A holin (ChiW) and an endopeptidase (ChiX) operate in tandem as components of a protein secretion system used by the gram-negative bacterium Serratia marcescens to secrete the chitinolytic machinery.
Abstract
Pathogenic bacteria adapt to their environment and manipulate the biochemistry of hosts by secretion of effector molecules. Serratia marcescens is an opportunistic pathogen associated with healthcare-acquired infections and is a prolific secretor of proteins, including three chitinases (ChiA, ChiB, and ChiC) and a chitin binding protein (Cbp21). In this work, genetic, biochemical, and proteomic approaches identified genes that were required for secretion of all three chitinases and Cbp21. A genetic screen identified a holin-like protein (ChiW) and a putative l-alanyl-d-glutamate endopeptidase (ChiX), and subsequent biochemical analyses established that both were required for nonlytic secretion of the entire chitinolytic machinery, with chitinase secretion being blocked at a late stage in the mutants. In addition, live-cell imaging experiments demonstrated bimodal and coordinated expression of chiX and chiA and revealed that cells expressing chiA remained viable. It is proposed that ChiW and ChiX operate in tandem as components of a protein secretion system used by gram-negative bacteria.
Introduction
Targeting of proteins to their sites of physiological function is an essential process in all cellular systems. Secretion of specific proteins completely out of the cell is a key process that bacteria use to adapt to and modulate their immediate environment (Trost et al., 2005). Six protein targeting and secretion systems have been identified in gram-negative bacteria, named type 1–type 6 (Economou et al., 2006). In general, protein secretion can occur either in a single step, in which substrates are secreted directly from cytoplasm to the cell exterior, or by a two-step process, in which substrates are first targeted to the periplasm and are then translocated across the outer membrane in a secondary transport event.
Serratia marcescens is an opportunistic pathogen thought to be a significant cause of healthcare-acquired infections (Mahlen, 2011). This bacterium is a prolific secretor of proteins, many of which include virulence factors such as phospholipases, proteases, and nucleases (Hines et al., 1988). In addition, S. marcescens secretes three extracellular chitinases (ChiA, ChiB, and ChiC) and one extracellular chitin binding protein (Cbp21). This chitinolytic machinery allows S. marcescens to use chitin as a carbon and nitrogen source for growth (Hejazi and Falkiner, 1997). Moreover, chitinases from bacterial pathogens, together with host chitinase-like proteins, are thought to have increasingly important roles in infection (Tran et al., 2011; Frederiksen et al., 2013), including in the inflammation process (Lee et al., 2011), where the bacterial chitinases and chitin binding proteins are believed to facilitate the initial attachment of bacteria onto host cells (Tran et al., 2011; Frederiksen et al., 2013).
The mechanism of secretion of the components of the S. marcescens chitinolytic machinery has not been reported. The S. marcescens ChiA and Cbp21 proteins are both predicted to be synthesized as precursors with N-terminal Sec signal peptides (Brurberg et al., 1996; Vaaje-Kolstad et al., 2005a), and as a result, it may have been assumed that they were externalized by a type 2 secretion system, in a similar fashion to the chitinase from Escherichia coli (Francetic et al., 2000). Interestingly, the primary sequences of the other chitinases (ChiB and ChiC) contain no canonical signal peptides or other obvious targeting signals (Brurberg et al., 1995; Suzuki et al., 1999), suggesting that these proteins may be secreted by an alternative route. Indeed, the homologous ChiC protein from Pseudomonas aeruginosa was previously implied to be secreted by an unknown mechanism, distinct from the then-characterized systems in that bacterium (Folders et al., 2001).
In this work, the molecular basis of chitinase secretion by S. marcescens was studied. A genetic screen initially identified a gene predicted to encode a holin-like integral membrane protein that was shown to be required for secretion of the entire chitinolytic machinery. The holin-like protein is encoded in an apparent four-cistron operon—chiWXYZ—that shares similarity with bacteriophage lysis cassettes. The chiX gene encodes a putative l-alanyl-d-glutamate endopeptidase that is also essential for chitinase secretion. The phenotype of a ΔchiW strain was rescued by expression of a modified version ChiX routed to the periplasm via the twin-arginine translocation (Tat) pathway, suggesting that ChiX normally operates in the periplasm and is targeted there by ChiW. Live-cell imaging experiments revealed bimodal and coordinated expression for chiX and chiA and demonstrated that cells expressing chitinase remained viable. Together, chiW and chiX form part of a genetic locus dedicated to chitin metabolism, and it is proposed that they represent components of a protein secretion system used by gram-negative bacteria.
Results
Chitinases are secreted proteins
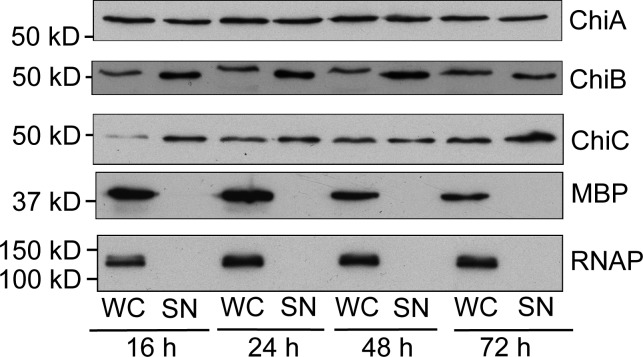
Initially, the subcellular localization of the S. marcescens chitinases was clarified (Fig. 1). S. marcescens was cultured aerobically in liquid medium before whole cells and cell-free supernatants were analyzed for the presence of the native chitinases by Western immunoblotting (Fig. 1). ChiA, ChiB, and ChiC were clearly detectable in the culture supernatant over the duration of the experiment (Fig. 1). In contrast, neither the cytoplasmic control protein RNA polymerase nor the normally periplasmic maltose binding protein could be detected in the extracellular milieu, even after an extended 72 h of growth (Fig. 1). It must be concluded that all three chitinases are deliberately secreted from the bacterial cell.
Figure 1.
S. marcescens chitinases are extracellular proteins. S. marcescens Db10 was grown aerobically in rich medium. At the time points indicated, an aliquot was taken and separated into culture supernatant (SN) and whole cells (WC) by centrifugation. Samples were analyzed for the presence of ChiA, ChiB, and ChiC by Western immunoblotting. The locations of a periplasmic control protein (maltose binding protein [MBP]) and a cytoplasmic control protein (RNA polymerase [RNAP]) are shown.
Identification of a locus required for ChiC secretion
Arguably the most well-studied secretion system is the general secretory pathway or type 2 secretion system (Korotkov et al., 2012). Inspection of the S. marcescens Db10/Db11 genome sequence (Iguchi et al., 2014) did not reveal any gene products that would encode typical components of a type 2 secretion system. Furthermore, chitinases ChiB and ChiC display no obvious targeting Sec-type sequences. Thus, to select for mutant strains defective in chitinase secretion, a genetic screen was designed. The initial focus was placed on ChiC and an S. marcescens mutant, JchiC (ΔchiA and ΔchiB), was constructed that would produce only ChiC. Random transposon mutagenesis was used to screen the JchiC (ΔchiA and ΔchiB) strain for mutants that could no longer degrade extracellular chitin. Eight transposon mutant strains devoid of chitinase activity were isolated (Fig. S1). To differentiate mutants defective in chiC expression from those defective in ChiC secretion, the transposon mutants were analyzed for the presence of ChiC in cellular and secreted/supernatant fractions by Western immunoblotting (Fig. S1). This approach revealed that three of the original transposon insertion mutants retained the ability to produce ChiC to normal levels but were apparently completely blocked in their ability to secret the enzyme into the culture supernatant (Fig. S1).
