Abstract
Pediatric tumors are relatively infrequent, but are often associated with significant lethality and lifelong morbidity. A major goal of pediatric cancer research has been to identify key drivers of tumorigenesis to eventually develop targeted therapies to enhance cure rate and minimize acute and long-term toxic effects. Here, we used genomic approaches to identify biomarkers and candidate drivers for fibrolamellar hepatocellular carcinoma (FL-HCC), a very rare subtype of pediatric liver cancer for which limited therapeutic options exist. In-depth genomic analyses of one tumor followed by immunohistochemistry validation on seven other tumors showed expression of neuroendocrine markers in FL-HCC. DNA and RNA sequencing data further showed that common cancer pathways are not visibly altered in FL-HCC but identified two novel structural variants, both resulting in fusion transcripts. The first, a 400 kb deletion, results in a DNAJB1-PRKCA fusion transcript, which leads to increased cAMP-dependent protein kinase (PKA) activity in the index tumor case and other FL-HCC cases compared with normal liver. This PKA fusion protein is oncogenic in HCC cells. The second gene fusion event, a translocation between the CLPTM1L and GLIS3 genes, generates a transcript whose product also promotes cancer phenotypes in HCC cell lines. These experiments further highlight the tumorigenic role of gene fusions in the etiology of pediatric solid tumors and identify both candidate biomarkers and possible therapeutic targets for this lethal pediatric disease.
INTRODUCTION
Hepatocellular carcinoma (HCC) is the primary cause of liver cancer and the third leading cause of cancer mortality, with more than 600 000 deaths annually. In the majority of cases, chronic hepatitis B virus and hepatitis C virus infections are responsible for the development of the disease, but other factors such as alcohol consumption and exposure to aflatoxin B1 contribute to cancer development (1,2). HCC is usually a cancer found in older adults, and primary hepatic carcinomas are rare in people younger than 20 years. In this age group, approximately two-thirds of pediatric cases are hepatoblastomas, an embryonal cancer thought to arise from hepatocyte precursor cells, and the remaining cases are primary HCCs. Unlike HCC in adults, the risk factors for pediatric HCC are poorly understood. Epidemiologic evidence indicates that underlying liver or metabolic disease is absent in most cases of pediatric HCC, and it is thought that most HCCs in children and young adults do not arise in the setting of histologically recognizable liver disease (3,4).
Fibrolamellar hepatocellular carcinoma (FL-HCC) is a very rare and distinct subtype of HCC that affects adolescents and young adults (5,6). FL-HCC develops in the setting of normal healthy livers posing the question as to why these tumors arise in the liver of young patients in the absence of known metabolic defects or viral infections (4). Furthermore, there are no known genetic predispositions to the development of FL-HCC, and there are no documented cases of it affecting more than one member of a family. Morphologically, FL-HCC tumors are characterized by a proliferation of pleomorphic malignant hepatocytes with large central nucleoli and abundant eosinophilic cytoplasm. The tumor cells are arranged in variably sized nests or cords set within a meshwork of lamellated collagen fibers. In the majority of cases, FL-HCC cells contain hyaline droplets and some isolated cells have round cytoplasmic inclusions known as ‘pale bodies’ (7,8). The background uninvolved liver parenchyma characteristically shows no increase in fibrosis. FL-HCCs express markers associated with biliary [Cytokeratin 7 (CK7) and MUC1] and hepatocytic (Heppar-1, Albumin and Glypican-3) differentiation, as well as hepatic progenitor/stem cells (CK19, CD10, CEA, EpCAM, CD133, CD44 and SALL4) (4,9–13). FL-HCCs also show elevated expression of epidermal growth receptor (EGFR) and Ras signaling as well as transforming growth factor β, compared with adult HCC (14–17). There is also some immunohistochemical evidence that the nuclear factor kappa B pathway is activated in FL-HCC (18), and that expression of the small heterodimer partner orphan nuclear receptor is decreased (19). But overall, the list of FL-HCC markers is still limited and the (epi)genetic alterations driving the growth of these tumors have remained unknown. There are also no serum biomarkers for early detection and no clear correlation between the expression of known markers and the survival rate of patients (reviewed in 3,4). FL-HCC is associated with better survival than HCC in adults, presumably due to the young age of the patients and the lack of cirrhosis, which makes more aggressive surgical therapy possible (20). However, few therapeutic options have been explored beyond surgery and little is known about the efficacy of chemotherapy for these patients. The 5-year survival rate ranges from 35 to 75% in patients treated with liver transplantation or resection. In cases where surgery is not possible, the survival drops to 12–14 months (compared with 8–9 months for adult HCC) (21–24).
The very low incidence rate of 0.02 per 100 000 (age-adjusted) has kept FL-HCC an understudied cancer (5). Indeed, a search through the pathology archives and tissue bank for Stanford University Hospital and Lucille Packard Children's Hospital revealed only eight cases (seven formalin-fixed paraffin-embedded and one fresh frozen tissue) for which the diagnosis was reproducible by an independent pathologist's review. We sought to gain a better understanding of the molecular mechanisms underlying FL-HCC initiation and progression, with the hope that it would allow for the development of better diagnosis and treatment strategies. To this end, we performed an in-depth molecular characterization of one FL-HCC tumor using transcriptome and whole genome sequencing (WGS). Similar to a recent study (25), we identified an intrachromosomal deletion resulting in a gene fusion product between DNAJB1 and PRKACA that results in activation of cAMP-dependent protein kinase (PKA) signaling; this fusion may play a driving role in FL-HCC initiation and progression. In addition, we found a translocation and gene fusion event between GLIS3 and CLPTM1L with oncogenic effects in liver cancer cells. These experiments identify novel potential therapeutic targets for FL-HCC.
RESULTS
A genome-wide RNA profiling analysis of a pediatric FL-HCC tumor identifies a neuroendocrine differentiation state
We performed an unbiased transcriptome sequencing analysis (RNA-seq) of a freshly resected FL-HCC tumor and paired normal liver tissue (Fig. 1A and Supplementary Material, Table S1). To investigate global patterns of differential gene expression, we performed gene set enrichment analysis (GSEA) and ingenuity pathway analysis (IPA). These analyses revealed that gene expression profiles in normal liver tissue were preferentially enriched for hepatic metabolism pathways, indicating that the tumor had decreased hepatic function, as would be expected (Supplementary Material, Table S2). Conversely, gene expression profiles in the tumor were enriched for pathways involved in protein synthesis, proliferation and extracellular matrix interactions, as would be expected for tumor cell populations (Supplementary Material, Table S2).
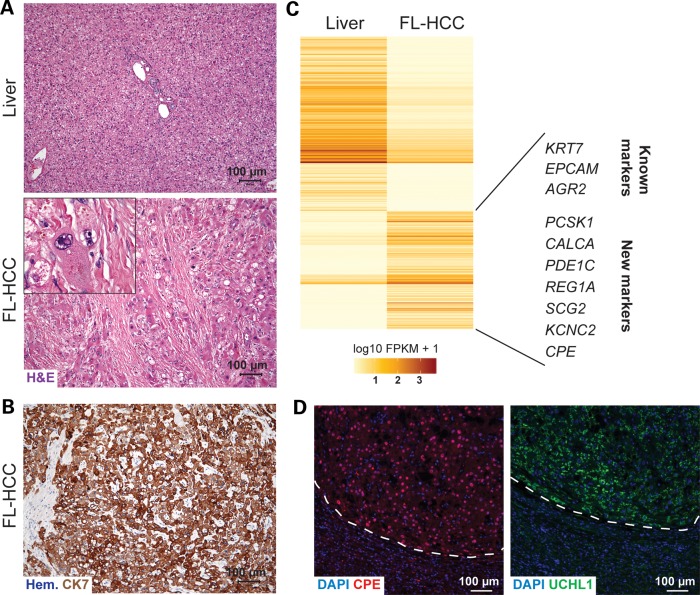
Figure 1.
Expression of neuroendocrine markers in FL-HCC. (A) Histopathology of the index case (H&E, hematoxylin and eosin staining) compared with the normal liver area. Indent, high magnification. Scale bar, 100 µm. (B) Representative image of an immunostaining analysis for the known FL-HCC marker Cytokeratin 7 (CK7, brown staining). The section was counterstained with hematoxylin (Hem.). Scale bar, 100 µm. (C) Heat map of the 543 significantly differentially expressed genes between tumor and normal liver tissue. Some known and novel genes upregulated in the tumor are shown, including neuroendocrine genes. (D) Immunofluorescence analysis of the neuroendocrine markers CPE and UCHL1. The tumor area (top) is demarked by a dashed line. The CPE signal is red, and the UCHL1 signal is green. DAPI marks the DNA in blue. Scale bar, 100 µm.
Further RNA-seq analysis with Cufflinks identified 543 significantly differentially expressed genes [false discovery rate (FDR)-adjusted P-values <0.05; Supplementary Material, Table S3]. Two hundred and nineteen genes were upregulated and 324 genes were downregulated in the tumor compared with normal liver (Fig. 1B). Among the 219 upregulated genes were genes coding for markers previously associated with FL-HCC, such as EpCAM, CK7, and AGR2 (Anterior Gradient Homolog 2) (4,10,26). Immunostaining for CK7 confirmed the expression of this protein in the tumor cells (Fig. 1C). Other less common FL-HCC markers (such as CK19, Neurotensin and Transcobalamin) were also upregulated in the tumor compared with control liver, although they did not pass the threshold for significance (Supplementary Material, Table S4). Of note, AFP (alpha-fetoprotein), a marker commonly associated with adult HCC, was expressed at very low, almost undetectable levels in the tumor. Taken together, these observations confirm the histopathological diagnosis of FL-HCC for this patient.
