Summary
Spx, a member of the ArsC (arsenate reductase) protein family, is conserved in Gram-positive bacteria, and interacts with RNA polymerase to activate transcription in response to toxic oxidants. In Bacillus anthracis str. Sterne, resistance to oxidative stress requires the activity of two paralogues, SpxA1 and SpxA2. Suppressor mutations were identified in spxA1 mutant cells that conferred resistance to hydrogen peroxide. The mutations generated null alleles of the saiR gene and resulted in elevated spxA2 transcription. The saiR gene resides in the spxA2 operon and encodes a member of the Rrf2 family of transcriptional repressors. Derepression of spxA2 in a saiR mutant required SpxA2, indicating an autoregulatory mechanism of spxA2 control. Reconstruction of SaiR-dependent control of spxA2 was accomplished in Bacillus subtilis, where deletion analysis uncovered two cis-elements within the spxA2 regulatory region that are required for repression. Mutations to one of the sequences of dyad symmetry substantially reduced SaiR binding and SaiR-dependent repression of transcription from the spxA2 promoter in vitro. Previous studies have shown that spxA2 is one of the most highly induced genes in a macrophage infected with B. anthracis. The work reported herein uncovered a key regulator, SaiR, of the Spx system of stress response control.
Keywords: Oxidative stress, Spx transcription regulator, Rrf2 family repressor, Bacillus anthracis
Introduction
Bacillus anthracis is the etiologic agent of anthrax. The most severe form of anthrax is caused by inhalation of B. anthracis spores. How spores are able to successfully germinate and survive the host’s innate immunity to ultimately cause the deadly disease are important for further understanding of B. anthracis pathogenesis (Hanna & Ireland, 1999). When encountering toxic oxidants such as those produced by professional phagocytes, bacteria upregulate the transcription of genes that function in detoxifying oxidative stressors and repairing the cellular damage that they cause (Imlay, 2008). The Spx transcription factor plays a pivotal role in combating oxidative stress in Gram-positive bacteria including Bacillus subtilis where investigation of Spx has been most extensively studied (Zuber, 2004). Additionally, the role of Spx in pathogenesis was uncovered in Streptococci (Chen et al., 2012, Kajfasz et al., 2010), which showed that paralogous Spx proteins are required for virulence. Spx, by interacting with the α subunit of RNA polymerase (RNAP) (Nakano et al., 2003b, Newberry et al., 2005, Lamour et al., 2009), recognizes the Spx element (5′-AGCA-3′) situated around −44 of the target promoters (Nakano et al., 2003a, Newberry et al., 2005, Lin et al., 2013, Nakano et al., 2010, Reyes & Zuber, 2008). Genes activated by the Spx-RNAP complex mostly function in alleviating oxidative stress (Nakano et al., 2003a, Rochat et al., 2012) and protein aggregation induced by thiol stress (Runde et al., 2014). Oxidants such as diamide and hydrogen peroxide cause two important post-translational changes in Spx, namely, disulfide bond formation between the conserved cysteines, which is required for full Spx-dependent induction of trxA (thioredoxin) and trxB (thioredoxin reductase) transcription (Nakano et al., 2005), and escape from the ClpXP-mediated proteolysis that results in an increase in Spx concentration (Nakano et al., 2003a). The regulated proteolysis of Spx in response to oxidants is mediated by the YjbH adaptor protein (Chan et al., 2012, Garg et al., 2009, Gohring et al., 2011, Larsson et al., 2007), which is a possible target of thiol stress control.
Unlike B. subtilis that carries a single spxA gene, B. anthracis has two Spx paralogues encoded by spxA1 and spxA2. The spxA2 gene is one of the most highly expressed B. anthracis genes in the infected macrophage (Bergman et al., 2007). Our previous study identified distinct roles of each paralogue in cell survival during treatments with different oxidants (Barendt et al., 2013). SpxA1 and SpxA2, together, confer diamide resistance, as only the double ΔspxA1ΔspxA2 mutant shows sensitivity to diamide. In contrast, the ΔspxA1 mutant, but not ΔspxA2 mutant, is sensitive to hydrogen peroxide. Nevertheless, inducible expression of ClpXP protease-resistant SpxA1 or SpxA2 is able to restore peroxide resistance in the ΔspxA1 mutant. This result suggests that both SpxA proteins are intrinsically capable of activating genes involved in peroxide resistance; however, either SpxA2 levels or its activity in the native form/context is not sufficient to activate genes that confer peroxide resistance. Here, we report the finding of a novel Rrf2-family transcriptional regulator, SaiR, which is conserved in the Bacillus cereus group and present evidence that SpxA2 is able to confer peroxide resistance in the B. anthracis ΔspxA1 mutant when SaiR is absent. The study revealed a transcriptional regulatory network governed by SpxA1, SpxA2, and SaiR that operates in the control of the oxidative stress response in B. anthracis.
Results
The ΔspxA1 mutant shows a conditional growth-defect phenotype
We have previously shown that the ΔspxA1 but not ΔspxA2 mutant is sensitive to hydrogen peroxide (Barendt et al., 2013). We show here that a null mutation in saiR (BAS3200) restores resistance to hydrogen peroxide in the ΔspxA1 mutant cells. This finding arose from an unexpected observation that the ΔspxA1 mutant (ORB8170), unlike the wild-type parent (7702) or ΔspxA2 mutant (ORB8438), was unable to grow on either tryptic soy broth (TSB, Neogen Co) agar or liquid media. However, the spxA1 mutant grew well on other media such as BHI (brain heart infusion), LB (lysogeny broth), and DS (Difco sporulation) media (data not shown). Interestingly, freshly prepared TSB medium supported the growth of the ΔspxA1 mutant, whereas TSB medium that was autoclaved and shelf-stored for more than five months (Aged TSB) did not support growth (Fig. 1A). In order to confirm that the growth defect of the ΔspxA1 mutant on aged TSB agar was caused by the lack of SpxA1, we carried out a complementation analysis. To this end, we used a previously isolated streptomycin-resistant variant of ΔspxA1 (ORB8398) and the ΔspxA1 mutant carrying Pspank(hy)-spxA1 (ORB8404) (Barendt et al., 2013). ORB8404 was constructed using ICEBs1-mediated conjugation (Auchtung et al., 2005). The streptomycin-resistant ΔspxA1 mutant did not grow on aged TSB agar, but the ΔspxA1 strain expressing the inducible copy of spxA1 grew on aged TSB (Fig. 1A). This result confirmed that the lack of SpxA1 is responsible for the observed growth defect phenotype.
Fig. 1. Growth phenotype of Bacillus anthracis 7702 and its mutant derivatives on fresh and aged TSB agar media.
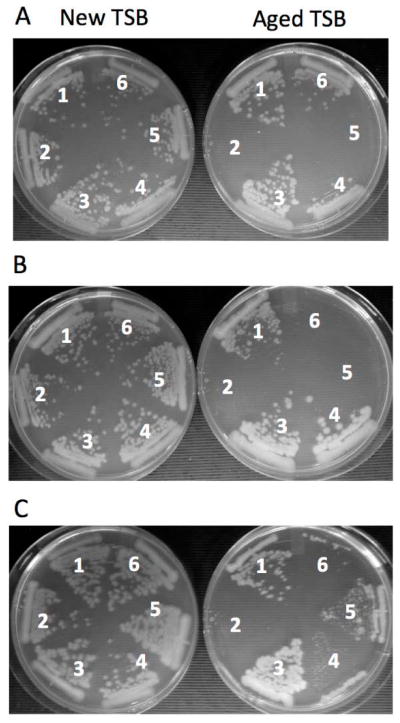
A. The ΔspxA1 strain is unable to grow on aged TSB unlike the wild-type parent and the ΔspxA2 mutant. Strains: 1, wt (7702 Sterne); 2, ΔspxA1 (ORB8170); 3, ΔspxA2 (ORB8438); 4, wtStrr (7702 SR1); 5, ΔspxA1Strr (ORB8398); 6, ΔspxA1Strr+spxA1DD (ORB8404).
B. The saiR mutations restore the growth of the ΔspxA1 mutant on aged TSB agar, but not of the ΔspxA1ΔspxA2 mutant. Strains: 1, wt (7702 Sterne); 2, ΔspxA1 (ORB8170); 3, ΔspxA1sup6 (ORB8492); 4, ΔspxA1ΔsaiR (ORB8606); 5, ΔspxA1ΔspxA2 (ORB8285); ΔspxA1ΔspxA2ΔsaiR (ORB8564).
C. The substitution of C96 in SaiR leads to a loss of repressor activity. Strains: 1, wt (7702 Sterne); 2, ΔspxA1 (ORB8170); 3, ΔspxA1ΔsaiR (ORB8606); 4, ΔspxA1ΔsaiR+saiR(C89S) (ORB8723); 5, ΔspxA1ΔsaiR+saiR(C96S) (ORB8724); 6, ΔspxA1ΔsaiR+saiR(wt) (ORB8725).
