Abstract
Cardiac muscle differentiation in vivo is guided by sequential growth factor signals, including endoderm-derived diffusible factors, impinging on cardiogenic genes in the developing mesoderm. Previously, by RNA interference in AB2.2 mouse embryonic stem cells (mESCs), we identified the endodermal transcription factor Sox17 as essential for Mesp1 induction in primitive mesoderm and subsequent cardiac muscle differentiation. However, downstream effectors of Sox17 remained to be proven functionally. In this study, we used genome-wide profiling of Sox17-dependent genes in AB2.2 cells, RNA interference, chromatin immunoprecipitation, and luciferase reporter genes to dissect this pathway. Sox17 was required not only for Hhex (a second endodermal transcription factor) but also for Cer1, a growth factor inhibitor from endoderm that, like Hhex, controls mesoderm patterning in Xenopus toward a cardiac fate. Suppressing Hhex or Cer1 blocked cardiac myogenesis, although at a later stage than induction of Mesp1/2. Hhex was required but not sufficient for Cer1 expression. Over-expression of Sox17 induced endogenous Cer1 and sequence-specific transcription of a Cer1 reporter gene. Forced expression of Cer1 was sufficient to rescue cardiac differentiation in Hhex-deficient cells. Thus, Hhex and Cer1 are indispensable components of the Sox17 pathway for cardiopoiesis in mESCs, acting at a stage downstream from Mesp1/2. Stem Cells 2014;32:1515–1526
Keywords: Cardiac development, Embryonic stem cells, Endoderm, Myogenesis, RNA interference
Introduction
The developmental restriction of primitive mesoderm to a cardiac muscle fate—whether in the embryo or in pluripotent cells—depends on signals from adjacent cell types, among them the developing endoderm 1–3. Notwithstanding differences in cardiac anatomy and even the body plan, the molecular cues responsible for cardiac induction are largely conserved across species, including bone morphogenetic proteins (BMPs), Activin and Nodal (a second branch of the BMP/transforming growth factor-β superfamily), fibroblast growth factors (FGFs), and Wnts. Previously, we demonstrated that an endoderm-associated Sry-box transcription factor, Sox17, was essential for cardiac specification in differentiating mouse embryonic stem cells (mESCs), depending on canonical Wnts and BMPs for its induction, and acting on cardiac myogenesis at least in part via cell-nonautonomous mechanisms upstream of Mesp1/2 4. Mesp1 and Mesp2, two closely related helix-loop-helix proteins, are among the earliest transcription factors that direct primitive mesoderm to a cardiovascular fate 5–8. A similar requirement for SOX17 was shown independently in human ESCs 9. However, the mechanism or mechanisms that communicate Sox17's effect on cardiac specification remain unproven.
A second endodermal transcription factor Hhex was contingent on Sox17 and is plausible as a candidate effector, although its functional role in ESCs has not been addressed 4. In Xenopus, a Wnt-activated Hhex pathway controls secreted signals for mesoderm patterning to a cardiac fate, acting in parallel with Nodal-dependent induction of Cerberus, a growth factor antagonist 10. Although Xenopus Cerberus is a broad-spectrum inhibitor of BMPs, Wnts, and Nodal, the mouse ortholog Cer1 inhibits Nodal and BMPs only 11.
What are the responsible effectors for the Sox17 pathway toward cardiac muscle differentiation in differentiating mESCs? Here, through genome-wide microarray analyses we identified Cer1, like Hhex 4, as a gene whose induction requires Sox17. We then used RNA interference to prove the requirement for Hhex and Cer1 in mesoderm patterning by ESCs, the requirement for Hhex in normal induction of Cer1, and the ability of ectopic Cer1 to rescue Hhex-deficient cells. We further show the direct binding of Sox17 to conserved sites in the Cer1 gene and demonstrate the induction of endogenous Cer1 by conditional expression of Sox17. Thus, Sox17 contributes to Cer1 expression both directly and, through Hhex, indirectly. Together, these results demonstrate an obligatory role for Hhex and Cer1 in differentiating ESCs, as mediators of the Sox17-dependent pathway for cardiac mesoderm formation.
Materials and Methods
Cell Culture
AB2.2 ESCs 12 were provided by Allan Bradley (Baylor College of Medicine) and DE14 Cripto−/− ESCs 13 by Eileen Adamson (Sanford-Burnham Medical Research Institute). Routinely, ESCs were cultivated in serum-containing medium as hanging droplets to form embryoid bodies, as previously detailed 4. ESCs were transduced with lentiviral vectors coexpressing enhanced green fluorescent protein (eGFP) with shRNA against the genes tested, or against firefly luciferase 4. Transduced cells were flow-sorted based on GFP fluorescence, grown as embryoid bodies, and transferred to tissue culture plates after 4.5 days 4. Expression profiling was performed after further culture for up to 10 days.
Where indicated, cells were plated directly as monolayers at 5 × 104 cells per milliliter using serum-free medium, containing 75% Iscove's modified Dulbecco's medium (Invitrogen, Carlsbad, CA; http://www.lifetechnologies.com), 25% Ham's medium F-12 (Invitrogen), 0.5× of supplements N2 and B27 (without retinoic acid) (Invitrogen), penicillin, streptomycin, 0.05% bovine serum albumin, 2 mM glutamine (Invitrogen), and 4.5 × 10−4 M 1-thioglycerol. Serum-free medium was replaced every 2 days. Recombinant Activin and Wnt3a were purchased from R&D Systems (Minneapolis, MN; http://www.rndsystems.com).
Microarray Analyses
Two independent samples were used for each condition, except where noted. Fluorescence intensities were captured with a GeneArray 2500 Scanner (Affymetrix, Santa Clara, CA; http://www.affymetrix.com). Differentially expressed genes were identified from log 2-expression data averaged over both replicates using a cross-sample variation >0.65 and a log 2-fold-change >1.2 across conditions for at least one time point. Genes were clustered according to their temporal structure following the procedure in Supporting Information Figure S1A.
RNA Interference
The modified pLL3.7 lentiviral vector (replacing the human cytomegalovirus promoter with the murine phosphoglycerate kinase promoter to drive GFP) and the Sox17 and firefly luciferase shRNA vectors were previously described 4. For Hhex and Cer1 shRNAs, the synthetic oligonucleotides and their reverse complement were annealed and ligated into the HpaI/XhoI sites of pLL3.7pgk, downstream from the murine U6 promoter. The two Hhex shRNAs target 5′-GCGTCTGGCCAAGATGTTA-3′ and 5′-GGTGCCTCTTTGGATCGTT-3′ in Hhex mRNA. The Cer1 shRNAs target 5′-GTCCAGAACAACCTTTGCT-3′, 5′-GATGGTGATGCAAGTAGAA-3′, and 5′-GGGATGTGGAAAGCGATCA-3′ in Cer1 mRNA.
For viral vector production, 20 µg of shRNA-encoding lentiviral backbone plasmid was transfected along with 15 µg of packaging vector psPAX2 and 10 µg of envelope vector pMD2.G (from Didier Trono, University of Geneva) into a 150 mm dish of 293T cells, using calcium phosphate. Culture supernatant containing lentiviral particles was collected 48 and 60 hours after transfection and combined. The pooled supernatant was first cleared of cell debris by centrifugation at 2,000 rpm (1,000g) for 5 minutes, and the subsequent supernatant subjected to ultracentrifugation at 25,000 rpm (82,705g) for 2 hours in a SW28 rotor (Beckman, Indianapolis, IN; https://www.beckmancoulter.com). The resulting pellet was resuspended in 136 µl phosphate-buffered saline, and the lentiviral vector stocks (>108 IU/ml) were stored at −80°C.
