Abstract
HPV-positive oropharyngeal carcinoma (OPC) patients have superior outcomes relative to HPV-negative patients, but the underlying mechanisms remain poorly understood. We conducted a proteomic investigation of HPV-positive (n = 27) and HPV-negative (n = 26) formalin-fixed paraffin-embedded OPC biopsies to acquire insights into the biological pathways that correlate with clinical behavior. Among the 2,633 proteins identified, 174 were differentially abundant. These were enriched for proteins related to cell cycle, DNA replication, apoptosis, and immune response. The differential abundances of cortactin and methylthioadenosine phosphorylase were validated by immunohistochemistry in an independent cohort of 29 OPC samples (p = 0.023 and p = 0.009, respectively). An additional 1,124 proteins were independently corroborated through comparison to a published proteomic dataset of OPC. Furthermore, utilizing the Cancer Genome Atlas, we conducted an integrated investigation of OPC, attributing mechanisms underlying differential protein abundances to alterations in mutation, copy number, methylation, and mRNA profiles. A key finding of this integration was the identification of elevated cortactin oncoprotein levels in HPV-negative OPCs. These proteins might contribute to reduced survival in these patients via their established role in radiation resistance. Through interrogation of Cancer Genome Atlas data, we demonstrated that activation of the β1-integrin/FAK/cortactin/JNK1 signaling axis and associated differential regulation of activator protein 1 transcription factor target genes are plausible consequences of elevated cortactin protein levels.
In the Western world, the overall decline in the incidence of head and neck squamous cell carcinoma (HNSCC)1 has been accompanied by an increase in oropharyngeal carcinoma (OPC), the most common HNSCC subtype (1, 2). This trend has been attributed to a concomitant decline in smoking and increase in oral human papillomavirus (HPV) infection rates (2). HPV has been reported to be associated with more than 60% of all OPCs in North America, with HPV16 being the predominant high-risk strain in HNSCC (2–5).
The process of tumorigenesis by oncogenic HPV is well characterized. It is driven by the viral oncoproteins E6 and E7, which in turn inactivate the tumor suppressor proteins p53 and pRb, respectively (6). E7 binds pRb, thereby releasing the E2F transcription factor from pRb, leading to subsequent overexpression of p16 (i.e. cyclin-dependent kinase inhibitor 2A (CDKN2A)) (3). p16 expression is lost in the majority of HPV-negative (HPV−) tumors; thus it has been well accepted as a surrogate marker for HPV positivity in OPC. Although in situ hybridization remains the gold standard for HPV detection because it allows direct visualization of HPV in tumor nuclei, p16 immunoexpression is an equivalent method for HPV status determination and is commonly utilized in clinical settings (5, 7).
HPV-positive (HPV+) and HPV− OPCs are distinct tumor entities with different clinical characteristics and molecular profiles (8). Unlike HPV− cancers, HPV+ OPCs are associated with a more favorable outcome and treatment response, younger age at diagnosis, and wild-type p53 status (3, 9, 10). This has been corroborated by our group in a Canadian cohort, where we demonstrated that the 3-year overall survival (OS) rates were 67% for HPV− OPC and 88% for HPV+ OPC (4, 5). Despite this differential outcome, OPCs are typically treated with radiation therapy with or without concurrent chemotherapy irrespective of tumor HPV status (4). Clinical trials are ongoing to determine the efficacy of treatment de-escalation for HPV+ OPC, with the expectation that this will reduce treatment-related toxicities without adversely affecting patient survival (11). However, further studies are required in order to unravel the biological mechanisms accounting for differences in outcome and therapeutic response between HPV+ and HPV− OPCs. An enhanced mechanistic understanding of these two distinct malignancies could aid in the development of tailored therapeutics, as well as biomarkers for treatment stratification.
Analytical protocols for the proteomic characterization of formalin-fixed paraffin-embedded (FFPE) patient materials have been recently developed, enabling utilization of this valuable resource (12, 13). When coupled with high-resolution mass spectrometry (MS) techniques, these approaches make it possible to obtain a quantitative snapshot of protein abundances across increasingly large sample cohorts, thereby providing novel insights into mechanisms of disease progression (14–16). In parallel, the efforts of the Cancer Genome Atlas (TCGA) have produced broad genomic characterization of ∼300 head and neck cancer (HNC) specimens, including OPCs, highlighting potential genomic alterations responsible for cancer initiation and progression.2 What is currently missing is an integration of TCGA genomic alterations with in-depth proteomic analyses (17). Because proteins and their functions are the ultimate mediators of biological activity, such integrative efforts could highlight important biological regulatory mechanisms unlikely to be discovered otherwise (17–20). Capitalizing on these developments, we undertook a comparative global proteomics investigation complemented by genomic analysis in order to identify differentially abundant proteins and, using these resources, explore the mechanisms underlying the distinct clinical behavior of HPV+ versus HPV− (HPV±) OPCs.
MATERIALS AND METHODS
Clinical Specimens
Institutional Research Ethics Board approval was obtained for this study (REB# 07–0521-CE). Primary diagnostic (i.e. pre-treatment) fully clinically annotated, non-metastatic FFPE OPC biopsies from a total of 82 patients were utilized (5-μm sections). The clinical characteristics of the patients are shown in Table I. The discovery and validation cohorts were group-matched for age, sex, stage, and treatment. Samples from the first cohort were utilized for MS-based proteomic discovery (n = 53; median follow-up time of 2.6 years; supplemental Table S1), and those from the second cohort were used for immunohistochemistry (IHC) validation (n = 29; median follow-up time of 2.4 years; supplemental Table S2). Both MS and IHC experiments were performed blinded, without knowledge of HPV status or clinical outcome.
Table I. Clinico-pathological characteristics of patients for tissue mass spectrometry and IHC studies.
| Clinical factors | Discovery cohort |
Validation cohort |
Discovery vs. validation |
||||
|---|---|---|---|---|---|---|---|
| HPV− (n = 26) | HPV+ (n = 27) | p value | HPV− (n = 17) | HPV+ (n = 12) | p value | p value | |
| Age (median, range) | 67 (33–80) | 67 (41–83) | 0.94 | 69 (43–93) | 69 (35–84) | 0.82 | 0.48 |
| Sex (male/female) | 22/4 | 20/7 | 0.50 | 15/2 | 12/0 | 0.50 | 0.12 |
| Stage | 0.08 | 0.26 | 0.99 | ||||
| I | 1 | 0 | 0 | 0 | |||
| II | 3 | 2 | 3 | 0 | |||
| III | 7 | 2 | 3 | 3 | |||
| IVA | 10 | 20 | 10 | 6 | |||
| IVB | 5 | 3 | 1 | 3 | |||
| Treatment | 0.20 | 1.00 | 0.44 | ||||
| RT | 18 | 14 | 12 | 8 | |||
| CRT | 8 | 13 | 5 | 4 | |||
Notes: RT, radiotherapy alone; CRT, chemo-radiotherapy.
Extraction and Digestion of Proteins from FFPE Tissue
Following macro-dissection of tissue sections to obtain >70% tumor cell content, tissues were de-paraffinized and rehydrated using sequential washes in xylene; 100%, 98%, and 70% ethanol; and water. The tumor sections were then incubated in a 100 mm ammonium bicarbonate, 20% acetonitrile, pH 8.5, buffer at 95 °C for 2 h followed by 65 °C for 1 h. The tissue was digested overnight at 37 °C with recombinant, proteomics-grade trypsin (Promega, Madison, WI). Undigested tissues were removed via centrifugation, and peptides were solid-phase extracted with C18 spin-columns (The Nest Group Inc., Southbrough, MA) based on the manufacturer's instructions, vacuum-concentrated, and reconstituted in Buffer A (0.1% formic acid, 5% acetonitrile). A BCA assay was performed to determine the peptide concentration, and 5-μg peptide aliquots were kept at −80 °C until analyzed via multi-dimensional protein identification technology (MudPIT).
MudPIT Analysis of FFPE Tissue Peptides
A fully automated five-cycle two-dimensional high-performance liquid chromatography sequence was set up as previously described (21). Peptides were loaded onto a 7-cm Kasil fritted pre-column (150-μm inner diameter) packed with 3.5 cm of 5-μm Magic C18 100-Å reversed-phase material (Michrom Bioresources Inc., Auburn, CA) followed by 3.5 cm of Luna 5-μm SCX 100-Å strong cation exchange resin (Phenomenex, Torrance, CA). Samples were loaded automatically from a 96-well microplate autosampler at 3 μl/min using the EASY-nLC system (Thermo Scientific). The pre-column was connected to a fused silica analytical column (8 cm long, 75-μm inner diameter) via a microsplitter tee (Thermo Scientific) to which a distal 2.0-kV spray voltage was applied. The analytical column was pulled to a fine electrospray emitter using a laser puller. For peptide separation on the analytical column, a water/acetonitrile gradient, controlled by the EASY-nLC (Thermo Scientific), was applied at an effective flow rate of 400 nL/min. Ammonium acetate salt bumps (8 μl) at concentrations of 100, 150, 200, and 500 mm were sequentially loaded, and peptides were eluted by a water/acetonitrile gradient as described previously (22). Sample analysis was performed on an LTQ Orbitrap XL (Thermo Scientific) using previously described instrument parameters (22).
