Abstract
Multiparental designs combined with dense genotyping of parents have been proposed as a way to increase the diversity and resolution of quantitative trait loci (QTL) mapping studies, using methods combining linkage disequilibrium information with linkage analysis (LDLA). Two new nested association mapping designs adapted to European conditions were derived from the complementary dent and flint heterotic groups of maize (Zea mays L.). Ten biparental dent families (N = 841) and 11 biparental flint families (N = 811) were genotyped with 56,110 single nucleotide polymorphism markers and evaluated as test crosses with the central line of the reciprocal design for biomass yield, plant height, and precocity. Alleles at candidate QTL were defined as (i) parental alleles, (ii) haplotypic identity by descent, and (iii) single-marker groupings. Between five and 16 QTL were detected depending on the model, trait, and genetic group considered. In the flint design, a major QTL (R2 = 27%) with pleiotropic effects was detected on chromosome 10, whereas other QTL displayed milder effects (R2 < 10%). On average, the LDLA models detected more QTL but generally explained lower percentages of variance, consistent with the fact that most QTL display complex allelic series. Only 15% of the QTL were common to the two designs. A joint analysis of the two designs detected between 15 and 21 QTL for the five traits. Of these, between 27 for silking date and 41% for tasseling date were significant in both groups. Favorable allelic effects detected in both groups open perspectives for improving biomass production.
Keywords: QTL detection, linkage disequilibrium information with linkage analysis (LDLA), allelic series, multiparental families, maize biomass production, MPP, multiparental populations, Multiparent Advanced Generation Inter-Cross (MAGIC)
MOST traits of agronomic interest present a continuous variation resulting from the sum of the effects of various quantitative trait loci (QTL). Mapping these QTL is a first step toward elucidating their molecular nature and offers important application perspectives for marker-assisted breeding. QTL mapping started in plants with segregating families derived from the cross of two inbred lines (Lander and Botstein 1989). However, such biparental designs address only a small portion of the diversity available (a maximum of two alleles can segregate at a given QTL) and the accuracy of QTL positions is usually poor. To overcome these limitations, Rebai and Goffinet (1993) and Charcosset et al. (1994) proposed models for joint QTL detection in several biparental families connected to each other by the use of common parental lines. When the number of parents is less than the number of families, connections can be taken into account to reduce the number of allelic effects to be estimated in the detection model. This increases power and accuracy of detection when QTL behave additively (see Blanc et al. 2006). However, such a model makes the assumption that each parental line carries a different allele, which limits its benefit when the number of parental lines is high relative to the number of families, a situation commonly encountered in breeding programs.
Recent advances in sequencing and genotyping technologies make it possible to genotype individuals for a large number of markers at reduced costs, so that one can expect to have markers closely linked to any QTL. This has paved the way toward association mapping, in which marker-trait associations are directly detected in populations composed of diverse inbred lines without the need to develop experimental segregating families. Association mapping, also often referred to as linkage disequilibrium (LD) mapping, has been widely used with success in the plant community (see for instance Bouchet et al. 2013 and Romay et al. 2013 for recent results of association mapping in maize). In this approach, it is important to use models accounting for potential underlying population structure and relatedness between individuals to prevent spurious QTL detection due to associations between loci that are not linked physically (Yu et al. 2006). As a consequence, the power to detect associations is low for causal polymorphisms correlated with the underlying population structure or when they are present in the population at a low frequency (Rincent et al. 2014). In addition, associations are generally tested at SNP (single nucleotide polymorphism) markers, which leads to the implicit assumption that the QTL are biallelic. These limitations can be alleviated by combining information coming from LD at the level of the parents and linkage within families, as first proposed for animal populations by Meuwissen and Goddard (2001). In this approach, referred to as linkage disequilibrium and linkage analysis (LDLA), dense genotyping of parents is used to detect identity by descent (IBD) at putative QTL, i.e., the fact that two individuals carry the same allele transmitted by a common ancestor. Different types of LDLA analyses have been proposed to account for the LD component. The simplest is to consider that parents carrying the same allele at a given marker are IBD (Yu et al. 2008; Liu et al. 2012) as done in association mapping. Haplotype-based approaches also have been proposed to group parental alleles and tested by simulations (for instance Jansen et al. 2003; Bink et al. 2012; Leroux et al. 2014). Advantages of LDLA have been shown experimentally in maize notably by using the nested association mapping (NAM) design developed in the United States (Yu et al. 2008; McMullen et al. 2009). This design consists of 25 biparental recombinant inbred line (RIL) populations derived from the cross of the inbred B73 with 25 diverse lines representing the diversity of maize (tropical, temperate, sweet corn, and popcorn lines). This design was studied with a linkage analysis model (Buckler et al. 2009; Kump et al. 2011; Tian et al. 2011) where QTL effects were nested within each family and each parental line was assumed to carry a different allele, and with LDLA through a genome-wide association mapping model (Kump et al. 2011; Tian et al. 2011) including allelic effects observed at individual SNP of the parents to identify IBD alleles. This design successfully led to the detection of numerous QTL and use of LDLA permitted in some cases to resolve QTL detection up to the gene level (Kump et al. 2011; Poland et al. 2011; Tian et al. 2011; Cook et al. 2012). Recently, Bardol et al. (2013) applied the haplotype-based approach of Leroux et al. (2014) to detect QTL in two data sets coming from an applied maize (Zea mays L.) breeding program and compared it to models considering each parental allele as different (linkage model) or considering that parents carrying the same allele at a given marker are IBD. Results showed that when parental lines are all issued from the same breeding program and related by pedigree, LDLA models were more powerful than linkage approaches. Bardol et al. (2013) also showed that the different ways of modeling allelic variation (either using haplotypes or single-marker information) had variable efficiencies depending on the QTL and trait considered and were therefore complementary. It is thus important to further evaluate the ability of diverse LDLA models to detect QTL in multiparental populations with different diversity levels.
The central line of the U.S. NAM (B73) is too late flowering for evaluation in Northern Europe and founder lines cover a very broad range of geographical origins, including even later tropical materials. This prevents the evaluation of the whole design for productivity traits in Northern European conditions and due to diversity of the lines it is difficult to use a single tester to investigate hybrid values. To overcome these limitations and expand the genetic pool investigated in maize QTL mapping studies, two parallel complementary NAM designs were developed within the European project CornFed. Each was derived from inbred lines representing the main diversity available for breeding in each of the two major heterotic groups (dent and flint) used in Northern Europe. Both designs were genotyped with a 50k SNP array (Ganal et al. 2011) and genotyping information was used to build individual population maps (Bauer et al. 2013). The two NAM designs were crossed with the central line of the opposite group to produce hybrids, which were analyzed for traits related to biomass production as described in Lehermeier et al. (2014). Increasing biomass production is of key interest in Northern Europe where maize has been extensively used for decades for silage and more recently for bioenergy production. To our knowledge no QTL mapping experiment has been carried out so far for traits related to biomass production in multiparental design assembling such large diversity. Note that both hybrid designs address variation compared to the same hypothetical reference hybrid (the one produced by crossing the two central lines), with each experimental hybrid of each group sharing on average 75% of its genome with the reference hybrid. In this context, effects of all segregating genotypes at a QTL (11 on the dent side and 12 on the flint side) are compared to a same genotype (having received alleles from two central lines). This makes this design particularly adapted for deciphering loci involved in genetic variation on the dent and flint sides for productivity traits.
This study aimed at comparing different methods of QTL detection in these two European NAM designs for five traits of agronomical interest for biomass production in maize: whole plant dry matter yield, whole plant dry matter content at harvest, female flowering, male flowering, and plant height. We compared a linkage approach with two LDLA approaches either considering haplotypic IBD or single-marker groupings. This allowed us to investigate the performance of the different LDLA approaches in two complementary heterotic groups in a more diverse context than a simple breeding program. A second important objective of this work was to compare the results of QTL detection conducted separately in the two heterotic groups or jointly for the whole design, to better understand the contribution of each group to trait variation.
Material and Methods
Plant material and phenotypic analysis
Two maize NAM designs composed of half-sib families from the two major heterotic groups (dent and flint) used for breeding in Europe were analyzed. The two designs are described in Bauer et al. (2013). In short, the dent and flint designs were respectively composed of 10 and 11 doubled haploid (DH) families, derived from the cross of respectively 10 and 11 diverse founder lines with a common central line: F353 for the dent and UH007 for the flint. F353 and UH007 represent very promising European lines created by public institutes in their respective heterotic groups. The parental lines were chosen to cover the diversity available within the two groups with a combination of ancestral and more recent material. From each cross, DH lines were generated, resulting in 919 lines for the dent and 1009 for the flint (Bauer et al. 2013) (Supporting Information, Table S1). For phenotypic evaluation (see below), the segregating DH lines of a given group were crossed with the central line of the other group. A total of 841 hybrids were produced for the dent group and 811 for the flint group (Lehermeier et al. 2014) (Table S1). The number of dent lines for which testcrossed progenies were phenotyped per family was 84 on average and varied between 53 and 104, depending on the family. For the flint group, the number of DH lines per family that were phenotyped for testcross values ranged from 17 to 133 with an average of 73. As the hybrids of each group were obtained by crossing DH lines with the central line of the other group, all the hybrids shared a large proportion of their genome and were expected to be heterozygotes F353/UH007 for 50% of their genome. Hybrids were evaluated in 2011 in four (dent) and six (flint) European locations. Five traits were considered: biomass dry matter yield (DMY, decitons per hectare, dt⋅ha−1) at the whole plant level, whole plant dry matter content (DMC, %) at harvest, days to tasseling (DtTAS, in days), and days to silking (DtSILK, in days) measured as the number of days from sowing until tasseling and silking, respectively. Field trial design is described in Lehermeier et al. (2014). Individual field plot measures were analyzed (Lehermeier et al. 2014) to compute for each hybrid the adjusted means over the different trials that were used in this study.
Genotyping and analysis of genotypic data
The 1928 DH lines and the 23 parental lines were genotyped with the Illumina MaizeSNP50 BeadChip containing 56,110 SNPs (Ganal et al. 2011). Markers with a call frequency <0.9, a GenTrainScore <0.7, a minor allele frequency (MAF) <0.01, or >10% missing values were discarded as in Lehermeier et al. (2014).
