Abstract
Homology-directed repair (HDR) of double-strand DNA breaks is a promising method for genome editing, but is thought to be less efficient than error-prone nonhomologous end joining in most cell types. We have investigated HDR of double-strand breaks induced by CRISPR-associated protein 9 (Cas9) in Caenorhabditis elegans. We find that HDR is very robust in the C. elegans germline. Linear repair templates with short (∼30–60 bases) homology arms support the integration of base and gene-sized edits with high efficiency, bypassing the need for selection. Based on these findings, we developed a systematic method to mutate, tag, or delete any gene in the C. elegans genome without the use of co-integrated markers or long homology arms. We generated 23 unique edits at 11 genes, including premature stops, whole-gene deletions, and protein fusions to antigenic peptides and GFP. Whole-genome sequencing of five edited strains revealed the presence of passenger variants, but no mutations at predicted off-target sites. The method is scalable for multi-gene editing projects and could be applied to other animals with an accessible germline.
Keywords: CRISPR, Cas9, genome editing, homology-directed repair, short homology arms, Caenorhabditis elegans
THE ultimate goal of genetic engineering is to rewrite the genome with precision and without extraneous modifications (e.g., marker insertion). The remarkable efficiency of CRISPR-associated protein 9 (Cas9) to induce double-strand breaks at defined locations has led to an explosion of new methods for genome engineering (see Carroll 2014 and Sander and Joung 2014 for review). Streptococcus pyogenes Cas9 is an endonuclease that is targeted to a specific DNA sequence by an associated guide RNA (Gasiunas et al. 2012; Jinek et al. 2012). In animal models, expression of Cas9/single-guide RNAs (sgRNA) complexes in zygotes creates double-strand breaks that can be repaired by nonhomologous end joining (NHEJ) or homology-dependent repair (HDR). NHEJ is an error-prone process that can create insertions, deletions, or mutations at the cut site. HDR, in contrast, is a precise mechanism that uses sequences from a homologous donor molecule to repair the break. If the donor molecule carries edits flanked by sequences homologous to the targeted locus (“homology arms”), the edits will be integrated as part of the repair process. In many systems, HDR is thought to be less efficient than NHEJ, requiring high concentrations of donor molecules or long homology arms to stimulate recombination (see Beumer et al. 2013 and Sander and Joung 2014 for review). Single-strand oligodeoxynucleotides (ssODNs) can be injected at high concentration, but their relatively small size (∼200 bp or less) limits the types of edits that can be introduced. Studies in zebrafish embryos have also shown that ssODN-templated HDR is often imprecise, involving at least one error-prone NHEJ-like step (Auer and Del Bene 2014). Plasmid donors can accommodate gene-sized edits and longer homology arms, but require cloning and often selection to facilitate the recovery of rare recombinants. The selection marker is integrated along with the edit and must be removed in a subsequent step. The requirement for selection can be bypassed by providing high levels of Cas9 and sgRNA, using genome-integrated transgenes [as reported in Drosophila (Port et al. 2014)] or RNA injections [as reported in mice (Yang et al. 2013)]. For gene-sized edits, these approaches, however, are still thought to require the construction of plasmids with long homology arms, which limits scalability. Linear PCR fragments with short (<100 bp) homology arms are easier to prepare and have been shown to support HDR in yeast and Drosophila tissue culture cells using selection for a co-integrated marker (DiCarlo et al. 2013; Bottcher et al. 2014). Our goal was to determine whether a similar approach could be developed in an animal, but with a high-enough frequency to bypass the need for selection to generate marker-free, gene-sized edits in a single step.
HDR has been used extensively in C. elegans to introduce edits near double-strand breaks created by Mos excision, TALENs, and, most recently, CRISPR/Cas9 (see Waaijers and Boxem 2014 for review). As in other systems, both plasmid donors and ssODNs have been used as repair templates. Using CRISPR/Cas9, plasmid donors with long homology arms (≥1 kb) have been used to insert GFP by coselection for a linked marker or by direct screening for GFP expression (Dickinson et al. 2013; Tzur et al. 2013; Kim et al. 2014). ssODNs have been used to introduce smaller, base-size edits near Cas9 cuts without selection (Zhao et al. 2014) or by screening worms co-edited at a second locus (Arribere et al. 2014; Zhang and Glotzer 2014). The Arribere et al. (2014) study reported that ssODN-templated HDR can be highly local and frequently gives rise to partial conversion events where edits >10 bases away from the cut site are not integrated (Arribere et al. 2014). Whether short homology arms can support gene-size edits has not yet been reported.
In this study, we demonstrate that, in C. elegans, short homology arms flanking Cas9 sites support robust and precise HDR regardless of the size of the edit. Based on this finding, we developed a systematic and scalable method to create marker-free mutations, insertions, and deletions at any locus. Unlike earlier approaches, our method uses the same 10-day protocol to mutate, tag or delete genes of interest, generates “clean” homozygous mutants with no co-integrated markers or footprints, and can be scaled up for systematic editing of multiple genes.
Materials and Methods
Protocol
We provide a detailed protocol in the Supporting Information File S1.
Whole-genome sequencing
Libraries were constructed on the Mondrian SP+ (Nugen) and sequenced on the HiSeq 2500 (Illumina). For each library, a minimum of 4.4 × 107 50-bp reads (22-fold genome coverage) were aligned to the reference genome WS220 (http://www.wormbase.org) using BFAST software (Homer et al. 2009). Potential off-target sites were predicted using the CRISPR Design Tool (http://crispr.mit.edu). Mutation screening was by visual inspection of the aligned data at predicted sites and flanking sequences (±35 bp). Potential insertion mutations were detected using split-end alignment (Smith 2011).