Protein Identification and Data Analysis
Raw data were converted to m/z XML files using ReAdW (v1.1) and searched against the Human UniProt database (version 2011/03; 20,227 sequences) using X! Tandem (CYCLONE v2011.12.01.1), OMSSA (v2.1.8), and MyriMatch (v2.1.97) search algorithms. The search was conducted with a fragment ion mass tolerance of ±0.40 Da and a parent ion tolerance of ±10 ppm. Complete tryptic digestion was assumed with one allowed missed cleavage site. Methionine oxidation was specified as a variable modification. For protein inference minimization, an in-house grouping scheme was applied, reporting only proteins with substantial peptide information (23). Target/decoy searches were performed to experimentally estimate the protein false discovery rate, which was determined to be 1.3%. Protein identifications with at least two unique tryptic peptides were considered (23, 24).
Protein Quantification via MS and Normalization
Adjusted spectral counts were determined in a manner previously described and used as a measure of relative protein abundance (25). To avoid division by 0, adjusted spectral count values of 0 were replaced by 0.1. The fold change (FC) for each protein was determined by calculating the ratio of averaged normalized adjusted spectral counts in HPV± samples.
Mass Spectrometry Raw Data
The raw mass spectrometry data associated with this manuscript have been submitted to a public repository (the Mass Spectrometry Interactive Virtual Environment, http://massive.ucsd.edu) for others to download. These data are associated with the identifier MassIVE ID MSV000078610 at the FTP site ftp://MSV000078610@massive.ucsd.edu.
Functional Enrichment Analysis
Protein Center Software (v3.12.10008) was used for Gene Ontology (GO) and KEGG Pathway enrichment analyses. The DAVID UCSB_TFBS annotation tool (v6.7) was used for transcription factor target enrichment analysis (26). Enrichment analyses were performed against all genes or identified proteins in a given study as the background. Significance was set at FDR < 0.05.
Data Integration with Published Proteomic Data and the Cancer Genome Atlas
For proteomic data integration, IPI accessions from Slebos et al. were mapped to their corresponding gene names and compared by linking both datasets by gene accessions using an in-house database (27). In cases where multiple proteins mapped to the same gene, the proteomic results were averaged.
For integrated -omics analysis, anonymized Level 3 processed TCGA results for the Head and Neck Provisional Data Release (2013_05_23 Firehose Analysis Run) were downloaded from the cBioPortal using the R (v3.0.1) API (cgds v1.1.29) on October 8, 2013 (28, 29). There were 310 total samples (295 complete), of which 306 had somatic point mutation and copy number alternation (CNA) data, 303 had RNA sequencing data, and 310 had methylation data. Types of data included functional mutation counts (missense, nonsense), copy number calls, RNA-sequencing normalized counts, Infinium 450 methylation values, and clinical attributes. Data processing protocols are described in the MAGE-TAB archives associated with the original data2 and in the TCGA bladder cancer marker paper (30).
Samples were limited to the oropharynx subsite (tonsil, oropharynx, or base of tongue) with information on HPV status (n = 32; 10 HPV+, 22 HPV−). HPV status was defined as any sample with evidence for reads aligned to the HPV viral sequence in DNA sequencing (exome, low-pass whole-genome sequencing) or the presence of E6/E7 viral transcripts in RNA sequencing. Normalized mRNA counts were calculated using RSEM (31). Copy number was based on the discretized log2 ratio of tumor to normal pre-processed probe intensity averaged across each gene (32). Methylation was based on the ratio of background corrected intensities for methylated versus unmethylated probes (β values) determined by interrogating the tumor independently. If more than one probe was assigned to a gene, the one with the strongest negative correlation with mRNA expression was selected.
The primary analysis was performed for selected genes that were identified as differentially abundant for proteins (n = 174). Analysis of AP-1 target genes (n = 23) was limited to mRNA counts. Data visualization used the lattice (v0.20–24) package for the R statistical environment. For qualitative assessment of correlation between proteomic and genomic profiles, concordance was defined as similar significance levels (p < 0.05 or p > 0.05) and equivalent directionality of relative changes of molecular profiles in HPV± OPCs.
Immunohistochemistry
Protein expression was evaluated on FFPE tumor sections using microwave antigen retrieval with 0.01 m citric acid (pH 6.0) or 10 mmol/l Tris, 1 mmol/l EDTA, pH 9.0, in combination with LSAB+ System-HRP (Dako, Burlington, Ontario, Canada). Mouse monoclonal anti-keratin 17 (1:20 dilution, Dako), rabbit polyclonal anti-MTAP (1:200 dilution, ProteinTech, Tucson, AZ), rabbit polyclonal anti-cortactin (1:25 dilution, Cell Signaling, Danvers, MA), and rabbit polyclonal anti-ICAM1 (1:250 dilution, Sigma-Prestige) antibodies were utilized. Primary antibody was omitted as a negative control.
Tumor cytoplasmic staining levels were scored according to intensity of immunoexpression, with 0, 1, 2, or 3 indicating negative, weak, moderate, or strong staining in tumor cells, respectively. Tumor cell plasma membrane staining was scored as a percentage of cells with staining, and the final score was calculated as a product of cytoplasmic and plasma membrane staining intensities. IHC scoring was performed blinded to knowledge of any clinico-pathological parameters or HPV status. Each tissue section was scored twice, and the scores were averaged.
Statistics
Patient OS was calculated from the date of diagnosis to death or the last follow-up time. Disease-free survival (DFS) was calculated from the date of diagnosis to death, recurrence, or the last follow-up time. Patient survival was estimated via the Kaplan–Meier method, and the log-rank test was used for comparison of OS and DFS rates among patient groups. The Cox proportional-hazard (PH) model was used to estimate hazard ratios and corresponding 95% confidence intervals. The non-parametric Kruskal–Wallis test was applied for group comparisons on continuous clinico-pathological variables, and Pearson's χ2 test or Fisher's exact test was used for categorical variables. Pearson's χ2 test was used to compare GO term classifications based on HPV status.
The Wilcoxon rank-sum test and log2(FC) were used to compare continuous genomic features (protein, mRNA, and methylation), whereas a proportion test and frequency difference were used to compare the significance and magnitude of discrete genomic features (CNAs and mutations). For these comparisons, significance was set at p < 0.05. A protein was considered to be significantly differentially abundant if it also had a |log2(FC)| value greater than 1. Correction for multiple testing was performed using the FDR method in order to determine the top differentially abundant proteins (|log2(FC)| ≥ 1; FDR < 0.05) and proteins for functional enrichment analysis (FDR < 0.05) (33).
Unsupervised Hierarchical Clustering
The globally normalized OPC protein expression dataset was filtered to include the top 10% of proteins with the highest variance. Unsupervised hierarchical clustering was performed with the hclust function (R software v3.0.2) using average linkage as the agglomeration method and Pearson's correlation as a measure of similarity.
RESULTS
Global Proteomic Profiling of HPV+ and HPV− OPCs
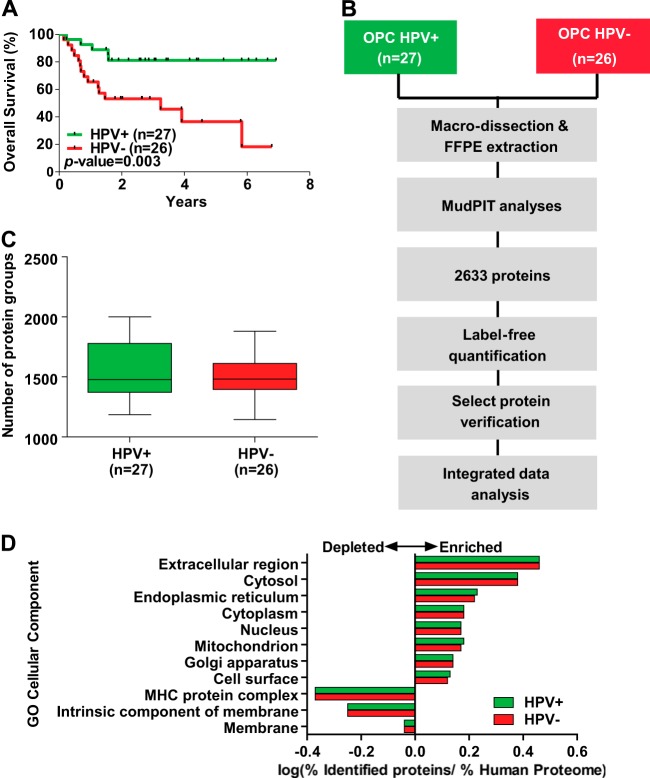
In order to obtain global protein expression profiles for OPCs, we conducted five-step MudPIT analyses on the 53 FFPE biopsy specimens serving as the discovery cohort, including 26 HPV− and 27 HPV+ OPCs. These specimens were deliberately selected to reflect the clinical behavior exemplified by the superior outcome of the HPV+ patient group while balancing for other clinico-pathological variables such as age, sex, tumor stage, and treatment (Table I; supplemental Table S1). The 3-year OS rates for HPV+ and HPV− patients were 81% and 46%, respectively (Fig 1A; p = 0.003). The workflow and experimental design of this study are shown in Fig. 1B. As a result of the MudPIT analyses, 2,633 protein groups were identified with high confidence (FDR = 1.3%), with an average of ∼1,500 protein groups identified in HPV+ and HPV− OPCs (Fig. 1C). The complete lists of protein identifications, normalized protein abundances, and peptide identifications are presented in supplemental Tables S3, S4, and S5. The identified proteins span all the major subcellular components; however, membrane proteins were underrepresented (Fig. 1D). There was no significant difference in relative protein distributions across subcellular components in HPV± OPCs (p value > 0.999, Pearson's χ2 test).
