Abstract
Drosophila melanogaster females commonly mate with multiple males establishing the opportunity for pre- and postcopulatory sexual selection. Traits impacting sexual selection can be affected by a complex interplay of the genotypes of the competing males, the genotype of the female, and compatibilities between the males and females. We scored males from 96 2nd and 94 3rd chromosome substitution lines for traits affecting reproductive success when mated with females from 3 different genetic backgrounds. The traits included male-induced female refractoriness, male remating ability, the proportion of offspring sired under competitive conditions and male-induced female fecundity. We observed significant effects of male line, female genetic background, and strong male by female interactions. Some males appeared to be “generalists” and performed consistently across the different females; other males appeared to be “specialists” and performed very well with a particular female and poorly with others. “Specialist” males did not, however, prefer to court those females with whom they had the highest reproductive fitness. Using 143 polymorphisms in male reproductive genes, we mapped several genes that had consistent effects across the different females including a derived, high fitness allele in Acp26Aa that may be the target of adaptive evolution. We also identified a polymorphism upstream of PebII that may interact with the female genetic background to affect male-induced refractoriness to remating. These results suggest that natural variation in PebII might contribute to the observed male–female interactions.
Key words: association testing, accessory gland proteins, adaptive evolution, coevolution, sperm competition, sexual conflict
It is well recognized that relations within and between the sexes form the basis of sexual selection. Recent research is also beginning to demonstrate that complex interactions between the sexes, such that certain male–female pairings have the highest reproductive success, contribute to variation in fitness and may promote sexual selection and reproductive isolation (Tregenza and Wedell 2000; Zeh and Zeh 2003; Snook et al. 2009). These male–female interactions fall along a continuum ranging from compete genetic incompatibilities between males and females of “true biological species” to cases where specific combinations of males and females have high fitness when together but the same males and females have reduced fitness when paired with other partners. The coevolution between sperm–egg recognition systems in marine invertebrates (Swanson and Vacquier 1998; Palumbi 1999; Evans and Marshall 2005) and self-incompatibility loci in plants (Castric and Vekemans 2004; Charlesworth et al. 2005) are 2 well-studied examples of these types of interactions. Incompatibilities exist in numerous other systems and efforts have often focused on understanding how mate choice and the potential for inbreeding avoidance might influence the evolution of these male–female interactions (Penn and Potts 1999; Mack et al. 2002; Nilsson et al. 2003; Birkhead et al. 2004; Evans and Marshall 2005; Marshall and Evans 2005; Panhuis and Nunney 2007; Clark et al. 2009; Pryke et al. 2010; Michalczyk et al. 2011; Sagga and Civetta 2011; Evans et al. 2013).
In Drosophila, extensive research has focused on hybrid incompatibilities between different species and several “speciation genes” have been identified (Coyne and Orr 1998; Tang and Presgraves 2009; Barbash 2010). More recently, researchers have focused on the role of male–female interactions impacting pre- and postcopulatory sexual selection between populations of a single species and even within individual populations. For example, strong male–female interactions have been observed between the Zimbabwe and cosmopolitan races of Drosophila melanogaster for mate preference such that the highest mating rate occurs when pairing individuals from the same race (Wu et al. 1995). Furthermore, a gametic incompatibility that results in reduced egg viability exists when mating Zimbabwe females to cosmopolitan males (Alipaz et al. 2001). There is also extensive evidence that male–female interactions exist between different cosmopolitan populations and within populations of both laboratory populations and lines derived from nature (Clark et al. 1999; Civetta and Clark 2000; Bjork et al. 2007; Svensson et al. 2009; Chow et al. 2010; Pischedda et al. 2012). These interactions occur for a variety of traits influencing pre- and postcopulatory sexual selection such as mating and remating rates, as well as sperm utilization and gamete viability. Clark (2002) suggested that male–female interactions might contribute to the maintenance of polymorphism within populations and Bjork et al. (2007) speculated that male–female interactions could have limited the response in an artificial selection experiment.
Although male–female interactions appear prevalent in D. melanogaster populations, with few exceptions, we know relatively little about the genes contributing to these interactions. One exception is that variation in desaturase-2, a gene involved with pheromone production, may be contributing to the interactions that determine mating preferences observed between Drosophila races (Fang et al. 2002; Greenberg et al. 2003). In another example, variation in the male produced seminal fluid protein, sex peptide, interacts with variation in the female produced sex peptide receptor, SPR, to impact male-induced refractoriness to remating in the females (Chow et al. 2010).
The purpose of this study is to provide a more thorough characterization of the role of natural genetic variation in driving genetic interactions between males and females for male reproductive traits that might be affected by sexual selection. Using 190 genetically distinct lines, we measured several male reproductive phenotypes when these males were mated with females from 3 different genetic backgrounds. We also used candidate gene association to investigate whether specific polymorphisms in male reproductive genes contribute to the observed male–female interactions.
Materials and Methods
Drosophila Lines
The experimental males were from 96 2nd chromosome substitution lines (Fiumera et al. 2005) and 94 3rd chromosome substitution lines (Fiumera et al. 2007). By backcrossing to appropriate balancer stocks, lines were created that are homozygous and isogenic but each line represents a unique 2nd or 3rd chromosome, respectively, that was segregating in a natural population from the Northeastern United States. These experimental males have dominant red eyes and females and tester males had recessive white eyes caused by cn bw mutations. Tester males were derived from the cn bw strain used in Fiumera et al. (2005, 2007), henceforth referred to as cnbw1. Females for the following experiments came from 1 of 6 cn bw strains; the original cnbw1 strain, a cnbw2 strain (introgressed into Canton-S), a cnbw3 strain (introgressed into Oregon-R), and 3 strains with the cn bw mutations introgressed into African genetic backgrounds (Pool and Aquadro 2006). Fly cultures were maintained on an agar–dextrose–yeast media (McGraw et al. 2007) and housed at 24 °C on a 12-h light/dark cycle. Virgins were collected using CO2 and housed in low density, single sex vials until 4–7 days.
Measures of Male Reproductive Fitness
Both the “offense” and “defense” components of sperm competition were measured for all the lines. The “defense” experiments score the reproductive fitness of an experimental male when they are the first male to mate to a doubly mated female and the “offense” experiments score the fitness when the experimental male is the second male to mate (see below). The overall experiment was conducted using an incomplete block design. Each block corresponded to a date when the first mating was completed. In each block, replicate males were tested from multiple lines, usually with multiple different female genetic backgrounds. Males were scored for each phenotype in at least 2 separate blocks. Males from every experimental line, however, may not have been mated to each genotype of female in every block. Overall matings were conducted in 6 different blocks for the 3rd chromosome lines and 7 different blocks for the 2nd chromosome lines. Data for some of the matings to the cnbw1 females were taken from Fiumera et al. (2005, 2007). These mating were conducted in multiple blocks and to effectively incorporate the block effects we completed additional blocks that had matings to all 3 genotypes of females simultaneously. Depending upon the phenotype, approximately 40% of the data came from Fiumera et al. (2005, 2007).
The “defense” experiments were conducted by tapping a single experimental male into vial 1 with a single female (of the appropriate genotype). Pairs were observed for mating approximately every 10min for 6h. After mating was completed, the male was removed to prevent double matings. Almost all virgin females mated and those that did not mate within this time period were discarded. On day 3, 2 virgin tester males were tapped into vial 1 with the female and vials were observed for mating every 10min for 6h and again males were removed after mating. Females that failed to remate were kept with the males for another 6h but were not observed. After the full 12h, any remaining males were discarded and females were transferred to vial 2. Females were allowed to oviposit in vial 2 for 3 days before being discarded. Once the progeny emerged, they were counted and eye color was used to assign paternity. The “offense” experiments were conducted similarly except the tester males were the first to mate and the experimental males were the second males to mate. At least 15 replicates (across the different blocks) were set up for each male–female combination. Females were discarded if they failed to produce progeny from the first male, failed to produce at least 5 progeny, died or escaped during the experiment. A small procedural difference as compared to Fiumera et al. (2005, 2007) should be noted. Here males were removed immediately after observed matings but in Fiumera et al. (2005, 2007) they were not. This change was made because, in a preliminary experiment, the cnbw3 females were frequently observed to mate multiple times within a few hours when left paired with the males (personal observation). The other females did not show this tendency. All females were ultimately given the same total amount of time to mate (if needed) and we removed males after mating as we felt it was most important to ensure that different females mated only once to a given male, as had been the case in Fiumera et al. (2005, 2007) for the cnbw1 females.
