Abstract
BRCA1 promotes homologous recombination-mediated DNA repair (HRR). However, HRR must be tightly regulated to prevent illegitimate recombination. We previously found that BRCA1 HRR function is regulated by the RAP80 complex, but the mechanism was unclear. We have now observed that PARP1 interacts with and poly-ADP-ribosylates (aka. PARsylates) BRCA1. PARsylation is directed at the BRCA1 DNA binding domain and down-modulates its function. Moreover, RAP80 contains a poly-ADP-ribose (PAR) interacting domain that binds PARsylated BRCA1 and helps to maintain the stability of PARP1-BRCA1-RAP80 complexes.
BRCA1 PARsylation is a key step in BRCA1 HRR control. When BRCA1 PARsylation is defective, it gives rise to excessive HRR and manifestations of genome instability. BRCA1 PARsylation and/or RAP80 expression is defective in a subset of sporadic breast cancer cell lines and patient-derived tumor xenograft (PDX) models. These observations are consistent with the possibility that such defects, when chronic, contribute to tumor development in BRCA1+/+ individuals.
Introduction
BRCA1 is a breast and ovarian cancer- suppressing gene and a major contributor to genome integrity control. The latter function is a major component of its tumor suppressing function (1, 2). Among its various DNA damage response functions, BRCA1 normally promotes error-free, homologous recombination-type, DNA damage repair (HRR). Defects in this pathway lead to DNA damage and genomic instability. Strong genetic and epidemiologic links exist between BRCA1 HRR function and its breast cancer suppression activity (3-6). Yet, how these phenomena are mechanistically connected is poorly understood.
Recent studies, including some from our group, showed that at least four, BRCA1-containing, nuclear protein complexes concentrate in DSB break - containing nuclear foci (e.g. ionizing radiation induced foci, or IRIF) and participate in these structures in the HR repair (HRR) pathway (7-11). One of them, the RAP80-BRCA1 complex, regulates the concentration in IRIF of two HRR-promoting (pro-HRR), BRCA1-containing protein complexes [i.e. the CtIP (aka. RBBP8) - BRCA1 and BACH1 (aka. BRIP1/FANCJ) - BRCA1 complexes]. BRCA1 employs this mechanism in a process that maintains a physiological amplitude of HR-mediated DSB repair. Loss of amplitude regulation (aka. tuning) after RAP80 depletion leads to excessive DSB end-resection and the type of chromosomal instability that, when chronic, is associated with breast and ovarian cancer development (12, 13).
Here, we report that PARP1 is a physiological, RAP80- and BRCA1- associated protein and that its ability to operate as a poly-ADP-ribosyl transferase (pADRT) supports proper HRR-tuning. More specifically, in this process PARP1 poly-ADP-ribosylates (aka. PARsylates) BRCA1, targeting its DNA binding domain and reducing its avidity for DNA. BRCA1 PARsylation is required for maintenance of the stability of the RAP80-BRCA1-PARP1 complex. Moreover, RAP80 contains a PAR-interacting domain (PID) that binds PARsylated BRCA1. This, in turn, enables fine-tuning of BRCA1 HRR function. A major outcome of this process is a BRCA1-driven contribution to chromosome integrity control.
Results
PARP1 is a partner of the RAP80-BRCA1 complex
Using crosslinking- assisted tag affinity purification (CATAP), we identified a number of novel binding partners of the tagged RAP80-BRCA1 complex in HeLa S3 cells (N= 95; Supplementary Fig. S1a and S1b, and see Supplementary information for a detailed description of the method). These proteins can be portrayed as a network of communicating polypeptides, based upon their gene ontology terms and their experimentally deciphered protein interaction properties (14). Among their interacting partners are proteins recently shown to be involved in cellular responses to DSBs, including SFPQ (15), CHD4 (16), and UBR5 (17) (Supplementary Fig. S1c).
Interestingly, PARP1 was identified as one such RAP80-BRCA1 partner (Supplementary Fig. S1c and S1d). Results of a gel filtration experiment showed that PARP1 was detected in a wide range of fractions including those containing BRCA1, RAP80, and ABRAXAS (ABRA1), another component of the RAP80 complex (Supplementary Fig. S1e). These results suggest that a fraction of the detected PARP1 is associated with the RAP80-BRCA1 complex.
We also detected an interaction between PARP1 and RAP80-BRCA1 by endogenous/endogenous coimmunoprecipitation (co-IP), performed in the absence of a cross-linking agent. As shown in Figs. 1a and 1b, endogenous PARP1 associated with endogenous BRCA1, RAP80 and ABRA1. PARP1 was also detected in endogenous BRCA1 IPs (Fig. 1c). Similar interactions between endogenous proteins were detected in co-IP experiments performed with other cell lines (e.g. U2OS, T98G and 293T cells). The same co-IP results were detected in cell lysates treated with ethidium bromide (EtBr), implying that the association between these proteins is not a result of nucleic acid bridging (Supplementary Fig. S1f) (18).
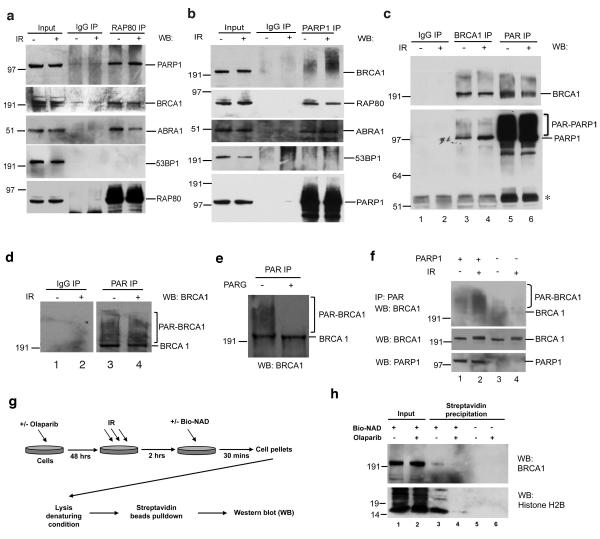
Figure 1. PARP1 is a partner of the RAP80-BRCA1 complex and promotes BRCA1 PARsylation.
(a) HeLa cells were exposed to 10 Gy IR or mock treated and lysed 4 hours later. Immunoprecipitation (IP) was performed with a rabbit polyclonal anti-RAP80 antibody or IgG control. The IPs were blotted, and the blots were probed with antibodies to PARP1, BRCA1, ABRA1, 53BP1, or RAP80. 5% of total cell lysate was blotted as an input control in all IP experiments. (b) HeLa cells were treated as described in (a). IPs were generated with a monoclonal anti-PARP1 antibody or IgG control, blotted, and the blots probed with antibodies to BRCA1, RAP80, ABRA1, 53BP1, or PARP1. (c) HeLa cells were treated as described in (a). IPs were generated with a rabbit polyclonal anti-BRCA1 antibody, or a monoclonal anti-PAR antibody, or IgG control, blotted, and the blots were probed with antibodies to BRCA1 or PARP1. The asterisk indicates the migration position of IgG heavy chain. (d) HeLa cells were treated as described in (a). IPs were generated with a rabbit polyclonal anti-PAR antibody or an IgG control, blotted, and the blots were probed with a monoclonal antibody to BRCA1. (e) HeLa cells were treated, and anti-PAR IPs were performed as described in (d). IPd proteins were eluted and then incubated with PARG or with buffer only. Proteins were then blotted and probed with a BRCA1 antibody. (f) Parp1−/− MEFs were stably reconstituted with human PARP1 cDNA or empty vector. Cells were treated with 10 Gy IR or mock treated and then lysed after 2 hours of recovery. IPs were performed with an anti-PAR antibody and then probed with an antibody directed against mouse Brca1. Input proteins were blotted directly and probed with antibodies recognizing PAR (a different antibody from that used in the PAR IP), mouse Brca1, or PARP1. (g and h) in vivo incorporation of biotinylated-NAD (bio-NAD). (g) A schematic diagram showing the experimental procedure (see details in Methods). (h) Western blots showing that BRCA1 and Histone H2B were detected in streptavidin precipitates while Olaparib pre-treatment blocked bio-NAD incorporation. 20 μg total proteins were loaded as an input control.
PARP1 promotes BRCA1 PARsylation
Interestingly, BRCA1 bands that smeared and migrated more slowly than normal BRCA1 p220 were detected in anti-PARP1 IPs (Fig. 1b). This suggested that the BRCA1 species that exist in complex with PARP1 are modified. To test whether these modified forms of BRCA1 represent poly-ADP-ribosylated (aka. PARsylated) BRCA1, we attempted to co-IP them with anti- poly-ADP-ribose (PAR) antibodies. Multiple BRCA1 bands that migrated more slowly than unmodified BRCA1 appeared in these IPs [Fig. 1c (lanes 5 and 6, a mouse monoclonal anti-PAR antibody was used in these IPs) and Fig. 1d (lanes 3 and 4, a rabbit polyclonal anti-PAR antibody was used in these IPs)]. Similar results were obtained when other cell lines were examined, including U2OS, T98G, 293T, MCF7 and telomerase-immortalized, primary mammary epithelial cells (iMECs), etc, and when three, different anti-BRCA1 antibodies were used (Supplementary Fig. S1g and data not shown).
Poly-ADP-ribose glycohydrolase (PARG) cleaves conjugated ADP-ribose polymers (19). In anti-PAR immunoprecipitates from a cell lysate that was exposed to purified PARG, the smear detected with an anti-BRCA1 antibody was greatly reduced (Fig. 1e). Thus, the modified, slowly-migrating, anti-PAR-reactive BRCA1 species are PARsylated.
To test whether PARP1 is the enzyme that catalyzes BRCA1 PARsylation, a Parp1−/− strain of mouse embryonic fibroblasts (MEFs) (20) was analyzed before and after reconstitution with a full-length human PARP1 cDNA (or with the empty vector as control). Much stronger signals of PARsylated mouse BRCA1 were consistently detected in PARP1-reconstituted than in control cells, and the difference was particularly strong after IR (Fig. 1f, lane 1 and 2). Thus, PARP1 likely catalyzes the vast majority of BRCA1 PARsylation in response to DNA damage. Some weak, anti-PAR- reactive BRCA1 bands were seen in vector-transfected cells, suggesting that other PARPs are responsible for a small fraction of BRCA1 PARsylation (Fig. 1f, lanes 3 and 4 of the top panel) (21, 22). However, these data indicate that BRCA1 is primarily PARsylated by PARP1.
BRCA1 promotes HR-mediated DNA repair in S and G2 phases, using undamaged sister chromatids as templates (23, 24). Thus, we asked whether cell cycle state influences BRCA1 PARsylation. In synchronized T98G cells irradiated at various time points after release from G0, PARsylated BRCA1 was apparent throughout the cell cycle (Supplementary Fig. S1h). However, we observed the most significant increase in PARsylated BRCA1 between 16 and 24 hrs after G1 release, when the largest proportion of cells were in S and G2 (i.e. S+G2 cells had increased from 11% to 70% of the total population) (Supplementary Fig. S1h, lane 4 and 5). From 24 to 32 hrs, when most cells had entered the next G1 (i.e. G1 increased from 30% to 65%), the PARsylated BRCA1 signal decreased significantly (Supplementary Fig. S1h, lane 6). Thus, BRCA1 is most heavily PARsylated during the period in which it exerts its HRR-related function(s) (i.e. S and G2).
Since PARP1 is abundant and stable, the possibility that the observed PARsylation of BRCA1 occurs after cell extraction was raised. However, adding 10 μM Olaparib (a PARP1 inhibitor with an IC50 of ~5 nM (25)) to the cell lysate did not affect the detection of BRCA1 PARsylation (Supplementary Fig. S1i).
Similarly, when permeabilized cells were briefly incubated (30 minutes) with a reaction buffer containing 12.5 μM biotinylated NAD (Bio-NAD) as substrates for PARsylation and then lysed under denaturing conditions (Fig. 1g), BRCA1 was detected in streptavidin- reactive precipitates (Fig. 1h, top panel, lane 3). As a positive control, histone H2B, a known substrate of PARP1 (26), was also found to label with biotin in this assay (Fig. 1h, bottom panel, lane 3). In contrast, pre-incubating cells with Olaparib abolished these PARsylation events (Fig. 1h, lane 4). These results further confirm that a fraction of endogenous BRCA1 is PARsylated in cells.
