Abstract
Population structure is a confounding factor in genome-wide association studies, increasing the rate of false positive associations. To correct for it, several model-based algorithms such as ADMIXTURE and STRUCTURE have been proposed. These tend to suffer from the fact that they have a considerable computational burden, limiting their applicability when used with large datasets, such as those produced by next generation sequencing techniques. To address this, nonmodel based approaches such as sparse nonnegative matrix factorization (sNMF) and EIGENSTRAT have been proposed, which scale better with larger data. Here we present a novel nonmodel-based approach, population structure inference using kernel-PCA and optimization (PSIKO), which is based on a unique combination of linear kernel-PCA and least-squares optimization and allows for the inference of admixture coefficients, principal components, and number of founder populations of a dataset. PSIKO has been compared against existing leading methods on a variety of simulation scenarios, as well as on real biological data. We found that in addition to producing results of the same quality as other tested methods, PSIKO scales extremely well with dataset size, being considerably (up to 30 times) faster for longer sequences than even state-of-the-art methods such as sNMF. PSIKO and accompanying manual are freely available at https://www.uea.ac.uk/computing/psiko.
Keywords: admixture inference, kernel-PCA, population structure, genome-wide association studies, Q-matrix
POPULATION stratification has been commonly used to investigate the structure of natural populations for some time and is also recognized as a confounding factor in genetic association studies (Knowler et al. 1988; Marchini et al. 2004). As a result, programs for detecting population stratification have become a standard tool for genetic analysis. Such approaches generally separate into two classes. Model-based approaches such as STRUCTURE (Pritchard et al. 2000) and the closely related ADMIXTURE approach (Alexander et al. 2009) are desirable in that they return a Q-matrix, which for each accession of the (marker) dataset indicates the proportion of its genotype that came from one of K ≥ 2 assumed founder populations. This biological interpretability of Q-matrices conveniently lends itself to a subsequent use in association studies. On the other hand, such approaches often suffer from long run times, particularly as dataset size increases. This problem is becoming particularly exacerbated with the increased use of next generation sequencing (NGS) and large SNP chips to develop marker datasets (International HapMap Consortium 2007; Kim et al. 2007). Conversely, nonmodel-based approaches such as Eigenstrat (Price et al. 2006), which uses principal component analysis (PCA), tend toward much shorter run times, making them more convenient when analyzing large marker sets. Unfortunately, EIGENSTRAT only returns principal components (PCs) of a dataset and not a Q-matrix. Some nonmodel-based approaches such as the recently introduced sparse nonnegative matrix factorization (sNMF) method (Frichot et al. 2014), have made advances regarding these issues and output a Q-matrix for use in association genetic analysis while significantly shortening run times. Like EIGENSTRAT, sNMF can be thought of as a feature extraction approach aimed at reducing the dimensionality of a high dimensional dataset. However, the matrices used by both approaches to achieve this reduction have different mathematical properties (Kim and Park 2007). Even so, sNMF still suffers from longer run times with increased number of markers.
In this article, we propose the novel PSIKO approach, which is linear-kernel PCA based. Like EIGENSTRAT, PSIKO returns significant principal components of a dataset. Contrary to EIGENSTRAT, however, it also generates Q-matrices, and these are of comparable quality to those produced by STRUCTURE, ADMIXTURE, and sNMF. In addition, PSIKO’s scaling properties are better than sNMF’s (and thus STRUCTURE’s and ADMIXTURE’s) when the dataset size increases, making it particularly attractive for large datasets.
We rigorously tested the performance of PSIKO using simulated datasets, designed to evaluate the effects of inbreeding, noise, missing data, and SNP pruning, while enabling us to compare run time and scaling properties in comparison to leading approaches such as STRUCTURE, ADMIXTURE, and sNMF.
Although we simulated a range of biologically motivated scenarios, as a more realistic test, we also assessed the performance of PSIKO for Q-matrix estimation from two biological datasets. The first of these was a relatively small diversity panel comprising 84 Brassica napus lines that had been previously used to perform associative transcriptomics of seed traits (Harper et al. 2012). This dataset is of particular interest as it could be considered to have a complex evolutionary history. B. napus is a relatively recently formed species, having arisen from spontaneous hybridization between B. rapa and B. oleracea as little as 10,000 years ago. It exhibits considerable phenotypic variation, includes spring, semi-winter, and winter ecotypes and has been cultivated as both vegetable and oilseed crops. The most intensive breeding occurred over the last 50–60 years to produce the most commonly used “canola type” oilseed rape cultivars with both low erucic acid and low glucosinolate content in the seed. Many of the lines in this biological dataset will have been included in these breeding programs and certain groups (such as the winter oilseed rape lines) may have a complex breeding history. Despite this, the wide diversity of accessions in the panel enabled 101,644 SNP markers to be discovered. Originally the population stratification of this set of accessions was analyzed using STRUCTURE before using the identified Q-matrix in a mixed linear association model (MLM). We decided to compare the Q-matrices from PSIKO to those of STRUCTURE as well as sNMF and ADMIXTURE, and determine how these Q-matrices affect the results of the MLM for the original seed oil traits.
On its own and in combination with PLINK’s sliding window SNP pruning procedure, we also tested the Q-matrices produced by PSIKO and the three other methods under investigation on a subset of the HapMap Phase 3 project dataset (International HapMap Consortium 2010). This dataset should provide a more standard random mating model than the Brassica dataset, while providing an excellent real-life example of the very large marker datasets that will become more common with the advances in sequencing technology.
Materials and Methods
In this section, we first provide an outline of PSIKO in terms of a two-step approach and then describe these two steps in detail. This also includes a brief description of kernel-PCA (Scholkopf et al. 1999) as its main underlying technique. We then present details on the simulation experiments and the real biological datasets that we used to assess the performance of PSIKO, where the former also includes behavior under noise, missing data, inbreeding, large datasets, and SNP pruning. A presentation of PSIKO in terms of pseudocode may be found in Supporting Information, File S1.