Consensus maps for the flint and the dent multipopulations were obtained following the same procedure. We considered for each consensus map the list of markers present in at least 1 of the 10 dent individual maps (respectively, 11 flint individual maps) from Bauer et al. (2013). The flint DH family resulting from the cross of EP44 and UH007 was not used due to small population size. For each marker of this list and for each individual genetic map, we computed the relative genetic position of this marker in this map by starting from its physical coordinate on the B73 genome assembly and converting it into a genetic coordinate with the spline-smoothing interpolating procedure described in Bauer et al. (2013). These genetic coordinates were then normalized between zero and one to obtain relative genetic positions. For the present study, each consensus map was built by computing the consensus relative genetic position of each marker as the average of its relative genetic positions in all individual maps involved, weighted by the numbers of individuals in the corresponding populations. Finally, the consensus genetic coordinate of each marker was obtained by multiplying its consensus relative genetic position by the genetic length of the consensus map, taken as the average of the genetic lengths of all maps, weighted by the numbers of individuals in the corresponding populations. The two consensus maps obtained are available at Maize GDB (http://maizegdb.org/cgi-bin/displayrefrecord.cgi?id=9024747, data available on 4th November 2014). A consensus map for the dent and flint multipopulations was built with the same procedure.
For the QTL detection we considered in the analysis only the PANZEA markers which were mapped on the consensus maps. PANZEA markers result from the alignment of sequences coming from resequencing data of the 27 lines used as parents of the U.S. NAM design (McMullen et al. 2009) and mapped against the B73 genome v. 2 (Gore et al. 2009). We discarded the other markers, mainly defined by comparing the sequences of the inbred lines B73 and Mo17, as they are known to create an ascertainment bias in diversity analyses (Ganal et al. 2011; Frascaroli et al. 2013).The dent and flint consensus genetics maps obtained were composed of, respectively, 21,878 and 20,406 PANZEA markers, corresponding respectively to 6808 and 7272 genetic positions on the consensus maps. The dent-flint consensus map was composed of 25,472 PANZEA markers, corresponding to 8124 genetic positions (Table 1).
Table 1. Number of mapped markers, length of the genetic map and linkage disequilibrium decay modeled with the Hill and Weir (1988) model for a r2 = 0.2 for the two groups dent and flint for each chromosome and for the whole genome.
| Dent |
Flint |
|||||
|---|---|---|---|---|---|---|
| Chromosome | Markers | Length (cM) | LD decay (cM) | Markers | Length (cM) | LD decay (cM) |
| 1 | 3287 | 184.5 | 0.96 | 2892 | 237.2 | 0.76 |
| 2 | 2402 | 137.9 | 2.51 | 2264 | 182.7 | 0.65 |
| 3 | 2480 | 151.0 | 1.99 | 2410 | 156.4 | 0.45 |
| 4 | 2528 | 134.6 | 1.47 | 2379 | 165.5 | 0.65 |
| 5 | 2405 | 136.6 | 0.45 | 2322 | 180.6 | 0.35 |
| 6 | 1695 | 119.9 | 1.47 | 1544 | 134.9 | 0.65 |
| 7 | 1820 | 128.9 | 1.37 | 1709 | 149.7 | 0.76 |
| 8 | 1992 | 125.6 | 1.47 | 1756 | 139.7 | 0.45 |
| 9 | 1699 | 118.5 | 0.96 | 1610 | 133.5 | 0.76 |
| 10 | 1570 | 105.8 | 1.89 | 1520 | 106.1 | 0.76 |
| Genome | 21878 | 1343.3 | 1.2 | 20406 | 1586.3 | 0.65 |
Clustering analysis of parental inbred lines
Clustering of the parental inbred lines was carried out with the R package “clusthaplo” (Leroux et al. 2014), separately on the dent and flint parents. This clustering was based on genomic similarities computed between each pair of individuals in a sliding window along the genome. To obtain insight into the length of the sliding window to use, we evaluated how fast LD between pairs of markers decays with the genetic distance. LD between pairs of markers was estimated for the 11 dent founder lines and for the 12 flint founder lines, according to Hill and Robertson (1968) as , with , where denotes the haplotype frequency of , the frequency of allele A at one marker locus, and the frequency of allele B at the other locus. The LD decay was estimated using the Hill and Weir (1988) model. The choice of the sliding window size was based on the LD decay observed in the dent and flint material considering the length in genetic distance needed to reach an r2 <0.2. Two values were chosen, 2 and 5 cM, each based on the LD decay observed for the flint and dent group, respectively. For facilitating comparisons between results obtained in the two groups, the clustering was carried out in each group using the two window sizes.
For each window size at each genotyped position, the similarity score between two parental lines i and j at a position t (center of the window) was calculated according to the formula described in Leroux et al. (2014) and used in Bardol et al. (2013). This formula is adapted from Li and Jiang (2005) and combined the number of alleles alike-in-state between the two lines inside the sliding window and the length of their longest common segment centered on t. Based on the similarity score curves obtained along each chromosome, a hidden Markov model (HMM) was used to determine at each position t if the two lines were similar and thus carried the same ancestral allele or not. After the clustering process, the number of ancestral alleles per position was plotted along chromosomes. We also computed similarities between inbred lines as the percentage of ancestral alleles shared over the genome and compared them with the similarities obtained from the SNP markers. A graphical representation of these similarities and a classification of the parental lines were carried out using the “heatmap” function in R (R Core Team 2013).
QTL detection
Analyses were first performed separately for each trait on the dent and flint multifamily designs, using their respective consensus map. Four statistical models were tested: one based on linkage analysis and three others combining linkage and LD information. All the models were multilocus models in which the significance of each QTL was tested, conditional on the inclusion of other QTL positions used as cofactors.
The first model corresponded to a conventional multifamily connected model. This model considered the connections between families through the sharing of the central inbred line and relied on the assumptions that each parental inbred line carried a different QTL allele and that each allelic effect was independent of the family
where y was the vector (N × 1) of the adjusted phenotypic means of the N individuals of the data set, J was a (N × P) matrix of 0 and 1 that linked each individual to the family it belonged to with P being the total number of families, μ was the column vector (P × 1) of family means, and Xq and Xc were (N × K) matrices with K being the number of parents. Each element (ranging from 0 to 2) of these matrices corresponded to the expected number of alleles of the parent k at QTL q and cofactor c for each individual, according to the genotyping information at the position of q and c when this information was available (i.e., when these positions correspond to markers polymorphic in the population the individual belong to) or at flanking markers otherwise. aq and ac were the column vectors (K × 1) of the additive intrafamily effects associated with QTL q and cofactor c, respectively. e was a column vector (N × 1) of the residuals of the model. This model will be further referred to as “connected.” Note that this model is close to the joint inclusive composite interval mapping (JCIM) model proposed by Buckler et al. (2009) and used on the U.S. NAM design.
The second and third models were LDLA multifamily connected models, which used the results of the clustering of parental alleles carried out with clusthaplo
where y, J, μ, Xq, Xc, and e were the same as described as in the previous model. Qq and Qc were (K × Aq) and (K × Ac) matrices with Aq and Ac being the number of ancestral alleles at QTL q and cofactor c. Each element (0 or 1) of these matrices linked the parental alleles at QTL q and cofactor c to the ancestral alleles identified by the clustering approach. hq and hc were column vectors (Aq × 1) and (Ac × 1) of the additive effects of the ancestral alleles associated with QTL q and cofactor c. Two models were considered, one based on the clustering approach using a window size of 2 cM and further referred to as “LDLA—2 cM,” and one based on the clustering approach using a window size of 5 cM and further referred to as “LDLA—5 cM.”
QTL detection using the three models described above were performed using the MCQTL_LD software (Jourjon et al. 2005) using an iterative composite interval QTL mapping method (iQTLm) (Charcosset et al. 2000). For these models, genotypic information of markers located at the same position of the consensus genetic map was concatenated to indicate which parental allele was transmitted. For missing data, MCQTL_LD software estimated the probability of transmission of each parental allele based on the information of flanking markers. At each tested position, the presence of a QTL was assessed based on the −log10 of the Fisher test P-value [−log10(P-value)]. Thresholds for considering a QTL as significant were computed for each trait and each data set using 5000 intrafamily permutations of the phenotypes for a type I risk of 10% across all families and total genome. In the iQTLm approach, the initial set of cofactors was chosen using a multiple regression with a forward selection of marker positions with a threshold equal to 80% of the QTL significance threshold value. At the end of the detection process, for the conventional connected model, confidence intervals at 95% were estimated on the basis of a 1 LOD unit fall. The confidence intervals were not estimated for the LDLA models as there is no established method proposed for these models.
The fourth model, referred to as single-marker LDLA model (“LDLA—1-marker”), considered that two parental lines carrying the same allele at a marker were IBD for this marker
y, J, μ, and e were as described in the previous model. Mq and Mc were (N × 2) matrices whose elements (0 or 1) corresponded to the genotyping information at QTL q and cofactor c for each individual. gq and gc were column vectors (2 × 1) of the additive effects of marker alleles associated with QTL q and cofactor c. This model can be viewed as a multilocus genome-wide association study with population structure controlled by family membership. It is equivalent to the association mapping model used to analyze the U.S. NAM design (Yu et al. 2008; Tian et al. 2011; Kump et al. 2011) except that in our model, dense marker genotyping information is directly available for the progenies and does not need to be inferred from the parental genotypes.
The analysis with the fourth model was performed in R (R Core Team 2013) using an R-script derived from the one used for the multilocus mixed model approach presented in Segura et al. (2012). We used a multilocus forward–backward stepwise linear regression model and selected the most appropriate model using the extended Bayesian information criterion (Segura et al. 2012). Loci of the selected model, which had P-values below the Bonferroni threshold for a genome-wide risk of 10%, were considered as QTL. For this model, imputation of the genotyping data for marker with missing data were done using the software BEAGLE (Browning and Browning 2009) family by family. Even if we considered the same type I error risk at the genome level as for other models, the threshold used for the LDLA—1-marker model was not obtained by permutations and is possibly more conservative than other models.
Analyses were then performed jointly for each trait on the two designs using the dent–flint consensus map. The model used corresponded to a conventional multifamily connected model except that all the dent and flint families were considered jointly. As the central line of the dent is used as tester in the flint design and reciprocally, the F353-UH007 genotype segregates against an alternative genotype in each population. This enabled us to connect allelic effects estimated in the two designs. QTL detection was performed using the MCQTL_LD software (Jourjon et al. 2005) following the same procedure as that used in group-specific QTL detection. Thresholds for considering a QTL as significant were computed for the joint data set for each trait using 5000 intrafamily permutations of the phenotypes for a type I risk of 10% across all families and total genome. To test whether effects were significant in a single group or in both groups, the effects of the QTL detected in the joint analysis were tested in each of the separate data sets. They were considered as significant if the −log10 of the Fisher test P-value was above the thresholds of the studied trait in the separate data set (estimated with the dent or flint consensus maps, respectively).
