Summary
Bistable flagellar and virulence gene expression generates specialized Salmonella subpopulations with distinct functions. Repressing flagellar genes allows Salmonella to evade caspase-1 mediated host defenses and enhances systemic colonization. By definition, bistability arises when intermediate states of gene expression are rendered unstable by the underlying genetic circuitry. We demonstrate sustained bistable fliC expression in virulent Salmonella 14028 and document dynamic control of the distribution, or single-cell census, of flagellar gene expression by the mutually repressing repressors YdiV and FliZ. YdiV partitions cells into the fliC-OFF subpopulation, while FliZ partitions cells into the fliC-HIGH subpopulation at late timepoints during growth. Bistability of ΔfliZ populations and ydiV-independent FliZ control of flagellar gene expression provide evidence that the YdiV-FliZ mutually repressing repressor circuit is not required for bistability. Repression and activation by YdiV and FliZ (respectively) can shape the census of fliC expression independently, and bistability collapses into a predominantly intermediate population in the absence of both regulators. Metered expression of YdiV and FliZ reveals variable sensitivity to these regulators and defines conditions where expression of FliZ enhances fliC expression and where FliZ does not alter the fliC census. Thus, this evolved genetic circuitry coordinates multiple layers of regulatory heterogeneity into a binary response.
Keywords: bistability, Salmonella, hysteresis, heterogeneity, flagella, stochastic
Introduction
Transcriptional responses at a promoter can be characterized as graded or binary (Biggar & Crabtree, 2001, Temme et al., 2008). In a graded response, all levels of expression from fully off to fully on are stable and the response to an activation signal is continuous, while a binary or switch-like response implies only two levels of activity, on and off. A switch-like and heterogeneous response within a population of single cells generates a bistable distribution, or census, of gene expression (Veening et al., 2008). Many cellular programs that direct construction of complex structures (flagella and other surface organelles), entail irreversible differentiation (sporulation), or risk a significant fitness cost in some other way (growth arrest leading to persistence, competence for DNA uptake) are controlled by genetic circuitry imparting bistable expression of key phenotypic determinants (Cummings et al., 2006, Stewart et al., 2011, Temme et al., 2008, Sturm et al., 2011, Chung et al., 1994, Bigger, 1944, Moyed & Bertrand, 1983, Smits et al., 2005).
Bistability requires a promoter signal-response function characterized by non-linear kinetics, positive feedback, and hysteresis (Veening et al., 2008, Ray et al., 2011). Non-linear kinetics coupled with positive feedback imparts high promoter sensitivity to supra-threshold signal concentrations, generating a strong, rapid transcriptional response. In hysteretic systems, distinct signal threshold levels initiate switching from OFF to ON versus ON to OFF. Hysteresis stabilizes cells in the OFF- or ON-state, pending a decisive change in signal concentration (Ray et al., 2011). Specific biochemical modules by which non-linearity, positive feedback and hysteresis are integrated into a genetic program can be transcriptional or non-transcriptional, including post-translational mechanisms such as phosphorylation, proteolytic degradation or physical sequestration (Chen & Arkin, 2012, Ray et al., 2011).
Our model system for evolved genetic circuitry conferring bistability is the Salmonella enterica serovar Typhimurium flagellar regulon. Host compartment-specific flagellar regulation is critical to Salmonella virulence. Flagellar gene expression promotes intestinal colonization by conferring motility (Stecher et al., 2008) and activating expression of invasion genes (Chubiz et al., 2010). Bistable flagellar and invasion gene expression generates distinct subpopulations that 1) invade the intestinal epithelium and activate inflammation, and 2) thrive on host products released into the gut lumen during inflammation, thereby outcompeting the intestinal microbiota and promoting transmission (Ackermann et al., 2008). In contrast, failure to repress flagellar genes in systemic tissues (where Salmonella live within phagocytes) is attenuating (Stewart et al., 2011, Miao et al., 2010). Salmonella signaling networks have evolved such that the flagellar regulon receives input from numerous environmental, physiological and metabolic pathways, many of which help Salmonella orient to host micro-environments (Bader et al., 2003, Adams et al., 2001, Spory et al., 2002, Lin et al., 2008, Wozniak et al., 2009, Clegg & Hughes, 2002, Lawhon et al., 2003, Mouslim & Hughes, 2014).
Salmonella flagella are constructed via the sequential expression of three classes of genes (figure 1). Class I comprises one operon, flhDC, encoding master regulatory proteins required to initiate class II expression. Cellular signaling pathways converge here to control the amount and activity of the FlhD4C2 complex, modulating its transcription (Mouslim & Hughes, 2014), translation (Wei et al., 2001, Jonas et al., 2010, Lawhon et al., 2003), activation (Takaya et al., 2006) and activity/stability (Tomoyasu et al., 2002, Saini et al., 2008, Wada et al., 2011a, Takaya et al., 2012, Li et al., 2012, Ahmad et al., 2013). Class II genes encode proteins of the hook/basal body (HBB), while class III genes encode the flagellar filament monomers (FliC and FljB) and proteins required for motor function and chemotaxis (Chevance & Hughes, 2008). Class II and III promoters also drive expression of multiple positive and negative regulators of the cascade (Wozniak et al., 2010), including the sigma factor (FliA) required for class III expression (Chevance & Hughes, 2008), the anti-sigma factor (FlgM) that sequesters FliA until HBB assembly is complete (Karlinsey et al., 2000, Hughes et al., 1993), and FliZ, an activator of class II promoters (Kutsukake et al., 1999).
Figure 1.
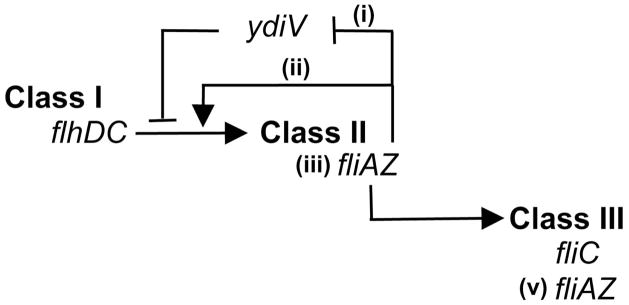
The flagellar three class regulatory cascade. Proteins encoded by flhDC, the sole class I operon, form a complex required to activate class II gene transcription. Class II genes encode structural proteins of the platform that anchors the flagellum within the bacterial cell envelope (the basal body) and of the flexible linker that connects the platform with the filament (the hook), as well as the regulators fliZ and fliA. FliZ enhances class II gene expression, forming a positive feedback loop. Two mechanisms have been proposed to account for this loop. FliZ represses the class II repressor ydiV (i); thus fliZ and ydiV are mutually repressing repressors with opposing effects upon class II transcription. FliZ has also been shown to augment the level of the class I protein FlhC (ii). FliA is the sigma factor required for transcription of class III genes, including the flagellin filament protein FliC. The fliAZ operon is transcribed from both a class II (iii) and a class III (v) promoter, thus forming another positive feedback loop (the sigma factor activates transcription of its own operon).
The three-class flagellar gene expression cascade includes biochemical features that generate positive feedback and ultrasensitivity (Ray et al., 2011) in other bistable systems, both natural (evolved) and synthetic. Complex genetic architecture comprising multiple positive and negative feedback loops (Kutsukake et al., 1999, Yamamoto & Kutsukake, 2006, Kalir et al., 2005), sequestration of the sigma factor FliA by the anti-sigma factor FlgM, and a system of mutually repressing repressors formed by FliZ and a protein encoded outside the regulon, YdiV (Wada et al., 2011b, Wozniak et al., 2009), are candidate mechanisms that might contribute to bistable expression of class II or III genes (Ray et al., 2011, Chen & Arkin, 2012, Dubnau & Losick, 2006). WT Salmonella populations comprise a subpopulation transcribing fliA and a subpopulation in which fliA transcription is post-translationally blocked by YdiV (Stewart et al., 2011), resulting in bistable fliA and fliC expression. YdiV binds FlhD, inhibits FlhD4C2 complex binding to DNA, and targets the complex to the ClpXP protease for degradation (Wada et al., 2011a, Takaya et al., 2012, Li et al., 2012). FliZ increases the level of class I proteins by an unknown mechanism, and represses ydiV transcriptionally. Thus YdiV and FliZ are mutually repressing repressors with opposing effects at the same important control point of flagellar gene expression, the FlhD4C2 complex. Repression of fliC by YdiV is required for full virulence (Stewart et al., 2011, Hisert et al., 2005). FliZ is also necessary for virulence, as a positive regulator of the Salmonella Pathogenicity Island-1 locus (Iyoda et al., 2001, Lucas et al., 2000, Chubiz et al., 2010). Whether FliZ’s role in flagellar regulation contributes to infection is unknown.
