Abstract
Background
Blood microRNAs (miRs) are a new promising area of disease research, but variability in miR measurements may limit detection of true-positive findings. Here, we measured sources of miR variability and determine whether repeated measures can improve power to detect fold-change differences between comparison groups.
Methods
Blood from healthy volunteers (N=12) was collected at three time points. The miRs were extracted by a method predetermined to give the highest miR-yield. Nine different miRs were quantified using different qPCR assays and analyzed using mixed models to identify sources of variability. A larger number of miRs from a publicly-available blood miR microarray dataset with repeated measures was used for a bootstrapping procedure to investigate effects of repeated-measures on power to detect fold-changes in miR expression for a theoretical case-control study.
Results
Technical variability in qPCR replicates was identified as a significant source of variability (p<0.05) for all nine miRs tested. Variability was larger in the TaqMan qPCR assays (SD = 0.15–0.61) versus the qScript qPCR assays (SD = 0.08–0.14). Inter- and intra- individual and extraction variability also contributed significantly for two miRs. The bootstrapping procedure demonstrated that repeated measures (20–50% of N) increased detection of a 2-fold change for ~10–45% more miRs.
Conclusion
Statistical power to detect small-fold changes in blood miRs can be improved by accounting for sources of variability using repeated measures and choosing appropriate methods to minimize variability in miR quantification.
Impact
This study demonstrates the importance of including repeated measures in experimental designs for blood miR research.
Keywords: blood miRNA, nuisance variability, repeated measures, extraction methods
Introduction
The use of microRNAs (miRs) as blood-based biomarkers is a new field of research for diagnostic and preventative medicine. A limitation of this field is the lack of statistical power to detect true differences between comparison groups, which can contribute to difficulties in validating results. Accounting for sources of variability in the experimental design may increase power in blood biomarker studies. Previously, we demonstrated that by controlling for technical variability in preparation of blood RNA for microarray analysis, we were able to improve power to detect small, yet significant, fold-changes in blood transcriptomic data (1). Here, we assess sources of inter- and intra-individual and technical variability for miRs found in blood samples and predict how repeated measures can improve power to detect fold-changes in miR expression.
MicroRNAs have been widely studied as biomarkers for a number of diseases. These small non-coding transcripts regulate translation of RNA by binding to the 3′ untranslated region of target RNA. Overall, miRs regulate 30–60% of RNA translation to protein usually by down-regulation of the transcript (2,3). Disease status, chemical exposures, and life-style factors have been linked to differences in expression of miRs observed between individuals (discussed in 4–6). However, as most reported miR expression fold-changes are small (~1.5–2-fold), it is difficult to replicate findings and discover true associations. Therefore, it is critical that important sources of variability are predetermined and controlled for in the experimental design, a priori.
Variability in RNA transcription within subjects over time has seldom been discussed in the literature, particularly for microRNAs (miRs). Several transcriptomic studies have shown limited fluctuation in blood RNAs when measured from healthy individuals over several weeks to months (7–11). The proportion of transcripts with high intra-individual variability was attributed to a small number of immunological genes (i.e., immunoglobulin) (9) or inseparable from technical variability due to poor experimental design (10,11). This evidence from transcriptomics suggests that there may be similarly small intra-individual variability for miR transcription, however, this has not been previously measured.
Other overlooked sources of variability include methods for miR quantification and extraction. For example, competing platforms for miR microarray and qPCR analysis have shown differences in sensitivity, which suggests variance in miR measurements (12–14). For processing of fresh blood samples, miRs studied in specific blood-partitions (i.e., plasma, red blood cells, platelets and leukocytes) have attributed certain miR expression in plasma and serum to contamination of red blood cells and platelets (15–18). Extraction of miRs can also introduce variability as systematic differences can depend on method or kit manufacturer (14,19–23). Most of these previous studies focused on samples obtained from cell lines and did not thoroughly compare miR yield obtained from primary cells.
Here, we hypothesized that there are important sources of inter-, intra- and technical variability in miRs extracted from primary human peripheral blood mononuclear cells (PBMCs). We calculated the contributions of these sources of variability using experimental data obtained by qPCR and compared this to estimates obtained from a miR microarray dataset from a previously published study. As PBMCs are a popular and non-invasive sample-type and can be affected in early stages of disease, it is important to improve methods of pre-analytical processing and measuring of PBMC biomarkers for future disease-related research.
Materials and Methods
Comparison of RNA extraction kits
Four kits were compared to each other for miR yield: miRNeasy kit (Qiagen, Valencia, CA), mirVana kit (Ambion / Life Technologies, Grand Island, NY), ZR-duet (Zymo Research Corporation, Irvine, CA), and Trizol (Life Technologies, Grand Island, NY), with the addition of 25nmoles–250nmoles of c. elegans oligos spike-ins (cel-39 and cel-54) to each sample during the cell lysis step. The AllPrep kit, which comprises of on-column extraction of both DNA and RNA, was also compared to the miRNeasy kit. Details are included in the Supplementary Methods.