Why is aged TSB medium not able to support the growth of the ΔspxA1 mutant? One possibility is that some nutritional components might undergo decomposition during prolonged storage. As SpxA1 is a transcriptional regulator that functions as an activator under oxidative stress conditions, we wondered whether aged TSB lacked a particular amino acid that becomes limited under oxidative stress conditions. Methionine and cysteine are susceptible to oxidation and the enzyme involved in biosynthesis of branched chain amino acids is inactivated in response to reactive oxygen species (ROS) (Boehm et al., 1976, Carlioz & Touati, 1986, Anjem & Imlay, 2012). Aged TSB supplemented with methionine, but not cysteine, cystine, nor a mixture of isoleucine/leucine/valine, supported growth of the ΔspxA1 mutant.
Mutations in the saiR (BAS3200) gene suppress the ΔspxA1 growth phenotype
The result that the aforementioned growth phenotype is specific to the ΔspxA1 mutant prompted us to further investigate the phenomenon. We hoped that the investigation would provide a clue to understanding the physiological role of the two Spx paralogous proteins in oxidative stress response. We first decided to isolate suppressor mutants that grow on aged TSB medium in the absence of spxA1. Three independently isolated mutants (ORB8476, 8478, and 8492) were chosen to identify the compensatory mutations by using whole genome sequencing (Tufts University Core Facility Genomics). Sequence analysis of the mutant genomes in comparison with the parental ΔspxA1 (ORB8170) genome identified SNP (single nucleotide polymorphism) and INDEL (base insertion/deletion) in BAS3200 (BA3453 according to the B. anthracis str. Ames nomenclature)(Table 1). The BAS3200 gene encodes a protein that belongs the Rrf2 family of transcriptional regulators. We named the BAS3200 gene saiR (suppressor of spxA1/regulator). As the mutations in saiR were found in the genomes of the three suppressor mutants, we determined whether all spxA1 suppressors have saiR mutations. Using PCR, the saiR gene was amplified from the remaining spxA1 suppressor strains and the PCR products were sequenced. As shown in Table 1, all suppressor mutants were found to carry mutations in saiR. These mutations include truncations and frameshifts, strongly suggesting that saiR loss-of-function mutations lead to the spxA1 suppressor phenotype.
Table 1.
Summary of spxA1 suppressor mutations
| strain | genotype | mutation | effect |
|---|---|---|---|
|
| |||
| ORB8476* | ΔspxA1 sup1 | BAS3200 (saiR) (C95A), cya (G2265T) | SaiR (A32E), adenylate cyclase (R755S) |
| ORB8477 | ΔspxA1 sup2 | BAS3200 (saiR) (T83A) | SaiR (I28N) |
| ORB8478* | ΔspxA1 sup3 | BAS3200 (saiR) (C86T), BAS3859 (C2237T) | SaiR (S29L), pyruvate carboxylase (T746M) |
| ORB8479 | ΔspxA1 sup4 | BAS3200 (saiR) (G179T) | SaiR (G60V) |
| ORB8480 | ΔspxA1 sup5 | BAS3200 (saiR) (G221A) | SaiR (W74 stop) |
| ORB8492* | ΔspxA1 sup6 | BAS3200 (saiR) (ΔT144) | SaiR (frameshift) |
| ORB8493 | ΔspxA1 sup7 | BAS3200 (saiR) (+T145) | SaiR (H49 stop) |
| ORB8494 | ΔspxA1 sup8 | BAS3200 (saiR) (G32A) | SaiR (S11N) |
| ORB8495 | ΔspxA1 sup9 | BAS3200 (saiR) (ΔACCCA104-108) | SaiR (frameshift) |
| ORB8496 | ΔspxA1 sup10 | BAS3200 (saiR) (ΔT201, ΔA204) | SaiR (L68 stop) |
| ORB8497 | ΔspxA1 sup11 | BAS3200 (saiR) (G181A) | SaiR (G61R) |
Mutations were identified by whole genome sequencing.
To confirm the assumption, we constructed an in-frame saiR deletion using a markerless allelic replacement technique developed by Janes and Stibitz (Janes & Stibitz, 2006), and introduced the ΔsaiR mutation into the wild-type, ΔspxA1, and ΔspxA1ΔspxA2 backgrounds. The ΔspxA1ΔsaiR strain (ORB8606) restored the growth on aged TSB agar, whereas the ΔspxA1ΔspxA2ΔsaiR strain (ORB8564) was unable to grow on the medium (Fig. 1B), indicating that the suppressor effect is indeed attributed to the lack of SaiR activity and the suppression by saiR requires SpxA2.
The saiR mutations restore hydrogen peroxide resistance in the ΔspxA1 strain
As with the growth defect on aged TSB, the ΔspxA1 strain is also more sensitive to hydrogen peroxide than the wild-type and ΔspxA2 strains, as previously reported (Barendt et al., 2013). Therefore, we examined whether the saiR mutations also suppress the peroxide sensitivity caused by ΔspxA1. Both a saiR spontaneous suppressor (sup6) mutation and the in-frame deletion conferred peroxide resistance in the ΔspxA1 strain to a similar level observed in the wild-type strain (Fig. S1). The saiR mutation did not increase peroxide resistance in the ΔspxA1ΔspxA2 background (data not shown), which is in good agreement with the saiR growth phenotype on aged TSB.
The ΔsaiR mutation results in elevated transcription of the spxA2 operon
We investigated the genome-wide extent of saiR transcriptional control in B. anthracis to understand how saiR restores the growth phenotype and peroxide resistance in the ΔspxA1 mutant. We also hoped that the study would provide insight into why SpxA2 is required for the compensatory effect of saiR. Microarray-based transcriptomic analysis was carried out using RNA isolated from the ΔspxA1 and ΔspxA1ΔsaiR strains. In addition, we compared the transcriptional profile between the wild-type and ΔsaiR strains to assess the influence of SaiR on the transcriptome when SpxA1 is present. The result showed that spxA2, BA3455, BA3454, and BA3452 were highly upregulated in the absence of saiR in either the spxA1+ or ΔspxA1 background (Table 2). These four genes, together with saiR, likely constitute an operon and a putative ρ-independent transcription terminator is present downstream of BA3452 (Fig. 2). In addition, the microarray analysis identified that BA0787 (BAS0749) encoding a major facilitator family protein was upregulated by the saiR mutation and the upregulation caused by the mutation was 7-fold higher in the spxA1 background than in the spxA1+ background (Table 2). These microarray results were validated by RT-qPCR (Table 2). Based on these results, we hypothesized that SaiR represses transcription of the spxA2 operon and the ΔsaiR mutation results in upregulation of spxA2, leading to the activation of SpxA1-controlled genes, some of which are required for resistance to hydrogen peroxide and the growth on aged TSB.
Table 2.
Genes upregulated by the saiR mutation
| Microarray (fold induction) | RT-qPCR (fold induction) | |||||
|---|---|---|---|---|---|---|
|
| ||||||
| locus | gene function | gene | ΔsaiR/wt | ΔspxA1ΔsaiR/ΔspxA1 | ΔsaiR/wt | ΔspxA1ΔsaiR/ΔspxA1 |
|
| ||||||
| BA3456 | transcription factor | spxA2 | 15.6 | 13.1 | 114.6±33.9 | 149.7±54.7 |
| BA3455 | hypothetical protein | 23.5 | 11.3 | 110.9±29.8 | 147.7±47.3 | |
| BA3454 | hypothetical protein | 30.1 | 22.8 | 111.8±25.6 | 132.4±40.5 | |
| BA3452 | Hypothetical protein | 49.2 | 44.6 | N/A | N/A | |
| BA0787 | major facilitator family | 2.0 | 13.9 | 1.6±0.2 | 3.8±1.4 | |
N/A: not applicable
Values in microarray are medians of triplicates and those in RT-qPCR are averages of triplicates with standard deviations. Locus is shown in B. anthracis Ames nomenclature used for microarray. BAS3453 is BAS3200 in B. anthracis Ames nomenclature.
Fig. 2.
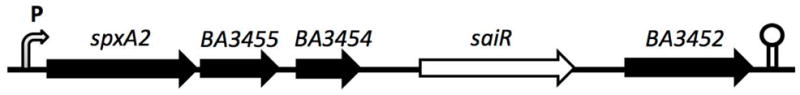
spxA2 and saiR reside within a five-gene operon in B. anthracis. spxA2 and saiR are likely transcribed from the spxA2 promoter together with three other genes shown using Ames nomenclature. The function of these three genes is unknown. The bent arrow marked with P indicates the operon promoter and the direction of transcription, and the lollipop indicates a putative ρ-independent transcription terminator.
