Abstract
The inhibitor of apoptosis protein Survivin regulates hematopoiesis, although its mechanisms of regulation of hematopoietic stem cells (HSCs) remain largely unknown. While investigating conditional Survivin deletion in mice, we found that Survivin was highly expressed in phenotypically defined HSCs and Survivin deletion in mice resulted in significantly reduced total marrow HSC and progenitor cells (HPC). Transcriptional analysis of Survivin−/− HSCs revealed altered expression of multiple genes not previously linked to Survivin activity. In particular, Survivin deletion significantly reduced expression of the Evi-1 transcription factor indispensable for HSC function, and the downstream Evi-1 target genes Gata2, Pbx1 and Sall2. The loss of HSCs following Survivin deletion and impaired long-term HSC repopulating function could be partially rescued by ectopic Evi-1 expression in Survivin −/− HSCs. These data demonstrate that Survivin partially regulates HSC function by modulating the Evi-1transcription factor and its downstream targets and identify new genetic pathways in HSCs regulated by Survivin.
INTRODUCTION
The endogenous inhibitor of apoptosis protein Survivin regulates apoptosis, cell division and cell cycle (1-3) and deregulated expression of Survivin is frequently observed in solid tumors and in leukemia cells (4, 5). Survivin is the fourth most highly expressed transcript in cancer cells (6), where its expression is commonly associated with a higher proliferative index, reduced apoptosis, resistance to chemotherapy and an increased rate of tumor recurrence. While early studies suggested that Survivin was expressed only during development and in neoplastic adult cells, it is now known that Survivin is expressed in normal tissues, including hematopoietic stem and progenitor cells (HSPC) (7, 8), T-cells (9-11), neutrophils (12), erythroid cells (13) and vascular endothelial cells (14, 15). In addition to its role during development, Survivin plays physiologic roles in proliferation and cell cycle control in mouse and human, particularly in highly proliferating cell systems such as hematopoiesis. Despite a better understanding of where and when Survivin is expressed, little is currently known about the mechanism of action of Survivin in normal adult cells or the growth and proliferative pathways in which it participates. We found that Survivin was highly expressed in primitive mouse hematopoietic stem cells (HSC) and conditional deletion caused hematopoietic collapse. To gain insight into Survivin function in hematopoiesis, particularly in HSC, we performed differential mRNA microarray analysis on mouse HSC shortly following conditional Survivin deletion. Survivin deletion was associated with a significant reduction in the expression of the transcription factor Evi-1 and its downstream target genes Gata2, Pbx1 and Sall2, transcription factors critical for normal hematopoiesis. Ectopic expression of Evi-1 in HSC significantly rescued the hematopoietic collapse due to the loss of Survivin in vivo. These studies provide the first mechanistic insight of the role of Survivin in regulating genetic pathways required for HSC function.
MATERIALS AND METHODS
Animals
SPF female C57Bl/6 mice between 6-8 weeks of age were purchased from the Jackson Laboratories (Bar Harbor, Maine). B6.SJL-PtrcAPep3B/BoyJ (B6.BoyJ) mice were bred at Indiana University. Mice in which the Survivin gene could be conditionally deleted by Tamoxifen (CreER-Survivinfl/fl) (16) or polyI:polyC (Mx1-Cre Survivinfl/fl) (17) were previously described. Littermate Survivinfl/fl mice without Cre were used as controls. PCR primers used to distinguish the wild-type, floxed or deleted Survivin alleles were prepared as described (9). The IACUC of Indiana University School of Medicine and Shimane University School of Medicine approved all experimental procedures.
Antibodies and cytokines
The following antibodies were purchased from BD Biosciences (San Diego, CA): anti-Fcγ-III/II receptor; APC-conjugated anti-mouse c-kit (clone 2B8); R-Phycoerythrin (PE)-conjugated anti-mouse CD3 (clone 143-2C11), GR-1 (clone RB6-8C5), B220 (clone RA3-6B2), Mac1 (clone M1/70) and Ter119 (clone Ter119); fluorescein isothiocyanate (FITC)-conjugated CD45.2 and PE-conjugated CD45.1; rat IgG2a; rat IgG2b and hamster IgG. Biotinylated lineage markers consisting of CD5, CD11b, Gr-1, 7-4 and Ter119 were purchased from Miltenyi Biotech (Auburn, CA). Recombinant human (rh) Flt3 ligand (FL), rh thrombopoietin (Tpo) and recombinant mouse stem cell factor (rmSCF) were purchased from R&D Systems (Minneapolis, MN). Recombinant murine GM-CSF (rmGM-CSF) was purchased from BioVision (Palo Alto, CA). Recombinant human erythropoietin was purchased from Amgen (Thousand Oaks, CA).
In vivo deletion of the Survivin gene
Survivinfl/fl and CreER-Survivinfl/fl mice received 5 mg Tamoxifen in corn oil (Sigma-Aldrich, St Louis, MO) intraperitoneally (i.p.) for 3 consecutive days, rested for 3 days, treated for an additional 3 days with Tamoxifen at the same dose and analyzed at 14 days following the final administration of Tamoxifen. Mx1-Cre Survivinfl/fl mice received 3 injections of 260 µg/mouse polyI:polyC (GE HealthCare Bioscience, Piscataway, NJ) i.p. every other day (days 1,3,5) and analyzed 14 days later. Red blood cells (RBC), platelets (PLT) and the absolute neutrophil count (ANC) were quantified using a Hemavet 950F hematology analyzer (Drew Scientific, Oxford, CT). Total nucleated cellularity was quantitated and marrow or spleen HSPC analyzed using flow cytometry and CFC analysis. For CFC analysis, cells were cultured in methylcellulose medium containing growth factors as described (18) . Colonies were scored after 7 days.
Retrovirus transduction and competitive HSC transplants
Bone marrow cells from Cre-Survivinfl/fl and Survivinfl/fl mice were transduced with ecotropic retrovirus generated using Phoenix Eco cells and MSCV (-) or mouse Evi-1 cDNA. Briefly, lineage-depleted marrow cells were pre-stimulated with 100 ng/ml of rhTpo, rmSCF and rhFL for 48 hours and were then infected with fresh retrovirus supernatant containing MSCV(-) or MSCV-Evi-1 (19) for 2 consecutive days. Transduced marrow cells from CreER-Survivinfl/fl and Survivinfl/fl mice were cultured in IMDM/10% FBS with 100 ng/ml of rhTpo, rmSCF and rhFL in the presence of 1 μM of 4OH-Tamoxifen (Sigma-Aldrich) for 7 days. Viable cells were enumerated by trypan blue exclusion and stained with antibodies against c-kit, Sca-1, lineage markers and CD34. For competitive transplantation assays, bone marrow cells from untreated Mx1-Cre Survivinfl/fl and Survivinfl/fl mice (CD45.2) were mixed with bone marrow cells from B6.BoyJ mice (CD45.1) and 1x106 cells transplanted into lethally irradiated (1050 cGy) C57Bl/6 mice (CD45.2). Transduced cells (CD45.2) were cultured in IMDM/10% FBS with 100 ng/ml of rhTpo, rmSCF and rhFL for 48 hrs, mixed with CD45.1 competitor bone marrow cells and 1x106 cells transplanted into lethally irradiated (1050 cGy) C57Bl/6 mice (CD45.2). At 8 weeks post-transplantation, Survivin gene deletion was induced as described above. The proportion of CD45.1 and CD45.2 cells in peripheral blood was determined monthly by flow cytometry.
