Abstract
Increasing evidences show that the actin cytoskeleton is a key parameter of the nuclear remodeling process in response to the modifications of cellular morphology. However, detailed information on the interaction between the actin cytoskeleton and the nuclear lamina was still lacking. We addressed this question by constraining endothelial cells on rectangular fibronectin-coated micropatterns and then using Structured Illumination Microscopy (SIM) to observe the interactions between actin stress fibers, nuclear lamina and LINC complexes at a super-resolution scale. Our results show that tension in apical actin stress fibers leads to deep nuclear indentations that significantly deform the nuclear lamina. Interestingly, indented nuclear zones are characterized by a local enrichment of LINC complexes, which anchor apical actin fibers to the nuclear lamina. Moreover, our findings indicate that nuclear indentations induce the formation of segregated domains of condensed chromatin. However, nuclear indentations and condensed chromatin domains are not irreversible processes and both can relax in absence of tension in apical actin stress fibers.
The cell nucleus is composed of two lipid bilayers, the outer and the inner nuclear membranes (ONM and INM), which connect at nuclear pores and delineate the perinuclear space. While the ONM is an extension of the rough endoplasmic reticulum, the INM adheres to the nuclear lamina, which is a meshwork of intermediate filaments composed of A- and B-type lamins. Extensive works have demonstrated that lamins contribute to nuclear stiffness1,2 and stability3. In addition, lamins are also believed to modulate transcription4 and have been speculated to mechanoregulate the genome5,6. Recently, Swift and colleagues have shown that the level of lamin-A scales with the tissue stiffness and enhances matrix-elasticity directed differentiation7.
Physical connections between the nuclear lamina and the peripheral cytoskeleton, called LInkers of Nucleoskeleton and Cytoskeleton (LINC) complexes, were recently uncovered8. They form through interactions between luminal domains of two families of transmembrane proteins of the nuclear envelope: Sun proteins and Nesprins9,10. This mechanical coupling of the nucleus to the cytoskeleton can potentially serve many purposes. Indeed, the interaction of the nucleus with the cytoskeleton is central to the nuclear positioning, which is involved in a variety of cellular processes, such as division, migration and mechanotransduction11.
The rapidly growing field of nuclear–cytoskeletal interactions has lead to numerous studies on the transduction of extracellular physical cues (e.g. force, geometry and stiffness) to the nucleus to regulate gene expression12,13,14,15,16,17. Among these works, some studies have revealed that the cytoskeleton maintains the nucleus in a prestressed state18 and demonstrated that the nucleus responds to external forces, emphasizing the role of physical links between the cytoskeleton and the nucleus19. Whereas an aligned actin filament structure has been shown to form a perinuclear actin cap20, the distinct mechanical role of apical and central actin stress fibers in the coordination between cell and nuclear shapes has been reported recently for endothelial cells21. Normal and lateral compressive loads exerted on the nucleus by apical and central stress fibers modulate nuclear homeostatic balance and internal chromatin structure, respectively21,22. Though a large number of studies report a coupling between active actin stress fibers and the cell nucleus, the molecular interactions of these links and the mechanical behavior of the nuclear sites under a compressive load have not been already reported.
To answer this question, high resolution imaging with Structured Illumination Microscopy (SIM) and confocal microscopy were carried out on Human Umbilical Vein Endothelial Cells (HUVECs) spread on rectangular micropatterns. We used immunostained HUVECs for actin, Lamin-A and C and Syne-2 to analyze the specific coupling between actin stress fibers and the nucleus and to identify the mechanical response of the nuclear lamina.
Results
The cellular morphology controls the nuclear shape through the reorganization of apical and central actin stress fibers
We used a microcontact printing technique (Fig. 1A) to functionalize polydimethylsiloxane (PDMS) culture substrates with fibronectin (FN) micropatterns23. Circular and rectangular (aspect ratio of 1:10) protein micropatterns were used to control the morphology of HUVECs. Uncoated regions were passivated with Pluronic F-127 and HUVECs were plated at low density to confine one single endothelial cell per FN island. The micropattern area was chosen to be 1600 μm2, which corresponds to the mean cell area observed on homogeneous FN-coated substrates. Single HUVECs were cultured on micropatterned substrates for 24 hours and confocal and SIM microscopy imagings were carried out on immunostained HUVECs.
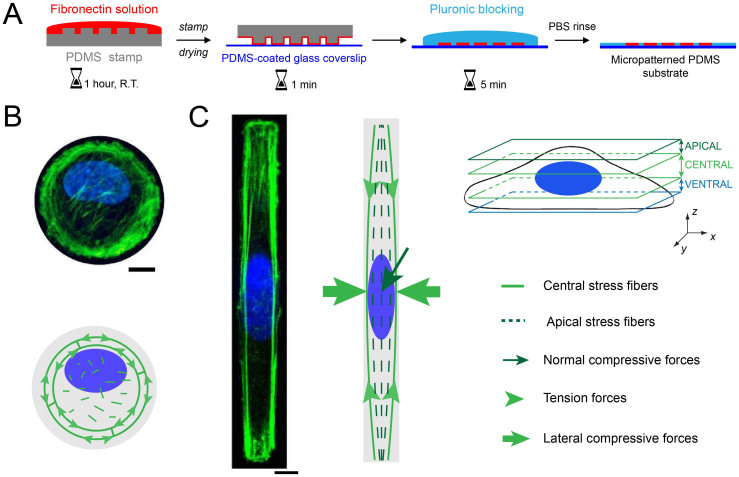
Figure 1. The control of the cell morphology by microcontact printing induces a modification of the nuclear shape.
