Abstract
The rapid rise in morbidity and mortality from drug-resistant pathogenic bacteria has generated elevated interest in combination therapy using antimicrobial agents. Antimicrobial peptides (AMPs) are a candidate drug class to advance the development of combination therapies. Although the literature is ambiguous, the generic membrane disrupting activity of AMPs could enable them to synergize with conventional small molecule antibiotics by increasing access to the cell and by triggering membrane damage mediators. We used a novel assay to measure interactions, expressed as fractional inhibitory concentration (FIC), between four conventional antibiotics in combination with four well-characterized, membrane permeabilizing AMPs, against three species of Gram negative and Gram positive bacteria, giving 40 total pair-wise measurements of FIC with statistical uncertainties. We chose a set of AMPs that are known to dramatically disrupt the membranes of both Gram negative and Gram positive bacteria. Yet none of the membrane permeabilizing antimicrobial peptides interacted synergistically with any of the conventional antibiotic drugs in any organism. Large-scale membrane disruption and permeabilization by AMPs is not sufficient to drive them to act synergistically with chemical antibiotics in either Gram negative or Gram positive microbes.
INTRODUCTION
One of the most significant and widespread problems in infectious disease treatment is the development of resistance to chemotherapeutics. Alarmingly, drug-resistant and multidrug-resistant bacteria, such as methicillin resistant Staphylococcus aureus (MRSA) or vancomycin resistant Enterococcus faecalis, have become commonplace in hospitals, leading to very significant morbidity and mortality from previously treatable bacterial infections (1). The rise of community acquired drug resistant infections has caused additional concern and has led some to question whether we have reached “the end of antibiotics”(2).
Because of the ineffectiveness of current antibiotic treatment options for drug-resistant bacterial infections, alternate classes of antibiotics are desperately needed and combination therapies must be seriously considered as a routine approach to antibacterial chemotherapy. Cationic antimicrobial peptides (AMP) that target the microbial membrane directly have long been a promising treatment alternative (3–8) for direct use, or for use in combination with other antibiotics. Yet, their potential is still mostly unfulfilled. The potential advantages of membrane permeabilizing antimicrobial peptides are significant. They have broad-spectrum μM activity against many strains of Gram negative and Gram positive bacteria, in vitro, including drug-resistant strains (4, 7, 9–20). Furthermore, resistance to AMPs is uncommon and is not readily selectable or inducible (21, 22), unlike the case for most chemical antibiotics. This is likely due to the fact that AMPs act on bacterial membranes globally, and not with a particular macromolecular component of the cell. The need to alter the architecture of the entire membrane to achieve resistance likely increases the overall fitness cost.
Multidrug combinations are sometimes used for the treatment of routine bacterial infections (23, 24) as well as tuberculosis (25). Thus it is reasonable to propose a combination therapy utilizing AMPs and chemical antibiotics because the mechanism of action of membrane permeabilizing antimicrobial peptides is so dramatically different from the chemical antibiotics. It has been proposed in the literature that AMPs could synergize with antibiotics increasing the permeability of bacterial membranes (26), including the outer membrane of Gram negative bacteria. It has also been suggested that AMPs could enhance the activity of bacterial murein hydrolases (27, 28) or other enzymes that decrease the integrity of the peptidoglycan layer, allowing antibiotics that affect cell wall synthesis (e.g. β-lactams) to act more efficiently. However, the data in the literature are ambiguous with some AMP-drug combinations reported to have synergy and some reported to not have synergy.
To measure antimicrobial synergy, one measures minimum inhibitory concentration (MIC) for growth in a broth dilution experiment. This is often done in a “checkerboard” type assay in which compounds A and B are serially diluted in the rows and columns of a multiwell plate, respectively, and the wells with no growth at the intersection between the effective concentrations of A and B are noted (29). While checkerboard synergy assays have been widely used, several authors have noted significant weaknesses in this approach (29–33), mainly arising from the fact that only one or a few wells in an entire plate actually provide information on synergy. This lack of statistical robustness causes the results to be sensitive to random experimental error. It also means that seemingly trivial differences in the method of interpreting checkerboard results can lead to very different conclusions, even for the same raw data (30, 31), an observation that we confirm below. These issues could explain why the literature on AMP-antibiotic synergy is ambiguous. For this work, we have developed a more robust synergy assay that circumvents many of the problems associated with checkerboard assays. This novel assay provides sensitive and statistically robust measurements of drug interactions with low sensitivity to random or systematic, day-to-day experimental error.
We use this novel assay to make an extensive set of statistically robust FIC measurements on all pairwise combinations of four well-characterized, membrane permeabilizing cationic antimicrobial peptides (AMP) and four classes of traditional chemical antibiotics against three species of bacteria, including both Gram positive and Gram negative species. In total, we measured 40 individual pairwise interactions. No statistically significant evidence for synergy or antagonism was detected in any combination; all pairs showed either simple additivity or independence of effects, suggesting that substantial membrane disruption caused by AMPs does not automatically lead to synergy, and does not fully explain the synergistic interactions observed for some peptide-drug pairs.
MATERIALS AND METHODS
Materials
Peptides were synthesized and purified by Biosynthesis, Inc. Purity and identity were independently verified by HPLC and MALDI mass spectrometry. The sequences of the peptide are as follows: *VAYR* = RRGWVLALYLRLYGRR; *ARVA = RRGWLALRLVLALY; VVRG = WVLVLRLGLY. Antibiotics were purchased from Sigma-Aldrich. Bacteria were obtained from ATCC and cultured/propagated according to standard protocols.
Measurement of MIC
Bacteria were grown to a log phase culture and then diluted to 4×105 in trypticase soy broth (TSB). Cells were added to each well of a plate containing serially diluted peptide, antibiotic or both. and the plates were incubated overnight at 37°C. Optical density measurements the next day showed a binary response, with almost all wells either opaque (OD600 ≥ 0.6) or transparent (OD600 ≤ 0.08). The rare wells with partial growth were considered to be not inhibited.
Fractional inhibitory concentration
FIC in a drug mixture is defined as
where MICs are measured for pure compounds and in mixtures of the two. FIC is not necessarily expected to be constant for all ratios of compounds A and B, however when the concentration ratio is similar to the MIC ratio, FIC values are relatively insensitive to small changes in the ratio between A and B. This is the regime where the experiments reported here are done.
