Abstract
We used a custom-designed microarray and quantitative PCR to characterize the rapid transcriptional response to long-term sensitization training in the marine mollusk Aplysia californica. Aplysia were exposed to repeated noxious shocks to one side of the body, a procedure known to induce a longlasting, transcription-dependent increase in reflex responsiveness that is restricted to the side of training. One hour after training, pleural ganglia from the trained and untrained sides of the body were harvested; these ganglia contain the sensory nociceptors which help mediate the expression of longterm sensitization memory. Microarray analysis from 8 biological replicates suggests that long-term sensitization training rapidly regulates at least 81 transcripts. We used qPCR to test a subset of these transcripts and found that 83% were confirmed in the same samples, and 86% of these were again confirmed in an independent sample. Thus, our new microarray design shows strong convergent and predictive validity for analyzing the transcriptional correlates of memory in Aplysia. Fully validated transcripts include some previously identified as regulated in this paradigm (ApC/EBP and ApEgr) but also include novel findings. Specifically, we show that long-term sensitization training rapidly upregulates the expression of transcripts which may encode Aplysia homologs of a C/EBPγ transcription factor, a glycine transporter (GlyT2), and a vacuolar-protein-sorting-associated protein (VPS36).
1. Introduction
The encoding of new memories triggers rapid changes in neuronal gene expression (Bailey, Bartsch, & Kandel, 1996; Tischmeyer & Grimm, 1999). This early wave of transcriptional change seems to set the stage for long-term memory maintenance, as blocking transcription during encoding impairs retention across a wide range of species and learning contexts (e.g. Chew, Mello, Nottebohm, Jarvis, & Vicario, 1995; Esdin, Pearce, & Glanzman, 2010; Hermitte, Pedreira, Tomsic, & Maldonado, 1999; Sangha, 2003; Watanabe et al., 2005). Thus, characterizing the immediate transcriptional response to memory encoding is an important step in understanding the long-term cellular and network changes that underlie long-term memory.
Here we use microarray and qPCR to characterize the rapid transcriptional response to long-term sensitization in the marine mollusk Aplysia californica. Aplysia have long served as an attractive model organism for studying the molecular mechanisms of memory. One particular focus has been long-term sensitization (Pinsker, Hening, Carew, & Kandel, 1973), a learning paradigm in which repeated exposure to a noxious stimulus produces a long-lasting, transcription-dependent increase in reflex responsiveness (Castellucci, Blumenfeld, Goelet, & Kandel, 1989). Sensitization of the tail-elicited siphon withdrawal reflex provides an especially attractive system for transcriptional analysis because 1) sensitization can be applied and expressed unilaterally (Scholz & Byrne, 1987), allowing for powerful within-subjects comparisons, 2) sensitization memory is known to depend at least in part on physiological changes in the VC nociceptors of the pleural ganglia (Cleary, Lee, & Byrne, 1998), providing a behaviorally-relevant target for transcriptional analysis, and 3) transcriptional and behavioral changes can be correlated at the level of individual animals, allowing exploration of individual differences in retention (Bonnick et al., 2012).
Previous research has shown that long-term sensitization training produces a rapid (within 1 hour) increase in the expression of six different transcripts in the pleural ganglia: ApC/EBP (GenBank: U00994; Alberini, Ghirardi, Metz, & Kandel, 1994), ApCREB1 (GenBank: NM_001256437; Bartsch, Casadio, Karl, Serodio, & Kandel, 1998), ApEgr (GenBank: KC608221; Cyriac et al., 2013), ApTBL-1 (GenBank: U57369; Liu et al., 1997), ApCalmodulin (GenBank: NM_001204580; Zwartjes et al., 1998), and an reductase-related protein (GenBank: NM_001204605; Zwartjes et al., 1998). To date, however, there have been no comprehensive efforts to characterize the rapid transcriptional response to long-term sensitization training (though see Castellucci, Kennedy, Kandel, & Goelet, 1988; Liu et al., 1997). Considerable effort, however, has gone into characterizing the transcriptional response of Aplysia neurons to exposure to serotonin (5-hydroxytryptamine, 5-HT), a neuromodulator released during sensitization training that is critical to its induction (Glanzman et al., 1989). For example, a microarray study in Aplysia kurodai has characterized the rapid transcriptional response to soaking whole animals in 200 μM 5-HT for 2 hours (Lee et al., 2008). Many others have examined changes in transcription or protein expression at different time points after 5-HT exposure in whole animals, ganglia, or cultured neurons (e.g. Barzilai, Kennedy, Sweatt, & Kandel, 1989; Monje et al., 2011). In a previous analysis of a small set of transcripts, however, we have shown that 5-HT and behavioral training do not always produce similar patterns of transcriptional change in Aplysia californica (Bonnick et al., 2012).
To facilitate characterization of learning-related transcriptional changes, we developed a new microarray design that leverages the entire known EST pool for Aplysia in a more compact format than previously available arrays. We find that this new platform has high convergent and predictive validity, and present 4 new transcripts which are strongly, consistently, and rapidly up-regulated by long-term sensitization training.
2. Materials and methods
We report how we determined our sample size, all data exclusions (if any), all manipulations, and all measures in these studies (Simmons, Nelson, & Simonsohn, 2012). All data for this project is posted to the Open Science Framework (https://osf.io/8pgfh/); the microarray data is also posted to GEO (Geo: GSE57458).
2.1 Animals
Animals (75–125g) were obtained from the RSMAS National Resource for Aplysia (Miami, FL) and maintained at 16°C in one of two 90-gallon aquariums with continuously circulating artificial sea water (Instant Ocean, Aquarium Systems Inc.). Animals were separately housed in rectangular colanders, fed dried seaweed twice a week, and maintained on a 12 hr light-dark cycle. 2 days prior to any experimental testing, animals were fed to satiation and then food deprived for the remainder of the experiment (Levenson, Byrne, & Eskin, 1999; Wainwright, Zhang, Byrne, & Cleary, 2002). To control for batch/shipment effects, animals from at least 2 different shipments were used for each experiment.
2.2 Long-term sensitization Training
A one-day long-term sensitization training protocol (Figure 1) was conducted similarly to Wainwright et al. (2002) but with two differences. First, we stimulated with a 60Hz biphasic square-wave pulse rather than true AC current, as this enabled the use of a constant-current stimulator (WPI High-Current Stimulus Isolation Unit, Model A385, Sarasota, FL) to provide more precise control over stimulus intensity. Second, we used a 90mA stimulus rather than a 60mA stimulus. We selected this intensity based on extensive pilot work for a previous set of experiments (Bonnick et al., 2012) where we observed that with our stimulator a 90mA stimulus produces more reliable elevations of both behavior and C/EBP expression.
Figure 1.