(A) Successive steps of the microcontact printing method used to create circular and rectangular micropatterns of fibronectin (in red) on a glass coverslip coated with a layer of polydimethylsiloxane (PDMS). Maximum intensity projections of confocal images for a single endothelial cell grown on an adhesive micropattern with (B) a circular and (C) a rectangular geometry. The actin cytoskeleton is stained in (B) and (C) with AlexaFluor 488 Phalloïdin (in green) and the nucleus is stained with Dapi (in blue). The drawings are schematic representations of the spatial organization of apical and central actin stress fibers in rounded and rectangular endothelial cells. Scale bars are 10 μm.
As we demonstrated recently21, geometrical constraints imposed on single HUVECs lead to a significant reorganization of central and apical actin fibers. Central actin fibers formed circular bundles at the periphery of circular-shaped cells (Fig. 1B), whereas they formed straight cables aligned with respect to the long cell axis in rectangular cells (Fig. 1C). While apical structures appeared as short segments randomly oriented on circular geometry, they formed thin parallel fibers aligned with respect to the long cell axis in rectangular geometries (Figs. S1A and S1B).
Nucleus of rectangular-shaped HUVECs was elongated with respect to the long cell axis (Fig. 1C), whereas it remained rounded in circular-shaped cells (Fig. 1B). Consistent with previous studies on other cell types, apical actin stress fibers were frequently observed in elongated HUVECs (~78% of cells for n = 24, Fig. S2A and S2B), while only less than ~10% of circular cells (n = 21) exhibited a network of apical actin fibers (Fig. 1B).
Concomitant with the emergence of apical actin filaments, the nuclear height of rectangular HUVECs decreased significantly with the average density of apical stress fibers (Fig. S2), as defined by the ratio of area between apical actin fibers and the nucleus. The regulation of the nuclear shape in response to cell shape changes is the result of the interplay between apical and central actin stress fibers that exert compressive forces on the nucleus. Central actin stress fibers exert lateral compressive forces on both sides of the nucleus in elongated cells, whereas apical actin fibers regulate the nuclear height by exerting normal compressive forces (Figs. S1C and S1D). Indeed, both actin structures are connected to focal adhesions that are located at the cell extremities in elongated cells21,22,24. We further investigated the behavior of nuclear lamina in response to these compressive forces.
Indentations of the nuclear lamina co-localize with apical actin stress fibers
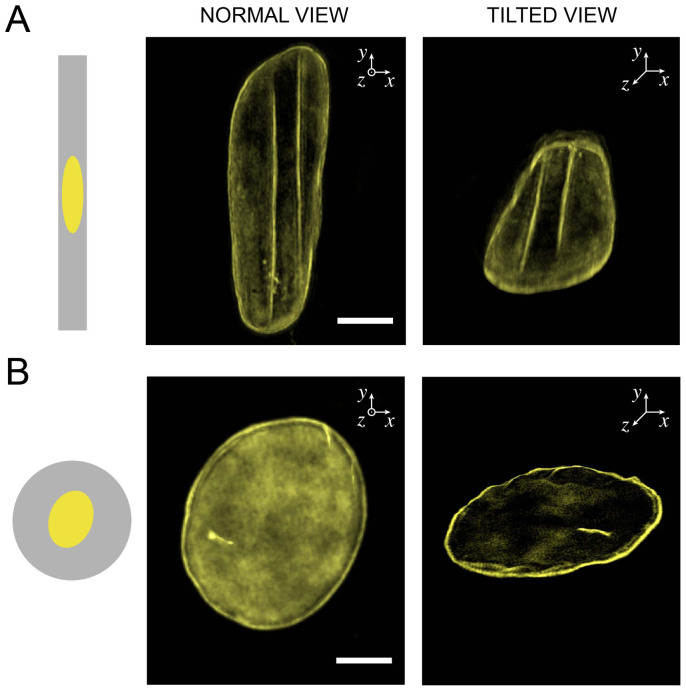
To investigate the behavior of the nuclear lamina in response to apical and central compressive forces, Structured Illumination Microscopy (SIM) in super-resolution mode was carried out on circular and rectangular-shaped HUVECs labeled for lamins A/C. SIM imaging of the nuclear lamina revealed mechanical indentations of the nuclear envelope of elongated cells (Fig. 2A), whereas the mechanical integrity of the nuclear membrane of circular-shaped cells remained intact (Fig. 2B). As shown in Fig. 2A and Movie S1, elongated nuclei were indented from their top side, leading to intense lines of fluorescence aligned with respect to the nuclear long axis. Since the global deformation of the nucleus is a consequence of compressive forces exerted by actin stress fibers, we investigated their role in the nuclear indentation.
Figure 2. The top of the nucleus in elongated cells presents linear deformations of the nuclear lamina.
Normal views (left column) and tilted views (right column) obtained from 3D-SIM imaging of the nuclear lamina (in yellow) of (A) a rectangular-shaped and (B) a circular-shaped endothelial cells. Scale bars are 5 μm.