BRCA1 PARsylation is directed at its DNA binding domain and controlled by an internal sequence
We attempted to determine which segment(s) of BRCA1 is (are) targeted by this modification. Given that mass spectrometry analyses of full-length BRCA1 were uninformative (cf. Supplementary information), we asked whether any of a collection of different GST- BRCA1 p220 fragments that, collectively, represent the entire protein are PARsylated by PARP1 in a cell- free assay (cf. Supplementary information). We found that the reaction primarily targeted one fragment, i.e. F3.7 = aa 501-744 (Supplementary Fig. S2a-f). This fragment is encoded by BRCA1 exon 11 (Fig. 2a). Of note, F3.7 contains sequences that support both BRCA1 nuclear localization (27, 28) and its DNA binding activity (29-32) (Fig. 2a).
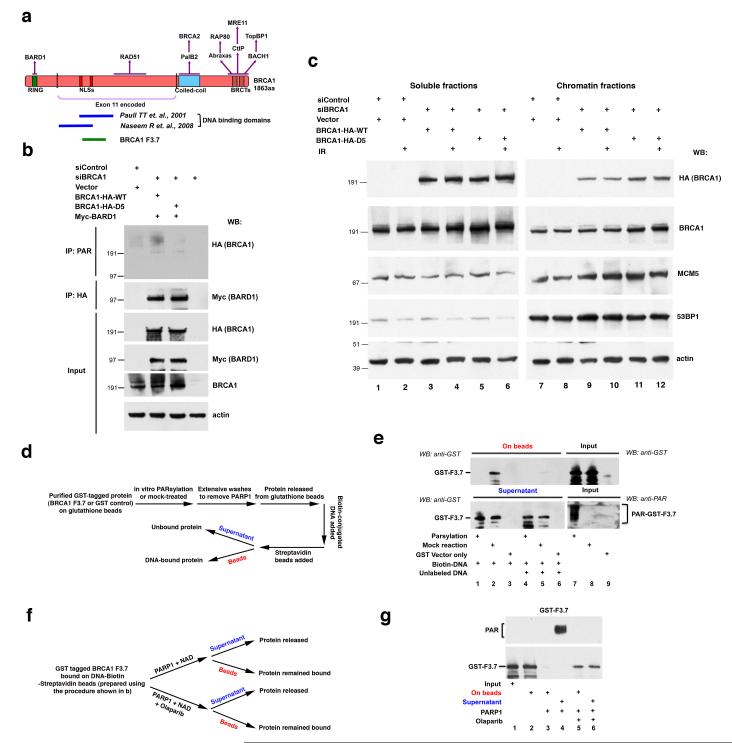
Figure 2. PARsylation modulates BRCA1 DNA binding activity.
(a) A schematic diagram showing elements of the domain structure of human BRCA1 p220. Domains are labeled under the protein diagram, while some of their respective binding partners are indicated above. The section indicated between the two dotted lines is encoded by BRCA1 exon 11. The dark blue lines indicate previously mapped regions required for BRCA1 DNA binding activity. The green line shows the location of BRCA1 fragment F3.7, which is targeted by PARsylation. (b) The BRCA1-D5 mutant is defective for PARsylation in cells. 293T cells were sequentially transfected with the indicated combinations of control siRNA or BRCA1 siRNA and BRCA1 siRNA-resistant WT or D5 mutant cDNA or BRCA1 siRNA only. 48 hours later, cells were irradiated with 10 Gy IR and lysed after 2 hours. IPs were performed with an anti-PAR or an anti-HA antibody (recognizing HA-tagged BRCA1). IPd proteins were blotted, and the blots were probed with HA or Myc antibody (recognizing Myc-tagged BARD1), respectively. Input samples were blotted and the blots were probed with HA, Myc, BRCA1 (recognizing endogenous and HA-tagged BRCA1) or Actin antibody. (c) The BRCA1-D5 mutant exhibits stronger association with chromatin fractions compared to BRCA1-WT. 293T cells transfected with either control or BRCA1 siRNA were subsequently transfected with vector, siRNA resistant- BRCA1-WT, or -BRCA1-D5 as indicated. 48 hours later, cells were irradiated with 10 Gy IR and collected after 4 hours. Cells were processed for extraction to separate soluble and chromatin fractions. Equivalent amounts of lysate protein derived from each fraction were blotted, and the blots were probed with HA, BRCA1 (endogenous), MCM5, 53BP1 or Actin antibody. (d and e) PARsylation of BRCA1 F3.7 abolished its DNA binding activity in vitro. (d) A schematic diagram showing the experimental procedure. For details, please see Methods. (e) DNA- bound (on beads) and unbound (supernatant) proteins were detected by Western blotting, using an anti-GST antibody (lane 1-6). Equivalent amounts of modified (PAR) and unmodified proteins to those used in the DNA binding assay were loaded as input samples (lane 7-9, upper panel). PARsylation was confirmed with an anti-PAR antibody (lane 7-9, lower panel). (f and g) PARP1-driven PARsylation released DNA -bound F3.7. (f) A schematic diagram showing the experimental procedure. For details, please see Methods. (g) Input (lane 1) and equivalent amount of DNA bound GST-F3.7 was mock treated (lane 2) or incubated with PARP1 in the absence (lanes 3 and 4) or presence of Olaparib (lanes 5 and 6). Released protein (supernatant, lanes 4 and 6) and protein still bound to DNA (on beads, lanes 2, 3 and 5) were blotted and probed with an anti-GST antibody. The presence of PARsylated proteins in the supernatant of PARP1-treated sample (lane 4) was detected by probing the same membrane with an anti-PAR antibody (upper panel).
We next asked whether BRCA1 PARsylation results in PAR being conjugated to a specific residue(s) in F3.7. However, after extensive analyses, it appeared that PAR was directed to a number of sites in the F3.7 region (see details in Supplementary Fig. S3a-d and Supplementary information). Such non-unique mono- or poly- ADP-ribose modifications - some within discrete/specific protein domains - have been described for other proteins [for reviews, see (33-35)].
Given that the F3.7 region rather than specific residues within it was targeted for PARsylation, we asked whether there is a dedicated F3.7 sequence that targets PARsylation to this region. We focused on 10, non-overlapping segments of F3.7, each composed of 8 residues. This is because, collectively, they encompass the most evolutionarily conserved region within this BRCA1 segment (36, 37) (Fig. 2a and Supplementary Fig. S3c, D1 – D10). The question was whether deletion of any of them from F3.7 affects its ability to be mono- and/or poly- ADP-ribosylated.
Repeatedly, internal deletion of the D5 segment, which contains BRCA1 p220 residues 611-618 (LRRKSSTR), inhibited both cell- free mono- and poly- ADP-ribosylation of otherwise intact F3.7 (Supplementary Fig. S3e). Moreover, MS analysis performed on peptides derived from Glu-C digestion of mono-ADP-ribosylated BRCA1 F3.7 (cf. Supplementary information) identified only unmodified D5 peptides, suggesting that the D5 sequence, itself, lacks ADP-ribosyl conjugating sites (data not shown).
More importantly, PARsylation of a full-length, wt BRCA1 polypeptide lacking D5 (BRCA1-D5), was also markedly diminished in cells (Fig. 2b). Although BRCA1-D5 was only minimally PARsylated, it is nuclear and appeared to interact normally with BARD1 (Fig.2b, second panel from the top). It also concentrated in IRIF in S/G2 phase cells, like endogenous, wt BRCA1 (Supplementary Fig. S4), and it interacted normally with the BRCA1 partners- CtIP, and BACH1 (please see below). Thus, the BRCA1-D5 mutant exhibited normal BRCA1 properties, including nuclear localization and interactions with important partner proteins. However, it cannot be properly PARsylated.
PARsylation modulates BRCA1 DNA binding activity
Since PARsylation targets a BRCA1 region that partially overlaps the previously mapped BRCA1 DNA binding domains (cf. Fig. 2a), we asked whether PARsylation influences BRCA1 DNA binding activity. Indeed, when two cell lines, each containing a single, genome-integrated I-SceI HRR repair reporter, were treated with Olaparib, the amount of endogenous BRCA1 associated with chromatin near the breakage site increased significantly, as measured by chromatin immunoprecipitation (ChIP) assay (Supplementary Fig. S5a and S5b). The avidity of BRCA1 for these two DSB-containing loci differed, even in the absence of Olaparib. Conceivably, this is due to the different location of each locus in the genome.
Although these results suggest strongly that PARsylation modulates BRCA1 DNA binding after DNA damage in cells, we could not exclude the possibility that global inhibition of PARsylation by Olaparib may disturb other factors, such as certain chromatin structures that might indirectly affect BRCA1 binding. We attempted to circumvent this complication by analyzing the non-PARsylatable BRCA1-D5 and its wt counterpart. Although we observed a small but reproducible increase in the amount of chromatin- associated BRCA1 in cells transiently expressing BRCA1-D5, compared to cells transiently expressing BRCA1-WT at comparable levels (Fig. 2c, compare lanes 11 and 12 with lanes 9 and 10), we found no consistent difference in chromatin-binding between BRCA1-WT or BRCA1-D5 in such cells by ChIP (data not shown). Since only a small fraction of BRCA1 is PARsylated at any time and this modification may turn over rapidly (cf. Fig. 1d and 1h), we may have been unable to detect the subtle and transient difference in chromatin binding between BRCA1-WT and BRCA1-D5 in response to a single DSB in these cells.
However, since BRCA1 preferentially binds four-way junction- structured DNA that exhibits certain features of a recombination intermediate in vitro (29, 32), we asked whether BRCA1 F3.7 manifests such an activity and whether it could be modulated by PARsylation. As shown in Fig. 2d and 2e, unmodified F3.7, which contains the previously designated BRCA1 DNA binding domains (29-32) bound Biotin-labeled, four-way junction structured DNA (Fig. 2e, lane 2, top panel). Binding was DNA sequence- as opposed to biotin- driven, since excessive amounts of the same, unlabeled DNA blocked biotinyl DNA binding to F3.7, likely by competition (Fig. 2e, lane 5, top panel). However, when F3.7 was PARsylated by PARP1 (Fig. 2e, lanes 1, 4 and 7), its DNA binding activity was completely abolished (Fig. 2e, lane 1, top panel). Thus, PARsylation prevented BRCA1 F3.7 binding to a four-way DNA junction structure ex vivo.
Next, we modified the same system to test whether PARP1-driven BRCA1 PARsylation triggers the release of BRCA1 that is already bound to DNA (Fig. 2f). As shown in Fig. 2g, incubating biotinyl DNA- bound BRCA1 F3.7 (Fig. 2g, lane 2, bottom panel) with purified PARP1 and NAD completely released F3.7 from DNA (Fig. 2g, lane 3, bottom panel), and the released F3.7 was clearly PARsylated (Fig. 2g, lane 4, top panel). However, a significant amount of F3.7 remained DNA-bound when PARP1 enzymatic activity was inhibited by Olaparib (Fig. 2g, lane 5, bottom panel), and GST did not bind four-way junction structured DNA (data not shown). Thus, PARsylation led to the release of BRCA1 F3.7 from a DNA substrate in vitro.
Stability of RAP80-BRCA1 complexes after DNA damage requires BRCA1 PARsylation and proper interactions between subunits
Since PARP1 is a RAP80-BRCA1 partner, we asked whether PARP1 enzymatic activity affects the integrity of RAP80-BRCA1 complexes. In cells exposed to a PARP inhibitor, Olaparib, post-IR BRCA1-RAP80 binding was significantly weakened, particularly at late time points after irradiation (Fig. 3a, lane 7-10). By contrast, Olaparib exposure had no effect on the ability of CtIP-, BACH1-, or RAD51- to co-IP with BARD1, a heterodimeric partner of BRCA1 (38) (Supplementary Fig. S6a), or on BRCA1 -BARD1 co-IP (Fig. 3b, lanes 5-8).
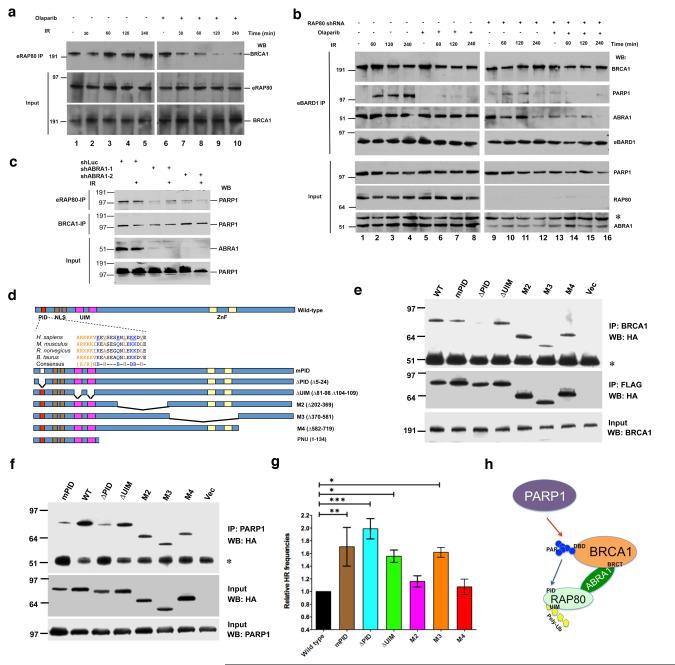
Figure 3. The interaction between PARsylated BRCA1 and RAP80 is required for maintaining BRCA1-RAP80-PARP1 complex integrity after DNA damage and normal HRR regulation.
