A nitrate transporter transports ABA and regulates primary root growth via an ABA-dependent nitrate signaling pathway.
Abstract
Elongation of the primary root during postgermination of Medicago truncatula seedlings is a multigenic trait that is responsive to exogenous nitrate. A quantitative genetic approach suggested the involvement of the nitrate transporter MtNPF6.8 (for Medicago truncatula NITRATE TRANSPORTER1/PEPTIDE TRANSPORTER Family6.8) in the inhibition of primary root elongation by high exogenous nitrate. In this study, the inhibitory effect of nitrate on primary root elongation, via inhibition of elongation of root cortical cells, was abolished in npf6.8 knockdown lines. Accordingly, we propose that MtNPF6.8 mediates nitrate inhibitory effects on primary root growth in M. truncatula. pMtNPF6.8:GUS promoter-reporter gene fusion in Agrobacterium rhizogenes-generated transgenic roots showed the expression of MtNPF6.8 in the pericycle region of primary roots and lateral roots, and in lateral root primordia and tips. MtNPF6.8 expression was insensitive to auxin and was stimulated by abscisic acid (ABA), which restored the inhibitory effect of nitrate in npf6.8 knockdown lines. It is then proposed that ABA acts downstream of MtNPF6.8 in this nitrate signaling pathway. Furthermore, MtNPF6.8 was shown to transport ABA in Xenopus spp. oocytes, suggesting an additional role of MtNPF6.8 in ABA root-to-shoot translocation. 15NO3−-influx experiments showed that only the inducible component of the low-affinity transport system was affected in npf6.8 knockdown lines. This indicates that MtNPF6.8 is a major contributor to the inducible component of the low-affinity transport system. The short-term induction by nitrate of the expression of Nitrate Reductase1 (NR1) and NR2 (genes that encode two nitrate reductase isoforms) was greatly reduced in the npf6.8 knockdown lines, supporting a role of MtNPF6.8 in the primary nitrate response in M. truncatula.
Nitrogen (N) is an inorganic nutrient that is essential for sustaining plant growth. The availability of N in soil is an important factor for plant establishment, crop yield, and quality. One of the main sources of N in the soil, available for plants, is nitrate (NO3−; Andrews and Lea, 2013; Andrews et al., 2013). In order to adapt to nitrate availability, plants have evolved two nitrate uptake systems: a low-affinity transporter system (LATS) and a high-affinity transporter system (HATS; Wang et al., 2012) that belong respectively to two families of proteins, named NITRATE TRANSPORTER1 (NRT1)/PEPTIDE TRANSPORTER (PTR) and NITRATE TRANSPORTER2 (NRT2; Nacry et al., 2013). According to the recent unified nomenclature, NPF is now the name to be used for the NRT1/PTR Family (Léran et al., 2014). Fifty-three putative NPF genes were predicted from the genome sequence of Arabidopsis (Arabidopsis thaliana) and 10 were characterized as nitrate transporters (Tsay et al., 1993; Huang et al., 1999; Chiu et al., 2004; Almagro et al., 2008; Lin et al., 2008; Fan et al., 2009; Li et al., 2010; Wang and Tsay, 2011; Hsu and Tsay, 2013). All of the NPF transporters are located in the plasma membrane. Most of these nitrate transporters are low-affinity nitrate transporters, except AtNPF6.3 (AtNRT1.1), which is a dual-affinity nitrate transporter (Liu et al., 1999). To date, only AtNPF6.3 and AtNPF4.6 (AtNRT1.2) have been shown to participate in the uptake of high nitrate concentrations by roots (Huang et al., 1999; Liu et al., 1999). In Arabidopsis, seven members of the NRT2 family were identified and characterized as high-affinity nitrate transporters. Most of these AtNRT2 transporters are located in the plasma membrane, except AtNRT2.7, which is located in the tonoplast (Chopin et al., 2007). AtNRT2.1 is the major NRT2 transporter that participates in high-affinity nitrate uptake, whereas AtNRT2.2 and AtNRT2.4 have been shown to play a minor role in nitrate uptake (Li et al., 2007; Kiba et al., 2012).
Developmental plasticity allows a plant to respond to nutrient availability by changing the architecture of the root system (Zhang and Forde, 2000; Linkohr et al., 2002). Nitrate is one of the major nutrients known to act as a signal molecule involved in the control of root architecture through the regulation of primary root growth and in the emergence and development of lateral roots (LRs), as part of an adaptive strategy (Zhang and Forde, 1998, 2000; Zhang et al., 1999; Tian et al., 2008; Vidal et al., 2010b; Celis-Arámburo et al., 2011). The molecular nature of nitrate sensing and signaling has been extensively studied in Arabidopsis in relation to the regulation of LR development. At high nitrate concentrations, a systemic response suppresses LR growth after initiation at the early LR emergence stage (Zhang et al., 1999). In addition, studies have shown that when a plant is subjected to nitrate deficiency, followed by the application of a high local nitrate concentration, LR growth is locally stimulated toward this most abundant source of nitrate (Zhang and Forde, 1998; Zhang et al., 1999). The localized effect of nitrate on LR growth was shown to be mediated by the dual-affinity nitrate transporter AtNPF6.3 (Remans et al., 2006) and Arabidopsis Nitrate Regulated1, a transcription factor of the MADS box family (Zhang and Forde, 1998). Furthermore, the finding that AtNPF6.3 transports nitrate and auxin (indoleacetic acid [IAA]) provides a model in which 1) under low nitrate conditions, AtNPF6.3 transports auxin away from the LR tip and the low content of auxin in the tip inhibits the outgrowth of LR; and 2) under high nitrate conditions, nitrate stimulates LR emergence by inhibiting AtNPF6.3-dependent auxin basipetal transport, leading to auxin accumulation in the LR tip (Krouk et al., 2010). In nonnodulated Medicago truncatula (i.e. in the absence of rhizobia), the modulation of shoot-to-root auxin transport in response to nitrate availability was shown to reduce the density of LR and this effect was mediated by the super numeric nodules gene (Jin et al., 2012).
The effect of nitrate on primary root growth has been less well studied compared with LR growth, although nitrate has been shown in several species to inhibit primary root growth through the alteration of hormone transport or signaling. In maize (Zea mays) seedlings, nitrate supply resulted in a decrease in auxin content in the phloem, thus decreasing auxin shoot-to-root translocation (Tian et al., 2008). In Arabidopsis, nitrate was shown to regulate primary root growth by a pathway involving the Auxin Receptor F-box3 (AFB3; Vidal et al., 2010a). Nitrate treatment induced the accumulation of the small RNA miR393, which was shown to specifically direct cleavage of AFB3 transcripts (Vidal et al., 2010a). In M. truncatula, nitrate-induced inhibition of primary root growth was altered in a mutant affected in the high-affinity nitrate transporter MtNPF1.7, also known as Lateral Root Organ Defective (LATD)/Numerous Infections and Polyphenolics (Harris and Dickstein, 2010; Yendrek et al., 2010; Bagchi et al., 2012). Because the root architecture phenotype of npf1.7 mutants was rescued by the application of exogenous abscisic acid (ABA; Liang et al., 2007), it is possible that the control of root architecture by nitrate in M. truncatula might involve an interaction with an ABA signaling pathway. Accordingly, the Arabidopsis low-affinity nitrate transporter AtNPF4.6 was shown to transport both nitrate (Huang et al., 1999) and ABA (Kanno et al., 2013). Three closely related proteins belonging to the NPF family (At1g27040/Arabidposis Abscisic Acid-Importing Transporter2 [AtAIT2]/AtNPF4.5, At3g25260/AtAIT3/AtNPF4.1, and At3g25280/AtAIT4/AtNPF4.2) were shown to transport ABA (Kanno et al., 2012).
In a previous study, we characterized the dual-affinity nitrate transporter MtNPF6.8 (MtNRT1.3) in M. truncatula (Morère-Le Paven et al., 2011; Léran et al., 2014). Based on genetic analyses and colocalization of MtNPF6.8 with a peak of a major quantitative trait locus (QTL) for primary root growth, we hypothesized that this transporter is involved in the regulation of primary root growth by nitrate during seedling establishment (Morère-Le Paven et al., 2011). In this study, we tested this hypothesis by studying the effect of nitrate on the growth of primary roots of npf6.8 knockdown lines generated by RNA interference (RNAi). Our findings support the hypothesis that MtNPF6.8 regulates primary root growth in response to nitrate availability by controlling the elongation of root cells. Our results also show that ABA is transported by MtNPF6.8 and is involved in nitrate inhibitory effects on primary root growth. Furthermore, analyses of short-term effects of nitrate (30 min) on nitrate-inducible genes support the proposal that MtNPF6.8 is involved in the primary response to nitrate in M. truncatula.
RESULTS
Subcellular Localization of the Dual-Affinity Nitrate Transporter MtNPF6.8
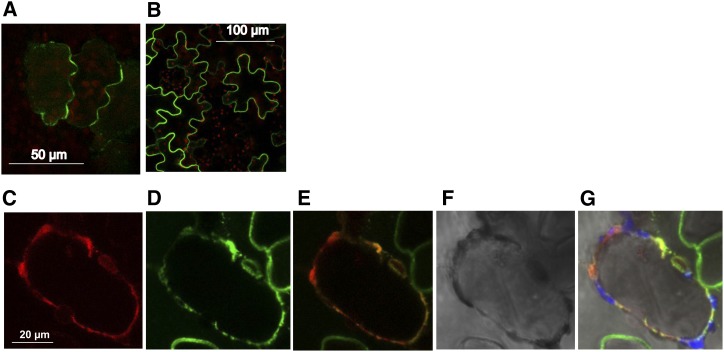
To determine the subcellular localization of MtNPF6.8, MtNPF6.8 was fused to GFP or to red fluorescent protein (RFP) under the control of the Cauliflower mosaic virus (CaMV) 35S promoter and was transiently expressed by agroinfiltration in M. truncatula or Nicotiana benthamiana leaf epidermal cells. Confocal microscopy observation of either M. truncatula (Fig. 1A) or N. benthamiana (Fig. 1B) leaf epidermal cells expressing MtNPF6.8:GFP strongly indicates a plasma membrane localization of MtNPF6.8. To further investigate the localization of MtNPF6.8, MtNPF6.8:RFP was detected in plasmolyzed N. benthamiana cells coexpressing a peptide Transmembrane Domain23 (TM23) fused to GFP (Fig. 1C). This peptide is specifically targeted to the plasma membrane (Brandizzi et al., 2002; Fig. 1D). Merged images of Figure 1, C and D, show that the green and red fluorescence overlapped, thus confirming the plasma membrane localization of MtNPF6.8 (Fig. 1E). A similar experiment was carried out with the tonoplast-specific protein tonoplast intrinsic protein1-1 (TIP1-1) fused to GFP (Boursiac et al., 2005); in this case, the green and red fluorescence did not overlap (data not shown).
Figure 1.
A and B, Subcellular localization of MtNPF6.8. MtNPF6.8-GFP fluorescence in leaf epidermal cells of M. truncatula (A) or N. benthamiana (B). C to F, Coexpression in N. benthamiana plasmolyzed leaf cells of MtNPF6.8-RFP (C) and the plasma membrane marker TM23:GFP (D). Near-perfect colocalization of coexpressed proteins is observed (E). A differential interference contrast image of the plasmolyzed cell highlighting the outline of the cell wall is shown (F). G, Overlay of C to F.