We start with remarking that we follow Engelhardt and Stephens (2010) to infer a SNP matrix from a dataset given in terms of a sequence of d ≥ 1 SNPs and n ≥ 1 accessions, that is, a d × n matrix whose entries are 0, 1, and 2. For this, we use a reference sequence and count for each locus of an accession the number of copies of the reference allele found at that locus. Such a reference sequence could, for example, be obtained as in Bancroft et al. (2011) or be one of the accessions present in the dataset.
Method outline
Given a dataset X in the form of a d × n SNP matrix, PSIKO aims to infer the number K of founders of X as well as significant PCs and a Q-matrix. It consists of two main steps: dimensionality reduction (step I) and population structure inference (step II). The purpose of step I is to infer significant principal components of X and also obtain an estimate for K. For this we use a combination of the Tracy–Widom test (Patterson et al. 2006) with a powerful PCA-based technique called linear-kernel PCA. Due to the centrality of that technique to PSIKO, we also present an outline of it in that step. The purpose of step II is to quickly find good estimates for the ancestry coefficients, that is, the entries of the Q-matrix. For this, we exploit the properties of a PCA-reduced dataset to cast the problem of inferring population structure within a least squares optimization framework.
Step I: Dimensionality reduction:
PCA is a popular dimensionality reduction method that allows one to reduce the number of variables of the input dataset X (given in terms of d), at the same time keeping as much variability in the data as possible. It has proven very useful in population genetics and found in Patterson et al. (2006) and Ma and Amos (2012) to exhibit desirable properties when applied to datasets containing admixed individuals. However, the inner workings of PCA imply that it does not scale well with an increasing number of SNPs. To overcome this problem and thus obtain a method that is applicable to large NGS datasets, we employ a special kind of PCA called kernel-PCA which is known to scale well for large numbers of variables (SNPs in our case) (Murphy 2012). Rather than carrying out a PCA analysis directly on a given dataset, in kernel-PCA that dataset is first projected to some new higher dimensional (unknown) feature space, and then classic PCA is applied to the resulting projection of the dataset. To overcome the problem that this projection may be difficult to compute, a technique called kernel trick is sometimes used. Due to its centrality to psiko, we next describe it within a kernel-PCA setting (Murphy 2012).
For X as above, we start with remarking that if it is centered as described in Price et al. (2006), then performing PCA on it reduces to finding an eigendecomposition of the d × d-dimensional sample covariance matrix XXT. Suppose W is the matrix of eigenvectors and Λ is the diagonal matrix of eigenvalues of such a decomposition. Then where U is the matrix of eigenvectors of the n × n-dimensional inner product matrix K = XTX and performing PCA on X is equivalent to carrying out an eigendecomposition of K.
Suppose X is projected into a higher dimensional space Φ via a map φ, and for all 1 ≤ i ≤ n, put φi := φ(xi). Then performing PCA on the projection of X is equivalent to carrying out an eigendecomposition of the inner product matrix KΦ = (〈φi, φj〉)1≤i,j≤n. Computing these inner products directly tends to be difficult as the projection φ is unknown. By replacing the inner products 〈φi, φj〉 of KΦ with the values κ(xi, xj) of a real-valued function κ on X × X called a kernel function, the kernel trick overcomes this problem by allowing for computation of said inner products without having to directly compute φ. Informally speaking, κ is a proxy for the inner product in Φ.
To obtain the required lower dimensional dataset, let UΦ denote the matrix of eigenvectors of KΦ in an eigendecomposition of KΦ, let ΛΦ denote the associated diagonal matrix of eigenvalues, and let x denote an accession of X. Then the kernel-PCA projection of x is where kx = (κ(x, x1), κ(x, x2), …, κ(x, xn)). The gain in speed of kernel-PCA over PCA (and thus the ability to cope with large NGS datasets) is an immediate consequence of the fact that computing KΦ requires O(n2d) operations and a further O(n3) is required for its eigendecomposition, [as opposed to O(d2n) and O(d3) for PCA for the corresponding tasks], which amounts to considerably fewer operations for kernel-PCA when d is much larger than n.
Then for step I we proceed as follows: We first perform a linear kernel-PCA for X, that is, we take the kernel function to be the inner product between accessions of X. Subsequent to this, we subject the resulting eigenvalues to the Tracy–Widom test to identify significant principal components (see, e.g., Peres-Neto et al. 2005) for a survey of attractive alternative approaches). This test has proven very popular in population genetics and relies on the fact that nonzero eigenvalues of a matrix follow a Tracy–Widom distribution. Checking whether an eigenvector is a significant principal component of that matrix or not then reduces to checking whether its associated eigenvalue passes a certain statistical significance test (Patterson et al. 2006).
Step II: Population structure inference:
Simulation studies indicate that a PCA-reduced dataset X obtained in step I can be represented in terms of a (K − 1)-dimensional simplex K−1, where K ≥ 2 [see, e.g., Figure 2 for examples for the case K = 3, and Patterson et al. (2006) and Ma and Amos (2012), where this phenomenon has also been observed for general K]. The vertices of such a simplex correspond to the putative founders of the dataset, that is, its nonadmixed accessions. The position of an accession relative to these vertices encodes the admixture proportion of that accession in the sense that it can be uniquely expressed as a convex combination of the vertices of that simplex. Put differently, with a1, a2, …, aK denoting the vertices of the simplex K−1 representing a dataset X found in step I, any of its accessions x can be expressed as
where, for all 1 ≤ i ≤ K, the quantity λi ≥ 0 is the genetic contribution of founder ai to x and Thus, the components of the ancestry vector λx = (λl)1≤l≤K of x can be thought of as the admixture coefficients of x and computing them is straight forward using standard arguments from linear algebra if the matrix A = (a1, a2, …, aK) of founders is known. If this is not the case then, by viewing the matrix AQ as an approximation of X, the matrix A (and thus the Q-matrix of X) can be inferred using least squares optimization. This boils down to minimizing, for a PCA-reduced SNP matrix X found in step I, the quantity
| (1) |
with respect to A and Q, where Q = (λx)x∈X, and is the Frobenius norm of a matrix B = (bij)1≤i≤k, 1≤j≤t. A detailed explanation on how Equation 1 is solved may be found in File S1.
