A strategy for the purification of forced POZ-domain heterodimers is described, and crystal structures of the heterodimeric POZ domains of Miz1/BCL6 and Miz1/NAC1 are reported.
Keywords: BTB domain, forced heterodimer, tethered heterodimer, transcriptional repressor, Miz1/BCL6, Miz1/NAC1
Abstract
The POZ domain is an evolutionarily conserved protein–protein interaction domain that is found in approximately 40 mammalian transcription factors. POZ domains mediate both homodimerization and the heteromeric interactions of different POZ-domain transcription factors with each other. Miz1 is a POZ-domain transcription factor that regulates cell-cycle arrest and DNA-damage responses. The activities of Miz1 are altered by its interaction with the POZ-domain transcriptional repressors BCL6 and NAC1, and these interactions have been implicated in tumourigenesis in B-cell lymphomas and in ovarian serous carcinomas that overexpress BCL6 and NAC1, respectively. A strategy for the purification of tethered POZ domains that form forced heterodimers is described, and crystal structures of the heterodimeric POZ domains of Miz1/BCL6 and of Miz1/NAC1 are reported. These structures will be relevant for the design of therapeutics that target POZ-domain interaction interfaces.
1. Introduction
Many eukaryotic transcription factors function as biological dimers, and individual members of transcription-factor networks may form distinct homodimeric and heterodimeric associations that influence the recognition of target gene DNA sequences and the recruitment of transcriptional co-regulators. The specificity and regulation of transcription-factor dimerization thereby play critical roles in controlling the gene expression patterns that govern proliferation and differentiation in many cell lineages.
The POZ [poxvirus and zinc finger, also known as BTB (bric-à-brac, tram-track and broad complex)] domain is a protein–protein interaction domain that is found at the N-terminus of approximately 40 mammalian transcription factors (Bardwell & Treisman, 1994 ▶; reviewed in Stogios et al., 2005 ▶). Many POZ-domain transcription factors (POZ-TFs) play roles in development and several have been implicated in specific human malignancies (reviewed in Kelly & Daniel, 2006 ▶). The POZ-domain fold comprises an α-helical core that is flanked by β-strands, and the transcription-factor POZ domains form highly stable obligate dimers that contain a central hydrophobic interface of α-helices, together with two β-sheet interfaces that each contain one strand contributed from each constituent monomer (Li et al., 1997 ▶). The POZ domains also mediate the recruitment of non-POZ partners such as transcriptional co-regulators, and most POZ-TFs interact with DNA via a zinc-finger domain that is well separated from the POZ domain by a flexible linker.
Miz1 (Myc-interacting zinc-finger protein) is a POZ-domain transcription factor (Peukert et al., 1997 ▶) that activates target genes via recruitment of the cofactors p300 (Staller et al., 2001 ▶) and nucleophosmin (Wanzel et al., 2008 ▶). Many Miz1 target genes play roles in cell proliferation (Seoane et al., 2001 ▶; Staller et al., 2001 ▶) and differentiation (Wu et al., 2003 ▶; Kime & Wright, 2003 ▶), and Miz1 activates the cell-cycle inhibitor gene cdkn1a in response to DNA damage (Herold et al., 2002 ▶; Seoane et al., 2002 ▶). Miz1 is also an important regulator of autophagy (Wolf et al., 2013 ▶) and of B-cell and T-cell development (Kosan et al., 2010 ▶; Möröy et al., 2011 ▶; Saba, Kosan, Vassen, Klein-Hitpass et al., 2011 ▶; Saba, Kosan, Vassen & Moroy, 2011 ▶). The transcriptional properties of Miz1 are modulated by its interaction with other transcription factors, and many of these associations are directly relevant in human malignancy. Miz1 acts as a repressor when complexed with the bHLH-LZ transcription factor c-Myc (Peukert et al., 1997 ▶), and the inappropriate repression of Miz1 target genes contributes to deregulated cell proliferation in c-Myc-overexpressing tumours (Hönnemann et al., 2012 ▶; van Riggelen et al., 2010 ▶); the interaction of Miz1 with c-Myc is mediated by residues located adjacent to its zinc-finger DNA-binding domain. Miz1 associates with other POZ-domain transcription factors via heteromeric POZ–POZ interactions, and these interactions are also important in normal development and in malignancy. Miz1 interacts with the POZ-domain transcriptional repressor BCL6 (B-cell lymphoma 6) in germinal centre B cells (Phan et al., 2005 ▶), leading to repression of the Miz1 target gene cdkn1a; this facilitates the proliferative expansion that normally takes place in spite of the natural DNA damage that accompanies the somatic hypermutation and class-switch recombination of immunoglobulin genes at this stage. However, the overexpression of BCL6 in diffuse large B-cell lymphoma (DLBCL) and follicular lymphoma leads to the inappropriate repression of Miz1 target genes and may contribute to malignancy. More recently, it has been recognized that Miz1 interacts with other POZ-domain transcription factors, including ZBTB4 (Weber et al., 2008 ▶) and NAC1 (nucleus accumbens-associated 1; Stead & Wright, 2014 ▶), and the inappropriate repression of Miz1-target genes may therefore also be relevant to oncogenesis in NAC1-overexpressing ovarian serous carcinomas (Nakayama et al., 2006 ▶). Although all transcription-factor POZ domains crystallize as homodimers (Ahmad et al., 1998 ▶, 2003 ▶; Ghetu et al., 2008 ▶; Stead et al., 2008 ▶, 2009 ▶; Rosbrook et al., 2012 ▶; Schubot et al., 2006 ▶; Li et al., 1999 ▶; Stogios et al., 2010 ▶, 2007 ▶), the Miz1 POZ domain also crystallized in a tetrameric form that resulted from the interaction of two classic POZ dimers (Stead et al., 2007 ▶). This observation raised the suggestion that heteromeric POZ-domain interactions might be mediated by a tetrameric association of different POZ dimers. Recent cross-linking studies have, however, indicated a heterodimeric interaction of the Miz1 and NAC1 POZ domains in transfected mammalian cells (Stead & Wright, 2014 ▶).