cDNA microarray and quantitative RT-PCR
Bone marrow cells were harvested from control Survivinfl/fl and CreER-Survivinfl/fl mice at 14 days post Survivin gene deletion, stained with anti-CD34, c-kit, Sca-1 and lineage markers, and CD34neg KSL cells were isolated by FACS. Sorted cells were immediately lysed and subjected to differential mRNA microarray analysis, which was performed by Miltenyi Biotec (Auburn, CA). Briefly, 250 ng of each cDNA was used as a template for Cy3 and Cy5 labeling. Equal amounts of corresponding Cy3- and Cy5-labeled cDNAs from Tamoxifen-treated Survivinfl/fl and CreER-Survivinfl/fl mice were combined and hybridized overnight (17 hours, 65°C) to Agilent Whole Mouse Genome Oligo Microarrays (44K). Fluorescence signals of the hybridized microarrays were detected using Agilent's DNA microarray scanner (Agilent, Palo Alto). The Agilent Feature Extraction Software (FES) was used to determine feature intensities and ratios (including background subtraction and normalization), rejects outliers and calculates statistical confidences (p-values). The differentially expressed genes were functionally classified according to their molecular function and role in biological processes, as defined by Gene Ontology (http://www.geneontology.org/) or the molecular pathways defined by the KEGG database (http://www.genome.jp/kegg/) using the DAVID program (the Database for Annotation, Visualization and Integrated Discovery; http://david.abcc.ncifcrf.gov/home.jsp) (20). The pathways and the molecular network regulated by Survivin were visualized using Cytoscape software (http://www.cytoscape.org/) (21). In separate experiments, total RNA isolated from the sorted control and Survivin-deleted CD34neg.KSL cells was used for quantitative RT-PCR using platinum SYBR Green qPCR SuperMix UDG (Invitrogen, Carlsbad, CA). Primers for the RT-PCR experiments are listed in Supplementary Table 1. Differences in mRNA were calculated using the ΔCT method and were normalized to HPRT, as previously described (7).
Statistics
The data are expressed as the mean ± standard error (SEM). Statistical differences between single agents and the control were determined using the two-tailed Student's t-test in Microsoft Excel ™ (Microsoft Corp, Seattle, WA). The correlation of gene expression with Survivin was determined by Pearson's correlation analysis using Microsoft Excel ™.
RESULTS
Survivin is expressed in mouse HSCs and disruption impairs hematopoiesis
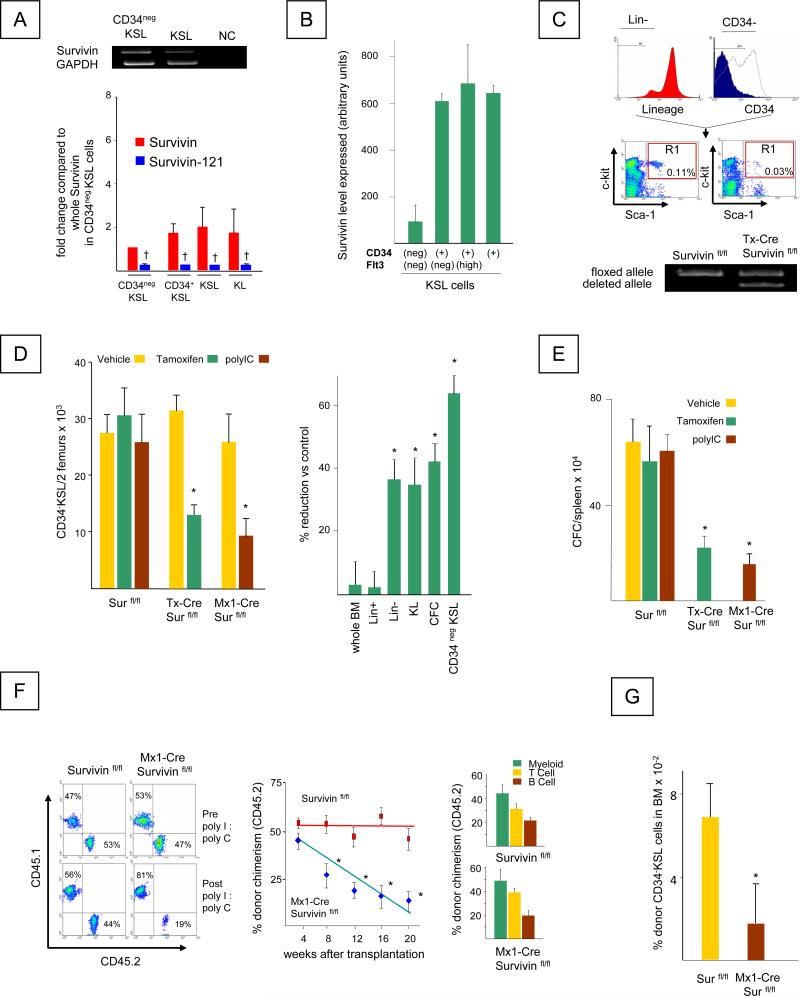
We previously demonstrated that Survivin is expressed in the human CD34+ cell population that contains HSC (7, 22), and is critical for CD34+ cell proliferation and survival (7, 8, 23). It was also shown that Survivin deletion in mice impairs hematopoiesis (17). However, the expression of Survivin in primitive mouse HSC and the consequences of Survivin deletion on HSC function have not been studied. We now show using RT-PCR analysis that mouse Survivin mRNA is expressed in the CD34neg KSL cell population that is enriched for long-term repopulating HSC and in the KSL cell population that contains HSC and HPC (Figure 1A-Upper Panel). In addition, the mouse Survivin-121 splice variant that also demonstrates anti-apoptotic activity (24) was also expressed in mouse HSC and HPC, but at significantly lower levels compared to full length Survivin (Figure 1A, Lower Panel). Consistent with the high expression of Survivin in CD34neg KSL cells, analysis of the public gene expression profile database GSE 4332 (www.ncbi.nlm.nih.gov/geo) showed that the Survivin signal was similarly expressed in mouse bone marrow Flt3 (CD135) neg CD34 neg KSL cells highly enriched for primitive HSC (25) (Figure 1B). Survivin was also detected in CD34 pos KSL cells that represent proliferating HPC, consistent with our previous demonstration of Survivin up regulation during cell cycle (7, 8).
Figure 1. Survivin is expressed in mouse HSC and disruption of Survivin impairs their function.
A. Left Panel, Top: RT-PCR for Survivin mRNAs in FACS sorted KSL and CD34negKSL bone marrow cells. GAPDH was used as an internal control. NC represents the negative control containing no template. Bottom: Quantitative analysis of wild-type Survivin and Survivin-121 in hematopoietic cell populations. † = significant reduction of Survivin-121 compared to full length Survivin in respective cell type, P<0.05.
B. Survivin expression in CD34neg Flt3 neg KSL, CD34pos Flt3 neg KSL, CD34pos Flt3 high KSL and CD34pos KSL cells from GSE 4322, (www.ncbi.nlm.nih.gov/geo).