Figure 2.
PCA reduced dataset under different simulation scenarios, each of which is represented by a separate panel. In each panel, the coordinate axes are the first two significant principal components; see text for details.
Simulated datasets and performance measure
In this section, we present an outline of how we generated the various types of datasets underpinning our simulation study for assessing PSIKO’s performance. In addition, we also briefly review the root mean squared error (RMSE) measure, which we use as assessment criterion. We start with providing details concerning our simulation study.
Simulated datasets generation:
We used the command line-based coalescent simulator msms (Ewing and Hermisson 2010) to first simulate founder allele frequencies and then used them to simulate admixture proportions and genotypes of admixed individuals. More precisely, we simulated K = 3, 4, …, 10 independent, randomly mating populations each of which comprised 100 individuals, where by an individual we mean a sequence composed of L loci evolved over a period of 10,000 generations (see File S1 for exact msms commands used). Here, the number of generations is biologically inspired and the number of individuals and the value K = 3 is based on Alexander et al. (2009). The values we chose for L were 13,262 [which is as in Alexander et al. (2009)] and, to shed light on the scalability of psiko, also 100,000, 250,000, and 2.5 million. We then used these individuals to calculate founder allele frequencies fk1, fk2, …, fkL for all 1 ≤ k ≤ K.
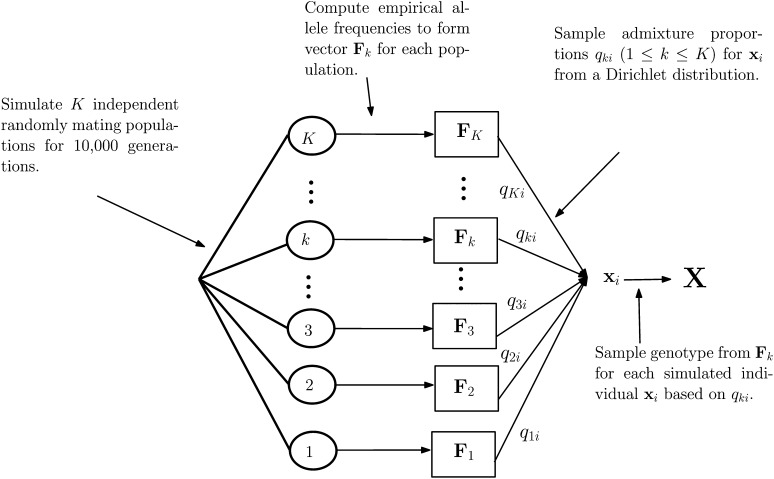
Once obtained, we simulated the genotype of an individual on a locus-by-locus basis using the following two-step process: For a locus l of an individual i, we first simulated the founder zl of l by sampling from a multinomial distribution with parameter the admixture proportions for individual i. The admixture proportions were sampled from a Dirichlet distribution and represent the contribution of each founder to the dataset. Subsequent to this, we simulated the genotype of individual i at locus l by sampling from a multinomial distribution with parameter the allele frequency of population zl at locus l (see Figure 1 for a summary of this two-step process).
Figure 1.
A summary of how the datasets underpinning our simulation experiments were generated. Each of the 1, 2, …, K encircled values indicates a founder population generated with the msms software. For all 1 ≤ k ≤ K, the vector Fk represents empirical allele frequencies computed for each of the K founder populations [i.e., Fk = (fk1, fk2, …, fkL)] and the values qki represent the proportion population k contributes to accession xi of the dataset X.
We repeated this process 1000 times to obtain an admixed dataset containing 1000 individuals.
Performance measure:
To assess the performance of the four approaches under consideration with regard to their ability to recover the known Q-matrix underlying a dataset, we used the RMSE between two Q-matrix and Q′, given by:
| (2) |
where n represents the number of individuals (1000 in our case), K represents the number of founders (K = 3, 4, …, 10 in our case), and and are the elements of and Q′ respectively, where 1 ≤ i ≤ n and 1 ≤ k ≤ K.
Parameter settings:
For all our simulation experiments we used ADMIXTURE and sNMF with their respective default settings, as suggested by their authors. For STRUCTURE, we used the following settings: We assumed admixed populations with independent allele frequencies. We set the length of the burn-in period to 2000 iterations and ran the program for an additional 2000 iterations after the burn-in period. All remaining parameters were used with default values. To ensure fairness in run time comparison between the above three methods and PSIKO, we only compared their run times for the ground truth value of K, thus ensuring that a single run of PSIKO was timed against a single run of all the other methods.
Biological datasets
To assess the performance of PSIKO with challenging biological datasets, we first performed a comparison of the Q-matrix provided by PSIKO to those estimated using STRUCTURE, ADMIXTURE and sNMF for a set of 84 diverse B. napus accessions as described in Harper et al. (2012). Over half of these accessions are winter oilseed rape (OSR) types (49), but the rest comprise diverse winter fodder types (5), spring OSR (14), Chinese semi-winter OSR (5), Japanese kale (2), Siberian kale (2), and Swede (7). Q-matrix estimations were compared directly and subsequently used to perform linear model association mapping following the method outlined in Harper et al. (2012). Briefly, the Q-matrices were used as covariates in general linear models (GLMs), and mixed linear models (MLMs), where a relatedness measure was included as a random effect for two seed oil traits, i.e., erucic acid and glucosinolate content using the program tassel (Bradbury et al. 2007). The results of these models were then compared to their P-value expectations. Results were presented as QQ plots showing observed against expected log10 P-values for each of the four stratification methods and each of the seed oil traits and association model types.
To also investigate psiko in a human population context, we applied it to a subset of the HapMap Phase 3 dataset (International HapMap Consortium 2010). That subset comprised 541 individuals spanning the groupings with the following sampling scenarios: African ancestry in Southwest United States (ASW), Yoruban in Ibadan, Nigeria, West Africa (YRI), Utah residents with Northern and Western European ancestry from the Centre d'Etude du Polymorphisme Humain collection (CEU) and Mexican ancestry in Los Angeles, California (MEX). Each individual was genotyped over 1,457,897 SNP loci. We remark in passing that the choice of dataset is as in Alexander et al. (2009) noting, however, that that article used an older version of the dataset and that those sequences had been pruned so that each comprised 13,298 genotyped SNP loci (Alexander et al. 2009). The general understanding of the dataset is that the ASW sample is admixed with ancestries from YRI and CEU and that MEX is admixed with ancestries from CEU and an unsampled founder population (Jakobsson et al. 2008; Li et al. 2008; Alexander et al. 2009). Therefore the number of founders for this dataset is expected to be three.