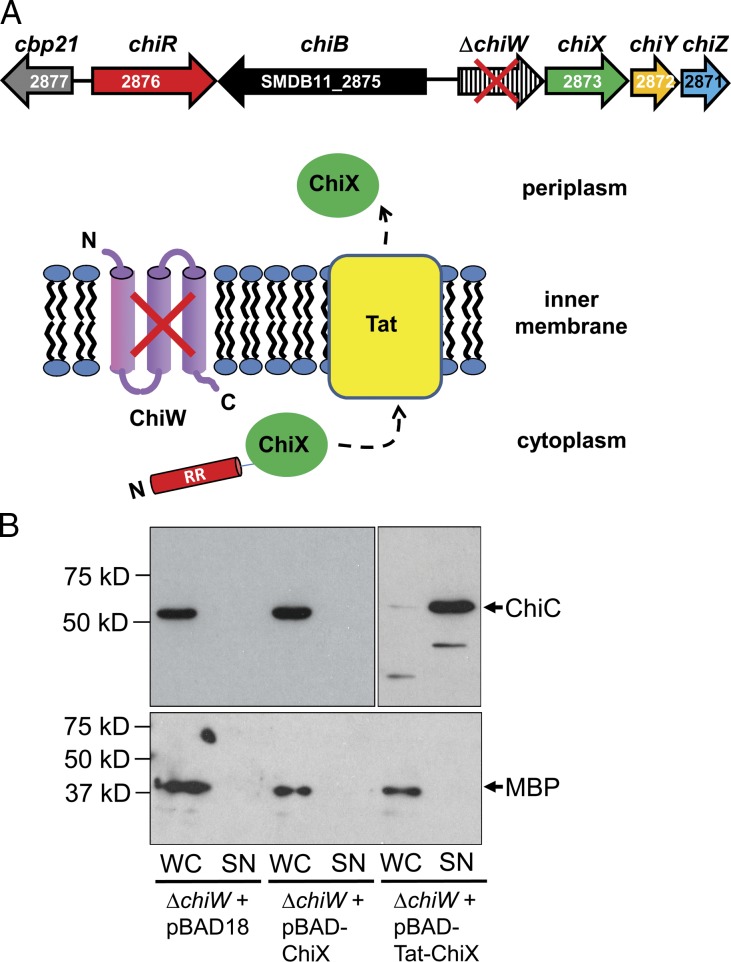
The locations of the transposon insertions in the three secretion-defective mutant strains were mapped with reference to the genome sequence of S. marcescens Db11 (Iguchi et al., 2014), which is a spontaneous streptomycin-resistant derivative of the Db10 strain used in this study. Two of the transposon insertions were mapped to different locations near the same gene (SMDB11_2874) and the third was located near SMDB11_2876, which had been previously identified as chiR (Fig. 2 A; Suzuki et al., 2001). The chiR gene encodes a transcriptional regulator of the LysR family thought to be important for expression of chitinase activity (Suzuki et al., 2001); however, SMDB11_2874 had not been previously studied. Further inspection of the S. marcescens Db11 genome sequence revealed that chiR and SMDB11_2874 are located adjacent to chiB on the S. marcescens chromosome (Fig. 2 A). The SMDB11_2874 gene is therefore part of a genetic locus dedicated to chitin metabolism and was named chiW (Fig. 2). The chiW gene encodes a predicted inner membrane protein of the phage holin-3 family. The archetypal member of this protein family is the bacteriophage λ holin S (Gründling et al., 2000). The chiW gene is located in an apparent four-cistron operon with a gene encoding a predicted metal-dependent l-alanyl-d-glutamate endopeptidase and two genes encoding homologues of bacteriophage spanins (Fig. 2), similar to a λ lysis cassette (Summer et al., 2007), and the operon was named chiWXYZ (Fig. 2).
Figure 2.
The genetics of chitinase secretion in S. marcescens. (A) A cartoon representing the organization of the S. marcescens Db10/11 chromosome around region 3,031,956–3,036,024 bp. The red arrows indicate the positions of three transposon insertion sites found to disrupt ChiC secretion. One was located 129 bp upstream of chiR (SMDB11_2876), one was 23 bp upstream of chiW (SMDB11_2874), and one was at position 165 within chiW. (B) Cartoon depicting the predicted subcellular localizations and topologies of the chiWXYZ gene products. ChiW is a member of the phage holin-3 family and is predicted to span the membrane three times with an N terminus–out, C terminus–in topology. The predicted endopeptidase ChiX does not have a classical signal peptide and is hypothesized to reach the periplasm via the ChiW holin. The ChiY protein is synthesized with a classical N-terminal type II signal peptide, which is typical of outer membrane lipoproteins. The ChiZ protein is predicted to have a single N-terminal transmembrane domain and span the membrane with an N terminus–in, C terminus–out topology. N, N terminus; C, C terminus.
Proteomics reveals all chitinolytic proteins require ChiW for secretion
Next, it was decided to take a whole systems approach to understand the involvement of ChiW in general protein secretion. First, a new deletion strain (JJH08p) was made that carried an unmarked complete deletion of chiW. Total protein extracts of culture supernatant (secretomes) were then prepared from the S. marcescens Db10 parental strain and the JJH08p chiW mutant. Label-free quantitative proteomics was then used to profile changes in protein abundance in the corresponding secretomes. Four biological replicates of secretome digests of both strains were analyzed by high-resolution mass spectrometry in a mass spectrometer (Orbitrap Velos Pro; Thermo Fisher Scientific), and data were processed through MaxQuant (Cox and Mann, 2008). Using strict filtering, 497 proteins (<1% false discovery rate) were identified, of which 351 showed good reproducibility and were quantified in at least two of the four replicates for each strain.
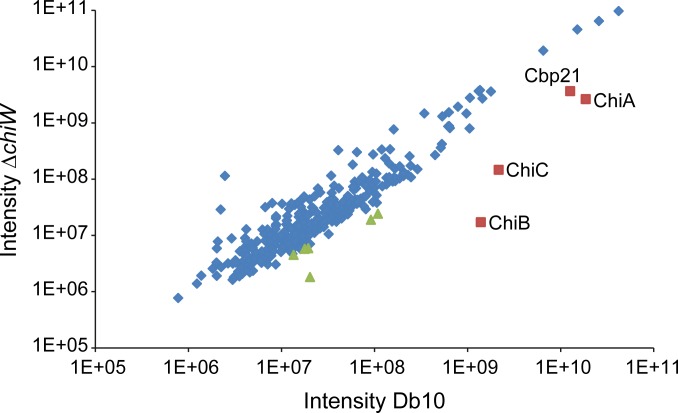
Although the relative levels of almost all proteins identified by this method remained unaffected by the chiW deletion, a subset of 10 proteins showed, with high confidence (P < 0.01), a decrease in abundance in the secretome of the mutant compared with the parent strain (Table 1). Strikingly, this group of 10 proteins included not only ChiC but also ChiA, ChiB, and Cbp21 (Table 1). In addition, these four chitinolytic proteins were by far the most abundant of the 10 whose extracellular levels were found to change in this experiment (Fig. 3, Table 1, and Fig. S3). Similarly, only a small number of mostly extracellular proteins, predominantly fimbrial subunits, were found to increase in abundance in the secretome of the chiW mutant (Table S1).
Table 1.
Extracellular proteins secreted in a ChiW-dependent manner identified by label-free mass spectrometry
| Identifier | Name | Sec signal peptide | Description | Intensity (relative intensitya) | Relative abundance Db10/ΔchiW | P-value | Unique peptides | Sequence coverage |
| % | ||||||||
| SMDB11_2875 | ChiB | No | Chitinase B | 8.5 × 109 (92) | 80.7 | 1.32 × 10−8 | 11 | 32 |
| SMDB11_0468 | ChiC | No | Chitinase C | 1.4 × 1010 (152) | 14.7 | 3.08 × 10−5 | 21 | 54 |
| SMDB11_3897 | − | No | Pirin | 1.3 × 108 (1.4) | 11.2 | 3.08 × 10−4 | 5 | 26 |
| SMDB11_4243 | ChiA | Yes | Chitinase A | 1.2 × 1011 (1,304) | 7.0 | 1.42 × 10−7 | 33 | 69 |
| SMDB11_1933 | TrpE | No | Anthranilate synthase | 5.5 × 108 (6) | 4.8 | 2.26 × 10−5 | 12 | 26 |
| SMDB11_4522 | MdhA | No | Malate dehydrogenase | 7.4 × 108 (8) | 3.4 | 1.94 × 10−5 | 9 | 36 |
| SMDB11_2877 | Cbp21 | Yes | Chitin binding protein | 6.7 × 1010 (728) | 3.4 | 0.0054 | 10 | 73 |
| SMDB11_2870 | MaeB | No | NADP-dependent malic enzyme | 1.3 × 108 (1.4) | 3.3 | 0.00032 | 11 | 18 |
| SMDB11_2390 | − | No | Putative heme oxygenase | 1.1 × 108 (1.2) | 3.1 | 0.0044 | 4 | 25 |
| SMDB11_3147 | ProS | No | Prolyl-tRNA synthetase | 9.2 × 107 (1) | 3.0 | 6.33 × 10−5 | 9 | 23 |
Proteins identified as present in the secretome of the S. marcescens parental strain (Db10) at an abundance greater than 3× higher than observed in the secretome of a strain (JJH08p) lacking the chiW gene (P < 0.01) are shown. Four biological replicates of each strain were analyzed. The most abundant proteins in this list are highlighted in bold. Minus signs indicate that these gene products have no name.
Relative intensity is calculated in relation to the intensity of least abundant protein in the list (prolyl-tRNA synthetase). Additional mass spectrometry data showing the proteins in the secretome of the ΔchiW strain, which are increased in abundance relative to the secretome of the Db10 parental strain, are shown in Table S1.
Figure 3.
Intensity scatter plot of proteome results. Label-free intensities of secretome proteins (mean of four biological replicates) of the ΔchiW (JJH08p) strain versus the secretome of the Db10 parental strain. Highly abundant proteins that were reduced significantly (P < 0.01) in the ΔchiW secretome are labeled red with the locations of the other, lower abundance, proteins shown in green (see Table 1). The three chitinases, ChiA, ChiB, and ChiC, as well as the chitin-binding protein, Cbp21, are the by far the most abundant of the proteins affected by the ΔchiW mutation.