Measurement of FIC
To the top row of a sterile 96-well plate we added 3xMIC of peptide in four columns, 3XMIC of antibiotic in four columns, and 1.5xMIC of each peptide and antibiotic mixed, in four columns. Then the top row was serially diluted row by row by a factor of 2/3. 4×105 bacterial cells/mL (as above) were added to each well and the plates were incubated overnight at 37°C. Optical density measurements the next day showed a binary response, with almost all wells either opaque (OD600 ≥ 0.6) or transparent (OD600 ≤ 0.08). Rare wells with partial growth or single microbial colonies were considered to be not inhibited.
Statistical analyses
The number of consecutive inhibited wells, starting from the highest concentration, rather than the actual drug concentration, was the raw data used in statistical calculations because well numbers are normally (Gaussian) distributed, while the concentrations calculated from them are not. For each set of experiments, we tabulated the number of unambiguously inhibited wells for peptide, for antibiotic and for the mixture. These values were entered into the program SynerStat, which calculates average dilution number (and standard deviation) for each column of peptide, antibiotic and mixture. (The analysis software is freely available from the authors upon request). FIC was calculated using a non-parametric, brute force numerical averaging of all the possible data combinations, giving a Gaussian-distributed mean FIC, SD and SE. Testing for rejection of potential outliers was done using Chauvenet’s criterion, but essentially no data points were rejected. The correct N is the equivalent number of unique experiments performed. In these experiments, there are 4 experiments for each 96 well plate. In all cases we used at least four plates (N≥16).
RESULTS AND DISCUSSION
Antimicrobial peptides, antibiotics and microorganisms
To test the hypothesis that membrane disruption will lead to synergistic interactions with chemical antibiotics, we use three well characterized peptides that were selected in a high-throughput screen for synthetic bilayer permeabilization(34). These cationic, interfacially active(35) peptides, referred to as *VAYR*, *ARVA and VVRG are 15, 12 and 9 residues long, respectively (see sequences above) with charges of +6, +4 and +2. They all bind selectively and strongly to microbes with partial β-sheet secondary structure and permeabilize their cytoplasmic membranes(15, 16). However some of the details of their actions vary. For example *VAYR* causes very rapid cytoplasmic membrane depolarization of E. coli (~20 sec). It also causes rapid entry of membrane impermeant SYTOX Green DNA binding dye into Gram positive and Gram negative bacteria within about 10 min. SYTOX Green is larger and more polar than most antibiotics(15) and thus is a good surrogate marker for antibiotic entry into cells. On the other hand, *ARVA does not cause membrane depolarization over the first 3 minutes, but by 5 minutes allows SYTOX Green into all bacteria tested. Interestingly VVRG, while broadly microbicidal, causes large scale permeabilization of Staph aureus membranes to SYTOX Green, but does not have the same effect in E. coli membranes. Despite the differences in apparent molecular mechanism, the peptide MIC values (which actually are minimum sterilizing concentrations) are all similarly in the low μM range. These AMPs have activity against all Gram positive and Gram negative microbes that we have tested, including drug resistant strains. They also have sterilizing activity against the pathogenic fungi Cryptococcus neoformans and Candida albicans (16, 34). Furthermore, these peptides have low cytolysis and cytotoxicity against mammalian cells(15).
The fourth peptide we tested was melittin, which belongs to a very different class. Melittin is a generic membrane permeabilizing (i.e. lytic) bee venom toxin of 26 residues that binds to and permeabilizes bacterial, eukaryotic and fungal membranes indiscriminately (36). Melittin is not a candidate AMP drug because it is highly toxic to human cells as well as bacteria. We use it here to assess whether an potently lytic class of membrane permeabilizing peptide can act synergistically with chemical antibiotics.
The chemical antibiotics tested were as follows: ampicillin (a β-lactam that inhibits cell wall synthesis), ciprofloxacin (a fluoroquinolone that inhibits DNA gyrase), streptomycin (an aminoglycoside that inhibits protein synthesis), and vancomycin (a glycopeptide that inhibits cell wall synthesis in Gram positive microbes). The site of action of ampicillin and vancomycin are outside of the cytoplasmic membrane, while ciprofloxacin and streptomycin act intracellularly. We used three strains of bacteria: E. coli (ATCC 25922) and Pseudomonas aeruginosa (ATCC 27853) as representative Gram negative organisms, and Staph. aureus (ATCC 25923) as a representative Gram positive microbe.
Weaknesses of the checkerboard assay
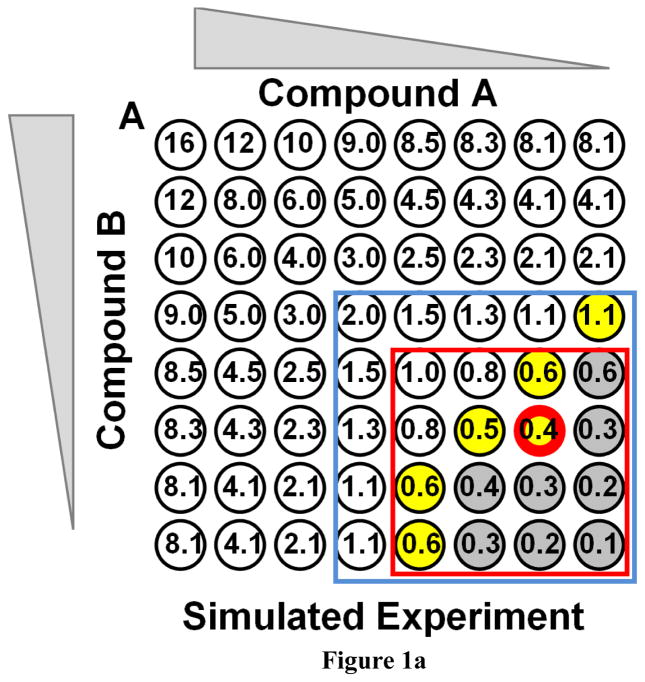
Mechanistic interactions between antibiotics are usually measured with some version of the broth dilution “checkerboard” assay. While this type of assay is convenient to perform, its weaknesses and statistical ambiguity have been well described in the literature (29–33). As an example, a simulated checkerboard antibiotic interaction assay is shown in Figure 1A. Hypothetical compound A is diluted by serial 2-fold steps horizontally and compound B is diluted in 2-fold steps vertically. Clear wells indicate the absence of growth (i.e. inhibition), while gray wells indicate growth. Yellow wells mark the last inhibited wells at the boundary between inhibition and growth. The numbers in the wells are the minimum FIC values that would be required to inhibit the microbe in that particular well in the array. The well bordered in red has the lowest FIC of any inhibited well. If there was no experimental variance, and if the drugs acted in an ideally additive manner (FIC=1), the red box would delineate the area that would not be inhibited. The blue box indicates wells that would not be inhibited if the drugs acted completely independently (FIC=2). In this simulated experiment, FIC has been set to 0.5, moderately synergistic, and a small, realistic amount of random experimental variation has been added
Figure 1.