Results
Bearing in mind that ADMIXTURE has been shown in Alexander et al. (2009) to be faster than STRUCTURE, FRAPPE (Tang et al. 2005), and INSTRUCT (Gao et al. 2007), and that the recently introduced fastSTRUCTURE approach (Raj et al. 2014) has run time comparable to ADMIXTURE (Raj et al. 2014), to asses psiko’s performance we only compared it against ADMIXTURE and sNMF and, due to its popularity, STRUCTURE. For this, we used a computing cluster with Intel Sandybridge Dual processor, 8 core E5-2670 2.6 GHz CPUs and 2 Gb of DDR3 memory at 1066 Mhz, with Intel Hyper-Threading disabled. We simulated different scenarios for how populations might have arisen. These simulation studies are similar in spirit to those performed in Alexander et al. (2009). Additionally we tested the methods on real biological examples. We start with describing the results of the simulation study, which also includes details on the parameters we varied and their ranges. We then present our findings for the biological datasets.
Simulated datasets
As outlined above (see Materials and Methods) the parameters we varied were the number K of founders and the respective Dirichlet distribution parameters for them. Since their choices depend on the values of K employed, we will detail them as part of a separate treatment of the cases K = 3 and K ≥ 4. Before detailing these cases, though, we remark that low values for the Dirichlet distribution parameters correspond to almost admixture-free populations, whereas values close to one correspond to heavily admixed populations. Thus, our simulation study allows us to assess the performances of the methods in question on highly admixed and highly nonadmixed populations. We start our discussion with remarking that the value for K was correctly recovered by all tested methods for each of the constructed simulated datasets.
For K = 3, and datasets with sequence length 13,262, we chose the same values for the three Dirichlet distribution parameters as in Alexander et al. (2009), resulting in six different simulation scenarios. Three of these scenarios were asymmetric, meaning that in each case at least one Dirichlet distribution parameter was different from the other two and the other three were symmetric, meaning that in each case all Dirichlet distribution parameters were the same. For each of the six scenarios, we generated 100 datasets, resulting in a total of 600 datasets. These we then analyzed with regard to their behavior under PSIKO (see below), and the average root mean square error for the Q-matrices found by each of the methods considered, where the average is taken over all 100 datasets of a scenario (see Materials and Methods). Furthermore, for each of the three sequence lengths, 100,000, 250,000, and 2.5 million, we generated 10 datasets as before using the symmetric Dir(1, 1, 1) parameter distribution. To assess the effect of SNP pruning, we also generated a further 100 datasets following a similar protocol (see below for details). Additionally, to test PSIKO’s robustness to deviations from our simulation model, we also simulate scenarios with noise, missing data, and inbreeding present.
Behavior of a dataset:
To investigate the behavior of PSIKO when applied to a dataset generated under each of the six scenarios, we randomly chose one dataset from each. Exploiting the observation that the number of founders of a dataset equals the number of significant principal components found for that dataset in step I of PSIKO plus one (see, e.g., Patterson et al. 2006), we depict each chosen dataset in terms of a panel containing a two-dimensional coordinate system whose axes are labeled by the two significant principal components found by PSIKO for that dataset (Figure 2). For each coordinate system that makes up that figure, its footer Dir(x, y, z) encodes the simulation scenario used to generate it in terms of the values x, y, and z for the three Dirichlet distribution parameters. For example, the footer Dir(0.2, 0.2, 0.5) of the leftmost coordinate system in the bottom row indicates that two of the three Dirichlet distribution parameters had value 0.2 and that the third one had value 0.5.
As expected (see also Patterson et al. 2006), each of the chosen datasets depicted in Figure 2 (after having applied PSIKO to them) corresponds to a 2-simplex with the dots inside the simplex representing the dataset’s accessions. PSIKO infers three founders for each dataset. We indicated them for each dataset in terms of three ellipses. These are clearly very close to the vertices of the simplex representing that dataset and thus the founders of that dataset. Also the figure suggests that the smaller the values for the Dirichlet distribution parameter are, the more the data points get pushed to the simplex’s vertices, which is again as expected. This holds not only for the asymmetric scenarios but also for the symmetric ones, where the data points get pushed away from the founder with the lowest value. An extreme case in this context is the asymmetric scenario corresponding to Dir(0.05, 0.05, 0.01) as it suggests that one of the founders (i.e., the one corresponding to Dirichlet distribution parameter value 0.01) had very little contribution to the represented dataset.
Average root mean square error:
We next turn our attention to assessing the estimated accuracy of PSIKO by measuring the average RMSE between the true and estimated Q-matrices under each one of the six simulation scenarios. For this we used the 600 datasets generated as described above as input to all four methods in question to obtain Q-matrix estimates from each of them. For each method and over all 100 datasets of a scenario, we then computed the average RMSE between the true and estimated Q-matrices. A summary of our results in terms of these averages is given in Table 1, which consists of six panels each of which corresponds to one of our six simulation scenarios. As can be readily observed, all methods seem to be performing similarly well under all simulation scenarios, with negligible differences between their estimates for the Q-matrices.