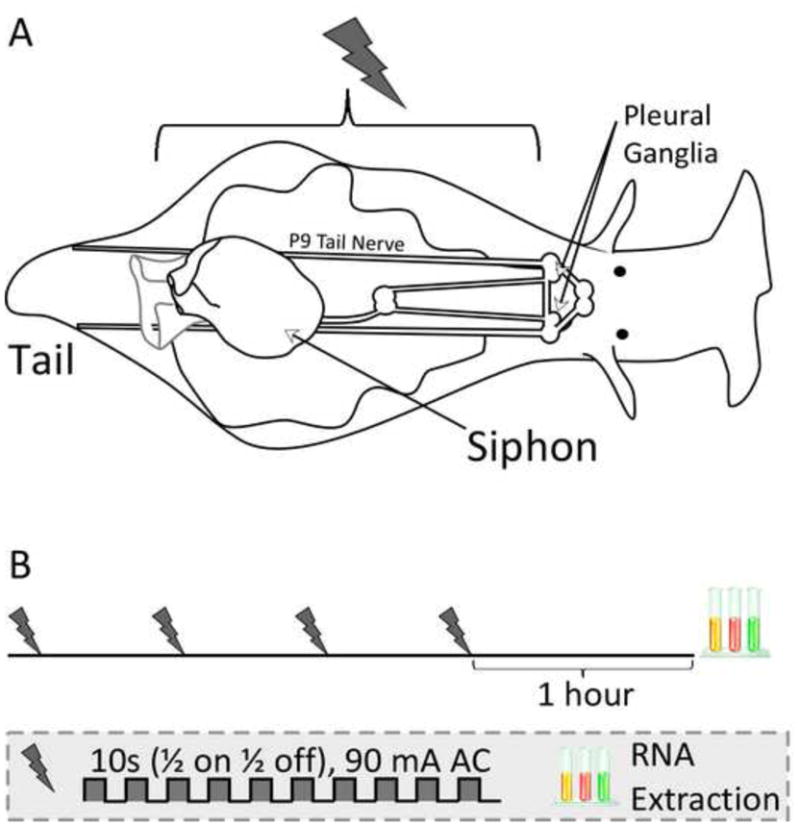
Long-term sensitization of the tail-elicited siphon withdrawal response. A) Noxious shocks to one side of the body alter the duration of the tail-elicited siphon withdrawal reflex (T-SWR) in Aplysia. The T-SWR is a defensive withdrawal of the siphon that is triggered by weak electrical stimulation to one side of the tail. The reflex is mediated by a simple 3-layer network. Stimulation of the tail is relayed in part by the VC nociceptors with axons in the P9 tail nerves and cell bodies in the pleural ganglia. The VCs excite a population of interneurons in the pedal ganglia with axons projecting to the abdominal ganglion. These interneurons activate motor neurons in the abdominal ganglia with axons in the siphon nerve, which cause contraction of the siphon. B) Experimental protocol. Long-term sensitization training consisted of 4 rounds of unilateral noxious shock every 30 minutes. This protocol reliably induces an increase in T-SWR duration lasting at least 24 hours on the side of training. One hour after training, pleural ganglia from the trained and untrained side were harvested separately for transcriptional analysis.
Training consisted of 4 rounds of noxious shock applied at 30 minute intervals to one side of the body with a hand-held electrode. Each round of shock consisted of 10 pulses of 500ms duration at a rate of 1hz and an amplitude of 90mA. During the course of each shock, the stimulating electrode was slowly moved from anterior (just behind neck) to posterior (just in front of tail) and back to cover nearly the entire surface of that side of the body. Side of training was counterbalanced within each experiment (left- and right-side training equalized for each experiment). One of the advantages of this training protocol is that both the induction and expression of sensitization is restricted to one side of the body (Antzoulatos & Byrne, 2007; Antzoulatos, Wainwright, Cleary, & Byrne, 2006; Cleary et al., 1998; Scholz & Byrne, 1987; Wainwright et al., 2002), enabling each animal to serve as its own control (trained vs. untrained sides).
2.3 Sample Size Determination
We compared gene expression from pleural ganglia on the trained vs. untrained side of the animal. For the microarray analysis, samples from two animals trained on opposite sides were pooled. This was done to decrease variability due to differential expression across sides. Pooling however was not used for the generalization experiment to ensure patterns of regulation were evident at the level of individual animals.
We collected 8 pairs of animals for microarray analysis. This enabled the entire experiment to be completed with a single microarray slide. This sample size is underpowered for subtly regulated transcripts (see discussion) but does meet the consensus recommendation of at least 5 biological replicates per group (Allison, Cui, Page, & Sabripour, 2006; Pavlidis, Li, & Noble, 2003; Tsai, Hsueh, & Chen, 2003).
We collected samples from an additional 12 animals to test for generalization to an independent sample with qPCR. However, RNA yields were very low in at least one sample from each of 3 animals, severely limiting the number of transcripts which could be measured in these animals. Thus, most results from this set are limited to the 9 high-yield animals. This sample size is sufficiently powered (>= 0.80) only for very large effects (d > 1.4). However, this is similar to effect size we have previously observed in this paradigm for strongly regulated transcripts such as ApC/EBP, ApCREB1, and ApEgr (Bonnick et al., 2012; Cyriac et al., 2013). Moreover, we also conduct an integrated analysis of qPCR measures across both experiments, providing a sample size of at least 17 and adequate power for d >= 1.
2.4 Isolation and Processing of Pleural Ganglia RNA
Animals were harvested 1 hour after training, the time point at which protein levels of the immediate-early gene C/EBP are most strongly up-regulated after training (Lyons, Collado, Khabour, Green, & Eskin, 2006).
To analyze transcription, pleural ganglia RNA was isolated. Briefly, animals were anesthetized with an injection of isotonic MgCl2 (50% of body weight), and an incision was then made along the ventral midline to expose the CNS. As dissection can alter gene expression (Alberini et al., 1994), we extracted ganglia rapidly (< 5 minutes per animal) and transferred them immediately to Trizol (Invitrogen, Carlsbad CA) for homogenization.
Tissue was homogenized using the Bullet Blender (NextAdvance, Averill Park, NY) and RNA extracted using Direct-Zol Mini RNA Kit (Zymo, Irvine, CA). Quantity and quality of RNA was assessed using the NanoDrop 1000 (Thermo Scientific, Wilmington, DE).
2.5 Microarray Design
Kandel, Moroz and associates (Moroz et al., 2006) have previously designed two microarray platforms for Aplysia californica based on an EST library—the Aplysia Discovery Array (ADA, GEO:GPL3635) and the Aplysia Annotated Array (AAA, GEO:GPL3636). These arrays have been estimated to provide 50–60% coverage of neurally-expressed transcripts (Moroz et al., 2006). In 2011, the AAA array was updated (updated AAA; GEO: GPL13815/GPL17112) to include ~ 2000 additional probes identified from 454 sequencing for kinesin-transported transcripts (Puthanveettil et al., 2013). Since the time these arrays were designed, however, the published library of Aplysia ESTs has grown from ~56k to ~215k. Thus, we developed a new microarray design to leverage this larger pool of ESTs. Because funding to develop and validate this array came in part from a grant from the Tellabs Foundation, we have named this new platform the Aplysia Tellabs Array (ATA: GEO: GPL18666).
To minimize redundancy from ESTs representing the same mRNA, we used the UniGene clustering of Aplysia ESTs (build 9, July 2011, ftp://ftp.ncbi.nih.gov/repository/UniGene/Aplysia_californica/). This build analyzed all 216,566 Aplysia californica ESTs and mRNAs which had been deposited with NCBI by July of 2011 and identified 24,709 distinct clusters. Of these, 7 consisted of short/repetitive sequences for which no suitable probes could be designed. For the remainder, we used the longest-length EST from each cluster as the target for probe design (Unigene targets). At the time of designing the array (January, 2012), an additional 35 mRNAs had been added to NCBI since the UniGene build. Of these, 15 did not have a strong match to any of the clusters in UniGene, so these were also included as targets for probe design (new mRNA targets). Agilent’s eArray platform was used to design a first- and second-best probe for each target (each Unigene cluster and each new mRNA target) with the complete target set used as the “genome” for determining probe specificity.
Because the UniGene clustering had included the ESTs used for the ADA and AAA designs, we expected that all the probes from these microarrays would be well-represented in our new design. Indeed, we found that the vast majority had perfect matches to elements within our target set. However, we found that 504 probes from the updated ADA design and 928 probes from the updated AAA design had matches of less the 58 nucleotides out of 60. To ensure comprehensive coverage, we directly included these probes in our new array design.
We selected Agilent’s 8×60k microarray platform. Both first- and second-best probes were spotted for all 24,702 Unigene targets and all 15 new mRNA targets. The probes selected from the updated AAA and ADA designs were spotted once. The remaining features (~9000) were filled with replicates of a random subset of first-best probes from the Unigene and new mRNA targets. Thus, with the exception of the probes from the AAA and ADA design, all targets were measured by at least 2 probes and about 37% were measured by 3.
