Abstract
Slowing the aging process can reduce the risk for multiple chronic diseases simultaneously. It is increasingly recognized that maintaining protein homeostasis (or proteostasis) is important for slowing the aging process. Since proteostasis is a dynamic process, monitoring it is not a simple task and requires use of appropriate methods. This review will introduce methods to assess protein and DNA synthesis using deuterium oxide (D2O), and how protein and DNA synthesis outcomes provide insight into proteostatic mechanisms. Finally, we provide a discussion on how these assessments of protein and DNA synthesis are “mechanistic” investigations and provide an appropriate framework for the further development of slowed aging treatments.
Keywords: Stable isotope, deuterium oxide, long-lived model, mitochondria, proliferation
1.1 Introduction
Long-lived models are ideal for studying slowed aging because it is presumed that slowed aging is what imparts long life. This mini-review will present a strategy for examining shared characteristics among long-lived models. We believe that shared characteristics between models are potentially indicative of globally applicable mechanistic insights for slowing the aging process.
The ability to maintain protein homeostasis (proteostasis) has become a key outcome in aging research (Austad, 2010; Balch et al., 2008; Perez et al., 2009; Salmon et al., 2009). Proteostasis refers to cellular control of the concentration, location, and conformation of individual proteins to achieve stability (Balch et al., 2008). Conceptually the idea of proteostasis is simple, protein synthesis (with accompanied folding and transport) must match the rate of protein degradation and the need to provision new daughter cells (Figure 1). It is thought that proteostasis promotes healthy aging through enhanced cellular stability and repair (Treaster et al., 2013). Whether or not increased proteostasis maintenance is an underlying mechanism of slowed aging is yet to be definitively determined, but investigations into proteostasis provide a contextual framework for studying the aging process. In order to study proteostasis, one needs to use appropriate methods. In this review, we show that in vivo heavy water labeling provides valuable insight into the connection between the aging process and changes in proteostasis.
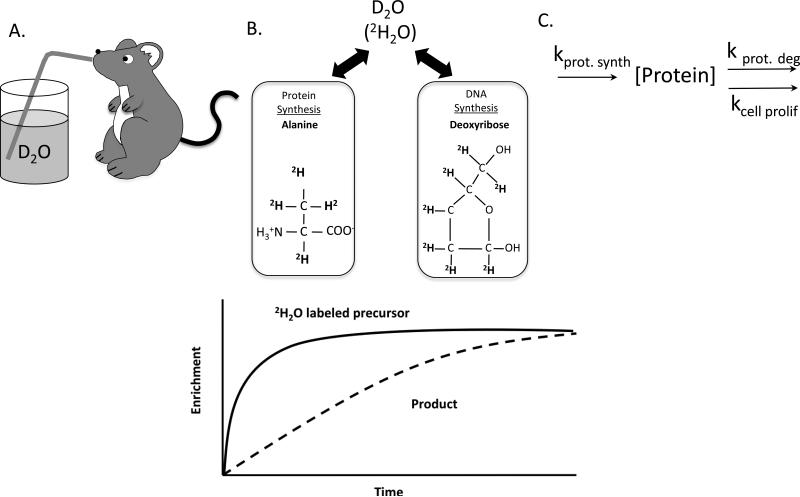
Figure 1. Using D2O for assessing synthesis of DNA and protein.
Following an intraperitoneal injection, D2O (2H2O) is administered via the animal's drinking water where it rapidly equilibrates with the body water pool (A). Incorporation of deuterium from body water into labile hydrogens of precursor building blocks and subsequent incorporation of those precursor species into stable C-H bonds allows monitoring of rates of newly synthesized proteins and DNA up until a period in which all are 100% new (B). The synthesis of new protein and DNA control protein concentration in vivo (C).
1.2 Discussion
1.2.1 Distinguishing cell growth, adaptation and repair to understand proteostasis
When a proliferative cell replicates it doubles its mass to have two equal sized daughter cells (Grebien et al., 2005). When a post-mitotic cell, such as a skeletal muscle cell, hypertrophies it recruits DNA from resident stem cells (Collins et al., 2005) to maintain a constant DNA to cytoplasm ratio (Allen et al., 1999; Pavlath et al., 1989). Therefore, in both proliferative and post-mitotic cells, new DNA, either from true DNA replication or DNA recruitment, is associated with increases in protein synthesis.