To help expand the limited number of markers currently available for FL-HCC, we searched for additional candidate biomarkers from the list of upregulated genes. The downstream effects analysis by IPA, which finds biological functions that are expected to be increased or decreased based on the observed gene expression changes, identified neurological disease and endocrine system disorders as among the top most significantly increased biological functions aside from those related to cancer (Supplementary Material, Table S5). Indeed, we noted that a number of the significantly upregulated genes are usually expressed in neural or neuroendocrine cells [such as genes coding for the p21-activated kinase 3 (PAK3), the carboxypeptidase (CPE), calcitonin-related polypeptide alpha (CALCA), the proprotein convertase subtilisin/kexin type 1 (PCSK1), and secretogranin 2 (SCG2); Supplementary Material, Table S4]. Given that neuroendocrine features have been previously noted in FL-HCC (10,27–29), we tested CPE and UCHL1 (ubiquitin thiolesterase, another commonly used neuroendocrine marker that was upregulated in FL-HCC with a P-value close to the significance threshold). CPE and UCHL1 showed stronger immunostaining in the FL-HCC tumor tissue compared with the adjacent normal liver tissue (Fig. 1D). Immunohistochemistry analysis on seven other FL-HCC sample tissues (Fig. 1A and Supplementary Material, Table S1) showed the expression of CPE in additional tumors (Supplementary Material, Fig. S1). These experiments confirm that at least a subset of FL-HCC cases have a neuroendocrine differentiation phenotype and suggest that specific neuroendocrine markers may help further distinguish FL-HCC from other HCC subtypes.
Genome-wide sequencing of a pediatric FL-HCC tumor highlights a low number of somatic mutations and a lack of mutations in classical cancer genes
The few studies aimed at understanding the genetics of FL-HCC tumorigenesis have analyzed the mutation status of candidate genes such as TP53, KRAS, EGFR and CTNNB1, and have found only a lack of mutations in the genes analyzed (4,14–16,30). To perform an in-depth characterization of the genetic changes in FL-HCC, we conducted WGS of both the FL-HCC tumor and paired normal tissue from the patient for which we had obtained fresh frozen tissue (Complete Genomics; Fig. 2A). 96.6 and 96.3% of the tumor and normal genomes, respectively, were sequenced with 77.7 and 73.2% of the genomes sequenced to a coverage of ≥30×. We conducted a two-step approach in identifying somatic mutations. First, to achieve maximum sensitivity, we screened for potential variants by using low thresholds in CGtools (default parameters) and identified 200 395 potential somatic small variants (indels, single-nucleotide variants and substitutions). This number decreased to 86 331 after applying additional quality control filters (minimum of 20× coverage in the normal genome and maximum of 1 read supporting the somatic allele in the normal genome to allow for sequencing error). Next, we determined the true somatic mutations by looking for validation in sequencing data generated by a different technology (to minimize sequencer-specific artifact). Using Illumina technology, we sequenced the cancer exome to an average depth of 66×. Filtering the WGS data against the exome sequencing data and the RNA-seq data, we confirmed 67 true somatic mutations in the cancer genome (with at least two supporting reads in either the RNA-seq or exome sequencing data; see Materials and Methods).
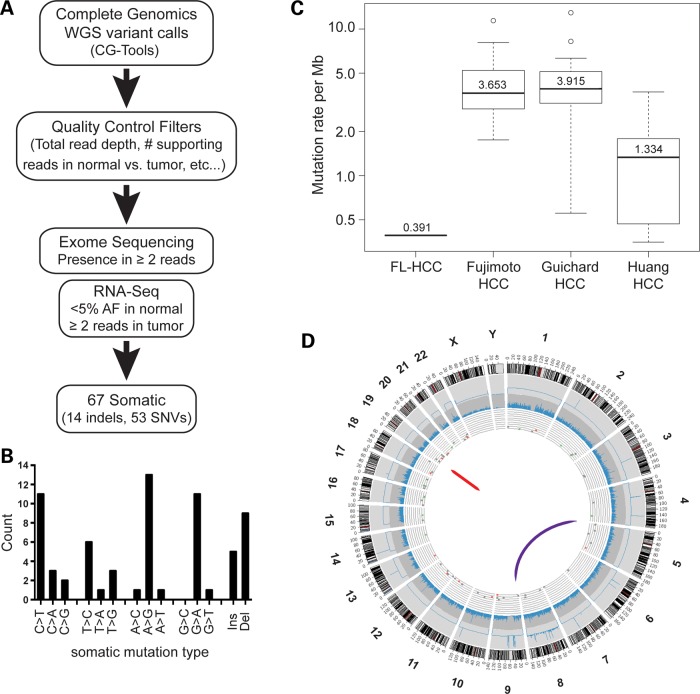
Figure 2.
WGS of a FL-HCC tumor and normal liver. (A) Flowchart describing the detection and validation of somatic small variants. (B) The majority of the somatic mutations in the tumor are transitions (A>G/T>C or C>T/G>A). (C) Overall mutation rate in the FL-HCC case sequenced compared with three studies of adult HCC (see text for detail). (D) Circos plot of the somatic changes in the tumor sample. From outermost to innermost layer: Track 1—human karyotype ideogram. Track 2—CNV as determined by Complete Genomics CNVcalls algorithm. Track 3—exome sequencing coverage over RefSeq coding regions. Track 4—scatterplot of somatic small variants (red, coding region; green, UTR), where y-axis is variant allele frequency. Track 5—large structural variants.
These 67 mutations were comprised of 5 insertions, 9 deletions and 53 single-nucleotide variants (Supplementary Material, Table S6). Unlike HCC cancer genomes, which have an overrepresentation of transversions (31,32), the majority of the variants identified in FL-HCC are transitions (A>G/T>C or C>T/G>A; Fig. 2B). Eleven of the somatic mutations were in coding regions resulting in two frameshifts, seven missense mutations, one synonymous alteration and one single amino acid deletion. Some of these mutations were predicted to have damaging or deleterious effects on protein function by SIFT and Provean scores, respectively (Supplementary Material, Table S7). However, none of the mutated genes in this list are known cancer genes nor do they have known involvement with cancer pathways. Compared with recent HCC genome sequencing studies (31–33), the overall mutation rate of FL-HCC is 3- to 10-fold lower and closer to the mutation rate of other pediatric cancers (34) (Fig. 2C).
The analysis of copy number variations and chromosomal events identifies two gene fusions in the FL-HCC genome
Data from traditional cytogenetic methods have suggested that chromosomes are largely stable in FL-HCC tumors, with some large chromosomal changes such as gains in chromosome arms 1q and 8q or loss of 18q (4,6,35,36). Copy number variation (CNV) analysis (CG Cancer Pipeline version 2.2) comparing the tumor genome with control liver detected a potential 89 somatic CNV amplifications; of which, 81 occurred on chromosome 8 (Fig. 2D). GSEA using positional gene sets (gene sets corresponding to human chromosomes and cytogenic bands) showed enrichment for gene sets corresponding to both arms of chromosome 8 in the tumor expression data (Supplementary Material, Table S8). Taken together, these observations suggest that the genome of this FL-HCC case is triploid for chromosome 8.
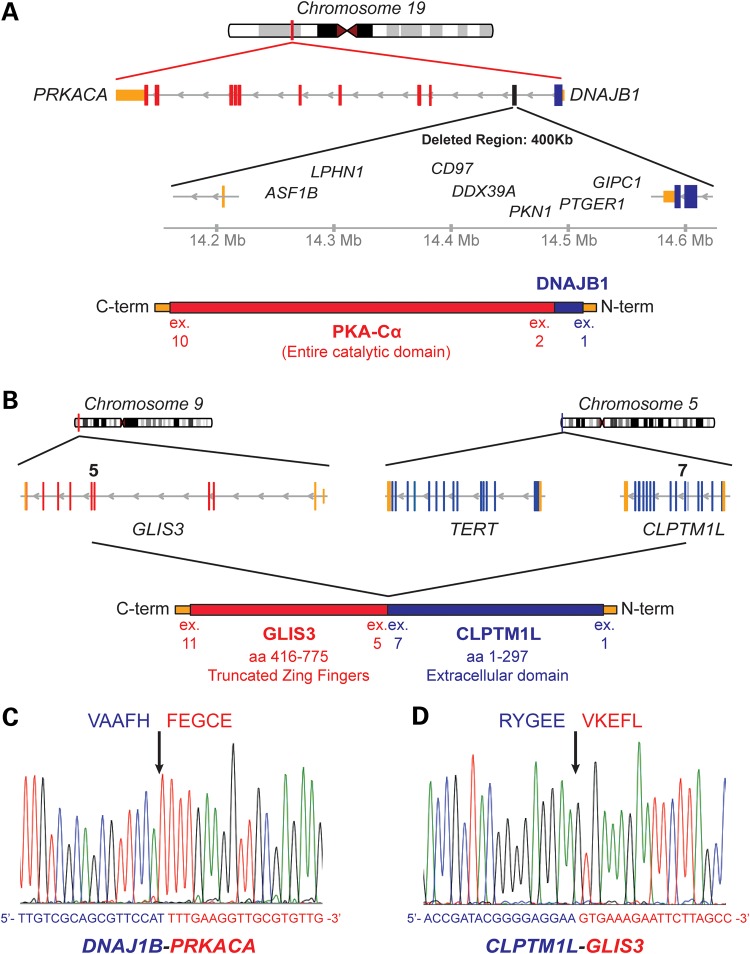
A recent study on adult HCC found an average of 20.8 genomic rearrangements per tumor by WGS (32). Here, using discordant mate-pairs and split-read mapping, we detected and validated only two large genomic rearrangements in the WGS data. The first rearrangement is an intrachromosomal deletion of ∼400 kb on chromosome 19. The second is a translocation between chromosome 5 and chromosome 9 (Fig. 2D). Both rearrangements resulted in fusion transcripts that were also detected in the RNA-seq expression data by TopHat-Fusion analysis (Supplementary Material, Table S9). The t(5;9) translocation fuses the N-terminus of CLPTM1L in frame to a part of the DNA-binding domain of GLIS3 (Fig. 3A). The large intrachromosomal deletion on chr19 fuses the N-terminal J domain of DNAJB1 in frame to the catalytic domain of PRKACA (PKA subunit Cα; Fig. 3B). Both fusions were validated by RT-PCR from tumor RNA and by subsequent Sanger sequencing (Fig. 3C and D).