Genetic dissection of the chiWXYZ locus
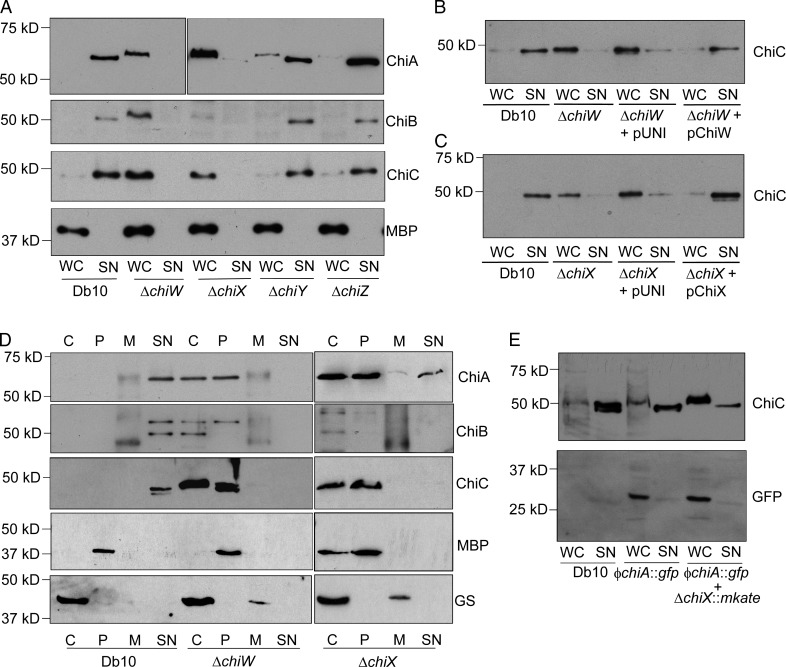
To understand the roles of all the members of the chiWXYZ locus, a bank of four in-frame deletion strains was constructed, and Western immunoblotting was then used to assess the localization of the three chitinases (Fig. 4 A). In line with the proteomics data (Fig. 3 and Table 1), deletion of the chiW gene deleteriously affected the extracellular levels of all three S. marcescens chitinases (Fig. 4 A). In addition to chiW, it was revealed that ChiX, a putative metal-dependent l-alanyl-d-glutamate endopeptidase, was also essential for secretion of the three known chitinases (Fig. 4 A). However, neither of the genes encoding the spanin-like proteins (chiY and chiZ) appeared individually essential for chitinase secretion (Fig. 4 A).
Figure 4.
S. marcescens chitinases are secreted in chiW- and chiX-dependent manner. (A) S. marcescens (Db10) together with JJH04w (ΔchiW), JJH05x (ΔchiX), JJH06y (ΔchiY), and JJH07z (ΔchiZ) were analyzed by Western immunoblotting using the specific antisera indicated. (B) Complementation of the ΔchiW strain. The JJH04w strain was transformed with pUNI-PROM or pUNI-ChiW (pChiW), and the localization of ChiC was analyzed. (C) Complementation of the ΔchiX strain. The JJH05x strain was transformed with pUNI-PROM or pUNI-ChiX (pChiX), and the localization of ChiC was analyzed. (D) S. marcescens (Db10) together with the JJH04w (ΔchiW) mutant and the JJH05x (ΔchiX) mutant were fractionated into cytoplasm (C), periplasm (P), total membranes (M), and culture supernatant (SN). Proteins were analyzed by Western immunoblotting using the antisera indicated. Controls were the periplasmic maltose binding protein (MBP) and the cytoplasmic glutamine synthetase (GS) enzyme. In all cases, the band marked by the single asterisk is the ChiA protein, which the polyclonal ChiB antiserum can also detect. (E) The FTG005 strain (ϕchiA::gfp) and the FTG006 strain (ϕchiA::gfp and ΔchiX::mKate) were grown in rich media before the localization of ChiC and GFP was analyzed by Western immunoblotting. WC, whole cell.
Both the ΔchiW and the ΔchiX mutant phenotypes could be complemented in trans (Fig. 4, B and C). The intact S. marcescens chiW and chiX genes were amplified by PCR and cloned under the control of the constitutive tat promoter from E. coli in plasmid pUNI-PROM (Jack et al., 2004). Transformation of the either the ΔchiW (Fig. 4 B) or ΔchiX mutant (Fig. 4 C) strain with the empty vector did not restore secretion of ChiC. However, plasmid-driven production of the holin-like protein in ΔchiW (Fig. 4 B) or the endopeptidase in ΔchiX (Fig. 4 C) rescued secretion of the ChiC chitinase.
Evidence that ChiW and ChiX operate in tandem to facilitate chitinase secretion
Although sequence analysis suggests ChiX could be an l-alanyl-d-glutamate endopeptidase, which would require a periplasmic localization to access peptidoglycan substrate, this protein is not synthesized with an obvious signal peptide. One hypothesis, consistent with the similarity of the chiWXYZ system to phage lysis cassettes, is that the role of the ChiW holin could be to allow ChiX access to the periplasm. To test this hypothesis, an experiment was designed in which ChiX was targeted to the periplasm via an alternative route, thus bypassing any requirement for ChiW (Fig. 5 A).
Figure 5.
Bypassing the holin: Tat-dependent rescue of chitinase secretion in a chiW mutant. (A) Experimental design. The JJH04w strain carries an in-frame deletion in chiW, encoding a holin-like membrane protein but retains an active Tat translocase in the inner membrane. A plasmid-encoded fusion between the S. marcescens SufI twin-arginine signal peptide and the S. marcescens ChiX protein will be targeted to the periplasm via Tat. N, N terminus; C, C terminus. (B) The JJH04w (ΔchiW) strain was transformed with empty vector (pBAD18), a vector encoding native ChiX (pBAD-ChiX), or a vector encoding the spSufI::ChiX fusion protein (pBAD-Tat-ChiX). All strains were grown aerobically in rich medium containing 0.02% (wt/vol) l-arabinose before being separated into culture supernatant (SN) and whole cell (WC) fractions. Samples were analyzed for the presence of ChiC and the periplasmic control protein (maltose binding protein [MBP]) by Western immunoblotting.
A set of plasmids was prepared, based on the l-arabinose–inducible pBAD18 system (Guzman et al., 1995), which allowed controlled production of ChiX, or ChiX fused to the Tat signal peptide of S. marcescens SufI (SMDB11_3496), in the S. marcescens host. The bacterial Tat pathway is a general protein export system for the transport of folded proteins across the cytoplasmic membrane and was chosen here because ChiX has a predicted metal-binding motif, as have most natural Tat substrates (Palmer and Berks, 2012). A ΔchiW strain, blocked for chitinase secretion, was transformed with the inducible plasmids (Fig. 5 B). Induction of chiX alone could not restore chitinase secretion to a ΔchiW strain (Fig. 5 B). Very interestingly, however, production of the spSufI::ChiX fusion was able to rescue secretion of ChiC in the ΔchiW strain (Fig. 5 B). It can be concluded from this that periplasmically located ChiX is required for chitinase secretion and that ChiW may normally play some role in targeting ChiX to this compartment.
Evidence for a two-step pathway for chitinase secretion
It is clear that genetic inactivation of ChiW and ChiX impaired chitinase secretion from the cell; however, it was of interest to determine the subcellular location of mislocalized enzymes in the mutant strains. To address this, the periplasmic, cytoplasmic, and total membrane fractions of the parental strain and the ΔchiW mutant were prepared, separated by SDS-PAGE, and analyzed by Western immunoblotting (Fig. 4 D). Probing with periplasmic and cytoplasmic control antisera established that the fractionation protocol had prepared samples of the ΔchiW strain periplasm that were free from cytoplasmic contamination (Fig. 4 D). Further inspection revealed that both ChiA and ChiC were located in the periplasm in the secretion-defective ΔchiW strain (Fig. 4 D). Similarly, fractionation of the ΔchiX strain revealed that ChiA and ChiC secretion was blocked at an outer membrane transport step because both proteins could be detected in the periplasm in the mutant background (Fig. 4 D). Taken altogether, these data suggest strongly that export of ChiA and ChiC to the periplasm is not adversely affected by either the chiW or chiX mutations but that secretion is completely blocked at the final outer membrane translocation stage. Surprisingly, ChiB appeared to behave differently in this experiment, where a periplasmic intermediate for this isoenzyme was not readily detectable in the mutant strains.
Evidence for bimodal and coordinated expression of chiA and chiX
Although the biochemical (Fig. 1, Fig. 4, and Fig. 5) and proteomic (Table 1, Table S1, Fig. 3, and Fig. S3) analyses described here showed negligible contamination of the extracellular milieu with periplasmic or cytoplasmic proteins, the obvious similarities between chiWXYZ and bacteriophage lysis cassettes means it is reasonable to question whether chitinase secretion by S. marcescens is associated with a cell lysis event. To address this directly, live single cell imaging experiments were designed.
To visualize expression of the chiWXYZ operon at the single cell level, a fluorescent reporter strain was constructed in which the chiX gene on the S. marcescens chromosome was replaced by the gene encoding the RFP mKate2 (Shcherbo et al., 2007). The JJH09 (ΔchiX::mKate) strain was grown for 16 h at 30°C in rich media before analysis by fluorescence microscopy (Fig. 6). The data clearly demonstrate bimodal chiX expression in the isogenic S. marcescens population, as both chiX ON cells (TRITC positive) and chiX OFF cells (TRITC negative) were apparent (Fig. 6). Calculations indicated that 0.95% of the cells per field of view (n > 4,500 cells) were TRITC (and thus chiX) positive (Fig. S4).
Figure 6.
