Abstract
In all organisms, chromosomal DNA must be compacted nearly three orders of magnitude to fit within the limited volume of a cell. However, chromosomes cannot be haphazardly packed, and instead must adopt structures compatible with numerous cellular processes, including DNA replication, chromosome segregation, recombination, and gene expression. Recent technical advances have dramatically enhanced our understanding of how chromosomes are organized in vivo and have begun to reveal the mechanisms and forces responsible. Here, we review the current arsenal of techniques used to query chromosome structure, focusing on (i) single-cell fluorescence microscopy approaches that directly examine chromosome structure and (ii) population-averaged biochemical methods that infer chromosome structure based on the interaction frequencies of different chromosomal loci. We describe the power of these techniques, highlighting the major advances they have produced while also discussing their limitations.
Introduction
How DNA is compacted and organized within the restricted volume of a cell remains a major unsolved problem in biology. Most bacterial chromosomes range from 2 to 8 Mbp in length. If fully stretched out, an individual chromosome would measure millimeters in length, yet it somehow fits within a cell just a few microns long. How do cells compact their chromosomes nearly three orders of magnitude, and how are chromosomes spatially arranged within cells? Studies to tackle these questions promise to reveal fundamental aspects of bacterial cell biology and, perhaps even more importantly, will impact our understanding of many other critical cellular processes involving the chromosome, including gene expression, DNA replication, chromosome segregation, DNA damage repair, recombination, the integration of horizontally-acquired DNA, and more.
An understanding of precisely how the genome is packaged and organized within cells has only recently begun to emerge, driven in large part by the advent of several new techniques for probing chromosome structure in vivo. Here, we review these techniques, including various microscopy-based methods for directly visualizing DNA loci [1-4], indirect methods based on measurements of recombination rates between loci, and new genomic technologies built around an assay called chromosome conformation capture [5-7]. We discuss the strengths and shortcomings of these different approaches in probing chromosomal organization. We highlight specific examples of how these methodologies have driven insights into chromosome biology; for more comprehensive reviews of bacterial chromosome structure, see references [8-10].
Microscopy and imaging of individual chromosomal loci
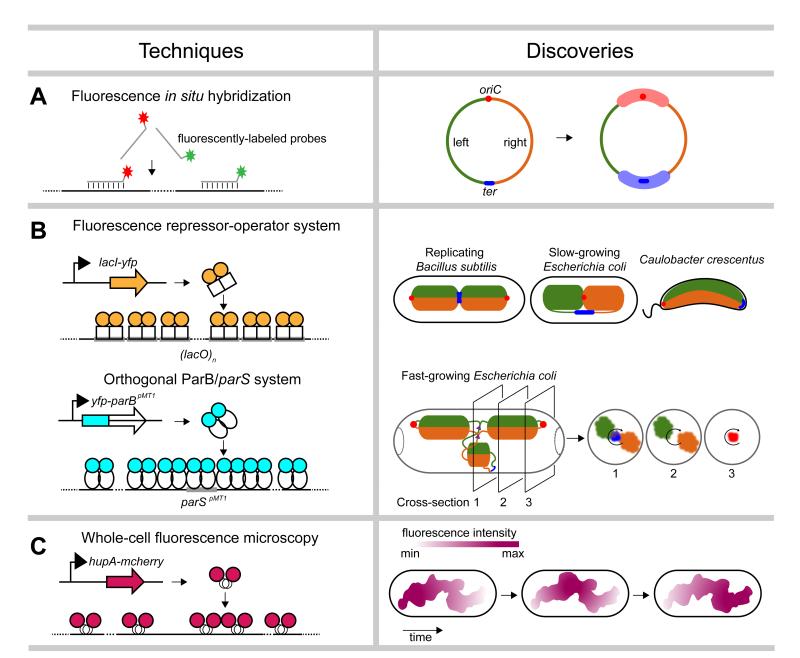
The spatial arrangement of chromosomes can be partially reconstructed by tracking the subcellular positions of specific loci using microscopy. One of the first methods developed for such visualizations was FISH, or fluorescence in situ hybridization. FISH involves the partial permeabilization of cells and subsequent addition of fluorescently-labeled DNA probes that hybridize to complementary regions of the chromosome (Figure 1a). Epi-fluorescence microscopy can then reveal the subcellular location of the labeled locus. For example, an early examination of 22 loci in Escherichia coli by FISH [11] revealed that loci are arranged within the cell in the same approximate order as they appear in the genome, implying a highly-organized chromosomal organization (Figure 1a). In addition, these studies indicated that the E. coli chromosome possesses two so-called macrodomains, called Ori and Ter, in which loci near the origin of replication (oriC) or terminus (dif), respectively, frequently co-localize (Figure 1a).