The capacity for cells to enzymatically repair proteins is low (Mary et al., 2004; Mortimore and Pösö, 1987; Poppek and Grune, 2005). Therefore to maintain proteostasis, damaged proteins are removed and replaced with new proteins (Poppek and Grune, 2005). Cells constantly integrate multiple signals about the intracellular and extracellular environment and synthesize new proteins in response to environmental stresses. This process is counter to increasing protein synthesis due to an increase in cell proliferation. In one case there are adaptive responses to prevent or replace protein damage to maintain proteostasis, whereas in the other, protein synthesis increases to equip the newly made cells. By this rationale, when examining protein synthesis in the context of increasing proteostatic mechanisms, one should consider both the rate of protein synthesis and cellular proliferation in order to account for how much protein is directed toward cell doubling versus how much is directed toward maintaining existing cellular structures.
1.2.3 Using D2O for assessing synthesis of DNA and protein
The concentration of each individual protein is the result of dynamic balance of synthesis and degradation rates. Assessing rates of synthesis of new protein and DNA is critical because simply measuring concentration masks ongoing cellular processes. For example, if protein synthesis increased by one hundred-fold and was matched by a one hundred-fold increase in breakdown, there would be no detectable change in protein concentration even though there were dramatic changes to proteostasis. Although it has been recognized previously that cell proliferation and protein synthesis are linked (Eden et al., 2011), few in vivo experimental approaches have been developed to allow measurement of these rates. During periods of cellular stress several post-transcriptional mechanisms such as selective translation (Zid et al., 2009), stress granules (Kimball et al., 2003), and micro-RNA (Adeli, 2011) determine which mRNA are translated to protein. Therefore, measurements of cellular signaling and mRNA content are not appropriate for assessing protein synthesis because they fail to account for post-transcriptional regulation, a concept we have previously discussed (Miller and Hamilton, 2012).
In vivo deuterium oxide (D2O) labeling allows for the simultaneous measurement of both protein (Busch et al., 2006; Drake et al., 2013; Miller et al., 2012; 2013; Robinson et al., 2011) and DNA synthesis (Drake et al., 2013; Miller et al., 2013; 2012; Neese et al., 2002; Robinson et al., 2011). For a detailed discussion of the calculations, assumptions, and limitations, the reader is referred to the following publications for protein (Busch et al., 2006) and DNA (Neese et al., 2002). To perform a study, D2O is introduced as an intraperitoneal injection (Gasier et al., 2009; Miller et al., 2013; Yuan et al., 2008) or as drinking water (Drake et al., 2013; Miller et al., 2013; 2012) to enrich the body water pool with deuterium (Figure 1). Equilibrium is rapidly established within the body water. Newly synthesized metabolites incorporate deuterium into stable C-H bonds, which allows identification of new molecules by mass spectrometry. A particular advantage of using D2O, compared to intravenous delivery of labeled amino acids, is that D2O can be used over prolonged periods, as opposed to acute 4-6 hr periods for labeled amino acids, to capture the synthesis of slowly synthesized species while an organism is free living. This ability to measure outcomes over the long-term is advantageous because the sum total of physiological effects over time, not just short snapshots that vary based on the prevailing metabolic state, can be assessed. When studying processes associated with aging and slowed aging in laboratory models and humans, the ability to capture data during prolonged free living allows the study of more subtle differences that accumulate over time.
1.2.4 What measuring proteostasis tells us about slowed aging
Long-lived models (i.e. models of slowed aging) are ideally suited for use in studies to identify the mechanisms responsible for aging and age-related declines. Caloric restriction without malnutrition (CR) (Masoro, 2005) and rapamycin treated mice (Harrison et al., 2009) are long-lived and demonstrate characteristics consistent with prolonged healthspan (Wilkinson et al., 2012). These models also share the characteristic of decreased mechanistic (formerly mammalian) target of rapamycin (mTOR) activity (Drake et al., 2013; Harrison et al., 2009; Miller et al., 2012), a key modulator of protein synthesis and cell cycling (Sengupta et al., 2010). However, as discussed below, decreased mTOR activity is not universal among long-lived models.