Lentiviral Vectors for Conditional Expression
To generate viral vectors for tetracycline-induced Sox17 expression, the tet transactivator-dependent vector plasmid pNLTREpitt-EGFP-ΔU3 14 was modified as follows. The SacII/BsrG1 fragment encoding eGFP was removed, the SacII/BsrG1 sites were blunted with T4 DNA polymerase, and a blunted BglII/NotI fragment of pIRES2-EGFP (Clontech, Mountain View, CA; http://www.clontech.com) containing IRES2-EGFP was inserted into the resulting sites. Conditional expression of this bicistronic vector (pNLTREpitt-IRES2-EGFP) is monitored by means of the Dox-induced eGFP fluorescence. A XhoI/NsiI fragment from pMSCVpuro (Clontech) was blunted and inserted into a blunted BamHI site of pNLrtTA2sM2, containing the rtTA2s-M2 tet transactivator sequence controlled by the constitutive human CMV-IE promoter; thus, cells harboring this vector, pNLrtTA2sM2-puro, are rendered subject to selection with puromycin. Wild-type Sox17 and the N3 and C1 truncation mutants were subcloned from pcDNA6.2 into the BamHI site of the pNLTREpitt-IRES2-EGFP, and lentiviruses were prepared as described above.
Conditional Expression of Sox17
ESCs were infected with pNLrtTA2sM2-puro then selected in ES culture media containing 1 µg/ml puromycin, from 3 to 5 days after infection. Surviving cells were infected with pNLTREpitt-IRES2-EGFP-derived vectors for wild-type or truncated Sox17 with a V5 epitope tag and then treated with 1 µg/ml Dox, after 3 days.
Chromatin Immunoprecipitation
Cyber-green PCR primers were designed for predicted Sox-binding elements 5′-(A/T)(A/T)CAA(A/T)−3′ 15 within the Cer1, Foxa1, and Foxa2 loci that are conserved in placental mammals (Supporting Information Table S3). Multispecies alignments and binding site detection were performed using Mulan and multi-TF in ECR Browser. Sox17 binding to the Cer1, Foxa1, and Foxa2 loci was determined using the EZ-ChIP kit (Millipore Corporation, Billerica, MA; http://www.millipore.com).
Results
Genome-Wide Expression Profiling of Sox17-Dependent Genes
For a transcriptome-wide assessment of Sox17-dependent genes, ESCs expressing Sox17 or luciferase shRNA were differentiated for up to 10 days by the embryoid body method, then were analyzed using Affymetrix microarrays (Supporting Information Fig. S1A and Table S1). Roughly 800 genes (4% of those tested) showed significant changes both across time and in response to Sox17 shRNA. The filtering thresholds were chosen guided by the change in five Sox17-dependent genes of known biological importance, which we had earlier identified by a candidate gene approach 4 (Foxa1, Mesp1, Nkx2−5, Mef2c, Myh6; Supporting Information Fig. S1B).
For the resulting overall set of Sox17-regulated genes, the significant gene ontology (GO) and GenMAPP terms are discussed below. Two GenMAPP categories requiring Sox17 were indicative of cell lineage, namely, striated muscle contraction (p = 1E-13) and smooth muscle contraction (p = 3E-05). Accordingly, many of the most affected GO Biological Process terms were related to cardiovascular development and function (Fig. 1A; 16 of the top 40 including: heart morphogenesis, p = 3E-17; heart development, p = 1E-14; vasculogenesis; p = 2E-10). Other highly dependent categories were related to more generic events (multicellular organismal development, p = 4E-30; regulation of transcription, DNA dependent, p = 3E-16; cell fate commitment, p = 5E-07), or, notably, endoderm development (p = 9E-05). Numerous pathways for growth factor signaling were Sox17-dependent (SMADs, p = 1E-06; Wnts, p = 5E-06; BMPs, p = 7E-05; transforming growth factor beta, p = 1E-03). As further steps to refine and visualize these results, the filtered genes were also clustered according to their dynamic profiles (Supporting Information Fig. S1C, S1D) and the temporal clusters then subjected to GO analysis (Supporting Information Fig. S2A). Cluster III, comprising transiently expressed genes with an onset between days 2 and 4, was notably enriched for the GO processes endoderm formation, ectoderm formation, embryonic heart tube morphogenesis, and heart looping (Supporting Information Figs. S2B, S3).
Figure 1.

Sox17-dependent genes in differentiating ESCs.(A): The top 20 GO biological process terms that were dysregulated in Sox17-deficient mouse ESCs. Those specific for cardiovascular development and function are highlighted (black), and a p-value of 1E-5 is noted for reference (Supporting Information Figs. S1, S2). (B): Heat map of gene expression levels for 126 Sox17-regulated genes from a curated gene set related to cardiac myogenesis (Supporting Information Table S2). Genes that fulfill the filtering criteria in Supporting Information Figure S1A are presented, grouped according to the temporal clusters obtained from the whole-transcriptome analysis (Supporting Information Fig. S1D). Functional annotations are shown at the right for transcription factors, extracellular/membrane proteins, and muscle-specific genes for cardiac contractility. For the complete set of affected genes refer Supporting Information Figure S3. Sox17, Hhex, and Cer1 are highlighted. Abbreviation: GO, gene ontology.
Next, the changes contingent on Sox17 were scrutinized using a manually curated set of 445 genes relevant to cardiac myogenesis, antecedent processes, and selected relatives for multigene families (Supporting Information Table S2). Significant dysregulation was observed in 28% of the genes, that is, enriched sevenfold compared to the unbiased 22K chipset (Fig. 1B). Endogenous Sox17 and its direct targets Foxa1 and Foxa2 were suppressed, as expected (preconditions for the knockdown experiment to be valid). Moreover, the genome-wide analysis and specific conditions chosen for data mining were sufficient to capture all the Sox17-dependent genes we had found previously by a limited candidate gene approach 4. In addition to just the representative markers Myh7 and Ryr2, the lack of Sox17 broadly downregulated the genes for diverse cardiac thick filament proteins (Mybpc3, Myh6, Myl2/3/4/7, Mylk3, Myom1, and Ttn), thin filament proteins (Actc1, Actn2, Tnnc1, Tnni1/3, Tnnt2, and Tpm1/2), Z disc proteins (Csrp3/Mlp), Nppa, and regulators of Ca2+ homeostasis (Atp2a2, Pln, and Srl).
With regard to cardiogenic transcription factors and their coactivators, the lack of Sox17 resulted in suppression of Foxc1/2, Gata4/5/6, Hopx, Irx3/5, Isl1, Smarcd3/Baf60c, and Smyd1/Bop, in addition to the three factors examined previously (Mesp1, Mef2c, and Nkx2–5). Unlike those mentioned above, Hand1/2 and Tbx20 were upregulated, concomitant with other genes for neural development, consistent with their additional roles, respectively, in neural crest and motoneurons 16. Thus, in unbiased genome-wide testing, Sox17 expression in ESCs was a prerequisite for the induction of highly diverse cardiogenic transcription factors and cardiac structural genes.
Demonstrated by induction of T, Eomes, Fgf8, Gsc, Cdx2, and Mixl1 to normal or increased levels, suppressing Sox17 did not prevent the induction of primitive mesoderm, mesendoderm, or the primitive streak (Fig. 1B, Supporting Information Fig. S3). In the case of Eomes, a direct activator of Mesp1 7,17, this may be due to loss of a known negative feedback loop 5. Several upregulated mesodermal genes were related to hematopoiesis (Etv2, Fli1, Hoxa9, Hoxb6, Hoxc8, Hba-a1/2, Hba-x, Hbb-bh1, and Hbb-y). Notably, however, key early markers of the multipotential cardiovascular progenitor cell were suppressed (Kdr/Flk1, Pdgfra) 18,19. Thus, taking these results together, genome-wide profiling substantiates that Sox17 specifically affects the direction of mesoderm toward a cardiovascular fate, not mesoderm formation per se 4.