We investigated the contribution of this mutually repressing repressor genetic circuit to fliC bistability, using flow cytometry to plot the single-cell census, or distribution, of fliC expression for WT and mutant populations. Bistable populations were maintained from 2–5 hours after backdilution, and the repressor YdiV maintained the fliC-OFF subpopulation throughout 5 hours of growth. FliZ was required for WT levels of fliC expression, however, bistable populations continued to form in a ΔfliZ strain, indicating that reciprocal repression by YdiV and FliZ is not crucial for fliC bistability. Instead, the individual roles of YdiV and FliZ are important, such that bistability collapses into a predominantly intermediate population in the absence of both regulators. Dose-dependent induction or metering of YdiV and FliZ production from separate exogenous promoters revealed underlying cell-to-cell heterogeneity in factors required for these regulators to modulate fliC expression. We propose that input from multiple pathways tuning transcription, translation, activation and stability of the FlhD4C2 complex generates population diversity in FlhD4C2 activity, determining which cells are locked into fliC-OFF mode and which activate fliC expression.
Results
The kinetic role of FliZ in fliC bistability
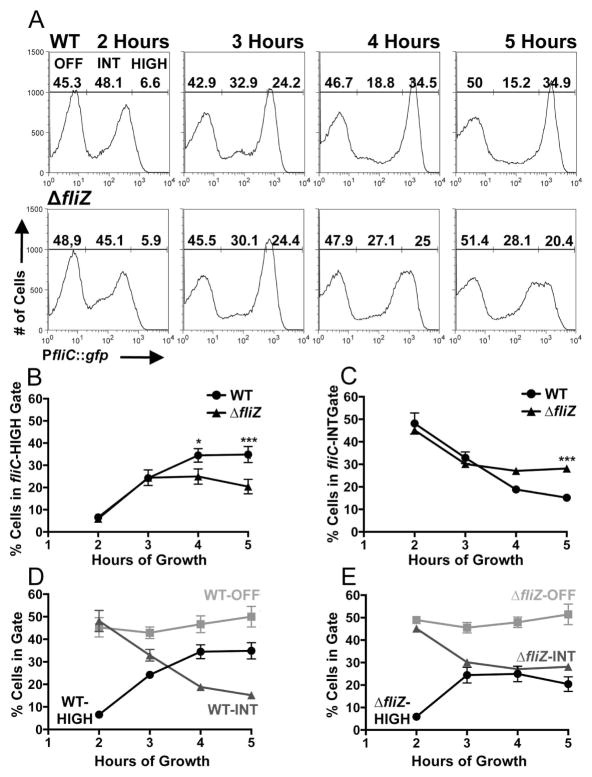
The census of fliC expression in populations of the virulent Salmonella enterica serovar Typhimurium strain 14028 was analyzed by flow cytometry, using a fliC-GFP transcriptional reporter (Cummings et al., 2006). (See figure 1 for an overview of flagellar regulation in Salmonella.) In order to quantitatively describe our observations, the range of fluorescence observed was divided into three gates: fliC-OFF, fliC-INT and fliC-HIGH (see figure S1 for a detailed description of gating). Percent gate occupation in these experiments depends upon fluorescent intensity of individual bacteria in the populations. When fluorescent proteins are used as reporters of bacterial gene transcription, fluorescent intensity is determined by the relative kinetics of promoter activity, dilution due to cell division, and proteolytic degradation. Proteolytic degradation is negligible in these experiments, due to the 9.5-day fluorescence half-life (Cummings et al., 2006) of the GFP (GFPmut3, (Cormack et al., 1996)) in this construct. Additionally, fluorescence of GFPmut3 is detectible within 8 minutes of transcriptional induction (Cormack et al., 1996). Thus, fluorescent intensity of Salmonella in these populations is a function of promoter activity and dilution due to growth. Similar growth curves and generation times for the four strain backgrounds used in this study (figure S2) indicate that comparisons of percent gate occupation can be made across strains at individual timepoints. Trends in gate occupation observed across timepoints for a single strain reflect changes in both promoter activity and growth rate during those intervals. Strains are in exponential growth phase between 1.5–3 hours after backdilution and post-exponential phase during the remainder of the timecourse (figure S2). Expression of a constitutive promoter is unimodal for all strain backgrounds used in this study (figures S3 and S4B).
Wild-type populations demonstrated well-defined bimodal fliC expression distributions at all timepoints tested (figure 2A, top panels, histograms from a representative experiment and average subpopulation percentages from three independent experiments). (See supplemental material for subpopulation percentages and mean fluorescent intensities from individual experiments, and tables 1 and S1 for strain specifics.) This kinetic analysis confirms that two stable states of fliC expression are maintained throughout the growth curve in Salmonella strain 14028, in contrast to strain LT2 (figure S4A) where flagellar class III expression becomes activated in all cells of the population during this timeframe (Saini et al., 2010). fliC expression increased over time, as cells were partitioned from the fliC-INT gate to the fliC-HIGH gate. The fliC-HIGH subpopulation grew significantly between 2–3 hours (p<0.001) and 3–4 hours (p<0.01), with corresponding decreases in the size of the fliC-INT subpopulation over both intervals (p<0.001).
Figure 2.
fliZ is required to maximize the fliC-HIGH subpopulation at late timepoints. A) A transcriptional fusion of the fliC promoter to gfp (pPfliC::gfp) was used with flow cytometry to take a census of fliC-expression in WT (BC2117) and ΔfliZ (BC2119) Salmonella populations as the cultures progressed through five hours of growth after backdilution (figure S2). B and C) Percentages of cells falling within the fliC-HIGH (B) and fliC-INT (C) gates for WT and the ΔfliZ mutant. D and E) Kinetic plots of subpopulation flux for WT (D) and ΔfliZ (E). Histograms in (A) depict the results of one representative experiment; associated subpopulation percentages in (A) are the means of three independent experiments. Line graphs in (B–D) represent the means with standard deviation of the three independent experiments. One-way ANOVA with Tukey’s multiple comparisons test was used to compare all strain/timepoint values. *p<0.05, **p<0.01, ***p<0.001.
Table 1.