The matings allowed several “defense” phenotypes to be scored. Male-induced refractoriness to remating (refractory) is the proportion of females that mate to the experimental male but fail to remate to the tester male. Among those females that do remate to the tester male (confirmed from the progeny of both males), P1′ is the proportion of offspring sired by the first male (i.e., the experimental male). For doubly mated females, male-induced fecundity (fec-V1) was estimated using those progeny in vial 1. Several “offense” phenotypes were also scored. The ability of a male to encourage an already mated female to remate with him (P2-mating) is the proportion of experimental males that mate to an already mated female. This measure of male reproductive performance is sometimes referred to as remating rate (Fiumera et al. 2005). For those males that were successful at getting the female to remate, P2′ is the proportion of offspring sired by the second male. For doubly mated females, male-induced fecundity was scored using the progeny from vial 2, after the experimental male mated, (fec-V2).
Sperm competition phenotypes were analyzed using linear or generalized linear models in R version 3.1.0 (R Core Team 2014). Measures of fecundity (fec-V1 and fec-V2) were analyzed using linear models (lm) and generalized linear models were used to analyze refractory, P2-mating, P1′ and P2′. Refractory and P2-mating utilized the quasibinomial distribution whereas P1′ and P2′ used the binomial distribution incorporating all the progeny from vial 2. The quasibinomial was used because of overdispersion as measured by the difference between the residual deviance and the residual degrees of freedom. Significance was tested according to the following model:
where where P ijkl is the phenotype, BLOCKi is the effect of the i th block, FEMALEj is the effect of the j th female genetic background, LINEk is the effect of the k th chromosome substitution line that the male came from and (FEMALE*LINE)jk is the interaction between the j th female genotype and the k th male chromosome substitution line. P values were corrected for over dispersion when necessary. The analysis was conducted separately for the 2nd and 3rd chromosome substitution lines. Line means, when males were mated to each of the different female genotypes, were calculated using the predict function in R (R Core Team 2014) and used in subsequent analyses.
“Specialist” and “Generalist” Males
We identified “specialist” and “generalist” males by testing for homogeneity of variances (Levene’s test) using the line means estimated for males from each line when mating to the 3 different female genotypes. A significant Levene’s test indicated that males from some lines performed equally well across all the female genetic backgrounds (“generalists”) whereas other males performed very well with some females but poorly when mated to others (“specialists”). We then chose males from 2 3rd chromosome lines that were identified as “specialists” for P2′ and males from 2 lines that were identified as “generalists” (see results) and tested if the “specialists” preferred to court the females with whom they have the highest fitness. Females were housed on colored food to allow identification (Wu et al. 1995). Six replicates of all 6 combinations of female genetic background and coloring were tested with each of the 4 male lines (36 total replicates per male line). All females were premated to cnbw1 males 48h prior to the preference tests because the specialist–generalist example was for an “offense” trait. Courtship vials consisted of 3 females (1 of each genetic background and of differing colors) and 1 experimental male. Each vial was observed at least every 15min, for approximately 1min, and which female the male was courting was recorded. Preference was scored as the genotype of female most frequently courted. Vials were watched for approximately 2.5h by which time female coloration began to fade. Male preference in courtship was tested using a 3 by 4 contingency table.
Candidate Gene Associations
The line means from the linear models were then used to test for associations between genotype and phenotype and test for interactions between polymorphisms in male reproductive genes and the female genetic background with whom he mated. A total of 70 polymorphisms in 10 male reproductive genes were scored in the 2nd chromosome substitution lines (Fiumera et al. 2005) and 73 polymorphisms in 13 male reproductive genes were scored in the 3rd chromosome substitution lines (Fiumera et al. 2007). FastPHASE (Scheet and Stephens 2006) was used to reconstruct missing data based on haplotypes but associations were tested using the single markers. Permutation tests conducted in MATLAB (The MathWorks Inc. 2010) were used to calculate experimentwise P values (Churchill and Doerge 1994). The model was P ijk = GMi + FEMALEj + (GM*FEMALE)ij + εijk where P ijk is the phenotype, GMi is the effect of the i th genetic marker, FEMALEj is the effect of the j th FEMALE and (GM*FEMALE)ij is the interaction between the male genetic marker and the female with whom he mated. Experimentwise P values were analyzed separately for each phenotype for both the 2nd and 3rd chromosome substitution lines.
Testing Effects of African Female Genetic Backgrounds
Based on results from this study (see below) and previous work (Fiumera et al. 2005), we tested the effects of females with African genetic backgrounds on the association between a serine to isoleucine polymorphism in Acp26Aa and P2′. We took a small number of the 2nd chromosome lines (4 with a serine allele and 4 with the isoleucine at the same position) and mated these males to 5 different female genetic backgrounds; cnbw2 and cnbw3 females derived from North American lines and cnbw5, cnbw6, and cnbw7 derived from African strains (Pool and Aquadro 2006).
Overall Sample Sizes
For the 2nd chromosome substitution lines, refractory was scored using 3075 females. Of those, 2587 produced progeny from both males and were used to estimate P1′ and fec-V1. P2-mating was scored using 4049 females and 2567 were used to estimate P2′ and fec-V2. For the 3rd chromosome substitution lines, refractory was scored using 3105 females and 2537 produced progeny from both males and were used to estimate P1′ and fec-V1. P2-mating was scored using 3322 females and 2614 were used to estimate P2′ and fec-V2. In total, 233 702 progeny (“defense”, 2nds), 311 292 progeny (“offense”, 2nds), 213 776 progeny (“defense”, 3rds) and 291 619 progeny (“offense”, 3rds) were counted for a total of 1 050 389 progeny. Of the 96 2nd chromosome lines, 83 had sperm competition phenotypes measured when mating to all 3 female genetic backgrounds and 85 were mated to 2 of the 3 females. Of the 94 3rd chromosome lines, 85 had sperm competition phenotypes measured when mating to all 3 females and 86 were mated to 2 of the 3 females for the “offense” phenotypes. For the “defense” phenotypes, 76 and 80 lines were mated to either 3 or 2 different female genetic backgrounds, respectively. In fulfillment of data archiving guidelines (Baker 2013), the data used in these analyses are available through Dryad (doi:http://dx.doi.org/doi:10.5061/dryad.2856m).
Results
Analysis of Male Reproductive Fitness
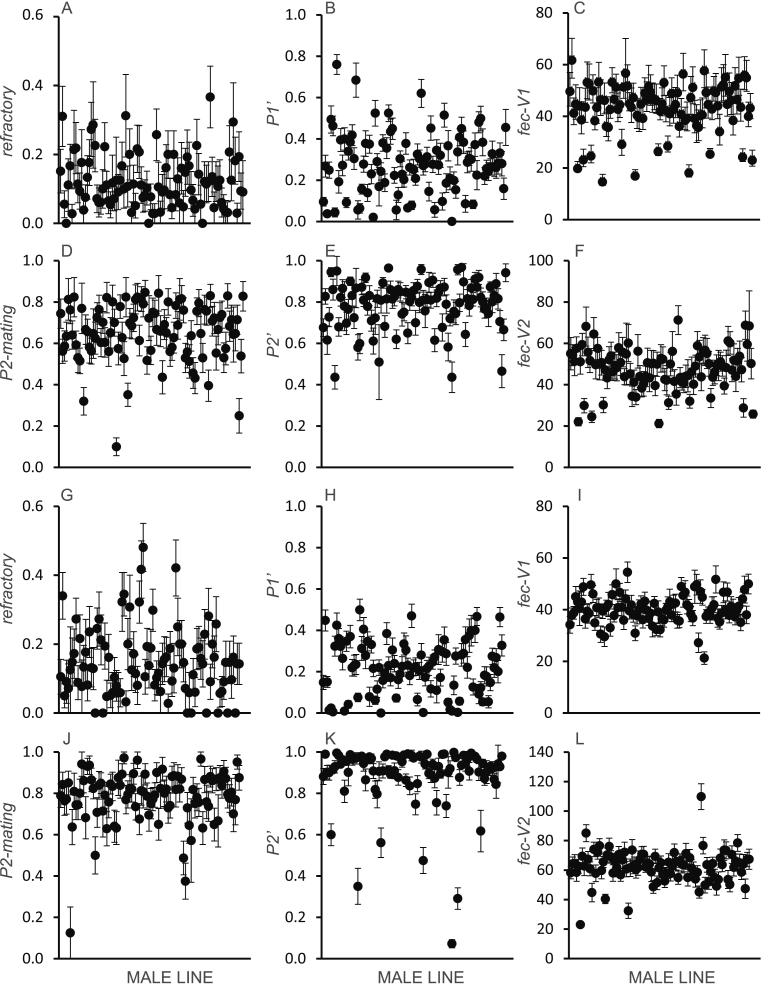
Male line was a significant predictor for all phenotypes, across the 3 female genetic backgrounds, in both the 2nd and 3rd chromosome lines indicating that there was a genetic component to the variation observed (Tables 1 and 2). Male-induced refractoriness ranged from 0 to 0.38 (average = 0.12) in the 2nds and from 0 to 0.49 (average = 0.16) in the 3rds. P1′ ranged from 0 to 0.78 (average = 0.18, 2nds) and 0 to 0.53 (average = 0.19, 3rds) while fec-V1 ranged from 15.4 to 68.7 (average = 52.3, 2nds) and 21.3 to 54.5 (average = 38.4, 3rds). P2-mating ranged from 0.10 to 0.86 (average = 0.65) in the 2nds and from 0.13 to 0.98 (average = 0.78) in the 3rds. P2′ ranged from 0.42 to 0.99 (average = 0.82, 2nds) and 0.07 to 1.00 (average = 0.95, 3rds) while fec-V2 ranged from 21.2 to 71.8 (average = 53.2, 2nds) and 23.0 to 109.8 (average = 66.4, 3rds). Means and standard errors for all phenotypes (using the predicted values from the linear models) are shown for males from different lines (Figure 1).