Structural studies of the BCL6 POZ domain (Ahmad et al., 2003 ▶; Ghetu et al., 2008 ▶) are guiding the design of targeted therapies for DLBCL, and crystal structures of the dimeric BCL6 POZ domain in complex with the co-repressors SMRT (silencing mediator for retinoid and thyroid hormone receptors) and BCoR (BCL6-interacting co-repressor) have been used to identify small molecules that block these interactions and that kill BCL6-positive B-cell lymphoma cells in vitro and in vivo (Cerchietti et al., 2009 ▶, 2010 ▶). No structures of heterodimeric POZ domains have been reported, and this may reflect difficulties in the production of pure heterodimeric forms. The crystallization of heterodimeric transcription factors is often hampered by the predominance of homodimers in bacterial co-expression systems, and alternative strategies are often used. For example, the purification of heterodimers of the bHLH-Zip protein Max with its bHLH-zip partners Myc or Mad was achieved using a semi-synthetic scheme for the production of chemoselectively ligated heterodimers (Nair & Burley, 2003 ▶).
In nature, flexible Gly-rich linkers sometimes separate the multiple domains of a single protein (Argos, 1990 ▶; Steinert et al., 1991 ▶), and synthetic Gly-rich linkers have been used in protein-engineering strategies to separate two binding partners within a chimeric protein (reviewed by Reddy Chichili et al., 2013 ▶). The increased proximity of the binding partners favours intramolecular over intermolecular interactions, and forced dimers therefore predominate. This approach has been used for the production of tethered homodimers and heterodimers both in bacteria and in mammalian cells. For example, a forced homodimer of the HIV protease was expressed in bacteria for structural and biochemical studies (Wlodawer et al., 1989 ▶; Cheng et al., 1990 ▶); the forced dimer was more stable than the natural protease dimer, and this strategy provided a means of studying the effect of monomer–dimer transitions on enzymatic activity and of producing asymmetric dimer mutants for structure–function studies. In mammalian cells, the ectopic expression of forced homodimers and heterodimers has been particularly useful for dissecting the interactions between family members of transcription-factor networks: forced homodimers of the transcription factor Nanog have been used to study the relative importance of dimers and monomers in stem-cell self-renewal and pluripotency (Wang et al., 2008 ▶), and tethered heterodimers have been used to analyse the role of the bHLH proteins MyoD and E47 in myogenesis (Neuhold & Wold, 1993 ▶).
Here, we describe a strategy for the production of heterodimeric POZ domains by using a Gly+Ser-rich tether to separate two different POZ domains within a chimeric protein, and we report the crystal structures of the heterodimeric POZ domains of Miz1/BCL6 and Miz1/NAC1.
2. Materials and methods
2.1. Cloning
Tethered POZ domains that formed forced heterodimers were expressed as GST-fusion proteins using the plasmid pGEX-6P-1 (GE Healthcare). The oligonucleotides 5′-AATTCAGATCTGGCGGAGGCTCGAGCGGTGGGAGCGGTACCG and 5′-TCGACGGTACCGCTCCCACCGCTCGAGCCTCCGCCAGATCTG were annealed and inserted between the EcoRI and SalI restriction-enzyme sites of pGEX-6P-1; this sequence encoded a polypeptide tether. A cDNA encoding human Miz1 residues 2–115 was inserted between the BamHI and BglII sites located at the 5′ end of this region, and cDNAs encoding human BCL6 residues 5–129 or human NAC1 residues 2–125 were inserted between the KpnI and SalI sites at the 3′ end. NAC1 contained the F98D mutation that enhances the solubility of the recombinant protein (Stead et al., 2009 ▶). The resulting plasmids encoded the Miz1 POZ domain joined to either the BCL6 or NAC1 POZ domain by a flexible in-frame polypeptide tether (Arg-Ser-Gly-Gly-Gly-Ser-Ser-Gly-Gly-Ser-Gly-Thr).
