Elmo–Dock complexes are involved not just in integrin-based adhesions but also in the initial formation of strong cadherin-based adhesions through the regulation of local Rho GTPase activity and actin remodeling.
Abstract
Cell–cell contact formation is a dynamic process requiring the coordination of cadherin-based cell–cell adhesion and integrin-based cell migration. A genome-wide RNA interference screen for proteins required specifically for cadherin-dependent cell–cell adhesion identified an Elmo–Dock complex. This was unexpected as Elmo–Dock complexes act downstream of integrin signaling as Rac guanine-nucleotide exchange factors. In this paper, we show that Elmo2 recruits Dock1 to initial cell–cell contacts in Madin–Darby canine kidney cells. At cell–cell contacts, both Elmo2 and Dock1 are essential for the rapid recruitment and spreading of E-cadherin, actin reorganization, localized Rac and Rho GTPase activities, and the development of strong cell–cell adhesion. Upon completion of cell–cell adhesion, Elmo2 and Dock1 no longer localize to cell–cell contacts and are not required subsequently for the maintenance of cell–cell adhesion. These studies show that Elmo–Dock complexes are involved in both integrin- and cadherin-based adhesions, which may help to coordinate the transition of cells from migration to strong cell–cell adhesion.
Introduction
Epithelial monolayers regulate and organize tissue structure and function (Bryant and Mostov, 2008). Epithelia formation requires coordination of cellular machinery that regulates cell adhesion to the ECM and other cells (Bryant and Mostov, 2008; Nelson, 2009). Focal adhesions (FAs) are integrin-based structures that bind to the ECM, and adherens junctions (AJs) are cadherin-based structures that regulate cell–cell adhesion (Niessen et al., 2011; Weber et al., 2011). These two well-studied adhesion complexes share several cellular links, including the actin cytoskeleton, Rho family GTPases, and other signaling proteins (Weber et al., 2011). However, remarkably little is known about how the transition between cell migration and intercellular adhesion is coordinated.
Rho family GTPases, comprising RhoA, Rac1, and Cdc42, play key roles at both FAs and AJs by regulating actin cytoskeleton dynamics, organization, and function (Tapon and Hall, 1997; McCormack et al., 2013). Rac1 has a central role in driving lamellipodia extension during cell migration (Côté and Vuori, 2007) and is transiently activated during initial cell–cell adhesion (Malliri et al., 2004; Yamada and Nelson, 2007; Kitt and Nelson, 2011). Rho family GTPases cycle between GTP- and GDP-bound states by the actions of guanine-nucleotide exchange factors (GEFs) and GTPase-activating proteins that spatially and temporally regulate GTPase activity. Much attention has been given to identifying GEFs that activate Rho GTPases at FAs and AJs. Tiam1, Tiam2, Trio, Asef, and ECT2 have Rac GEF activity and are implicated in the maintenance of cell–cell contacts (McCormack et al., 2013). However, the Rac GEF that regulates Rac activation during initial cell–cell contact formation (Yamada and Nelson, 2007) remains elusive. At FAs, an equally complex picture has emerged with the Rac GEFs α-Pix, β-Pix, Trio, Vav2, Tiam1, and Dock1 implicated in cell migration (Marignani and Carpenter, 2001; Medley et al., 2003; Rosenberger et al., 2003; Nayal et al., 2006; Côté and Vuori, 2007; O’Toole et al., 2011).
Recently, we reported a genome-wide screen of Drosophila melanogaster S2 cells for proteins required for cadherin-based cell–cell adhesion in suspension culture that was designed to exclude proteins involved in integrin-based cell spreading, adhesion, and migration. We identified Elmo2, a component of an Elmo–Dock complex (Toret et al., 2014). This was surprising because the Elmo–Dock complex has a well-established role downstream of integrins in cell–ECM spreading and migration pathways (Meller et al., 2005; Côté and Vuori, 2007). An Elmo–Dock complex consists of a scaffolding component (Elmo protein) and a Rac GEF catalytic component (Dock protein), both of which are required for full Rac GEF activity of the complex (Brugnera et al., 2002). At FAs, activated RhoG opens Elmo and activates Dock (Katoh and Negishi, 2003; Côté and Vuori, 2007; Patel et al., 2010).
That different Elmo–Dock complexes may be involved in integrin- and cadherin-based cell adhesion places the complex in a unique position to provide novel insight into how these different adhesion pathways might be regulated during cell–cell interactions. Here, we show that a specific Elmo–Dock complex is transiently recruited to early cell–cell contacts where it is required for the proper reorganization of E-cadherin, F-actin, and Rho GTPase activities and thereby initiates strong cell–cell adhesion.
Results and discussion
Knockdown of Dock1 and Elmo2 slows the formation of initial cadherin-mediated cell–cell adhesion
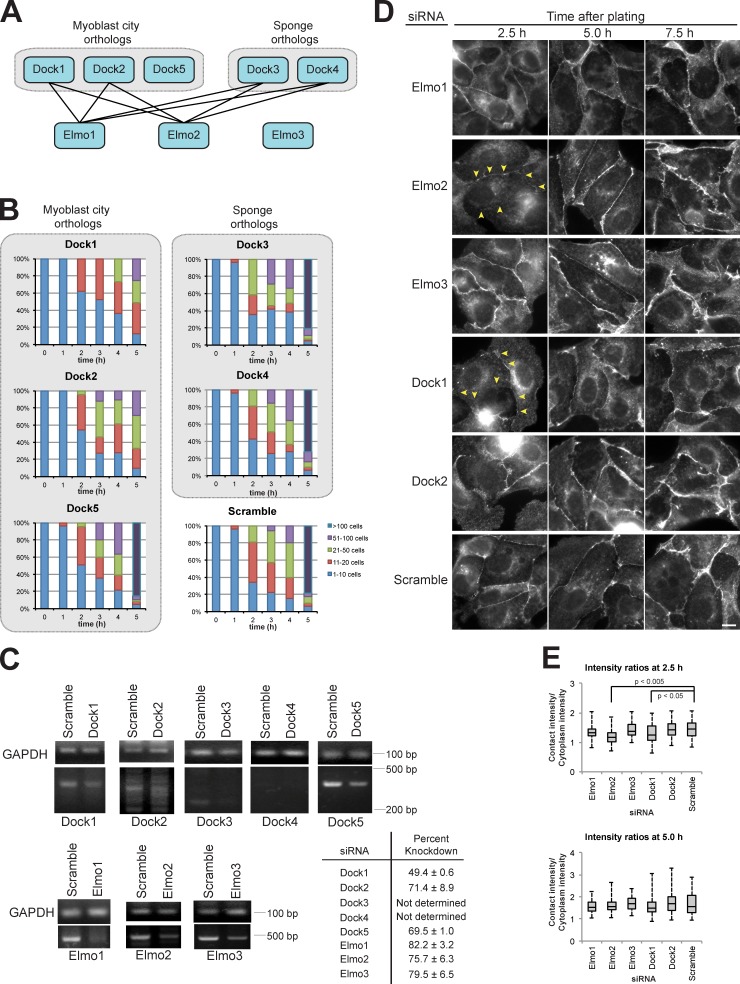
Elmo2 is essential for rapid cadherin-mediated cell–cell adhesion (Toret et al., 2014), but it is unclear whether this function requires a Dock protein. There are five Dock protein orthologues in mammals that bind to, or contain, N-terminal SH3 domains that could bind to Elmo2 (Fig. 1 A; Meller et al., 2005; Côté and Vuori, 2007). To test whether Dock proteins were required for Ca2+-dependent cell–cell adhesion, we measured cell–cell adhesion in MDCK cells treated with two independent siRNAs to each orthologue.
