Abstract
Peroxisomes lack their own genetic material and must therefore import proteins encoded by genes in the nucleus. Amino acids within these proteins serve as targeting signals: they direct the delivery of the proteins to the organelle. The majority of soluble proteins destined for the peroxisomal matrix utilize a type 1 peroxisomal targeting signal (PTS1): a C-terminal tripeptide that follows the pattern small/basic/hydrophobic. We have discovered two new C-terminal tripeptides that target proteins to peroxisomes in Arabidopsis thaliana. The tripeptides PSL and KRR do not fit the major PTS1 consensus but cause green fluorescent protein to accumulate in peroxisomes of stably transformed Arabidopsis. We have identified forty-one proteins in the Arabidopsis genome that also bear these tripeptides at their C-termini and may therefore be peroxisomal.
Keywords: Peroxisome, PTS1, Arabidopsis thaliana
Introduction
Peroxisomes are single membrane bounded organelles found in nearly all eukaryotes (Schluter et al., 2006). In plants, the dominant functions of peroxisomes are photorespiration and β-oxidation of fatty acids. Peroxisomes also play important roles in a number of other metabolic pathways including synthesis of the hormones jasmonic acid and auxin (reviewed in Hu et al., 2012). Recently, proteomic studies of purified peroxisomes have identified still more enzyme activities in these organelles (reviewed in Reumann, 2011). Another approach to the discovery of novel peroxisomal enzymes and functions is bioinformatics-based: the identification of peroxisomal proteins based on genome data. This strategy is made possible by the fact that peroxisomes lack their own genome and are forced to import nuclear-encoded proteins. Proteins destined for the peroxisomes carry one of a number of signaling peptides that target them to the organelle. Bioinformatic approaches depend on the ability of computer algorithms to identify these signaling peptides in amino acid sequences.
The main import pathway for peroxisomal matrix proteins utilizes the type 1 peroxisomal targeting signal (PTS1). The PTS1 consists of a C-terminal tripeptide. In plants, major PTS1s direct protein accumulation to peroxisomes regardless of upstream sequences and follow the consensus [SA][RK][LM]> (where > denotes the C-terminus of the protein) (Reumann, 2004). Minor PTS1s direct peroxisomal accumulation only when certain upstream enhancing sequences are present. Allowing for these minor PTS1s, the overall PTS1 consensus broadens to [SAPC][RKNMSLH][LMIVY]> (Lingner et al., 2011).
Other peroxisomal proteins utilize a number of less well characterized pathways. Some soluble proteins use a type 2 peroxisomal targeting signal (PTS2) which consists of an N-terminal nonapeptide that follows the consensus R[ILQ]X5HL (Kato et al., 1998). A membrane peroxisomal targeting signal (mPTS) consisting of an internal stretch of five amino acids heavy in basic residues has also been proposed for integral membrane proteins (Dyer et al., 1996; Mullen et al., 2000). An internal PTS1 (QKL) has been identified within pumpkin catalase (Kamigaki et al., 2003). Finally, a “piggyback” method has been reported that allows proteins to be imported into peroxisomes by protein-protein interaction with another protein carrying its own peroxisomal targeting signal (Lee et al., 1997).
Here, we report in vivo verification of two new PTS1 sequences that can target green fluorescent protein (GFP) to peroxisomes. These C-terminal tripeptides are not predicted to be PTS1s by existing prediction algorithms. Peroxisomal targeting by these sequences suggests that forty-one Arabidopsis proteins containing the same C-terminal tripeptides may also be peroxisomal. Incorporating these newly confirmed PTS1s into prediction algorithms may improve future bioinformatic investigations of peroxisome biology.
Material and methods
Plant material and growth conditions
Transgenic Arabidopsis thaliana lines were obtained from the Arabidopsis Biological Resource Center (Columbus, OH, USA). Seeds were surface sterilized in 10% sodium hypochlorite and 0.2% Tween-20 and then stratified for 2–4 days at 4°C in darkness. Seedlings were germinated on sterile media containing Murashige and Skoog basal medium (Murashige et al., 1962), Gamborg’s vitamins (Gamborg et al., 1968), 3% (w/v) sucrose, and 0.85% (w/v) agar. Mature plants were grown on Miracle Grow potting soil (Scotts Miracle-Grow, Marysville, OH, USA).