Figure 3.
WGS of a FL-HCC tumor and normal liver. (A) Schematic representation of the fusion between the DNAJB1 and PRKACA genes. The 400 kb deletion on chr19 results in the N-terminal J domain of DNAJB1 fused to the Cα subunit of PKA (PRKACA). The 400 kb deletion also results in the loss of eight genes: ASF1B, CD97, GIPC1, LOC100130932, LPHN1, PKN1 and PTGER1. (B) Schematic representation of the t(5;9) translocation between GLIS3 and CLPTM1L resulting in an in-frame chimeric protein. (C and D) Sanger sequencing chromogram of the RT-PCR product around the fusion point.
Taken together, these observations confirmed the relatively stable nature of the FL-HCC genome and led us to investigate further the possible tumorigenic role of the two fusion proteins.
The DNAJB1-PRKACA fusion codes for an active and oncogenic form of the PKA enzyme
DNAJB1 codes for a heat shock protein (also named HSP40) and PRKACA for the Cα catalytic subunit of the PKA. This same DNAJB1-PRKACA fusion was recently identified as a recurrent event in a study of 15 cases of FL-HCC, strongly suggesting it is an initiating event in the disease (25). Our study provides independent support for this previous finding. An immunoblot analysis on tumor extracts using an antibody directed against PKA-Cα showed a protein at the predicted size for the fusion product, slightly larger than PKA-Cα (Fig. 4A). Interestingly, the levels of the fusion protein were higher than PKA-Cα levels from the wild-type PRKACA allele (Fig. 4A). This is most likely, in part, due to the higher DNAJB1-PRKACA fusion transcript levels compared with the PRKACA transcript levels, as evidenced by the relatively higher number of fusion junction spanning reads (276) compared with the number of exon1–exon2 wild-type PRKACA spanning reads (11) (data not shown).
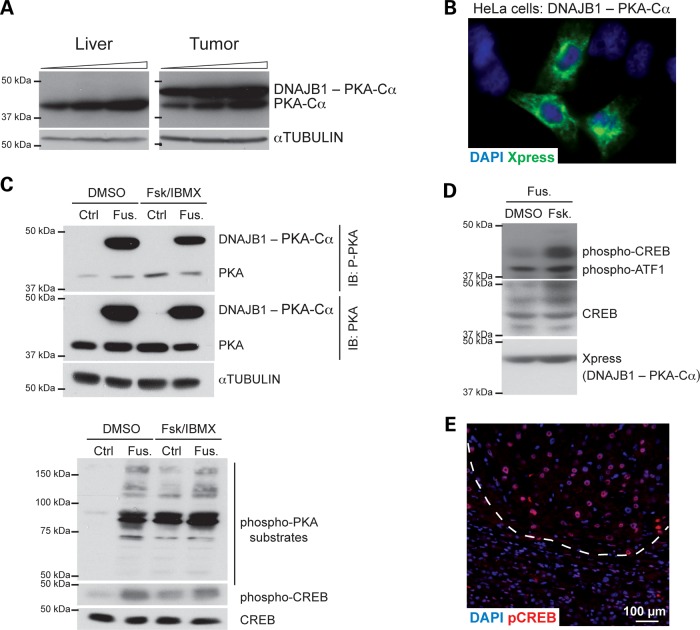
Figure 4.
The DNAJB1-PRKACA fusion codes for an active form of PKA. (A) Immunoblot analysis of PKA and the fusion protein (PKA antibody) in normal liver and tumor material. Increasing amounts of protein extracts were loaded. Tubulin serves as a loading control. (B) Immunofluorescence analysis of the tagged fusion protein (antibody against the Xpress tag) following transfection in HeLa cells. (C) Immunoblot analysis from 293T cells stably infected with an empty vector (Ctrl) or a vector expressing the fusion protein (Fus.); the cells were treated with vehicle (DMSO) or 50 µM Forskolin and 100 µM 3-Isobutyl-1-methylxanthine (IBMX) (Fsk/IBMX) for 30 min. Immunoblot was performed for PKA, phospho-PKA, CREB, phospho-CREB and an antibody that recognizes phosphorylated PKA substrates. The same extracts were used in the two immunoblots. Tubulin serves as a loading control. (D) Immunoblot analysis for the indicated antibodies as in (C), 293T cells were treated with vehicle or 10 µM Forskolin for 1 h. (E) Immunofluorescence analysis of phospho-CREB (red signal). The tumor area (top) is demarked by a dashed line. DAPI marks the DNA in blue. Scale bar, 100 µm.
We were not able to generate a cell line or a xenograft from the patient tumor (data not shown), hampering functional studies with the endogenous fusion protein. To circumvent this limitation, we transiently expressed a tagged form of the fusion protein in human cancer cells. We found that the fusion protein localized mostly in the cytosol of HeLa cells (Fig. 4B), as would be expected given the largely cytosolic localization of both DNAJB1 (37) and PKA-Cα (38) under normal conditions. When transfected into 293T cells, the fusion protein was phosphorylated at a site in PKA-Cα often associated with kinase activity (Serine 133) even in the absence of activators of adenylyl cyclase [Forskolin and 3-Isobutyl-1-methylxanthine (IBMX)] (Fig. 4C, top panel); adenylyl cyclase activation leads to PKA activation in cells by increasing the intracellular levels of the PKA activator cAMP. Accordingly, we detected increased phosphorylation of cAMP response element-binding protein (CREB) and other PKA substrates in these cells even in the absence of Forskolin and IBMX treatment (Fig. 4C, bottom panel). Furthermore, treatment with Forskolin alone induced a greater increase in CREB phosphorylation in cells expressing the fusion protein (Fig. 4D). These experiments suggest that the fusion protein is constitutively more active than wild-type PKA-Cα, but can be further induced with signals that normally activate PKA in cells. We also found increased levels of phospho-CREB in the tumor compared with adjacent normal liver on tissue sections (Fig. 4D), suggesting that the fusion protein is active in tumor cells in vivo. Accordingly, the upstream regulator analysis by IPA identified CREB as a candidate activated regulator of the gene signatures that were significantly different in the tumor compared with normal liver (Supplementary Material, Table S10). Increased phospho-CREB levels were detected in the additional FL-HCC samples, validating the observations by Honeyman et al. that the fusion is present in most if not all cases of FL-HCC (25) (Supplementary Material, Fig. S2).
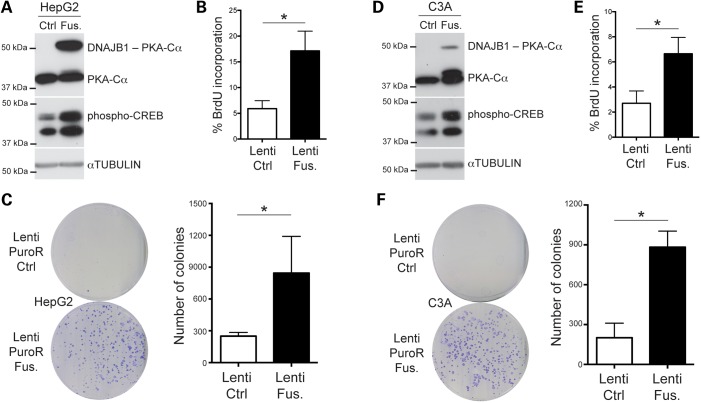
Intriguingly, our initial attempts to obtain cancer cells stably expressing high levels of the PKA fusion by transfection failed; similarly, we could not obtain Hep3B cells (derived from a case of well-differentiated HCC in an adolescent) stably expressing the fusion protein after lentiviral infection and selection (data not shown). However, we were able to stably infect several other HCC cell lines (SNU-449, HepG2 and C3A) using the same lentiviral vector. In HepG2 cells (derived from an HCC case in an adolescent), expression of the fusion protein induced increased levels of phospho-CREB (Fig. 5A) and led to an increased BrdU incorporation, a marker of DNA replication (Fig. 5B). In addition, expression of the PKA fusion protein enhanced the ability of these cells to form colonies when plated a low density, a feature of transformed cells (Fig. 5C). In C3A cells (a clonal expansion of HepG2 cells with far more differentiated features), selected cells could only express low levels of the fusion protein (Fig. 5D), but BrdU incorporation was nevertheless still increased compared with the empty vector control (Fig. 5E) and the ability to form colonies at low density was also significantly enhanced (Fig. 5F). Expression of the PKA fusion protein in SNU-449 cells also led to increased phospho-CREB and significant BrdU incorporation, but there was only a non-significant trend in the colony-forming ability of these cells when they expressed the fusion protein, presumably because these cells have a higher basal proliferation rate and naturally form more colonies in this assay (data not shown).
Figure 5.
The DNAJB1-PRKACA fusion is oncogenic in HCC cells. (A and D) Immunoblot analysis of the fusion (Fus.) protein following lentiviral infection in human HCC cells compared with cells infected with an empty control (Ctrl) vector (A, HepG2 and D, C3A). The top portion of each blot shows PKA and the fusion protein (PKA antibody). The middle panel shows immunoblot analysis for phospho-CREB. The lower blot shows Tubulin as a loading control. (B and E) Quantification of BrdU incorporation in HepG2 (B) and C3A (E) cells stably expressing the fusion protein compared with an empty control virus (n = 3 independent experiments). (C and F) Representative images (left) of 10 cm plates from colony-formation assays from HepG2 (C) and C3A (F) cells stably infected with a lentivirus expressing the fusion protein compared with an empty control virus (both selected for puromycin resistance) and quantification (right) (n ≥ 3 independent experiments). *P < 0.05 (paired t-test).