(a) HeLa cells stably expressing FLAG/HA-tagged RAP80 (eRAP80) were incubated with 30 nM Olaparib for 48 hours and then irradiated with 10 Gy IR. Cells were lysed 30 min, 60 min, 120 min or 240 min post-IR. IPs were performed with antibodies directed against FLAG (recognizing eRAP80) and then probed with anti-BRCA1. Input samples were blotted and probed with an anti-HA (recognizing eRAP80) or an anti-BRCA1 antibody. (b) HeLa cells that stably express FLAG/HA-tagged BARD1 (eBARD1) and an shRNA containing either a RAP80-targeting or a control sequence were generated. Where indicated, these cells were incubated with 30 nM Olaparib or DMSO for 48 hours. Cells were then irradiated with 10 Gy IR and lysed 60 min, 120 min or 240 min later. IPs were performed with anti-FLAG antibody (recognizing eBARD1) and then probed with antibodies directed against BRCA1, PARP1 or ABRA1. Input proteins were blotted and probed with antibodies against PARP1, RAP80 or ABRA1. The asterisk in the ABRA1 blot indicates the migration position of a non-specific band detectable by this antibody. (c) HeLa cells stably expressing eRAP80 were infected with a lentivirus encoding an shRNA directed at Luciferase (shLuc) or ABRA1 (shABRA-1 or shABRA-2). 48 hours later, cells were irradiated with 10 Gy IR or mock treated. Cells were collected 4 hours after IR. IPs were performed with antibodies directed at FLAG (recognizing eRAP80) or BRCA1 and then probed with anti-PARP1 antibody. Input samples were blotted and probed with an anti-ABRA1 or an anti-PARP1 antibody. (d) RAP80 contains a PAR interacting domain. Schematic diagrams indicate the domain structure of human RAP80. PID, poly-ADP-ribose interacting domain; NLS, nuclear localization sequence; UIM, ubiquitin interacting motif; and ZnF, Zinc finger domain. The sequences of the PID domain in four mammalian species are shown. The conserved K/R-rich cluster, hydrophobic, and basic amino acids are indicated in different colors. Seven conserved amino acids (underlined) were mutated to alanine [A] in the mPID mutant. Structures of six N-terminal GST or C-terminal FLAG/HA-tagged RAP80 mutants and an N-terminal T7-tagged RAP80 PNU fragment are shown below. (e) 293T cells were transiently transfected with the RAP80-FLAG/HA vectors described in (d) and, 72 hours later, cells were irradiated with 10 Gy IR. Cells were lysed 3 hours later, and IPs were generated with an anti- BRCA1 antibody, and the IPd proteins were blotted and the blots probed with an anti-HA (top panel and middle panels) antibody. The asterisk indicates the position of IgG heavy chain. 15 μg of protein from each lysate were loaded onto each gel lane, and the subsequent blots were probed with an anti-BRCA1 antibody (bottom panel). (f) 293T cells were treated as in (e). IPs were performed with an anti-PARP1 antibody. IPd proteins were blotted and probed with an anti-HA antibody to recognize epitope-tagged RAP80 mutants (top panel). The asterisk indicates the migration position of IgG heavy chain. 15 μg of protein from each lysate were loaded onto each gel lane, and the subsequent blots were probed with anti-HA (middle panel) or anti-PARP1 antibody (bottom panel). (g) A graph summarizing relative HRR frequencies of cells over-expressing wild-type RAP80 or various RAP80 mutants. DR-GFP HRR assays were performed as described in methods. The abundance of GFP-positive (HRR) cells over-expressing wild-type RAP80 was normalized to one. Experiments were performed in triplicate, and error bars indicate standard deviation. *, p<0.05; **, p<0.01; ***, p<0.001. (h) A schematic diagram showing the relationship between PARP1, BRCA1, ABRA1 and RAP80 in the HRR-tuning complex. Red arrow indicates that PARP1 promotes BRCA1 PARsylation. The blue arrow indicates the binding of PAR to the RAP80 PID domain. BRCA1 BRCT and DNA-binding (DBD) domains, and RAP80 UIM domain are also indicated in the diagram.
In keeping with this observation, the interaction between the non-PARsylatable BRCA1 mutant, BRCA1-D5, and RAP80 was mostly lost after DNA damage. However, its interaction with CtIP or BACH1 was unaffected (cf. Supplementary Fig. S6b, compare lane 7 with lane 3). Thus, PARP1-driven BRCA1 PARsylation is required for stable BRCA1-RAP80 complex formation, but not for BRCA1-CtIP or -BACH1 binding.
Just as Olaparib disrupted the interaction between PARP1 and BARD1 (Fig. 3b, compare lanes 5-8 with lanes 1-4), RAP80 depletion (by shRNA transduction) also disrupted the association between BARD1 and PARP1 (Fig. 3b, lanes 9-12). As expected, the interaction between BRCA1 and BARD1 was unaffected by either treatment (Fig. 3b, top panel). Thus, BRCA1 PARsylation and RAP80 are, individually required for PARP1-BRCA1/BARD1-RAP80 complex stability.
Earlier studies showed that ABRA1 bridges RAP80 and BRCA1 and that the interaction between BRCA1 and ABRA1 does not require RAP80 either before and shortly after IR (i.e. < 2 hrs) (10, 39, 40). However, at a late time point (4 hrs) after IR, we found that ABRA1 binding to BRCA1/BARD1 was weakened in RAP80- depleted cells (Fig. 3b, lane 9-12). Moreover, when cells were exposed to both RAP80 shRNA and Olaparib, the interaction between ABRA1 and BRCA1/BARD1 was further impaired (Fig. 3b, lane 13-16). Thus, RAP80 and PARsylation are also both required for stable BRCA1-ABRA1 interaction.
We then depleted ABRA1 from cells and examined its effects on RAP80-BRCA1/BARD1-PARP1 complex stability. Under this condition, the interaction between RAP80 and PARP1 was significantly weakened, but the interaction between BRCA1 and PARP1 was not affected (Fig. 3c). Thus, the association between RAP80 and PARP1, while indirect, is largely mediated through the BRCA1-ABRA1 complex.
These data reinforce the view that, the overall integrity of RAP80-ABRA1-BRCA1/BARD1-PARP1 complexes is influenced by specific interactions between subunits, PAR, and PARsylated BRCA1. Parenthetically, since BRCA1 was still PARsylated in RAP80- or ABRA1-depleted cells (Supplementary Fig. S6c and data not shown), RAP80 and ABRA1 are not required for BRCA1 PARsylation per se.
Only a small fraction of the RAP80 within RAP80-BRCA1 complexes co-IPd with PARsylated BRCA1 in an IP-re-IP experiment (Supplementary Fig. S6d, lanes 5 and 6). This is in keeping with the knowledge that only a small fraction of BRCA1 in cells is PARsylated. Since rapid turnover is a common fate for DNA damage-induced ADP-ribose polymers (41), we speculate that BRCA1 PARsylation is a transient modification subject to rapid turnover.
RAP80 possesses a functioning PAR interacting domain that is required for normal HRR tuning
Some proteins that functionally communicate with PARP1 and/or PARsylated proteins contain specific domains that bind PAR. These PAR-interacting units (PIDs) play critical roles in performing and/or regulating PARsylation -related biological processes (33, 42). One such PID contains a stretch of 19-21 amino acids that begins with a cluster of basic residues followed by a pattern of hydrophobic amino acids interspersed with basic residues (43-45). We identified such a 21-residue region at the N-terminus of RAP80. It is well conserved in mouse, rat, and bovine RAP80 (Fig. 3d).
To test whether this motif behaves as a bona fide PID, tagged RAP80 mutants were generated (Fig. 3d). The PAR binding activity of purified GST-tagged, full-length wild-type RAP80 and of comparable amounts of these mutant derivatives (Supplementary Fig. S7a) was tested in a slot-blot binding assay. GST-WT RAP80 bound purified PAR polymers, while GST did not (Supplementary Fig. S7b). The PAR-binding capacity of mPID or ΔPID was significant weaker than that of WT (Supplementary Fig. S7b), suggesting that RAP80 can bind PAR, mediated at least in part through its PID. Histone H1, which also contains a PID, served as a positive control (46).
We next asked whether the RAP80 PID could interact directly with PARsylated BRCA1. Since full-length RAP80 binds BRCA1 protein through multiple domains in vitro (data not shown), we tested the binding of a RAP80 fragment that retained only the PID, NLS and UIM motifs (Fig. 3d, fragment PNU) to PARsylated and unmodified BRCA1 F3.7 (i.e. the BRCA1 segment targeted by PARsylation). Mutant fragments containing an mPID mutation, ΔPID or a UIM deletion (ΔUIM) were also tested. The results indicate that the RAP80 PID but not its UIM domain is required for efficient binding to PARsylated BRCA1 F3.7. Moreover, PARsylation of BRCA1 F3.7 was required for this interaction (Supplementary Fig. S7c). Thus, the RAP80 PID facilitates the binding of PARsylated BRCA1 in the RAP80-BRCA1 complex.
Since PARsylation supports the integrity of RAP80-BRCA1 complexes after DNA damage (cf. Fig. 3a and Supplementary Fig. S6b), we asked whether the RAP80 PID contributes to this function. Consistent with the finding that the RAP80 PID is required for binding to PARsylated BRCA1 F3.7 in vitro (Supplementary Fig. S7c), HA-tagged mPID and ΔPID were defective in their association with intact BRCA1 in cells after IR by comparison with WT and other RAP80 mutants (Fig. 3e). Moreover, mPID or ΔPID was also defective in binding to endogenous PARP1 in cells (Fig. 3f). This implies that the PID is required for the stable association of RAP80 and PARP1 (through BRCA1, cf. Fig. 3c). The interaction defect was not associated with failure of these mutants to concentrate in IRIF, since mPID and ΔPID concentrated there normally, while, as expected, ΔUIM failed to do so (8-10, 47) (Supplementary Fig. S7d).
Thus, the RAP80 PID domain, likely through an interaction with PARsylated BRCA1, contributes to overall RAP80-BRCA1-PARP1 complex stability after DNA damage. That said, it is not required for RAP80 concentration in IRIF.
Since a defective PID cannot sustain a normal interaction between RAP80 and either BRCA1 or PARP1 (c.f. Fig. 3e and 3f), we asked whether expressing RAP80 mPID or ΔPID interferes with RAP80-driven HRR-regulation function. Indeed, expression of each resulted in significantly higher HRR frequencies in U2OS cells that contain a single, integrated DR-GFP HRR reporter (U2OS-DR-GFP) (48, 49). As controls for non-specific effects, two other deletion mutants that map outside of the PID sequence, M2 and M4, had no such effect (Fig. 3g). These data imply that RAP80-PAR binding is required to sustain proper HRR regulation.
ΔUIM expression also led to higher HRR frequencies. This result implies that RAP80-polyubiquitin binding, which tethers RAP80 in post-damage foci, is also required for it to function in HRR regulation (Fig. 3g).
ABRA1 serves as another bridge between RAP80 and BRCA1 (10, 39, 40). Thus, we wondered whether this interaction was also important for HRR regulation. The RAP80 sequences that are required for ABRA1 binding map to two, partially overlapping regions within aa 235 – 400 of RAP80 (10, 39). Our RAP80 M2 and M3 mutants cover this region (Fig. 3d). Although the M3 mutant was functional for PAR binding in vitro (cf. Supplementary Fig. S7b), its binding to BRCA1 was considerably weaker than WT (Fig. 3e), presumably due to its defective interaction with ABRA1. Consistent with this observation, expression of the M3 mutant also resulted in increased HRR (Fig. 3g).