Western blotting
Transgenic worms were lysed by freeze–thaw lysis in 1× M9 with 2.5% SDS. For embryonic lysates, 50 μl of packed embryos were resuspend in lysis buffer (2% SDS, 10% glycerol, 65 mM Tris–HCl, pH 7.5, protease inhibitors). Embryos were lysed using a Misonix Sonicator 3000 with total of 30 sec of sonication (15 sec on, 45 sec off at power level 2). Samples were run on a polyacrylamide gel and transferred overnight to a PVDF membrane. The membrane was blocked with 5% nonfat milk in PBS-Tween, washed, and probed with the following antibodies: anti-V5 HRP (R961-25, Invitrogen, 1/1000) for 2 hr at room temperature and anti-FLAG HRP (2044-S, Cell Signaling Technology, 1/1000).
Immunofluorescence
For staining, embryos were freeze-cracked on poly-l-lysine-coated slides and fixed in −20° methanol for 15 min and −20° acetone for 10 min. Samples were blocked in PBS-Triton-BSA for 30 min and stained with anti-FLAG M2 (1/500, Sigma F1804) and anti-PGL-1 (K76, 1/15) overnight at 4°. Primary antibody was detected using appropriate fluorescent secondary antibodies, mounted, and imaged. N2 worms were used as negative control.
In gel TetraCys tag detection
Transgenic worms were transferred in PBS containing protease inhibitors and freeze-thawed to lyse in NP40 buffer containing protease inhibitors. The lysate was processed using the Lumio Green detection kit (Invitrogen) following the manufacturer’s instructions, run on polyacrylamide gel, and imaged.
Results
Insertion of premature stop codons and small protein tags using ssODNs
To test the robustness of HDR using short homology arms, we first established a systematic protocol to generate small insertions/deletions using ssODNs. We designed ssODNs and sgRNAs (Jinek et al. 2012) to target Cas9 to eight different loci in five genes using genomic sequence information available on WormBase (sgRNA and ssODN sequences used for each experiment are listed in Table S1, Table S2, and Table S3). The ssODNs contained homology arms (43–100 bases in length; average of ∼60 bases) flanking the insertion/deletion placed directly at the Cas9/sgRNA site (Figure 1A, strategy 1; Figure S1, A and B) or up to 27 bases away (Figure 1A, strategy 2). In strategy 2, we also included silent mutations in the ssODNs to prevent recutting (Figure 1A). We also embedded a restriction site in the insertion to help identification of the edits by PCR (Figure S1 and Figure S2). We co-injected the ssODNs with a plasmid coding for Cas9 and the sgRNA (Dickinson et al. 2013) (see Figure 2 for protocol outline). We also included in the injection mix a plasmid coding for a visible marker. pRF4 is maintained extrachromosomally and causes a roller phenotype (Rol) in the F1’s that inherit it (Mello et al. 1991). We used this marker to identify injected mothers that incorporated the DNAs in germ-cell nuclei, as evidenced by the appearance of Rol progeny in their broods, and screened only these “marked broods.” We screened all F1 progeny (Rol and non-Rol) laid within ∼24–48 hr after gonad injection. The F1’s (singly or in pools of two) were allowed to lay eggs overnight and processed the next day by PCR and restriction digest. Four days later, 8–16 F2’s derived from positive F1 pools were processed in the same way to confirm germline transmission and isolate animals homozygous for the edit. Edits frequencies were calculated based on the number of positive PCR reactions divided by the total number of F1’s screened (Table 1 and Table S1).
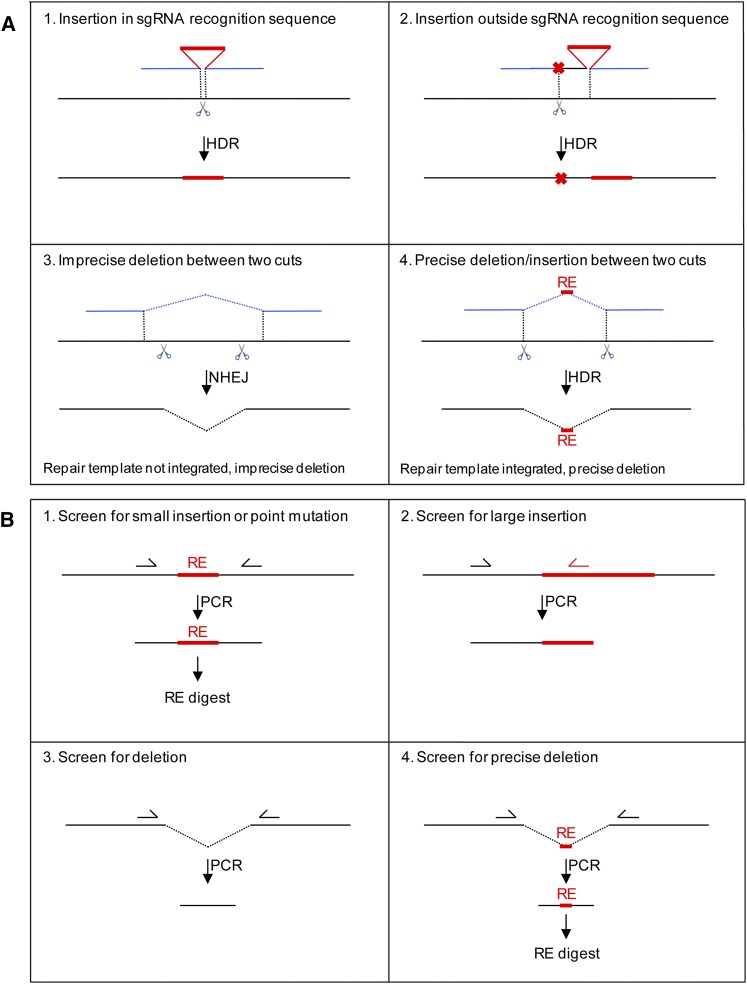
Figure 1.
