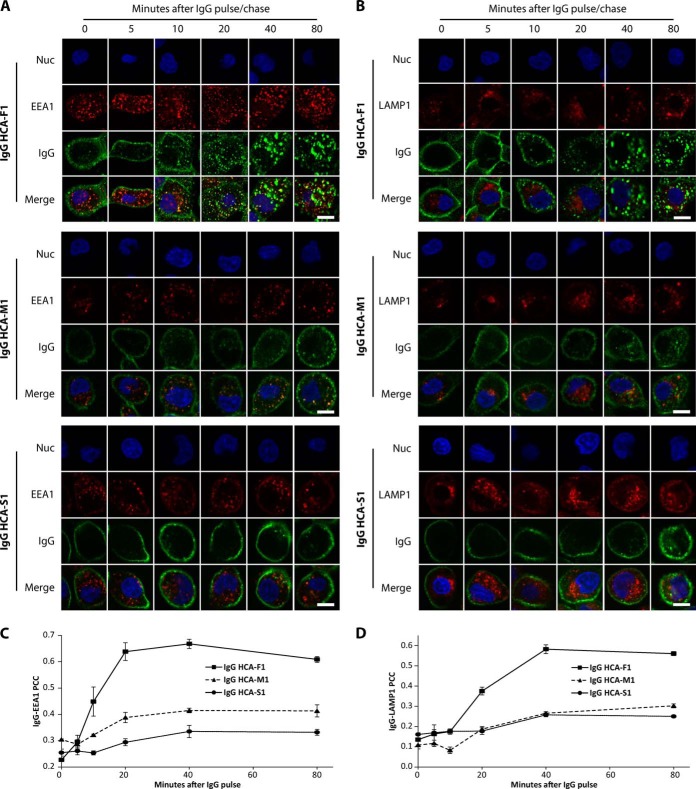
We next utilized immunolabeling against the endocytic markers EEA1 and LAMP1 to examine the colocalization of IgGs HCA-F1, HCA-M1, and HCA-S1 with early endosomes and lysosomes over varying time intervals. All of the IgGs bound to the surface of DU145 cells almost immediately after administration (Fig. 5A, 5B). IgG HCA-F1 fluorescence increased in intensity over time in punctate-like structures at the expense of cell surface fluorescence (Fig. 5A, 5B). IgG HCA-F1 addition also led to increased numbers of EEA1-labeled punctate structures when compared with either IgGs HCA-M1 or -S1 (Fig. 5A). Antibody colocalization with both organelles was quantitated via PCC analysis across all time points. IgG HCA-F1 possessed significantly higher PCC values at earlier time points for both EEA1 and LAMP1 when compared with either IgG HCA-M1 or HCA-S1 (Fig. C, D). IgG HCA-F1 did not significantly colocalize with caveolin-2 or clathrin heavy chain, especially at earlier time points (supplemental Fig. S6). Furthermore, the HCA-F1 scFv-Fc fusion also bound, internalized, and colocalized with both EEA1 and LAMP1 within DU145 cells in the same fashion as its IgG counterpart (supplemental Fig. S7).
Fig. 5.
Kinetics of antibody internalization and subcellular localization. DU145 cells were incubated with three different IgGs (HCA-F1, HCA-M1, or HCA-S1) at 10 μg/ml for 15 min at 4 °C and then chased with complete DMEM/10% FBS for indicated time periods. Cells were then fixed, permeabilized, and immunolabeled against human IgG (green) and A, early endosomes (EEA1, red) or B, lysosomes (LAMP1, red). Nuclei were stained with Hoechst 33342 (blue). Scale bar: 20 μm. Pearson's correlation coefficients between immunolabeled C, EEA1 or D, LAMP1 and immunolabeled IgG were averaged from a minimum of 30 cells. Error bars denote S.E. of n = 3.
IgG HCA-F1 Internalizes via Macropinocytosis
To confirm antibody internalization via macropinocytosis, we studied antibody internalization with and without inhibitors of macropinocytosis. Previous studies have demonstrated that cytochalasin D and ethylisopropylamiloride (EIPA) both inhibit macropinocytosis (9, 23–25). DU145 cells pretreated with cytochalasin D, EIPA, or DMSO for 30 min were chased with IgG HCA-F1 in the presence of drug or DMSO. Both cytochalasin D and EIPA significantly inhibited IgG HCA-F1 internalization into DU145 cells (Fig. 6A). Measurements of internalized, immunolabeled IgG HCA-F1 fluorescence showed that both cytochalasin D and EIPA decreased endocytosed IgG HCA-F1 by >50% when compared with DMSO control (Fig. 6B).