Figure 1.
A specific Elmo–Dock complex machinery is essential for cell–cell contact formation. (A) Schematic showing mammalian Dock and Elmo protein orthologues with known interactions (Meller et al., 2005; Côté and Vuori, 2007). (B) Quantification of hanging drop assays for the indicated siRNAs in which the cells were binned into cluster classes: 1–10, 11–20, 21–50, 51–100, or >100 cells (Toret et al., 2014). The percentage of cells in each category is shown for each time point. The data shown are from a representative experiment of three repeats in which ∼5 × 104 cells were analyzed for each time point. (C) RT-PCR analysis of transcript levels for the indicated genes of interest. Dashes indicate molecular mass standards. Percentage of knockdown for each siRNA was calculated by taking the mean from three experiments. (D) E-cadherin immunofluorescence for the indicated siRNA-treated cells at different times after cell plating. Yellow arrowheads indicate reduced E-cadherin staining at cell–cell contacts. Bar, 5 µm. (E) Box plot quantification of the ratio of E-cadherin fluorescence intensity at a region of cell–cell contact normalized to the intensity of an equal region of the cytoplasm underlying the contact (n = 67 for each condition). Whiskers show minimum and maximum values, horizontal lines show medians, and boxes show 1st and 3rd quartiles. P-values were determined by unpaired t test for the indicated samples.
We used a hanging drop assay in which MDCK cells in suspension form cadherin-dependent cell aggregates in a time-dependent manner in the absence of cell–substrate (ECM) adhesion and cell migration pathways (Benjamin et al., 2010); each condition was performed in more than three independent experiments. Knockdown of Dock1 expression by specific siRNAs resulted in the failure of cells to form large aggregates of >100 cells by 5 h compared with the scramble siRNA control in which large cell aggregates with strong cell–cell adhesion formed within 4–5 h (Fig. 1 B). Depletion of Dock2 expression had a similar, albeit weaker, cell–cell adhesion phenotype compared with Dock1 knockdown (Fig. 1 B). However, siRNA knockdown of Dock3, 4, or 5 had no effect on cell–cell adhesion in this assay; all formed large (>100 cell) aggregates within 5 h similar to the scramble siRNA control (Fig. 1 B). Note that we detected little to no expression of endogenous Dock3 and Dock4 in MDCK cells (Fig. 1 C), but the effect of siRNA knockdown was analyzed in case low levels of protein expression were required for cell–cell adhesion. To exclude off-target effects, cells were transfected with a second, independent siRNA for each gene, and hanging drop assays were performed. This analysis confirmed that cell–cell adhesion was strongly disrupted in cells depleted of Dock1 and mildly disrupted after Dock2 knockdown, and no effect was observed with Dock3–5 siRNAs (Fig. S1 A).
Knockdown of gene transcripts (RT-PCR) and protein (Western blot) levels were measured (Fig. 1 C and Fig. S1 C). Dock1 siRNA concentration was titrated to generate ∼50% knockdown of transcript levels (Fig. 1 C and Fig. S1 B) and protein expression (Fig. S1 C) because higher levels of Dock1 depletion resulted in cell lethality (unpublished data). Knockdown of Elmo1–3 and Dock 2 and 5 transcript and protein levels was 70–80% in three independent experiments (Fig. 1 C and Fig. S1, B and C).
We examined E-cadherin localization in MDCK cells treated with two independent siRNAs to different components of an Elmo–Dock complex (Fig. 1, D and E; and Fig. S1, D and E) to investigate the defect in cell–cell adhesion. Cells were plated on collagen-coated coverslips, allowed to form cell–cell contacts, and then fixed and processed for immunofluorescence microscopy at different times after plating and, hence, cell–cell contact formation. Depletion of either Dock1 or Elmo2 resulted in significantly reduced E-cadherin staining intensity at cell–cell contacts after 2.5 h (Fig. 1, D and E; and Fig. S1, D and E), compared with the scramble siRNA control. However, E-cadherin staining intensity at cell–cell contacts 5 and 7.5 h after induction of cell–cell adhesion was similar in cells treated with either Elmo2, Dock1, or scramble siRNAs (Fig. 1, D and E). Treatment with siRNAs specific for Elmo1 or Elmo3 had little or no effect on E-cadherin localization or staining intensity at any time during cell–cell adhesion in agreement with earlier observations (Toret et al., 2014). Knockdown of Dock2, which partially reduced the rate and amount of cell aggregation (Fig. 1 B and Fig. S1 A), had little or no quantifiable effect on E-cadherin staining at cell–cell contacts compared with knockdown of Dock1 (Fig. 1, C and D; and Fig. S1, D and E), and consequently, Dock2 was not examined further here. Collectively, these results indicate that Elmo2 and Dock1 have similar functions and may act in a complex in early cadherin-mediated cell–cell contacts.
Elmo2–Dock1 depletion perturbs E-cadherin accumulation and spreading at cell–cell contacts
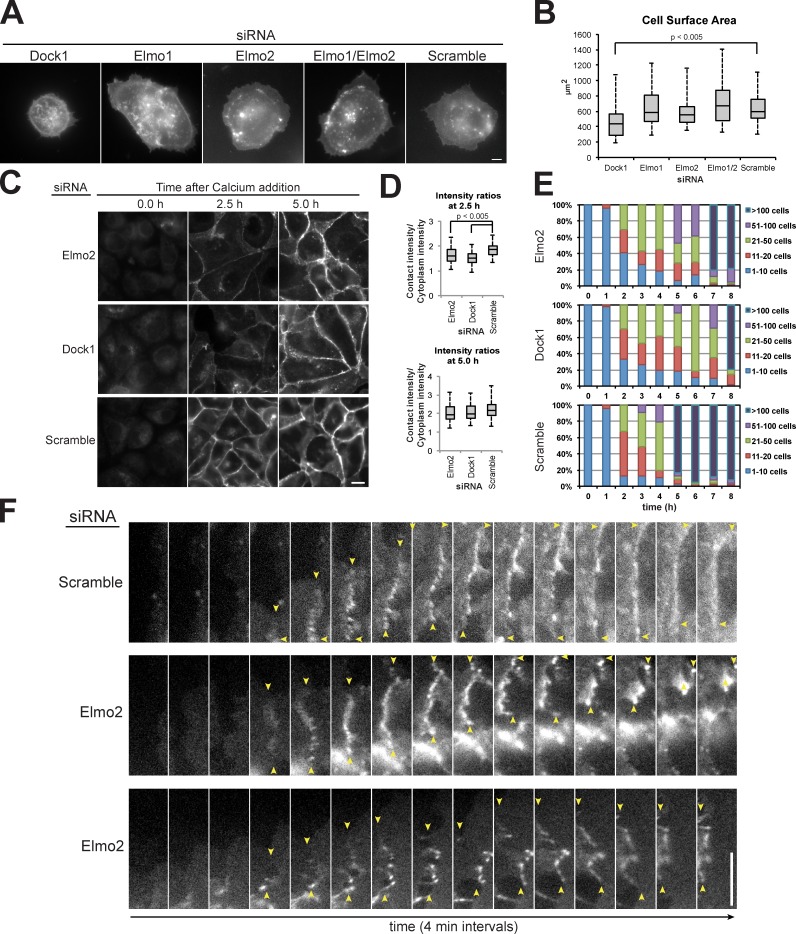
The Elmo–Dock complex has an established role in cell spreading and migration on the ECM (Côté and Vuori, 2007). Thus, we were concerned that the defect in E-cadherin accumulation at early cell–cell contacts in Dock1 and Elmo2 siRNA-treated cells (Fig. 1 C) might have been caused by reduced cell spreading and, hence, cell–cell collisions leading to AJ formation. We measured the area that MDCK cells spread on ECM after depletion of Elmo–Dock complex components (Fig. 2 A). siRNA knockdown of Dock1 resulted in cells that were defective in cell spreading, consistent with a previous study (Katoh and Negishi, 2003). siRNA knockdown of either Elmo2 or Elmo1 or a double knockdown of Elmo1 and Elmo2 had no measurable effect on cell spreading (Fig. 2, A and B). A previous study reported that expression of dominant-negative Elmo mutants reduced cell spreading (Katoh and Negishi, 2003). However, the effects of overexpression of Elmo mutants that antagonize Elmo–Dock complex function may not be comparable to siRNA knockdown of Elmo orthologues. Alternatively, depletion of all three Elmo orthologues may be necessary to achieve an ECM spreading defect in MDCK cells. Nevertheless, the defects in cadherin localization and cell–cell adhesion observed in the Elmo2 depletion were not linked to a defect in cell spreading.
