Abstract
Developing improved methods for breast cancer risk prediction could facilitate the targeting of interventions to women at highest risk, thereby reducing mortality, while sparing low-risk women the costs and inconvenience of unnecessary testing and procedures. However, currently available risk assessment tools fall short of achieving accurate individual risk prediction, precluding implementation of this approach. Improving these tools will require the identification of new methods of assessing risk and increasing the accuracy of existing risk indicators. We review four emerging topics that may have importance for breast cancer risk assessment: etiological heterogeneity, genetic susceptibility, mammographic breast density and assessment of breast involution.
Keywords: Breast Cancer, Risk, Genetics, Mammographic Density, Involution, Etiology, Epidemiology
Introduction
Accurate estimation of breast cancer risk could enable the identification of high-risk women who might be most likely to benefit from specific interventions, while allowing low-risk women to safely avoid unnecessary screening and procedures. Successful application of this strategy could help reduce breast cancer mortality and limit false positive screening tests.
Breast cancer risk models are generally useful in predicting the number of cancers that will develop in populations, but cannot identify which individuals will develop cancer [1]. Thus, risk models are useful for determining sample sizes needed for adequate statistical power in clinical trials, but have less value for patient management. Accordingly, discovering new breast cancer risk factors and refining the assessment of established ones is important for improving risk assessment. In this review, we discuss four emerging topics in breast cancer risk prediction (Table 1): 1) etiological heterogeneity; 2) genetic susceptibility; 3) mammographic breast density and 4) molecular histology: involution of normal breast tissue.
Table 1.
Emerging topics in breast cancer risk prediction: Evidence and opportunities
| Emerging Topic | Sources of Evidence | Potential Translation | Future Directions |
|---|---|---|---|
Etiological Heterogeneity
|
|
|
|
Genetic susceptibility
|
|
|
|
Mammographic Breast Density (MBD)
|
|
|
|
Breast Involution
|
|
|
|
Etiological Heterogeneity
Breast cancer can be classified into distinctive, clinically relevant molecular subtypes based on mRNA profiling [2–5]. Detailed molecular characterization of breast cancer has revealed increasing biological diversity, which has been matched by a growing recognition that risk factor associations vary by tumor subtype.
Molecular epidemiological studies using immunohistochemistry for tumor subtyping show that reproductive risk factors are more strongly linked to estrogen receptor (ER)-positive or progesterone receptor (PR)-positive cancers than to receptor negative tumors [6, 7]. These relationships are compatible with the probable importance of cumulative exposure to sex-steroid hormones in the pathogenesis of ER-positive breast cancer [8].
In contrast to cancers that are ER-positive/PR-positive and human epidermal growth factor receptor 2 (HER 2) negative (“luminal” molecular subtype), triple-negative (TN) breast cancers are less strongly related to reproductive factors [9]. In particular, basal-like cancers (a subset of TN tumors) are clinically aggressive and associated with early onset, African American race and BRCA1 germline mutations [10–13]. Data suggest that basal-like cancers may underlie many of the differences in risk factor associations between ER-positive and ER-negative cancers [9].
Nulliparity
Nulliparity is associated with increased breast cancer risk overall, but does not seem to increase risk of TN tumors [7, 9], and may even be protective [10, 14, 15]. In a pooled analysis including up to 35,568 cases within the Breast Cancer Association Consortium (BCAC), women diagnosed with ER-negative breast cancers were less likely to be nulliparous than women diagnosed with ER-positive tumors (P=3×10−6), and the frequency of nulliparity was lowest among TN tumors (13% among women with TN cancers vs. 17% among women with luminal cancers). In a subset of 12 population-based BCAC studies, nulliparity was associated with increased risk of luminal tumors but not TN cancers [9]. Similarly, nulliparity was not associated with risk for TN tumors in another population-based case-control study not included within the BCAC analysis [16]. Moreover, nulliparity was associated with decreased risk of TN cancer in the Women’s Health Initiative (WHI) cohort [14] and decreased risk of ER-negative/PR-negative breast cancer in the Black Women’s Health Study [15], and the Carolina Breast Cancer Study [10], which included a high percentage of African American women. In addition, increasing age at first birth, an established breast cancer risk factor, was not associated with increased risk for TN tumors in several studies [9, 14, 16]
Breastfeeding
Breastfeeding for long durations has been associated with a modest reduction in breast cancer risk in two meta-analyses [17, 18], one of which included prospective data. However, some studies have found that breastfeeding is associated specifically with a substantial risk reduction for ER-negative/PR-negative or basal-like tumors, but not for ER-positive cancers [10, 15], whereas another did not find significant protection for any tumor type [14]. In addition, data suggest that breastfeeding eliminates the association between high parity and increased risk of ER-negative/PR-negative or basal-like breast cancers [10, 15, 19]. Limited data also suggest that breastfeeding may be particularly protective for women with a family history of breast cancer [20] and carriers of germline BRCA1 mutations [21].
Age at menarche
Most investigations suggest that early age at menarche is more strongly associated with increased risk of hormone receptor positive than receptor negative cancers [6, 7, 9, 14], with one investigation finding the strongest relationship with PR status [9]. However, data suggest that early age at menarche is not significantly different between women with TN tumors versus women with luminal tumors [9].
Obesity
Premenopausal obesity is protective for breast cancer, whereas postmenopausal obesity is associated with increased risk [22, 23]. Obesity may produce several potentially pro-carcinogenic effects, related to sex-steroid hormones, growth factors and inflammation [24]. In one report, waist-to-hip ratio (WHR), a measure of central obesity, was related to increased risk for TN tumors, among both premenopausal women (Odds ratio (OR)=1.8, 95% confidence interval (CI)=1.0, 3.4, Ptrend=0.07) and postmenopausal women (OR=2.7, 95% CI=1.3, 5.4, Ptrend=0.006) [10]. Obesity has been related to increased risk for TN tumors in two studies of predominantly white women [19, 25] and a similar suggestion was found in the pooled BCAC analysis [9].
Other risk factors
Menopausal hormone therapy (MHT) use has been associated with increased risk of ER-positive but not ER-negative tumors [26–28], whereas reported associations between oral contraceptive use and breast cancer subtypes are more variable [14, 16, 29]. A positive family history increases risk, irrespective of ER status [9, 30], though risks may be greatest for basal-like cancers [9].
Etiological heterogeneity: risk prediction and future directions
Data suggest that the Gail Model provides more accurate risk prediction of ER-positive breast cancer than ER-negative tumors [31]. The development of a risk model for ER-positive breast cancer could enable more specific identification of women most likely to benefit from chemoprevention with endocrine agents, which may only reduce risk for hormone-dependent tumors. By analogy, models could be developed to identify women at high-risk for other specific molecular subtypes of breast cancer that could be prevented with targeted interventions. However, implementation of multiple risk models would be complex compared with use of an omnibus risk prediction model for all tumor types.