Expression of MtNPF6.8 in Seedlings
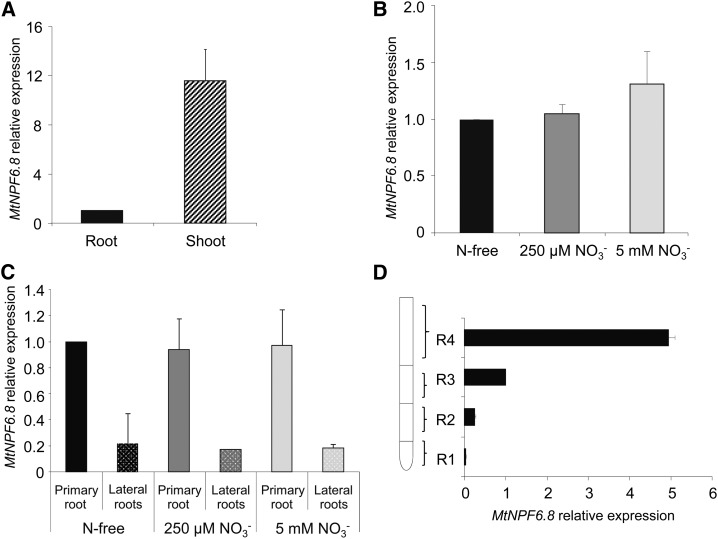
Expression of MtNPF6.8 was determined by quantitative reverse transcription (RT)-PCR in shoots and roots in 10-d-old seedlings grown on a 5 mm NO3−-supplied medium (Fig. 2A). The expression of MtNPF6.8 was 12 times higher in the shoots than in the roots at this stage of development. Expression of MtNPF6.8 was also examined in the root system taken as a whole (Fig. 2B) and in the primary root and the LR separately (Fig. 2C), in 10-d-old seedlings grown on either an N-free or an NO3−-supplied (250 μm or 5 mm) medium. Expression of MtNPF6.8 was not affected by nitrate supply at this stage of development (Fig. 2B); however, expression was 5- to 10-fold higher in the primary roots than that in the LR (Fig. 2C). The pattern of expression of MtNPF6.8 along the primary root was examined in 6-d-old seedlings grown on 5 mm NO3−. The primary root was cut into four sections starting with the tip and three 0.5-cm-long sections R1, R2 (elongation zone), and R3 (elongation zone), and section R4 consisted of the rest of the root, of 2 to 3 cm in length (Fig. 2D). Expression of MtNPF6.8 was barely detectable in the root tip (R1), whereas expression increased along the root and was much higher in the mature parts of the root.
Figure 2.
MtNPF6.8 expression determined by quantitative RT-PCR in 10-d-old seedlings of M. truncatula. A, MtNPF6.8 relative expression in roots or shoots of seedlings grown on 5 mm nitrate. B and C, MtNPF6.8 relative expression in the entire root system (B) or in the primary root and the LR (C) in seedlings grown on either an N-free or an NO3−-supplied (250 μm or 5 mm) medium. D, MtNPF6.8 relative expression from four sequential sections of the primary root starting with the tip: R1, R2, and R3 (0.5 cm long) and the root remainder R4. The R3 section was used as the calibrator. Seedlings were grown on 5 mm nitrate. Error bars represent the sd of three independent biological replicates.
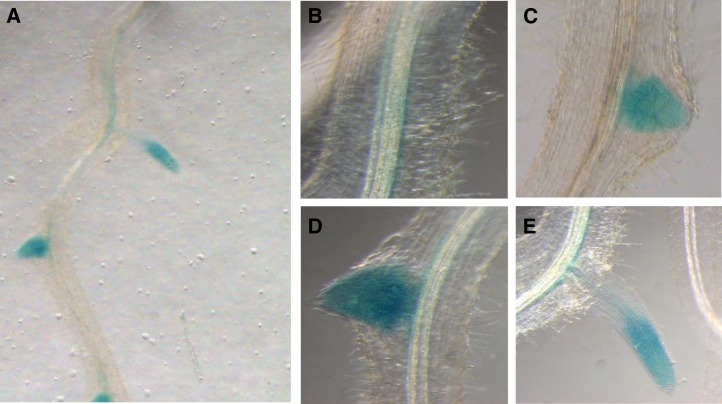
To elucidate the expression pattern in more detail, roots expressing the GUS reporter gene under the control of the MtNPF6.8 promoter were regenerated using Agrobacterium rhizogenes (Fig. 3). GUS staining suggests that MtNPF6.8 is expressed in the vascular system all along the primary root in the pericycle region (Fig. 3, A and B) and in the LR primordia and the LR tip (Fig. 3, C–E). An apparent discrepancy appears between the levels of expression of MtNPF6.8 in the primary root and the LR as determined by quantitative RT-PCR (Fig. 2C) and GUS staining (Fig. 3). This finding may indicate that the quantitative RT-PCR quantitation of MtNPF6.8 in the LR is underestimated when the whole LR is used for RNA extraction while GUS staining shows that the expression is concentrated in the tips.
Figure 3.
Histochemical localization of GUS activity in transgenic M. truncatula roots expressing the GUS reporter gene under the control of the MtNPF6.8 promoter. A, M. truncatula root expressing pMtNPF6.8:GUS stained histologically for GUS activity. B to E, Enlarged views of the primary root and of different developmental stages of LR: primary root near the shoot-root section (B), LR prior to emergence (C), LR immediately after emergence (D), and mature LR (E).
Generation of npf6.8 Knockdown Lines by RNAi for Functional Characterization of MtNPF6.8 in Planta
To investigate the physiological function of MtNPF6.8 in M. truncatula, plants were constructed using RNAi technology. A 401-bp sense and antisense fragment of MtNPF6.8, located in the third and fourth exons (Supplemental Fig. S1A), was inserted into a plasmid and placed under the control of the CaMV 35S promoter. After transformation of the wild-type M. truncatula R108 genotype, homozygous plants of the T2 generation were selected. Three independent transformants were retained for further analyses. To evaluate the efficiency of RNAi-mediated silencing, the expression of MtNPF6.8 was determined by quantitative RT-PCR in 10-d-old seedlings of wild-type and transgenic RNAi plants grown on 5 mm NO3−. Compared with the wild type, the expression of MtNPF6.8 was reduced by 85.7% (npf6.8-2), 77.7% (npf6.8-3), and 81.5% (npf6.8-5) in roots and 73.5% (npf6.8-2), 57% (npf6.8-3), and 81.4% (npf6.8-5) in shoots (Supplemental Fig. S1B). Expression of two putative nitrate transporters, Medtr4g101380 and Medtr5g012290, which possess the greatest sequence identity (60% and 59.6%, respectively) with MtNPF6.8, exhibited no difference between the wild type and the RNAi plants (Supplemental Fig. S1C), indicating that the RNAi fragment was specific for MtNPF6.8.
Nitrate Influx and N Status in the Wild Type and npf6.8 Knockdown Lines
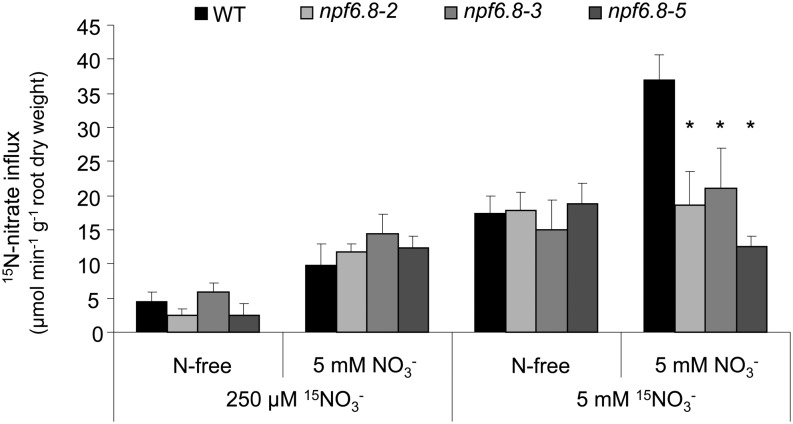
To elucidate the role of MtNPF6.8 in nitrate influx, the wild type and npf6.8 knockdown lines were germinated for 2 d on N-free or NO3−-supplied (5 mm) medium and transferred for 8 d to a hydroponic system with the same N regime. Nitrate influx through the HATS and LATS was measured by feeding the seedlings with 250 µm or 5 mm 15NO3− for 5 min (Fig. 4). Plants grown on N-free medium allow for measuring activities of both constitutive HATS and LATS, whereas the NO3− influxes determined in plants acclimatized to nitrate are the result of both constitutive and inducible HATS and LATS systems of nitrate transport (Siddiqi et al., 1990; Huang et al., 1996; Faure-Rabasse et al., 2002).
Figure 4.
Nitrate uptake by the M. truncatula wild type and npf6.8 knockdown lines. Seedlings were grown in hydroponic solution containing N-free or 5 mm nitrate for 10 d. Root 15NO3− influx was assayed by 5-min labeling in complete nutrient solutions containing 15NO3− (99 atom percentage 15N) at the concentration indicated. Values are the mean of four biological replicates ± se. P < 0.05 compared with the wild type for each condition (ANOVA). WT, Wild type.
Nitrate influx measured by feeding seedlings with 250 µm 15NO3− indicated that neither constitutive nor inducible HATS were significantly affected by the silencing of MtNPF6.8 (Fig. 4). Nitrate influx measured by feeding N-starved seedlings with 5 mm 15NO3− showed similar constitutive LATS activity in the wild type and npf6.8 knockdown lines. However, when LATS activity was measured in seedlings acclimatized to 5 mm NO3−, the influx of 15NO3− was 2-fold higher in the wild type but remained unchanged in the npf6.8 knockdown lines. This result suggests the involvement of MtNPF6.8 in the increase in LATS activity (Fig. 4).
To investigate whether the decrease of nitrate influx due to MtNPF6.8 silencing affected N status, the NO3−, NH4+, amino acid, and protein contents of the roots and shoots were determined in the wild type and npf6.8 knockdown lines grown for 10 d on 250 µm or 5 mm NO3−-supplied medium (Table I). There was no significant difference in these biochemical parameters between the wild type and any of the three npf6.8 knockdown lines, irrespective of the nitrate supply.
Table I. NO3−, NH4+, amino acid, and soluble protein contents in roots or shoots of M. truncatula 10-d-old seedlings of the wild type and of three npf6.8 knockdown lines, grown on 250 µm- or 5 mm NO3−-supplied medium.