C. Top: Gating criteria for isolation of CD34neg KSL cells. Bottom: RT-PCR analysis for floxed and deleted Survivin alleles in Survivinfl/fl and CreER-Survivinfl/fl mice following Tamoxifen treatment. The data are representative of 3 independent experiments.
D. Left panel: The absolute number of CD34negKSL cells per 2 femurs in Survivinfl/fl and CreER-Survivinfl/fl and Mx1-Cre Survivinfl/fl mice before and after Survivin deletion, determined by multiplying the absolute total nucleated cell number in 2 femurs by the percentage of CD34negKSL cells. Data represent the mean ± SEM from 3 independent experiments. *P<0.05. Right panel: The percent reduction of total nucleated marrow cells, lineage+ cells (Lin+), lineage- (Lin-) cells, c-kit+ lin- (KL) cells, CFC (CFU-GM, BFU-E, CFU-GEMM) and CD34negKSL cells in bone marrow obtained from Tamoxifen treated CreER-Survivinfl/fl and Survivinfl/fl mice. Data are the mean ± SEM of 3 independent experiments. *P<0.05
E. Mean total CFC/spleen ± SEM, in spleen of 3 individual mice, each assayed individually. *P<0.05
F. Left panel: Representative staining for nucleated peripheral blood CD45.1 and CD45.2 cells in recipient C57Bl/6 mice transplanted with donor (CD45.2) (Survivinfl/fl or Mx1-Cre Survivinfl/fl) and competitor cells (CD45.1) at a ratio of 1:1. Peripheral blood was harvested. Peripheral blood chimerism in the recipients receiving Survivinfl/fl marrow cells (left plots) or Mx1-Cre Survivinfl/fl marrow cells (right plots) at 4 weeks post-transplant prior to polyI:polyC injection (top) and at 20 weeks, 16 weeks after polyI:polyC injection (bottom).
Center panel: Donor (CD45.2) chimerism in the recipients transplanted with Survivinfl/fl marrow cells (red squares) or Mx1-Cre Survivinfl/fl marrow cells (blue diamonds) at 4 weeks post-transplantation prior to polyI:polyC treatment and at 4 week intervals thereafter. *P<0.05 (N=10).
Right panel: Tri-lineage analysis for donor derived (CD45.2) T, B and myeloid cells in recipient mice at 20 weeks post-transplant. Mean ± SEM; N= 10 mice/group, each assayed individually).
G. The frequency of CD34negKSL cells in the bone marrow of recipient mice was determined at 24 weeks post transplantation (20 weeks post polyI:polyC treatment). *P<0.05 (N=10)
Peripheral blood white blood cells (WBC), red blood cells, marrow and spleen cellularity or the proportion or total number of lineage-committed cells (Gr-1+, Mac-1+, B220+, CD4+ and CD8+) in the bone marrow or peripheral blood of CreER-Survivinfl/fl mice were within normal ranges at 14 days after Survivin deletion with the exception of a significant reduction in platelet count (Table 1). In contrast, Survivin deletion significantly decreased the frequency and the absolute number of primitive CD34neg KSL cells containing HSC (61±5% reduction compared to Survivinfl/fl mice receiving the same treatment) (Figure 1C, D) and also reduced total bone marrow linneg cells (37±6%), KL cells (35±8%) and total CFC (44±6%) (Figure 1D). Similar to bone marrow, total CFC in the spleen of Tamoxifen treated CreER-Survivinfl/fl mice were decreased by 62±12% (Figure 1E).
Table 1.
Effects of Tamoxifen treatment on cells in peripheral blood, bone marrow and spleens of control Survivin fl/fl and Cre-ER Survivin fl/fl mice
| Peripheral Blood | ||||||
|---|---|---|---|---|---|---|
| Group | WBCa | RBCb | Plta | |||
| Survivin fl/fl | 5.6±0.5 | 9.4±0.6 | 1607±103 | |||
| Cre-ER Survivin fl/fl | 6.4±1.6 | 9.8±0.6 | 1041±196* | |||
| Mac-1a | GR-1a | Ter199a | CD4a | CD8a | B220a | |
|---|---|---|---|---|---|---|
| Survivin fl/fl | 1.9±0.2 | 2.0±0.3 | 1.2±0.3 | 1.2±0.1 | 0.5±0.04 | 1.1±0.1 |
| Cre-ER Survivin fl/fl | 2.1±0.5 | 2.4±0.6 | 2.9±0.9 | 1.3±0.3 | 0.5±0.2 | 2.6±1.3 |
| Bone Marrow | ||||||
|---|---|---|---|---|---|---|
| TNCc | Mac-1d | GR-1d | Ter199d | |||
| Survivin fl/fl | 3.2±0.4 | 62±3 | 61±3 | 24±10 | ||
| Cre-ER Survivin fl/fl | 3.1±0.2 | 31±4 | 61±4 | 28±6 | ||
| Spleen | ||||||
|---|---|---|---|---|---|---|
| TNCe | ||||||
| Survivin fl/fl | 207±21 | |||||
| Cre-ER Survivin fl/fl | 174±13 | |||||
Control and Survivin fl/fl and Cre-ER Survivin fl/fl mice were treated with Tamoxifen as described in the Method section. CBC and bone marrow and spleen cellularity were determined using a Hemavet hematology analyzer and flow cytometry at 14 days after gene deletion.
×103 per MicroL
×106 per MicroL
×107/2 femurs
% of total marrow cells
×106/spleen
P<0.05
To further evaluate the functional role of Survivin in HSC regulation; we performed long-term competitive repopulating assays. Lethally irradiated C57Bl/6 mice were transplanted with 1x106 cells consisting of congenic B6.BoyJ (CD45.1) marrow cells plus marrow cells (CD45.2) from Survivinfl/fl or Mx1-Cre Survivinfl/fl mice at ratios from 1:1 to 0.5:1. At 1 month post-transplant, the levels of CD45.2/CD45.1 chimerism observed (1:1 ratio shown) were at the expected ratios and were equivalent in all transplant recipients (Figure 1F; Left panels)(Supplemental Figure 1A, 1B). Following Survivin deletion, a significant and progressive reduction in donor CD45.2 chimerism derived from Mx1-Cre Survivinfl/fl bone marrow cells was observed (Figure 1F, Center Panel)(Supplemental Figure 1A, 1B) as previously described (17). Analysis of T, B and myeloid cells in peripheral blood showed equal reduction in all lineages (Figure 1F, Right Panel). In addition to reduced contribution to chimerism, the absolute number of Survivin gene-deleted donor derived CD34negKSL cells in the recipient mice marrow 20 weeks post-transplant (16 weeks post polyI:polyC treatment) was also significantly lower compared to donor cells derived from Survivin fl/fl control mice (Figure 1G). These data confirm a significant role for Survivin in regulation of hematopoiesis, indicating that it is essential for HSC maintenance and function.