For each analysis, variances explained by each QTL (partial ) were defined as the ratio between the sum of squares associated with the QTL effect in the model including the other detected QTL and the residual sums of squares of a linear model considering only the family effects. Total percentage of variance explained by the detected QTL () was defined as the ratio between the sum of squares of all the detected QTL and the residual sums of squares of a linear model considering only the effects of the families. All the R2 were adjusted by the number of degrees of freedom of the considered models (Charcosset and Gallais 1996). Differences in effects among pairs of alleles at a given QTL were tested a posteriori using a t-test (α = 5%). For facilitating comparisons between models and the interpretation of the QTL results, the allelic effect of the central lines were set to zero, and the other allelic effects were estimated accordingly.
Comparison of the positions of the QTL detected separately in the two groups and in the joint analysis was based on the results of the connected model. QTL detected in each separate group and on the joint data set were projected on the dent–flint consensus map using BioMercator v. 4.2 (Sosnowski et al. 2012). A QTL was considered common for a trait when the confidence intervals of the QTL after projection were overlapping.
Results
Analysis of parental linkage disequilibrium and parental clustering
The average genetic distance to reach a LD below r2 =0.2 was 1.2 and 0.65 cM for the dent and flint groups, respectively (Table 1). This distance varied according to the chromosome between 0.45 cM (chromosome 5) and 2.51 cM (chromosome 2) for the dent group and 0.35 cM (chromosome 5) and 0.76 cM (chromosomes 1, 7, 9, 10) for the flint group. The two different sliding window sizes that we considered for computing the similarity score with clusthaplo approximately correspond to two times the distance beyond which LD becomes negligible for all the chromosomes. Note that 2 cM was the minimum window size that we could consider since the HMM-based clustering approach did not converge for smaller window sizes.
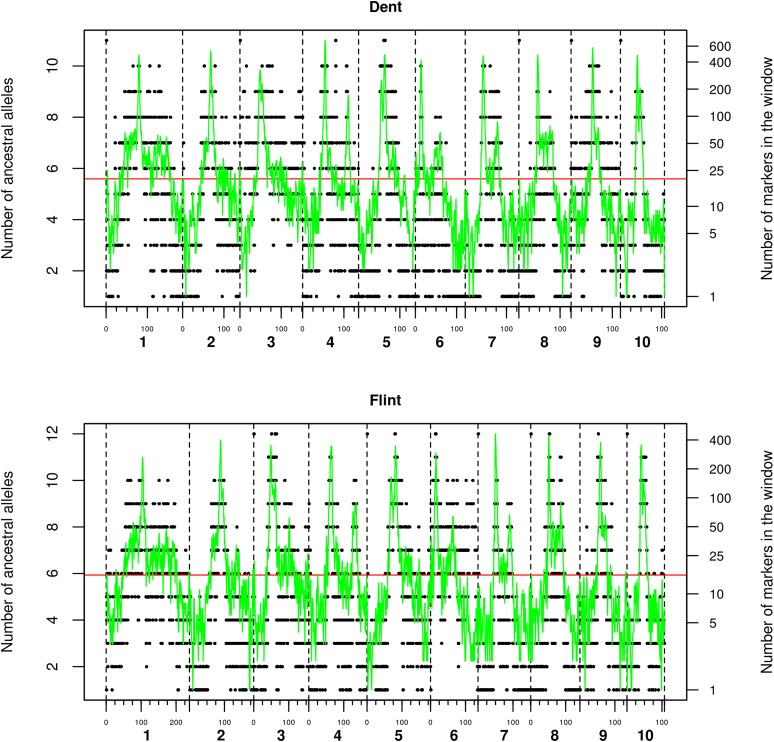
The 5-cM sliding window size led to a higher number of ancestral alleles than the 2 cM one for the two designs. For dent, the average number of ancestral alleles along the genome was 5.6 per genetic position for the 2-cM sliding window size and 6.5 for the 5-cM window. For flint, the average number of ancestral alleles was 5.9 per genetic position for the 2-cM sliding window size and 7.2 for the 5-cM window. It has to be noted that the number of ancestral alleles varied along the genome. For both window sizes, clustering was more important in telomeric than in centromeric regions, where quite often the number of ancestral alleles equaled the number of parental lines (Figure 1).
Figure 1.
Number of ancestral alleles along the genome after clustering with clusthaplo using a 2-cM sliding window size and number of markers in the 2-cM sliding window along the genome for the dent design (6808 unique positions on the genome—1343.3 cM in total) and the flint design (7272 unique positions on the genome—1586.3 cM in total). The black points correspond to the number of ancestral alleles. The green line corresponds to the number of markers in the 2-cM sliding window along the genome. Horizontal red lines correspond to the average number of ancestral alleles along the whole genome. The vertical black dotted lines correspond to the limits of each chromosome.
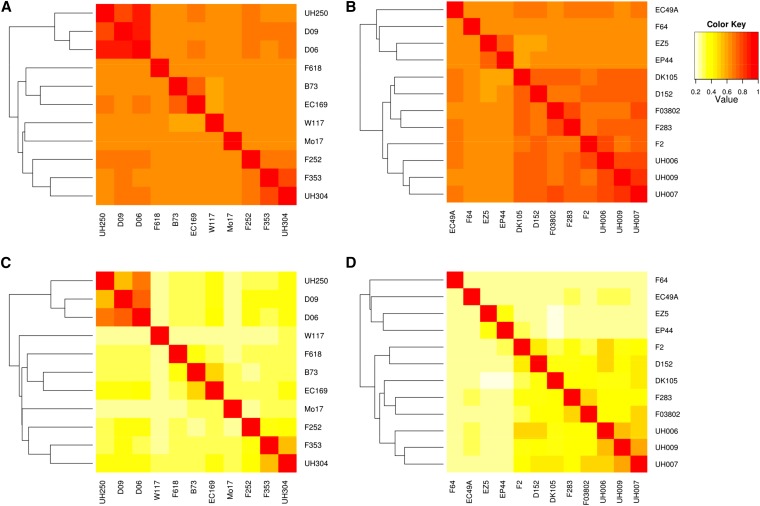
For both sliding window sizes, similarities between the parental inbred lines estimated based on ancestral alleles sharing showed a structured pattern (Figure 2). Within the dent group, pairs of lines involving (i) UH250, D09, and D06 and (ii) F353 and UH304 shared the same ancestral alleles for >47% of the genetic positions for both sizes of sliding window. In the flint group, with the 5-cM window, closest pairs of lines involved UH006, UH007, and UH009. With the 2-cM window size, this expanded to F03802, D152, and F2. The classifications of parental lines based on single markers were globally consistent with those based on ancestral alleles, at least for grouping the most similar lines. Only positions of inbred lines that showed low levels of similarities with the other lines slightly changed in the dendrogram depending on the allele definition considered. In the dent group, three related lines, UH250, D09, and D06, are clearly separated from a nonstructured group among which only F353 (the central line of the dent design) and UH304 were related. In the flint group, similarities separated a subgroup composed of F64, EC49A, EZ5, and EP44 from the other lines that appeared to be more closely related to each other. In this subgroup, UH009 and UH006 are both related to UH007, the central line of the flint design.
Figure 2.
Similarities between the dent (left) and the flint parental lines (right), computed based on direct marker genotyping (top) and on ancestral allele sharing (using clusthaplo and a 2-cM window size) (bottom). Yellow corresponds to a low similarity, red corresponds to a high similarity (color scale on the top-right corner). Lines were ordered according to their position in the dendogram (on the top and on the left of each graph) obtained by a hierarchical clustering based on similarities. (A) Similarities between the dent parental lines computed based on direct marker genotyping. (B) Similarities between the flint parental lines computed based on direct marker genotyping. (C) Similarities between the dent parental lines computed based on ancestral allele sharing (using clusthaplo and a 2-cM window size). (D) Similarities between the flint parental lines computed based on ancestral allele sharing (using clusthaplo and a 2-cM window size).
Comparison of the thresholds used in the QTL detection models
For the separate data sets analyses, threshold values [−log10(P-value)] were higher for the LDLA models than for the linkage model (Table S2). For LDLA models, the threshold increased as the size of the considered window decreased. This suggests that reducing the size of the window decreases the dependence between tests. For every model, threshold values were lower for DMC and higher for DtSILK and DtTAS (except for the conventional connected model for the flint group). This might be due to heterogeneity of within-family variances for some traits. For instance, for DtSILK, for the dent data set, genetic variances varied from 0.95 to 4.93 (see Lehermeier et al. 2014 for an estimation of these variances). As for the separate data sets thresholds, for the joint data set, threshold values for the connected model were lower for DMC and higher for DtSILK and DtTAS.
Comparison of the QTL detected with the different models in the dent and flint designs
For a given trait and group, the number of detected QTL varied according to the model (Table 2, Table S3, Table S4, Table S5, Table S6, Table S7, Table S8, Table S9, and Table S10). Between 5 (for DMY with LDLA—5 cM and LDLA—1-marker models) and 16 (for DMC with LDLA—2 cM model) QTL were detected in the dent design and between 7 (for DMC with LDLA—1-marker model) and 16 QTL (for DtSILK and DtTAS with LDLA—1-marker model) in the flint design.
Table 2. Number of QTL detected (Nb) and adjusted percentage of variance explained by the detected QTL (R2) for the five traits in the two separate data sets for each model and for the joint data set for the connected model.
| DMC |
DMY |
DtSILK |
DtTAS |
PH |
Total |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Nb | R2 (%) | Nb | R2 (%) | Nb | R2 (%) | Nb | R2 (%) | Nb | R2 (%) | Nb | R2 (%) | |
| Dent | ||||||||||||
| Connected | 12 | 51.4 | 8 | 32.7 | 11 | 52.3 | 7 | 41.2 | 14 | 57.1 | 52 | 46.9 |
| LDLA—5 cM | 15 | 51.1 | 5 | 22.5 | 12 | 53.7 | 11 | 49.2 | 13 | 54.1 | 56 | 46.1 |
| LDLA—2 cM | 16 | 53.6 | 6 | 23.4 | 12 | 53.2 | 9 | 45.1 | 12 | 49.5 | 55 | 45.0 |
| LDLA—1-marker | 12 | 37.4 | 5 | 18.6 | 11 | 43.2 | 7 | 33.3 | 10 | 36.4 | 45 | 33.8 |
| Flint | ||||||||||||
| Connected | 8 | 46.0 | 11 | 48.6 | 15 | 69.3 | 12 | 65.3 | 9 | 52.3 | 55 | 56.3 |
| LDLA—5 cM | 11 | 49.2 | 10 | 41.9 | 14 | 67.5 | 13 | 61.1 | 10 | 51.7 | 58 | 54.3 |
| LDLA—2 cM | 8 | 42.1 | 12 | 45.3 | 11 | 62.0 | 14 | 62.2 | 11 | 51.9 | 56 | 52.7 |
| LDLA—1-marker | 7 | 36.1 | 11 | 39.0 | 16 | 61.7 | 16 | 58.0 | 9 | 41.9 | 59 | 47.3 |
| Joint | ||||||||||||
| Connected | 18 | 54.6 | 16 | 45.5 | 15 | 59.7 | 17 | 61.4 | 21 | 61.2 | 87 | 56.5 |
We also indicated the total number of QTL detected over the traits and the average percentage of variance explained (“Total” column).