One of the conserved cysteine residues is essential for SaiR repressor activity
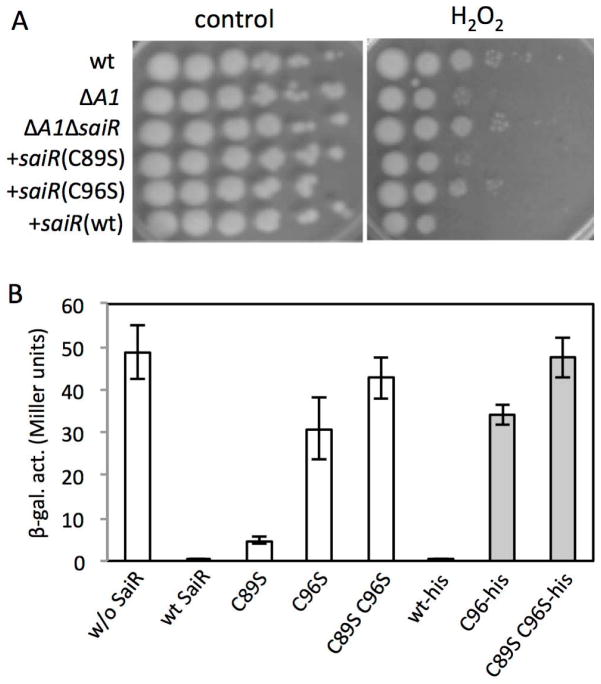
Members of the Rrf2 family of transcription regulators such as IscR, NsrR, and CymR have a C-terminal region that serves a sensory function by interacting with effector ligands. IscR and NsrR coordinate an Fe-S cluster (Isabella et al., 2009, Schwartz et al., 2001, Tucker et al., 2008, Yukl et al., 2008, Kommineni et al., 2010) and CymR forms a complex with CysK, O-acetylserine-thiol-lyase (Tanous et al., 2008). The effectors often enhance DNA-binding activity of the repressors and play pivotal roles in sensing signals such as Fe, NO, and O-acetyl serine (thus intracellular cysteine levels). The C-terminal domain of NsrR contains three conserved cysteine residues that coordinate the Fe-S cluster. The corresponding region in SaiR has two cysteines that are fully conserved in the orthologues from the B. cereus group (B. anthracis, B. cereus, and Bacillus thuringiensis), and other Bacilli including Bacillus firmus, Bacillus methanolicus, and Geobacillus thermoglucosidasius (Fig. S2). As posttranslational modifications of cysteine are known to play roles in redox signaling and oxidative stress response (Antelmann & Helmann, 2011), we determined whether one or both cysteines function in modulating DNA-binding activity. To test the possibility, the C89 and C96 residues were substituted with serine, and the mutant and wild-type alleles of saiR were introduced into the ΔspxA1ΔsaiR strain (ORB8611) using ICEBs1 conjugation system (Auchtung et al., 2005). We examined the sensitivity of these conjugants to hydrogen peroxide in comparison with that of the wild-type, ΔspxA1, and ΔspxA1ΔsaiR strains (Fig. 3A). The result showed that ΔspxA1ΔsaiR cells producing the wild-type or C89S SaiR exhibit hydrogen peroxide sensitivity similar to the ΔspxA1 mutant, indicating that the C89S mutation hardly affects SaiR repressor activity. In contrast, the sensitivity was comparable between the SaiR(C96S)-complementing strain and the parental ΔspxA1ΔsaiR strain, which suggests that the C96S mutation leads to the loss of SaiR activity. Growth phenotype experiments on aged TSB agar also confirmed that C96 plays a more important role than C89 for SaiR transcription repressing activity (Fig. 1C).
Fig. 3. Cys96 in SaiR is required for repressor activity.
A. H2O2 sensitivity assays were carried out as previously described. Five μl of 10-fold serial dilutions of cells (left to right) were spotted onto LB agar with and without 0.44 mM H2O2. The saiR genes, which encode the wild-type, C89S, and C96S protein, were introduced into the ICEBs1 element of the ΔspxA1ΔsaiR mutant. Strains: wt (7702SR1); ΔspxA1 (ORB8398); ΔspxA1ΔsaiR (ORB8611); ΔspxA1ΔsaiR +saiR(wt) (ORB8725); ΔspxA1ΔsaiR +saiR(C89S) (ORB8723); ΔspxA1ΔsaiR +saiR(C96S) (ORB8724).
B. SaiR represses spxA2 transcription in B. subtilis. Open bars show spxA2-lacZ expression in the absence of SaiR (ORB8884), in the presence of wild-type SaiR (ORB8820), SaiR(C89S) (ORB8825), SaiR(C96S) (ORB8826), and SaiR(C89S C96S) (ORB9055). The spxA2 promoter contains the region between −247 and +268 relative to the spxA2 transcription start site. Gray bars show that the his6 tag has no effect on the activity of SaiR. Strains: wild-type saiR-his6 (ORB8985); saiR(C96S)-his6 (ORB9047); saiR(C89S C96S)-his6 (ORB9064).
The spxA2 promoter carries two cis-acting regions required for SaiR repression of spxA2
The results above suggested that SaiR directly or indirectly interacts with the spxA2 promoter and represses transcription under nonstress conditions. We examined whether the repressive effect of SaiR on spxA2 transcription could be reconstituted in B. subtilis, which does not carry the saiR gene. A 5′-RACE experiment using RNA purified from the B. anthracis 7702 identified the spxA2 transcription start site at 220 bp upstream of the translational start site. The spxA2 fragment (−247 to +268 relative to the transcription start site) was fused to a promoter-less lacZ gene (Experimental procedures). The transcriptional lacZ fusion was integrated into the thrC locus, while an IPTG-inducible allele of saiR was placed at the amyE locus in B. subtilis 168. spxA2 was transcribed in B. subtilis that lacks saiR, whereas transcription was highly repressed in the strain carrying saiR (Fig. 3B). The repression is specific to spxA2 and saiR, as SaiR did not show any significant effect on transcription of B. subtilis trxB, a gene belonging to the Spx regulon (Nakano et al., 2003a) (data not shown). In addition, integration of the empty vector plasmid pDR111 (Britton et al., 2002) into the amyE locus did not repress spxA2 transcription (data not shown). The SaiR(C89S) mutant showed a 10-fold reduction in repressor activity compared to the wild-type SaiR, whereas the C96S mutation nearly abolished repression of spxA2 transcription. Furthermore, spxA2 transcription is fully active in the C89S C96S SaiR mutant (Fig. 3B). In order to eliminate the possibility that C96S and C89S C96S SaiR proteins are unstable in B. subtilis, we carried out western blot analysis. To this end, we constructed B. subtilis strains that produce the wild-type and the two mutant SaiR proteins tagged with 6xHis at the C-termini. To verify that the tagged SaiR is functional, we introduced saiR-his6 at the amyE locus in B. subtilis carrying spxA2-lacZ. spxA2 transcription in each strain carrying various saiR-his6 was similar compared to the expression in the strain with the corresponding untagged saiR gene (Fig. 3B). Western blot analysis of SaiR-His6 using anti-His tag antibody showed that the C96 and C89S/C96S SaiR, like the wild-type protein, were produced in B. subtilis (Fig. S3). We concluded that the conserved cysteine residues, C96 in particular, play an important role in SaiR repressor activity.
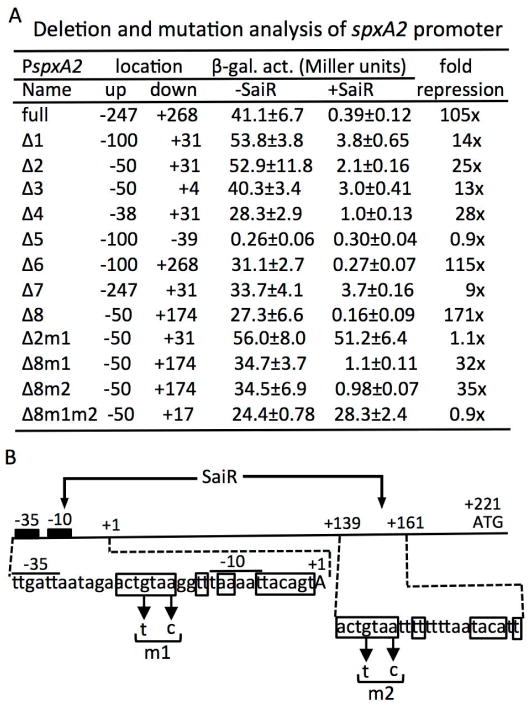
In order to localize the cis-regulatory region of SaiR in the spxA2 promoter, we constructed a series of promoter deletion mutations and examined the effect of SaiR on the promoter activity (Fig. 4A). The spxA2 (−247 to +268) was used as the full-length promoter. The Δ1 (−100 to +31) promoter exhibited β-galactosidase activity similar to the full-length promoter in the absence of SaiR; however, repression by SaiR was 14-fold, much weaker than repression by the full promoter (105-fold). Three successive deletions (Δ2, Δ3, and Δ4) retained promoter activity, which was also partially repressed by SaiR, indicating that the region between −38 and +4 contains a cis site targeted directly or indirectly by SaiR repression (SaiR site 1). As expected from the 5′-RACE result, the Δ5 fragment (−100 to −39) was unable to promote lacZ expression. As SaiR represses more strongly the full-length promoter than the deleted (Δ2 to Δ4) promoters, we next determined whether the 5′ or 3′ region that is missing in the deleted promoters (from Δ1, Δ2 and Δ4) is required for full repression. The Δ6 (−100 to +268) promoter restored the full-length level repression, while the Δ7 (−247 to +31) fragment did not, demonstrating that the region downstream from +31 contains another SaiR target site (SaiR site 2). The minimum region that is fully susceptible to SaiR repression was localized between −50 and +174, as deduced from the observed expression levels of the Δ8 promoter mutant.