Genetic susceptibility
Women who have a family history of breast cancer are personally at increased risk, suggesting the importance of genetic factors in the etiology of the disease [32]. Furthermore, risk varies with the number of affected relatives, the closeness of their relationship (i.e. first or second degree) and the relatives’ ages at diagnosis [32]. Risk is similar for women with an affected maternal as compared to an affected paternal relative. A meta-analysis of 74 studies showed that women who have a first-degree relative with breast cancer are at approximately 2-fold increased risk, whereas those with an affected second-degree relative were at approximately 1.5-fold increased risk [32]. Having a relative diagnosed by age 40 years increases risk fivefold whereas having a relative diagnosed at age 60 years or older increases risk by 40%. In contrast to a positive family history, a negative family history provides limited risk information because 80% to 90% breast cancers occur among women without an affected close relative. Increasingly, genetic research has focused on identifying specific markers of risk, including uncommon variants conferring large or moderate risk, and common variants conferring small increases in risk.
Genetic risk factors for breast cancer: Genes with high or moderate penetrance
An estimated 57% of women with BRCA1 mutations and 49% of those with BRCA2 mutations will develop breast cancer by age 70 years [33]. Cancers among BRCA2 mutation carriers differ from those among BRCA1 carriers in that they more close resemble the range of cancer subtypes that occur in the general population (i.e. predominantly ER-positive) and are diagnosed at older ages. Other high penetrance mutations linked to elevated breast cancer risk are discussed elsewhere, including: TP53 in Li-Fraumeni syndrome [34], PTEN in Cowden syndrome [35, 36], and STK11/LKB1 in Peutz-Jegher syndrome [37]. Mutations in CDH1 (which encodes e-cadherin) have been linked to increased risk, especially for lobular cancers [38].
Mutations with moderate penetrance (ATM, CHEK2, BRIP1, and PALB2) [39–42] confer a two- to four-fold increase in breast cancer risk and are quite rare in the general population. CHEK2 mutations may confer susceptibility specifically to luminal breast cancer subtypes, including tumors with lobular histology [43–45]. Clear links between mutations in ATM, PALB2, and BRIP1 and breast cancers with specific molecular or pathologic characteristics have not been established. Efforts to identify additional moderately penetrant susceptibility mutations are ongoing.
Common variants of low penetrance
A prior report based on 13 common low penetrant genetic variants suggested that these factors explained about 8.3% of familial risk, as compared with 5% for moderately penetrant genes and 22% for highly penetrant mutations [46]. Genome-wide association studies (GWAS) have identified over 30 common low penetrant variants as of this writing, the majority of which are single nucleotide polymorphism (SNP) markers, which confer breast cancer risks ranging from 1.04–1.40 per-allele. SNPs have been associated with important breast cancer features, such as ER, PR or HER2 status, grade, and histology [47–54].
Genetic susceptibility: risk prediction and future directions
Analyses suggest that adding known SNPs to breast cancer risk models produces only modest improvements in risk prediction [55]. In addition, strong gene-gene or gene-environment interactions that would identify subsets of high-risk women have not been found [56–58], nor have variants been strongly linked to breast cancer outcomes [59]. Research on the functional consequences of SNPs on carcinogenic processes is in its infancy, and the identification of SNPs in non-coding regions of the genome raises intriguing questions about the mechanisms that mediate the risk associated with of these variants. Identification of variants related to ER-negative breast cancers and early onset tumors is a research priority. A web-based computer program for assessing risk among patients with a family history of breast and/or ovarian cancer has been developed (BOADICEA) [60]. Finally, a polygenic model that combines multiple common susceptibility variants to predict risk has been developed, which could inform future public health recommendations about screening women based on level of risk [54, 61, 62].
Mammographic breast density
Mammographic breast density (MBD) reflects the tissue composition of the breast: high MBD corresponds to a greater percentage of fibroglandular tissue relative to fat, whereas low MBD is related to a higher percentage of fat relative to non-fatty tissue. Although high MBD is related to some breast cancer risk factors such as nulliparity, a positive family history, and MHT use, many cross-sectional and prospective studies have consistently demonstrated that high MBD is a strong and independent breast cancer risk factor, conferring relative risks (RRs) of 4- to 5-fold when comparing women with highest to lowest MBD (reviewed in [63]). Dense areas on a mammogram, which appear white (Figure 1B), may mask tumors, leading to delayed detection; however, high MBD is related to long-term prospective increases in tumor incidence, independent of its effects on detection [64]. Given that elevated MBD is the strongest risk factor for non-familial breast cancer apart from age and gender [65, 66], and that many women have high MBD [66], assessment of MBD represents a potentially useful breast cancer risk assessment tool.
Figure 1.
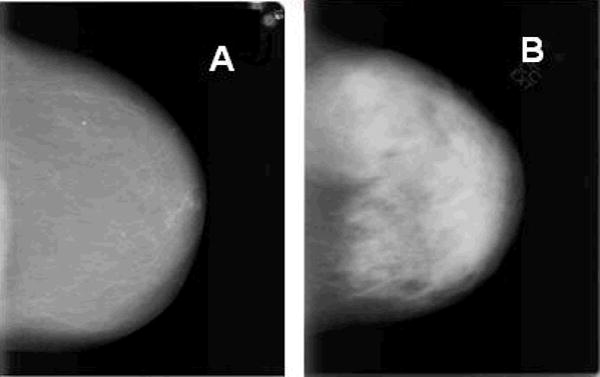
Digitized mammograms from NCI Polish Breast Cancer Study participants where A and B represent breasts of low and high mammographic breast density (MBD), respectively.
Little is known about the biology of high MBD and why it is related to breast cancer risk. MBD has a substantial heritable component; it is estimated that ~2/3 of the variance in density is genetically determined [67], suggesting that density acts at early stages in carcinogenesis. As described by Boyd et al. [68], density at young ages may be a key risk marker because it reflects the number of undifferentiated cells that are vulnerable to carcinogenic insults before the differentiating effects of pregnancy and age-related involution.
Methods for assessing mammographic breast density
Methods for assessing MBD have become increasingly more quantitative, reproducible and automated. MBD can be estimated visually as the extent of dense tissue in mammograms (e.g., Wolfe’s parenchymal patterns and the American College of Radiology Breast Imaging-Reporting and Data System (BI-RADS)) or quantitatively as an absolute area or as a percentage of total breast area using planimetry or computer-assisted methods, which are more reliable. Recently, techniques to assess radiologically dense tissue as a volume have been developed (reviewed in [69]).