Strategies for HDR and PCR screening. (A) Donor design strategies. Black lines represent genomic DNA, red lines represent inserted sequence, and blue lines are homology arms on template DNA. Scissors denote the sgRNA recognition sequence (pairing site + PAM). Red crosses indicate mutations in the repair template in sequences corresponding to the sgRNA recognition sequence. See text for strategy descriptions. Note that, for strategies 1 and 2, only insertions are shown in the schematics, but it is possible to simultaneously insert and delete sequences as described in Figure S1. In strategy 3, the homology arms in the donor template do not extend to the cuts and, as a result, NHEJ is preferred over HDR, and the donor template is not used. This strategy can be used without a template to generate an imprecise deletion between two distant sgRNA cut sites. (B) PCR screening strategies: see text and Supporting Information for details. Half-arrows denote primers, RE: restriction enzyme site. Strategy 1: Amplify region with primers flanking the insertion and digest with a restriction enzyme whose site is embedded in the insertion. See the protocol for sequences coding for antigenic tags that contain convenient restriction sites. This strategy works best with small F1 pools (2 F1’s). Strategy 2: Amplify with a primer specific for the insertion and a primer specific for the gene of interest. This approach could also be used for small insertions, but, in practice, when using ssODNs to generate small insertions, we have observed nonspecific products likely due to perduring ssODNs in the F1’s. Strategy 3: Amplify with primers flanking the deletion. This approach is suitable for pools of eight F1’s since the smaller deletion band is favored in the PCR. Strategy 4: Amplify with primers flanking the deletion and digest with the restriction enzyme the site of which is embedded at the junction (RE). This digest is used to distinguish HDR events from NHEJ events.
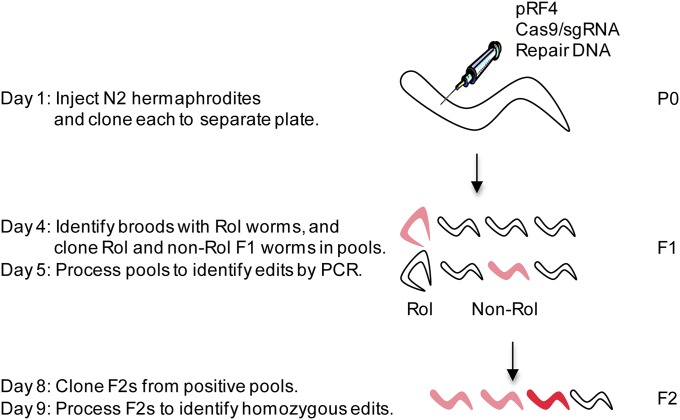
Figure 2.
Experimental outline. On day 1, ∼40 young adult hermaphrodites are injected and allowed to self-fertilize (one injected hermaphrodite per plate). The injections deliver the Cas9/sgRNA and pRF4 plasmids directly into the syncytial oogenic germline (sperm are formed at an earlier stage of development and stored away from the site of injection). The Cas9/sgRNA plasmid is presumed to be expressed shortly after injection in oogenic germ cells, as it contains promoters predicted to be active in that tissue (Dickinson et al. 2013). The pRF4 plasmid encodes a mutated collagen which, when expressed in F1’s, causes the worms to roll (Rol phenotype). This marker is used to identify broods derived from mothers that were successfully injected as evidenced by their transmission of the pRF4 plasmid to the next generation. On day 4 after injection, Rol and non-Rol F1’s from plates with Rol animals (“marked broods”) are screened directly for GFP expression or transferred to new plates in pools of two or eight for PCR screening. On day 5 (after F1’s have laid eggs on plates), F1 pools are screened by PCR for the desired genome edits (light red). On day 8, 8–24 F2’s from each positive F1 pool are transferred singly to new plates and allowed to self-fertilize. On day 9, F2’s are screened by PCR for homozygous genome edits (dark red).
Table 1. Summary of experiments.
| Experiment | Strategy (Figure 1A) | Edit | Gene | sgRNAs | No. of P0’sa | No. of PCRsb | No. of F1’sc | No. of editsd | % editse | No. of precise edits/no. of sequenced edits | Verification of expression |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Insertion of premature stop | |||||||||||
| 1 | 1 | Insertion-STOP | K08F4.2 | APa4-2 | 18 | 277 | 554 | 41 | 7.4 | 5/5 | — |
| 2 | 1 | Insertion/deletion-STOP | K08F4.2 | APs1 | 3 | 95 | 178 | 13 | 7.3 | 2/2 | — |
| 3 | 1 | Insertion/deletion-STOP | K08F4.2 | APs4 | 6 | 141 | 267 | 1 | 0.4 | — | — |
| 4 | 1 | Insertion/deletion-STOP | nos-2 | SL225; SL232 | 3 | 194 | 267 | 3 | 1.1 | 3/3 | — |
| Insertion of small protein tag | |||||||||||
| 5 | 2 | TetraCys at C terminus | K08F4.2 | APs6 | 11 | 188 | 376 | 0 | 0.0 | — | — |
| 6 | 2 | TetraCys at C terminus | K08F4.2 | APs5 | 5 | 143 | 286 | 5 | 1.7 | 2/5 | In gel detection |
| 7 | 2 | Myc, 3× Flag, V5, His, HA at C terminus | K08F4.2 | APs5 | 9 | 179 | 316 | 3 | 0.9 | 1/3 | WB/IF |
| 8 | 2 | V5, 3× Flag, OLLAS at N terminus | mex-5 | JS129; JS130 | 11 | 235 | 412 | 12 | 2.9 | 3/4 | f |
