Abstract
Many neurodegenerative diseases, such as frontotemporal dementia (FTD), are associated with behavioral deficits, but the anatomical and molecular bases remain poorly understood. Here we show that forebrain-specific expression of FTD-associated mutant CHMP2B causes several age-dependent neurodegenerative phenotypes, including social behavioral impairments. The social deficits were accompanied by a change in AMPA receptor (AMPAR) composition, leading to imbalance between Ca2+-permeable and -impermeable AMPARs. Expression of most AMPAR subunits was regulated by the brain-enriched microRNA (miR-124), whose abundance was markedly decreased in the superficial layers of cerebral cortex of FTD mice. We found similar changes in miR-124 and AMPAR levels in the frontal cortex and iPSC-derived neurons of subjects with behavioral variant FTD. Moreover, miR-124 expression in the medial prefrontal cortex decreased AMPAR levels and partially rescued behavioral deficits. Knockdown of Gria2 also alleviated social impairments in FTD mice. Our results identify a novel mechanism involving miR-124 and AMAPRs in regulating social behavior in FTD and suggest a potential therapeutic avenue.
Keywords: AMPA receptor, CHMP2B, frontotemporal dementia, microRNA, neurodegeneration, social behavior
Different neurodegenerative diseases are characterized by progressive dysfunction and loss of specific neuronal populations, often resulting in various behavioral abnormalities1-3. The molecular mechanisms of these impairments, and the neural circuits involved, are poorly understood. Frontotemporal dementia (FTD) is the second most common cause of dementia before 65 years of age. More than 50% of subjects with FTD have the behavioral variant, characterized by marked changes in personality and social behavior4. FTD has a strong genetic component; about 40% of subjects have a family history of the disease5. FTD is linked clinically, pathologically, and molecularly to amyotrophic lateral sclerosis6,7.
Mutations in several seemingly unrelated genes cause familial FTD6,7. However, the clinical outcome in subjects with FTD carrying different mutations is often similar, suggesting that the same neural circuits are affected. Mutations in charged multivesicular body protein 2B (CHMP2B) are especially interesting, as they are implicated not only in FTD and amyotrophic lateral sclerosis8,9 but also recently in early-onset Alzheimer’s disease10. CHMP2B encodes a component of the endosomal sorting complexes required for transport-III (ESCRT-III), which functions in the endosomal-lysosomal and autophagy pathways11,12. The mutation that causes FTD results in a C-terminally truncated CHMP2B (CHMP2BIntron5)9 and a number of studies suggest a gain-of toxic function unique to the CHMP2BIntron5 isoform13-16.
Although the molecular basis of the social deficits associated with FTD is unknown, microRNAs (miRNAs), a class of small noncoding RNAs, are important contributors to neurodegeneration17,18. In that regard, miR-124 is an attractive candidate because it is evolutionary conserved and one of the most abundant miRNAs in the brain. Although miR-124 has well-established functions during neuronal development19, its specific roles in neurodegeneration are poorly understood17,18. Here, through a novel mouse model of FTD with the CHMP2BIntron5 mutation and studies on iPSC-derived human neurons and brain tissues of behavioral variant FTD (bvFTD), we uncovered unexpected contributions of miR-124 and AMPA receptors (AMPARs) to FTD-associated social impairments.
RESULTS
A novel mouse model of FTD exhibits deficits in sociability
To examine the molecular basis of behavioral abnormalities in FTD, we generated a novel transgenic mouse model expressing, under the control of the tetracycline promoter, CHMP2BIntron5 (Fig. 1a). When crossed with a reporter line (TetO-GFP), CamkIIa-tTA drives GFP expression starting around E18 in forebrain neurons (Supplementary Fig. 1a). We then crossed CaMKIIa-tTA mice with different CHMP2BWT or CHMP2BIntron5 founder lines. Transgene expression pattern in tTA:CHMP2BWT and tTA:CHMP2BIntron5 mice was very similar to that of the reporter GFP (Supplementary Fig. 1b).
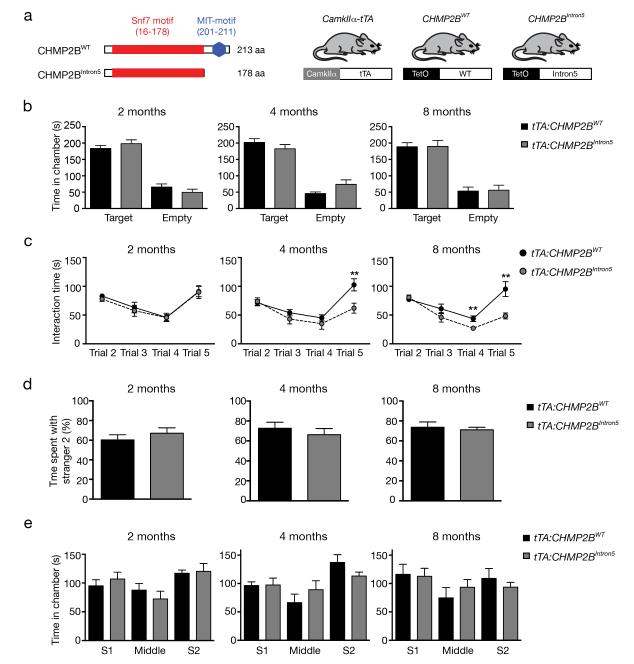
Figure 1.
Age-dependent deficits in sociability in tTA:CHMP2BIntron5 mice. (a) Schematic representation of the wildtype and mutant CHMP2B proteins and the transgenic mouse strains used. (b) Social recognition, namely total time spent in the target chamber and empty chamber in Trial 2 at different ages, is measured in tTA:CHMP2BIntron5 mice (n = 11 mice per genotype, p > 0.6 by two-sided t test). (c) Sociability of tTA:CHMP2BWT and tTA:CHMP2BIntron5 mice are measured based on interaction time (n = 11 mice per genotype. **: P < 0.01 by two-sided t test). (d) Proportion of interaction time spent with Stranger 2 in Trial 5 was measured as an indicator for social memory and novelty in tTA:CHMP2BWT and tTA:CHMP2BIntron5 mice (n = 11 mice per genotype; P > 0.5 by two-sided t test). All values are mean ± s.e.m. (e) Time spent in each chamber during Trial 5 was quantified as the exploration patterns (n = 11 mice per genotype; P > 0.5 by two-sided t test). All values are mean ± s.e.m.
The extent of transgene expression was modest at both the mRNA level (Supplementary Fig. 1c) and the protein level (Supplementary Fig. 1e) in tTA:CHMP2BWT and tTA:CHMP2BIntron5 mice. We selected tTA:CHMP2BWT line 3 and tTA:CHMP2BIntron5 line 3 for further analysis in most experiments because of their equal transgene expression at the mRNA level (Supplementary Fig. 1c) and because CHMP2BIntron5 and CHMP2BWT proteins are equally stable15. Moreover, a doxycycline-containing diet repressed transgene transcription (Supplementary Fig. 1d).
To determine whether our mice have disease-relevant behavioral deficits, we first used the resident-intruder test20. Although tTA:CHMP2BIntron5 mice performed similarly to tTA:CHMP2BWT mice at the beginning of the test, their interaction times at the last two time points decreased (Supplementary Fig. 2a), suggesting specific defects in this social behavior. To further characterize the social behavioral deficits, we used a modified version of the three-chamber social paradigm21. In Trial 2, both tTA:CHMP2BWT and tTA:CHMP2BIntron5 mice spent equally more time in the target chamber that contains Stranger 1 than in the empty chamber (Fig. 1b), indicating that CHMP2BIntron5 expression does not perturb the ability to recognize a conspecific mouse.
To quantify sociability, we measured the time test mice spent in close interaction with social partners across trials. In Trial 2, both tTA:CHMP2BWT and tTA:CHMP2BIntron5 mice showed equal sociability, which gradually decreased in later trials, suggesting progressive loss of social interest in the familiar mouse (Fig. 1c). However, tTA:CHMP2BIntron5 mice had a more pronounced, age-dependent decrease in sociability, particularly in Trial 5, when both social partners were present (Fig. 1c). We also observed a similar age-dependence in the elevated plus maze (Supplementary Fig. 2c). In Trial 5, the proportion of time tTA:CHMP2BIntron5 mice spent interacting with Stanger 2 was similar to that of tTA:CHMP2BWT mice, suggesting that they distinguished Stranger 1 from Stranger 2 equally well (Fig. 1d). Moreover, time spent in each chamber during Trial 5 did not differ between tTA:CHMP2BIntron5 and tTA:CHMP2BWT mice (Fig. 1e), suggesting that social behavior but not exploration pattern is specifically compromised in mutant mice. Moreover, in a novel object recognition task in which exploration can be assessed independently of social cues, the times exploring identical objects during the familiarization phase as well as the familiar and novel object during the test phase were similar in tTA:CHMP2BIntron5 and tTA:CHMP2BWT mice (Supplementary Fig. 2d), confirming the specificity of the social deficits.
The selective effect of CHMP2BIntron5 is also supported by the lack of major deficiency in locomotion (Supplementary Fig. 2e). Moreover, it is unlikely that the social deficits were due to perturbed olfaction, as we did not observe gross morphological defects in the olfactory bulb (Supplementary Fig. 3a,b). Moreover, tTA:CHMP2BIntron5 showed no differences from tTA:CHMP2BWT mice at 8 months of age in the time spent with either two supposedly preferred scents (cinnamon and vanilla) or an aversive scent (2-methyl butyrate) (Supplementary Fig. 2f), suggesting that their olfactory discrimination is not disrupted.
Thus, the FTD mice have a selective impairment in sociability. Moreover, seven of 36 old tTA:CHMP2BIntron5 mice (age 14-20 months) had cutaneous lesions compatible with excessive grooming, suggesting an obsessive-compulsive-like behavior (Supplementary Fig. 2b). Together, these results indicate that expression of CHMP2BIntron5 in the mouse forebrain causes age-dependent behavioral deficits that recapitulate some of the clinical symptoms of bvFTD.