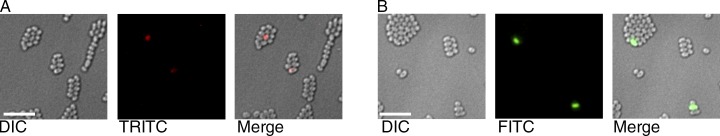
The expression of the chiX and chiA genes exhibit a bimodal distribution. (A) Representative still frames of the JJH09 (ΔchiX::mKate) strain. The ΔchiX::mKate cells were chosen to represent the range of expression values observed. (B) Representative still frames of the chiA::gfp strain (FTG005). In each case, cells were grown for 16 h in rich media, and images were taken using a 100×, 1.4 NA lens (Olympus) and a camera (CoolSNAP HQ). Bars, 10 µm.
To visualize expression of a secreted substrate, expression of the chitinase gene chiA was monitored at the single-cell level using a transcriptional reporter strain. A modified version of S. marcescens strain was constructed, FTG005, which contained a promoterless gfp gene (encoding GFP) positioned 23 base pairs downstream of the chiA termination codon on the bacterial chromosome. The FTG005 (ϕchiA-gfp) strain was grown for 16 h at 30°C and before analysis by fluorescence microscopy (Fig. 6). The data clearly demonstrate bimodal chiA expression in the isogenic S. marcescens population, as both chiA ON cells (FITC positive) and chiA OFF cells (FITC negative) were apparent (Fig. 6). Calculations indicated that 0.76% of the cells per field of view (n > 7,500 cells) were FITC (and thus chiA) positive (Fig. S4).
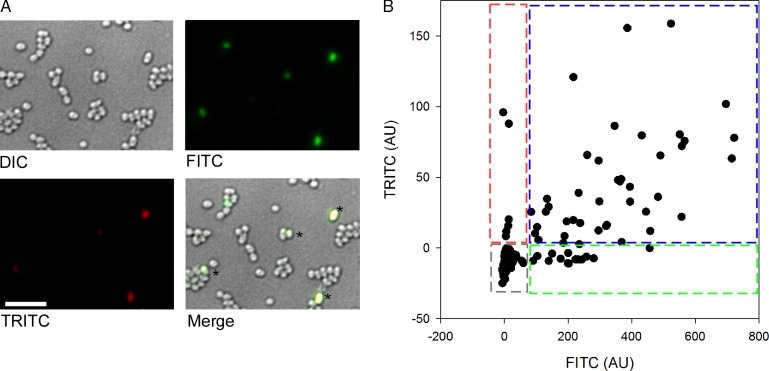
Next, a dual-fusion strain was constructed that would report both chiA (GFP) and chiX (mKate) expression simultaneously. Analysis of the FTG006 (ϕchiA-gfp and ΔchiX::mKate) strain established that bimodal expression of both chiX and chiA was evident from this experiment (Fig. 7). Most interestingly, however, the microscopy also revealed that expression from the distinct chiX and chiA operons was frequently coordinated within a single cell (Fig. 7).
Figure 7.
Coordinated expression of chiA and chiX. Representative data for chiA and chiX expression as followed via GFP and mKate fluorescence, respectively, for strain FTG006 (ΔchiX::mkate and ϕchiA::gfp). (A) Representative still frames of the FTG006 strain. Cells were grown for 16 h in rich media, and images were taken using a 100×, 1.4 NA lens (Olympus) and a camera (CoolSNAP HQ). Asterisks highlight cells that exhibit both red and green fluorescence and so appear yellow upon merging. Bar, 10 µm. (B) The background level for both channels was calculated as the mean value of S. marcescens Db10 parent strain grown under the same conditions, plus two times the standard deviation. This value was subtracted from the fluorescence values obtained for strain FTG006. TRICT values in arbitrary units (AU) are plotted on the y axis, and FITC (arbitrary units) is plotted in the x axis. 500 cells are represented, and the boxed areas on the chart highlight cells with TRICT only (red), FITC only (green), and TRICT with FITC (blue) expression. The cell population analyzed was biased toward analysis of the fluorescent cells to allow the ability of coexpression to be viewed.
Expression of chiA does not predispose cells to lysis
Having established that expression of chiX and chiA is tightly coordinated, it was now possible to use GFP-linked expression of chiA to monitor the viability of cells within the subpopulation known to be secreting chitinases. This is because the FTG005 strain produces GFP linked to chiA expression but also possesses an intact copy of active chiX, which is essential for the extracellular release of chitinases.
First, a biochemical approach was taken. Cells harboring the chiA::gfp fusion were analyzed for chitinase secretion activity and the localization of coexpressed GFP (Fig. 4 E). In this case, coordinately expressed GFP was found in the whole cells by Western immunoblotting only, rather than in the culture supernatant, whereas chitinase was secreted (Fig. 4 E).
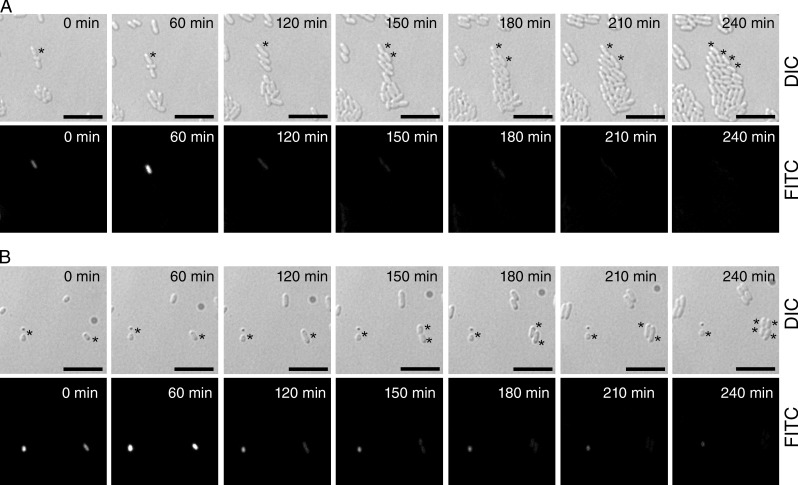
Next, the fate of the FTG005 (ϕchiA-gfp) population expressing chiA and gfp was followed using time-lapse microscopy (Fig. 8 and Videos 1 and 2). FTG005 (ϕchiA-gfp) cells were grown for 18 h in minimal media and were then diluted and monitored at 15-min intervals under the differential interference contrast (DIC; light microscopy) and FITC (green fluorescence) channels while growing on agarose slides. Growth was followed over a 4-h period and showed unequivocally that fluorescent FTG005 (ϕchiA-gfp) cells were capable of dividing (twice during this time course) and differentiating into a nonfluorescent cell population (Fig. 8 and Videos 1 and 2). From a total of 77 fluorescent cells observed, 55 were noted to undergo cell division (∼71%). Furthermore, none of the remaining 22 cells (∼29%) exhibited an obvious lysis event but appeared to adopt a quiescent (nondividing) state and were either subsumed within the developing microcolony (not depicted) or remained dormant (Fig. 8 and Video 2). It can be concluded that these behaviors are incompatible with chiA expression, representing a terminal differentiation state that automatically results in lysis.
Figure 8.
Expression of chiA gene is not concurrent with a lysis event. Representative still frames (separate DIC and FITC channels) taken from time-lapse videos for strain FTG005 (ϕchiA::gfp) demonstrating the two fates of GFP-positive cells upon time-lapse imaging. The S. marcescens FTG005 (ϕchiA::gfp) strain was grown for 18 h in minimal media before being diluted and applied to a microscope slide. (A) FITC-positive cells divide and propagate. The asterisks highlight a single cell growing and dividing, twice, into daughter cells. (B) A subpopulation of FITC-positive cells remains fluorescent but does not divide even over an extended period of time. Progression from left to right is with respect to time, as noted on the micrograph (minutes). The left-hand asterisks highlight a quiescent cell that does not divide. The right-hand asterisks highlight a single cell growing and dividing, twice, into daughter cells. Bars, 10 µm. Videos of these images (merged) are available as Videos 1 and 2.
Discussion
This work has identified three genes—chiW (SMDB11_2874), chiX (SMDB11_2873), and chiR (SMDB11_2876)—that are essential for chitinase secretion by S. marcescens. All three are part of a locus that includes chiB and cbp21 and so is dedicated to chitin metabolism. There are no adjacent genes encoding phage tail proteins or transposases, which suggests that these genes are of proteobacterial origin and not associated with a prophage. The chiR gene was originally identified by a genetic screen designed to identify strains with reduced extracellular chitinase activity (Suzuki et al., 2001), whereas chiW and chiX have not been previously implicated in chitinase secretion.
Evidence presented in this work points to a two-step chitinase secretion process in which the chitinolytic machinery is first targeted to the periplasm and then transported across the outer membrane. Western analysis placed both ChiA and ChiC in the periplasm of the ΔchiW and the ΔchiX strains (Fig. 4 D). In support of this model, proteomic analysis of secreted ChiA and Cbp21 revealed that their Sec signal peptides were correctly processed during secretion, implying translocation to the periplasm via Sec and processing via LepB, a membrane-bound enzyme with its active site exposed to the periplasm. Moreover, structural studies of ChiA and Cbp21 reveal that these proteins contain disulfide bonds (Papanikolau et al., 2001; Vaaje-Kolstad et al., 2005b), which are formed in the periplasm by the DsbAB oxidoreductase system (Goemans et al., 2014), reinforcing the need for periplasmic intermediates during the secretion of these enzymes.