Table 1. For K = 3, average RMSEs between true and estimated Q-matrices for simulated datasets.
| Asymmetric | Dir(0.2, 0.2, 0.5) | Dir(0.2, 0.2, 0.05) | Dir(0.05, 0.05, 0.01) |
|---|---|---|---|
| PSIKO | 0.008 | 0.007 | 0.005 |
| ADMIXTURE | 0.008 | 0.005 | 0.002 |
| sNMF | 0.008 | 0.005 | 0.002 |
| STRUCTURE | 0.053 | 0.022 | 0.021 |
| Symmetric | Dir(1, 1, 1) | Dir(0.5, 0.5, 0.5) | Dir(0.1, 0.1, 0.1) |
| PSIKO | 0.011 | 0.009 | 0.004 |
| ADMIXTURE | 0.018 | 0.01 | 0.004 |
| sNMF | 0.02 | 0.013 | 0.005 |
| STRUCTURE | 0.015 | 0.016 | 0.03 |
Longer sequences:
As can be readily observed from Table 2, PSIKO is faster than snmf for each of the three sequence lengths used i.e., 100,000, 250,000, and 2.5 million (Materials and Methods). [Since it has been shown in Frichot et al. (2014) that ADMIXTURE is slower than sNMF, we only compared PSIKO against sNMF.] In fact, as the length of the sequences grows, so too does the difference in run time between PSIKO and sNMF with that difference being significantly in favor of PSIKO. A possible reason for this might be that PSIKO is based on kernel-PCA, which is known to scale very well with the number of variables of a dataset which, in our case, is the number of SNPs i.e., the sequence length (see also Materials and Methods). This behavior seems to suggest that PSIKO scales better than sNMF with increasing sequence length making it highly attractive for population structure estimation from the very large datasets that are becoming increasingly more common in modern, whole-genome studies.
Table 2. Summarized relative run times of sNMF and PSIKO as averages over all 30 datasets.
| Sequence length | 100,000 | 250,000 | 2,500,000 |
|---|---|---|---|
| PSIKO | 8 sec | 11 sec | 1 min 25 sec |
| sNMF | 55.5 sec | 1 min 40 sec | 22 min 28 sec |
i.e., 10 datasets for each symmetric Dirichlet distribution parameter setting given in Table 1.
SNP pruning:
A popular way to turn a large SNP dataset into a dataset of more manageable size is to employ linkage disequilibrium (LD) (Purcell et al. 2007), which is essentially a measure of how frequently SNPs get transmitted together. This technique, however, has the potential to remove relevant information thus introducing bias to a dataset. To test the robustness of the three methods with regards to this, we proceeded as follows. For K = 3 we used msms to simulate 100 datasets each comprising 1000 individuals and 1 million SNPs per individual. From the resulting sequences we then randomly removed 90% of SNPs and then ran PSIKO, ADMIXTURE, and sNMF on the resulting 100 datasets. We found that the average RMSE was below 0.025 for all of PSIKO, sNMF, and ADMIXTURE, corresponding to at most a 2.5% error in ancestry estimates. Once again, all of the tested methods correctly inferred K = 3. The average run times were 3 sec for PSIKO, 7 sec for sNMF, and 30 sec for ADMIXTURE.
Larger values for K:
Due to the combinatorial explosion caused by asymmetric Dirichlet parameter distributions for increasing values of K, we only considered symmetric Dirichlet distribution parameters for higher values of K, that is, for K ranging between 4 and 10. For each of these values for K, we chose the same values for the Dirichlet distribution parameters as for the symmetric Dirichlet distribution parameters for K = 3 i.e., all 1, all 0.5, and all 0.1.
We found that the performance of each of the methods is comparable for all of the resulting 2100 datasets (see File S1). It is worth noting, however, that the run time of PSIKO is much faster than that of ADMIXTURE (and hence also STRUCTURE), and slightly faster than that of sNMF, with sNMF taking on average 7 sec to complete processing each dataset, PSIKO taking on average 4 sec to complete, and ADMIXTURE taking on average 55 sec to complete.
Noise:
Due to the possibility of complex evolutionary processes such as hybridization having confounded the coalescent signal in a dataset, we also tested the robustness of PSIKO for noisy datasets. These we obtained by employing a parameter p that governs the amount of noise that we allowed a dataset’s sequences to contain. More precisely, we started with a dataset obtained for K = 3 and Dirichlet distribution parameters Dir(1, 1, 1) (see Materials and Methods for details), and then, for every one of its sequences, flipped on a locus-by-locus basis the allele of that locus with probability p. Using this modification process we generated 100 noisy datasets for 1000 accessions at 13,262 loci with noise level p set to 0.01, 0.05, 0.1, and 0.15, corresponding to 1, 5, 10, and 15% noise, respectively.
As can be readily seen, the difference in the average RMSE between the estimated and true Q-matrix for each approach in question under each of the aforementioned noise levels is marginal (Table 3), suggesting that all methods are equally robust under the considered simulation scenarios with the observed differences being marginal.
Table 3. Average RMSE between true and estimated Q-matrix for Dir(1, 1, 1) for each approach under each noise level p.
| p | 0.01 | 0.05 | 0.1 | 0.15 |
|---|---|---|---|---|
| PSIKO | 0.011 | 0.012 | 0.013 | 0.015 |
| sNMF | 0.016 | 0.012 | 0.012 | 0.02 |
| ADMIXTURE | 0.018 | 0.013 | 0.013 | 0.019 |
Missing data:
Reflecting the fact that even with current NGS technology, missing data are still a problem (Harper et al. 2012), we also assessed the robustness of PSIKO for this type of data. To obtain such datasets, we proceeded as in the previous data experiment only now instead of flipping a locus allele state with probability p, we set it to a missing value character with probability p. More precisely, for K = 3 and Dirichlet distribution parameters Dir(1, 1, 1), we generated 100 datasets for 1000 accessions each of which was 13,262 loci long (Materials and Methods). We set the missing value character probability p to 0.1 and 0.2, corresponding to 10 and 20% missing data, respectively. Using again the average RMSE as assessment criterion, we present our findings in Table 4.
Table 4. Average RMSE between true and estimated Q-matrix for Dir(1, 1, 1) for each approach under each missing value probability character p.
| p | 0.1 | 0.2 |
|---|---|---|
| PSIKO | 0.012 | 0.012 |
| sNMF | 0.013 | 0.012 |
| ADMIXTURE | 0.019 | 0.021 |
As can be readily seen, even with large proportions of data missing, all three methods perform equally well with only marginal differences, a fact that was also observed for sNMF and ADMIXTURE in Frichot et al. (2014).