In addition to Foxa1/2, discussed above, multiple markers of early endoderm (Cldn6, Dpp4, Epcam, Foxg1, Nr2f1, Rhox5, and Sparcl1), definitive endoderm (Cd24a, Cxcr4, Foxa1/2, and Kitl), and visceral endoderm (Afp, Cited1, Dab2, Fxyd3, Hesx, and Ttr) were inhibited, the latter notably including Cer1 (Fig. 1B; Supporting Information Fig. S3, cluster III). Several of the endodermal genes cited are reportedly direct targets of Sox17 by chromatin immunoprecipitation (ChIP) and whole-genome promoter tiling arrays 20, although whether Sox17 directly activates Cer1 is unsubstantiated. Other Sox17-dependent genes included two related F box family members, Sox18 and Sox7, with which Sox17 can be redundant 21,22. Given these genes' sequence similarity, we confirmed that the Sox17 shRNAs have no promiscuous effects on cotransfected Sox18 4 or Sox7 (Supporting Information Fig. S4A). Thus, under the conditions tested, lack of Sox17 downregulates the redundant family members that are required in concert with Sox17 during embryogenesis.
Because the action of Sox17 in mesoderm patterning is cell-nonautonomous 4, we next tested for operation of a secretory pathway, as opposed to ones requiring cell-cell contact. Doxycycline- (Dox-) dependent “inducer” ESCs bearing Sox17 gain-of-function mutations were able to upregulate Nkx2–5 and Myh6 in wild-type “responder” ESCs, across a semipermeable membrane (Fig. 2A). Thus, Sox17 is sufficient to promote cardiac muscle differentiation from ESCs via one or more soluble signals. In Xenopus embryos, the endodermal genes that most conclusively regulate secreted signals for cardiac specification by primitive mesoderm—the stage regulated by Sox17—are Hhex (whose relevant target is unknown) acting in parallel with Cer1, induced by Wnt antagonists and Nodal, respectively 10,23. Using the transmembrane induction assay, Hhex, like Sox17, was sufficient to upregulate Nkx2–5 and Myh6 in responder cells (Fig. 2A). As a prelude to more detailed investigation of these two putative effectors and their potential relationship, we substantiated our microarray finding that Cer1, like Hhex, depended on Sox17 using quantitative real-time RT-PCR (QRT-PCR; Fig. 2B).
Figure 2.
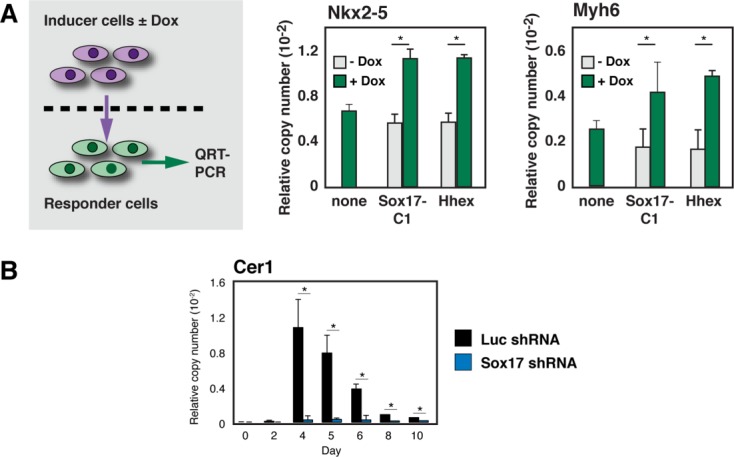
Sox17 regulates secreted signals for cardiac differentiation. (A): (Left) Schematic cartoon of the experimental design. For details of the Sox17 gain-of-function mutation refer Figure 4D and Supporting Information Figure S5B–S5D. (Right) QRT-PCR results for Nkx2–5 and Myh6. (B): Corroboration by QRT-PCR of Cer1 induction as contingent on Sox17. *, p < .05 versus control cells; n ≥ 3. Abbreviation: QRT-PCR, quantitative real-time RT-PCR.
Hhex and Cer1 Act in Series for Mesoderm Patterning to a Cardiac Fate in Differentiating ESCs
To test the requirement for Hhex and Cer1 by RNA interference, preparatory studies confirmed the shRNAs' effect on cotransfected Hhex, these sequences were retested as recombinant lentiviruses, and a block to endogenous Hhex was confirmed (Supporting Information Fig. S4B). The consequences of Hhex shRNA shown by QRT-PCR strongly resembled those of blocking Sox17 (Fig. 3A): (a) suppression of cardiac structural genes (Myh6 and Ryr2), (b) suppression of cardiogenic transcription factors (Nkx2–5, Myocd, and Mef2c), (c) lack of interference with downregulation of stemness factors (Oct4 and Sox2), and (d) failure to inhibit T, implicating one or more steps later than the formation of primitive mesoderm.
Figure 3.
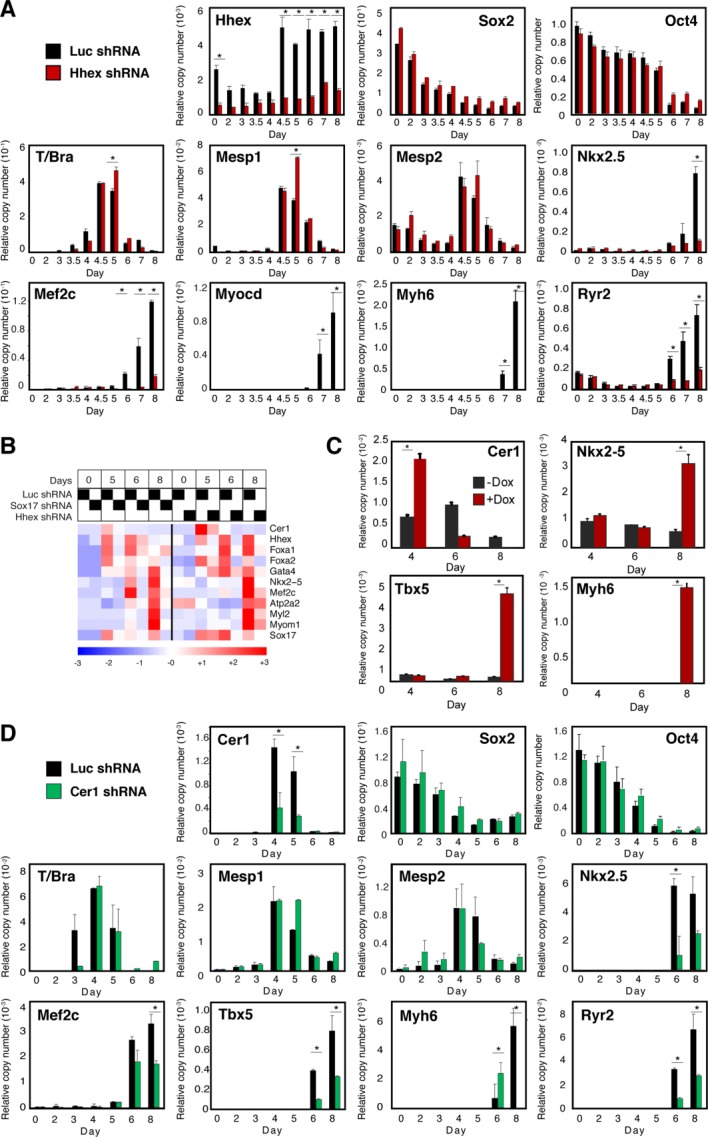
The Sox17-dependent genes Hhex and Cer1 are important for cardiac myogenesis in differentiating ESCs. (A–C): Hhex and (D) Cer1 shRNA suppressed the respective cognate genes in differentiating ESCs and inhibited the induction of cardiac transcription factors and structural genes, acting at a stage subsequent to induction of Mesp1/2. *, p < .05 versus control cells; n ≥ 3. (A, D): Results are shown for the most potent of the shRNAs tested, measured by effectiveness against the endogenous transcripts. For each gene, qualitatively similar results were obtained using at least two independent shRNAs. (B): Partial comparison of the microarray findings with Hhex and Sox17 shRNAs, illustrating the shared impairment of Cer1, cardiac transcription factors, and cardiac structural genes. In addition, a potential positive feedback loop between Hhex and Sox17 is noted. n = 2 for Hhex shRNA; n = 1 for the Luc shRNA controls. (C): Ectopic Cer1 expression rescues cardiac differentiation in Hhex-knock down ESCs. Cer1 was encoded by a tetO-regulated lentiviral vector, and was induced on day 3 by doxycycline. Gene expression was assayed by QRT-PCR. *, p < .05 versus control cells; n ≥ 3.