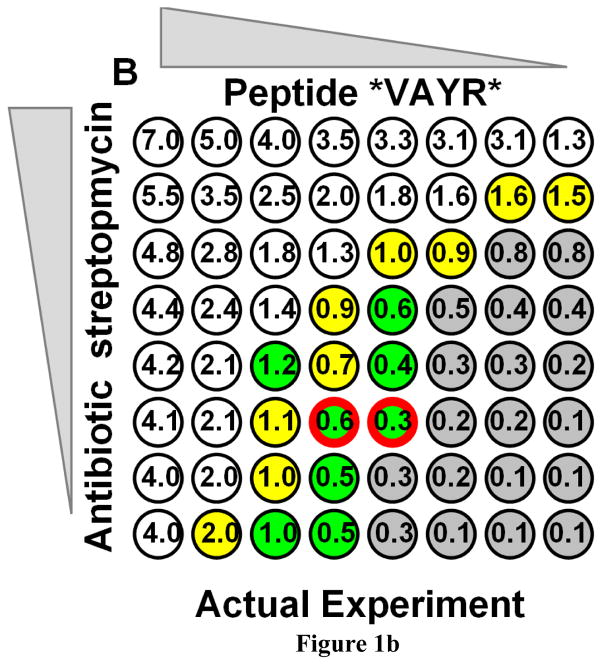
Checkerboard synergy assays. A: A simulated checkerboard synergy assay for antibiotic interactions. Both hypothetical compounds are diluted serially from 8 x MIC by a factor of 2 each step. Shown is a single result from a simulation to which a small random experimental error was added. Colorless wells are inhibited. Gray wells are not inhibited. Yellow marks the last inhibited wells at the boundary. The numbers in the wells are the minimum FIC values that would be required for inhibition of that well. The red bordered well is the single lowest FIC observed for an inhibited well. B: An actual checkerboard assay which was performed twice. *ARVA and streptomycin were serially diluted by two-fold from 15 μM each and used to inhibit Staph aureus. The color coding is the same as above except that green wells were inhibited in only one of two repeats.
In this work we use the simple mathematical definition of FIC < 1 for synergy, 1 ≤ FIC ≤ 2 for additivity to independence and FIC > 2 for antagonism. However we note that many authors follow more conservative guidelines and use the following values: Synergy: FIC ≤ 0.5; Additivity: 0.5 < FIC ≤ 2; Indifference: 2 < FIC ≤ 4; Antagonism FIC > 4. In the experiments in Figure 1A, if there is synergy between the drugs (FIC < 1) then additional wells inside the red box will be inhibited. The FIC values of the last inhibited wells at the interface between inhibition and no inhibition carry the information used to calculate FIC. However, this highlights one of the weaknesses of the checkerboard assay because there are at least four ways to interpret such an experimental result, as shown by White and colleagues(30, 32, 33). These various approaches to determining FIC from a checkerboard assay give very different answers with respect to synergy, even when applied to the same raw data (30–33). While the true FIC value lies between the last inhibited and first non-inhibited well, the boundary values vary significantly at the interface because the “slope” of the FIC change per row/column varies throughout the plate matrix. In the example shown in Figure 1A, to which we have added a small random experimental variance, the boundary FIC values range from 1.1 to 0.4. To analyze a checkerboard assay result, one can average all of the FIC values at the boundary (although there are several different ways to identify boundary wells(30)) which will provide what might seem like a statistically robust number, but one that will be far from a true random sampling of a parent population and thus does not actually have any statistical meaning. The average FIC will always be an overestimate of the true FIC. Alternately, researchers sometimes note the single well with the smallest FIC value that was inhibited. This approach will give a statistically weak number that is highly sensitive to experimental error (with N=1 for an entire 96-well plate), but one that may be closer to the true value. Finally, FIC values from checkerboard assays can be significantly affected by small systematic errors in the MIC values which are used to calculate FIC. While some authors measure MIC on the same plate as the checkerboard, reducing systematic errors due to day-to-day variations, others rely on previous MIC measurements to calculate FIC. Given the exponential nature of serial dilutions, small variations in MIC can lead to large systematic errors in FIC.
The weaknesses of the checkerboard assay are further exemplified by the actual checkerboard assay data shown in Figure 1B. In this case, we performed two independent checkerboard assays with an antimicrobial peptide and a chemical antibiotic against Staph. aureus under nominally identical conditions. Colorless wells in Figure 1B were inhibited in both plates, green wells were inhibited in only 1 of the 2 plates, and gray wells were not inhibited in either plate. Notice here again, the boundary wells (in yellow/green) range widely from FIC = 2.0 to 0.3. If we average the common boundary wells observed in both plates we obtain a minimum FIC of 1.2 +/− 0.4, indicating simple additivity. But these are not really independent measurements, thus the uncertainty has little physical meaning. If we average the set of boundary wells with the lowest FIC observed in either plate, then the minimum FIC = 0.8 +/− 0.5. If we average the lowest FIC single wells observed in each plate (red bordered wells in Figure 1B), we get FIC = 0.45, indicating synergy. But, since the SD is undefined in this case, we do not know the error in FIC value without analyzing additional individual plates. Thus, as discussed above(30–33), the very same data can have widely different interpretations in checkerboard assays (FIC for this one data set ranges from 1.2 to 0.45) and there is no universal consensus on how such assays should be interpreted. (Note: Using the new assay described here, we obtain FIC = 1.5 +/− 0.2 (SD, N=16) for this pair of compounds against Staph. aureus, see below).