Fig. 6.
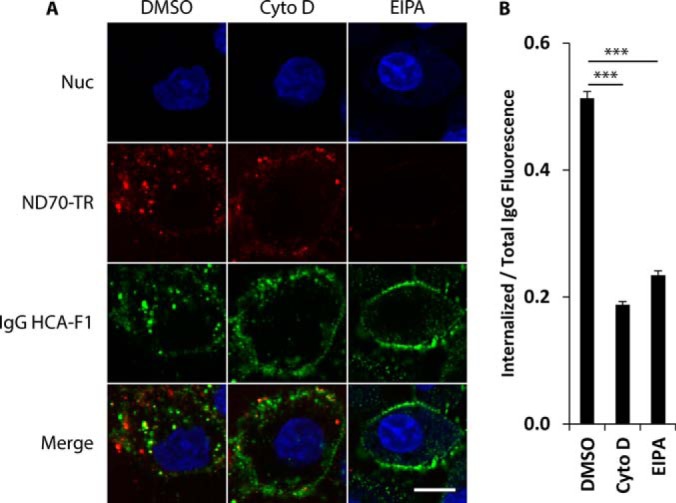
Macropinocytosis inhibitors prevent internalization of IgG HCA-F1. DU145 cells were pretreated with 50 μg/ml cytochalasin D, 7.5 μg/ml EIPA, or DMSO (control) for 30 min at 37 °C followed by co-incubation with 10 μg/ml IgG HCA-F1 and ND70-TR (red) in the presence of cytochalasin D, EIPA, or DMSO in complete DMEM/10% FBS for 40 min at 37 °C. Cells were then immunolabeled for human IgG (green). Nuclei were stained with Hoechst 33342 (blue). A, Individual confocal Z-slices of representative cells. CytoD: cytochalasin D. Scale bar: 20 μm. B, The percentage of internalized IgG HCA-F1 was quantitated by measuring the ratio of internalized, cytosolic IgG HCA-F1 fluorescence over total cell IgG HCA-F1 fluorescence, analyzing >15 cells over three independent experiments. *** indicates p value of <0.001 using two-tailed student's t test assuming unequal variance. Error bars represent S.E. with n = 3.
EphA2 Identified as Antigen Target for Macropinocytosing IgG HCA-F1
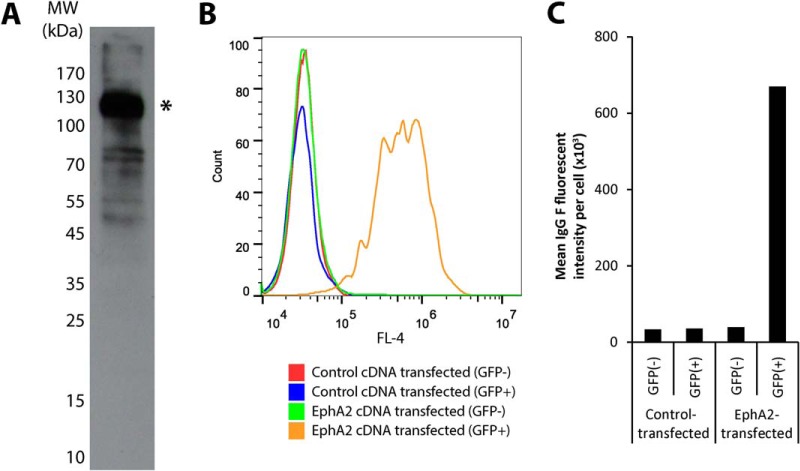
We next sought to determine the target antigen bound by the rapidly internalizing macropinocytosing IgG HCA-F1. We surface-biotinylated DU145 cells, prepared cell lysates, and performed immunoprecipitation with HCA-F1 scFv-Fc immobilized to agarose beads. Immunoprecipitation products underwent parallel SDS-PAGE and immunoblotting. Immunoblotting results with streptavidin-conjugated horseradish peroxidase (HRP) showed a dominant band at ∼110 kDa (Fig. 7A). After excising the corresponding band from the Coomassie-stained gel, the extracted protein gel slice underwent trypsin-digestion and analysis via tandem mass spectrometry. The results identified a transmembrane protein, ephrin type-A receptor 2 (EphA2), as the target antigen (supplemental Table S2. Peptide identification is shown in supplemental Table S3). For an independent verification, we ectopically expressed human EphA2 cDNA in Chinese hamster ovary (CHO) cells and found that IgG HCA-F1 bound strongly to these cells but not CHO cells transfected with a control cDNA (Fig. 7B, C).