2.2. Protein expression and purification
Plasmids were transformed into Escherichia coli BL21 (DE3) pLysS cells and plated onto Luria–Bertani agar plates containing 34 µg ml−1 chloramphenicol and 100 µg ml−1 ampicillin. A single colony was cultured overnight at 310 K with shaking in 2TY supplemented with the same antibiotics. 84 ml of the overnight culture was diluted into 12 l 2TY supplemented with 100 µg ml−1 ampicillin and the cells were grown at 310 K with shaking to an optical density at 600 nm of ∼0.7. Expression of recombinant protein was induced by the addition of isopropyl β-d-1-thiogalactopyranoside to a final concentration of 0.1 mM and the culture was incubated at 289 K for 16 h. Cells were harvested by centrifugation and resuspended in phosphate-buffered saline (PBS) containing 5 mM dithiothreitol (DTT).
Cells were lysed using a cell disruptor (Constant Systems) and Triton X-100 was added to a final concentration of 0.1%. The lysate was clarified by centrifugation at 39 000g for 20 min and the supernatant was passed through a 1.2 µm filter. Fusion proteins were bound to Super-Glu glutathione resin (Generon) equilibrated with binding buffer (PBS, 0.1% Triton X-100, 5 mM DTT) for 1 h at 277 K. Nonspecifically bound proteins were removed from the resin by alternating washes with binding buffer and with binding buffer supplemented with 1 M NaCl. The glutathione resin was further rinsed with 20 mM Tris–HCl, 75 mM NaCl, 5 mM DTT pH 7.5, and the GST tag was removed by cleavage with PreScission protease overnight at 277 K. POZ domains were further purified by size-exclusion chromatography on a pre-equilibrated HiLoad Superdex 75 26/60 column (GE Healthcare) in 20 mM Tris–HCl, 150 mM NaCl, 5 mM DTT, 5% glycerol pH 7.5. Purified proteins were concentrated to 6 mg ml−1 using Vivaspin 20 centrifugal concentrators (Sartorius). Protein concentration was determined using the Bradford reagent. The yield of purified protein was approximately 1 mg per litre of bacterial culture.
2.3. Protein crystallization
Crystallization trials were set up in Corning 3552 96-well plates using a variety of commercially available sparse-matrix crystallization screens.
Crystals of the Miz1/BCL6 heterodimeric POZ domain were obtained in 8.25% polyethylene glycol (PEG) 3350, 4.95% 2-propanol, 0.2 M ammonium citrate pH 4.5 using a protein concentration of 3.5 mg ml−1. Optimization of this condition was carried out by hanging-drop vapour diffusion in 24-well plates using dichlorodimethylsilane-treated coverslips. Crystals with average dimensions of 100 × 70 × 70 µm were grown by mixing 2.5 µl protein solution at 3.5 mg ml−1 with 2.5 µl reservoir solution (4% PEG 3350, 3% 2-propanol, 0.2 M ammonium citrate pH 4.5) and incubating at 291 K for 5 d. Crystals were harvested using a nylon cryo-loop and transferred for 30 s to a 5 µl drop of mother liquor supplemented with 15% glycerol before being flash-cooled at 100 K.
Crystals of the Miz1/NAC1 heterodimeric POZ domain were obtained in 0.33% PEG 4000, 0.33 M ammonium citrate pH 5.5 using a protein concentration of 7 mg ml−1. Optimization of this condition was carried out by sitting-drop vapour diffusion using 24-well Intelli-Plates (Art Robbins). Crystals with average dimensions of 200 × 100 × 100 µm were grown by mixing 3 µl protein at 3.5 mg ml−1 with 2.5 µl reservoir solution (0.2% PEG 4000, 0.1 M sodium citrate pH 5.6) and incubating at 291 K for 5 d. Crystals were harvested as above using mother liquor supplemented with 25% glycerol.
2.4. Data processing
X-ray data were collected on Diamond Light Source beamline I04 and were processed, reduced and scaled with XDS (Kabsch, 2010 ▶) and AIMLESS (Evans & Murshudov, 2013 ▶) as part of the xia2 pipeline (Winter, 2010 ▶). The structure of the Miz1/BCL6 POZ-domain heterodimer was solved by molecular replacement using Phaser (McCoy et al., 2007 ▶) with the Miz1 and BCL6 POZ-domain monomers as search models [Miz1, PDB entry 3m52, chain A residues 2–50 (Stogios et al., 2010); BCL6, PDB entry 1r28, chain A residues 7–128 (Ahmad et al., 2003 ▶)]. Phaser located a Miz1/BCL6 POZ-domain heterodimer that resembled the structures of reported POZ-domain dimers; this was then used as a model in Phaser to find all four copies of the heterodimeric POZ domain in the asymmetric unit. Density modification was carried out using Parrot (Zhang et al., 1997 ▶) and the model was built using ARP/wARP (Cohen et al., 2008 ▶).
The structure of the Miz1/NAC1 heterodimer was solved by molecular replacement using the Miz1/BCL6 POZ-domain heterodimer as the search model; the asymmetric unit contained two Miz1/NAC1 POZ-domain heterodimers.
The models of the heterodimeric Miz1/BCL6 and Miz1/NAC1 POZ domains underwent iterative refinement in REFMAC5 (Murshudov et al., 2011 ▶) and model building using Coot (Emsley et al., 2010 ▶). Stereochemistry was analysed with MolProbity (Chen et al., 2010 ▶).