Strains and plasmids used in this study
| Strains | ||
| BC2117 | Salmonella Typhimurium 14028 fljBA::FRT pPfliC::gfp, carbR | This Study |
| BC2118 | Salmonella Typhimurium 14028 fljBA::FRT ydiV::FRT pPfliC::gfp, carbR | This Study |
| BC2119 | Salmonella Typhimurium 14028 fljBA::FRT fliZ::FRT pPfliC::gfp, carbR | This Study |
| BC2675 | Salmonella Typhimurium 14028 fljBA::FRT ydiV::FRT fliZ::FRT pPfliC::gfp, carbR | This Study |
| BC2264 | Salmonella Typhimurium 14028 fljBA::FRT FlhC::3XFLAG, kanR | Stewart et al., 2011 |
| BC2265 | Salmonella Typhimurium 14028 fljBA::FRT ydiV::FRT FlhC::3XFLAG, kanR | Stewart et al., 2011 |
| BC2266 | Salmonella Typhimurium 14028 fljBA::FRT fliZ::FRT FlhC::3XFLAG, kanR | This Study |
| BC2638 | Salmonella Typhimurium 14028 fljBA::FRT ydiV::FRT fliZ::FRT FlhC::3XFLAG, kanR | This Study |
| BC2307 | Salmonella Typhimurium 14028 fljBA::FRT ydiV::FKF::pKG136, kanR | This Study |
| BC3276 | Salmonella Typhimurium 14028 fljBA::FRT fliZ::FRT ydiV::FKF::pKG136 pJN105, kanR gentR | This Study |
| BC3277 | Salmonella Typhimurium 14028 fljBA::FRT fliZ::FRT ydiV::FKF::pKG136 pfliZ, kanR gentR | This Study |
| BC3314 | Salmonella Typhimurium 14028 fljBA::FRT fliZ::FRT ydiV T-POP, pfliZ pPfliC::gfp, tetR gentR carbR | This Study |
| Plasmids | ||
| pPfliC::gfp | pSRB1, PfliC::gfp construct in pBR322 vector | Cummings et al., 2006 |
| pJN105 | Vector, arabinose-inducible expression of inserts (ParaBAD) | Newman et al., 1999 |
| pfliZ | Salmonella Typhimurium 14028 fliZ cloned into pJN105 | This Study |
In order to define the role of FliZ in shaping the census of fliC expression in a bistable population, fliC expression in a ΔfliZ mutant was next examined. ΔfliZ populations also proved bistable throughout the timecourse (figure 2A, bottom panels, histograms from a representative experiment and average subpopulation percentages from three independent experiments). When kinetic plots of subpopulation sizes were compared, the population distributions of fliC expression for WT and the ΔfliZ mutant were indistinguishable at 2 and 3 hours (figures 2B,C). At 4 hours, however, the WT fliC-HIGH subpopulation was larger (figure 2B) than that of the ΔfliZ mutant, and this difference increased between 4 and 5 hours. The opposite trend was observed when the sizes of the fliC-INT subpopulations were compared (figure 2C), whereby at 5 hours the WT fliC-INT subpopulation was smaller than that of the ΔfliZ mutant. The average mean fluorescent intensity (MFI, measured in arbitrary fluorescence units, AFU) of the WT fliC-HIGH subpopulations was significantly higher than the average MFI of the ΔfliZ mutant fliC-HIGH subpopulations (at 5 hours: WT fliC-HIGH MFI=1588 AFU and ΔfliZ mutant fliC-HIGH MFI=1415 AFU, p<0.001), consistent with the observation that the peak of fliC-expressing cells for ΔfliZ mutant is shifted toward lower levels of expression, spanning the junction between the fliC-INT and fliC-HIGH gates. Individual plots of the population distributions for WT (figure 2D) and the ΔfliZ mutant (figure 2E) highlight this fliZ- and time-dependent partitioning of cells from the fliC-INT subpopulation to the fliC-HIGH subpopulation.
Thus, in a bistable strain, FliZ increases the probability that an individual cell will maximize fliC expression, in a growth-phase dependent manner. Intriguingly, many cells are able to maximize fliC expression in the absence of FliZ, revealing biochemical heterogeneity in these well-mixed, homogeneously grown cultures.
FliZ controls flagellar gene expression via ydiV-dependent and -independent mechanisms
Two mechanisms have been proposed to account for FliZ-mediated upregulation of flagellar genes. Whether these activities constitute a single pathway or distinct pathways was unknown. FliZ and YdiV participate in a genetic circuit of mutually repressing repressors (figure 1), wherein FliZ repression of ydiV is predicted to increase flagellar class II gene expression because YdiV inhibits binding of the FlhD4C2 complex to DNA and targets the complex for ClpXP-mediated proteolysis (Takaya et al., 2012, Wada et al., 2011a). Additionally, FliZ increases the level of the class I protein FlhC concurrently with fliC transcription (Saini et al., 2008); specifics of this pathway have not been elucidated, and ydiV-dependence of the finding was not tested. We investigated whether the impact of either mechanism varied with time in our system.
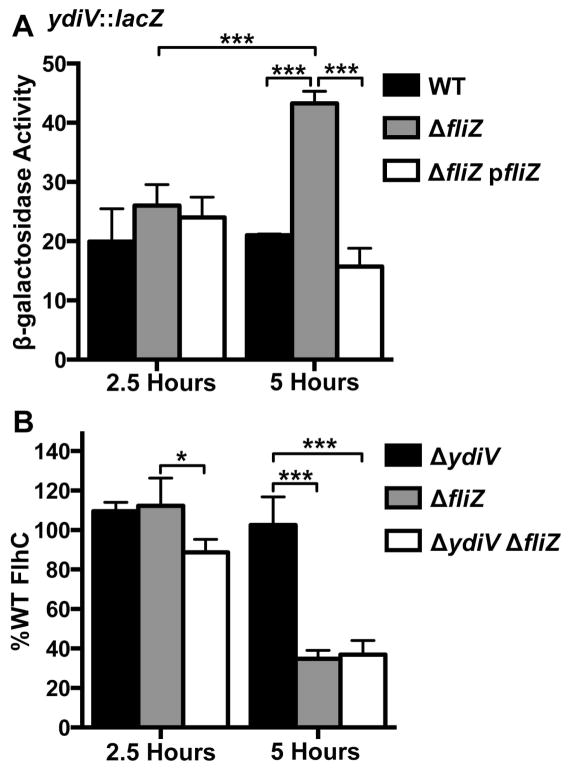
A β-galactosidase reporter fusion was used to measure ydiV transcription in the presence and absence of FliZ. In WT cells, ydiV was transcribed at similar levels at both 2.5 and 5 hours (figure 3A, compare black bars across timepoints, difference is not significant), and there were no significant differences between the WT, ΔfliZ, and ΔfliZ pfliZ strains at 2.5 hours. In contrast, the temporal difference in ydiV transcription for the ΔfliZ mutant was highly significant (figure 3A, compare grey bars across timepoints), and at 5 hours ydiV was more highly transcribed in fliZ than in the WT strain (compare black bar to grey bar). Complementation with exogenous FliZ was significant at 5 hours (compare grey bar to white bar). These data demonstrate that FliZ represses ydiV to a greater extent as cells approach stationary phase (figure S2).
Figure 3.
fliZ impacts flagellar regulators via a mechanism that is independent of the mututally repressing repressors genetic circuit. A) A transcriptional, chromosomal β-galactosidase fusion to the ydiV promoter was used to characterize the kinetics of FliZ-mediated ydiV repression. 0.01% L-arabinose (to induce ΔfliZ transcription from ParaBAD in the complementation strain, ΔfliZ pfliZ) was added to all samples 2.5 hours before cells were lysed and β-galactosidase activity was measured. One-way ANOVA with Tukey’s multiple comparisons test was used to compare all strain/timepoint values. (WT: BC2307, ΔfliZ: BC3276, ΔfliZ pfliZ: BC3277) B) FliZ increases the amount of FlhC by a ydiV-independent mechanism. A 3XFLAG tagged version of the class I protein FlhC was detected by western blot, normalized to DnaK expression, and adjusted to %WT expression by timepoint. One-sample t-tests with hypothetical value of 100 were used to compare mutant FlhC levels to WT. One-way ANOVA with Tukey’s multiple comparisons test was used to compare FlhC values within timepoints. (WT (not shown, used to compute %WT values): BC2264, ΔydiV: BC2265, ΔfliZ: BC2266, ΔydiV ΔfliZ: BC2638) Bars in (A) and (B) represent the mean plus standard deviation of three independent experiments. *p<0.05, **p<0.01, ***p<0.001.
FlhC levels were measured by western blot and are reported for the ΔydiV, ΔfliZ, and ΔydiV ΔfliZ mutants as the percentage of WT FlhC at 2.5 or 5 hours (figure 3B). FliZ did not regulate FlhC at 2.5 hours (figure 3B, none of the mutants differed significantly from WT which is set at 100%). However, at 5 hours, FliZ was required to stabilize WT FlhC levels (figure 3B, FlhC in ΔfliZ mutant is 34.8% of WT, p=0.0014, and FlhC in ΔydiV ΔfliZ mutant is 37% of WT, p=0.0041). Thus, both the kinetic pattern of ydiV repression by FliZ (figure 3A) and FlhC regulation by FliZ (figure 3B) support our fliC transcription timecourse, in which FliZ increases fliC expression at later timepoints during growth (figure 2A-E).