Table 1.
Summary of male reproductive phenotypes for the 2nd chromosome lines
| Phenotype | Terma | Degrees of freedom for term | Degrees of freedom remainingb | P value |
|---|---|---|---|---|
| Refractory | FEMALE | 2 | 3066 | 1.0×10−4 |
| MALE | 95 | 2971 | 8.2×10−9 | |
| FEMALE*MALE | 166 | 2805 | 6.5×10−10 | |
| P1ʹ | FEMALE | 2 | 2317 | 0.819 |
| MALE | 95 | 2317 | <2.2×10−16 | |
| FEMALE*MALE | 166 | 2317 | <2.2×10−16 | |
| Fec-V1 | FEMALE | 2 | 2317 | 7.6×10−10 |
| MALE | 95 | 2317 | 1.7×10−4 | |
| FEMALE*MALE | 166 | 2317 | 3.2×10−7 | |
| P2-mating | FEMALE | 2 | 4040 | <2.2×10−16 |
| MALE | 95 | 3945 | <2.2×10−16 | |
| FEMALE*MALE | 166 | 3779 | 1.7×10−5 | |
| P2ʹ | FEMALE | 2 | 2296 | 9.1×10−4 |
| MALE | 95 | 2296 | <2.2×10−16 | |
| FEMALE*MALE | 166 | 2296 | 8.6×10−13 | |
| Fec-V2 | FEMALE | 2 | 2296 | <2.2×10−16 |
| MALE | 95 | 2296 | 3.8×10−5 | |
| FEMALE*MALE | 166 | 2296 | 5.2×10−8 |
aBLOCK, with 6 degrees of freedom, was highly significant for all the traits measured.
bRemaining degrees of freedom is the residual degrees of freedom from the glm model (family = quasibinomial or binomial) for refractory, P2-mating, P1ʹ and P2ʹ or the error degrees of freedom for the lm models for Fec-V1 and Fec-V2.
Table 2.
Summary of male reproductive phenotypes for the 3rd chromosome lines
| Phenotype | Terma | Degrees of freedom for term | Degrees of freedom remainingb | P value |
|---|---|---|---|---|
| Refractory | FEMALE | 2 | 3097 | 1.0×10−4 |
| MALE | 93 | 3004 | 8.2×10−9 | |
| FEMALE*MALE | 155 | 2849 | 6.5×10−10 | |
| P1ʹ | FEMALE | 2 | 2282 | 0.819 |
| MALE | 93 | 2282 | <2.2×10−16 | |
| FEMALE*MALE | 154 | 2282 | <2.2×10−16 | |
| Fec-V1 | FEMALE | 2 | 2282 | 7.6×10−10 |
| MALE | 93 | 2282 | 1.7×10−4 | |
| FEMALE*MALE | 154 | 2282 | 3.2×10−7 | |
| P2-mating | FEMALE | 2 | 3314 | <2.2×10−16 |
| MALE | 93 | 3221 | <2.2×10−16 | |
| FEMALE*MALE | 170 | 3051 | 1.7×10−5 | |
| P2ʹ | FEMALE | 2 | 2342 | 9.1×10−4 |
| MALE | 93 | 2342 | <2.2×10−16 | |
| FEMALE*MALE | 169 | 2342 | 8.6×10−13 | |
| Fec-V2 | FEMALE | 2 | 2342 | <2.2×10−16 |
| MALE | 93 | 2342 | 3.8×10−5 | |
| FEMALE*MALE | 169 | 2342 | 5.2×10−8 |
aBLOCK, with 5 degrees of freedom, was highly significant for all the traits measured.
bRemaining degrees of freedom is the residual degrees of freedom from the glm model (family = quasibinomial or binomial) for refractory, P2-mating, P1ʹ and P2ʹ or the error degrees of freedom for the lm models for Fec-V1 and Fec-V2.
Figure 1.
Differences among the genetic lines for male reproductive phenotypes. Means and standard errors (across all 3 female genetic backgrounds) are shown for each phenotype for males from the 2nd (A–F) and 3rd (G–L) chromosome lines.
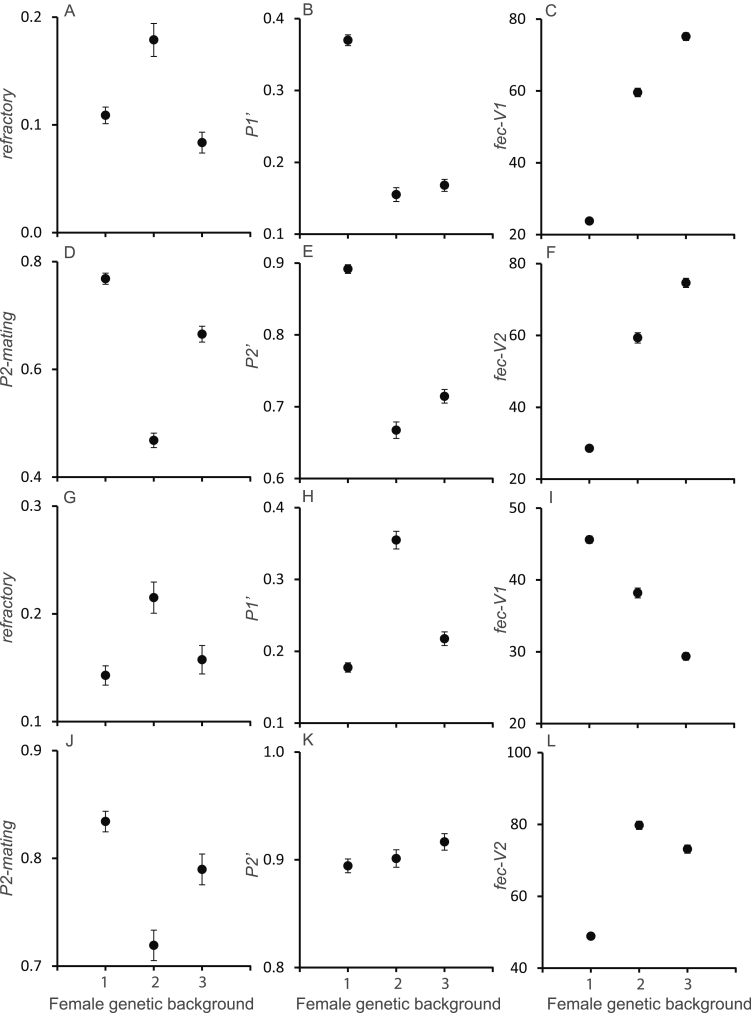
Female genetic background also was a significant predictor, across all male lines, for all phenotypes except P1′ when mating with males from the 2nd chromosome lines (Tables 1 and 2). This indicates that the female genetic background affected the reproductive success of the males. On average, male-induced female refractoriness was 0.12 (0.11, 0.18, and 0.08 for cnbw1, cnbw2, and cnbw3 females, respectively) when mating with males from the 2nd chromosome lines and 0.16 (0.14, 0.21, and 0.14) with the 3rds. Similarly ordered, average P1′ was 0.17 (0.35, 0.09, 0.11) and 0.18 (0.12, 0.31, 0.16); average fecundity of vial 1 was 52.3 offspring (23.7, 61.1, 75.5) and 38.4 offspring (45.6, 38.9, 29.4); average P2-mating was 0.65 (0.77, 0.49, 0.69) and 0.78 (0.83, 0.72, 0.79); average P2′ was 0.82 (0.94, 0.72, 0.76) and 0.95 (0.95, 0.94, 0.95); and average fecundity of vial 2 was 53.2 (28.5, 59.8, 74.6) and 66.4 (48.8, 79.3, 72.6). Means and standard errors for all phenotypes (using the predicted values from the linear models) are shown for the different female genetic backgrounds (Figure 2).