Clinical relevance of mammographic breast density
High MBD is clinically important because women with such breasts are more likely to develop interval cancers (undetected by screening mammography). Furthermore, most studies have found that high MBD increases risk for both ER-positive and ER-negative cancers [70–73]. MBD and its associations with risk also do not appear to differ between carriers of high penetrant mutations, such as BRCA1/2, as compared to non-carriers [74, 75]. Therefore, understanding the mechanisms that mediate the risk related to high MBD may increase our knowledge about etiological factors that contribute to most subtypes of breast cancer and could facilitate the development of prevention strategies with broad impact.
High MBD has been linked to increased risk of breast cancer recurrence (reviewed in [76]). In addition, MBD may reflect the breast cancer risk associated with use of exogenous hormones. Specifically, limited data suggest that women whose MBD declines after receiving endocrine agents for chemoprevention [77] or adjuvant treatment [78] are more likely to benefit from these medications than those whose MBD does not fall. In contrast, elevated MBD in the context of MHT or oral contraceptive use may be associated with increased breast cancer risk [79]. Thus, MBD is potentially a strong, modifiable “biosensor” of breast cancer risk, which may have utility in multiple populations and in different clinical settings.
Mammographic breast density: risk prediction and future directions
Several studies [80–83] have suggested that adding MBD to the Gail model may improve breast cancer risk prediction modestly and efforts to incorporate MBD in newer risk models are ongoing [84]. Of those studies incorporating MBD into the Gail model, three [80, 81, 83] evaluated the addition of BI-RADS density categories and the fourth [82] used a quantitative measure of MBD as assessed by planimetry. These results show modest but consistent improvements in risk prediction by adding MBD to existing risk prediction models [80–82]. However, the potential gains in risk prediction that might be realized by using automated, quantitative measures of density obtained through full field digital mammography or other emerging technologies have not been fully explored. In addition, data related to MBD and breast cancer risk in non-White populations are limited. Finally, elevated MBD may produce its strongest effect among young women who are below the age of initiation of mammographic screening, but who might benefit from preventive interventions [66]. Evaluating density without exposing young women to ionizing radiation is critical, and these approaches have not been implemented in clinical practice.
Subjective visual assessment of MBD is often performed clinically, but this approach has only moderate inter-rater reliability [85, 86] and assigns most women to two of four possible categories, thereby providing limited risk discrimination. Measurement of MBD is limited by properties of mammography, including: 1) two-dimensional representation; 2) need for compression, which distorts tissue architecture and varies between examinations; and 3) ionizing radiation exposure, which poses cancers risks.
It is hoped that more accurate and precise measurement of MBD is achievable through technological advances, which will increase its clinical utility. Methods for measuring breast density as a volume and in specific regions of the breast using digital mammography with density phantoms [87] and other breast imaging modalities are rapidly evolving [69]. Non-ionizing technologies, such as MRI and ultrasound tomography, may be ideally suited for assessing volumetric density in young or high risk women or in situations where it is desirable to perform more frequent measurements [88]. These evolving technologies may also offer further opportunities for increasing accuracy in measurement and identifying stronger risk associations.
Molecular histology: involution of normal breast tissue
“Molecular histology” may be defined as the sum total of all microscopic and molecular characteristics of the breast [89]. With aging, the breast undergoes dramatic structural and compositional changes, including a reduction in epithelium, followed successively by stromal and then adipose tissue replacement [90–92]. Recent studies suggest that analysis of terminal duct lobular units (TDLUs), the benign structures from which most breast cancers arise, may be useful for breast cancer risk prediction [92–94]. TDLU involution begins prior to menopause, progresses with aging, and varies substantially among women, reflecting differences in reproductive history and other factors [90, 91, 93]. Thus, TDLU involution could represent a global measure of risk, which reflects the interaction of genetic and environmental risk factors over time.
The number of TDLUs in the breast, like MBD, declines with aging, although breast cancer risk increases [95]. As has been proposed for MBD [88], this apparent paradox may be reconciled by viewing these factors as proxies for exposure to carcinogenic influences, culminating during critical periods of heightened susceptibility to malignant transformation, like prior to a first birth. From this perspective, women with less TDLU involution are postulated to have had more at-risk epithelium over their lifetimes or during vulnerable periods.
TDLU involution may be viewed as a reduction in the number of TDLUs or simplification of the structure of individual TDLUs, manifested as a shorter diameter or a reduced number of acini (functional subunits of TDLUs). A retrospective analysis of 8,736 benign breast biopsies found that TDLU involution was absent in 18.6%, partial in 59.5% and complete in 21.9% [93]. The percentage of biopsies with complete TDLU involution increased with age, reaching 53.1% among women aged 70 years or older. Having given birth, a strong family history of breast cancer, use of menopausal hormones or having proliferative breast disease was associated with less TDLU involution. Preliminary analyses suggest that many of these relationships hold in normal breast tissues donated by volunteers (unpublished observation).
Compared with population-based rates, Milanese et al. reported that women with TDLU involution categorized as “none” had increased breast cancer risk (RR=1.88, 95% CI=1.59, 2.21), as did women with “partial” involution (RR=1.47, 95% CI=1.33, 1.61); risk for women with “complete” involution was close to unity [93]. Levels of TDLU involution stratified women’s breast cancer risk irrespective of other factors including family history, parity, age at first birth biopsy and the presence of hyperplasia or atypical hyperplasia. Similar but less significant relationships with risk were reported in a smaller case-control comparison nested within another cohort [94].
Data indicated that levels of TDLU involution are similar throughout both breasts [96], suggesting that TDLU involution represents a global marker of risk. Breasts that contain more TDLUs (less involution) are associated with high MBD [95].
Molecular histology: risk prediction and future directions
The degree of TDLU involution is generally inversely related to factors that increase breast cancer risk. However, these associations raise questions as to whether TDLU involution represents an independent risk marker. In one analysis, both MBD and TDLU involution were significantly associated with breast cancer risk after mutual adjustment. Compared with women who had low MBD and complete involution, women who had high MBD and no involution had a RR=4.08 (95% CI=1.72, 9.68) [97]. Limited data also suggest that TLDU involution levels in tissues surrounding breast cancers may represent a marker of etiological heterogeneity. Specifically, TDLUs associated with basal-like cancers showed significantly less involution than those surrounding luminal cancers in age-adjusted analyses [98].
Molecular histology generally, and TDLU analysis specifically, may offer an objective method for breast cancer risk assessment that reflects cumulative effects of exposures on the target organ. The approach bypasses concerns related to imperfect recall of medical history and may reflect important effects of unknown risk factors and interactions among factors, both genetic and environmental [89]. Despite subjectivity, data suggest that visual and computer-assisted assessment of TDLU characteristics yield reproducible results [98, 99] (and unpublished data). A limitation of TDLU analysis is that it requires access to tissue; however, breast biopsies are common and precede a cancer diagnosis among many women. Another significant challenge is that when TDLUs are not identified in a biopsy of limited dimensions, it is difficult to judge whether this represents complete involution (i.e. most TDLUs have been replaced by stroma throughout the breast) or sampling of a non-representative area.