Overall, our new design incorporates all the available sources of Aplysia ESTs and mRNAs and should have even better CNS coverage than the combined ADA and AAA platforms. With clustering, however, we were able to distill down to a single-chip design.
We compiled annotations for our design from several sources. First, we collected the annotations for each UniGene cluster. Second, we included annotations to the EST or mRNA that serves as an exemplar for each cluster. Third, we blasted exemplars against the predicted gene models from the recently released third draft of the Aplysia Genome (GenBank: GCA_000002075.2) and included the automated annotations from these gene models wherever matches were very strong (>20% coverage an e < 1*10−10; 50% of targets had a strong match). For reporting results in text, we refer to each probe by the GenBank accession number for the EST or mRNA that served as the exemplar for designing the probe. These records are directly linked to their UniGene clusters. Supplemental Table 1 provides the accession number for each target included on the array.
2.6 Microarray Processing
Processing was completed by Mogene Inc. (St. Louis, MO). A two-color approach was used with each array hybridized to a trained (Cy5) or untrained (Cy3) sample. Sample integrity was determined by Bioanalyzer RNA 6000, Pico total RNA protocol. 300ng of total RNA was amplified and labeled with Cy3 or Cy5 using the Agilent Quick Amp Two-Color Labeling Kit. Dye incorporation and yield was determined by Nanodrop. Samples were hybridized to the microarray slide at 65C and 10rpm for 17 hours. Slides were scanned on an Agilent C scanner at 3um resolution. Data was extracted using Agilent Feature Extraction software, v. 11.5. All labeling and post-labeling processing was carried out in an ozone regulated environment, monitored at < 5ppb.
2.7 Microarray Data Analysis
Microarray data was analyzed using limma (Smyth, 2005) from the Bioconductor suite of tools (Gentleman et al., 2004) for R (Ihaka & Gentleman, 1996). Our processing script is posted on the Open Science Framework. Median expression values were analyzed (Zahurak et al., 2007). These were corrected for background using the normexp+offset algorithm recommended for Agilent microarrays by Ritchie et al. (2007). An offset of 30 was selected based on inspection of MA Plots (supplemental Figure 1) and based on maximizing correlations with qPCR measurements from the same material (a set of measurements for 10 transcripts).
Expression was normalized within each array using the loess function (Smyth & Speed, 2003). Where multiple probes were used to measure the same EST or mRNA, these were then averaged. Finally, trained and untrained expression were compared using an empirical Bayes-moderated t-test (Smyth, 2004). Statistical significance was calculated using Benjamini-Hochberg correction for multiple comparisons to maintain a 5% overall false-discovery rate (Benjamini & Hochberg, 1995).
Rather than test for significant regulation and then use a fold-change threshold to evaluate practical significance, we used the treat function from limma (McCarthy & Smyth, 2009). This function allows testing of statistical significance against different null hypotheses. We tested against the standard null hypothesis of no regulation (Ho: Mean Fold Change <> 1; note that a ratio of 1 is produced when both trained and untrained samples have the same levels of gene expression). In addition, we tested against mean fold changes of 1.1, 1.25, and 1.5-fold change in either direction, each representing progressively more stringent criteria for regulation.
2.8 Reverse-Transcription Quantitative PCR (qPCR)
RNA was reverse transcribed using oligo(dT) primes and RevertAid First Strand cDNA Synthesis Kit (Fermentas, Glen Burnie MD).
Quantitative PCR was conducted using Sybr Green and the MyIQ real time PCR system (Bio-Rad, Los Angeles CA). Primers were validated for correct PCR efficiency and are listed in Supplemental Table 2. qPCR samples were analyzed in duplicate and the relative amounts of each transcript were determined using the ddCT method and the Bio-rad IQ5 gene expression analysis (Bio-Rad, Los Angeles CA). All qPCR expression levels were normalized to levels of histone H4, a transcript which is stable during long-term sensitization training (Bonnick et al., 2012).
To determine the effects of sensitization training on gene expression, a fold-change score was calculated for each animal as the ratio of trained to untrained expression. For all analyses, fold change scores were log transformed (base 2). This ensures equal weight to both up- and down-regulated measures and maintains consistency with microarray analysis. Changes from control were tested using a one-sample t test against an expected value of 0 for the null hypothesis (0 represents no change for log-transformed fold-change scores). Adjustments for multiple comparisons were not made for qPCR analyses.
For ease of interpretation, fold-change scores are plotted in raw format on a log scale and are reported in text in raw format as mean fold-change (MFC) with 95% confidence intervals in brackets. Cohen’s d is reported as an estimate of effect size, calculated so that positive values indicate increased expression on the trained side.
3. Results
3.1 Positive transcriptional controls confirm the efficacy of LTS training
To characterize the rapid transcriptional response to long-term sensitization training, we conducted a microarray analysis on 8 pairs of animals from pleural ganglia harvested 1 hour after long-term sensitization training (Figure 1). These ganglia were selected because they contain the cell bodies of the VC nociceptors (Illich & Walters, 1997; Walters, Byrne, Carew, & Kandel, 1983) which are thought to serve as an important site for the neural plasticity underlying behavioral sensitization (Cleary et al., 1998). In addition, the pleural ganglia contain a variety of other cell types. This includes a set of interneurons which inhibit the VCs (Buonomano, Cleary, & Byrne, 1992; Mackey et al., 1987), motor neurons controlling the opaline-gland (Tritt & Byrne, 1980) and mucus secretion (Rayport, Ambron, & Babiarz, 1983), and a number of additional cell clusters and types which have not been fully characterized (Fredman & Jahan-Parwar, 1979).
Although animals were sacrificed before long-term sensitization could be observed, we have previously shown (Bonnick et al., 2012) that this training protocol consistently results in unilateral long-term sensitization training, with 13 out of 13 animals showing an increase in T-SWR duration from before training to 24 hours after training on the side of training.
To further ensure the efficacy of the treatment, we used qPCR to measure the expression of three transcripts known to be rapidly up-regulated by long-term sensitization training: ApC/EBP, ApEgr, and ApCREB1 (Figure 2, circles). We found significant up-regulation of ApC/EBP in the trained samples from 8 out of 8 pairs tested (Mean Fold Change (MFC) = 4.5, 95% CI [2.5, 8.2], d = 2.1, t(7) = 5.9, p = 0.0006). The same pattern was observed for ApEgr (MFC = 2.0 [1.4, 3.0], d = 1.6, t(7) = 4.4, p = 0.003). For ApCREB1, 7 of 8 samples were responsive. Due to low power, however, this yielded a non-significant result (MFC = 1.15 [0.7, 1.8], d = 0.27, t(7) = 0.75, p = 0.48). This one non-response is likely due to normal variability in ApCREB1 expression as we observed strong regulation in a second sample (see below).
Figure 2.