1.2.4.1 Caloric restriction
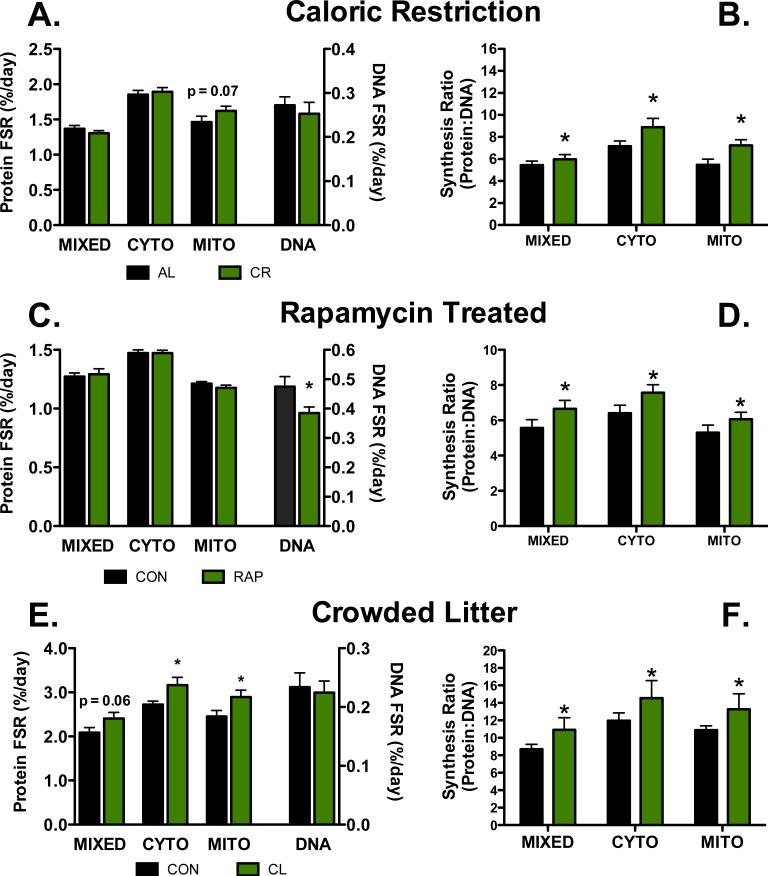
CR is a fascinating aging intervention because some evidence suggests that mitochondrial biogenesis is maintained or increased during CR (Nisoli, 2005), while others have speculated that decreased protein synthesis leads to lifespan extension (Kapahi, 2010; Tavernarakis, 2008). Given the complex mechanisms of post-transcriptional regulation employed during activation of pathways associated with energetic stress, we thought that it is possible that protein synthetic responses were not truly captured with previous methods. Using D2O we measured protein synthesis, including mitochondrial protein synthesis, both acutely (4 hours) and over the long-term (6 weeks) in liver, heart, and skeletal muscle of CR animals compared to ad libitum fed controls (CON) (Miller et al., 2013; 2012). We found that almost uniformly among the three tissues, protein synthesis was maintained in CR mice, including in the mitochondrial fraction (Miller et al., 2013; 2012) (Figure 2A). Although the finding that there are no differences in protein synthesis between CR and CON appears fairly unremarkable, when considered in relation to DNA synthesis a different picture emerges (Figure 2B). Unchanged rates of protein synthesis during CR directly conflicts with the idea that a decrease in protein synthesis is a mechanism of slowed aging. Interestingly, because DNA synthesis rates were lower in CR animals compared to CON, the ratio of protein synthesis to DNA synthesis is greater in CR compared to CON. The combination of slowed DNA and unchanged protein synthesis, suggests a greater proportion of protein synthesis is dedicated to maintaining existing cellular structures rather than cell proliferation.
Figure 2. Representative protein synthesis, DNA synthesis, and protein synthesis:DNA synthesis in three models of slowed aging.
(A) Synthetic rates of DNA and protein in three fractions of skeletal muscle (MIXED = myofibrillar, CYTO = cytoplasmic, MITO = Mitochondrial) in lifelong calorically restricted mice (CR) and ad libitum fed controls (AL). Although there is a trend for greater mitochondrial protein synthesis in CR, protein synthesis rates are largely similar between CR and AL (data reproduced from (Miller et al., 2013; 2012)). (B) When the data are expressed as a synthesis ratio, it is clear that there is increase protein synthesis dedicated to proteostasis in CR animals as compared to AL. (C) Synthetic rates of DNA and three fractions of heart in chronically fed rapamycin treated (RAP) and age matched controls (CON). There are no differences between RAP and CON for protein synthetic rates, although DNA synthesis is lower in RAP compared to CON. (D) When the data are expressed as a synthesis ratio, there is increased protein synthesis dedicated to maintaining existing protein structures in RAP compared to CON (data reproduced from (Drake et al., 2013)). (E) Synthetic rates of DNA and three fractions of skeletal muscle in crowded litter mice (CL) and normal litter sized controls (CON). As opposed to CR and RAP models, there is greater protein synthesis in CL compared to CON and no difference in DNA synthesis (data reproduced from (Drake et al., 2014)). (F) Despite qualitative differences in synthesis rates in the CL model compared to CR and RAP, the ratio of protein and DNA synthesis rates still imply maintained proteostasis. Data are means ± SEM, n = 4-12/group. *p<0.05 compared to CON for the same fraction.