Improving criteria for visually categorizing TDLU involution and developing image analysis tools for quantifying TDLU metrics may increase the utility of this approach for risk prediction [99]. It is unclear which metrics of TDLU involution best predict breast cancer risk. Given that increased expression of ER in benign epithelium has been associated with elevated breast cancer risk [100], it is hoped that molecular analysis of TDLUs might be useful for risk prediction, if valid techniques can be developed.
Conclusion
Developing improved approaches for breast cancer risk assessment offers a potential means of targeting interventions to women most likely to benefit, which could lead to reduced mortality, lowered costs and more efficient screening. We explore four possible approaches for achieving that objective: accounting for etiological heterogeneity; testing for genetic susceptibility factors, measurement of MBD and assessment of TDLU involution. These topics reflect the overall complexity of breast cancer risk prediction, given the enormous diversity among women and the breast cancers that they develop. Future research will reveal whether understanding the heterogeneity of this complex disease represents a gateway to progress or a challenge to clinical translation.
Acknowledgments
Funding
This work was supported (in part) by the Intramural Research Program of the National Institutes of Health, National Cancer Institute.
Contributor Information
Gretchen L. Gierach, Email: gierachg@mail.nih.gov, National Cancer Institute, Division of Cancer Epidemiology and Genetics, Hormonal and Reproductive Epidemiology Branch, 6120 Executive Blvd., Rockville, MD 20852-7234; Phone: 301-594-5635; Fax: 301-402-0916.
Xiaohong R. Yang, Email: royang@mail.nih.gov, National Cancer Institute, Division of Cancer Epidemiology and Genetics, Genetic Epidemiology Branch, 6120 Executive Blvd., Rockville, MD 20852-7234; Phone: 301-594-7804.
Jonine D. Figueroa, Email: figueroaj@mail.nih.gov, National Cancer Institute, Division of Cancer Epidemiology and Genetics, Hormonal and Reproductive Epidemiology Branch, 6120 Executive Blvd., Rockville, MD 20852-7234; Phone: 301-402-3654; Fax: 301-402-0916.
Mark E. Sherman, Email: shermanm@mail.nih.gov, National Cancer Institute, Division of Cancer Epidemiology and Genetics, Hormonal and Reproductive Epidemiology Branch, 6120 Executive Blvd., Rockville, MD 20852-7234; Phone: 301-594-7661; Fax: 301-402-0916.
References
* Denotes annotated reference of importance
** Denotes annotated reference of outstanding importance
- *1.Cummings SR, Tice JA, Bauer S, Browner WS, Cuzick J, Ziv E, Vogel V, Shepherd J, Vachon C, Smith-Bindman R, et al. Prevention of breast cancer in postmenopausal women: approaches to estimating and reducing risk. Journal of the National Cancer Institute. 2009;101(6):384–398. doi: 10.1093/jnci/djp018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, et al. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 3.Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(19):10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486(7403):346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Althuis MD, Fergenbaum JH, Garcia-Closas M, Brinton LA, Madigan MP, Sherman ME. Etiology of hormone receptor-defined breast cancer: a systematic review of the literature. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2004;13(10):1558–1568. [PubMed] [Google Scholar]
- 7.Ma H, Bernstein L, Pike MC, Ursin G. Reproductive factors and breast cancer risk according to joint estrogen and progesterone receptor status: a meta-analysis of epidemiological studies. Breast cancer research : BCR. 2006;8(4):R43. doi: 10.1186/bcr1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Henderson BE, Feigelson HS. Hormonal carcinogenesis. Carcinogenesis. 2000;21(3):427–433. doi: 10.1093/carcin/21.3.427. [DOI] [PubMed] [Google Scholar]
- 9.Yang XR, Chang-Claude J, Goode EL, Couch FJ, Nevanlinna H, Milne RL, Gaudet M, Schmidt MK, Broeks A, Cox A, et al. Associations of breast cancer risk factors with tumor subtypes: a pooled analysis from the Breast Cancer Association Consortium studies. Journal of the National Cancer Institute. 2011;103(3):250–263. doi: 10.1093/jnci/djq526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Millikan RC, Newman B, Tse CK, Moorman PG, Conway K, Dressler LG, Smith LV, Labbok MH, Geradts J, Bensen JT, et al. Epidemiology of basal-like breast cancer. Breast cancer research and treatment. 2008;109(1):123–139. doi: 10.1007/s10549-007-9632-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carey LA, Perou CM, Livasy CA, Dressler LG, Cowan D, Conway K, Karaca G, Troester MA, Tse CK, Edmiston S, et al. Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. JAMA : the journal of the American Medical Association. 2006;295(21):2492–2502. doi: 10.1001/jama.295.21.2492. [DOI] [PubMed] [Google Scholar]
- 12.Turner NC, Reis-Filho JS. Basal-like breast cancer and the BRCA1 phenotype. Oncogene. 2006;25(43):5846–5853. doi: 10.1038/sj.onc.1209876. [DOI] [PubMed] [Google Scholar]
- 13.Livasy CA, Karaca G, Nanda R, Tretiakova MS, Olopade OI, Moore DT, Perou CM. Phenotypic evaluation of the basal-like subtype of invasive breast carcinoma. Mod Pathol. 2006;19(2):264–271. doi: 10.1038/modpathol.3800528. [DOI] [PubMed] [Google Scholar]
- 14.Phipps AI, Chlebowski RT, Prentice R, McTiernan A, Wactawski-Wende J, Kuller LH, Adams-Campbell LL, Lane D, Stefanick ML, Vitolins M, et al. Reproductive history and oral contraceptive use in relation to risk of triple-negative breast cancer. Journal of the National Cancer Institute. 2011;103(6):470–477. doi: 10.1093/jnci/djr030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Palmer JR, Boggs DA, Wise LA, Ambrosone CB, Adams-Campbell LL, Rosenberg L. Parity and lactation in relation to estrogen receptor negative breast cancer in African American women. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2011;20(9):1883–1891. doi: 10.1158/1055-9965.EPI-11-0465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ma H, Wang Y, Sullivan-Halley J, Weiss L, Marchbanks PA, Spirtas R, Ursin G, Burkman RT, Simon MS, Malone KE, et al. Use of four biomarkers to evaluate the risk of breast cancer subtypes in the women’s contraceptive and reproductive experiences study. Cancer research. 2010;70(2):575–587. doi: 10.1158/0008-5472.CAN-09-3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Breast cancer and breastfeeding: collaborative reanalysis of individual data from 47 epidemiological studies in 30 countries, including 50302 women with breast cancer and 96973 women without the disease. Lancet. 2002;360(9328):187–195. doi: 10.1016/S0140-6736(02)09454-0. [DOI] [PubMed] [Google Scholar]