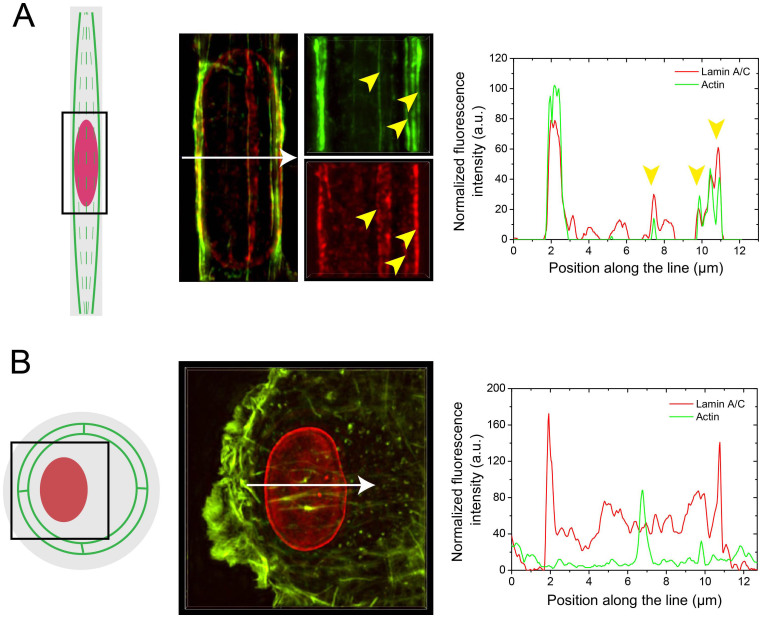
A detailed observation by SIM microscopy of elongated cells labeled for both actin and lamins A/C reveals the presence of fluorescence lines oriented with respect to the long nuclear axis. The overlay of the intensity profiles presented in Fig. 3A shows that most of the intense lamin signals correspond to the presence of apical actin filaments, suggesting that large nuclear deformations co-localize with apical actin filaments. Apical actin filaments were observed to be thinner structures (~100–150 nm in diameter) than central actin stress fibers (~300–500 nm in diameter), which were localized on both sides of the nucleus. The low diameter of apical fibers may explain that few of them were characterized by a discontinous fluorescence signal that did not permit to reveal a colocalization with lamin A/C structures. In contrary to rectangular cells, circular-shaped cells presented a very low amount of apical actin characterized by a weak fluorescence level (Fig. 3B). In addition, no co-localization between F-actin and lamin A/C was observed on rounded cells (Fig. 3B).
Figure 3. Nuclear lamina deformations correspond to the localization of apical stress fibers.
(A) Maximum intensity projections obtained from 3D-SIM microscopy of the actin microfilaments (in green) and the nuclear lamina (in red) of a single endothelial cell spread on a FN-coated rectangular micropattern. Yellow arrows highlight a co-localization of the lines of lamina deformation with the apical and lateral stress fibers surrounding the nucleus. The normalized fluorescence intensity of actin (emission at 488 nm) and lamin A/C (emission at 561 nm) were measured along the white arrow of the merged view. (B) Maximum intensity projections obtained from 3D-SIM microscopy of the actin microfilaments (in green) and the nuclear lamina (in red) of a single endothelial cell spread on a FN-coated circular micropattern. The normalized fluorescence intensity of actin (emission at 488 nm) and lamin A/C (emission at 561 nm) were measured along the white arrow. White arrows in (A) and (B) represent 13 μm.
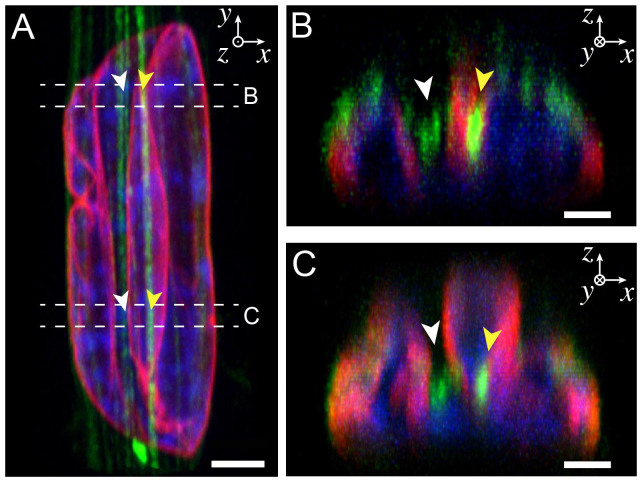
In order to confirm these observations, we imaged the deformed nucleus and actin filaments of a rectangular endothelial cells (aspect ratio 1:10) by confocal microscopy. Orthogonal views showed clearly the presence of straight apical actin fibers that crossed the nucleus and formed deep valleys by deforming the lamina (Fig. 4A). Cross-sections on different zones of the nucleus (Figs. 4B and 4C, Supplementary Movies S2 and S3) indicated that actin stress fibers were localized at the valley floors formed by the lamina deformations, suggesting that apical actin filaments applied important normal compressive forces. One can note that the diameter of the apical stress fibers involved in deep nuclear indentations (>50% of the nuclear height) was two or three times larger (~300–500 nm in diameter) than other apical filaments, and thus directly comparable to the diameter of central actin structures.
Figure 4. Apical actin fibers are located at the bottom of indentation sites that form deep valleys in the nucleus.
(A) Normal confocal view (XY) of an elongated nucleus indented by straight apical actin fibers. Lamins A/C are in red, DNA in blue and actin in green. Two cross-sectional views (XZ) obtained from the top (B) and the bottom (C) parts showed actin fibers localized at the bottom of indentation sites (white arrows) and others embedded in the nucleus (yellow arrows). Apical actin fibers exert important compressive forces that lead to large deformations of the nuclear envelope and the formation of deep valleys. The scale bars correspond to (A) 3 μm and (B, C) 1.5 μm.
Taken together, these results suggest that the reorganization of the actin cytoskeleton in response to cell shape changes leads to the formation of parallel apical and central actin fibers that surround elongated nuclei in rectangular shaped-cells (1:10 aspect ratio). Apical actin structures control the height of the deformed nuclei and accumulated tension in thicker apical actin stress fibers leads to deep indentations of the nuclear lamina localized at the top of the nucleus.
LINC complexes anchor apical and central actin fibers to the nuclear lamina and are enriched at nuclear indentation sites
We investigated whether these nuclear deformations were associated with a modification of the mechanical link tethering actin filaments and lamins A/C. To do this, we labeled LINC complexes, which are involved in the anchoring of both nuclear membranes and nuclear lamina to the actin cytoskeleton. LINC is primarily composed of Suns and Nesprins, which are also called Synes, for synaptic nuclear envelopes. We labeled therefore Nesprin-2, which is the first protein to localize to the outer nuclear envelope25 and connects the nuclear lamina to the actin cytoskeleton.