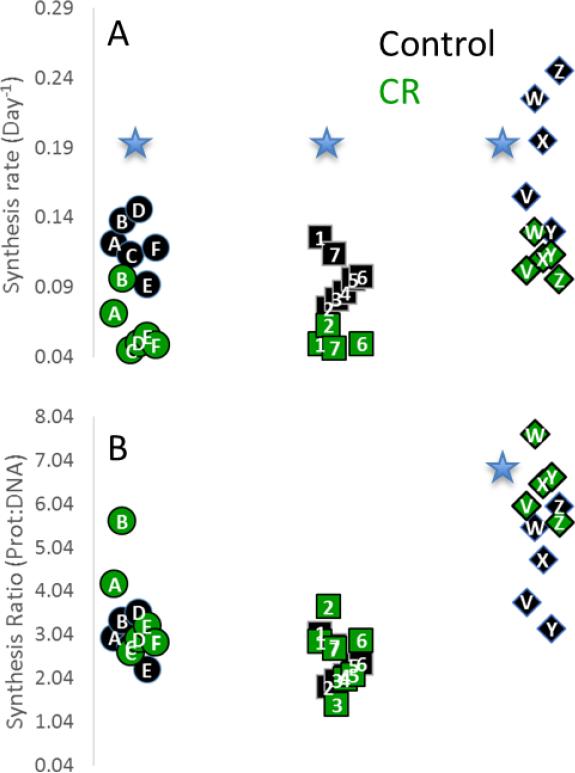
These data are suggestive of increases in proteostatic maintenance and quality control. We expect that changes in the synthesis of individual proteins and enzymes will give further insight into the CR-dependent cellular response. Indeed, we measured the synthesis rates of several hundred individual proteins in the liver (Price et al., 2012), and found that a majority of these proteins were synthesized more slowly during CR. We observed that many proteins which were functionally related seemed to be regulated together (Figure 3), in keeping with our previous observations (Price et al., 2010). Interestingly, in the liver although the synthesis rates of many proteins were significantly different the protein:DNA ratio was often the same between controls and CR. A few selected protein ontologies, like the urea cycle (Figure 3B) showed an increase in protein:DNA ratio suggestive of increased proteostatic maintenance. This indicates that the cell may adapt to CR by enacting a specific program of proteostasis instead of global changes in metabolism. The application of this technique to other tissues and models may shed light on mechanisms of increased proteostasis.
Figure 3.
(A) Individual protein components of ATP Synthase (· ·), Ribosome (· ·), and urea cycle (◇) are synthesized significantly more slowly (٭, p<0.05) during CR. (B) Comparison of DNA:Protein synthesis rate ratios suggests that synthesis of urea cycle proteins is maintained at higher comparative rate in CR compared to AL controls.
1.2.4.2 Rapamycin treatment
Chronic administration of the mTORC1 inhibitor rapamycin has been shown to extend mean and maximal life span in heterogeneous mice (Harrison et al., 2009). Our comparison of protein synthesis and DNA synthesis over a 4-week period in heterogeneous mice treated for 12 weeks with rapamycin (Drake et al., 2013) showed that skeletal muscle experienced a decrease in protein synthesis rates in mixed (nuclei, plasma membrane, and contractile proteins) and cytosolic fractions, while synthesis rates of mitochondrial proteins were maintained (Figure 2C). In both the skeletal muscle and heart, there was a decrease in DNA synthesis. In a subsequent calculation of protein synthesis to DNA synthesis ratios, it became apparent that like CR, there is an increase in proteostatic mechanisms in rapamycin treated mice compared to CON (Figure 2D).
1.2.4.3 Crowded litter
The crowded litter model (CL) is a newly described long-lived model (Steinbaugh et al., 2012; Sun et al., 2009). In the CL model, litter size is increased by 50% and this increase in litter size presumably imposes transient caloric restriction during a critical developmental period until the pups are weaned and subsequently given free access to food. Even though the CL mice are given ad libitum access to food for the remainder of their lives, mean and maximal lifespan are increased (Sun et al., 2009).
Recently we characterized protein and DNA synthetic rates in the long-lived CL mouse (Drake et al., 2014). Surprisingly, we found that compared to normal litter sized controls (CON), CL mice rates of protein synthesis that were uniformly greater in heart, liver, and skeletal muscle in all sub-fractions examined (Drake et al., 2014)(Figure 2E). In addition, contrary to other long-lived models, DNA synthesis was relatively unchanged. Importantly, when the protein synthesis to DNA synthesis was calculated, it again appeared that there was greater activation of proteostatic mechanisms in the CL mice compared to CON (Figure 2F). Finally, activation of ribosomal protein subunit S6 (rpS6), a marker of mTOR activation, was greater in CL compared to CON (Drake et al., 2014). Therefore, although a decrease in mTOR activity observed in both the CR and rapamycin treated mice, the CL model illustrates that the resultant improvement in proteostasis is likely a more important outcome than decreased mTOR activity itself.