Fig. 1.
Global proteomic analysis of OPCs. A, Kaplan–Meier estimates of overall survival among OPC patients in the discovery cohort according to HPV status. B, outline of the experimental workflow for in-depth proteomic analysis of FFPE OPC tissues. C, number of protein groups identified in HPV+ and HPV− OPCs. D, cellular component Gene Ontology (GO) term enrichment analysis for proteins identified in HPV+ and HPV− OPCs (p > 0.999, Pearson's χ2 test). Select terms with FDR < 0.05 are shown.
Among the 2,633 protein groups identified, 159 were significantly up-regulated and 15 were significantly down-regulated in HPV± OPCs (Fig. 2A). The surrogate marker for HPV positivity, CDKN2A, was the most significantly up-regulated protein in HPV± OPCs (FC = 10.17, p < 0.0001). CDKN2A was detected in 12 of 27 (44%) HPV+ tumors, but it was not detected in any of the HPV− samples (Fig. 2B).
Fig. 2.
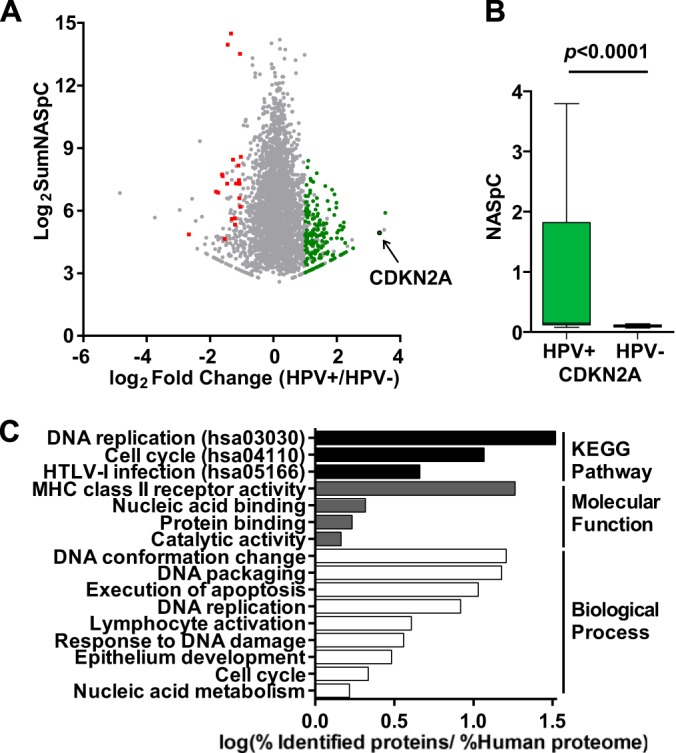
Differentially expressed proteins in HPV+ versus HPV− OPCs. A, protein ratios as a function of protein abundances of the 2,633 protein groups identified. Fifteen proteins were significantly more abundant in HPV− OPCs (red data points), and 159 proteins were significantly more abundant in HPV+ OPCs (green data points). The arrow denotes CDKN2A. B, significantly higher CDKN2A protein abundance denoted in HPV+ relative to HPV− OPCs (p < 0.0001, Wilcoxon rank sum test). C, significantly enriched (FDR < 0.05) KEGG Pathway, Biological Process, and Molecular Function GO terms among the differentially abundant proteins.
Functional Term Enrichment Analysis
In order to determine the biological functions that were enriched among the differentially abundant proteins, KEGG Pathway and GO term enrichment analyses were performed. Significantly enriched categories included proteins involved in cell cycle, DNA replication, execution of apoptosis, and lymphocyte activation, among others (Fig. 2C). Cell cycle/DNA replication proteins included those induced by the E2F transcription factors: mini-chromosome maintenance proteins (MCM2/4/5/6), proliferating cell nuclear antigen, flap structure specific endonuclease 1, and p16, as well as the 14–3-3 family protein (YWHAB) and structural maintenance chromosome protein (SMC1A), among others (34). Examples of proteins involved in apoptosis included p16, DNA fragmentation factor subunit α, and high-mobility group protein 2. Lastly, lymphocyte activation proteins were enriched in HPV+ OPC, consisting of MHC class II proteins (HLA-DRA and HLA-DRB1/3/5), c-SRC kinase, and ICAM1, among others. It should be noted that numerous proteins belonged to more than one category (supplemental Table S6).
Comparative Analysis with a Published OPC Proteome
As a means of verifying the protein abundance patterns observed in the current study, we compared our protein profiles to results from a study of two small pools of frozen HPV+ and HPV− OPC tissues (10 samples per pool) (27). A total of 1,626 protein groups were identified in both studies; >1,000 proteins were uniquely identified in our work, and ∼600 were observed in the pooled analysis (27) (Fig. 3A; supplemental Table S7). Because a different analytical method was utilized by Slebos et al. (27) and raw data were unavailable, it was not possible to quantify the correlation between protein ratios across the two studies (supplemental Fig. S1A). However, by comparing the overlap of proteins present at significantly higher or lower abundance in HPV± OPC and those with unchanged abundance, we found that 1,124/1,626 (∼70%) of the co-detected proteins demonstrated the same trend in expression in both studies. Specifically, the relative expression patterns of 1,124 (from 1,626) commonly identified proteins were similarly unchanged, up- or down-regulated, based on p < 0.05 (supplemental Figs. S1B–S1D). Furthermore, of our 34 most differentially abundant proteins after multiple-testing adjustment (see “Materials and Methods”), 29 were also detected by Slebos et al. (27), of which 12 (41%) proteins were concordantly expressed and 12 additional proteins (∼41%) exhibited similar trends in relative abundance, corroborating our observations (Fig. 3B) (27).
Fig. 3.
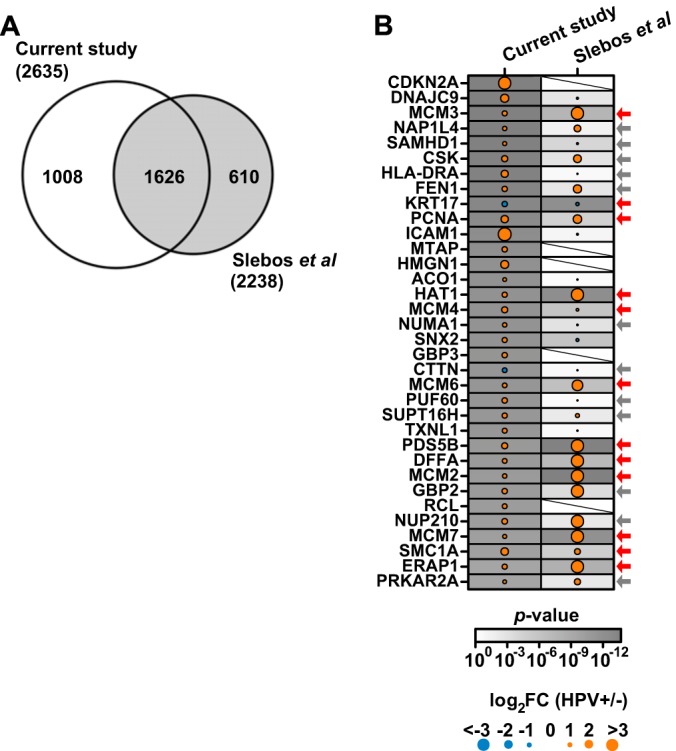
Comparison of relative protein abundance and protein functions in the current study with the published pooled OPC proteome (27). A, overlap of protein group identifications between the two OPC proteome studies. B, protein ratios of the top most significantly differentially abundant proteins in the current study and in the study by Slebos et al. (27). Arrows denote the 24 proteins with similar relative abundance trends in both studies, with red arrows highlighting the 12 proteins that were significantly differentially abundant.