Data are expressed as the mean ± se. ND indicates not detected.
| Components | Plant Grown on 250 µm NO3− |
Plant Grown on 5 mm NO3− |
||||||
|---|---|---|---|---|---|---|---|---|
| Wild Type | npf6.8-2 | npf6.8-3 | npf6.8-5 | Wild Type | npf6.8-2 | npf6.8-3 | npf6.8-5 | |
| Root NO3− content (µg g−1 root dry weight) | ND | ND | ND | ND | 8.17 ± 1.06 | 8.83 ± 2.28 | 8.88 ± 0.85 | 6.74 ± 1.59 |
| Shoot NO3− content | ND | ND | ND | ND | 0.76 ± 0.69 | 0.35 ± 0.05 | 0.53 ± 0.31 | 1.10 ± 0.44 |
| Root NH4+ content (µmol g−1 root fresh weight) | 1.19 ± 0.24 | 1.39 ± 0.30 | 1.41 ± 0.32 | 1.46 ± 0.29 | 1.27 ± 0.41 | 1.02 ± 0.01 | 1.10 ± 0.10 | 1.23 ± 0.10 |
| Shoot NH4+ content | 0.68 ± 0.06 | 0.74 ± 0.09 | 0.83 ± 0.09 | 1.08 ± 0.19 | 0.73 ± 0.12 | 0.55 ± 0.09 | 0.58 ± 0.04 | 0.66 ± 0.19 |
| Root amino acid content (µmol g−1 root fresh weight) | 4.96 ± 1.88 | 4.30 ± 0.76 | 4.66 ± 0.78 | 4.96 ± 1.15 | 7.01 ± 0.39 | 6.42 ± 1.25 | 5.75 ± 1.35 | 8.18 ± 0.29 |
| Shoot amino acid content | 10.49 ± 4.17 | 7.54 ± 0.65 | 10.05 ± 1.88 | 12.55 ± 2.03 | 12.03 ± 1.96 | 8.52 ± 2.96 | 10.13 ± 4.06 | 15.26 ± 3.38 |
| Root soluble protein content (µg g−1 root fresh weight) | 2.90 ± 0.31 | 2.63 ± 0.03 | 2.19 ± 0.19 | 2.54 ± 0.02 | 2.53 ± 0.09 | 2.53 ± 0.07 | 2.53 ± 0.36 | 2.94 ± 0.14 |
| Shoot soluble protein content | 6.27 ± 1.04 | 6.67 ± 1.20 | 6.39 ± 1.63 | 6.77 ± 0.87 | 5.02 ± 0.90 | 7.44 ± 1.52 | 6.94 ± 1.24 | 7.16 ± 0.93 |
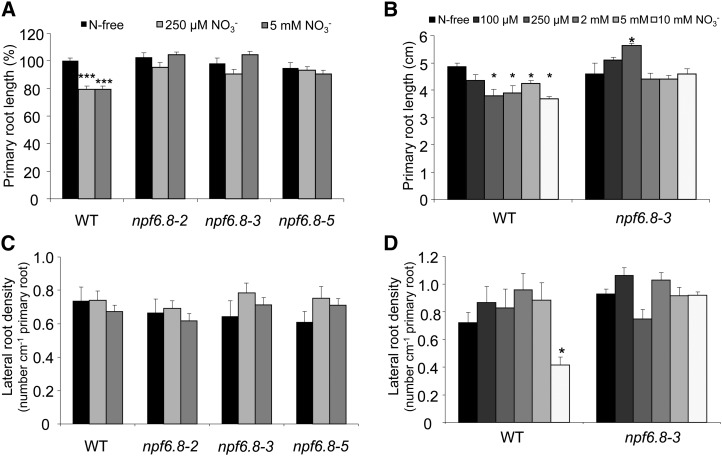
The Effect of Nitrate on Root System Architecture Is Abolished in npf6.8 Knockdown Lines
In a previous study, on the basis of a quantitative genetic approach using a QTL determination, we hypothesized that MtNPF6.8 might be involved in the control of primary root length (Morère-Le Paven et al., 2011). To test this hypothesis, the three npf6.8 knockdown lines and the wild type were grown for 10 d on an N-free or NO3−-supplied (250 µm or 5 mm) medium.
The length of the primary root was similar in the wild type and npf6.8 knockdown lines, when the seedlings were grown on N-free medium (Fig. 5A). When the seedlings were fed with either 250 µm or 5 mm NO3−, the wild type exhibited a reduction of 20% in primary root length compared with growth in the N-free medium, whereas no effect was observed in any of the npf6.8 knockdown lines (Fig. 5A). Because the three npf6.8 knockdown lines showed a similar response to nitrate, only one (the npf6.8-3 line) was used in a dose-dependent study of the effect of nitrate on the growth of the primary root compared with the wild type. With the exception of the treatment with 100 µm NO3−, there was a similar inhibitory effect on the growth of the primary root of the wild type at all of the concentrations tested from 250 µm up to 10 mm NO3− (Fig. 5B). The growth of the primary root of the npf6.8-3 knockdown line was insensitive to increasing nitrate concentrations (Fig. 5B), thus strengthening the idea that the inhibitory effect of nitrate requires a functional MtNPF6.8.
Figure 5.
Effect of nitrate treatment on root architecture. A to D, Primary root length (A and B) and LR density (C and D) in the wild type or npf6.8 knockdown lines. Seedlings were grown on N-free medium for 24 h and then transferred on vertical plates for 10 d on medium containing nitrate at the indicated concentration. Primary root length was determined by image analysis (A and B), and LR density was calculated as the number of LRs per centimeter of primary root length (C and D). Means represent average values ± se of 32 seedlings of four biological replicates (A and C) and 16 seedlings of two biological replicates (B and D). Asterisks indicate significant differences compared with the untreated control (N-free) according to a one-way ANOVA (one asterisk indicates P < 0.05, whereas three asterisks indicate P < 0.001). WT, Wild type.
LR density was compared between the wild type and npf6.8 knockdown lines grown for 10 d on an N-free or NO3−-supplied (250 µm or 5 mm) medium. At these concentrations, no significant effect of nitrate on the LR density was observed in the wild type and npf6.8 knockdown lines (Fig. 5C). However, a 10 mm NO3− concentration led to a significant decrease in the density of the LR in the wild type, whereas no effect was observed in the npf6.8-3 knockdown line (Fig. 5D), indicating that LR density of the npf6.8-3 knockdown line was insensitive to 10 mm NO3−.
The overall growth of the seedlings was examined in terms of fresh weight (roots or shoots). The fresh weight of the roots (primary root and LR) was significantly reduced in the wild-type seedlings fed with either 250 µm or 5 mm NO3−, compared with those grown on the N-free medium (Supplemental Fig. S2A). However, the fresh weight of the roots of the three npf6.8 knockdown lines was not affected by nitrate supply. The fresh weight of the shoots of the wild type and the three npf6.8 knockdown lines was not affected by nitrate treatments (Supplemental Fig. S2B).
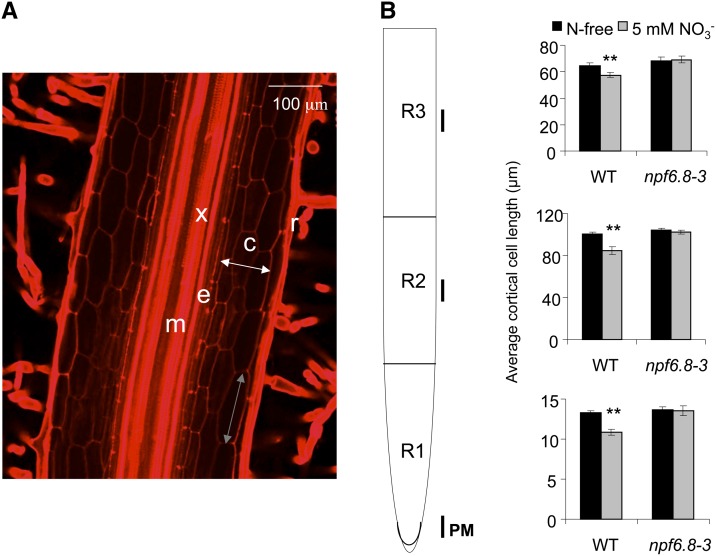
The Inhibitory Effect of Nitrate on Cell Elongation Is Abolished in npf6.8 Knockdown Lines
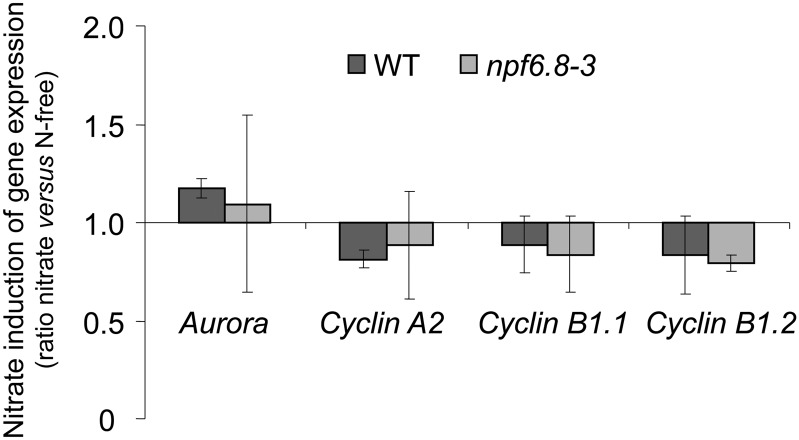
The effect of nitrate on primary root growth was further investigated by measuring cell elongation. Plants were grown on N-free or 5 mm NO3−-supplied medium and the length of the root cortical cells was measured in 6-d-old seedlings in three different root sections (R1, R2, and R3), by staining with propidium iodide and observation under a confocal microscope (Fig. 6A). In wild-type plants, the length of cortical cells was decreased by 20% in NO3−-fed seedlings compared with seedlings grown on N-free medium regardless of the root section (Fig. 6B). Interestingly, no difference in the length of the cortical cells was observed between the npf6.8-3 knockdown line seedlings grown on N-free and 5 mm NO3−-supplied medium. To check whether MtNPF6.8 was also involved in cell division, expression of four marker genes that encode proteins of cell division was measured (Aurora, Cyclin A2, Cyclin B1.1-like, and Cyclin B1.2-like). No difference in the expression of these cell division marker genes was observed between the wild type and the npf6.8-3 knockdown line (Fig. 7).
Figure 6.
Effect of nitrate treatment on the cortical cell length of primary root. Seedlings of the wild type and the npf6.8-3 knockdown line were germinated on N-free medium for 24 h and then transferred on vertical plates on N-free or 5 mm nitrate for 6 d. A, Roots were stained with propidium iodide and observed under a confocal microscope. B, Cortical cell length was then measured on the 1-mm part indicated by vertical black lines (primary meristem [PM] region) of three sequential sections of the primary root starting with the tip: root sections R1 (0.7 cm long), R2 (0.7 cm long), and R3 (the root remainder) by image analysis. Results are the means ± se of 6 to 12 replicates. The mean was modeled in a linear model (two asterisks indicate P < 0.01) with genotype and condition as quantitative factors and taking genotype and condition interactions into account. c, Cortex; e, endoderm; m, medulla; r, rhizoderm; WT, wild type; x, xylem.
Figure 7.
Effect of nitrate treatment on the expression of cell division marker genes. Seedlings were grown as described in the legend for Figure 6. Expression of cell division marker genes (Aurora, Cyclin A2, Cyclin B1.1, and Cyclin B1.2) was determined by quantitative RT-PCR in response to nitrate in the primary root tip. Each value represents the mean ± se of three independent biological experiments. Gene numbers are as follows: Aurora (TC901033), Cyclin A2 (Medtr2g102520), Cyclin B1.1 (Medtr5g088980), and Cyclin A2 (Medtr2g102520). WT, Wild type.