Fig. 4. The spxA2 promoter contains two cis-sites required for SaiR repression.
A. Deletion and mutational analysis of the spxA2 promoter. Endpoints of the promoter fused to lacZ are shown in numbers relative to the transcriptional start site. Each strain was grown in LB and cells were harvested around OD600≈0.4 to measure β-galactosidase activities. Experiments were repeated at least three times using independent isolates obtained from strain construction and the averages are shown with standard deviations.
B. A schematic map of the spxA2 promoter region. The location of the transcription site, the core promoter, and the translation start site are marked as +1, −10 and −35, and ATG, respectively. The nucleotide sequences of the two SaiR sites are shown below the map. Boxed nucleotides constitute a dyad symmetry sequence in site 1 and a partial dyad symmetry sequence in site 2. Arrows show the base substitutions introduced in site 1 (m1) and site 2 (m2).
Members of the Rrf2-family of transcription factors are known to bind as dimers to DNA containing sequences exhibiting dyad symmetry (Shepard et al., 2011). We searched for such sequences in the region between −38 and +4 where SaiR site 1 was likely residing. The sequence ACTGTAAN2TTtAAN2TTACAGT (the lowercase t represents the centre of the symmetry) was detected in the core promoter region that overlaps with the promoter −10 element (Fig. 4B). Based on the deletion analysis, we assumed that SaiR site 2 exists between +31 and +174. A sequence of partial dyad symmetry (ACTGTAAN2TTtN5TACANT) was found in the region between +139 and +161. The sequence of the left half in SaiR site 2 shows a perfect match with the corresponding region of SaiR site 1, whereas the right half has a 5/9 match to site 1.
In order to confirm that the two cis sites are involved in SaiR repression, base substitutions were introduced in the putative control elements having dyad symmetry. When substitutions of two conserved bases in SaiR site 1 were introduced in the Δ2 (−50 to +31) promoter, the resultant Δ2m1 promoter was fully resistant to SaiR-dependent repression. In contrast, the m1 mutation in the Δ8 (−50 to +174, Fig. 4A) promoter that carries the two SaiR sites resulted in a partial loss of repression (see Δ8m1, Fig. 4A). Similarly, the same base substitutions in SaiR site 2 led to 35-fold repression by SaiR (see Δ8m2). In contrast, SaiR was unable to repress the Δ8m1m2 promoter, which supports the conclusion that both SaiR sites are required for full repression by SaiR. Binding of SaiR to the downstream site might function as a roadblock to RNAP elongation or in DNA loop formation together with its binding to the upstream, SaiR site 1.
SaiR directly controls spxA2 transcription
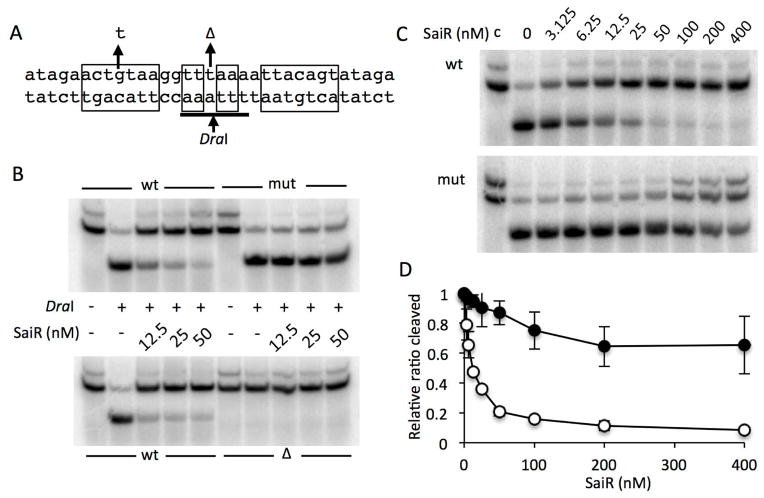
The in vivo spxA2 promoter analysis clearly demonstrated that SaiR repression is dependent on the core promoter (−23 to −1) region as well as the downstream but untranslated (+139 to +157) region. We next examined whether SaiR directly interacts with the dyad-symmetry motifs to repress transcription. As SaiR-His6 was shown to be functional as described above, we overproduced the SaiR-His6 in E. coli and purified as described in Experimental procedures. EMSA (electrophoretic mobility shift assay) was initially attempted to determine whether SaiR binds to the motifs identified by in vivo transcription study. In some cases, we obtained high-affinity binding of SaiR to the wild-type SaiR site 1 in a sequence-specific manner, but we were not always able to reproduce the results. We assumed that the SaiR-DNA complex might be easily dissociated during electrophoresis. Therefore, we conducted restriction enzyme protection assays, which do not involve gel electrophoresis of protein-DNA complex. As shown in Fig. 5A, there is a DraI recognition site in the centre of SaiR site 1. If SaiR interacts with the dyad symmetry sequence, SaiR likely causes reduced accessibility of DraI to the recognition site. As expected, increasing concentrations of SaiR protected the wild-type DNA from the cleavage by DraI; whereas, the mutant DNA was susceptible to DraI cleavage (Fig. 5B). Deleting the nucleotide at the centre resulted in the loss of DraI site (Fig. 5A), thus becoming insensitive to DraI (Fig. 5B), confirming that the digestion of the wild-type and mutant DNA is specific to DraI. In addition, SaiR-dependent protection is not targeted directly to DraI, as SaiR did not protect DraI recognition sites other than the spxA2 DNA (Fig. S4). A titration experiment indicated that 6 nM (3 nM as dimer) SaiR provides 50% protection from cleavage of the wild-type DNA (Figs. 5C and D). With the mutant DNA bearing the single base substitution that reduces symmetry, 50% inhibition of DraI cleavage was observed at concentrations higher than 200 nM, but full inhibition was not attained in the range of concentrations (up to 400nM) used in the experiment. Based on these results, we concluded that SaiR binds to the sequence of dyad symmetry constituting SaiR site 1.
Fig. 5. SaiR directly interacts with the spxA2 core promoter.
A. The probes used for DraI-digestion protection assay. The sequence from −28 to +5 (relative to the transcriptional start site) of the spxA2 promoter is shown. The nucleotide sequences in rectangular boxes indicate the dyad symmetry required for SaiR repression. The arrows above the sequence show a base substitution and deletion used in the protection assay. The DraI recognition sequence is underlined and digestion site is marked by the arrow below the sequence.
B. The wild-type probe and the probe carrying the base substitution (mut) and deletion (Δ) were incubated without or with various concentrations of SaiR-His6, followed by DraI digestion. The results are representative of four independent experiments.
C. Titration of SaiR for protection against DraI digestion. All lanes except those marked with c were from reactions treated with DraI as described in Experimental procedures.
D. Results in triplicate of reactions shown in Fig. 5C were used to quantify band intensities using Image J. First, intensity of digested band in each lane was divided by the sum of digested and undigested band intensities in the corresponding lane (ratio cleaved). The relative ratio cleaved was calculated by dividing the ratio cleaved in each lane by the ratio obtained from the lane without SaiR. Symbols: open circles, the wild-type probe; closed circles, the probe with the base substitution.
As the in vivo study showed that C96 plays an important role in SaiR repressor activity, we examined whether the C96S mutation or C89S C96S mutation results in reduced DNA binding using the DraI digestion protection assay. The result showed that both mutant proteins retain DNA binding activity similar to the wild-type protein (data not shown). Based on these results, we concluded that SaiR directly binds to the SaiR sites to repress spxA2 transcription and the C96 (or C89 and C96) residue may interact with a ligand or small protein, which was lost during purification process of SaiR.