- 18.Bernier MO, Plu-Bureau G, Bossard N, Ayzac L, Thalabard JC. Breastfeeding and risk of breast cancer: a metaanalysis of published studies. Human reproduction update. 2000;6(4):374–386. doi: 10.1093/humupd/6.4.374. [DOI] [PubMed] [Google Scholar]
- 19.Gaudet MM, Press MF, Haile RW, Lynch CF, Glaser SL, Schildkraut J, Gammon MD, Douglas Thompson W, Bernstein JL. Risk factors by molecular subtypes of breast cancer across a population-based study of women 56 years or younger. Breast cancer research and treatment. 2011;130(2):587–597. doi: 10.1007/s10549-011-1616-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stuebe AM, Willett WC, Xue F, Michels KB. Lactation and incidence of premenopausal breast cancer: a longitudinal study. Archives of internal medicine. 2009;169(15):1364–1371. doi: 10.1001/archinternmed.2009.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kotsopoulos J, Lubinski J, Salmena L, Lynch HT, Kim-Sing C, Foulkes WD, Ghadirian P, Neuhausen SL, Demsky R, Tung N, et al. Breastfeeding and the risk of breast cancer in BRCA1 and BRCA2 mutation carriers. Breast cancer research : BCR. 2012;14(2):R42. doi: 10.1186/bcr3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ursin G, Longnecker MP, Haile RW, Greenland S. A meta-analysis of body mass index and risk of premenopausal breast cancer. Epidemiology. 1995;6(2):137–141. doi: 10.1097/00001648-199503000-00009. [DOI] [PubMed] [Google Scholar]
- 23.Lahmann PH, Hoffmann K, Allen N, van Gils CH, Khaw KT, Tehard B, Berrino F, Tjonneland A, Bigaard J, Olsen A, et al. Body size and breast cancer risk: findings from the European Prospective Investigation into Cancer And Nutrition (EPIC) International journal of cancer Journal international du cancer. 2004;111(5):762–771. doi: 10.1002/ijc.20315. [DOI] [PubMed] [Google Scholar]
- 24.Brown KA, Simpson ER. Obesity and breast cancer: mechanisms and therapeutic implications. Front Biosci (Elite Ed) 2012;4:2515–2524. doi: 10.2741/e562. [DOI] [PubMed] [Google Scholar]
- 25.Phipps AI, Malone KE, Porter PL, Daling JR, Li CI. Body size and risk of luminal, HER2-overexpressing, and triple-negative breast cancer in postmenopausal women. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2008;17(8):2078–2086. doi: 10.1158/1055-9965.EPI-08-0206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rosenberg LU, Einarsdottir K, Friman EI, Wedren S, Dickman PW, Hall P, Magnusson C. Risk factors for hormone receptor-defined breast cancer in postmenopausal women. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2006;15(12):2482–2488. doi: 10.1158/1055-9965.EPI-06-0489. [DOI] [PubMed] [Google Scholar]
- 27.Bao PP, Shu XO, Gao YT, Zheng Y, Cai H, Deming SL, Ruan ZX, Su Y, Gu K, Lu W, et al. Association of hormone-related characteristics and breast cancer risk by estrogen receptor/progesterone receptor status in the shanghai breast cancer study. Am J Epidemiol. 2011;174(6):661–671. doi: 10.1093/aje/kwr145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li CI, Malone KE, Porter PL, Weiss NS, Tang MT, Cushing-Haugen KL, Daling JR. Relationship between long durations and different regimens of hormone therapy and risk of breast cancer. JAMA : the journal of the American Medical Association. 2003;289(24):3254–3263. doi: 10.1001/jama.289.24.3254. [DOI] [PubMed] [Google Scholar]
- 29.Dolle JM, Daling JR, White E, Brinton LA, Doody DR, Porter PL, Malone KE. Risk factors for triple-negative breast cancer in women under the age of 45 years. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2009;18(4):1157–1166. doi: 10.1158/1055-9965.EPI-08-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Phipps AI, Buist DS, Malone KE, Barlow WE, Porter PL, Kerlikowske K, Li CI. Family history of breast cancer in first-degree relatives and triple-negative breast cancer risk. Breast cancer research and treatment. 2011;126(3):671–678. doi: 10.1007/s10549-010-1148-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chlebowski RT, Anderson GL, Lane DS, Aragaki AK, Rohan T, Yasmeen S, Sarto G, Rosenberg CA, Hubbell FA. Predicting risk of breast cancer in postmenopausal women by hormone receptor status. Journal of the National Cancer Institute. 2007;99(22):1695–1705. doi: 10.1093/jnci/djm224. [DOI] [PubMed] [Google Scholar]
- 32.Pharoah PD, Day NE, Duffy S, Easton DF, Ponder BA. Family history and the risk of breast cancer: a systematic review and meta-analysis. International journal of cancer Journal international du cancer. 1997;71(5):800–809. doi: 10.1002/(sici)1097-0215(19970529)71:5<800::aid-ijc18>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 33.Chen S, Parmigiani G. Meta-analysis of BRCA1 and BRCA2 penetrance. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2007;25(11):1329–1333. doi: 10.1200/JCO.2006.09.1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mai PL, Malkin D, Garber JE, Schiffman JD, Weitzel JN, Strong LC, Wyss O, Locke L, Means V, Achatz MI, et al. Li-Fraumeni syndrome: report of a clinical research workshop and creation of a research consortium. Cancer genetics. 2012;205(10):479–487. doi: 10.1016/j.cancergen.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liaw D, Marsh DJ, Li J, Dahia PL, Wang SI, Zheng Z, Bose S, Call KM, Tsou HC, Peacocke M, et al. Germline mutations of the PTEN gene in Cowden disease, an inherited breast and thyroid cancer syndrome. Nature genetics. 1997;16(1):64–67. doi: 10.1038/ng0597-64. [DOI] [PubMed] [Google Scholar]
- 36.Hemel D, Domchek SM. Breast cancer predisposition syndromes. Hematology/oncology clinics of North America. 2010;24(5):799–814. doi: 10.1016/j.hoc.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 37.Hemminki A. The molecular basis and clinical aspects of Peutz-Jeghers syndrome. Cellular and molecular life sciences : CMLS. 1999;55(5):735–750. doi: 10.1007/s000180050329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schrader KA, Masciari S, Boyd N, Wiyrick S, Kaurah P, Senz J, Burke W, Lynch HT, Garber JE, Huntsman DG. Hereditary diffuse gastric cancer: association with lobular breast cancer. Familial cancer. 2008;7(1):73–82. doi: 10.1007/s10689-007-9172-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Renwick A, Thompson D, Seal S, Kelly P, Chagtai T, Ahmed M, North B, Jayatilake H, Barfoot R, Spanova K, et al. ATM mutations that cause ataxia-telangiectasia are breast cancer susceptibility alleles. Nature genetics. 2006;38(8):873–875. doi: 10.1038/ng1837. [DOI] [PubMed] [Google Scholar]