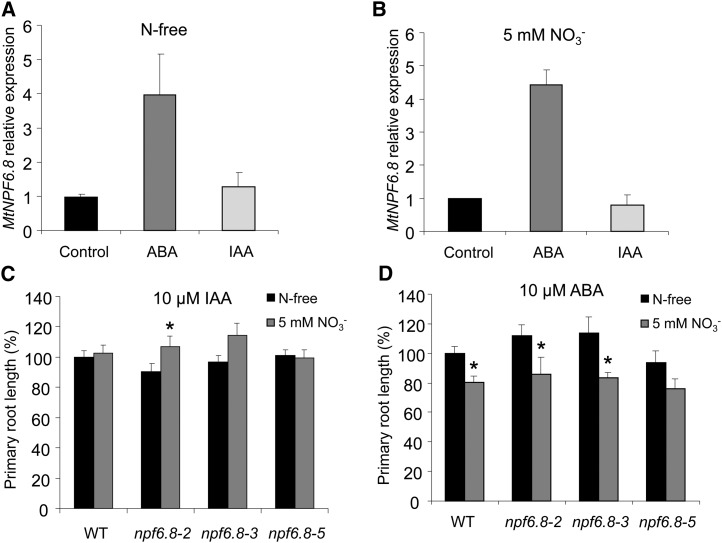
ABA Treatment Restores the Inhibitory Effect of Nitrate on Primary Root Length
As far as IAA and ABA were proposed to play a role in mediating the nitrate regulatory effect on LR development (Signora et al., 2001; De Smet et al., 2006; Walch-Liu et al., 2006; Zhang et al., 2007), we have investigated whether these hormones interact with nitrate signaling pathway via MtNPF6.8 in M. truncatula. For this purpose, we first examined the regulation of MtNPF6.8 expression by both hormones. The expression of MtNPF6.8 was determined in 10-d-old seedlings grown on N-free medium and then treated with 10 µm IAA or ABA in the presence or absence of 5 mm NO3− for 5 h. The results showed that MtNPF6.8 expression is induced by ABA but is insensitive to IAA regardless of the exogenous nitrate content (Fig. 8, A and B). Second, we investigated the effect of nitrate on primary root growth in the wild type and in the three npf6.8 knockdown lines in IAA- or ABA-treated seedlings. In the npf6.8 knockdown lines, the inhibitory effect of nitrate on primary root growth was not restored by exogenous IAA; in the wild type, it seems that the effect of IAA abolished the effect of nitrate, leading to similar growth of the primary root in the presence and absence of nitrate (Fig. 8C). Unlike IAA, ABA treatment did not hamper the effect of nitrate in the wild-type seedlings in which ABA-/nitrate-treated seedlings showed a decrease of 20% in the primary root length compared with the nitrate-treated seedlings (Fig. 8D), a nitrate inhibitory effect of the same magnitude as observed in seedlings nontreated with phytohormones (Fig. 5A). More interestingly, in the absence of a functional MtNPF6.8 in the mutants, ABA rescues the inhibitory effect of nitrate on primary root elongation (Fig. 8D).
Figure 8.
Combined effect of nitrate and ABA or IAA on MtNPF6.8 expression and on primary root length. A and B, MtNPF6.8 expression in root determined by quantitative RT-PCR. Ten-day-old seedlings of M. truncatula were grown on N-free medium and exposed to 10 µm IAA or ABA during 5 h without (A) and with (B) 5 mm nitrate. Error bars represent the sd of three independent biological replicates. C and D, Combined effect of nitrate and hormonal treatment on primary root length in the wild type or npf6.8 knockdown lines. C and D, Seedlings were grown on medium for 24 h and then transferred on vertical plates for 10 d on medium containing N-free or 5 mm nitrate with 10 µm IAA (C) or ABA (D). Primary root length was determined by image analysis. Means represent average values ± se of 16 seedlings of two biological replicates. Asterisks indicate significant differences compared with the untreated control (N-free) according to a one-way ANOVA (asterisk indicates P < 0.05). WT, Wild type.
MtNPF6.8 Transports ABA in Xenopus spp. Oocytes
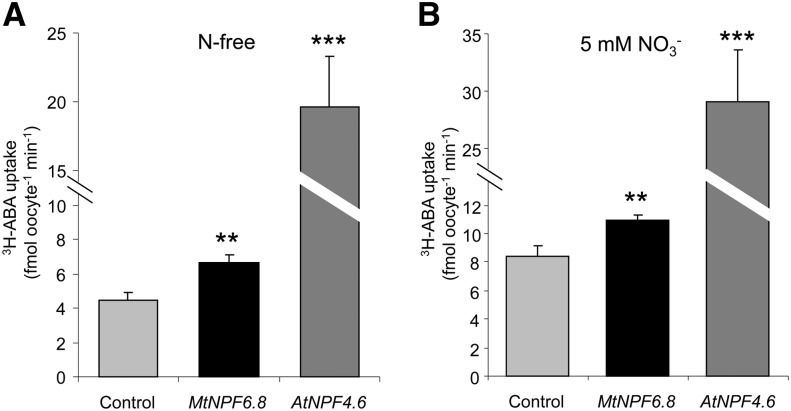
The reestablishment of the inhibitory effect of nitrate on primary root growth by exogenous ABA in npf6.8 knockdown lines and the sensitivity of MtNPF6.8 expression to exogenous ABA prompted us to investigate whether MtNPF6.8 would transport ABA. For this purpose, we used Xenopus spp. oocytes as a heterologous expression system and AtNPF4.6 as a positive control (Kanno et al., 2012). Oocytes were assessed for ABA transport activity using [3H]ABA at 1 µm in the assay medium with or without 5 mm NO3−. [3H]ABA uptake by MtNPF6.8 in oocytes was lower but still significant compared with that mediated by AtNPF4.6 (Fig. 9A). In addition, injection of MtNPF6.8 and AtNPF4.6 complementary RNA (cRNA) resulted in a significant increase of [3H]ABA uptake in oocytes similarly in the absence or presence of nitrate (Fig. 9). These results show that MtNPF6.8 is able to transport ABA or to facilitate ABA transport in a heterologous system.
Figure 9.
ABA uptake activities in MtNPF6.8-cRNA- or AtNPF4.6-cRNA-injected Xenopus spp. oocytes. Injected or control oocytes were incubated with 1 µm [3H]ABA at pH 5.5. Each data point is a mean ± se for eight oocytes. Asterisks indicate significant differences compared with the control according to a one-way ANOVA (two asterisks indicate P < 0.01, whereas three asterisks indicate P < 0.001). Similar results were obtained with oocytes isolated from three different frogs.
Expression of Nitrate-Inducible Genes NR1, NR2, and GS2 in npf6.8 Knockdown Lines
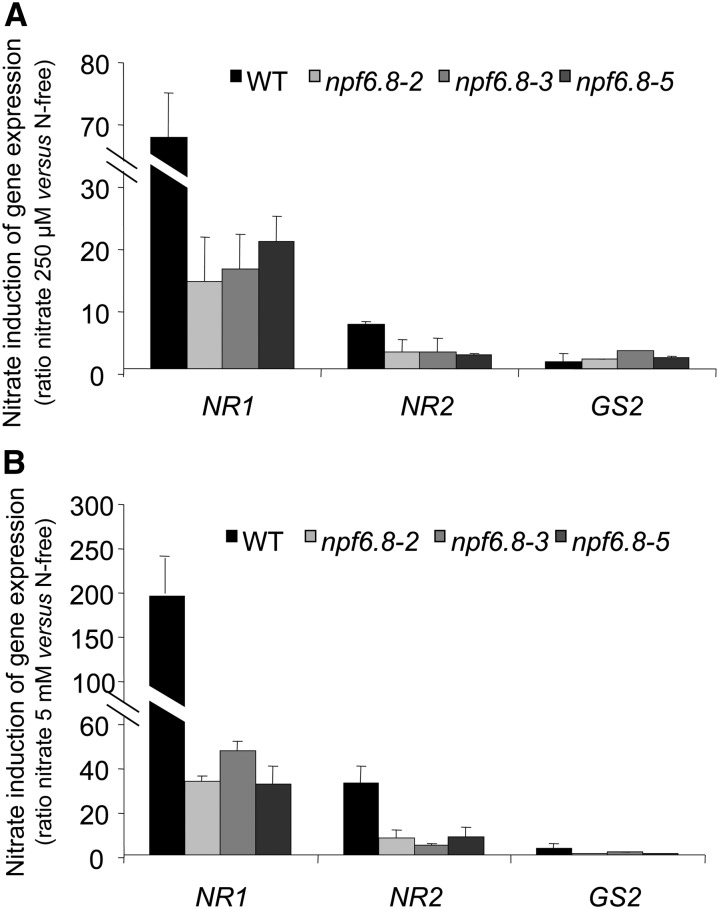
In Arabidopsis, over 1,000 genes display rapid responses to nitrate at low concentration, the primary response to nitrate (Wang et al., 2003). To further characterize the role of MtNPF6.8 in nitrate signaling, the expression of three nitrate-inducible genes (two genes encoding nitrate reductase NR1 and NR2 and a gene encoding a plastidic Gln synthetase GS2) was analyzed in the wild type and in npf6.8 knockdown lines to determine whether MtNPF6.8 is involved in the primary nitrate response. The wild type and the three npf6.8 knockdown lines were grown on N-free medium and transferred to a 250 μm (Fig. 10A) or 5 mm (Fig. 10B) NO3−-supplied medium for 30 min. The data show that the induction of NR1 and NR2 by nitrate was reduced at least by 60% in all of the npf6.8 knockdown lines compared with the wild type, in both 250 μm and 5 mm NO3−-supplied medium. GS2 expression was not significantly induced by 250 μm NO3− (Fig. 10A), whereas it showed a 4-fold increase by 5 mm NO3− in the wild type (Fig. 10B). Although no difference was observed between the wild type and npf6.8 knockdown lines at 250 μm NO3−, the induction of GS2 by 5 mm NO3− was reduced by at least 40% in the npf6.8 knockdown lines (Fig. 10B). Altogether, these data suggest that MtNPF6.8 takes part in the transcriptional regulation of nitrate-inducible genes in response to nitrate.
Figure 10.
Nitrate induction of the expression of nitrate reductase genes (NR1 and NR2) and Gln synthetase gene (GS2) in roots. A and B, Seedlings of the wild type and the npf6.8 knockdown line were grown on vertical plates on N-free for 6 d and then transferred to N-free or 250 µm nitrate (A) and to N-free or 5 mm nitrate (B) for 30 min. The root mRNA level was determined by quantitative RT-PCR. Nitrate induction was calculated using N-free medium as the control condition. Error bars represent ± sd of biological replicates (n = 3). Gene numbers are as follows: NR1 (Medtr3g073180), NR2 (Medtr5g059820), and GS2 (Medtr2g021250). WT, Wild type.