Therefore, in addition to maintaining the integrity of RAP80-ABRA1-BRCA1/BARD1-PARP1 complexes, these results imply that the contribution to HRR regulation by RAP80 also requires its intact PID- and ABRA1-BRCA1 interactions as well as normal UIM-poly-ubiquitin binding (Fig. 3h).
BRCA1 PARsylation is required for normal HRR regulation
Consistent with the observation that Olaparib led to increased BRCA1 chromatin association at sites near a single DSB (cf. Supplementary Fig. S5), the magnitude of HR repair of this DSB increased after PARP1 depletion or exposure to 30 nM Olaparib (Supplementary Fig. S8a-c). A similar increase was observed in a mouse ES cell line containing an independent HRR reporter (Supplementary Fig. S8d). At this Olaparib concentration, the inhibitor blocked PARP1 enzymatic activity effectively but did not cause detectable DNA damage, cell cycle changes, or toxicity (Supplementary Fig. S8e and S8f).
Two studies previously reported that PARP1 inactivation or inhibition did not affect the outcomes of I-SceI induced HRR (50, 51). To address the apparent difference in our observations, we compared the effects of four, different PARP inhibitors - 1,5-dihydroxyisoquinoline (ISQ), which was used in one of the two studies noted above (50), Olaparib, Veliparib, and BMN-673. BMN-673 is the most potent and specific PARP1 inhibitor known to date with a IC50 against PARP1 of ~0.5 nM (52). In this experiment, we employed a U2OS- cell line that allows one to measure both I-SceI –induced short track (STGC) –type and long track gene conversion (LTGC) –type HRR (Fig. 4a). The former is a faithful reflection of error-free HRR, and the latter could give rise to non-allelic, error-prone recombination events (53, 54).
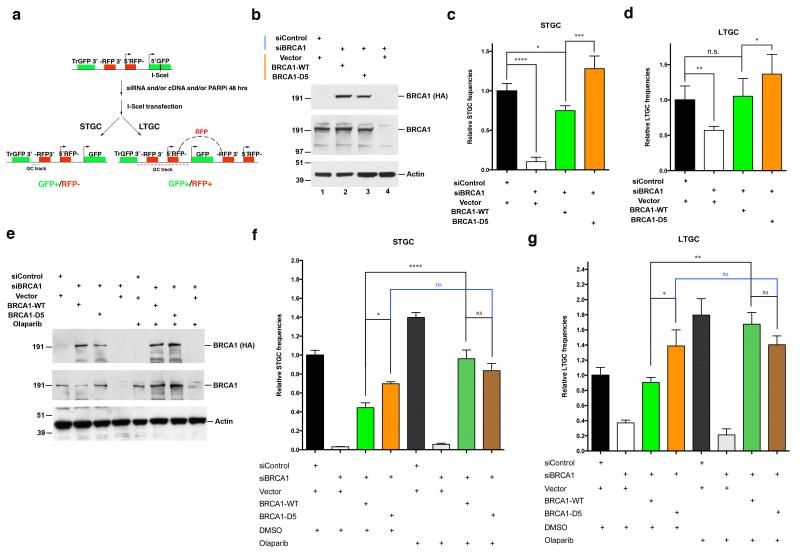
Figure 4. BRCA1 PARsylation is required for normal HRR tuning.
(a) A schematic diagram showing the steps in the I-SceI induced DSB HR repair assay, using cells containing a single copy of the SCR reporter that can measure both STGC and LTGC. Tr-GFP: a 5′ truncated GFP fragment; -RFP3′: 3′ exon of the RFP cassette; 5′RFP-: 5′ exon of the RFP cassette; 5′GFP: A 3′ truncated GFP cassette containing the I-SceI recognition sequence. STGC results in a smaller gene conversion track (green dashed line) and only restores the GFP cassette. LTGC results in a longer gene conversion track and duplications (red dashed line) that leads to a functional RFP cassette by restoring the correct order of RFP exons (black dashed line above the GFP cassette on the right). (b-d) Effects of BRCA1-WT and BRCA1-D5 on STGC and LTGC. U2OS cells containing the SCR reporter were sequentially transfected with BRCA1 siRNA and siRNA-resistant BRCA1-WT or BRCA1-D5. STGC and LTGC assays were performed as described in Methods. (b) Western blots showing depletion of endogenous BRCA1 and expression of BRCA1-WT or BRCA1-D5 in these reporter-containing cells. 25 μg protein was loaded in each lane, and the blot was probed with antibodies directed against HA (detects HA-tagged BRCA1-WT or BRCA1-D5), BRCA1, or Actin. (c) Results of STGC assay. The STGC frequencies of cells transfected with control siRNA plus vector were normalized to one. (d) Results of LTGC assay. The LTGC frequencies of cells transfected with control siRNA and vector were normalized to one. (e-g) Effects of Olaparib on STGC and LTGC in cells depleted of BRCA1 or expressing endogenous BRCA1, or depleted of endogenous BRCA1 and expressing ectopic BRCA1-WT or BRCA1-D5. (e) Western blots showing depletion of endogenous BRCA1 and re-expression of BRCA1-WT or BRCA1-D5 in cells. These cells were either treated with DMSO or 30 nM Olaparib for 48 hours before being transfected with an I-SceI expressing plasmid. 25 μg of protein were loaded in each lane, and the blot was probed with HA (detects HA-tagged BRCA1-WT or BRCA1-D5), BRCA1 or Actin antibodies. (f) Results of STGC assay. The STGC frequencies of cells transfected with control siRNA and vector were normalized to one. (g) Results of LTGC assay. The LTGC frequencies of cells transfected with control siRNA and vector were normalized to one. *, p<0.05; **, p<0.01; ***, p<0.001; ****, p<0.0001; n.s., not significant.
When cells were exposed to each of these inhibitors over a range of relatively low concentrations (i.e. 5 - 40 times the IC50 for PARP1 for each inhibitor-Supplementary Fig. S8g), all four stimulated HRR (both STGC and LTGC) to various extents (Supplementary Fig. S8h and S8i). However, at higher doses, ISQ either had no effect (at 75 μM) or actually inhibited HRR (at 300 μM) (Supplementary Fig. S8h and S8i). The ISQ concentration used in the earlier report was 600 μM (50). These data suggest the presence of an off-target effect by a compound (ISQ) with significantly lower avidity for PARP1 than the other inhibitors. Thus, the effect of PARP inhibitors on HRR is dose-dependent. Within a range that elicited what are likely to be on-target effects, HRR stimulation was a constant finding.
To further determine the effect of BRCA1 PARsylation on HRR, and to avoid potential confounding effects of PARP1 inhibition or depletion, we analyzed cells that transiently expressed HA-tagged BRCA1-WT or identically tagged, PARsylation-defective BRCA1-D5. Each was expressed at levels comparable to that of endogenous BRCA1. Moreover, the mRNA for both had been rendered BRCA1 sh/siRNA- resistant. Expression of endogenous BRCA1 was suppressed by shRNA and siRNA, respectively, followed by a comparison of the frequencies of HR- mediated repair of a single I-SceI induced DSB in these cells and controls (Fig. 4b).
Surprisingly, RNAi- resistant BRCA1-D5 expression in BRCA1- depleted cells led to significantly higher HRR frequencies (for both STGC and LTGC) than those observed in both control cells and cells re-expressing BRCA1-WT (Fig. 4c and 4d, compare column 3 and 4). In addition, Olaparib treatment led to an enhancement of both STGC and LTGC in cells expressing BRCA1-WT, but not in cells expressing BRCA1-D5 (Fig. 4e - g). This implies that the observed HRR stimulation caused by Olaparib within its on-target concentration range is mostly contributed by a failure of proper BRCA1 PARsylation.
These data indicate that BRCA1 PARsylation is required for normal HRR amplitude regulation. We hypothesize that the hyper-HRR phenotype associated with PARP1 inhibition is a product of defective BRCA1 PARsylation, since it was the only major difference observed between BRCA1-D5 and BRCA1-WT-driven HHR function in these experiments.
BRCA1 PARsylation is required for maintaining chromosome integrity in response to DNA damage
Recent studies demonstrated that the chromosomal rearrangements observed after IR exposure in BRCA1-deficient cells are largely a product of combining inactive HRR with overactive, end joining- mediated, and erroneous repair of DSBs (55, 56). Since cells, in which endogenous, wild-type BRCA1 was replaced by the non-PARsylatable mutant, BRCA1-D5, actually revealed higher HRR activity (cf. Fig. 4c and 4d), we asked whether there were fewer rearranged chromosomes induced by DNA damage in these cells compared to cells expressing comparable amounts of BRCA1-WT (Supplementary Fig. S9a).
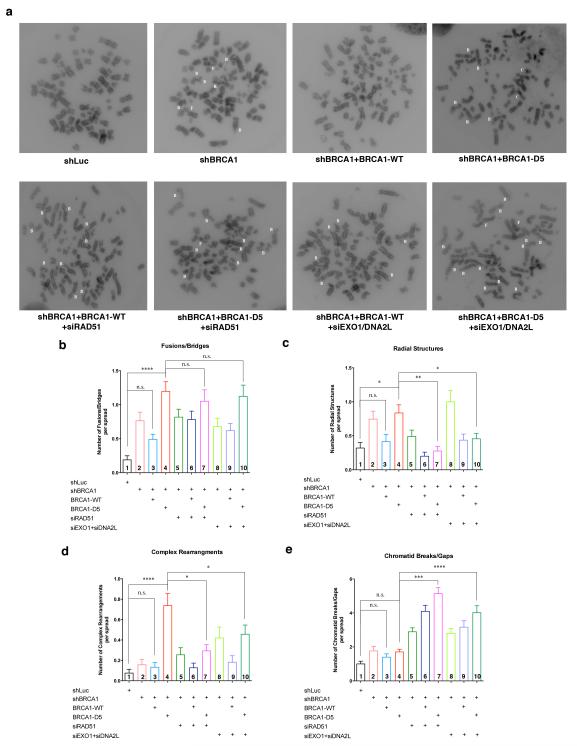
To test this possibility we irradiated cells and then analyzed them for chromosomal abnormalities. Each damaged or rearranged chromosome present in a series of cell spreads was assigned to one of four categories: (1) Fusions/Bridges (F), in which two chromatids from different chromosomes appear to be connected; (2) Radial structures (R), in which chromosomes are composed of three- or four-armed structures; (3) Complex rearrangements (C), in which three or more chromosomes are connected to form complex structures that cannot be assigned to any of the other three types; and (4) Chromatid breaks/gaps (B) (Fig. 5a).
Figure 5. Defective BRCA1 PARsylation results in HRR-dependent chromosome rearrangements after IR.
(a) Representative metaphase spreads of U2OS cells: transduced with control shRNA (shLuc), shBRCA1, treansduced with shBRCA1 and re-expressing BRCA1-WT or BRCA1-D5, or cells transfected with RAD51 siRNA or EXO1/DNA2L siRNA in addition to the previous treatment. Cells were irradiated with 150 rad and collected at 8 hours later for chromosome analysis. White letters in each spread indicates the nature of aberrant chromosomes present: B – chromatid breaks/gaps; F – fusions/bridges; R – radial structures; C – complex rearrangements. (b-e) Bar graphs showing frequencies of these various aberrant chromosomes in cells treated with indicated combinations of shRNA, siRNA and/or cDNAs. (b) Frequencies of fusions/bridges, (c) frequencies of radial structures, (d) frequencies of complex rearrangements, and (e) frequencies of chromatid breaks/gaps. At least 50 metaphase spreads were counted for each category of cells. Statistical analyses were performed using Kruskal-Wallis tests. Results of selected multiple comparisons are shown. ****, p < 0.0001; ***, p < 0.001; **, p < 0.01; *, p < 0.05; n.s., not significant.
Compared with cells transduced with control (shLuc) shRNA, cells in which endogenous BRCA1 was depleted by shBRCA1 (Supplementary Fig. S9a) displayed significantly higher frequencies of fusions/bridges (Fig. 5b, column 2) and radial structures (Fig. 5c, column 2). But there were only slight increases in complex rearrangements (Fig. 5d, column 2) and breaks/gaps (Fig. 5e, column 2) in these cells.
Re-introduction of shRNA-resistant BRCA1-WT expression in these endogenous BRCA1-depleted cells (Supplementary Fig. S9a) partially reduced the frequencies of fusion/bridges (Fig. 5b, column 3) and radial structures (Fig. 5c, column 3).