- 40.Meijers-Heijboer H, van den Ouweland A, Klijn J, Wasielewski M, de Snoo A, Oldenburg R, Hollestelle A, Houben M, Crepin E, van Veghel-Plandsoen M, et al. Low-penetrance susceptibility to breast cancer due to CHEK2(*)1100delC in noncarriers of BRCA1 or BRCA2 mutations. Nature genetics. 2002;31(1):55–59. doi: 10.1038/ng879. [DOI] [PubMed] [Google Scholar]
- 41.Rahman N, Seal S, Thompson D, Kelly P, Renwick A, Elliott A, Reid S, Spanova K, Barfoot R, Chagtai T, et al. PALB2, which encodes a BRCA2-interacting protein, is a breast cancer susceptibility gene. Nature genetics. 2007;39(2):165–167. doi: 10.1038/ng1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Seal S, Thompson D, Renwick A, Elliott A, Kelly P, Barfoot R, Chagtai T, Jayatilake H, Ahmed M, Spanova K, et al. Truncating mutations in the Fanconi anemia J gene BRIP1 are low-penetrance breast cancer susceptibility alleles. Nature genetics. 2006;38(11):1239–1241. doi: 10.1038/ng1902. [DOI] [PubMed] [Google Scholar]
- 43.Huzarski T, Cybulski C, Domagala W, Gronwald J, Byrski T, Szwiec M, Woyke S, Narod SA, Lubinski J. Pathology of breast cancer in women with constitutional CHEK2 mutations. Breast cancer research and treatment. 2005;90(2):187–189. doi: 10.1007/s10549-004-3778-2. [DOI] [PubMed] [Google Scholar]
- 44.Nagel JH, Peeters JK, Smid M, Sieuwerts AM, Wasielewski M, de Weerd V, Trapman-Jansen AM, van den Ouweland A, Bruggenwirth H, van IJWF, et al. Gene expression profiling assigns CHEK2 1100delC breast cancers to the luminal intrinsic subtypes. Breast cancer research and treatment. 2012;132(2):439–448. doi: 10.1007/s10549-011-1588-x. [DOI] [PubMed] [Google Scholar]
- 45.Domagala P, Wokolorczyk D, Cybulski C, Huzarski T, Lubinski J, Domagala W. Different CHEK2 germline mutations are associated with distinct immunophenotypic molecular subtypes of breast cancer. Breast cancer research and treatment. 2012;132(3):937–945. doi: 10.1007/s10549-011-1635-7. [DOI] [PubMed] [Google Scholar]
- 46.Mavaddat N, Antoniou AC, Easton DF, Garcia-Closas M. Genetic susceptibility to breast cancer. Molecular oncology. 2010;4(3):174–191. doi: 10.1016/j.molonc.2010.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Broeks A, Schmidt MK, Sherman ME, Couch FJ, Hopper JL, Dite GS, Apicella C, Smith LD, Hammet F, Southey MC, et al. Low penetrance breast cancer susceptibility loci are associated with specific breast tumor subtypes: findings from the Breast Cancer Association Consortium. Human molecular genetics. 2011;20(16):3289–3303. doi: 10.1093/hmg/ddr228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Figueroa JD, Garcia-Closas M, Humphreys M, Platte R, Hopper JL, Southey MC, Apicella C, Hammet F, Schmidt MK, Broeks A, et al. Associations of common variants at 1p11.2 and 14q24.1 (RAD51L1) with breast cancer risk and heterogeneity by tumor subtype: findings from the Breast Cancer Association Consortium. Human molecular genetics. 2011;20(23):4693–4706. doi: 10.1093/hmg/ddr368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Milne RL, Benitez J, Nevanlinna H, Heikkinen T, Aittomaki K, Blomqvist C, Arias JI, Zamora MP, Burwinkel B, Bartram CR, et al. Risk of estrogen receptor-positive and -negative breast cancer and single-nucleotide polymorphism 2q35-rs13387042. Journal of the National Cancer Institute. 2009;101(14):1012–1018. doi: 10.1093/jnci/djp167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Haiman CA, Chen GK, Vachon CM, Canzian F, Dunning A, Millikan RC, Wang X, Ademuyiwa F, Ahmed S, Ambrosone CB, et al. A common variant at the TERT-CLPTM1L locus is associated with estrogen receptor-negative breast cancer. Nature genetics. 2011;43(12):1210–1214. doi: 10.1038/ng.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lambrechts D, Truong T, Justenhoven C, Humphreys MK, Wang J, Hopper JL, Dite GS, Apicella C, Southey MC, Schmidt MK, et al. 11q13 is a susceptibility locus for hormone receptor positive breast cancer. Human mutation. 2012;33(7):1123–1132. doi: 10.1002/humu.22089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fletcher O, Johnson N, Orr N, Hosking FJ, Gibson LJ, Walker K, Zelenika D, Gut I, Heath S, Palles C, et al. Novel breast cancer susceptibility locus at 9q31.2: results of a genome-wide association study. Journal of the National Cancer Institute. 2011;103(5):425–435. doi: 10.1093/jnci/djq563. [DOI] [PubMed] [Google Scholar]
- 53.Ghoussaini M, Fletcher O, Michailidou K, Turnbull C, Schmidt MK, Dicks E, Dennis J, Wang Q, Humphreys MK, Luccarini C, et al. Genome-wide association analysis identifies three new breast cancer susceptibility loci. Nature genetics. 2012;44(3):312–318. doi: 10.1038/ng.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Antoniou AC, Wang X, Fredericksen ZS, McGuffog L, Tarrell R, Sinilnikova OM, Healey S, Morrison J, Kartsonaki C, Lesnick T, et al. A locus on 19p13 modifies risk of breast cancer in BRCA1 mutation carriers and is associated with hormone receptor-negative breast cancer in the general population. Nature genetics. 2010;42(10):885–892. doi: 10.1038/ng.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Park JH, Gail MH, Greene MH, Chatterjee N. Potential usefulness of single nucleotide polymorphisms to identify persons at high cancer risk: an evaluation of seven common cancers. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2012;30(17):2157–2162. doi: 10.1200/JCO.2011.40.1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Campa D, Kaaks R, Le Marchand L, Haiman CA, Travis RC, Berg CD, Buring JE, Chanock SJ, Diver WR, Dostal L, et al. Interactions between genetic variants and breast cancer risk factors in the breast and prostate cancer cohort consortium. Journal of the National Cancer Institute. 2011;103(16):1252–1263. doi: 10.1093/jnci/djr265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Travis RC, Reeves GK, Green J, Bull D, Tipper SJ, Baker K, Beral V, Peto R, Bell J, Zelenika D, et al. Gene-environment interactions in 7610 women with breast cancer: prospective evidence from the Million Women Study. Lancet. 