Verification of Select Markers via Immunohistochemistry
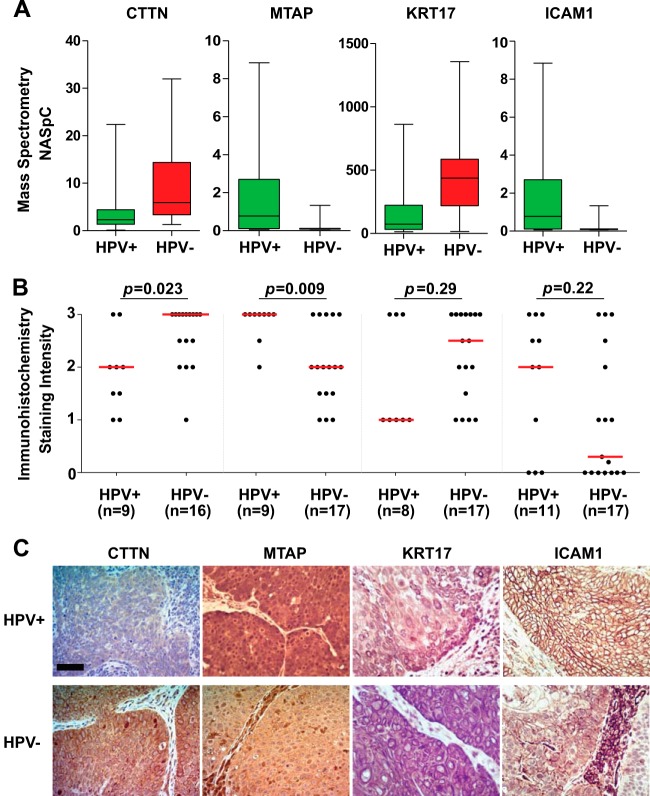
Four proteins, cortactin (CTTN), methylthioadenosine phosphorylase (MTAP), cytokeratin 17, and ICAM1, were selected for IHC verification in an independent cohort of pre-treatment OPC biopsies (n = 29, Fig. 4; supplemental Table S2). This selection was based on relative abundance in HPV± OPCs from the discovery cohort (Fig. 4A), concordance with the published proteome or genomic profiles from TCGA, and reported biological function. The validation cohort was age-, sex-, stage-, and treatment-matched to the discovery cohort, as well as between HPV+ and HPV− OPCs (Table I). Two proteins, CTTN and MTAP, were confirmed to be differentially abundant (p = 0.023 and p = 0.009, respectively; Wilcoxon rank-sum test), whereas ICAM1 and cytokeratin 17 exhibited similar relative expression (i.e. higher in HPV+ or HPV−) in comparison with the discovery cohort (p = 0.22 and p = 0.29, respectively; Wilcoxon rank-sum test) (Fig. 4B). Representative IHC images are shown in Fig. 4C.
Fig. 4.
Verification of selected differentially expressed proteins by IHC on pre-treatment OPC biopsies. A, relative abundances of selected proteins in the discovery cohort. B, IHC staining intensities in HPV+ and HPV− OPCs. The Wilcoxon rank-sum test was used to compare median immunoexpression levels (red lines). For a few specimens, IHC staining could not be evaluated because of the absence of tumor cells in the given tissue section; this explains the variations in the number of samples. C, representative IHC images. Scale bar, 50 μm.
Examining mRNA Abundance Downstream of Cortactin
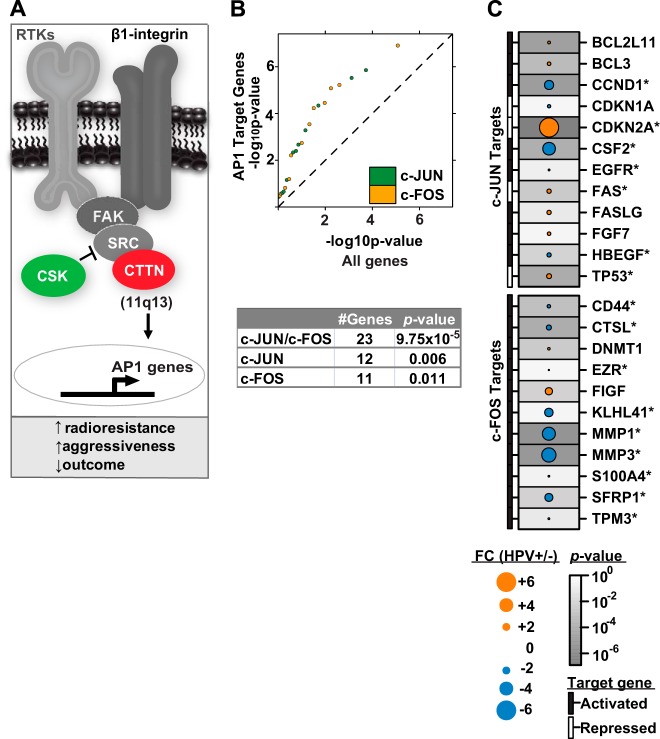
Building on the observation that cortactin is down-regulated in HPV+ versus HPV− OPC and its reported biological role in tumor radiation resistance (35), we sought evidence of differential regulation of cortactin-mediated radiation resistance based on HPV status. We hypothesized that selective activation of the β1-integrin/FAK/cortactin/JNK1 (MAPK8) signaling axis in HPV− OPC would be associated with induction of AP-1 transcription factor target genes, thereby leading to potential radiation resistance (Fig. 5A) (35, 36). Thus, relative mRNA abundance of established AP-1 target genes was examined in HPV± OPCs from the TCGA HNC dataset. Upon phosphorylation by activated JNK1, c-JUN acts in concert with c-FOS to stimulate the transcription of target genes; thus we specifically examined the target genes regulated by the oncogenic c-JUN and c-FOS proteins of the AP-1 complex (reviewed in Ref. 36). These reported AP-1 target genes were significantly enriched among the differentially abundant mRNAs in HPV± OPCs (qq-plot slope = 1.43; p = 9.8 × 10−5, 0.006, and 0.011 in Kolmogorov–Smirnov (KS) tests for c-FOS/c-JUN, c-JUN, and c-FOS targets, respectively) (Fig. 5B). Among the 23 genes examined, 16 (70%) demonstrated the expected relative mRNA abundance levels, and 11 (48%) of these were also significantly differentially abundant based on HPV status. Notably, transcriptional target genes both repressed and activated by AP-1 demonstrated expected alterations in relative mRNA expression levels (Fig. 5C). Prominent examples of the former included CDKN2A, TP53, and apoptosis-mediating surface antigen FAS, whereas examples of the latter included CD44 antigen, cathepsin L, matrix metalloproteinases (MMP1 and MMP3), and cyclin D1. In a second and unbiased attempt to examine AP-1 target gene enrichment, transcription factor enrichment analysis was performed using the DAVID UCSC_TFBS enrichment tool on two sets of differentially abundant mRNA transcripts from the TCGA HNC dataset: (i) in HPV+ versus HPV− OPCs and (ii) in OPCs with high versus low CTTN mRNA abundance. AP-1 target genes were significantly enriched (FDR < 0.0001) among these differentially abundant mRNAs, whether they were stratified according to HPV status or CTTN mRNA abundance. Taken together, these results point toward differential regulation of AP-1 target genes based on HPV status.
Fig. 5.
Model for radioresistance mediated by cortactin in HPV− OPC. A, potential signaling pathway downstream of β1-integrin that results in cortactin protein activation and subsequent induction of radiation resistance in HPV− OPC. Alternatively, CTTN signaling may occur via receptor tyrosine kinases (RTKs) (35, 52). CTTN (red) and c-SRC kinase (CSK, green) were identified among the top differentially abundant proteins during our proteomic discovery. Increased relative CTTN protein expression in HPV− OPCs via 11q13 amplification along with a corresponding decrease in the negative regulator CSK potentially leads to increased radioresistance in HPV− OPC. We hypothesize that this occurs via induction of downstream signaling (e.g. JNK1 phosphorylation) leading to induction of AP-1 transcription factor target genes. B, known AP-1 target transcripts are enriched among the differentially abundant mRNAs in HPV± OPCs (36). The table summarizes the p values based on target genes that are reported to be regulated by c-JUN, c-FOS, or both. C, relative mRNA abundance levels of reported AP-1 target genes in HPV± OPCs. Target gene activation or repression by AP-1 (c-FOS or c-JUN) is indicated. Genes with expected relative mRNA abundance levels are denoted by an asterisk.
Prognostic Significance of Cortactin in OPC
The prognostic significance of cortactin was evaluated through estimation of OS and DFS using the Kaplan–Meier method and Cox PH regression analyses. High cortactin protein levels were associated with significantly lower OS and DFS rates in OPC patients (supplemental Figs. S2A and S2B, supplemental Table S9). In addition, patients with high tumor cortactin protein levels had a 5-fold increase in the risk of disease progression or death (hazard ratio = 5.05 (95% confidence interval, 1.80–14.15), p = 0.002) (supplemental Figs. S2A and S2B, supplemental Table S9). Cortactin levels were associated with several clinico-pathological variables including smoking and drinking, sex, T-stage, and HPV status (supplemental Table S10). It was not possible to assess the independent prognostic utility of cortactin in a multivariate model for OS or DFS given the small number of events in the present cohort. Nonetheless, our findings validate the reported association of cortactin with OPC patient outcome in a univariate analysis model (37).