For the dent group, the LDLA—1-marker model detected fewer QTL over all traits (45 QTL in total) and explained the smaller percentage of variance (33.8% on average). In this group, the LDLA models using clusthaplo information detected more QTL (56 in total for the LDLA—5 cM, 55 for the LDLA—2 cM) than the conventional connected model (52 QTL in total). This advantage of the LDLA models in terms of number of QTL detected was found for DMC, DtSILK, and DtTAS. In contrast, for DMY and PH the connected model detected more QTL. Even if more QTL were detected on average with the LDLA models, the connected model explained a higher percentage of variance (46.9%) than the other models.
For the flint group, the LDLA—1-marker model detected more QTL (59 QTL in total) but explained a smaller percentage of variance (47.3% on average) than the other models. In this group, the conventional connected model detected the smallest number of QTL (55 in total). The LDLA models using clusthaplo information detected an intermediate number of QTL (58 and 56 for the LDLA—5 cM and LDLA—2 cM models, respectively). The ranking of the models in terms of number of detected QTL varied depending on the trait. For instance, the two LDLA models using clusthaplo information detected more QTL than the conventional connected model for DtTAS, PH, DMC (with the LDLA—5 cM model only), and for DMY (with the LDLA—2 cM model only). For the flowering traits, the LDLA—1-marker model detected more QTL than the other models. As for the dent group, the connected model explained a higher percentage of variance (56.3%) compared to the other models even if it did not detect a higher number of QTL.
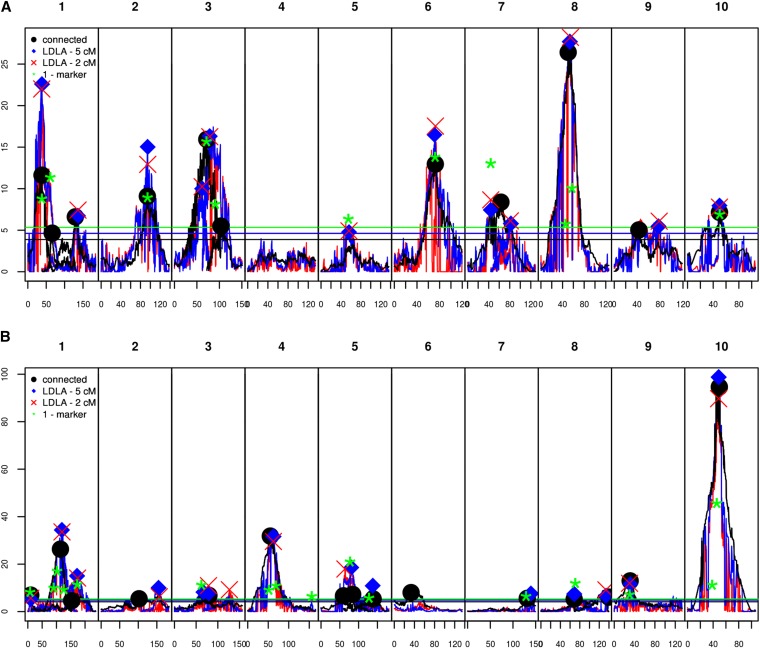
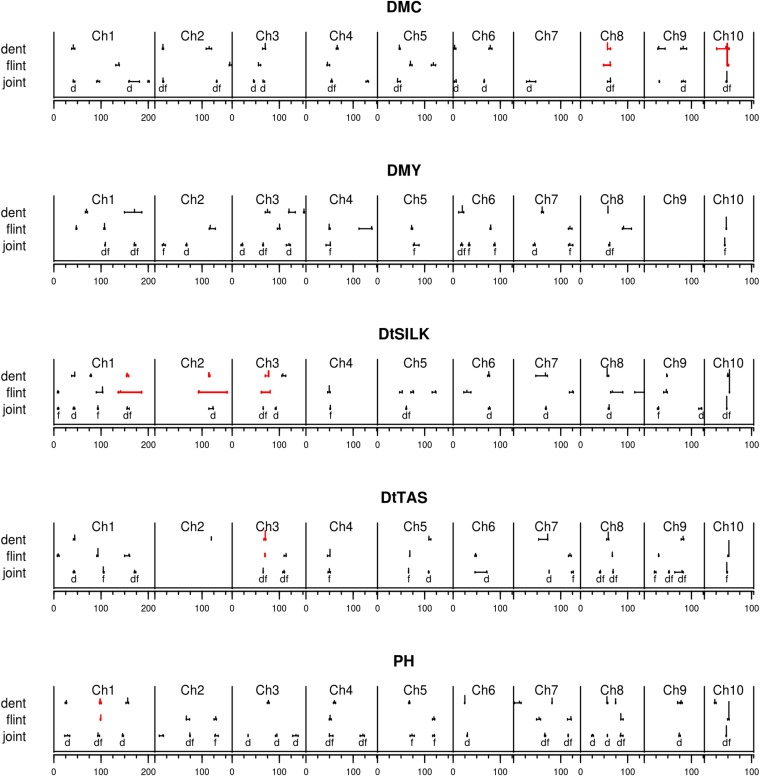
One can note that the −log10(P-values) curves showed relatively noisy patterns along the genome, especially for the LDLA models (Figure 3, Figure S1, Figure S2, Figure S3, and Figure S4). However, curves displaying evolution of –log10(P-values) along the genome were globally highly consistent across models and all models detected the same major QTL (Figure 3, Figure S1, Figure S2, Figure S3, and Figure S4). This was true even in cases when they detected a different number of QTL on the same chromosome. For instance, in the flint design, for DMC, all models detected a major QTL at 45–46 cM on chromosome 10 but two models detected other QTL in the region without challenging the position of the major QTL: the LDLA—2 cM model at 69.9 cM and the LDLA—1-marker model at 68.9 cM (Figure S1, Table S3, Table S4, Table S5, and Table S6).
Figure 3.
Results of the QTL detection with each model for DtSILK for (A) the dent design and (B) the flint design. The −log10(P-values) of the connected model are represented by black lines, the QTL positions of the connected models by black dots. The −log10(P-values) of the LDLA—5 cM model are represented by blue lines and the QTL positions by blue diamonds. The −log10(P-values) of the LDLA—2 cM model are represented by red lines and the QTL positions by red crosses. The −log10(P-values) of the QTL detected by the LDLA—1-marker model are represented by green stars. Horizontal lines correspond to the threshold values of the different models.
Considering the QTL that were detected by different models, the ranking of the models according to their −log10(P-value) varied with the QTL. For instance, for the QTL detected with all models for DtSILK in the dent group at 70–74 cM on chromosome 6, the highest –log10(P-value) was found with the LDLA—2 cM model (17.5) and the lowest with the connected model (13) (Figure 3). In contrast, for the QTL detected with all models for DMY in the dent group on chromosome 6 at 14–17 cM, the highest −log10(P-value) was found with the connected model (14.9) (Figure S2 and Table S7) and the lowest with the LDLA—2 cM model (13.3) (Table S9).
Allelic effect series and comparison of the different allelic models for the major QTL detected for female flowering time
Visualization of allelic effects of the connected model through heat maps (Figure S5, Figure S6, Figure S7, Figure S8, Figure S9, Figure S10, Figure S11, Figure S12, Figure S13, and Figure S14) illustrated a continuous range of effects for all QTL. The central line had an intermediate value for most of the loci in both designs. Each parental line carried alleles with either positive or negative effects compared to the central line. LDLA models are expected to outperform the connected model if the clustering process correctly identifies underlying allelic series at QTL. To get further insight into this point, we compared allelic effects estimated by the different models for the two major DtSILK QTL found in this study.
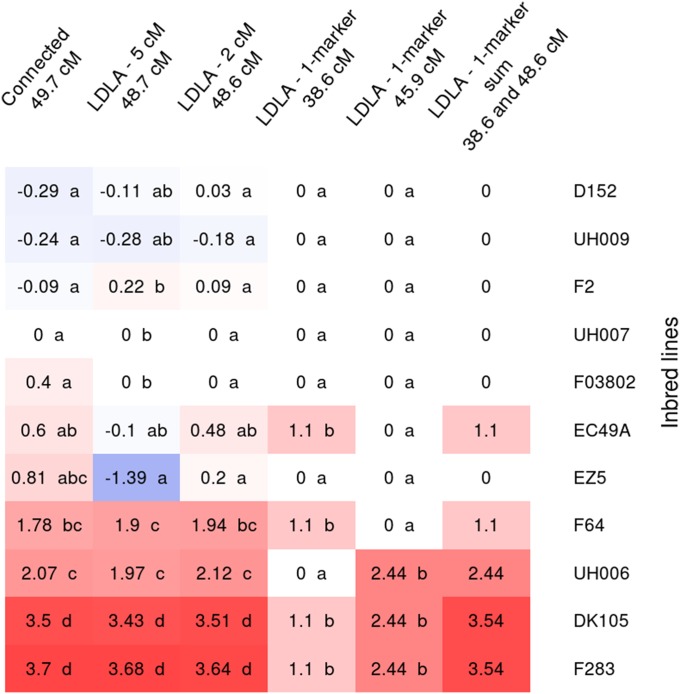
The allelic effects of the DtSILK major QTL detected in the flint group on chromosome 10 at 38–50 cM clearly showed an allelic series (Figure 4). The four models detected QTL in this region but at slightly different positions. For the QTL detected with the connected model, at least three classes of effects were identified based on t-tests. F283 and DK105 carried a late allele (3.7 and 3.5 days compared to UH007), UH006 an intermediate allele (2.07 days), and D152, UH009, F2, UH007, and F03802 an early allele (between −0.29 and 0.4 days), the three other parental lines showing effects between the early and the intermediate classes. For the QTL detected with the LDLA—5 cM and LDLA—2 cM models, allelic effects were globally consistent with those found for the QTL detected with the connected model except for EZ5, which had the earliest allele with the LDLA—5 cM model. Note that the family derived from this parent was one of the smallest of the design. The LDLA—1-marker model detected two QTL in this region: one at position 45.9 cM (close to the position of the QTL found with the other models) and one, of smaller effect, 7 cM apart at the position 38.6 cM. For the marker detected at position 45.9 cM, the late allele (2.44 days) was shared by F283, DK105 and UH006, which also carried the latest alleles according to the other models. All the other lines shared the same early allele (0 days). For the marker detected at position 38.6 cM, the late allele (1.1 days) was shared by DK105, F283 (the lines carrying the latest alleles in the other models), EC49A, and F64 (which carried alleles classified as intermediate). All the other lines shared the early allele (0 days). So, when considered jointly, these two markers account for the allelic series observed for the QTL detected with the other models: DK105 and F283 carrying the late alleles at the two markers, UH006 carrying the late allele for the marker with the strongest effect and the early allele for the other marker, EC49A and F64 carrying the late allele at the marker with the smallest effect and the early allele for the other one, and D152, UH009, F2, UH007, and F03802 carrying at both markers the early alleles. The two QTL detected with the LDLA—1-marker model individually explained 2.2 and 11.1% of the variance for the marker at positions 38.6 and 45.9 cM, respectively, but they jointly explained 26.8% of the variance, only slightly less than the variance explained by the QTL detected with the other models (between 27.5 and 28.2%).