2010;375(9732):2143–2151. doi: 10.1016/S0140-6736(10)60636-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Milne RL, Gaudet MM, Spurdle AB, Fasching PA, Couch FJ, Benitez J, Arias Perez JI, Zamora MP, Malats N, Dos Santos Silva I, et al. Assessing interactions between the associations of common genetic susceptibility variants, reproductive history and body mass index with breast cancer risk in the breast cancer association consortium: a combined case-control study. Breast cancer research : BCR. 2010;12(6):R110. doi: 10.1186/bcr2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fasching PA, Pharoah PD, Cox A, Nevanlinna H, Bojesen SE, Karn T, Broeks A, van Leeuwen FE, van’t Veer LJ, Udo R, et al. The role of genetic breast cancer susceptibility variants as prognostic factors. Human molecular genetics. 2012;21(17):3926–3939. doi: 10.1093/hmg/dds159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cunningham AP, Antoniou AC, Easton DF. Clinical software development for the Web: lessons learned from the BOADICEA project. BMC medical informatics and decision making. 2012;12(1):30. doi: 10.1186/1472-6947-12-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pharoah PD. Genetic susceptibility, predicting risk and preventing cancer. Recent results in cancer research Fortschritte der Krebsforschung Progres dans les recherches sur le cancer. 2003;163:7–18. doi: 10.1007/978-3-642-55647-0_2. discussion 264–266. [DOI] [PubMed] [Google Scholar]
- 62.Pharoah PD, Antoniou AC, Easton DF, Ponder BA. Polygenes, risk prediction, and targeted prevention of breast cancer. The New England journal of medicine. 2008;358(26):2796–2803. doi: 10.1056/NEJMsa0708739. [DOI] [PubMed] [Google Scholar]
- 63.Boyd NF, Lockwood GA, Byng JW, Tritchler DL, Yaffe MJ. Mammographic densities and breast cancer risk. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 1998;7(12):1133–1144. [PubMed] [Google Scholar]
- 64.Vachon CM, van Gils CH, Sellers TA, Ghosh K, Pruthi S, Brandt KR, Pankratz VS. Mammographic density, breast cancer risk and risk prediction. Breast cancer research : BCR. 2007;9(6):217. doi: 10.1186/bcr1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Byrne C, Schairer C, Wolfe J, Parekh N, Salane M, Brinton LA, Hoover R, Haile R. Mammographic features and breast cancer risk: effects with time, age, and menopause status. Journal of the National Cancer Institute. 1995;87(21):1622–1629. doi: 10.1093/jnci/87.21.1622. [DOI] [PubMed] [Google Scholar]
- 66.Boyd NF, Guo H, Martin LJ, Sun L, Stone J, Fishell E, Jong RA, Hislop G, Chiarelli A, Minkin S, et al. Mammographic density and the risk and detection of breast cancer. The New England journal of medicine. 2007;356(3):227–236. doi: 10.1056/NEJMoa062790. [DOI] [PubMed] [Google Scholar]
- 67.Kelemen LE, Sellers TA, Vachon CM. Can genes for mammographic density inform cancer aetiology? Nature reviews Cancer. 2008;8(10):812–823. doi: 10.1038/nrc2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Boyd N, Martin L, Chavez S, Gunasekara A, Salleh A, Melnichouk O, Yaffe M, Friedenreich C, Minkin S, Bronskill M. Breast-tissue composition and other risk factors for breast cancer in young women: a cross-sectional study. The lancet oncology. 2009;10(6):569–580. doi: 10.1016/S1470-2045(09)70078-6. [DOI] [PubMed] [Google Scholar]
- 69.Yaffe MJ. Mammographic density. Measurement of mammographic density. Breast cancer research : BCR. 2008;10(3):209. doi: 10.1186/bcr2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ziv E, Tice J, Smith-Bindman R, Shepherd J, Cummings S, Kerlikowske K. Mammographic density and estrogen receptor status of breast cancer. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2004;13(12):2090–2095. [PubMed] [Google Scholar]
- 71.Ma H, Luo J, Press MF, Wang Y, Bernstein L, Ursin G. Is there a difference in the association between percent mammographic density and subtypes of breast cancer? Luminal A and triple-negative breast cancer. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2009;18(2):479–485. doi: 10.1158/1055-9965.EPI-08-0805. [DOI] [PubMed] [Google Scholar]
- 72.Olsen AH, Bihrmann K, Jensen MB, Vejborg I, Lynge E. Breast density and outcome of mammography screening: a cohort study. British journal of cancer. 2009;100(7):1205–1208. doi: 10.1038/sj.bjc.6604989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yaghjyan L, Colditz GA, Collins LC, Schnitt SJ, Rosner B, Vachon C, Tamimi RM. Mammographic breast density and subsequent risk of breast cancer in postmenopausal women according to tumor characteristics. Journal of the National Cancer Institute. 2011;103(15):1179–1189. doi: 10.1093/jnci/djr225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gierach GL, Loud JT, Chow CK, Prindiville SA, Eng-Wong J, Soballe PW, Giambartolomei C, Mai PL, Galbo CE, Nichols K, et al. Mammographic density does not differ between unaffected BRCA1/2 mutation carriers and women at low-to-average risk of breast cancer. Breast cancer research and treatment. 2010;123(1):245–255. doi: 10.1007/s10549-010-0749-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mitchell G, Antoniou AC, Warren R, Peock S, Brown J, Davies R, Mattison J, Cook M, Warsi I, Evans DG, et al. Mammographic density and breast cancer risk in BRCA1 and BRCA2 mutation carriers. Cancer research. 2006;66(3):1866–1872. doi: 10.1158/0008-5472.CAN-05-3368. [DOI] [PubMed] [Google Scholar]
- 76.Maskarinec G, Woolcott CG, Kolonel LN. Mammographic density as a predictor of breast cancer outcome. Future Oncol. 2010;6(3):351–354. doi: 10.2217/fon.10.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cuzick J, Warwick J, Pinney E, Duffy SW, Cawthorn S, Howell A, Forbes JF, Warren RM. Tamoxifen-induced reduction in mammographic density and breast cancer risk reduction: a nested case-control study. Journal of the National Cancer Institute. 2011;103(9):744–752. doi: 10.1093/jnci/djr079. [DOI] [PubMed] [Google Scholar]
- 78.Kim J, Han W, Moon HG, Ahn SK, Shin HC, You JM, Han SW, Im SA, Kim TY, Koo HR, et al. Breast density change as a predictive surrogate for response to adjuvant endocrine therapy in hormone receptor positive breast cancer. Breast cancer research : BCR. 2012;14(4):R102. doi: 10.1186/bcr3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Boyd NF, Melnichouk O, Martin LJ, Hislop G, Chiarelli AM, Yaffe MJ, Minkin S. Mammographic density, response to hormones, and breast cancer risk. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2011;29(22):2985–2992. doi: 10.1200/JCO.2010.33.7964. [DOI] [PubMed] [Google Scholar]