Inbreeding:
The assumption of random mating is frequently violated in natural populations. To test the robustness of psiko under these circumstances, we also simulated datasets where inbreeding is present. To do this, we first simulated K = 3 independently mating populations as in the noise experiment. For each population 1 ≤ k ≤ 3 and each locus l in such a population, we then computed the empirical allele frequencies fkl (Materials and Methods). Subsequent to this and following Frichot et al. (2014), we used a preset value for the inbreeding coefficient FIS (i.e., FIS = 0.25 and FIS = 1) to compute genotype frequencies gkl at locus l in population k. Using the Dirichlet distribution parameters Dir(1, 1, 1), we then applied the same simulation protocol as above (see Materials and Methods for details), with gkl taking the place of fkl. For each value of FIS, we simulated 100 datasets comprising 1000 individuals each with 13,262 genotyped SNP positions.
As can be seen in Table 5, all methods are equally robust to inbreeding being present in the dataset, although psiko seems to be slightly more accurate than sNMF and ADMIXTURE (see also (Frichot et al. 2014) where a similar trend was observed for sNMF and ADMIXTURE).
Table 5. Average RMSE between true and estimated Q-matrix for Dir(1, 1, 1) for each approach under each value for the inbreeding coefficient FIS.
| FIS | 0.25 | 1 |
|---|---|---|
| PSIKO | 0.016 | 0.017 |
| sNMF | 0.026 | 0.027 |
| ADMIXTURE | 0.022 | 0.026 |
Biological datasets
To further assess PSIKO, we also subjected it to the test of two biological datasets, one of which is an oilseed rape dataset that was originally studied in Harper et al. (2012) and the other is from the HapMap 3 project (see Materials and Methods for a brief description of each). We compared our findings with that of ADMIXTURE and sNMF, again using the average RMSE as an assessment measure.
Oilseed rape dataset:
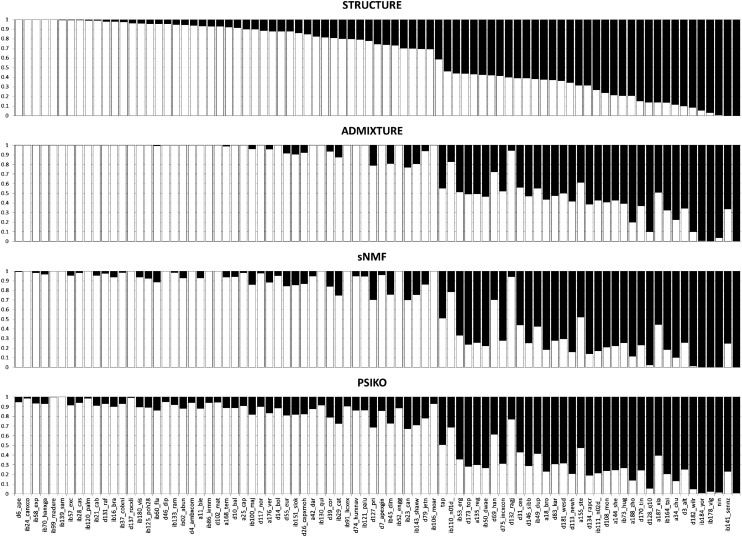
Two of the four methods tested predicted two population clusters (i.e., K = 2). ADMIXTURE predicted three population clusters, while sNMF predicted five clusters. For the purposes of comparing the four models equally, we elected to use the Q-matrices generated for K = 2 from each of the programs. Similarly and as recommended by their respective authors, we ran all programs with their default parameter values. Additionally, we ran STRUCTURE with a burn-in period of 20,000 iterations, followed by another 20,000 iterations. Direct comparison of the four obtained Q-matrices (Figure 3) indicates great similarity, particularly between ADMIXTURE, sNMF, and PSIKO.
Figure 3.
Q-matrix plots for the 84 line Brassica napus dataset comparing the performance of psiko to other leading methods. The proportion of alleles belonging to each of the clusters is shown by respective white bars (cluster 1) or black bars (cluster 2).
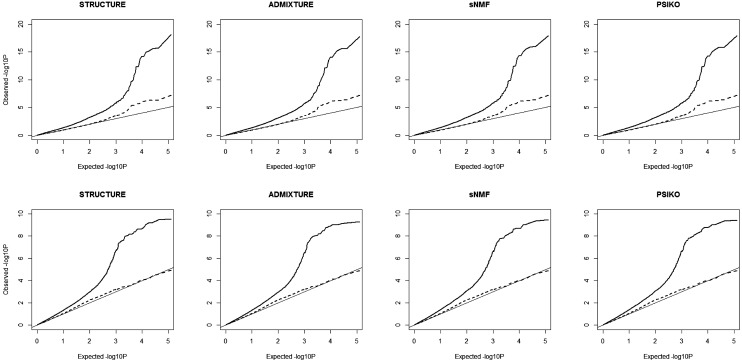
The results of the association mapping using each of the four matrices were very similar (Figure 4). As expected, incorporating the relatedness matrix as a random effect in a MLM reduced the supposed type I error rate. For the erucic acid trait, the residual error was minimized by the MLM/STRUCTURE model, and for the seed glucosinolates trait the residual error was minimized by the MLM/PSIKO model. It is worth noting, however, that the difference between the Q-matrices was not enough to alter identification of markers in close proximity to the major causative loci (see Harper et al. 2012 for details).
Figure 4.
QQ plots illustrating population structure corrections using the four methods in genome-wide association studies (GWAS) analysis of two traits in the 84 lines Brassica napus panel, erucic acid (A) and seed glucosinolate content (B). The expected −log10 P (x-axis) are plotted against those observed (y-axis) from either a general linear model (solid lines) using population structure correction only and a mixed linear model (dashed lines) with population structure and relatedness corrections. The diagonal line is a guide for the perfect fit to the expected −log10 P-values.
HapMap 3 dataset:
Given the size of the dataset and thus the prohibitively long run time of STRUCTURE, we only investigated it with ADMIXTURE, sNMF, and PSIKO (again with all parameter values set to default). Since there are no trait data available, we measured the difference between any two of the three returned Q-matrices in terms of their RMSE and their R2 correlation coefficient.