| 9 | 1 | Myc, V5 at N terminus | swan-1 | CSD35 | 1 | 19 | 19 | 5 | 26.3 | 3/3 | WB/IF |
| 10 | 2 | 3× Flag at C terminus | pgl-1 | TL001; TL002 | 6 | 134 | 134 | 1 | 0.8 | 0/1 | — |
| 11 | 2 | 3× Flag at N terminus | nos-2 | SL225; SL232 | 11 | 370 | 781 | 19 | 2.4 | 6/8 | WB/IF |
| 12 | 2 | V5 at N terminus | mbk-2 | HS516 | 2 | 48 | 48 | 1 | 2.1 | 1/1 | WB/IF |
| Insertion of GFP | |||||||||||
| 13 | 1 | GFP at sgRNA site near C terminus | K08F4.2 | APs5 | 12 | 108 | 741 | 30 | 4.0 | 4/4 | GFP expression |
| 14 | 2 | GFP at C terminus | K08F4.2 | APs5 | 20 | 179 | 1134 | 10 | 0.9 | — | GFP expression |
| 15 | 2 | GFP at C terminus | fbf-2 | APs12; APa13 | 17 | 134 | 832 | 3 | 0.4 | — | GFP expression |
| 16 | 1 | GFP at C terminus | mes-2 | sg16 | 18 | 150 | 738 | 5 | 0.7 | — | GFP expression |
| 17 | 1 | GFP at C terminus | lin-15b | sg23; sg25 | 5 | 51 | 336 | 2 | 0.6 | — | GFP expression |
| 18 | 2 | GFP at C terminus | deps-1 | sg6; sg21 | 11 | 83 | 487 | 7 | 1.4 | — | GFP expression |
| 19 | 1 | GFP at C terminus | mex-6 | sg3; sg18 | 10 | 90 | 666 | 8 | 1.2 | — | GFP expression |
| 20 | 2 | GFP at C terminus | glh-1 | sg4 | 5 | 54 | 402 | 0 | 0.0 | — | — |
| 21 | 1 | GFP at C terminus | htp-3 | sg5 | 1 | 12 | 84 | 0 | 0.0 | — | — |
| Deletion | |||||||||||
| 22 | 3 | ORF deletion (1.6 kb) | K08F4.2 | (APs1; APs4); (APs5; APs6) | 16 | 117 | 675 | 22 | 3.3 | 0/3 | — |
| 23 | 3 | ORF deletion (2.6 kb) | mbk-2 | HS516; (JS117; JS118) | 11 | 66 | 108 | 3 | 2.8 | — | f |
| 24 | 3 | Operon deletion (6 kb) | swan-1/2 | CSD35; (CSD53; CSD54) | 11 | 86 | 318 | 5 | 1.6 | 0/2 | — |
| 25 | 4 | ORF deletion/NheI insertion | K08F4.2 | APs1; APs5 | 11 | 84 | 528 | 20 | 3.8 | 2/2 | — |
| 26 | 1 | ORF deletion/NheI insertion | K08F4.2 | APs5 | 15 | 73 | 475 | 0 | 0.0 | — | — |
WB, Western blot; IF, immunofluorescence; GFP expression, GFP fluorescence in live animals in a pattern expected for the targeted ORF (see Figure 3).
Number of injected hermaphrodites whose broods were screened.
Total number of F1 pools that were screened by PCR.
Number of F1’s screened.
Number of positive PCRs.
Number of positive PCRs divided by the number of F1’s screened. The assumption is that each positive pool contains only one positive F1 animal. This may be an underestimate for the GFP experiments where each pool contained eight F1’s. Indeed, in some pools, we observed a higher frequency of edits among F2’s than expected if the pool contained only one edited F1.
Edits are maternal-effect lethal (V5-tagged MEX-5 and deletion in mbk-2 locus). Interestingly, 2/2 independent V5-MEX-5 edits are maternal-effect lethal as is a mex-5(0) mutant (Schubert et al. 2000). In contrast, the FLAG-MEX-5 and OLLAS-MEX-5 edits were viable. The ability to generate fusions to different tags in the same experiment will be useful to determine the best tagging strategy for each gene and avoid those tags that interfere with normal protein function.
Edit frequencies ranged widely (Table 1) and did not appear to correlate with insert size. For example, using the same sgRNAs to target the nos-2 locus, we recovered insertion of a premature stop (12 bp) and a FLAG tag (66 bp) at similar frequencies (1 and 2.4%) (experiments 4 and 11, Table 1). In another experiment, we co-injected three ssODNs carrying insertions of two different sizes (42, 42, and 66 nucleotides, experiment 8) and recovered all three edits. We observed, however, significant variability in sgRNA efficiency. In two cases where we used the same ssODN with two different sgRNAs, we obtained different edit frequencies [0.4 vs. 7.3% (experiments 3/2) and 0 vs. 1.7% (experiments 5/6) Table 1].
Sequencing of PCR-amplified regions in F2 worms confirmed correct insertion for 26 of 35 independent edits. Interestingly, all 9 incorrect edits were obtained using strategy 2 (25 edits), where the insert is placed at a distance from the sgRNA/Cas9 site. The 9 incorrect edits contained mutations, deletions, or insertions around the cut site and/or the insertion and, in one case (experiment 6, Table 1), a single base change in the tag sequence. In contrast, 13 of 13 edits obtained using strategy 1 (insertion directly in the sgRNA site) were error-free. These observations suggest that HDR is more robust when the homology arms directly flank the cut site. Analyses of lines established for a subset of edits (Table 1) confirmed that the tags were expressed as protein fusions of the expected size (Figure 3, A and B) and in the expected cells (Figure 3C).
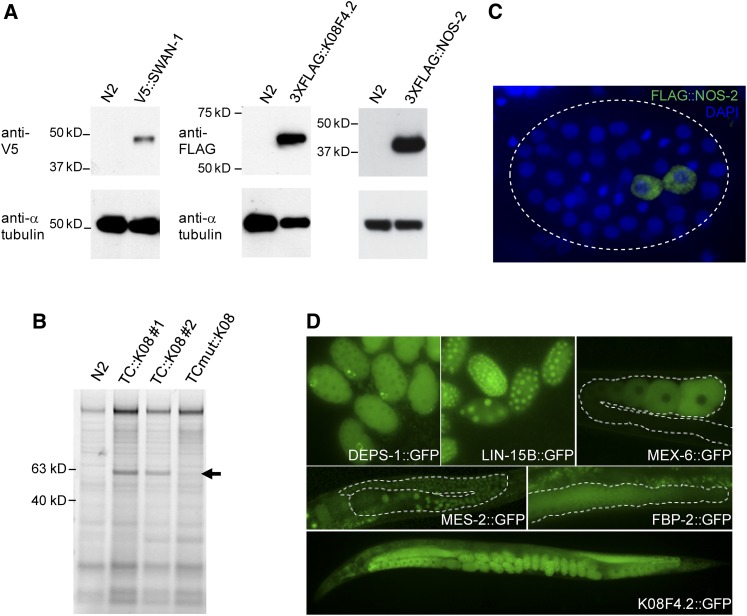
Figure 3.