Fig. 7.
EphA2 identified as target antigen bound by macropinocytosing antibody IgG HCA-F1. A, Immunoprecipiation of the target antigen from surface-biotinylated Du145 whole cell lysates using scFv HCA-F1-Fc fusion immobilized onto a solid matrix. The immunoprecipitation product was run on SDS-PAGE and subjected to Western blot analysis using streptavidin-HRP to locate the position of membrane proteins. The dominant band, denoted by “*,” represents the approximate region from which the corresponding SDS-PAGE gel was extracted for mass spectrometry analysis. B, Binding to ectopically expressed EphA2. Chinese hamster ovarian (CHO) cells were co-transfected with pEGFP-N2 (to label transfected cells) and pCMV6 expression constructs bearing either human EphA2 or Lgr5 (control). Cells were then incubated with IgG HCA-F1, followed by immunolabeling using anti-human Fc AlexaFluor® 647. Cells were gated for GFP expression and plotted for AlexaFluor® 647 fluorescence (FL4). C, Plot of MFI values as analyzed by FACS. IgG HCA-F1 binds specifically to ectopically expressed EphA2, confirming the target identification.
Neither IgG HCA-M1 nor -S1 binds to EphA2 as they did not bind to cells expressing EphA2 such as benign prostatic hyperplasia (BPH-1) cells that IgG HCA-F1 bound (supplemental Fig. S8). We screened our current cell-binding phage antibody collection and found no additional EphA2 binders within this pool (supplemental Fig. S9). Nonetheless additional anti-EphA2 antibodies could still exist in the LCM selection output, which could be uncovered by expanding the scope of the screening in the future.
Receptor-dependent Macropinocytosis of the Anti-EphA2 IgG
As EphA2 is widely overexpressed by cancer cells (26, 27), we next examined whether IgG HCA-F1 is capable of binding to other cancer cell lines and internalizing via macropinocytosis. We analyzed the binding of IgG HCA-F1 to five human cancer cell lines (prostate cancer DU145, breast cancer MDA-MB-231, lung cancer A549, cervical cancer HeLa, epidermoid carcinoma A431) and two noncancer cell lines (Hs27 and BPH-1) by FACS. IgG HCA-F1 binding was higher for all five cancer cell lines when compared with the noncancer cell lines (supplemental Fig. S10). IgG HCA-F1 did not bind to the LNCaP line that does not express EphA2 (Fig. 8A), demonstrating the receptor-dependent nature of this type of cell entry. To assess binding to cross-species epitopes, we also performed FACS analysis of IgG HCA-F1 on a mouse melanoma cell line B16F10 and observed binding, which suggests that IgG HCA-F1 binds to an EphA2 epitope conserved across species (data not shown). To investigate the specificity of internalization, we co-incubated IgG HCA-F1 and ND70-TR, over the aforementioned panel of both cancer and noncancer cell lines. Confocal imaging using equal exposure times confirmed that IgG HCA-F1 bound strongly to cancer cell lines when compared with noncancer cell lines (supplemental Fig. S11A). Internalized IgG HCA-F1 was quantified by measuring mean fluorescent intensities of IgG HCA-F1 within individual, confocal slices of cytosolic areas of cells. Quantitation of internalized IgG HCA-F1 across all cell lines revealed that cancer cell lines possess greater amounts of internalized IgG HCA-F1 when compared with noncancer cell lines (supplemental Fig. S11B).
Fig. 8.