Among the few differences from Sox17-deficient ESCs 4, Hhex-deficient ones showed little or no loss of Mesp1/2. Thus, these results suggest that Sox17 acts on Mesp gene induction, whereas Hhex mediates a later stage. In preliminary microarray analyses, further cardiogenic transcription factors and cardiac structural genes were suppressed by Hhex shRNA, and, notably, like Sox17, Hhex was required for the normal induction of Cer1 (Fig. 3B). Conversely, forced expression of Cer1 was sufficient to rescue cardiac differentiation in Hhex-deficient ESCs, as shown by Nkx2–5, Tbx5, and Myh6 expression (Fig. 3C). Thus, Cer1 stands out among the plausible candidates to explain the impact of Sox17, Hhex, or both on cardiac myogenesis.
As done for Hhex, we selected shRNAs against Cer1 and proved their efficacy in transfected 293T cells (Supporting Information Fig. S4B). After lentiviral delivery into ESCs, each shRNA inhibited endogenous Cer1, cardiogenic transcription factors, and cardiac structural genes (Fig. 3D), identical to the results obtained in Hhex-deficient cells. Like Hhex, Cer1 shRNAs did not suppress the tested markers of “stemness” (Oct4 and Sox2), primitive mesoderm (T), or precardiac mesoderm (Mesp1/2). Thus, Cer1—like Hhex—acts at a later stage than the conversion of primitive mesoderm to Mesp-expressing mesoderm and, consequently, later than Sox17. We saw no difference between shRNA-mediated silencing of Hhex versus Cer1, based on the candidate genes studied in both backgrounds. These results do not exclude differences that could emerge from broader surveys or genome-wide profiling. The combined knockdown of Hhex and Cer1 was similar qualitatively and quantitatively to that of either alone (not shown), consistent with the ability of Cer1 to rescue Hhex-deficient cells under these conditions. The lack of any additive effect suggests that Hhex and Cer1 act in series, an interpretation supported strongly by the Cer1 rescue experiment.
Sox17 Couples the Activin/Nodal Pathway to Hhex, Cer1, and Cardiac Specification
In all the experiments above, the requirement for a Sox17-Hhex-Cer1 circuit was demonstrated in differentiating embryoid bodies after aggregation in serum-containing medium, that is, conditions that are spontaneous but biochemically undefined. To ascertain whether this Sox17-Hhex-Cer1 circuit might be essential even if an exogenous differentiating signal were provided, we first tested for induction of these genes in serum-free monolayer culture 24 containing 25 ng/ml Activin 25 (Fig. 4A). Along with transient induction of T on day 4, Activin induced sustained expression of Sox17, Hhex, and Cer1. As expected, Activin was sufficient to provoke a cardiomyocyte phenotype, denoted by Nkx2–5, Tbx5, and Myh6 at day 7. Thus, in addition to the later expression of cardiac markers, Activin elicited the prior induction of Sox17, Hhex, and Cer1.
Figure 4.
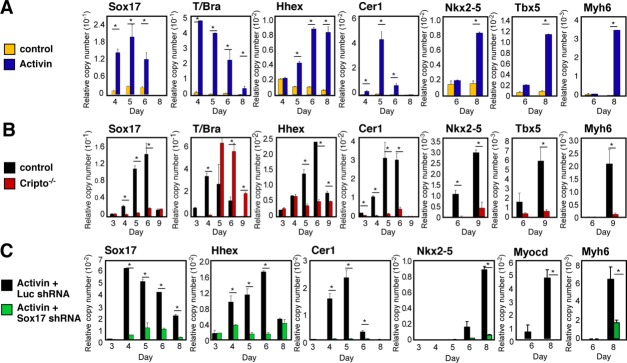
Sox17 mediates the Activin/Nodal pathway for cardiac myogenesis. (A): Induction of Sox17, Hhex, Cer1, and cardiac genes in Activin-treated embryonic stem cells (ESCs). (B): The Nodal receptor Cripto is essential for induction of the endoderm-associated Sox17-Hhex-Cer1 pathway. (C): Sox17 shRNA recapitulates the Cripto-deficient phenotype in Activin-treated ESCs. Cells were grown in monolayer culture for panels (A) and (C), and as embryoid bodies for panel (B). *, p < .05 versus control cells; n ≥ 3.
Conversely, to assess whether the Nodal/Activin pathway was required to induce the Sox17-Hhex-Cer1 module, we used homologous-null ESCs lacking the coreceptor Cripto. As reported 26, cardiac transcription factors (Nkx2–5 and Tbx5) and Myh6 were not induced in the absence of Cripto (Fig. 4B). Similarly, the lack of Cripto reduced Sox17, Hhex, and Cer1 each by 80%–90% (Fig. 4B). By contrast, T was expressed at levels even higher than in wild-type cells, although delayed by 1 day. Thus, in addition to canonical Wnts and BMPs 4, a third signal is essential to confer Sox17 induction, namely the Nodal/Activin cascade.
To verify whether Sox17 is essential for the induction of Hhex and Cer1 by exogenous Activin, we next compared control and knockdown ESCs in the serum-free monolayer cultures (Fig. 4C). Corresponding to the requirement for Sox17 in embryoid bodies (Fig. 1B; Supporting Information Fig. S3, cluster III), suppressing Sox17 likewise prevented the induction of Hhex and Cer1 by recombinant Activin. Despite forced stimulation of the Activin/Nodal pathway, suppressing Sox17 resulted in the failure of cardiac myocyte differentiation, measured here by Nkx2–5, Myocd, and Myh6 (Fig. 4C). Together, these complementary results clearly position Sox17 upstream from both Hhex and Cer1, mammalian counterparts of the endodermal signals for cardiac myogenesis in Xenopus, and likely explain the absence of Cer1 seen in Cripto-deficient ESCs 27.
Sox17 Binds and Activates Endogenous Cer1
To distinguish whether Sox17 confers expression of Cer1 only indirectly, via Hhex, or also acts on Cer1 directly, we performed Sox17 ChIP assays, using (A/T)(A/T)CAA(A/T) sites conserved across the mouse, canine, rhesus, and human Cer1 genes 28, guided by the (A/T)(A/T)CAA(A/T)G consensus binding site for the Sox family 15 and the ATTGT core site for Sox17 itself 20 (Fig. 5A; Supporting Information Table S3). Epitope-tagged Sox17 was transduced into ESCs using a tetracycline-inducible lentiviral system, to obviate potential confounding effects of constitutive expression. Negative controls were randomly selected regions lacking this motif, remote from predicted binding sites. Predicted Sox sites from the Foxa1 and Foxa2 loci (Fig. 6A) also were assayed, as these genes are proven direct targets of Sox17 20. In concordance with identification of Cer1 as a potential Sox17 target by ChIP-chip, albeit along with 1,800 other genes 20, we specifically confirmed Sox17 binding at each of the predicted sites we tested from the upstream 6 kbp, with enrichment at least equal to that for the sites in Foxa1/2 (Fig. 5B). Thus, predicted binding sites in the upstream region bind Sox17 efficiently.
Figure 5.