Figure 1. Single-cell microscopic techniques.
A summary of microscopic techniques (left) used to probe the spatial organization of bacterial chromosomes, with example discoveries (right). (a) Fluorescence in situ hybridization. The origin of replication (oriC) is shown as a red dot and the terminus (ter) as a blue line. The left and right arms of the chromosome are colored in green and orange, respectively. The Ori and Ter macrodomains are indicated by thick opaque red and blue lines, respectively. (b) Fluorescent repressor-operator systems. Proposed patterns of chromosome organization in replicating Bacillus subtilis, slow-growing Escherichia coli, and Caulobacter crescentus based on traditional fluorescent repressor-operator systems (top). The proposed spatial organization of replicating chromosomes in fast-growing E. coli cells, based on studies using an orthogonal ParB/parS system is also shown (bottom). Cross-sections show the radial distribution of the left and right chromosomal arms as well as oriC, ter, and replication forks (purple triangles). (c) Imaging of fluorescently-tagged nucleoid-associated proteins. Reconstruction of the nucleoid of slow-growing E. coli cells expressing HupA-mCherry indicated a coiled-shaped structure of no particular handedness. Nucleoid intensities, summarized from Z projections, also showed rapid fluxes of density along the length of the nucleoid.
Although FISH allows the visualization of loci without modification of the genome, it requires the fixing and permeabilization of cells, treatments that may alter chromosome conformation. Moreover, because FISH requires cell fixation, it cannot be used to track the dynamic movement of chromosomal loci in living cells. These limitations were overcome in part through the development of fluorescent repressor-operator systems (FROS) in which an array (~240 copies) of operator sites (e.g. lacO, tetO or λOL1) is inserted near a locus of interest and cells are engineered to express a cognate, fluorescently-tagged DNA-binding protein (e.g. LacI, TetR, or λCI) [4,12,13]. The binding of multiple fluorescent proteins to an operator array produces a discrete focus detectable by fluorescence microscopy that can be tracked over time in living cells (Figure 1b).
The use of fluorescent repressor-operator systems has produced significant advances in our understanding of chromosome organization and dynamics in bacteria. Early studies of Bacillus subtilis [14,15] analyzed four different loci in replicating cells and revealed that for a given chromosome, the origin of replication resides at a cell quarter position while the terminus lies at mid-cell with the left and right arms in between (Figure 1b). Similar analyses performed on slow-growing E. coli cells revealed a strikingly different overall organization with the origin and terminus near mid-cell and the left and right chromosomal arms in opposite halves of the cell (Figure 1b) [2,16]. A global investigation of chromosome organization in Caulobacter crescentus using the LacI-lacO system examined the subcellular positioning of 112 different loci [17]. This study demonstrated that, as in E. coli, the spatial positions of loci within the cell recapitulate the genetic map with the replication origin at one cell pole and the terminus at the opposite pole (Figure 1b). Further, loci moved to their final subcellular positions coincident with their replication during the cell cycle. This study solidified the notion that bacterial chromosomes are highly organized, both spatially and temporally.
Although FROS has revolutionized the study of bacterial chromosome organization and been applied now to many different species [18-21], it has two important limitations. First, it requires the insertion of a large (~8 kbp) array into the genome, which can be difficult to engineer and, more importantly, can potentially perturb local genome architecture. The integrated operator arrays are also sometimes unstable, with recombination between operators leading to smaller arrays and diminished signal. Although not yet widely used, super-resolution microscopy can eliminate the need for large arrays; cells harboring arrays of just three tetO or lacO operators yield detectable signal [22,23].