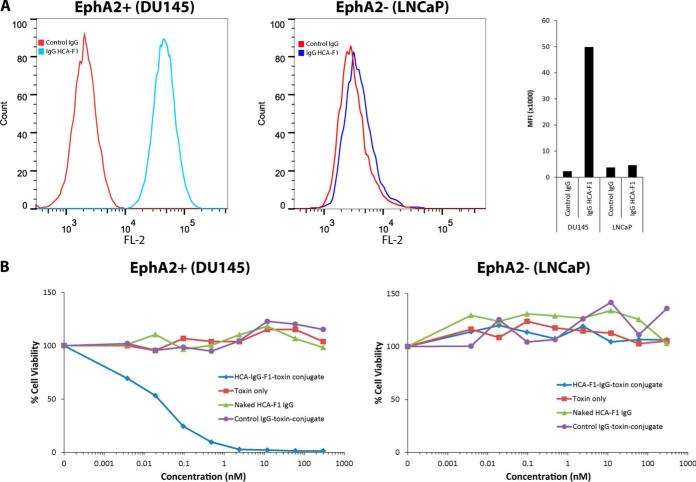
Functional internalization assay using IgG HCA-F1-toxin conjugates. A, FACS analysis showing EphA2-positive (DU145) and EphA2-negative (LNCaP, control) cells. IgG HCA-F1 was incubated with the cells and binding was detected with anti-human Fc. MFI values are shown in the far right panel. B, IgG HCA-F1 was conjugated to saporin and incubated with target (DU145) and control (LNCaP) cells. Controls: toxin only and IgG HCA-F1 only. Cell viability was measured 4 days later using the CCK-8 assay.
Antibody-toxin Conjugate Exhibits Potent Cytotoxicity In Vitro
To obtain functional evidence for internalization, we investigated whether an IgG HCA-F1-based antibody-toxin conjugate could lead to targeted killing of tumor cells. We created an IgG HCA-F1-toxin conjugate by first modifying IgG HCA-F1 with amine-reactive biotin, followed by attachment of streptavidin-conjugated saporin, a highly potent ribosome-inactivating protein toxin. Saporin lacks a chain required for cell insertion and is thus nontoxic by itself. We incubated the antibody-toxin conjugates at varying concentrations with both DU145 (EphA2 positive) and LNCAP (EphA2-negative) cells and examined cell viability after 4 days. The IgG HCA-F1-toxin conjugate exhibited potent cytotoxicity against DU145 cells (EC50 about 19 pm) but not on control LNCaP cells (Fig. 8B), demonstrating functionally a receptor-dependent internalization mechanism. Toxin conjugated to a control nonbinding human IgG did not kill tumor cells, neither did toxin alone nor naked HCA-F1 IgG. These studies provide functional evidence for rapid internalization by our anti-EphA2 antibody IgG HCA-F1 and demonstrate potential for the development of targeted therapeutics against EphA2-positive tumors.
DISCUSSION
Recent studies suggest that macropinocytosis is a rapid and efficient cellular internalization pathway that is up-regulated selectively by tumor cells (9, 28). Exploring this pathway for targeted therapy development has the potential of improving potency and selectivity for tumor targeting agents. Although studies have been done previously to identify internalizing antibodies from phage antibody display libraries (13–15, 29, 30), no method has been developed to identify macropinocytosing antibodies. In this study, we developed an HCA-based high throughput method to identify macropinocytosing antibodies from phage antibody display libraries. Following conversion into full-length human IgG1s, we determined by confocal microscopy that one of the antibodies, IgG HCA-F1, rapidly internalizes via macropinocytosis and colocalizes with early endosome and lysosome markers. The microscopic internalization studies were confirmed by functional internalization assays based on the plant toxin saporin that lacks an internalization mechanism on its own. The rapid internalization of the HCA-F1 IgG resulted in potent cytotoxicity of antibody-toxin conjugate against a broad panel of tumor cells expressing the target antigen, demonstrating functionally that this antibody is efficiently internalized by target cells.
Previous methods to select and screen for internalizing phage antibodies have utilized low pH, high salt wash buffers in an attempt to strip away surface-bound phage antibodies (14, 15, 29). Although this approach has been at least partially successful, strong binding high affinity phage antibodies may be resistant to even these harsh conditions. Indeed, when we tested strong binding phage antibodies on fixed cells that are incapable of internalization, we found that binding was resistant to low pH, high salt washes. In addition, we found that analysis of patterns of cell-associated phage that were generated by nonconfocal HCA instruments was not sufficient to determine if the phage is internalized. Many heterogeneous patterns were observed, and it was difficult to reliably associate any of the patterns with internalization, let alone macropinocytosis. Thus, our new methods based on multimarker microscopic HCA establish an effective means for the identification of internalizing and furthermore macropinocytosing antibodies from phage display libraries.