SSM structure superpositions were calculated in CCP4mg (McNicholas et al., 2011 ▶) and contact residues were obtained using the PISA server (Krissinel & Henrick, 2007 ▶). Illustrations of protein structures were prepared using CCP4mg.
3. Results
3.1. Bacterial expression of tethered POZ domains
To obtain crystal structures of heterodimeric POZ domains, we expressed chimeric proteins that contained the Miz1 POZ domain joined to either the BCL6 or the NAC1 POZ domain by a Gly+Ser-rich linker that was predicted to form a flexible hydrophilic tether. The Miz1 POZ domain was placed at the N-terminus of the chimeric proteins, as reasoned from the structures of homodimeric POZ domains. The BCL6 and NAC1 POZ domains each contain six α-helices and five β-strands, with the N-terminal β1 of each chain interacting with β5 of the opposite chain; the N-terminus of each chain is therefore close to the C-terminus of the opposite chain in the domain-swapped homodimer. The Miz1 POZ domain lacks a β1 strand, and the N- and C-termini of the opposite chains of the homodimer are further apart. It was therefore anticipated that the formation of forced heterodimers would be more favoured if the Miz1 POZ domain were positioned at the N-terminus rather than at the C-terminus of the chimeric proteins.
The tethered Miz1/BCL6 and Miz1/NAC1 POZ domains eluted at the same volume as classic POZ-domain homodimers when purified by size-exclusion chromatography.
3.2. Crystal structures of heterodimeric Miz1/BCL6 and Miz1/NAC1 POZ domains
The tethered POZ domains crystallized readily, and structures were determined by molecular replacement using the constituent POZ-domain monomers as search models; structures of Miz1/BCL6 and of Miz1/NAC1 were determined to 2.2 and to 2.3 Å resolution, respectively (Table 1 ▶ and Fig. 1 ▶).
Table 1. Data collection and processing.
Values in parentheses are for the outer shell.
| Miz1/BCL6 | Miz1/Nac1 | |
|---|---|---|
| PDB entry | 4u2m | 4u2n |
| Crystal parameters | ||
| Space group | P212121 | P21212 |
| Unit-cell parameters (, ) | a = 91.85, b = 97.88, c = 119.88, = = = 90 | a = 109.09, b = 115.19, c = 58.29, = = = 90 |
| Data collection | ||
| Resolution range () | 58.472.23 (2.292.23) | 50.932.30 (2.362.30) |
| Wavelength () | 0.97949 | 0.97949 |
| R merge † (%) | 4.6 (31.1) | 3.0 (66.3) |
| R p.i.m. ‡ (%) | 2.5 (17.3) | 1.7 (37.0) |
| I/(I) | 19 (4.8) | 24 (2.4) |
| Total No. of reflections | 215105 (14633) | 133634 (10182) |
| No. of unique reflections | 52962 (3738) | 33315 (2420) |
| Multiplicity | 4.1 (3.9) | 4.0 (4.2) |
| Completeness (%) | 99.3 (96.2) | 99.7 (99.7) |
| Refinement | ||
| Resolution () | 2.23 | 2.30 |
| R work § (%) | 21.1 | 23.8 |
| R free ¶ (%) | 24.0 | 28.6 |
| R.m.s.d.†† bond angles () | 1.502 | 1.623 |
| R.m.s.d.†† bond lengths () | 0.015 | 0.017 |
| No. of non-H protein atoms | 7187 | 3581 |
| No. of water molecules | 163 | 30 |
| Average B factor (2) | 47.0 | 66.0 |
| Ramachandran favoured‡‡ (%) | 98.2 | 98.5 |
| Ramachandran outliers‡‡ (%) | 0 | 0 |
R
merge = 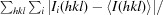
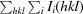 , where Ii(hkl) is the intensity of the ith measurement of reflection hkl and I(hkl) is the mean intensity of all i measurements.
, where Ii(hkl) is the intensity of the ith measurement of reflection hkl and I(hkl) is the mean intensity of all i measurements.
R
p.i.m. = 
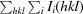 , where Ii(hkl) is the intensity of the ith measurement of the reflection hkl, I(hkl) is the mean intensity of all i measurements and N(hkl) is the multiplicity.
, where Ii(hkl) is the intensity of the ith measurement of the reflection hkl, I(hkl) is the mean intensity of all i measurements and N(hkl) is the multiplicity.
R
work = 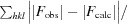
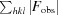 , where F
obs and F
calc are the observed and calculated structure factors, respectively.
, where F
obs and F
calc are the observed and calculated structure factors, respectively.
R free is the same as R work but calculated using a random 5% of the data that was excluded from refinement.
R.m.s.d. is the deviation from ideal values.
Ramachandran analysis was called out in MolProbity (Chen et al., 2010 ▶).