Blood sample collection from volunteers
In order to calculate sources of inter- and intra-individual and technical variability, we measured miR expression in PBMCs of healthy volunteers over an 8-month time period. A sample size of N=12 healthy subjects were included in the study (exclusion criteria for volunteer subjects were chronic illness or pregnancy at the time of blood draws). Samples were obtained at three time points, roughly 2–4 months apart. On the day of collection, blood samples were processed to isolate PBMCs. Data collection for this study was approved by the Internal Review Board within University of California Berkeley’s Human Research Protection Program. Informed consent was obtained from all participants.
PBMCs were isolated from fresh whole blood collected in EDTA tubes using the standard Ficoll gradient protocol (24). Upon isolation of the PBMCs, they were immediately washed in PBS, pelleted, and resuspended in aliquots of RNA Protect Cell reagent (Qiagen, Valencia, CA) and frozen at −80°C until further use. At a later time, PBMC samples were thawed and RNA was extracted by the miRNeasy kit (Qiagen) as described in the Supplementary Methods.
Real-time PCR quantification
Probe-based miRNA TaqMan Assays (LifeTechnologies, Grand Island, NY), or PerfeCTa SYBER Green-based qScript microRNA Assays (Quanta BioSciences, Gaithersburg, MD) were used to quantify miR targets of interest. Reaction volumes were proportionately scaled-down from the initial protocols (see Supplementary Methods). Six miRs (miR-30d, let-7d, miR-185, miR-130a, miR-451, miR-342-3p) were chosen based on overlap between miRs expressed in the miR microarray dataset (used for some of the simulation studies) (25) and miRs differentially expressed in PBMCs of Type II diabetics (26). One miR (cel-39) was used as an exogenous control for elution variability. Two small RNAs (SNU6 and RNU48) and miR-16, frequently used for normalization of miR expression, were included as well.
Statistical analysis of qPCR results
We used a mixed-effects model to determine contributions of different sources of variability in miR expression for the healthy volunteer blood samples; time-point, within-extraction batch, between-extraction batch, and qPCR replicate variability.
| (1) |
The fixed effect coefficient, β1, represents a change in miR Cq value per unit change in RNA concentration for each sample, which we used as precaution against any effects that were not accounted for by using the same input (μg) of RNA for each RT-PCR reaction. The random effects terms are defined as the change in expression (Cq value) from baseline levels for each index. Thus, Yijklm represents the Cq value for the ith (i= 1…13) individual at the jth (j= 1, 2, 3) time point, in the kth (k=1, 2) extraction batch, for the lth (l= 1, 2) within-extraction replicate, for the mth (m =1, 2, 3) sample for a given individual on the nth (n = 1, 2, 3) plate. Thus, β0im is the random effects term for the qPCR replicate for a given individual at a given time point. Lastly the term, eijklmn, is defined as the ‘residual variability,’ which includes differences from plate-to-plate and other unaccounted for sources of variability. ANOVA tests were run for each small RNA model to determine which random effects terms were significant (p<0.05).
Estimating minimum detectable fold-changes based on qPCR data
We observed several measurable sources of variability for two miRs from the qPCR experiment, miR-185 and miR-451. These two miRs were used to determine if repeated measures would improve detection of fold-changes in a theoretical study. The constraints for the theoretical study were a sample size of N= 75 vs. 75 subjects in two comparison groups (e.g., disease vs. healthy controls) under two experimental designs conditions. Study 1 had no repeated measures and Study 2 had four repeated measures for 50% of the subjects for each of the four modeled sources of variability based on our empirical qPCR data— seasonal, between-batch, within-batch and qPCR replicate. The estimates of the parameters of interest, which is the minimum detectable fold-change in the mean level of the miRNAs, are computed using the variance values attributed to the four different sources of variability from the empirical study data of N=12 subjects.
Therefore, in order to obtain estimates of variability for the parameters of interest associated with repeating the empirical study, a clustered bootstrap procedure (using 100 bootstrap samples) was used. This procedure provides both a point estimate and confidence intervals for these fold-change estimates. Each bootstrap sample consists of data associated with 12 subjects drawn with replacement from the N=12 subjects of the empirical study. The data associated with a bootstrap sample was used to estimate the variances attributed to the four sources of variability using linear mixed models (27). The distribution of the minimum detectable fold-changes over the 100 bootstrap samples was used to estimate the confidence intervals of this parameter of interest.