Survivin modulates the expression of genes associated with divergent molecular functions in mouse HSC
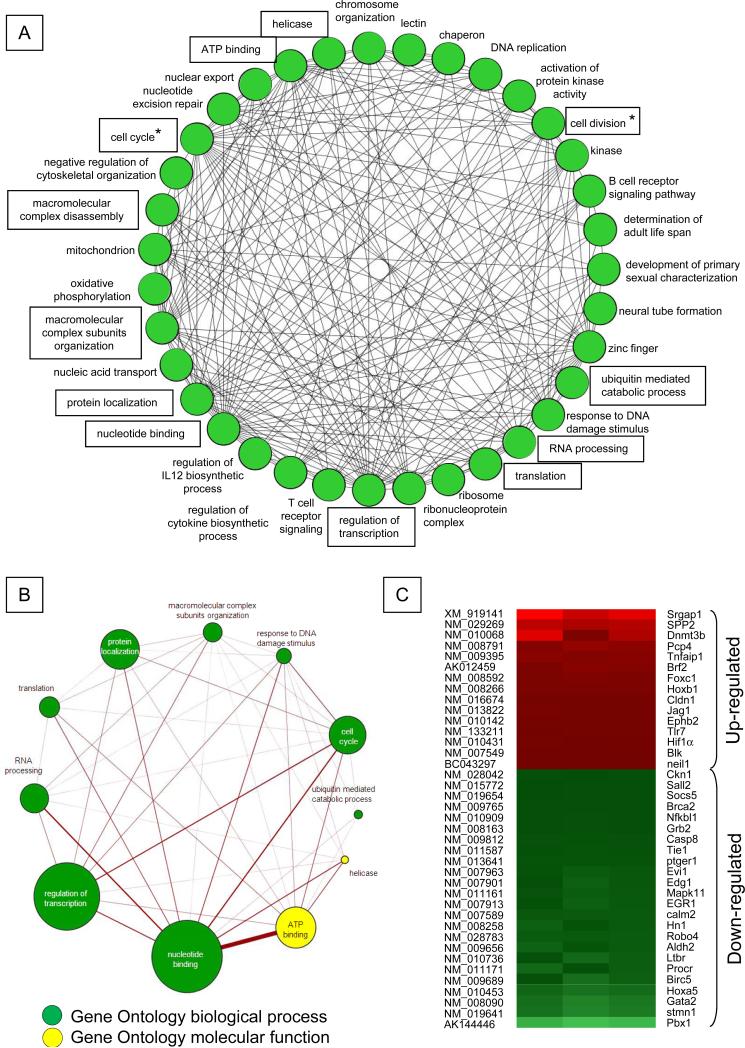
To identify downstream pathways regulated by Survivin in HSC, mRNA expression in FACS sorted CD34negKSL cells before and after Survivin deletion were compared using cDNA microarrays. In two separate experiments, we found 1,653 transcripts that were differentially regulated in CD34neg KSL cells upon Survivin deletion, 965 down-regulated and 688 up-regulated. The full list of Survivin regulated genes has been deposited in the public domain. Birc5 (Survivin) mRNA was reduced by an average 10.4±0.2-fold (P<0.001), confirming the efficacy of Survivin deletion. Significantly regulated genes (>/< 2-fold) were classified into 35 clusters according to the Gene Ontology Database (Figure 2A). The 11 functional categories containing the greatest number of genes among the 35 groups are boxed (Figure 2A) and shown separately in Figure 2B. The circles in Figure 2 indicate individual functions, and the lines between functional groups represent the presence of shared genes. Survivin disruption altered the expression of genes regulating cell cycle and cell division, as expected based on the known mechanism of action studies (shown with asterisks in Figure 2A). However, analysis of gene functional categories revealed that Survivin deletion also affected the expression of genes associated with transcription, protein localization, translation, nucleotide binding and RNA processing, functions not previously linked to Survivin activity (Figure 2A, B). Comparison of our microarray data with the list of previously reported mouse HSC-associated genes (26) identified a number of these genes that were differentially modulated by Survivin depletion (Figure 2C). Expression of several transcription factors associated with embryogenesis, hematopoiesis and cell fate determination, including the homeobox transcription factors HoxA5 and Hoxb1, HIF1α, Evi-1, Foxc1, Pbx1, Gata2 and Sall2 were altered as a consequence of Survivin deletion in CD34neg KSL cells. To validate the microarray data, QRT-PCR analysis was performed on pairs of CD34neg KSL cells from Survivinfl/fl and CreER-Survivinfl/fl mice (Table 2) following Survivin deletion. An 8.5-fold reduction in Evi-1 transcripts as well as transcripts of several downstream Evi-1 target genes was observed , including Gata2, which regulates HSC proliferation/self-renewal (27, 28); Pbx1(29), that maintains HSCs quiescence (30); and Sall2 that affects committed myeloid progenitor cell differentiation and myeloid cell maturation (31, 32). The Foxc1 transcript was up regulated by Survivin deletion, consistent with a reported inverse relationship with Pbx1 (33). In addition to the fact that Gata2, Pbx1 and Sall2 are transcriptional targets of Evi-1, analysis of genes in the public mouse KSL cell gene expression database (GSE 2031, www.ncbi.nlm.nih.gov/geo) (34) showed a strong correlation between Survivin (Birc5) and Evi-1(r=0.35, N=44, P<0.05) and similar to mouse HSC, Survivin showed a positive correlation (r=78) with Evi-1 in highly purified human HSC (Gene expression Omnibus:www.ncbi.nlm.nih.gov/geo (35).
Figure 2. Functional gene networks regulated by Survivin in mouse CD34negKSL cells.
A. Genes significantly regulated by Survivin in three independent microarray analyses (P<0.05) were functionally classified according to the biological process and molecular function listed in the Gene Ontology database, and the analysis was performed using DAVID software (20) and visualized using the Cytoscape program (21). Genes for which annotations could not be assigned were excluded. A circle (node) indicates individual functions, and the line (edge) indicates the presence of shared genes between each functional group. Functional categories are boxed and also shown in Figure 2B. Asterisks indicate functions related to the cell cycle and cell division for which Survivin has been implicated.
B. The panel shows the 11 functional categories containing the greatest number of genes among the 35 groups. The size of each circle represents the number of genes involved in each functional category, and the thickness of the line indicates the number of genes shared between given functions. Green or yellow circles represent functions defined by biological process or molecular function, respectively.
C. Heat map for mouse HSC-specific genes (26) found to be differentially modulated by Survivin depletion in CD34negKSL cells.
Table 2.
Genes modulated by Survivin deletion and implicated in HSC function
| Gene symbol | Fold change by array | Fold change by RT-PCR | Gene name [GenBank Accession number] |
|---|---|---|---|
| Birc5 | −10.5 | −20.0 | Survivin [NM_009689] |
| Evi-1 | −4.4 | −8.5 | Ecotropic viral integration site [NM_007963] |
| Gata2 | −14.4 | −11.1 | GATA binding protein 2 [NM_008090] |
| Pbx1 | −33.7 | −12.5 | pre B-cell leukemia transcription factor 1 [AK144446] |
| Sall2 | −2.2 | −33.3 | sal-like 2 [NM_015772] |
| Hoxa5 | −10.9 | −3.8 | homeo box A5 [NM_010453] |
| Egr1 | −4.8 | −2.5 | early growth response 1 [NM_007913] |
| Tie1 | −2.9 | −4.6 | tyrosine kinase receptor 1 [NM_011587] |
| Robo4 | −5.3 | −6.4 | roundabout homolog 4 (Drosophila) [NM_028783] |
| Hif1α | 2.3 | 4.1 | hypoxia inducible factor 1, alpha subunit [NM_010431] |
| FoxC1 | 4.5 | ND | Forkhead box C1 [NM_008592] |
Selected genes whose expression was affected by Survivin deletion in mouse CD34negKSL cells. Microarray data represent the average of three independent experiments, and the fold change is shown. Quantitative RT-PCR data are the average for three separate experiments using the ΔΔCt method with the fold change compared to HPRT (P <0.05). ND: not determined.