Integrative -Omics Analysis of OPC
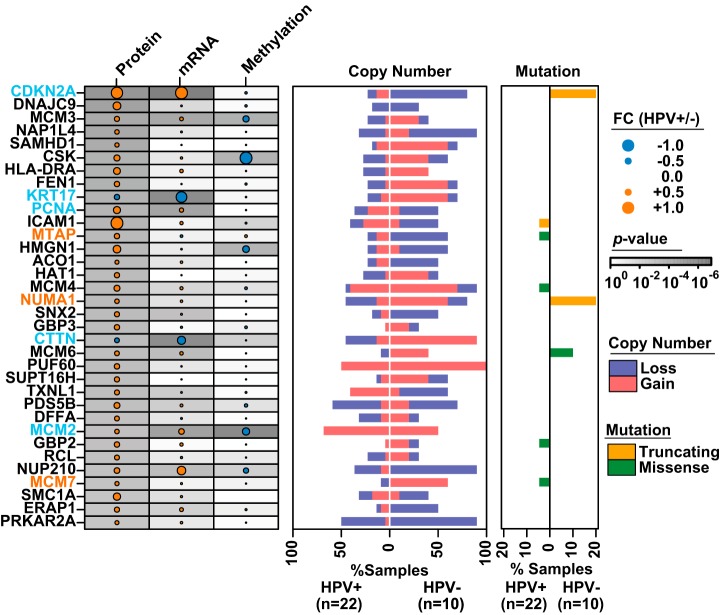
In order to identify potential mechanisms of protein regulation, we conducted a central dogma analysis based on all available -omics data by integrating our differentially abundant proteins with mutation, CNA, methylation, and mRNA profiles from the TCGA HNC study (Fig. 6; supplemental Table S8). Significant differences in gene mutations correlating with differential protein abundance were not observed, likely because of insufficient power, given the low number of patient samples examined. However, CDKN2A and nuclear mitotic apparatus protein 1 appeared to exhibit a trend toward increased mutation frequencies in HPV− OPC (p = 0.17, proportion test) (Fig. 6). Despite being derived from different cohorts of patients, 44/174 of the differentially abundant proteins were associated with corresponding changes in CNAs. In addition, 29 of 38 genes with statistically significant changes in methylation between HPV+ and HPV− OPC showed inverse changes in relative protein abundance. Additionally, 65/174 of the differentially abundant proteins were associated with corresponding changes in mRNA abundance. Importantly, a number of genes displayed corresponding changes across their entire molecular profiles, with CDKN2A, CTTN, cytokeratin 17, MCM2, and proliferating cell nuclear antigen serving as noteworthy examples. In contrast, relative abundance levels of numerous potentially functionally relevant proteins would not have been predicted or prioritized for follow-up investigations based on genomic data alone. As one prominent example, MTAP was identified and verified as expressed at significantly higher levels in HPV± OPC at the protein level using MS and IHC, respectively; however, mRNA, methylation, and mutation profiles did not serve as reliable predictors for relative MTAP protein levels. Copy number alterations revealed that deletion in HPV− OPC and, to a lesser degree, amplification in HPV+ OPC might serve as putative mechanisms for differential MTAP regulation. As a second example, significantly higher nuclear mitotic apparatus protein 1 levels in HPV+ versus HPV− OPCs were associated with a complex genomic profile: truncating mutations in two HPV− OPC specimens, significant amplifications in HPV− OPCs, and no significant changes in methylation status yet significantly higher mRNA levels in HPV+ OPC. Even though relative mRNA abundance served as a reliable predictor for relative protein abundance for nuclear mitotic apparatus protein 1, because it was not among the top differentially expressed mRNAs, it would not have been prioritized for further follow-up in a genomics-driven study, despite potential functional significance (38). As a third example, MCM7 protein was significantly more abundant in HPV± OPC (Fig. 3B); however, at the genomic level it was significantly amplified in HPV− OPC and showed no significant changes in mRNA, methylation, or mutation status, which would have rendered the genomic profiling unhelpful in predicting relative MCM7 protein abundance.
Fig. 6.
Integrated -omics analysis of OPC via integration with TCGA data. The top differentially abundant proteins among HPV± OPCs and the corresponding mRNA expression, methylation, copy number, and mutational changes in the OPC subset of the TCGA HNC study. FC values were normalized to the maximum across features for visualization. Light blue font denotes examples of genes with concordance across multiple -omic profiles; orange font denotes examples of genes for which relative protein abundance would have been difficult to predict based on genomic profiles alone.
Proteomic Subtypes of OPC
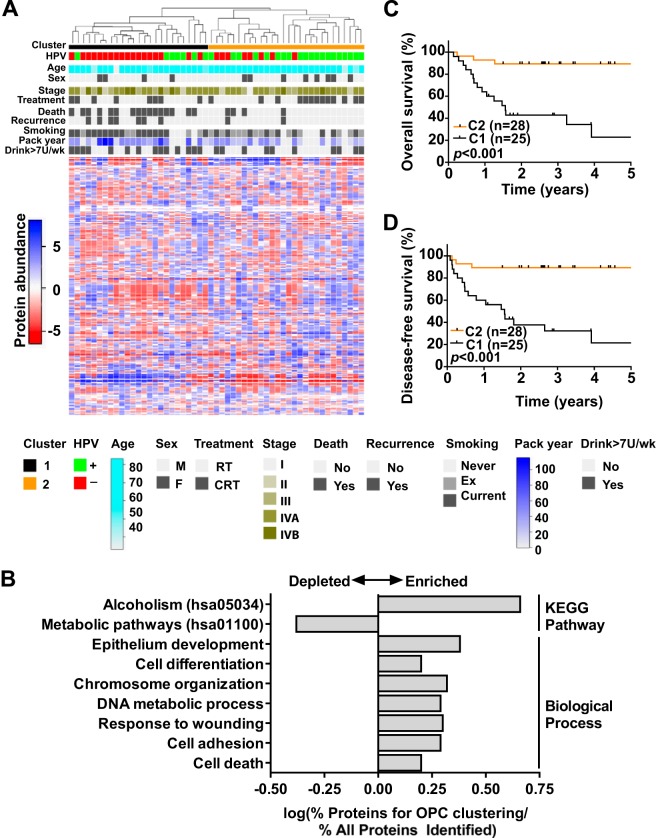
In order to examine whether proteomic profiles can be used to identify newly prognostic subtypes of OPC, we performed unsupervised hierarchical clustering on the 53 OPCs of the proteomic discovery cohort, revealing two main subgroups (C1, n = 25; C2, n = 28) (Fig. 7A). The subgroups differed significantly in HPV status (p = 0.0024, Fisher's exact test) but were otherwise balanced for clinico-pathological variables such as age, sex, and clinical stage (supplemental Table S10). However, C1 was marginally significantly associated with a higher proportion of smokers (p = 0.051, Fisher's exact test) and higher median smoking pack years (p = 0.061, Kruskal–Wallis test) (supplemental Table S10). The top most variable proteins used to delineate OPC subgroups were enriched for various functional categories, such as the alcoholism pathway (KEGG), cell differentiation, DNA metabolism, and cell adhesion (GO biological process categories), among others (Fig. 7B).
Fig. 7.
Proteomic subtypes of OPC. A, unsupervised hierarchical clustering of 53 OPCs based on globally normalized adjusted spectral counts of the top 10% of proteins (n = 263) with the highest variance. B, significantly enriched (FDR < 0.05) KEGG Pathway and GO Biological Process terms for proteins used for cluster analysis. Select categories are shown. C, five-year overall survival in OPC patients according to proteomic clusters. D, five-year disease-free survival in OPC patients according to proteomic clusters. Comparison of the survival rates between patient groups was based on the log-rank test.
Next, the prognostic significance of the proteomic subgroups was evaluated through estimation of OS and DFS using the Kaplan–Meier method and Cox PH regression analyses. C2 patients were found to experience significantly better OS and DFS than C1 patients; the corresponding 3-year rates of OS were 89% versus 26% (p < 0.001, Kaplan–Meier log-rank test), and the 3-year DFS rates were 89% versus 27% (p < 0.001, Kaplan–Meier log-rank test) (Figs. 7C and 7D, supplemental Table S11). Accordingly, C2 patients had a ∼7-fold reduction in the risk of disease progression or death relative to C1 patients (hazard ratio = 0.14 (95% confidence interval, 0.05–0.42), p < 0.001, Cox PH model) (supplemental Table S11). These associations remained significant in a multivariate model adjusted for HPV status, suggesting that proteomic subgroups might provide independent prognostic value in OPC (supplemental Table S11).