We next determined that SaiR, by binding to the dyad symmetry, represses spxA2 transcription in vitro. The linear DNA templates carry the spxA2 promoter with and without the m1/m2 base substitutions (Δ8 and Δ8m1m2 in Fig. 4B). The template was expected to produce a 174 base run-off transcript. Addition of 16 nM SaiR resulted in reduced transcript synthesis from the wild-type promoter and transcription was almost completely repressed by 64 nM SaiR (Fig. 6). The mutant promoter is more resistant to SaiR repression, with transcript observed in reactions containing 64 nM SaiR. SaiR(C96S) was able to repress spxA2 transcription in vitro to a degree similar to the wild-type protein (data now shown), which is consistent with the result that showed no effect of C96S on DNA binding activity in vitro.
Fig. 6. SaiR represses spxA2 transcription in vitro.
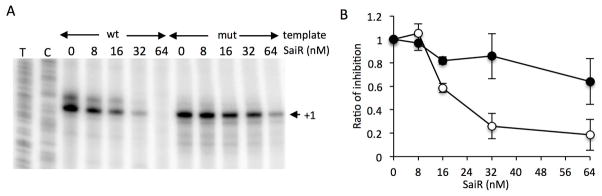
A. The wild-type and mutant (m1m2) templates were incubated with 25 nM RNAP at 37 C for 10 min with increasing concentrations of SaiR-His6. Transcription was initiated by the addition of nucleotide mixtures including radioactive UTP as described in Experimental procedures. T and C sequencing ladders are also shown. The results are representative of three independent experiments.
B. The intensity of the corresponding bands was quantified with ImageJ and is shown as the ratio of transcript level in the presence and absence of SaiR for each template. The values are averages of three independent experiments with standard deviations. Symbols: open circles, the wild-type template; closed circles, the mutant template.
spxA2 transcription is upregulated in response to disulfide stress
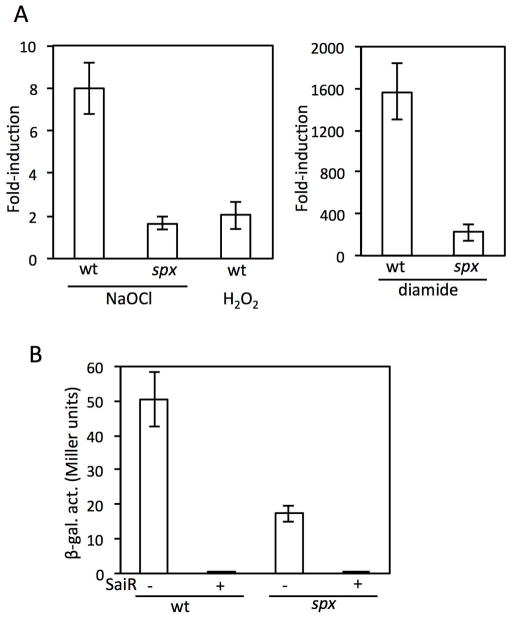
The two conserved cysteines in SaiR are important in spxA2 repression under nonstress conditions in vivo, but data from DNA binding and transcription studies in vitro with SaiR cysteine mutant proteins did not support the in vivo results. In hope of understanding how SaiR activity is regulated, we examined conditions where spxA2 transcription is derepressed in the presence of SaiR. As SpxA2 activates genes involved in oxidative stress response and spxA2 transcription is induced in the infected macrophage (Bergman et al., 2007), we first tested the effect of oxidizing agents that are produced in macrophages; namely, hypochlorite, hydrogen peroxide, and NO. spxA2-lacZ was not induced in response to NO in B. subtilis ORB8820 cells that produce SaiR (data not shown). RT-qPCR showed that spxA2 transcription is induced around 8-fold in response to 0.8 or 2 mM NaOCl treatment for 10 min, whereas only 2-fold induction was detected with 1 mM hydrogen peroxide (Fig. 7A and data not shown for 0.8 mM NaOCl). ORB8820 carries B. subtilis spx and spxA2 transcription is 3-fold higher in the B. subtilis spx+ strain than spx mutant (Fig. 7B). NaOCl was known to evoke Spx-controlled disulfide stress response (Chi et al., 2011). Hence, the observed spxA2 upregulation in response to NaOCl could be due to increased activity/concentrations of B. subtilis Spx, the loss of SaiR activity, or both. To eliminate the ambiguity, we carried out RT-qPCR to measure the spxA2 induction in the spx mutant cells in response to NaOCl. The results showed a significant decrease in the fold-induction in the absence of Spx, indicating that the induction caused by NaOCl is largely (if not solely) attributed to Spx response. Although diamide is a synthetic substance and not produced in macrophages, it is a potent thiol-oxidizing agent. As NaOCl is known to elicit disulfide stress (Chi et al., 2011, Gray et al., 2013), we wondered whether diamide also increases spxA2 transcription. When spx+ cells were treated with 0.5 mM diamide, spxA2 transcription was induced over 1000-fold, and more importantly, 150-fold induction was observed even in the absence of Spx. This result indicates that thiol oxidation first triggers SaiR inactivation and Spx further activates spxA2 transcription in response to disulfide stress response.
Fig. 7. Transcription of spxA2 is upregulated by B. subtilis Spx in vivo in the absence of SaiR and transcription is induced in response to oxidants in the presence of SaiR.
A. RT-qPCR of spxA2 in B. subtilis that produces SaiR. The wild-type (ORB8820) and spx mutant (ORB9050) cells were grown in LB. At OD600=0.3–0.4, cells were treated with NaOCl (final concentration of 2 mM), H2O2 (1 mM), or diamide (0.5 mM) for 10 min. RNA purification and cDNA preparation were previously described (Barendt et al., 2013). The fold-induction was shown as averages of biological triplicates with standard deviations after standardized with rpoB as described in Experimental procedures.
B. spxA2(−247/+268)-lacZ was measured in the wild-type strain in the absence (ORB8827) and presence (ORB8820) of the saiR gene. The lacZ expression was also measured in the strain lacking B. subtilis spx in the absence (ORB9049) and presence (ORB9050) of saiR.
Purified SaiR protein is not inactivated by responding to disulfide stress
RT-qPCR results showed that spxA2 transcription is induced in B. subtilis that produces SaiR in response to oxidants. We examined whether SaiR directly senses diamide using the DraI protection assay described earlier. Unexpectedly, incubation of SaiR with diamide, as well as with hydrogen peroxide, did not affect SaiR DNA-binding activity; in contrast, 50 μM NaOCl led to dissociation of SaiR from spxA2 DNA carrying the SaiR box 1 (Fig. S5A). Furthermore, the single and double cysteine mutants of SaiR still responded to NaOCl (Fig. S5B). In these experiments, SaiR was treated with the oxidants at room temperature for 5 min before the addition of the spxA2 DNA followed by DraI digestion. When SaiR was incubated with the DNA before NaOCl addition, SaiR did not dissociate from spxA2 (Fig S5C). These results suggested that diamide likely weakens the interaction between SaiR and the effector and that NaOCl does not target cysteine residues in SaiR to weaken its DNA binding activity.
Discussion
The most important finding of this study is that elevated spxA2 transcription due to inactivation of the SaiR repressor is a prerequisite for SpxA2-dependent oxidative stress resistance. This study began with a serendipitous finding that the spxA1 mutant is unable to grow on aged TSB but that the growth is restored by the addition of methionine. We assume that prolonged storage of the medium at room temperature might generate an oxidative condition, which leads to conversion of methionine to methionine sulfoxide (Met-SO). Although B. anthracis is generally a methionine auxotroph, the amount of methionine in aged TSB is sufficient to support the growth if SpxA1 is produced. An alternative but not mutually exclusive possibility is that Met-SO generated over time in TSB could be transported by the MetNPQ ABC-type transporter (Hullo et al., 2004), and possibly reduced back to methionine by methionine sulfoxide reductases (Msr) in cells producing SpxA1. MsrA and MsrB catalyze the reduction of the S-epimer and the R-epimer of Met-SO, respectively [reviewed in (Ezraty et al., 2005)]. The msrAB operon belongs to the Spx regulon in B. subtilis (You et al., 2008). The B. anthracis genome contains msrA1 and msrA2, the latter of which codes for bifunctional methionine sulfoxide reductase A/B protein. Our previous microarray data showed that SpxA1DD and SpxA2DD activate transcription of msrA2 but not msrA1. Taken together, the spxA1 mutant likely has a low MsrA2 activity that does not generate methionine from methionine sulfoxide, hence the spxA1 mutant is unable to grow on aged TSB.