Figure 2.
Dock1 and Elmo2 are essential for early E-cadherin recruitment to cell–cell contacts. (A) Basal focal plane of Lyn-GFP expression in cells treated with indicated siRNAs and fixed 90 min after plating on collagen. (B) Box plot quantification of Lyn-GFP fluorescence intensity for an equal surface area in indicated siRNA-treated cells (n = 40 for each condition). Whiskers show minimum and maximum values, horizontal lines show medians, and boxes show 1st and 3rd quartiles. P-values were determined by unpaired t test for the indicated samples. (C) E-cadherin immunofluorescence in indicated siRNA-treated cells at different times after calcium addition to induce cell–cell adhesion. (D) Box plot quantification of the ratio of E-cadherin fluorescence intensity at a region of cell–cell contact normalized to the intensity of an equal region of the cytoplasm underlying the contact (n = 100 for each condition). P-values were determined by unpaired t test for the indicated samples. (E) Quantification of hanging drop assays for the indicated siRNAs in which the cells were binned into cluster classes: 1–10, 11–20, 21–50, 51–100, or >100 cells. The percentage of cells in each category is shown for each time point. The data shown are from a representative experiment from three repeats in which ∼5 × 104 cells were analyzed for each time point. (F) Montages of individual frames from Video 1 of cells expressing E-cadherin–GFP treated with scramble or Elmo2 siRNA. Yellow arrows indicate the boundary of the cell–cell contact. Bars, 5 µm.
To circumvent any effect of cell spreading on cell–cell adhesion, we examined E-cadherin distribution in cells plated at ultra-high density in media containing 5 µM Ca2+ to prevent cadherin- but not integrin-mediated adhesions; this condition caused cells to pack tightly in the absence of cadherin contacts with minimal spreading on the substrate. Formation of cadherin-mediated cell–cell contacts was induced by the addition of 1.8 mM Ca2+ to the medium, and cells were fixed at 0, 2.5, and 5 h after Ca2+ addition and processed for E-cadherin immunofluorescence (Fig. 2, C and D). siRNA-mediated depletion of Dock1 or Elmo2 reduced the amount of E-cadherin recruited to contacts between closely opposed cells after 2.5 h. However, 5 h after induction of Ca2+-dependent cell–cell adhesion, the intensity of E-cadherin staining was high at cell–cell contacts in Dock1- and Elmo2-depleted cells and similar to that at cell–cell contacts in scramble siRNA control cells (Fig. 2, C and D). This ∼2.5-h delay in the accumulation of E-cadherin in cells depleted of either Elmo2 or Dock1 correlated with the delay in the formation of large cell aggregates (>100 cells) by those cells measured in the hanging drop assay; few large aggregates were formed initially compared with the control (5 h), but they formed normally by 7–8 h, an ∼2.5-h delay (Fig. 2 E). Thus, Elmo–Dock complex depletion slows initial E-cadherin recruitment and strengthening of cell–cell adhesion (<2.5 h), but it is not required subsequently to maintain E-cadherin at cell–cell contacts and strong cell–cell adhesion (>5 h).
Previous studies showed that E-cadherin engagement results in reduction of membrane lamellipodia that may enable the conversion of weak initial cell–cell adhesion to strong adhesion (Ehrlich et al., 2002; Yamada and Nelson, 2007; Xue et al., 2013). We speculated that delayed E-cadherin recruitment and cell–cell adhesion by Elmo–Dock complex depletion might be caused by abnormal plasma membrane dynamics that perturbed initial cell–cell adhesion. Because Dock1 depletion reduced cell spreading on a collagen substrate (Fig. 2, A and B), we could not examine cell–cell adhesion in isolation, and therefore, we focused on the effects of Elmo2 depletion during the initial moments of cadherin-mediated cell–cell contact formation. In the scramble siRNA control, E-cadherin first localized to puncta at initial cell–cell contacts and then spread along the plasma membrane as the contact expanded laterally (Fig. 2 F and Video 1), as described previously for MDCK cells (Adams et al., 1998). In cells depleted of Elmo2, E-cadherin localized in small puncta at initial cell–cell contacts, similar to the scramble siRNA control, but these contacts were unstable and collapsed concomitantly with abnormal dynamics and organization of the E-cadherin puncta (Fig. 2 F and Video 1). Unstable cell–cell contacts and abnormal E-cadherin dynamics were observed at 88% (n = 17) of newly forming cell–cell contacts in Elmo2-depleted cells but not in control cells (n = 14). These results indicate that expression of the Elmo2–Dock1 complex reduces lamellipodia dynamics at newly forming cell–cell contacts, thereby allowing the sequential formation of local E-cadherin puncta along the plasma membrane that stabilizes the lateral expansion of the contact between cells. Subsequently, maintenance of E-cadherin at cell–cell contacts and strong cell–cell adhesion become independent of the Elmo2–Dock1 complex.
The Elmo2–Dock1 complex localizes to early cell–cell contacts
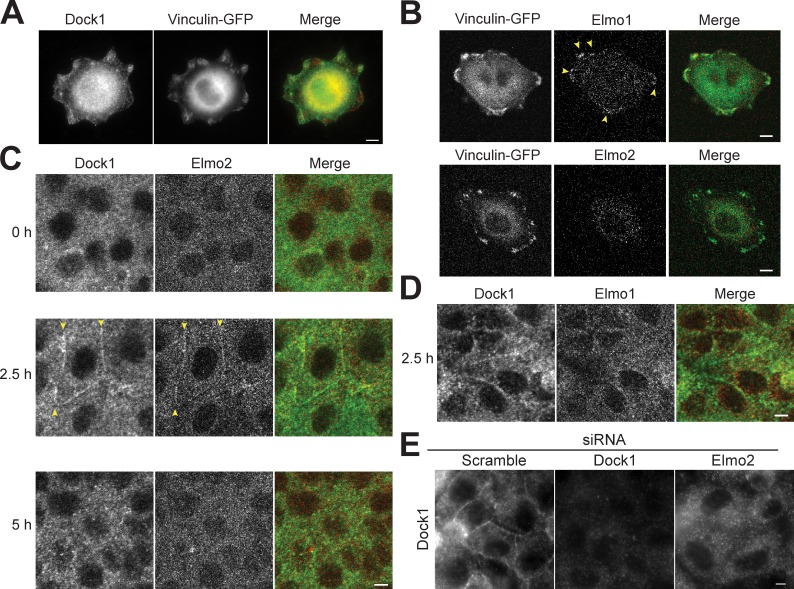
Because the Elmo2–Dock1 complex appears to play a critical role in the initial, but not later, phase of E-cadherin–mediated cell–cell adhesion, we sought to examine whether the complex is transiently localized to initial cell–cell contacts but not matured contacts. In single MDCK cells, Dock1 (Fig. 3 A) and Elmo1 (Fig. 3 B) localized to FAs marked by vinculin-GFP (Fig. 3, A and B), consistent with earlier observations (Brugnera et al., 2002). However, Elmo2 was diffuse in the cytoplasm and was not detected at FAs (Fig. 3 B). This is in contrast to a study of Elmo2 at FAs in MDA-MB-231 cells (Margaron et al., 2013) and may be caused by differences in cell types.