The limitations of operator arrays can also be partially overcome by using ParB/parS systems derived from plasmid and chromosome partitioning systems. ParB proteins typically bind a single cognate parS site (~140-280 bp) and initiate the polymerization and spreading of additional ParB proteins on nearby DNA. Thus, insertion of a single parS site into a genome of interest and expression of a fluorescently-tagged ParB can enable the visualization of specific loci. ParB/parS derived from plasmids or chromosomes of different organisms, i.e. orthogonal ParB/parS systems, avoid interference with an endogenous ParB/parS. Further, the use of orthogonal ParB/parS systems with different recognition elements can enable the simultaneous examination of multiple loci in individual cells. The spatial organization of E. coli chromosomes was recently investigated during fast growth using ParB/parS systems from plasmid pMT1 and phage P1 to probe the positioning of 13 different loci. The left and right arms of replicating chromosomes were observed to form separate but parallel running structures in the outer shell of the nucleoid in each cell half (Figure 1b) [3]. In contrast, the replication forks, the origin of replication and the terminus were found at the center of the nucleoid (Figure 1b) [3]. The notion that different regions of the chromosome prefer different shells of the nucleoid, or positions along the short axis of a rod-shaped cells, had not been appreciated previously.
A second limitation of fluorescent repressor-operator systems is the tight binding (Kd ~ 1 nM) of TetR and LacI to multiple operator sites or the spreading of ParB around parS, which can produce a roadblock that impedes DNA replication and results in replication fork collapse or DNA stress [24,25]. Additionally, multimerization between the DNA-binding domains of repressors or between the fluorescent proteins fused to repressors can artificially increase cohesion of sister DNA loci [26]. This artifact can be partially reduced by using a variant of ParB (Nielsen, personal communication) or a fluorescent protein derivative with diminished propensity for multimerization [26].
Global organization revealed by imaging DNA and DNA-binding proteins
Another limitation of fluorescent repressor-operator systems is that only 2-3 loci can be visualized at a time. Assessing global properties of chromosome structure requires a different set of techniques. One common method involves imaging of cells stained with dyes such as 4′,6-diamidino-2-phenylindole (DAPI) that bind DNA non-specifically, thereby revealing the overall cellular distribution and gross structure of bacterial chromosomes in vivo. DAPI staining has demonstrated that the chromosomes of many bacteria, including E. coli and B. subtilis, do not fill the volume of the cell whereas in others, such as C. crescentus, staining occurs throughout the cell. In the former cases, co-staining for other proteins indicates that the nucleoids of these cells often exclude ribosomes, setting up a partial separation of transcription and translation [27]. In B. subtilis, DAPI-staining has also revealed major changes to chromosome morphology upon entry to sporulation, with the nucleoid transitioning from a globular state to an extended axial filament, which may facilitate pumping of one chromosome into the forespore [28].
Like FISH, DAPI staining involves fixed cells, precluding analyses of living cells. Thus, many labs have begun imaging fluorescently-tagged proteins that bind throughout the chromosome, including RNA polymerase subunits and nucleoid-associated proteins (NAPs) which, like histones in eukaryotes, often play important roles in structuring the chromosome [1,29,30]. The subcellular distribution of these proteins serves as a reasonable proxy for the chromosome and has driven significant new insights. One recent study of E. coli used an mCherry fusion to the nucleoid-associated protein HupA to image chromosomes at high spatial and temporal resolution (Figure 1c). This work revealed an overall helical structure of the E. coli nucleoid, although with no handedness preference (Figure 1c) [1]. This study also revealed the dynamic nature of the E. coli nucleoid. Even in non-replicating G1 cells, nucleoid density waves fluxed back and forth along the longitudinal axis of the cell (Figure 1c) [1]. In replicating cells, sister chromosomes segregated end-to-end in sequential, discontinuous pulses [1], with periodic (~20 min intervals) accumulation and relief of intra-nucleoid tethers. The nature of such tethers and the mechanisms responsible for the dynamic motions observed are currently unknown but may be critical to chromosome segregation.
Although many NAPs, including HU, Fis, and StpA, are relatively evenly distributed across the E. coli genome [23], super-resolution imaging studies have suggested that some are not. For instance, H-NS appears to form discrete subcellular structures that may help to establish or reinforce chromosome structure, but the precise function of H-NS in genome organization remains unclear [23]. Still in its infancy, super-resolution imaging promises to provide important new insights into chromosome organization in the coming years.
Inferring chromosome organization by recombination frequencies and Hi-C
Although fluorescence microscopy-based methods have been the mainstay of chromosome biology, these approaches are limited in spatial resolution, even using super-resolution techniques. An alternative class of approaches involves the inference of chromosome structure through assays that report on the frequency of collisions between different loci. Collision, or contact, frequencies in a population of cells will strongly reflect interlocus distances, although they are not always equivalent.