Taken together, these observations suggest that expression of the DNAJB1-PRKACA fusion may have toxic effects when expressed at high levels in the wrong cellular contexts, but also provide evidence that this fusion protein is oncogenic in certain HCC cell lines.
The CLPTM1L-GLIS3 fusion codes for an oncogenic protein in HCC cell lines
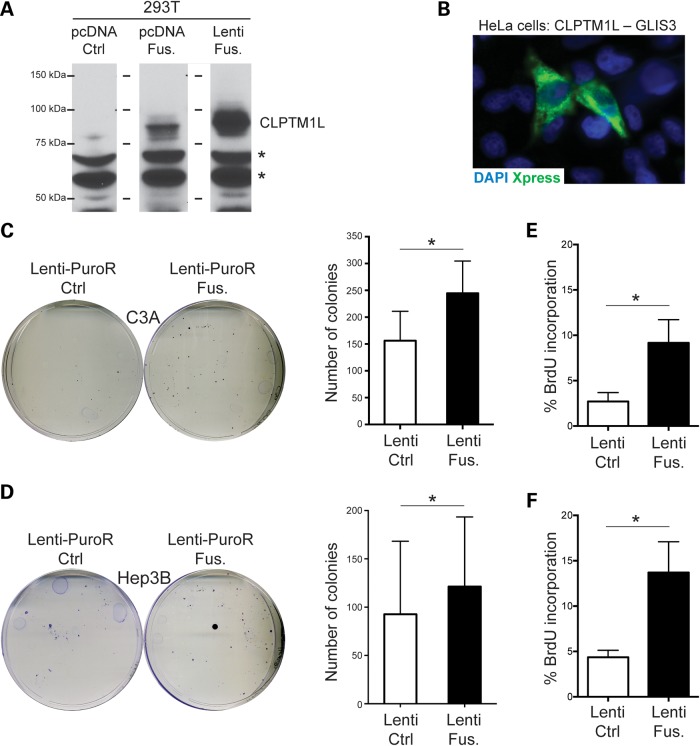
CLPTM1L is located adjacent to TERT on chromosome 5p. Multiple studies have associated this locus with an increased cancer risk (39). We found no evidence of the reciprocal t(9;5) translocation, suggesting that TERT is not in an abnormal location in the FL-HCC genome. GLIS3 mutations are responsible for a rare syndrome involving hepatic fibrosis, neonatal diabetes, congenital hypothyroidism, congenital glaucoma and polycystic kidneys (40). To investigate if this fusion protein could participate in the development of FL-HCC in this patient, we cloned a tagged form of the CLPTM1L-GLIS3 cDNA into an expression vector and transfected this construct into mammalian cells. The fusion protein was detected at the predicted size by immunoblot in 293T cells (Fig. 6A). We also found that the fusion protein was localized mostly in the cytosol of HeLa cells and not at the membrane (where CLPTM1L would be expected to localize based on its transmembrane domain) or in the nucleus (where GLIS3 would be expected to localize based on its nuclear localization signal and DNA-binding zinc finger domains) (Fig. 6B). We were unable to detect the fusion protein by immunostaining on tumor sections or by immunoblot on tumor extracts, presumably because of the low quality of the available commercial antibodies for these two proteins (these antibodies also did not readily recognize the transfected tagged fusion protein—data not shown).
Figure 6.
The CLPTM1L-GLIS3 fusion is oncogenic in HCC cells. (A) Immunoblot analysis of the fusion protein following transient transfection in 293T cells (antibody against CLPTM1L). Non-specific bands (asterisks) serve as a loading control. pcDNA Ctrl is the vector control for pcDNA-Fus.; the right lane is from cells transfected with a lentivirus to stably express the fusion protein (same lentivirus used in C–F). (B) Immunofluorescence analysis of the tagged fusion protein (Xpress tag) following transfection of pcDNA-Fus. in HeLa cells. (C and D) Representative images (left) of 10 cm plates from colony-formation assays from C3A (C) and Hep3B (D) cells stably infected with a lentivirus expressing the fusion protein compared with an empty control virus (both selected for puromycin resistance) and quantification (right) (n ≥ 3 independent experiments). (E and F) Quantification of BrdU incorporation in C3A (E) and Hep3B (F) cells stably expressing the fusion protein compared with an empty control virus (n = 3 independent experiments). *P < 0.05 (paired t-test).
In the absence of available FL-HCC cell lines, we sought to express the fusion protein in human HCC cells using lentiviral infection. We verified that the selected cells expressed the fusion transcript (data not shown), and measured the ability of these cells to form colonies when plated at low density compared with control cells. Expression of the fusion protein had no significant effect on the ability of fast-growing SNU-449 and HepG2 cells to form colonies at low density (data not shown). In contrast, expression of the CLPTM1L-GLIS3 cDNA in slower-growing C3A and Hep3B cells (from an 8-year-old HCC patient) enhanced the colony-forming ability of these cells to form colonies at low density (Fig. 6C and D). Accordingly, expression of the CLPTM1L-GLIS3 fusion protein led to a significant increase in BrdU incorporation in these cells (Fig. 6E and F). These pro-tumorigenic effects of the CLPTM1L-GLIS3 fusion in juvenile HCC cells suggest that this fusion protein may have contributed to the development of FL-HCC in the tumor of this patient.
DISCUSSION
In children and adolescents, HCC is a rare disease and its development is often unexplained. Here, we sought to investigate in depth one case of FL-HCC, an exceedingly rare subtype of pediatric liver cancer for which no etiological factor was known. We identified two gene fusion events that may contribute to the development of cancer in this patient and others.
Increasing evidence indicates that pediatric tumors often develop with many fewer alterations than adult tumors, possibly because children with cancer have not been exposed to the same level of carcinogens, both in terms of time and absolute amount, as their adult counterparts (34,41,42). In addition, pediatric tumors may arise from stem/progenitor cell populations that are endowed with an inherently greater self-renewal and proliferative potential, owing to their relative similarities with embryonic cells (43). One striking example is retinoblastoma, a tumor type in which the only recurrent gene-altering events are in the RB1 tumor suppressor gene (44). Other examples include pediatric cancers initiated and driven by simple gene fusion events; strikingly, these gene fusions are specific for different cancer types, suggesting that they are oncogenic only in certain contexts (45–56). Interestingly, these oncogenic events are often not obviously accompanied by loss of function of tumor suppressors such as RB or p53, raising the thus far untested possibility that the cell of origin for these pediatric tumors does not undergo senescence in response to this specific oncogenic stress.
In the case of FL-HCC, this work and recent work from Honeyman et al. (25) suggest that a fusion protein with the catalytic domain of the PKA enzyme may be a driving oncogenic force. This DNAJB1-PRKACA fusion has not been described in any other cancer types and how it promotes the development of cancer specifically in the livers of children remains unknown. Our data suggest that one target of PKA in FL-HCC cells could be the transcription factor CREB, but the functional relevance of CREB phosphorylation in this context remains unknown and it is likely that the DNAJB1-PKA-Cα has multiple targets in cancer cells.
Interestingly, a human syndrome associated with 19p13.12 microdeletions has been described (57). This deletion encompasses the genes present in the chromosomal region that is lost as a result of the intrachromosomal deletion that gives rise to the DNAJB1-PRKACA fusion, but these patients are not known to develop FL-HCC. These observations support the idea that the DNAJB1-PKA-Cα fusion protein is a key driver of cancer in FL-HCC, but it will be important in the future to determine if haploinsufficiency of one or more of these deleted loci may contribute to cancer development in FL-HCC patients.
This work and previous work indicate that FL-HCC tumors often express neuroendocrine markers (10,27–29). Our recent study in neuroendocrine small cell lung carcinoma (58) and previous experiments in neuroendocrine prostate cancer cells (59,60) and in adrenal tumors (61–63) suggest that PKA activity may be important in the growth of neuroendocrine tumor cells. This further raises the question of the cell of origin of FL-HCC. There are no known neuroendocrine cell types residing in the liver, but it is possible that FL-HCC arises upon activation of PKA from embryonic liver progenitors with neuroendocrine features; conversely, liver progenitors may acquire neuroendocrine features upon PKA activation and may retain these features as tumors progress. Resolving this issue will likely require the development of genetically engineered mouse models for FL-HCC.
Previous observations have underscored the lack of mutations in cancer pathways in FL-HCC (4,29,64), including the recent study from Honeyman et al. (25). While the activation of PKA signaling may be a driving event in FL-HCC, one would still expect to identify second hits during tumor progression, including possible epigenetic changes such as alterations in DNA methylation patterns (65) or mutations in the mitochondrial genome (66). The CLPTM1L-GLIS3 fusion identified in our analysis is an interesting candidate. CLPTM1L may regulate the risk of developing cancer (39,67), although its cellular functions remain largely unknown. GLIS3 is a transcription factor implicated in liver and pancreas development, including neuroendocrine cells; GLIS3 is mutated in a human syndrome associated with multiple organ defects, including the liver (40). We re-analyzed the RNA-seq data recently published by Honeyman et al. with 11 cases of FL-HCC (25): our analysis confirmed the presence of the DNAJB1-PRKACA fusion transcript in all the samples, but failed to identify fusion events involving either CLPTM1L or GLIS3 (Supplementary Material, Table S11). Thus, it is possible that the CLPTM1L-GLIS3 fusion is a rare event in FL-HCC, possibly unique to this patient. Nevertheless, other genetic or epigenetic events may have similar effects in FL-HCC cells, and further studies of the effects of this second fusion on the growth of tumor populations may provide novel insights into FL-HCC biology. Our present work also does not exclude that loss of one GLIS3 allele may be oncogenic.