A robust assay for antibiotic interactions
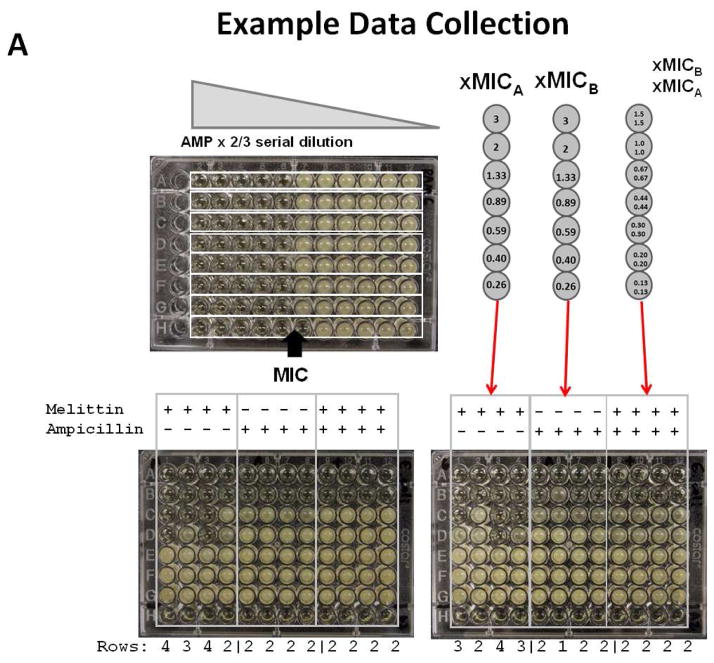
The assay we developed for this work was designed for increased statistical power in a multiwell plate-based measurement of FIC, and for circumventing the weaknesses of the checkerboard assay. To perform the assay, first the MIC of the two compounds are measured independently, as in Figure 2A. While these MIC determinations are used to design the assay plates, they are not used to calculate FIC. Next, 96-well assay plates are designed with three sets of four columns, where each set of columns contains compound A only, compound B only, or a mixture of compounds A and B. Compounds A and B are serially diluted from 3 x MIC down by a factor of 2/3. As such, the third row of each column has each compound present above the expected MIC (1.3 x MIC) and the fourth row is below the expected MIC (0.9 x MIC). In the absence of day-today experimental variance, the third row will be the last inhibited row for the pure compounds. Normal experimental variance for 2/3 serial dilution can be up to +/− 1 row, especially for peptides (see Figure 2). If the compounds are ideally additive, (FIC=1) the third row of mixture will also be the last inhibited row. If there is synergy, the mixture will inhibit more rows than the pure compounds. An average of one additional row will be inhibited for FIC=0.67, two additional rows will be inhibited for FIC = 0.44 and three additional rows will be inhibited for FIC = 0.3. If there is antagonism, the mixture will inhibit fewer rows than the pure compounds. Example plates for this assay are shown in Figure 2. After overnight incubation, inhibited wells have no growth and have low optical density. Non-inhibited wells have stationary phase growth and high optical density. Intermediate optical densities are very rare, thus the identification of inhibited wells is straightforward.
Figure 2.
Example assay plates from the novel antibiotic interaction assay we developed here. Top: Simple MIC measurement for a peptide antibiotic against Staph aureus. Each row is a repeat of a 2/3 fold serial dilution of melittin from 15 μM. Colorless wells have no growth (and no detectable CFUs). Opaque wells have stationary phase growth after overnight incubation. MIC is calculated by averaging the number of wells sterilized and then converting the mean to concentration. Bottom: Two interaction assay plates for the peptide melittin and ampicillin against Staph aureus. The first two sets of columns contain either peptide or antibiotic diluted by 2/3 serially from 3 x MIC. The third set of four columns contains a mixture of peptide and antibiotic diluted by 2/3 from 1.5 x MIC for each. Clear wells have no growth after overnight incubation. Opaque wells have stationary phase growth after overnight incubation. All calculations are done using the number of consecutive wells that were inhibited from the highest concentration.
By comparing pure compounds and mixtures side by side, this assay design eliminates the effect of errors or day-to-day fluctuation in MIC. In a typical MIC assay, there are many sources of variation that can lead to random or systematic errors in inhibition by at least one row in a 2/3 serial dilution assay. In a checkerboard assay, uncertainties or fluctuation in MIC can dominate the measurement of drug interaction because they directly enter into the calculation of FIC. In the new assay, the effect of systematic experimental errors is greatly reduced because we obtain FIC values directly using multiple side-by-side measurements of MIC using pure compounds and mixtures. Neither errors in the a priori knowledge of MIC, nor systematic errors in the measurement of MIC affect the determination of FIC in our assay.
We note here that the conditions where FIC is most sensitive to interactions is when both compounds are present in a ratio that is equal to the ratio of their MIC values. One potential disadvantage of this assay compared to the checkerboard assay is that the latter simultaneously explores multiple ratios of the two compounds being tested. However, large variations in synergy at ratios that are far from the ratio of pure MICs are extremely rare. To more accurately measure the MIC and FIC, we use small serial dilutions of 2/3. Finally, each plate contains four measurements of each compound’s MIC and four measurements of the mixture’s MIC. Thus four measurements of FIC are made on each plate by directly comparing mixtures with individual compounds on the same plate.
Statistical analysis
Another potentially significant source of error in synergy assays (actually in all serial dilution assays) arises from the fact that the inhibitory concentration values obtained in repeats of serial dilution experiments do not have a Gaussian probability distribution, and thus are not subject to Gaussian-based statistical analyses (Student’s t-tests, standard ANOVA etc.). Non-parametric analyses are required to analyze non-Gaussian data except at large N when the distribution of means approaches a Gaussian distribution. The statistical calculations that we perform here (e.g. MIC calculation) are done using the number of consecutive wells inhibited as the raw numerical data, (instead of the antibiotic concentration). The former do have normal (Gaussian) probability distribution (p-value for the Kolmogorov-Smirnov test for normality > 0.1). Each 96-well synergy plate (Figure 2) provides four measurements, each, of the number of wells inhibited for compound A and B and for the mixture. While repeat measurements on one plate are not necessarily expected to be statistically independent, we found by ANOVA that variations in calculated FIC values within plates are indistinguishable from the variation between plates measured on separate days, so they are effectively independent measurements. Each FIC measurement presented here is done using at least four plates total, prepared over at least two separate days from new stocks.
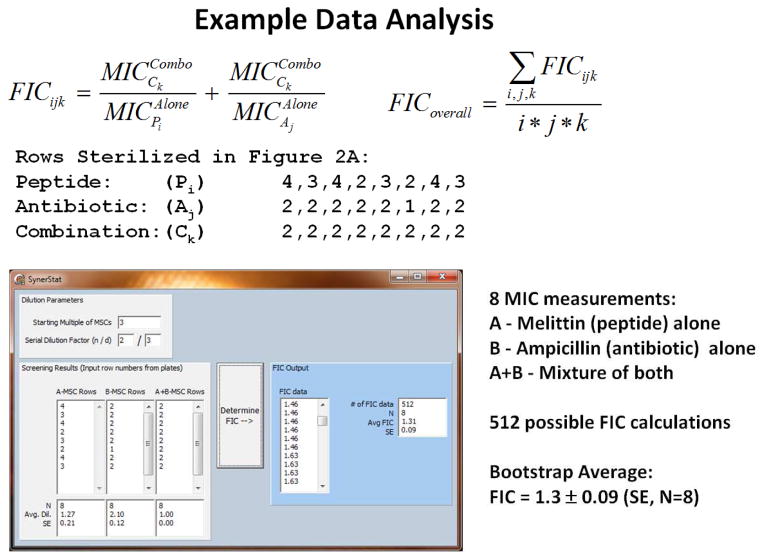
We use a numerical (non-parametric) bootstrap averaging of all possible FIC values using the individual values of wells inhibited from all experimental plates combined. This is shown in Figure 3. For calculation of p-values, the effective sample number, N, is 4 for each plate. Given the redundancy of MIC measurements in our assay, four independent plates provides data at N=16 from which we can obtain statistically robust measurements of FIC, standard deviation and standard error of the mean for each combination.