A simulation procedure was used to estimate the parameters of interest for Study 1 and Study 2 given the variance estimates from a bootstrap sample, bs. For each miR within a given study design, data for 100 studies were simulated (assuming a normal distribution) using the bs variances estimates. The standard deviation of the miR expression across these 100 studies provides an estimate of the standard error (SEi,bs) of the mean level of the miR in a bs sample for a given study design. This standard error was then used to estimate the minimum detectable fold-change, FCi,bs, with 80% power (corresponding to a 5% family-wise error rate) by the following equation:
| (2) |
Where
| (3) |
Estimating minimum detectable fold-changes based on previously published data
To expand upon our findings of variability for individually-tested miRs by qPCR, we examined a publicly-available microarray dataset that measured hundreds of blood miRs simultaneously at several time-points for each subject by Honda et al (25). The study looked at the effects of chronic academic stress on miR levels in whole blood of medical students by obtaining measurements two months before, two days before, and one month after an exam for medical practitioners. The GSE49677 series from the Gene Expression Omnibus (28) Agilent-021827 Human miR Microarray (V3) is used in this study. The low expressed miRs with mean intensity levels <20 were filtered out as indicated in the study (25) and 143 miRs remained. The levels of these miRs across the four subjects and three time points are normalized using Cyclic Loess (29).
Variability estimates from the academic stress data were used to determine whether repeated measures would increase detection of differentially expressed blood miRs between two comparison groups (e.g., disease versus healthy controls). We calculated estimates of inter-individual variability from this study and assumed that the residual (unexplained) variability in blood miR levels was due to other sources. For ease of exposition, we assumed the residual to be time-point (e.g., seasonal) variability, although it is likely composed of multiple sources.
For our theoretical replicate design simulation study, our sample size was 75 vs. 75 subjects in each of two comparison groups (e.g., disease vs. controls). A total of 2000 markers were evaluated for purposes of multiple testing under realistic omic-level conditions. We assumed that the sample collection for the subjects in the two groups occurred at two different time points. Therefore, seasonal-effects on miR levels were not blocked in these experimental designs. We varied the proportion of subjects with repeated measures and the number of repeated measures per subject for each of the seven proposed designs shown in Table 1. We used a clustered bootstrap method, similar to the one described for the simulation of qPCR data, in order to predict minimum detectable fold-changes in the mean levels for the 143 miRNAs under the seven theoretical experimental design conditions. Like before, we again provide confidence intervals for our parameters of interest. These confidence intervals are based on repeating the academic stress experiment (25) with four subjects. The academic stress experiment provides estimates of inter-individual and residual variability. Residual variability is assumed to be attributed to seasonal variation.
Table 1.
Summary table of experimental designs used for simulations of miR microarray data
| Design | N1 | N2 | Number of subjects with repeated measures (%) | number of repeated measures |
|---|---|---|---|---|
| 0 | 75 | 75 | 0 (0) | 0 |
|
| ||||
| 1A | 75 | 75 | 30 (20) | 1 |
| 1B | 75 | 75 | 30 (20) | 4 |
|
| ||||
| 2A | 75 | 75 | 75 (50) | 1 |
| 2B | 75 | 75 | 75 (50) | 4 |
|
| ||||
| 3A | 75 | 75 | 150 (100) | 1 |
| 3B | 75 | 75 | 150 (100) | 4 |
Results and Discussion
Comparing methods of miR extraction
We evaluated miR extraction procedures to find the most efficient and accurate method for our downstream applications. We presumed that lower Cq values for a given extraction method would be a proxy for both greater overall yield of all miRs and lower technical variability (i.e. between and within a given batch of extractions). We compared four methods; miRNeasy, miRVana, Trizol, and Zymo-Duet (which extracts both RNA and DNA). The miRNeasy kit had the lowest Cq value for all small RNAs tested (see Supplementary Results and Supplementary Figure S1A) which is supported by similar previous findings (14). The miRNeasy kit also slightly out-performed the AllPrep kit (Qiagen) (Supplementary Figure S1B) and was thus chosen as the extraction method for the downstream measurements of miR variability in the volunteer blood samples.
Measuring sources of variability in miR from qPCR data
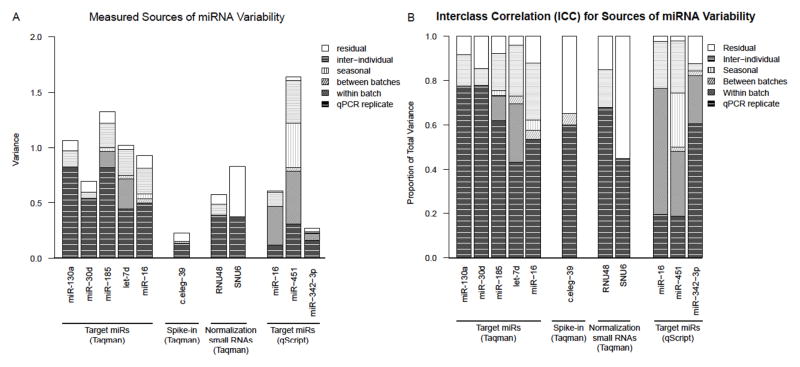
We measured several sources of variability for nine chosen miRs from 12 volunteer subject PBMC samples obtained at three time points over the course of several months. We included extraction replicates to account for both between- and within-extraction batch variability. Our results were analyzed using mixed-effects models. The variance attributed to each source of variability for a given miR is shown in the stacked bar graph (Figure 1A and 1B), and the significance of each term in the model is reported in Table 2.
Figure 1. qPCR measurements of sources of blood miRNA variability.