From a personalized medicine perspective, the identification of the CLPTM1L-GLIS3 fusion does not offer a direct therapeutic target. In contrast, inhibitors of PKA could provide novel targeted therapies in FL-HCC patients, especially if these inhibitors could be specifically delivered to the tumor cells in the liver. In that respect, the RNA-seq data that we generated here could in the future not only help identify additional biomarkers for FL-HCC, but could also help find cell surface markers that may serve as specific delivery sites for drugs.
MATERIALS AND METHODS
WGS and processing
Appropriate IRB approval (IRB-21796) was obtained from Stanford University to collect and study archived pathology tissue samples obtained from patients aged 45 years and younger with the diagnosis of HCC or FL-HCC. Thirty-two patients were identified. Patients with hepatitis, increased background parenchymal fibrosis or tumor morphology atypical of FL-HCC were excluded. Eight patients remained for the study population. Informed consent was obtained from case 8 (Supplementary Material, Table S1), and tissue was collected from both the tumor and distant non-affected normal liver tissue.
The genomes of both the tumor and normal tissue were sequenced at Complete Genomics using their DNA nanoball array and combinatorial probe-anchor ligation sequencing technology (68). The Complete Genomics Cancer Pipeline (version 2.2) was used for sequence alignment, read mapping and initial data analysis. Briefly, reads are initially mapped to Human Genome Build 37, then areas which appear to contain variation undergo a local de novo assembly algorithm to refine initial mappings and both call and score differences between the reference genome and either the tumor or the normal genome (68).
All the sequencing data from this study (raw DNA and RNA sequencing files) are available on database of genotypes and phenotypes or can be obtained by contacting the corresponding author.
Small variant detection
Potential small variants (single nucleotide polymorphisms, indels and substitutions) were derived from the masterVar files provided by Complete Genomics. These include potential somatic small variants that were identified with the CallDiff method (CGtools) within the CGAtools v1.6.0 package (www.completegenomics.com/analysis-tools/cgatools/). From these initial calls generated by the default parameters (200 395 potential somatic changes), only calls which had a minimum of 20× coverage in the normal genome and <2 reads supporting the somatic allele in the normal genome were considered. Then to this set of 86 331 sequence changes, we filtered for true positives by looking for corresponding support in either the RNA-seq reads of the Whole Exome Sequencing reads. A true somatic mutation is called if either of two conditions were satisfied: (1) it was present in at least two reads of the Whole Exome Sequencing (Illumina) or (2) it was present in at least two reads of the RNA-seq for the tumor and had a <5% allele frequency in the normal RNA-seq data (to account for sequencing error). This identified 69 somatic mutations, 2 of which were discarded because they occurred at non-fully called loci and were thus most likely germline variants that were undercalled in the normal genome. Variants were annotated using data and tools provided by UCSC, HGNC, PROVEAN (provean.jcvi.org), SIFT (sift.jcvi.org/), dbSNP (www.ncbi.nlm.nih.gov/SNP), Annovar (www.openbioinformatics.org/annovar) and CG.
CNV detection
We used is CallCNVs (Complete Genomics), where genome-wide coverage is calculated, corrected for GC bias, and then smoothed and averaged in 2 kb windows. The tumor sample coverage was then normalized against the normal sample coverage and then segmented and scored using Hidden Markov Models. Results were visualized within IGV (Broad Institute).
Structural variant detection
JunctionDiff and Junction2Event tools (CGAtools v1.6.0) from Complete Genomics were used to compare the tumor and normal genomes. The somatic events were filtered based on the following exclusion and inclusion criteria. Exclusion criteria: somatic events annotated as artifacts or complex, frequency in CG Baseline Genome Set >0, or overlap with known underrepresented repeats. Inclusion criteria: at least 10 supporting discordant mate pairs and the junction sequence are resolved or evidence of the somatic event in sequencing data generated from other platforms (e.g. transcriptome sequencing).
Whole exome sequencing and processing
Library construction and targeted sequence capture and enrichment were performed by Centrillion Biosciences using Agilent's SureSelect Human All Exon Kit v4. Briefly, 1 µg genomic DNA was analyzed for quality control on the Agilent Bioanalyzer, then exome enrichment was performed using the Agilent's SureSelect Human All Exon Kit v4 kit. Post-library validation was performed using the Agilent DNA 1000 Kit and the Illumina Library Quantification Kit (Kapa Biosystems). Clustering was performed on the Illumina cBot using Illumina TruSeq PE Cluster kit v3, and sequencing was done on Illumina HiSeq 2000 using the Illumina TruSeq SBS Kit v3-HS to generate 2 × 100 pair end reads. Reads were mapped to UCSC hg19 genome using burrows-wheeler aligner (bio-bwa.sourceforge.net). PCR duplicates were removed by Picard (picard.sourceforge.net). Reads then underwent local realignment and base recalibration by genome analysis toolkit (www.broadinstitute.org/gatk/). Samtools (samtools.sourceforge.net) mpileup was used to generate a pileup file that was compared with Complete Genomics somatic variant calls by custom perl scripts. The targeted regions were sequenced to an average depth of 115.7×, with ∼99.9% of the targeted regions covered by ≥1×, ∼98.1% covered by >10×, ∼88.9% covered by >30× and ∼75.8% covered by >50×. Bedtools (https://github.com/arq5x/bedtools2) was used for coverage calculations in conjunction with Agilent's Human All Exon v4 target bed file.
Whole transcriptome sequencing (RNA-seq) and processing
RNA was extracted from the tumor and nonadjacent normal liver using TRIzol, followed by DNAse treatment to remove genomic DNA contamination and purified using a Qiagen RNEasy column. RNA was assessed for quality using an Agilent 2100 Bioanalyzer. The RNA-Seq library construction and sequencing were performed by Centrillion Biosciences. Briefly, after passing quality control, 1 µg RNA underwent rRNA depletion with the Epicentre Ribo-Zero Magnetic rRNA Removal Kit (Human/Mouse/Rat). The library was prepared using the Epicentre ScriptSeq v2 RNA-Seq Library Preparation Kit, and post-library validation was performed using the Agilent DNA 1000 Kit and the Illumina Library Quantification Kit (Kapa Biosystems). Clustering was performed on Illumina cBot using the Illumina TruSeq PE Cluster Kit v3 and then sequenced by the Illumina HiSeq 2000 using the Illumina TruSeq SBS Kit v3-HS to generate 2 × 100 paired end reads. Reads were mapped to the reference genome hg19 using Tophat (tophat.cbcb.umd.edu) (with parameters –r 100 and –mate-std-dev 50). Cufflinks (cufflinks.cbcb.umd.edu) cuffdiff was used to determine differential gene expression between the tumor and normal transcriptomes using a hg19 reference transcript annotation file, refFlat.gtf, downloaded from the UCSC genome browser. CummeRbund (compbio.mit.edu/cummeRbund) was then used to visualize the data and generate the heat map. For fusion transcript discovery, TopHat-Fusion was used (tophat v2.0.6 with –fusion-search option using bowtie1 -r 100 --mate-std-dev 50 and tophat-fusion-post using --num-fusion-reads 1 --num-fusion-pairs 2 --num-fusion-both 5).
Comparison of somatic mutation rates
Previously published data for HCC were used (31–33). Mutation rates for Guichard et al., Huang et al. and this study were calculated by counting somatic small variant mutations (silent, missense, indels and those affecting splice sites) occurring in the exome and dividing by the total exome targeted bases covered. Mutation rates for Fujimoto et al. were calculated by counting all somatic small variant mutations and dividing by the size of the hg19 genome (2897.3 Mb excluding N bases).
Gene set enrichment analysis
To investigate global patterns of differential gene expression and to determine if there were gene expression features significantly enriched in either the tumor or normal transcriptome, we performed GSEA using java GSEA downloaded from http://www.broadinstitute.org/gsea/index.jsp. Mean fragments per kilobase of exon per million fragments mapped (FPKM) expression values from cuffdiff (for genes which passed the test status for comparison) were used to generate a GCT file that was then fed into GSEA. Gene sets were downloaded from MSigDB. A cutoff value of FDR q-value of <0.25 was used.
Circos plot
Circos plot was generated by Circos (v. 0.62) (available at http://circos.ca/software/). Variant allele frequency was calculated as the number of mutant allele calls over the total number of mutant and reference base calls. To visualize the exome sequencing coverage across all protein coding regions, a BED file was generated from the UCSC genome browser containing all RefSeq coding exons and then edited to remove overlapping regions. This BED file, spanning 34.9 Mb or ∼1% of the genome, was then the reference file for Bedtools' bedCoverage to calculate the coverage for each feature. The result was then displayed as a histogram in the Circos plot.
Ingenuity pathway analysis
A list of differentially expressed genes in addition to the log2(fold change) and the associated P-values and q-values were derived from cuffdiff output of the RNA-seq data—where log2(fold change) = log2([tumor mean FPKM]/[normal mean FPKM]). This served as the input for IPA software (www.ingenuity.com). Core analysis was then performed on all genes whose q-value of <0.25 using the default settings with the exception that an additional filter (all tissues and cell lines) was selected. Overrepresented disease and biological functions in the downstream effects analysis were considered significant if P-value <0.05 and the absolute activation Z-score was >2. For upstream regulator analysis, we filtered for transcriptional regulators and considered them significant if the overlap P-value was <0.05 and the absolute activation Z-score was >2. Both types of analyses are based on prior knowledge of expected effects between genes and biological functions (for downstream effects analysis) or transcriptional regulators and their target genes (for upstream regulatory analysis) from IPA's Ingenuity Knowledge Base.