1.2.5 Why are studies of proteostasis mechanistic?
It is easy to dismiss the measurement of protein and DNA synthesis as largely descriptive and not mechanistic. However, when investigating the end result of cellular processes, the measurement of synthesis rates is perhaps one of the more insightful mechanistic outcomes. Protein concentration is controlled through the dynamic balance of synthesis versus degradation and cell proliferation (Figure 1) and is poorly correlated with mRNA concentration. For example, aerobic exercise, a known modulator of the aging process (Short et al., 2005), activates both the transcriptional coactivator peroxisome proliferator-activated receptor-coactivator-1α (PGC-1α) (Pilegaard et al., 2003) and AMP-activated protein kinase (AMPK) (Winder and Hardie, 1996). Whereas PGC-1α activation triggers increases in the transcription of nuclear and mitochondrial coded mRNA for mitochondrial protein synthesis, AMPK inhibits protein translation through its interactions with mTOR (Mounier et al., 2011). Therefore, without a measurement of protein synthesis, it is difficult to determine whether the newly transcribed mRNA was actually translated into new protein. Also, the manipulation of gene targets can have multiple unknown downstream effects (pleiotropy), by monitoring protein synthesis we measure the final action in the cascade of events. Measuring protein synthesis rates in the context of DNA synthesis rates provides novel insight into proteostatic mechanisms. These measurements are perhaps even more discerning when using in vivo models of slowed aging since long-lived models have complex systemic changes that might not be captured during short term analysis of signaling pathways or protein concentration.
To date, our data strongly support the concept that slowed aging is associated with improved proteostasis. Importantly, they also illustrate the importance of using the assessment of synthetic rates as a mechanistic outcome. The inhibition of mTOR has been the focus of much research, because of its potential to slow the aging process (Flynn et al., 2013; Wilkinson et al., 2012; Wu et al., 2013; Ye et al., 2013). However, our findings in CL indicate that lifespan extension can happen independently of a decrease in mTOR activity (Drake et al., 2014). This lifespan extension independent of a decrease in mTOR activity is seen in other longevity treatments such as branched chain amino acid supplementation (D'Antona et al., 2010). It is yet to be determined which proteins are most important for slowed aging although we hypothesize that these are proteins involved in mitochondrial respiration and stress resistance. Future studies could be enhanced by interrogating mechanisms of proteostasis control as a key regulator of lifespan, as well as the determination of protein synthetic “signatures” of slowed aging.
1.3 Conclusions
By recognizing that proteostasis is a key feature of slowed aging, maintaining proteostasis can then be a mechanistic target to slow the aging process. Monitoring cellular control of individual proteins via protein synthesis is critical to this research because it represents the sum of the highly regulated transcriptional and post-transcriptional processes. However, to properly assess the contribution of the increase in protein synthesis to proteostasis, one must also consider the making of new cells to determine how much protein synthesis is dedicated to new cells versus replacing of existing proteins. It is our goal to further characterize protein synthetic responses in long-lived models in order to identify methods of proteostatic control that contribute to slow aging.
Highlights.
Proteostasis is a dynamic process requiring appropriate methods for monitoring.
Labeling with 2H2O can capture proteostatic mechanisms.
The Protein:DNA synthesis rate is indicative of proteostatic processes.
Examining proteostatic processes is mechanistic for studies of slowed aging.
Acknowledgements
We acknowledge Dr. Richard Miller for his contributions to the original work. The work the led to this review was funded in part by 1K01AG031829-01A1, P30AG013283, and DARPA N66001-10-c-2134.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Bibliography
- Adeli K. Translational control mechanisms in metabolic regulation: critical role of RNA binding proteins, microRNAs, and cytoplasmic RNA granules. AJP: Endocrinology and Metabolism. 2011;301:E1051–64. doi: 10.1152/ajpendo.00399.2011. doi:10.1152/ajpendo.00399.2011. [DOI] [PubMed] [Google Scholar]