To determine whether FliZ requires ydiV in order to augment the amount of FlhC protein, FlhC levels between the ΔydiV single mutant and the ΔydiV ΔfliZ double mutant were compared (figure 3B, compare black bars to white bars within timepoints). FliZ represses ydiV transcription (Wada et al., 2011b, Wozniak et al., 2009). If this were the sole mechanism by which FliZ increased FlhC, the amount of FlhC should be identical between ΔydiV and ΔydiV ΔfliZ. FliZ significantly increases the amount of FlhC in a ΔydiV background at 5 hours (FlhC in ΔydiV=102.5% WT, FlhC in ΔydiV ΔfliZ=37% WT, p<0.001). Therefore, FliZ also plays a ydiV-independent role in flagellar gene regulation.
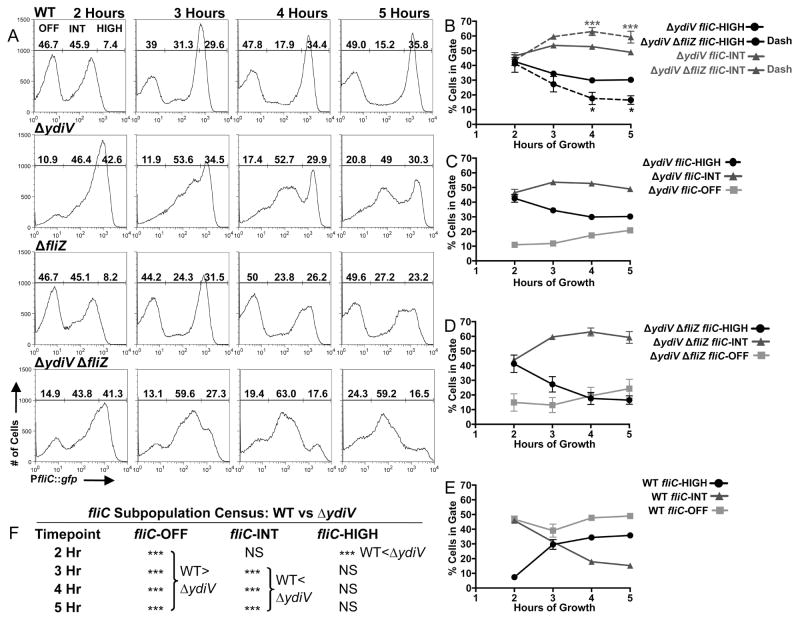
We hypothesized that if FliZ can boost fliC expression by a ydiV-independent mechanism, ΔydiV single and ΔydiV ΔfliZ double mutant populations would have distinct patterns of fliC expression (figure 4A, second and fourth rows, histograms from a representative experiment and average subpopulation percentages from three independent experiments). Indeed, the percentage of cells in the fliC-HIGH gate was significantly larger for ΔydiV than for ΔydiV ΔfliZ when distributions of fliC expression were compared at 4 and 5 hours (figure 4B, black circles). In contrast, the fliC-INT subpopulations were significantly larger for ΔydiV ΔfliZ than for ΔydiV (figure 4B, dark grey triangles). Thus, fliZ significantly shapes these distributions even in the absence of ydiV.
Figure 4.
Inversion of bistability in the absence of fliZ and ydiV. A) Flow cytometry census of fliC expression for WT (BC2117) and the ΔydiV (BC2118), ΔfliZ (BC2119), and ΔydiV ΔfliZ (BC2675) mutants. B) Comparisons of fliC-HIGH and fliC-INT subpopulations for the ΔydiV and ΔydiV ΔfliZ mutants. Black asterisks (bottom of figure) reference comparisons between population percentages in the fliC-HIGH gate, dark grey asterisks (top of figure) reference comparisons between population percentages in the fliC-INT gate. There were no significant differences between the ΔydiV and ΔydiV ΔfliZ strains in the sizes of the fliC-OFF subpopulations. C, D and E) Kinetics of subpopulation flux for strains ΔydiV (C), ΔydiV ΔfliZ (D), and WT (E, from this set of experiments, distinct from WT data represented in figure 2). F) Significance of comparisons between WT and the ΔydiV mutant. Histograms in (A) depict the results of one representative experiment; associated subpopulation percentages in (A) are the means of three independent experiments. Line graphs in (B–E) represent the means with standard deviation of the three independent experiments. One-way ANOVA with Tukey’s multiple comparisons test was used to compare all strain/timepoint values. *p<0.05, **p<0.01, ***p<0.001.
The changing role of YdiV during growth
At 2 hours, there were significantly more fliC-HIGH cells and significantly fewer fliC-OFF cells for the ΔydiV mutant compared to WT (figures 4A,C,E,F), while the fliC-INT subpopulations did not significantly differ in size. Thus YdiV partitions cells from the fliC-HIGH gate to the fliC-OFF gate early during growth.
This kinetic analysis of fliC expression in the ΔydiV mutant revealed significant ydiV-independent fliC repression at 4 and 5 hours. This manifested as 1) a drop in the percentage of fliC-HIGH cells across time for ΔydiV (figure 4C, black circles, compare 2 hours to 5 hours, p<0.001) and 2) an increase in the percentage of fliC-OFF cells across time (figure 4C, light grey squares, compare 2 hours to 5 hours, p<0.05). Furthermore, the average MFI for cells in the fliC-INT gate fell significantly from 2 hours (246.7 AFU) to 5 hours (144.7 AFU, p<0.01) for the ΔydiV mutant, indicating a shift to a lower level of fliC expression for this subpopulation. From 3 hours on, significant differences in the fliC expression distributions between WT and the ΔydiV mutant occurred in the fliC-OFF and fliC-INT subpopulations (figures 4A,C,E,F), while the sizes of the fliC-HIGH subpopulations did not significantly differ. These data demonstrate a changing role for YdiV during growth whereby YdiV partitions cells into fliC-OFF from fliC-HIGH at 2 hours, and from fliC-INT at later timepoints (3, 4 and 5 hours).
The census of fliC expression is inverted in the absence of YdiV and FliZ
We predicted that deleting both ydiV and fliZ would invert the WT census of fliC expression as cells approach stationary phase, since YdiV is critical to maintain the fliC-OFF subpopulation, and FliZ is required to maximize fliC expression under these conditions. In WT distributions, intermediate states of fliC expression are unstable compared to the fliC-OFF and fliC-HIGH states (figures 2A,D and 4A,E). This observation suggested the possibility that intermediate states of fliC expression might be strongly favored in the absence of YdiV-mediated repression and FliZ-mediated enhancement. This proved to be true at late timepoints (4 and 5 hours, figures 4A (bottom panels),D), when the fliC-INT subpopulation of ΔydiV ΔfliZ distributions contained a significantly higher percentage of cells than either the fliC-OFF or fliC-HIGH gates, which did not differ from one another (at 4 hours, fliC-INT compared to fliC-OFF, p<0.001, and to fliC-HIGH, p<0.001; at 5 hours, fliC-INT compared to fliC-OFF, p<0.001, and to fliC-HIGH, p<0.001).
Multiple layers of heterogeneity shape the fliC expression census
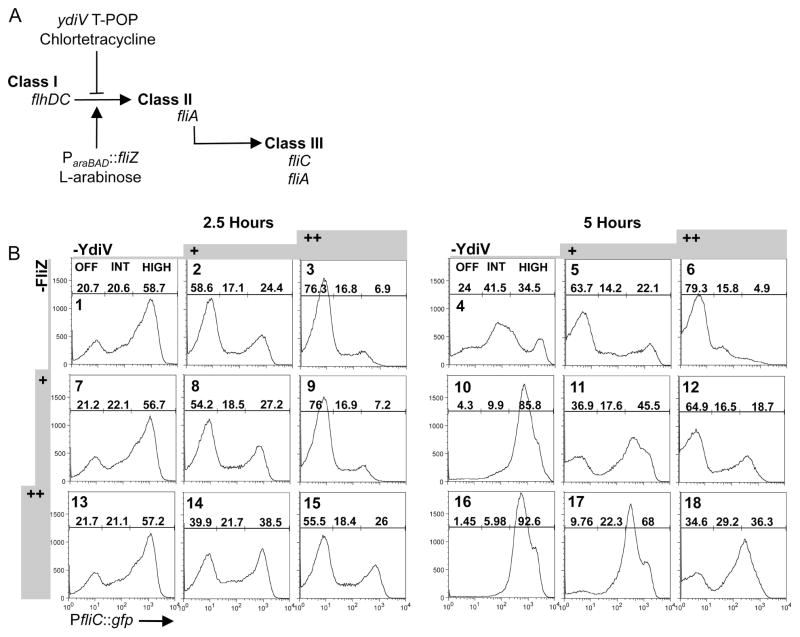
We hypothesized that modulating production of YdiV and FliZ would reveal the extent to which fliC expression can be controlled by these two regulators. Accordingly, a system with which to study the sensitivity of fliC transcription to perturbation by YdiV and FliZ was constructed. Regulator production is metered from separate exogenous promoters (figure 5A), YdiV according to the concentration of chlortetracycline, and FliZ according to the concentration of L-arabinose. Neither promoter inherently produces bistable populations in the presence or absence of inducer (figure S5). Bimodal distributions of cells in many panels of figure 5B evince switch-like, heterogeneous control of fliC expression in the metered strain.