DISCUSSION
MtNPF6.8, a Nitrate Transporter Essential for the Inducible Component of the LATS
MtNPF6.8 was previously shown to be a dual-affinity nitrate transporter by expression in Xenopus spp. oocytes and determination of the kinetics of 15NO3 uptake (Morère-Le Paven et al., 2011). In this study, MtNPF6.8 was characterized further in planta by an investigation into the subcellular localization, gene expression, and contribution to the nitrate transport system.
Expression of MtNPF6.8:GFP and coexpression of MtNPF6.8:RFP and TM23, a plasma membrane peptide marker fused to GFP, in both M. truncatula and N. benthamiana leaf epidermal cells unequivocally demonstrated the plasma membrane localization of MtNPF6.8 (Fig. 1). This localization is identical to that of all of the NPF transporters studied to date (Wang et al., 2012).
Investigation of the spatiotemporal expression of MtNPF6.8 in M. truncatula indicated important differences compared with that of the known dual-affinity nitrate transporter AtNPF6.3 in Arabidopsis. Indeed, MtNPF6.8 is constitutively expressed in the roots (Fig. 2) and, as previously observed, it is not a nitrate-inducible gene (Morère-Le Paven et al., 2011), whereas AtNPF6.3 is a nitrate-inducible gene (Tsay et al., 1993). In earlier work, we detected a slight inhibitory effect of nitrate on MtNPF6.8 expression in the radicles of 2- to 3-d-old seedlings when 5 mm NO3− was administered for 5 or 24 h (Morère-Le Paven et al., 2011). However, in this study, using seedlings grown either on nitrate-free or nitrate-supplied medium for at least 10 d, the inhibitory effect of nitrate on MtNPF6.8 expression was not observed in the primary roots. This result strengthens the concept of constitutive expression of MtNPF6.8, similar to that observed for other genes encoding low-affinity transporters LeNRT1.1 (Lauter et al., 1996), AtNPF4.6 (Huang et al., 1999), and OsNPF8.9 (OsNRT1; Lin et al., 2000), or the high-affinity transporter MtNPF1.7 (Yendrek et al., 2010; Bagchi et al., 2012). Unlike AtNPF6.3, MtNPF6.8 expression was not confined to nascent organs. In the roots, MtNPF6.8 was expressed in the pericycle region and interestingly in the LR primordia and tips (Fig. 3).
The expression of MtNPF6.8 also showed differences with the expression of another NPF gene, MtNPF1.7, identified in M. truncatula (Yendrek et al., 2010). MtNPF6.8 showed a lower expression in the primary root tip and the young LR compared with the expression in the elongation zone of the primary root, and the highest expression was observed in the mature parts of the primary root and in the shoot (Fig. 2). Unlike MtNPF6.8, MtNPF1.7 showed a 5-fold higher expression in the primary root tip versus the remainder of the root system (Yendrek et al., 2010). Whereas MtNPF1.7 was expressed in nodules (Yendrek et al., 2010), MtNPF6.8 was very weakly expressed or was not expressed in nodules (Morère-Le Paven et al., 2011; Cabeza et al., 2014). These results suggest that MtNPF6.8 and MtNPF1.7 might have different roles in M. truncatula.
The expression of MtNPF6.8 in the root system and plasma membrane localization of the encoded protein would suggest a contribution of MtNPF6.8 to the exogenous nitrate uptake and/or loading in the xylem for long-distance transport. To study this contribution, nitrate influx through both the HATS and LATS was studied in seedlings of both 10-d-old wild-type and npf6.8 knockdown lines that had been previously grown on N-free or 5 mm nitrate-supplied medium. The absence of expression of MtNPF6.8 affected only the inducible component of the LATS (Fig. 4). This result indicates that although MtNPF6.8 has the properties of a dual-affinity nitrate transporter, it does not make a contribution to either the constitutive or inducible components of the HATS. The npf6.8 knockdown lines exhibited the same rate of 15NO3 influx through the LATS irrespective of whether seedlings were grown on an N-free medium or a high nitrate supply. This result suggests that MtNPF6.8 does not contribute to the constitutive component of the LATS. It appears, however, that almost all of the inducible component of the LATS requires a functional MtNPF6.8 as either a direct contributor to nitrate transport or as an intermediate in nitrate sensing and induction of inducible low-affinity nitrate transporters in M. truncatula, or both. To our knowledge, this is the first time that the knockdown of one gene encoding a nitrate transporter affected the entire inducible component of the nitrate uptake, rather than induced a partial decrease of inducible LATS activity as observed for other mutants or underexpressors affected in nitrate transport (Nacry et al., 2013).
The analysis of N status of the plants grown on a high-nitrate regime showed that in the long term, the npf6.8 knockdown lines did not exhibit symptoms of N deficiency (Table I). This would indicate that the absence of the low-affinity uptake component in the npf6.8 knockdown lines is compensated by the higher root biomass (Supplemental Fig. S2) without ruling out a decrease in nitrate efflux.
MtNPF6.8 Is Involved in Nitrate Signaling and Primary Root Growth Regulation
During the postgermination phase, after radicle protrusion, primary root elongation is a developmental event that is required for the seedling to colonize the soil in order to rapidly access mineral nutrients and water. In our previous study, using quantitative genetics and QTL determination, we showed that the growth of the primary root of M. truncatula is a multigenic trait responsive to nitrate availability in the rhizosphere (Morère-Le Paven et al., 2011). Although in some studies, as with Arabidopsis, high nitrate stimulated primary root growth (Walch-Liu and Forde, 2008), it is generally well established that uniform high nitrate availability in the root zone results in a decrease in primary root growth (Scheible et al., 1997). Primary root elongation of 4-d-old maize seedlings was inhibited by treatments with 5 to 10 mm NO3− for 7 d (Tian et al., 2008); primary root growth of Capsicum chinense was inhibited in 10-d-old seedlings exposed for 4 to 6 d to nitrate concentrations ranging between 0.1 and 10 mm NO3− (Celis-Arámburo et al., 2011). Inhibition of primary root elongation was also observed in Arabidopsis grown either on vertical plates for 18 d on 1 mm NO3− (Linkohr et al., 2002), or in a hydroponic system for 14 d and then treated for 3 d with 5 mm NO3− (Vidal et al., 2010a). In addition, the dual-affinity nitrate transporter AtNPF6.3 of Arabidopsis was shown to function as a nitrate sensor involved in nitrate signaling. As a result of the coincidence of MtNPF6.8 with the peak of a major QTL involved in the control of primary root elongation, we hypothesized that MtNPF6.8 is a reliable candidate to mediate the primary root elongation response to nitrate in M. truncatula. This hypothesis is supported in this study by the finding that the inhibitory effect of nitrate on primary root elongation was abolished in the npf6.8 knockdown lines, indicating that the effect of nitrate required a functional MtNPF6.8 (Fig. 5). To further investigate the mechanisms underlying the inhibitory effect of nitrate on primary root elongation, the effect on cortical cell size was determined in various regions of the primary root of the wild type and the npf6.8-3 knockdown line of M. truncatula. As expected, the length of the cortical cells was decreased by approximately 20% in the roots of the wild-type seedlings that had been supplied with 5 mm NO3−, compared with those growing on an N-free medium (Fig. 6). By contrast, the size of the cortical cells of the npf6.8-3 knockdown line was very similar irrespective of whether seedlings were grown in the presence or absence of 5 mm NO3−. Analysis of the expression of several genes involved in the cell cycle showed that they were not affected by nitrate supply (Fig. 7), either in the wild type or in the npf6.8-3 knockdown line, indicating that only cell elongation is affected by the mutation. This compares with the localized control of LR growth by nitrate via AtNPF6.3 that results in accelerated meristematic cell division (Mounier et al., 2014). Altogether, these results support the assumption that in M. truncatula, nitrate regulates primary root growth by controlling the elongation of cortical cells, probably independently of its metabolic role (Table I), and that MtNPF6.8 is involved in this signaling cascade.
ABA Acts Downstream of MtNPF6.8 in the Nitrate Inhibitory Effect on Primary Root Growth
ABA treatment (10 μm) of M. truncatula wild-type seedlings severely inhibited primary root and LR growth (Liang and Harris, 2005) by inhibiting cell elongation (Zhang et al., 2014). Because nitrate induced the inhibition of primary root growth with the inhibition of cortical cell elongation (Figs. 5 and 6) and because the expression of the gene encoding MtNPF6.8 was strongly stimulated by exogenous ABA (Fig. 8, A and B), we checked for a potential involvement of ABA in the nitrate signaling pathway. This involvement is evidenced by the fact that the inhibitory effect of nitrate on primary root growth was restored in npf6.8 knockdown lines by exogenous ABA (10 μm; Fig. 8D), showing for the first time that ABA acts downstream of a nitrate transporter (MtNPF6.8) in the nitrate signaling pathway for the regulation of root growth in M. truncatula. The abolition of nitrate-induced inhibition of primary root growth in the npf1.7 mutant affected in ABA sensitivity (Yendrek et al., 2010) strongly supports our assumption, thereby indicating that nitrate signaling also requires a functional MtNPF1.7. Interestingly, MtNPF1.7 was found to encode a high-affinity nitrate transporter (Bagchi et al., 2012). Analysis of the npf1.7 mutant showed that MtNPF1.7 functions downstream of ABA to regulate root meristem organization and activity (Liang et al., 2007). The integration of our results together with the literature (Liang and Harris, 2005; Liang et al., 2007; Yendrek et al., 2010) allowed us to propose a model in which the nitrate signaling pathway for the regulation of primary root growth is ABA dependent and involves two nitrate transporters: MtNPF6.8 acts upstream of ABA and MtNPF1.7 acts downstream of ABA (Fig. 11). Whether MtNPF6.8 and MtNPF1.7 are directly or indirectly connected along the nitrate/ABA signaling pathway is still to be unraveled.
Figure 11.
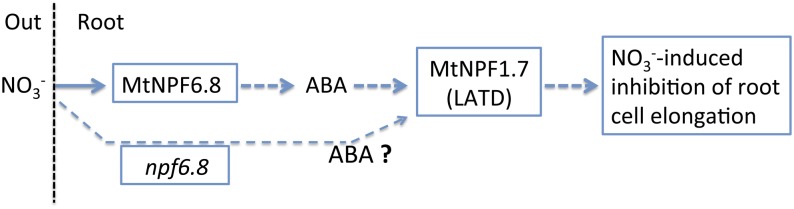
Schematic representation of a model of ABA-dependent nitrate signaling pathway for the regulation of primary root growth. This model includes two nitrate transporters: Nitrate-induced inhibition of primary root elongation was abolished in mutants knocked down in either MtNPF6.8 (this work) or MtNPF1.7 (Yendrek et al., 2010). In this work, the rescue of nitrate-induced inhibition of primary root growth by exogenous ABA in the npf6.8 mutant shows that MtNPF6.8 acts upstream of ABA, whereas MtNPF1.7 (LATD) was shown to act downstream of ABA (Liang et al., 2007). The rescue of nitrate inhibitory effect by exogenous ABA in the npf6.8 mutant suggests that nitrate is sensed at another site than MtNPF6.8. MtNPF1.7, a high-affinity nitrate transporter, is a good candidate for the integration of both nitrate and ABA signals.