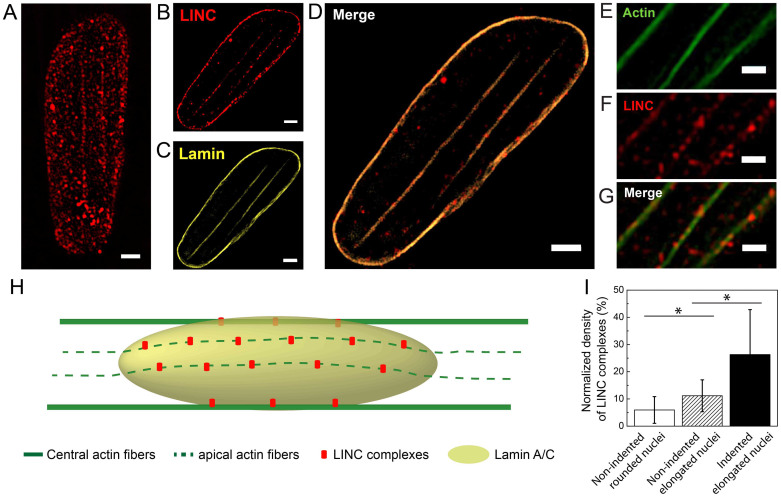
To investigate whether apical actin stress fibers were co-localized with Syne-2 sites, we analyzed extended depth of focus images of LINC complexes labeled with a Syne-2 antibody (Fig. 5A). The plane of focus was lowered in SIM mode to exhibit only the Syne-2 fluorescence signal corresponding to the depth of indentation sites. In this deeper plane of focus, a large amount of Syne-2 signal disappeared to exhibit only discrete patches of Syne-2 organized along straight lines (Fig. 5B), which appeared to be localized as lamina indentation sites (Figs. 5C and 5D). Interestingly, the observation in SIM mode of a double immunofluorescence staining for F-actin (Fig. 5E) and Syne-2 (Fig. 5F) revealed a large co-localization (Fig. 5G) of Syne-2 with apical and central actin fibers. Taken together, these observations suggest that apical actin filaments are anchored to the lamina indentation sites through discrete nesprin-2 patches, as depicted in the schematic view presented in Fig. 5H.
Figure 5. The density of LINC complexes is enriched at the sites of nuclear lamina indentations.
(A) SIM images of LINC complexes labeled with Syne-2 antibody (red) in a deformed nucleus. A focal plane in SIM images located in the central zone was selected to show (B) LINC complexes fluorescence signals at the bottom of (C) lamin indentations sites. (D) The merged image of lamin and LINC complexes from the same focal plane showed that both signals spatially co-localized. (E) Apical actin filaments and (F) LINC complexes observed in SIM showed (G) a strong co-localization on the merged image. (H) Schematic representation of the spatial organization of actin microfilaments (green), nuclear lamina (yellow) and LINC complexes (red) in a rectangular-shaped endothelial cell. (I) Evolution of the normalized density of LINC complexes for rounded and non-indented nuclei (white bar, n = 12), for non-indented elongated nuclei (dashed bar, n = 11) and for elongated nuclei with deep indentations (black bar, n = 14). Bars are mean ± SD. *p < 0.05, unpaired Student's test. Scale bars correspond to 1 μm.
We next assessed whether the presence of tensed apical filaments can affect quantitatively the density of LINC patches. To do this, we normalized the total LINC area obtained from extended depth of focus images by the total nuclear area for different nuclear types. Non-indented rounded nuclei in non-patterned cells were characterized by a mean density of LINC complexes of 5.9 ± 4.9% (Fig. 5I), whereas the LINC density was two times higher (11.2 ± 5.9%) for elongated but non-indented nuclei in rectangular cells. Interestingly, we found that the LINC density increased up to 26.3 ± 16.5% for elongated cells with indented nuclei (Fig. 5I). Our results suggest therefore that the presence of compressive central and apical actin fibers increased the density of LINC complexes in deformed nuclei. In addition, indentation domains were characterized by a significant enrichment of LINC complexes (~135%), as demonstrated by the large co-localization of apical actin fibers with LINC patches (Fig. 5G).
To further confirm these observations, we performed nuclear compression experiments (Fig. S2) and we quantified the evolution of the mean area and the number of LINC complexes (Figs. S3A and S3B, respectively). Artificial compression of the nucleus was obtained by placing a glass coverslip of 22 mm in diameter in the culture dish over endothelial cells, as described recently26. The glass coverslip was coated with Pluronic F-127 to avoid cellular adhesions and then placed during 6 hours on top of circular micropatterned endothelial cells cultured for 20 hours. The weight of the coverslip (m = 0.136 g) was chosen to apply a gentle normal pressure, without damaging the cell nucleus as described previously27. Confocal imaging of compressed cells demonstrated a flattening of the nucleus (Fig. S2), whereas the cellular morphology remained perfectly rounded. The quantification of the mean area (Fig. S3A) and the number (Fig. S3B) of LINC complexes did not show statistically significant differences for compressed nuclei. Interestingly, the number of LINC complexes per actin fiber area increased for elongated indented nuclei (Fig. S3B), whereas it remained constant for rounded cells compressed by a coverslip. Taken together, these data strengthen the hypothesis of a LINC complex enrichment at specific nuclear indentation sites where actin filaments are anchored to nuclear lamina.
Nuclear indentations induce reversible modifications of the chromatin structure
Considering the large nuclear deformations caused by indentations of actin filaments, the following questions arise: (i) is the chromatin structure affected by indentations and (ii) are indentations irreversible deformations of the nucleus? To answer these questions, we first observed by confocal microscopy the cross-sections of a DAPI-stained nucleus of rectangular cells that presented indented sites (Movie S4). As shown in Fig. 6A, indented nuclei presented the formation of rich domains of chromatin with a high level of condensation (higher fluorescence intensity). In contrary, the chromatin was homogeneously localized to the periphery of non-indented nuclei (Movie S5), which is in agreement with the rationale that chromatin fibers are attached to the nuclear envelope28. The three-dimensional organization of chromatin fibers depends on the stability of lamina structure and nuclear indentations can lead to the formation of high-condensed chromatin domains.