Figure 4.
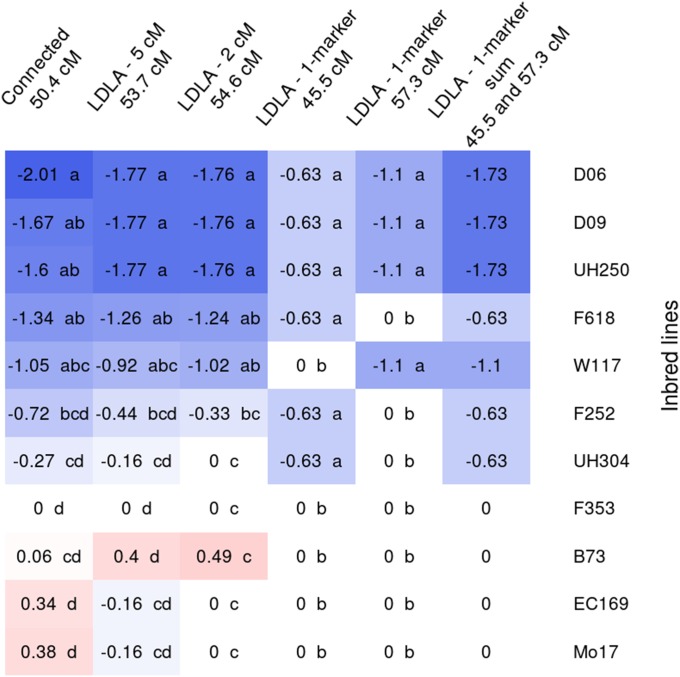
Allelic effects for the different flint lines for the QTL detected on chromosome 10 at 38–50 cM for DtSILK with all the QTL detection models. Allelic effects are estimated in contrast to the central line allelic effect (UH007), which was set to zero. The same letter was given to allelic effects not significantly different at a 5% risk level. Alleles with intermediate effects may be attributed to more than one letter. The last column corresponds to the joint effect of the two QTL detected in the region with LDLA—1-marker model. Allelic effects estimated for EP44 were not shown because the population where it segregates was too small (17 individuals) to obtain a reliable estimation. Inbred lines are ranked according to their allelic effects obtained with the connected model.
The allelic effects of the DtSILK QTL detected in the dent group, on chromosome 8 at 45–58 cM also clearly showed an allelic series and the same type of pattern (Figure 5). With the connected model, allelic effects showed a continuous variation and at least two classes of alleles could be identified. Four inbred lines (D06, D09, UH250, and F618) carried early alleles compared to the group consisting of F353 (central line), EC169, and Mo17. The other parental alleles were not clearly classified but had intermediate effects. In this chromosome region, the two LDLA models based on ancestral allele clustering both identified a QTL. With both window sizes, D06, D09, and UH250, which carried the earliest alleles in the connected model, were attributed to the same ancestral allele with an early effect (−1.77 with LDLA—5 cM and −1.76 with LDLA—2 cM compared to F353). Mo17, EC169 (the two lines with latest allelic effects in the connected model), UH304, and F353 were attributed to the same or to different ancestral alleles depending on the window size but in both cases their allelic effects were equal or close to zero. With these models, B73 was attributed the latest effect (0.4 or 0.49) but this effect was not significantly different from zero. The other lines had allelic effects consistent with the effects estimated with the connected model. Two QTL were detected in this region with the LDLA—1-marker model: one at 45.5 cM and the other at 57.3 cM, on either side of the QTL detected with the other models. D06, D09, and UH250, which carried the earliest allele of the connected model and were attributed to the same early ancestral allele with LDLA—2 cM and LDLA—5 cM models, carried the early allele at both QTL. Mo17, EC169, B73, and F353, the lines with the latest allelic effects with the other models, carried the late allele at both QTL. The other lines, which had intermediate allelic effects with the other models, carried the late allele at one QTL and the early allele at the other QTL. Thus, marker effects at these two QTL jointly mimic the allelic series identified by the other models. The two QTL detected with the LDLA—1-marker individually explained 1.5 and 2.9% of the variance but they jointly explained 7.9% of the variance, which is only slightly less than the other models (8.9% for the LDLA—5 cM and LDLA—2-cM models and 9.6% for the connected model).
Figure 5.
Allelic effects for the different dent lines for the QTL detected on chromosome 8 at 45–58 cM for DtSILK with all the models. Allelic effects are estimated in contrast to the central line allelic effect (F353), which was set to zero. The same letter was given to allelic effects not significantly different at a 5% risk level. Alleles with intermediate effects may be attributed to more than one letter. The last column corresponds to the joint effect of the two QTL detected in the region with LDLA—1-marker model. Inbred lines are ranked according to their allelic effects obtained with the connected model.
Comparison of the QTL detected in the two heterotic groups analyzed individually and jointly
In total, for the connected model, 52 QTL were detected in the dent design for all traits and 55 in the flint design (Table 2). More QTL were found in the dent than in the flint design for DMC and PH, whereas the reverse was observed for DtSILK, DtTAS, and DMY.
Based on overlap of their confidence intervals, when comparing results obtained in the two separate data sets, only seven QTL were common between the two groups. Two of these QTL were for DMC (chromosomes 8 and 10), three for DtSILK (chromosomes 1, 2 and 3), one for DtTAS (chromosome 3), and one for PH (chromosome 1). No common QTL were found for DMY. In addition, some chromosome regions carried QTL detected in the two groups but not for the same trait (Figure 6).
Figure 6.
QTL projection on the flint–dent consensus map of the QTL detected in the dent data set, the flint data set, and the joint data set for DMC, DMY, DtSILK, DtTAS, and PH. Each QTL is displayed by one horizontal line bound by two vertical lines representing the confidence region and a vertical line proportional to the QTL adjusted R2 symbolizing the QTL position. QTL common to dent and flint according to the overlap of their confidence region on the dent–flint consensus map are represented in red. For the QTL detected in the joint analysis, the letters d and f written below the QTL indicate that the QTL was significant when tested in the dent or flint data set respectively.
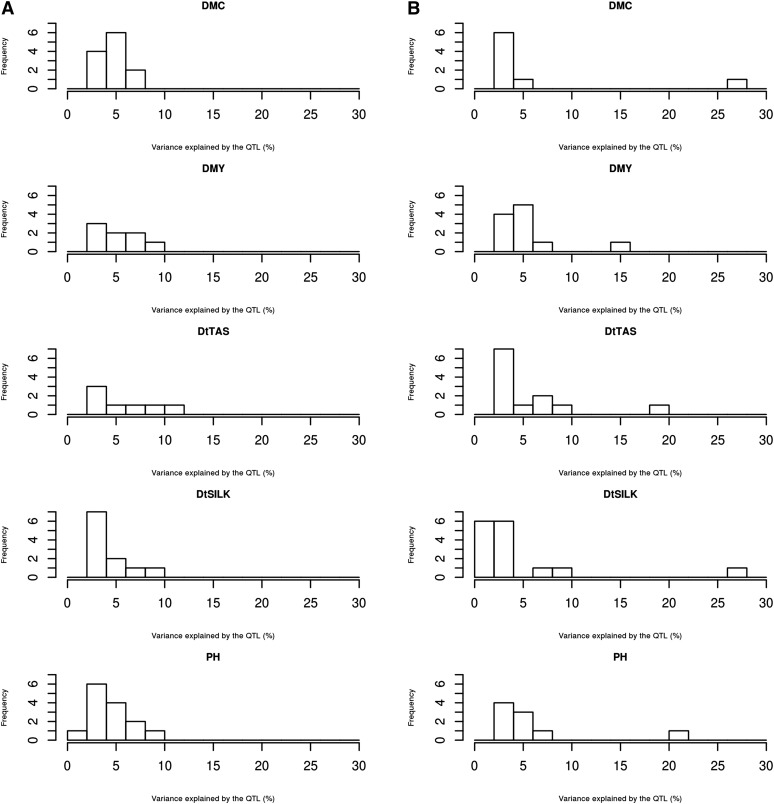
The distribution of QTL effects (in terms of R2) differed in the two groups (Figure 7). In the dent group, all the QTL had low to medium effect (R2 < 10%). The QTL with the biggest effect was detected on chromosome 3 at 63 cM for DtTAS and explained 10.4% of the variance (Table S7). A QTL was also detected at this position for DMC but with a smaller effect. The second biggest QTL was detected on chromosome 8 at position 50 cM for DtSILK and explained 9.4% of the genetic variance. This region was also detected for the other traits but with smaller effects. On the contrary, in the flint group, one region located on chromosome 10 around position 44–50 cM showed a major effect on all the traits (Table S3). Depending on the trait considered, this region explained between 14% of the variation for DMY and 27.5% for DtSILK. All the other QTL detected in this group showed milder effects with R2 < 10%. It is interesting to note that the QTL that exhibited a strong effect in one group (the QTL detected on chromosome 10 in the flint group and the QTL detected on chromosome 3 and 8 in the dent group) did not have such a strong effect in the other group for the same traits.
Figure 7.