As described earlier, cells reconstituted with the non-PARsylatable BRCA1-D5 revealed a higher HRR amplitude than those reconstituted with BRCA1-WT (cf. Fig. 4c and 4d). Thus, we expected to find that BRCA1-D5- expressing cells revealed even fewer damaged chromosomes than BRCA1-WT-reconstituted expressing cells. Surprisingly, BRCA1-D5 expressing cells revealed significantly higher frequencies of fusion/bridges (Fig. 5b, column 4), radial structures (Fig. 5c, column 4), and complex rearrangements (Fig. 5d, column 4) than those of cells expressing BRCA1-WT. Indeed, the frequencies of complex rearrangements in BRCA1-D5 expressing cells were 5-7 fold higher than those in control, BRCA1-depleted cells either before or after BRCA1-WT re-expression (Fig. 5d, compare column 4 with columns 1, 2 and 3). By contrast, there was no difference in the frequencies of breaks/gaps between BRCA1-WT and BRCA1-D5 re-expressing cells (Fig. 5e, column 4).
Given these observations, we hypothesized that certain forms of chromosome rearrangement observed in cells expressing BRCA1-D5 might be the results of aberrant HR-dependent repair. To test this, we cotransfected cells with siRNAs targeting RAD51 or EXO1+DNA2L. RAD51 is the prime recombinase required for performing homology search and strand exchange reactions in HR (57). EXO1 and DNA2L are two major nucleases that facilitate the generation of long, single-stranded DNA segments needed for HRR (58).
Importantly, depletion of either RAD51 or EXO1+DNA2L reduced the levels of both radial structures (Fig. 5c, compare columns 7 and 10 with column 4) and complex rearrangements (Fig. 5d, compare columns 7 and 10 with column 4) in BRCA1-D5- expressing cells. This suggests that these two types of chromosome aberration in cells expressing BRCA1-D5 were dependent upon key aspects of the HRR mechanism.
Transfecting cells with RAD51 or EXO1+DNA2L siRNAs resulted in the blocking of HRR downstream of BRCA1-CtIP (59, 60). This would result in arrested repair of those initial IR-induced chromosomal breaks that would have been repaired by HRR. Therefore, we expected to observe higher accumulations of chromosomal breaks/gaps in cells expressing BRCA1-D5 than in control, BRCA1-depleted, or BRCA1-WT re-expressing cells, when RAD51 or EXO1+DNA2L were depleted. This was indeed the case (Fig. 5e, columns 7 and 10). This result further suggests that hyperactive, aberrant HRR is responsible for the radial structures and complex rearrangements seen in cells expressing BRCA1-D5.
In these experiments, RAD51 depletion elicited a much stronger inhibitory effect on HRR than that of EXO1+DNA2L depletion, since other nucleases could compensate for some lost function of EXO1 and DNA2L (Supplementary Fig. S9b). Moreover, RAD51 function is considered essential for HRR. This may help to explain why RAD51 depletion led to a more extensive reduction in radial structures and complex rearrangements than depletion of EXO1+DNA2L (Fig. 5c and 5d).
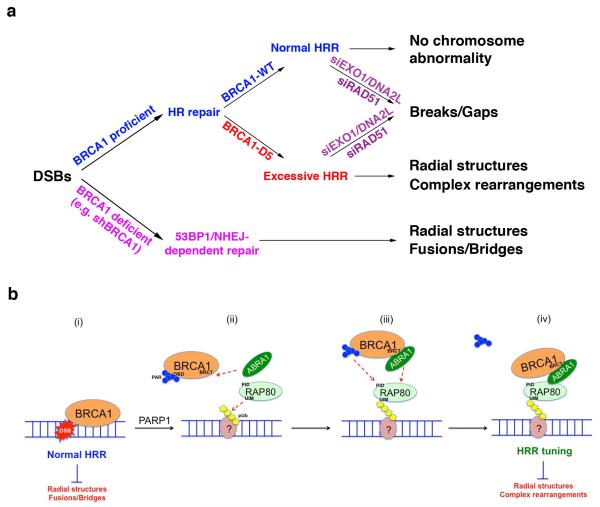
In summary, these observations indicate that defective BRCA1 PARsylation, observed in cells expressing BRCA1-D5 in place of wt BRCA1, fosters the development of chromosome aberrations after DNA damage. Thus, loss of BRCA1 function, which leads to HRR deficiency and a preference for NHEJ- type repair, and deregulated, excessive BRCA1 HRR activity, can both elicit deleterious, mutagenic chromosome instability (Fig. 6a and 6b).
Figure 6. Models showing the effects of BRCA1 PARsylation on genome integrity control after DNA damage.
(a) A schematic diagram showing the development of NHEJ-dependent or HRR-dependent chromosome damage and rearrangements in cells depleted of BRCA1 or in cells expressing either wild-type (BRCA1-WT) or non-PARsylatable BRCA1 (BRCA1-D5), respectively. (b) A schematic diagram showing the role of timely PARP1-driven BRCA1 PARsylation in HRR tuning and genome integrity control. (i) BRCA1 normally promotes HRR by interacting with damaged DNA and recombination intermediates. Normal HRR suppresses the development of radial structures and fusions/bridges. (ii) PARP1-driven BRCA1 PARsylation releases BRCA1 from DNA by targeting its DNA binding domain (DBD). ABRA1 interacts with BRCA1 BRCT domain and RAP80 binds to poly-ubiquitin chains (pUb) formed at or near the site of DNA damage (indicated by a circle with “?” since the nature of the chromatin structure involved remains unclear). The sequence of these events is unknown and it is possible that they occur concurrently. (iii) RAP80 interacts with PARsylated BRCA1 through its PID and also through its interaction with ABRA1. (iv) PAR is removed after the formation of stable RAP80-ABRA1-BRCA1 complexes. The successful completion of these steps ensures proper HRR amplitude control (i.e. tuning) and prevents the development of radial structures and complex chromosome rearrangements.
Finally, RAD51 or EXO1+DNA2L depletion had no significant impact on the frequencies of fusions/bridges in cells expressing BRCA1-D5, which remained significantly higher than other groups of cells (Fig. 5b). We speculate that a BRCA1-D5 function other than promoting hyper- HRR is responsible for this abnormal phenotype. This is consistent with the fact that BRCA1 engages in repair processes other than HRR and that BRCA1-D5 may participate in these and yet other deregulated repair processes (61, 62)
The BRCA1 PARsylation-RAP80 pathway is defective in a subset of breast cancer cells and tumors
Since BRCA1 is a breast cancer gene, we investigated whether BRCA1 is PARsylated in normal, human mammary and breast cancer cells. Here we examined a panel of 13 well-established sporadic breast cancer cell lines along with iMECs, a telomerase- immortalized normal, primary mammary epithelial cell line (Fig. 7a). Among the 13 breast cancer cell lines, 9 were characterized as triple-negative, basal-like (TNBC, i.e. ER-, PR- and HER2 not amplified) subtype. Two of them contain disease-producing mutations in the BRCA1 gene. The remaining 4 were of the luminal subtype (63, 64).
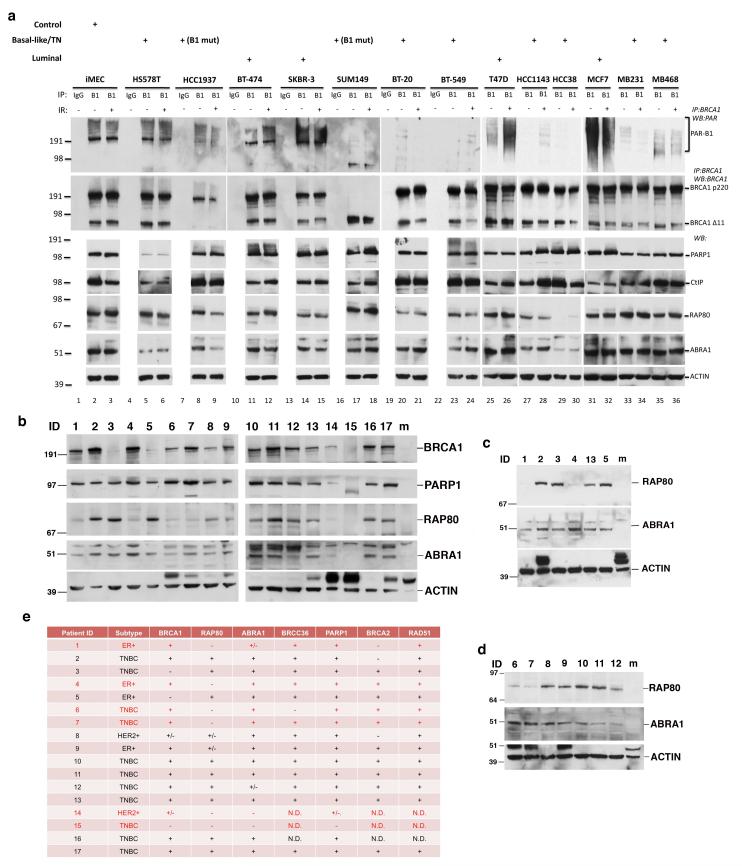
Figure 7. BRCA1 PARsylation and/or RAP80 expression is suppressed in a subset of breast cancer cell lines and tumors.
(a) Results of IP-Western blots searching for BRCA1 PARsylation in a panel of normal breast and breast cancer cell lines (the pathological subtype of each cell line is indicated above the relevant blot; B1-mut: cell lines that contain pathological BRCA1 mutations). Cells were irradiated with 10 Gy IR or mocked treated and collected 8 hours later for analysis. IPs were performed using a rabbit polyclonal anti-BRCA1 antibody. Immunoprecipitated proteins were blotted and probed with a monoclonal anti-PAR antibody (top panels) or a monoclonal anti-BRCA1 antibody (the second row of panels). 20 μg of protein extract from each cell line were blotted and probed with antibodies recognizing PARP1, CtIP, RAP80, ABRA1 and Actin, respectively. (b-d) Results of Western blots detecting BRCA1, PARP1, RAP80, ABRA1 and ACTIN expression in subsets of tumor samples from PDX models. 20 μg of protein extract from each tumor were blotted and probed with antibodies recognizing the above noted proteins. “m” indicates lanes that were loaded with extracts from mouse breast tissues. Please note that mouse Brca1, Rap80 and Abra1 were not detectable by the antibodies used, which specifically recognize human proteins. Results shown were obtained in three, different experiments using independent snap-frozen tumor samples (see Supplementary Methods). (e) Table summarizing the results of protein expression analysis by Western blotting of tumor samples from 17 PDX models. +, positive; −, negative; +/−, weaker expression than other samples; N.D., not determined.
We evaluated the status of BRCA1 PARsylation in these cell lines before and after IR by IP-Western assay (Fig. 7a). Surprisingly, although comparable levels of intact BRCA1 p220 were detected at in all but the two BRCA1 mutant lines (HCC1937 and SUM149) (Fig. 7a), the levels of PARsylated BRCA1 were significantly lower or even undetectable in 6 lines, all of which were derived from sporadic TNBC (Fig. 7a, top panel). This suggests the existence of certain defects in the generation and/or maintenance of BRCA1 PARsylation in a subset of breast cancer lines that express wild-type BRCA1.
We next asked whether similar defects in BRCA1 PARsylation and/or RAP80 expression are also present in primary human breast tumors. However, since the only way to detect BRCA1 PARsylation is IP-Western blotting, which requires large amounts of freshly prepared protein lysates, we were unable to obtain results of satisfactory quality from primary tumor samples.
Therefore, we turned to an analysis of patient-derived xenograft (PDX) breast cancer tumors, which have been shown to resemble, pathologically and genetically, the primary tumors from which they were derived (65-67). While the status of BRCA1 PARsylation remained undetermined due to high background signals (likely from host mouse tissues), we repeatedly detected defects in RAP80 and/or ABRA1 expression in some of these tumors (Fig. 7b-d). A summary of these observations is provided in table (Fig. 7e).
Discussion
We have identified PARP1 as a physiological partner of the RAP80-BRCA1 complex. PARP1 PARsylates BRCA1, a pivotal contributor to HR-mediated DSB repair (HRR). Although PARP1-driven BRCA1 PARsylation is not required for HRR function, per se, it represents a critical step in its regulation (aka tuning). BRCA1 PARsylation is targeted to a polypeptide segment within its DNA-binding domain and is controlled by a short oligopeptide sequence, D5, that resides near the center of this segment. PARsylation modulates BRCA1 DNA binding activity in vitro and is required for normal HRR tuning. It is required to suppress the development of a hyper-HRR state. Importantly, following DNA damage, hyper-recombination resulting from defective BRCA1 PARsylation constitutes a genome-destabilizing force (cf. Fig. 6b).