The saiR gene is located in the spxA2 operons of B. cereus and B. thuringiensis and both SaiR-binding sites are conserved in the spxA2 promoter of these bacteria (Fig. S6B), which strongly suggests that SaiR plays a pivotal role in the control of SpxA2-dependent transcriptional activation among the B. cereus group of bacteria. The spxA2 operon genes were highly upregulated by the saiR mutation regardless of the presence or absence of spxA1. On the other hand, ΔsaiR led to a moderate upregulation of BA0787 only in the absence of spxA1. We found that the BA0787 promoter contains a sequence with partial homology to the SaiR consensus binding-site (Fig. S6B). The sequence overlaps with a putative −35 sequence (or extended −10 element) and the left half-site has a 7/9 match to the consensus sequence and the right half-site showed a 4/9 match. In addition, the putative SaiR-binding sequence is also well conserved in B. cereus and B. thuringiensis (Fig. S6B), further supporting the direct role of SaiR in BA0787 transcription.
Our previous work identified genes induced by the Spx paralogues (Barendt et al., 2013). While this transcriptomic analysis was informative, it was carried out using IPTG-inducible spx paralogues that encode protease-resistant proteins, which likely circumvented stress-responsive transcriptional and proteolytic control. In fact, the study showed the essential role in peroxide resistance of SpxA1, but not SpxA2, in a saiR+ background; nevertheless, protease-resistant SpxA2 restores peroxide resistance in spxA1 mutant cells (Barendt et al., 2013). The results suggested that SpxA2 is capable of activating genes that function in peroxide resistance if the protein is present in sufficient levels. The study presented herein confirmed the assumption and uncovered SaiR-dependent spxA2 repression, which explains the previous peroxide sensitivity results. As with B. subtilis Spx, SpxA2 activity is probably stimulated through disulfide-bond formation of the conserved cysteines, which could be generated only under the conditions where SaiR-dependent spxA2 repression is relieved either by the saiR mutation or a loss of SaiR activity.
The Rrf2 family transcription repressors are known to undergo modulation of DNA-binding activity in response to environmental stress (Shepard et al., 2011). Previous transcriptome studies by Hanna and co-workers showed that transcription of the putative spxA2 operon (spxA2-BA3455-BA3454-saiR-BA3452) is activated in the last 3 h (called wave 5) of the 8 h life cycle of B. anthracis grown in vitro (Bergman et al., 2006). In contrast, spxA1 transcription was upregulated in the first 2 h (wave 1) after germination. Interestingly, the same group observed that members of the spxA2 operon are among the most highly upregulated B. anthracis genes in infected macrophages (RAW264.7 cells) compared to cultures in vitro (Bergman et al., 2007).
Taken together, it is likely that SaiR is inactivated in macrophages. We have tested whether any oxidant produced in macrophages is able to induce spxA2 transcription in the presence of SaiR. The results showed that NaOCl moderately induces spxA2 among oxidants tested. A weak induction with hydrogen peroxide is in good agreement with the result of peroxide sensitivity assays indicating that spxA1 mutant is sensitive to hydrogen peroxide unless saiR is mutated (compare the peroxide sensitivity between ΔspxA1 and ΔspxA1ΔsaiR in Fig. 3A). In comparison, a synthetic thiol-oxidizing reagent, diamide, induced spxA2 to much higher levels and substantial induction was detected even in the absence of Spx. The result argues that oxidation of thiol(s) is a key to inactivation of SaiR repressor activity. However, it is unlikely that thiol oxidation of the conserved cysteines is sufficient for the loss of SaiR activity because of the following reasons. First, diamide did not affect DNA-binding activity of purified SaiR in vitro. Second, substitutions of the conserved cysteines did not show any effect on in vitro transcription nor DNA binding, despite the observation that the mutation showed a dramatic effect on spxA2 transcription in vivo. We purified SaiR from B. subtilis as well as E. coli, under both aerobic and anaerobic conditions, to obtain the protein that reflects in vivo activity but neither approach has been successful. We currently hypothesize that a ligand/co-repressor (a metabolite or protein) that interacts with the C-terminal half of SaiR is lost during purification. Even in the absence of such a ligand, SaiR interacts with the SaiR-binding sites in vitro, but perhaps with lower affinity than what is achieved in vivo (Figs. 5 and 6), which explains why the SaiR-DNA complex was unstable during electrophoresis in EMSA experiments. In addition, SaiR lacking the ligand/co-repressor is unable to respond to the signal. In this scenario, C96 (and C89 to a lesser extent) is essential for interaction with the ligand/co-repressor that increases DNA binding activity. Diamide, by oxidizing C89/C96 or thiol(s) in the interacting partner, causes ligand/co-repressor dissociation. We are currently not certain whether the effect of NaOCl on SaiR activity in vitro has physiological relevance. One possibility is that once SaiR is released from the ligand, NaOCl might oxidize SaiR. If it is the case, the oxidized residue leading to target release is not cysteine, as the cysteine SaiR mutants still respond to NaOCl (Fig. S5B).
In conclusion, this study presented evidence that SaiR is a key player in the control of SpxA2-dependent transcription activation and studies are underway to identify the effector ligand/co-repressor of SaiR and the accompanying environmental/metabolic signal that controls SaiR activity.
Experimental procedures
Bacterial strains
B. anthracis strains used in this study are derivatives of the 7702 Sterne strain (pXO1+ pXO2−) and listed in Table S1. B. subtilis strains are derivatives of either JH642 or 168 as listed in Table S1. Plasmids and oligonucleotides are listed in Tables S2 and S3, respectively. A marker-less in-frame deletion of saiR was constructed using the method described by Janes and Stibitz (Janes & Stibitz, 2006). In constructing the saiR deletion, the codons specifying the first seven amino acids were fused in-frame to those for the last six amino acids.
Identification of spxA1 suppressor mutations
Genomic DNA was isolated from the parental ΔspxA1 (ORB8170) strain and three suppressor mutants. Sequence of genomic DNA was determined by Tufts University Core Facility Genomics. The input DNA (1μg per sample) was sheared by sonication to 200–300bp. The sheared DNA was then used as input for library preparation using TruSeq DNA Sample Preparation Kit (Illumina). The resulting fragment library was quantified and sequenced with HiSeq 2000 using SE50 format. The reads were then aligned to the reference genome, followed by SNP/Indel calling using CLC Genomics Workbench (CLC bio, Qiagen).
Transcriptomic and RT-qPCR analyses
RNA was isolated from 7702 (wild type), ORB8170 (ΔspxA1), ORB8571 (ΔsaiR), and ORB8606 (ΔspxA1ΔsaiR) strains grown in LB until OD600 reached around 0.4 to 0.5. RNA was isolated using glass beads/phenol method (Igo & Losick, 1986) and microarray analysis was carried out as described in a previous study (Barendt et al., 2013). The arrays used for hybridization contained 70-mers that were designed to hybridize to the 3′-end of transcripts. The microarray result was sorted and listed in Table S4 and the original data was deposited to NCBI GEO (accession ID: GSE57851).
RT-qPCR was performed using two independently isolated RNA samples that were used in microarray experiments and two additional RNA preparations. Normalization was conducted using gatB transcript, thoe level of which is largely constant during growth (Reiter et al., 2011), as described before (Barendt et al., 2013), and the ratio of induction was calculated with the normalized values. Oligonucleotide pairs spxA2-F3/spxA2-R3 and gatB-F1/gatB-R1 were used for spxA2 and gatB RT-qPCR, respectively, and the sequences of these oligonucleotides are reported in a publication of a previous study (Barendt et al., 2013). Other oligonucleotides used for qPCR of BA0787, BA3454, and BA3455 are listed in Table S3.
Induction of spxA2 transcription in response to oxidants was examined using B. subtilis spx+ (ORB8820) and spx (ORB9050) strains carrying spxA2-lacZ and saiR. The B. subtilis strains were grown until OD600=0.3–0.4, and treated with 2 mM (or 0.8 mM) NaOCl, 1 mM H2O2, or 0.5 mM diamide for 10 min. RNA was isolated from untreated and treated cells, and the induction ratio was calculated after normalization with the induction ratio of the rpoB transcript. Oligonucleotides used for qPCR are listed in Table S3.
Complementation analysis using the wild-type, C89S-, and C96S-SaiR
SaiR complementation analysis was performed using the B. subtilis ICEBs1 system (Auchtung et al., 2005). The saiR gene was amplified by PCR and the resultant fragment was cloned downstream of the IPTG-inducible Pspank(hy) promoter in pDR111 (Britton et al., 2002) to generate pSS9. The fragment containing Pspank(hy)-saiR and the lacI gene was isolated from pSS9 and subcloned into pJMA402, an ICEBs1 conjugation system vector plasmid, resulting in pMMN850. The cysteine substitution mutations were generated using two-step site-directed PCR mutagenesis and a similar procedure used for the pMMN850 construction was applied to generate pMMN851(C89S) and pMMN852(C96S). Introduction of the wild-type and mutant saiR alleles into the B. anthracis genome was carried out by mating streptomycin-resistant B. anthracis with B. subtilis strains carrying the saiR genes in recombinant ICEBs1.
Determination of the spxA2 transcription start site
RNA was purified as described above from the 7702 parental strain and 5′-RACE was carried out using 5′-RACE system (Invitrogen) per manufacture’s instructions.