Figure 6. Indentation of the nuclear lamina induces chromatin segregation, which is a reversible process.
(A) 3D confocal cross-section views (XZ) of indented (left column) and normal (right column) nuclei show the formation of rich chromatin domains within an indented nucleus. DNA is in blue and actin filaments in green. DAPI images were digitized in 256 bits and colour coded for clarity. Highly condensed domains show higher fluorescence intensity with respect to the less condensed ones. (B) Schematic representation of the cell detachment process by addition of accutase in the culture medium. The focal adhesion disassembly leads to the cell detachment followed by a progressive loss of the actin cytoskeletal tension and the rounding of the cell. (C) Successive normal views in epifluorescence mode of the nuclear relaxation process induced by the cell detachment. The histone 2B of the nucleus were labeled with green fluorescent protein (GFP) and accutase was added at t = 2 sec. Green arrows indicate the initial indentations of the elongated nucleus due to normal forces applied by apical stress fibers. The red dashed ellipse shows the initial outline of the nucleus (t = 0 sec). The relaxation of the nucleus shows that nuclear indentations are reversible and non-plastic deformations. Scale bars represent 2 μm for (A) and 5 μm for (C).
We then followed by epifluorescence microscopy the relaxation dynamics of a deformed nucleus that was indented by apical stress fibers. Nuclear histones 2B were labeled with green fluorescent protein (GFP) and de-adhesion of elongated endothelial cells (aspect ratio 1:10) was initiated by adding accutase in the culture medium (see Methods for additional information). As we have shown previously, rectangular-shaped endothelial cells grown on FN-coated micropatterns exhibit focal adhesions that are located at both extremities of cells21. The spatial organization of focal adhesions connected to straight actin fibers that are elongated with respect to the long-cell axis results in an anisotropic force contraction dipole with two opposite contractile forces that are localized at both cell extremities. We used accutase, which is a marine-origin enzyme with proteolytic and collagenolytic activities for the detachment of primary cells, to dissociate the specific anchorages to the substrate (Fig. 6B). Consequently, both cell extremities retracted to dissipate the tension accumulated in the actin fibers and the cell relaxed. As a consequence, compressive lateral and normal forces exerted on the nucleus by the actin network were removed and the deformed nucleus started to relax during the cell detachment process.
By recording the fluorescence of the nucleus labeled with H2B-GFP, we observed that indentation lines disappeared after ~30 seconds, while the nucleus was still significantly deformed (Fig. 6C and Movie S5). Then the relaxation process continued and the nucleus was fully relaxed after 180 seconds (Fig. 6C). Interestingly, relaxed nuclei adopted a rounded morphology, which was characterized by a homogeneous H2B-GFP fluorescent signal, demonstrating that chromatin adopted a decondensed state in relaxed nuclei. Taken together, these observations suggest that the deep nuclear indentations induced by apical actin fibers are not plastic deformations of the lamina and furthermore the modifications of the chromatin structure associated with nuclear indentations are reversible.
Discussion
Previous studies have shown that cell shape changes can regulate the nuclear morphology through a spatial reorganization of the actin network20,21,22,29. Apical and central actin filaments have been observed to exert compressive forces on the nucleus that permit to adapt its morphology21. Interestingly, recent works have demonstrated that the tension in apical actin structures is related to the formation of specific focal adhesion sites that are fundamentally distinct from conventional focal adhesions associated with basal or dorsal actin filaments24. Consistent with a recent study on fibroblasts22, we show that elongated nuclei observed in rectangular-shaped endothelial cells exhibit important nuclear lamina deformations. Co-localization of actin filaments with nuclear lamina suggests that the highly ordered apical actin structures that form arches above the nucleus apply normal compressive forces, resulting in the formation of indentation sites. Nuclear indentations have been reported previously by Martini and colleagues during the nucleokinesis process of migrating neurons30. Interestingly, it appeared that the nuclear indentation in migrating neurons was a consequence of the forces exerted by actomyosin on the rear of the nucleus. As illustrated in Fig. 4, cross-sectional views of indented nuclei in endothelial cells show that actin stress fibers originated from the apical region can deeply indent the nucleus to finally be localized within the nucleus in the ventral cellular domain. In addition to these observations, our results show that the relative density of apical actin in elongated cells is inversely proportional to the nuclear height (Fig. S4), confirming that apical actin structures apply a normal compressive load on the nucleus. This mechanical picture is in good agreement with 4D traction force investigations on endothelial31 and dyctiostelium cells32, reporting that the cell nucleus experiences a pushing down force.
Apical actin stress fibers are directly connected to the nuclear lamina through LINC complexes. Using super resolution microscopy, we have shown that the density of LINC complexes is two times higher at the nuclear indentation sites (Fig. 5I), demonstrating that apical stress fibers are firmly anchored to the bottom of the indentation sites. These results suggest that deformed nuclei indented by actin stress fibers cannot escape the cage formed by the actin network. Our findings of enriched coupling between apical actin and nucleus at the sites of lamina indentations suggest that apical actin structures maintain the stability in nuclear positioning, which is central to important cellular functions such as cell polarity, division, differentiation or motility33. Furthermore our findings suggest a force-dependent LINC complexes recruitment to actin stress fibers, as observed previously for vinculin in focal adhesions34,35. In contrary to rectangular cells, the absence of apical actin cytoskeleton in rounded cells (Fig. 3B) does not permit to form a mechanical coupling with the nuclear lamina (Fig. 5I), which may explain the manifestation of nuclear rotation in rounded cells36.