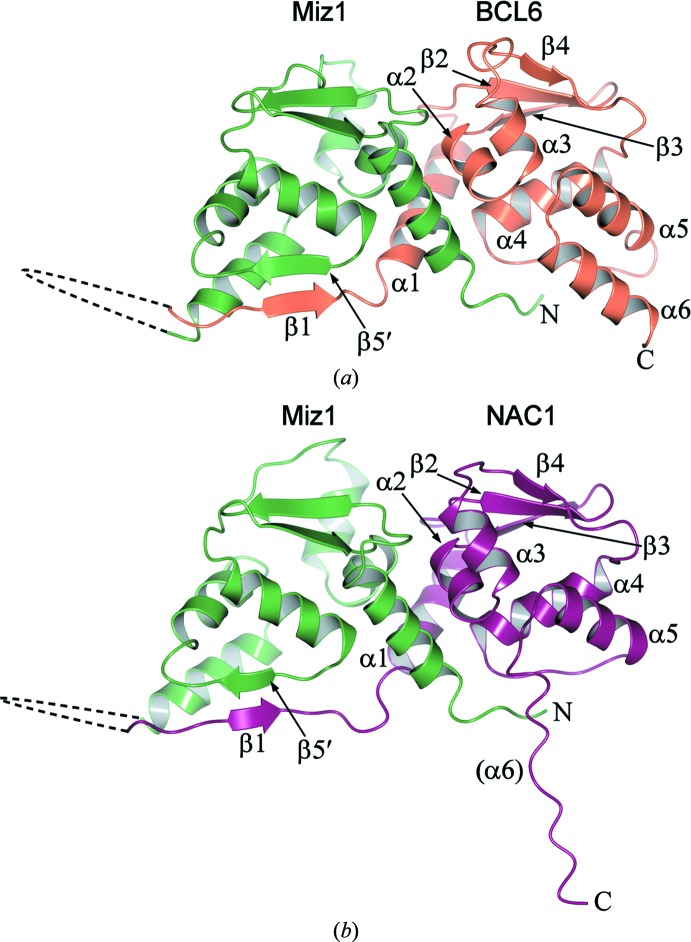
Figure 1.
Structures of heterodimeric POZ domains. Ribbon representations of (a) the Miz1/BCL6 (PDB entry 4u2m, chain D) and (b) the Miz1/NAC1 (PDB entry 4u2n, chain B) heterodimeric POZ domains. The Miz1 residues are coloured green and the BCL6 and NAC1 residues are coloured orange and purple, respectively. Secondary-structure elements of BCL6 and of NAC1 are shown, together with the β5 strand of Miz1; the unstructured region that corresponds to NAC1 α6 is indicated in parentheses. The dashed lines denote the linkers between the POZ domains; the electron density in these regions was poor and the residues were not modelled in the structures.
The tethered POZ domains formed forced heterodimers that resemble the overall structure of reported POZ-domain homodimers. The heterodimeric POZ domain of Miz1/BCL6 superposed well with the homodimeric POZ domains of Miz1 and BCL6 (r.m.s.d. values of 2.0 and 1.14 Å2, respectively), and the heterodimeric POZ domain of Miz1/NAC1 superposed well with the Miz1 and NAC1 homodimers (r.m.s.d. values of 1.86 and 1.66 Å2, respectively; Figs. 2 ▶ a and 2 ▶ b). The α6 region of the NAC1 chain was unstructured in the heterodimeric Miz1/NAC1 POZ domain, as observed in the NAC1 homodimer. The N-terminal β1 strands of the BCL6 and NAC1 POZ domains interact with β5 of the opposite chain in the domain-swapped homodimers, and although the Miz1 POZ domain lacks a β1 strand, its β5 region interacts with β1 of BCL6 or NAC1 in the heterodimeric POZ complexes.
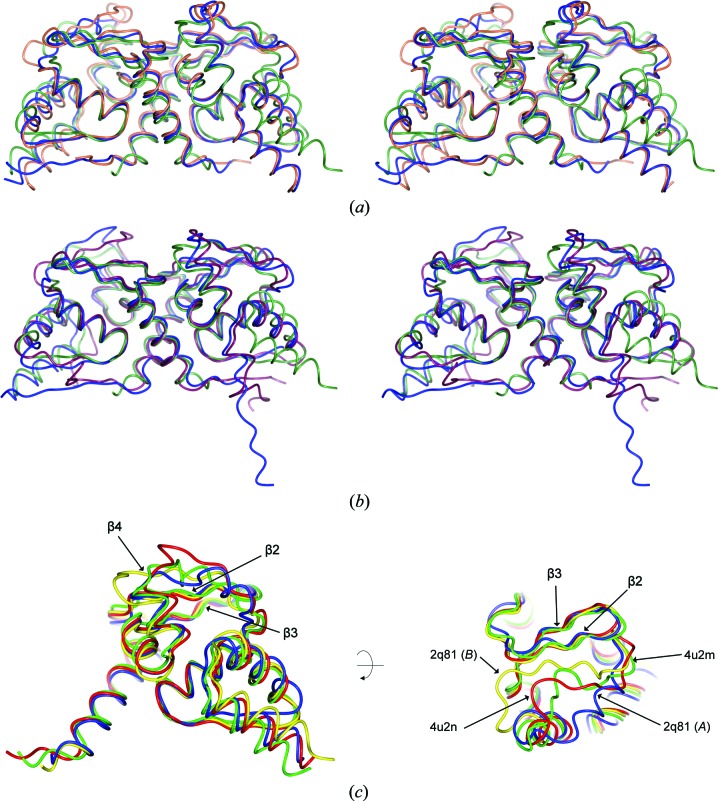
Figure 2.