Expression of tagged proteins. (A) Western blot using whole-worm or embryonic lysates and commercially available anti-V5 and anti-FLAG antibodies. (B) TC::K08F4.2 detection using Lumio Green. Arrows show a band of the expected size for two independent TetraCys (TC) edits of K08F4.2; the band is absent in N2 (wild-type) animals and also in a third independent edit of K08F4.2 that contains an inactivating mutation in the TC tag (C to Y). (C) Immunofluorescence image of a fixed embryo stained with DAPI and anti-FLAG antibody. FLAG-NOS-2 is present in the two primordial germ cells, Z2 and Z3, reflecting the wild-type distribution of NOS-2 at this stage of development. (D) Fluorescence pictures of live embryos (deps-1, lin-15), germline (mex-6, fbf-2), and whole animals (mes-2, K08F4.2) expressing the indicated GFP fusion proteins. Dashed lines outline the gonad boundary.
To assess the potential for off-target effects, we performed whole-genome sequencing and variant analysis of five edited lines obtained from two different sgRNA/repair ssODN combinations (experiments 1 and 12, Table 1 and Figure S3A) plus two wild-type (N2) populations from which the edited lines were derived. No mutations were observed in the 7 (experiment 1, Table 1) or 13 (Exp. 12, Table 1) predicted off-target sites with sequence similarity to their respective sgRNAs (Figure S3, B and C). We also screened for extraneous insertions of the repair ssODNs or Cas9 and Rol plasmid sequences within the genomes of the edited strains. No insertion events beyond the targeted edit sites were detected. We conclude that ssODNs with short (>60 nt) homology arms can be used to create insertions (largest tested: 66 nt) at or near Cas9 sites without also causing random insertions in the genome or mutations in predicted off-target sites. However, we did observe a number of variants (mostly single-nucleotide polymorphisms) unique to the edited strains (Figure S3, D and E). Potential sources of such “passenger variants” in the edited lines are addressed below (see Discussion).
Insertion of GFP using PCR fragments with short homology arms
To test whether short homology arms can support the integration of larger edits, we used PCR fragments as repair templates, as is standard in yeast (Horecka and Davis 2014), except that we did not use a selection marker. We first attempted to insert GFP (864 bases) using an sgRNA used in experiment 6 to insert the small protein tag TetraCys (18 bases) at the C terminus of K08F4.2. We amplified GFP with PCR primers designed to contain 59/59-nt homology arms that extended from the cut site (strategy 1, experiment 13, Table 1) or arms designed to position GFP precisely before the stop codon, 27 bases away from the cut site (experiment 14, Table 1). In the latter, we included in the repair template mutations in the sgRNA pairing sequence to prevent recutting (strategy 2 in Figure 1A, Table 1, and Table S3). We screened F1’s laid over a 48-hr period after injection in pools of eight worms and identified edits at an estimated minimum frequency of 4% (experiment 13) and 0.9% (experiment 14). Remarkably, these frequencies were comparable to the frequency (1.7%) observed for the insertion of the much smaller TetraCys tag using the same sgRNA.
To test whether this approach is robust and can be used at other loci, we designed sgRNAs to target the C termini of seven other genes. Where possible, we used sgRNA sites that overlapped the stop codon (strategy 1 in Figure 1A). Alternatively, we chose sgRNAs close (<30 bases) to the stop codon and used silent mutations to prevent recutting (strategy 2 in Figure 1A). We obtained GFP insertions for five of the seven genes attempted at estimated minimum frequencies ranging from 0.4 to 1.4% (experiments 15–21, Table 1). Twenty-four F2’s derived from positive F1’s were screened singly by PCR or by visual inspection for GFP fluorescence to recover and propagate the edits. All derived strains (each started from a single homozygous F2/3) showed stable GFP expression in the expected pattern (Figure 3D, Table S5).
In one of the two experiments that failed, only one brood (84 F1’s) was screened, which may have been too few (experiment 21). In the other failed experiment, a large number of F1’s were screened (402 F1’s), raising the possibility that the sgRNA may have been inefficient (experiment 20). We found that it is possible to use two sgRNAs with overlapping recognition sites in the same experiment (experiments 15, 17, and 19), which could help avoid low-efficiency sgRNAs.
We conclude that small homology arms are sufficient to support GFP insertion at most loci, provided that an efficient sgRNA site can be identified within ∼30 bases of the desired site of insertion. We have not yet tested whether insertions could be created at an even greater distance from the sgRNA site.
Precise gene-sized deletions using ssODNs
Using TALENs, Lo et al. (2013) reported the isolation of small precise deletions (<100 bases) templated by ssODNs. To test whether ssODNs could also be used to create gene-size deletions, we attempted to delete an entire ORF, using an ssODN with 67- and 57-base homology arms that fused the START and STOP codons of K08F4.2. We co-injected this ssODN with four sgRNAs with cut sites at the 5′ and 3′ ends of the K08F4.2 ORF (experiment 22, Table 1 and Figure S1). Unlike in the insertion experiments described above, both homology arms of the ssODN were separated from the cut sites by 10–31 bases (strategy 3 in Figure 1A and Figure S1C). We obtained 22 deletions (frequency 3.3%). The deletions, however, were imprecise as evidenced by their varied sizes and sequencing results (Table 1), suggestive of NHEJ repair. NHEJ repair of two cuts separated by 53 bp was reported previously (Cho et al. 2013). We confirmed that large deletions can be created directly by NHEJ alone, using sgRNA pairs targeting the 5′ and 3′ ends of mbk-2 and the swan-1/swan-2 operon (experiments 23 and 24, Table 1). Sequencing of the deletion breakpoints revealed small insertion/deletions consistent with error-prone NHEJ (data not shown).