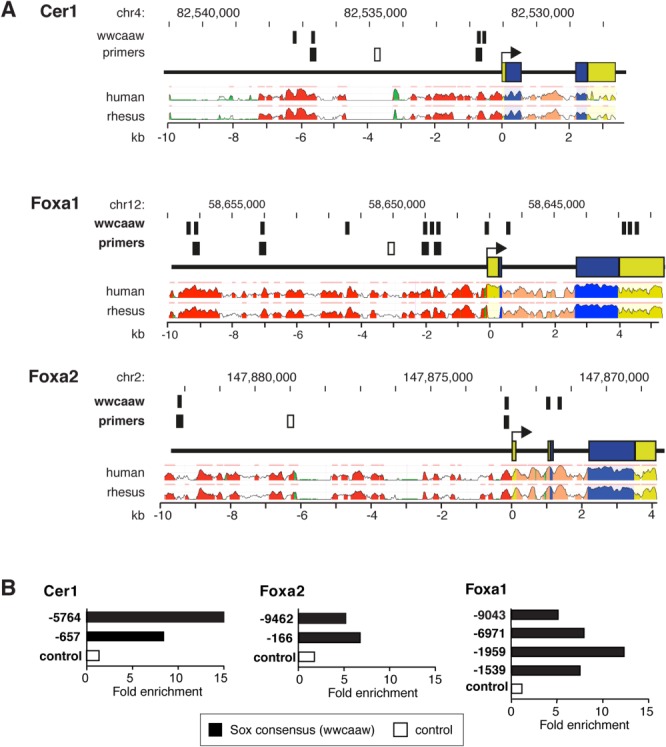
Sox17 binds to evolutionarily conserved Sox sites in the Cer1 upstream region. (A): Predicted Sox17 binding sites in the Cer1, Foxa1, and Foxa2 loci. Primers corresponding to predicted binding sites versus irrelevant control regions are indicated in black and white, respectively. Conservation profiles are shown for the human and rhesus orthologs (range, 50%–100%). Pink bars above the profiles denote regions of conservation with the mouse genome; blue, coding exons; yellow, untranslated regions; salmon, introns; red, intergenic regions; green, transposons and simple repeats. (B): Chromatin immunoprecipitation, assayed by quantitative PCR, shown as the fold enrichment for the indicated regions (V5 antibody, normalized for nonspecific precipitation by nonimmune IgG). Black, predicted Sox binding sites; white, irrelevant regions. For Foxa1 and Foxa2 refer Figure 6A.
Figure 6.
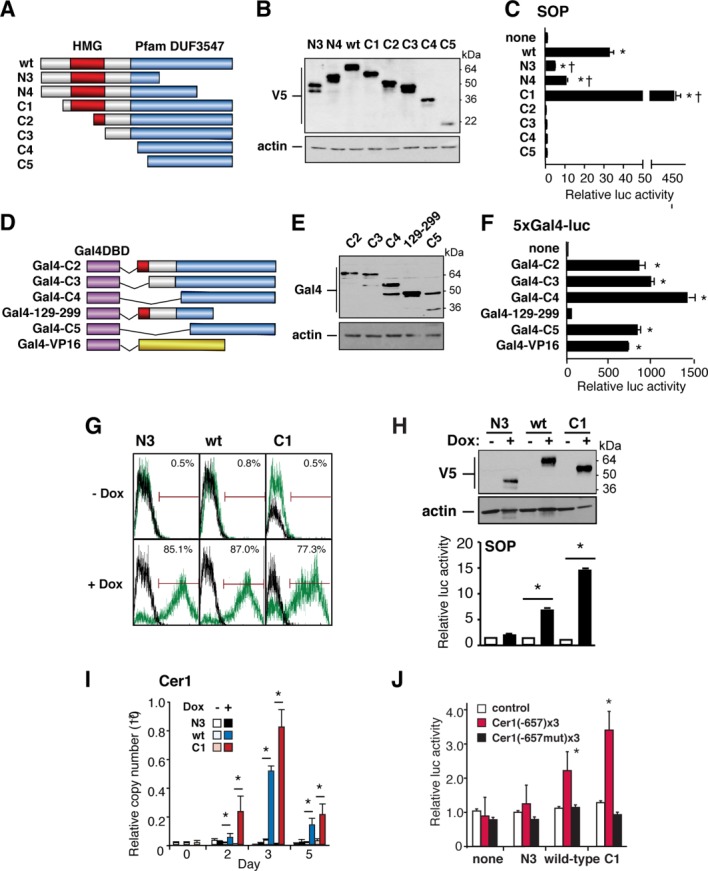
Sox17 activates Cer1. (A–F): Mapping the Sox17 transactivation domain. (A) Schematic representation of the Sox17 deletion mutants. Domain of unknown function 3547 (Pfam 24.0) designates the conserved C-terminal region of F group Sox proteins. (B): Western blot analysis of the constructs in 293T cells. (C): Sox-dependent reporter gene activity (SOP-FLASH) in 293T cells, in the presence of cotransfected Sox17 expression vectors. Deletion of the N terminus (C1) increases the transcriptional activity. Deletions of the conserved C-terminal domain (N3, N4) attenuate transactivation. (D): Schematic representation of the GAL4DBD-Sox17 fusion proteins. (E): Western blot analysis of the constructs in 293T cells, using antibody to Gal4. (F): Reporter gene activity (5xGal4-luc) in 293T cells, induced by GAL4-Sox17 vectors. Deletion of the Sox17 C-terminal domain (GAL4 129–299) cripples transactivation. Other constructs showed activity equal to or greater than that of GAL4-VP16. (G, H): Doxycycline-dependence of the Sox17 vectors, measured in AB2.2 cells by flow cytometry (G), Western blotting (H, above), and transactivation of SOP (H, below). (I): Wild-type Sox17 and the C1 truncation both induce endogenous Cer1. n ≥ 3; *, p < .05 versus control embryonic stem cells. (J): The multimerized Sox17 site at −657 of the Cer1 locus mediates Dox-dependent, sequence-specific trans-activation. n = 6; *, p < .01 versus the absence of Dox.
To test whether exogenous Sox17 suffices to induce Cer1, we first mapped the transactivation domain of mouse Sox17, based on the structural organization of XSox17β and other Sox proteins 29 (Fig. 6A, 6B). Each construct was cotransfected into 293T cells with the Sox-dependent luciferase reporter SOP, containing a concatamer of CTTTGTT (an inverse of AACAAAG) 30 (Fig. 6C). Activation was obtained only with wild-type Sox17 or an N-terminal truncation that retains both the DNA binding domain (HMG box) and C-terminus (“C1”). C1 was 14-fold more potent than wild-type Sox17, suggesting the presence of auto-inhibitory elements in the N terminus. None of the constructs activated the inactive control reporter, NOP.
To confirm the putative function of this C-terminal activation domain, we constructed chimeric expression vectors encoding the Sox17 truncations in frame with the Gal4 DNA-binding domain (DBD; Fig. 6D, 6E). Tested using a Gal4-dependent luciferase reporter gene, four of the five fusion proteins—all those preserving the distal C-terminus—were at least as potent as the Gal4-VP16 control, but the C-terminal residues 129–299 were inactive, similar to the Gal4DBD alone (Fig. 6F). Thus, the Sox17 trans-activation domain is located in the distal C-terminus.
On this basis, wild-type Sox17, the N-terminal portion N3, and the C-terminal portion C1 were compared for their ability to induce endogenous Cer1, using Dox-dependent lentiviral vectors. Flow cytometry for eGFP 1 day after Dox administration demonstrated that roughly 80% of the cells were successfully induced (Fig. 6G), versus 0.5%–0.8% for cells without Dox. All three Sox17 proteins were induced efficiently, without discernible leak (Fig. 6H, upper panel). In agreement with their respective activity toward the SOP reporter (Fig. 6H, lower panel), viruses encoding wild-type Sox17 and C1 conferred Dox-dependent induction of endogenous Cer1 (16–120-fold; Fig. 6I), with C1 (lacking the auto-inhibitory domain) being even more potent than the wild-type protein and N3 (lacking the activation domain) being altogether ineffective.