The function of BRCA1 in supporting HRR has long been considered an important suppressor of breast and ovarian cancer (2). This notion was strengthened by the observation that chromosome instability and cancer development arising when BRCA1 function is absent from cells are mainly the result of an inability to activate normal DSB end-processing by BRCA1 and/or by certain of its partner proteins that are required for HRR (55, 56).
However, recent observations indicate that excessive BRCA1 activity and/or failure to regulate such activity is also associated with genomic instability (12, 13, 68). Importantly, as shown here, BRCA1 PARsylation, which requires a built-in, cis-acting BRCA1 element (i.e. D5), contributes to the BRCA1 genome integrity control system. Given the strong connection between its genome integrity control and its tumor suppression function, it is likely that D5 function contributes to BRCA1 tumor suppressing activity.
The exact action by the D5 sequence that results in the licensing of BRCA1 PARsylation remains unknown. The D5 sequence is not the direct target of poly-ADP-ribose (PAR) coupling (cf. Supplementary Fig. S3), and its integrity is not required for the DNA binding activity of full length BRCA1 in the absence of PARsylation (data not shown). A mutant D5 sequence also failed to interfere with BRCA1 recruitment to sites of DNA damage (i.e. nuclear foci) or its interactions with known BRCA1 partner proteins that perform important steps in HRR (cf. Supplementary Fig. S4 and Fig. S6). Therefore, it is possible that the role of this sequence is to maintain a critical structural conformation of BRCA1 that is required for proper and timely PARsylation and subsequent modulation of BRCA1 DNA binding activity. Yet other possibilities exist. However, due to the location of the D5 sequence within the exon 11-encoded region, which is neither functionally nor structurally well characterized, we are currently unable to test the conformation hypothesis.
Unexpectedly, hyper-recombination resulting from defects in BRCA1 PARsylation was associated with the appearance of radial structures and complex chromosome rearrangements shortly after DNA damage. In contrast to the radial structures observed in BRCA1-depleted cells, which are dependent on 53BP1 and the NHEJ pathway (55, 56), those observed in BRCA1-D5 expressing cells were hyper HRR -dependent. The complex rearrangements observed in BRCA1-D5 cells were similarly hyper HRR-dependent (cf. Fig. 5 and 6a). Since these rearranged chromosomes emerged within one cell cycle, they might represent products of unresolved recombination intermediates.
PARP1 is an abundant enzyme that likely participates in multiple pathways, which, directly or indirectly, affect the outcome of HRR. Two previous reports, both of which also used I-SceI-based HRR reporters, concluded that PARP1 is not involved in the repair of I-SceI induced DSBs (50, 51) - in contrast to what was reported here. In keeping with the apparent off-target effects triggered by very high concentrations of ISQ (cf. Supplementary Fig. S8g-i), differences in experimental conditions may have contributed to this discrepancy. In addition, the immortalized Parp1−/− MEF cells used in one of the above-noted studies (51) retained detectable, albeit low level BRCA1 PARsylation activity, probably due to compensatory effects of other PARP enzymes (21) (cf. Fig. 1f). Therefore, a possible explanation for the lack of effect on HRR in these Parp1−/− MEFs is that other PARPs, through an emerging state of adaptation, were able to support normal HRR regulation at a single I-SceI induced DSB in these cells. Whatever the case, the demonstration that ISQ, at concentrations much closer to its IC50, enhanced HRR, much like three, more specific PARP inhibitors (Supplementary Fig. S8h and S8i), reinforces the view that PARP1 functions, at least in part, by negatively regulating BRCA1-driven HRR function.
Thus, we speculate that preventing the formation of proper RAP80-BRCA1-PARP1 complexes and blocking BRCA1 PARsylation, e.g. with a suitable PARP1 inhibiting agent or by replacing endogenous BRCA1 with a non-PARsylable mutant, will translate into a major breakdown in HRR control. Since HRR control is also a suppressor of radial and complex chromosomal rearrangements, this effect is likely to be accompanied by overt chromosomal instability (cf. Fig. 6). Such an outcome would be expected in any proliferating cell, such as certain hematopoietic progenitor cells in the bone marrow. Sufficient chronic genome disorder could, in time, nurture elements of a neoplastic phenotype.
Multiple PARP inhibitors are being tested in clinical trials as a novel class of therapeutic agents. BRCA1- or BRCA2- deficient cancer cells are hypersensitive to PARP inhibitors due to their inability to repair PARP inhibitor-induced DNA damage or trapped PARP-DNA complexes in the absence of efficient HRR (69-72). Currently, PARP inhibitors are under clinical investigations for use in other cancers that synthesize fully functional BRCA1, such as ovarian cancers, either as single agent or in combination with other therapeutic agents (73, 74). However, our results suggest that the use of PARP inhibitors in a BRCA1-proficient background requires caution, since chronic hyper-recombination in conjunction with sporadic or therapeutic DNA damage may result in deleterious genomic instability in non-tumor cells.
Given the value of HRR tuning in genome stability control, one wonders whether it also operates in breast cancer suppression, since BRCA1 PARsylation defects were noted in some sporadic TNBC cell lines (cf. Fig. 7a). If so, PARP1+RAP80-driven regulation of the amplitude of BRCA1 HRR function would be a physiological process that, if defective, might be clinically relevant.
Recent studies suggest that certain mutations or polymorphisms in the RAP80, ABRA1, or MERIT40/NBA1 (another member of the RAP80 complex) genes are associated with increased susceptibility to breast and/or ovarian cancer (75-80). In addition, the genomic region containing the RAP80 gene is not infrequently lost in TNBCs, which commonly exhibit gross manifestations of genomic instability (81). RAP80 and/or ABRA1 expression was also suppressed in 6 out of 17 of our PDX breast cancer models, of which 3 were TNBC in origin (cf. Fig. 7e). Finally, cells of RAP80 or ABRA1 knockout mice experience a breakdown in genome integrity control, and these mice are more cancer -prone than wt controls (82-84). These observations, together with our findings, suggest that a deficiency in RAP80 complex function and/or BRCA1 PARsylation contributes to the genomic instability that drives cancer development.
Methods
(For additional Methods, please see supplementary information.)
Cell culture
All cells were cultivated at 37°C in a humidified incubator in an atmosphere containing 10% CO2. U2OS, HeLa, 293T, T98G, and MEF cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS). HeLa S3 cells were grown in Joklik’s Minimum Essential Medium (Sigma) supplemented with 7% newborn calf serum at 37°C. Mouse ES cells were grown in ES medium (Invitrogen) on either MEF feeder cells or gelatinized plates supplemented with LIF, and cultured at 37°C in a humidified incubator in an atmosphere containing 5% CO2. iMEC cells were grown in MEBM medium (Lonza Walkersville) supplemented with mammalian epithelium growth factors. Breast cancer cell lines were cultured according to the guidelines provided by ATCC or the suppliers. No authentication of these cell lines was performed by the authors.
I-SceI recombination assay
U2OS cells containing a single copy of the DR-GFP reporter (U2OS-DR) were employed using methods that were described previously (49). Experiments using U2OS or mouse ES cell lines, each containing a single copy of the SCR reporter were performed as described (85). Two versions of the SCR reporter were used in experiments. One detects LTGC events by forming blasticidin (BSD) resistant colonies, while the other measures LTGC events by counting cells with RFP fluorescence. Briefly, for the STGC assay, cells were transfected with pCBASce (I-SceI) plasmid and collected 72 hours later. GFP positive cell populations (i.e. STGC) were detected and measured by flow cytometry. For the LTGC assay, after pCBASce transfection, cells were either plated in 60 mm plates at 5 × 105 cell/plate and then selected with 10 μg/ml blasticidin (BSD) beginning 48 hours later for 12 days, or measured by flow cytometry for RFP positive cells. In all experiments, a plasmid expressing DsRed was transfected in parallel to determine transfection efficiency. To determine plating efficiency (PE), cells were plated in 60 mm plates at 5000 cells/plate but were not selected with BSD. In the case of BRCA1- depleted and -reconstituted cells, since their ability to form colonies was compromised, the rates of cell survival after BSD selection were determined using the CellTiter-Glo method (Promega) following the manufacturer’s protocol.
PARG treatment
Immunoprecipitated proteins were eluted by incubating beads with elution buffer (0.1 M Glycine pH 2.5, 0.1 M NaCl). The eluted protein-containing solution was diluted with 2X PARG reaction buffer (100 mM KH2PO4, 100 mM KCl, 0.2 mg/ml BSA, 0.2% Triton X-100). Recombinant PARG (Trevigen) was added to a final concentration of 10 ng/ml, and the reaction mixture was incubated at room temperature for 60 minutes.
PARP inhibitor
The PARP inhibitors, Olaparib (AZD2281), Veliparib (ABT-888), ISQ and BMN-673, were dissolved in Dimethyl sulfoxide (DMSO) and stored at -20oC. When employed in tissue culture experiments, each was further diluted in tissue culture medium. In most experiments, the final concentration of Olaparib in the medium was 30 nM, and control cells were incubated with medium containing the same concentration of the solvent, DMSO (final concentration: 0.003%).
Biotinylated-NAD incorporation
HeLa cells growing in plates were washed twice with PBS and then incubated with PARP reaction buffer (56 mM HEPES, pH = 8.0, 28 mM KCl, 2mM MgCl2, 0.01% digitonin, 12.5 μM biotinylated-NAD (Trevigen), supplemented with 100 nM Olaparib or 0.01% DMSO) for 30 minutes at 37oC. Cells were then collected and lysed in pre-heated denaturing lysis buffer [400 mM NaCl, 0.5 mM EDTA, 50 mM Tris-HCl pH 7.5, 0.5% NP-40, 70 mM ß-Mercaptoethanol, 1.5 mM MgCl2, 10% Glycerol, 5 mM NEM, 1 μM ADP-HPD (a PARG inhibitor) and protease inhibitors] and heated for 10 min at 95 °C. After incubating for 30 min at 4 oC, lysates were cleared by centrifugation and then incubated with Streptavidin M-280 Dynal beads (Invitrogen) to trap Biotin- containing proteins according to manufacturer’s guidelines.
In vitro ADP-ribosylation assay
Bead-bound GST-tagged BRCA1 fragments of defined sequence were incubated with 10 pmoles purified human PARP1 (Enzo, ALX-201-063-C020) or PARP3 (Enzo, ALX-201-252-C010) in reaction buffer (50 mM Tris-HCl pH 7.5, 4 mM MgCl2, 250 μM DTT, 20 mM NaCl, 100 μM NAD, and activated DNA (Trevigen)) at room temperature for 20 minutes. For reactions containing Bio-NAD, 75 μM NAD and 25 μM Bio-NAD were combined and added in reaction buffer solution. Beads were then washed 3 times with washing buffer (55 mM Tris-HCl pH 7.5, 1 mM EDTA, 200 mM NaCl, 1 mM DTT, 1% Triton X-100) to stop the reaction. Beads were washed once more with 50 mM Tris-HCl pH 8.0 before GST-tagged proteins were eluted and analyzed.
DNA binding assay
Oligonucleotides (sequences available upon request) were labelled using a Biotin 3′ End DNA Labelling Kit following manufacturer’s instructions (Pierce/Thermo). They were then annealed to generate Biotin-conjugated four-way junction DNA structures. GST-F3.7, while still bound to glutathione beads, was PARsylated in vitro by PARP1, as described above, or mock treated. After the reaction, beads were washed with GST binding buffer (55 mM Tris-HCl, pH7.5, 1 mM EDTA, 200 mM NaCl, 1% Triton X-100, 1mM DTT and protease inhibitors) four times before GST-F3.7 or GST was eluted. GST -F3.7 PARsylated by PARP1 or mock treated, or GST, treated similarly, was then incubated with Biotinyl four-way junction DNA in DNA binding buffer (10 mM Tris-HCl, pH = 8.0, 60 mM KCl, 20 μg/ml BSA, 0.1% Triton X-100, 400 μM MgCl2) for 30 minutes at room temperature. 10 μl Streptavidin M-280 Dynal beads (Invitrogen) were then added to each reaction mixture and incubated at room temperature for 10 minutes. Supernatants containing unbound proteins were collected, and the beads were washed 4 times with DNA binding buffer before being processed for further analyses.
For PARsylation-driven binding release assays, Biotinyl four-way junction DNA- bound F3.7 on Streptavidin beads was incubated with PARP1 in PARP reaction buffer (see above), in the presence and absence of 100 nM Olaparib. After the reaction, the supernatant (containing any released protein) and beads (containing remaining bound protein) were collected and analyzed.