To obtain a deletion with a precise fusion point, we modified the design of the ssODN targeting K08F4.2 to contain (1) 80- to 51-base homology arms that precisely flanked the sgRNA cut sites and (2) a restriction site inserted at the cut site (strategy 4 in Figure 1A and Figure S1D). This time, we obtained 22 correctly sized deletions, 20 of which contained the edited restriction site (3.8%, experiment 25, Table 1). We repeated the same experiment, omitting the sgRNA on one side of the deletion, and failed to obtain any deletions (experiment 26, Table 1 and Figure S1E). We conclude that large deletions with precise breakpoints (including insertions at the fusion point) can be obtained using two sgRNAs that target the ends of the deletion and one ssODN with homology arms that closely flanks the sgRNA sites (strategy 4).
Edit frequency is highest in marked broods, fluctuates from brood to brood, and does not increase with longer homology arms
In all the experiments described above, we screened only the broods of injected mothers that segregated Roller animals (“marked broods”) for genome edits. Because the Rol marker is on a different plasmid than Cas9/sgRNA, we investigated whether nonmarked broods might also contain edits. We repeated experiment 25 to generate a precise deletion in K08F4.2 and screened F1’s from 16 marked and 16 nonmarked broods in pools of one to eight. We identified 12 deletions from 295 F1’s from marked broods (estimated 4% efficiency) and 0 deletions from 332 F1’s from nonmarked broods. We conclude that screening only marked broods enriches for broods with edits, as expected.
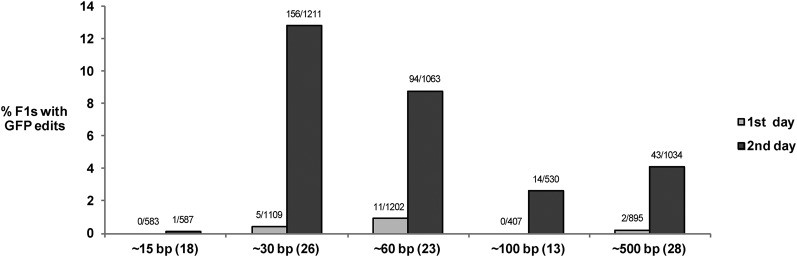
To determine the optimal length for homology arms, we repeated experiment 13 using the same PCR fragment (GFP flanked by 59/59-bp homology arms) and with new PCR fragments with shorter and longer arms (Figure 4). We separated the F1’s laid in the first and second 24 hr after injection and screened each F1 directly for GFP expression by live fluorescence microscopy. We found most GFP+ edits among the F1’s laid on the second day after injections; 33/33-bp homology arms gave the highest frequency of GFP+ edits and 15-/19-bp arms gave the lowest (12.8 and 0.1% on the second day). Longer arms did not increase, and in fact appeared to decrease, edit frequency (Figure 4). Overall, edit frequency for 59/59-bp homology arms (4.6%) was comparable to that found by PCR screening (4%, Table 1, experiment 13), strongly suggesting that all edits express GFP already in the first generation.
Figure 4.
Effect of arm length on edit frequency for a GFP PCR fragment. Graph showing the percentage of GFP+ F1’s among marked broods derived from hermaphrodites injected with Cas9/sgRNA APs5 as in experiment 13, except that the GFP PCR fragment contained homology arms of different lengths. Exact sizes were 15/19, 33/33, 59/59, 99/101, and 504/502 bp. The number of GFP+/ total F1’s scored is indicated on each bar, and the number of broods scored for each experiment are shown in parentheses below. The data shown here for the ∼30- and ∼60-bp homology arms are also shown broken down by broods in Table S4.
Edit frequency varied greatly between broods. For example, on the second day after injection using 33/33-bp homology arms, ∼40% of broods yielded no edits and 20% of broods gave 20–60% edits (“jackpot broods,” Table S4). We also observed jackpot broods when using ssODNs (example shown in Figure S2), as also reported by Arribere et al. 2014. In the one experiment using ssODNs where we separated F1’s laid on the first and second day (experiment 2, Table 1), we did not note a difference in edit frequency between the two egg-laying periods (data not shown).
We conclude that GFP edits can be obtained with homology arms as short as 33 bp, that longer arms do not increase edit frequencies, and that edits are distributed unevenly between broods.
Discussion
In this study, we demonstrate that linear templates with short homology arms (∼30–60 bp) support robust integration of both small and large (gene size) edits in C. elegans. The efficiency of HDR (typical range: 0.4–7% edits across all F1’s and as high as 60% in jackpot broods) is high enough to bypass the need for selection and is not affected significantly by edit size. HDR efficiency is affected, however, by the distance between the homology arms and the cut site(s). We obtained best results when both homology arms were homologous to sequences immediately flanking the cut site (strategy 1). To create a small deletion or an insertion at a distance from the cut site, it is possible to use repair templates with one homology arm flanking the cut and the other at a distance, but this approach yielded more imprecise edits (strategy 2, maximum distance tested 27 bp). This strategy was also used by Lo et al. (2013) to create a 77-bp deletion using a single, TALEN-induced cut. We were not able, however, to generate a 1.6-kb deletion using this single-cut strategy, possibly because the distance was too large between the cut site and the distant homology arm (experiement 26). We were able to make such a deletion, however, using a two-cut strategy (strategy 4, experiment 25). When using two cuts, we found again that it is critical that the homology arms in the ssODN extend close to the cut sites. Separation of both homology arms from the cut sites by as few as 10–31 bp favors an error-prone NHEJ-like mechanism where the repair template is not used (experiment 22). In fact, it is possible to create large imprecise deletions by NHEJ using two cuts and no repair template (strategy 3, largest deletion attempted: 6 kb). We conclude that, for precise edits, it is best to design repair templates with homology arms that extend as close to the cut site(s) as possible at least on one side of the cut. Since it is possible to use multiple cut sites and to simultaneously insert and delete sequences, this requirement still allows for many different types of edits.