To determine whether Sox17 can specifically activate the binding sites verified in the Cer1 locus, the inducible expression vectors were then also tested against Cer1 luciferase reporter genes. Construction was based on the SOP reporter, containing instead a 3× multimer of the Cer1 proximal or distal Sox17 binding site, and using each in its wild-type or mutationally inactivated form (−657, Fig. 6J; −6017, not shown). Both wild-type Sox17 and the gain-of-function mutation C1 evoked Dox-dependent, sequence-specific trans-activation via the multimerized proximal site. No induction was seen if the Sox17 binding motif was mutated, a minimal promoter was tested, or N3 was used, lacking the trans-activation domain. As the distal Sox17 site was activated by C1, but not wild-type Sox17, its activation should be interpreted more cautiously, even though sequence-specific. Together with our evidence that Sox17 specifically binds the Cer1 promoter, these gain-of-function studies indicate that Sox17 may directly drive Cer1 transcription, at least in part through the proximal Sox17 site. None of the Sox17 expression vectors upregulated endogenous Hhex (data not shown), suggesting that Sox17 is required but not sufficient for Hhex induction.
Discussion
We and others have shown previously that Sox17 is essential for cardiac mesoderm specification in ESCs, working through a cell nonautonomous mechanism for patterning the primitive mesoderm, whose effectors were suppositional 4,9. As summarized schematically in Figure 7, these findings establish, that (a) Sox17 integrates three convergent pathways for cardiac differentiation, requiring Nodal/Activin for its induction, beyond just Wnts and BMPs 4; (b) the cell nonautonomous requirement for Sox17 is mediated by secreted signals, not cell contact; (c) Sox17 is essential for induction of an endodermal growth factor inhibitor Cer1, in addition to the endodermal gene Hhex, a Sox17-dependent transcription factor 4; (d) Sox17 is required for the induction of Hhex, Cer1, and cardiac differentiation even using exogenous Activin as a defined stimulus; (e) Cer1 and Hhex are essential in ESCs for mesoderm patterning to cardiac muscle, akin to their obligatory role in Xenopus; (f) Hhex, in addition, was required for normal induction of Cer1; (g) exogenous Cer1 was sufficient to rescue Hhex-deficient cells; (h) Sox17 binds consensus sites in the Cer1 locus and activates sequence-specific transcription of Cer1 reporter genes.
Figure 7.
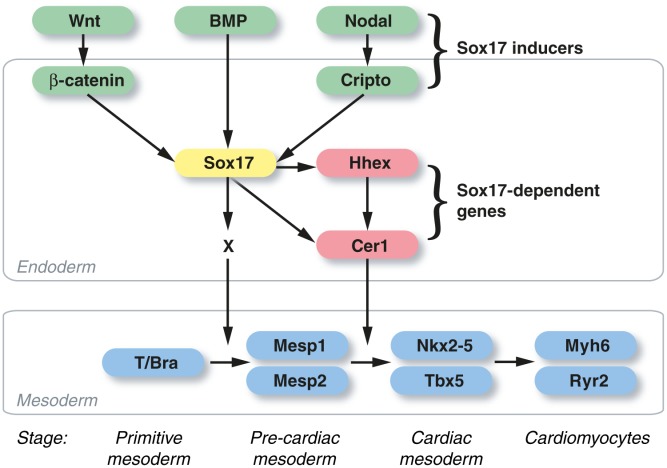
The Sox17-Hhex-Cer1 pathway for heart induction in differentiating mouse embryonic stem cells (ESCs). Sox17 expression is contingent on input from β-catenin-dependent Wnts, BMPs, and Nodal. Its induction in the endoderm is mandatory for a cell nonautonomous signal (X) that activates Mesp1 and Mesp2, the essential first step directing the primitive mesoderm toward cardiac muscle specification (stages noted below the mesoderm compartment). Two Sox17-dependent endodermal genes, Hhex and Cer1, act in series downstream from Mesp1/2 to trigger the induction of cardiogenic transcription factors, such as Nkx2–5 and Tbx5, which denote and execute the cardiac muscle lineage decision. Sox17 activates Cer1 both directly, via sequence-specific binding and trans-activation, and indirectly, via Hhex. Forced expression of Cer1 can reduce the lack of cardiac muscle differentiation in Hhex-deficient ESCs.
Sox17 is best known for its function in the developing endoderm 29,31–34, but, in addition, is required in fetal and neonatal hematopoietic stem cells and can convert adult hematopoietic progenitors to fetal-like hematopoietic stem cells 35. Sox17, Sox18, and Sox7 have nonredundant functions in hematopoiesis 36, yet seeming redundancy in cardiovascular development, which has complicated defining their contribution in vivo 21,22. A combined deletion of all three F box family members remains to be reported, and in zebrafish, the ortholog sox17 itself lacks the conserved β-catenin binding site, whereas endoderm formation relies instead on a unique fourth F group gene, casanova/sox32 37,38. Loss of Hhex in mice is sufficient for severe cardiac structural malformations including hypoplasia of the right ventricle and abnormal development of compact myocardium, along with atrioventricular cushion, septal, outflow tract, and vascular defects 39.
Like our studies of Sox17-deficient ESCs created by RNA interference, investigations of Sox17-null ESCs observe phenotypes (in the latter case, an essential role in extraembryonic mesoderm) 20 that are not manifest in Sox17-deficient embryos 31,40. It was speculated that the difference reflects compensatory changes in networks controlling intrauterine development, illustrated by the persistence of Gata4 and Sox7, two proven Sox17 targets, in embryos lacking Sox17 20. Another potential basis is the effect of mouse strain 21,41. Alternatively, either the greater spatial precision of instructive signals in the embryo than in embryoid bodies, differences in coexpressed factors 42, or the trans-placental rescue of soluble factors in heterozygous mothers, might make differentiating ESCs a more sensitized system for disruption of cell nonautonomous circuits. Illustrated by genetic modifiers as well as transplacental rescue in the case of TGFβ1 deficiency 43,44, two or even more of these mechanisms might coexist.
Several features of this Sox17-Hhex-Cer1 circuit bear mention. First, although all three genes act on mesoderm pattern to a cardiac fate, Hhex and Cer1 act at a later stage than Sox17 itself and are dispensable for the induction of Mesp1/2. This signifies the operation of as-yet-unidentified effectors of Sox17 beyond just Hhex and Cer1 in endoderm that dictate this initial step. Indeed, factors apart from Cer1 contribute to cardiac myogenesis in mESCs stimulated by endoderm from embryonal carcinoma cells, in part via contact-dependent signals, or by conditioned medium from extraembryonic endoderm 45. Although we show that Cer1 expression requires Hhex, and is sufficient to rescue Hhex-deficient cells, these results do not altogether exclude the possibility that Hhex might also function in parallel with Cer1, the epistatic relation in Xenopus. Potential paracrine effectors of Sox17 shRNA at the time of Mesp1 induction include increased expression of Nodal (Fig. 2B), whose upregulation can impair Mesp1 expression 46. Interestingly, Eomes, a pivotal inducer of Mesp1, was expressed despite the lack of Sox17; however, its ability to drive Mesp1 expression is known to be blocked by high levels of Activin 17.
Second, the molecular mechanisms whereby Sox17 and Hhex both activate Cer1 are expected to differ, with Sox17 binding the Cer1 promoter directly as shown here, but Hhex, a homeodomain transcriptional repressor related to Antennapedia, perhaps more likely inhibiting a key inhibitor 47. Interestingly, in addition to its role as a trans-activator, Sox17 can function as a repressor of Wnt/β-catenin signals 4,48, its mode of action at later stages of heart development in an endocardial pathway for FGF expression 49.