A. Proportions of inter-individual, intra-individual, and technical variability were estimated for N=12 subjects using a mixed-effects model of qPCR data from seven target miRs (miR-16, miR-342-3p, miR-30d, miR-185, let7d, miR-130a, miR-451), two endogenous control small RNAs (RNU48 and snRNA U6) and one exogenous spike-in (cel-39). Technical variability includes variability within- and between-extraction batches as well as plate-to-plate variability. B. Interclass correlation (ICC) for each source of variability was calculated as the proportion of total variance for each miR.
Table 2.
Variance terms and P values for sources of variability
| miRNA Target | Random effect |
Residual |
Total | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| qPCR replicate |
Interindividual |
Seasonal |
Between batches |
Within batch |
||||||||
| Variance | P value | Variance | P value | Variance | P value | Variance | P value | Variance | P value | Variance | ||
| miR130a | 0.826 | <0.001 | 0.149 | 0.387 | 0.000 | 1.000 | 0.000 | 1.000 | 0.000 | 1.000 | 0.089 | 1.064 |
| miR30d | 0.541 | <0.001 | 0.053 | 0.560 | 0.000 | 1.000 | 0.000 | 1.000 | 0.000 | 1.000 | 0.100 | 0.693 |
| miR185 | 0.820 | <0.001 | 0.222 | 0.153 | 0.031 | 0.910 | 0.000 | 1.000 | 0.149 | 1.000 | 0.102 | 1.324 |
| let-7d | 0.441 | <0.001 | 0.234 | 0.103 | 0000 | 1.000 | 0.036 | 0.770 | 0.271 | 0.647 | 0.040 | 1.021 |
| miR16 (TaqMan) | 0.499 | <0.001 | 0.237 | <0.05 | 0.045 | 0.706 | 0.036 | 0.888 | 0.000 | 1.000 | 0.113 | 0.929 |
| c.eleg-39 | 0.137 | <0.001 | 0.000 | 1.000 | 0.000 | 1.000 | 0.011 | 0.815 | 0.000 | 1.000 | 0.080 | 0.228 |
| RNU48 | 0.388 | <0.001 | 0.097 | 0.264 | 0.000 | 1.000 | 0.000 | 0:213 | 0.000 | 1.000 | 0.086 | 0.571 |
| SNU6 | 0.375 | <0.001 | 0.000 | 1.000 | 0.000 | 1.000 | 0.000 | 1.000 | 0.000 | 1.000 | 0.458 | 0.833 |
| miR16 (qScript) | 0.119 | <0.001 | 0.129 | <0.001 | 0.000 | 1.000 | 0.000 | 1.000 | 0.346 | <0.05 | 0.014 | 0.607 |
| miR451 | 0.308 | <0.001 | 0.386 | <0.001 | 0.401 | <0.05 | 0.029 | 0.691 | 0.482 | 0.073 | 0.031 | 1.640 |
| miR342-3p | 0.164 | <0.001 | 0.009 | 0.535 | 0.000 | 1.000 | 0.006 | 1.000 | 0.058 | 1.000 | 0.033 | 0.271 |
NOTE: The variability for each random-effect term in the model is reported, as well as P values based on ANOVA tests for each term of each modeled miRNA.
An ANOVA test for each of the random effects in each miR model indicated the significant variability terms (p<0.05). For all endogenous and exogenous small RNAs tested, a significant proportion of variability was due to replicate qPCR reactions for a given sample. For miR-16 and miR-451, inter-individual variability was also significant. Furthermore, for the qScript SYBR Green assays, miR-451 showed a significant “time point” effect while miR-16 showed a significant batch-replicate effect. This is the first evidence to suggest time-point and extraction variability in miR expression. Other transcriptomics studies have also found differences in RNA expression over the course of 1-day to several weeks (7,8) while longer time-points were inconclusive due to confounding of technical variability (10,11).
The residual variability present for all miRs may be due to several sources. Covariate information was not included in this model, such as age, race, gender and BMI, which may contribute to the residual variability. There also may be unmeasured technical variability in the processing of samples. For example, Ficoll separation of PBMCs is not 100% efficient, so miRs such as miR-16 and miR-451, both known to be highly-expressed in red blood cells (15)), could contribute to variability of these miR expression levels. SNU6 and the exogenous miR, cel-39, demonstrated the highest proportions of residual variability (Figure 1B). As expected, biological and technical variability contributed minimally to total variability and overall variability for the spike-in cel-39, and it had the lowest total variability of all miRs tested. For SNU6, the source of residual variability remains unknown, but perhaps this small RNA is not ideal to use for normalization of target miRs in future studies if sources of variability are not representative of other miRs.