Figure 3.
Example data analysis for determining FIC. Top: Each column in a plate (see Fig. 2) provides a single unique MIC measurement. For any combination of peptide, antibiotic and combination columns, a single FIC can be calculated from the number of wells inhibited. The overall FIC average and standard deviation from a related set of assay plates can be calculated using a bootstrap approach that is based on the calculation of FIC for all possible combinations of experimental values. Middle: The table of values (number of rows sterilized) for the two example plates in Figure 2. In this example, the peptide is melittin (m) and the antibiotic is ampicillin. Bottom: The program SynerStat (freely available from the authors upon request) is used to calculate all possible individual FIC values using the number of rows inhibited. From the list of all possible FIC values, the overall average FIC is calculated along with SD and SE. For statistical calculations, the effective N is the number of columns assayed for each compound.
Lack of synergy between membrane permeabilizing AMPs and chemical antibiotics
Using the new assay, we measured the FIC and standard deviations for 40 peptide-drug-microbe combinations available from the list above. In Table 1, we show the MIC values for each compound, alone. The peptide MICs are all in the low μM range. Antibiotic MICs are mostly in the low μM range with a few exceptions. Ciprofloxacin has sub micromolar MIC values. Ampicillin, on the other hand, has low activity against the strain of Pseudomonas we used, requiring mM concentrations for inhibition. Vancomycin is not active against Gram negative microbes.
Table 1.
Minimum sterilization concentration (MIC, in μM) of tested antibiotics and antimicrobial peptides against bacteria.
| E. coli | S. aureus | P. aeruginosa | |
|---|---|---|---|
| Ampicillin | 18.3 ± 2.0 | 14.8 ± 2.7 | 1.4 ± 0.31 |
| Ciprofloxacin | 0.05 ± 0.01 | 1.8 ± 0.2 | 1.0 ± 0.2 |
| Streptomycin | 3.2 ± 1.8 | 8.9 ± 1.9 | 8.9 ± 1.8 |
| Vancomycin | N/A2 | 86.8 ± 8.0 | N/A2 |
| Melittin (26 aa) | 2.7 ± 0.6 | 1.5 ± 0.4 | 2.3 ± 0.5 |
| *VAYR* (15 aa) | 4.3 ± 2.2 | 5.8 ± 1.3 | 7.6 ± 1.5 |
| *ARVA (12 aa) | 3.9 ± 1.3 | 4.1 ± 0.8 | 7.8 ± 3.2 |
| VVRG (9 aa) | 2.9 ± 1.6 | 4.8 ± 0.8 | 7.2 ± 1.4 |
MIC of Ampicillin vs. P. aeruginosa is shown in mM;
Gram negative strains were not susceptible to Vancomycin.
In all cases uncertainties are standard deviations and N=16.
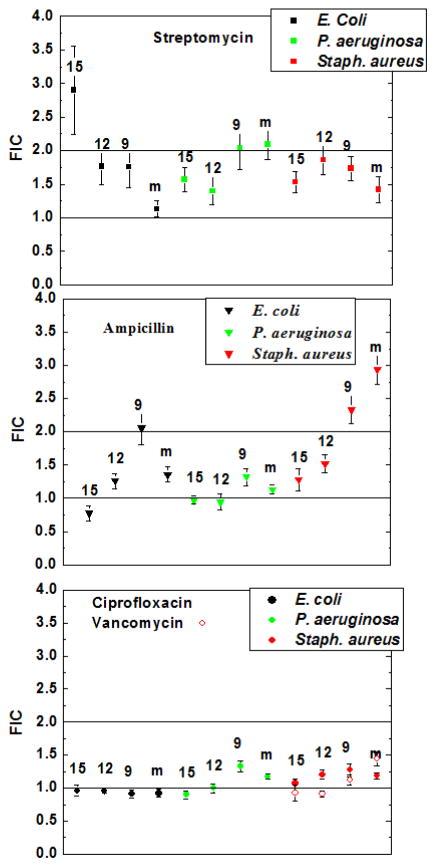
The 40 interactions are expressed as FIC values +/− SD in Figure 4 A, B and C. For each combination, at least four plates were analyzed, giving at least 16 independent measurements of the individual MIC, combined MIC and FIC. The results in Figure 4 A, B and C show that there was no detectable synergy between any pair. While a few measurements are below the mathematical cutoff of 1 for synergy, the statistical concept of multiple parallel comparisons indicates that this is likely a Type I error and not a statistically significant effect. A few measurements are above the mathematical cutoff of 2 for independence, suggesting antagonism, but again the differences are not statistically significant in the context of 40 comparisons.
Figure 4.
Measured fractional inhibitory concentrations (FIC) for all of the combinations of peptide, antibiotic and microbe studied in this work. In each case, at least four plates were analyzed giving a minimum N=16. Error bars are standard deviations calculated from the raw data. Horizontal lines on the plates indicate mathematically ideal independence (FIC=2) and additivity (FIC=1). Each panel shows data for a single drug, with vancomycin added to the third panel. Colors indicate different organisms and the markings above the points indicate the peptide: 15:the 15-residue peptide *VAYR*; 12: the 12-residue peptide *ARVA; 9: the 9-residue peptide VVRG; m: the 26 residue bee venom peptide melittin.
Comparison to published results
There are mixed reports in the literature of synergy between various AMPs and conventional antibiotics (26, 28, 37–44). The aggregated results of some representative large studies are given in Table 2. Synergy, defined by most authors as FIC < 0.5, is reported in roughly 1 in 5 experiments against Gram positive bacteria (mostly Staph. aureus) and in roughly 1 in 3 experiments against Gram negative bacteria (mostly E. coli and Pseudomonas aeruginosa). Many of the reported synergistic FIC values are between 0.3 and 0.5, slightly lower than the common threshold, thus statistical uncertainties are critical for assessment of the effects. Yet, most synergy studies, including the studies in Table 2, are striking in their complete lack of stated statistical uncertainties. Taken together, the lack of statistical uncertainties, and the large errors in FIC that can arise from small experimental errors (see above), means that most published FIC measurements for AMP/antibiotic interactions have unknown confidence intervals and unknown statistical significance. Further adding to the ambiguity, the large number of multiple comparisons in most synergy studies significantly increases the probability of type I statistical errors (i.e. false positives), an effect that can be accounted for, as we have done here, only if experimental uncertainties are known quantitatively.