Figure 3.
Dock1 and Elmo2 localize to early cell–cell contacts. (A) Basal focal plane epifluorescence image of single MDCK cells expressing vinculin-GFP and stained with Dock1 antibodies. (B) Basal focal plane confocal image of single MDCK cells expressing vinculin-GFP and stained with Elmo1 and Elmo2 antibodies. (C) Confocal images of confluent MDCK monolayers stained with Dock1 and Elmo2 antibodies at different times after calcium addition and induction of cell–cell adhesion. (D) Confocal imaging of confluent MDCK monolayers stained with Dock1 and Elmo2 antibodies 2.5 h after calcium addition. (E) Epifluorescence images of confluent MDCK monolayer stained with Dock1 and Elmo1 antibodies 2.5 h after calcium addition in cells treated with the indicated siRNAs. Yellow arrowheads indicate the boundary of the cell–cell contact. Bars, 5 µm.
We next analyzed Elmo–Dock complex localization during initial cell–cell contact formation in confluent cell monolayers using the calcium switch assay (Fig. 2, C and D). At 0 h, before induction of cell–cell adhesion, both Dock1 and Elmo2 localized as puncta in the cytoplasm of confluent monolayers (Fig. 3 C). At 2.5 h, both Elmo2 and Dock1 staining appeared at initial cell–cell contacts (Fig. 3 C), coincident with E-cadherin accumulation at cell–cell contacts (Fig. 2 C). However, at 5 h after induction of cell–cell adhesion, both Elmo2 and Dock1 were not localized at cell–cell contacts (Fig. 3 C). Quantitation of Elmo2 and Dock1 colocalization showed that Elmo2 was detected at 97.8% (n = 45) of Dock1-positive cell–cell contacts. In contrast, Elmo1 was only detected at 14.4% (n = 41) of Dock1-positive cell–cell contacts (Fig. 3 D). Significantly, siRNA depletion of Elmo2 inhibited the recruitment of Dock1 to cell–cell contacts, although the cytoplasmic pool of Dock1 was unaffected (Fig. 3 E). Thus, Elmo2 may recruit Dock1 to cell–cell contacts, as suggested for the organization of an Elmo–Dock complex at FAs (Katoh and Negishi, 2003; Côté and Vuori, 2007). Collectively, these data indicate that Elmo2 and Dock1 transiently colocalize at initial E-cadherin–mediated cell–cell contacts in MDCK cells and that Elmo2 is required to recruit Dock1 to those sites.
The Elmo2–Dock1 complex regulates actin dynamics at cell–cell contacts
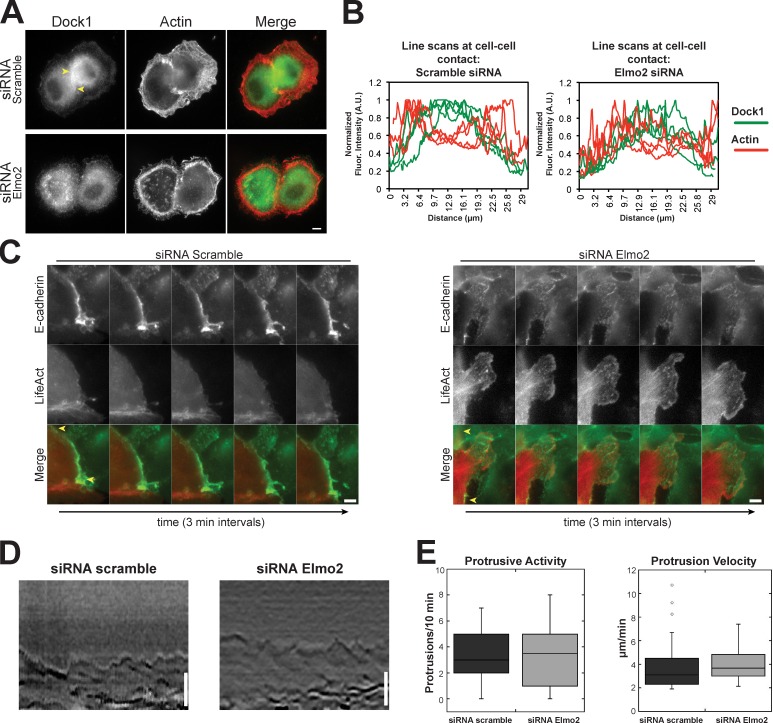
To further explore Elmo2–Dock1 complex function during cell–cell contact formation, we analyzed actin dynamics during cell–cell interactions in pairs of MDCK cells. Previous studies showed that the cortical actin bundle present in migratory cells is reorganized underneath the spreading cell–cell contact (Adams et al., 1998; Yamada and Nelson, 2007). In scramble siRNA-treated cells, Dock1 was enriched at the plasma membrane in the middle of the expanding cell–cell contact (Fig. 4 A, between arrowheads). This corresponds to the region in which the cortical bundle of actin had undergone dispersion and reorganization, whereas the cortical actin bundle appeared contiguous around the rest of the plasma membrane outside the area of cell–cell adhesion (Fig. 4, A and B). In cells that were depleted of Elmo2, Dock1 was not localized to the cell–cell contact, and the cortical actin bundle appeared to completely circumscribe the cell even in areas of close cell–cell apposition (Fig. 4, A and B). Thus, loss of Elmo2 and Dock1 localization at initial cell–cell contacts appears to result in a lack of reorganization of the cortical actin bundle at that site. Thus, the Elmo2–Dock1 complex may play a role in localized actin reorganization at initial cell–cell contacts.
Figure 4.
Dock1 and Elmo2 regulate actin dynamics at cell–cell contacts. (A) Cells treated with the indicated siRNAs were fixed 1 h after plating on collagen and imaged by epifluorescence for Dock1 and actin distributions. Yellow arrowheads indicate the boundary of the cell–cell contact. (B) Quantification of Dock1 and actin fluorescence intensities in a 1-pixel-wide line along four cell contacts. A.U., arbitrary unit. (C) Montage of images from a video of live cells expressing E-cadherin–GFP and LifeAct-RFP and treated with scramble or Elmo2 siRNAs. Yellow arrows indicate the boundary of the cell–cell contact. (D) Representative kymographs of membrane protrusions in cells treated scramble or Elmo2 siRNAs. 2-pixel-wide kymographs were compiled over 10 min. (E) Number of protrusions over a 10-min interval (left) and mean protrusion velocity (right) were measured in cells expressing scramble or Elmo2 siRNA. 10 cells with 33 protrusions (siRNA scramble) and 10 cells with 37 protrusions (siRNA Elmo2) were quantified. Results are presented in a box and whisker format, in which the ends of the box mark the upper and lower quartiles, the horizontal line in the box indicates the median, and the whiskers outside the box extend to the highest and lowest value within 1.5× the interquartile range. Outliers are indicated by dots. Bars, 5 µm.