Our studies showed that there are major differences in internalization kinetics between an antibody in soluble form and on phage, which must be taken into consideration for screening design. For example, when tested in full-length IgG or scFv-Fc fusion forms, the highly active macropinocytosing antibody HCA-F1 starts internalization almost immediately and completes the process in 40–80 min, whereas the same antibody in phage format does so in 8 h. The large size of the phage particle may have slowed down the internalization process considerably regardless of how rapidly the antibody internalizes in soluble forms. In addition, although in soluble forms different antibodies utilized disparate internalization pathways, in phage forms they seem to converge into the macropinocytosis pathway. This is not entirely surprising considering the size of the phage particle. Nonetheless, despite the generally retarded rate and the near uniform route of phage particle internalization (phage macropinocytosis), the kinetic differences in phage antibody internalization are a function of the underlying scFv, with HCA-F1-like phage internalizing in 8 h, HCA-M1-like in 24 h and HCA-S1-like >24 h. These kinetics differences allowed us to develop screening schemes to uncover rapidly internalizing antibodies such as the macropinocytosing antibody HCA-F1. In this context, we would like to re-emphasize that HCA screening using phage directly is convenient and compatible with the high throughout format, but the result must be verified using antibodies in soluble forms. Either scFv-Fc fusions or full IgGs can be used for secondary screening or validation study as we have not found significant differences between these two forms of soluble antibodies with regard to internalization patterns. We normally produce and test the scFv-Fc fusion molecules first because of ease in cloning and production, followed by conversion of scFvs into human IgG molecules for bioassays.
Another peculiar feature of phage internalization is revealed by our organelle-labeling experiment. Although phage antibodies are seen to colocalize to lysosomes, they could not be readily seen entering cells via the early endosomal pathway. We reconciled this observation by hypothesizing that large, macropinocytosed phage particles may be trafficking via endosomes distinct from traditional coated vesicle-formed endosomes, which has been previously observed (8). For IgG however, we were able to observe that the phage-derived IgG could internalize via macropinocytosis toward the lysosomal organelles via the endosomal pathway.
EphA2 is known to be expressed by various tumor cells and play roles in tumor invasion and metastasis (26). EphA2 also interacts with Kaposi's sarcoma-associated herpesvirus and regulates viral entry into endothelial cells by macropinocytosis (31). Several groups have developed anti-EphA2 antibodies (32–34), and it appears that different epitopes mediate different rates of internalization (32, 35). No phage antibody library selection scheme has been developed previously that allows for selection of macropinocytosing antibodies binding to EphA2 or other antigens. Our unbiased screening has uncovered an antibody that binds to EphA2 and is rapidly internalized by the macropinocytosis pathway, thereby creating novel agents against this receptor.
EphA2 has been the target for many forms of cancer therapy development. Nanoparticles conjugated with anti-EphA2 antibodies have been used for siRNA delivery (36, 37). In addition, an anti-EphA2 antibody auristatin E conjugate was tested in a phase I trial for solid tumor treatment (33, 38). This particular anti-EphA2 antibody-auristatin E-conjugate showed unacceptably high toxicity at sub-therapeutic doses (38). Given that different EphA2 epitopes distinctly influence the kinetics and pathway of internalization, it is possible that the aforementioned setback with the anti-EphA2 antibody-auristatin E-conjugate is an isolated phenomenon relating to the particular antibody used. In any event, anti-EphA2 antibodies can be utilized to deliver payloads other than auristatin. As such there still could be further development of an anti-EphA2 antibody-based therapeutic in the future. Our anti-EphA2 antibody is internalized by the tumor selective macropinocytosis pathway. It remains to be determined if our antibody has a different potency/toxicity profile than those previously reported anti-EphA2 antibodies. Given that the macropinocytosing epitope bound by our HCA-F1 antibody is conserved across species, any targeted therapeutics developed from this antibody can be tested in small rodents to obtain meaningful toxicology profiles. Finally, we have only screened a limited fraction of our LCM-selection output and identified one anti-EphA2 binder. Work is underway to expand the scope of the screening to identify additional macropinocytosing anti-EphA2 antibodies.
We have previously developed an LCM-based selection strategy to enrich for phage antibodies binding to tumor cells in situ residing in their tissue microenvironment as opposed to cell line artifacts (1). In this report we further screened the LCM selection output using our HCA-based method and identified novel macropinocytosing human antibodies targeting clinically relevant tumor antigens. Integrating LCM and HCA into phage antibody display library selection thus allows identification of novel antibodies that target true tumor antigens expressed by tumor cells residing in their tissue microenvironment and enter target cells via tumor selective pathways such as macropinocytosis. Targeted therapeutics based on these novel antibodies have the potential to improve potency in tumor killing and reduce toxicity on normal tissues, thus widening the therapeutic window and improving effectiveness of such antibody-targeted therapeutics.