Table 2.
Representative synergy results from the literature.
| Study | # of AMPs | # of Antibiotics | # Gram + sp(st) | FIC < 0.5 | # Gram – sp(st) | FIC <0.5 |
|---|---|---|---|---|---|---|
| This study, 2014 | 4 | 4 | 1 | 0/24 | 2 | 0/16 |
| Niu, 2013 (37) | 1 | 9 | 2 | 2/15 | 1 | 1/6 |
| Yenugu,2010 (38) | 2 | 10 | -- | -- | 1 | 20/20 |
| Sánchez,2010 (39) | 3 | 12 | -- | -- | 1 | 63/81 |
| van der Linden,2009(40) | 2 | 2 | 1 | 1/4 | 1 | 0/4 |
| Ulvatne 2001(41) | 6 | 8 | 1 | 1/40 | 1 | 9/40 |
| Giacometti 2000a(42) | 5 | 8 | 1(6) | 48/240 | 2(6) | 30/150 |
| Giacometti 2000b(28) | 5 | 8 | -- | -- | 1(3) | 15/120 |
| Giacometti 2000c(26) | 3 | 10 | -- | -- | 1(2) | 30/60 |
| Zhang,1999(43) | 9 | 2 | -- | -- | 2(4) | 26/72 |
| Scott, 1999(44) | 20 | 4 | -- | -- | 1 | 16/61 |
The number of bacterial species (sp) and strains (st) are shown, along with the number of combinations that are synergistic over the total tested, where synergy is defined empirically by most authors as FIC<0.5.
To enable a direct comparison between published values obtained with checkerboard assays and values obtained using the assay described here, we examined the literature for a peptide/antibiotic combination that was consistently reported to display synergy. In three papers published at nearly the same time by one laboratory, it was reported that the amphibian AMP magainin II interacted synergistically with all tested β-lactam antibiotics (26, 28, 42), but not with other antibiotics. Magainin II has a mechanism of action that may be similar to melittin but with greater selectivity for anionic microbial membranes (45). However unlike melittin, which we showed here does not act synergistically with chemical antibiotics, magainin was reported to have FIC values between 0.15 and 0.3 for interaction with about 10 different β-lactams in multiple strains of 5 different species of Gram positive and Gram negative bacteria. Strengthening the validity of the reported synergy between magainin and β-lactam interactions, in the same papers, synergy was not observed between magainin II and other classes of antibiotics. Nor was synergy observed between other AMPs, including the bovine neutrophil AMP indolicidin, and β-lactam antibiotics.
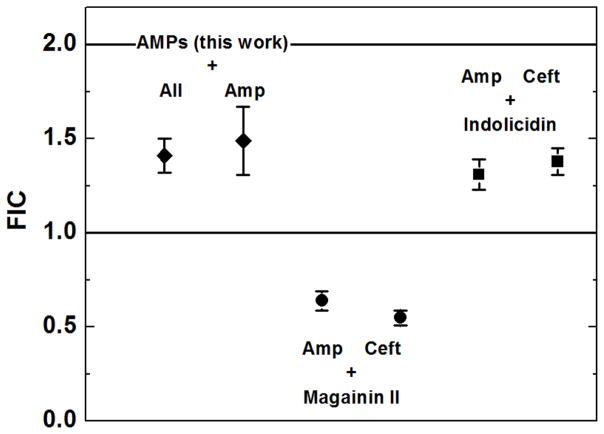
To make this direct comparison, we used the assay described here to measure the synergy between the peptides magainin II and indolicidin in the presence of either of two β-lactam antibiotics, ampicillin and ceftazidime. The results are shown in Figure 5, where we compare average FIC values of our peptides with the four new measurements. Qualitatively, these measurements agree well with the published results. Magainin II interacts synergistically with both β-lactam antibiotics, relative to the mathematical cutoff of FIC=1 for synergy (p<0.0001). In the magainin II assay plates (as in Figure 2), the combination columns showed sterilization in one or two additional rows, compared to the pure compounds. Indolicidin, like the other peptides we studied here, does not interact synergistically with β-lactam antibiotics (p>0.05). By ANOVA, both of the magainin II-antibiotic FIC values are significantly different from the FICs of the peptides measured here, against ampicillin, and from the FICs for indolicidin against the two β-lactams (p>0.001). At the same time, indolicidin’s two FIC values are not distinguishable from the average of the peptide-antibiotic combinations described here (p>0.05).
Figure 5.
Comparison of fractional inhibitory concentrations (FIC) against E. coli. The two points derived from this work are an average +/− SE (N=40) for all FIC measurements in Figure 4, or an average +/− SE (N=12) for all FIC using the β-lactam ampicillin. The magainin II and indolicidin points are FIC values +/− SE (N=16) for the combination of peptide and the β-lactams, ampicillin or ceftazidime. FICs were measured as described above. The lines at FIC=1 and 2 represent the mathematical boundaries for synergy and antagonism, respectively.
Mechanistic interpretation
In this work we tested the hypothesis that membrane disruption will lead to generic synergistic interactions between cationic antimicrobial peptides and chemical antibiotics. The literature is ambiguous with respect to addressing this hypothesis and the lack of statistical robustness in most of the literature makes interpretation difficult. To address this hypothesis, we developed a novel assay for antibiotic interaction that provides an unambiguous and statistically robust measurement of FIC for drug pairs in a relatively small number of microwell plates. Furthermore, we selected four antimicrobial peptides that are very well characterized with respect to bacterial membrane disruption , including knowledge of time-course and effective “pore size” (15, 16, 34, 46, 47). Our results show that bacterial membrane disruption by itself does not automatically give rise to synergy with any class of chemical antibiotic, including those that act intracellularly. The once example of synergy we observed here, which others have also observed, between magainin II and β-lactam antibiotics is unusual, suggesting a unique mechanism that, for the moment, remains unknown.
Highlights.