- Allen DL, Roy RR, Edgerton VR. Myonuclear domains in muscle adaptation and disease. Muscle Nerve. 1999;22:1350–1360. doi: 10.1002/(sici)1097-4598(199910)22:10<1350::aid-mus3>3.0.co;2-8. doi:10.1002/(SICI)1097-4598(199910)22:10<1350::AID-MUS3>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Austad SN. Methusaleh's Zoo: how nature provides us with clues for extending human health span. J. Comp. Pathol. 2010;142(Suppl 1):S10–21. doi: 10.1016/j.jcpa.2009.10.024. doi:10.1016/j.jcpa.2009.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balch WE, Morimoto RI, Dillin A, Kelly JW. Adapting proteostasis for disease intervention. Science. 2008;319:916–919. doi: 10.1126/science.1141448. doi:10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- Busch R, Kim Y-K, Neese RA, Schade-Serin V, Collins M, Awada M, Gardner JL, Beysen C, Marino ME, Misell LM, Hellerstein MK. Measurement of protein turnover rates by heavy water labeling of nonessential amino acids. Biochim Biophys Acta. 2006;1760:730–744. doi: 10.1016/j.bbagen.2005.12.023. doi:10.1016/j.bbagen.2005.12.023. [DOI] [PubMed] [Google Scholar]
- Collins CA, Olsen I, Zammit PS, Heslop L, Petrie A, Partridge TA, Morgan JE. Stem cell function, self-renewal, and behavioral heterogeneity of cells from the adult muscle satellite cell niche. Cell. 2005;122:289–301. doi: 10.1016/j.cell.2005.05.010. doi:10.1016/j.cell.2005.05.010. [DOI] [PubMed] [Google Scholar]
- D'Antona G, Ragni M, Cardile A, Tedesco L, Dossena M, Bruttini F, Caliaro F, Corsetti G, Bottinelli R, Carruba MO, Valerio A, Nisoli E. Branched-chain amino acid supplementation promotes survival and supports cardiac and skeletal muscle mitochondrial biogenesis in middle-aged mice. Cell Metabolism. 2010;12:362–372. doi: 10.1016/j.cmet.2010.08.016. doi:10.1016/j.cmet.2010.08.016. [DOI] [PubMed] [Google Scholar]
- Drake JC, Bruns DR, Peelor FF, Biela LM, Miller RA, Hamilton KL, Miller BF. Long-lived crowded litter mice have an age-dependent increase in protein synthesis to DNA synthesis ratio and mTORC1-substrate phosphorylation. AJP: Endocrinology and Metabolism. 2014 doi: 10.1152/ajpendo.00256.2014. doi:10.1152/ajpendo.00256.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake JC, Peelor FF, Biela LM, Watkins MK, Miller RA, Hamilton KL, Miller BF. Assessment of mitochondrial biogenesis and mTORC1 signaling during chronic rapamycin feeding in male and female mice. The Journals of Gerontology Series A: Biological Sciences and Medical Sciences. 2013;68:1493–1501. doi: 10.1093/gerona/glt047. doi:10.1093/gerona/glt047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden E, Geva-Zatorsky N, Issaeva I, Cohen A, Dekel E, Danon T, Cohen L, Mayo A, Alon U. Proteome half-life dynamics in living human cells. Science. 2011;331:764–768. doi: 10.1126/science.1199784. doi:10.1126/science.1199784. [DOI] [PubMed] [Google Scholar]
- Flynn JM, O'Leary MN, Zambataro CA, Academia EC, Presley MP, Garrett BJ, Zykovich A, Mooney SD, Strong R, Rosen CJ, Kapahi P, Nelson MD, Kennedy BK, Melov S. Late life rapamycin treatment reverses age-related heart dysfunction. Aging Cell. 2013 doi: 10.1111/acel.12109. doi:10.1111/acel.12109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasier HG, Riechman SE, Wiggs MP, Previs SF, Fluckey JD. A comparison of 2H2O and phenylalanine flooding dose to investigate muscle protein synthesis with acute exercise in rats. AJP: Endocrinology and Metabolism. 2009;297:E252–E259. doi: 10.1152/ajpendo.90872.2008. doi:10.1152/ajpendo.90872.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grebien F, Dolznig H, Beug H, Mullner EW. Cell size control: new evidence for a general mechanism. Cell Cycle. 2005;4:418–421. doi: 10.4161/cc.4.3.1523. [DOI] [PubMed] [Google Scholar]
- Harrison DE, Strong R, Sharp ZD, Nelson JF, Astle CM, Flurkey K, Nadon NL, Wilkinson JE, Frenkel K, Carter CS, Pahor M, Javors MA, Fernandez E, Miller RA. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 2009;460:392–395. doi: 10.1038/nature08221. doi:10.1038/nature08221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapahi P. Protein synthesis and the antagonistic pleiotropy hypothesis of aging. Adv. Exp. Med. Biol. 2010;694:30–37. doi: 10.1007/978-1-4419-7002-2_3. [DOI] [PubMed] [Google Scholar]