Given the widely accepted fact that the number of founders for this particular dataset is three, all three methods were run with K = 3. They all found strikingly similar Q-matrices. More precisely, the RMSE between any two matrices was never larger than 0.02 (corresponding to an ∼2% difference) and the R2 correlation coefficient was always >0.99 (suggesting that they are almost perfectly correlated). However, there was a discrepancy between the methods with regard to estimating the number of founders for the dataset with PSIKO and ADMIXTURE returning K = 3, whereas sNMF returned K = 4. The very fast run time of 48 sec for PSIKO (as compared to ADMIXTURE, whose run time was 5212 sec, equating to 1 hr and 27 min and sNMF, whose run time was 17 min and 18 sec) is strikingly apparent with this large-scale dataset.
Since mapping information is available for this dataset, which can be used for LD-based SNP-pruning purposes, we also investigated the performance of PSIKO, sNMF, and ADMIXTURE when the sequences are pruned. More precisely, we used the sliding window-based SNP-pruning approach implemented in PLINK (Purcell et al. 2007) (with default settings) to obtain a pruning of the HapMap 3 dataset. We found that PSIKO, sNMF, and ADMIXTURE all correctly infer the widely accepted number of three founders for that dataset, and that the RMSE between any pair of estimated Q-matrices is never >0.02 (i.e., a 2% disagreement), suggesting that all tested methods yield very similar results (data not shown). However, PSIKO took 21 sec to complete. Using K = 3 as input, sNMF took 6 min and ADMIXTURE took 36 min. Additionally, we found that the SNP pruning took 52 min to terminate, resulting in a 52-min overhead in the total running time of each method for this experiment. This is in stark contrast to the 48 sec it took PSIKO to analyze the complete, unpruned dataset.
Discussion
Population structure is a confounding factor in population association studies, hampering our understanding of how, for example, agronomically important traits have been selected for in crop plants or how diseases might have spread throughout a population (Price et al. 2006). It is therefore important to be able to correct for it and this entails gaining insight into a dataset’s Q-matrix as well as the number of its founders. Popular software packages such as STRUCTURE, FRAPPE, INSTRUCT, and ADMIXTURE infer both. Many of them are based on sophisticated models and rely on assumptions such as satisfying Hardy–Weinberg equilibrium. However, if the dataset in question violates such assumptions or is very large, as would be the case for NGS datasets, these approaches tend to suffer from long run times. To address the issue, among others, of scalability, the sNMF approach has been proposed (Frichot et al. 2014). Unlike STRUCTURE and ADMIXTURE, it is not model based and uses sophisticated algorithmic techniques to ensure fast run times on large datasets.
Here, we propose the novel and fast PSIKO approach for population structure inference. By combining linear kernel-PCA with a quick-to-solve optimization problem, it couples the fast run time and robustness of PCA with the biological interpretability of Q-matrices obtained from model-based approaches such as STRUCTURE and ADMIXTURE. This allows quick estimation of the Q-matrix underpinning a marker dataset as well as the number of founders of that dataset. Due to PCA’s few underlying assumptions, PSIKO is widely applicable and generally has a very low run time, at the same time producing results that are comparable in quality with those obtained by ADMIXTURE, STRUCTURE, and sNMF.
To assess the performance of PSIKO with regard to Q-matrix estimation and inference of founder number, we rigorously tested it on both simulated and real biological datasets. In our simulation studies, we varied the number of founders for a dataset as well as the admixture scenarios for generating a dataset. To help ensure biological relevance, we based our choices for the range of these parameters on those made in Alexander et al. (2009). Across a wide range of simulation scenarios, we found that PSIKO provides Q-matrix estimates that are very close to the estimates for the respective datasets produced by STRUCTURE, ADMIXTURE, and sNMF where closeness is measured in terms of the root mean squared error between two matrices (Alexander et al. 2009). Our missing data, noise, and inbreeding experiments suggest that PSIKO as well as ADMIXTURE and sNMF handle these types of data extremely well. However for large datasets, PSIKO seems to be superior, even if such a dataset is pruned based on, e.g., linkage disequilibrium.
The first of our biological datasets comprises 84 oilseed rape accessions, representing some seven crop types, genotyped over 101,644 SNP loci. The second comprises 541 human samples from differing geographic regions, genotyped at 1,457,897 SNP loci. For each dataset, we found that the Q-matrix estimates generated by PSIKO were very close to those produced by ADMIXTURE and sNMF for that dataset, using the same measure of closeness as in our simulation study. However, it is worth pointing out that independent of whether the dataset had been pruned or not, PSIKO’s run time was only a fraction of that of ADMIXTURE, especially on the human dataset, and was also considerably faster than sNMF.
Although great effort has been put into the development of powerful tools for deriving the number K of founders of a population dataset, inferring that number is still a formidable statistical and computational problem. For example, finding that number using STRUCTURE can be a very time-consuming task due to the fact that it has to be run on a range of different values for K, each of which might take a long time to complete. Even for newer methods such as ADMIXTURE or sNMF, finding the optimal value of K relies on running the methods for a range of values of K. In PSIKO, we exploit the behavior of the eigenvalues returned by linear-kernel PCA for a dataset to infer K. Due to the algorithmic internals of PCA, this can be done quickly. We are also motivated by a study in Patterson et al. (2006) as well as numerous simulation studies that indicate that the number of founders of a dataset equals the number of significant principal components for that dataset plus one. Our simulation studies as well as our two real biological examples suggest that PSIKO holds great promise for this.
The speed of PSIKO is similar to that of sNMF for smaller datasets and is faster than that of ADMIXTURE. While for small datasets the differences in speed between PSIKO and sNMF are negligible, with increasing sequence length PSIKO proves to be significantly faster than sNMF and implicitly also ADMIXTURE. We therefore argue that PSIKO could be a very attractive tool for analyzing the larger datasets that arise from NGS technologies. For smaller datasets (<50 K SNPs), the differences between the three methods are not as clear cut, and the user should choose whichever method would suit their particular dataset best.