AMPAR composition is altered in the cortex of tTA:CHMP2BIntron5 mice
To investigate the cellular and molecular bases of the behavioral deficits, we found that at 8 months of age, forebrain structures of tTA:CHMP2BIntron5 mice were similar to those of tTA:CHMP2BWT mice (Supplementary Fig. 3a,b) and showed no obvious neuronal loss (Supplementary Fig. 3c,d). However, tTA:CHMP2BInton5 mice had some FTD-like histopathological features, including astrogliosis, ubiquitin deposits, and an increase of p62 in insoluble fraction (Supplementary Fig. 4a-d). In addition, in pyramidal neurons in the superficial layers (II and III) of the medial prefrontal cortex (mPFC)—which are most vulnerable in subjects with FTD22,23—spine number and density were greater in tTA:CHMP2BInton5 than in tTA:CHMP2BWT mice (Supplementary Fig. 3e,f). This increase was largely due to “thin” spines, which are considered immature24-26. We did not observe spine changes in hippocampal CA3 pyramidal neurons (Supplementary Fig. 3f), consistent with a greater susceptibility of certain neuronal populations in the mPFC.
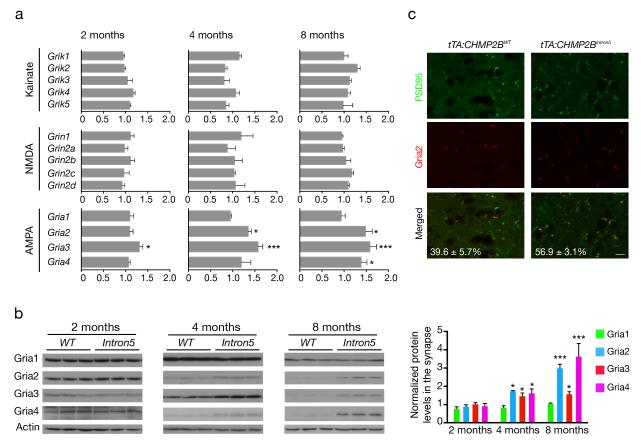
To further examine synaptic defects, we first analyzed glutamate receptor levels and composition. qRT-PCR analysis of the various NMDA and kainate subunits showed no apparent differences between tTA:CHMP2BIntron5 and tTA:CHMP2BWTmice (Fig. 2a). However, in tTA:CHMP2BIntron5 mice, mRNAs encoding AMPAR subunits Gria2, Gria3, and Gria4 but not Gria1 were upregulated at 4 and 8 months of age (Fig. 2a), correlating with the age-dependent defects in sociability. Then we analyzed postsynaptic density (PSD) fractions from the cortex of tTA:CHMP2BIntron5 and tTA:CHMP2BWT mice. Interestingly, NMDA subunits and Gria1 were maintained at similar levels in tTA:CHMP2BIntron5 mice at all time points examined, whereas Gria2, Gria3, and Gria4 levels were increased substantially in mice aged 4 months and older (Fig. 2b and Supplementary Fig. 5a). Moreover, tTA:CHMP2BIntron5 mice at 8 months of age had a higher number of Gria2+PSD95+ (Fig. 2c) and Gria4+PSD95+ (Supplementary Fig. 5b) puncta in the mPFC, confirming the increased synaptic content of these subunits in situ. Thus, an age-dependent dysregulation of AMPAR subunit composition correlates with the onset of behavioral deficits in tTA:CHMP2BIntron5 mice.
Figure 2.
Biochemical characterization of glutamate receptor subunits in the cortex of tTA:CHMP2BIntron5 mice. (a) Quantitative RT-PCR analysis of mRNA levels of different glutamate receptor subunits (kainate, NMDA, and AMPA) in tTA:CHMP2BIntron5 and tTA:CHMP2BWT mice (n = 3 mice per genotype; *: P < 0.05, ***: P < 0.001 by two-sided t test). (b) Western blot analysis of the expression levels of AMPAR subunits in the PSD fractions of tTA:CHMP2BWT and tTA:CHMP2BIntron5 mice of different ages (n = 3 mice per genotype, *: P < 0.05, ***: P < 0.001 by two-sided t test). (c) Double-immunostaining analysis and quantification of PSD95-positive (green) and Gria2-positive (red) puncta in the mPFC (layers II and III) of 8-month-old mice (n = 3 mice per genotype, P < 0.02 by two-sided t test). Scale bar, 200 μm. All values are mean ± s.e.m.
AMPAR function in the mPFC is altered in tTA:CHMP2BIntron5 mice
We next sought to link altered postsynaptic AMPARs to social defects in tTA:CHMP2BIntron5 mice. In 8-month old tTA:CHMP2BWT mice, intraperitoneal injection of NBQX (50 mg kg−1), a general AMPAR antagonist27, did not modify sociability (Fig. 3a). In 8-month-old tTA:CHMP2BIntron5 mice, however, NBQX increased sociability, suggesting that an abnormal AMPAR activity might underlie the social defects in these mice (Fig. 3a). Injections of saline or the NMDA receptor antagonist AP5 (10 mg kg−1) did not affect sociability (data not shown) in either tTA:CHMP2BIntron5 or tTA:CHMP2BWT mice. Thus, these results, together with the changes in AMPAR composition in the cortex of tTA:CHMP2BIntron5 mice (Fig. 2), suggest that impaired social behaviors result from postsynaptic perturbation of synaptic AMPARs.
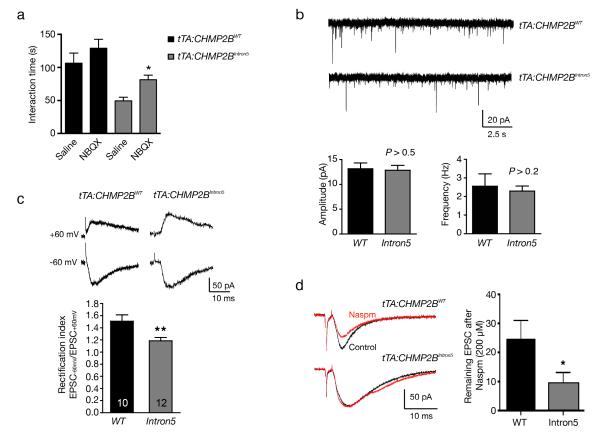
Figure 3.
Functional consequences of altered AMPAR composition in tTA:CHMP2BIntron5 mice. (a) Behavioral consequences of AMPAR inhibitor NBQX in tTA:CHMP2BWT and tTA:CHMP2BIntron5 mice at 8 months of age (n = 10 for all groups except n = 13 for tTA:CHMP2BWT saline; *: P < 0.05 by two-sided Kruskal-Wallis test corrected for multiple comparisons). (b) Whole-cell patch-clamp recordings of mEPSCs in pyramidal neurons in the mPFC. The amplitude and frequency of these miniature events were compared between tTA:CHMP2BWT mice (n = 11) and tTA:CHMP2BIntron5 mice (n = 12, P > 0.1 by two-sided t test). (c) The rectification index, calculated as EPSC-60/EPSC+60, for AMPAR-mediated EPSCs, was measured (n = 10 for tTA:CHMP2BWT mice and n = 12 for tTA:CHMP2BIntron5 mice; **: P < 0.01 by two-sided t test). (d) Decreased sensitivity of evoked EPSCs to Naspm (200 μM) inhibition in pyramidal neurons expressing CHMP2BIntron5. Representative EPSC traces at −60 mV before (Control) and after Naspm application are shown on the left (n = 8 mice for tTA:CHMP2BWT and n = 9 for tTA:CHMP2BIntron5. *: P < 0.05 by two-sided t test). Mice used in b-d were 15-16 weeks of age. All values are mean ± s.e.m.
To further investigate AMPAR alterations at the functional level in FTD mice, we examined the electrophysiological properties of synapses in layers II and III made on apical dendrites of layer V pyramidal neurons in the mPFC in acute brain slices from 15-16-week-old mice. The amplitude or frequency of AMPAR-mediated miniature excitatory postsynaptic currents (AMPAR-mEPSCs) was not altered in tTA:CHMP2BIntron5 mice (Fig. 3b), suggesting that the presynaptic release mechanism or the overall synaptic strength of these neurons at the resting level was largely intact in the mutant mice.
Most AMPARs in principal excitatory neurons contain Gria2 and hence are Ca2+-impermeable28. However, some AMPARs lack Gria229, causing these receptors to be Ca2+-permeable and exhibit an inwardly rectifying current-voltage relationship due to voltage-dependent blockade by intracellular polyamines30,31. Since Gria2 expression was increased in tTA:CHMP2BIntron5 mice (Fig. 2), one would expect a functional increase in the proportion of Gria2-containing AMPARs in mutant synapses. To test this hypothesis, we recorded AMPAR-mediated EPSCs in response to stimuli delivered to layer II and III at −60 and +60 mV and calculated the rectification index, EPSC−60mV/EPSC+60mV. Synapses of tTA:CHMP2BIntron5 mice had a lower rectification index than those of tTA:CHMP2BWT mice (Fig. 3c), suggesting an increased proportion of Gria2-containing receptors. Moreover, at a concentration of 200 μM, Naspm (a specific inhibitor of Ca2+-permeable AMPARs)32 partially blocked AMPAR-mediated currents in tTA:CHMP2BWT neurons, indicating the existence of a substantial amount of AMPARs lacking Gria2 under control conditions. In contrast, EPSCs of tTA:CHMP2BIntron5 neurons were less sensitive to inhibition by Naspm (Fig. 3d), again indicating an altered AMPAR subunit composition characterized by an enrichment of Gria2-containing Ca2+-impermeable receptors. Thus, our data suggest that Gria2-lacking AMPARs exist in mPFC neurons of adult tTA:CHMP2BWT mice and that these native Ca2+-permeable receptors are replaced by Gria2-containing, Ca2+-impermeable AMPARs in tTA:CHMP2BIntron5 mice.