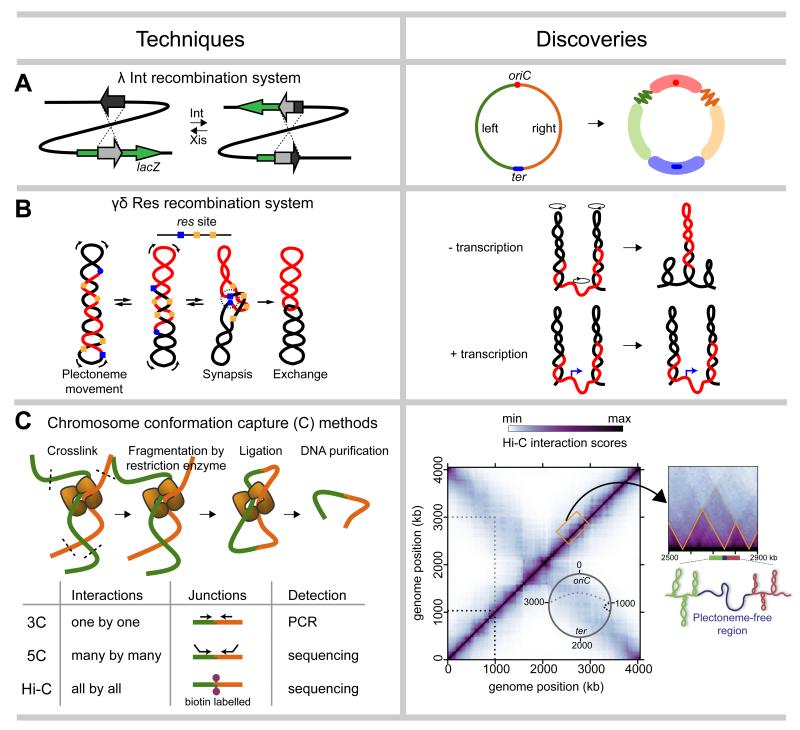
One contact-frequency method uses site-specific recombination systems. Typically, two DNA elements, such as λ attP and attB, the sites recognized by λ integrase, are engineered at different chromosomal loci and the frequency of recombination between them is measured following induction of the recombinase (Figure 2a). This approach was used to measure recombination rates between att sites inserted at various loci around the E. coli genome (Figure 2a), confirming and extending the existence of macrodomains discovered initially by FISH [31]. Loci within a given macrodomain recombined at higher frequencies with other loci in the same macrodomain.
Figure 2. Population-averaged biochemical methods based on locus-locus collision frequencies.
As in Figure 1, a summary of techniques (left) used to probe the spatial organization of bacterial chromosomes, with example discoveries (right). (a) λ Int recombination. Recombination between attP and attB sites causes the disruption of a reporter gene lacZ and creation of two new attL and attR sites. The direction of recombination is controlled by Int and Xis protein. The schematic of the E. coli genome shows oriC and ter as a red dot and a blue line, respectively. The left and right arms of the chromosome are colored green and orange. Four macrodomains: Ori, Ter, Left and Right are shown as thick opaque red, blue, green and orange lines, respectively. Two additional unstructured regions are shown as fuzzy green and orange lines. (b) γσ Res recombination. The slithering of supercoiled plectoneme brings γσ res sites (blue and orange squares or circles) together for synapsis, and subsequent excision of intervening DNA (red). The γσ Res system has shown that high levels of transcription (blue arrows) inhibits supercoil diffusion (black circles with arrowheads, preventing synapsis between flanking loci. (c) Genome-wide chromosome conformation capture assays (5C and Hi-C) have revealed the architecture of the C. crescentus chromosome. A Hi-C heat map uses colors to indicate the frequency of interactions between locus pairs across the genome. The main diagonal captures interactions within chromosomal arms (black dashed line) and the opposite diagonal indicates interactions between arms (grey dashed lines). The Hi-C map also indicates (see zoomed region) the presence of chromosomal interaction domains (CID) (orange triangles). A plectoneme-free region (blue) formed through high gene expression at domain boundaries spatially insulates DNA in flanking domains (green and red).
A complementary system involves site-specific resolvases derived from transposon Tn3. Whereas λ Int can report on long-distance interactions, these resolvase systems, such as γσ Res, are well-suited to probing shorter-range contacts as they rely on negative supercoiling and plectoneme slithering to bring resolution (res) sites together for recombination rather than random collisions (Figure 2b). Studies using the γσ Res system in Salmonella typhimurium revealed the maximal length of a plectoneme, i.e. the longest distance over which res site can be brought together, to be ~100 kb [32]. This work further showed that barriers to plectoneme diffusion appear stochastically distributed within the chromosome [33], although regions of high transcription often formed major barriers to supercoil diffusion, suggesting that the chromosome may be partitioned somewhat deterministically (Figure 2b) [34,35].