In summary we propose a novel, nonmodel-based method for inferring population structure. It exploits the advantages of linear kernel-PCA to quickly and accurately describe a SNP dataset’s population stratification. It is much (up to 300 times) faster than classical, model-based approaches while outputs match those of state-of-the-art methods such as sNMF. Its superior speed for large datasets makes it particularly attractive for datasets generated by NGS approaches.
Supplementary Material
Acknowledgments
The authors thank the referees for their helpful suggestions and comments. A.-A.P. thanks the Norwich Research Park (NRP) for financial support through provision of an NRP studentship. This work was supported by UK Biotechnology and Biological Sciences Research Council [BBSRC BB/H004351/1 (Integrated Biorefining Research and Technology Club), BB/E017363/1, ERAPG08.008] and UK Department for Environment, Food, and Rural Affairs (Defra IF0144). The research presented in this article was carried out on the High Performance Computing Cluster supported by the Research and Specialist Computing Support service at the University of East Anglia.
Footnotes
Supporting information is available online at http://www.genetics.org/lookup/suppl/doi:10.1534/genetics.114.171314/-/DC1.
Communicating editor: G. A. Churchill
Literature Cited
- Alexander D. H., Novembre J., Lange K., 2009. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19: 1655–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bancroft I., Morgan C., Fraser F., Higgins J., Wells R., et al. , 2011. Dissecting the genome of the polyploid crop oilseed rape by transcriptome sequencing. Nat. Biotechnol. 29: 762–766. [DOI] [PubMed] [Google Scholar]
- Bradbury P. J., Zhang Z., Kroon D. E., Casstevens T. M., Ramdoss Y., et al. , 2007. Tassel: software for association mapping of complex traits in diverse samples. Bioinformatics 23: 2633–2635. [DOI] [PubMed] [Google Scholar]
- Engelhardt B. E., Stephens M., 2010. Analysis of population structure: a unifying framework and novel methods based on sparse factor analysis. PLoS Genet. 6: e1001117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing G., Hermisson J., 2010. MSMS: a coalescent simulation program including recombination, demographic structure and selection at a single locus. Bioinformatics 26: 2064–2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frichot E., Mathieu F., Trouillon T., Bouchard G., François O., 2014. Fast inference of admixture coefficients using sparse non-negative matrix factorization algorithms. Genetics 196: 973–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H., Williamson S., Bustamante C. D., 2007. A Markov chain Monte Carlo approach for joint inference of population structure and inbreeding rates from multilocus genotype data. Genetics 176: 1635–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper A. L., Trick M., Higgins J., Fraser F., Clissold L., et al. , 2012. Associative transcriptomics of traits in the polyploid crop species Brassica napus. Nat. Biotechnol. 30: 798–802. [DOI] [PubMed] [Google Scholar]
- International HapMap Consortium , 2007. A second generation human haplotype map of over 3.1 million SNPs. Nature 449: 851–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International HapMap Consortium , 2010. Integrating common and rare genetic variation in diverse human populations. Nature 467: 52–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsson M., Scholz S. W., Scheet P., Gibbs J. R., VanLiere J. M., et al. , 2008. Genotype, haplotype and copy-number variation in worldwide human populations. Nature 451: 998–1003. [DOI] [PubMed] [Google Scholar]
- Kim H., Park H., 2007. Sparse non-negative matrix factorizations via alternating non-negativity-constrained least squares for microarray data analysis. Bioinformatics 319: 1495–1502. [DOI] [PubMed] [Google Scholar]
- Kim S., Plagnol V., Hu T. T., Toomajian C., Clark R. M., et al. , 2007. Recombination and linkage disequilibrium in Arabidopsis thaliana. Nat. Genet. 39: 1151–1155. [DOI] [PubMed] [Google Scholar]
- Knowler W. C., Williams R. C., Pettitt D. J., Steinberg A. G., 1988. Gm3;5,13,14 and type 2 diabetes mellitus: an association in american indians with genetic admixture. Am. J. Hum. Genet. 43: 520526. [PMC free article] [PubMed] [Google Scholar]
- Li J. Z., Absher D. M., Tang H., Southwick A. M., Casto A. M., et al. , 2008. Worldwide human relationships inferred from genome-wide patterns of variation. Science 319: 1100–1104. [DOI] [PubMed] [Google Scholar]
- Ma J., Amos C. I., 2012. Principal components analysis of population admixture. PLoS ONE 7: e40115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchini J., Cardon L., Phillips M., Donnelly P., 2004. The effects of human population structure on large genetic association studies. Nat. Genet. 36: 512–517. [DOI] [PubMed] [Google Scholar]
- Murphy K. P., 2012. Machine Learning: A Probabilistic Perspective, MIT Press, Cambridge, MA. [Google Scholar]
- Patterson N., Price A. L., Reich D., 2006. Population structure and eigenanalysis. PLoS Genet. 2: e190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peres-Neto P. R., Jackson D. A., Somers K. M., 2005. How many principal components? Stopping rules for determining the number of non-trivial axes revisited. Comput. Stat. Data Anal. 49: 974–997. [Google Scholar]
- Price A. L., Patterson N. J., Plenge R. M., Michael E. W., Shadick N. A., et al. , 2006. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38: 904–909. [DOI] [PubMed] [Google Scholar]
- Pritchard J. K., Stephens M., Donnelly P., 2000. Inference of population structure using multilocus genotype data. Genetics 155: 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M. A., et al. , 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81: 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj, A., M. Stephens, and J. K. Pritchard, 2014 fastSTRUCTURE: variational inference of population structure in large SNP data sets. Genetics 197: 573–589. [DOI] [PMC free article] [PubMed]
- Scholkopf, B., A. Smola, and K.-R. Mller, 1999 Kernel principal component analysis, pp. 327–352 in Advances in Kernel Methods: Support Vector Learning, MIT Press, Cambridge, MA. [Google Scholar]
- Tang H., Peng J., Wang P., Risch N. J., 2005. Estimation of individual admixture: analytical and study design considerations. Genet. Epidemiol. 28: 289–301. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.