Membrane permeabilizing antimicrobial peptides could synergize with antibiotics
Commonly used checkerboard assay for synergy is not statistically robust
A novel assay provides robust quantitation of synergy
A set of membrane permeabilizing peptides do not act synergistically with antibiotics
Acknowledgments
We thank Ramesh Rathinakumar for his insights during the initiation of this work and Deepa Roy for performing some early checkerboard assays. We thank Bill Walkenhorst for many insightful conversations. We also thank Thomas C. Freeman for many insightful conversations, and for writing the data analysis software “Synerstat”. Funded by NIH GM60000.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- 1.D'Agata EM. Rapidly rising prevalence of nosocomial multidrug-resistant, Gram-negative bacilli: a 9-year surveillance study. Infect Control Hosp Epidemiol. 2004;25:842–846. doi: 10.1086/502306. [DOI] [PubMed] [Google Scholar]
- 2.Srinivasan A. Hunting the nightmare bacteria. PBS Broadcasting; 2014. [Google Scholar]
- 3.White SH, Wimley WC, Selsted ME. Structure, function, and membrane integration of defensins. Cur Opinion Struc Biol. 1995;5:521–527. doi: 10.1016/0959-440x(95)80038-7. [DOI] [PubMed] [Google Scholar]
- 4.Hancock RE, Sahl HG. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat Biotechnol. 2006;24:1551–1557. doi: 10.1038/nbt1267. [DOI] [PubMed] [Google Scholar]
- 5.Yount NY, Yeaman MR. Multidimensional signatures in antimicrobial peptides. Proc Natl Acad Sci. 2004;101:7363–7368. doi: 10.1073/pnas.0401567101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yeaman MR, Yount NY. Mechanisms of antimicrobial peptide action and resistance. Pharmacol Rev. 2003;55:27–55. doi: 10.1124/pr.55.1.2. [DOI] [PubMed] [Google Scholar]
- 7.Easton DM, Nijnik A, Mayer ML, Hancock RE. Potential of immunomodulatory host defense peptides as novel anti-infectives. Trends Biotechnol. 2009;27:582–590. doi: 10.1016/j.tibtech.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gardy JL, Lynn DJ, Brinkman FS, Hancock RE. Enabling a systems biology approach to immunology: focus on innate immunity. Trends Immunol. 2009;30:249–262. doi: 10.1016/j.it.2009.03.009. [DOI] [PubMed] [Google Scholar]
- 9.Hamill P, Brown K, Jenssen H, Hancock RE. Novel anti-infectives: is host defence the answer? Curr Opin Biotechnol. 2008;19:628–636. doi: 10.1016/j.copbio.2008.10.006. [DOI] [PubMed] [Google Scholar]
- 10.Jenssen H, Hamill P, Hancock RE. Peptide antimicrobial agents. Clin Microbiol Rev. 2006;19:491–511. doi: 10.1128/CMR.00056-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marr AK, Gooderham WJ, Hancock RE. Antibacterial peptides for therapeutic use: obstacles and realistic outlook. Curr Opin Pharmacol. 2006;6:468–472. doi: 10.1016/j.coph.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 12.Selsted ME, Ouellette AJ. Mammalian defensins in the antimicrobial immune response. Nat Immunol. 2005;6:551–557. doi: 10.1038/ni1206. [DOI] [PubMed] [Google Scholar]
- 13.Tang YQ, Yeaman MR, Selsted ME. Antimicrobial peptides from human platelets. Infect Immun. 2002;70:6524–6533. doi: 10.1128/IAI.70.12.6524-6533.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ouellette AJ, Selsted ME. Enteric defensins. Cur Opin Gastroenterology. 1997;13:494–499. [Google Scholar]
- 15.Rathinakumar R, Wimley WC. High-throughput discovery of broad-spectrum peptide antibiotics. FASEB J. 2010;24:3232–3238. doi: 10.1096/fj.10-157040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rathinakumar R, Walkenhorst WF, Wimley WC. Broad-spectrum antimicrobial peptides by rational combinatorial design and high-throughput screening: The importance of interfacial activity. J Am Chem Soc. 2009;131:7609–7617. doi: 10.1021/ja8093247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rausch JM, Marks JR, Rathinakumar R, Wimley WC. Beta-sheet pore-forming peptides selected from a rational combinatorial library: mechanism of pore formation in lipid vesicles and activity in biological membranes. Biochemistry. 2007;46:12124–12139. doi: 10.1021/bi700978h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang Y, Wiradharma N, Xu K, Ji Z, Bi S, Li L, Yang YY, Fan W, Pollard JE, Snarr J, Chaudhary V, Jennings JD, Shaw H, Christiansen B, Wright J, Jia W, Bishop RE, Savage PB. Cationic amphiphilic alpha-helical peptides for the treatment of carbapenem-resistant Acinetobacter baumannii infection. Biomaterials. 2012;33:8841–8847. doi: 10.1016/j.biomaterials.2012.08.026. [DOI] [PubMed] [Google Scholar]
- 19.Arias CA, Murray BE. Antibiotic-resistant bugs in the 21st century--a clinical super-challenge. N Engl J Med. 2009;360:439–443. doi: 10.1056/NEJMp0804651. [DOI] [PubMed] [Google Scholar]
- 20.Schwab U, Gilligan P, Jaynes J, Henke D. In vitro activities of designed antimicrobial peptides against multidrug-resistant cystic fibrosis pathogens. Antimicrob Agents Chemother. 1999;43:1435–1440. doi: 10.1128/aac.43.6.1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pollard JE, Snarr J, Chaudhary V, Jennings JD, Shaw H, Christiansen B, Wright J, Jia W, Bishop RE, Savage PB, Yang YY, Fan W. In vitro evaluation of the potential for resistance development to ceragenin CSA-13. J Antimicrob Chemother. 2012 doi: 10.1093/jac/dks276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Daniels R. Surviving the first hours in sepsis: getting the basics right (an intensivist's perspective) J Antimicrob Chemother. 2011;66(Suppl 2):ii11–ii23. doi: 10.1093/jac/dkq515. [DOI] [PubMed] [Google Scholar]
- 23.Ball P. Conclusions: the future of antimicrobial therapy - Augmentin and beyond. Int J Antimicrob Agents. 2007;30(Suppl 2):S139–S141. doi: 10.1016/j.ijantimicag.2007.08.016. [DOI] [PubMed] [Google Scholar]
- 24.Tangden T. Combination antibiotic therapy for multidrug-resistant Gram-negative bacteria. Ups J Med Sci. 2014 doi: 10.3109/03009734.2014.899279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zumla AI, Gillespie SH, Hoelscher M, Philips PP, Cole ST, Abubakar I, McHugh TD, Schito M, Maeurer M, Nunn AJ. New antituberculosis drugs, regimens, and adjunct therapies: needs, advances, and future prospects. Lancet Infect Dis. 2014;14:327–340. doi: 10.1016/S1473-3099(13)70328-1. [DOI] [PubMed] [Google Scholar]