Further reinforcing the results above, C2 OPCs remained associated with superior OS and DFS upon stratification of HPV+ and HPV− OPCs according to proteomic subgroups (supplemental Figs. S2C–S2F, supplemental Table S12). Specifically among HPV+ OPCs, C2 tumors were associated with significantly higher DFS rates than C1 OPCs, with corresponding 3-year DFS estimates of 90% versus 43% (p = 0.034, Kaplan–Meier log-rank test) (supplemental Fig. S2F). Concordant associations were observed using Cox PH models, where C2 OPCs were associated with significantly lower risks of disease progression or death than C1 OPCs (supplemental Table S12). Cortactin protein levels were significantly lower in C2 than in C1 OPCs (supplemental Figs. S2G and S2H). Collectively, these findings point toward the presence of two proteomic subtypes of OPC that are strongly linked to HPV status, with distinct prognoses and expression of disease-aggressiveness markers.
DISCUSSION
We conducted integrative proteomic profiling on a richly annotated cohort of OPCs in order to acquire insight into OPC progression in relation to HPV status. MudPIT analysis of 53 FFPE OPCs yielded 2,633 protein identifications, establishing this as one of the largest FFPE tumor studies employing MS-based proteomics to date. We observed that similar to genomic and transcriptomic profiles, the proteomes of HPV+ and HPV− OPCs harbor distinct differences, with p16 demonstrating the most significant change. Because of its relatively low abundance and small molecular size, p16 protein was identified in only about half of the HPV+ OPCs; however, with the latest generation of MS instrumentation, this detection bias will likely be overcome in the future.3 HPV proteins were not detected, perhaps because some were lost during viral integration and were no longer expressed (6). Alternatively, they might have been of low relative abundance or undetectable as a result of the tissue fixation process.
The proteomic analysis of primary pre-treatment OPCs herein provides further evidence supporting numerous key hypotheses that have been explored previously to elucidate biological mechanisms associated with differential HPV status (8, 39–41). First, functional pathway enrichment analyses corroborated previous reports that differences in the cell cycle regulatory system, the process most well documented in the literature, differentially drive HPV+ versus HPV− OPC (39). Among the cell cycle proteins, several E2F transcription factor targets were enriched in HPV+ OPC, consequent to E7-induced loss of pRB.
As a second notable mechanism, post-therapy enhanced immune response in HPV-associated OPC has been postulated to play a potentially critical role in the superior outcomes of these patients (42, 43). Consistent with this hypothesis, we identified significant differences in expression profiles of immune response proteins in HPV± OPCs. Of particular interest was the apparent enrichment of nine proteins involved in lymphocyte activation in HPV+ OPC (supplemental Table S6), many of which participate in antigen processing and presentation for immune recognition. A recent report described an association between higher levels of tumor infiltrating lymphocytes and HPV positivity and improved outcome (40). Our study does suggest the influence of the tumor microenvironment, particularly related to the tumor infiltrating lymphocytes in HPV± OPC, which secrete cytokines and can stimulate the expression of several identified proteins (e.g. ICAM1 and MHC class II antigens HLA-DRA, HLA-DRB1, and HLA-DRB3). The specific biological roles of these proteins in HPV+ OPC progression and modulation of treatment response clearly require further investigation.
Third, the activation of residual p53 in HPV+ tumor models has been shown to induce apoptosis following radiation, thereby potentially leading to a favorable therapeutic response (41). This is in contrast to HPV− tumors, which were largely associated with inactivating p53 mutations. We and others have previously demonstrated that a greater proportion of HPV+ OPC samples harbor p53 immunoexpression (3, 5, 10). In the current study, we also observed elevated TP53 levels in HPV+ OPC, which might be inversely associated with the activation of the cortactin-mediated radiation resistance pathway (Figs. 5A and 5C), and we propose that the relationship between TP53 and CTTN should be further explored in this context.
Cortactin is an oncoprotein located on chromosome 11q13 along with cyclin D1, a site that is frequently amplified in HNSCC (44). In accordance with our findings, high tumor CTTN expression has been shown to correlate with increased aggressiveness and poor outcome in many cancers, but most frequently in HNSCC (37, 45). A recent pre-clinical study reported tumor radiosensitization via abrogation of the β1-integrin/FAK/cortactin/JNK1 signaling axis by administration of an anti-β1-integrin antibody in vitro and in vivo, making this pathway a therapeutically attractive target for investigation in OPC (35). To the best of our knowledge, we have identified and verified for the first time that cortactin protein expression is significantly higher in HPV− than in HPV+ OPC. Only one other study with parallel findings described higher CTTN mRNA levels in HPV− than in transcriptionally active (i.e. positive for E6 and E7 mRNA) HPV+ OPC (46).
The association between CTTN amplification and differential regulation of AP-1 target genes suggests that this pathway has increased activity in HPV− OPC; thus, we propose that differences in CTTN signaling might partially account for the increased radioresistance of HPV− OPC (Fig. 5A). Along the same pathway, higher expression of an SRC signaling inhibitor (c-SRC kinase) in HPV+ OPC, observed in the current proteomic study, might also contribute to enhanced radioresistance (47), though this remains to be validated in mechanistic studies. An anticipated challenge for future investigation of the described pathways is the identification of appropriate HPV+ and HPV− OPC model systems (cell lines and animal models) that faithfully recapitulate the molecular biology and treatment response profiles observed clinically.
We have also shown that tumor MTAP levels were significantly higher in HPV± OPC. MTAP, along with p16, is located at 9p21, a commonly deleted chromosomal region in HNSCC (48, 49). Tumor MTAP loss or decreased levels are associated with poor patient survival (48). Deficient MTAP levels could be exploited therapeutically in HPV− OPC, as these deficient cells are more chemosensitive and can be selectively targeted with purine synthesis inhibitors or toxic purine and pyrimidine analogues (e.g. 5-fluorouracil and 6-thioguanine) when combined with methylthioadenosine (the cleavage substrate for MTAP) (48, 50).
As biology is ultimately driven by the expression of functional proteins, the integration of protein expression profiles identified herein with TCGA genomics data might provide a vital link between cancer genomics and clinical phenotypes. Observed concordance between CNA, methylation, and mRNA profiles and relative protein abundance strengthened the confidence in protein level changes and provided multidimensional insight into the regulation of these proteins. In addition, although numerous proteins had parallel evidence of dysregulation across the central dogma, relative protein abundances were frequently difficult to predict based on mutation, CNA, methylation, or mRNA data alone, underscoring the importance of proteomic studies. There are numerous causes for discordance across various molecular profiles, including inter- and intrapatient tumor heterogeneity, different assays used for HPV status determination, and true differences in molecular regulation.
Lastly, two prognostic subgroups of OPC were identified in our analyses. The observation of significant differences in risk of disease progression following multivariate analysis suggests that these subgroups capture biological variation that cannot be accounted for by differences in HPV status alone. Furthermore, the identification of a subset of HPV+ OPCs with “atypical” behavior characterized by relatively worse outcome and potentially increased aggressiveness, as indicated by higher tumor cortactin levels, presents the possibility of identifying patients at greater risk for disease progression. This is an intriguing finding in light of the currently unmet need to establish criteria (e.g. clinico-pathological or molecular biomarkers) for the selection of patients with HPV-associated OPC toward de-escalated treatment (4). Previous investigation of HNC microarray gene expression data supports the presence of two HPV+ subgroups that were classified as mesenchymal and classical types. The former was associated with worse patient outcomes and featured an increased abundance of epithelial-to-mesenchymal transition markers (51). Although we did not observe significant differences in the expression of epithelial-to-mesenchymal markers between the subgroups (data not shown), the enrichment of cell differentiation and cell adhesion protein groups supports the presence of two biologically distinct phenotypes with varying disease aggressiveness.
In conclusion, our study demonstrates complex interplay among several key pathways that might be associated with the differences in the clinical phenotypes of HPV+ and HPV− OPC and reveals additional biological variation in OPC to HPV, rendering the interpretation of the molecular biology of this disease challenging. In addition, because of the intricacy of genomic, epigenetic, and transcriptomic alterations in HPV± cancers, comparative proteomics will play an important role in clarifying OPC biology, as proteins are the functional cellular units and represent the final products of such alterations. Our study points toward pathways and highlights proteins that differ as a function of HPV status that would not have been detected through genomic investigations alone, allowing for the identification of potential therapeutic targets that could improve outcomes for HPV− OPC. Further evaluations will be needed to investigate these proteins, and the differential regulation of these signaling pathways, both in the presence or absence of HPV and in the context of the relevant treatment modalities.
Supplementary Material
Footnotes
Author contributions: L.S., A.H., W.X., P.C.B., F.L., and T.K. designed research; L.S., D.W., A.H., W.S., and M.L. performed research; A.H., W.S., I.W., J.W., B.O., J.C.I., P.C.B., F.L., and T.K. contributed new reagents or analytic tools; L.S., D.W., A.H., W.S., S.S., A.I., V.I., S.H., and W.X. analyzed data; L.S., D.W., P.C.B., F.L., and T.K. wrote the paper.