Cloning of fusion transcripts
RT-PCR verification of fusion transcripts was done by primers targeting either side of the breakpoint, amplification of the subsequent product and Sanger sequencing. The entire DNAJB1-PRKACA fusion transcript was amplified in one PCR reaction with primers targeting the ATG start codon of DNAJB1 (DNAJB1-PRKACA-cDNA-full-F1) and the stop codon of PRKACA (DNAJB1-PRKACA-cDNA-full-R1). Product was then cloned into pCDNA4-HisMax-TOPO vector and subsequently sequenced for confirmation. The CLPTM1L-GLIS3 fusion transcript was amplified in three separate PCRs: the first PCR amplified a 5′ fragment starting with the ATG start codon of CLPTM1L and the second PCR amplified the 3′ fragment containing the stop codon of GLIS3. Both fragments contained an overlap of ∼500 bp that included the breakpoint. The fragments were then used as the template for the final third reaction, which used the forward primer of the first reaction and the reverse primer of the second reaction to obtain the full-length transcript (∼2.2 kb). Primers are provided in Supplementary Material, Table S12.
Similar forward and reverse primers were used to clone the full transcripts (from start to stop codon) of CLPTM1L-GLIS3 and DNAJB1-PRKACA out of the pCDNA4-HisMax-TOPO plasmids into the multiple cloning site of the pLEX-MCS lentiviral vector system. Sanger sequencing was used to verify the plasmids.
Cell culture
293T and HeLa cells were grown in DMEM with 10% serum. HCC cells lines were purchased from the ATCC collection and were grown in RPMI with 10% serum. 293T cells and HeLa cells were transfected using the calcium phosphate method. Lentiviral supernatants were produced by transfecting 293T cells with pLEX-MCS (Open Biosystems) and pLEX- CLPTM1L-GLIS3 plasmids together with packaging plasmids (pCMV Δ8.9 and pMD VSV-G). Transduced cells were selected with puromycin. For colony-formation assays, pLEX-MCS and pLEX- CLPTM1L-GLIS3 stably infected HCC cell lines were seeded at 1000 cells/10 cm plate. Plates were stained with Crystal Violet 10 days after plating: plates were washed with PBS and then fixed for 10′ in 4% formaldehyde at room temperature (RT). After a rinse in PBS, 0.1% Crystal Violet was added for 30 min at RT. Excess staining was removed under tap water and plates were let to dry. Stained colonies were counted manually. BrdU incorporation (10 µM) was performed for 30 min before fixation and analysis by FACS, as described before (69). Statistical significance was assayed by paired Student's t-test (*P-value <0.05). Mean and standard error are shown.
RNA and protein analysis
RNA was extracted from frozen cell pellets with TRIzol (Invitrogen). Expression levels were measured using Taqman or SYBR green by quantitative PCR. For immunoblotting, cells were homogenized using lysis buffer containing 1% NP40, 50 mm HEPES-KOH pH 7.8, 150 mm NaCl, 10 mm EDTA and a cocktail of protease and phosphate inhibitors. The antibodies used were phospho-CREB (Ser133), CREB, phospho-PKA C (Thr197), PKA C and phospho-(Ser/Thr) PKA substrate (all purchased from Cell Signaling). Loading was verified with staining of total protein with Ponceau and antibodies against anti-alpha-Tubulin (Sigma T9026).
Immunostaining
For paraffin sections, normal and tumor tissue chunks were fixed in 4% paraformaldehyde and then transferred to 70% ethanol prior to processing and embedding. For immunofluorescence on paraffin sections, antigen retrieval was performed using Trilogy (Cell Marque) for 15 min in a pressure cooker. Sections were then briefly washed in PBS + 0.1% Tween (PBST), and then blocked for 1 h at RT in PBST with 10% serum and 1% bovine serum albumin (BSA). Sections were next incubated with primary antibodies in block solution overnight at 4°C, washed in PBST, incubated with fluorescent secondary antibodies diluted to 1 : 500 in block solution for 1 h at RT in the dark, then washed with PBST and mounted with ProLong Gold (Life Technologies). Dilutions for primary antibodies used: 1 : 800 for phospho-CREB (Ser133, Cell Signaling); 1 : 500 UCHL1 (Sigma-Aldrich) and 1 : 100 CPE (BD Biosciences). For immunocytochemistry, HeLa cells were grown on 10 mm coverslips in 24-well plates and then transfected with the pCDNA4-HisMax-TOPO-CLPTM1L-GLIS3 plasmid, the pCDNA4-HisMax-TOPO-DNAJB1-PRKACA plasmid or a control GFP plasmid. Cells were fixed 48 h later for 10 min at RT in 4% paraformaldehyde, then washed in PBS and permeabilized for 10 min in 0.2% Triton X-100 in PBS. Coverslips were incubated with primary antibody anti-Xpress (1 : 500 dilution, Life Technologies) diluted in PBST + 10% serum + 1% BSA for 1 h at RT, washed with PBST, then incubated with secondary antibody diluted in PBST + 10% serum + 1% BSA for 1 h at RT, then washed with PBST and rinsed with PBS before being mounted on a slide with ProLong Gold. Immunohistochemical staining was performed on paraffin-embedded tissue of FL-HCC and normal background liver parenchyma from each patient using standard techniques. Each case was evaluated for protein expression of CPE (polyclonal, Santa Cruz Biotechnology), CREB (polyclonal, Abcam), phospho-CREB (polyclonal, Abcam) and CK7 (OV-TL 12/30, Dako).
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by the National Institutes of Health (CA114102 to J.S.) and the Lucile Packard Foundation for Children's Health (grant UL1 RR025744). J.S. is the Harriet and Mary Zelencik Scientist in Children's Cancer and Blood Diseases. L.X. was funded by the Stanford Medical Scholars program and the Howard Hughes Medical Institute Medical Fellows program.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank members of the Sage laboratory for helpful comments throughout the course of this study, especially Jing Lim and Pawel Mazur, as well as Dedeepya Vaka, Cuiping Pan and Jason Reuter for advice with the bioinformatics analysis, Gary Mantalas for his help with library construction, Janet Bueno for her help with tissue collection and Kathleen Sakamato for helpful suggestions with the manuscript.
Conflict of Interest statement. None declared.
REFERENCES
- 1.El-Serag H.B., Rudolph K.L. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557–2576. doi: 10.1053/j.gastro.2007.04.061. [DOI] [PubMed] [Google Scholar]
- 2.Arzumanyan A., Reis H.M., Feitelson M.A. Pathogenic mechanisms in HBV- and HCV-associated hepatocellular carcinoma. Nat. Rev. Cancer. 2013;13:123–135. doi: 10.1038/nrc3449. [DOI] [PubMed] [Google Scholar]
- 3.Klein W.M., Molmenti E.P., Colombani P.M., Grover D.S., Schwarz K.B., Boitnott J., Torbenson M.S. Primary liver carcinoma arising in people younger than 30 years. Am. J. Clin. Pathol. 2005;124:512–518. doi: 10.1309/TT0R7KAL32228E99. [DOI] [PubMed] [Google Scholar]
- 4.Ward S.C., Waxman S. Fibrolamellar carcinoma: a review with focus on genetics and comparison to other malignant primary liver tumors. Semin. Liver Dis. 2011;31:61–70. doi: 10.1055/s-0031-1272835. [DOI] [PubMed] [Google Scholar]
- 5.El-Serag H.B., Davila J.A. Is fibrolamellar carcinoma different from hepatocellular carcinoma? A US population-based study. Hepatology. 2004;39:798–803. doi: 10.1002/hep.20096. [DOI] [PubMed] [Google Scholar]
- 6.Kakar S., Burgart L.J., Batts K.P., Garcia J., Jain D., Ferrell L.D. Clinicopathologic features and survival in fibrolamellar carcinoma: comparison with conventional hepatocellular carcinoma with and without cirrhosis. Mod. Pathol. 2005;18:1417–1423. doi: 10.1038/modpathol.3800449. [DOI] [PubMed] [Google Scholar]
- 7.Craig J.R., Peters R.L., Edmondson H.A., Omata M. Fibrolamellar carcinoma of the liver: a tumor of adolescents and young adults with distinctive clinico-pathologic features. Cancer. 1980;46:372–379. doi: 10.1002/1097-0142(19800715)46:2<372::aid-cncr2820460227>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 8.Liu S., Chan K.W., Wang B., Qiao L. Fibrolamellar hepatocellular carcinoma. Am. J. Gastroenterol. 2009;104:2617–2624. doi: 10.1038/ajg.2009.440. quiz 2625. [DOI] [PubMed] [Google Scholar]
- 9.Yamashita T., Forgues M., Wang W., Kim J.W., Ye Q., Jia H., Budhu A., Zanetti K.A., Chen Y., Qin L.X., et al. EpCAM and alpha-fetoprotein expression defines novel prognostic subtypes of hepatocellular carcinoma. Cancer Res. 2008;68:1451–1461. doi: 10.1158/0008-5472.CAN-07-6013. [DOI] [PubMed] [Google Scholar]