Third, our study defines the pathway coupling Sox17 to Cer1 and thus complements a recent investigation elegantly mapping the effectors of Cer1, including inhibition of BMPs and Nodal (growth factors that are essential to the induction of Sox17 in the endoderm, at an earlier stage), thereby leading to induction of Baf60c, the cardiomyogenic component of the SWItch/Sucrose NonFermentable (SWI/SNF) chromatin remodeling complex (42). Consistent with the latter report, Baf60c was markedly suppressed by the absence of Sox17 (Supporting Information Fig. S2, Cluster V), as predicted from the resulting defect in Cer1. Taken together with the latter findings, this study represents a further advance in the genetic dissection of endodermal signals for cardiac muscle creation. Because Sox17 is also expressed by cardiac-resident stem cells 50, it will be intriguing to assess their function in adult cardiac progenitor cells and especially the context of cardiac self-repair.
Conclusion
We report a genome-wide analysis of Sox17-dependent genes in differentiating ESCs, substantiate through unbiased expression profiling that Sox17 controls cardiac myocyte creation at the stage of mesoderm patterning, and prove the requirement for two Sox17-dependent genes, Hhex and Cer1, acting in series.
Acknowledgments
We thank A. Bradley, J. Brickman, H. Clevers, E. De Robertis, L. Van Parijs, J. Reiser, and D. Trono for reagents; S. Dimmeler, D. Garry, K. Niederreither, M. Noseda, and M. Sano for discussions; M. Ramirez, N. Jiang, the Baylor Flow Cytometry Core, and the Baylor Microarray Core Facility for expert assistance. This work was supported by grants from the British Heart Foundation (CH/08/002, RE/08/002), European Commission (223372), Fondation Leducq (04 CVD 03), Medical Research Council (G0901467), and National Institutes of Health (P01 HL49953) to M.D.S. Y.L. and R.J.S. are currently affiliated with the Department of Biology & Biochemistry, University of Houston, Houston, TX. R.K. is currently affiliated with Department of Regenerative Medicine and Advanced Cardiac Therapeutics, Keio University School of Medicine, Tokyo 160–8582, Japan.
Author Contributions
Y.L.: conception and design, collection and assembly of data, data analysis and interpretation, and manuscript writing; R.K.: conception and design, collection and assembly of data, and data analysis and interpretation; T.W.L., R.J.S., and M.B.: data analysis and interpretation and manuscript writing; T.S.: collection and assembly of data and data analysis and interpretation; O.T.: collection and assembly of data; G.M.: provision of study material; M.D.S.: conception and design, financial support, data analysis and interpretation, manuscript writing, and final approval of manuscript. Y.L. and R.K. contributed equally to this article.
Disclosure of Potential Conflicts of Interest
The authors indicate no potential conflicts of interest.
Supporting Information
See www.StemCells.com for supporting information available online.
References
- 1.Murry CE, Keller G. Differentiation of embryonic stem cells to clinically relevant populations: Lessons from embryonic development. Cell. 2008;132:661–680. doi: 10.1016/j.cell.2008.02.008. [DOI] [PubMed] [Google Scholar]
- 2.Noseda M, Peterkin T, Simoes FC, et al. Cardiopoietic factors: Extracellular signals for cardiac lineage commitment. Circ Res. 2011;108:129–152. doi: 10.1161/CIRCRESAHA.110.223792. [DOI] [PubMed] [Google Scholar]
- 3.Mercola M, Ruiz-Lozano P, Schneider MD. Cardiac muscle regeneration: Lessons from development. Genes Dev. 2011;25:299–309. doi: 10.1101/gad.2018411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu Y, Asakura M, Inoue H, et al. Sox17 is essential for the specification of cardiac mesoderm in embryonic stem cells. Proc Natl Acad Sci USA. 2007;104:3859–3864. doi: 10.1073/pnas.0609100104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bondue A, Lapouge G, Paulissen C, et al. Mesp1 acts as a master regulator of multipotent cardiovascular progenitor specification. Cell Stem Cell. 2008;3:69–84. doi: 10.1016/j.stem.2008.06.009. [DOI] [PubMed] [Google Scholar]
- 6.Lindsley RC, Gill JG, Murphy TL, et al. Mesp1 coordinately regulates cardiovascular fate restriction and epithelial-mesenchymal transition in differentiating ESCs. Cell Stem Cell. 2008;3:55–68. doi: 10.1016/j.stem.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Costello I, Pimeisl IM, Drager S, et al. The T-box transcription factor Eomesodermin acts upstream of Mesp1 to specify cardiac mesoderm during mouse gastrulation. Nat Cell Biol. 2011;13:1084–1091. doi: 10.1038/ncb2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan SS, Shi X, Toyama A, et al. Mesp1 patterns mesoderm into cardiac, hematopoietic, or skeletal myogenic progenitors in a context-dependent manner. Cell Stem Cell. 2013;12:587–601. doi: 10.1016/j.stem.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stefanovic S, Abboud N, Desilets S, et al. Interplay of Oct4 with Sox2 and Sox17: A molecular switch from stem cell pluripotency to specifying a cardiac fate. J Cell Biol. 2009;186:665–673. doi: 10.1083/jcb.200901040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Foley AC, Mercola M. Heart induction by Wnt antagonists depends on the homeodomain transcription factor Hex. Genes Dev. 2005;19:387–396. doi: 10.1101/gad.1279405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Belo JA, Bachiller D, Agius E, et al. Cerberus-like is a secreted BMP and nodal antagonist not essential for mouse development. Genesis. 2000;26:265–270. [PubMed] [Google Scholar]
- 12.Matzuk MM, Finegold MJ, Su JG, et al. Alpha-inhibin is a tumour-suppressor gene with gonadal specificity in mice. Nature. 1992;360:313–319. doi: 10.1038/360313a0. [DOI] [PubMed] [Google Scholar]
- 13.Xu C, Liguori G, Adamson ED, et al. Specific arrest of cardiogenesis in cultured embryonic stem cells lacking Cripto-1. Dev Biol. 1998;196:237–247. doi: 10.1006/dbio.1998.8862. [DOI] [PubMed] [Google Scholar]
- 14.Pluta K, Luce MJ, Bao L, et al. Tight control of transgene expression by lentivirus vectors containing second-generation tetracycline-responsive promoters. J Gene Med. 2005;7:803–817. doi: 10.1002/jgm.712. [DOI] [PubMed] [Google Scholar]
- 15.Mertin S, McDowall SG, Harley VR. The DNA-binding specificity of SOX9 and other SOX proteins. Nucleic Acids Res. 1999;27:1359–1364. doi: 10.1093/nar/27.5.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takeuchi JK, Mileikovskaia M, Koshiba-Takeuchi K, et al. Tbx20 dose-dependently regulates transcription factor networks required for mouse heart and motoneuron development. Development. 2005;132:2463–2474. doi: 10.1242/dev.01827. [DOI] [PubMed] [Google Scholar]
- 17.van den Ameele J, Tiberi L, Bondue A, et al. Eomesodermin induces Mesp1 expression and cardiac differentiation from embryonic stem cells in the absence of Activin. EMBO Rep. 2012;13:355–362. doi: 10.1038/embor.2012.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kattman SJ, Huber TL, Keller GM. Multipotent flk-1+ cardiovascular progenitor cells give rise to the cardiomyocyte, endothelial, and vascular smooth muscle lineages. Dev Cell. 2006;11:723–732. doi: 10.1016/j.devcel.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 19.Prall OW, Menon MK, Solloway MJ, et al. An Nkx2–5/Bmp2/Smad1 negative feedback loop controls heart progenitor specification and proliferation. Cell. 2007;128:947–959. doi: 10.1016/j.cell.2007.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Niakan KK, Ji H, Maehr R, et al. Sox17 promotes differentiation in mouse embryonic stem cells by directly regulating extraembryonic gene expression and indirectly antagonizing self-renewal. Genes Dev. 2010;24:312–326. doi: 10.1101/gad.1833510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hosking B, Francois M, Wilhelm D, et al. Sox7 and Sox17 are strain-specific modifiers of the lymphangiogenic defects caused by Sox18 dysfunction in mice. Development. 2009;136:2385–2391. doi: 10.1242/dev.034827. [DOI] [PubMed] [Google Scholar]