MiR-124 regulates AMPAR levels and is decreased in the cortex of tTA:CHMP2BIntron5 mice
We next examined mechanisms of the upregulation of AMPARs in tTA:CHMP2BIntron5 mice. We turned our attention to miRNAs, as they control gene expression at the posttranscriptional level33 and are essential regulators of synaptic function34. Furthermore, each miRNA can regulate multiple mRNA targets that may contribute to complex processes, such as sociability. We examined the expression levels of a number of brain-enriched miRNAs in the cortex in tTA:CHMP2BIntron5 mice at one month and at one year of age. At both time points, many miRNAs were expressed in the cortex at similar levels in tTA:CHMP2BIntron5 mice and tTA:CHMP2BWT mice (Supplementary Fig. 6b); however, at one year, some miRNAs were downregulated in tTA:CHMP2BIntron5 mice, especially miR-124, which is one of the most abundant miRNAs in the brain (Supplementary Fig. 6a,b). miR-124 downregulation was specific to the expression of the FTD-causing CHMP2BIntron5, as these miRNAs remained unchanged in the cerebellum, where the transgene was not expressed (Supplementary Fig. 6a). Importantly, miR-124 was also downregulated to a lesser extent in the cortex of two other independent transgenic lines expressing lower level of CHMP2BIntron5 (Supplementary Fig. 6c). In contrast, variable levels of CHMP2BWT expression in the other mouse lines did not affect miR-124 abundance (Supplementary Fig. 6c). Moreover, when CHMP2BIntron5 expression was turned off in tTA:CHMP2BIntron5 mice fed a diet containing doxycycline (Supplementary Fig. 1d), their cortical miR-124 expression was identical to that in tTA:CHMP2BWT mice fed a regular diet or a diet containing doxycycline (Supplementary Fig. 7a). These results support a specific correlation between CHMP2BIntron5 expression and miR-124 reduction.
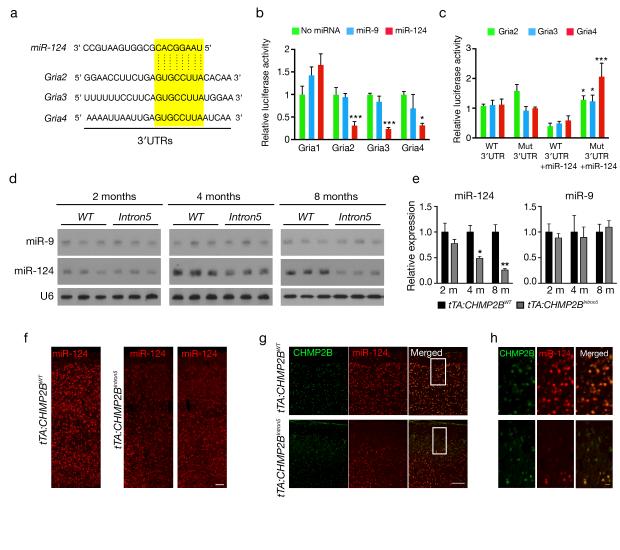
Computer algorithms (Targetscan, PicTar, miRanda) indicate that three AMPAR subunits, Gria2, Gria3, and Gria4, are potential targets of miR-124 (Fig. 4a). Thus, downregulation of miR-124 might contribute to the upregulation of these AMPARs in tTA:CHMP2BIntron5 mice. Indeed, in transfected HEK293 cells, miR-124 markedly suppressed the expression of the reporter gene with the 3′UTRs of Gria2, Gria3, and Gria4 but not of Gria1 (Fig. 4b). Their expression was not suppressed when we mutated the miR-124 binding site (Fig. 4c). Similarly, in the cortex of tTA:CHMP2BIntron5 mice, miR-9 levels remained unchanged, whereas miR-124 was downregulated in an age-dependent manner (Fig. 4d,e), correlating with the age-dependent increase in Gria2, Gria3, and Gria4 levels (Fig. 2) and the social behavioral deficits (Fig. 1).
Figure 4.
MiR-124 targets AMPAR subunits, and its expression is reduced in the cortex of tTA:CHMP2BIntron5 mice. (a) Alignment of potential binding sites for miR-124 in the 3′UTRs of Gria2, Gria3, and Gria4 mRNAs. (b) Direct interaction between 3′UTRs of Gria2, Gria3, and Gria4 mRNAs and miR-124 (n > 4 assays, *: P < 0.05, ***: P < 0.001 by two-sided t test). (c) The effects of miR-124 on the expression of luciferase-containing subunit 3′UTRs in which we mutated the miR-124 binding site. n = 4 assays, *: P < 0.05, **: P < 0.001 by two-sided t test. (d) Northern blot analysis of miR-9 and miR-124 expression levels in the cortex of tTA:CHMP2BWT and tTA:CHMP2BIntron5 mice at different ages. (e) Quantification of the northern blots in panel d (n = 3 mice per group; *: P < 0.05, **: P < 0.01 by two-sided t test). (f) Fluorescence in situ hybridization of miR-124 in brain sections of tTA:CHMP2BWT and tTA:CHMP2BIntron5 mice at 8 months of age. (g) Double-fluorescence in situ hybridization staining of miR-124 (red) in CHMP2BIntron5-expressing cells (green) in the cortex at 8 months of age. Scale bar, 150 μm in f and g. (h) Higher magnification of the boxed area in G. Scar bar, 20 μm. Values are mean ± s.e.m. in b and c and mean ± s.d. in e.
Consistent with previous studies35, miR-124 was widely expressed in all layers of the cortex of 2-month-old mice (Supplementary Fig. 7b). In contrast, miR-124 expression was markedly decreased in the cortex of tTA:CHMP2BIntron5 mice at 8 months of age, and this decrease seemed to be most pronounced in the layers II and III (Fig. 4f). Most neurons expressing miR-124 at low levels in the superficial layers were also strongly positive for CHMP2BIntron5 (Fig. 4g,h). Thus, expression of CHMP2BIntron5 triggers a progressive loss of miR-124 that is greater in superficial layers.
miR-124 and AMPARs are dysregulated in a subset of subjects with behavioral variant FTD
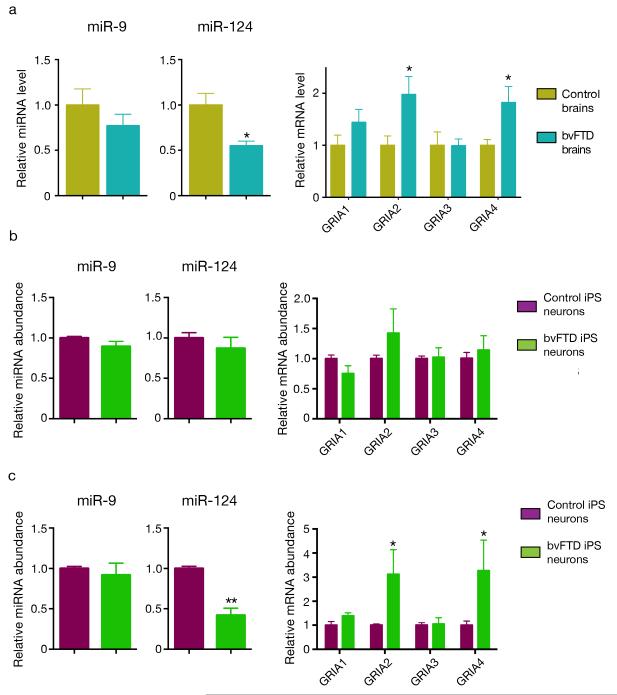
To confirm that our findings in mice are relevant to human disease, we next examined miR-124 and AMPAR levels in subjects with FTD. Since FTD is a heterogeneous disorder, we focused on the frontal cortex, a brain region critical for social behavior, in a subset of subjects with the behavioral variant (bvFTD), whose clinical presentation is closest to the phenotypes in our mice. In samples from two brain banks, we found a decrease in miR-124, but not miR-9, and a concomitant upregulation of GRIA2 and GRIA4 in the frontal cortex of five bvFTD cases compared with five age-matched controls (Fig. 5a).
Figure 5.
miR-124 and AMPAR levels are altered in the frontal cortex of subjects with bvFTD and in cortical neurons derived from iPSC lines of three subjects with bvFTD. (a) Quantification of the abundance of miR-124, miR-9, and AMPAR transcripts by RT-PCR in the frontal cortex of subjects with bvFTD and controls. We normalized the content of AMPAR mRNAs against the geometric mean of four different neuronal-specific reference genes (MAP2, Enolase 2, GAP43, and PSD95). *: P < 0.05 by Mann-Whitney test. (b) Quantification of miR-124, miR-9, and AMPAR transcripts in 2-week old human neurons derived from 3 iPSC lines of two control individuals and 4 iPSC lines from three subjects with bvFTD36,37. P > 0.1 by two-sided t test). (c) The levels of miR-124, miR-9, and AMPAR transcripts were measured again in these human neurons at 8 weeks (*: P < 0.05, **: P < 0.01, ***: P < 0.001 by two-sided t test). All values are mean ± s.e.m.
Then we took advantage of induced pluripotent stem cell lines derived from three subjects with bvFTD we established previously36,37. The levels of miR-124 and AMPARs in 2-week-old neurons derived from four iPSC lines of three subjects with bvFTD were the same as those in controls (Fig. 5b). However, in 8-week old neurons of subjects with bvFTD, miR-124 was substantially reduced and some AMPAR subunit mRNAs were upregulated (Fig. 5c). This change is not due to culture condition since the percentages of different neuronal subtypes remained the same as those in cultures derived from controls (Supplementary Fig. 8a). As controls, the miR-9 level was not decreased (Fig. 5c) and we did not detect a change in NMDA or kainate receptor transcripts or neuronal subtype-specific markers (Supplementary Fig. 8a,b).