Figure 2.
Differences among the 3 different female genetic backgrounds for phenotypes affecting reproductive success. Means and standard errors (across all male lines) are shown for each phenotype for mating to males from the 2nd (A–F) and 3rd (G–L) chromosome lines.
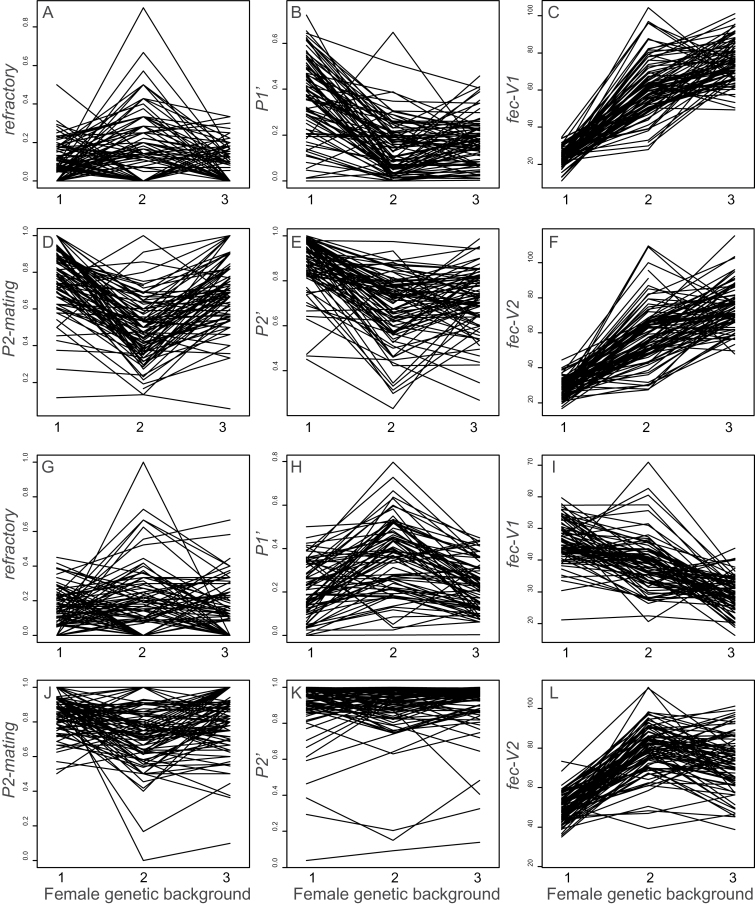
There were highly significant interactions between the male line and the genetic background of the females. Significant interactions were detected for all the phenotypes measured in both the 2nd and 3rd chromosome lines (Tables 1 and 2; Figure 3) indicating that the reproductive success of the male varied depending upon the female with whom he mated. In some cases, the interactions appear to be driven primarily by the rank order of male lines changing depending upon the female with whom they mate (e.g., P1′ and P2′ in Figure 3B,E,H,K). In other cases, the interactions appear to be driven more by certain genotypes of females showing greater variability among the male lines (e.g., fec-V1 Figure 3C,F).
Figure 3.
Male by female interactions affect sperm competition phenotypes. Interaction plots are shown for all the measured phenotypes between the 3 female genetic backgrounds (cnbw1, cnbw2, and cnbw3) and the 2nd chromosome males (A–F) or the 3rd chromosome males (G–L).
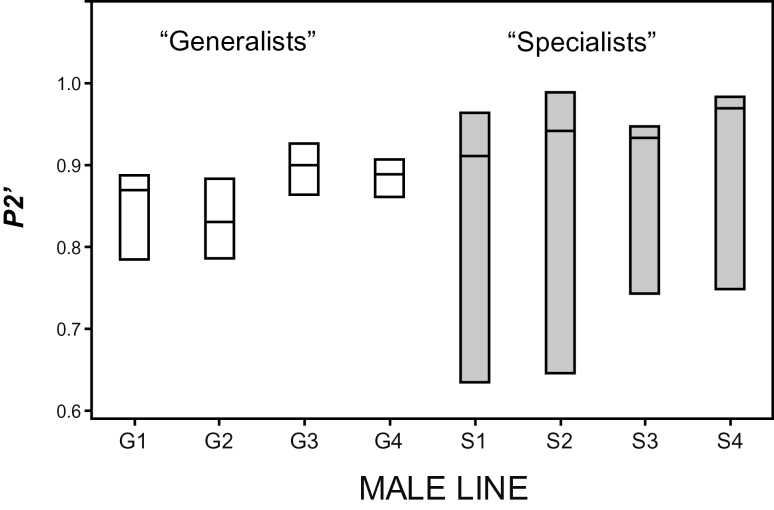
To investigate the potential for “generalist” versus “specialist” males we tested for homogeneity of variances (Levene’s test) using the line means derived from mating the chromosome substitution males to the 3 different females. We detected a significant deviation from homogeneity of variances (P < 0.001) in line means for the proportion of offspring sired when the 3rd chromosome males were the second males to mate, P2′ (Figure 4). Some males did equally average when mating to any female (“generalists”) while some males did particularly well or poorly depending upon the female with whom they mated (“specialists”). We then tested whether “specialist” males tended to preferentially court the females with whom they have higher reproductive success. There was no evidence, however, that males showed differential courtship (χ 2 = 3.42, d.f. = 6, P = 0.75).
Figure 4.
Box plots of generalist versus specialist males for the proportion of offspring sired by the second male to mate (P2′). Males from some 3rd chromosome lines (G1–G4) show consistent patterns of sperm use when mating to different females and can be termed “generalist” males. Other males (S1–S4) show high variance in P2′ when mating to different females and can be termed “specialist” males.
We also investigated genetic correlations among the male reproductive traits. Several of the sperm competition phenotypes consistently showed positive correlations (Supplementary Tables 1–3 online). For example, P1′ and fecundity in vial 1 (fec-V1) were positively correlated in all 6 comparisons (i.e., both the 2nd and 3rd chromosome lines when mating to all 3 females). P1′ and P2′ were significantly positively correlated in 5 of the comparisons while P2′ and fecundity in vial 2 (fec-V2) were positively correlated in the 3rd chromosome lines for all 3 females. When considering all 3 female genetic backgrounds simultaneously, however, several male reproductive traits were negatively correlated (Supplementary Table 4 online). One example is P1′ and fec-V1. Within each female genetic background P1′ and fec-V1 are positively correlated but because the female’s differ in mean P1′ and fec-V1 this creates a significant negative correlation observed when considering all the data simultaneously (Supplementary Figure 1 online) and highlights the utility of investigating the role of genetic diversity in both males and females simultaneously.
Associations Between Genotype and Phenotype
We identified 11 associations between genotypes at male reproductive genes and phenotypes affecting sexual selection that conferred consistent effects across the 3 females (strict experimentwise P value < 0.05, Table 3). These associations were with 8 different genes and 5 of the measured male reproductive traits; we did not identify any associations with fec-V1. Many of the traits (e.g., P1′, P2-mating, P2′, and fec-V2) showed associations with polymorphisms in multiple genes and each association explained only a small proportion of the phenotypic variation (adjusted r 2 never exceeded 0.09). In addition, polymorphisms in the same gene occasionally associate with multiple traits. For example, different markers in CG14560 associate with refractory and P1′ while the same marker in CG6168 associates with both P2′ and fecundity in vial 2.
Table 3.