- 22.Sakamoto Y, Hara K, Kanai-Azuma M, et al. Redundant roles of Sox17 and Sox18 in early cardiovascular development of mouse embryos. Biochem Biophys Res Commun. 2007;360:539–544. doi: 10.1016/j.bbrc.2007.06.093. [DOI] [PubMed] [Google Scholar]
- 23.Foley AC, Korol O, Timmer AM, et al. Multiple functions of Cerberus cooperate to induce heart downstream of Nodal. Dev Biol. 2007;303:57–65. doi: 10.1016/j.ydbio.2006.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yasunaga M, Tada S, Torikai-Nishikawa S, et al. Induction and monitoring of definitive and visceral endoderm differentiation of mouse ES cells. Nat Biotechnol. 2005;23:1542–1550. doi: 10.1038/nbt1167. [DOI] [PubMed] [Google Scholar]
- 25.Kattman SJ, Witty AD, Gagliardi M, et al. Stage-specific optimization of activin/nodal and BMP signaling promotes cardiac differentiation of mouse and human pluripotent stem cell lines. Cell Stem Cell. 2011;8:228–240. doi: 10.1016/j.stem.2010.12.008. [DOI] [PubMed] [Google Scholar]
- 26.Parisi S, D'Andrea D, Lago CT, et al. Nodal-dependent Cripto signaling promotes cardiomyogenesis and redirects the neural fate of embryonic stem cells. J Cell Biol. 2003;163:303–314. doi: 10.1083/jcb.200303010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Farina A, D'Aniello C, Severino V, et al. Temporal proteomic profiling of embryonic stem cell secretome during cardiac and neural differentiation. Proteomics. 2011;11:3972–3982. doi: 10.1002/pmic.201100063. [DOI] [PubMed] [Google Scholar]
- 28.Loots GG, Ovcharenko I, Pachter L, et al. rVista for comparative sequence-based discovery of functional transcription factor binding sites. Genome Res. 2002;12:832–839. doi: 10.1101/gr.225502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sinner D, Rankin S, Lee M, et al. Sox17 and beta-catenin cooperate to regulate the transcription of endodermal genes. Development. 2004;131:3069–3080. doi: 10.1242/dev.01176. [DOI] [PubMed] [Google Scholar]
- 30.Medina PP, Castillo SD, Blanco S, et al. The SRY-HMG box gene, SOX4, is a target of gene amplification at chromosome 6p in lung cancer. Hum Mol Genet. 2009;18:1343–1352. doi: 10.1093/hmg/ddp034. [DOI] [PubMed] [Google Scholar]
- 31.Kanai-Azuma M, Kanai Y, Gad JM, et al. Depletion of definitive gut endoderm in Sox17-null mutant mice. Development. 2002;129:2367–2379. doi: 10.1242/dev.129.10.2367. [DOI] [PubMed] [Google Scholar]
- 32.Spence JR, Lange AW, Lin SC, et al. Sox17 regulates organ lineage segregation of ventral foregut progenitor cells. Dev Cell. 2009;17:62–74. doi: 10.1016/j.devcel.2009.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang P, Rodriguez RT, Wang J, et al. Targeting SOX17 in human embryonic stem cells creates unique strategies for isolating and analyzing developing endoderm. Cell Stem Cell. 2011;8:335–346. doi: 10.1016/j.stem.2011.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chng SC, Ho L, Tian J, et al. ELABELA: A hormone essential for heart development signals via the apelin receptor. Dev Cell. 2013;27:672–680. doi: 10.1016/j.devcel.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 35.He S, Kim I, Lim MS, et al. Sox17 expression confers self-renewal potential and fetal stem cell characteristics upon adult hematopoietic progenitors. Genes Dev. 2011;25:1613–1627. doi: 10.1101/gad.2052911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Serrano AG, Gandillet A, Pearson S, et al. Contrasting effects of Sox17- and Sox18-sustained expression at the onset of blood specification. Blood. 2010;115:3895–3898. doi: 10.1182/blood-2009-10-247395. [DOI] [PubMed] [Google Scholar]
- 37.Alexander J, Rothenberg M, Henry GL, et al. Casanova plays an early and essential role in endoderm formation in zebrafish. Dev Biol. 1999;215:343–357. doi: 10.1006/dbio.1999.9441. [DOI] [PubMed] [Google Scholar]
- 38.Chung MI, Ma AC, Fung TK, et al. Characterization of Sry-related HMG box group F genes in zebrafish hematopoiesis. Exp Hematol. 2011;39:986–998 e985. doi: 10.1016/j.exphem.2011.06.010. [DOI] [PubMed] [Google Scholar]
- 39.Hallaq H, Pinter E, Enciso J, et al. A null mutation of Hhex results in abnormal cardiac development, defective vasculogenesis and elevated Vegfa levels. Development. 2004;131:5197–5209. doi: 10.1242/dev.01393. [DOI] [PubMed] [Google Scholar]
- 40.Kim I, Saunders TL, Morrison SJ. Sox17 dependence distinguishes the transcriptional regulation of fetal from adult hematopoietic stem cells. Cell. 2007;130:470–483. doi: 10.1016/j.cell.2007.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pfister S, Jones VJ, Power M, et al. Sox17-dependent gene expression and early heart and gut development in Sox17-deficient mouse embryos. Int J Dev Biol. 2011;55:45–58. doi: 10.1387/ijdb.103158sp. [DOI] [PubMed] [Google Scholar]
- 42.Cai W, Albini S, Wei K, et al. Coordinate Nodal and BMP inhibition directs Baf60c-dependent cardiomyocyte commitment. Genes Dev. 2013;27:2332–2344. doi: 10.1101/gad.225144.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Letterio JJ, Geiser AG, Kulkarni AB, et al. Maternal rescue of transforming growth factor-beta 1 null mice. Science. 1994;264:1936–1938. doi: 10.1126/science.8009224. [DOI] [PubMed] [Google Scholar]
- 44.Bonyadi M, Rusholme SAB, Cousins FM, et al. Mapping of a major genetic modifier of embryonic lethality in TGF beta 1 knockout mice. Nat. Genet. 1997;15:207–211. doi: 10.1038/ng0297-207. [DOI] [PubMed] [Google Scholar]
- 45.Brown K, Doss MX, Legros S, et al. eXtraembryonic ENdoderm (XEN) stem cells produce factors that activate heart formation. PLoS One. 2010;5:e13446. doi: 10.1371/journal.pone.0013446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Powers SE, Taniguchi K, Yen W, et al. Tgif1 and Tgif2 regulate Nodal signaling and are required for gastrulation. Development. 2010;137:249–259. doi: 10.1242/dev.040782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zamparini AL, Watts T, Gardner CE, et al. Hex acts with β-catenin to regulate anteroposterior patterning via a Groucho-related co-repressor and Nodal. Development. 2006;133:3709–3722. doi: 10.1242/dev.02516. [DOI] [PubMed] [Google Scholar]
- 48.Zorn AM, Barish GD, Williams BO, et al. Regulation of Wnt signaling by Sox proteins: XSox17 alpha/beta and XSox3 physically interact with beta-catenin. Mol Cell. 1999;4:487–498. doi: 10.1016/s1097-2765(00)80200-2. [DOI] [PubMed] [Google Scholar]
- 49.Zhang Y, Li S, Yuan L, et al. Foxp1 coordinates cardiomyocyte proliferation through both cell-autonomous and nonautonomous mechanisms. Genes Dev. 2010;24:1746–1757. doi: 10.1101/gad.1929210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Martin CM, Meeson AP, Robertson SM, et al. Persistent expression of the ATP-binding cassette transporter, Abcg2, identifies cardiac SP cells in the developing and adult heart. Dev Biol. 2004;265:262–275. doi: 10.1016/j.ydbio.2003.09.028. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
See www.StemCells.com for supporting information available online.