Figure 5.
Multi-layered heterogeneity regulates fliC expression. A) Schematic of our metered system (BC3314). YdiV and/or FliZ are expressed with the addition of chlortetracycline (+1.6 or ++3.1μg ml−1) (T-POP) and/or L-arabinose (+0.01 or ++0.1%) (ParaBAD), respectively, and fliC transcription is measured using the GFP reporter fusion. B) FliZ’s impact upon fliC transcription remains time-dependent, and heterogeneity in fliC expression is maintained in the metered system. Populations were grown in media +/− chlortetracycline for 2.5 or 5 hours, and L-arabinose was added 2.5 hours before harvesting the cells at both timepoints.
Metered induction of YdiV reduced fliC expression, partitioning cells into the fliC-OFF subpopulation. Without FliZ (figure 5B, panels 1–3 and 4–6, histograms and subpopulation percentages from a single experiment), fliC transcriptional responses to YdiV induction were similar at the two timepoints tested. At 2.5 hours, titration of YdiV into the system increases the percentage of the population in the fliC-OFF gate from 20.7% (panel 1) to 58.6% (2) to 76.3% (3). At 5 hours, the fliC-OFF population rose from 24% (panel 4), to 63.7% (5) and 79.3% (6).
At 2.5 hours, no YdiV-independent role of FliZ was observed (figure 5B, compare panels 1,7 and 13). FliZ boosted fliC expression only when ydiV was concurrently induced (figure 5B, columns 2 and 3). In the context of high YdiV expression, strong induction of fliZ increased the fliC-HIGH subpopulation from 6.9% (panel 3) to 26% (15) and reduced the fliC-OFF subpopulation from 76.3% (panel 3) to 55.5% (15). The fliC-INT subpopulation remained nearly constant at 16.8–18.4% (figure 5B, panels 3,9 and 15).
At 5 hours, in the absence of YdiV and FliZ, the largest subpopulation is fliC-INT at 41.5% (figure 5B, panel 4). This mirrors the inverted census of fliC expression observed for the ΔydiV ΔfliZ strain in figure 4A (fourth row, 4 and 5 hours). At 5 hours, as at 2.5, metered YdiV expression partitioned cells into the fliC-OFF subpopulation at all strengths of FliZ induction (figure 5B, compare panels 4 to 6, 10 to 12 and 16 to 18). However, in contrast to observations at 2.5 hours, metered induction of FliZ now partitioned cells into fliC-HIGH in the presence and absence of YdiV (figure 5B, compare fliC-HIGH subpopulations in panels 4–6, where FliZ is not induced, to fliC-HIGH subpopulations in panels 10–12 and 16–18). This demonstrates that FliZ is able to counter both YdiV-dependent and –independent fliC repression at 5 hours. Thus, the impact of FliZ upon the fliC census is enhanced at 5 hours compared to 2.5 hours under conditions of metered expression. These data are consistent with the requirement for FliZ to maintain both WT fliC expression at late timepoints (figure 2A-E) and WT levels of FlhC in a ydiV-independent manner at 5 hours (figure 3B).
Partitioning of 92.6% of cells into the fliC-HIGH gate with strong fliZ induction at 5 hours (figure 5B, panel 16) shows that most cells in the population contained all cellular factors required for fliC expression except for FliZ at this time. Intriguingly, though fliZ was induced throughout the population (figure S5) in our metered strain, the ability of FliZ to counter YdiV-dependent (figure 5B, panels 11,12,17,18) and –independent (figure 5B, panels 10,16) fliC repression varies from cell to cell. Similarly, at both timepoints, the addition of increasing amounts of YdiV partitions some cells from fliC-ON (fliC-INT + fliC-HIGH) to the fliC-OFF subpopulation, but cell-to-cell sensitivity to the repressor varies (compare column 1 data to columns 2 and 3 for both timepoints). By titrating expression levels of ydiV and fliZ, we reveal heterogeneity in cellular factors that potentiate the impact of these two important regulators upon fliC expression.
Discussion
YdiV and FliZ destabilize intermediate states of fliC expression
Previously, we showed that the virulent Salmonella strain 14028 demonstrates bistable fliC expression at a single timepoint (Stewart et al., 2011), and the present data show that bistable distributions of fliC expression in 14028 persist through 5 hours of growth. Using these kinetic analyses, we define the roles of two flagellar regulators in subpopulation flux and the maintenance of fliC bistability. The activator FliZ partitions cells from intermediate to high levels of fliC expression at late timepoints. This finding is supported by a study of flagellar gene expression dynamics in the transiently bimodal strain LT2, showing that a class III gene was expressed at a lower intensity at late timepoints in a ΔfliZ mutant (Saini et al., 2010). YdiV partitions cells from fliC-HIGH (at early timepoints) or fliC-INT (at later timepoints) into the fliC-OFF subpopulation. Thus, both regulators contribute to the instability of intermediate states of expression, organizing the population into tightly defined fliC-OFF and fliC-HIGH subpopulations.
Individual roles of YdiV and FliZ shape fliC bistability, while reciprocal repression is dispensible
FliZ and YdiV are mututally repressing repressors, a genetic feature demonstrated to contribute to phenotypic heterogeneity in other systems. The positive feedback loops formed when FliZ increases class I protein levels or decreases ydiV transcription were candidate hysteretic mechanisms for fliC regulation. Positive feedback loops could lock cells into the fliC-ON state by increasing the level of a positive regulator or decreasing the level of a negative regulator of flagellar class II gene expression. Here we demonstrate bistability in the absence of FliZ (figures 2A and 4A). Therefore, neither coordinated downregulation of ydiV by FliZ as flagellar gene expression increases nor the positive feedback loop formed by FliZ’s impact upon class I proteins is required for cells to activate fliC expression or persist in fliC-ON mode across time. Additionally, the present work shows that these two activities of FliZ constitute distinct pathways. FliZ represses ydiV, and increases both class I protein levels (figure 3B) and fliC expression in the absence of ydiV (figure 4A,B). These regulators therefore play important individual roles in fliC expression, and bistable population distributions continue to form in the absence of the mututally repressing repressor circuit.
FliZ activity is temporally regulated
The impact of FliZ upon fliC expression was most evident at 5 hours in both the WT (figure 2A,B,C) and engineered backgrounds (figure 5B). At the population level, this suggests temporal fluctuations in average concentrations of cellular factors required for FliZ to activate fliC expression, most likely regulators of class I. Transcription of class I genes is tightly controlled by growth-phase specific expression of multiple positive and negative regulators (Mouslim & Hughes, 2014). One positive transcriptional regulator of flhDC, HilD, is itself more highly expressed as growth progresses (Mouslim & Hughes, 2014, Kröger et al., 2013) and is a master activator of Salmonella Pathogenicity Island-1 transcription (Schechter et al., 1999). HilD is post-translationally activated by FliZ (Chubiz et al., 2010), and therefore represents a potential growth-phase specific mechanism by which FliZ could upregulate flhDC transcription coordinately with SPI-1 genes. However, control of class I proteins by FliZ is reported to be predominantly post-translational (Saini et al., 2008), and availability of cellular factors impacting translation, activation, and stability of FlhD4C2 may also fluctuate with growth phase. These hypotheses will be tested in future studies.