- 26.Giacometti A, Cirioni O, Del Prete MS, Barchiesi F, Fortuna M, Drenaggi D, Scalise G. In vitro activities of membrane-active peptides alone and in combination with clinically used antimicrobial agents against Stenotrophomonas maltophilia. Antimicrob Agents Chemother. 2000;44:1716–1719. doi: 10.1128/aac.44.6.1716-1719.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lebel G, Piche F, Frenette M, Gottschalk M, Grenier D. Antimicrobial activity of nisin against the swine pathogen Streptococcus suis and its synergistic interaction with antibiotics. Peptides. 2013;50C:19–23. doi: 10.1016/j.peptides.2013.09.014. [DOI] [PubMed] [Google Scholar]
- 28.Giacometti A, Cirioni O, Barchiesi F, Scalise G. In-vitro activity and killing effect of polycationic peptides on methicillin-resistant Staphylococcus aureus and interactions with clinically used antibiotics. Diagn Microbiol Infect Dis. 2000;38:115–118. doi: 10.1016/s0732-8893(00)00175-9. [DOI] [PubMed] [Google Scholar]
- 29.Greco WR, Bravo G, Parsons JC. The search for synergy: a critical review from a response surface perspective. Pharmacol Rev. 1995;47:331–385. [PubMed] [Google Scholar]
- 30.Bonapace CR, Bosso JA, Friedrich LV, White RL. Comparison of methods of interpretation of checkerboard synergy testing. Diagn Microbiol Infect Dis. 2002;44:363–366. doi: 10.1016/s0732-8893(02)00473-x. [DOI] [PubMed] [Google Scholar]
- 31.Hsieh MH, Yu CM, Yu VL, Chow JW. Synergy assessed by checkerboard. A critical analysis. Diagn Microbiol Infect Dis. 1993;16:343–349. doi: 10.1016/0732-8893(93)90087-n. [DOI] [PubMed] [Google Scholar]
- 32.Bonapace CR, White RL, Friedrich LV, Bosso JA. Evaluation of antibiotic synergy against Acinetobacter baumannii: a comparison with Etest, time-kill, and checkerboard methods. Diagn Microbiol Infect Dis. 2000;38:43–50. doi: 10.1016/s0732-8893(00)00163-2. [DOI] [PubMed] [Google Scholar]
- 33.White RL, Burgess DS, Manduru M, Bosso JA. Comparison of three different in vitro methods of detecting synergy: time-kill, checkerboard, and E test. Antimicrob Agents Chemother. 1996;40:1914–1918. doi: 10.1128/aac.40.8.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rathinakumar R, Wimley WC. Biomolecular engineering by combinatorial design and high-throughput screening: small, soluble peptides that permeabilize membranes. J Am Chem Soc. 2008;130:9849–9858. doi: 10.1021/ja8017863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wimley WC. Describing the mechanism of antimicrobial peptide action with the interfacial activity model. ACS Chem Biol. 2010;5:905–917. doi: 10.1021/cb1001558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dempsey CE. The actions of melittin on membranes. Biochim Biophys Acta. 1990;1031:143–161. doi: 10.1016/0304-4157(90)90006-x. [DOI] [PubMed] [Google Scholar]
- 37.Niu M, Li X, Gong Q, Wang C, Qin C, Wang W, Chen P. Expression of 4kD scorpion defensin and its in vitro synergistic activity with conventional antibiotics. World J Microbiol Biotechnol. 2013;29:281–288. doi: 10.1007/s11274-012-1181-4. [DOI] [PubMed] [Google Scholar]
- 38.Yenugu S, Narmadha G. The human male reproductive tract antimicrobial peptides of the HE2 family exhibit potent synergy with standard antibiotics. J Pept Sci. 2010;16:337–341. doi: 10.1002/psc.1246. [DOI] [PubMed] [Google Scholar]
- 39.Sanchez-Gomez S, Japelj B, Jerala R, Moriyon I, Fernandez AM, Leiva J, Blondelle SE, Andra J, Brandenburg K, Lohner K, Martinez dT. Structural features governing the activity of lactoferricin-derived peptides that act in synergy with antibiotics against Pseudomonas aeruginosa in vitro and in vivo. Antimicrob Agents Chemother. 2011;55:218–228. doi: 10.1128/AAC.00904-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van der Linden DS, Short D, Dittmann A, Yu PL. Synergistic effects of ovine-derived cathelicidins and other antimicrobials against Escherichia coli O157:H7 and Staphylococcus aureus 1056 MRSA. Biotechnol Lett. 2009;31:1265–1267. doi: 10.1007/s10529-009-0010-9. [DOI] [PubMed] [Google Scholar]
- 41.Ulvatne H, Karoliussen S, Stiberg T, Rekdal O, Svendsen JS. Short antibacterial peptides and erythromycin act synergically against Escherichia coli. J Antimicrob Chemother. 2001;48:203–208. doi: 10.1093/jac/48.2.203. [DOI] [PubMed] [Google Scholar]
- 42.Giacometti A, Cirioni O, Del Prete MS, Paggi AM, D'Errico MM, Scalise G. Combination studies between polycationic peptides and clinically used antibiotics against Gram-positive and Gram-negative bacteria. Peptides. 2000;21:1155–1160. doi: 10.1016/s0196-9781(00)00254-0. [DOI] [PubMed] [Google Scholar]
- 43.Zhang L, Benz R, Hancock RE. Influence of proline residues on the antibacterial and synergistic activities of alpha-helical peptides. Biochemistry. 1999;38:8102–8111. doi: 10.1021/bi9904104. [DOI] [PubMed] [Google Scholar]
- 44.Scott MG, Yan H, Hancock RE. Biological properties of structurally related alpha-helical cationic antimicrobial peptides. Infect Immun. 1999;67:2005–2009. doi: 10.1128/iai.67.4.2005-2009.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Matsuzaki K, Sugishita K-I, Harada M, Fujii N, Miyajima K. Interactions of an antimicrobial peptide, magainin 2, with outer and inner membranes of Gram-negative bacteria. Biochim Biophys Acta. 1997;1327:119–130. doi: 10.1016/s0005-2736(97)00051-5. [DOI] [PubMed] [Google Scholar]
- 46.Krauson AJ, He J, Wimley WC. Determining the mechanism of membrane permeabilizing peptides: Identification of potent, equilibrium pore-formers. Biochim Biophys Acta. 2012;1818:1625–1632. doi: 10.1016/j.bbamem.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wiedman G, Herman K, Searson P, Wimley WC, Hristova K. The electrical response of bilayers to the bee venom toxin melittin: evidence for transient bilayer permeabilization. Biochim Biophys Acta. 2013;1828:1357–1364. doi: 10.1016/j.bbamem.2013.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]