Distribution of the percentage of variance explained by the QTL detected in (A) the dent design and (B) the flint design, with the connected model and for the five traits.
Eighty-seven QTL were detected in total with the joint analysis, which is less than the sum of the QTL found in the two separate data sets (107) (Table 2 and Table S11). For each trait, the number of QTL detected with the joint analysis was equal or superior to that detected in each single data set analysis. For DMC and PH, QTL detected with the joint analysis explained a larger fraction of variance than the one explained in the separate data sets analysis. On the contrary, for DMY, DtSILK, and DtTAS, more variance was explained in the flint data set analysis than in the joint analysis.
QTL found in the joint analysis were generally found at the same position or close to QTL detected in one or both separate analyses (Figure 6). In some cases, they were detected between two QTL detected in a single data set analysis (for instance, QTL on chromosome 5 for DtSILK), or between one QTL detected in the dent data set and one detected in the flint data set (QTL at 130 cM on chromosome 2 for DMC). In some cases, no QTL was detected with the joint analysis although QTL were detected in the separate data sets (for instance flint QTL at 9 cM on chromosome 1 or dent QTL on chromosome 2 for DtTAS). Other QTL were detected only with the joint analysis (and not close to or between two QTL detected with the separate analysis), as the one detected for DMC on chromosome 7.
When testing the effects of these 87 QTL in the separate data sets, 30 were significant in both data sets, 52 in a single data set only, and 5 in none of the data sets (Table 3). So the number of QTL with effect in both data set varied between 27% for DtSILK and 41% for DtTAS.
Table 3. Number of QTL detected for the five traits in the joint data set for the connected model and found significant in each separate data set, in both separate data sets and in none of the separate data set.
| DMC | DMY | DtSILK | DtTAS | PH | Total | |
|---|---|---|---|---|---|---|
| Significant in the whole data set (Nb) | 18 | 16 | 15 | 17 | 21 | 87 |
| Significant in the dent data set (Nb) | 14 | 9 | 11 | 11 | 17 | 62 |
| Significant in the flint data set (Nb) | 6 | 12 | 8 | 13 | 11 | 50 |
| Significant in both data sets (Nb) | 6 | 5 | 4 | 7 | 8 | 30 |
| Non significant in both data sets (Nb) | 4 | 0 | 0 | 0 | 1 | 5 |
Concerning the seven QTL found common when comparing the dent and flint separate analyses, the joint analysis always found a QTL in the region nearby (not necessarily with overlapping of the confidence regions but really close). Except for the QTL found on chromosome 2 for DtSILK, these QTL were significant in both groups.
Discussion
Our study aimed at comparing genetic determinism of biomass related traits in two complementary flint and dent genetic pools that are often used to produce commercial hybrids in Northern Europe. To do so, a new NAM DH population was developed for each group. Both NAM populations display intermediate levels of diversity compared to the U.S. NAM design and classical elite breeding programs. Data from each design were analyzed with four models: a connected model where parents are assumed to carry different alleles, an LDLA model based on single-marker information close to the one successfully used for the U.S. NAM design, and two LDLA models based on ancestral allele modeling previously used with success by Leroux et al. (2014) and Bardol et al. (2013). In addition, data of the two designs were analyzed jointly with the connected model, considering that the central line of one design was used as tester in the other design and reciprocally.
Linkage disequilibrium and clustering of parental alleles
The haplotype clustering approach of Leroux et al. (2014) requires the definition of a window size according to genetic map units (centimorgans, cM). We defined it based on the estimation of the LD extent at the level of the parental lines. This showed that LD decreased below r2 = 0.2 after ∼1 and 2 cM in the flint and dent parental lines, respectively. Although estimated with only 11 and 12 inbred lines, for the dent and flint group respectively, these values were consistent with the LD extent observed for these groups by van Inghelandt et al. (2011). Based on this result, we considered two window sizes for the parental clustering, one of 2 cM, more adapted to the flint group and one of 5 cM, more adapted to the dent group. Note that a 1 cM window was also considered but the HMM approach did not converge with the R version we used for this study. These values are smaller than the 10-cM window size used in Bardol et al. (2013) to analyze a multiparental design derived from highly related founders.
In both flint and dent groups, the clustering process identified on average six and seven ancestral alleles per position for the 2- and 5-cM window sizes, respectively. The percentage of genome detected as IBD was in agreement with the marker-based similarities between inbred line pairs and pedigree information. These results showed that among dent lines, there were two groups of related lines: (i) D09, D06, and UH250, which came from the breeding program of the University of Hohenheim, and (ii) UH304 and F353, which share a common Iodent background (Bauer et al. 2013). For the flint, there was a separation between EC49A, EZ5, EP44 (the three lines with Spanish origin), and F64 (Argentinean origin) and all the other lines.
The number of ancestral alleles detected after clustering with clusthaplo varied along the genome, first at the local level, from one position to the next. This results in a variation in model dimension along the genome that certainly explains the erratic pattern of the –log10(P-values) curves of the LDLA models (see below). Beyond this local variation we observed that on average more ancestral alleles were detected in the centromeric than in the telomeric regions. This result is probably related to the higher number of marker loci per centimorgan in centromeric regions than in telomeric ones. It may be also related to a higher divergence between lines in centromeric regions. The similarity score used in clusthaplo is expected to be robust against the difference of marker density inside the sliding windows (Leroux et al. 2014). Our results suggest, however, that we reached the limits in this robustness. As most of the lines were not closely related, the size of IBD segments was expected to be limited, which made them difficult to detect. Visual inspection of the graphs of IBD segments (results not shown) indeed revealed that the segments were in general shorter than in Bardol et al. (2013) except for related lines such as D06 and D09. The method implemented in the clusthaplo software should therefore be adapted to cope with more diverse sets of lines than the one considered in Leroux et al. (2014), possibly by reducing window sizes in regions of the genome where marker density is high and local LD is low relative to the genetic map.
Adapting the method to cope with populations with limited LD also raises issues regarding the genetic map to be considered for the clustering process. Bauer et al. (2013) showed that even if the individual maps of the families of a given group had globally consistent order, putative inversions were found in some areas. This is in agreement with recent studies that showed copy-number variations (Springer et al. 2009; Swanson-Wagner et al. 2010), chromosomal inversions, or translocations between the different maize lines. Ganal et al. (2011) also suggested that some regions of the physical map of B73 v. 2 are not correctly assembled. This may have affected our consensus maps since information from the physical map was used for positioning the markers and this may have affected the clustering process. It appears thus important to further evaluate the properties of the clustering approach when using denser genotyping data and also evaluate its potential interest in the context of the rapid emergence of sequencing data that may enable a more direct identification of conserved haplotypes between inbred lines.
Comparison of the different QTL detection models
The highest total number of QTL was detected by one of the three LDLA models in both designs. We noted, however, different trends for the two designs. For the dent, LDLA—2 cM and LDLA—5 cM detected very similar numbers of QTL (55 and 56, respectively), more than for both the connected and LDLA—1-marker models (52 and 45, respectively). Note that Bardol et al. (2013) also found that in an elite dent breeding pool, the LDLA method based on ancestral alleles detected on average more QTL than the LDLA—1-marker model. Our results suggest that the genotyping data and window sizes used for clusthaplo were well suited for LDLA models for the dent design. For the flint design, the connected model detected fewer QTL (55) than the LDLA—5 cM, the LDLA—2 cM and the 1-marker model (58, 56, and 59 respectively), but differences between models were small on average This suggests that the available density of genotyping data and/or window size we could use with the HMM approach were not necessarily optimal for this design. Interestingly, although the connected model was globally outperformed by LDLA models in terms of number of QTL detected, it explained a higher percentage of variance than the other models for nearly all the traits. Conversely, the LDLA—1-marker model explained a smaller percentage of variance even when detecting more QTL. As the estimations of the percentages of variation explained were adjusted for the number of parameters, this cannot be due to model overfitting. One can thus hypothesize that a large part of the QTL showed allelic series that are not completely accounted for by local similarities or single-marker information. This is consistent with Würschum et al. (2012) who compared by simulation different models for joint linkage association mapping. They concluded that, even if the single SNP model was more powerful in terms of detection, the model considering one allele per parent was better adapted to estimate QTL effects in case of multiallelic series, corroborating experimental results of Liu et al. (2011).
Globally, LDLA models and linkage analysis detected QTL in the same chromosome regions although fine comparison of QTL positions was complicated by the relatively noisy pattern of the LDLA −log10(P-value) curves. We noted that the number of QTL in a given genomic region could either be the same or vary across models. In cases when a single QTL position is detected by all models, one can assume that variation is most likely due to a single QTL with two alleles well reflected by a single biallelic marker. On the opposite, a variable number of QTL across models suggests a more complex situation with linkage between several QTL or allelic series at a single QTL. This can be exemplified by the DtSILK QTL detected on chromosome 8 in the dent design. In this region, the LDLA—1-marker model detected two QTL 12 cM apart and located on both sides of the single QTL detected with the connected model. This suggests that either the two marker loci were needed to account for the allelic series at a single QTL or conversely that the connected model failed at distinguishing the two underlying QTL due to limited recombination in DH families.
The different models thus showed variable efficiency depending on the trait and region considered, which highlights complementarities of different allele coding methods in deciphering allelic series in genetic studies.
Comparison between the QTL detected in the two heterotic groups and evolutionary interpretation
Similar numbers of QTL were detected in the two groups with the separate data set analyses, showing that both can contribute genetic variation useful for breeding in Northern Europe. Less than 15% of the QTL was common between the dent and flint design when comparing the positions of the QTL detected in the separate data set analyses. This is consistent with the long time divergence between the dent and flint heterotic groups: >500 years (Tenaillon and Charcosset 2011). Part of this low value can be due to power issues. Indeed the joint analysis enabled us to detect additional QTL compared to single group analysis and among the detected QTL with the joint analysis, 34% on average were significant in both groups. However, some QTL detected in individual designs disappeared in the joint analysis, which suggests that they were really specific of one group and that variation within the other group diminished power at these QTL in the joint analysis. Some of the QTL detected in the joint analysis were found at an intermediate position between the positions of design-specific QTL. This may correspond to a gain in precision but one cannot exclude that these QTL might also correspond to an artifact “ghost” QTL between actual QTL.
Note that in addition to the common QTL, some chromosome regions had an effect in both designs but for different traits. These QTL could be pleiotropic QTL for which effects on some traits were not detected in one of the designs, due to a lack of power, diversity, etc.