The remaining miRs did not show significant contributions of variability from the other measured sources. This may be because 1) the sample size was too small too assign statistical significance or 2) the scaled-down volumes for the TaqMan assay were not sufficient to measure these effects. Comparing miR-16 measurements in both assays, the variance term for the qPCR replicates in the model was much smaller for the SYBR Green assay than the TaqMan assay (0.119 vs. 0.499) and are plotted for each individual in Supplementary Figure S2A and B. Additionally, three significant random-effects terms were found with the SYBR Green assay for miR-16, while only two were significant using the TaqMan kit, providing further evidence that perhaps other sources of variability could be unveiled if qPCR replicate variance was reduced. A less rigorous comparison of the two assays has been made in a previous study, however, the opposite results were found (13). Our results on the performance of miR SYBR Green-based qPCR are supported by a very recent study that examined miR expression analysis of qScript and several other platforms in much greater detail (30). As many studies use TaqMan-based assays, our results could help explain the lack of reproducibility reported between studies examining miRs in the same tissue for the same disease in similar populations.
Estimating effects of repeated measures from qPCR data
We used the estimates of inter-individual, intra-individual, and technical variability for two of the miRs (miR-185 and miR-451) for further analysis in a theoretical study of N= 75 vs. 75 subjects. We calculated the minimum detectable fold-change with 80% power in a study with no repeated measures (Study 1) versus four repeated measures for each of the following; between-batch, within-batch, time point and qPCR replicates for 50% of the subjects in each group (Study 2). The minimum fold-change estimate for miR-451 decreased with repeated measures from 3.77 (95% CI [1.75, 5.16]) to 2.38 (95% CI [0.97, 3.36]) and the minimum fold-change estimate for miR-185 decreased from 4.18 (95% CI [2.67, 6.46]) to 2.4 (95% CI [1.71, 3.74]) (Table 3). For miR-185, a marginally significant (90% CI) decrease in fold-change was observed with repeated measures from Study 1 to Study 2. Our estimate of detectable differences in fold-change for miR-185 is similar to previous findings that showed a 1.82-fold difference between disease conditions measured in PBMC samples (26).
Table 3. Estimations of minimum detectable fold-changes for 2 study designs.
The estimates of variability obtained from the empirical qPCR study of miR-451 and miR-185 were used to determine the minimum detectable fold-change with 80% statistical power for a theoretical study (N=75 vs 75) of two miRs. The mean fold-change, standard error (SE) and 95% CI and 90% CI are reported given no replicates (Study 1) versus a study given five extraction batches, five within-batch replicates, five time point replicates, and five qPCR replicates for 50% of the subjects (Study 2).
| miR-451 | miR-185 | |||
|---|---|---|---|---|
| Study 1 | Study 2 | Study 1 | Study 2 | |
| mean | 3.77 | 2.38 | 4.18 | 2.4 |
| SE | 0.95 | 0.53 | 1.2 | 0.67 |
| 95% CI | (1.75, 5.16) | (0.97, 3.36) | (2.67, 6.46) | (1.71, 3.74) |
| 90% CI | (1.88, 5.11) | (1.07, 3.20) | (3.69, 6.37) | (2.11, 3.68) |
Estimating variability in miR from previously published data
We expanded our investigation of repeated measures to examine more miRs simultaneously, as is currently done in omics-level studies. We used a previously published miR microarray dataset on four medical students over three time points to estimate the variability in 143 miRs (25). The inter-individual variability of each miR from our empirical study (miR-342, miR-451, miR-16, miR-185, miR-30d, let-7d, miR-130a) was compared to results obtained from Honda et al. There was no significant correlation between the two estimates (data not shown). This lack of correlation may be explained by differences in expression variability in each of the sample types (PBMCs versus whole blood), as high expression for some of these miRs has been reported in red blood cells (15), or by the small sample sizes used to estimate variability for the qPCR data and the Honda et al. dataset.
Estimating effects of repeated measures using simulated miR microarray data
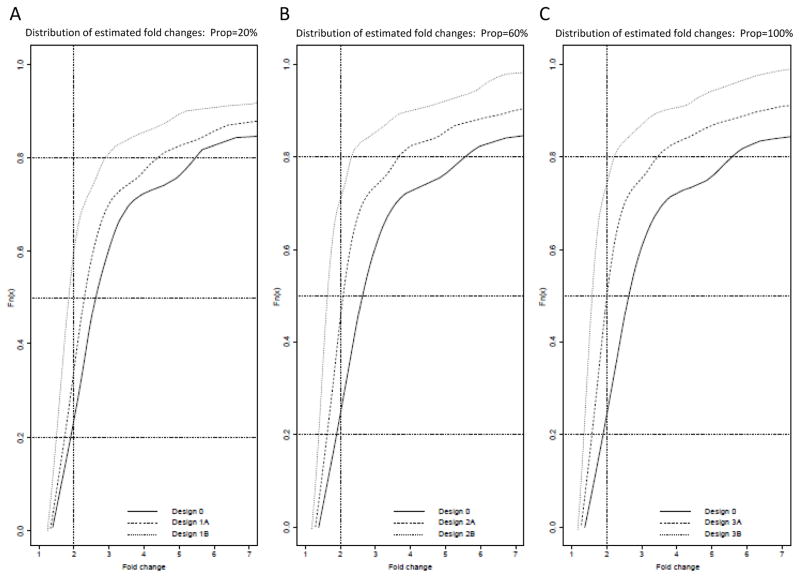
From the Honda et al. dataset, we simulated data for a theoretical study to demonstrate benefits of including repeated measures. We ran the analyses with 20%, 50% and 100% of the subjects randomly chosen for repeat sampling, and tested a total of seven different experimental designs summarized in Table 1. The cumulative distribution over the 143 miRs for the minimum detectable fold-change across the 100 bootstrap samples is plotted for each of the seven replicate designs (Figure 2A–C). Without repeated measures, a ≥2-fold change could be detected in ~24% of miRs. Inclusion of repeated measures for 20% of the samples improved the detection rate to 34% and 59% for Designs 1A and 1B, respectively (Figure 2A). When repeated measures were included for 50% of the samples, the detection rate for a ≥2-fold change improved further to 46% and 69% for Designs 2A and 2B, respectively (Figure 2B). Only a minimal increase in detection rate was gained beyond this when performing repeated measures on 100% of the samples in Designs 3A and 3B (Figure 2C).