Supplementary Material
Acknowledgments
We thank Drs. Christopher R. Behrens, NamKyung Lee, and Daniel Sherbenou for helpful discussions.
Footnotes
Author contributions: K.D.H., S.M.B., and B.L. designed research; K.D.H., S.M.B., Y.Z., and Y.S. performed research; K.D.H., S.M.B., Y.Z., and Y.S. contributed new reagents or analytic tools; K.D.H., S.M.B., Y.Z., Y.S., and B.L. analyzed data; K.D.H. and B.L. wrote the paper.
* This work was supported by the National Institutes of (R01 CA118919, R01 CA129491 and R01 CA171315) and the Center for Mass Spectrometry and Proteomics at the University of Minnesota for mass spectrometry analysis.
 This article contains Supplemental Figs. S1 to S11, Tables S1 to S3 and Movies S1 and S2.
This article contains Supplemental Figs. S1 to S11, Tables S1 to S3 and Movies S1 and S2.
1 The abbreviations used are:
- HCA
- High content analysis
- ScFv
- single chain variable fragment
- PCC
- Pearson's correlation coefficient
- CFU
- Colony forming unit
- MFI
- mean fluorescence intensity
- EEA
- early endosomal antigen
- LAMP
- lysosomal-associated membrane protein
- IgG
- immunoglobulin G
- ND70-TR
- Texas Red-conjugated neutral dextran 70 kDa
- FBS
- Fetal bovine serum
- HEK
- human embryonic kidney
- LCM
- Laser capture microdissection
- EphA2
- ephrin type-A receptor 2
- HRP
- horseradish peroxidase
- EC50
- half maximal effective concentration
- MAbs
- monoclonal antibodies.
REFERENCES
- 1. Ruan W., Sassoon A., An F., Simko J. P., Liu B. (2006) Identification of clinically significant tumor antigens by selecting phage antibody library on tumor cells in situ using laser capture microdissection. Mol. Cell. Proteomics. 5, 2364–2373 [DOI] [PubMed] [Google Scholar]
- 2. Austin C. D., De Maziere A. M., Pisacane P. I., van Dijk S. M., Eigenbrot C., Sliwkowski M. X., Klumperman J., Scheller R. H. (2004) Endocytosis and sorting of ErbB2 and the site of action of cancer therapeutics trastuzumab and geldanamycin. Mol. Biol. Cell 15, 5268–5282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Burris H. A., 3rd, Tibbitts J., Holden S. N., Sliwkowski M. X., Lewis Phillips G. D. (2011) Trastuzumab emtansine (T-DM1): a novel agent for targeting HER2+ breast cancer. Clin. Breast Cancer 11, 275–282 [DOI] [PubMed] [Google Scholar]
- 4. Sievers E. L., Senter P. D. (2013) Antibody-drug conjugates in cancer therapy. Annu. Rev. Med. 64, 15–29 [DOI] [PubMed] [Google Scholar]
- 5. Behrens C. R., Liu B. (2013) Methods for site-specific drug conjugation to antibodies. MAbs. 6, 46–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sutherland M. S., Sanderson R. J., Gordon K. A., Andreyka J., Cerveny C. G., Yu C., Lewis T. S., Meyer D. L., Zabinski R. F., Doronina S. O., Senter P. D., Law C. L., Wahl A. F. (2006) Lysosomal trafficking and cysteine protease metabolism confer target-specific cytotoxicity by peptide-linked anti-CD30-auristatin conjugates. J. Biol. Chem. 281, 10540–10547 [DOI] [PubMed] [Google Scholar]
- 7. Erickson H. K., Park P. U., Widdison W. C., Kovtun Y. V., Garrett L. M., Hoffman K., Lutz R. J., Goldmacher V. S., Blattler W. A. (2006) Antibody-maytansinoid conjugates are activated in targeted cancer cells by lysosomal degradation and linker-dependent intracellular processing. Cancer Res. 66, 4426–4433 [DOI] [PubMed] [Google Scholar]
- 8. Hewlett L. J., Prescott A. R., Watts C. (1994) The coated pit and macropinocytic pathways serve distinct endosome populations. J. Cell Biol. 124, 689–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Commisso C., Davidson S. M., Soydaner-Azeloglu R. G., Parker S. J., Kamphorst J. J., Hackett S., Grabocka E., Nofal M., Drebin J. A., Thompson C. B., Rabinowitz J. D., Metallo C. M., Vander Heiden M. G., Bar-Sagi D. (2013) Macropinocytosis of protein is an amino acid supply route in Ras-transformed cells. Nature 497, 633–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hayward S. W., Dahiya R., Cunha G. R., Bartek J., Deshpande N., Narayan P. (1995) Establishment and characterization of an immortalized but nontransformed human prostate epithelial cell line: BPH-1. In Vitro Cell Dev. Biol. Anim. 31, 14–24 [DOI] [PubMed] [Google Scholar]