Our results provide evidence for a modification of the nuclear homeostatic balance through the coupling between apical stress fibers and the nuclear lamina via LINC complexes. Alterations of the homeostatic balance impair the chromatin assembly by forming domains of segregated condensed chromatin that may affect gene expression and/or DNA replication37,38. Interestingly, our findings have shown that indentations of nucleus by apical stress fibers and the associated modifications of the chromatin architecture are reversible processes. The slow recovery observed in our experiments reflects the viscoelastic signature of nucleus, in agreement with previous reports39. The relaxation time scale of nuclear indentations is of particular importance for understanding the dynamics of the chromatin remodeling in response to the constant reorganization of the apical actin stress fibers22. Future work will use high-resolution imaging to observe the influence of nuclear indentations on chromatin/lamin interactions and stability of DNA repair foci. Indeed, recent findings suggest a mechanism wherein the dynamic structural meshwork formed by A-type lamins may spatially anchor the genome to compartmentalize DNA transactions within the mammalian cell nucleus40,41.
Methods
Microcontact printing
Circular and rectangular (aspect ratio of 1:10) microfeatures of 1600 μm2 were drawn with the Clewin software and generated to a silicon master by deep reactive-ion etching (FH Vorarlberg University of Applied Sciences, Microtechnology, Dornbirn, Austria). The silicon surface was passivated under vacuum with vapors of fluorosilane (tridecafluoro-1,1,2,2-tetrahydrooctyl-1-trichlorosilane) for 30 min, and then molded with polydimethylsiloxane, PDMS, (Sylgard 184 Silicone Elastomer Kit; Dow Corning, Midland, MI). After 4 h of curing at 60°C, the PDMS layer was peeled off and stamps of 1 cm2 were cut manually. The structured surface of the PDMS stamps were first oxidized in an UV/O3 cleaner for 8 min and then inked for 1 h at room temperature with a 25 μg.ml−1 fibronectin (FN) solution from human plasma. FN-coated stamps were dried under filtered nitrogen and gently deposited on a flat PDMS-coated glass coverslip for 15 seconds, as described previously42,43. Uncoated regions were blocked by incubating micropatterned coverslips for 5 min in a 1% solution of Pluronic F-12744.
Cell culture
Primary human umbilical vein endothelial cells, HUVECs (Cells Applications, San Diego, CA) were grown in complete endothelial cell growth medium (Cells Applications, San Diego, CA) supplemented with 1% antibiotics and antimycotics and maintained at 37°C in a humidified atmosphere with 5% CO2. Cells between passages 2 and 8 were cultured on microprinted PDMS coverslips at a concentration of about 15,000 cells ml−1.
Immunohistochemistry and labeling
After 24 h in culture on micropatterned substrates, HUVECs were fixed and permeabilized with 4% paraformaldehyde, 0.05% Triton X-100 in Phosphate buffered saline (PBS) for 15 min at 37°C and then washed three times in PBS. Fixed cells were incubated for 30 min at room temperature in a blocking solution (FBS 5%, BSA 1%) and labeled for F-actin (Alexa Fluor 488 Phalloidin 1:200, Molecular Probes), nuclear lamina (anti-lamin A/C antibody produced in mouse 1:200, Sigma-Aldrich SAB4200236) and LINC-complex (anti Syne-2 antibody produced in rabbit 1:100, Sigma-Aldrich HPA003435) for 45 min at 37°C in PBS. After three successive washes in PBS, cells were incubated for 45 min at 37°C with an anti-mouse antibody produced in goat and labeled with tetramethylrhodamine (Molecular Probes, Invitrogen, T2762), an anti-mouse antibody produced in rabbit and labeled with fluorescein isothiocyanate (Sigma-Aldrich, F9137) or an anti-rabbit antibody produced in goat an labeled with tetramethylrhodamine (Sigma-Aldrich, T6778). All secondary antibodies were used in a 1:200 concentration in PBS. Slides were mounted in Slow Fade Gold Antifade (Molecular Probes).
Living cell relaxation
To observe nuclear DNA in living cells, HUVECs were transduced with the BacMam 2.0 CellLight Histone 2B-GFP according to the manufacturer's protocol (Molecular Probes). Routine culture flasks were incubated with approximately 20 BacMam particles/cell for at least 24 hours. Labeled cells were then detached and grown on rectangular FN micropatterns (1:10 aspect ratio) for 24 hours and finally transferred to an inverted confocal microscope equipped with a temperature and CO2 level controller. The GFP fluorescence signal was recorded during the relaxation of the elongated cells, which was induced by the addition of 1 ml of accutase after a gentle PBS wash.
Structured illumination microscopy and confocal imaging
Immunostained preparations were observed in Structured Illumination Microscopy (SIM) performed on an Eclipse Ti inverted microscope equipped with a Nikon Plan Apo ×100 TIRF objective (NA 1.49, oil immersion) and an Andor DU-897X-5254 camera. Z-step size was set to 0.120 um, which is well within the Nyquist criterion. For each focal plane, 15 images (5 phases, 3 angles) were captured with the NIS-Elements software. SIM image processing, reconstruction and analysis were carried out using the N-SIM module of the NIS-Element Advanced Research software. Confocal imaging was performed with an inverted Nikon Eclipse Ti-E motorized microscope (Nikon C1 scanhead; Nikon, Japan) equipped with Plan Apo ×60 and ×100 (NA 1.45, oil immersion) objectives, two lasers (Ar ion 488 nm; HeNe, 543 nm) and a modulable diode (408 nm). Confocal images were acquired with NIS Elements Advanced Research 3.0 software (Nikon) by using small Z-depth increments between focal sections (0.150 μm).