Figure 2. Cumulative distributions of minimum detectable fold-changes in miRs for bootstrap procedure using repeated measures.
Smallest fold-changes detected for the 143 miRNAs (with 80% statistical power) are plotted under the seven experimental design conditions (for N= 75 vs. 75 subjects), which vary in proportion of repeated measures, A) 20%, B) 50%, C) 100%, and number of repeated measures per subject (n1=0, n1=1 or n1=4). Fold-changes are reported with 80% power for simulations with 100 bootstraps for 4 unique subjects each with 3 time point measurements (25). The vertical line in each figure is for purposes of comparing distributions at a 2-fold change in miR expression.
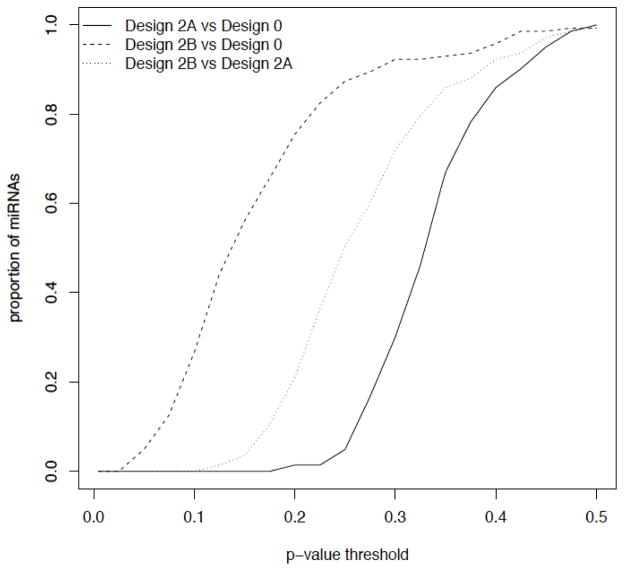
The estimates of 95% confidence intervals (based on bootstrapping) for these cumulative distribution curves overlapped under each design scenario (Supplementary Figure S3A–C), providing inconclusive evidence of statistically significant improvements of one design over another. To investigate this further, we compared each of the three designs to each other within the 50% repeated-measure parameter (Design 0 vs. 2A vs. 2B) and looked at a range of confidence intervals (i.e., p-values) for the minimum detectable fold-change of miRs in one study versus another. The proportion of miRs with detectable fold-change differences in one design versus another at a given p-value is plotted for each pair of designs (Figure 3). From this analysis, Design 2B (with four repeated measures) shows lower detectable fold-changes for ~20% of the miRs (at p<0.10) than Design 0 (with no repeated measures). Similar comparisons for Design 1B vs. Design 0 showed very few miRs with lower detectable fold-changes, whereas Design 0 vs. Design 3B showed lower detectable fold-changes in ~40% of miRs (at p<0.10) (see Supplementary Results and Supplementary Figure S4A and B, respectively).
Figure 3. Comparison of 50% repeated measure designs for detection of significant fold-changes in miRs.
Designs 0, 2A and 2B were compared to each other to calculate the proportion of the 143 miRNAs for which two designs’ confidence intervals do not overlap at a given p-value. (Designs differ by number of repeated measures for each subject.)
While inferences can be made from the simulations of this miR microarray dataset, there are still several limitations of this study. First, we assumed that the remaining variability after accounting for inter-individual differences is due to intra-individual variability over time, however, technical variability from time-point-to-time-point are included too. Our estimates of intra-individual variability may be higher than expected due to the lack of technical variability measurements. Also, changes in study participants’ stress levels during the collection time points (25) might also lead to over-estimates of intra-individual variability. Note that in order to mimic realistic omic-level conditions, the theoretical experimental design for this study was limited to N= 75 vs. 75 subjects, and we used 2000 measured endpoint markers (including the 143 miRs) for purposes of multiple-hypothesis testing. Altering these parameters by including a larger (or smaller) number of subjects and/or a greater (or reduced) proportion of repeated measures would shift all three curves to the left (or right) (see Figure 2). Thus, we consider the improvements of incorporating repeated measures observed herein to underestimate those expected for detecting smaller fold-changes in molecular epidemiological studies with larger sample sizes.