Microscopy
Live samples of rosette leaves and dark-grown seedlings were mounted on slides in water. Fluorescence of GFP-cDNA fusion proteins was documented by laser scanning confocal microscopy using a Leica HC PL APO 63× objective (N.A. = 1.4). GFP was excited with 488nm light and fluorescence was detected between 500nm and 600nm. Colocalization of GFP-cDNA fusions and peroxisome-targeted mCherry was documented by wide-field epifluorescence microscopy using 40× (N.A. = 0.95) and 100× (N.A. = 1.4) Zeiss Plan Apochromat objectives. GFP fluorescence was detected with a Zeiss 38 filter set: excitation (470/40nm), dichroic (495nm), emission (525/50nm). mCherry fluorescence was detected with a Chroma 49306 filter set: excitation (580/25nm), dichroic (600nm), emission (625/30nm).
Contrast of micrographs was enhanced by adjusting maximum and minimum grey levels with Photoshop (Adobe Systems, San Jose, CA, USA). Grey levels were adjusted linearly to preserve relative brightness of different structures.
Transient expression by Agrobacterium tumefaciens infiltration
Rosette leaves of Arabidopsis lines CS84743 and CS84812 were injected with Agrobacterium strain GV3101 containing the vector described by Nelson et al. (2007) as px-rk (CD3-983). This vector causes high-level expression of mCherry fluorescent protein fused to the PTS1 SKL>. Transformed leaves were observed 24–48 hours after infection.
Molecular biology
Genomic DNA was isolated from mature rosette leaves using a PureLink Plant Total DNA Purification Kit (Invitrogen, Carlsbad, CA, USA). GFP-cDNA inserts were amplified with the pEGAD forward and reverse primers described by Cutler et al. (2000). PCR products were cloned into the pJET1.2 (Fermentas, Pittsburg, PA, USA) or pCR-BluntII-TOPO (Invitrogen) cloning vectors. Cloned PCR products were sequenced by Sanger sequencing (Laragen, Culver City, CA, USA).
Results
Our experiments began while investigating a pool of transgenic Arabidopsis created by Sean Cutler and David Ehrhardt and colleagues (2000). These researchers had generated a library of Arabidopsis cDNAs that were fused at their 5′ ends to the coding region for GFP (GFP-cDNA). These GFP-cDNA fusions were then inserted into Arabidopsis under the control of the constitutive CaMV 35S promoter. The vast majority of transgenic plants recovered showed GFP fluorescence in the nucleus and cytosol (Cutler et al., 2000). This result was expected since lack of signal peptides by GFP causes it to accumulate in the cytosol and its small size allows it to diffuse through the nuclear pore complex. More interestingly, Cutler and colleagues also observed several classes of plants that showed GFP accumulation in unique structures. These structures did not appear to be any of the membrane bounded organelles commonly found in plant cells. Cutler and colleagues called one of these structural classes Q-balls: GFP fluorescence appeared as a small ring with a bright sphere attached. They identified eight independent transgenic lines as belonging to the Q-ball class.
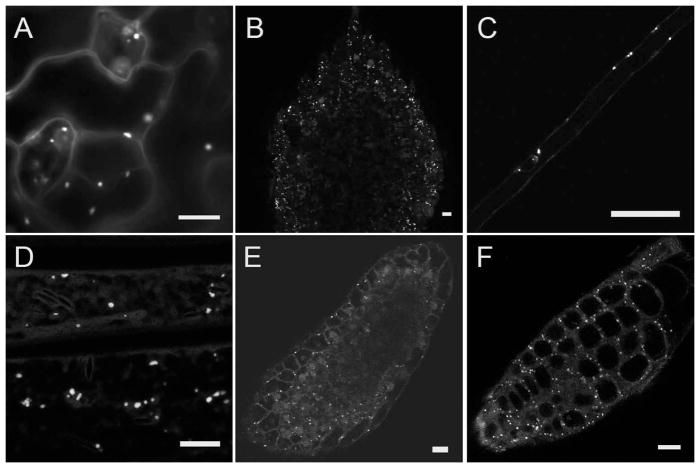
Curious about these novel Q-ball structures, we further examined these eight transgenic lines. However, observation of two of the Q-ball lines, CS84743 and CS84812, showed only circular structures about 1μm in diameter (Fig. 1). These structures were present in all organs investigated. Multiple circular structures were observed within each cell and some were motile (data not shown). A low level of GFP-cDNA fluorescence was also detected in the cytosol and nuclei of these cells. The fluorescent structures in Arabidopsis lines CS84743 and CS84812 appear identical to fluorescently labeled peroxisomes from previous experiments in plants (Mano et al., 1999; Mathur et al., 2002). Mathur and colleagues investigated matrix-targeted GFP in Arabidopsis peroxisomes and observed circular structures with a diameter between 0.8 and 1.9μm, similar to ours (Mathur et al., 2002). These same authors also observed movement of peroxisomes and showed that motility was actin-dependent. The fusion proteins in lines CS84743 and CS84812 appeared matrix-targeted and not membrane-localized because fluorescent protein fusions to peroxisomal membrane proteins appear as rings around the periphery of the organelle (Reumann et al., 2007), a conformation not observed in these lines. Finally, overexpression of peroxisome-targeted GFP often results in accumulation in the cytosol, possibly due to overloading of the cytosolic carrier proteins that shepherd proteins to peroxisomes (Reumann et al., 2007).