The S. marcescens ChiB and ChiC proteins do not display obvious Sec signal peptides. Proteomic analysis in this work established that the ChiB protein was not proteolytically processed during secretion. However, ChiB does contain a disulfide bond (Vaaje-Kolstad et al., 2004), which implies that a periplasmic intermediate must be a feature of its biosynthesis. Surprisingly, the ChiB protein was not detected in the periplasm of the chiW and chiX strains. It seems unlikely that ChiB is exported to the periplasm via ChiW because no periplasmic intermediate was obvious even in the ΔchiX strain that has an intact chiW gene (Fig. 4 D). Instead, it seems more likely that ChiB is rapidly degraded when mistargeted to the periplasm. The secreted version of S. marcescens ChiC was also found to be intact in the proteomic experiments and therefore not processed during secretion. However, previous work on P. aeruginosa ChiC suggested that secretion of this enzyme was accompanied by an unusual processing at the N terminus (Folders et al., 2001). Indeed, in some instances, the electrophoretic mobility of S. marcescens ChiC appeared different in whole cell and supernatant fractions (Fig. 4 D), and this can also be observed in some ChiA (Fig. 4 A) and ChiB (Fig. 1 and Fig. 4 A) samples. The basis of these electrophoretic shifts will require further investigation because the proteomic analysis of the secreted version of these enzymes did not reveal any processing events. The mode of initial export of ChiC to the periplasm remains to be established; however, it is clear that ChiW is not involved in this step (Fig. 4 D). It is possible that ChiC is a Sec-dependent protein but that it lacks an obvious signal peptide, perhaps analogous to SodA from Rhizobium leguminosarum (Krehenbrink et al., 2011).
The role of a holin in protein secretion
ChiW is a holin-like protein encoded within a putative four-gene operon that is itself located within a wider genetic locus dedicated to chitin utilization (Fig. 2). The ChiW protein shares sequence identity with bona fide phage holin proteins and is predicted to be an inner membrane protein with three transmembrane domains. A canonical phage or prophage holin would be expected to be involved in catalyzing cell lysis. Thus, because the evidence presented in this work suggests that cell lysis is not the major facilitator of chitinase secretion, ChiW is termed a holin-like protein in this study.
ChiW shares 32% overall sequence identity, and 46% similarity, with the canonical λ S holin. ChiW is clearly a member of the holin family (Reddy and Saier, 2013) but shows a high degree of divergence from phage sequences. Indeed, it is worth noting that of 52 potential holin families, there are 12 holin families predicted to be of proteobacterial origin, which includes ChiW, that have not been extensively characterized (Reddy and Saier, 2013). One key difference is that, unlike most holins encoded by phage or prophage, the S. marcescens ChiW protein has a single translation start (Fig. S2). The λ S holin, for example, has two translation initiation codons that result in active λ S105 and anti-holin λ S107 forms (White et al., 2010). ChiW therefore probably has no anti-holin activity associated with it. Indeed, this itself may point to a different modus operandi compared with the canonical λ holin S.
If the S. marcescens ChiW holin-like protein were to behave in the same way as the λ S holin, the possibility would be raised that the chitinases may not be actively secreted but instead released into the extracellular milieu via an altruistic cell lysis event induced by ChiW allowing ChiX, and possibly other hydrolytic enzymes, access to the periplasm. However, the evidence presented in this work allows an argument to be made against a crude cell lysis model. Neither Western analysis, nor the sensitive and wider ranging label-free proteomics experiments, could identify a lysis event associated with the chitinase-positive S. marcescens parent strain. Here, 497 different proteins were identified in the extracellular milieu of both parental and chiW strains. These included many previously described secreted proteins, as well as a large number of proteins known or predicted to be cytoplasmic or membrane-bound proteins, but that can be detected and identified even at very low levels here using this very sensitive technique. For a ChiWX-triggered general lysis model to be correct, it would be expected that all 497 proteins would be equally decreased in the mutant strain secretome compared with the parent strain, and so after normalization of the data, no significant differences would be expected to be observed. In reality, 10 proteins behaved very differently from the remaining 487 and are clearly reduced in the secretome of the holin-deficient strain (Table 1), with only four of those (the chitinolytic enzymes) being normally very highly abundant in the parental strain secretome (Fig. 3). Furthermore, live-cell imaging techniques revealed that chiA was coexpressed with the chiWXYZ operon in a subpopulation of cells (Fig. 7) and that cells coexpressing chiA and gfp did not release GFP into the growth media as would be expected in a lysis event (Fig. 4 D) but were instead able to grow and divide (Fig. 8). This provides strong evidence that, although bimodal, chitinase expression is not concomitant with an altruistic lysis event. Taken altogether, these data clearly point to a specific role in chitinase secretion for the ChiW holin-like protein.
Holins have been implicated in protein secretion in some other systems, but not without controversy. The S. marcescens NucE protein (SMDB11_0177) was suggested to be a holin required for secretion of the Sec-dependent NucA nuclease (Berkmen et al., 1997). However, subsequent deletion of nucE was found to have no effect on nuclease secretion (Strych et al., 1999). Similarly, one recent study suggested a holin-like protein (TcdE) was dedicated to the nonlytic toxin secretion by Clostridium difficile (Govind and Dupuy, 2012), whereas alternative research concluded that tcdE was not involved in toxin secretion (Olling et al., 2012).
The role of an endolysin in chitinase secretion
The ChiX protein is predicted to be a 15-kD soluble protein that has been classified in some databases as a d-ala-d-ala carboxypeptidase of the VanY family but actually shares very little sequence identity with genuine VanY-like proteins. Instead, ChiX is closely related to the Bacillus subtilis CwlK protein, which has been characterized as an l-alanyl-d-glutamate endopeptidase (Fukushima et al., 2007). Unlike CwlK, ChiX contains no obvious targeting signals, but its coexpression with a gene encoding a holin-like protein (ChiW) suggested that it could be targeted to the periplasm via its partner holin. Experimental evidence presented in this work supports that model because a ΔchiW mutant phenotype can be complemented by a Tat-targeted ChiX protein (Fig. 5).
The important role of the chiX gene in chitinase secretion in S. marcescens is clear; however, the precise biochemical function of ChiX protein remains to be determined. Although bacteriophage-encoded endolysins are often closely linked with cell lysis events when they operate in partnership with holins, evidence is now emerging of endolysins being involved in the specific nonlytic secretion of proteins. For example, recently an N-acetyl-β-d-muramidase (TtsA) from Salmonella enterica serovar Typhi, similar to phage endolysins, was found to be involved in typhoid toxin secretion across the bacterial outer membrane (Hodak and Galán, 2013). It is possible that ChiX is similarly involved in remodeling the peptidoglycan to facilitate protein secretion. Indeed, many apparently homologous enzymes have been identified encoded within bacterial genomes that may have such specialized roles, because it is believed that the periplasmic peptidoglycan layer presents a permeability barrier to all but the smallest of proteins (Scheurwater et al., 2008). Endogenous endolysins may be required to remodel the peptidoglycan to allow, for example, cell division to proceed or for large transmembrane protein complexes, such as secretion systems, to be assembled (Koraimann, 2003; Scheurwater et al., 2008).
A proposed scheme for chitinase secretion
Collectively, the multidisciplinary approaches used in this study allow an initial model to be proposed for chitinase secretion by S. marcescens. A small subpopulation of cells coexpress chitinase genes together with chiWXYZ. The chitinases produced are most likely first targeted to the periplasm by the Sec pathway, where ChiA and Cbp21 are further proteolytically processed and ChiA, ChiB, and Cbp21 have disulfide bonds correctly inserted. ChiW is a holin-like protein that meanwhile integrates into the inner membrane and, by an as-yet-unexplored mechanism for this system, allows the periplasmic targeting of ChiX, which is predicted to be a metal-dependent l-alanyl-d-glutamate endopeptidase. The role of ChiX is most likely to remodel the peptidoglycan by removing cross-links between the glycan strands, and this activity seems critical for the correct functioning of the final outer membrane secretion step.
The role for a bacterial holin-like protein and a peptidoglycan hydrolase in protein transport across the outer membrane is an intriguing one. It is very likely that these proteins will have to operate in partnership with other, as-yet-unidentified, outer membrane-associated proteins. In addition, ChiA, ChiB, ChiC, and Cbp21 may contain a common signal that allows specific secretion of this subset of proteins above all others.
Materials and methods
Mutant and plasmid construction, transposon mutagenesis, and genetic screening
Deletion strains of S. marcescens were constructed by allelic exchange using the suicide vector pKNG101 (Kaniga et al., 1991), and they are listed in Table 2. All in-frame deletions were constructed to preserve surrounding regulatory elements; however, note that strain JJH08p is devoid of chiW and also lacks the initiation codon of chiX and therefore exhibits a ΔchiW and chiX genotype. S. marcescens strains carrying fluorescent transcriptional markers were constructed using pKNG101. For strain JJH09 (ΔchiX::mKate), a DNA fragment containing a ribosome binding site and the mkate gene (Shcherbo et al., 2007) was amplified from a plasmid supplied by T. Norman (Harvard University, Cambridge, MA) and inserted into a ΔchiX allele prepared in pKNG101. For strain FTG005 (ϕchiA::gfp), a 1,000-bp fragment of DNA covering the 3′ end of chiA was amplified by PCR and cloned into pKNG101. Then, an EcoRV restriction site 35 bp downstream of the chiA termination codon was used to insert gfp-mut2 amplified from pNW704 (Marlow et al., 2014) to give an ϕchiA::gfp that was then transferred to the chromosome of S. marcescens Db10.