We next investigated actin dynamics at cell–cell contacts in control cells and cells depleted of Elmo2 using live cells expressing LifeAct-RFP. In scramble siRNA control cells, the cortical actin bundle dissolved beneath the plasma membrane along the cell–cell contact (Fig. 4 C and Video 2), as expected in MDCK cells (see also Fig. 4 A in fixed cells; Adams et al., 1998; Yamada and Nelson, 2007). Dissolution of the cortical actin bundle and dampened membrane activity along cell–cell contacts were found in 91.1% of 45 cell–cell contacts among 15 imaged scramble siRNA control cells expressing LifeAct-RFP. In Elmo2 siRNA-treated cells, actin at cell–cell contacts was highly dynamic similar to that in lamellipodia-like structures (Fig. 4 C and Video 2); this organization of actin was very different to that in scramble siRNA control cells. In addition, actin cables of unknown origin protruded into the protrusion from deep in the cytoplasm. Highly dynamic membranes along contacts that had protruding actin cables was found in 70.5% of 44 cell–cell contacts among 15 imaged Elmo2-treated cells expressing LifeAct-RFP.
Actin dynamics in lamellipodia at the plasma membrane outside areas of cell–cell contacts appear similar in scramble and Elmo2 siRNA-treated cells; there was an actin-rich leading edge and a cortical bundle of actin in the lamella (Fig. S2 and Video 2). Kymograph analysis showed that the plasma membrane from both scramble siRNA control and Elmo2 siRNA-treated cells exhibited similar protrusive activity and velocity (Fig. 4, D and E). These results indicate that actin dynamics and lamellipodia activity at the plasma membrane outside the area of cell–cell contact were unperturbed in Elmo2 knockdown cells. Thus, the Elmo2–Dock1 complex appears to play a key role in actin dynamics and reorganization at newly formed cell–cell contacts and not at other sites on the plasma membrane. Abnormal actin and membrane dynamics in the absence of the Elmo2–Dock1 complex may perturb the formation of cell–cell adhesions.
The Elmo2–Dock1 complex regulates Rho family GTPase activity at initial cell–cell contacts
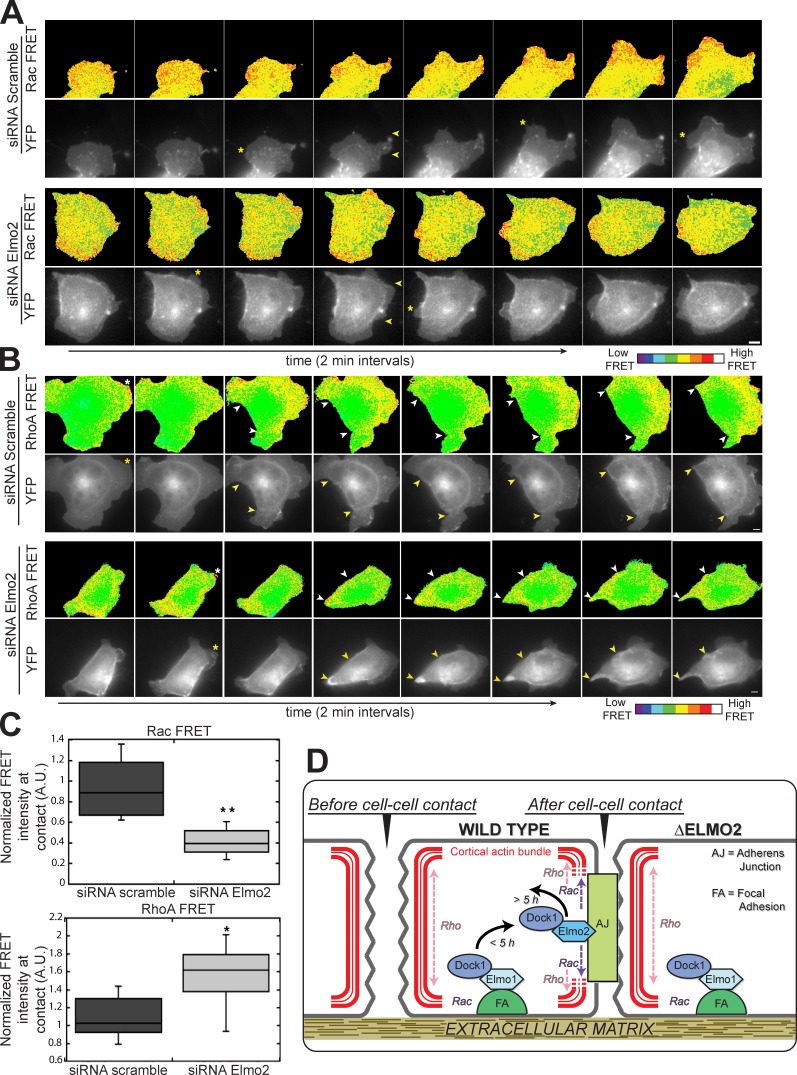
Given the importance of actin cytoskeleton rearrangements at initial cell–cell contacts (Adams et al., 1998; Zhang et al., 2005; Yamada and Nelson, 2007) and the known Rac GEF activity of the Elmo–Dock complex (Côté and Vuori, 2007), we next sought to test whether expression of Elmo2 and Dock1 was essential for Rac activity at cell–cell contacts. Cells were transfected with a Rac fluorescence resonance energy transfer (FRET) probe to localize Rac activity (Itoh et al., 2002; Yoshizaki et al., 2003; Yamada and Nelson, 2007). In scramble siRNA control cells, Rac activity was restricted to plasma membrane protrusions and at newly forming cell–cell contacts (Fig. 5 A), as reported previously (Yamada and Nelson, 2007); in these cells, 98% (n = 51) of lamellipodia and 80% (n = 10) of cell–cell contacts had increased Rac activity.
Figure 5.
An Elmo–Dock complex influences GTPase activity at cell–cell contacts. (A) Montage of images from movies of live-cells expressing the Rac FRET sensor and treated with scramble or Elmo2 siRNA. Asterisks indicate Rac-positive lamella, and yellow arrows indicate the boundary of the cell–cell contact. (B) Montage of images from videos of live cells expressing the RhoA FRET sensor and treated with scramble or Elmo2 siRNAs. Asterisks indicate RhoA-positive lamellipodia. Yellow and white arrowheads mark expanding cell–cell contacts in black/white and color images, respectively. (C) Mean Rac (top) and RhoA (bottom) FRET pixel intensity in scramble and Elmo2 siRNA-treated cells. Mean pixel intensity per unit area was quantified for 8–10 cell–cell contacts per condition. Results are presented in a box and whisker format, in which the ends of the box mark the upper and lower quartiles, the horizontal line in the box indicates the median, and the whiskers outside the box extend to the highest and lowest value within 1.5× the interquartile range: *, P < 0.01; **, P < 0.0002 (Mann–Whitney test). A.U., arbitrary unit. (D) Model of Elmo–Dock complex function during cadherin–integrin cross talk. Bars, 5 µm.
In Elmo2 siRNA-treated cells, the distribution of Rac activity was different from that in the scramble siRNA control cells: Rac activity was less focused at cell–cell contacts and appeared more ubiquitous throughout the cell and plasma membrane (Fig. 5 A). Although Rac FRET was less focused at cell–cell contacts, high activity was still observed in membrane protrusions, indicating that the FRET probe localized at sites of increased actin dynamics as expected (Fig. 5 A). In Elmo2 siRNA-treated cells, 84% (n = 32) of lamellipodia and 20% (n = 25) of cell–cell contacts had increased Rac FRET activity.