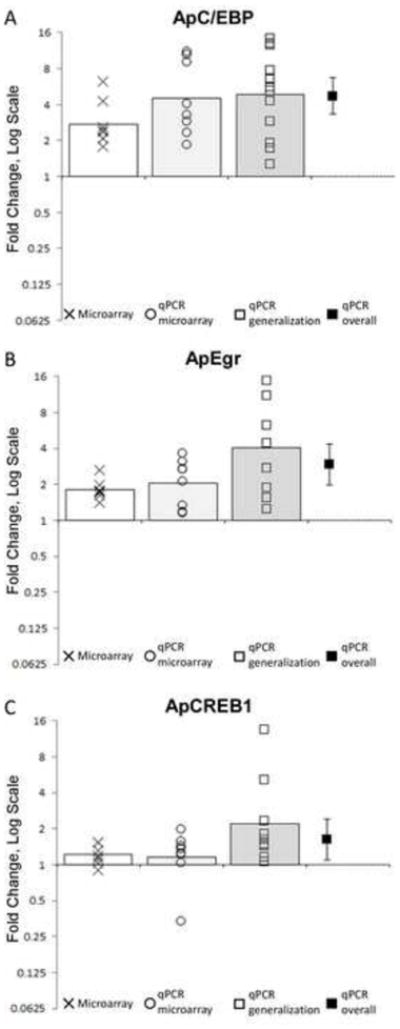
Training-induced change in gene expression in positive controls: A) ApC/EBP, B) ApEgr, and C) ApCREB1. Each panel shows data from the microarray analysis (X symbols, n = 8 pairs of animals), qPCR on the same samples (circles, n = 8 pairs of animals) and qPCR from a second, independent sample (squares, n = 9–12 animals). Bars represent means for each analysis. The black square shows the overall average across all qPCR samples, with the error bar representing the 95% confidence interval. Where the confidence interval does not include 0 the transcript is significantly regulated at p < 0.05. Every comparison for every transcript is statistically significant at p < 0.05 except for CREB-1 in both analyses of the original sample.
Despite the failure of ApCREB1 levels to reach statistical significance, we conclude that the samples collected show transcriptional changes indicative of successful LTS training.
3.2 Microarray analysis indicates 81 strongly regulated transcripts
Microarray analysis indicated statistically significant regulation for 1,494 transcripts, even with adjustment for multiple comparisons (Table 1). However, this standard (H0: MFC <> 1) selects transcripts which are consistently regulated, but without regard to the magnitude of regulation. Testing for regulation that can be statistically distinguished from at least a 1.1 fold change revealed only 81 transcripts—we consider these transcripts to be strongly regulated, both because the statistical evidence for regulation is more convincing and because their average fold change is likely to be meaningful (MFC in this group = 1.5). Adopting a more stringent null hypothesis further reduced this list, with only 10 transcripts showing regulation significantly greater than 1.25 in either direction. A clear bias towards up-regulation is evident. This may be because the time point examined (1 hour after training) is too short for down-regulation of mRNA production to substantially affect the pool of available transcript. A complete list of all transcripts and the highest-fold change exceeded with confidence is given in Supplementary Table 1.
Table 1.
Summary of microarray results. This table tallies the number of transcripts that show mean fold changes (MFC) that can be statistically differentiated from 1, 1.1, and 1.25 in either direction (a MFC of 1 represents no regulation as this is the ratio formed when trained and control tissues have the same level of expression for a transcript). Statistical significance was assessed with an empirical Bayes-moderated t-test with Benjamini-Hochberg adjustment for multiple comparisons to maintain an overall 5% falsediscovery rate.
| Criterion | Total | Up | Down |
|---|---|---|---|
| Not significant | 24597 | ||
| MFC > 1 | 1494 | 974 | 520 |
| MFC > 1.1 | 81 | 81 | 0 |
| MFC > 1.25 | 10 | 10 | 0 |
| Total | 26091 |
As predicted based on our positive controls, probes for ApEgr and ApC/EBP were flagged as regulated by at least 1.25 fold (Figure 2, marked by X symbols). As expected based on the qPCR data, probes for ApCREB1 did not emerge as strongly regulated in this sample (though see below).
In contrast to Liu et al. (1997), we did not find that training increases the expression of ApTBL-1, a tolloid/BMP-1-like protein (MFC = 0.97). In addition, we did not replicate previous findings (Zwartjes et al., 1998) of up-regulation of a reductase-related protein (MFC = 1.02) nor ApCalmodulin (MFC = 1.08). This is not surprising, however, as we analyzed a different timepoint (immediate in prior work versus 1 hour after training in this study) and a different tissue (sensory neurons only in prior work versus entire pleural ganglia in this study).
3.3 qPCR confirms good convergent validity of microarray results
Does the microarray platform we designed provide valid measures of regulation? To find out, we used qPCR to measure the expression of 27 different transcripts in the exact same samples as used in the microarray analysis. We selected 6 transcripts predicted to be strongly regulated (> 1.1 fold change in either direction), 8 predicted to be at least somewhat regulated (> 1 fold change), and 13 predicted to be not at all regulated (we include in these totals the positive controls ApEgr, ApCREB1, and ApC/EBP already described).
We compared the mean fold change scores obtained from microarray with those obtained via qPCR (Figure 3). We observed fairly good convergent validity for an EST-based array (Figure 2), with a strong linear correlation between log fold change measures from qPCR and microarray (r = 0.77, 95% CI [0.56, 0.89], transcripts = 27, p = 0.000004). Note that the slope of the regression line between these measures is less than 1 (m = 0.61), which indicates some regression to the mean.
Figure 3.
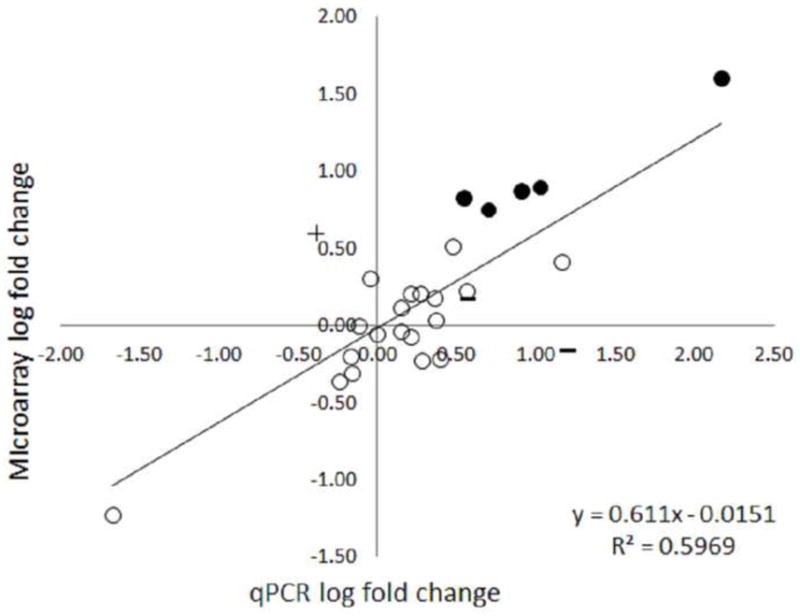
Convergent validity of the new developed Aplysia Tellabs Array platform. Fold-change scores for 27 transcripts were compared when measured by microarray or qPCR. Each measurement is the average log-fold change (base 2) across 8 pairs of animals. Symbols indicate the degree of convergence in the assessment of statistical significance. Circles represent matches in assessment of statistical significance between microarray (> 1.1 fold change in either direction) and qPCR with both techniques indicating either non-significant (open circles) or significant (filled circles). False positives are marked with an + (significant in microarray but not in qPCR) and false negatives are marked with a – (significant in qPCR but not microarray).
We also assessed how well microarray and qPCR converged on the assessment of statistically significant regulation. Of 6 strongly regulated transcripts identified from microarray (> 1.1 fold), 5 showed statistically significant regulation in qPCR, a true-positive rate of 83%. Of the 7 predicted to be only somewhat regulated by microarray, 2 reached statistical significance, and the rest failed to validate (though 1 reached p < 0.1). Thus, as expected, adopting a lower standard for regulation produced a much lower true-positive rate (29%). It should be kept in mind, though, that this study is adequately powered only for large effects, so some transcripts which did not reach statistical significance may actually be regulated (e.g. ApCREB1; see below). Finally, we tested 13 transcripts which did not show statistically significant regulation with the microarray. Of these, 11 confirmed as not-regulated, but 2 showed statistically significant up-regulation (data not shown). This gives a true-negative rate of 82%, but note than neither of these transcripts generalized to an independent sample (see below).
3.4 The majority of confirmed microarray results generalize to an independent sample
To confirm that results from this data set would generalize to an independent sample, we trained an additional 12 animals and harvested their pleural ganglia 1 hour after training. We used qPCR to examine convergent validity in this sample.