In summary, miRs have great potential as reliable blood biomarkers of early effects of the disease state. In measuring miRNA expression, we concluded that variability due to the qPCR reaction replicates generally outweighs other measured sources of variability. In the future, it would be advantageous to either troubleshoot qPCR reaction conditions to reduce this variance (e.g., increase reaction volumes, modify reaction temperatures, etc.) or increase the number of replicates per sample to account for this source of variability. Previous publications on small-fold changes in blood-miR analyzed by qPCR should be viewed with skepticism in light of this finding. Additionally, methods of extraction and miR quantification must be rigorously tested to maximize yield and interpretable results, as performance can vary by kit and assay. For unavoidable sources of variability, block experimental designs or repeated measures should be implemented. Identifying sources of variability in future omics-level experimental designs and estimating power a priori using these described methods can save precious resources, funding, and time for molecular epidemiological studies.
Supplementary Material
Acknowledgments
Financial support: This work was supported by grant # P42ES004705 from National Institute of Environmental Health Sciences Superfund Research Program (M.T.S.) and a fellowship from the National Science Foundation (S.I.D.).
Footnotes
Conflict of interest disclosure: The authors declare no competing financial interests.
References Cited
- 1.McHale CM, Zhang L, Lan Q, Vermeulen R, Li G, Hubbard AE, et al. Global Gene Expression Profiling of a Population Exposed to a Range of Benzene Levels. Environ Health Perspect. 2010 Dec 13;119:628–34. doi: 10.1289/ehp.1002546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Friedman RC, Farh KK-H, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009 Jan;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005 Jan 14;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 4.Lu M, Zhang Q, Deng M, Miao J, Guo Y, Gao W, et al. An Analysis of Human MicroRNA and Disease Associations. PLoS ONE. 2008 Oct 15;3:e3420. doi: 10.1371/journal.pone.0003420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang Q, Qiu C, Yang J, Wu Q, Cui Q. miREnvironment Database: providing a bridge for microRNAs, environmental factors and phenotypes. Bioinformatics. 2011 Dec 1;27:3329–30. doi: 10.1093/bioinformatics/btr556. [DOI] [PubMed] [Google Scholar]
- 6.Alegria-Torres JA, Baccarelli A, Bollati V. Epigenetics and lifestyle. Epigenomics. 2011 Jun;3:267–77. doi: 10.2217/epi.11.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Whitney AR, Diehn M, Popper SJ, Alizadeh AA, Boldrick JC, Relman DA, et al. Individuality and variation in gene expression patterns in human blood. Proc Natl Acad Sci. 2003 Feb 18;100:1896–901. doi: 10.1073/pnas.252784499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Radich JP, Mao M, Stepaniants S, Biery M, Castle J, Ward T, et al. Individual-specific variation of gene expression in peripheral blood leukocytes. Genomics. 2004 Jun;83:980–8. doi: 10.1016/j.ygeno.2003.12.013. [DOI] [PubMed] [Google Scholar]
- 9.Eady JJ. Variation in gene expression profiles of peripheral blood mononuclear cells from healthy volunteers. Physiol Genomics. 2005 May 24;22:402–11. doi: 10.1152/physiolgenomics.00080.2005. [DOI] [PubMed] [Google Scholar]
- 10.McLoughlin K, Turteltaub K, Bankaitis-Davis D, Gerren R, Siconolfi L, Storm K, et al. Limited dynamic range of immune response gene expression observed in healthy blood donors using RT-PCR. Mol Med. 2006;12:185. doi: 10.2119/2006-00018.McLoughlin. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karlovich C, Duchateau-Nguyen G, Johnson A, McLoughlin P, Navarro M, Fleurbaey C, et al. A longitudinal study of gene expression in healthy individuals. BMC Med Genomics. 2009 Jun 7;2:33. doi: 10.1186/1755-8794-2-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Git A, Dvinge H, Salmon-Divon M, Osborne M, Kutter C, Hadfield J, et al. Systematic comparison of microarray profiling, real-time PCR, and next-generation sequencing technologies for measuring differential microRNA expression. RNA. 2010 May;16:991–1006. doi: 10.1261/rna.1947110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Redshaw N, Wilkes T, Whale A, Cowen S, Huggett J, Foy CA. A comparison of miRNA isolation and RT-qPCR technologies and their effects on quantification accuracy and repeatability. BioTechniques. 2013 Mar;54:155–64. doi: 10.2144/000114002. [DOI] [PubMed] [Google Scholar]