Although activation of Rac is localized to forming cell–cell contacts, RhoA activity appears to be absent from sites of cell–cell contacts and instead is concentrated at the edge of the expanding contact at sites of actomyosin contractility (Yamada and Nelson, 2007). RhoA activity in scramble siRNA control and Elmo2 siRNA-treated cells was localized using a RhoA FRET probe (Itoh et al., 2002; Yoshizaki et al., 2003; Yamada and Nelson, 2007). In control cells, 94% of lamellipodia (n = 31) were enriched for RhoA activity. Increased RhoA FRET activity was detected in 13% (n = 9) of cells forming cell–cell interactions, but the majority of the activity generally localized to the plasma membrane outside the area of the cell–cell contact (Fig. 5 B), as reported previously (Yamada and Nelson, 2007). In Elmo2-depleted cells, high RhoA FRET activity was observed in 88% (n = 33) of lamellipodia and, in contrast to control siRNA cells, 85% (n = 9) of newly forming cell–cell contacts (Fig. 5 B). The overall distribution of RhoA activity was also different from that in control scramble siRNA-treated cells: high RhoA activity was present around the entire periphery of the cell during cell–cell contact formation. Generally, Elmo2 siRNA-treated cells displayed lower Rac FRET activity and higher RhoA FRET activity at newly forming cell–cell contacts compared with control cells (Fig. 5 C). RhoA and Rac activity are often inversely correlated (Burridge and Wennerberg, 2004), and activation of Rac generally antagonizes RhoA activity (Sander et al., 1999; Nimnual et al., 2003). Although the mechanisms that drive this inverse activity relationship remain to be established, Elmo2 may regulate the activities of both Rac and Rho GTPases, directly or indirectly, during cell–cell contact formation (Fig. 5 D).
In summary, this study reveals a novel pathway involving an Elmo–Dock complex in cadherin-mediated cell–cell adhesion, in addition to the known role of the complex in integrin-based adhesion (Fig. 5 D; Meller et al., 2005; Côté and Vuori, 2007). In MDCK cells, the Elmo1–Dock1 complex appears to be the predominant form of the complex localized at integrin-based FAs. During cadherin-mediated cell–cell adhesion, Elmo2 localizes to cell–cell contacts and is required to recruit Dock1 to those sites, and presumably, together they act as a functional unit to locally regulate Rac activity. That the Elmo2–Dock1 complex is required for and transiently localizes during the formation of initial cell–cell adhesions suggests that it is the major Rac GEF that functions transiently (<2.5 h) during initial cell–cell contact (Fig. 5 D). Other Rac GEFs that have been demonstrated to act on cell–cell adhesions (McCormack et al., 2013) may function subsequently to complete and stabilize cell–cell adhesions. At present, it is unclear how Elmo2 is recruited to cell–cell contacts. RhoG, Arf6, and Arl4 have been implicated in the recruitment and activation of Elmo proteins at FAs (Katoh and Negishi, 2003; Santy et al., 2005; Patel et al., 2011). A similar mechanism may exist at cell–cell contacts. Perhaps different GTPases control the recruitment of Elmo1 versus Elmo2 to different sites on the plasma membrane (integrin adhesions vs. cadherin adhesions). Thus, different combinations of Elmo and Dock protein orthologues in complex may regulate cell transitions between a migratory state and a cell–cell adhesion state by localized control of Rho GTPases at those sites (Fig. 5 D). Further work is needed to understand these mechanisms.
Materials and methods
Cell culture
MDCK G type II cells were grown in DMEM with 1 g/liter sodium bicarbonate, 10% fetal bovine serum (Atlas Biologicals), penicillin, streptomycin, and kanamycin.
siRNA and plasmids
For siRNA treatment, two rounds of 10 µg siRNA (one round of 5 µg for Dock1) were transfected (Lipofectamine 2000; Invitrogen) for 18-h periods, and cells were analyzed after a further 24-h recovery. Dock and Elmo (Toret et al., 2014) protein orthologue siRNA oligonucleotides were designed by Thermo Fisher Scientific (DOCK1 #1, 5′-CUUUAGAGCUCAUGAAAUA-3′; DOCK1 #2, 5′-CAGCAAACAUCAAGAGAUA-3′; DOCK2 #1, 5′-GAAGAAAUAUCGAGAACAU-3′; DOCK2 #2, 5′-CAUCCAAGGUUCAAGAAUA-3′; DOCK3 #1, 5′-GGAUGAUAAUACAGAGAAA-3′; DOCK3 #2, 5′-CCACGGAGGAGGAGAAAUA-3′; DOCK4 #1, 5′-CGGGAAACAUGGAGGGAAA-3′; DOCK4 #2, 5′-GCAUAAGUGUGAAGAGAAU-3′; DOCK5 #1, 5′-CCAAGAUAGUGGAGAGCAA-3′; DOCK5 #2, 5′-GGGCAGAGGAGAUGAACAA-3′; ELMO1 #1, 5′-AACAAGACCUGGAAGGAAA-3′; ELMO1 #2, 5′-GGAAGGAUAUGAUGAGUGA-3′; ELMO2 #1, 5′-AAGAAAGGAUGAUGACCAA-3′; ELMO2 #2, 5′-GAGCAGACGCGCAGUGAUA-3′; ELMO3#1, 5′-GGAGAAGGGCUCAGGGAAA-3′; and ELMO3 #2, 5′-GCAUCCAGCUGUUGAAUAA-3′). Raichu FRET probes for Rac1 (1,026×; a sequential fusion of four proteins), YFP-CRIB (Cdc42/Rac interactive binding) domain [p21-activated kinase]-Rac1-CFP, expressed from a cytomegalovirus [CMV] promoter in a pCAGGS backbone), and RhoA (1,298×; a sequential fusion of four proteins, YFP-RBD domain[PKN]-RhoA-CFP, expressed from a CMV promoter in a pCAGGS backbone) were variants of published probes that contained Venus instead of YFP (http://www.fret.lif.kyoto-u.ac.jp/e-phogemon/vector.htm) and were gifts from M. Matsuda (Kyoto University, Kyoto, Japan). The LifeAct-RFP plasmid (first 17 aa of yeast Abp140p fused to mRFPruby expressed under the CMV promoter in a pEGFP-N1 backbone) was previously described (Riedl et al., 2008).
RT-PCR
Total RNA was extracted from siRNA-treated cells using the RNeasy kit (QIAGEN) according to the manufacturer’s instructions. 1 µg of RNA was used to generate cDNA using cDNA synthesis kit (iScript; Bio-Rad Laboratories). PCR was performed using specific primers to measure the levels of each gene of interest. The following primer sequences were used in these studies: Dock1 forward (F), 5′-ATTCGACACAAGCCGCTGAA-3′; Dock1 reverse (R), 5′-AAATCACATGCTCCAGCCGT-3′; Dock2 F, 5′-GTACAAGCTGGGCCAGAACA-3′; Dock2 R, 5′-TCTGCAGCACACTCCATCAG-3′; Dock3 F, 5′-TCTGTTCTGTCCTCGTCCCA-3′; Dock3 R, 5′-TTGGCAGCTGTCCATTCTCC-3′; Dock4 F, 5′-GCCAGAGCTTCCCCTTTGTT-3′; Dock4 R, 5′-CCTTCAACGGAGACCTCCCA-3′; Dock5 F, 5′-GGCTTCGTCAGGTGCTAGAG-3′; Dock5 R, 5′-AGGAGCAATCTCGGTGGAGT-3′; Elmo1 F, 5′-AGTGGCACCGAACGATACCAGA-3′; Elmo1 R, 5′-GCACGTACAGCTGGTGTGCCAT-3′; Elmo2 F, 5′-TGGGAACCGCCGAAGGCAAG-3′; Elmo2 R, 5′-AGCTGCTGGGCTCCTTGGGT-3′; Elmo3 F, 5′-CGCACTCTGGCCCTGAAGCC-3′; and Elmo3 R, 5′-CAGCCGAGTCTGCTCGCTGC-3′.