We again confirmed the efficacy of training by measuring transcripts known to be regulated (Figure 2, Squares). As expected, ApC/EBP was up-regulated in 12 out of 12 animals tested (MFC = 4.9 [2.9, 8.2], d = 1.9, t(11) = 6.7, p = 0.00003). However, 3 animals yielded insufficient RNA for further qPCR analysis, so only the remaining 9 were used to measure predictive validity. Our other positive controls showed significant regulation in this set. ApEgr was up-regulated on the trained side of 9 out of 9 animals (MFC = 4.0 [2.0, 7.9], d = 1.6, t(8) = 4.75, p = 0.001). ApCREB1 was also up-regulated in all samples (MFC = 2.19 [1.17, 4.13], d = 0.96, t(8) = 2.87, p = 0.02). Note that when integrating results from both sets of samples there is a clear overall regulation of ApCREB1, with 16 of 17 samples showing higher expression on the trained side (MFC = 1.6 [1.1, 2.4], d = 0.63, t(16) = 2.6, p =0.02).
We tested 5 additional transcripts which had shown significant regulation by both microarray and qPCR in the original sample (Figure 4). We found that 4 of these generalized to the new sample. EB339038, which is predicted to encode a vacuolar-protein-sorting-associated-like protein, was strongly up-regulated in 9 of 9 samples (MFC = 3.1 [1.4, 6.8], d = 1.1, t(5) = 3.35, p = 0.01). EB289383, predicted to encode a glycine transporter2-like protein was also up-regulated in 9 of 9 samples tested (MFC = 2.5 [1.5, 4.1], d = 1.35, t(8) = 4.1, p = 0.003). Similarly, GD219501.1, which maps to an uncharacterized protein, was up-regulated in 7 of 9 samples tested (MFC = 2.0 [1.2, 3.2], d = 1.1, t(8) = 3.29, p = 0.01). Finally, EB233406, which is predicted to encode a C/EBPγ-like transcription factor, was up-regulated in 6 of 8 samples tested (MFC = 2.8 [1.2, 6.6], d = 0.98, t(7) = 2.76, p = 0.03). The other transcript, 7.UF_CU.8090.C2, did not show statistically significant regulation in this second sample. We also tested the two false-negative transcripts (not significant in microarray but significant in qPCR) but these did not generalize (data not shown).
Figure 4.
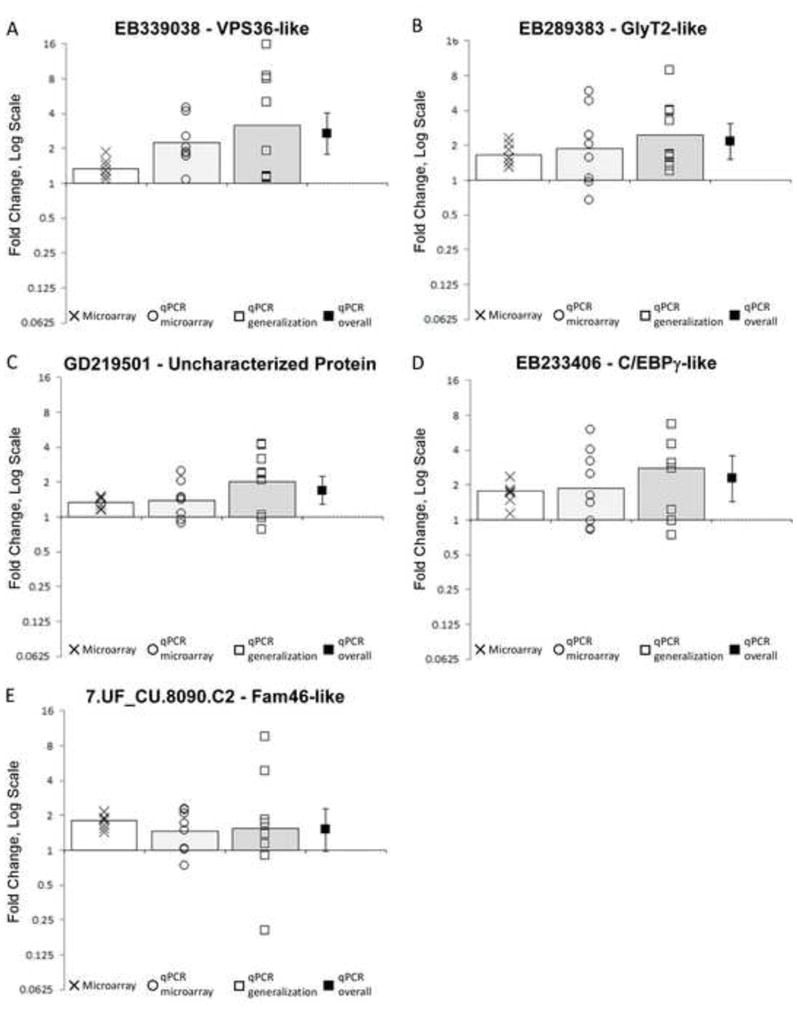
Training-induced changes in gene expression in novel transcripts. These are results from the transcripts which were significantly regulated according to both microarray and qPCR in the original sample. Each panel shows data from the microarray analysis (Xs, n = 8 pairs of animals), qPCR on the same samples (circles, n = 8 pairs of animals) and qPCR from a second, independent sample (squares, n = 9–12 animals). Bars represent means for each analysis. The black square shows the overall average across all qPCR samples, with the error bar representing the 95% confidence interval. Where the confidence interval does not include 0 the transcript is significantly regulated at p < 0.05. Every comparison for every transcript is statistically significant at p < 0.05 except 7.UF_CU.8090.C2 in panel E which does not reach statistical significance in the second sample nor in the overall qPCR analysis.
Overall, our microarray results show strong predictive validity. Of the 7 transcripts which were significantly regulated in the original sample by both microarray and qPCR, 6 were confirmed in an independent sample, a true positive rate of 86% (this total includes ApEgr and ApC/EBP as well as the 5 novel transcripts tested). Moreover, these results nearly double the pool of known transcripts exhibiting a rapid change in expression after long-term sensitization training.
4. Discussion
We have used microarray and qPCR to characterize the rapid transcriptional response to long-term sensitization training in the pleural ganglia, a component of the Aplysia CNS which contains the VC nociceptors thought to play a critical role in the expression of sensitization memory.
We developed a new microarray design for this project to provide a more compact but comprehensive platform for characterizing changes in gene expression in Aplysia. For an EST-based array, we found strong convergent (83%) and predictive validity (86%) for this new design. By comparison, the EST-based array for Aplysia kurodai has shown only 38% (5 of 13) convergence with qPCR data from the same samples (Lee et al., 2008). This difference could be due to disparate design strategies. In addition, measures of convergent validity are likely to depend in part on the achieving reliable measurement through adequate sample size. We used 8 biological replicates rather than the 2–3 which have typically been used for microarray analysis in Aplysia (e.g. Kadakkuzha et al., 2013, analysis of age-related changes in isolated R15 neurons; Lee et al., 2008, analysis of serotonin-related changes in pleural ganglia). A strong consensus has now emerged that microarray analysis requires at least 5 biological replicates per group to produce reliable results (Allison et al., 2006; Pavlidis et al., 2003; Tsai et al., 2003).