To further confirm this finding, we surveyed published genome-wide studies of miRNAs on frontal cortex samples from subjects with FTD38. Re-analysis of the data showed a decrease in miR-124 level in the frontal cortex of subjects with progranulin deficiency (FTD-GRN) (Supplementary Fig. 9a). Another published study, of a small number of subjects, also showed a trend toward a decrease39. Similarly, re-analysis of a published transcriptome profiling data40 revealed an increase in some AMPAR subunits, but not of some NMDA receptor subunits, in the frontal cortex but not the cerebellum of six subjects with FTD-GRN (Supplementary Fig. 9b,c). These data are relevant to our study since most subjects with FTD-GRN belong to the behavioral variant41. Together, these findings suggest that dysregulation of miR-124 and AMPARs discovered in our mouse model also occurs in the frontal cortex in a subset of subjects with bvFTD.
MiR-124 expression and Gria2 knockdown in the mPFC partially rescue social deficits in tTA:CHMP2BIntron5 mice
To determine whether miR-124 downregulation contributes to FTD-associated social deficits in tTA:CHMP2BIntron5 mice, we engineered an adeno-associated vector (AAV) to express miR-124 in vitro (Supplementary Fig. 10a) and in vivo (Fig. 6a). The mPFC controls most social behaviors in mice42, and we detected synaptic defects in that region in tTA:CHMP2BIntron5 mice (Fig. 2). Therefore, we injected an AAV driving miR-124 expression or a control AAV-GFP vector into the mPFC of 7-month-old mice.
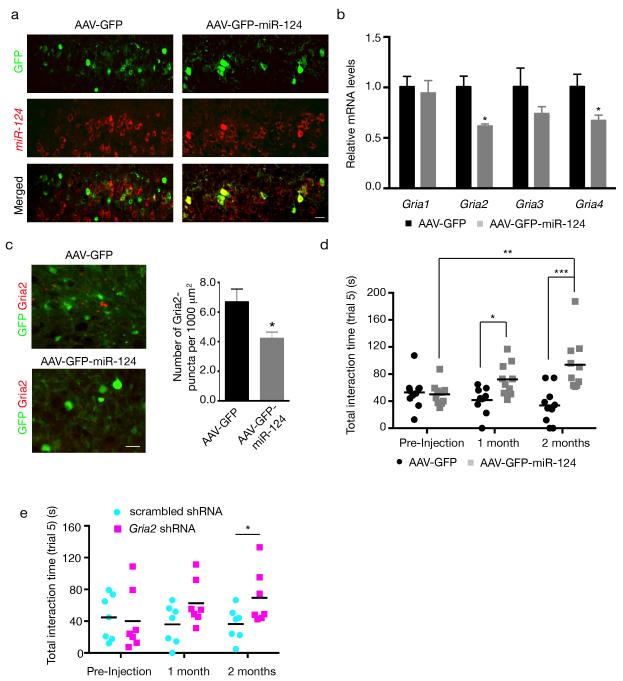
Figure 6.
miR-124 expression and Gria2 knockdown in the mPFC partially rescue social deficits in tTA:CHMP2BIntron5 mice. (a) Double staining for GFP and miR-124 in tTA:CHMP2BIntron5 mice (8 months) that received an intracranial injection of AAV-GFP (left panels) or AAV-GFP-miR-124 (right panels). To facilitate assessment of differences in miR-124 levels, we took pictures at the periphery of the injection site so infected and noninfected cells are present in the same field. miR-124 levels were similar in AAV-GFP-infected cells and noninfected neighboring cells. In contrast, AAV-GFP-miR-124 induced a robust increase in miR-124 expression. (b) qRT-PCR analysis of Gria transcripts in the mPFC of tTA:CHMP2BIntron5 mice 2 months after AAV injection (n = 5 AAV-GFP-infected mice and n = 4 AAV-GFP-miR-124-infected mice; *: P < 0.05 by two-sided t test). (c) Quantification of the number of Gria2-positive puncta in tTA:CHMP2BIntron5 mice 2 months after intracranial injection of AAV-GFP or AAV-GFP-miR-124 (n = 3 mice per genotype, *: P < 0.05 by two-sided t test). (d) Quantification of miR-124-dependent rescue of social deficits in tTA:CHMP2BIntron5 mice 1 and 2 months after bilateral injection. Sociability was significantly greater in mice injected with AAV-GFP-miR-124 in than in those injected with the control vector (n = 10 per AAV vector; *: P < 0.05, **: P < 0.01, ***: P < 0.001 by two-sided repeated-measures two-way ANOVA corrected for multiple comparison, Bonferroni test). (e) Quantification of the rescue effects of in vivo downregulation of Gria2 expression on social deficits in tTA:CHMP2BIntron5 mice (n = 7 per group; *: P < 0.05, by two-sided repeated-measures two-way ANOVA followed by Fisher’s LSD test). Scar bar, 20 μm. All values are mean ± s.e.m.
We first observed that AAV-mediated miR-124 expression reduced mRNA levels of Gria2 and Gria4 in the mPFC of tTA:CHMP2BIntron5 mice (Fig. 6b). The decrease was not due to the surgical procedure, since mRNA levels did not differ in the occipital cortex of the same mice (Supplementary Fig. 10b). Although we could not obtain enough material from these experiments for PSD fractionation, we did find a decrease in the number and the intensity of synaptic Gria2 puncta in brain sections (Fig. 6c). More importantly, AAV-GFP injection did not improve the sociability of tTA:CHMP2BIntron5 mice at either 1 or 2 months (Fig. 6d). In contrast, one month after injection of AAV-GFP-miR-124, the mice showed a substantial increase in sociability in Trial 5. These mice showed a further improvement at 2 months that was significantly better than the pre-injection level (P < 0.001), suggesting that the deficits were reversed (Fig. 6d). Finally, the time spent with Stranger 2 did not differ in AAV-GFP mice and those injected with AAV-GFP-miR-124 (Supplementary Fig. 10c), arguing for a role of miR-124 in specifically regulating sociability but not novelty recognition.
To further demonstrate that a decrease in Ca2+-impermeable AMPARs contributes to the social deficits in tTA:CHMP2BIntron5 mice, we constructed lentiviral vectors expressing scrambled short-hairpin RNA (shRNA) or shRNA directed against mouse Gria2 (Supplementary Fig. 11a). After lentivirus injection into the mPFC of 7-month-old tTA:CHMP2BIntron5 mice, downregulation of Gria2 expression partially rescued the social deficits (Fig. 6e). Interestingly, recognition of the novel mouse (Stranger 2) during this trial was not affected (Supplementary Fig. 11b), suggesting that Gria2 levels are critical for sociability but not all aspects of social behavior. Together, these results support the notion that alterations in AMPARs composition in the mPFC contribute to social defects in tTA:CHMP2BIntron5 mice.
DISCUSSION
In a novel mouse model of FTD harboring the CHMP2BIntron5 mutation, we found that downregulation of miR-124 causes a dysregulation in AMPAR composition and a selective impairment in sociability. AAV-mediated ectopic expression of miR-124 in the mPFC partially restored the levels of AMPAR subunits and rescued the social behavioral deficits. Knockdown of Gria2 also alleviated the social deficits in FTD mice, suggesting a key role for Ca2+-impermeable AMPARs. Similar alterations in miR-124 and AMPAR levels were also found in human cortical neurons derived from iPSCs of subjects with bvFTD and in the frontal cortex of a subset of subjects with bvFTD. Thus, the mechanisms uncovered in our mouse model of FTD may have general implications for understanding behavioral abnormalities in human subjects with the disease.
Marked changes in social behavior such as social withdrawal and obsessive-compulsive behaviors are common in subjects with bvFTD4,43. Social deficits similar to those in our tTA:CHMP2BIntron5 mice were also reported in progranulin happloinsufficient mice44, an independent FTD model. In addition, a number of FTD histopathological hallmarks were present in our mice and previously published mouse models14,44, suggesting that the anatomical and behavioral abnormalities in subjects with FTD can be reproduced in mice. Despite widespread forebrain expression of mutant CHMP2BIntron5 protein in our FTD mice, only sociability was impaired at an early age, indicating that specific circuits are particularly vulnerable. Consistent with this notion, neurons in some regions of the human prefrontal cortex are most susceptible in FTD45. Moreover, ectopic miR-124 expression or Gria2 knockdown in this cortical area partially rescued the social behavioral deficits.
Our findings suggest that AMPAR composition is regulated by miR-124 and altered in FTD mice. More importantly, we also found miR-124 downregulation and a concomitant increase in AMPARs levels in the frontal cortex and iPSC-derived cortical neurons from subjects with bvFTD. AMPARs have been implicated in social behaviors. For instance, Gria3 knockout mice show increased aggression46. Modulating AMPAR activity in the mPFC can result in both upward and downward movement in the social hierarchy42. Our electrophysiological analysis suggested that FTD-related social behaviors are linked to an increase in Ca2+-impermeable AMPARs at excitatory synapses of PFC pyramidal neurons. These abnormally inserted receptors may interfere with synaptic Ca2+ signaling or impair synaptic efficacy and plasticity47, leading to structural alterations in dendritic spines and behavioral deficits in social interaction. We speculate these molecular changes may also contribute to the early behavioral abnormalities in subjects with FTD as well.
MiRNAs are deregulated in many neurodegenerative disorders18. The exact molecular link between CHMP2BIntron5 mutation and miR-124 reduction remains to be elucidated. However, previous works implicated a role of ESCRTs in the miRNA pathway48,49, raising the possibility that CHMP2BIntron5 mutation could affect miR-124 stability. Only six of 91 neurotransmitter receptor subunit mRNAs are predicted targets of miR-124, including three of four known AMPAR subunits (Supplementary Table 1). Thus, miR-124 may play a unique role in fine-tuning glutamate neurotransmission by controlling the expression and composition of AMPARs, consistent with the inverse correlation between miR-124 and Gria2 levels reported here and by others50. Moreover, our finding that ectopic expression of miR-124 or Gria2 knockdown partially rescues behavioral deficits in early stages of FTD may suggest a potentially beneficial therapeutic approach before neuronal cell loss becomes apparent and irreversible.