Although powerful, the throughput of recombination-based systems for probing chromosome structure is limited and dependent on an ability to insert the relevant sites throughout a genome of interest. However, a related class of approaches based on the technique chromosome conformation capture (3C) enables much greater throughput and has recently driven major advances in understanding chromosome structure in bacteria and eukaryotes [7,36]. For 3C, cells are typically treated with formadehyde to crosslink proteins to DNA and DNA to DNA, thereby preserving the conformation of the chromosome (Figure 2c). The chromosome is then restriction digested, followed by a ligation reaction under dilute conditions, which favors the joining of loci that were crosslinked together. These ligation events fuse loci that were in close spatial proximity when formaldehyde was added. Ligated junctions are then isolated for analysis. The original 3C assay uses PCR to query a junction of interest; 5C uses multiplexed PCR and sequencing of many different junctions; Hi-C involves the incorporation of a biotinylated nucleotide prior to ligation, a subsequent streptavidin-based precipitation of ligated regions, and next-generation sequencing to query all possible junctions (Figure 2c).
Both 5C and Hi-C reveal the identities of many interacting loci and the frequency of each interaction. The resolution of these techniques depends on the distribution and number of restriction sites and on the depth of sequencing performed. Although 3C-based methods do not require any genetic modifications, and are thus potentially applicable to any organism, success can hinge on the restriction enzyme used and the nature of the cell lysate produced following formaldehyde treatment. Because the initial steps of 3C-based methods are performed with a crude cell lysate, endogenous exonucleases or restriction enzyme inhibitors can significantly impact experimental outcomes.
The first 5C performed on bacteria was for C. crescentus swarmer cells that harbor a single circular chromosome [6]. These data revealed that loci typically interacted most strongly with other loci within ~100 kb in primary sequence on the same chromosome arm, with weaker interactions occuring with loci at the same approximate position on the opposite arm of the chromosome (Figure 2c). This pattern reinforced FISH and FROS data showing that the Caulobacter origin is anchored at one pole of the cell with the two arms running in parallel down the long axis of the cell. These 5C data were also used to model the global configuration of the chromosome by (i) assuming that interaction frequencies reflect physical distance between loci and then (ii) searching for overall conformations that satisfy these inferred spatial constraints. The resulting ensemble of models suggested that the two chromosomal arms may adopt a helical shape with the two arms gently twisted around one another [6], reminiscent of structures observed microscopically in E. coli [1,30], B. subtilis [37], and B. bacteriovorus [38].
This 5C study queried 169×170 locus interactions. The recent application of Hi-C to Caulobacter expanded the number of interactions queried to 700×700 and 2025×2025 interactions using BglII and NcoI, respectively [5]. The increase in interactions examined produced contact maps with significantly higher resolution and, consequently, revealed new features of the Caulobacter chromosome. In particular, Hi-C revealed ~23 chromosomal interaction domains (CID), regions of the genome within which loci interact more frequently with each other than with loci in other domains (Figure 2c) [5]. CIDs range in size from 30-400 kb with a mean of ~166 kb, and many appeared nested within larger domains that could correspond to macrodomains (Figure 2c). Importantly, because Hi-C signal comes from a population of cells, the CIDs documented are likely present in most or all cells; additional domains could be present in individual cells, but escape detection by Hi-C.
Domain boundaries frequently coincide with very highly-expressed genes, suggesting a potential causal relationship. Indeed, treating cells with rifampicin, which inhibits RNA polymerase, demonstrated that active gene expression is necessary for boundary formation. Further, insertion of a highly-expressed gene, rsaA, within a domain was sufficient to induce a new boundary [5]. Highly-transcribed genes likely form plectoneme-free regions that are less compacted than neighboring regions. A polymer model of the chromosome suggested that plectoneme-free regions produce boundaries because their decompaction physically separates flanking domains and because they block the diffusion of supercoils, a primary driver of short-range interactions.