* This study was funded by a grant from the Canadian Institutes of Health Research (CIHR) to T.K., P.C.B., and F.F.L. (MOP-114896), as well as by the Ontario Institute for Cancer Research through funding provided by the Government of Ontario to P.C.B. L.S. was supported by the CIHR Canada Graduate Scholarship (CGS) Master's Award, the Ontario Graduate Scholarship, and the CIHR Terry Fox Foundation Strategic Training Initiative for Excellence in Radiation Research in the 21st Century. T.K. is supported by the Canada Research Chair Program. F.F.L. is the Dr. Mariano Elia Chair for Head & Neck Oncology. P.C.B. is supported by a Terry Fox Research Institute New Investigator Award and a CIHR New Investigator Award. We gratefully acknowledge the philanthropic support from the Wharton Family, Joe's Team, and Gordon Tozer. Support is also provided by the Campbell Family Institute for Cancer Research and the Ministry of Health and Long-term Planning.
 This article contains supplemental material.
This article contains supplemental material.
2 The Cancer Genome Atlas, accepted for publication.
3 L. Sepiashvili and T. Kislinger, unpublished data.
1 The abbreviations used are:
- HNSCC
- head and neck squamous cell carcinoma
- AP-1
- activator protein 1
- CDKN2A
- cyclin-dependent kinase inhibitor 2A
- CNA
- copy number alternation
- CTTN
- cortactin
- DFS
- disease-free survival
- FC
- fold change
- FDR
- false discovery rate
- FFPE
- formalin-fixed paraffin-embedded
- GO
- Gene Ontology
- HNC
- head and neck cancer
- HPV
- human papillomavirus
- HPV−
- HPV-negative
- HPV+
- HPV-positive
- HPV±
- HPV+ versus HPV−
- ICAM
- intercellular adhesion molecule
- IHC
- immunohistochemistry
- MTAP
- methylthioadenosine phosphorylase
- MudPIT
- multi-dimensional protein identification technology
- OPC
- oropharyngeal carcinoma
- OS
- overall survival
- PH
- proportional hazard
- TCGA
- the Cancer Genome Atlas.
REFERENCES
- 1. Chaturvedi A. K., Engels E. A., Anderson W. F., Gillison M. L. (2008) Incidence trends for human papillomavirus-related and -unrelated oral squamous cell carcinomas in the United States. J. Clin. Oncol. 26, 612–619 [DOI] [PubMed] [Google Scholar]
- 2. Habbous S., Chu K. P., Qiu X., La Delfa A., Harland L. T., Fadhel E., Hui A., Perez-Ordonez B., Weinreb I., Liu F. F., Waldron J., O'Sullivan B., Goldstein D., Xu W., Huang S. H., Liu G. (2013) The changing incidence of human papillomavirus-associated oropharyngeal cancer using multiple imputation from 2000 to 2010 at a Comprehensive Cancer Centre. Cancer Epidemiol. 37, 820–829 [DOI] [PubMed] [Google Scholar]
- 3. Leemans C. R., Braakhuis B. J., Brakenhoff R. H. (2011) The molecular biology of head and neck cancer. Nat. Rev. Cancer 11, 9–22 [DOI] [PubMed] [Google Scholar]
- 4. O'Sullivan B., Huang S. H., Siu L. L., Waldron J., Zhao H., Perez-Ordonez B., Weinreb I., Kim J., Ringash J., Bayley A., Dawson L. A., Hope A., Cho J., Irish J., Gilbert R., Gullane P., Hui A., Liu F. F., Chen E., Xu W. (2013) Deintensification candidate subgroups in human papillomavirus-related oropharyngeal cancer according to minimal risk of distant metastasis. J. Clin. Oncol. 31, 543–550 [DOI] [PubMed] [Google Scholar]
- 5. Shi W., Kato H., Perez-Ordonez B., Pintilie M., Huang S., Hui A., O'Sullivan B., Waldron J., Cummings B., Kim J., Ringash J., Dawson L. A., Gullane P., Siu L., Gillison M., Liu F. F. (2009) Comparative prognostic value of HPV16 E6 mRNA compared with in situ hybridization for human oropharyngeal squamous carcinoma. J. Clin. Oncol. 27, 6213–6221 [DOI] [PubMed] [Google Scholar]
- 6. zur Hausen H. (2002) Papillomaviruses and cancer: from basic studies to clinical application. Nat. Rev. Cancer 2, 342–350 [DOI] [PubMed] [Google Scholar]
- 7. El-Naggar A. K., Westra W. H. (2012) p16 expression as a surrogate marker for HPV-related oropharyngeal carcinoma: a guide for interpretative relevance and consistency. Head Neck 34, 459–461 [DOI] [PubMed] [Google Scholar]
- 8. Slebos R. J., Yi Y., Ely K., Carter J., Evjen A., Zhang X., Shyr Y., Murphy B. M., Cmelak A. J., Burkey B. B., Netterville J. L., Levy S., Yarbrough W. G., Chung C. H. (2006) Gene expression differences associated with human papillomavirus status in head and neck squamous cell carcinoma. Clin. Cancer Res. 12, 701–709 [DOI] [PubMed] [Google Scholar]
- 9. Gillison M. L., Koch W. M., Capone R. B., Spafford M., Westra W. H., Wu L., Zahurak M. L., Daniel R. W., Viglione M., Symer D. E., Shah K. V., Sidransky D. (2000) Evidence for a causal association between human papillomavirus and a subset of head and neck cancers. J. Natl. Cancer Inst. 92, 709–720 [DOI] [PubMed] [Google Scholar]
- 10. Braakhuis B. J., Snijders P. J., Keune W. J., Meijer C. J., Ruijter-Schippers H. J., Leemans C. R., Brakenhoff R. H. (2004) Genetic patterns in head and neck cancers that contain or lack transcriptionally active human papillomavirus. J. Natl. Cancer Inst. 96, 998–1006 [DOI] [PubMed] [Google Scholar]
- 11. Lui V. W., Grandis J. R. (2012) Primary chemotherapy and radiation as a treatment strategy for HPV-positive oropharyngeal cancer. Head Neck Pathol. 6 Suppl 1, S91–S97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Magdeldin S., Yamamoto T. (2012) Toward deciphering proteomes of formalin-fixed paraffin-embedded (FFPE) tissues. Proteomics 12, 1045–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ralton L. D., Murray G. I. (2011) The use of formalin fixed wax embedded tissue for proteomic analysis. J. Clin. Pathol. 64, 297–302 [DOI] [PubMed] [Google Scholar]
- 14. Patel V., Hood B. L., Molinolo A. A., Lee N. H., Conrads T. P., Braisted J. C., Krizman D. B., Veenstra T. D., Gutkind J. S. (2008) Proteomic analysis of laser-captured paraffin-embedded tissues: a molecular portrait of head and neck cancer progression. Clin. Cancer Res. 14, 1002–1014 [DOI] [PubMed] [Google Scholar]
- 15. Naidoo K., Jones R., Dmitrovic B., Wijesuriya N., Kocher H., Hart I. R., Crnogorac-Jurcevic T. (2012) Proteome of formalin-fixed paraffin-embedded pancreatic ductal adenocarcinoma and lymph node metastases. J. Pathol. 226, 756–763 [DOI] [PubMed] [Google Scholar]
- 16. Wisniewski J. R., Ostasiewicz P., Dus K., Zielinska D. F., Gnad F., Mann M. (2012) Extensive quantitative remodeling of the proteome between normal colon tissue and adenocarcinoma. Mol. Syst. Biol. 8, 611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ellis M. J., Gillette M., Carr S. A., Paulovich A. G., Smith R. D., Rodland K. K., Townsend R. R., Kinsinger C., Mesri M., Rodriguez H., Liebler D. C. (2013) Connecting genomic alterations to cancer biology with proteomics: the NCI Clinical Proteomic Tumor Analysis Consortium. Cancer Discov. 3, 1108–1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Crick F. (1970) Central dogma of molecular biology. Nature 227, 561–563 [DOI] [PubMed] [Google Scholar]
- 19. Zhang B., Wang J., Wang X., Zhu J., Liu Q., Shi Z., Chambers M. C., Zimmerman L. J., Shaddox K. F., Kim S., Davies S. R., Wang S., Wang P., Kinsinger C. R., Rivers R. C., Rodriguez H., Townsend R. R., Ellis M. J., Carr S. A., Tabb D. L., Coffey R. J., Slebos R. J., Liebler D. C., and National Cancer Institute Clinical Proteomics Tumor Analysis Consortium (2014) Proteogenomic characterization of human colon and rectal cancer. Nature 513, 382–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Alfaro A. A., Sinha A., Kislinger T., Boutros P. C. (2014) Onco-proteogenomics: cancer proteomics joins forces with genomics. Nat. Methods, 11, 1107–1113 [DOI] [PubMed] [Google Scholar]
- 21. Sepiashvili L., Hui A., Ignatchenko V., Shi W., Su S., Xu W., Huang S. H., O'Sullivan B., Waldron J., Irish J. C., Perez-Ordonez B., Liu F. F., Kislinger T. (2012) Potentially novel candidate biomarkers for head and neck squamous cell carcinoma identified using an integrated cell line-based discovery strategy. Mol. Cell. Proteomics 11, 1404–1415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Taylor P., Nielsen P. A., Trelle M. B., Horning O. B., Andersen M. B., Vorm O., Moran M. F., Kislinger T. (2009) Automated 2D peptide separation on a 1D nano-LC-MS system. J. Proteome Res. 8, 1610–1616 [DOI] [PubMed] [Google Scholar]