Associations between genotype and sperm competition phenotypes
| Phenotype | Gene | Polymorphisma | Adj. r 2b | Polymorphism type |
|---|---|---|---|---|
| Refractory | CG14560 | SNP 1173 | 0.058 | Upstream |
| P1′ | Acp33A | SNP 2125 | 0.033 | Downstream |
| CG14560 | SNP 1474 | 0.090 | Upstream | |
| P2-mating | CG31872 | SNP 4238 | 0.031 | Synonymous |
| Acp36DE | SNP 3566 | 0.027 | Synonymous | |
| P2′ | Acp26Aa | SNPs 2201 and 2202 | 0.089 | together forms SER:ISO |
| CG31872 | SNP 3824 | 0.004c | synonymous | |
| Acp62F | SNP 1016 | 0.060 | upstream | |
| CG6168 | SNP 1583 | 0.065 | synonymous | |
| Fec-V2 | CG6168 | SNP 1583 | 0.020 | Synonymous |
| Acp70A | SNP 1624 | 0.014 | TRP:GLY |
Note: All reported associations at experimentwise P < 0.05.
aTranscription start site was given the arbitrary notation of position 1500 (Fiumera et al 2005 2007).
bAdjusted r 2 is based on model with only the male genetic marker and does not include female genetic background. When multiple genetic markers associate, the one with the highest adjusted r 2 is reported.
cMarker not significant in model without the effect of female genetic background.
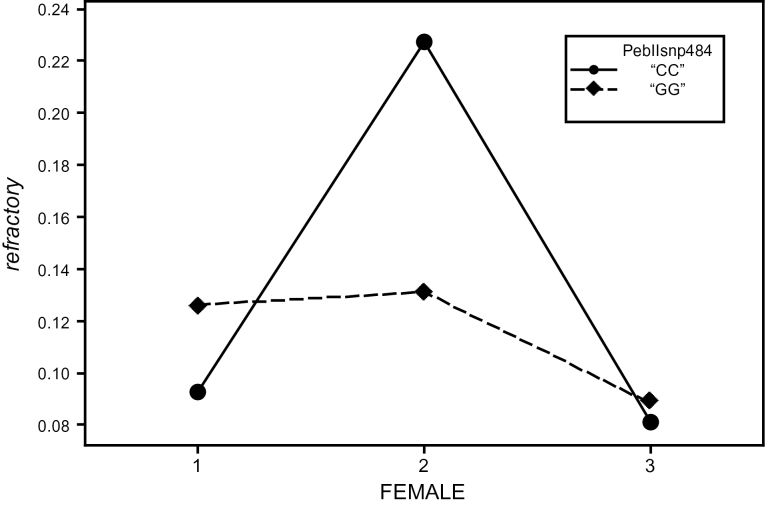
We also tested for genetic interactions between the male genotype and the female genetic background. The strongest mapped interaction was for male-induced female refractoriness with a marker upstream of PEBII (SNP484). Although it failed to meet the strict experimentwise P value < 0.05 (experimentwise P = 0.15 and markerwise P = 0.0015), an FDR analysis (Storey and Tibshirani 2003) estimates a q-value of 0.07 indicating this is unlikely to be a false positive. Males homozygous for “G” are much better at inducing refractoriness in cnbw2 females but worse with cnbw1 females as compared to males homozygous for “C”; there appears to be little difference between the different genotypes when mating to cnbw3 females (Figure 5). Given the reduced power that is expected for a higher order interaction term, these results suggest that natural variation in PEBII contributes to male–female genetic interactions.
Figure 5.
Variation in PebII associates with male-induced female refractoriness and contributes to male by female interactions. Interaction plot showing the ability of males homozygous for the “G” allele or the “C” allele at PebIIsnp484 to induce refractoriness to remating (refractory) in the 3 different female genetic backgrounds.
Effects of Genetic Backgrounds from Africa
A serine to isoleucine polymorphism in Acp26Aa strongly associated with P2′ across all 3 female genetic backgrounds derived from North America and males with the serine allele consistently sired a larger proportion of offspring (Table 3). We then tested this association when the males were mated to females derived from African genetic backgrounds. Again, there was a significant effect of male genotype at Acp26Aa (F 1,354 = 88.4; P < 2.2×10−16) with the serine allele showing higher fitness in all the female genetic backgrounds from both locations. Interestingly, there was a significant effect of female location (F 1,354 = 16.25; P < 6.8×10–5) with the African females being more likely to use the sperm of the second male. There was also a significant effect of male line nested within Acp26Aa genotype (F 6,354 = 2.84; P = 0.01) but neither an effect of female genetic background nested within female location (F 3,354 = 1.1; P = 0.35) nor an interaction between the male genotype at Acp26Aa and the female location (F 1,354 = 0.068; P = 0.794). Interestingly, the high fitness serine allele appears to be a new, derived mutation that is polymorphic in D. melanogaster (Figure 6) as the close relatives of D. melanogaster all appear to be monomorphic for isoleucine based on at least 9 available sequences for each species in GenBank.
Figure 6.

Neighbor-joining tree for Acp26Aa alleles. Four randomly selected serine and isoleucine alleles from D. melanogaster were sequenced and one representative allele from GenBank was selected to represent D. simulans, D. sechellia, and D. mauritiana. Tree was constructed in Mega 5 (Tamura et al 2011) using a partial coding sequencing of 804 base pairs that was aligned with Clustal W (Larkin et al 2007). D. yakuba, and further outgroups were excluded due to alignment difficulties.
Discussion
In this study, we investigated the potential for male–female interactions to influence sexual selection in Drosophila melanogaster. We used males from 190 chromosome extraction lines (96 2nd and 94 3rd chromosome lines) and mated them to females from 3 different genetic backgrounds. Our results provide strong evidence that natural genetic variation in males, the female genetic background and the interaction between the male and female are important factors influencing sexual selection. Interestingly, our results suggest that certain male lines may be “specialized” on certain female genetic backgrounds while other males may be more “generalists”. Furthermore, we were able to map several polymorphisms in male reproductive genes that had consistent effects across the different female genetic backgrounds and have identified a role for PebII contributing to male by female interactions for induced refractoriness to remating.
This study expanded our understanding of the role of male–female interactions influencing sexual selection in D. melanogaster. We observed highly significant male–female interactions for all investigated phenotypes (both “offense” and “defense”) when our 3 female genetic backgrounds mated to males from either the 2nd or 3rd chromosome substitution lines. Clark et al. (1999), using a set of 6 male and female lines, identified male–female interactions for P1′ but not P2′ while measures of fecundity or mating rate where not reported. Chow et al. (2010), focusing on male variation on chromosome 3 and female variation on the X chromosome, identified significant male–female interactions for the “defense” phenotypes when the focal male is the first male to mate (i.e., P1, refractory and fecundity) but did not investigate the “offense” phenotypes.
Interestingly, how the observed interactions were manifested vary across the different phenotypes. For example, our observed interactions in P1′ and P2′ appear to be driven by the relative success of males changing across the different female genetic backgrounds (Figure 3B,E,H,K). In contrast, the interactions in male-induced female fecundity appears to be caused by cnbw2 females showing very high variability when mating to males from different 2nd chromosome lines, while the other female genetic backgrounds show relatively less variation among the different males (Figure 3C,F). Male–female interactions are important from the perspective of mapping the genes contributing to sexual selection (see below) but also from an evolutionary perspective. Complex male–female interactions have the potential to produce frequency dependent selection (Svensson et al. 2005; Clark et al. 2009; Svensson et al. 2009) which can significantly impact the consequences of selection and affect the rate and/or direction of evolution (Heino et al. 1998).
Through association testing, we believe that natural variation in PEBII contributes to male–female interactions for male-induced female refractoriness. The relative success of males homozygous for an upstream G to C polymorphism changes depending upon the genetic background of his partner. PEBII is a major component of the mating plug in Drosophila melanogaster (Lung and Wolfner 2001; Bretman et al. 2010) and mating plugs (or their components) have been shown to facilitate sperm storage (Polak et al. 1998; Neubaum and Wolfner 1999; Chapman et al. 2000; Bloch Qazi and Wolfner 2003) and inhibit female remating in Drosophila (Polak et al. 2001; Bretman et al. 2010). Bretman et al. (2010) used RNAi to demonstrate that the transfer of PEBII increases male-induced refractoriness to remating after 4h; females mated to knockdown males were more likely to remate. Interestingly, we observed an interaction of PebII and the female genetic background and it would be interesting to test if the knockdown of PebII shows evidence for male–female interactions.