Chromosome analysis
Cells were treated with indicated siRNA or drugs, or transfected with indicated cDNA for 48 hours and then exposed to 150 rad ionizing radiation. 30 ng/ml colcemid was added to culture 5 hours after IR and cells were incubated for additional 3 hours, collected, and then prepared for analysis of metaphase spreads. Spreads were stained with 4′,6-diamidino-2-phenylindole, dihydrochloride (DAPI).
PDX breast cancer tumor samples
PDX tumor models were established as described (5). Patient IDs were de-identified to protect confidentiality. Tumor samples were snap-frozen in liquid nitrogen immediately after surgery. Frozen tumor samples were pulverized using a pre-chilled metal pestle while remained submerged in liquid nitrogen. Then the vials containing these pulverized samples were slowly removed from the liquid nitrogen bath. Liquid nitrogen evaporated and pulverized tumor tissues remained in the vials and were ready for further processing and analysis. Appropriate amounts of NET-N 400 lysis buffer (400 mM NaCl, 0.5 mM EDTA, 20 mM Tris-HCl pH 7.5, 0.5% NP-40, 1.5 mM MgCl2, 10% Glycerol, 5 mM NEM, 1 μM ADP-HPD and protease inhibitors) were added to achieve desired a protein concentration in lysates.
Supplementary Material
Significance.
We propose a model that describes how BRCA1 functions to both support and restrict HR. BRCA1 PARsylation is a key event in this process, failure of which triggers hyper-recombination and chromosome instability. Thus, hyper-functioning BRCA1 can elicit genomic abnormalities similar to those observed in the absence of certain BRCA1 functions.
Acknowledgements
We wish to thank Drs. Zhao-Qi Wang and Ya Wang for providing Parp1 knockout MEF cells, and Dr. Andrea Richardson for providing human breast cancer samples. We are grateful to Astra-Zeneca/Kudos for providing a generous initial supply of Olaparib. We also want to thank Drs. Guillaume Adelmant, Jean-Bernard Lazaro, and members of the Livingston Group for numerous helpful discussions and technical advice. This work was supported by grants from the National Cancer Institute, the Breast Cancer Research Foundation, and the Susan G Komen Foundation for the Cure to D.M.L. and by an NCI SPORE grant in breast cancer research to Dana-Farber/Harvard Cancer Center. Y.H. was supported by a US Department of Defense postdoctoral fellowship award (W81XWH-09-1-0632).
Footnotes
Conflict of interest statement: The authors disclose no potential conflicts of interest.
References
- 1.Scully R, Livingston DM. In search of the tumour-suppressor functions of BRCA1 and BRCA2. Nature. 2000;408:429–32. doi: 10.1038/35044000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Walsh T, King MC. Ten genes for inherited breast cancer. Cancer Cell. 2007;11:103–5. doi: 10.1016/j.ccr.2007.01.010. [DOI] [PubMed] [Google Scholar]
- 3.Jasin M. Homologous repair of DNA damage and tumorigenesis: the BRCA connection. Oncogene. 2002;21:8981–93. doi: 10.1038/sj.onc.1206176. [DOI] [PubMed] [Google Scholar]
- 4.Moynahan ME, Chiu JW, Koller BH, Jasin M. Brca1 controls homology-directed DNA repair. Mol Cell. 1999;4:511–8. doi: 10.1016/s1097-2765(00)80202-6. [DOI] [PubMed] [Google Scholar]
- 5.Chen JJ, Silver D, Cantor S, Livingston DM, Scully R. BRCA1, BRCA2, and Rad51 operate in a common DNA damage response pathway. Cancer Res. 1999;59:1752s–6s. [PubMed] [Google Scholar]
- 6.Venkitaraman AR. Cancer susceptibility and the functions of BRCA1 and BRCA2. Cell. 2002;108:171–82. doi: 10.1016/s0092-8674(02)00615-3. [DOI] [PubMed] [Google Scholar]
- 7.Greenberg RA, Sobhian B, Pathania S, Cantor SB, Nakatani Y, Livingston DM. Multifactorial contributions to an acute DNA damage response by BRCA1/BARD1-containing complexes. Genes Dev. 2006;20:34–46. doi: 10.1101/gad.1381306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sobhian B, Shao G, Lilli DR, Culhane AC, Moreau LA, Xia B, et al. RAP80 targets BRCA1 to specific ubiquitin structures at DNA damage sites. Science. 2007;316:1198–202. doi: 10.1126/science.1139516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim H, Chen J, Yu X. Ubiquitin-binding protein RAP80 mediates BRCA1-dependent DNA damage response. Science. 2007;316:1202–5. doi: 10.1126/science.1139621. [DOI] [PubMed] [Google Scholar]
- 10.Wang B, Matsuoka S, Ballif BA, Zhang D, Smogorzewska A, Gygi SP, et al. Abraxas and RAP80 form a BRCA1 protein complex required for the DNA damage response. Science. 2007;316:1194–8. doi: 10.1126/science.1139476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu X, Chen J. DNA damage-induced cell cycle checkpoint control requires CtIP, a phosphorylation-dependent binding partner of BRCA1 C-terminal domains. Mol Cell Biol. 2004;24:9478–86. doi: 10.1128/MCB.24.21.9478-9486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coleman KA, Greenberg RA. The BRCA1-RAP80 complex regulates DNA repair mechanism utilization by restricting end resection. J Biol Chem. 2011;286:13669–80. doi: 10.1074/jbc.M110.213728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu Y, Scully R, Sobhian B, Xie A, Shestakova E, Livingston DM. RAP80-directed tuning of BRCA1 homologous recombination function at ionizing radiation-induced nuclear foci. Genes Dev. 2011;25:685–700. doi: 10.1101/gad.2011011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee I, Blom UM, Wang PI, Shim JE, Marcotte EM. Prioritizing candidate disease genes by network-based boosting of genome-wide association data. Genome Res. 2011;21:1109–21. doi: 10.1101/gr.118992.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rajesh C, Baker DK, Pierce AJ, Pittman DL. The splicing-factor related protein SFPQ/PSF interacts with RAD51D and is necessary for homology-directed repair and sister chromatid cohesion. Nucleic Acids Res. 2011;39:132–45. doi: 10.1093/nar/gkq738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Polo SE, Kaidi A, Baskcomb L, Galanty Y, Jackson SP. Regulation of DNA-damage responses and cell-cycle progression by the chromatin remodelling factor CHD4. EMBO J. 2010;29:3130–9. doi: 10.1038/emboj.2010.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gudjonsson T, Altmeyer M, Savic V, Toledo L, Dinant C, Grofte M, et al. TRIP12 and UBR5 Suppress Spreading of Chromatin Ubiquitylation at Damaged Chromosomes. Cell. 2012;150:697–709. doi: 10.1016/j.cell.2012.06.039. [DOI] [PubMed] [Google Scholar]
- 18.Lai JS, Herr W. Ethidium bromide provides a simple tool for identifying genuine DNA-independent protein associations. Proc. Natl. Acad. Sci. U.S.A. 1992;89:6958–62. doi: 10.1073/pnas.89.15.6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hatakeyama K, Nemoto Y, Ueda K, Hayaishi O. Purification and characterization of poly(ADP-ribose) glycohydrolase. Different modes of action on large and small poly(ADP-ribose) J Biol Chem. 1986;261:14902–11. [PubMed] [Google Scholar]
- 20.Wang ZQ, Auer B, Stingl L, Berghammer H, Haidacher D, Schweiger M, et al. Mice lacking ADPRT and poly(ADP-ribosyl)ation develop normally but are susceptible to skin disease. Genes Dev. 1995;9:509–20. doi: 10.1101/gad.9.5.509. [DOI] [PubMed] [Google Scholar]
- 21.Shieh WM, Ame JC, Wilson MV, Wang ZQ, Koh DW, Jacobson MK, et al. Poly(ADP-ribose) polymerase null mouse cells synthesize ADP-ribose polymers. J Biol Chem. 1998;273:30069–72. doi: 10.1074/jbc.273.46.30069. [DOI] [PubMed] [Google Scholar]
- 22.Ame JC, Rolli V, Schreiber V, Niedergang C, Apiou F, Decker P, et al. PARP-2, A novel mammalian DNA damage-dependent poly(ADP-ribose) polymerase. J Biol Chem. 1999;274:17860–8. doi: 10.1074/jbc.274.25.17860. [DOI] [PubMed] [Google Scholar]
- 23.Scully R, Chen J, Ochs RL, Keegan K, Hoekstra M, Feunteun J, et al. Dynamic changes of BRCA1 subnuclear location and phosphorylation state are initiated by DNA damage. Cell. 1997;90:425–35. doi: 10.1016/s0092-8674(00)80503-6. [DOI] [PubMed] [Google Scholar]
- 24.Bekker-Jensen S, Lukas C, Kitagawa R, Melander F, Kastan MB, Bartek J, et al. Spatial organization of the mammalian genome surveillance machinery in response to DNA strand breaks. J Cell Biol. 2006;173:195–206. doi: 10.1083/jcb.200510130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Menear KA, Adcock C, Boulter R, Cockcroft XL, Copsey L, Cranston A, et al. 4-[3-(4-cyclopropanecarbonylpiperazine-1-carbonyl)-4-fluorobenzyl]-2H-phth alazin-1-one: a novel bioavailable inhibitor of poly(ADP-ribose) polymerase-1. J Med Chem. 2008;51:6581–91. doi: 10.1021/jm8001263. [DOI] [PubMed] [Google Scholar]
- 26.Ogata N, Ueda K, Hayaishi O. ADP-ribosylation of histone H2B. Identification of glutamic acid residue 2 as the modification site. J Biol Chem. 1980;255:7610–5. [PubMed] [Google Scholar]
- 27.Thakur S, Zhang HB, Peng Y, Le H, Carroll B, Ward T, et al. Localization of BRCA1 and a splice variant identifies the nuclear localization signal. Mol. Cell. Biol. 1997;17:444–52. doi: 10.1128/mcb.17.1.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen CF, Li S, Chen Y, Chen PL, Sharp ZD, Lee WH. The nuclear localization sequences of the BRCA1 protein interact with the importin-alpha subunit of the nuclear transport signal receptor. J Biol Chem. 1996;271:32863–8. doi: 10.1074/jbc.271.51.32863. [DOI] [PubMed] [Google Scholar]
- 29.Paull TT, Cortez D, Bowers B, Elledge SJ, Gellert M. Direct DNA binding by Brca1. Proc Natl Acad Sci U S A. 2001;98:6086–91. doi: 10.1073/pnas.111125998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Naseem R, Sturdy A, Finch D, Jowitt T, Webb M. Mapping and conformational characterization of the DNA-binding region of the breast cancer susceptibility protein BRCA1. Biochem J. 2006;395:529–35. doi: 10.1042/BJ20051646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Simons AM, Horwitz AA, Starita LM, Griffin K, Williams RS, Glover JN, et al. BRCA1 DNA-binding activity is stimulated by BARD1. Cancer Res. 2006;66:2012–8. doi: 10.1158/0008-5472.CAN-05-3296. [DOI] [PubMed] [Google Scholar]
- 32.Sturdy A, Naseem R, Webb M. Purification and characterisation of a soluble N-terminal fragment of the breast cancer susceptibility protein BRCA1. J Mol Biol. 2004;340:469–75. doi: 10.1016/j.jmb.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 33.Hassa PO, Haenni SS, Elser M, Hottiger MO. Nuclear ADP-ribosylation reactions in mammalian cells: where are we today and where are we going? Microbiol Mol Biol Rev. 2006;70:789–829. doi: 10.1128/MMBR.00040-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krishnakumar R, Kraus WL. The PARP side of the nucleus: molecular actions, physiological outcomes, and clinical targets. Mol Cell. 2010;39:8–24. doi: 10.1016/j.molcel.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hassa PO, Hottiger MO. The diverse biological roles of mammalian PARPS, a small but powerful family of poly-ADP-ribose polymerases. Front Biosci. 2008;13:3046–82. doi: 10.2741/2909. [DOI] [PubMed] [Google Scholar]
- 36.Ramirez CJ, Fleming MA, Potter JD, Ostrander GK, Ostrander EA. Marsupial BRCA1: conserved regions in mammals and the potential effect of missense changes. Oncogene. 2004;23:1780–8. doi: 10.1038/sj.onc.1207292. [DOI] [PubMed] [Google Scholar]
- 37.Fleming MA, Potter JD, Ramirez CJ, Ostrander GK, Ostrander EA. Understanding missense mutations in the BRCA1 gene: an evolutionary approach. Proc. Natl. Acad. Sci. U.S.A. 2003;100:1151–6. doi: 10.1073/pnas.0237285100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu LC, Wang ZW, Tsan JT, Spillman MA, Phung A, Xu XL, et al. Identification of a RING protein that can interact in vivo with the BRCA1 gene product. Nat Genet. 1996;14:430–40. doi: 10.1038/ng1296-430. [DOI] [PubMed] [Google Scholar]