- 23. Gortzak-Uzan L., Ignatchenko A., Evangelou A. I., Agochiya M., Brown K. A., St Onge P., Kireeva I., Schmitt-Ulms G., Brown T. J., Murphy J., Rosen B., Shaw P., Jurisica I., Kislinger T. (2008) A proteome resource of ovarian cancer ascites: integrated proteomic and bioinformatic analyses to identify putative biomarkers. J. Proteome Res. 7, 339–351 [DOI] [PubMed] [Google Scholar]
- 24. Sodek K. L., Evangelou A. I., Ignatchenko A., Agochiya M., Brown T. J., Ringuette M. J., Jurisica I., Kislinger T. (2008) Identification of pathways associated with invasive behavior by ovarian cancer cells using multidimensional protein identification technology (MudPIT). Mol. Biosyst. 4, 762–773 [DOI] [PubMed] [Google Scholar]
- 25. Fermin D., Basrur V., Yocum A. K., Nesvizhskii A. I. (2011) Abacus: a computational tool for extracting and pre-processing spectral count data for label-free quantitative proteomic analysis. Proteomics 11, 1340–1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dennis G., Jr., Sherman B. T., Hosack D. A., Yang J., Gao W., Lane H. C., Lempicki R. A. (2003) DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 4, P3. [PubMed] [Google Scholar]
- 27. Slebos R. J., Jehmlich N., Brown B., Yin Z., Chung C. H., Yarbrough W. G., Liebler D. C. (2013) Proteomic analysis of oropharyngeal carcinomas reveals novel HPV-associated biological pathways. Int. J. Cancer 132, 568–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cerami E., Gao J., Dogrusoz U., Gross B. E., Sumer S. O., Aksoy B. A., Jacobsen A., Byrne C. J., Heuer M. L., Larsson E., Antipin Y., Reva B., Goldberg A. P., Sander C., Schultz N. (2012) The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gao J., Aksoy B. A., Dogrusoz U., Dresdner G., Gross B., Sumer S. O., Sun Y., Jacobsen A., Sinha R., Larsson E., Cerami E., Sander C., Schultz N. (2013) Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6, pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cancer Genome Atlas (2014) Comprehensive molecular characterization of urothelial bladder carcinoma. Nature 507, 315–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li B., Dewey C. N. (2011) RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cancer Genome Atlas (2008) Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455, 1061–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Benjamini Y., Hochberg Y. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57, 289–300 [Google Scholar]
- 34. Ren B., Cam H., Takahashi Y., Volkert T., Terragni J., Young R. A., Dynlacht B. D. (2002) E2F integrates cell cycle progression with DNA repair, replication, and G(2)/M checkpoints. Genes Dev. 16, 245–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Eke I., Deuse Y., Hehlgans S., Gurtner K., Krause M., Baumann M., Shevchenko A., Sandfort V., Cordes N. (2012) beta(1)Integrin/FAK/cortactin signaling is essential for human head and neck cancer resistance to radiotherapy. J. Clin. Invest. 122, 1529–1540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Eferl R., Wagner E. F. (2003) AP-1: a double-edged sword in tumorigenesis. Nat. Rev. Cancer 3, 859–868 [DOI] [PubMed] [Google Scholar]
- 37. Dumitru C. A., Bankfalvi A., Gu X., Eberhardt W. E., Zeidler R., Lang S., Brandau S. (2013) Neutrophils activate tumoral CORTACTIN to enhance progression of orohypopharynx carcinoma. Front. Immunol. 4, 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nguyen C. L., Munger K. (2009) Human papillomavirus E7 protein deregulates mitosis via an association with nuclear mitotic apparatus protein 1. J. Virol. 83, 1700–1707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lechner M., Frampton G. M., Fenton T., Feber A., Palmer G., Jay A., Pillay N., Forster M., Cronin M. T., Lipson D., Miller V. A., Brennan T. A., Henderson S., Vaz F., O'Flynn P., Kalavrezos N., Yelensky R., Beck S., Stephens P. J., Boshoff C. (2013) Targeted next-generation sequencing of head and neck squamous cell carcinoma identifies novel genetic alterations in HPV+ and HPV- tumors. Genome Med. 5, 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ward M. J., Thirdborough S. M., Mellows T., Riley C., Harris S., Suchak K., Webb A., Hampton C., Patel N. N., Randall C. J., Cox H. J., Jogai S., Primrose J., Piper K., Ottensmeier C. H., King E. V., Thomas G. J. (2013) Tumour-infiltrating lymphocytes predict for outcome in HPV-positive oropharyngeal cancer. Br. J. Cancer 110, 489–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kimple R. J., Smith M. A., Blitzer G. C., Torres A. D., Martin J. A., Yang R. Z., Peet C. R., Lorenz L. D., Nickel K. P., Klingelhutz A. J., Lambert P. F., Harari P. M. (2013) Enhanced radiation sensitivity in HPV-positive head and neck cancer. Cancer Res. 73, 4791–4800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Andersen A. S., Koldjaer Solling A. S., Ovesen T., Rusan M. (2013) The interplay between HPV and host immunity in head and neck squamous cell carcinoma. Int. J. Cancer 134, 2755–2763 [DOI] [PubMed] [Google Scholar]
- 43. Guerrera I. C., Quetier I., Fetouchi R., Moreau F., Vauloup-Fellous C., Lekbaby B., Rousselot C., Chhuon C., Edelman A., Lefevre M., Nicolas J. C., Kremsdorf D., Lacau Saint Guily J., Soussan P. (2014) Regulation of interleukin-6 in head and neck squamous cell carcinoma is related to papillomavirus infection. J. Proteome Res. 13, 1002–1011 [DOI] [PubMed] [Google Scholar]
- 44. Rodrigo J. P., Garcia-Carracedo D., Garcia L. A., Menendez S., Allonca E., Gonzalez M. V., Fresno M. F., Suarez C., Garcia-Pedrero J. M. (2009) Distinctive clinicopathological associations of amplification of the cortactin gene at 11q13 in head and neck squamous cell carcinomas. J. Pathol. 217, 516–523 [DOI] [PubMed] [Google Scholar]
- 45. Clark E. S., Brown B., Whigham A. S., Kochaishvili A., Yarbrough W. G., Weaver A. M. (2009) Aggressiveness of HNSCC tumors depends on expression levels of cortactin, a gene in the 11q13 amplicon. Oncogene 28, 431–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Jung A. C., Briolat J., Millon R., de Reynies A., Rickman D., Thomas E., Abecassis J., Clavel C., Wasylyk B. (2010) Biological and clinical relevance of transcriptionally active human papillomavirus (HPV) infection in oropharynx squamous cell carcinoma. Int. J. Cancer 126, 1882–1894 [DOI] [PubMed] [Google Scholar]
- 47. Okada M., Nada S., Yamanashi Y., Yamamoto T., Nakagawa H. (1991) CSK: a protein-tyrosine kinase involved in regulation of src family kinases. J. Biol. Chem. 266, 24249–24252 [PubMed] [Google Scholar]
- 48. Bertino J. R., Waud W. R., Parker W. B., Lubin M. (2011) Targeting tumors that lack methylthioadenosine phosphorylase (MTAP) activity: current strategies. Cancer Biol. Ther. 11, 627–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhang H., Chen Z. H., Savarese T. M. (1996) Codeletion of the genes for p16INK4, methylthioadenosine phosphorylase, interferon-alpha1, interferon-beta1, and other 9p21 markers in human malignant cell lines. Cancer Genet. Cytogenet. 86, 22–28 [DOI] [PubMed] [Google Scholar]
- 50. Lubin M., Lubin A. (2009) Selective killing of tumors deficient in methylthioadenosine phosphorylase: a novel strategy. PLoS One 4, e5735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zuo Z., Khattri A., Keck M. K., Cohen E. E. W., Lingen M. W., Vokes E. E., Seiwert T. Y. (2014) Validation of two biologically distinct HPV-associated head and neck cancer subtypes and correlation with E5 expression. J. Clin. Oncol. 32 32:5s, 2014 (suppl; abstr 6031) [Google Scholar]
- 52. MacGrath S. M., Koleske A. J. (2012) Cortactin in cell migration and cancer at a glance. J. Cell Sci. 125, 1621–1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.