Model of YdiV-dependent hysteresis in fliC heterogeneity
Metered expression (figure 5B) revealed that sensitivity to YdiV and FliZ varies from cell to cell within populations. The presence of a YdiV-dependent fliC-OFF subpopulation in the WT census (figures 2A,4A, top panels) suggests variable sensitivity, but could also be explained by a variable distribution of YdiV across the population. In the metered strain, however, variable responses occurred with uniform induction of these regulators (figure S5). We observed that addition of FliZ organized a population with broadly heterogeneous fliC expression into a single fliC-HIGH peak (figure 5B, compare panel 4 to panel 16), demonstrating that these cells contained all factors required for fliC expression, and suggesting that post-translationally increasing class I protein levels (figure 3B) was critical.
We propose a model in which variable sensitivity to YdiV and FliZ is predicated upon apparently stochastic cell-to-cell differences (Elowitz et al., 2002) in the amount and activity of the FlhD4C2 complex. Production, activity and stability of FlhD4C2 are tuned by many regulators, in accordance with information flow through diverse signaling pathways (RcsB (Wang et al., 2007), RflM (Singer et al., 2013), SlyA (Mouslim & Hughes, 2014), LrhA (Mouslim & Hughes, 2014), RtsB (Ellermeier & Slauch, 2003), FimZ (Clegg & Hughes, 2002), HilD (Singer et al., 2014), CsrA (Lawhon et al., 2003), DnaK (Takaya et al., 2006), ClpX (Tomoyasu et al., 2002), HNS (Kutsukake, 1997), cAMP-CRP (Komeda et al., 1976), and others). Inputs from multiple cellular systems include simultaneous positive and negative regulation (Mouslim & Hughes, 2014). Cumulatively, small cell-to-cell variations in regulatory input from multiple pathways could generate population heterogeneity in the net level of activated FlhD4C2 complex. Cells low in activated FlhD4C2 would require more FliZ to activate fliC expression, and cells with higher concentrations of activated FlhD4C2 would require more YdiV to switch off fliC expression.
YdiV is required for a stable fliC-OFF peak (figure 4A), and for fliC repression and Salmonella virulence in host systemic tissues (Stewart et al., 2011). Li et al. recently proposed a model based upon work in E. coli whereby up to four YdiV molecules can bind to FlhD in the FlhD4C2 ring (Li et al., 2012). The FlhD4C2 conformation was unaffected by the addition of one or two molecules of YdiV, but the third and fourth distorted the ring, abrogating DNA binding and flagellar class II transcription. Depending upon the strength of the association, YdiV4-FlhD4C2 could act as a dead-end complex (Igoshin et al., 2008, Ray et al., 2011). High concentrations of YdiV are also reported to mediate degradation of FlhD4C2 by ClpXP (Takaya et al., 2012). Dead-end complex formation and/or YdiV-driven FlhD4C2 degradation could impart hysteresis. Such cells without sufficient FlhD4C2 would be locked into fliC-OFF mode until the relative concentrations of factors preventing and promoting flagellar gene expression shift decisively, perhaps due to unequal partitioning to daughter cells during division. Thus, we provide evidence that coordinated input from a landscape of heterogeneously distributed cellular factors shapes the binary output of this evolved genetic circuit.
Experimental Procedures
Bacterial Strains and Growth Conditions
Strains and plasmids used in this study are listed in tables 1 and S1. Mutants were generated in the Salmonella enterica serovar Typhimurium 14028 background as previously described (Datsenko & Wanner, 2000). Strains express only fliC flagellin due to deletion of fljBA (Bonifield & Hughes, 2003). The FlhC::3XFLAG construct was a gift from the Rao lab (Saini et al., 2008), and the ydiV::FKF::pKG136 and T-POP::ydiV (ydiV240::Tn10dTc(del-25)) constructs were gifts from the Hughes lab (Wozniak et al., 2009). Constructs were transferred between Salmonella strains by P22 transduction (Maloy et al., 1996). Salmonella were grown at 37°C with shaking at 225 rpm in Miller Luria Broth (Product #244620, Difco) with appropriate antibiotics, backdiluted 1:100 after overnight incubation, and grown to indicated timepoints. Carbenicillin, kanamycin and gentamicin were added at 100μg/ml, 50μg/ml and 20μg/ml, respectively. Chlortetracycline hydrochloride (Sigma #26430-5G, autoclaved with liquid media) and L-arabinose were added at the indicated concentrations. Growth curves were generated (figure S2) by using a Beckman Coulter Multisizer 4 to measure the density of bacterial cultures every 30 minutes from backdilution through five hours of growth. Generation times were calculated as follows (Fuchs & Kroger, 1999): tg=(t-t0)/[(log(Nt/N0))*3.3], where tg=generation time, N=number of cells at time t, and N0=number of cells at time t0.
Construction of pfliZ and pgfp
The fliZ coding sequence from Salmonella 14028 and the gfp coding sequence from pDW5 (Cummings et al., 2006) were amplified using primers GAATTCGTATAGACTACCAGGAGTTC and GAGCTCGACTCTGCTACATCTTATGC, or GAATTCGATTTAAGAAGGAGATATAC and GAGCTCCAGGTCTGGACATTTATTTG, respectively. PCR products were cloned into pCR2.1, sequenced, digested out with EcoRI and SacI, and ligated into EcoRI/SacI digested pJN105 (Newman & Fuqua, 1999). For both plasmids, L-arabinose induces transcription of the insert from ParaBAD.
Flow Cytometry
Samples were diluted into 0.5ml 0.2% sodium azide in filtered PBS. Unless otherwise noted (Figures S3-S5), 100,000 events were collected on a Becton Dickinson FACScan using the following parameters: FSC-H E01 log, SSC-H 400 log, SSC threshold of 129, FL1 750 log, FL2 and FL3 150 log. Data were analyzed using FlowJo (Tree Star).
Western Blotting
At indicated timepoints cells were pelleted and resuspended in PBS. Concentrations were determined using a Beckman Coulter Multisizer 4 and adjusted to 5×108 cells/ml. Proteins from 5×106 cells per sample were separated by SDS-PAGE electrophoresis on a 4–20% gel and transferred to nitrocellulose overnight at 30V. FlhC detected was normalized to DnaK detected for each lane and to FlhC/DnaK detected for the WT strain at the specified timepoint within that experiment. Primary antibodies used were α-3XFLAG (F3165, Sigma-Aldrich) and α-DnaK (SPA-880, Assay Designs). Goat α-Mouse secondary antibody (926-32280, 800CW) was purchased from Li-Cor. Western blots were visualized and proteins were quantified on the Li-Cor Odyssey CLx Imaging System.
β-galactosidase Activity
β-galactosidase activities were determined as described previously (Slauch & Silhavy, 1991) with the following modifications. At the indicated timepoints, bacterial cultures were centrifuged and resuspended in Z-buffer. Concentrations were determined using a Beckman Coulter Multisizer 4 and adjusted to 108 cells/ml. β-galactosidase activity of 100μl of cell suspension was measured in triplicate in 96-well plate format by the kinetic method of (Slauch & Silhavy, 1991) on a Molecular Devices SpectraMax M3 and is reported as [μmoles of ortho-nitrophenyl formed/minute/ml of cell suspension]·2×108.
Statistics
All statistical comparisons were performed using GraphPad Prism version 6.
Supplementary Material
Acknowledgments
This work was supported by NIH grant U19 AI 090882A and a University of Washington Cellular and Molecular Biology Training Grant.
Footnotes
The authors declare no conflict of interest.