Table 2.
S. marcescens strains used and constructed in this work
| Strain | Relevant genotype | Reference |
| Db10 | Nonpigmented wild strain | Flyg et al., 1980 |
| JJH01 | As Db10 ΔchiA (SMDB11_4243) | This study |
| JJH02 | As Db10 ΔchiB (SMDB11_2875) | This study |
| JJH03 | As Db10 ΔchiC (SMDB11_0468) | This study |
| JchiA | As Db10 ΔchiB, ΔchiC | This study |
| JchiB | As Db10 ΔchiA, ΔchiC | This study |
| JchiC | As Db10 ΔchiA, ΔchiB | This study |
| Nochi | As Db10 ΔchiA, ΔchiB, ΔchiC | This study |
| JJH04w | As Db10 ΔchiW (SMDB11_2874) | This study |
| JJH05x | As Db10 ΔchiX (SMDB11_2873) | This study |
| JJH06y | As Db10 ΔchiY (SMDB11_2872) | This study |
| JJH07z | As Db10 ΔchiZ (SMDB11_2871) | This study |
| JJH08p | As Db10 ΔchiW, chiX | This study |
| FTG005 | As Db10 ϕchiA::gfp | This study |
| JJH09 | As Db10 ΔchiX::mkate | This study |
| FTG006 | As Db10 ϕchiA::gfp, ΔchiX::mkate | This study |
Transposon mutagenesis of the JchiC (chiC+, ΔchiA, and ΔchiB) strain was performed using E. coli donor strain SM10 λpir (pUTmini-Tn5Sm/Sp) as outlined previously (de Lorenzo et al., 1990). JchiC colonies that contained Tn5 were patched onto Luria Bertani (LB) agar plates supplemented with 2% (wt/vol) colloidal chitin prepared in house. Transposon insertion sites were mapped using single primer–specific PCR in which genomic DNA was digested with XhoI and PstI and then cloned into pBluescript followed by a round of PCR with vector- and Tn5-specific primers. PCR products were sequenced with Tn5 primers, and the positions of the Tn5 insertions were located by analysis of the S. marcescens Db11 genome at the Wellcome Trust Sanger Institute, for which Db10 is the direct parent.
For complementation analysis, the intact chiW and chiX genes were amplified by PCR and cloned into the vector pUNI-PROM (AmpR) that allows constitutive gene expression (Jack et al., 2004). S. marcescens was transformed by electroporation, and transformants were isolated on solid LB media supplemented with 200 µg/ml carbenicillin. For protein secretion analysis, the transformants were grown aerobically in liquid LB medium supplemented with 50 µg/ml carbenicillin at 30°C.
For the holin bypass experiments, the chiX gene was amplified by PCR and cloned as an engineered XbaI–SphI fragment into pBAD18 (KanR) that allows regulated expression with l-arabinose (Guzman et al., 1995) to give plasmid pBAD-ChiX. To attach a Tat signal peptide onto the N terminus of ChiX, the pBAD18 derivative pSC144 (English et al., 2012) was modified such that a SacI–XbaI fragment encoding an OmpA signal peptide was replaced by a PCR product encoding the initial 30 amino acids of S. marcescens SufI (SMDB11_3496) to give plasmid pSC143. Next, chiX (minus its initiation codon) was amplified and cloned as an XbaI–SphI fragment, resulting in pBAD-Tat-ChiX encoding an spSufI::ChiX fusion protein.
Protein production and secretion analysis
SMDB11_0468 (ChiC), SMDB11_2875 (ChiB), and SMDB11_4243 (ChiA) were overproduced in E. coli as C-terminally His-tagged proteins. Purified proteins were used to raise rabbit polyclonal antisera (Eurogentec). For Western immunoblotting, samples were separated by SDS-PAGE electroblotted onto polyvinylidene difluoride sheets (EMD Millipore). The in-house α-ChiA, α-ChiB, and α-ChiC sera were raised in rabbits against purified, recombinant, hexa-His–tagged S. marcescens chitinases produced in E. coli and were each used at a 1:20,000 dilution. The anti–glutamine synthetase antiserum was raised in rabbits against purified, recombinant, E. coli glutamine synthetase as previously described (Javelle et al., 2004) and was used at 1:10,000 dilution. Commercially available antibodies purchased for this work were the α-MBP (E. coli maltose binding protein) mouse monoclonal antibody (New England Biolabs, Inc.), which was used at a 1:20,000 dilution, and the α-RNAP (E. coli RNA polymerase) mouse monoclonal antibody (NeoClone), which was used at a 1:40,000 dilution. Immunodetection was performed using a HRP-conjugated goat anti–mouse or goat anti–rabbit IgG (Bio-Rad Laboratories) used at 1:20,000 dilution together with a chemiluminescent substrate (EMD Millipore). For small scale protein secretion assays, strains were grown in 5-ml volumes at 30°C. The OD600 for each sample was normalized before whole cell and supernatant fractions were collected by centrifugation. Culture supernatants were subjected to ultrafiltration through a 0.2-µm device (EMD Millipore).
Proteomic sample preparation and mass spectrometry
S. marcescens Db10 and JJH08p (ΔchiW and chiX) secreted protein samples were prepared for label-free mass spectrometry by growth in 50 ml minimal media at 30°C for 16 h. 2.6 ml of a 6.1-N trichloroacetic acid solution (∼100% wt/vol) was added to 40 ml of culture supernatant, and proteins were precipitated overnight at 4°C. The protein precipitates were then washed four times in 1 ml of 80% acetone (−20°C) before being dried at room temperature for 45 min. Samples were resuspended in 500 µl of 20 mM Tris-HCl, pH 8.0, 4% (wt/vol) SDS, and 1 mM Tris(2-carboxyethyl)phosphine and alkylated by addition of 5 mM iodoacetamide for 20 min in the dark at room temperature. After quenching with 5 mM DTT, samples were treated using the Filter-Aided Sample Prep protocol (Manza et al., 2005; Wiśniewski et al., 2009) and digested using trypsin (sequencing grade; Promega). Approximately 0.5 µg of each digest was injected in an interleaved manner onto a 2-cm × 100-µm trap column and separated on a 15-cm × 75-µm reversed-phase column (PepMap C18; Thermo Fisher Scientific) on a Rapid Separation LC (UltiMate 3000; Dionex). Peptides were eluted by a linear 2-h gradient of 95% A/5–35% B (A: 0.1% [wt/vol] formic acid; B: 80% [vol/vol] acetonitrile and 0.08% [wt/vol] formic acid) at 300 nl/min into a LTQ Orbitrap Velos Pro. Data were acquired using a data-dependent top 20 method, dynamically choosing the most abundant precursor ions from the survey scan (350–1,650 Th, 60,000 resolution, and automatic gain control target value of 106). Precursors above the threshold of 500 counts were isolated within a 2 Th window and fragmented by collision-induced dissociation in the LTQ Velos using a normalized collision energy of 35% and an activation time of 10 ms. Dynamic exclusion was defined by a list size of 500 features and exclusion duration of 60 s. Lock mass was used and set to 445.120025 for ions of polydimethylcyclosiloxane (Ritorto et al., 2013).
Label-free quantitation was performed using MaxQuant v1.3.0.5 (Cox and Mann, 2008). Mass spectrometric runs of four biological replicates of Db10 and JJH08p were searched against a combined database of S. marcescens containing 4,720 sequences and a list of common contaminants in proteomics experiments using the following settings: enzyme trypsin/P, allowing for two missed cleavages, fixed modifications were set to carbamidomethyl, and variable modifications were set to acetyl (protein N terminus) and oxidation. Mass spectrometry/mass spectrometry tolerance was set to 0.5 D, and precursor tolerance was set to 6 ppm. Peptide and protein false discovery rate was set to 0.01, minimal peptide length was 7, and one unique peptide was required. Requantify and retention time alignment (2 min) were enabled.
Subcellular fractionation
Preparation of cytoplasmic, total membrane, periplasmic, and secreted (culture supernatant) fractions was as follows. Initially, 5-ml cultures were grown overnight in LB medium. The cells were harvested by centrifugation and then suspended in 1 ml of 30 mM Tris-HCl, pH 7.8, 40% (wt/vol) glucose, and 2 mM EDTA. Cell suspensions were incubated at 30°C for 10 min before being harvested by centrifugation and immediately resuspended in 1 ml ice-cold distilled water. After incubation on ice for 10 min, the resultant spheroplasts were harvested by centrifugation, and the supernatant was retained as the periplasmic fraction. Spheroplasts were suspended in 50 mM Tris-HCl, pH 7.8, and 0.2 M Na2CO3 and lysed by sonication. Total membranes were recovered by ultracentrifugation and resuspended in 50 mM Tris-HCl, pH 7.8, by homogenization.