Hi-C has also driven new insights into how NAPs contribute to chromosome organization in vivo. Hi-C analysis of cells lacking HU showed that it specifically bolsters DNA interactions at short-length scales but without impacting CID formation, consistent with biophysical studies indicating that HU can bend and compact DNA in vitro [5]. In contrast, Hi-C analysis indicated that the structural maintenance of chromosomes (SMC) protein, a homolog of eukaryotic cohesin and condensin [39,40], does not impact short-range interactions, and instead promotes the colinearity of chromosomal arms. Whether SMC physically tethers the arms together or somehow acts locally on individual arms to keep them in register remains unclear. Collectively, these Hi-C studies are providing important new insight into how specific proteins and other factors, such as supercoiling, contribute to chromosome organization at different length-scales.
Concluding remarks
Recent advances in microscopy and the advent of new genomic techniques such as Hi-C have opened new windows into the organization and structure of chromosomes inside bacterial cells. The combination of population-averaged biochemical methods with single-cell microscopy studies, as well as classical molecular genetic studies, is revealing both global and local patterns of organization. Despite great progress, there is still much to be learned, and we anticipate that efforts to improve the spatial and temporal resolution of both imaging techniques and Hi-C approaches will spur new insights and answer unsolved questions about the structure, dynamics, and functioning of bacterial chromosomes.
Highlights.
Bacterial chromosomes are highly organized spatially and temporally
Recent technical advances dramatically enhance studies of chromosome biology
Microscopy approaches reveal chromosome organization at the single-cell level
Genome-wide methods such as Hi-C generate high resolution maps of chromosomes
Acknowledgements
This work is supported by NIH grant R01GM082899 (M.T.L) and by the Gordon and Betty Moore Foundation and a Life Sciences Research Foundation fellowship (T.B.K.L.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended readings
- 1.Fisher JK, Bourniquel A, Witz G, Weiner B, Prentiss M, Kleckner N. Four-dimensional imaging of E. coli nucleoid organization and dynamics in living cells. Cell. 2013;153:882–895. doi: 10.1016/j.cell.2013.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2**.Nielsen HJ, Ottesen JR, Youngren B, Austin SJ, Hansen FG. The Escherichia coli chromosome is organized with the left and right chromosome arms in separate cell halves. Mol Microbiol. 2006;62:331–338. doi: 10.1111/j.1365-2958.2006.05346.x. Early use of an orthogonal ParB/parS system to label and track DNA loci in live cells. Revealed a strikingly different organization of chromosomes in slow-growing E. coli compared to other rod-shaped bacteria.
- 3*.Youngren B, Nielsen HJ, Jun S, Austin S. The multifork Escherichia coli chromosome is a self-duplicating and self-segregating thermodynamic ring polymer. Genes Dev. 2014;28:71–84. doi: 10.1101/gad.231050.113. Examined the chromosome conformation of fast-growing E. coli cells using ParB/parS systems and showed that different parts of the chromosome reside in different shells of the nucleoid.
- 4.Lau IF, Filipe SR, Soballe B, Okstad OA, Barre FX, Sherratt DJ. Spatial and temporal organization of replicating Escherichia coli chromosomes. Mol Microbiol. 2003;49:731–743. doi: 10.1046/j.1365-2958.2003.03640.x. [DOI] [PubMed] [Google Scholar]
- 5**.Le TB, Imakaev MV, Mirny LA, Laub MT. High-resolution mapping of the spatial organization of a bacterial chromosome. Science. 2013;342:731–734. doi: 10.1126/science.1242059. First use of Hi-C to produce a high-resolution DNA interaction map, revealing ~23 chromosomal interaction domains (CID). Demonstrated that highly-expressed genes are critical to domain boundary formation, and illustrated the power of Hi-C for investigating the in vivo functions and roles of SMC and NAPs.
- 6*.Umbarger MA, Toro E, Wright MA, Porreca GJ, Bau D, Hong SH, Fero MJ, Zhu LJ, Marti-Renom MA, McAdams HH, et al. The three-dimensional architecture of a bacterial genome and its alteration by genetic perturbation. Mol Cell. 2011;44:252–264. doi: 10.1016/j.molcel.2011.09.010. Used genome-wide chromosome conformation capture (5C) to investigate bacterial chromosome structure. Together with live imaging and computational modeling, produced 3D models of the Caulobacter chromosome showing intertwined arms.
- 7*.Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. The original report describing 3C for determining the interaction frequency of two DNA loci..