Figure 1.
GFP-cDNA fusion proteins in Arabidopsis lines CS84743 and CS84812 accumulate in circular structures about 1μm in diameter. Live samples of lines CS84743 (A–C) and CS84812 (D–F) were observed by epifluorescence and laser scanning confocal microscopy. Fluorescent structures were observed in all organs examined, including leaf epidermis (A), cotyledons (B), root hairs (C), etiolated hypocotyl epidermis (D), leaf primordia (E), and root tips (F). Faint GFP fluorescence is sometimes visible in nuclei and in the cortical cytoplasm. Scale bars = 10μm.
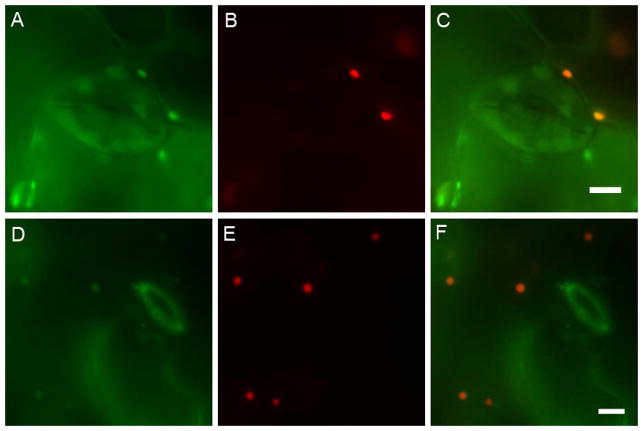
To test the hypothesis that these transgenic lines contained GFP-cDNA-labeled peroxisomes, mCherry fluorescent protein carrying an established PTS1 (SKL>) (Nelson et al., 2007) was transiently expressed in these plants. Colocalization was performed by infiltrating Agrobacterium carrying the peroxisome-targeted mCherry into mature leaves of lines CS84743 and CS84812. In successfully transformed epidermal cells, the green fluorescence of the GFP-cDNA fusions overlapped the red fluorescence of the peroxisome-targeted mCherry (Fig. 2). The overlap between the two fusion proteins confirms that GFP-cDNA fusions are targeting to the matrix of peroxisomes. Agroinfiltration resulted in only a fraction of cells expressing the mCherry protein in peroxisomes. However, this served as an internal control for the fluorescence filter sets: cells not transformed with mCherry-PTS1 (but still carrying the GFP-cDNA transgenes) did not show any signal in the mCherry channel, confirming that the mCherry filter set is specific for mCherry and does not allow bleed-through of GFP fluorescence. Likewise, the GFP filter set does not detect mCherry fluorescent protein (Supplementary Figure 1). All cells containing both GFP-cDNA and mCherry-PTS1 signals showed a 100% overlap of the two fluorophores.
Figure 2.
Peroxisome-targeted mCherry fluorescent protein colocalizes with GFP-cDNA fusion proteins in Arabidopsis lines CS84743 and CS84812. Rosette leaves of Arabidopsis lines CS84743 (A–C) and CS84812 (D–F) were infiltrated with Agrobacterium containing a peroxisome-targeted mCherry fluorescent protein. Epifluorescence images of leaf epidermis show fluorescence from GFP-cDNA fusions (A,D), peroxisome-targeted mCherry fluorescent protein (B,E), and an overlay of the two fluorophores (C,F). GFP-labeled organelles in cells that were not transformed with the peroxisome-targeted mCherry showed fluorescence only in the GFP and merged channels (arrows). Scale bars= 5μm.