To our knowledge, this is the first comprehensive examination of transcriptional changes immediately following long-term sensitization. Our findings suggest that the pool of regulated transcripts is much larger than the set of 6 previously identified transcripts. Specifically, our microarray analysis identifies 81 transcripts showing fairly strong regulation after training (statistically greater than a 1.1 fold change). If our convergent (83% true-positive rate) and predictive validity (86% true-positive rate) results are representative, the expected number of strongly and reliably regulated transcripts is predicted to be ~58. This number may be larger, however, given that the microarray is based on an EST library that may not have full coverage of all CNS-expressed transcripts. In addition, even with a sample size of 8 animals, only very strongly regulated transcripts could be reliably detected; many weakly regulated transcripts could have been missed. Thus, there may be considerable complexity in the transcriptional response to long-term sensitization memory. The reasons for this complexity are not clear—it could allow different responses in different cell types, provide parallel signaling pathways to implement different phases of memory, and/or provide redundancy to ensure proper encoding. It is notable that a similar number of regulated transcripts (27) has been reported immediately following serotonin exposure in Aplysia kurodai, a treatment which mimics some aspects of long-term sensitization training (Lee et al., 2008, though note this report used a >2 fold change criterion).
We have specifically confirmed learning-related regulation in 4 novel ESTs, showing that they are consistently up-regulated not only in the original sample but also in a completely independent sample. Confirmation in an independent sample is an important follow-up for microarray analysis, as this is a primary predictor of generalizability to other contexts (Siontis, Patsopoulos, & Ioannidis, 2010).
One EST, EB233406 aligns to a gene model (GenBank: XM_005105406) predicted to encode a CCAAT/enhancer-binding protein gamma-like gene (C/EBPγ also known as Ig/EBP). In other species, C/EBPγ is a ubiquitously expressed member of the C/EBP transcription factor family (Lekstrom-Himes & Xanthopoulos, 1998). C/EBPγ is distinguished within this family by lacking a transcriptional activation domain, and is thus thought to function as a transdominant negative inhibitor of other C/EBPs (Cooper, Henderson, Artandi, Avitahl, & Calame, 1995). It is expressed in the mammalian CNS (Roman, Platero, Shuman, & Calame, 1990) but to date, no nervous system role has been described for C/EBPγ. Through its ability to heterodimerize with C/EBP isoforms, an Aplysia C/EBPγ could act as a modulator of ApC/EBP (Alberini et al., 1994), which is known to play an essential role in the long-term synaptic facilitation thought to underlie the expression of LTS memory. Further exploration of this hypothesis is planned.
A second EST, EB289383 aligns to a gene model (GenBank: XM_005092349) predicted to encode a sodium- and chloride-dependent glycine transporter 2 -like protein (GlyT2). Glycine transporters 1 and 2 belong to the large family of Na+/Cl−-dependent transporters and mediate glycine uptake into glia and neurons, respectively (Kristensen et al., 2011). Interestingly, it has been reported that inhibition of GlyT2 in mice helps ameliorate increased pain sensitivity after nerve damage (Morita et al., 2008). This is consistent with the large body of work showing overlap in the mechanisms of long-term sensitization and hyperalgesia (e.g. Walters, 1987; Weragoda & Walters, 2007). We are now testing if pharmacological inhibition of GlyT2 can block the induction or maintenance of long-term sensitization memory.
A third EST, EB339038 aligns to a gene model (GenBank: XM_005112280) predicted to encode a vacuolar protein-sorting-associated 36-like protein (VPS36). In other species, VPS36 is a subunit of the endosomal sorting complexes required for transport-II (ESCRT-II) (Saksena & Emr, 2009). ESCRT complexes are known to regulate protein trafficking from endosomes to lysosomes and it is well established that aberrant endosomal trafficking in neurons can be linked to a number of neurodegenerative diseases (Lee & Gao, 2012).
Finally, the EST GD219501 aligns to a gene model (GenBank: XM_005113453) of an uncharacterized protein. We are currently exploring the function of this protein.
We have yet to further investigate the majority of the other transcripts identified as regulated in our microarray screen. In the set of 81 regulated significantly greater than 1.1-fold, 2 were already known and we have conducted qPCR validation on an additional 6, leaving 73 without further exploration at this point. Of these, 50 are annotated only as “transcribed locus” or “uncharacterized locus” (see Supplemental Table 1). Additional work will be required not only to validate these transcripts but to determine their function. Of the 23 with annotations, some are particularly notable. For example several transcripts encode proteins that may be related to transcription. These include EB344520 (Predicted: INO80 complex subunit E-like, important for chromatin remolding), EB325811 (Predicted: elongator complex protein 4-like), and GD232035 (Predicted: DNA-directed RNA polymerases I and III subunit RPAC2-like). Another group of transcripts encodes proteins that may be related to metabolism and growth. This includes EB248521 (Predicted: insulin-like growth factor-binding protein), EB265745 (Predicted: 3-hydroxy-3-methylglutaryl-coenzyme A reductase-like), GD224855 (Predicted: bifunctional purine biosynthesis protein PURH-like), and EB349048 (Predicted: Tob1-like). Finally, one transcript (FF065849, predicted: ubiquitin thioesterase) may have a role in protein degradation and turn-over. This is consistent with previous research in Aplysia (Hegde et al., 1997; Mohamed, Yao, Fioravante, Smolen, & Byrne, 2005) and other animals (Jarome & Helmstetter, 2013) which has implicated the ubiquitin-proteosome system in learning and memory.
One limitation of this work is that it was conducted with tissue harvested from the entire pleural ganglia. The VC nociceptors are a prominent landmark in these ganglia, but many other cell bodies are also located there, some of which may not contribute to the expression of long-term sensitization (e.g. the opaline-gland motor neurons described by Tritt & Byrne, 1980). The inclusion of unrelated material decreases signal to noise and thus the sensitivity of transcriptional analysis. For that reason, we have recently begun to focus on the VC cluster and plan to perform experiments with this more specific cell population.
Supplementary Material
Highlights.
We developed a new Aplysia microarray with high convergent and predictive validity.
Long-term sensitization training rapidly regulates 81 transcripts in pleural ganglia.
Regulated transcripts include putative homologs of C/EBPgamma, GLYT2, and VPS36.