- 8.Wang X, Montero Llopis P, Rudner DZ. Organization and segregation of bacterial chromosomes. Nat Rev Genet. 2013;14:191–203. doi: 10.1038/nrg3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reyes-Lamothe R, Wang X, Sherratt D. Escherichia coli and its chromosome. Trends Microbiol. 2008;16:238–245. doi: 10.1016/j.tim.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 10.Reyes-Lamothe R, Nicolas E, Sherratt DJ. Chromosome replication and segregation in bacteria. Annu Rev Genet. 2012;46:121–143. doi: 10.1146/annurev-genet-110711-155421. [DOI] [PubMed] [Google Scholar]
- 11**.Niki H, Yamaichi Y, Hiraga S. Dynamic organization of chromosomal DNA in Escherichia coli. Genes Dev. 2000;14:212–223. First report describing macrodomains in the E. coli chromosome based on FISH studies.
- 12.Fekete RA, Chattoraj DK. A cis-acting sequence involved in chromosome segregation in Escherichia coli. Mol Microbiol. 2005;55:175–183. doi: 10.1111/j.1365-2958.2004.04392.x. [DOI] [PubMed] [Google Scholar]
- 13.Robinett CC, Straight A, Li G, Willhelm C, Sudlow G, Murray A, Belmont AS. In vivo localization of DNA sequences and visualization of large-scale chromatin organization using lac operator/repressor recognition. J Cell Biol. 1996;135:1685–1700. doi: 10.1083/jcb.135.6.1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Teleman AA, Graumann PL, Lin DC, Grossman AD, Losick R. Chromosome arrangement within a bacterium. Curr Biol. 1998;8:1102–1109. doi: 10.1016/s0960-9822(98)70464-6. [DOI] [PubMed] [Google Scholar]
- 15.Webb CD, Teleman A, Gordon S, Straight A, Belmont A, Lin DC, Grossman AD, Wright A, Losick R. Bipolar localization of the replication origin regions of chromosomes in vegetative and sporulating cells of B. subtilis. Cell. 1997;88:667–674. doi: 10.1016/s0092-8674(00)81909-1. [DOI] [PubMed] [Google Scholar]
- 16**.Wang X, Liu X, Possoz C, Sherratt DJ. The two Escherichia coli chromosome arms locate to separate cell halves. Genes Dev. 2006;20:1727–1731. doi: 10.1101/gad.388406. Using fluorescent repressor-operator systems, this paper (along with ref. 2) reported the general pattern of chromosome organization for slow-growing E. coli.
- 17*.Viollier PH, Thanbichler M, McGrath PT, West L, Meewan M, McAdams HH, Shapiro L. Rapid and sequential movement of individual chromosomal loci to specific subcellular locations during bacterial DNA replication. Proc Natl Acad Sci U S A. 2004;101:9257–9262. doi: 10.1073/pnas.0402606101. Used fluorescent repressor-operators systems to show that 112 different chromosomal loci reside at specific locations and in the same approximate order along the long axis of the cell as in the genome sequence. Newly-replicated DNA loci segregate sequentially to their final locations in the growing cell.
- 18.Vallet-Gely I, Boccard F. Chromosomal organization and segregation in Pseudomonas aeruginosa. PLoS Genet. 2013;9:e1003492. doi: 10.1371/journal.pgen.1003492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harms A, Treuner-Lange A, Schumacher D, Sogaard-Andersen L. Tracking of chromosome and replisome dynamics in Myxococcus xanthus reveals a novel chromosome arrangement. PLoS Genet. 2013:e1003802. doi: 10.1371/journal.pgen.1003802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jain IH, Vijayan V, O’Shea EK. Spatial ordering of chromosomes enhances the fidelity of chromosome partitioning in cyanobacteria. Proc Natl Acad Sci U S A. 2012;109:13638–13643. doi: 10.1073/pnas.1211144109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fiebig A, Keren K, Theriot JA. Fine-scale time-lapse analysis of the biphasic, dynamic behaviour of the two Vibrio cholerae chromosomes. Mol Microbiol. 2006;60:1164–1178. doi: 10.1111/j.1365-2958.2006.05175.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hensel Z, Weng X, Lagda AC, Xiao J. Transcription-factor-mediated DNA looping probed by high-resolution, single-molecule imaging in live E. coli cells. PLoS Biol. 2013;11:e1001591. doi: 10.1371/journal.pbio.1001591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang W, Li GW, Chen C, Xie XS, Zhuang X. Chromosome organization by a nucleoid-associated protein in live bacteria. Science. 