- 11. O'Connell D., Becerril B., Roy-Burman A., Daws M., Marks J. D. (2002) Phage versus phagemid libraries for generation of human monoclonal antibodies. J. Mol. Biol. 321, 49–56 [DOI] [PubMed] [Google Scholar]
- 12. Liu B., Marks J. D. (2000) Applying phage antibodies to proteomics: selecting single chain Fv antibodies to antigens blotted on nitrocellulose. Anal. Biochem. 286, 119–128 [DOI] [PubMed] [Google Scholar]
- 13. Zhu X., Bidlingmaier S., Hashizume R., James C. D., Berger M. S., Liu B. (2010) Identification of internalizing human single-chain antibodies targeting brain tumor sphere cells. Mol. Cancer Ther. 9, 2131–2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. An F., Drummond D. C., Wilson S., Kirpotin D. B., Nishimura S. L., Broaddus V. C., Liu B. (2008) Targeted drug delivery to mesothelioma cells using functionally selected internalizing human single-chain antibodies. Mol. Cancer Ther. 7, 569–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liu B., Conrad F., Cooperberg M. R., Kirpotin D. B., Marks J. D. (2004) Mapping tumor epitope space by direct selection of single-chain Fv antibody libraries on prostate cancer cells. Cancer Res. 64, 704–710 [DOI] [PubMed] [Google Scholar]
- 16. Smith K., Garman L., Wrammert J., Zheng N. Y., Capra J. D., Ahmed R., Wilson P. C. (2009) Rapid generation of fully human monoclonal antibodies specific to a vaccinating antigen. Nat. Protoc. 4, 372–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Liu B., Conrad F., Roth A., Drummond D. C., Simko J. P., Marks J. D. (2007) Recombinant full-length human IgG1s targeting hormone-refractory prostate cancer. J. Mol. Med. 85, 1113–1123 [DOI] [PubMed] [Google Scholar]
- 18. Conrad F., Zhu X., Zhang X., Chalkley R. J., Burlingame A. L., Marks J. D., Liu B. (2009) Human antibodies targeting cell surface antigens overexpressed by the hormone refractory metastatic prostate cancer cells: ICAM-1 is a tumor antigen that mediates prostate cancer cell invasion. J. Mol. Med. 87, 507–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Keller A., Nesvizhskii A. I., Kolker E., Aebersold R. (2002) Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74, 5383–5392 [DOI] [PubMed] [Google Scholar]
- 20. Nesvizhskii A. I., Keller A., Kolker E., Aebersold R. (2003) A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75, 4646–4658 [DOI] [PubMed] [Google Scholar]
- 21. Schnatwinkel C., Christoforidis S., Lindsay M. R., Uttenweiler-Joseph S., Wilm M., Parton R. G., Zerial M. (2004) The Rab5 effector Rabankyrin-5 regulates and coordinates different endocytic mechanisms. PLoS Biol. 2, E261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Veithen A., Amyere M., Van Der Smissen P., Cupers P., Courtoy P. J. (1998) Regulation of macropinocytosis in v-Src-transformed fibroblasts: cyclic AMP selectively promotes regurgitation of macropinosomes. J. Cell Sci. 111, 2329–2335 [DOI] [PubMed] [Google Scholar]
- 23. Gold S., Monaghan P., Mertens P., Jackson T. (2010) A clathrin independent macropinocytosis-like entry mechanism used by bluetongue virus-1 during infection of BHK cells. PLoS One. 5, e11360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Veithen A., Cupers P., Baudhuin P., Courtoy P. J. (1996) v-Src induces constitutive macropinocytosis in rat fibroblasts. J. Cell Sci. 109, 2005–2012 [DOI] [PubMed] [Google Scholar]