References
- Ackermann M, Stecher B, Freed NE, Songhet P, Hardt WD, Doebeli M. Self-destructive cooperation mediated by phenotypic noise. Nature. 2008;454:987–990. doi: 10.1038/nature07067. [DOI] [PubMed] [Google Scholar]
- Adams P, Fowler R, Kinsella N, Howell G, Farris M, Coote P, O’Connor CD. Proteomic detection of PhoPQ- and acid-mediated repression of Salmonella motility. Proteomics. 2001;1:597–607. doi: 10.1002/1615-9861(200104)1:4<597::AID-PROT597>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- Ahmad I, Wigren E, Le Guyon S, Vekkeli S, Blanka A, El Mouali Y, et al. The EAL- like protein STM1697 regulates virulence phenotypes, motility and biofilm formation in Salmonella typhimurium. Mol Microbiol. 2013;90:1216–1232. doi: 10.1111/mmi.12428. [DOI] [PubMed] [Google Scholar]
- Bader MW, Navarre WW, Shiau W, Nikaido H, Frye JG, McClelland M, et al. Regulation of Salmonella typhimurium virulence gene expression by cationic antimicrobial peptides. Mol Microbiol. 2003;50:219–230. doi: 10.1046/j.1365-2958.2003.03675.x. [DOI] [PubMed] [Google Scholar]
- Biggar SR, Crabtree GR. Cell signaling can direct either binary or graded transcriptional responses. EMBO J. 2001;20:3167–3176. doi: 10.1093/emboj/20.12.3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigger J. Treatment of Staphylococcal Infections with Penicillin by Intermittent Sterilization. The Lancet. 1944;244:497–500. [Google Scholar]
- Bonifield HR, Hughes KT. Flagellar phase variation in Salmonella enterica is mediated by a posttranscriptional control mechanism. J Bacteriol. 2003;185:3567–3574. doi: 10.1128/JB.185.12.3567-3574.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Arkin AP. Sequestration-based bistability enables tuning of the switching boundaries and design of a latch. Mol Syst Biol. 2012;8:620. doi: 10.1038/msb.2012.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevance FF, Hughes KT. Coordinating assembly of a bacterial macromolecular machine. Nat Rev Microbiol. 2008;6:455–465. doi: 10.1038/nrmicro1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chubiz JE, Golubeva YA, Lin D, Miller LD, Slauch JM. FliZ regulates expression of the Salmonella pathogenicity island 1 invasion locus by controlling HilD protein activity in Salmonella enterica serovar typhimurium. J Bacteriol. 2010;192:6261–6270. doi: 10.1128/JB.00635-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung JD, Stephanopoulos G, Ireton K, Grossman AD. Gene expression in single cells of Bacillus subtilis: evidence that a threshold mechanism controls the initiation of sporulation. J Bacteriol. 1994;176:1977–1984. doi: 10.1128/jb.176.7.1977-1984.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clegg S, Hughes KT. FimZ is a molecular link between sticking and swimming in Salmonella enterica serovar Typhimurium. J Bacteriol. 2002;184:1209–1213. doi: 10.1128/jb.184.4.1209-1213.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormack BP, Valdivia RH, Falkow S. FACS-optimized mutants of the green fluorescent protein (GFP) Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- Cummings LA, Wilkerson WD, Bergsbaken T, Cookson BT. In vivo, fliC expression by Salmonella enterica serovar Typhimurium is heterogeneous, regulated by ClpX, and anatomically restricted. Mol Microbiol. 2006;61:795–809. doi: 10.1111/j.1365-2958.2006.05271.x. [DOI] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubnau D, Losick R. Bistability in bacteria. Mol Microbiol. 2006;61:564–572. doi: 10.1111/j.1365-2958.2006.05249.x. [DOI] [PubMed] [Google Scholar]
- Ellermeier CD, Slauch JM. RtsA and RtsB coordinately regulate expression of the invasion and flagellar genes in Salmonella enterica serovar Typhimurium. J Bacteriol. 2003;185:5096–5108. doi: 10.1128/JB.185.17.5096-5108.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elowitz MB, Levine AJ, Siggia ED, Swain PS. Stochastic gene expression in a single cell. Science. 2002;297:1183–1186. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- Fuchs G, Kroger A. Growth and Nutrition. In: Lengler JW, Drews G, Schlegel HG, editors. Biology of the Prokaryotes. Georg Thieme Verlag; Stuttgart, Germany: 1999. [Google Scholar]
- Hisert KB, MacCoss M, Shiloh MU, Darwin KH, Singh S, Jones RA, et al. A glutamate-alanine-leucine (EAL) domain protein of Salmonella controls bacterial survival in mice, antioxidant defence and killing of macrophages: role of cyclic diGMP. Mol Microbiol. 2005;56:1234–1245. doi: 10.1111/j.1365-2958.2005.04632.x. [DOI] [PubMed] [Google Scholar]
- Hughes KT, Gillen KL, Semon MJ, Karlinsey JE. Sensing structural intermediates in bacterial flagellar assembly by export of a negative regulator. Science. 1993;262:1277–1280. doi: 10.1126/science.8235660. [DOI] [PubMed] [Google Scholar]
- Igoshin OA, Alves R, Savageau MA. Hysteretic and graded responses in bacterial two-component signal transduction. Mol Microbiol. 2008;68:1196–1215. doi: 10.1111/j.1365-2958.2008.06221.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyoda S, Kamidoi T, Hirose K, Kutsukake K, Watanabe H. A flagellar gene fliZ regulates the expression of invasion genes and virulence phenotype in Salmonella enterica serovar Typhimurium. Microb Pathog. 2001;30:81–90. doi: 10.1006/mpat.2000.0409. [DOI] [PubMed] [Google Scholar]
- Jonas K, Edwards AN, Ahmad I, Romeo T, Romling U, Melefors O. Complex regulatory network encompassing the Csr, c-di-GMP and motility systems of Salmonella Typhimurium. Environ Microbiol. 2010;12:524–540. doi: 10.1111/j.1462-2920.2009.02097.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalir S, Mangan S, Alon U. A coherent feed-forward loop with a SUM input function prolongs flagella expression in Escherichia coli. Mol Syst Biol. 2005;1:2005.0006. doi: 10.1038/msb4100010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlinsey JE, Tanaka S, Bettenworth V, Yamaguchi S, Boos W, Aizawa SI, Hughes KT. Completion of the hook-basal body complex of the Salmonella typhimurium flagellum is coupled to FlgM secretion and fliC transcription. Mol Microbiol. 2000;37:1220–1231. doi: 10.1046/j.1365-2958.2000.02081.x. [DOI] [PubMed] [Google Scholar]
- Komeda Y, Suzuki H, Ishidsu JI, Iino T. The role of cAMP in flagellation of Salmonella typhimurium. Mol Gen Genet. 1976;142:289–298. doi: 10.1007/BF00271253. [DOI] [PubMed] [Google Scholar]
- Kröger C, Colgan A, Srikumar S, Händler K, Sivasankaran SK, Hammarlöf DL, et al. An infection-relevant transcriptomic compendium for Salmonella enterica Serovar Typhimurium. Cell Host Microbe. 2013;14:683–695. doi: 10.1016/j.chom.2013.11.010. [DOI] [PubMed] [Google Scholar]
- Kutsukake K. Autogenous and global control of the flagellar master operon, flhD, in Salmonella typhimurium. Mol Gen Genet. 1997;254:440–448. doi: 10.1007/s004380050437. [DOI] [PubMed] [Google Scholar]
- Kutsukake K, Ikebe T, Yamamoto S. Two novel regulatory genes, fliT and fliZ, in the flagellar regulon of Salmonella. Genes Genet Syst. 1999;74:287–292. doi: 10.1266/ggs.74.287. [DOI] [PubMed] [Google Scholar]
- Lawhon SD, Frye JG, Suyemoto M, Porwollik S, McClelland M, Altier C. Global regulation by CsrA in Salmonella typhimurium. Mol Microbiol. 2003;48:1633–1645. doi: 10.1046/j.1365-2958.2003.03535.x. [DOI] [PubMed] [Google Scholar]
- Li B, Li N, Wang F, Guo L, Huang Y, Liu X, et al. Structural insight of a concentration-dependent mechanism by which YdiV inhibits Escherichia coli flagellum biogenesis and motility. Nucleic Acids Res. 2012;40:11073–11085. doi: 10.1093/nar/gks869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin D, Rao CV, Slauch JM. The Salmonella SPI1 type three secretion system responds to periplasmic disulfide bond status via the flagellar apparatus and the RcsCDB system. J Bacteriol. 2008;190:87–97. doi: 10.1128/JB.01323-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas RL, Lostroh CP, DiRusso CC, Spector MP, Wanner BL, Lee CA. Multiple factors independently regulate hilA and invasion gene expression in Salmonella enterica serovar typhimurium. J Bacteriol. 2000;182:1872–1882. doi: 10.1128/jb.182.7.1872-1882.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maloy SR, Stewart VJ, Taylor RK. Genetic Analysis of Pathogenic Bacteria. New York: Cold Spring Harbor Laboratory Press; 1996. [Google Scholar]