- Kimball SR, Horetsky RL, Ron D, Jefferson LS, Harding HP. Mammalian stress granules represent sites of accumulation of stalled translation initiation complexes. Am. J. Physiol., Cell Physiol. 2003;284:C273–84. doi: 10.1152/ajpcell.00314.2002. doi:10.1152/ajpcell.00314.2002. [DOI] [PubMed] [Google Scholar]
- Mary J, Vougier S, Picot CR, Perichon M, Petropoulos I, Friguet B. Enzymatic reactions involved in the repair of oxidized proteins. Experimental Gerontology. 2004;39:1117–1123. doi: 10.1016/j.exger.2004.06.008. [DOI] [PubMed] [Google Scholar]
- Masoro EJ. Overview of caloric restriction and ageing. Mechanisms of Ageing and Development. 2005;126:913–922. doi: 10.1016/j.mad.2005.03.012. doi:10.1016/j.mad.2005.03.012. [DOI] [PubMed] [Google Scholar]
- Miller BF, Hamilton KL. A perspective on the determination of mitochondrial biogenesis. AJP: Endocrinology and Metabolism. 2012;302:E496–9. doi: 10.1152/ajpendo.00578.2011. doi:10.1152/ajpendo.00578.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller BF, Robinson MM, Bruss MD, Hellerstein M, Hamilton KL. A comprehensive assessment of mitochondrial protein synthesis and cellular proliferation with age and caloric restriction. Aging Cell. 2012;11:150–161. doi: 10.1111/j.1474-9726.2011.00769.x. doi:10.1111/j.1474-9726.2011.00769.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller BF, Robinson MM, Reuland DJ, Drake JC, Peelor FF, Bruss MD, Hellerstein MK, Hamilton KL. Calorie restriction does not increase short-term or long-term protein synthesis. The Journals of Gerontology Series A: Biological Sciences and Medical Sciences. 2013;68:530–538. doi: 10.1093/gerona/gls219. doi:10.1093/gerona/gls219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimore GE, Pösö AR. Intracellular protein catabolism and its control during nutrient deprivation and supply. Annu. Rev. Nutr. 1987;7:539–564. doi: 10.1146/annurev.nu.07.070187.002543. doi:10.1146/annurev.nu.07.070187.002543. [DOI] [PubMed] [Google Scholar]
- Mounier R, Lantier L, Leclerc J, Sotiropoulos A, Foretz M, Viollet B. Antagonistic control of muscle cell size by AMPK and mTORC1. Cell Cycle. 2011;10:2640–2646. doi: 10.4161/cc.10.16.17102. [DOI] [PubMed] [Google Scholar]
- Neese RA, Misell LM, Turner S, Chu A, Kim J, Cesar D, Hoh R, Antelo F, Strawford A, McCune JM, Christiansen M, Hellerstein MK. Measurement in vivo of proliferation rates of slow turnover cells by 2H2O labeling of the deoxyribose moiety of DNA. Proc. Natl. Acad. Sci. U.S.A. 2002;99:15345–15350. doi: 10.1073/pnas.232551499. doi:10.1073/pnas.232551499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nisoli E. Calorie Restriction Promotes Mitochondrial Biogenesis by Inducing the Expression of eNOS. Science. 2005;310:314–317. doi: 10.1126/science.1117728. doi:10.1126/science.1117728. [DOI] [PubMed] [Google Scholar]
- Pavlath GK, Rich K, Webster SG, Blau HM. Localization of muscle gene products in nuclear domains. Nature. 1989;337:570–573. doi: 10.1038/337570a0. doi:10.1038/337570a0. [DOI] [PubMed] [Google Scholar]
- Perez VI, Buffenstein R, Masamsetti V, Leonard S, Salmon AB, Mele J, Andziak B, Yang T, Edrey Y, Friguet B, Ward W, Richardson A, Chaudhuri A. Protein stability and resistance to oxidative stress are determinants of longevity in the longest-living rodent, the naked mole-rat. Proceedings of the National Academy of Sciences. 2009;106:3059–3064. doi: 10.1073/pnas.0809620106. doi:10.1073/pnas.0809620106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilegaard H, Saltin B, Neufer PD. Exercise induces transient transcriptional activation of the PGC-1 gene in human skeletal muscle. The Journal of Physiology. 2003;546:851–858. doi: 10.1113/jphysiol.2002.034850. doi:10.1113/jphysiol.2002.034850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poppek D, Grune T. Protein Repair and Degradation. 2005:177–201. doi:10.1007/b101151. [Google Scholar]
- Price JC, Guan S, Burlingame A, Prusiner SB, Ghaemmaghami S. Analysis of proteome dynamics in the mouse brain. Proceedings of the National Academy of Sciences. 2010;107:14508–14513. doi: 10.1073/pnas.1006551107. doi:10.1073/pnas.1006551107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price JC, Khambatta CF, Li KW, Bruss MD, Shankaran M, Dalidd M, Floreani NA, Roberts LS, Turner SM, Holmes WE, Hellerstein MK. The effect of long term calorie restriction on in vivo hepatic proteostatis: a novel combination of dynamic and quantitative proteomics. Molecular & Cellular Proteomics. 