The spxA2 promoter analysis
Repression of spxA2 transcription by SaiR was reconstituted in a heterologous system using B. subtilis 168. Various regions of the spxA2 promoter were amplified by PCR and cloned into a promoter-probe vector, pDG793 (Guérout-Fleury et al., 1996) to generate transcriptional lacZ fusions. The resulting plasmids were used to transform B. subtilis 168, which leads to the integration of the lacZ fusions into the thrC locus. Base substitutions of the spxA2 promoter were performed using PCR-based site-directed mutagenesis and the mutated promoters were cloned in pDG793. The wild-type saiR gene amplified from 7702 genomic DNA was cloned in pDR111 that carries an IPTG-inducible promoter. The mutant alleles of saiR, which bear substitutions of Cys89, Cys96, and Cys89/96 with serine, were generated by site-directed mutagenesis and cloned in pDR111. These plasmids were used to transform B. subtilis to generate the strains carrying the wild-type and mutant saiR genes at the amyE locus. Although a promoter-less saiR was placed downstream of the IPTG-inducible Pspank(hy) promoter, repression was observed even in the absence of IPTG due to the leaky IPTG-inducible promoter. Hence, all experiments using Pspank(hy)-saiR in this study were carried out without IPTG, except otherwise stated.
Purification of SaiR protein
SaiR carrying a C-terminal 6xhistidine tag (SaiR-His6) was produced and purified from E. coli. The saiR gene was amplified from pMMN850 and the PCR product was cloned in pET23a to generate pMMN856. To produce SaiR(C89S) and SaiR(C96S), pMMN857 and pMMN858 were constructed similarly using pMMN851 and pMMN852 as templates, respectively. The saiR gene with the C89S C96S mutation was generated by two-step PCR-based site-directed mutagenesis using pSS10 as template and cloned into pET23a to construct pMMN881. E. coli BL21(DE3)pLysS carrying each plasmid was used to overproduce SaiR-His6 and mutant derivatives. Purification was carried out using Ni-NTA resin (Thermo Scientific) according to manufacturer’s protocol. To determine whether SaiR-His6 is functional, saiRhis6 was amplified using pMMN856, pMMN858, and pMMN881 as template and the resultant fragment was cloned in pDR111 to generate amyE-integration plasmids, pMMN880, pYA1, and pYA3, respectively. SaiR repressor activity was tested using the full-length spxA2-lacZ, as described above.
Restriction enzyme protection assay
DNA carrying the wild-type SaiR site 1 was generated by annealing complementary oligonucleotides oMN14-666 and oMN14-667. The resultant double stranded DNA corresponds to the region between −28 and +5 in the spxA2 promoter. Similarly, DNA carrying the base substitution (C-22T) or deletion of −12T was generated by annealing of oMN14-668/oMN14-669 and oMN14-670/oMN14-671, respectively. The sequences of these oligonucleotides are listed in Table S3. One of the oligonucleotides (oMN14-666, oMN14-668, or oMN14-669) labeled with T4 polynucleotide kinase and γ-32P ATP was annealed with the unlabeled complementary oligonucleotide. Unincorporated γ-32P ATP was removed by nucleotide removal kit (Qiagen). The labeled DNA was mixed with various concentrations of SaiR-His6 in 9 μl of 1x CutSmart buffer (50 mM potassium acetate, 20 mM Tris-acetate, 10 mM magnesium acetate, 100 μg ml−1 BSA, pH 7.9: New England BioLabs) and the mixture was incubated at room temperature for 20 min. After the addition of 1 μl of freshly diluted DraI (0.2 units), digestion was carried out at room temperature for 30 min before heat inactivation at 65°C for 20 min. DNA was resolved on a pre-run 15% native polyacrylamide gel in TAE buffer.
In vitro transcription
In vitro transcription was carried out using the wild-type and mutant spxA2 promoter fragments. The wild-type template was PCR amplified from pMMN874 using 793up and oMN13-638 and the mutant template was generated using 793up and oMMN14-661 with pMMN879 as template. The wild-type and mutant templates were identical with Δ8 and Δ8 m1m2 (in Fig. 4A), respectively, except that an extra 79 bp originating from pDG793 was adjacent to −50 of spxA2. The expected transcript size was 174 bases. A reaction mixture (20 μl total) contained 40 mM Tris-HCl (pH 8.0), 10 mM MgCl2, 5% glycerol, 0.1 mM EDTA, 1 mM DTT, 0.1 mg ml−1 bovine serum albumin, 10 nM template DNA, 25 nM RNAP, and increasing concentrations of SaiR. B. subtilis His-tagged RNAP and σA protein previously purified (Lin et al., 2013) were preincubated on ice for 30 min (the final concentration used was 25 nM and 125 nM, respectively). The reaction mixture was incubated at 37°C for 10 min, and transcription was started by the addition of GTP, CTP, ATP (20 μM each), UTP (1 μM), and 5 μCi of α-32P UTP. The reaction was stopped after 10 min, precipitated with ethanol, and products were resolved on a pre-run 6% polyacrylamide-urea gel. Sequence ladders were generated using Thermo Sequenase Cycle Sequencing Kit (Affimetrix) with the labeled downstream oligonucleotide (oMN13-638) and pMMN874.
Supplementary Material
Acknowledgments
This work is supported by National Institutes of Health Grants AI092313 and GM045898 (to P.Z.) and in part by National Science Foundation Grant MCB1157424 (to M.M.N.). S.M.B. is supported in part by NIH 5T32AI007472 and Y.M.N. was supported Equity Summer Internship Program by Center for Diversity & Inclusion at OHSU. We thank Dr. Boris Belitsky for helpful technical advice.
References
- Anjem A, Imlay JA. Mononuclear iron enzymes are primary targets of hydrogen peroxide stress. J Biol Chem. 2012;287:15544–15556. doi: 10.1074/jbc.M111.330365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antelmann H, Helmann JD. Thiol-based redox switches and gene regulation. Antioxidants & redox signaling. 2011;14:1049–1063. doi: 10.1089/ars.2010.3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auchtung JM, Lee CA, Monson RE, Lehman AP, Grossman AD. Regulation of a Bacillus subtilis mobile genetic element by intercellular signaling and the global DNA damage response. Proc Natl Acad Sci USA. 2005;102:12554–12559. doi: 10.1073/pnas.0505835102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barendt S, Lee H, Birch C, Nakano MM, Jones M, Zuber P. Transcriptomic and phenotypic analysis of paralogous spx gene function in Bacillus anthracis Sterne. Microbiology Open. 2013;2:695–714. doi: 10.1002/mbo3.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergman NH, Anderson EC, Swenson EE, Janes BK, Fisher N, Niemeyer MM, et al. Transcriptional profiling of Bacillus anthracis during infection of host macrophages. Infect Immun. 2007;75:3434–3444. doi: 10.1128/IAI.01345-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergman NH, Anderson EC, Swenson EE, Niemeyer MM, Miyoshi AD, Hanna PC. Transcriptional profiling of the Bacillus anthracis life cycle in vitro and an implied model for regulation of spore formation. J Bacteriol. 2006;188:6092–6100. doi: 10.1128/JB.00723-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm DE, Vincent K, Brown OR. Oxygen and toxicity inhibition of amino acid biosynthesis. Nature. 1976;262:418–420. doi: 10.1038/262418a0. [DOI] [PubMed] [Google Scholar]
- Britton RA, Eichenberger P, Gonzalez-Pastor JE, Fawcett P, Monson R, Losick R, Grossman AD. Genome-wide analysis of the stationary-phase sigma factor (sigma-H) regulon of Bacillus subtilis. J Bacteriol. 2002;184:4881–4890. doi: 10.1128/JB.184.17.4881-4890.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlioz A, Touati D. Isolation of superoxide dismutase mutants in Escherichia coli: is superoxide dismutase necessary for aerobic life? EMBO J. 1986;5:623–630. doi: 10.1002/j.1460-2075.1986.tb04256.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CM, Garg S, Lin AA, Zuber P. Geobacillus thermodenitrificans YjbH recognizes the C-terminal end of Bacillus subtilis Spx to accelerate Spx proteolysis by ClpXP. Microbiology. 2012;158:1268–1278. doi: 10.1099/mic.0.057661-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Ge X, Wang X, Patel JR, Xu P. SpxA1 involved in hydrogen peroxide production, stress tolerance and endocarditis virulence in Streptococcus sanguinis. PLoS One. 2012;7:e40034. doi: 10.1371/journal.pone.0040034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi BK, Gronau K, Mader U, Hessling B, Becher D, Antelmann H. S-bacillithiolation protects against hypochlorite stress in Bacillus subtilis as revealed by transcriptomics and redox proteomics. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.M111.009506. M111 009506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezraty B, Aussel L, Barras F. Methionine sulfoxide reductases in prokaryotes. Biochim Biophys Acta. 2005;1703:221–229. doi: 10.1016/j.bbapap.2004.08.017. [DOI] [PubMed] [Google Scholar]