- 10.Ward S.C., Huang J., Tickoo S.K., Thung S.N., Ladanyi M., Klimstra D.S. Fibrolamellar carcinoma of the liver exhibits immunohistochemical evidence of both hepatocyte and bile duct differentiation. Mod. Pathol. 2010;23:1180–1190. doi: 10.1038/modpathol.2010.105. [DOI] [PubMed] [Google Scholar]
- 11.Ross H.M., Daniel H.D., Vivekanandan P., Kannangai R., Yeh M.M., Wu T.T., Makhlouf H.R., Torbenson M. Fibrolamellar carcinomas are positive for CD68. Mod. Pathol. 2011;24:390–395. doi: 10.1038/modpathol.2010.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oikawa T., Kamiya A., Zeniya M., Chikada H., Hyuck A.D., Yamazaki Y., Wauthier E., Tajiri H., Miller L.D., Wang X.W., et al. Sal-like protein 4 (SALL4), a stem cell biomarker in liver cancers. Hepatology. 2013;57:1469–1483. doi: 10.1002/hep.26159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Van Eyken P., Sciot R., Brock P., Casteels-Van Daele M., Ramaekers F.C., Desmet V.J. Abundant expression of cytokeratin 7 in fibrolamellar carcinoma of the liver. Histopathology. 1990;17:101–107. doi: 10.1111/j.1365-2559.1990.tb00679.x. [DOI] [PubMed] [Google Scholar]
- 14.Kannangai R., Vivekanandan P., Martinez-Murillo F., Choti M., Torbenson M. Fibrolamellar carcinomas show overexpression of genes in the RAS, MAPK, PIK3, and xenobiotic degradation pathways. Hum. Pathol. 2007;38:639–644. doi: 10.1016/j.humpath.2006.07.019. [DOI] [PubMed] [Google Scholar]
- 15.Buckley A.F., Burgart L.J., Kakar S. Epidermal growth factor receptor expression and gene copy number in fibrolamellar hepatocellular carcinoma. Hum. Pathol. 2006;37:410–414. doi: 10.1016/j.humpath.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 16.Cieply B., Zeng G., Proverbs-Singh T., Geller D.A., Monga S.P. Unique phenotype of hepatocellular cancers with exon-3 mutations in beta-catenin gene. Hepatology. 2009;49:821–831. doi: 10.1002/hep.22695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Orsatti G., Hytiroglou P., Thung S.N., Ishak K.G., Paronetto F. Lamellar fibrosis in the fibrolamellar variant of hepatocellular carcinoma: a role for transforming growth factor beta. Liver. 1997;17:152–156. doi: 10.1111/j.1600-0676.1997.tb00798.x. [DOI] [PubMed] [Google Scholar]
- 18.Li W., Tan D., Zenali M.J., Brown R.E. Constitutive activation of nuclear factor-kappa B (NF-kB) signaling pathway in fibrolamellar hepatocellular carcinoma. Int. J. Clin. Exp. Pathol. 2010;3:238–243. [PMC free article] [PubMed] [Google Scholar]
- 19.Wilczek E., Szparecki G., Lukasik D., Koperski L., Winiarska M., Wilczynski G.M., Wasiutynski A., Gornicka B. Loss of the orphan nuclear receptor SHP is more pronounced in fibrolamellar carcinoma than in typical hepatocellular carcinoma. PLoS ONE. 2012;7:e30944. doi: 10.1371/journal.pone.0030944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Njei B., Konjeti V.R., Ditah I. Prognosis of patients with fibrolamellar hepatocellular carcinoma versus conventional hepatocellular carcinoma: a systematic review and meta-analysis. Gastrointest. Cancer Res. 2014;7:49–54. [PMC free article] [PubMed] [Google Scholar]
- 21.Stevens W.R., Johnson C.D., Stephens D.H., Nagorney D.M. Fibrolamellar hepatocellular carcinoma: stage at presentation and results of aggressive surgical management. AJR Am. J. Roentgenol. 1995;164:1153–1158. doi: 10.2214/ajr.164.5.7717223. [DOI] [PubMed] [Google Scholar]
- 22.Katzenstein H.M., Krailo M.D., Malogolowkin M.H., Ortega J.A., Qu W., Douglass E.C., Feusner J.H., Reynolds M., Quinn J.J., Newman K., et al. Fibrolamellar hepatocellular carcinoma in children and adolescents. Cancer. 2003;97:2006–2012. doi: 10.1002/cncr.11292. [DOI] [PubMed] [Google Scholar]
- 23.Ang C.S., Kelley R.K., Choti M.A., Cosgrove D.P., Chou J.F., Klimstra D., Torbenson M.S., Ferrell L., Pawlik T.M., Fong Y., et al. Clinicopathologic characteristics and survival outcomes of patients with fibrolamellar carcinoma: data from the fibrolamellar carcinoma consortium. Gastrointest. Cancer Res. 2013;6:3–9. [PMC free article] [PubMed] [Google Scholar]
- 24.Chun Y.S., Zimmitti G. Fibrolamellar variant of hepatocellular carcinoma. Recent Results Cancer Res. 2013;190:101–110. doi: 10.1007/978-3-642-16037-0_7. [DOI] [PubMed] [Google Scholar]
- 25.Honeyman J.N., Simon E.P., Robine N., Chiaroni-Clarke R., Darcy D.G., Lim I.I., Gleason C.E., Murphy J.M., Rosenberg B.R., Teegan L., et al. Detection of a recurrent DNAJB1-PRKACA chimeric transcript in fibrolamellar hepatocellular carcinoma. Science. 2014;343:1010–1014. doi: 10.1126/science.1249484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vivekanandan P., Micchelli S.T., Torbenson M. Anterior gradient-2 is overexpressed by fibrolamellar carcinomas. Hum. Pathol. 2009;40:293–299. doi: 10.1016/j.humpath.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Collier N.A., Weinbren K., Bloom S.R., Lee Y.C., Hodgson H.J., Blumgart L.H. Neurotensin secretion by fibrolamellar carcinoma of the liver. Lancet. 1984;1:538–540. doi: 10.1016/s0140-6736(84)90934-6. [DOI] [PubMed] [Google Scholar]
- 28.Wang J.H., Dhillon A.P., Sankey E.A., Wightman A.K., Lewin J.F., Scheuer P.J. ‘Neuroendocrine’ differentiation in primary neoplasms of the liver. J. Pathol. 1991;163:61–67. doi: 10.1002/path.1711630111. [DOI] [PubMed] [Google Scholar]
- 29.Malouf G.G., Job S., Paradis V., Fabre M., Brugieres L., Saintigny P., Vescovo L., Belghiti J., Branchereau S., Faivre S., et al. Transcriptional profiling of pure fibrolamellar hepatocellular carcinoma reveals an endocrine signature. Hepatology. 2014;59:2228–2237. doi: 10.1002/hep.27018. [DOI] [PubMed] [Google Scholar]
- 30.Ding S.F., Delhanty J.D., Bowles L., Dooley J.S., Wood C.B., Habib N.A. Infrequent chromosome allele loss in fibrolamellar carcinoma. Br. J. Cancer. 1993;67:244–246. doi: 10.1038/bjc.1993.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guichard C., Amaddeo G., Imbeaud S., Ladeiro Y., Pelletier L., Maad I.B., Calderaro J., Bioulac-Sage P., Letexier M., Degos F., et al. Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nat. Genet. 2012;44:694–698. doi: 10.1038/ng.2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fujimoto A., Totoki Y., Abe T., Boroevich K.A., Hosoda F., Nguyen H.H., Aoki M., Hosono N., Kubo M., Miya F., et al. Whole-genome sequencing of liver cancers identifies etiological influences on mutation patterns and recurrent mutations in chromatin regulators. Nat. Genet. 2012;44:760–764. doi: 10.1038/ng.2291. [DOI] [PubMed] [Google Scholar]
- 33.Huang J., Deng Q., Wang Q., Li K.Y., Dai J.H., Li N., Zhu Z.D., Zhou B., Liu X.Y., Liu R.F., et al. Exome sequencing of hepatitis B virus-associated hepatocellular carcinoma. Nat. Genet. 2012;44:1117–1121. doi: 10.1038/ng.2391. [DOI] [PubMed] [Google Scholar]
- 34.Vogelstein B., Papadopoulos N., Velculescu V.E., Zhou S., Diaz L.A., Jr., Kinzler K.W. Cancer genome landscapes. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marchio A., Pineau P., Meddeb M., Terris B., Tiollais P., Bernheim A., Dejean A. Distinct chromosomal abnormality pattern in primary liver cancer of non-B, non-C patients. Oncogene. 2000;19:3733–3738. doi: 10.1038/sj.onc.1203713. [DOI] [PubMed] [Google Scholar]
- 36.Wilkens L., Bredt M., Flemming P., Kubicka S., Klempnauer J., Kreipe H. Cytogenetic aberrations in primary and recurrent fibrolamellar hepatocellular carcinoma detected by comparative genomic hybridization. Am. J. Clin. Pathol. 2000;114:867–874. doi: 10.1309/BMTT-JBPD-D13H-1UVD. [DOI] [PubMed] [Google Scholar]
- 37.Hattori H., Liu Y.C., Tohnai I., Ueda M., Kaneda T., Kobayashi T., Tanabe K., Ohtsuka K. Intracellular localization and partial amino acid sequence of a stress-inducible 40-kDa protein in HeLa cells. Cell Struct. Funct. 1992;17:77–86. doi: 10.1247/csf.17.77. [DOI] [PubMed] [Google Scholar]
- 38.Dyachok O., Isakov Y., Sagetorp J., Tengholm A. Oscillations of cyclic AMP in hormone-stimulated insulin-secreting beta-cells. Nature. 2006;439:349–352. doi: 10.1038/nature04410. [DOI] [PubMed] [Google Scholar]
- 39.Rafnar T., Sulem P., Stacey S.N., Geller F., Gudmundsson J., Sigurdsson A., Jakobsdottir M., Helgadottir H., Thorlacius S., Aben K.K., et al. Sequence variants at the TERT-CLPTM1L locus associate with many cancer types. Nat. Genet. 2009;41:221–227. doi: 10.1038/ng.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Senee V., Chelala C., Duchatelet S., Feng D., Blanc H., Cossec J.C., Charon C., Nicolino M., Boileau P., Cavener D.R., et al. Mutations in GLIS3 are responsible for a rare syndrome with neonatal diabetes mellitus and congenital hypothyroidism. Nat. Genet. 2006;38:682–687. doi: 10.1038/ng1802. [DOI] [PubMed] [Google Scholar]