Survivin Regulates HSC Function through Evi-1
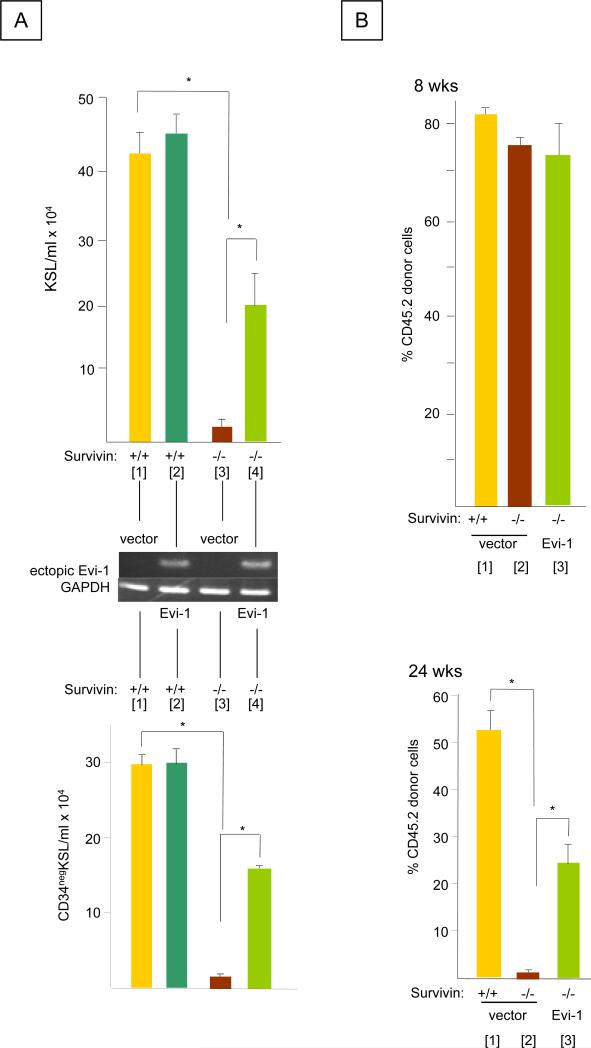
Differential expression of transcription factors upon Survivin deletion was an unexpected result, since a direct role for Survivin in regulating mammalian gene transcription has not been reported. However, Bir1, the C. elegans homologue of Survivin, can function as a transcriptional regulator (36) and Survivin has been shown to affect the expression of multiple genes in cancer cells (37-39). Since a strong correlation was seen between Survivin deletion and reduction in Evi-1 as well as Evi-1 downstream targets genes, we focused on evaluating a potential Survivin/Evi-1 functional axis in regulating HSC function. We hypothesized that restoration of Evi-1 signaling in HSC would restore or protect impaired hematopoiesis resulting from Survivin deletion. Survivin deletion in bone marrow cells from CreER-Survivinfl/fl mice transduced with retroviruses containing MSCV vector resulted in an almost total reduction in the number of KSL cells (Figure 3A Top) and CD34neg KSL cells (Figure 3A Bottom; Survivin in +/+ vector [Bar 1] vs. Survivin −/− vector [Bar 3]). In contrast, significantly more KSL and CD34neg KSL cells were observed when Survivin was deleted in bone marrow cells ectopically expressing Evi-1 (Figure 3, N=3, P<0.05; Survivin −/− vector [Bar 3] vs. Survivin −/− Evi-1[Bar 4]). The enhancing effect of ectopic Evi-1 in Survivin deleted bone marrow cells was not observed in control Survivinfl/fl bone marrow cells (Survivin +/+ vector [Bar 1] vs. Survivin +/+Evi-1 [Bar 2].
Figure 3. Effect of ectopic Evi-1 expression on the in vitro proliferation of CD34negKSL cells and the long-term repopulating activity of HSC lacking Survivin.
A. Bone marrow cells from CreER-Survivinfl/fl and control Survivinfl/fl were retrovirally transduced with MSCV (-) or MSCV containing human Evi-1 (19) and cultured with 100 ng/ml each of rhTpo, rmSCF and rhFL in 10% FBS/IMDM containing 1μM of 4OH-Tamoxifen for 7days. Top panel: KSL cells and Bottom panel: CD34negKSL cells were quantified by cell enumeration and flow cytometry (* P<0.05, N=3). Middle panel: Expression of ectopic human Evi-1 and GAPDH (as an internal control) determined by RT-PCR.
B. Peripheral blood chimerism in (CD45.1) recipients transplanted with (CD45.2) Mx1-Cre Survivinfl/fl cells expressing vector control or Evi-1or Survivinfl/fl marrow cells. PolyI:polyC was injected (i.p.) at 8 weeks post-transplantation. The proportion of CD45.2 cells before polyI:polyC injection (8 week, Top panel) and 24 weeks (Bottom panel) after polyI:polyC treatment are shown (*P<0.05 N=10).
Next, marrow cells from Survivinfl/fl or Mx1-Cre Survivinfl/fl mice (CD45.2) were transduced with MSCV vector or human Evi-1 cDNA and subsequently transplanted along with competitor marrow cells (CD45.1) into lethally irradiated congenic mice. At 8 weeks post-transplantation prior to induction of Survivin gene deletion, there were no significant differences in peripheral blood chimerism between Evi-1 and vector control treatments (Figure 3B; Top Panel). However, upon deletion of Survivin selectively in donor cells, a significant reduction of CD45.2 donor cell chimerism was observed over time as expected (Figure 3B Bottom Survivin +/+ [Bar 1] vs. Survivin −/− [Bar 2]). In contrast, significantly higher chimerism was observed for donor cells lacking Survivin but expressing ectopic Evi-1, compared to Survivin-deleted cells (Figure 3B Survivin −/− vector [Bar 2] vs. Survivin −/− Evi-1[Bar 3]). The competitive repopulating units (CRU) of the donor cells expressing Evi-1 was elevated by 17±2-fold in comparison to cells lacking Survivin and without Evi-1 (P<0.0001). None of the recipient mice developed signs of leukemia or died during a 6-month follow up period.
DISCUSSION
Survivin is expressed during development and elevated re-expression is a hallmark of neoplastic cells, associated with neoplastic growth, high proliferative index and resistance to chemotherapy (1, 2). Increasing evidence also indicates that Survivin plays physiological roles in normal cell division, including hematopoiesis (3), however little is known regarding signaling or functional pathways through which Survivin regulates normal cell growth.
While we have previously shown that Survivin is expressed in normal human CD34+ stem and progenitor cells and is required for cell cycle entry and progression, and survival (7, 22, 23), and others have demonstrated that conditional gene deletion in mice alters steady-state hematopoiesis (17), a direct functional role in HSC, and downstream mechanistic targets have not been identified. While basal Survivin expression is low in mouse HSC compared to less quiescent hematopoietic populations, as we previously described for human HSCs (7, 23), the catastrophic effect of Survivin deletion became highly notable upon HSC stress due to transplantation. This suggests that Survivin activity or molecular networks that associate with Survivin in HSC critically modulate HSC fate/function. We now demonstrate that Survivin is essential for normal long-term repopulating HSC function and identify differentially expressed genes associated with HSC function, as well as a number of biological pathways not previously linked to Survivin activity. Surprisingly, Survivin deletion significantly down-regulated the expression of Evi-1, a transcriptional factor selectively expressed in HSC and indispensable for HSC function (40). Over expression of Evi-1 in HSC compensated, for the impaired HSC function caused by Survivin deletion.