Immunofluorescence and Western blotting
MDCK cells, plated on collagen-coated cover glass, were fixed in 100% methanol (−20°C) or 3.8% paraformaldehyde. Antibodies used in immunofluorescence experiments were E-cadherin/mouse (Decma [Sigma-Aldrich]; rr1 [Developmental Studies Hybridoma Bank]; Ozawa et al., 1989), Elmo2/goat (ab2240; Abcam), Elmo1/goat (ab2239; Abcam), Dock1/rabbit (ab97325; Abcam), actin/mouse (MAB1501R; EMD Millipore), and GAPDH/mouse (ab8245; Abcam). For Western blotting, all whole cell lysates collected from dishes were washed once with PBS (4°C), and then, cells were scraped in 4× Laemmli buffer (4°C). Quantification of blots was performed using ImageJ (National Institutes of Health).
Hanging drop assay
The assay was performed as previously described (Toret et al., 2014). MDCK cells were plated at low density, and cells were trypsinized, centrifuged, and resuspended at a density of 2.5 × 105 cells/ml. 20-µl drops of the cell suspension were placed on 35-mm culture dish lids, which were inverted on top of the dish that contained medium, and incubated for different times at 37°C. At each time point, drops were triturated 10 times through a 20-µl pipette, and 4 µl of 16% PFA was added. The entire sample was mounted on a slide, observed at 10× magnification, and scored by eye.
Imaging and FRET analysis
All live-cell imaging was performed at 37°C in DMEM (without phenol red) with 1 g/liter sodium bicarbonate, 25 mM Hepes, 10% fetal bovine serum, penicillin, streptomycin, and kanamycin. Epifluorescence imaged used a 63×/1.4 NA Plan Apochromat lens (Carl Zeiss) or 100×/1.4 NA Plan Apochromat lens (Carl Zeiss), and images were captured with AxioVision LE64 software (Carl Zeiss) and a camera (AxioCam MRm; Carl Zeiss). Time-lapse imagining of GFP, RFP, CFP, and YFP signals used Plan Apochromat 63×/1.4 NA or 100×/1.4 NA objectives, and images were captured with SlideBook software (Intelligent Imaging Innovations) and a camera (CoolSNAP HQ; Roper Scientific). Confocal imaging used a 63×/1.4 NA Plan Apochromat lens (Carl Zeiss) and a laser-scanning confocal system (LSM 510 Meta; Carl Zeiss). All images were analyzed with ImageJ. FRET analysis was performed using PixFRET (Feige et al., 2005) with a bleed-though correction of 0 (Hodgson et al., 2010). Rac and RhoA FRETs at cell–cell contacts were measured by calculating the mean pixel intensity within a small region along the newly formed contact. Values were normalized by dividing by the area measured.
Lamellipodia kymograph analysis
MDCK cells treated with scramble or Elmo2 siRNAs were plated on collagen-coated 35-mm glass-bottom dishes (MatTek Corporation) 24 h after transfection. Differential interference contrast images were captured every 10 s over a 10-min time course (see previous paragraph for live cell imaging). Kymographs were generated by generating a time-lapse montage of a single 2-pixel-wide frame rectangle (perpendicular to the cell edge) for each frame of the video (ImageJ). Protrusion activity was defined as the number of peaks extending >0.5 µm and persisting for >30 s formed in 10 min. Protrusion velocity was defined as the rate of membrane extension (mean slope of peaks).
Online supplemental material
Fig. S1 shows results of hanging drop assays, RT-PCRs, and E-cadherin localizations for alternate Dock and Elmo siRNA constructs. Fig. S2 is a montage representation of lamellipodia shown in Video 2. Video 1 shows E-cadherin dynamics at cell–cell contacts in control and Elmo2-depleted cells. Video 2 shows actin dynamics at cell–cell contacts in control and Elmo2-depleted cells. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201406135/DC1.
Supplementary Material
Acknowledgments
C.P. Toret and C. Collins were supported by National Institutes of Health, National Research Service Award (T32 CA09151) from the National Cancer Institute. This work was supported by a grant from the National Institutes of Health (GM35527) to W.J. Nelson.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- AJ
- adherens junction
- CMV
- cytomegalovirus
- FA
- focal adhesion
- FRET
- fluorescence resonance energy transfer
- GEF
- guanine-nucleotide exchange factor
References
- Adams C.L., Chen Y.T., Smith S.J., and Nelson W.J.. 1998. Mechanisms of epithelial cell–cell adhesion and cell compaction revealed by high-resolution tracking of E-cadherin–green fluorescent protein. J. Cell Biol. 142:1105–1119 10.1083/jcb.142.4.1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin J.M., Kwiatkowski A.V., Yang C., Korobova F., Pokutta S., Svitkina T., Weis W.I., and Nelson W.J.. 2010. αE-catenin regulates actin dynamics independently of cadherin-mediated cell–cell adhesion. J. Cell Biol. 189:339–352 10.1083/jcb.200910041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brugnera E., Haney L., Grimsley C., Lu M., Walk S.F., Tosello-Trampont A.-C., Macara I.G., Madhani H., Fink G.R., and Ravichandran K.S.. 2002. Unconventional Rac-GEF activity is mediated through the Dock180-ELMO complex. Nat. Cell Biol. 4:574–582. [DOI] [PubMed] [Google Scholar]
- Bryant D.M., and Mostov K.E.. 2008. From cells to organs: building polarized tissue. Nat. Rev. Mol. Cell Biol. 9:887–901 10.1038/nrm2523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burridge K., and Wennerberg K.. 2004. Rho and Rac take center stage. Cell. 116:167–179 10.1016/S0092-8674(04)00003-0 [DOI] [PubMed] [Google Scholar]
- Côté J.-F., and Vuori K.. 2007. GEF what? Dock180 and related proteins help Rac to polarize cells in new ways. Trends Cell Biol. 17:383–393 10.1016/j.tcb.2007.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich J.S., Hansen M.D.H., and Nelson W.J.. 2002. Spatio-temporal regulation of Rac1 localization and lamellipodia dynamics during epithelial cell-cell adhesion. Dev. Cell. 3:259–270 10.1016/S1534-5807(02)00216-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feige J.N., Sage D., Wahli W., Desvergne B., and Gelman L.. 2005. PixFRET, an ImageJ plug-in for FRET calculation that can accommodate variations in spectral bleed-throughs. Microsc. Res. Tech. 68:51–58 10.1002/jemt.20215 [DOI] [PubMed] [Google Scholar]
- Hodgson L., Shen F., and Hahn K.. 2010. Biosensors for characterizing the dynamics of Rho family GTPases in living cells. Curr. Protoc. Cell Biol. 46:14.11.1–14.11.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh R.E., Kurokawa K., Ohba Y., Yoshizaki H., Mochizuki N., and Matsuda M.. 2002. Activation of rac and cdc42 video imaged by fluorescent resonance energy transfer-based single-molecule probes in the membrane of living cells. Mol. Cell. Biol. 22:6582–6591 10.1128/MCB.22.18.6582-6591.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh H., and Negishi M.. 2003. RhoG activates Rac1 by direct interaction with the Dock180-binding protein Elmo. Nature. 424:461–464 10.1038/nature01817 [DOI] [PubMed] [Google Scholar]