Live-cell imaging and image processing
For static images, cells were grown in LB aerobically overnight at 30°C with shaking at 220 rpm. Next, 5 ml of fresh LB was inoculated with 25 µl of the starter culture and grown for 16 h at 30°C with shaking at 220 rpm. Cells were harvested by centrifugation, washed in 5 ml PBS, and suspended in 10 ml PBS. Each microscope slide was prepared as follows. A 125 µl Gene Frame (AB-0578; ABgene House) was attached to a standard microscope slide (super premium; VWR International). The Gene Frame was next filled with molten minimal media (0.2% wt/vol glucose) solidified with 1% agarose and covered firmly with a standard microscope slide, to flatten the agarose surface. When the agarose had sufficiently cooled and solidified, the upper slide was carefully removed. After inoculation, the cell suspension was allowed to dry, after which the Gene Frame was sealed with a number 1.5 coverslip (22 × 22 mm; VWR International). Images were acquired using a wide-field microscope (DeltaVision Core; Applied Precision) mounted on an inverted stand (IX71; Olympus) with a 100×, 1.4 NA lens (Olympus) and electron-multiplying charge-coupled device camera (Cascade2 512; Photometrics). Datasets (512 × 512 pixels with 13 z sections spaced by 0.2 µm) were acquired with DIC and fluorescence optics. DIC images were acquired with an light-emitting diode transmitted light source (Applied Precision) at 32% intensity and exposure times between 25 and 50 ms. Fluorescence from mKate2 was imaged using a 100-W mercury lamp and a TRITC filter set (excitation of 555/28 nm; emission of 617/73 nm) with an exposure time of 300 ms. Fluorescence from GFP was imaged using a 100-W mercury lamp and an FITC filter set (excitation of 490/20 nm; emission of 528/38 nm) with an exposure time of 200 ms.
For time-lapse microscopy, cells were grown in an identical manner except they were subcultured into minimal media (0.2% wt/vol glucose) and grown aerobically at 30°C with shaking at 220 rpm for 18 h before being diluted to OD600 of 0.01 in minimal media. Agarose microscope slides were prepared, exactly as described for collecting static images, and incubated at 30°C in a temperature-controlled environmental chamber (Weather Station; Applied Precision) to allow the cells to equilibrate on the agarose pads for 3 h. Time-lapse imaging of microcolony development and gene expression was performed using a wide-field microscope (DeltaVision Core) mounted on an inverted stand (IX71) with a 60×, 1.4 NA lens (Olympus) and a camera (CoolSNAP HQ; Photometrics) with DIC and fluorescence optics. For each experiment, 12 independent fields, each containing one or two cells, were manually identified, and their xyz positions were stored in the microscope-control software (softWoRx; Applied Precision). Datasets (512 × 512 pixels with 2 × 2 binning and 12 z sections spaced by 1 µm) were acquired every 15 min for ≤12 h. GFP was imaged using a 100-W mercury lamp and a FITC filter set (excitation of 490/20 nm; emission of 528/38 nm) with an exposure time of 50 ms. DIC images were acquired with an light-emitting diode transmitted light source (Applied Precision) at 32% intensity and exposure times between 25 and 50 ms. Postacquisition datasets were rendered and analyzed using OMERO software (Open Microscopy Environment). During analysis, the threshold used to define activation of the transcriptional reporter was set as a fluorescence intensity value greater than two standard deviations above the mean background fluorescence of the wild-type strain Db10.
Online supplemental material
Fig. S1 shows that directed and transposon mutagenesis identifies genes required for in vivo chitinolytic activity by S. marcescens. Fig. S2 shows the basic bioinformatics analysis of the ChiW holin-like protein. Fig. S3 shows the additional analysis of the proteomics data, demonstrating the reproducibility of the experiment. Fig. S4 shows the fluorescence data collected and used to calculate the different levels of expression of chiA and chiX within a population of cells. Table S1 shows analysis of the label-free quantitative proteomic data to reveal secreted proteins that are increased in abundance in the secretome of the ΔchiW strain as compared with the parental strain. Video 1 shows time-lapse microscopy of an S. marcescens strain expressing GFP transcriptionally coupled to chiA. Video 2 shows the same S. marcescens strain expressing GFP under time-lapse microscopy, but this time, it demonstrates a small subset of fluorescent cells that are quiescent in that they do not divide over the timescale of the experiment. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201404127/DC1. Additional data are available in the JCB DataViewer at http://dx.doi.org/10.1083/jcb.201404127.dv.
Supplementary Material
Acknowledgments
We thank Dr. Sarah Murdoch (Dundee) for valuable help and constant technical advice throughout the course of this project. We would like to thank Dr. Graeme Ball (Dundee) for preparing the video files, and we are also grateful to Dr. Sam Swift and the light microscopy team at the College of Life Sciences, Dundee, for expert assistance.
This work was funded by a Wellcome Trust PhD studentship (089693/Z/09/Z) and a Medical Research Council PhD studentship award to the College of Life Sciences, Dundee, which supported J.J. Hamilton and R.A. Owen, respectively. V.L. Marlow was supported by the Biotechnology and Biological Sciences Research Council via a research grant (BB/C520404/1) awarded to N.R. Stanley-Wall. M.d.A.A. Costa was supported by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior PhD program. S.J. Coulthurst was funded by a Royal Society of Edinburgh/Scottish Government Personal Research Fellowship, and T. Palmer was the recipient of a Royal Society Wolfson Research Merit Award. The microscopy facilities in the College of Life Sciences, Dundee, are supported by the Wellcome Trust Strategic Award (grant number 083524/Z/07/Z) and the Medical Research Council Next Generation Optical Microscopy award (grant number MR/K015869/1).
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- DIC
- differential interference contrast
- LB
- Luria Bertani
- Tat
- twin-arginine translocation
References
- Berkmen M., Benedik M.J., and Bläsi U.. 1997. The Serratia marcescens NucE protein functions as a holin in Escherichia coli. J. Bacteriol. 179:6522–6524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brurberg M.B., Eijsink V.G., Haandrikman A.J., Venema G., and Nes I.F.. 1995. Chitinase B from Serratia marcescens BJL200 is exported to the periplasm without processing. Microbiology. 141:123–131 10.1099/00221287-141-1-123 [DOI] [PubMed] [Google Scholar]
- Brurberg M.B., Nes I.F., and Eijsink V.G.. 1996. Comparative studies of chitinases A and B from Serratia marcescens. Microbiology. 142:1581–1589 10.1099/13500872-142-7-1581 [DOI] [PubMed] [Google Scholar]
- Cox J., and Mann M.. 2008. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26:1367–1372 10.1038/nbt.1511 [DOI] [PubMed] [Google Scholar]
- de Lorenzo V., Herrero M., Jakubzik U., and Timmis K.N.. 1990. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J. Bacteriol. 172:6568–6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Economou A., Christie P.J., Fernandez R.C., Palmer T., Plano G.V., and Pugsley A.P.. 2006. Secretion by numbers: Protein traffic in prokaryotes. Mol. Microbiol. 62:308–319 10.1111/j.1365-2958.2006.05377.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- English G., Trunk K., Rao V.A., Srikannathasan V., Hunter W.N., and Coulthurst S.J.. 2012. New secreted toxins and immunity proteins encoded within the Type VI secretion system gene cluster of Serratia marcescens. Mol. Microbiol. 86:921–936 10.1111/mmi.12028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flyg C., Kenne K., and Boman H.G.. 1980. Insect pathogenic properties of Serratia marcescens: phage-resistant mutants with a decreased resistance to Cecropia immunity and a decreased virulence to Drosophila. J. Gen. Microbiol. 120:173–181. [DOI] [PubMed] [Google Scholar]
- Folders J., Algra J., Roelofs M.S., van Loon L.C., Tommassen J., and Bitter W.. 2001. Characterization of Pseudomonas aeruginosa chitinase, a gradually secreted protein. J. Bacteriol. 183:7044–7052 10.1128/JB.183.24.7044-7052.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francetic O., Belin D., Badaut C., and Pugsley A.P.. 2000. Expression of the endogenous type II secretion pathway in Escherichia coli leads to chitinase secretion. EMBO J. 19:6697–6703 10.1093/emboj/19.24.6697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederiksen R.F., Paspaliari D.K., Larsen T., Storgaard B.G., Larsen M.H., Ingmer H., Palcic M.M., and Leisner J.J.. 2013. Bacterial chitinases and chitin-binding proteins as virulence factors. Microbiology. 159:833–847 10.1099/mic.0.051839-0 [DOI] [PubMed] [Google Scholar]
- Fukushima T., Yao Y., Kitajima T., Yamamoto H., and Sekiguchi J.. 2007. Characterization of new L,D-endopeptidase gene product CwlK (previous YcdD) that hydrolyzes peptidoglycan in Bacillus subtilis. Mol. Genet. Genomics. 278:371–383 10.1007/s00438-007-0255-8 [DOI] [PubMed] [Google Scholar]