Conditional Evi-1 knock-down in mice has been shown to reduce marrow KSL and CD34negKSL cells without affecting differentiation of mature blood cells (31). We also observed marginal but significant reduction in platelets in both the Evi-1 (31) and Survivin conditional knockout mice (Table 2). The embryonic lethality of Survivin knockout mice is also consistent with the Evi-1 phenotype (41). These findings suggest that Survivin regulates hematopoiesis similarly to Evi-1. In addition to Evi-1, Survivin knock-down reduced the expression of Gata2, Pbx1 and Sall2 in HSC, which are transactivated by Evi-1 and necessary for normal hematopoietic function (31). The shared regulation of these genes by Evi-1 and Survivin is consistent with the reduction of Evi-1 expression following Survivin deletion. In contrast, Evi-1 deletion had no effect on Survivin expression in mouse HSC (31)(GSE11557); suggesting Survivin is upstream of Evi-1. Importantly, marrow cells derived from conditional Survivin−/− mice expressing ectopic Evi-1 showed significantly enhanced numbers of CD34negKSL cells and competitive repopulating activity in comparison to cells in which Survivin was ablated without Evi-1 expression, indicating that Survivin regulates HSC function at least in part through modulation of Evi-1 and that repression of Evi-1 expression is not secondary to the cell cycle alteration and/or apoptosis resulting from Survivin ablation in HSC.
Ectopic expression of Evi-1 only partially rescued the hematopoietic deficits caused by Survivin deletion, suggesting that Evi-1 is necessary but not sufficient for full Survivin activity. Evi-1 can repress transcription of PTEN, which regulates HSC function (42) and correlates with Survivin expression in CD34+CD38− cells in patients with AML (43). Our microarray data showed that Survivin deletion up-regulated PTEN mRNA expression by ~1.4-3.0 fold, however, it did not reach statistical significance. Despite the parallel change in Gata2 expression following Survivin deletion, ectopic Gata2 failed to compensate for the hematopoietic defects impaired by Survivin deletion (not shown). This however, is consistent with previous reports showing that the forced expression of Gata2 blocks hematopoiesis (44). Recent studies indicate that Survivin regulates not only cell division/cell cycle but also intracellular signaling (45). Our microarray data suggests that Survivin impacts multiple signaling pathways including transcription, protein localization and translation in CD34negKSL cells. While some gene alterations may represent changes in cell cycle and/or apoptosis upon Survivin deletion, these findings support the concept that Survivin is involved in divergent HSC cellular functions. Functional grouping informatics analysis revealed that genes associated with transcription were significantly enriched among those regulated by Survivin. Although there is no evidence that Survivin directly regulates gene transcription, Bir1, the C. elegans homologue of Survivin, has been shown to regulate transcription (36), Survivin expression affects transcription in cancer cells (37-39) and transgenic expression of Survivin alters expression of multiple genes in the bladder (46). A transcription factor binding analysis revealed several binding sequences for transcriptional factor Sp1 on the 5’ non-coding region of the mouse Evi-1 gene (GRCm38.p2 C57BL/6J; NC_00069). Survivin has been shown to modulate DNA binding activity of Sp1 by enhancing its phosphorylation (37). These findings suggest that Sp1 may be involved in the transcriptional regulation of Evi-1 expression by Survivin.
A previous report has indicated that induced Survivin deletion in Mx1-Cre Survivinfl/fl mice led to bone marrow ablation, loss of HSPC and rapid mortality (17). Although we observed a significant reduction in CD34negKSL cells in conditional CreER-Survivinfl/fl mice, all mice survived for at least 6 months. This difference may relate to either the incomplete deletion of the Survivin gene by Tamoxifen or to strain differences. Alternatively, the induction of interferons, which are known inhibitors of hematopoiesis (47, 48), by treatment with polyI:polyC (49, 50) in vivo, may have synergistically inhibited HSC in the previous study (17). Nevertheless, although the effect of Survivin gene deletion was not identical, both model systems clearly demonstrated the catastrophic consequences of Survivin deletion on HSC function.
In summary, Survivin regulates HSC proliferation through Evi-1. In addition to Evi-1, genes associated with transcription, cell cycle and those with divergent functions may function as effectors of Survivin activity in HSC. Presently, it is not clear if Survivin regulates genes differentially in normal versus leukemic HSC. This could highlight beneficial pathways for treatments aimed to protect normal HSC and therapeutically leukemic HSC.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Susan Rice and Denessa Lucket for cell sorting. This work was supported by a Biomedical Research Grant from the Indiana University School of Medicine, Research Support Funds Grants from Indiana University and Purdue University, Indianapolis, Research Support Funds from Mochida Memorial Foundation for Medical and Pharmaceutical Research, the Sankyo Biomedical Research Foundation and Mitsubishi Pharma Research Foundation, the Japan Leukaemia Research Fund, an AstraZeneca Research Grant, the Naito Memorial Foundation, a Grant-in-Aid for Scientific Research (B) (20390298) from the Japan Society for the Promotion of Science (to SF) and US Public Health Service grants (HL69669, HL079654 and HL096305) from the National Institutes of Health (to LMP). EMC is an adjunct scientist with the Canadian Blood Services and holds a Canada Research Chair in Endothelial Biology and a CSL-Behring Research Chair. The authors disclose no conflicts of interest associated with this work.
S.F. directed the projects, designed the research, performed experiments, analyzed the data and wrote the manuscript. J.H., performed experiments, analyzed data and wrote the manuscript. P.S., J.S., M.A. and S.Y performed experiments and analyzed the data. E.M.C and G.N. prepared materials. L.M.P directed the project, performed experiments, interpreted the data and wrote the manuscript.
Footnotes
Supplementary information is available at Leukemia's website.
The authors have no competing financial interests is relation to this work.