Male genotype had a highly significant effect on all the measured phenotypes affecting sexual selection. This is consistent with numerous studies investigating the role of male genotype on measures of sperm competition (Clark et al. 1995; Hughes 1997; Civetta and Clark 2000; Chippindale and Rice 2001; Sawby and Hughes 2001; Fiumera et al. 2005, 2006; Hughes and Leips 2006; Fiumera et al. 2007; Civetta et al. 2008; Chow et al. 2010; Greenspan and Clark 2011). Interestingly, our results indicate that some males might be “generalists”, having similar reproductive success across multiple female genetic backgrounds while others might be “specialists” such that they do very well when mating with some females and relatively poorly when mating with others (Figure 4). Although this concept is most commonly applied to the species level (Devictor et al. 2010), numerous studies have found evidence for “generalists” and “specialists” within species (Bolnick et al. 2003). Our data, however, is possibly the first example of male–female specialization. Such intraspecific differences in specialization could result in a form of frequency dependent selection (Abrams et al. 1993) that could potentially contribute to reproductive isolation and genetic differentiation (Via 1999). Sexual conflict (Parker 2006) could also be impacted by the presence of “generalist” and “specialist” males, even if the males do not necessarily show a preference for the different females.
We were successful at mapping genes that contribute to phenotypes affecting male reproductive success across the 3 female genetic backgrounds (Table 3). Several of the associations were consistent with our previous mapping efforts, including associations between Acp33A and CG14560 and P1′, as well as between CG6168 and Acp26Aa (see below) and P2′. Many of the associations we identified, when considering the 3 genetic backgrounds of females, differed from those identified when only incorporating data from a single genetic background (Clark et al. 1995; Fiumera et al. 2005, 2007). Some of the differences may not be biologically relevant due to marginal significance in one or the other analysis. The others, however, highlight the importance of studying variation in both males and females simultaneously. For example, certain polymorphisms might only matter when mating to a specific genotype of female and thus the relative frequency of the female genotypes in a population will have a large effect on the relative fitness of the alternative alleles and the potential for selection to operate. Although we only surveyed 3 female genetic backgrounds, we demonstrated that female genotype plays an important role in sexual selection and is consistent with previous work on females (Clark and Begun 1998; Clark et al. 1999; Lawniczak and Begun 2005; Lew et al. 2006; Giardina et al. 2011; Chow et al. 2013; Lüpold et al. 2013).
Several associations are supported by known roles of these proteins. For example, we identified an association with an amino acid polymorphism in Acp70A and fecundity in vial 2 (fec-V2). Several studies have shown that that Acp70A influences egg production, deposition, and ovulation (Soller et al. 1997, 1999; Chapman et al. 2003; Liu and Kubli 2003). This study also identified a radical serine to isoleucine amino acid polymorphism in Acp26Aa that associates with the proportion of offspring sired by the second male to male (P2′) and is consistent with the results from Fiumera et al. (2005) using only the cnbw1 females. The serine allele is favored across the 3 different North American female genetic backgrounds that were investigated and also the 3 African genetic backgrounds. The favored serine allele appears to be derived in D. melanogaster as its close relatives, D. simulans, D. sechellia, and D. mauritiana, all appear to be monomorphic for isoleucine at this locus (based on at least 9 sequences from each species available in GenBank). This strongly suggests that the serine allele is a new mutation that has arisen in D. melanogaster, is favored due to its impact on postcopulatory sexual selection, and is increasing in frequency. Ultimately it may become fixed as an amino acid substitution and these results strongly suggest that sexual selection is driving the rapid evolution observed in Acp26Aa (Tsaur and Wu 1997; Tsaur et al. 1998; Aguadé 1999). We do not, however, observe reduced genetic diversity on the serine haplotypes (not shown) suggesting that if a sweep is in progress it may have started from standing genetic variation (Przeworski et al. 2005).
Furthermore, when considering all 3 genetic backgrounds simultaneously, we observed a positive correlation between a male’s ability to acquire mates (P2-mating) and the proportion of offspring that male sires (P2′). A positive covariance between male mating success and the number of offspring sired is one condition predicted to be necessary for polyandrous mating to enhance sexual selection among males (Shuster et al. 2013). Several previous studies have identified positive correlations between different components of male reproductive performance (Clark et al. 1995; Fiumera et al. 2005, 2007) suggesting that a common molecular mechanism might underlie the different traits. Other studies have identified negative correlations between P1 and P2, suggesting that tradeoffs exist (Fricke et al. 2010). Fricke et al. (2010), however, tested P1 and P2 using the same male such that P2 was measured after the male had previously mated. If males are able to strategically allocate sperm and/or seminal fluids (Wigby et al. 2009; Bretman et al. 2011; Lüpold et al. 2011; Garbaczewska et al. 2013; Moatt et al. 2014), then high allocation to the first mating could result in reduced allocation to the second mating and thus a tradeoff in performance between successive matings. Questions of strategic allocation, depletion and replenishment of sperm and seminal fluid proteins are exciting areas of current research.
Caveats and Future Directions
The potentials and pitfalls of association testing are well known (Cowperthwaite et al. 2010; Weir and Laurie 2010) but a few aspects do warrant comment. First, it is always prudent to remember that association testing does not demonstrate causation and whenever feasible it is important to attempt to verify the observed associations through additional studies. Second, a lack of association does not imply the gene is not important for the phenotype under study. It may simply indicate that the particular variants that are segregating (or segregating at a high enough frequency to detect associations) do not contribute to that particular trait. In addition, the observation of male–female interactions highlights how external factors (such as the mating partner or the environment) can affect the observed associations. It is also important to recognize that our measure of sperm competitive ability, the proportion of offspring sired (P2′) is a composite measure that includes both differential fertilization success and differential offspring viability (Gilchrist and Partridge 1997; Droge-Young et al. 2012) and thus we cannot determine the underlying mechanisms for the differences in the proportion of offspring sired. Drosophila is rich in shared genomic resources for studying the genetic basis for complex trait variation, including the DGRP collection (Mackay et al. 2012), the Drosophila Population Genetics program (Langley et al. 2012), the Drosophila Synthetic Population Resource (King et al. 2012a; King et al. 2012b) and the Global Diversity Lines (Greenberg et al. 2011). These resources will be invaluable as we to aim to further dissect the genetic and environmental contributions to sexual selection.
Supplementary Material
Supplementary material can be found at http://www.jhered.oxfordjournals.org/.
Funding
National Science Foundation (DEB-0743125 to A.C.F and A.G.C.); National Institutes of Health (R01 HD059060 to A.G.C.).
Supplementary Material
Acknowledgments
Assistance with the Drosophila experiments was provided by M. Eng, S. Fecht, C. Hwang, T. Giardina, M. Leib, M. Leung, J. Lin, F. Ooi, B. Patel, M Rubino, M. Sausner, M. Savoury, G. Tollin, N. Ulrich, S. Vayalumkal, J. Yahav, and R. Zhang.