- Garg SK, Kommineni S, Henslee L, Zhang Y, Zuber P. The YjbH protein of Bacillus subtilis enhances ClpXP-catalyzed proteolysis of Spx. J Bacteriol. 2009;191:1268–1277. doi: 10.1128/JB.01289-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gohring N, Fedtke I, Xia G, Jorge AM, Pinho MG, Bertsche U, Peschel A. New role of the disulfide stress effector YjbH in beta-lactam susceptibility of Staphylococcus aureus. Antimicrob Agents Chemother. 2011;55:5452–5458. doi: 10.1128/AAC.00286-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray MJ, Wholey WY, Jakob U. Bacterial responses to reactive chlorine species. Annu, Rev, Microbiol. 2013;67:141–160. doi: 10.1146/annurev-micro-102912-142520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guérout-Fleury AM, Frandsen N, Stragier P. Plasmids for ectopic integration in Bacillus subtilis. Gene. 1996;180:57–61. doi: 10.1016/s0378-1119(96)00404-0. [DOI] [PubMed] [Google Scholar]
- Hanna PC, Ireland JA. Understanding Bacillus anthracis pathogenesis. Trends Microbiol. 1999;7:180–182. doi: 10.1016/s0966-842x(99)01507-3. [DOI] [PubMed] [Google Scholar]
- Hullo MF, Auger S, Dassa E, Danchin A, Martin-Verstraete I. The metNPQ operon of Bacillus subtilis encodes an ABC permease transporting methionine sulfoxide, D- and L-methionine. Res Microbiol. 2004;155:80–86. doi: 10.1016/j.resmic.2003.11.008. [DOI] [PubMed] [Google Scholar]
- Igo MM, Losick R. Regulation of a promoter that is utilized by minor forms of RNA polymerase holoenzyme in Bacillus subtilis. J Mol Biol. 1986;191:615–624. doi: 10.1016/0022-2836(86)90449-3. [DOI] [PubMed] [Google Scholar]
- Imlay JA. Cellular defenses against superoxide and hydrogen peroxide. Annu Rev Biochem. 2008;77:755–776. doi: 10.1146/annurev.biochem.77.061606.161055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isabella VM, Lapek JD, Jr, Kennedy EM, Clark VL. Functional analysis of NsrR, a nitric oxide-sensing Rrf2 repressor in Neisseria gonorrhoeae. Mol Microbiol. 2009;71:227–239. doi: 10.1111/j.1365-2958.2008.06522.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janes BK, Stibitz S. Routine markerless gene replacement in Bacillus anthracis. Infect Immun. 2006;74:1949–1953. doi: 10.1128/IAI.74.3.1949-1953.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajfasz JK, Rivera-Ramos I, Abranches J, Martinez AR, Rosalen PL, Derr AM, et al. Two Spx proteins modulate stress tolerance, survival, and virulence in Streptococcus mutans. J Bacteriol. 2010;192:2546–2556. doi: 10.1128/JB.00028-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kommineni S, Yukl E, Hayashi T, Delepine J, Geng H, Moënne-Loccoz P, Nakano MM. Nitric oxide-sensitive and -insensitive interaction of Bacillus subtilis NsrR with a ResDE-controlled promoter. Mol Microbiol. 2010;78:1280–1293. doi: 10.1111/j.1365-2958.2010.07407.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamour V, Westblade LF, Campbell EA, Darst SA. Crystal structure of the in vivo-assembled Bacillus subtilis Spx/RNA polymerase alpha subunit C-terminal domain complex. J Struct Biol. 2009;168:352–356. doi: 10.1016/j.jsb.2009.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson JT, Rogstam A, von Wachenfeldt C. YjbH is a novel negative effector of the disulphide stress regulator, Spx, in Bacillus subtilis. Mol Microbiol. 2007;66:669–684. doi: 10.1111/j.1365-2958.2007.05949.x. [DOI] [PubMed] [Google Scholar]
- Lin AA, Walthers D, Zuber P. Residue substitutions near the redox center of Bacillus subtilis Spx affect RNA polymerase interaction, redox control and Spx-DNA contact at a conserved cis-acting element. J Bacteriol. 2013;195:3967–3978. doi: 10.1128/JB.00645-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano MM, Lin A, Zuber CS, Newberry KJ, Brennan RG, Zuber P. Promoter recognition by a complex of Spx and the C-terminal domain of the RNA polymerase alpha subunit. PLoS One. 2010;5:e8664. doi: 10.1371/journal.pone.0008664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano S, Erwin KN, Ralle M, Zuber P. Redox-sensitive transcriptional control by thiol/disulphide switch in the global regulator, Spx. Mol Microbiol. 2005;55:498–510. doi: 10.1111/j.1365-2958.2004.04395.x. [DOI] [PubMed] [Google Scholar]
- Nakano S, Kuster-Schock E, Grossman AD, Zuber P. Spx-dependent global transcriptional control is induced by thiol-specific oxidative stress in Bacillus subtilis. Proc Natl Acad Sci USA. 2003a;100:13603–13608. doi: 10.1073/pnas.2235180100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano S, Nakano MM, Zhang Y, Leelakriangsak M, Zuber P. A regulatory protein that interferes with activator-stimulated transcription in bacteria. Proc Natl Acad Sci USA. 2003b;100:4233–4238. doi: 10.1073/pnas.0637648100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newberry KJ, Nakano S, Zuber P, Brennan RG. Crystal structure of the Bacillus subtilis anti-alpha, global transcriptional regulator, Spx, in complex with the alpha C-terminal domain of RNA polymerase. Proc Natl Acad Sci USA. 2005;102:15839–15844. doi: 10.1073/pnas.0506592102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter L, Kolsto AB, Piehler AP. Reference genes for quantitative, reverse-transcription PCR in Bacillus cereus group strains throughout the bacterial life cycle. J Microbiol Methods. 2011;86:210–217. doi: 10.1016/j.mimet.2011.05.006. [DOI] [PubMed] [Google Scholar]
- Reyes DY, Zuber P. Activation of transcription initiation by Spx: formation of transcription complex and identification of a cis-acting element required for transcriptional activation. Mol Microbiol. 2008;69:765–779. doi: 10.1111/j.1365-2958.2008.06330.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochat T, Nicolas P, Delumeau O, Rabatinova A, Korelusova J, Leduc A, et al. Genome-wide identification of genes directly regulated by the pleiotropic transcription factor Spx in Bacillus subtilis. Nucleic Acids Res. 2012;40:9571–9583. doi: 10.1093/nar/gks755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz CJ, Giel JL, Patschkowski T, Luther C, Ruzicka FJ, Beinert H, Kiley PJ. IscR, an Fe-S cluster-containing transcription factor, represses expression of Escherichia coli genes encoding Fe-S cluster assembly proteins. Proc Natl Acad Sci USA. 2001;98:14895–14900. doi: 10.1073/pnas.251550898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepard W, Soutourina O, Courtois E, England P, Haouz A, Martin-Verstraete I. Insights into the Rrf2 repressor family--the structure of CymR, the global cysteine regulator of Bacillus subtilis. The FEBS journal. 2011;278:2689–2701. doi: 10.1111/j.1742-4658.2011.08195.x. [DOI] [PubMed] [Google Scholar]
- Tanous C, Soutourina O, Raynal B, Hullo MF, Mervelet P, Gilles AM, et al. The CymR regulator in complex with the enzyme CysK controls cysteine metabolism in Bacillus subtilis. J Biol Chem. 2008;283:35551–35560. doi: 10.1074/jbc.M805951200. [DOI] [PubMed] [Google Scholar]
- Tucker NP, Hicks MG, Clarke TA, Crack JC, Chandra G, Le Brun NE, et al. The transcriptional repressor protein NsrR senses nitric oxide directly via a [2Fe-2S] cluster. PLoS ONE. 2008;3:e3623. doi: 10.1371/journal.pone.0003623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You C, Sekowska A, Francetic O, Martin-Verstraete I, Wang Y, Danchin A. Spx mediates oxidative stress regulation of the methionine sulfoxide reductases operon in Bacillus subtilis. BMC Microbiol. 2008;8:128. doi: 10.1186/1471-2180-8-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yukl ET, Elbaz MA, Nakano MM, Moënne-Loccoz P. Transcription factor NsrR from Bacillus subtilis senses nitric oxide with a 4Fe-4S cluster. Biochemistry. 2008;47:13084–13092. doi: 10.1021/bi801342x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber P. Spx-RNA polymerase interaction and global transcriptional control during oxidative stress. J Bacteriol. 2004;186:1911–1918. doi: 10.1128/JB.186.7.1911-1918.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.