ONLINE METHODS
Mice
All procedures involving mice were approved by the University of Massachusetts Medical School Institutional Animal Care and Use Committee. We cloned CHMP2BWT and CHMP2BIntron5 into the NotI and SalI sites of pTRE-Tight-BI-ZsGreen1 mammalian expression vector (Clontech). We linearized pTRE-CHMP2BWT vector with BsrBI, and purified DNA for microinjection. Since no adequate restriction enzymes were present on the pTRE-CHMP2BIntron5 plasmid, we amplified the cassette by PCR (TRESacIIFwd 5′-CCGCGGCCACCTGACGTCG GCAGT GAA-3′ and TREApaIRV 5′-GGGCCCCGAGTCAGTGAGCGAGGAAGCTC-3′) and subcloned into pCR2.1 TOPO (Invitrogen). We linearized this construct with SacII and ApaI enzymes, and purified DNA for microinjection. For both CHMP2BWT and CHMP2BIntron5 constructs, ZsGreen coding sequence was lost after linearization. Injected eggs were transferred to female recipients, and F1 litters analyzed for transgene insertion by PCR with two specific sets of primers (forward primer: 5′-GCTCGTTTAGTGAACCGTCAG-3′; reverse primer: 5′-AGCTC GGGGCAGTGAAAA-3′ or forward primer 5′-ATGGCGTCCCTCTTCAAGAAGA AAAG-3′ and reverse primer 5′-GGGCCCCGAGTCAGTGAGCGAGGAAGCTC-3′).
We maintained germline-transmitting founders (three for CHMP2BWT and three for CHMP2BIntron5) as hemizygous lines by backcrossing to wildtype C57BL/6 mice. To drive transgene expression in the forebrain, we crossed CHMP2BWT and CHMP2BIntron5 transgenic mice with CaMKIIα-tTA mice (Jackson Laboratory 003010). We used both male and female double-transgenic offspring for all experiments.
Behavioral tests
We conducted all experiments between 9 am and 5 pm. We housed mice under specific-pathogen-free conditions in an animal facility with a regular 12-h light and dark cycle (light on at 7:00 am) under constant conditions (21 ± 1 °C; 60% humidity). Food and water were supplied ad libitum.
We conducted all behavioral studies using both male and female mice and analyzed data blind to genotypes. For all tasks, each mouse was selected randomly and then tested on the same day. We tested the first cohort of mice for sociability in the three-chamber social approach task. 2 d after the social test, we tested subjects in the open field and, 2 d later, in the elevated plus maze. We used independent cohorts of mice at 8 months of age for the other tests; these mice were not subjected to any other task.
Open field
We tested general exploratory locomotion in a novel open-field environment in 15 min sessions in a 40 × 40 × 40 cm white Plexiglas arena. The mouse was placed in the center of the arena and allowed to freely move while being recorded by an overhead camera. We analyzed behavior with an automated tracking system (Ethovision software, Noldus). The total distance traversed in the arena and the time spent in the center vs. the perimeter were automatically calculated.
Elevated plus maze
The elevated plus maze (Med Associates) consisted of two open arms and two closed arms situated opposite each other and separated by a 6 cm square center platform. Each runway is 6 cm wide × 35 cm long. The open arms have lips 0.5 cm high to prevent mice from slipping off the arm, and the closed arms are surrounded on three sides by 20 cm high walls. The floors and walls are black polypropylene. For each test, the mouse was placed in the center square facing the open arms and allowed to explore freely for 5 min. The number of entries in the open and closed arms and the times spent in the open arms, closed arms, and the center square were quantified by automated analysis of beam breaks.
Three-chamber social task
We evaluated sociability, social recognition, and response to social novelty by using the three-chamber task with minor modifications. Test mice were housed individually for 1 week before the test. We habituated the mice in the testing room for at least 1 min before the start of behavioral tasks. The social test apparatus consisted of a white Plexiglas box with removable floor and partitions dividing the box into three chambers with 5 cm openings between chambers. Target subjects (Stranger 1 and Stranger 2) were 10-14 week old males. We used wire cages to contain the stranger mice. We placed a paper cup on the top of the cage to prevent the test mice from climbing on the top of the wire cage. This task was carried out in five trials of 5 min each. After each trial, we returned the mouse to his home cage for 15 min.
Trial 1: the test mouse is placed to the middle chamber and left to explore the arena containing the empty wire cages for 5 min (habituation).
Trials 2–4: the mouse is placed in the middle chamber, but an unfamiliar mouse (Stranger 1) is placed into a wire cage in one of the side-chambers (the wire cage in the other side-chamber remains empty) (sociability, social learning acquisition).
Trial 5: a novel stranger mouse (Stranger 2) is placed in the previously empty wire cage and again the test mouse is left to explore for 5 min (sociability, social recognition memory).
We recorded and analyzed all the trials post hoc. We measured the time spent in each chamber and the time spent in close interaction with stranger mice for each trial. The following parameters were considered: (1) social recognition, defined as the ability to identify a conspecific (Stranger 1 versus the empty cage) in Trial 2 (spending more time in the side-chamber containing Stranger 1); (2) social novelty, defined as the ability to discriminate between a novel mouse (Stranger 2) and a familiar mouse (Stranger 1) in Trial 5 (spending more time interacting with Stranger 2); and (3) sociability, which reflects the motivation of the test mouse to spontaneously interact with target mice in Trials 2-5.
Social interaction in home cage (resident-intruder test)
We assessed social interaction in the home cage by standard protocols52. Briefly, we housed individual test mouse for 1 week before the task. Then, they were habituated for 30 min to the test room. In this task, a mouse in his home cage is allowed to freely roam in the absence of the cage top for 1 min. A novel juvenile (4-week-old) male intruder is then placed in the opposite corner as the resident subject and allowed to roam freely for 5 min. The task is recorded, and total physical interaction between the two mice is quantified visually; social interaction is scored as the time during which the resident mouse actively explores the intruder. We did not observe fighting, biting, or attacking in this task.
Olfactory testing
After habituation to empty cages with no bedding, we tested mice for odor discrimination or sensitivity, as described53. In the discrimination task, mice are challenged with a filter paper embedded with an attractive scent (vanilla or cinnamon), an aversive scent (2-methyl butyrate), or a neutral scent (water). For the sensitivity task, dilutions of the same scent (vanilla) are used. The mice are videotaped, and the time spent sniffing the filter paper during a 3 min period is calculated post hoc.
Novel object recognition task
We performed the object recognition task as described54 following the no-habituation paradigm. The testing set-up was the same as for open field experiment. We placed mice in the testing room for at least 30 min for habituation. Then, mice were placed into the set-up containing two identical plastic sample objects (cube) placed 5 cm from one of the walls and ~12 cm apart. The mice were allowed to explore the environment for 10 min, during which we recorded their movements. After this familiarization session, we cleaned the objects and open-field with 70% ethanol to eliminate any olfactory cues, and returned the mice to their home cage for 2 min. We then placed the mice again in the open field, but this time a novel object of similar size and complexity (cylinder instead of a cube) replaced one of the objects present during the familiarization session. The mice were allowed to explore the environment for 10 min (test session), after which we returned them to their cages. For both sessions, we placed mice into the open field, with the head positioned opposite the objects. Object exploration, defined as the duration of time in which the head of the mouse faced less than 2 cm from the object, was measured during the first 5 min of each session. We calculated the recognition index as the percentage of time spent exploring the novel object versus the total time spent exploring the objects.
Drug injections
We diluted NBQX and AP5 (both from Tocris) in 0.9% saline solution and intraperitoneally injected them 10 min before the start of the test. Doses were 50 mg kg−1 for NBQX and 10 mg kg−1 for AP5. We injected at least 10 mice per genotype with each drug, but each mouse received only a single injection. We quantified the total interaction time with both Stranger 1 and Stranger 2 (Trial 5). To repress transgene expression, we fed mice with a diet containing doxycycline (200 mg kg−1) (Bioserv). To ensure early repression, we maintained breeder mice on a doxycycline-containing diet. After weaning and genotyping, we fed mice with this diet until 8 months of age.
Golgi staining
We processed mouse brains for morphological assessment with the FD Rapid Golgi Stain Kit (FD NeuroTechnologies) according to the manufacturer’s protocol. We submerged mouse brains in impregnation solution for 14 d and then flash frozen in isopentane at −70 °C. Next, we cut 200 μm thick cryosections with a cryostat (Leica), mounted on gelatin-coated slides, rinsed in dH2O, and incubated in developing solution for 10 min. Sections were dehydrated with increasing concentrations of ethanol, cleared with xylene, and coverslipped with Permount mounting medium.
We examined the slides with a Nikon inverted microscope at the imaging core facility (University of Massachusetts Medical School). We took image stacks of 40-120 μm segments of apical dendrites on pyramidal neurons of layer II-III of the mPFC or in the CA3 region with a 100X oil-immersion lens (N.A. 1.45). For spine analysis, we quantified 15-25 dendritic segments (n = 3 mice per genotype). Total dendritic length was 1897 μm for tTA:CHMP2BIntron5 mice and 1702 μm for tTA:CHMP2BWT mice. All image stacks were first deconvolved (with the iterative three-dimensional (3D) deconvolution plug-in for Image J) using a point-spread function (PSF) as a reference (PSF plug-in for ImageJ). We imported images into NeuronStudio for 3D analysis of spine density and spine size4 and measured spines in 3D from the z-stacks. We calculated the density by dividing the total number of spines per 100 μm of dendrite. A spine was labeled thin if its head was < 0.6 μm in diameter and had a maximum length of at least twice the head diameter. We classified a spine as mushroom if its head diameter was > 0.6 μm. Stubby spines were those lacking a neck. We classified other spines as “other”.