The edit efficiencies that we report here are within the range reported for similar experiments in Drosophila and zebrafish using ssODNs or PCR fragments (and plasmids) with long (>800 bp) homology arms (Auer and Del Bene 2014; Beumer and Carroll 2014). In zebrafish embryos, the majority of repair events are imprecise and involve an NHEJ-like mechanism on at least one side of the edit (Auer and Del Bene 2014). The practice in C. elegans of injecting the repair template and Cas9/sgRNA plasmid directly into the meiotic (oogenic) germline syncytium may help favor HDR. It is likely that most edits were generated in the oogenic germline of the injected hermaphrodites, since most edits were heterozygous in the F1 generation and were transmitted to the F2 generation in the expected 1:2:1 ratio. We speculate that high rates of HDR could also be obtained in other animals by targeting meiotic germ cells instead of embryonic cells where NHEJ dominates (Auer and Del Bene 2014).
We sequenced the genomes of five edited lines made using two different sgRNA/ssODN combinations and observed no mutations at predicted off-target sites, as also reported by Chiu et al. (2013). We also observed no insertions of the ssODNs outside of the targeted loci, confirming specificity. However, we did observe several variants (from 1 to 18 per strain, predominantly SNPs) in the edited strains that were not detected in the wild-type populations (N2) used for injections. The source of these variants is unclear. One possibility is that the variants observed in the edited strains were derived from rare alleles present in the wild-type population that became fixed during edit isolation. Each edited line was founded by a single hermaphrodite and underwent at least one additional round of clonal isolation. These putative rare alleles may have been lost from the wild-type strains during propagation or may have remained present but at a frequency below the level of detection. Both the wild-type and edited strains were passaged several times between the day of injection and the time of harvest for sequencing, providing an opportunity for genetic drift. The proposed model of fixation of rare parental variants is most consistent with the data. We observed the same variants in independent edited lines obtained from the same injected hermaphrodite (Figure S3: AP-1 and AP-3) and different variants in edited lines derived from different injected hermaphrodites (Figure S3: AP-1/AP-3 vs. AP-2 vs. YW-1/YW-2). Alternatively or additionally, some of the variants could have been caused by a sequence nonspecific mutagenic effect of Cas9 and/or the ssODNs. Although further analyses are needed to distinguish between these possibilities, our findings so far indicate that (1) passenger mutations can become fixed in the edited lines and that (2) it is advisable to isolate at least two independent edits (from different injected mothers) to avoid possible background effects.
Based on these observations, we created a simple method for genome editing in C. elegans. Our approach differs in several respects from previous methods (Waaijers and Boxem 2014). First, our method is versatile, allowing users to follow four different strategies (Figure 1A) and the same protocol (Figure 2, File S1) to tag, mutate, or delete their gene of interest. Second, our method does not use co-integrated markers and thus generates marker-free edits and does not require a specific genetic background or time-consuming selection schemes. Because injected DNAs form stable extrachromosomal arrays in C. elegans (Stinchcomb et al. 1985), selection-based approaches must also include counterselection against such arrays (Chen et al. 2013). The selection marker integrates with the edit and must be removed in an additional step using flanking recombination sites, which can leave a footprint (Dickinson et al. 2013). Third, by relying on direct screening of F1’s, edits are identified 5 days after injection, compared to 2 weeks or more when using selection markers, although PCR screening requires more hands-on time. Fourth, genome edits are identified in heterozygous animals in the first generation after editing, ensuring the recovery of both viable and lethal alleles (footnote f in Table 1). Finally, the use of short homology arms does not reduce edit frequency even for gene-sized insertions like GFP. PCR fragments with longer arms (up to 500 bp) do not exhibit higher edit rates (Figure 4). Also, using plasmids with 1-kb homology arms and the same marker plasmid that we used here (pRF4-Rol), Kim et al. (2014) reported 1–10% GFP edits among Rol F1’s. Similarly, using 60-bp homology arms, we obtained 3 –11% GFP edits among Rol F1’s (Table S1). Importantly, we also found significant numbers of GFP edits among unmarked F1’s, especially in the second day after injection (9–13%, Figure 4 and Table S4). Unmarked F1’s are more numerous than marked F1’s and therefore require fewer injections to generate. The ability to recover edits in unmarked F1’s also reduces exposure to the Cas9/sgRNA plasmid, which likely is co-inherited with the pRF4 marker plasmid in the Rol F1’s (Mello et al. 1991).
The use of short homology arms also offers several technical advantages. ssODNs and PCR fragments require no cloning, making our approach scalable. We successfully designed repair templates targeting 17 unique sequences in 11 different genes using genome sequence information available in WormBase, suggesting that, even when relying on microhomology, polymorphisms that could interfere with HDR are not an issue at least in the common C. elegans lab strain Bristol (N2). The oligo-based design of the templates also facilitates the incorporation of helpful modifications in the homology arms and/or the insertion. These modifications include restriction sites to facilitate screening and mutations in sgRNA sites to prevent recutting after editing. This is particularly useful when using multiple sgRNAs to target a single site to reduce the chance of choosing inefficient sgRNAs, since it is advisable to mutate each site in the repair template to prevent recutting (Kim et al. 2014). Short homology arms also greatly simplify PCR screening by making it possible to use primers close to the insert without risking amplification of non-integrated templates that might persist in F1’s. Finally, when making GFP fusions, short homology arms can avoid the inclusion of promoter sequences, which could drive expression directly from the template. In this way, GFP edits can be identified directly by visual inspection of F1 animals.
When using PCR to identify edits, the most labor-intensive part of the protocol is the handling and processing of the F1’s. We used two strategies to minimize this step. First, we used a dominant episomal marker (pRF4) to identify successfully injected mothers and screened only their progeny. This approach cut by ∼50% the number of F1’s that need to be screened. Second, we processed the F1’s in pools. We used pools of two F1’s for microedits that require restriction digestion and pools of eight F1’s for larger edits that can be detected directly by PCR. One hundred to 200 pools can be processed in 2 days, and edits are easily isolated from the pool in the next generation by individual screening of 8–24 F2’s. Our observations indicate that edits are distributed highly unevenly among broods, with ∼15% of injected hermaphrodites generating broods with 20% or more edited F1’s (“jackpot broods,” Table S4). Identification of these jackpot broods before F1 screening would substantially reduce workload. Recently developed Co-CRISPR methods should make it possible to identify jackpot broods by selecting for broods containing edits at a second locus with a visible phenotype (Arribere et al. 2014; Kim et al. 2014; Zhang and Glotzer 2014). Particularly promising is the recent report of Arribere et al. (2014) who showed that single-base edits can be recovered in as high as 80% of F1’s selected for editing at a second locus with a dominant phenotype.