REFERENCES
- 1.Altieri DC. Validating survivin as a cancer therapeutic target. Nat Rev Cancer. 2003 Jan;3(1):46–54. doi: 10.1038/nrc968. [DOI] [PubMed] [Google Scholar]
- 2.Altieri DC. Survivin, cancer networks and pathway-directed drug discovery. Nat Rev Cancer. 2008 Jan;8(1):61–70. doi: 10.1038/nrc2293. [DOI] [PubMed] [Google Scholar]
- 3.Fukuda S, Pelus LM. Survivin, a cancer target with an emerging role in normal adult tissues. Mol Cancer Ther. 2006 May;5(5):1087–98. doi: 10.1158/1535-7163.MCT-05-0375. [DOI] [PubMed] [Google Scholar]
- 4.Carter BZ, Milella M, Altieri DC, Andreeff M. Cytokine-regulated expression of survivin in myeloid leukemia. Blood. 2001 May 1;97(9):2784–90. doi: 10.1182/blood.v97.9.2784. [DOI] [PubMed] [Google Scholar]
- 5.Adida C, Recher C, Raffoux E, Daniel MT, Taksin AL, Rousselot P, et al. Expression and prognostic significance of survivin in de novo acute myeloid leukaemia. Br J Haematol. 2000 Oct;111(1):196–203. doi: 10.1046/j.1365-2141.2000.02328.x. [DOI] [PubMed] [Google Scholar]
- 6.Velculescu VE, Madden SL, Zhang L, Lash AE, Yu J, Rago C, et al. Analysis of human transcriptomes. Nat Genet. 1999 Dec;23(4):387–8. doi: 10.1038/70487. [DOI] [PubMed] [Google Scholar]
- 7.Fukuda S, Foster RG, Porter SB, Pelus LM. The antiapoptosis protein survivin is associated with cell cycle entry of normal cord blood CD34(+) cells and modulates cell cycle and proliferation of mouse hematopoietic progenitor cells. Blood. 2002 Oct 1;100(7):2463–71. doi: 10.1182/blood.V100.7.2463. [DOI] [PubMed] [Google Scholar]
- 8.Fukuda S, Mantel CR, Pelus LM. Survivin regulates hematopoietic progenitor cell proliferation through p21WAF1/Cip1-dependent and -independent pathways. Blood. 2004 Jan 1;103(1):120–7. doi: 10.1182/blood-2003-05-1756. [DOI] [PubMed] [Google Scholar]
- 9.Xing Z, Conway EM, Kang C, Winoto A. Essential role of survivin, an inhibitor of apoptosis protein, in T cell development, maturation, and homeostasis. J Exp Med. 2004 Jan 5;199(1):69–80. doi: 10.1084/jem.20031588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Song J, So T, Cheng M, Tang X, Croft M. Sustained survivin expression from OX40 costimulatory signals drives T cell clonal expansion. Immunity. 2005 May;22(5):621–31. doi: 10.1016/j.immuni.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 11.Okada H, Bakal C, Shahinian A, Elia A, Wakeham A, Suh WK, et al. Survivin loss in thymocytes triggers p53-mediated growth arrest and p53-independent cell death. J Exp Med. 2004 Feb 2;199(3):399–410. doi: 10.1084/jem.20032092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Altznauer F, Martinelli S, Yousefi S, Thurig C, Schmid I, Conway EM, et al. Inflammation-associated cell cycle-independent block of apoptosis by survivin in terminally differentiated neutrophils. J Exp Med. 2004 May 17;199(10):1343–54. doi: 10.1084/jem.20032033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gurbuxani S, Xu Y, Keerthivasan G, Wickrema A, Crispino JD. Differential requirements for survivin in hematopoietic cell development. Proc Natl Acad Sci U S A. 2005 Aug 9;102(32):11480–5. doi: 10.1073/pnas.0500303102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blanc-Brude OP, Mesri M, Wall NR, Plescia J, Dohi T, Altieri DC. Therapeutic targeting of the survivin pathway in cancer: initiation of mitochondrial apoptosis and suppression of tumor-associated angiogenesis. Clin Cancer Res. 2003 Jul;9(7):2683–92. [PubMed] [Google Scholar]
- 15.Mesri M, Morales-Ruiz M, Ackermann EJ, Bennett CF, Pober JS, Sessa WC, et al. Suppression of vascular endothelial growth factor-mediated endothelial cell protection by survivin targeting. Am J Pathol. 2001 May;158(5):1757–65. doi: 10.1016/S0002-9440(10)64131-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fukuda S, Singh P, Moh A, Abe M, Conway EM, Boswell HS, et al. Survivin mediates aberrant hematopoietic progenitor cell proliferation and acute leukemia in mice induced by internal tandem duplication of Flt3. Blood. 2009 Jul 9;114(2):394–403. doi: 10.1182/blood-2008-11-188714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leung CG, Xu Y, Mularski B, Liu H, Gurbuxani S, Crispino JD. Requirements for survivin in terminal differentiation of erythroid cells and maintenance of hematopoietic stem and progenitor cells. J Exp Med. 2007 Jul 9;204(7):1603–11. doi: 10.1084/jem.20062395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pelus LM, Bian H, King AG, Fukuda S. Neutrophil-derived MMP-9 mediates synergistic mobilization of hematopoietic stem and progenitor cells by the combination of G-CSF and the chemokines GRObeta/CXCL2 and GRObetaT/CXCL2delta4. Blood. 2004 Jan 1;103(1):110–9. doi: 10.1182/blood-2003-04-1115. [DOI] [PubMed] [Google Scholar]
- 19.Buonamici S, Li D, Chi Y, Zhao R, Wang X, Brace L, et al. EVI1 induces myelodysplastic syndrome in mice. J Clin Invest. 2004 Sep;114(5):713–9. doi: 10.1172/JCI21716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang dW Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 21.Cline MS, Smoot M, Cerami E, Kuchinsky A, Landys N, Workman C, et al. Integration of biological networks and gene expression data using Cytoscape. Nat Protoc. 2007;2(10):2366–82. doi: 10.1038/nprot.2007.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fukuda S, Pelus LM. Regulation of the inhibitor-of-apoptosis family member survivin in normal cord blood and bone marrow CD34(+) cells by hematopoietic growth factors: implication of survivin expression in normal hematopoiesis. Blood. 2001 Oct 1;98(7):2091–100. doi: 10.1182/blood.v98.7.2091. [DOI] [PubMed] [Google Scholar]
- 23.Fukuda S, Pelus LM. Elevation of Survivin levels by hematopoietic growth factors occurs in quiescent CD34+ hematopoietic stem and progenitor cells before cell cycle entry. Cell Cycle. 2002 Sep;1(5):322–6. [PubMed] [Google Scholar]
- 24.Conway EM, Pollefeyt S, Cornelissen J, DeBaere I, Steiner-Mosonyi M, Ong K, et al. Three differentially expressed survivin cDNA variants encode proteins with distinct antiapoptotic functions. Blood. 2000 Feb 15;95(4):1435–42. [PubMed] [Google Scholar]
- 25.Rossi DJ, Bryder D, Zahn JM, Ahlenius H, Sonu R, Wagers AJ, et al. Cell intrinsic alterations underlie hematopoietic stem cell aging. Proc Natl Acad Sci U S A. 2005 Jun 28;102(26):9194–9. doi: 10.1073/pnas.0503280102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ivanova NB, Dimos JT, Schaniel C, Hackney JA, Moore KA, Lemischka IR. A stem cell molecular signature. Science. 2002 Oct 18;298(5593):601–4. doi: 10.1126/science.1073823. [DOI] [PubMed] [Google Scholar]