The effect of auxin was also studied. It appears that exogenously supplied IAA (10 μm) alleviated the inhibitory effect of nitrate in the wild type, but no significant effect was observed in the npf6.8 knockdown lines neither on N-free nor on nitrate-supplied medium (Fig. 8C). It is interesting to note that exogenous auxin counteracting the nitrate inhibitory effect on root growth in the wild type is consistent with the hypothesis of a systemic effect of nitrate via the inhibition of the long-distance transport of auxin from shoot to root (Zhang et al., 1999; Walch-Liu et al., 2006; Krouk et al., 2011).
MtNPF6.8 Transports ABA in Xenopus spp. Oocytes
Experiments in Xenopus spp. oocytes showed that, in addition to nitrate, MtNPF6.8 transports ABA, although at a low rate (Fig. 9). Interestingly, MtNPF6.8, a dual-affinity nitrate transporter that belongs to the same clade as the dual-affinity nitrate transporter AtNPF6.3, transports ABA like AtNPF4.6 that belongs to a different clade. This finding supports the assumption that, among NPF transporters, sequence homologies do not correlate with substrate selectivity (Léran et al., 2014).
At this stage, how the ABA transport activity of MtNPF6.8 fits within our signaling model (Fig. 11) still needs more investigation. On one hand, it seems unlikely that MtNPF6.8 mediates the inhibitory effect of nitrate on primary root growth through its ABA transport activity because ABA rescued this phenotype in the decreased MtNPF6.8 background of the npf6.8 knockdown lines. Furthermore, ABA transport activity in Xenopus spp. oocytes was similar in the presence or absence of nitrate in the external medium indicating that there was no competition for the transport between the two substrates as observed for ABA and nitrate transport by the Arabidopsis transporter AtNPF4.6 (Kanno et al., 2013). On the other hand, the localization of the expression of MtNPF6.8 in the pericycle region (Fig. 3) suggests that MtNPF6.8 may have a role in the redistribution of ABA within the root via retrieval of ABA from the xylem sap. As a consequence, underexpression of MtNPF6.8 would lead to lower ABA levels in the root and prevent ABA-mediated repression of root elongation that could be rescued by exogenous ABA application.
MtNPF6.8 Is Involved in the Primary Nitrate Response
In order to strengthen our assumption that MtNPF6.8 is involved in the nitrate signaling, the role of MtNPF6.8 in nitrate-inducible events was determined in the npf6.8 knockdown lines and compared with the wild type (Fig. 10). Two genes encoding two nitrate reductase isoforms (NR1 and NR2) proved to be highly sensitive to nitrate signaling in M. truncatula, because they were induced by up to 200- and 30-fold, respectively, in primary roots after 30 min of treatment with 5 mm NO3−. To a lesser extent, these two NR genes were also induced by nitrate at a lower concentration (250 μm NO3−). Nitrate induction of NR genes is strongly impaired in the npf6.8 knockdown lines; this result was also obtained at the 250-µm supply, where no defect in root nitrate uptake was recorded in these lines (Fig. 4). This suggests that MtNPF6.8 regulates the nitrate response of NR genes independently of its role in nitrate transport, thereby supporting a direct signaling role. In addition, the GS2 gene, which encodes a plastidic Gln synthetase and is less sensitive to nitrate signaling (induced by 4-fold), also showed reduced expression in all three npf6.8 knockdown lines after 30 min on 5 mm NO3−-supplied medium. These data indicate that MtNPF6.8 is very likely to play a role in the response to nitrate signaling in M. truncatula.
MATERIALS AND METHODS
Seed Germination and Seedling Growth Conditions
The R108 Medicago truncatula line was used in this study. Seed germination was performed as previously described (Morère-Le Paven et al., 2011), using N-free modified Murashige and Skoog medium (MS) containing 3 mm CaCl2, 1.5 mm MgSO4, 1.25 mm KH2PO4/K2HPO4, 5 mm KCl, and complete micronutrients (Murashige and Skoog, 1962). This medium was complemented with KNO3 as a sole N source at the concentration indicated for each individual experiment. The K+ concentration was adjusted to 5 mm by the addition of KCl in all media with KNO3 concentrations lower than 5 mm. For phenotypic and gene expression analyses, eight plants were transferred 24 h after germination on filter paper with 7 mL of the appropriate solution in 12-cm square transparent plates. Plates were placed at a 45° angle at 22°C with a 16-h photoperiod (120 µE m2 s−2) in a growth chamber. Primary root length was scored every 24 h by marking the plate covers, and corresponding images were analyzed using ImageJ software (http://rsbweb.nih.gov/ij/). For primary nitrate response analysis, plants were transferred 7 d after germination for 30 min onto N-free-modified (5 mm KCl-), 250 µm KNO3-modified, or 5 mm KNO3-modified MS medium.
Subcellular Localization of MtNPF6.8
For the subcellular localization of MtNPF6.8, transient expression of MtNPF6.8 fused to GFP or RFP in M. truncatula or Nicotiana benthamiana leaves was performed using Gateway cloning technology (Invitrogen). The MtNPF6.8 open reading frame (ORF) was previously obtained and cloned into a pGEM-HEJUELvector (Morère-Le Paven et al., 2011). The MtNPF6.8 ORF was amplified by PCR from the recombinant plasmid using the primer pair named MtNPF6.8-ORF (Supplemental Table S1) and Pfu DNA polymerase (Promega). Reactions were carried out in an iCycler (Bio-Rad) with a standard protocol (Melting temperature 58°C). The PCR products were purified using Genelute minus Ethidium Bromide spin columns (Sigma-Aldrich), following the manufacturer’s instructions. The MtNPF6.8 ORF was subcloned into pENTR/D-TOPO using a pENTR directional TOPO cloning kit (Invitrogen), as described by the manufacturer. The constructs were checked by DNA sequencing (GATC Biotec). For expression of the recombinant MtNPF6.8 fused to GFP or RFP, the insert was transferred into pK7FWG2 or pB7RWG2, respectively, using the Gateway LR Clonase II enzyme mix (Invitrogen). Recombinant plasmids 35S:NPF6.8:GFP/RFP were then transferred into Agrobacterium tumefaciens (strain EHA101) by electroporation. Positive clones were grown in Luria-Bertani medium supplemented with spectinomycin and rifampicin, until reaching an absorbance of 1 at 600 nm. This was followed by centrifugation at 3,000g for 20 min and resuspension in water to an absorbance of 0.2 to 0.4 at 600 nm. Suspensions were used to infiltrate leaves of M. truncatula or N. benthamiana and were incubated for 2 d. For colocalization experiments, two markers were used: TIP1-1, a marker protein of the tonoplast (Boursiac et al., 2005), and TM23, a marker peptide of the plasma membrane (Brandizzi et al., 2002). A. tumefaciens containing a recombinant plasmid 35S:NPF6.8:RFP was infiltrated into leaves of N. benthamiana together with either A. tumefaciens containing a recombinant plasmid 35S:TIP1-1:GFP or A. tumefaciens containing a recombinant plasmid 35S:TM23:GFP. For these experiments, cells were plasmolyzed by incubating leaf samples in 1 m mannitol for 30 min in vacuo, prior to observation. Cortical regions of leaf epidermal cells were observed by confocal microscopy, as described by Renard et al. (2011).
Histochemical Assay
A 2-kb genomic fragment containing the promoter, ATG, first intron, and partial first exon of MtNPF6.8 was isolated by PCR using Pfu DNA polymerase (Promega) using gene-specific primers (ProNPF6.8-For and ProNPF6.8-Rev; Supplemental Table S1). Reactions were carried out in an iCycler (Bio-Rad) with a standard protocol (Melting temperature 60°C). The PCR products were purified and subcloned into pENTR/D-TOPO as described above. The constructs were checked by DNA sequencing (GATC Biotech). For expression of the MtNPF6.8 promoter fused to the GUS reporter gene, the insert was transferred into pKGWFS7 using the Gateway LR Clonase II enzyme mix (Invitrogen). Recombinant plasmid pMtNPF6.8:GUS was then transferred into Agrobacterium rhizogenes ARqua1 (Smr-derivative strain of A4T; Gonzalez-Rizzo et al., 2006) by electroporation and used for M. truncatula root transformation. The transgenic roots were obtained using the protocol described by Boisson-Dernier et al. (2001). After 30 h of germination, the primary root was cut and seedlings were brought into contact with A. rhizogenes. After 3 weeks of growth, transgenic roots were stained for β-galactosidase activity as described by Gonzalez-Rizzo et al. (2006). Observations were performed using a SZX16 Research Stereo Microscope (Olympus) equipped with an Olympus DP71 camera and a SDFPLAPO1 XPF objective.
Generation of Stable npf6.8 Knockdown Lines by RNAi
To obtain the npf6.8 knockdown lines, a 401-bp complementary DNA fragment of the MtNPF6.8 gene was amplified with a primer pair named MtNPF6.8-fragment (Supplemental Table S1). This PCR product was introduced into the entry vector pCR8/GW/TOPO by the TOPO Cloning Reaction (Gateway System; Invitrogen) and transferred into the pFRB plasmid pFGC5941 by recombination with the Gateway LR Clonase Enzyme Mix. pFRB-NPF6.8 contained the sense and antisense sequences of the 401-bp fragment under the control of the CaMV 35S promoter. Transformation of the R108 line of M. truncatula by the vector using A. tumefaciens strain EHA105 and in vitro culture was performed according to Trinh et al. (1998). Six plants were regenerated. After a first selection of positive lines (T0 generation) by PCR using a specific primer pair named MtNPF6.8-specific (Supplemental Table S1), seedlings of the T1 generation were examined for a 3:1 segregation. Only seedlings with an RNAi construction (either heterozygous or homozygous) were kept to obtain the T2 generation. Thirty-five to 45 seedlings of the T2 generation were analyzed by PCR for selection of RNAi MtNPF6.8 homozygotes. Expression of MtNPF6.8 in homozygous npf6.8 lines was determined by quantitative RT-PCR to detect the efficiency of targeted knockdown of the RNAi fragment using the specific primer pair MtNPF6.8-RNAi (Supplemental Table S1). Three homozygous lines (T2) were retained for further analysis (npf6.8-2, npf6.8-3, and npf6.8-5).
RNA Extraction, Reverse Transcription, and Real-Time Quantitative PCR
Total RNA extraction, reverse transcription, and real-time quantitative PCR were performed as described by Charrier et al. (2012), with the following modifications: 2 µg of RNA was treated for 3 min with 200 U of DNase I (Invitrogen). DNase was denatured for 8 min at 70°C before reverse transcription. Each measurement was carried out with at least three independent biological replicates, using a triplicate PCR reaction for determining cycle threshold values. The ratio was calculated using an equation with normalization by mean of two endogenous reference genes (MtRPB1 and MtUbiquitin10).