2012;11:1801–1814. doi: 10.1074/mcp.M112.021204. doi:10.1074/mcp.M112.021204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MM, Turner SM, Hellerstein MK, Hamilton KL, Miller BF. Long-term synthesis rates of skeletal muscle DNA and protein are higher during aerobic training in older humans than in sedentary young subjects but are not altered by protein supplementation. FASEB J. 2011;25:3240–3249. doi: 10.1096/fj.11-186437. doi:10.1096/fj.11-186437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmon AB, Leonard S, Masamsetti V, Pierce A, Podlutsky AJ, Podlutskaya N, Richardson A, Austad SN, Chaudhuri AR. The long lifespan of two bat species is correlated with resistance to protein oxidation and enhanced protein homeostasis. The FASEB Journal. 2009;23:2317–2326. doi: 10.1096/fj.08-122523. doi:10.1096/fj.08-122523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta S, Peterson TR, Sabatini DM. Regulation of the mTOR complex 1 pathway by nutrients, growth factors, and stress. Molecular Cell. 2010;40:310–322. doi: 10.1016/j.molcel.2010.09.026. doi:10.1016/j.molcel.2010.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Short KR, Bigelow ML, Kahl J, Singh R, Coenen-Schimke J, Raghavakaimal S, Nair KS. Decline in skeletal muscle mitochondrial function with aging in humans. Proc. Natl. Acad. Sci. U.S.A. 2005;102:5618–5623. doi: 10.1073/pnas.0501559102. doi:10.1073/pnas.0501559102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbaugh MJ, Sun LY, Bartke A, Miller RA. Activation of genes involved in xenobiotic metabolism is a shared signature of mouse models with extended lifespan. AJP: Endocrinology and Metabolism. 2012;303:E488–95. doi: 10.1152/ajpendo.00110.2012. doi:10.1152/ajpendo.00110.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Sadighi Akha AA, Miller RA, Harper JM. Life-span extension in mice by preweaning food restriction and by methionine restriction in middle age. The Journals of Gerontology Series A: Biological Sciences and Medical Sciences. 2009;64:711–722. doi: 10.1093/gerona/glp051. doi:10.1093/gerona/glp051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavernarakis N. Ageing and the regulation of protein synthesis: a balancing act? Trends in Cell Biology. 2008;18:228–235. doi: 10.1016/j.tcb.2008.02.004. doi:10.1016/j.tcb.2008.02.004. [DOI] [PubMed] [Google Scholar]
- Treaster SB, Ridgway ID, Richardson CA, Gaspar MB, Chaudhuri AR, Austad SN. Superior proteome stability in the longest lived animal. AGE. 2013 doi: 10.1007/s11357-013-9597-9. doi:10.1007/s11357-013-9597-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson JE, Burmeister L, Brooks SV, Chan C-C, Friedline S, Harrison DE, Hejtmancik JF, Nadon N, Strong R, Wood LK, Woodward MA, Miller RA. Rapamycin slows aging in mice. Aging Cell –. 2012 doi: 10.1111/j.1474-9726.2012.00832.x. doi:10.1111/j.1474-9726.2012.00832.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winder WW, Hardie DG. Inactivation of acetyl-CoA carboxylase and activation of AMP-activated protein kinase in muscle during exercise. Am. J. Physiol. Endocrinol. Metab. 1996;270:E299–E304. doi: 10.1152/ajpendo.1996.270.2.E299. [DOI] [PubMed] [Google Scholar]
- Wu JJ, Liu J, Chen EB, Wang JJ, Cao L, Narayan N, Fergusson MM, Rovira II, Allen M, Springer DA, Lago CU, Zhang S, Dubois W, Ward T, deCabo R, Gavrilova O, Mock B, Finkel T. Increased Mammalian Lifespan and a Segmental and Tissue-Specific Slowing of Aging after Genetic Reduction of mTOR Expression. CellReports. 2013 doi: 10.1016/j.celrep.2013.07.030. doi:10.1016/j.celrep.2013.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye L, Widlund AL, Sims CA, Lamming DW, Guan Y. Rapamycin doses sufficient to extend lifespan do not compromise muscle mitochondrial content or endurance. Aging (Albany NY) 2013 doi: 10.18632/aging.100576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan CL, Sharma N, Gilge DA, Stanley WC, Li Y, Hatzoglou M, Previs SF. Preserved protein synthesis in the heart in response to acute fasting and chronic food restriction despite reductions in liver and skeletal muscle. AJP: Endocrinology and Metabolism. 2008;295:E216–E222. doi: 10.1152/ajpendo.00545.2007. doi:10.1152/ajpendo.00545.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zid BM, Rogers AN, Katewa SD, Vargas MA, Kolipinski MC, Lu TA, Benzer S, Kapahi P. 4E-BP extends lifespan upon dietary restriction by enhancing mitochondrial activity in Drosophila. Cell. 2009;139:149–160. doi: 10.1016/j.cell.2009.07.034. doi:10.1016/j.cell.2009.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]