References
- Abrams PA, Matsuda H, Harada Y. 1993. Evolutionarily unstable fitness maxima and stable fitness minima of continuous traits. Evol Ecol. 7:465–487. [Google Scholar]
- Aguadé M. 1999. Positive selection drives the evolution of the Acp29AB accessory gland protein in Drosophila. Genetics. 152:543–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alipaz JA, Wu CI, Karr TL. 2001. Gametic incompatibilities between races of Drosophila melanogaster . Proc Roy Soc Lond Series B Biol Sci. 268:789–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker CS. 2013. Journal of heredity adopts joint data archiving policy. J Hered. 104:1. [DOI] [PubMed] [Google Scholar]
- Barbash DA. 2010. Ninety years of Drosophila melanogaster hybrids. Genetics. 186:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkhead TR, Chaline N, Biggins JD, Burke T, Pizzari T. 2004. Nontransitivity of paternity in a bird. Evolution. 58:416–420. [PubMed] [Google Scholar]
- Bjork A, Starmer WT, Higginson DM, Rhodes CJ, Pitnick S. 2007. Complex interactions with females and rival males limit the evolution of sperm offence and defence. Proc Biol Sci. 274:1779–1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloch Qazi MC, Wolfner MF. 2003. An early role for the Drosophila melanogaster male seminal protein Acp36DE in female sperm storage. J Exp Biol. 206(Pt 19):3521–3528. [DOI] [PubMed] [Google Scholar]
- Bolnick DI, Svanbäck R, Fordyce JA, et al. 2003. The ecology of individuals: incidence and implications of individual specialization. Am Nat. 161:1–28. [DOI] [PubMed] [Google Scholar]
- Bretman A, Gage MJG, Chapman T. 2011. Quick-change artists: male plastic behavioural responses to rivals. Trends Ecol Evol. 26:467–473. [DOI] [PubMed] [Google Scholar]
- Bretman A, Lawniczak MK, Boone J, Chapman T. 2010. A mating plug protein reduces early female remating in Drosophila melanogaster. J Insect Physiol. 56:107–113. [DOI] [PubMed] [Google Scholar]
- Castric V, Vekemans X. 2004. Plant self-incompatibility in natural populations: a critical assessment of recent theoretical and empirical advances. Mol Ecol. 13:2873–2889. [DOI] [PubMed] [Google Scholar]
- Chapman T, Bangham J, Vinti G, et al. 2003. The sex peptide of Drosophila melanogaster: female post-mating responses analyzed by using RNA interference. Proc Natl Acad Sci USA. 100:9923–9928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman T, Neubaum DM, Wolfner MF, Partridge L. 2000. The role of male accessory gland protein Acp36DE in sperm competition in Drosophila melanogaster. Proc Roy Soc Lond B Biol Sci. 267:1097–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth D, Vekemans X, Castric V, Glémin S. 2005. Plant self-incompatibility systems: a molecular evolutionary perspective. New Phytol. 168:61–69. [DOI] [PubMed] [Google Scholar]
- Chippindale AK, Rice WR. 2001. Y chromosome polymorphism is a strong determinant of male fitness in Drosophila melanogaster. Proc Natl Acad Sci USA. 98:5677–5682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow CY, Wolfner MF, Clark AG. 2010. The genetic basis for male x female interactions underlying variation in reproductive phenotypes of Drosophila. Genetics. 186:1355–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow CY, Wolfner MF, Clark AG. 2013. Large neurological component to genetic differences underlying biased sperm use in Drosophila. Genetics. 193:177–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill GA, Doerge RW. 1994. Empirical threshold values for quantitative trait mapping. Genetics. 138:963–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civetta A, Clark AG. 2000. Chromosomal effects on male and female components of sperm precedence in Drosophila. Genet Res. 75:143–151. [DOI] [PubMed] [Google Scholar]
- Civetta A, Rosing KR, Fisher JH. 2008. Differences in sperm competition and sperm competition avoidance in Drosophila melanogaster . Anim Behav. 75:1739–1746. [Google Scholar]
- Clark AG. 2002. Sperm competition and the maintenance of polymorphism. Heredity (Edinb). 88:148–153. [DOI] [PubMed] [Google Scholar]
- Clark AG, Aguadé M, Prout T, Harshman LG, Langley CH. 1995. Variation in sperm displacement and its association with accessory gland protein loci in Drosophila melanogaster. Genetics. 139:189–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark AG, Begun DJ. 1998. Female genotypes affect sperm displacement in Drosophila. Genetics. 149:1487–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark AG, Begun DJ, Prout T. 1999. Female x male interactions in Drosophila sperm competition. Science. 283:217–220. [DOI] [PubMed] [Google Scholar]
- Clark NL, Gasper J, Sekino M, et al. 2009. Coevolution of interacting fertilization proteins. PLoS Genet. 5, 10.1371/journal.pgen.1000570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowperthwaite MC, Mohanty D, Burnett MG. 2010. Genome-wide association studies: a powerful tool for neurogenomics. Neurosurg Focus. 28:E2. [DOI] [PubMed] [Google Scholar]
- Coyne JA, Orr HA. 1998. The evolutionary genetics of speciation. Philos Trans R Soc Lond B Biol Sci. 353:287–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devictor V, Clavel J, Julliard R, et al. 2010. Defining and measuring ecological specialization. J Appl Ecol. 47:15–25. [Google Scholar]
- Droge-Young EM, Manier MK, Lüpold S, Belote JM, Pitnick S. 2012. Covariance among premating, post-copulatory and viability fitness components in Drosophila melanogaster and their influence on paternity measurement. J Evol Biol. 25:1555–1563. [DOI] [PubMed] [Google Scholar]
- Evans JP, Marshall DJ. 2005. Male-by-female interactions influence fertilization success and mediate the benefits of polyandry in the sea urchin Heliocidaris erythrogramma. Evolution. 59:106–112. [PubMed] [Google Scholar]
- Evans JP, Rosengrave P, Gasparini C, Gemmell NJ. 2013. Delineating the roles of males and females in sperm competition. Proc Roy Soc B Biol Sci. 280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang S, Takahashi A, Wu CI. 2002. A mutation in the promoter of desaturase 2 is correlated with sexual isolation between Drosophila behavioral races. Genetics. 162:781–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiumera AC, Dumont BL, Clark AG. 2005. Sperm competitive ability in Drosophila melanogaster associated with variation in male reproductive proteins. Genetics. 169:243–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiumera AC, Dumont BL, Clark AG. 2006. Natural variation in male-induced ‘cost-of-mating’ and allele-specific association with male reproductive genes in Drosophila melanogaster. Philos Trans R Soc Lond B Biol Sci. 361:355–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiumera AC, Dumont BL, Clark AG. 2007. Associations between sperm competition and natural variation in male reproductive genes on the third chromosome of Drosophila melanogaster. Genetics. 176:1245–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fricke C, Martin OY, Bretman A, Bussière LF, Chapman T. 2010. Sperm competitive ability and indices of lifetime reproductive success. Evolution. 64:2746–2757. [DOI] [PubMed] [Google Scholar]
- Garbaczewska M, Billeter JC, Levine JD. 2013. Drosophila melanogaster males increase the number of sperm in their ejaculate when perceiving rival males. J Insect Physiol. 59:306–310. [DOI] [PubMed] [Google Scholar]
- Giardina TJ, Beavis A, Clark AG, Fiumera AC. 2011. Female influence on pre- and post-copulatory sexual selection and its genetic basis in Drosophila melanogaster. Mol Ecol. 20:4098–4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist AS, Partridge L. 1997. Heritability of pre-adult viability differences can explain apparent heritability of sperm displacement ability in Drosophila melanogaster. Proc R Soc Lond Series B Biol Sci. 264:1271–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg AJ, Hackett SR, Harshman LG, Clark AG. 2011. Environmental and genetic perturbations reveal different networks of metabolic regulation. Mol Syst Biol. 7563, 10.1038/msb.2011.96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg AJ, Moran JR, Coyne JA, Wu CI. 2003. Ecological adaptation during incipient speciation revealed by precise gene replacement. Science. 302:1754–1757. [DOI] [PubMed] [Google Scholar]
- Greenspan L, Clark AG. 2011. Associations between variation in X chromosome male reproductive genes and sperm competitive ability in Drosophila melanogaster. Int J Evol Biol. 2011:214280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heino M, Metz JAJ, Kaitala V. 1998. The enigma of frequency-dependent selection. Trends Ecol Evol. 13:367–370. [DOI] [PubMed] [Google Scholar]
- Hughes KA. 1997. Quantitative genetics of sperm precedence in Drosophila melanogaster . Genetics. 145:139–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes KA, Leips J. 2006. Quantitative trait locus analysis of male mating success and sperm competition in Drosophila melanogaster. Evolution. 60:1427–1434. [PubMed] [Google Scholar]