- 41.Stratton M.R., Campbell P.J., Futreal P.A. The cancer genome. Nature. 2009;458:719–724. doi: 10.1038/nature07943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mitelman F., Johansson B., Mertens F. The impact of translocations and gene fusions on cancer causation. Nat. Rev. Cancer. 2007;7:233–245. doi: 10.1038/nrc2091. [DOI] [PubMed] [Google Scholar]
- 43.Marshall G.M., Carter D.R., Cheung B.B., Liu T., Mateos M.K., Meyerowitz J.G., Weiss W.A. The prenatal origins of cancer. Nat. Rev. Cancer. 2014;14:277–289. doi: 10.1038/nrc3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang J., Benavente C.A., McEvoy J., Flores-Otero J., Ding L., Chen X., Ulyanov A., Wu G., Wilson M., Wang J., et al. A novel retinoblastoma therapy from genomic and epigenetic analyses. Nature. 2012;481:329–334. doi: 10.1038/nature10733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Westbrook C.A., Hooberman A.L., Spino C., Dodge R.K., Larson R.A., Davey F., Wurster-Hill D.H., Sobol R.E., Schiffer C., Bloomfield C.D. Clinical significance of the BCR-ABL fusion gene in adult acute lymphoblastic leukemia: a Cancer and Leukemia Group B Study (8762) Blood. 1992;80:2983–2990. [PubMed] [Google Scholar]
- 46.Thirman M.J., Gill H.J., Burnett R.C., Mbangkollo D., McCabe N.R., Kobayashi H., Ziemin-van der Poel S., Kaneko Y., Morgan R., Sandberg A.A., et al. Rearrangement of the MLL gene in acute lymphoblastic and acute myeloid leukemias with 11q23 chromosomal translocations. N. Engl. J. Med. 1993;329:909–914. doi: 10.1056/NEJM199309233291302. [DOI] [PubMed] [Google Scholar]
- 47.Davis R.J., D'Cruz C.M., Lovell M.A., Biegel J.A., Barr F.G. Fusion of PAX7 to FKHR by the variant t(1;13)(p36;q14) translocation in alveolar rhabdomyosarcoma. Cancer Res. 1994;54:2869–2872. [PubMed] [Google Scholar]
- 48.Sorensen P.H., Lessnick S.L., Lopez-Terrada D., Liu X.F., Triche T.J., Denny C.T. A second Ewing's sarcoma translocation, t(21;22), fuses the EWS gene to another ETS-family transcription factor, ERG. Nat. Genet. 1994;6:146–151. doi: 10.1038/ng0294-146. [DOI] [PubMed] [Google Scholar]
- 49.Golub T.R., Barker G.F., Bohlander S.K., Hiebert S.W., Ward D.C., Bray-Ward P., Morgan E., Raimondi S.C., Rowley J.D., Gilliland D.G. Fusion of the TEL gene on 12p13 to the AML1 gene on 21q22 in acute lymphoblastic leukemia. Proc. Natl. Acad. Sci. USA. 1995;92:4917–4921. doi: 10.1073/pnas.92.11.4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Barr F.G., Nauta L.E., Davis R.J., Schafer B.W., Nycum L.M., Biegel J.A. In vivo amplification of the PAX3-FKHR and PAX7-FKHR fusion genes in alveolar rhabdomyosarcoma. Hum. Mol. Genet. 1996;5:15–21. doi: 10.1093/hmg/5.1.15. [DOI] [PubMed] [Google Scholar]
- 51.Knezevich S.R., Garnett M.J., Pysher T.J., Beckwith J.B., Grundy P.E., Sorensen P.H. ETV6-NTRK3 gene fusions and trisomy 11 establish a histogenetic link between mesoblastic nephroma and congenital fibrosarcoma. Cancer Res. 1998;58:5046–5048. [PubMed] [Google Scholar]
- 52.Raza-Egilmez S.Z., Jani-Sait S.N., Grossi M., Higgins M.J., Shows T.B., Aplan P.D. NUP98-HOXD13 gene fusion in therapy-related acute myelogenous leukemia. Cancer Res. 1998;58:4269–4273. [PubMed] [Google Scholar]
- 53.Rubnitz J.E., Pui C.H., Downing J.R. The role of TEL fusion genes in pediatric leukemias. Leukemia. 1999;13:6–13. doi: 10.1038/sj.leu.2401258. [DOI] [PubMed] [Google Scholar]
- 54.Argani P., Lui M.Y., Couturier J., Bouvier R., Fournet J.C., Ladanyi M. A novel CLTC-TFE3 gene fusion in pediatric renal adenocarcinoma with t(X;17)(p11.2;q23) Oncogene. 2003;22:5374–5378. doi: 10.1038/sj.onc.1206686. [DOI] [PubMed] [Google Scholar]
- 55.Kleinman C.L., Gerges N., Papillon-Cavanagh S., Sin-Chan P., Pramatarova A., Quang D.A., Adoue V., Busche S., Caron M., Djambazian H., et al. Fusion of TTYH1 with the C19MC microRNA cluster drives expression of a brain-specific DNMT3B isoform in the embryonal brain tumor ETMR. Nat. Genet. 2014;46:39–44. doi: 10.1038/ng.2849. [DOI] [PubMed] [Google Scholar]
- 56.Nowell P.C., Emanuel B.S., Finan J.B., Erikson J., Croce C.M. Chromosome rearrangements in oncogenesis. Microbiol. Sci. 1984;1:223–228. [PubMed] [Google Scholar]
- 57.Bonaglia M.C., Marelli S., Novara F., Commodaro S., Borgatti R., Minardo G., Memo L., Mangold E., Beri S., Zucca C., et al. Genotype-phenotype relationship in three cases with overlapping 19p13.12 microdeletions. Eur. J. Hum. Genet. 2010;18:1302–1309. doi: 10.1038/ejhg.2010.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jahchan N.S., Dudley J.T., Mazur P.K., Flores N., Yang D., Palmerton A., Zmoos A.F., Vaka D., Tran K.Q., Zhou M., et al. A drug repositioning approach identifies tricyclic antidepressants as inhibitors of small cell lung cancer and other neuroendocrine tumors. Cancer Discov. 2013;3:1364–1377. doi: 10.1158/2159-8290.CD-13-0183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Deeble P.D., Cox M.E., Frierson H.F., Jr., Sikes R.A., Palmer J.B., Davidson R.J., Casarez E.V., Amorino G.P., Parsons S.J. Androgen-independent growth and tumorigenesis of prostate cancer cells are enhanced by the presence of PKA-differentiated neuroendocrine cells. Cancer Res. 2007;67:3663–3672. doi: 10.1158/0008-5472.CAN-06-2616. [DOI] [PubMed] [Google Scholar]
- 60.Park M.H., Lee H.S., Lee C.S., You S.T., Kim D.J., Park B.H., Kang M.J., Heo W.D., Shin E.Y., Schwartz M.A., et al. p21-Activated kinase 4 promotes prostate cancer progression through CREB. Oncogene. 2013;32:2475–2482. doi: 10.1038/onc.2012.255. [DOI] [PubMed] [Google Scholar]
- 61.Goh G., Scholl U.I., Healy J.M., Choi M., Prasad M.L., Nelson-Williams C., Kuntsman J.W., Korah R., Suttorp A.C., Dietrich D., et al. Recurrent activating mutation in PRKACA in cortisol-producing adrenal tumors. Nat. Genet. 2014;46:613–617. doi: 10.1038/ng.2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cao Y., He M., Gao Z., Peng Y., Li Y., Li L., Zhou W., Li X., Zhong X., Lei Y., et al. Activating hotspot L205R mutation in PRKACA and adrenal Cushing's syndrome. Science. 2014;344:913–917. doi: 10.1126/science.1249480. [DOI] [PubMed] [Google Scholar]
- 63.Sato Y., Maekawa S., Ishii R., Sanada M., Morikawa T., Shiraishi Y., Yoshida K., Nagata Y., Sato-Otsubo A., Yoshizato T., et al. Recurrent somatic mutations underlie corticotropin-independent Cushing's syndrome. Science. 2014;344:917–920. doi: 10.1126/science.1252328. [DOI] [PubMed] [Google Scholar]
- 64.Kakar S., Chen X., Ho C., Burgart L.J., Sahai V., Dachrut S., Yabes A., Jain D., Ferrell L.D. Chromosomal changes in fibrolamellar hepatocellular carcinoma detected by array comparative genomic hybridization. Mod. Pathol. 2009;22:134–141. doi: 10.1038/modpathol.2008.178. [DOI] [PubMed] [Google Scholar]
- 65.Trankenschuh W., Puls F., Christgen M., Albat C., Heim A., Poczkaj J., Fleming P., Kreipe H., Lehmann U. Frequent and distinct aberrations of DNA methylation patterns in fibrolamellar carcinoma of the liver. PLoS ONE. 2010;5:e13688. doi: 10.1371/journal.pone.0013688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vivekanandan P., Daniel H., Yeh M.M., Torbenson M. Mitochondrial mutations in hepatocellular carcinomas and fibrolamellar carcinomas. Mod. Pathol. 2010;23:790–798. doi: 10.1038/modpathol.2010.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Haiman C.A., Chen G.K., Vachon C.M., Canzian F., Dunning A., Millikan R.C., Wang X., Ademuyiwa F., Ahmed S., Ambrosone C.B., et al. A common variant at the TERT-CLPTM1L locus is associated with estrogen receptor-negative breast cancer. Nat. Genet. 2011;43:1210–1214. doi: 10.1038/ng.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Drmanac R., Sparks A.B., Callow M.J., Halpern A.L., Burns N.L., Kermani B.G., Carnevali P., Nazarenko I., Nilsen G.B., Yeung G., et al. Human genome sequencing using unchained base reads on self-assembling DNA nanoarrays. Science. 2010;327:78–81. doi: 10.1126/science.1181498. [DOI] [PubMed] [Google Scholar]
- 69.Viatour P., Somervaille T.C., Venkatasubrahmanyam S., Kogan S., McLaughlin M.E., Weissman I.L., Butte A.J., Passegue E., Sage J. Hematopoietic stem cell quiescence is maintained by compound contributions of the retinoblastoma gene family. Cell Stem Cell. 2008;3:416–428. doi: 10.1016/j.stem.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.