In summary, we have found that short homology arms stimulate HDR at high-enough efficiency in the C. elegans germline to create marker-free, gene-sized edits in a single step. The scalability of our method should make it possible to produce precise ORF deletions and reporter (e.g., GFP) fusions for every gene in C. elegans, a first for an animal model. There is no reason, a priori, to think that a similar approach could not be applied to other organisms with an accessible germline, thereby expanding the versatility and applicability of this exciting new era of genome engineering.
Supplementary Material
Acknowledgments
We thank Craig Mello for sharing results prior to publication, Mario de Bono for reagents, Bram Lambrus for sgRNA cloning assistance, Dominique Rasoloson for strain archival, and Andrew Folkmann and Dan Dickinson for discussion and comments on the manuscript. This work was supported by National Institutes of Health (NIH) grant R01HD37047, by the Intramural Research Program of the NIH, and the National Institute of Diabetes and Digestive and Kidney Diseases. G.S. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Supporting information is available online at http://www.genetics.org/lookup/suppl/doi:10.1534/genetics.114.170423/-/DC1.
Communicating editor: O. Hobert
Literature Cited
- Arribere J. A., Bell R. T., Fu B. X., Artiles K. L., Hartman P. S., et al. , 2014. Efficient marker-free recovery of custom genetic modifications with CRISPR/Cas9 in Caenorhabditis elegans. Genetics (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auer T. O., Del Bene F., 2014. CRISPR/Cas9 and TALEN-mediated knock-in approaches in zebrafish. Methods 69: 142–150. [DOI] [PubMed] [Google Scholar]
- Beumer K. J., Carroll D., 2014. Targeted genome engineering techniques in Drosophila. Methods 68: 29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beumer K. J., Trautman J. K., Mukherjee K., Carroll D., 2013. Donor DNA utilization during gene targeting with zinc-finger nucleases. G3 (Bethesda) 3: 1717–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottcher R., Hollmann M., Merk K., Nitschko V., Obermaier C., et al. , 2014. Efficient chromosomal gene modification with CRISPR/cas9 and PCR-based homologous recombination donors in cultured Drosophila cells. Nucleic Acids Res. 42: e89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll D., 2014. Genome engineering with targetable nucleases. Annu. Rev. Biochem. 83: 409–439. [DOI] [PubMed] [Google Scholar]
- Chen C., Fenk L. A., de Bono M., 2013. Efficient genome editing in Caenorhabditis elegans by CRISPR-targeted homologous recombination. Nucleic Acids Res. 41: e193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu H., Schwartz H. T., Antoshechkin I., Sternberg P. W., 2013. Transgene-free genome editing in Caenorhabditis elegans using CRISPR-Cas. Genetics 195: 1167–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S. W., Lee J., Carroll D., Kim J. S., Lee J., 2013. Heritable gene knockout in Caenorhabditis elegans by direct injection of Cas9-sgRNA ribonucleoproteins. Genetics 195: 1177–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiCarlo J. E., Norville J. E., Mali P., Rios X., Aach J., et al. , 2013. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res. 41: 4336–4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson D. J., Ward J. D., Reiner D. J., Goldstein B., 2013. Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nat. Methods 10: 1028–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasiunas G., Barrangou R., Horvath P., Siksnys V., 2012. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc. Natl. Acad. Sci. USA 109: E2579–E2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homer N., Merriman B., Nelson S. F., 2009. BFAST: an alignment tool for large scale genome resequencing. PLoS ONE 4: e7767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horecka J., Davis R. W., 2014. The 50:50 method for PCR-based seamless genome editing in yeast. Yeast 31: 103–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J. A., et al. , 2012. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337: 816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., Ishidate T., Ghanta K. S., Seth M., Conte D., Jr, et al. , 2014. A co-CRISPR strategy for efficient genome editing in Caenorhabditis elegans. Genetics 197: 1069–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello C. C., Kramer J. M., Stinchcomb D., Ambros V., 1991. Efficient gene transfer in C.elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 10: 3959–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Port F., Chen H. M., Lee T., Bullock S. L., 2014. Optimized CRISPR/Cas tools for efficient germline and somatic genome engineering in Drosophila. Proc. Natl. Acad. Sci. USA 111: E2967–E2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander J. D., Joung J. K., 2014. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat. Biotechnol. 32: 347–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert C. M., Lin R., de Vries C. J., Plasterk R. H., Priess J. R., 2000. MEX-5 and MEX-6 function to establish soma/germline asymmetry in early C. elegans embryos. Mol. Cell 5: 671–682. [DOI] [PubMed] [Google Scholar]
- Smith H. E., 2011. Identifying insertion mutations by whole-genome sequencing. Biotechniques 50: 96–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinchcomb D. T., Shaw J. E., Carr S. H., Hirsh D., 1985. Extrachromosomal DNA transformation of Caenorhabditis elegans. Mol. Cell. Biol. 5: 3484–3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzur Y. B., Friedland A. E., Nadarajan S., Church G. M., Calarco J. A., et al. , 2013. Heritable custom genomic modifications in Caenorhabditis elegans via a CRISPR-Cas9 system. Genetics 195: 1181–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waaijers S., Boxem M., 2014. Engineering the Caenorhabditis elegans genome with CRISPR/Cas9. Methods 68: 381–388. [DOI] [PubMed] [Google Scholar]
- Yang H., Wang H., Shivalila C. S., Cheng A. W., Shi L., et al. , 2013. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell 154: 1370–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, D., and M. Glotzer, 2014 Efficient site-specific editing of the C. elegans genome. bioRxiv DOI: 10.1101/007344.
- Zhao P., Zhang Z., Ke H., Yue Y., Xue D., 2014. Oligonucleotide-based targeted gene editing in C. elegans via the CRISPR/Cas9 system. Cell Res. 24: 247–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.