- 39.Kim H, Huang J, Chen J. CCDC98 is a BRCA1-BRCT domain-binding protein involved in the DNA damage response. Nat Struct Mol Biol. 2007;14:710–5. doi: 10.1038/nsmb1277. [DOI] [PubMed] [Google Scholar]
- 40.Liu Z, Wu J, Yu X. CCDC98 targets BRCA1 to DNA damage sites. Nat Struct Mol Biol. 2007;14:716–20. doi: 10.1038/nsmb1279. [DOI] [PubMed] [Google Scholar]
- 41.Alvarez-Gonzalez R, Althaus FR. Poly(ADP-ribose) catabolism in mammalian cells exposed to DNA-damaging agents. Mut Res. 1989;218:67–74. doi: 10.1016/0921-8777(89)90012-8. [DOI] [PubMed] [Google Scholar]
- 42.Schreiber V, Dantzer F, Ame JC, de Murcia G. Poly(ADP-ribose): novel functions for an old molecule. Nat Rev Mol Cell Biol. 2006;7:517–28. doi: 10.1038/nrm1963. [DOI] [PubMed] [Google Scholar]
- 43.Pleschke JM, Kleczkowska HE, Strohm M, Althaus FR. Poly(ADP-ribose) binds to specific domains in DNA damage checkpoint proteins. J Biol Chem. 2000;275:40974–80. doi: 10.1074/jbc.M006520200. [DOI] [PubMed] [Google Scholar]
- 44.Stilmann M, Hinz M, Arslan SC, Zimmer A, Schreiber V, Scheidereit C. A nuclear poly(ADP-ribose)-dependent signalosome confers DNA damage-induced IkappaB kinase activation. Mol Cell. 2009;36:365–78. doi: 10.1016/j.molcel.2009.09.032. [DOI] [PubMed] [Google Scholar]
- 45.Haince JF, Kozlov S, Dawson VL, Dawson TM, Hendzel MJ, Lavin MF, et al. Ataxia telangiectasia mutated (ATM) signaling network is modulated by a novel poly(ADP-ribose)-dependent pathway in the early response to DNA-damaging agents. J Biol Chem. 2007;282:16441–53. doi: 10.1074/jbc.M608406200. [DOI] [PubMed] [Google Scholar]
- 46.Panzeter PL, Realini CA, Althaus FR. Noncovalent interactions of poly(adenosine diphosphate ribose) with histones. Biochemistry. 1992;31:1379–85. doi: 10.1021/bi00120a014. [DOI] [PubMed] [Google Scholar]
- 47.Yan J, Kim YS, Yang XP, Li LP, Liao G, Xia F, et al. The ubiquitin-interacting motif containing protein RAP80 interacts with BRCA1 and functions in DNA damage repair response. Cancer Res. 2007;67:6647–56. doi: 10.1158/0008-5472.CAN-07-0924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rouet P, Smih F, Jasin M. Introduction of double-strand breaks into the genome of mouse cells by expression of a rare-cutting endonuclease. Mol Cell Biol. 1994;14:8096–106. doi: 10.1128/mcb.14.12.8096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xia B, Sheng Q, Nakanishi K, Ohashi A, Wu J, Christ N, et al. Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2. Mol Cell. 2006;22:719–29. doi: 10.1016/j.molcel.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 50.Schultz N, Lopez E, Saleh-Gohari N, Helleday T. Poly(ADP-ribose) polymerase (PARP-1) has a controlling role in homologous recombination. Nucleic Acids Res. 2003;31:4959–64. doi: 10.1093/nar/gkg703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang YG, Cortes U, Patnaik S, Jasin M, Wang ZQ. Ablation of PARP-1 does not interfere with the repair of DNA double-strand breaks, but compromises the reactivation of stalled replication forks. Oncogene. 2004;23:3872–82. doi: 10.1038/sj.onc.1207491. [DOI] [PubMed] [Google Scholar]
- 52.Shen Y, Rehman FL, Feng Y, Boshuizen J, Bajrami I, Elliott R, et al. BMN 673, a novel and highly potent PARP1/2 inhibitor for the treatment of human cancers with DNA repair deficiency. Clin Can Res. 2013;19:5003–15. doi: 10.1158/1078-0432.CCR-13-1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Puget N, Knowlton M, Scully R. Molecular analysis of sister chromatid recombination in mammalian cells. DNA Repair. 2005;4:149–61. doi: 10.1016/j.dnarep.2004.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nagaraju G, Odate S, Xie A, Scully R. Differential regulation of short- and long-tract gene conversion between sister chromatids by Rad51C. Mol Cell Biol. 2006;26:8075–86. doi: 10.1128/MCB.01235-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bouwman P, Aly A, Escandell JM, Pieterse M, Bartkova J, van der Gulden H, et al. 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nat Struc Mol Biol. 2010;17:688–95. doi: 10.1038/nsmb.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bunting SF, Callen E, Wong N, Chen HT, Polato F, Gunn A, et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell. 2010;141:243–54. doi: 10.1016/j.cell.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Baumann P, Benson FE, West SC. Human Rad51 protein promotes ATP-dependent homologous pairing and strand transfer reactions in vitro. Cell. 1996;87:757–66. doi: 10.1016/s0092-8674(00)81394-x. [DOI] [PubMed] [Google Scholar]
- 58.Zhu Z, Chung WH, Shim EY, Lee SE, Ira G. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell. 2008;134:981–94. doi: 10.1016/j.cell.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bhattacharyya A, Ear US, Koller BH, Weichselbaum RR, Bishop DK. The breast cancer susceptibility gene BRCA1 is required for subnuclear assembly of Rad51 and survival following treatment with the DNA cross-linking agent cisplatin. J Biol Chem. 2000;275:23899–903. doi: 10.1074/jbc.C000276200. [DOI] [PubMed] [Google Scholar]
- 60.Tomimatsu N, Mukherjee B, Deland K, Kurimasa A, Bolderson E, Khanna KK, et al. Exo1 plays a major role in DNA end resection in humans and influences double-strand break repair and damage signaling decisions. DNA Repair. 2012;11:441–8. doi: 10.1016/j.dnarep.2012.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pathania S, Nguyen J, Hill SJ, Scully R, Adelmant GO, Marto JA, et al. BRCA1 is required for postreplication repair after UV-induced DNA damage. Mol Cell. 2011;44:235–51. doi: 10.1016/j.molcel.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schlacher K, Wu H, Jasin M. A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell. 2012;22:106–16. doi: 10.1016/j.ccr.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–27. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Elstrodt F, Hollestelle A, Nagel JH, Gorin M, Wasielewski M, van den Ouweland A, et al. BRCA1 mutation analysis of 41 human breast cancer cell lines reveals three new deleterious mutants. Cancer Res. 2006;66:41–5. doi: 10.1158/0008-5472.CAN-05-2853. [DOI] [PubMed] [Google Scholar]
- 65.Ding L, Ellis MJ, Li S, Larson DE, Chen K, Wallis JW, et al. Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature. 2010;464:999–1005. doi: 10.1038/nature08989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.DeRose YS, Wang G, Lin YC, Bernard PS, Buys SS, Ebbert MT, et al. Tumor grafts derived from women with breast cancer authentically reflect tumor pathology, growth, metastasis and disease outcomes. Nat Med. 2011;17:1514–20. doi: 10.1038/nm.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Landis MD, Lehmann BD, Pietenpol JA, Chang JC. Patient-derived breast tumor xenografts facilitating personalized cancer therapy. Breast Cancer Res. 2013;15:201. doi: 10.1186/bcr3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dimitrov SD, Lu D, Naetar N, Hu Y, Pathania S, Kanellopoulou C, et al. Physiological modulation of endogenous BRCA1 p220 abundance suppresses DNA damage during the cell cycle. Genes Dev. 2013;27:2274–91. doi: 10.1101/gad.225045.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Helleday T. The underlying mechanism for the PARP and BRCA synthetic lethality: clearing up the misunderstandings. Mol Oncol. 2011;5:387–93. doi: 10.1016/j.molonc.2011.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Murai J, Huang SY, Das BB, Renaud A, Zhang Y, Doroshow JH, et al. Trapping of PARP1 and PARP2 by Clinical PARP Inhibitors. Cancer Res. 2012;72:5588–99. doi: 10.1158/0008-5472.CAN-12-2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–7. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 72.Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–21. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 73.Ledermann J, Harter P, Gourley C, Friedlander M, Vergote I, Rustin G, et al. Olaparib maintenance therapy in platinum-sensitive relapsed ovarian cancer. N Engl J Med. 2012;366:1382–92. doi: 10.1056/NEJMoa1105535. [DOI] [PubMed] [Google Scholar]
- 74.Liu JF, Tolaney SM, Birrer M, Fleming GF, Buss MK, Dahlberg SE, et al. A Phase 1 trial of the poly(ADP-ribose) polymerase inhibitor olaparib (AZD2281) in combination with the anti-angiogenic cediranib (AZD2171) in recurrent epithelial ovarian or triple-negative breast cancer. Eur J Cancer. 2013;49:2972–8. doi: 10.1016/j.ejca.2013.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rebbeck TR, Mitra N, Domchek SM, Wan F, Friebel TM, Tran TV, et al. Modification of BRCA1-Associated Breast and Ovarian Cancer Risk by BRCA1-Interacting Genes. Cancer Res. 2011;71:5792–805. doi: 10.1158/0008-5472.CAN-11-0773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Antoniou AC, Wang X, Fredericksen ZS, McGuffog L, Tarrell R, Sinilnikova OM, et al. A locus on 19p13 modifies risk of breast cancer in BRCA1 mutation carriers and is associated with hormone receptor-negative breast cancer in the general population. Nat Genet. 2010;42:885–92. doi: 10.1038/ng.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bolton KL, Tyrer J, Song H, Ramus SJ, Notaridou M, Jones C, et al. Common variants at 19p13 are associated with susceptibility to ovarian cancer. Nat Genet. 2010;42:880–4. doi: 10.1038/ng.666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nikkila J, Coleman KA, Morrissey D, Pylkas K, Erkko H, Messick TE, et al. Familial breast cancer screening reveals an alteration in the RAP80 UIM domain that impairs DNA damage response function. Oncogene. 2009;28:1843–52. doi: 10.1038/onc.2009.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Solyom S, Aressy B, Pylkas K, Patterson-Fortin J, Hartikainen JM, Kallioniemi A, et al. Breast cancer-associated Abraxas mutation disrupts nuclear localization and DNA damage response functions. Sci Transl Med. 2012;4:122ra23. doi: 10.1126/scitranslmed.3003223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Solyom S, Patterson-Fortin J, Pylkas K, Greenberg RA, Winqvist R. Mutation screening of the MERIT40 gene encoding a novel BRCA1 and RAP80 interacting protein in breast cancer families. Breast Cancer Res Treat. 2010;120:165–8. doi: 10.1007/s10549-009-0453-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Weigman VJ, Chao HH, Shabalin AA, He X, Parker JS, Nordgard SH, et al. Basal-like Breast cancer DNA copy number losses identify genes involved in genomic instability, response to therapy, and patient survival. Breast Cancer Res Treat. 2012;133:865–80. doi: 10.1007/s10549-011-1846-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wu J, Liu C, Chen J, Yu X. RAP80 protein is important for genomic stability and is required for stabilizing BRCA1-A complex at DNA damage sites in vivo. J Biol Chem. 2012;287:22919–26. doi: 10.1074/jbc.M112.351007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yin Z, Menendez D, Resnick MA, French JE, Janardhan KS, Jetten AM. RAP80 is critical in maintaining genomic stability and suppressing tumor development. Cancer Res. 2012;72:5080–90. doi: 10.1158/0008-5472.CAN-12-1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Castillo A, Paul A, Sun B, Huang TH, Wang Y, Yazinski SA, et al. The BRCA1-Interacting Protein Abraxas Is Required for Genomic Stability and Tumor Suppression. Cell Rep. 2014;8:807–17. doi: 10.1016/j.celrep.2014.06.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Xie A, Puget N, Shim I, Odate S, Jarzyna I, Bassing CH, et al. Control of sister chromatid recombination by histone H2AX. Mol Cell. 2004;16:1017–25. doi: 10.1016/j.molcel.2004.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.