- Kitt K.N., and Nelson W.J.. 2011. Rapid suppression of activated Rac1 by cadherins and nectins during de novo cell-cell adhesion. PLoS ONE. 6:e17841 10.1371/journal.pone.0017841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malliri A., van Es S., Huveneers S., and Collard J.G.. 2004. The Rac exchange factor Tiam1 is required for the establishment and maintenance of cadherin-based adhesions. J. Biol. Chem. 279:30092–30098 10.1074/jbc.M401192200 [DOI] [PubMed] [Google Scholar]
- Margaron Y., Fradet N., and Côté J.-F.. 2013. ELMO recruits actin cross-linking family 7 (ACF7) at the cell membrane for microtubule capture and stabilization of cellular protrusions. J. Biol. Chem. 288:1184–1199 10.1074/jbc.M112.431825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marignani P.A., and Carpenter C.L.. 2001. Vav2 is required for cell spreading. J. Cell Biol. 154:177–186 10.1083/jcb.200103134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormack J., Welsh N.J., and Braga V.M.M.. 2013. Cycling around cell-cell adhesion with Rho GTPase regulators. J. Cell Sci. 126:379–391 10.1242/jcs.097923 [DOI] [PubMed] [Google Scholar]
- Medley Q.G., Buchbinder E.G., Tachibana K., Ngo H., Serra-Pagès C., and Streuli M.. 2003. Signaling between focal adhesion kinase and trio. J. Biol. Chem. 278:13265–13270 10.1074/jbc.M300277200 [DOI] [PubMed] [Google Scholar]
- Meller N., Merlot S., and Guda C.. 2005. CZH proteins: a new family of Rho-GEFs. J. Cell Sci. 118:4937–4946 10.1242/jcs.02671 [DOI] [PubMed] [Google Scholar]
- Nayal A., Webb D.J., Brown C.M., Schaefer E.M., Vicente-Manzanares M., and Horwitz A.R.. 2006. Paxillin phosphorylation at Ser273 localizes a GIT1–PIX–PAK complex and regulates adhesion and protrusion dynamics. J. Cell Biol. 173:587–589 10.1083/jcb.200509075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson W.J.2009. Remodeling epithelial cell organization: transitions between front-rear and apical-basal polarity. Cold Spring Harb. Perspect. Biol. 1:a000513 10.1101/cshperspect.a000513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niessen C.M.C., Leckband D., and Yap A.S.A.. 2011. Tissue organization by cadherin adhesion molecules: dynamic molecular and cellular mechanisms of morphogenetic regulation. Physiol. Rev. 91:691–731 10.1152/physrev.00004.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimnual A.S., Taylor L.J., and Bar-Sagi D.. 2003. Redox-dependent downregulation of Rho by Rac. Nat. Cell Biol. 5:236–241 10.1038/ncb938 [DOI] [PubMed] [Google Scholar]
- O’Toole T.E., Bialkowska K., Li X., and Fox J.E.B.. 2011. Tiam1 is recruited to β1-integrin complexes by 14-3-3ζ where it mediates integrin-induced Rac1 activation and motility. J. Cell. Physiol. 226:2965–2978 10.1002/jcp.22644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozawa M., Baribault H., and Kemler R.. 1989. The cytoplasmic domain of the cell adhesion molecule uvomorulin associates with three independent proteins structurally related in different species. EMBO J. 8:1711–1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel M., Margaron Y., Fradet N., Yang Q., Wilkes B., Bouvier M., Hofmann K., and Côté J.-F.. 2010. An evolutionarily conserved autoinhibitory molecular switch in ELMO proteins regulates Rac signaling. Curr. Biol. 20:2021–2027 10.1016/j.cub.2010.10.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel M., Chiang T.-C., Tran V., Lee F.-J.S., and Côté J.-F.. 2011. The Arf family GTPase Arl4A complexes with ELMO proteins to promote actin cytoskeleton remodeling and reveals a versatile Ras-binding domain in the ELMO proteins family. J. Biol. Chem. 286:38969–38979 10.1074/jbc.M111.274191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedl J., Crevenna A.H., Kessenbrock K., Yu J.H., Neukirchen D., Bista M., Bradke F., Jenne D., Holak T.A., Werb Z., et al. 2008. Lifeact: a versatile marker to visualize F-actin. Nat. Methods. 5:605–607 10.1038/nmeth.1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberger G., Jantke I., Gal A., and Kutsche K.. 2003. Interaction of αPIX (ARHGEF6) with β-parvin (PARVB) suggests an involvement of αPIX in integrin-mediated signaling. Hum. Mol. Genet. 12:155–167 10.1093/hmg/ddg019 [DOI] [PubMed] [Google Scholar]
- Sander E.E., ten Klooster J.P., van Delft S., van der Kammen R.A., and Collard J.G.. 1999. Rac downregulates Rho activity: reciprocal balance between both GTPases determines cellular morphology and migratory behavior. J. Cell Biol. 147:1009–1022 10.1083/jcb.147.5.1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santy L.C., Ravichandran K.S., and Casanova J.E.. 2005. The DOCK180/Elmo complex couples ARNO-mediated Arf6 activation to the downstream activation of Rac1. Curr. Biol. 15:1749–1754 10.1016/j.cub.2005.08.052 [DOI] [PubMed] [Google Scholar]
- Tapon N., and Hall A.. 1997. Rho, Rac and Cdc42 GTPases regulate the organization of the actin cytoskeleton. Curr. Opin. Cell Biol. 9:86–92 10.1016/S0955-0674(97)80156-1 [DOI] [PubMed] [Google Scholar]
- Toret C.P., D’Ambrosio M.V., Vale R.D., Simon M.A., and Nelson W.J.. 2014. A genome-wide screen identifies conserved protein hubs required for cadherin-mediated cell–cell adhesion. J. Cell Biol. 204:265–279 10.1083/jcb.201306082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber G.F., Bjerke M.A., and DeSimone D.W.. 2011. Integrins and cadherins join forces to form adhesive networks. J. Cell Sci. 124:1183–1193 10.1242/jcs.064618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue B., Krishnamurthy K., Allred D.C., and Muthuswamy S.K.. 2013. Loss of Par3 promotes breast cancer metastasis by compromising cell-cell cohesion. Nat. Cell Biol. 15:189–200 10.1038/ncb2663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada S., and Nelson W.J.. 2007. Localized zones of Rho and Rac activities drive initiation and expansion of epithelial cell–cell adhesion. J. Cell Biol. 178:517–527 10.1083/jcb.200701058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizaki H., Ohba Y., Kurokawa K., Itoh R.E., Nakamura T., Mochizuki N., Nagashima K., and Matsuda M.. 2003. Activity of Rho-family GTPases during cell division as visualized with FRET-based probes. J. Cell Biol. 162:223–232 10.1083/jcb.200212049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Betson M., Erasmus J., Zeikos K., Bailly M., Cramer L.P., and Braga V.M.M.. 2005. Actin at cell-cell junctions is composed of two dynamic and functional populations. J. Cell Sci. 118:5549–5562 10.1242/jcs.02639 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.