AAV production and injections
We constructed AAV particles (serotype 9) as described56. As a control, we use an AAV-GFP vector so that no exogenous miRNA is expressed (limiting potentially off-target effects). We constructed AAV-GFP-miR-124 by inserting 500 bp of mouse pri-miR-124-1 into the 3′UTR of the GFP cassette of the control vector. Lentivirus expressing short hairpin RNA (shRNA) were from the University of Massachusetts RNAi core facility, which houses the complete collection of mouse lentiviral shRNA libraries from Open Biosystems57. We packed lentivirus particles as described58,59.
tTA:CHMP2BIntron5 and tTA:CHMP2BWT mice were deeply anesthetized with ketamine (100 mg kg−1) and xylazine (10 mg kg−1) and placed in a stereotaxic frame. The coordinates according to the mouse brain atlas of Paxinos & Franklin were: AP, 2.43 mm; ML, ± 0.28 mm; DV, −1.81mm relative to the bregma, angled 14° toward the midline in the coronal plane. We injected high-titer vector (0.8-1.2 × 1013; 1.5 μl) bilaterally. At the end of the behavioral experiments, we sectioned the brains of these mice to verify the accuracy and extent of transduction. We analyzed the data from mice with correct injections in the target area. To calculate transduction efficiency, we stained brains from mice infected with AAVs or lentivirus (n = 6 per vector) with antibody against GFP before microscopic examination. For each mouse, we quantified the area transduced on four 20-μm-thick sections (bregma +2.2 to +1.9 mm) using Image J. The mean area of transduction per slice was 0.43 ± 0.095 mm2 and 0.46 ± 0.087 mm2 for AAVs and lentivirus, respectively.
Electrophysiology
We conducted all experiments in accordance with the National Institutes of Health “Guidelines for the Care and Use of Laboratory Animals” and an approved animal protocol from the Harvard Medical Area Standing Committee on Animals.
We used tTA:CHMP2BWT and tTA:CHMP2BIntron5 mice at 15-16 weeks of age. The brains were rapidly removed and placed in ice-cold artificial CSF (ACSF) containing 126 mM NaCl, 2.5 mM KCl, 2.5 mM CaCl2, 1.2 mM MgCl2, 25 mM NaHCO3, 1.2 mM NaH2PO4, and 25 mM D-glucose. ACSF was saturated with 95% O2 and 5% CO2. We cut coronal cortical slices (300 μm) containing the mPFC with a Leica VT1200 Vibratome, and collected slices into a tissue incubator (Harvard Apparatus) and allowed them to recover for 1 h in a 32 °C water bath. We then incubated slices in oxygenated ACSF for 1 h at room temperature (21-23 °C) and transferred to a recording chamber continuously perfused with oxygenated ACSF.
We did whole-cell voltage-clamp recordings of individual layer V pyramidal neurons using an Axoclamp 2B or a Multiclamp 700B amplifier (Molecular Devices) under infrared differential interference contrast microscopy. We identified pyramidal neurons by their morphology and delivered presynaptic stimuli (0.033 Hz, 200 μs) with a concentric bipolar electrode (FHC) placed at layer II and III of the mPFC cortex. For recordings of evoked EPSCs and miniature EPSCs (mEPSCs), neurons were voltage clamped at −60 mV unless indicated otherwise. We recorded evoked EPSCs every 30 s. The superfusion medium contained picrotoxin (100 μM) to block GABAA receptor-mediated synaptic responses. We added tetrodotoxin (1 μM, Sigma) and AP5 (2-amino-5-phosphonopentanoic acid, 50 μM, Abcam Biochemicals) during recordings of mEPSCs. We filled recording pipettes with solution containing 142 mM Cs-gluconate, 8 mM NaCl, 10 mM HEPES, 0.4 mM EGTA, 2.5 mM QX-314 [N-(2,6-dimethylphenylcarbamoylmethyl)triethylammonium bromide], 2 mM Mg-ATP, and 0.25 mM GTP-Tris, pH 7.25. For all recordings, temperature was maintained at 32°C with a temperature controller (Warner Instruments). For the Naspm (1-naphthylacetyl spermine trihydrochloride) sensitivity assay, after stable EPSCs were obtained for 10 min as baseline, we delivered the drug to the bath for 10-15 min with a gravity-driven perfusion system (Harvard Apparatus). We acquired data with a Digidata 1440A and pClamp software (version 10.2; Molecular Devices). Signals were filtered at 1 kHz, digitized at 10-20 kHz, and analyzed with Clampfit. We analyzed mEPSCs with Mini Analysis 6 (Synaptosoft).
In situ hybridization and immunofluorescence
Mice were deeply anesthetized with ketamine (100 mg kg−1) and xylazine (10 mg kg−1) and perfused intracardially with ice-cold PBS and then with 4% paraformaldehyde. Brains were extracted, postfixed in the same fixative overnight at 4 °C, transferred to a 30% sucrose solution for cryoprotection, frozen, and stored at −80 °C. We sectioned brain samples with a standard cryostat.
We used standard protocols for in situ hybridization and immunofluorescence60. For detection of miR-124, we purchased digoxigenin-labeled LNA probes from Exiqon. For transgene expression, a biotin- or digoxigenin-labeled RNA probe was synthesized with gene-specific PCR primers (forward 5′-AAATAATACGACTCACTATAGGGAGATTCTTGATGA AATTGGAATTGAAAT-3′ and reverse 5′-GCAGTGAAAAAAATGCTTTATTTGT-3′) and cDNA templates from transgenic mouse brain. Probes were hybridized overnight at 55 °C, and the slides were incubated with horseradish peroxidase-conjugated antidigoxigenin antibody (Roche; 1:500) and steptavidin linked to the peroxidase (Roche). FITC or Cy3 TSA Plus kit (Perkin-Elmer) were used for final detection.
For immunofluorescence, we diluted primary antibodies in PBS containing 10% donkey serum (Sigma), 3% bovine albumin (Sigma), and 0.3% Triton X-100 and incubated overnight at 4 °C. Corresponding donkey anti-rabbit or anti-mouse Alexa 488 or 555 (Invitrogen) was used for secondary detection. The primary antibodies were anti-NeuN (Milipore, MAB377 1:1,000), PSD95 (NeuroMab, 75-028; 1:5,000), GFAP (Dako, Z0334; 1:2,000), Gria2 (Abcam, ab52932; 1:2,000), Gria4 (Abcam, ab77407; 1:1,000), ubiquitin (Enzo Life Sciences, BML-PW8810; 1;1000) and anti-GFP (Life Technologies, A11122; 1:3,000).
Human brain samples
We obtained five FTD cases (3 men and 2 women, mean age at death 68 ± 6 years) and five age-matched normal controls (2 men and 3 women, mean age at death 79 ± 5 years) from the Mayo Clinic Jacksonville and the UCSF Neurodegenerative Disease Brain Bank. Informed consent was obtained for postmortem studies. Sex and postmortem interval (PMI) to autopsy were not different between cases and controls. FTD cases had sporadic behavioral variant FTD (bvFTD) due to FTLD-TDP, Type B. Control brains had no evidence of neurological disease either clinically or neuropathologically. We used frozen tissue (100-150 mg) from gray matter of the middle frontal gyrus for RNA extraction and subsequent RT-PCR analysis.
Image acquisition and processing
For immunofluorescence and in situ hybridization, we acquired images with an Eclipse C1 confocal microscope and software (Nikon). We quantified the number of NeuN neurons from fluorescence images of four tTA:CHMP2BWT mice and four tTA:CHMP2BIntron5 mice at 8 months of age; observers were blinded to genotype. For NeuN counting, we counted three to four sections of the mPFC with the Image J cell-counter plug-in.
For Gria puncta quantification, we used confocal z-stacks (159 × 159 × 10 μm). We analyzed three to four sections (n = 3 mice per genotype) or two sections (n = 6 mice per AAV vector). Following z-projection images, we quantified puncta using Particle Analyzer plug-in. We carried out GFAP coverage quantification as previously described14.
Northern blot
We extracted RNA from specific brain regions with Trizol reagent (Invitrogen) according to the manufacturer’s instructions. We loaded 5–10 μg total RNA into a urea-containing 12.5% acrylamide gel (Sequagel-Ureagel, National Diagnostics) and electrophoresed in standard TBE (1x) buffer. We then transferred RNA to a positively charged nylon membrane (Amersham) and hybridized overnight with digoxigenin-labeled LNA probes (Exiqon). We used anti-digoxigenin antibody linked to alkaline phosphatase (Roche, 11 093 274 910; 1:10,000) and CDP-star reagent (Roche) for detection.
Western blot
We obtained cortical extracts61 and cortical PSD fractions21 as described. PSD or cortical samples (20 μg) were subjected to SDS-PAGE and probed with specific primary antibodies. After incubation with the appropriate secondary antibody linked to horseradish peroxidase (Jackson Immunoresearch), West Pico Signal Chemoluminescent Substrate (Thermo) was used for detection. Antibodies used in these experiments included rabbit antibodies against CHMP2B (Abcam, ab 33174; 1:2,000), Gria1 (Abcam, ab109450; 1:5,000), Gria2 (Abcam, ab52932; 1:2,000), Gria3 (Abcam, ab78366; 1:1,000), Gria4 (Abcam, ab77407; 1:1,000), Grin1 (Upstate Biotechnology, 05-4320; 1:1,000), Grin2a (Upstate Biotechnology, 07-632; 1:2,000), Grin2b (Upstate Biotechnology, 0 -600; 1:1,000), p62 (Cell Signaling Technology, 5114; 1:2,000), and β-actin (Sigma, A-5316; 1:10,000) as a loading control.
Luciferase assays
We cultured HEK cells in Dulbecco’s modified Eagle’s medium containing 10% fetal calf serum and split the day before transfection to achieve ~50% confluence at the time of transfection. We amplified 3′UTRs from Gria1, Gria2, Gria3, and Gria4 transcripts by PCR from mouse brain cDNA and cloned downstream from the renilla luciferase coding sequence of the psicheck2 vector (Promega). We obtained 3′UTRs containing mutated miR-124 binding sites (WT binding site GUGCCUA; mutated binding site GUCGAAAA) by PCR with specific primers including the mutated nucleotide sequence. We transfected cells with a plasmid containing an AMPAR subunit 3′UTR together with a vector driving the expression of miR-124 or miR-9 or an empty vector (pSuper, Oligoengine). For transfection, we used Fugene (Roche) according to the manufacturer’s instructions. After 24 h, we lysed the cells and measured luciferase activity with the Dual Luciferase Reporter Assay (Promega), and normalized results to firefly luciferase activity.