- 27.Tsai FY, Keller G, Kuo FC, Weiss M, Chen J, Rosenblatt M, et al. An early haematopoietic defect in mice lacking the transcription factor GATA-2. Nature. 1994 Sep 15;371(6494):221–6. doi: 10.1038/371221a0. [DOI] [PubMed] [Google Scholar]
- 28.Rodrigues NP, Janzen V, Forkert R, Dombkowski DM, Boyd AS, Orkin SH, et al. Haploinsufficiency of GATA-2 perturbs adult hematopoietic stem-cell homeostasis. Blood. 2005 Jul 15;106(2):477–84. doi: 10.1182/blood-2004-08-2989. [DOI] [PubMed] [Google Scholar]
- 29.Shimabe M, Goyama S, Watanabe-Okochi N, Yoshimi A, Ichikawa M, Imai Y, et al. Pbx1 is a downstream target of Evi-1 in hematopoietic stem/progenitors and leukemic cells. Oncogene. 2009 Dec 10;28(49):4364–74. doi: 10.1038/onc.2009.288. [DOI] [PubMed] [Google Scholar]
- 30.Ficara F, Murphy MJ, Lin M, Cleary ML. Pbx1 regulates self-renewal of long-term hematopoietic stem cells by maintaining their quiescence. Cell Stem Cell. 2008 May 8;2(5):484–96. doi: 10.1016/j.stem.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goyama S, Yamamoto G, Shimabe M, Sato T, Ichikawa M, Ogawa S, et al. Evi-1 is a critical regulator for hematopoietic stem cells and transformed leukemic cells. Cell Stem Cell. 2008 Aug 7;3(2):207–20. doi: 10.1016/j.stem.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 32.Chai L. The role of HSAL (SALL) genes in proliferation and differentiation in normal hematopoiesis and leukemogenesis. Transfusion. 2011 Nov;51(Suppl 4):87S–93S. doi: 10.1111/j.1537-2995.2011.03371.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Berry FB, O'Neill MA, Coca-Prados M, Walter MA. FOXC1 transcriptional regulatory activity is impaired by PBX1 in a filamin A-mediated manner. Mol Cell Biol. 2005 Feb;25(4):1415–24. doi: 10.1128/MCB.25.4.1415-1424.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bystrykh L, Weersing E, Dontje B, Sutton S, Pletcher MT, Wiltshire T, et al. Uncovering regulatory pathways that affect hematopoietic stem cell function using ‘genetical genomics’. Nat Genet. 2005 Mar;37(3):225–32. doi: 10.1038/ng1497. [DOI] [PubMed] [Google Scholar]
- 35.Notta F, Doulatov S, Laurenti E, Poeppl A, Jurisica I, Dick JE. Isolation of single human hematopoietic stem cells capable of long-term multilineage engraftment. Science. 2011 Jul 8;333(6039):218–21. doi: 10.1126/science.1201219. [DOI] [PubMed] [Google Scholar]
- 36.Kostrouchova M, Kostrouch Z, Saudek V, Piatigorsky J, Rall JE. BIR-1, a Caenorhabditis elegans homologue of Survivin, regulates transcription and development. Proc Natl Acad Sci U S A. 2003 Apr 29;100(9):5240–5. doi: 10.1073/pnas.0730770100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Asanuma K, Tsuji N, Endoh T, Yagihashi A, Watanabe N. Survivin enhances Fas ligand expression via up-regulation of specificity protein 1-mediated gene transcription in colon cancer cells. J Immunol. 2004 Mar 15;172(6):3922–9. doi: 10.4049/jimmunol.172.6.3922. [DOI] [PubMed] [Google Scholar]
- 38.Takizawa BT, Uchio EM, Cohen JJ, Wheeler MA, Weiss RM. Downregulation of survivin is associated with reductions in TNF receptors' mRNA and protein and alterations in nuclear factor kappa B signaling in urothelial cancer cells. Cancer Invest. 2007 Dec;25(8):678–84. doi: 10.1080/07357900701600954. [DOI] [PubMed] [Google Scholar]
- 39.Balkhi MY, Christopeit M, Chen Y, Geletu M, Behre G. AML1/ETO-induced survivin expression inhibits transcriptional regulation of myeloid differentiation. Exp Hematol. 2008 Nov;36(11):1449–60. doi: 10.1016/j.exphem.2008.05.008. [DOI] [PubMed] [Google Scholar]
- 40.Kataoka K, Sato T, Yoshimi A, Goyama S, Tsuruta T, Kobayashi H, et al. Evi1 is essential for hematopoietic stem cell self-renewal, and its expression marks hematopoietic cells with long-term multilineage repopulating activity. J Exp Med. 2011 Nov 21;208(12):2403–16. doi: 10.1084/jem.20110447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Uren AG, Wong L, Pakusch M, Fowler KJ, Burrows FJ, Vaux DL, et al. Survivin and the inner centromere protein INCENP show similar cell-cycle localization and gene knockout phenotype. Curr Biol. 2000 Nov 2;10(21):1319–28. doi: 10.1016/s0960-9822(00)00769-7. [DOI] [PubMed] [Google Scholar]
- 42.Yoshimi A, Goyama S, Watanabe-Okochi N, Yoshiki Y, Nannya Y, Nitta E, et al. Evi1 represses PTEN expression and activates PI3K/AKT/mTOR via interactions with polycomb proteins. Blood. 2011 Mar 31;117(13):3617–28. doi: 10.1182/blood-2009-12-261602. [DOI] [PubMed] [Google Scholar]
- 43.Carter BZ, Qiu Y, Huang X, Diao L, Zhang N, Coombes KR, et al. Survivin is highly expressed in CD34(+)38(-) leukemic stem/progenitor cells and predicts poor clinical outcomes in AML. Blood. 2012 Jul 5;120(1):173–80. doi: 10.1182/blood-2012-02-409888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Persons DA, Allay JA, Allay ER, Ashmun RA, Orlic D, Jane SM, et al. Enforced expression of the GATA-2 transcription factor blocks normal hematopoiesis. Blood. 1999 Jan 15;93(2):488–99. [PubMed] [Google Scholar]
- 45.Altieri DC. Survivin and IAP proteins in cell-death mechanisms. Biochem J. 2010 Sep 1;430(2):199–205. doi: 10.1042/BJ20100814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Salz W, Eisenberg D, Plescia J, Garlick DS, Weiss RM, Wu XR, et al. A survivin gene signature predicts aggressive tumor behavior. Cancer Res. 2005 May 1;65(9):3531–4. doi: 10.1158/0008-5472.CAN-04-4284. [DOI] [PubMed] [Google Scholar]
- 47.Broxmeyer HE, Lu L, Platzer E, Feit C, Juliano L, Rubin BY. Comparative analysis of the influences of human gamma, alpha and beta interferons on human multipotential (CFU-GEMM), erythroid (BFU-E) and granulocyte-macrophage (CFU-GM) progenitor cells. J Immunol. 1983 Sep;131(3):1300–5. [PubMed] [Google Scholar]
- 48.Pelus LM, Ottmann OG, Nocka KH. Synergistic inhibition of human marrow granulocyte-macrophage progenitor cells by prostaglandin E and recombinant interferon-alpha, -beta, and -gamma and an effect mediated by tumor necrosis factor. J Immunol. 1988 Jan 15;140(2):479–84. [PubMed] [Google Scholar]
- 49.Magee WE, Griffith MJ. The liver as a site for interferon production in response to poly I:poly C. Life Sci II. 1972 Nov 22;11(22):1081–6. doi: 10.1016/0024-3205(72)90216-0. [DOI] [PubMed] [Google Scholar]
- 50.Manetti R, Annunziato F, Tomasevic L, Gianno V, Parronchi P, Romagnani S, et al. Polyinosinic acid: polycytidylic acid promotes T helper type 1-specific immune responses by stimulating macrophage production of interferon-alpha and interleukin-12. Eur J Immunol. 1995 Sep;25(9):2656–60. doi: 10.1002/eji.1830250938. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.