- 25. West M. A., Bretscher M. S., Watts C. (1989) Distinct endocytotic pathways in epidermal growth factor-stimulated human carcinoma A431 cells. J. Cell Biol. 109, 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wykosky J., Debinski W. (2008) The EphA2 receptor and ephrinA1 ligand in solid tumors: function and therapeutic targeting. Mol. Cancer Res. 6, 1795–1806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tandon M., Vemula S. V., Mittal S. K. (2011) Emerging strategies for EphA2 receptor targeting for cancer therapeutics. Expert. Opin. Ther. Targets. 15, 31–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Reyes-Reyes E. M., Teng Y., Bates P. J. (2010) A new paradigm for aptamer therapeutic AS1411 action: uptake by macropinocytosis and its stimulation by a nucleolin-dependent mechanism. Cancer Res. 70, 8617–8629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Poul M. A., Becerril B., Nielsen U. B., Morisson P., Marks J. D. (2000) Selection of tumor-specific internalizing human antibodies from phage libraries. J. Mol. Biol. 301, 1149–1161 [DOI] [PubMed] [Google Scholar]
- 30. Rudnick S. I., Lou J., Shaller C. C., Tang Y., Klein-Szanto A. J., Weiner L. M., Marks J. D., Adams G. P. (2011) Influence of affinity and antigen internalization on the uptake and penetration of Anti-HER2 antibodies in solid tumors. Cancer Res. 71, 2250–2259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chakraborty S., Veettil M. V., Bottero V., Chandran B. (2012) Kaposi's sarcoma-associated herpesvirus interacts with EphrinA2 receptor to amplify signaling essential for productive infection. Proc. Natl. Acad. Sci. U. S. A. 109, E1163–E1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ansuini H., Meola A., Gunes Z., Paradisi V., Pezzanera M., Acali S., Santini C., Luzzago A., Mori F., Lazzaro D., Ciliberto G., Nicosia A., La Monica N., Vitelli A. (2009) Anti-EphA2 antibodies with distinct in vitro properties have equal in vivo efficacy in pancreatic cancer. J. Oncol. 2009, 951917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jackson D., Gooya J., Mao S., Kinneer K., Xu L., Camara M., Fazenbaker C., Fleming R., Swamynathan S., Meyer D., Senter P. D., Gao C., Wu H., Kinch M., Coats S., Kiener P. A., Tice D. A. (2008) A human antibody-drug conjugate targeting EphA2 inhibits tumor growth in vivo. Cancer Res. 68, 9367–9374 [DOI] [PubMed] [Google Scholar]
- 34. Zhou Y., Zou H., Zhang S., Marks J. D. (2010) Internalizing cancer antibodies from phage libraries selected on tumor cells and yeast-displayed tumor antigens. J. Mol. Biol. 404, 88–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Peng L., Oganesyan V., Damschroder M. M., Wu H., Dall'Acqua W. F. (2011) Structural and functional characterization of an agonistic anti-human EphA2 monoclonal antibody. J. Mol. Biol. 413, 390–405 [DOI] [PubMed] [Google Scholar]
- 36. Shen H., Rodriguez-Aguayo C., Xu R., Gonzalez-Villasana V., Mai J., Huang Y., Zhang G., Guo X., Bai L., Qin G., Deng X., Li Q., Erm D. R., Aslan B., Liu X., Sakamoto J., Chavez-Reyes A., Han H. D., Sood A. K., Ferrari M., Lopez-Berestein G. (2013) Enhancing chemotherapy response with sustained EphA2 silencing using multistage vector delivery. Clin. Cancer Res. 19, 1806–1815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tanaka T., Mangala L. S., Vivas-Mejia P. E., Nieves-Alicea R., Mann A. P., Mora E., Han H. D., Shahzad M. M., Liu X., Bhavane R., Gu J., Fakhoury J. R., Chiappini C., Lu C., Matsuo K., Godin B., Stone R. L., Nick A. M., Lopez-Berestein G., Sood A. K., Ferrari M. (2010) Sustained small interfering RNA delivery by mesoporous silicon particles. Cancer Res. 70, 3687–3696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Annunziata C. M., Kohn E. C., LoRusso P., Houston N. D., Coleman R. L., Buzoianu M., Robbie G., Lechleider R. (2013) Phase 1, open-label study of MEDI-547 in patients with relapsed or refractory solid tumors. Invest. New Drugs. 31, 77–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.