Quantitative RT-PCR
We extracted RNA with the miRNeasy kit (Qiagen) according to the manufacturer’s instructions and digested with DNase for 30 min on-column. We performed first-strand synthesis with 500 ng of total RNA, random hexamers, and TaqMan reverse transcription reagents (Applied Biosystems). Reactions without reverse transcription were always included.
For quantitative PCR, we designed and tested specific primers (Supplementary Table 2) at different cDNA dilutions, and calculated their efficiency. We only used primers showing 95–105% efficiency for further analysis. We performed real-time quantitative PCR with a StepOnePlus system (Applied Biosystems). We carried out reactions (in triplicate) with SYBR Green PCR Master Mix (Applied Biosystems). Each SYBR Green reaction (total volume, 20 μl) contained 1 μl of cDNA as template and each primer at 0.25 μM. Controls without template DNA (reverse transcription minus reaction) were always negative. We incubated he reactions at 95 °C for 10 min to activate the HotStar Taq polymerase followed by 40 cycles at 95 °C for 15 s (denaturation) and at 60 °C for 1 min (annealing and extension). We used β-Actin or GADPH as internal control. Because SYBR Green indiscriminately binds to double-stranded DNA, other products in the PCR such as primer dimers may be detected along with the target gene. To verify that the SYBR Green dye detected only one PCR product, we subjected the samples to the heat dissociation protocol after the final cycle of PCR to check for the presence of only one peak.
Given the limited amount of human samples available, we used TaqMan miRNAs assays to quantify miRNA abundance in brain tissues and iPS-derived neurons (Applied Biosystems, assay 001182 for miR-124 and assay 000583 for miR-9). We extracted RNA as described for mouse samples. Then, we used specific primers for each miRNA for both reverse transcription and subsequent TaqMan quantitative PCR as specified by the manufacturer. We used U6 as internal reference gene for those experiments.
Mouse cortical cultures and iPSC-derived neuronal cultures
To test the efficiency of Gria2 shRNAs, we prepared neuronal cortical cultures from newborn mice as described10. 2 d after plating, 250 μl of lentiviral supernatant was added to the medium. We allowed cells to grow for 3 d and then lysed cells for protein extraction, and assessed Gria2 levels by western blot.
For iPSC-derived neuronal cultures, we used four different lines from three subjects with bvFTD. Two of these lines carry the progranulin S116X mutation36; the other two lines were derived from two subjects with bvFTD caused by C9ORF72 repeat expansion37. We used two iPSC lines from one healthy subject36 as controls and performed neuronal differentiation as described36,37.
Statistical analysis
We performed all statistical analyses with Prism GraphPad 6.0. We compared tTA:CHMP2BWT and tTA:CHMP2BIntron5 mice for each behavioral task. We used the Student’s t test to detect genotype differences in electrophysiological experiments, qPCR, NeuN counting, biochemical analysis, and some behavioral tests. For shRNA efficiency, we used one-way ANOVA. We also used repeated-measure two-way ANOVA to detect differences in the AAV injections. We used Bonferroni’s or Fisher’s LSD test after ANOVA results and analyzed the results of antagonist injections with the Kruskal-Wallis test corrected for multiple comparisons. We used Mann-Whitney test for comparisons of brain human samples.
Supplementary Material
ACKNOWLEDGMENTS
We thank S. Ordway and Gao lab members for comments, Y. Li and A. Philbrook for help with some experiments, Andrew Tapper for sharing behavioral test equipment, the Digital Light Microscopy Core at the University of Massachusetts Medical School (UMMS) for assistance with Golgi staining, the UMMS Viral Vector core for help with AAV vectors, and University of California San Francisco (UCSF) Neurodegenerative Disease Brain Bank for some human brain tissues. We also thank R. Rademakers for genotyping some human samples in a previous work51 that we used in the current study, and A. Chen-Plotkin for sharing published array data38,40. This work was supported by a UMMS startup fund (F.-B.G.), The Consortium for Frontotemporal Dementia Research (W.W.S.), and the US National Institutes of Health (NS057553, NS066586 and NS079725 to F.-B.G., DA032283 to W.D.Y., MH086509 to S.A., AG023501, AG19724 to W.W.S., AG016574 to D.W.D. and L.P.
Footnotes
Accession Numbers. Microarray and deep sequencing data39,40 are available at the NCBI Gene Expression Omnibus database under series accession numbers GDS3459 and GPL10999.
AUTHOR CONTRIBUTIONS
EG, KL, SA, and HZ performed most experiments. HR and WDY carried out the electrophysiology analysis and wrote the relevant sections. JV, DS, and JJ generated the transgenic mouse lines. LP, DWD, and WWS provided brain tissues from controls and subjects with FTD. MJ and SA assisted with behavioral tests. EG and FBG analyzed the data and wrote the manuscript. FBG conceived and supervised the project.
REFERENCES
- 1.Dickson DW. Parkinson’s disease and parkinsonism: neuropathology. Cold Spring Harb. Perspect. Med. 2012;2:a009258. doi: 10.1101/cshperspect.a009258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goldstein LH, Abrahams S. Changes in cognition and behaviour in amyotrophic lateral sclerosis: nature of impairment and implications for assessment. Lancet Neurol. 2013;12:368–380. doi: 10.1016/S1474-4422(13)70026-7. [DOI] [PubMed] [Google Scholar]
- 3.Serrano-Pozo A, Frosch MP, Masliah E, Hyman BT. Neuropathological alterations in Alzheimer disease. Cold Spring Harb. Perspect. Med. 2011;1:a006189. doi: 10.1101/cshperspect.a006189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Neary D, Snowden J, Mann D. Frontotemporal dementia. Lancet Neurol. 2005;4:771–780. doi: 10.1016/S1474-4422(05)70223-4. [DOI] [PubMed] [Google Scholar]
- 5.Loy CT, Schofield PR, Turner AM, Kwok JB. Genetics of dementia. Lancet. 2014;383:828–840. doi: 10.1016/S0140-6736(13)60630-3. [DOI] [PubMed] [Google Scholar]
- 6.Gendron TF, Belzil VV, Zhang YJ, Petrucelli L. Mechanisms of toxicity in C9FTLD/ALS. Acta Neuropathol. 2014;127:359–376. doi: 10.1007/s00401-013-1237-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ling SC, Polymenidou M, Cleveland DW. Converging mechanisms in ALS and FTD: disrupted RNA and protein homeostasis. Neuron. 2013;79:416–438. doi: 10.1016/j.neuron.2013.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cox LE, et al. Mutations in CHMP2B in lower motor neuron predominant amyotrophic lateral sclerosis (ALS) PLoS One. 2010;5:e9872. doi: 10.1371/journal.pone.0009872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Skibinski G, et al. Mutations in the endosomal ESCRTIII-complex subunit CHMP2B in frontotemporal dementia. Nat. Genet. 2005;37:806–808. doi: 10.1038/ng1609. [DOI] [PubMed] [Google Scholar]
- 10.Hooli BV, et al. Rare autosomal copy number variations in early-onset familial Alzheimer’s disease. Mol. Psychiatry. 2013;19:676–681. doi: 10.1038/mp.2013.77. [DOI] [PubMed] [Google Scholar]
- 11.Henne WM, Buchkovich NJ, Emr SD. The ESCRT pathway. Dev. Cell. 2011;21:77–91. doi: 10.1016/j.devcel.2011.05.015. [DOI] [PubMed] [Google Scholar]
- 12.Hurley JH, Hanson PI. Membrane budding and scission by the ESCRT machinery: it’s all in the neck. Nat. Rev. Mol. Cell. Biol. 2010;11:556–566. doi: 10.1038/nrm2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Belly A, et al. CHMP2B mutants linked to frontotemporal dementia impair maturation of dendritic spines. J. Cell Sci. 2010;123:2943–2954. doi: 10.1242/jcs.068817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ghazi-Noori S, et al. Progressive neuronal inclusion formation and axonal degeneration in CHMP2B mutant transgenic mice. Brain. 2012;135:819–832. doi: 10.1093/brain/aws006. [DOI] [PubMed] [Google Scholar]
- 15.Lee JA, Beigneux A, Ahmad ST, Young SG, Gao FB. ESCRT-III dysfunction causes autophagosome accumulation and neurodegeneration. Curr. Biol. 2007;17:1561–1567. doi: 10.1016/j.cub.2007.07.029. [DOI] [PubMed] [Google Scholar]
- 16.Lee JA, Gao FB. Inhibition of autophagy induction delays neuronal cell loss caused by dysfunctional ESCRT-III in frontotemporal dementia. J. Neurosci. 2009;29:8506–8511. doi: 10.1523/JNEUROSCI.0924-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abe M, Bonini NM. MicroRNAs and neurodegeneration: role and impact. Trends Cell Biol. 2013;23:30–36. doi: 10.1016/j.tcb.2012.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gascon E, Gao FB. Cause or effect: Misregulation of microRNA pathways in neurodegeneration. Front. Neurosci. 2012;6:48. doi: 10.3389/fnins.2012.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao FB. Context-dependent functions of specific microRNAs in neuronal development. Neural Dev. 2010;5:25. doi: 10.1186/1749-8104-5-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yizhar O, et al. Neocortical excitation/inhibition balance in information processing and social dysfunction. Nature. 2011;477:171–178. doi: 10.1038/nature10360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peca J, et al. Shank3 mutant mice display autistic-like behaviours and striatal dysfunction. Nature. 2011;472:437–442. doi: 10.1038/nature09965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rademakers R, Neumann M, Mackenzie IR. Advances in understanding the molecular basis of frontotemporal dementia. Nat. Rev. Neurol. 2012;8:423–434. doi: 10.1038/nrneurol.2012.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seelaar H, Rohrer JD, Pijnenburg YA, Fox NC, van Swieten JC. Clinical, genetic and pathological heterogeneity of frontotemporal dementia: a review. J. Neurol. Neurosurg. Psychiatry. 2011;82:476–486. doi: 10.1136/jnnp.2010.212225. [DOI] [PubMed] [Google Scholar]