When comparing the single data set analyses, QTL common to flint and dent designs were observed for DMC, DtSILK, DtTAS, and PH. It is interesting to note that no common QTL was observed for DMY. With the joint analysis, the percentage of QTL significant in both data sets was smaller for DtSILK and DMY (27 and 31%, respectively) than for the other traits (33% for DMC to 41% for DtTAS). For traits subjected to directional selection such as DMY, several alleles must have been fixed over time but there is no reason that the same alleles were fixed in both groups, especially considering that selection for hybrid value certainly favored fixation of complementary alleles in each group (Schön et al. 2010; Larièpe et al. 2012). This may explain why only few common QTL or QTL significant in both groups were detected for DMY. On the contrary, for traits for which a stabilizing selection is performed, the same polymorphisms are more likely to be maintained in both groups. This is the case for PH and DtTAS and also indirectly for DMC since DMC at harvest of a genotype depends on its precocity and its drying speed. Interestingly, common DMC QTL between groups and most of the DMC QTL detected with the joint analysis and significant in both data sets were detected in regions also carrying QTL for flowering time (DtSILK or DtTAS).
The few common QTL between dent and flint groups that we detected could explain the low predictive abilities of the prediction between dent and flint in genomic selection (Meuwissen et al. 2001; Jannink et al. 2010) when dent are in the estimation set and flint in the test set and vice versa (Lehermeier et al. 2014). The presence of a major effect QTL in the flint group might also partly explain this result.
Overview of detected QTL and comparison with literature studies
For the single data set analyses, between 20 QTL for DMY and 28 QTL for DtSILK were detected in total over the two groups when considering the model that detected the highest number of QTL. For the joint analysis, between 15 QTL for DtSILK and 21 QTL for PH were detected.
For DtSILK, although high, the number of detected QTL is less than the one reported for the U.S. NAM design (39 QTL detected with the multiple family joint stepwise model, 52 with JCIM) (Buckler et al. 2009; Li et al. 2011). This is also less than the total number of QTL estimated through metaanalysis for flowering time (62 and 59 in Chardon et al. 2004 and Salvi et al. 2009, respectively). QTL detected in our study explained a smaller proportion of the variance (for the connected model the detected QTL explained 52.3%, for the dent design, 59.7% for the joint analysis, and 69.3%, for the flint design of the within family variability) than the one detected on the U.S. NAM design (89%) (Buckler et al. 2009; Li et al. 2011). Similar trends were observed for male flowering (DtTAS). In our study, all QTL explained 10% or less of variation, with the exception of the main QTL found in the flint design on chromosome 10 (45–50 cM with the connected model). In the joint analysis, this QTL was significant for female flowering when tested in both data sets, whereas for male flowering it was significant only in the flint data set. This QTL was also found by Blanc et al. (2006) and is close to the ZmCCT gene, which was fine mapped as a major flowering time QTL by Ducrocq et al. (2009) and validated by Coles et al. (2011). In the flint design, for the connected model, this QTL explained 18.7 and 27.5% of male and female flowering time, respectively. In the joint analysis, it explained 12 and 15.2% of male and female flowering time, respectively. This value is higher than that reported for the same region in the U.S. NAM (1.1% for male flowering and 1.3% for female flowering with joint linkage stepwise model in Buckler et al. 2009) and in Blanc et al. (2006) (18% for female flowering). These differences can be explained by the fact that several lines in our flint design share a late allele and possibly suggest that the expression of the effect of this QTL is amplified in early flowering backgrounds compared to the later U.S. NAM background. In the dent design analyzed separately, the most significant DtSILK QTL was found on chromosome 8. This QTL does not seem to be located in the region where two major flowering time QTL, vgt1 and vgt2 (ZCN8), have been fine mapped (Salvi et al. 2007; Bouchet et al. 2013). It seems to be close to an area where other studies also found QTL for flowering time (Ducrocq et al. 2008; Salvi et al. 2009; Bouchet et al. 2013).
For plant height (PH), we detected in total 25 QTL, which explained 55.0 and 57.1% of the variation for the flint and dent designs, respectively. With the joint analysis, we detected 21 QTL, which explained 61.2% of the variation. A recent study (Peiffer et al. 2014) based on the U.S. NAM and IBM family (Lee et al. 2002) reported 89 family-nested markers detected with an adaptation of JCIM and 277 associations through a joint-linkage-assisted genome-wide association study (Tian et al. 2011). Except the QTL found on chromosome 10 in the flint design and that likely corresponds to a pleiotropic effect of a major flowering time QTL, no QTL explained >10% of the variation, in the separate or joint data sets. As in Peiffer et al. (2014), none of the QTL detected in this study seem to be located in the vicinity of known candidate genes for plant height.
For DMY, with the separate analyses, we detected in total 20 QTL, which is lower than the number of QTL detected for the other traits. With the joint analysis, we detected 16 QTL, which is one of the lowest number of QTL detected. This may be explained by the lower heritability of this trait and the fact that variation for this trait may involve numerous QTL of small effects that are difficult to detect. For DMC, we detected in total 27 QTL with the separate analyses and 18 with the joint analysis. Only few studies address QTL detection for biomass yield and dry matter content, mainly in biparental populations (e.g., Lübberstedt et al. 1998; Méchin et al. 2001; Barriere et al. 2010; Barriere et al. 2012). They reported only limited number of QTL and are not easily comparable with our results. Our study, which led to the detection of many QTL in a multiparental context, therefore represents a large advance toward understanding the genetics of biomass yield.
Thus globally, although high compared to the number of QTL indentified in biparental populations, the number of QTL detected in this study appears lower than those detected in most comprehensive designs and meta-analysis. Several explanations can be given for this result. First, compared with the U.S. NAM design, our experimental designs explore less diversity and included fewer individuals (841 and 811 DH lines for the dent and flint designs, respectively, compared to 5000 RILs for the U.S. NAM design). Moreover, as DH lines were used instead of RILs, the number of recombination events in our designs is expected to be two times lower per family. This certainly affected the power and resolution of our designs for deciphering trait variation even with LDLA models. One cannot exclude that QTL detected in our study may indeed correspond to clusters of linked QTL that could have been individually detected using a higher number of individuals, a higher number of markers and progenies exhibiting more crossovers (Huang et al. 2010). The main specificity of our study compared to the U.S. NAM design was that the different families were evaluated through their testcross progeny to evaluate traits related to biomass production at usual productivity levels. Under the hypothesis of additivity, the genetic variance is expected to be four times lower for testcross value than for per se value. In addition, the two central lines of each group that were used as testers for the other group belong to two complementary heterotic pools, so one expects to observe some dominance effects between the flint and the dent alleles at QTL. Such dominance effects may have masked part of the variability in each group. Despite these limitations, as progenies were evaluated based on testcross performance, the QTL detected in this study directly reflect the genetic variation present in each of the two main heterotic groups that is useful for breeding in European conditions.
Supplementary Material
Acknowledgments
Results have been achieved within the framework of the Transnational Cooperation within the PLANT-KBBE Initiative CornFed, with funding from the Federal Ministry of Education and Research (BMBF, Germany), Agence Nationale de la Recherche (ANR, France), and Ministry of Science and Innovation (MICINN, Spain). Part of this research was funded by the Federal Ministry of Education and Research (BMBF, Germany) within the Agro-ClustEr Synbreed-Synergistic plant and animal breeding (FKZ 0315528A).
Footnotes
Supporting information is available online at http://www.genetics.org/lookup/suppl/doi:10.1534/genetics.114.169367/-/DC1.
Communicating editor: B. S. Yandell
Literature Cited
- Bardol N., Ventelon M., Mangin B., Jasson S., Loywick V., et al. , 2013. Combined linkage and linkage disequilibrium QTL mapping in multiple families of maize (Zea mays L.) line crosses highlights complementarities between models based on parental haplotype and single locus polymorphism. Theor. Appl. Genet. 126: 2717–2736. [DOI] [PubMed] [Google Scholar]
- Barriere Y., Mechin V., Denoue D., Bauland C., Laborde J., 2010. QTL for yield, earliness, and cell wall quality traits in topcross experiments of the F838 × F286 early maize RIL progeny. Crop Sci. 50: 1761–1772. [Google Scholar]
- Barriere Y., Méchin V., Lefevre B., Maltese S., 2012. QTLs for agronomic and cell wall traits in a maize RIL progeny derived from a cross between an old Minnesota13 line and a modern Iodent line. Theor. Appl. Genet. 125: 531–549. [DOI] [PubMed] [Google Scholar]
- Bauer E., Falque M., Walter H., Bauland C., Camisan C., et al. , 2013. Intraspecific variation of recombination rate in maize. Genome Biol. 14: R103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bink M. C. A. M., Totir L. R., ter Braak C. J. F., Winkler C. R., Boer M. P., et al. , 2012. QTL linkage analysis of connected populations using ancestral marker and pedigree information. Theor. Appl. Genet. 124: 1097–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G., Charcosset A., Mangin B., Gallais A., Moreau L., 2006. Connected populations for detecting quantitative trait loci and testing for epistasis: an application in maize. Theor. Appl. Genet. 113: 206–224. [DOI] [PubMed] [Google Scholar]
- Bouchet S., Servin B., Bertin P., Madur D., Combes V., et al. , 2013. Adaptation of maize to temperate climates: mid-density genome-wide association genetics and diversity patterns reveal key genomic regions, with a major contribution of the Vgt2 (ZCN8) locus. PLoS ONE 8: e71377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning B. L., Browning S. R., 2009. A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am. J. Hum. Genet. 84: 210–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckler E. S., Holland J. B., Bradbury P. J., Acharya C. B., Brown P. J., et al. , 2009. The genetic architecture of maize flowering time. Science 325: 714–718. [DOI] [PubMed] [Google Scholar]