Total Soluble Protein, Amino Acid, Nitrate, and Ammonium Extraction and Analysis
For total soluble protein, amino acid, nitrate, and ammonium extraction, roots and shoots from five plants were separately excised from the wild type and npf6.8 knockdown lines, which were grown 10 d in transparent plates, as previously described. Total soluble protein was extracted from samples in 25 mm Tris-HCl, pH 7.6, with 1 mm MgCl2, 1 mm EDTA, 0.1% (v/v) β-mercaptoethanol, and 1% (w/v) polyvinylpolypyrrolidone. The resulting homogenate was centrifuged (14,000g for 30 min at 4°C). The protein content of the soluble fractions was determined using the Bradford reagent and bovine serum albumin as a standard (Bradford, 1976). Each measurement was carried out with four biological replicates, using a triplicate colorimetric reaction. Total amino acids and ammonium were extracted from samples with ethanol and water fractions and were measured by HPLC using the same method as described by Planchet et al. (2011). The amounts of all amino acids determined by HPLC were summed, in order to determine the total amount of amino acids. Measurements were repeated three times with independent biological samples. For the nitrate content analysis, roots and shoots were dried for 48 h at 60°C. Samples were ground, and 10 to 20 mg of powder was extracted in 500 µL of ultrapure water for 1 h at 100°C. The extract was centrifuged at 14,000g for 8 min, and the nitrate content of the supernatant was determined following the method of Cataldo et al. (1975). Each measurement was carried out with five biological replicates, using a triplicate colorimetric reaction.
Nitrate Influx Analysis
For analysis of the nitrate influx, eight plants of each genotype were grown for 2 d after germination on a plate with N-free modified MS medium or complemented with 5 mm KNO3, as previously described. Plants were then transferred to a hydroponic system for 8 d in the same nutrient solutions. Individual plants were rinsed in a 15 mm CaCl2 solution and the total root system was exposed to a modified MS medium containing 250 µm or 5 mm of K15NO3 for 5 min. The 15N-exposed plants were then immediately washed three times in cold 15 mm CaCl2. Roots were dried for 48 h at 60°C, and the retained 15N was determined using a C/N analyzer linked to an isotope ratio mass spectrometer (EA3000; EuroVector) coupled to a mass spectrometer (IsoPrime; Elementar).
Propidium Iodide Coloration
For confocal microscopy observations, seedlings were harvested 6 d after germination. Roots were removed and immediately immersed in the fixing solution (3:1 [v/v] absolute ethanol:acetic acid and 0.1% [v/v] Tween 20) for 24 h. The roots were incubated in 50% (v/v) ethanol for 1 h, and then in 70% (v/v) ethanol for 24 h. Prior to staining, primary roots were first rinsed in ultrapure water and incubated for 30 min in 1% (w/v) SDS and 200 mm NaOH. Second, the roots were rinsed in ultrapure water and incubated for 40 min in 1% (w/v) periodic acid. Finally, the roots were rinsed in ultrapure water and immersed for 8 min in a 1% (w/v) propidium iodide solution and stored in 40% (v/v) glycerol with 24 m chloral hydrate at 4°C for a minimum of 24 h until analysis. Observations were performed under a Nikon A1S1 confocal laser microscope (Nikon Instruments) equipped with argon-ion (488 nm) and diode (561 nm) lasers. Propidium iodide staining was detected with the emission light set at 520 to 720 nm.
ABA Uptake Assay in Xenopus spp. Oocytes
Oocytes were isolated and injected with 50 ng of MtNPF6.8 mRNA or AtNPF4.6 mRNA, as previously described (Morère-Le Paven et al., 2011). Control oocytes were either not injected or were injected with 50 nL of water. Oocytes were incubated for 20 min in a 250-µL solution containing 230 mm mannitol, 0.3 mm CaCl2, and 10 mm MES-Tris pH 5.5, containing 1 µm [3H]ABA (130 nm [3H]ABA from PerkinElmer, diluted with 870 nm cold ABA from Sigma-Aldrich). They were then washed four times with standard oocyte saline buffer (100 mm NaCl, 2 mm KCl, 1.8 mm CaCl2, 1 mm MgCl2, and 5 mm HEPES, pH 7.5) at 4°C, containing 5 µm cold ABA. Each oocyte was lysed with 50 µL of 2% (w/v) SDS and 2 mL of Ultima Gold (PerkinElmer) scintillating solution was added. Incorporated radioactivity was measured by a liquid scintillation analyzer (Tri-Carb 2100TR; Packard).
Statistical Analysis
All statistical tests were performed using R statistical software (version 2.13.0; http://cran.r-project.org). For all data, appropriate statistical tests were carried out and are described in the legend of each figure.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers Aurora (TC901033), Cyclin A2 (Medtr2g102520), Cyclin B1.1 (Medtr5g088980), Cyclin A2 (Medtr2g102520), NR1 (Medtr3g073180), NR2 (Medtr5g059820), and GS2 (Medtr2g021250).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Characterization of npf6.8 knockdown lines.
Supplemental Figure S2. Effect of nitrate treatment on fresh weight seedlings in the wild type or in npf6.8.
Supplemental Table S1. Sequences of primers used for PCR.
Supplementary Material
Acknowledgments
We thank Dr. Philippe Nacry and Peter Lea (Biochimie et Physiologie Moléculaire des Plantes, Montpellier, France) for critical reading of the article and helpful comments, Dr. Nadine Paris (Biochimie et Physiologie Moléculaire des Plantes, Montpellier, France) for the gift of recombinant plasmid containing the 35S:TM23:GFP plasma membrane marker, Dr. Benoit Lacombe (Biochimie et Physiologie Moléculaire des Plantes, Montpellier, France) for the gift of recombinant plasmid containing AtNPF4.6, Dr. François Barbier (Institut de Recherche en Horticulture et Semences, Angers, France) for the propidium iodide coloration advice, Mayeul Milien (Institut de Recherche en Horticulture et Semences, Angers, France) for histochemical assay, Patrice Chiron (Laboratoire Récepteurs et Canaux Ioniques Membranaires, Angers, France) for technical assistance, and Dr. Romain Berruyer (Institut de Recherche en Horticulture et Semences, Angers, France) for helpful discussions on statistical analysis.
Glossary
- ABA
abscisic acid
- LATS
low-affinity transporter system
- HATS
high-affinity transporter system
- LR
lateral root
- IAA
indoleacetic acid
- QTL
quantitative trait locus
- RNAi
RNA interference
- RFP
red fluorescent protein
- CaMV
Cauliflower mosaic virus
- cRNA
complementary RNA
- MS
Murashige and Skoog
- ORF
open reading frame
Footnotes
This work was supported by the Qualité des Semences program funded by Région Pays de Loire and by the CROISSANCE RACINAIRE program funded by Structure Fédérative de Recherche 4207 Qualité et Santé du Végétal.
The online version of this article contains Web-only data.
References
- Almagro A, Lin SH, Tsay YF (2008) Characterization of the Arabidopsis nitrate transporter NRT1.6 reveals a role of nitrate in early embryo development. Plant Cell 20: 3289–3299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews M, Lea PJ (2013) Our nitrogen ‘footprint’: the need for increased crop nitrogen use efficiency. Ann Appl Biol 163: 165–169 [Google Scholar]
- Andrews M, Raven JA, Lea PJ (2013) Do plants need nitrate? The mechanisms by which nitrogen form affects plants. Ann Appl Biol 163: 174–199 [Google Scholar]
- Bagchi R, Salehin M, Adeyemo OS, Salazar C, Shulaev V, Sherrier DJ, Dickstein R (2012) Functional assessment of the Medicago truncatula NIP/LATD protein demonstrates that it is a high-affinity nitrate transporter. Plant Physiol 160: 906–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisson-Dernier A, Chabaud M, Garcia F, Bécard G, Rosenberg C, Barker DG (2001) Agrobacterium rhizogenes-transformed roots of Medicago truncatula for the study of nitrogen-fixing and endomycorrhizal symbiotic associations. Mol Plant Microbe Interact 14: 695–700 [DOI] [PubMed] [Google Scholar]
- Boursiac Y, Chen S, Luu DT, Sorieul M, van den Dries N, Maurel C (2005) Early effects of salinity on water transport in Arabidopsis roots: molecular and cellular features of aquaporin expression. Plant Physiol 139: 790–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Brandizzi F, Frangne N, Marc-Martin S, Hawes C, Neuhaus JM, Paris N (2002) The destination for single-pass membrane proteins is influenced markedly by the length of the hydrophobic domain. Plant Cell 14: 1077–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabeza R, Koester B, Liese R, Lingner A, Baumgarten V, Dirks J, Salinas-Riester G, Pommerenke C, Dittert K, Schulze J (2014) An RNA sequencing transcriptome analysis reveals novel insights into molecular aspects of the nitrate impact on the nodule activity of Medicago truncatula. Plant Physiol 164: 400–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cataldo DA, Haroon M, Shrader LS, Youngs VL (1975) Rapid colorimetric determination of nitrate in plant tissue by nitration of salicylic acid. Commun Soil Sci Plant Anal 6: 71–80 [Google Scholar]
- Celis-Arámburo TdeJ, Carrillo-Pech M, Castro-Concha LA, Miranda-Ham MdeL, Martínez-Estévez M, Echevarría-Machado I (2011) Exogenous nitrate induces root branching and inhibits primary root growth in Capsicum chinense Jacq. Plant Physiol Biochem 49: 1456–1464 [DOI] [PubMed] [Google Scholar]
- Charrier A, Planchet E, Cerveau D, Gimeno-Gilles C, Verdu I, Limami AM, Lelièvre E (2012) Overexpression of a Medicago truncatula stress-associated protein gene (MtSAP1) leads to nitric oxide accumulation and confers osmotic and salt stress tolerance in transgenic tobacco. Planta 236: 567–577 [DOI] [PubMed] [Google Scholar]
- Chiu CC, Lin CS, Hsia AP, Su RC, Lin HL, Tsay YF (2004) Mutation of a nitrate transporter, AtNRT1:4, results in a reduced petiole nitrate content and altered leaf development. Plant Cell Physiol 45: 1139–1148 [DOI] [PubMed] [Google Scholar]
- Chopin F, Orsel M, Dorbe MF, Chardon F, Truong HN, Miller AJ, Krapp A, Daniel-Vedele F (2007) The Arabidopsis ATNRT2.7 nitrate transporter controls nitrate content in seeds. Plant Cell 19: 1590–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smet I, Zhang H, Inzé D, Beeckman T (2006) A novel role for abscisic acid emerges from underground. Trends Plant Sci 11: 434–439 [DOI] [PubMed] [Google Scholar]
- Fan SC, Lin CS, Hsu PK, Lin SH, Tsay YF (2009) The Arabidopsis nitrate transporter NRT1.7, expressed in phloem, is responsible for source-to-sink remobilization of nitrate. Plant Cell 21: 2750–2761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faure-Rabasse S, Le Deunff E, Lainé P, Macduff JH, Ourry A (2002) Effects of nitrate pulses on BnNRT1 and BnNRT2 genes: mRNA levels and nitrate influx rates in relation to the duration of N deprivation in Brassica napus L. J Exp Bot 53: 1711–1721 [DOI] [PubMed] [Google Scholar]