- King EG, Macdonald SJ, Long AD. 2012. Properties and power of the Drosophila synthetic population resource for the routine dissection of complex traits. Genetics. 191:935–U522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King EG, Merkes CM, McNeil CL, et al. 2012b. Genetic dissection of a model complex trait using the Drosophila Synthetic Population Resource. Genome Res. 22:1558–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langley CH, Stevens K, Cardeno C, et al. 2012. Genomic variation in natural populations of Drosophila melanogaster. Genetics. 192:533–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics. 23:2947–2948. [DOI] [PubMed] [Google Scholar]
- Lawniczak MK, Begun DJ. 2005. A QTL analysis of female variation contributing to refractoriness and sperm competition in Drosophila melanogaster. Genet Res. 86:107–114. [DOI] [PubMed] [Google Scholar]
- Lew TA, Morrow EH, Rice WR. 2006. Standing genetic variance for female resistance to harm from males and its relationship to intralocus sexual conflict. Evolution. 60:97–105. [PubMed] [Google Scholar]
- Liu H, Kubli E. 2003. Sex-peptide is the molecular basis of the sperm effect in Drosophila melanogaster . Proc Natl Acad Sci USA. 100:9929–9933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lung O, Wolfner MF. 2001. Identification and characterization of the major Drosophila melanogaster mating plug protein. Insect Biochem Mol Biol. 31:543–551. [DOI] [PubMed] [Google Scholar]
- Lüpold S, Manier MK, Ala-Honkola O, Belote JM, Pitnick S. 2011. Male Drosophila melanogaster adjust ejaculate size based on female mating status, fecundity, and age. Behav Ecol. 22:184–191. [Google Scholar]
- Lüpold S, Pitnick S, Berben KS, et al. 2013. Female mediation of competitive fertilization success in Drosophila melanogaster. Proc Natl Acad Sci USA. 110:10693–10698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mack PD, Hammock BA, Promislow DE. 2002. Sperm competitive ability and genetic relatedness in Drosophila melanogaster: similarity breeds contempt. Evolution. 56:1789–1795. [DOI] [PubMed] [Google Scholar]
- Mackay TF, Richards S, Stone EA, et al. 2012. The Drosophila melanogaster Genetic Reference Panel. Nature. 482:173–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall DJ, Evans JP. 2005. Does egg competition occur in marine broadcast-spawners? J Evol Biol. 18:1244–1252. [DOI] [PubMed] [Google Scholar]
- McGraw LA, Fiumera AC, Ramakrishnan M, et al. 2007. Larval rearing environment affects several post-copulatory traits in Drosophila melanogaster . Biol Lett. 3:607–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalczyk Ł, Millard AL, Martin OY, et al. 2011. Inbreeding promotes female promiscuity. Science. 333:1739–1742. [DOI] [PubMed] [Google Scholar]
- Moatt JP, Dytham C, Thom MDF. 2014. Sperm production responds to perceived sperm competition risk in male Drosophila melanogaster. Physiol Behav. 131:111–114. [DOI] [PubMed] [Google Scholar]
- Neubaum DM, Wolfner MF. 1999. Mated Drosophila melanogaster females require a seminal fluid protein, Acp36DE, to store sperm efficiently. Genetics. 153:845–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson T, Fricke C, Arnqvist G. 2003. The effects of male and female genotype on variance in male fertilization success in the red flour beetle (Tribolium castaneum). Behav Ecol Sociobiol. 53:227–233. [Google Scholar]
- Palumbi SR. 1999. All males are not created equal: fertility differences depend on gamete recognition polymorphisms in sea urchins. Proc Natl Acad Sci USA. 96:12632–12637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panhuis TM, Nunney L. 2007. Insight into post-mating interactions between the sexes: relatedness suppresses productivity of singly mated female Drosophila melanogaster . J Evol Biol. 20:1988–1997. [DOI] [PubMed] [Google Scholar]
- Parker GA. 2006. Sexual conflict over mating and fertilization: an overview. Philos Trans Roy Soc Lond B Biol Sci. 361:235–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penn DJ, Potts WK. 1999. The evolution of mating preferences and major histocompatibility complex genes. Am Nat. 153:145–164. [DOI] [PubMed] [Google Scholar]
- Pischedda A, Stewart AD, Little MK. 2012. Male × female interaction for a pre-copulatory trait, but not a post-copulatory trait, among cosmopolitan populations of Drosophila melanogaster . PLoS One. 7:e31683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polak M, Starmer WT, Barker JSF. 1998. A mating plug and male mate choice in Drosophila hibisci Bock. Anim Behav. 56:919–926. [DOI] [PubMed] [Google Scholar]
- Polak M, Wolf LL, Starmer WT, Barker JSF. 2001. Function of the mating plug in Drosophila hibisci Bock. Behav Ecol Sociobiol. 49:196–205. [Google Scholar]
- Pool JE, Aquadro CF. 2006. History and structure of sub-Saharan populations of Drosophila melanogaster . Genetics. 174:915–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryke SR, Rollins LA, Griffith SC. 2010. Females use multiple mating and genetically loaded sperm competition to target compatible genes. Science. 329:964–967. [DOI] [PubMed] [Google Scholar]
- Przeworski M, Coop G, Wall JD. 2005. The signature of positive selection on standing genetic variation. Evolution. 59:2312–2323. [PubMed] [Google Scholar]
- R Core Team. 2014. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna: (Austria: ). http://www.R-project.org. [Google Scholar]
- Sagga N, Civetta A. 2011. Male-female interactions and the evolution of postmating prezygotic reproductive isolation among species of the virilis subgroup. Int J Evol Biol. 2011:485460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawby R, Hughes KA. 2001. Male genotype affects female longevity in Drosophila melanogaster . Evolution. 55:834–839. [DOI] [PubMed] [Google Scholar]
- Scheet P, Stephens M. 2006. A fast and flexible statistical model for large-scale population genotype data: applications to inferring missing genotypes and haplotypic phase. Am J Hum Genet. 78:629–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuster SM, Briggs WR, Dennis PA. 2013. How multiple mating by females affects sexual selection. Philos Trans Roy Soc B Biol Sci. 368, 10.1098/Rstb.2012.0046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snook RR, Chapman T, Moore PJ, Wedell N, Crudgington HS. 2009. Interactions between the sexes: new perspectives on sexual selection and reproductive isolation. Evol Ecol. 23:71–91. [Google Scholar]
- Soller M, Bownes M, Kubli E. 1997. Mating and sex peptide stimulate the accumulation of yolk in oocytes of Drosophila melanogaster . Eur J Biochem. 243:732–738. [DOI] [PubMed] [Google Scholar]
- Soller M, Bownes M, Kubli E. 1999. Control of oocyte maturation in sexually mature Drosophila females. Dev Biol. 208:337–351. [DOI] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R. 2003. Statistical significance for genomewide studies. Proc Natl Acad Sci USA. 100:9440–9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensson EI, Abbott J, Hardling R. 2005. Female polymorphism, frequency dependence, and rapid evolutionary dynamics in natural populations. Am Nat. 165:567–576. [DOI] [PubMed] [Google Scholar]
- Svensson EI, Abbott JK, Gosden TP, Coreau A. 2009. Female polymorphisms, sexual conflict and limits to speciation processes in animals. Evol Ecol. 23:93–108. [Google Scholar]
- Swanson WJ, Vacquier VD. 1998. Concerted evolution in an egg receptor for a rapidly evolving abalone sperm protein. Science. 281:710–712. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, et al. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 28:2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang S, Presgraves DC. 2009. Evolution of the Drosophila nuclear pore complex results in multiple hybrid incompatibilities. Science. 323:779–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The MathWorks Inc. 2010. Matlab. Natick: (MA: ): The MathWorks Inc. [Google Scholar]
- Tregenza T, Wedell N. 2000. Genetic compatibility, mate choice and patterns of parentage: invited review. Mol Ecol. 9:1013–1027. [DOI] [PubMed] [Google Scholar]
- Tsaur SC, Ting CT, Wu CI. 1998. Positive selection driving the evolution of a gene of male reproduction, Acp26Aa, of Drosophila: II. Divergence versus polymorphism. Mol Biol Evol. 15:1040–1046. [DOI] [PubMed] [Google Scholar]
- Tsaur SC, Wu CI. 1997. Positive selection and the molecular evolution of a gene of male reproduction, Acp26Aa of Drosophila. Mol Biol Evol. 14:544–549. [DOI] [PubMed] [Google Scholar]
- Via S. 1999. Reproductive isolation between sympatric races of pea aphids. I. Gene flow restriction and habitat choice. Evolution. 53:1446–1457. [DOI] [PubMed] [Google Scholar]
- Weir BS, Laurie CC. 2010. Characterizing allelic association in the genome era. Genet Res (Camb). 92:461–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigby S, Sirot LK, Linklater JR, et al. 2009. Seminal fluid protein allocation and male reproductive success. Curr Biol. 19:751–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CI, Hollocher H, Begun DJ, et al. 1995. Sexual isolation in Drosophila melanogaster: a possible case of incipient speciation. Proc Natl Acad Sci USA. 92:2519–2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeh JA, Zeh DW. 2003. Toward a new sexual selection paradigm: polyandry, conflict and incompatibility (Invited article). Ethology. 109:929–950. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.