Superpositions of heterodimeric POZ domains. (a) Stereoimage of the superpositions of the Miz1/BCL6 heterodimeric POZ domain (blue; PDB entry 4u2m, chain D) with the homodimeric POZ domains of Miz1 (green; PDB entry 2q81 chains A and B) and of BCL6 (orange; PDB entry 1r28). (b) Stereoimage of the superpositions of the Miz1/NAC1 heterodimeric POZ domain (blue; PDB entry 4u2n, chain B) with the homodimeric POZ domains of Miz1 (green; PBD entry 2q81 chains A and B) and of NAC1 (purple; PDB entry 3ga1, Stead et al., 2009 ▶). (c) Superposition of Miz1 POZ-domain monomers from PDB entries 2q81 chain A (blue), 2q81 chain B (yellow), 4u2m chain D residues 2–115 (green) and 4u2n chain B residues 2–115 (red).
In most POZ-domain homodimers, the secondary-structure elements β2, β3 and β4 form a three-stranded β-sheet that is located at the ‘top’ of the molecule; however, in Miz1 the β4 region is displaced relative to the β2 and β3 strands in the crystal structures of Miz1 POZ-domain homodimers and tetramers. Each constituent dimer of the Miz1 POZ-domain tetramer (PDB entry 2q81; Stead et al., 2007 ▶) contains one ‘normal’ and one ‘displaced’ β4 region (Fig. 2 ▶ c, chain B and chain A, respectively), and the position of these displaced β4 strands was similar to that subsequently reported in a structure of the Miz1 POZ homodimer (PDB entry 3m52; Stogios et al., 2010 ▶). The Miz1 β4 region of the heterodimeric Miz1/BCL6 and Miz1/NAC1 POZ domains is located between the previously described ‘normal’ and ‘displaced’ positions, indicating that this region is flexible (Fig. 2 ▶ c).
The dimerization interfaces of the heterodimeric POZ domains resemble those of POZ-domain homodimers, comprising the central hydrophobic interface of α-helices together with the strand-exchanged β-sheet interface (β1–β5′; Fig. 1 ▶) that is found on one side only of the POZ-domain heterodimers. Several of the residues involved in hydrogen-bond interactions at the interface of the Miz1 homodimer (PDB entry 3m52) are involved in similar interactions in the Miz1/BCL6 and Miz1/NAC1 heterodimers. For example, Phe3 and His6 of Miz1 α1 each interact with α4 residues of the opposite chain in the Miz1 homodimer and in both the Miz1/BCL6 and Miz1/NAC1 heterodimers. Other common interactions involve the Miz1 α1 residue Gln17, which interacts with α2 of the opposite chain, and the Miz1 α4 residues Ala80 and Leu82, which interact with α1.
4. Conclusion
The interaction of the Miz1 POZ domain with the POZ-domain transcriptional repressors BCL6 and NAC1 leads to the repression of Miz1 target genes and is relevant in human malignancy. The transcriptional co-repressors BCoR and SMRT bind to the lateral groove of the BCL6 POZ domain and make contacts with residues from both chains of the BCL6 POZ homodimer; notably, these residues are not conserved in the Miz1 chain of the Miz1/BCL6 heterodimer. The production of a forced POZ-domain heterodimer of Miz1/BCL6 will enable a direct experimental analysis of its co-repressor interactions, and these structures will inform the design of therapeutic inhibitors. The network of heteromeric interactions between POZ-domain transcription factors has not been extensively studied and the structures of forced POZ-domain heterodimers will reveal features that determine the specificity of these associations.
Supplementary Material
PDB reference: Miz1/NAC1 POZ domains, 4u2n
PDB reference: Miz1/BCL6 POZ domains, 4u2m
Acknowledgments
We thank the staff of beamline I04 at Diamond Light Source for advice on X-ray data collection. This work was funded by Yorkshire Cancer Research (grant reference L334).