2011;333:1445–1449. doi: 10.1126/science.1204697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang X, Lesterlin C, Reyes-Lamothe R, Ball G, Sherratt DJ. Replication and segregation of an Escherichia coli chromosome with two replication origins. Proc Natl Acad Sci U S A. 2011;108:E243–250. doi: 10.1073/pnas.1100874108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bernard R, Marquis KA, Rudner DZ. Nucleoid occlusion prevents cell division during replication fork arrest in Bacillus subtilis. Mol Microbiol. 2010;78:866–882. doi: 10.1111/j.1365-2958.2010.07369.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mirkin EV, Chang FS, Kleckner N. Protein-mediated chromosome pairing of repetitive arrays. J Mol Biol. 2014;426:550–557. doi: 10.1016/j.jmb.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mascarenhas J, Weber MH, Graumann PL. Specific polar localization of ribosomes in Bacillus subtilis depends on active transcription. EMBO Rep. 2001;2:685–689. doi: 10.1093/embo-reports/kve160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Balassa G. The genetic control of spore formation in bacilli. Curr Top Microbiol Immunol. 1971;56:99–192. doi: 10.1007/978-3-642-65241-7_4. [DOI] [PubMed] [Google Scholar]
- 29.Dillon SC, Dorman CJ. Bacterial nucleoid-associated proteins, nucleoid structure and gene expression. Nat Rev Microbiol. 2010;8:185–195. doi: 10.1038/nrmicro2261. [DOI] [PubMed] [Google Scholar]
- 30.Hadizadeh Yazdi N, Guet CC, Johnson RC, Marko JF. Variation of the folding and dynamics of the Escherichia coli chromosome with growth conditions. Mol Microbiol. 2012;86:1318–1333. doi: 10.1111/mmi.12071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31**.Valens M, Penaud S, Rossignol M, Cornet F, Boccard F. Macrodomain organization of the Escherichia coli chromosome. EMBO J. 2004;23:4330–4341. doi: 10.1038/sj.emboj.7600434. Employed the λ Int recombination system in E. coli to reveal four macrodomains and two less-structured (NS) regions, extending the prior discovery of the Ori and Ter macrodomains by FISH.
- 32.Higgins NP, Yang X, Fu Q, Roth JR. Surveying a supercoil domain by using the gamma delta resolution system in Salmonella typhimurium. J Bacteriol. 1996;178:2825–2835. doi: 10.1128/jb.178.10.2825-2835.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stein RA, Deng S, Higgins NP. Measuring chromosome dynamics on different time scales using resolvases with varying half-lives. Mol Microbiol. 2005;56:1049–1061. doi: 10.1111/j.1365-2958.2005.04588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34**.Booker BM, Deng S, Higgins NP. DNA topology of highly transcribed operons in Salmonella enterica serovar Typhimurium. Mol Microbiol. 2010;78:1348–1364. doi: 10.1111/j.1365-2958.2010.07394.x. The γδ Res recombination system was used to show that a region with high rates of transcription can form a barrier to plectoneme diffusion.
- 35.Deng S, Stein RA, Higgins NP. Organization of supercoil domains and their reorganization by transcription. Mol Microbiol. 2005:1511–1521. doi: 10.1111/j.1365-2958.2005.04796.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dekker J, Marti-Renom MA. Exploring the three-dimensional organization of genomes: interpreting chromatin interaction data. Nat Rev Genet. 2013;14:390–403. doi: 10.1038/nrg3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Berlatzky IA, Rouvinski A, Ben-Yehuda S. Spatial organization of a replicating bacterial chromosome. Proc Natl Acad Sci U S A. 2008;105:14136–14140. doi: 10.1073/pnas.0804982105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Butan C, Hartnell LM, Fenton AK, Bliss D, Sockett RE, Subramaniam S, Milne JL. Spiral architecture of the nucleoid in Bdellovibrio bacteriovorus. J Bacteriol. 2011;193:1341–1350. doi: 10.1128/JB.01061-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schwartz MA, Shapiro L. An SMC ATPase mutant disrupts chromosome segregation in Caulobacter. Mol Microbiol. 2011;82:1359–1374. doi: 10.1111/j.1365-2958.2011.07836.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jensen RB, Shapiro L. The Caulobacter crescentus smc gene is required for cell cycle progression and chromosome segregation. Proc Natl Acad Sci U S A. 1999;96:10661–10666. doi: 10.1073/pnas.96.19.10661. [DOI] [PMC free article] [PubMed] [Google Scholar]