- 14.Ach RA, Wang H, Curry B. Measuring microRNAs: comparisons of microarray and quantitative PCR measurements, and of different total RNA prep methods. BMC Biotechnol. 2008;8:69. doi: 10.1186/1472-6750-8-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pritchard CC, Kroh E, Wood B, Arroyo JD, Dougherty KJ, Miyaji MM, et al. Blood Cell Origin of Circulating MicroRNAs: A Cautionary Note for Cancer Biomarker Studies. Cancer Prev Res (Phila Pa) 2012 Mar 1;5:492–7. doi: 10.1158/1940-6207.CAPR-11-0370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheng HH, Yi HS, Kim Y, Kroh EM, Chien JW, Eaton KD, et al. Plasma Processing Conditions Substantially Influence Circulating microRNA Biomarker Levels. PLoS ONE. 2013 Jun 7;8:e64795. doi: 10.1371/journal.pone.0064795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kirschner MB, Kao SC, Edelman JJ, Armstrong NJ, Vallely MP, van Zandwijk N, et al. Haemolysis during Sample Preparation Alters microRNA Content of Plasma. PLoS ONE. 2011 Sep 1;6:e24145. doi: 10.1371/journal.pone.0024145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kirschner MB, Edelman JJB, Kao SC-H, Vallely MP, van Zandwijk N, Reid G. The Impact of Hemolysis on Cell-Free microRNA Biomarkers. Front Genet [Internet] 2013 May 24; doi: 10.3389/fgene.2013.00094. [cited 2014 May 12];4. Available from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3663194/ [DOI] [PMC free article] [PubMed]
- 19.Mraz M, Malinova K, Mayer J, Pospisilova S. MicroRNA isolation and stability in stored RNA samples. Biochem Biophys Res Commun. 2009 Dec 4;390:1–4. doi: 10.1016/j.bbrc.2009.09.061. [DOI] [PubMed] [Google Scholar]
- 20.Masotti A, Caputo V, Da Sacco L, Pizzuti A, Dallapiccola B, Bottazzo GF. Quantification of Small Non-Coding RNAs Allows an Accurate Comparison of miRNA Expression Profiles. J Biomed Biotechnol [Internet] 2009 doi: 10.1155/2009/659028. [cited 2014 May 12];2009. Available from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2735750/ [DOI] [PMC free article] [PubMed]
- 21.Eikmans M, Rekers NV, Anholts JDH, Heidt S, Claas FHJ. Blood cell mRNAs and microRNAs: optimized protocols for extraction and preservation. Blood. 2013 Mar 14;121:e81–89. doi: 10.1182/blood-2012-06-438887. [DOI] [PubMed] [Google Scholar]
- 22.Eldh M, Lotvall J, Malmhall C, Ekstrom K. Importance of RNA isolation methods for analysis of exosomal RNA: evaluation of different methods. Mol Immunol. 2012 Apr;50:278–86. doi: 10.1016/j.molimm.2012.02.001. [DOI] [PubMed] [Google Scholar]
- 23.Remakova M, koda M, Faustova M, Vencovsky J, Novota P. Validation of RNA extraction procedures focused on micro RNA expression analysis. Folia Biol (Praha) 2013;59:47–50. [PubMed] [Google Scholar]
- 24.Kanof ME, Smith PD, Zola H. Current Protocols in Immunology [Internet] John Wiley & Sons, Inc; 2001. Isolation of Whole Mononuclear Cells from Peripheral Blood and Cord Blood. [cited 2013 Dec 5]. Available from: http://onlinelibrary.wiley.com/doi/10.1002/0471142735.im0701s19/abstract. [DOI] [PubMed] [Google Scholar]
- 25.Honda M, Kuwano Y, Katsuura-Kamano S, Kamezaki Y, Fujita K, Akaike Y, et al. Chronic Academic Stress Increases a Group of microRNAs in Peripheral Blood. PloS One. 2013;8:e75960. doi: 10.1371/journal.pone.0075960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karolina DS, Armugam A, Tavintharan S, Wong MTK, Lim SC, Sum CF, et al. MicroRNA 144 Impairs Insulin Signaling by Inhibiting the Expression of Insulin Receptor Substrate 1 in Type 2 Diabetes Mellitus. PLoS ONE [Internet] 2011 Aug 1; doi: 10.1371/journal.pone.0022839. [cited 2012 Nov 6];6. Available from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3148231/ [DOI] [PMC free article] [PubMed]
- 27.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986 Sep;7:177–88. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 28.Barrett T, Edgar R. Gene expression omnibus: microarray data storage, submission, retrieval, and analysis. Methods Enzymol. 2006;411:352–69. doi: 10.1016/S0076-6879(06)11019-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ballman KV, Grill DE, Oberg AL, Therneau TM. Faster cyclic loess: normalizing RNA arrays via linear models. Bioinforma Oxf Engl. 2004 Nov 1;20:2778–86. doi: 10.1093/bioinformatics/bth327. [DOI] [PubMed] [Google Scholar]
- 30.Mestdagh P, Hartmann N, Baeriswyl L, Andreasen D, Bernard N, Chen C, et al. Evaluation of quantitative miRNA expression platforms in the microRNA quality control (miRQC) study. Nat Methods. 2014 Aug;11:809–15. doi: 10.1038/nmeth.3014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.