References
- Ahmad, K. F., Engel, C. K. & Privé, G. G. (1998). Proc. Natl Acad. Sci. USA, 95, 12123–12128. [DOI] [PMC free article] [PubMed]
- Ahmad, K. F., Melnick, A., Lax, S., Bouchard, D., Liu, J., Kiang, C.-L., Mayer, S., Takahashi, S., Licht, J. D. & Privé, G. G. (2003). Mol. Cell, 12, 1551–1564. [DOI] [PubMed]
- Argos, P. (1990). J. Mol. Biol. 211, 943–958. [DOI] [PubMed]
- Bardwell, V. J. & Treisman, R. (1994). Genes Dev. 8, 1664–1677. [DOI] [PubMed]
- Cerchietti, L. C. et al. (2010). Cancer Cell, 17, 400–411. [DOI] [PMC free article] [PubMed]
- Cerchietti, L. C., Yang, S. N., Shaknovich, R., Hatzi, K., Polo, J. M., Chadburn, A., Dowdy, S. F. & Melnick, A. (2009). Blood, 113, 3397–3405. [DOI] [PMC free article] [PubMed]
- Chen, V. B., Arendall, W. B., Headd, J. J., Keedy, D. A., Immormino, R. M., Kapral, G. J., Murray, L. W., Richardson, J. S. & Richardson, D. C. (2010). Acta Cryst. D66, 12–21. [DOI] [PMC free article] [PubMed]
- Cheng, Y.-S. E., Yin, F. H., Foundling, S., Blomstrom, D. & Kettner, C. A. (1990). Proc. Natl Acad. Sci. USA, 87, 9660–9664. [DOI] [PMC free article] [PubMed]
- Cohen, S. X., Ben Jelloul, M., Long, F., Vagin, A., Knipscheer, P., Lebbink, J., Sixma, T. K., Lamzin, V. S., Murshudov, G. N. & Perrakis, A. (2008). Acta Cryst. D64, 49–60. [DOI] [PMC free article] [PubMed]
- Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. (2010). Acta Cryst. D66, 486–501. [DOI] [PMC free article] [PubMed]
- Evans, P. R. & Murshudov, G. N. (2013). Acta Cryst. D69, 1204–1214. [DOI] [PMC free article] [PubMed]
- Ghetu, A. F., Corcoran, C. M., Cerchietti, L., Bardwell, V. J., Melnick, A. & Privé, G. G. (2008). Mol. Cell, 29, 384–391. [DOI] [PMC free article] [PubMed]
- Herold, S., Wanzel, M., Beuger, V., Frohme, C., Beul, D., Hillukkala, T., Syvaoja, J., Saluz, H. P., Haenel, F. & Eilers, M. (2002). Mol. Cell, 10, 509–521. [DOI] [PubMed]
- Hönnemann, J., Sanz-Moreno, A., Wolf, E., Eilers, M., Elsässer, H. P. & Roemer, K. (2012). PLoS One, 7, e34885. [DOI] [PMC free article] [PubMed]
- Kabsch, W. (2010). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Kelly, K. F. & Daniel, J. M. (2006). Trends Cell Biol. 16, 578–587. [DOI] [PubMed]
- Kime, L. & Wright, S. C. (2003). Biochem. J. 370, 291–298. [DOI] [PMC free article] [PubMed]
- Kosan, C., Saba, I., Godmann, M., Herold, S., Herkert, B., Eilers, M. & Möröy, T. (2010). Immunity, 33, 917–928. [DOI] [PubMed]
- Krissinel, E. & Henrick, K. (2007). J. Mol. Biol. 372, 774–797. [DOI] [PubMed]
- Li, X., Lopez-Guisa, J. M., Ninan, N., Weiner, E. J., Rauscher, F. J. & Marmorstein, R. (1997). J. Biol. Chem. 272, 27324–27329. [DOI] [PubMed]
- Li, X., Peng, H., Schultz, D. C., Lopez-Guisa, J. M., Rauscher, F. J. & Marmorstein, R. (1999). Cancer Res. 59, 5275–5282. [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst. 40, 658–674. [DOI] [PMC free article] [PubMed]
- McNicholas, S., Potterton, E., Wilson, K. S. & Noble, M. E. M. (2011). Acta Cryst. D67, 386–394. [DOI] [PMC free article] [PubMed]
- Möröy, T., Saba, I. & Kosan, C. (2011). Semin. Immunol. 23, 379–387. [DOI] [PubMed]
- Murshudov, G. N., Skubák, P., Lebedev, A. A., Pannu, N. S., Steiner, R. A., Nicholls, R. A., Winn, M. D., Long, F. & Vagin, A. A. (2011). Acta Cryst. D67, 355–367. [DOI] [PMC free article] [PubMed]
- Nair, S. K. & Burley, S. K. (2003). Cell, 112, 193–205. [DOI] [PubMed]
- Nakayama, K., Nakayama, N., Davidson, B., Sheu, J. J.-C., Jinawath, N., Santillan, A., Salani, R., Bristow, R. E., Morin, P. J., Kurman, R. J., Wang, T.-L. & Shih, I.-M. (2006). Proc. Natl Acad. Sci. USA, 103, 18739–18744. [DOI] [PMC free article] [PubMed]
- Neuhold, L. A. & Wold, B. (1993). Cell, 74, 1033–1042. [DOI] [PubMed]