Image analysis
Confocal and SIM images were acquired and analyzed with the NIS-Elements Advanced Research 3.0 software. Three-dimensional normal views were obtained in maximum intensity projection mode. Unless otherwise stated, orthogonal views were obtained in maximum intensity projection mode and cropped to represent a one micron-length of the nucleus. LINC complexes quantifications were obtained after the thresholding and the binarization of SIM slides or Extended Depth of Focus images for actin co-localization or quantification on the whole nuclear projected area, respectively.
Nuclear compression experiments
We used circular glass coverslips of 22 mm in diameter, 170 μm thick and 0.136 grams to apply a homogeneous pressure on the nucleus. After 20 hours in culture on FN-coated circular micropatterns, a glass coverslip was placed over HUVECs during 6 hours. The top side of glass coverslips was coated with a 1% solution of pluronic F-127 to avoid cellular adhesion and exerted a normal pressure proportional to its weight.
Data presentation and statistics
Results are presented as the means ± Standard Deviation (SD). Student's two-tailed t-test was performed in Origin 8.0 where two groups were compared. Values of p < 0.05 were considered statistically significant.
Author Contributions
M.V. and S.G. designed the experiments, wrote the manuscript and prepared figures. J.B.B. and M.V. performed SIM experiments. M.R., T.G., J.L. and M.V. performed microprinting, immunohistochemistry experiments, confocal microscopy imaging, nuclear compression and relaxation experiments. All authors reviewed and accepted the manuscript.
Supplementary Material
Supplementary Information
Movie S1
Movie S2
Movie S3
Movie S4
Movie S5
Acknowledgments
The authors gratefully acknowledge Philippe Baert (Nikon Belux Instruments) and the Nikon Imaging Center of the Curie Institute (Paris, France). This work was supported by the Fonds National de la Recherche Scientifique – FNRS under Grants “Nanomotility” FRFC n°2.4622.11 and “TIRF Microscopy” n°1.5013.11F. MR is a Research Fellow of the Fonds National de la Recherche Scientifique – FNRS. T.G. and J.L. doctoral fellowships are supported by the Foundation for Training in Industrial and Agricultural Research (FRIA). The Mechanobiology & Soft Matter group belongs to the French research consortium GDR 3070 CellTiss. Published with the financial support of the Fonds National de la Recherche Scientifique – FNRS.
References
- Pajerowski J. D., Dahl K. N., Zhong F. L., Sammak P. J. & Discher D. E. Physical plasticity of the nucleus in stem cell differentiation. Proc Natl. Acad. Sci. U.S.A. 104, 15619–15624 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammerding J. et al. Lamins A and C but not lamin B1 regulate nuclear mechanics. J. Biol. Chem. 281, 25768–25780 (2006). [DOI] [PubMed] [Google Scholar]
- De Vos W. H. et al. Repetitive disruptions of the nuclear envelope invoke temporary loss of cellular compartmentalization in laminopathies. Hum. Mol. Genet. 20, 4175–4186 (2011). [DOI] [PubMed] [Google Scholar]
- Shimi T. et al. The A- and B-type nuclear lamin networks: microdomains involved in chromatin organization and transcription. Genes Dev. 22, 3409–3421 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N., Tytell J. D. & Ingber D. E. Mechanotransduction at a distance: Mechanically coupling the extracellular matrix with the nucleus. Nat. Rev. Mol. Cell Biol. 10, 75–82 (2009). [DOI] [PubMed] [Google Scholar]
- Shivashankar G. V. Mechanosignaling to the cell nucleus and gene regulation. Annu. Rev. Biophys. 40, 361–378 (2011). [DOI] [PubMed] [Google Scholar]
- Swift J. et al. Nuclear Lamin-A Scales with Tissue Stiffness and Enhances Matrix-Directed Differentiation. Science 341, 1240104 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crisp M. et al. Coupling of the nucleus and cytoplasm: role of the LINC complex. J. Cell. Biol. 2006, 172, 41–53 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGee M. D., Rillo R., Anderson A. S. & Starr D. A. UNC-83 is a KASH protein required for nuclear migration and is recruited to the outer nuclear membrane by a physical interaction with the SUN protein UNC-84. Mol Biol Cell. 17, 1790–1801 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosa B. A., Rothballer A., Kutay U. & Schwartz T. U. LINC complexes form by binding of three KASH peptides to domain interfaces of trimeric SUN proteins. Cell 149, 1035–1047 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardi M. L. et al. The interaction between nesprins and sun proteins at the nuclear envelope is critical for force transmission between the nucleus and cytoskeleton. J. Biol. Chem. 286, 26743–26753 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel V. & Sheetz M. Local force and geometry sensing regulate cell functions. Nat. Rev. Mol. Cell Biol. 7, 265–275 (2006). [DOI] [PubMed] [Google Scholar]
- Engler A. J., Sen S., Sweeney H. L. & Discher D. E. Matrix elasticity directs stem cell lineage specification. Cell 126, 677–689 (2006). [DOI] [PubMed] [Google Scholar]
- Chen C. S. Mechanotransduction - a field pulling together? J. Cell Sci. 121, 3285–3292 (2008). [DOI] [PubMed] [Google Scholar]
- Geiger B., Spatz J. P. & Bershadsky A. D. Environmental sensing through focal adhesions. Nat. Rev. Mol. Cell Biol. 10, 21–33 (2009). [DOI] [PubMed] [Google Scholar]