- Miao EA, Leaf IA, Treuting PM, Mao DP, Dors M, Sarkar A, et al. Caspase-1-induced pyroptosis is an innate immune effector mechanism against intracellular bacteria. Nat Immunol. 2010;11:1136–1142. doi: 10.1038/ni.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouslim C, Hughes KT. The Effect of Cell Growth Phase on the Regulatory CrossTalk between Flagellar and Spi1 Virulence Gene Expression. PLoS Pathog. 2014;10:e1003987. doi: 10.1371/journal.ppat.1003987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyed HS, Bertrand KP. hipA, a newly recognized gene of Escherichia coli K-12 that affects frequency of persistence after inhibition of murein synthesis. J Bacteriol. 1983;155:768–775. doi: 10.1128/jb.155.2.768-775.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman JR, Fuqua C. Broad-host-range expression vectors that carry the L-arabinose-inducible Escherichia coli araBAD promoter and the araC regulator. Gene. 1999;227:197–203. doi: 10.1016/s0378-1119(98)00601-5. [DOI] [PubMed] [Google Scholar]
- Ray JC, Tabor JJ, Igoshin OA. Non-transcriptional regulatory processes shape transcriptional network dynamics. Nat Rev Microbiol. 2011;9:817–828. doi: 10.1038/nrmicro2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini S, Brown JD, Aldridge PD, Rao CV. FliZ Is a posttranslational activator of FlhD4C2-dependent flagellar gene expression. J Bacteriol. 2008;190:4979–4988. doi: 10.1128/JB.01996-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini S, Koirala S, Floess E, Mears PJ, Chemla YR, Golding I, et al. FliZ induces a kinetic switch in flagellar gene expression. J Bacteriol. 2010;192:6477–6481. doi: 10.1128/JB.00751-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schechter LM, Damrauer SM, Lee CA. Two AraC/XylS family members can independently counteract the effect of repressing sequences upstream of the hilA promoter. Mol Microbiol. 1999;32:629–642. doi: 10.1046/j.1365-2958.1999.01381.x. [DOI] [PubMed] [Google Scholar]
- Singer HM, Erhardt M, Hughes KT. RflM functions as a transcriptional repressor in the autogenous control of the Salmonella Flagellar master operon flhDC. J Bacteriol. 2013;195:4274–4282. doi: 10.1128/JB.00728-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer HM, Kühne C, Deditius JA, Hughes KT, Erhardt M. The Salmonella Spi1 virulence regulatory protein HilD directly activates transcription of the flagellar master operon flhDC. J Bacteriol. 2014;196:1448–1457. doi: 10.1128/JB.01438-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slauch JM, Silhavy TJ. cis-acting ompF mutations that result in OmpR-dependent constitutive expression. J Bacteriol. 1991;173:4039–4048. doi: 10.1128/jb.173.13.4039-4048.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits WK, Eschevins CC, Susanna KA, Bron S, Kuipers OP, Hamoen LW. Stripping Bacillus: ComK auto-stimulation is responsible for the bistable response in competence development. Mol Microbiol. 2005;56:604–614. doi: 10.1111/j.1365-2958.2005.04488.x. [DOI] [PubMed] [Google Scholar]
- Spory A, Bosserhoff A, von Rhein C, Goebel W, Ludwig A. Differential regulation of multiple proteins of Escherichia coli and Salmonella enterica serovar Typhimurium by the transcriptional regulator SlyA. J Bacteriol. 2002;184:3549–3559. doi: 10.1128/JB.184.13.3549-3559.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stecher B, Barthel M, Schlumberger MC, Haberli L, Rabsch W, Kremer M, Hardt WD. Motility allows S. Typhimurium to benefit from the mucosal defence. Cell Microbiol. 2008;10:1166–1180. doi: 10.1111/j.1462-5822.2008.01118.x. [DOI] [PubMed] [Google Scholar]
- Stewart MK, Cummings LA, Johnson ML, Berezow AB, Cookson BT. Regulation of phenotypic heterogeneity permits Salmonella evasion of the host caspase-1 inflammatory response. Proc Natl Acad Sci U S A. 2011;108:20742–20747. doi: 10.1073/pnas.1108963108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm A, Heinemann M, Arnoldini M, Benecke A, Ackermann M, Benz M, et al. The cost of virulence: retarded growth of Salmonella Typhimurium cells expressing type III secretion system 1. PLoS Pathog. 2011;7:e1002143. doi: 10.1371/journal.ppat.1002143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaya A, Matsui M, Tomoyasu T, Kaya M, Yamamoto T. The DnaK chaperone machinery converts the native FlhD2C2 hetero-tetramer into a functional transcriptional regulator of flagellar regulon expression in Salmonella. Mol Microbiol. 2006;59:1327–1340. doi: 10.1111/j.1365-2958.2005.05016.x. [DOI] [PubMed] [Google Scholar]
- Takaya A, Erhardt M, Karata K, Winterberg K, Yamamoto T, Hughes KT. YdiV: a dual function protein that targets FlhDC for ClpXP-dependent degradation by promoting release of DNA-bound FlhDC complex. Mol Microbiol. 2012;83:1268–1284. doi: 10.1111/j.1365-2958.2012.08007.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temme K, Salis H, Tullman-Ercek D, Levskaya A, Hong SH, Voigt CA. Induction and relaxation dynamics of the regulatory network controlling the type III secretion system encoded within Salmonella pathogenicity island 1. J Mol Biol. 2008;377:47–61. doi: 10.1016/j.jmb.2007.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomoyasu T, Ohkishi T, Ukyo Y, Tokumitsu A, Takaya A, Suzuki M, et al. The ClpXP ATP-dependent protease regulates flagellum synthesis in Salmonella enterica serovar typhimurium. J Bacteriol. 2002;184:645–653. doi: 10.1128/JB.184.3.645-653.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veening JW, Smits WK, Kuipers OP. Bistability, epigenetics, and bet-hedging in bacteria. Annu Rev Microbiol. 2008;62:193–210. doi: 10.1146/annurev.micro.62.081307.163002. [DOI] [PubMed] [Google Scholar]
- Wada T, Morizane T, Abo T, Tominaga A, Inoue-Tanaka K, Kutsukake K. EAL domain protein YdiV acts as an anti-FlhD4C2 factor responsible for nutritional control of the flagellar regulon in Salmonella enterica Serovar Typhimurium. J Bacteriol. 2011a;193:1600–1611. doi: 10.1128/JB.01494-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada T, Tanabe Y, Kutsukake K. FliZ acts as a repressor of the ydiV gene, which encodes an anti-FlhD4C2 factor of the flagellar regulon in Salmonella enterica serovar typhimurium. J Bacteriol. 2011b;193:5191–5198. doi: 10.1128/JB.05441-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Zhao Y, McClelland M, Harshey RM. The RcsCDB signaling system and swarming motility in Salmonella enterica serovar typhimurium: dual regulation of flagellar and SPI-2 virulence genes. J Bacteriol. 2007;189:8447–8457. doi: 10.1128/JB.01198-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei BL, Brun-Zinkernagel AM, Simecka JW, Prüss BM, Babitzke P, Romeo T. Positive regulation of motility and flhDC expression by the RNA-binding protein CsrA of Escherichia coli. Mol Microbiol. 2001;40:245–256. doi: 10.1046/j.1365-2958.2001.02380.x. [DOI] [PubMed] [Google Scholar]
- Wozniak CE, Lee C, Hughes KT. T-POP array identifies EcnR and PefI-SrgD as novel regulators of flagellar gene expression. J Bacteriol. 2009;191:1498–1508. doi: 10.1128/JB.01177-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak CE, Chevance FF, Hughes KT. Multiple promoters contribute to swarming and the coordination of transcription with flagellar assembly in Salmonella. J Bacteriol. 2010;192:4752–4762. doi: 10.1128/JB.00093-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto S, Kutsukake K. FliT acts as an anti-FlhD2C2 factor in the transcriptional control of the flagellar regulon in Salmonella enterica serovar typhimurium. J Bacteriol. 2006;188:6703–6708. doi: 10.1128/JB.00799-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.