- Peukert, K., Staller, P., Schneider, A., Carmichael, G., Hänel, F. & Eilers, M. (1997). EMBO J. 16, 5672–5686. [DOI] [PMC free article] [PubMed]
- Phan, R. T., Saito, M., Basso, K., Niu, H. & Dalla-Favera, R. (2005). Nature Immunol. 6, 1054–1060. [DOI] [PubMed]
- Reddy Chichili, V. P., Kumar, V. & Sivaraman, J. (2013). Protein Sci. 22, 153–167. [DOI] [PMC free article] [PubMed]
- Riggelen, J. van, Müller, J., Otto, T., Beuger, V., Yetil, A., Choi, P. S., Kosan, C., Möröy, T., Felsher, D. W. & Eilers, M. (2010). Genes Dev. 24, 1281–1294. [DOI] [PMC free article] [PubMed]
- Rosbrook, G. O., Stead, M. A., Carr, S. B. & Wright, S. C. (2012). Acta Cryst. D68, 26–34. [DOI] [PubMed]
- Saba, I., Kosan, C., Vassen, L., Klein-Hitpass, L. & Moroy, T. (2011). J. Immunol. 187, 2982–2992. [DOI] [PubMed]
- Saba, I., Kosan, C., Vassen, L. & Moroy, T. (2011). Blood, 117, 3370–3381. [DOI] [PubMed]
- Schubot, F. D., Tropea, J. E. & Waugh, D. S. (2006). Biochem. Biophys. Res. Commun. 351, 1–6. [DOI] [PubMed]
- Seoane, J., Le, H.-V. & Massagué, J. (2002). Nature (London), 419, 729–734. [DOI] [PubMed]
- Seoane, J., Pouponnot, C., Staller, P., Schader, M., Eilers, M. & Massagué, J. (2001). Nature Cell Biol. 3, 400–408. [DOI] [PubMed]
- Staller, P., Peukert, K., Kiermaier, A., Seoane, J., Lukas, J., Karsunky, H., Möröy, T., Bartek, J., Massagué, J., Hänel, F. & Eilers, M. (2001). Nature Cell Biol. 3, 392–399. [DOI] [PubMed]
- Stead, M. A., Carr, S. B. & Wright, S. C. (2009). Acta Cryst. F65, 445–449. [DOI] [PMC free article] [PubMed]
- Stead, M. A., Rosbrook, G. O., Hadden, J. M., Trinh, C. H., Carr, S. B. & Wright, S. C. (2008). Acta Cryst. F64, 1101–1104. [DOI] [PMC free article] [PubMed]
- Stead, M. A., Trinh, C. H., Garnett, J. A., Carr, S. B., Baron, A. J., Edwards, T. A. & Wright, S. C. (2007). J. Mol. Biol. 373, 820–826. [DOI] [PubMed]
- Stead, M. A. & Wright, S. C. (2014). Biosci. Rep. 34, 227–235. [DOI] [PMC free article] [PubMed]
- Steinert, P. M., Mack, J. W., Korge, B. P., Gan, S.-Q., Haynes, S. R. & Steven, A. C. (1991). Int. J. Biol. Macromol. 13, 130–139. [DOI] [PubMed]
- Stogios, P. J., Chen, L. & Privé, G. G. (2007). Protein Sci. 16, 336–342. [DOI] [PMC free article] [PubMed]
- Stogios, P. J., Cuesta-Seijo, J. A., Chen, L., Pomroy, N. C. & Privé, G. G. (2010). J. Mol. Biol. 400, 983–997. [DOI] [PubMed]
- Stogios, P. J., Downs, G. S., Jauhal, J. J., Nandra, S. K. & Privé, G. G. (2005). Genome Biol. 6, r82. [DOI] [PMC free article] [PubMed]
- Wang, J., Levasseur, D. N. & Orkin, S. H. (2008). Proc. Natl Acad. Sci. USA, 105, 6326–6331. [DOI] [PMC free article] [PubMed]
- Wanzel, M., Russ, A. C., Kleine-Kohlbrecher, D., Colombo, E., Pelicci, P. G. & Eilers, M. (2008). Nature Cell Biol. 10, 1051–1061. [DOI] [PubMed]
- Weber, A., Marquardt, J., Elzi, D., Forster, N., Starke, S., Glaum, A., Yamada, D., Defossez, P. A., Delrow, J., Eisenman, R. N., Christiansen, H. & Eilers, M. (2008). EMBO J. 27, 1563–1574. [DOI] [PMC free article] [PubMed]
- Winter, G. (2010). J. Appl. Cryst. 43, 186–190.
- Wlodawer, A., Miller, M., Jaskólski, M., Sathyanarayana, B. K., Baldwin, E., Weber, I. T., Selk, L. M., Clawson, L., Schneider, J. & Kent, S. B. (1989). Science, 245, 616–621. [DOI] [PubMed]
- Wolf, E., Gebhardt, A., Kawauchi, D., Walz, S., von Eyss, B., Wagner, N., Renninger, C., Krohne, G., Asan, E., Roussel, M. F. & Eilers, M. (2013). Nature Commun. 4, 2535. [DOI] [PMC free article] [PubMed]
- Wu, S., Cetinkaya, C., Munoz-Alonso, M. J., von der Lehr, N., Bahram, F., Beuger, V., Eilers, M., Leon, J. & Larsson, L. G. (2003). Oncogene, 22, 351–360. [DOI] [PubMed]
- Zhang, K. Y. J., Cowtan, K. & Main, P. (1997). Methods Enzymol. 277, 53–64. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: Miz1/NAC1 POZ domains, 4u2n
PDB reference: Miz1/BCL6 POZ domains, 4u2m