Acknowledgments
This work was supported by a grant from the Tellabs foundation and by NIH Grant R15MH090998-01.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errorsmaybe discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
6. Works Cited
- Alberini CM, Ghirardi M, Metz R, Kandel ER. C/EBP is an immediate-early gene required for the consolidation of long-term facilitation in Aplysia. Cell. 1994;76(6):1099–114. doi: 10.1016/0092-8674(94)90386-7. [DOI] [PubMed] [Google Scholar]
- Allison DB, Cui X, Page GP, Sabripour M. Microarray data analysis: from disarray to consolidation and consensus. Nature Reviews Genetics. 2006;7(1):55–65. doi: 10.1038/nrg1749. [DOI] [PubMed] [Google Scholar]
- Antzoulatos EG, Byrne JH. Long-term sensitization training produces spike narrowing in Aplysia sensory neurons. The Journal of Neuroscience. 2007;27(3):676–83. doi: 10.1523/JNEUROSCI.4025-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antzoulatos EG, Wainwright ML, Cleary LJ, Byrne JH. Long-term sensitization training primes Aplysia for further learning. Learning & Memory. 2006;13(4):422–5. doi: 10.1101/lm.230306. [DOI] [PubMed] [Google Scholar]
- Bailey CH, Bartsch D, Kandel ER. Toward a molecular definition of long-term memory storage. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(24):13445–52. doi: 10.1073/pnas.93.24.13445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartsch D, Casadio A, Karl KA, Serodio P, Kandel ER. CREB1 encodes a nuclear activator, a repressor, and a cytoplasmic modulator that form a regulatory unit critical for long-term facilitation. Cell. 1998;95(2):211–23. doi: 10.1016/s0092-8674(00)81752-3. [DOI] [PubMed] [Google Scholar]
- Barzilai a, Kennedy TE, Sweatt JD, Kandel ER. 5-HT modulates protein synthesis and the expression of specific proteins during long-term facilitation in Aplysia sensory neurons. Neuron. 1989;2(6):1577–86. doi: 10.1016/0896-6273(89)90046-9. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society. Series B. 1995;57(1):289–300. doi: 10.2307/2346101. [DOI] [Google Scholar]
- Bonnick K, Bayas K, Belchenko D, Cyriac A, Dove M, Lass J, Calin-Jageman RJ. Transcriptional changes following long-term sensitization training and in vivo serotonin exposure in Aplysia californica. PloS One. 2012;7(10):e47378. doi: 10.1371/journal.pone.0047378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buonomano DV, Cleary LJ, Byrne JH. Inhibitory neuron produces heterosynaptic inhibition of the sensory-to-motor neuron synapse in Aplysia. Brain Research. 1992;577(1):147–50. doi: 10.1016/0006-8993(92)90548-n. [DOI] [PubMed] [Google Scholar]
- Castellucci VF, Blumenfeld H, Goelet P, Kandel ER. Inhibitor of protein synthesis blocks long-term behavioral sensitization in the isolated gill-withdrawal reflex of Aplysia. Journal of Neurobiology. 1989;20(1):1–9. doi: 10.1002/neu.480200102. [DOI] [PubMed] [Google Scholar]
- Castellucci VF, Kennedy TE, Kandel ER, Goelet P. A quantitative analysis of 2-D gels identifies proteins in which labeling is increased following long-term sensitization in Aplysia. Neuron. 1988;1(4):321–328. doi: 10.1016/0896-6273(88)90080-3. [DOI] [PubMed] [Google Scholar]
- Chew SJ, Mello C, Nottebohm F, Jarvis E, Vicario DS. Decrements in auditory responses to a repeated conspecific song are long-lasting and require two periods of protein synthesis in the songbird forebrain. Proceedings of the National Academy of Sciences of the United States of America. 1995;92(8):3406–10. doi: 10.1073/pnas.92.8.3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleary LJ, Lee WL, Byrne JH. Cellular correlates of long-term sensitization in Aplysia. The Journal of Neuroscience. 1998;18(15):5988–98. doi: 10.1523/JNEUROSCI.18-15-05988.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper C, Henderson A, Artandi S, Avitahl N, Calame K. Ig/EBP (C/EBPγ) is a transdominant negative inhibitor of C/EBP family transcriptional activators. Nucleic Acids Research. 1995;23(21):4371–4377. doi: 10.1093/nar/23.21.4371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyriac A, Holmes G, Lass J, Belchenko D, Calin-Jageman RJ, Calin-Jageman IE. An Aplysia Egr homolog is rapidly and persistently regulated by long-term sensitization training. Neurobiology of Learning and Memory. 2013;102:43–51. doi: 10.1016/j.nlm.2013.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esdin J, Pearce K, Glanzman DL. Long-term habituation of the gill-withdrawal reflex in aplysia requires gene transcription, calcineurin and L-type voltage-gated calcium channels. Frontiers in Behavioral Neuroscience. 2010;4(November):181. doi: 10.3389/fnbeh.2010.00181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredman SM, Jahan-Parwar B. Intra- and interganglionic synaptic connections in the CNS of Aplysia. Brain Research Bulletin. 1979;4(3):393–406. doi: 10.1016/s0361-9230(79)80017-9. [DOI] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Zhang J. Bioconductor: open software development for computational biology and bioinformatics. Genome Biology. 2004;5(10):R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glanzman DL, Mackey SL, Hawkins RD, Dyke AM, Lloyd PE, Kandel ER. Depletion of serotonin in the nervous system of Aplysia reduces the behavioral enhancement of gill withdrawal as well as the heterosynaptic facilitation produced by tail shock. The Journal of Neuroscience. 1989;9(12):4200–13. doi: 10.1523/JNEUROSCI.09-12-04200.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegde AN, Inokuchi K, Pei W, Casadio A, Ghirardi M, Chain DG, Schwartz JH. Ubiquitin C-Terminal Hydrolase Is an Immediate-Early Gene Essential for Long-Term Facilitation in Aplysia. Cell. 1997;89(1):115–126. doi: 10.1016/S0092-8674(00)80188-9. [DOI] [PubMed] [Google Scholar]
- Hermitte G, Pedreira ME, Tomsic D, Maldonado H. Context shift and protein synthesis inhibition disrupt long-term habituation after spaced, but not massed, training in the crab Chasmagnathus. Neurobiology of Learning and Memory. 1999;71(1):34–49. doi: 10.1006/nlme.1998.3858. [DOI] [PubMed] [Google Scholar]
- Ihaka R, Gentleman R. R: A Language for Data Analysis and Graphics. Journal of Computational and Graphical Statistics. 1996;5(3):299–314. doi: 10.1080/10618600.1996.10474713. [DOI] [Google Scholar]
- Illich P, Walters E. Mechanosensory neurons innervating Aplysia siphon encode noxious stimuli and display nociceptive sensitization. The Journal of Neuroscience. 1997;17(1):459–469. doi: 10.1523/JNEUROSCI.17-01-00459.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarome TJ, Helmstetter FJ. The ubiquitin-proteasome system as a critical regulator of synaptic plasticity and long-term memory formation. Neurobiology of Learning and Memory. 2013 doi: 10.1016/j.nlm.2013.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadakkuzha BM, Akhmedov K, Capo TR, Carvalloza AC, Fallahi M, Puthanveettil SV. Age-associated bidirectional modulation of gene expression in single identified R15 neuron of Aplysia. BMC Genomics. 2013;14(1):880. doi: 10.1186/1471-2164-14-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristensen AS, Andersen J, Jørgensen TN, Sørensen L, Eriksen J, Loland CJ, Gether U. SLC6 neurotransmitter transporters: structure, function, and regulation. Pharmacological Reviews. 2011;63(3):585–640. doi: 10.1124/pr.108.000869. [DOI] [PubMed] [Google Scholar]
- Lee, Choi S-L, Kim T-H, Lee J-A, Kim HK, Kim H, Kaang B-K. Transcriptome analysis and identification of regulators for long-term plasticity in Aplysia kurodai. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(47):18602–7. doi: 10.1073/pnas.0808893105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JA, Gao FB. Neuronal Functions of ESCRTs. Experimental Neurobiology. 2012;21(1):9–15. doi: 10.5607/en.2012.21.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lekstrom-Himes J, Xanthopoulos KG. Biological Role of the CCAAT/Enhancer-binding Protein Family of Transcription Factors. Journal of Biological Chemistry. 1998;273(44):28545–28548. doi: 10.1074/jbc.273.44.28545. [DOI] [PubMed] [Google Scholar]