- Jaalouk D. E. & Lammerding J. Mechanotransduction gone awry. Nat. Rev. Mol. Cell Biol. 10, 63–73 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilian K. A., Bugarija B., Lahn B. T. & Mrksich M. Geometric cues for directing the differentiation of mesenchymal stem cells. Proc. Natl. Acad. Sci. U.S.A. 107, 4872–4877 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazumder A., Roopa T., Basu A., Mahadevan L. & Shivashankar G. V. Dynamics of chromatin decondensation reveals the structural integrity of a mechanically prestressed nucleus. Biophys. J. 95, 3028–3035 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergani L., Grattarola M. & Nicolini C. Modifications of chromatin structure and gene expression following induced alterations of cellular shape. Int. J. Biochem. Cell Biol. 36, 1447–1461 (2004). [DOI] [PubMed] [Google Scholar]
- Khatau S. B. et al. A perinuclear actin cap regulates nuclear shape. Proc. Natl. Acad. Sci. U.S.A. 106, 19017–19022 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versaevel M., Grevesse T. & Gabriele S. Spatial coordination between cell and nuclear shape within micropatterned endothelial cells. Nat. Commun. 3, 671 (2012). [DOI] [PubMed] [Google Scholar]
- Li Q., Kumar A., Makhija E. & Shivashankar G. V. The regulation of dynamic mechanical coupling between actin cytoskeleton and nucleus by matrix geometry. Biomaterials. 35, 961–969 (2014). [DOI] [PubMed] [Google Scholar]
- Versaevel M., Grevesse T., Riaz M., Lantoine J. & Gabriele S. Micropatterning hydroxy-PAAm hydrogels and Sylgard 184 silicone elastomers with tunable elastic moduli. Methods Cell Biol. 121, 33–48 (2014). [DOI] [PubMed] [Google Scholar]
- Kim D. H. et al. Actin cap associated focal adhesions and their distinct role in cellular mechanosensing. Sci. Rep. 2, 555 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhen Y. Y., Libotte T., Munck M., Noegel A. A. & Korenbaum E. NUANCE, a giant protein connecting the nucleus and actin cytoskeleton. J. Cell Sci. 115, 3207–3222 (2002). [DOI] [PubMed] [Google Scholar]
- Vishavkarma R. et al. Role of Actin Filaments in Correlating Nuclear Shape and Cell Spreading. PLoS ONE 9, e107895 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Berre M., Aubertin J. & Piel M. Fine control of nuclear confinement identifies a threshold deformation leading to lamina rupture and induction of specific genes. Integr Biol 4, 1406–1414 (2012). [DOI] [PubMed] [Google Scholar]
- Woollam D. H., Millen J. W. & Ford E. H. Points of attachment of pachytene chromosomes to the nuclear membrane in mouse spermatocytes. Nature 213, 298–299 (1967). [DOI] [PubMed] [Google Scholar]
- Versaevel M., Grevesse T., Riaz M. & Gabriele S. Cell confinement: putting the squeeze on the nucleus. Soft Matter 9, 6665–6676 (2013). [Google Scholar]
- Martini F. J. & Valdeolmillos M. Actomyosin Contraction at the Cell Rear Drives Nuclear Translocation in Migrating Cortical Interneurons. J. Neurosci. 30, 8660–8670 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hur S. S., Zhao Y., Li Y.-S., Botvinick E. & Chien S. Live cells exert 3-dimensional traction forces on their substrata. Cell. Mol. Bioeng. 2, 425–436 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delanoë-Ayari H., Rieu J. P. & Sano M. 4D Traction Force Microscopy Reveals Asymmetric Cortical Forces in Migrating Dictyostelium Cells. Phys. Rev. Lett. 105, 248103 (2010). [DOI] [PubMed] [Google Scholar]
- Luxton G. W., Gomes E. R., Folker E. S., Vintinner E. & Gundersen G. G. Linear arrays of nuclear envelope proteins harness retrograde actin flow for nuclear movement. Science 329, 956–959 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbraith C. G., Yamada K. M. & Sheetz M. P. The relationship between force and focal complex development. J Cell Biol. 159, 695–705 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riveline D. et al. Focal contacts as mechanosensors: externally applied local mechanical force induces growth of focal contacts by an mDia1-dependent and ROCK-independent mechanism. J Cell Biol. 153, 1175–1185 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A., Maitra A., Sumit M., Ramaswamy S. & Shivashankar G. V. Actomyosin contractility rotates the cell nucleus. Sci. Rep. 4, 3871 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S., Marcel N., Sarin A. & Shivashankar G. V. Role of actin dependent nuclear deformation in regulating early gene expression. PLoS ONE 7, e53031 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain N., Venkatesan Iyer K., Kumar A. & Shivashankar G. V. Cell geometric constraints induce modular gene-expression patterns via redistribution of HDAC3 regulated by actomyosin contractility. Proc. Natl. Acad. Sci. U.S.A. 110, 11349–11354 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya D., Talwar S., Mazumder A. & Shivashankar G. V. Spatio-temporal plasticity in chromatin organization in mouse cell differentiation and during drosophila embryogenesis. Biophys. J. 96, 3832–3839 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman R. D., Gruenbaum Y., Moir R. D., Shumaker D. K. & Spann T. P. Nuclear lamins: building blocks of nuclear architecture. Genes Dev 16, 533–547 (2002). [DOI] [PubMed] [Google Scholar]
- Mahen R. et al. A-Type Lamins Maintain the Positional Stability of DNA Damage Repair Foci in Mammalian Nuclei. PLoS ONE 8, e61893 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grevesse T., Versaevel M., Circelli G., Desprez S. & Gabriele S. A simple route to functionalize polyacrylamide gels for the independent tuning of mechanotransduction cues. Lab Chip. 13, 777–780 (2013). [DOI] [PubMed] [Google Scholar]
- Grevesse T., Versaevel M. & Gabriele S. Preparation of hydroxy-PAAm hydrogels for decoupling the effects of mechanotransduction cues. J. Vis. Exp. 28, e51010 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriele S., Benoliel A.-M., Bongrand P. & Théodoly O. Microfluidic Investigation Reveals Distinct Roles for Actin Cytoskeleton and Myosin II Activity in Capillary Leukocyte Trafficking. Biophys. J. 96, 4308–4318 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information
Movie S1
Movie S2
Movie S3
Movie S4
Movie S5