- Charcosset A., Gallais A., 1996. Estimation of the contribution of quantitative trait loci (QTL) to the variance of a quantitative trait by means of genetic markers. Theor. Appl. Genet. 93: 327–333. [DOI] [PubMed] [Google Scholar]
- Charcosset, A., M. Causse, L. Moreau, and A. Gallais, 1994 Investigation into effect of genetic background on QTL expression using three connected maize recombinant inbred lines (RIL) populations, pp. 75–84 in Biometrics in plant breeding: applications of molecular markers: Proceedings of the 9th Meeting of the Eucarpia Section Biometrics in Plant Breeding, edited by J. W. van Oijen and J. Jansen. Wageningen, The Netherlands. [Google Scholar]
- Charcosset, A., B. Mangin, L. Moreau, L. Combes, M.-F. Jourjon et al., 2000 Heterosis in maize investigated using connected RIL populations, pp. 89–98 in Quantitative Genetics and Breeding Methods: The Way Ahead, edited by A. Gallais, C. Dillman, and I. Goldringer. INRA, Paris, France. [Google Scholar]
- Chardon F., Virlon B., Moreau L., Falque M., Joets J., et al. , 2004. Genetic architecture of flowering time in maize as inferred from quantitative trait loci meta-analysis and synteny conservation with the rice genome. Genetics 168: 2169–2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coles N. D., Zila C. T., Holland J. B., 2011. Allelic effect variation at key photoperiod response quantitative trait loci in maize. Crop Sci. 51: 1036–1049. [Google Scholar]
- Cook J. P., McMullen M. D., Holland J. B., Tian F., Bradbury P., et al. , 2012. Genetic architecture of maize kernel composition in the nested association mapping and inbred association panels. Plant Physiol. 158: 824–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducrocq S., Madur D., Veyrieras J.-B., Camus-Kulandaivelu L., Kloiber-Maitz M., et al. , 2008. Key impact of Vgt1 on flowering time adaptation in maize: evidence from association mapping and ecogeographical information. Genetics 178: 2433–2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducrocq S., Giauffret C., Madur D., Combes V., Dumas F., et al. , 2009. Fine mapping and haplotype structure analysis of a major flowering time quantitative trait locus on maize chromosome 10. Genetics 183: 1555–1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frascaroli E., Schrag T. A., Melchinger A. E., 2013. Genetic diversity analysis of elite European maize (Zea mays L.) inbred lines using AFLP, SSR, and SNP markers reveals ascertainment bias for a subset of SNPs. Theor. Appl. Genet. 126: 133–141. [DOI] [PubMed] [Google Scholar]
- Ganal M. W., Durstewitz G., Polley A., Bérard A., Buckler E. S., et al. , 2011. A large maize (Zea mays L.) SNP genotyping array: development and germplasm genotyping, and genetic mapping to compare with the B73 reference genome. PLoS ONE 6: e28334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gore M. A., Chia J.-M., Elshire R. J., Sun Q., Ersoz E. S., et al. , 2009. A first-generation haplotype map of maize. Science 326: 1115–1117. [DOI] [PubMed] [Google Scholar]
- Hill W. G., Robertson A., 1968. Linkage disequilibrium in finite populations. Theor. Appl. Genet. 38: 226–231. [DOI] [PubMed] [Google Scholar]
- Hill W. G., Weir B. S., 1988. Variances and covariances of squared linkage disequilibria in finite populations. Theor. Popul. Biol. 33: 54–78. [DOI] [PubMed] [Google Scholar]
- Huang X., Wei X., Sang T., Zhao Q., Feng Q., et al. , 2010. Genome-wide association studies of 14 agronomic traits in rice landraces. Nat. Genet. 42: 961–967. [DOI] [PubMed] [Google Scholar]
- Jannink J. L., Lorenz A. J., Iwata H., 2010. Genomic selection in plant breeding: from theory to practice. Brief. Funct. Genomics 9: 166–177. [DOI] [PubMed] [Google Scholar]
- Jansen, R. C., J.-L. Jannink, and W. D. Beavis, 2003 Mapping quantitative trait loci in plant breeding populations: use of parental haplotype sharing. Crop Sci. 43: 829–834. [Google Scholar]
- Jourjon M.-F., Jasson S., Marcel J., Ngom B., Mangin B., 2005. MCQTL: multi-allelic QTL mapping in multi-cross design. Bioinformatics 21: 128–130. [DOI] [PubMed] [Google Scholar]
- Kump K. L., Bradbury P. J., Wisser R. J., Buckler E. S., Belcher A. R., et al. , 2011. Genome-wide association study of quantitative resistance to Southern leaf blight in the maize nested association mapping population. Nat. Genet. 43: 163–168. [DOI] [PubMed] [Google Scholar]
- Lander E. S., Bolstein D. 1989. Mapping mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics 121: 185–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larièpe A., Mangin B., Jasson S., Combes V., Dumas F., et al. , 2012. The genetic basis of heterosis: multiparental quantitative trait loci mapping reveals contrasted levels of apparent overdominance among traits of agronomical interest in maize (Zea mays L.). Genetics 190: 795–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M., Sharopova N., Beavis W. D., Grant D., Katt M., et al. , 2002. Expanding the genetic map of maize with the intermated B73 × Mo17 (IBM) population. Plant Mol. Biol. 48: 453–461. [DOI] [PubMed] [Google Scholar]
- Lehermeier C., Krämer N., Bauer E., Bauland C., Camisan C., et al. , 2014. Usefulness of multi-parental populations of maize (Zea mays L.) for genome-based prediction of testcross performance. Genetics 198: 3–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroux D., Rahmani A., Jasson S., Ventelon M., Louis F., et al. , 2014. Clusthaplo: a plug-in for MCQTL to enhance QTL detection using ancestral alleles in multi-cross design. Theor. Appl. Genet. 127: 921–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Bradbury P., Ersoz E., Buckler E. S., Wang J., 2011. Joint QTL linkage mapping for multiple-cross mating design sharing one common parent. PLoS ONE 6: e17573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Jiang T., 2005. Haplotype-based linkage disequilibrium mapping via direct data mining. Bioinformatics 21: 4383–4393. [DOI] [PubMed] [Google Scholar]
- Liu W., Gowda M., Steinhoff J., Maurer H. P., Würschum T., et al. , 2011. Association mapping in an elite maize breeding population. Theor. Appl. Genet. 123: 847–858. [DOI] [PubMed] [Google Scholar]
- Liu W., Reif J. C., Ranc N., Porta G. D., Würschum T., 2012. Comparison of biometrical approaches for QTL detection in multiple segregating families. Theor. Appl. Genet. 125: 987–998. [DOI] [PubMed] [Google Scholar]
- Lübberstedt T., Melchinger A. E., Fahr S., Klein D., Dally A., et al. , 1998. QTL mapping in testcrosses of flint lines of maize. III. Comparison across populations for forage traits. Crop Sci. 38: 1278–1289. [Google Scholar]
- McMullen M. D., Kresovich S., Villeda H. S., Bradbury P., Li H., et al. , 2009. Genetic properties of the maize nested association mapping population. Science 325: 737–740. [DOI] [PubMed] [Google Scholar]
- Méchin V., Argillier O., Hebert Y., Guingo E., Moreau L., et al. , 2001. Genetic analysis and QTL mapping of cell wall digestibility and lignification in silage maize. Crop Sci. 41: 690–697. [Google Scholar]
- Meuwissen T. H., Goddard M. E., 2001. Prediction of identity by descent probabilities from marker-haplotypes. Genet. Sel. Evol. 33: 605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuwissen T. H., Hayes B. J., Goddard M. E., 2001. Prediction of total genetic value using genome-wide dense marker maps. Genetics 157: 1819–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiffer J. A., Romay M. C., Gore M. A., Flint-Garcia S. A., Zhang Z., et al. , 2014. The genetic architecture of maize height. Genetics 196: 1337-1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poland J. A., Bradbury P. J., Buckler E. S., Nelson R. J., 2011. Genome-wide nested association mapping of quantitative resistance to northern leaf blight in maize. Proc. Natl. Acad. Sci. USA 108: 6893–6898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team , 2013. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- Rebai A., Goffinet B., 1993. Power of tests for QTL detection using replicated progenies derived from a diallel cross. Theor. Appl. Genet. 86: 1014–1022. [DOI] [PubMed] [Google Scholar]
- Rincent R., Moreau L., Monod H., Kuhn E., Melchinger A. E., et al. , 2014. Recovering power in association mapping panels with variable levels of linkage disequilibrium. Genetics 197: 375–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romay M. C., Millard M. J., Glaubitz J. C., Peiffer J. A., Swarts K. L., et al. , 2013. Comprehensive genotyping of the USA national maize inbred seed bank. Genome Biol. 14: R55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvi S., Sponza G., Morgante M., Tomes D., Niu X., et al. , 2007. Conserved noncoding genomic sequences associated with a flowering-time quantitative trait locus in maize. Proc. Natl. Acad. Sci. USA 104: 11376–11381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvi S., Castelletti S., Tuberosa R., 2009. An updated consensus map for flowering time QTLs in maize. Maydica 54: 501–512. [Google Scholar]
- Schön C., Dhillon B., Utz H., Melchinger A., 2010. High congruency of QTL positions for heterosis of grain yield in three crosses of maize. Theor. Appl. Genet. 120: 321–332. [DOI] [PubMed] [Google Scholar]
- Segura V., Vilhjálmsson B. J., Platt A., Korte A., Seren Ü., et al. , 2012. An efficient multi-locus mixed-model approach for genome-wide association studies in structured populations. Nat. Genet. 44: 825–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosnowski O., Charcosset A., Joets J., 2012. BioMercator V3: an upgrade of genetic map compilation and quantitative trait loci meta-analysis algorithms. Bioinformatics 28: 2082–2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer N. M., Ying K., Fu Y., Yeh C. T., Jia Y., et al. , 2009. Maize inbreds exhibit high levels of copy number variation (CNV) and presence/absence variation (PAV) in genome content. PLoS Genet. 11: e1000734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson-Wagner R. A., Eichten S. R., Kumari S., Tiffin P., Stein J. C., et al. , 2010. Pervasive gene content variation and copy number variation in maize and its undomesticated progenitor. Genome Res. 20: 1689–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenaillon M. I., Charcosset A., 2011. A European perspective on maize history. C. R. Biol. 334: 221–228. [DOI] [PubMed] [Google Scholar]
- Tian F., Bradbury P. J., Brown P. J., Hung H., Sun Q., et al. , 2011. Genome-wide association study of leaf architecture in the maize nested association mapping population. Nat. Genet. 43: 159–162. [DOI] [PubMed] [Google Scholar]
- van Inghelandt D., Reif J. C., Dhillon B. S., Flament P., Melchinger A. E., 2011. Extent and genome-wide distribution of linkage disequilibrium in commercial maize germplasm. Theor. Appl. Genet. 123: 11–20. [DOI] [PubMed] [Google Scholar]
- Würschum T., Liu W., Gowda M., Maurer H. P., Fischer S., et al. , 2012. Comparison of biometrical models for joint linkage association mapping. Heredity 108: 332–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Pressoir G., Briggs W. H., Vroh Bi I., Yamasaki M., et al. , 2006. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat. Genet. 38: 203–208. [DOI] [PubMed] [Google Scholar]
- Yu J., Holland J. B., McMullen M. D., Buckler E. S., 2008. Genetic design and statistical power of nested association mapping in maize. Genetics 178: 539–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.