- Goemans C., Denoncin K., and Collet J.F.. 2014. Folding mechanisms of periplasmic proteins. Biochim. Biophys. Acta. 1843:1517–1528 10.1016/j.bbamcr.2013.10.014 [DOI] [PubMed] [Google Scholar]
- Govind R., and Dupuy B.. 2012. Secretion of Clostridium difficile toxins A and B requires the holin-like protein TcdE. PLoS Pathog. 8:e1002727 10.1371/journal.ppat.1002727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gründling A., Bläsi U., and Young R.. 2000. Biochemical and genetic evidence for three transmembrane domains in the class I holin, λ S. J. Biol. Chem. 275:769–776 10.1074/jbc.275.2.769 [DOI] [PubMed] [Google Scholar]
- Guzman L.M., Belin D., Carson M.J., and Beckwith J.. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121–4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hejazi A., and Falkiner F.R.. 1997. Serratia marcescens. J. Med. Microbiol. 46:903–912 10.1099/00222615-46-11-903 [DOI] [PubMed] [Google Scholar]
- Hines D.A., Saurugger P.N., Ihler G.M., and Benedik M.J.. 1988. Genetic analysis of extracellular proteins of Serratia marcescens. J. Bacteriol. 170:4141–4146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodak H., and Galán J.E.. 2013. A Salmonella Typhi homologue of bacteriophage muramidases controls typhoid toxin secretion. EMBO Rep. 14:95–102 10.1038/embor.2012.186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iguchi A., Nagaya Y., Pradel E., Ooka T., Ogura Y., Katsura K., Kurokawa K., Oshima K., Hattori M., Parkhill J., et al. . 2014. Genome evolution and plasticity of Serratia marcescens, an important multidrug-resistant nosocomial pathogen. Genome Biol. Evol. 6:2096–2110 10.1093/gbe/evu160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack R.L., Buchanan G., Dubini A., Hatzixanthis K., Palmer T., and Sargent F.. 2004. Coordinating assembly and export of complex bacterial proteins. EMBO J. 23:3962–3972 10.1038/sj.emboj.7600409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javelle A., Severi E., Thornton J., and Merrick M.. 2004. Ammonium sensing in Escherichia coli. Role of the ammonium transporter AmtB and AmtB-GlnK complex formation. J. Biol. Chem. 279:8530–8538 10.1074/jbc.M312399200 [DOI] [PubMed] [Google Scholar]
- Kaniga K., Delor I., and Cornelis G.R.. 1991. A wide-host-range suicide vector for improving reverse genetics in gram-negative bacteria: inactivation of the blaA gene of Yersinia enterocolitica. Gene. 109:137–141 10.1016/0378-1119(91)90599-7 [DOI] [PubMed] [Google Scholar]
- Koraimann G.2003. Lytic transglycosylases in macromolecular transport systems of Gram-negative bacteria. Cell. Mol. Life Sci. 60:2371–2388 10.1007/s00018-003-3056-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korotkov K.V., Sandkvist M., and Hol W.G.. 2012. The type II secretion system: biogenesis, molecular architecture and mechanism. Nat. Rev. Microbiol. 10:336–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krehenbrink M., Edwards A., and Downie J.A.. 2011. The superoxide dismutase SodA is targeted to the periplasm in a SecA-dependent manner by a novel mechanism. Mol. Microbiol. 82:164–179 10.1111/j.1365-2958.2011.07803.x [DOI] [PubMed] [Google Scholar]
- Lee C.G., Da Silva C.A., Dela Cruz C.S., Ahangari F., Ma B., Kang M.J., He C.H., Takyar S., and Elias J.A.. 2011. Role of chitin and chitinase/chitinase-like proteins in inflammation, tissue remodeling, and injury. Annu. Rev. Physiol. 73:479–501 10.1146/annurev-physiol-012110-142250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahlen S.D.2011. Serratia infections: from military experiments to current practice. Clin. Microbiol. Rev. 24:755–791 10.1128/CMR.00017-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manza L.L., Stamer S.L., Ham A.J., Codreanu S.G., and Liebler D.C.. 2005. Sample preparation and digestion for proteomic analyses using spin filters. Proteomics. 5:1742–1745 10.1002/pmic.200401063 [DOI] [PubMed] [Google Scholar]
- Marlow V.L., Porter M., Hobley L., Kiley T.B., Swedlow J.R., Davidson F.A., and Stanley-Wall N.R.. 2014. Phosphorylated DegU manipulates cell fate differentiation in the Bacillus subtilis biofilm. J. Bacteriol. 196:16–27 10.1128/JB.00930-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olling A., Seehase S., Minton N.P., Tatge H., Schröter S., Kohlscheen S., Pich A., Just I., and Gerhard R.. 2012. Release of TcdA and TcdB from Clostridium difficile cdi 630 is not affected by functional inactivation of the tcdE gene. Microb. Pathog. 52:92–100 10.1016/j.micpath.2011.10.009 [DOI] [PubMed] [Google Scholar]
- Palmer T., and Berks B.C.. 2012. The twin-arginine translocation (Tat) protein export pathway. Nat. Rev. Microbiol. 10:483–496. [DOI] [PubMed] [Google Scholar]
- Papanikolau Y., Prag G., Tavlas G., Vorgias C.E., Oppenheim A.B., and Petratos K.. 2001. High resolution structural analyses of mutant chitinase A complexes with substrates provide new insight into the mechanism of catalysis. Biochemistry. 40:11338–11343 10.1021/bi010505h [DOI] [PubMed] [Google Scholar]
- Reddy B.L., and Saier M.H. Jr. 2013. Topological and phylogenetic analyses of bacterial holin families and superfamilies. Biochim. Biophys. Acta. 1828:2654–2671 10.1016/j.bbamem.2013.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritorto M.S., Cook K., Tyagi K., Pedrioli P.G., and Trost M.. 2013. Hydrophilic strong anion exchange (hSAX) chromatography for highly orthogonal peptide separation of complex proteomes. J. Proteome Res. 12:2449–2457 10.1021/pr301011r [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheurwater E., Reid C.W., and Clarke A.J.. 2008. Lytic transglycosylases: bacterial space-making autolysins. Int. J. Biochem. Cell Biol. 40:586–591 10.1016/j.biocel.2007.03.018 [DOI] [PubMed] [Google Scholar]
- Shcherbo D., Merzlyak E.M., Chepurnykh T.V., Fradkov A.F., Ermakova G.V., Solovieva E.A., Lukyanov K.A., Bogdanova E.A., Zaraisky A.G., Lukyanov S., and Chudakov D.M.. 2007. Bright far-red fluorescent protein for whole-body imaging. Nat. Methods. 4:741–746 10.1038/nmeth1083 [DOI] [PubMed] [Google Scholar]
- Strych U., Dai W., and Benedik M.J.. 1999. The NucE and NucD lysis proteins are not essential for secretion of the Serratia marcescens extracellular nuclease. Microbiology. 145:1209–1216 10.1099/13500872-145-5-1209 [DOI] [PubMed] [Google Scholar]
- Summer E.J., Berry J., Tran T.A., Niu L., Struck D.K., and Young R.. 2007. Rz/Rz1 lysis gene equivalents in phages of Gram-negative hosts. J. Mol. Biol. 373:1098–1112 10.1016/j.jmb.2007.08.045 [DOI] [PubMed] [Google Scholar]
- Suzuki K., Taiyoji M., Sugawara N., Nikaidou N., Henrissat B., and Watanabe T.. 1999. The third chitinase gene (chiC) of Serratia marcescens 2170 and the relationship of its product to other bacterial chitinases. Biochem. J. 343:587–596 10.1042/0264-6021:3430587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K., Uchiyama T., Suzuki M., Nikaidou N., Regue M., and Watanabe T.. 2001. LysR-type transcriptional regulator ChiR is essential for production of all chitinases and a chitin-binding protein, CBP21, in Serratia marcescens 2170. Biosci. Biotechnol. Biochem. 65:338–347 10.1271/bbb.65.338 [DOI] [PubMed] [Google Scholar]
- Tran H.T., Barnich N., and Mizoguchi E.. 2011. Potential role of chitinases and chitin-binding proteins in host-microbial interactions during the development of intestinal inflammation. Histol. Histopathol. 26:1453–1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trost M., Wehmhöner D., Kärst U., Dieterich G., Wehland J., and Jänsch L.. 2005. Comparative proteome analysis of secretory proteins from pathogenic and nonpathogenic Listeria species. Proteomics. 5:1544–1557 10.1002/pmic.200401024 [DOI] [PubMed] [Google Scholar]
- Vaaje-Kolstad G., Houston D.R., Rao F.V., Peter M.G., Synstad B., van Aalten D.M., and Eijsink V.G.. 2004. Structure of the D142N mutant of the family 18 chitinase ChiB from Serratia marcescens and its complex with allosamidin. Biochim. Biophys. Acta. 1696:103–111 10.1016/j.bbapap.2003.09.014 [DOI] [PubMed] [Google Scholar]
- Vaaje-Kolstad G., Horn S.J., van Aalten D.M., Synstad B., and Eijsink V.G.. 2005a. The non-catalytic chitin-binding protein CBP21 from Serratia marcescens is essential for chitin degradation. J. Biol. Chem. 280:28492–28497 10.1074/jbc.M504468200 [DOI] [PubMed] [Google Scholar]
- Vaaje-Kolstad G., Houston D.R., Riemen A.H., Eijsink V.G., and van Aalten D.M.. 2005b. Crystal structure and binding properties of the Serratia marcescens chitin-binding protein CBP21. J. Biol. Chem. 280:11313–11319 10.1074/jbc.M407175200 [DOI] [PubMed] [Google Scholar]
- White R., Tran T.A., Dankenbring C.A., Deaton J., and Young R.. 2010. The N-terminal transmembrane domain of λ S is required for holin but not antiholin function. J. Bacteriol. 192:725–733 10.1128/JB.01263-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiśniewski J.R., Zougman A., Nagaraj N., and Mann M.. 2009. Universal sample preparation method for proteome analysis. Nat. Methods. 6:359–362 10.1038/nmeth.1322 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.