- 80.Barlow WE, White E, Ballard-Barbash R, Vacek PM, Titus-Ernstoff L, Carney PA, Tice JA, Buist DS, Geller BM, Rosenberg R, et al. Prospective breast cancer risk prediction model for women undergoing screening mammography. Journal of the National Cancer Institute. 2006;98(17):1204–1214. doi: 10.1093/jnci/djj331. [DOI] [PubMed] [Google Scholar]
- 81.Tice JA, Cummings SR, Ziv E, Kerlikowske K. Mammographic breast density and the Gail model for breast cancer risk prediction in a screening population. Breast cancer research and treatment. 2005;94(2):115–122. doi: 10.1007/s10549-005-5152-4. [DOI] [PubMed] [Google Scholar]
- 82.Chen J, Pee D, Ayyagari R, Graubard B, Schairer C, Byrne C, Benichou J, Gail MH. Projecting absolute invasive breast cancer risk in white women with a model that includes mammographic density. Journal of the National Cancer Institute. 2006;98(17):1215–1226. doi: 10.1093/jnci/djj332. [DOI] [PubMed] [Google Scholar]
- 83.Cecchini R, Costantino JP, Cauley JA, Cronin WM, Wickerham DL, Bandos H, Weissfeld JL, Wolmark N. Baseline mammographic breast density and the risk of invasive breast cancer in postmenopausal women participating in the NSABP Study of Tamoxifen and Raloxifene (STAR) Cancer Prev Res (Phila) 2012 doi: 10.1158/1940-6207.CAPR-12-0273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tice JA, Cummings SR, Smith-Bindman R, Ichikawa L, Barlow WE, Kerlikowske K. Using clinical factors and mammographic breast density to estimate breast cancer risk: development and validation of a new predictive model. Annals of internal medicine. 2008;148(5):337–347. doi: 10.7326/0003-4819-148-5-200803040-00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ciatto S, Houssami N, Apruzzese A, Bassetti E, Brancato B, Carozzi F, Catarzi S, Lamberini MP, Marcelli G, Pellizzoni R, et al. Categorizing breast mammographic density: intra- and interobserver reproducibility of BI-RADS density categories. Breast. 2005;14(4):269–275. doi: 10.1016/j.breast.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 86.Kerlikowske K, Grady D, Barclay J, Frankel SD, Ominsky SH, Sickles EA, Ernster V. Variability and accuracy in mammographic interpretation using the American College of Radiology Breast Imaging Reporting and Data System. Journal of the National Cancer Institute. 1998;90(23):1801–1809. doi: 10.1093/jnci/90.23.1801. [DOI] [PubMed] [Google Scholar]
- 87.Shepherd JA, Kerlikowske K, Ma L, Duewer F, Fan B, Wang J, Malkov S, Vittinghoff E, Cummings SR. Volume of mammographic density and risk of breast cancer. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2011;20(7):1473–1482. doi: 10.1158/1055-9965.EPI-10-1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Boyd NF, Martin LJ, Bronskill M, Yaffe MJ, Duric N, Minkin S. Breast tissue composition and susceptibility to breast cancer. Journal of the National Cancer Institute. 2010;102(16):1224–1237. doi: 10.1093/jnci/djq239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sherman ME, Figueroa JD, Henry JE, Clare SE, Rufenbarger C, Storniolo AM. The Susan G. Komen for the Cure Tissue Bank at the IU Simon Cancer Center: a unique resource for defining the “molecular histology” of the breast. Cancer Prev Res (Phila) 2012;5(4):528–535. doi: 10.1158/1940-6207.CAPR-11-0234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hutson SW, Cowen PN, Bird CC. Morphometric studies of age related changes in normal human breast and their significance for evolution of mammary cancer. Journal of clinical pathology. 1985;38(3):281–287. doi: 10.1136/jcp.38.3.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cowan df. Involution of the breast in women aged 50 to 104 years: a histopathological study of 102 cases. Surg Pathol. 1989;2:323–333. [Google Scholar]
- 92.Henson DE, Tarone RE. Involution and the etiology of breast cancer. Cancer. 1994;74(1 Suppl):424–429. doi: 10.1002/cncr.2820741330. [DOI] [PubMed] [Google Scholar]
- 93.Milanese TR, Hartmann LC, Sellers TA, Frost MH, Vierkant RA, Maloney SD, Pankratz VS, Degnim AC, Vachon CM, Reynolds CA, et al. Age-related lobular involution and risk of breast cancer. Journal of the National Cancer Institute. 2006;98(22):1600–1607. doi: 10.1093/jnci/djj439. [DOI] [PubMed] [Google Scholar]
- 94.Baer HJ, Collins LC, Connolly JL, Colditz GA, Schnitt SJ, Tamimi RM. Lobule type and subsequent breast cancer risk: results from the Nurses’ Health Studies. Cancer. 2009;115(7):1404–1411. doi: 10.1002/cncr.24167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ghosh K, Hartmann LC, Reynolds C, Visscher DW, Brandt KR, Vierkant RA, Scott CG, Radisky DC, Sellers TA, Pankratz VS, et al. Association between mammographic density and age-related lobular involution of the breast. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2010;28(13):2207–2212. doi: 10.1200/JCO.2009.23.4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Vierkant RA, Hartmann LC, Pankratz VS, Anderson SS, Radisky D, Frost MH, Vachon CM, Ghosh K, Distad TJ, Degnim AC, et al. Lobular involution: localized phenomenon or field effect? Breast cancer research and treatment. 2009;117(1):193–196. doi: 10.1007/s10549-008-0082-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ghosh K, Vachon CM, Pankratz VS, Vierkant RA, Anderson SS, Brandt KR, Visscher DW, Reynolds C, Frost MH, Hartmann LC. Independent association of lobular involution and mammographic breast density with breast cancer risk. Journal of the National Cancer Institute. 2010;102(22):1716–1723. doi: 10.1093/jnci/djq414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yang XR, Figueroa JD, Falk RT, Zhang H, Pfeiffer RM, Hewitt SM, Lissowska J, Peplonska B, Brinton L, Garcia-Closas M, et al. Analysis of terminal duct lobular unit involution in luminal A and basal breast cancers. Breast cancer research : BCR. 2012;14(2):R64. doi: 10.1186/bcr3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.McKian KP, Reynolds CA, Visscher DW, Nassar A, Radisky DC, Vierkant RA, Degnim AC, Boughey JC, Ghosh K, Anderson SS, et al. Novel breast tissue feature strongly associated with risk of breast cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2009;27(35):5893–5898. doi: 10.1200/JCO.2008.21.5079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Khan SA, Rogers MA, Obando JA, Tamsen A. Estrogen receptor expression of benign breast epithelium and its association with breast cancer. Cancer research. 1994;54(4):993–997. [PubMed] [Google Scholar]