- Gonzalez-Rizzo S, Crespi M, Frugier F (2006) The Medicago truncatula CRE1 cytokinin receptor regulates lateral root development and early symbiotic interaction with Sinorhizobium meliloti. Plant Cell 18: 2680–2693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris JM, Dickstein R (2010) Control of root architecture and nodulation by the LATD/NIP transporter. Plant Signal Behav 5: 1365–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PK, Tsay YF (2013) Two phloem nitrate transporters, NRT1.11 and NRT1.12, are important for redistributing xylem-borne nitrate to enhance plant growth. Plant Physiol 163: 844–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang NC, Chiang CS, Crawford NM, Tsay YF (1996) CHL1 encodes a component of the low-affinity nitrate uptake system in Arabidopsis and shows cell type-specific expression in roots. Plant Cell 8: 2183–2191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang NC, Liu KH, Lo HJ, Tsay YF (1999) Cloning and functional characterization of an Arabidopsis nitrate transporter gene that encodes a constitutive component of low-affinity uptake. Plant Cell 11: 1381–1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J, Watt M, Mathesius U (2012) The autoregulation gene SUNN mediates changes in root organ formation in response to nitrogen through alteration of shoot-to-root auxin transport. Plant Physiol 159: 489–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno Y, Hanada A, Chiba Y, Ichikawa T, Nakazawa M, Matsui M, Koshiba T, Kamiya Y, Seo M (2012) Identification of an abscisic acid transporter by functional screening using the receptor complex as a sensor. Proc Natl Acad Sci USA 109: 9653–9658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno Y, Kamiya Y, Seo M (2013) Nitrate does not compete with abscisic acid as a substrate of AtNPF4.6/NRT1.2/AIT1 in Arabidopsis. Plant Signal Behav 8: e26624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiba T, Feria-Bourrellier AB, Lafouge F, Lezhneva L, Boutet-Mercey S, Orsel M, Bréhaut V, Miller A, Daniel-Vedele F, Sakakibara H, et al. (2012) The Arabidopsis nitrate transporter NRT2.4 plays a double role in roots and shoots of nitrogen-starved plants. Plant Cell 24: 245–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krouk G, Lacombe B, Bielach A, Perrine-Walker F, Malinska K, Mounier E, Hoyerova K, Tillard P, Leon S, Ljung K, et al. (2010) Nitrate-regulated auxin transport by NRT1.1 defines a mechanism for nutrient sensing in plants. Dev Cell 18: 927–937 [DOI] [PubMed] [Google Scholar]
- Krouk G, Ruffel S, Gutiérrez RA, Gojon A, Crawford NM, Coruzzi GM, Lacombe B (2011) A framework integrating plant growth with hormones and nutrients. Trends Plant Sci 16: 178–182 [DOI] [PubMed] [Google Scholar]
- Lauter FR, Ninnemann O, Bucher M, Riesmeier JW, Frommer WB (1996) Preferential expression of an ammonium transporter and of two putative nitrate transporters in root hairs of tomato. Proc Natl Acad Sci USA 93: 8139–8144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Léran S, Varala K, Boyer JC, Chiurazzi M, Crawford N, Daniel-Vedele F, David L, Dickstein R, Fernandez E, Forde B, et al. (2014) A unified nomenclature of NITRATE TRANSPORTER 1/PEPTIDE TRANSPORTER family members in plants. Trends Plant Sci 19: 5–9 [DOI] [PubMed] [Google Scholar]
- Li JY, Fu YL, Pike SM, Bao J, Tian W, Zhang Y, Chen CZ, Zhang Y, Li HM, Huang J, et al. (2010) The Arabidopsis nitrate transporter NRT1.8 functions in nitrate removal from the xylem sap and mediates cadmium tolerance. Plant Cell 22: 1633–1646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Wang Y, Okamoto M, Crawford NM, Siddiqi MY, Glass AD (2007) Dissection of the AtNRT2.1:AtNRT2.2 inducible high-affinity nitrate transporter gene cluster. Plant Physiol 143: 425–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y, Harris JM (2005) Response of root branching to abscisic acid is correlated with nodule formation both in legumes and nonlegumes. Am J Bot 92: 1675–1683 [DOI] [PubMed] [Google Scholar]
- Liang Y, Mitchell DM, Harris JM (2007) Abscisic acid rescues the root meristem defects of the Medicago truncatula latd mutant. Dev Biol 304: 297–307 [DOI] [PubMed] [Google Scholar]
- Lin CM, Koh S, Stacey G, Yu SM, Lin TY, Tsay YF (2000) Cloning and functional characterization of a constitutively expressed nitrate transporter gene, OsNRT1, from rice. Plant Physiol 122: 379–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SH, Kuo HF, Canivenc G, Lin CS, Lepetit M, Hsu PK, Tillard P, Lin HL, Wang YY, Tsai CB, et al. (2008) Mutation of the Arabidopsis NRT1.5 nitrate transporter causes defective root-to-shoot nitrate transport. Plant Cell 20: 2514–2528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linkohr BI, Williamson LC, Fitter AH, Leyser HM (2002) Nitrate and phosphate availability and distribution have different effects on root system architecture of Arabidopsis. Plant J 29: 751–760 [DOI] [PubMed] [Google Scholar]
- Liu KH, Huang CY, Tsay YF (1999) CHL1 is a dual-affinity nitrate transporter of Arabidopsis involved in multiple phases of nitrate uptake. Plant Cell 11: 865–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morère-Le Paven MC, Viau L, Hamon A, Vandecasteele C, Pellizzaro A, Bourdin C, Laffont C, Lapied B, Lepetit M, Frugier F, et al. (2011) Characterization of a dual-affinity nitrate transporter MtNRT1.3 in the model legume Medicago truncatula. J Exp Bot 62: 5595–5605 [DOI] [PubMed] [Google Scholar]
- Mounier E, Pervent M, Ljung K, Gojon A, Nacry P (2014) Auxin-mediated nitrate signalling by NRT1.1 participates in the adaptive response of Arabidopsis root architecture to the spatial heterogeneity of nitrate availability. Plant Cell Environ 37: 162–174 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Nacry P, Bouguyon E, Gojon A (2013) Nitrogen acquisition by roots: physiological and developmental mechanisms ensuring plant adaptation to a fluctuating resource. Plant Soil 370: 1–29 [Google Scholar]
- Planchet E, Rannou O, Ricoult C, Boutet-Mercey S, Maia-Grondard A, Limami AM (2011) Nitrogen metabolism responses to water deficit act through both abscisic acid (ABA)-dependent and independent pathways in Medicago truncatula during post-germination. J Exp Bot 62: 605–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remans T, Nacry P, Pervent M, Filleur S, Diatloff E, Mounier E, Tillard P, Forde BG, Gojon A (2006) The Arabidopsis NRT1.1 transporter participates in the signaling pathway triggering root colonization of nitrate-rich patches. Proc Natl Acad Sci USA 103: 19206–19211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renard M, Alkhalfioui F, Schmitt-Keichinger C, Ritzenthaler C, Montrichard F (2011) Identification and characterization of thioredoxin h isoforms differentially expressed in germinating seeds of the model legume Medicago truncatula. Plant Physiol 155: 1113–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheible WR, Gonzalez-Fontes A, Lauerer M, Muller-Rober B, Caboche M, Stitt M (1997) Nitrate acts as a signal to induce organic acid metabolism and repress starch metabolism in tobacco. Plant Cell 9: 783–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqi MY, Glass AD, Ruth TJ, Rufty TW (1990) Studies of the uptake of nitrate in barley: I. Kinetics of 13NO3 influx. Plant Physiol 93: 1426–1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signora L, De Smet I, Foyer CH, Zhang H (2001) ABA plays a central role in mediating the regulatory effects of nitrate on root branching in Arabidopsis. Plant J 28: 655–662 [DOI] [PubMed] [Google Scholar]
- Tian Q, Chen F, Liu J, Zhang F, Mi G (2008) Inhibition of maize root growth by high nitrate supply is correlated with reduced IAA levels in roots. J Plant Physiol 165: 942–951 [DOI] [PubMed] [Google Scholar]
- Trinh TH, Ratet P, Kondorosi E, Durand P, Kamaté K, Bauer P, Kondorosi A (1998) Rapid and efficient transformation of diploid Medicago truncatula and Medicago sativa spp. falcata lines improved in somatic embryogenesis. Plant Cell Rep 17: 345–355 [DOI] [PubMed] [Google Scholar]
- Tsay YF, Schroeder JI, Feldmann KA, Crawford NM (1993) The herbicide sensitivity gene CHL1 of Arabidopsis encodes a nitrate-inducible nitrate transporter. Cell 72: 705–713 [DOI] [PubMed] [Google Scholar]
- Vidal EA, Araus V, Lu C, Parry G, Green PJ, Coruzzi GM, Gutiérrez RA (2010a) Nitrate-responsive miR393/AFB3 regulatory module controls root system architecture in Arabidopsis thaliana. Proc Natl Acad Sci USA 107: 4477–4482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal EA, Tamayo KP, Gutierrez RA (2010b) Gene networks for nitrogen sensing, signaling, and response in Arabidopsis thaliana. Wiley Interdiscip Rev Syst Biol Med 2: 683–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walch-Liu P, Forde BG (2008) Nitrate signalling mediated by the NRT1.1 nitrate transporter antagonises L-glutamate-induced changes in root architecture. Plant J 54: 820–828 [DOI] [PubMed] [Google Scholar]
- Walch-Liu P, Ivanov II, Filleur S, Gan Y, Remans T, Forde BG (2006) Nitrogen regulation of root branching. Ann Bot (Lond) 97: 875–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Okamoto M, Xing X, Crawford NM (2003) Microarray analysis of the nitrate response in Arabidopsis roots and shoots reveals over 1,000 rapidly responding genes and new linkages to glucose, trehalose-6-phosphate, iron, and sulfate metabolism. Plant Physiol 132: 556–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YY, Hsu PK, Tsay YF (2012) Uptake, allocation and signaling of nitrate. Trends Plant Sci 17: 458–467 [DOI] [PubMed] [Google Scholar]
- Wang YY, Tsay YF (2011) Arabidopsis nitrate transporter NRT1.9 is important in phloem nitrate transport. Plant Cell 23: 1945–1957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yendrek CR, Lee YC, Morris V, Liang Y, Pislariu CI, Burkart G, Meckfessel MH, Salehin M, Kessler H, Wessler H, et al. (2010) A putative transporter is essential for integrating nutrient and hormone signaling with lateral root growth and nodule development in Medicago truncatula. Plant J 62: 100–112 [DOI] [PubMed] [Google Scholar]
- Zhang C, Bousquet A, Harris JM (2014) Abscisic acid and LATERAL ROOT ORGAN DEFECTIVE/NUMEROUS INFECTIONS AND POLYPHENOLICS modulate root elongation via reactive oxygen species in Medicago truncatula. Plant Physiol 166: 644–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Forde BG (1998) An Arabidopsis MADS box gene that controls nutrient-induced changes in root architecture. Science 279: 407–409 [DOI] [PubMed] [Google Scholar]
- Zhang H, Forde BG (2000) Regulation of Arabidopsis root development by nitrate availability. J Exp Bot 51: 51–59 [PubMed] [Google Scholar]
- Zhang H, Jennings A, Barlow PW, Forde BG (1999) Dual pathways for regulation of root branching by nitrate. Proc Natl Acad Sci USA 96: 6529–6534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Rong H, Pilbeam D (2007) Signalling mechanisms underlying the morphological responses of the root system to nitrogen in Arabidopsis thaliana. J Exp Bot 58: 2329–2338 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.