- 24.Bourne J, Harris KM. Do thin spines learn to be mushroom spines that remember? Curr. Opin. Neurobiol. 2007;17:381–386. doi: 10.1016/j.conb.2007.04.009. [DOI] [PubMed] [Google Scholar]
- 25.Kasai H, Matsuzaki M, Noguchi J, Yasumatsu N, Nakahara H. Structure-stability-function relationships of dendritic spines. Trends Neurosci. 2003;26:360–368. doi: 10.1016/S0166-2236(03)00162-0. [DOI] [PubMed] [Google Scholar]
- 26.Kasai H, Fukuda M, Watanabe S, Hayashi-Takagi A, Noguchi J. Structural dynamics of dendritic spines in memory and cognition. Trends Neurosci. 2010;33:121–129. doi: 10.1016/j.tins.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 27.Namba T, Morimoto K, Sato K, Yamada N, Kuroda S. Antiepileptogenic and anticonvulsant effects of NBQX, a selective AMPA receptor antagonist, in the rat kindling model of epilepsy. Brain Res. 1994;638:36–44. doi: 10.1016/0006-8993(94)90630-0. [DOI] [PubMed] [Google Scholar]
- 28.Lu W, et al. Subunit composition of synaptic AMPA receptors revealed by a single-cell genetic approach. Neuron. 2009;62:254–268. doi: 10.1016/j.neuron.2009.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rozov A, Sprengel R, Seeburg PH. GluA2-lacking AMPA receptors in hippocampal CA1 cell synapses: evidence from gene-targeted mice. Front. Mol. Neurosci. 2012;5:22. doi: 10.3389/fnmol.2012.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cull-Candy S, Kelly L, Farrant M. Regulation of Ca2+-permeable AMPA receptors: synaptic plasticity and beyond. Curr. Opin. Neurobiol. 2006;16:288–297. doi: 10.1016/j.conb.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 31.Liu SJ, Zukin RS. Ca2+-permeable AMPA receptors in synaptic plasticity and neuronal death. Trends Neurosci. 2007;30:126–134. doi: 10.1016/j.tins.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 32.Noh KM, et al. Blockade of calcium-permeable AMPA receptors protects hippocampal neurons against global ischemia-induced death. Proc. Natl. Acad. Sci. U. S. A. 2005;102:12230–12235. doi: 10.1073/pnas.0505408102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 34.Siegel G, Saba R, Schratt G. microRNAs in neurons: manifold regulatory roles at the synapse. Curr. Opin. Genet. Dev. 2011;21:491–497. doi: 10.1016/j.gde.2011.04.008. [DOI] [PubMed] [Google Scholar]
- 35.Deo M, Yu JY, Chung KH, Tippens M, Turner DL. Detection of mammalian microRNA expression by in situ hybridization with RNA oligonucleotides. Dev. Dyn. 2006;235:2538–2548. doi: 10.1002/dvdy.20847. [DOI] [PubMed] [Google Scholar]
- 36.Almeida S, et al. Induced pluripotent stem cell models of progranulin-deficient frontotemporal dementia uncover specific reversible neuronal defects. Cell Rep. 2012;2:789–798. doi: 10.1016/j.celrep.2012.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Almeida, et al. Modeling key pathological features of frontotemporal dementia with C9ORF72 repeat expansion in iPSC-derived human neurons. Acta Neuropathol. 47:115–123. doi: 10.1007/s00401-013-1149-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen-Plotkin AS, et al. TMEM106B, the risk gene for frontotemporal dementia, is regulated by the microRNA-132/212 cluster and affects progranulin pathways. J. Neurosci. 2012;32:11213–11227. doi: 10.1523/JNEUROSCI.0521-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hebert SS, Wang WX, Zhu Q, Nelson PT. A study of small RNAs from cerebral neocortex of pathology-verified Alzheimer’s disease, dementia with Lewy bodies, hippocampal sclerosis, frontotemporal lobar dementia, and non-demented human controls. J. Alzheimers. Dis. 2013;35:335–348. doi: 10.3233/JAD-122350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen-Plotkin AS, et al. Variations in the progranulin gene affect global gene expression in frontotemporal lobar degeneration. Hum. Mol. Genet. 2008;17:1349–1362. doi: 10.1093/hmg/ddn023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Swieten JC, Heutink P. Mutations in progranulin (GRN) within the spectrum of clinical and pathological phenotypes of frontotemporal dementia. Lancet Neurol. 2008;7:965–974. doi: 10.1016/S1474-4422(08)70194-7. [DOI] [PubMed] [Google Scholar]
- 42.Wang F, et al. Bidirectional control of social hierarchy by synaptic efficacy in medial prefrontal cortex. Science. 2011;334:693–697. doi: 10.1126/science.1209951. [DOI] [PubMed] [Google Scholar]
- 43.Piguet O, Hornberger M, Mioshi E, Hodges JR. Behavioural-variant frontotemporal dementia: diagnosis, clinical staging, and management. Lancet Neurol. 2011;10:162–172. doi: 10.1016/S1474-4422(10)70299-4. [DOI] [PubMed] [Google Scholar]
- 44.Filiano AJ, et al. Dissociation of frontotemporal dementia-related deficits and neuroinflammation in progranulin haploinsufficient mice. J. Neurosci. 2013;33:5352–5361. doi: 10.1523/JNEUROSCI.6103-11.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim EJ, et al. Selective frontoinsular von Economo neuron and fork cell loss in early behavioral variant frontotemporal dementia. Cereb. Cortex. 2012;22:251–259. doi: 10.1093/cercor/bhr004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Adamczyk A, et al. GluA3-deficiency in mice is associated with increased social and aggressive behavior and elevated dopamine in striatum. Behav. Brain Res. 2012;229:265–272. doi: 10.1016/j.bbr.2012.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bezprozvanny I, Hiesinger PR. The synaptic maintenance problem: membrane recycling, Ca2+ homeostasis and late onset degeneration. Mol. Neurodegener. 2013;8:23. doi: 10.1186/1750-1326-8-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gibbings DJ, Ciaudo C, Erhardt M, Voinnet O. Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nat. Cell Biol. 2009;11:1143–1149. doi: 10.1038/ncb1929. [DOI] [PubMed] [Google Scholar]
- 49.Lee YS, et al. Silencing by small RNAs is linked to endosomal trafficking. Nat. Cell Biol. 2009;11:1150–1156. doi: 10.1038/ncb1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dutta R, et al. Hippocampal demyelination and memory dysfunction are associated with increased levels of the neuronal microRNA miR-124 and reduced AMPA receptors. Ann. Neurol. 2013;73:637–645. doi: 10.1002/ana.23860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Prudencio M, et al. Misregulation of human sortilin splicing leads to the generation of a nonfunctional progranulin receptor. Proc. Natl. Acad. Sci. U. S. A. 2012;109:2151–21515. doi: 10.1073/pnas.1211577110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Winslow JT. Mouse social recognition and preference. Curr. Protoc. Neurosci. 2003;22:8.16.1–8.16.16. doi: 10.1002/0471142301.ns0816s22. [DOI] [PubMed] [Google Scholar]
- 53.Witt RM, Galligan MM, Despinoy JR, Segal R. Olfactory behavioral testing in the adult mouse. J. Vis. Exp. 2009;23:949. doi: 10.3791/949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Leger M, et al. Object recognition test in mice. Nat. Protoc. 2013;8:2531–2537. doi: 10.1038/nprot.2013.155. [DOI] [PubMed] [Google Scholar]
- 55.Rodriguez A, Ehlenberger DB, Dickstein DL, Hof PR, Wearne SL. Automated three-dimensional detection and shape classification of dendritic spines from fluorescence microscopy images. PLoS One. 2008;3:e1997. doi: 10.1371/journal.pone.0001997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mueller C, Ratner D, Zhong L, Esteves-Sena M, Gao G. Production and discovery of novel recombinant adeno-associated viral vectors. Curr Protoc Microbiol. 2012;26:14D.1.1–14D.1.21. doi: 10.1002/9780471729259.mc14d01s26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chang K, Elledge SJ, Hannon GJ. Lessons from Nature: microRNA-based shRNA libraries. Nat. Methods. 2006;3:707–714. doi: 10.1038/nmeth923. [DOI] [PubMed] [Google Scholar]
- 58.Gao GP, Sena-Esteves M. Introducing Genes into Mammalian Cells: Viral Vectors. In: Green MR, Sambrook J, editors. Molecular Cloning, Vol 2: A Laboratory Manual. Cold Spring Harbor Laboratory Press; New York: 2012. pp. 1209–1313. [DOI] [PubMed] [Google Scholar]
- 59.Kutner RH, Zhang XY, Reiser J. Production, concentration and titration of pseudotyped HIV-1-based lentiviral vectors. Nat. Protoc. 2009;4:495–505. doi: 10.1038/nprot.2009.22. [DOI] [PubMed] [Google Scholar]
- 60.Gascon E, et al. Hepatocyte growth factor-Met signaling is required for Runx1 extinction and peptidergic differentiation in primary nociceptive neurons. J. Neurosci. 2010;30:12414–12423. doi: 10.1523/JNEUROSCI.3135-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gascon E, Vutskits L, Jenny B, Durbec P, Kiss JZ. PSA-NCAM in postnatally generated immature neurons of the olfactory bulb: a crucial role in regulating p75 expression and cell survival. Development. 2007;134:1181–1190. doi: 10.1242/dev.02808. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.