- Levenson J, Byrne JH, Eskin A. Levels of serotonin in the hemolymph of Aplysia are modulated by light/dark cycles and sensitization training. The Journal of Neuroscience. 1999;19(18):8094–103. doi: 10.1523/JNEUROSCI.19-18-08094.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu QR, Hattar S, Endo S, MacPhee K, Zhang H, Cleary LJ, Eskin A. A developmental gene (Tolloid/BMP-1) is regulated in Aplysia neurons by treatments that induce long-term sensitization. The Journal of Neuroscience. 1997;17(2):755–64. doi: 10.1523/JNEUROSCI.17-02-00755.1997. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/8987797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons LC, Collado MS, Khabour O, Green CL, Eskin A. The circadian clock modulates core steps in long-term memory formation in Aplysia. The Journal of Neuroscience. 2006;26(34):8662– 71. doi: 10.1523/JNEUROSCI.2307-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackey SL, Glanzman DL, Small Sa, Dyke aM, Kandel ER, Hawkins RD. Tail shock produces inhibition as well as sensitization of the siphon-withdrawal reflex of Aplysia: possible behavioral role for presynaptic inhibition mediated by the peptide Phe-Met-Arg-Phe-NH2. Proceedings of the National Academy of Sciences of the United States of America. 1987;84(23):8730–4. doi: 10.1073/pnas.84.23.8730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy DJ, Smyth GK. Testing significance relative to a fold-change threshold is a TREAT. Bioinformatics (Oxford, England) 2009;25(6):765–71. doi: 10.1093/bioinformatics/btp053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamed Ha, Yao W, Fioravante D, Smolen PD, Byrne JH. cAMP-response elements in Aplysia creb1, creb2, and Ap-uch promoters: implications for feedback loops modulating long term memory. The Journal of Biological Chemistry. 2005;280(29):27035–43. doi: 10.1074/jbc.M502541200. [DOI] [PubMed] [Google Scholar]
- Monje FJ, Birner-Gruenberger R, Darnhofer B, Divisch I, Pollak DD, Lubec G, Hüttinger B. Proteomics reveals selective regulation of proteins in response to memory-related serotonin stimulation in Aplysia californica ganglia. Proteomics. 2011;12(3):1–25. doi: 10.1002/pmic.201100418. [DOI] [PubMed] [Google Scholar]
- Morita K, Motoyama N, Kitayama T, Morioka N, Kifune K, Dohi T. Spinal antiallodynia action of glycine transporter inhibitors in neuropathic pain models in mice. The Journal of Pharmacology and Experimental Therapeutics. 2008;326(2):633–45. doi: 10.1124/jpet.108.136267. [DOI] [PubMed] [Google Scholar]
- Moroz LL, Edwards JR, Puthanveettil SV, Kohn AB, Ha T, Heyland A, Kandel ER. Neuronal transcriptome of aplysia: neuronal compartments and circuitry. Cell. 2006;127(7):1453–67. doi: 10.1016/j.cell.2006.09.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlidis P, Li Q, Noble WS. The effect of replication on gene expression microarray experiments. Bioinformatics. 2003;19(13):1620–1627. doi: 10.1093/bioinformatics/btg227. [DOI] [PubMed] [Google Scholar]
- Pinsker HM, Hening WA, Carew TJ, Kandel ER. Long-term sensitization of a defensive withdrawal reflex in Aplysia. Science. 1973;182(4116):1039–42. doi: 10.1126/science.182.4116.1039. [DOI] [PubMed] [Google Scholar]
- Puthanveettil SV, Antonov I, Kalachikov S, Rajasethupathy P, Choi YB, Kohn AB, Kandel ER. A strategy to capture and characterize the synaptic transcriptome. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(18):7464–9. doi: 10.1073/pnas.1304422110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayport SG, Ambron RT, Babiarz J. Identified cholinergic neurons R2 and LPl1 control mucus release in Aplysia. Journal of Neurophysiology. 1983;49(4):864–76. doi: 10.1152/jn.1983.49.4.864. [DOI] [PubMed] [Google Scholar]
- Ritchie ME, Silver J, Oshlack A, Holmes M, Diyagama D, Holloway A, Smyth GK. A comparison of background correction methods for two-colour microarrays. Bioinformatics (Oxford, England) 2007;23(20):2700–7. doi: 10.1093/bioinformatics/btm412. [DOI] [PubMed] [Google Scholar]
- Roman C, Platero JS, Shuman J, Calame K. Ig/EBP-1: a ubiquitously expressed immunoglobulin enhancer binding protein that is similar to C/EBP and heterodimerizes with C/EBP. Genes & Development. 1990;4(8):1404–1415. doi: 10.1101/gad.4.8.1404. [DOI] [PubMed] [Google Scholar]
- Saksena S, Emr SD. ESCRTs and human disease. Biochemical Society Transactions. 2009;37(Pt 1):167–72. doi: 10.1042/BST0370167. [DOI] [PubMed] [Google Scholar]
- Sangha S. Intermediate and long-term memories of associative learning are differentially affected by transcription versus translation blockers inLymnaea. Journal of Experimental Biology. 2003;206(10):1605–1613. doi: 10.1242/jeb.00301. [DOI] [PubMed] [Google Scholar]
- Scholz KP, Byrne JH. Long-term sensitization in Aplysia: biophysical correlates in tail sensory neurons. Science. 1987;235(4789):685–7. doi: 10.1126/science.2433766. [DOI] [PubMed] [Google Scholar]
- Simmons JP, Nelson LD, Simonsohn U. A 21 Word Solution. SSRN Electronic Journal. 2012:1–4. doi: 10.2139/ssrn.2160588. [DOI] [Google Scholar]
- Siontis KCM, Patsopoulos Na, Ioannidis JPa. Replication of past candidate loci for common diseases and phenotypes in 100 genome-wide association studies. European Journal of Human Genetics : EJHG. 2010;18(7):832–7. doi: 10.1038/ejhg.2010.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Statistical Applications in Genetics and Molecular Biology. 2004;3(1):Article 3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- Smyth GK. Limma: linear models for microarray data. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. 2005:397–420. doi: 10.1007/0-387-29362-0_23. [DOI] [Google Scholar]
- Smyth GK, Speed T. Normalization of cDNA microarray data. Methods. 2003;31(4):265–273. doi: 10.1016/S1046-2023(03)00155-5. [DOI] [PubMed] [Google Scholar]
- Tischmeyer W, Grimm R. Activation of immediate early genes and memory formation. Cellular and Molecular Life Sciences : CMLS. 1999;55(4):564–74. doi: 10.1007/s000180050315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tritt SH, Byrne JH. Motor controls of opaline secretion in Aplysia californica. Journal of Neurophysiology. 1980;43(3):581–94. doi: 10.1152/jn.1980.43.3.581. [DOI] [PubMed] [Google Scholar]
- Tsai CA, Hsueh H, Chen JJ. Estimation of false discovery rates in multiple testing: application to gene microarray data. Biometrics. 2003;59(4):1071–81. doi: 10.1111/j.0006-341x.2003.00123.x. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/14969487. [DOI] [PubMed] [Google Scholar]
- Wainwright ML, Zhang H, Byrne JH, Cleary LJ. Localized neuronal outgrowth induced by long-term sensitization training in aplysia. The Journal of Neuroscience. 2002;22(10):4132–41. doi: 10.1523/JNEUROSCI.22-10-04132.2002. 20026347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters ET. Site-specific sensitization of defensive reflexes in Aplysia: a simple model of longterm hyperalgesia. The Journal of Neuroscience : The Official Journal of the Society for Neuroscience. 1987;7(2):400–7. doi: 10.1523/JNEUROSCI.07-02-00400.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters ET, Byrne JH, Carew TJ, Kandel ER. Mechanoafferent neurons innervating tail of Aplysia. I. Response properties and synaptic connections. Journal of Neurophysiology. 1983;50(6):1522–42. doi: 10.1152/jn.1983.50.6.1522. [DOI] [PubMed] [Google Scholar]
- Watanabe H, Takaya T, Shimoi T, Ogawa H, Kitamura Y, Oka K. Influence of mRNA and protein synthesis inhibitors on the long-term memory acquisition of classically conditioned earthworms. Neurobiology of Learning and Memory. 2005;83(2):151–7. doi: 10.1016/j.nlm.2004.11.003. [DOI] [PubMed] [Google Scholar]
- Weragoda RMS, Walters ET. Serotonin induces memory-like, rapamycin-sensitive hyperexcitability in sensory axons of aplysia that contributes to injury responses. Journal of Neurophysiology. 2007;98(3):1231–9. doi: 10.1152/jn.01189.2006. [DOI] [PubMed] [Google Scholar]
- Zahurak M, Parmigiani G, Yu W, Scharpf RB, Berman D, Schaeffer E, Cope L. Pre-processing Agilent microarray data. BMC Bioinformatics. 2007;8:142. doi: 10.1186/1471-2105-8-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwartjes RE, West H, Hattar S, Ren X, Noel F, Nunez-Regueiro M, Eskin a. Identification of specific mRNAs affected by treatments producing long-term facilitation in Aplysia. Learning & Memory. 1998;4(6):478–495. doi: 10.1101/lm.4.6.478. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