Once it was demonstrated that the GFP-cDNA fusions were accumulating in peroxisomes, the amino acids responsible for peroxisomal targeting were investigated. PCR analysis of genomic DNA from each transgenic line revealed a single GFP-cDNA transgene in each plant (data not shown). Cloning and sequencing of the PCR products revealed the GFP coding region followed by a short linker region and the appended cDNA sequences. The linker region is as described by Cutler et al. (2000): 10 Alanines (Ala10) followed by Glu Phe (EF) encoded by the EcoRI restriction site used to join the linker to the cDNA. Both CS84743 and CS84812 contained cDNAs that were fused to GFP out-of-frame: the fusion proteins did not contain the amino acid sequences specified by the original genes.
The GFP-cDNA fusion protein in Line CS84743 consists of the 4 amino acid sequence IKRR appended to GFP (GFP-Ala10-EF-IKRR). The nucleotide sequence of the cDNA is significantly longer and database analysis indicates that it is not from Arabidopsis: it is a contaminating sequence of human DNA; specifically, from the MAGI3 gene on human chromosome 1. The sequence is a fragment from the first intron of the human gene, so the amino acids fused to GFP do not represent the amino acids of the MAGI3 protein. KRR> is not a known or predicted PTS1. Only the R at the −2 position (counting from the c-terminal end) matches the consensus of a major PTS1 as defined by Reumann et al. (2004). The PredPlantPTS1 algorithm, which considers the 14 C-terminal amino acids of a protein, does not identify the cDNA sequence as a PTS1 either (Reumann et al., 2012). Indeed, of the 8 Ala and EF that are included in the PredPlantPTS1 analysis, only the Ala at −11 gives a positive score for peroxisomal targeting (data not shown). Based on our reading of the proteomic studies conducted in plants, KRR> has never been found on a protein purified from peroxisomes of Arabidopsis (Fukao et al., 2002; Fukao et al., 2003; Reumann et al., 2007; Eubel et al., 2008; Reumann et al., 2009), soybean (Arai et al., 2008), or spinach (Babujee et al., 2010). Furthermore, the KRR> tripeptide has never been validated as a PTS1 by other experimental methods (Reumann et al., 2012).
The GFP-cDNA fusion protein in Line CS84812 consists of the 4 amino acid sequence KPSL appended to GFP (GFP-Ala10-EF-KPSL). As with the previous line, the nucleotide sequence of the cDNA is much longer. Here, the cDNA is derived from the overlapping untranslated regions (UTRs) of two Arabidopsis genes: the 3′UTR of an Arabidopsis Na+/H+ antiporter (AT3G19490) and the 5′UTR of Arabidopsis D-3-phosphoglycerate dehydrogenase (AT3G19480). The PSL tripeptide falls within the consensus of a minor PTS1. In addition, the K at the −4 position has been shown to improve targeting of minor PTS1s (Mullen et al., 1997; Reumann, 2004; Lingner et al., 2011). However, when the entire C-terminal sequence is evaluated by the PredPlantPTS1 algorithm it is not predicted to be peroxisomal (Lingner et al., 2011). Again, all but one of the 10 amino acids upstream of KPSL are negative contributors to peroxisomal targeting. The PSL> tripeptide has never been identified in proteomic studies of Arabidopsis (Fukao et al., 2002; Fukao et al., 2003; Reumann et al., 2007; Eubel et al., 2008; Reumann et al., 2009), soybean (Arai et al., 2008a), or spinach (Babujee et al., 2010). Furthermore, the PSL tripeptide has never been validated as a plant PTS1 by other experimental methods (Reumann et al., 2012).
The discovery that KRR> and PSL> can serve as PTS1s suggested that other proteins bearing the same C-terminal tripeptides might also be peroxisome-targeted. A search of the Arabidopsis genome revealed twenty-seven proteins that share the KRR> tripeptide and fourteen proteins that share the PSL> tripeptide (Table 1). None of these proteins are predicted to be peroxisomal by the PredPlantPTS1 algorithm (Lingner et al., 2011). In addition, none of these proteins have functions that are associated with known major peroxisomal activities. However, some of the proteins on the list bear further investigation on the chance that they are actually peroxisome-targeted. The presence of the kinases and kinase inhibitor mirrors reports from several proteomics experiments: other kinases/phosphatases have been found in peroxisomes and some peroxisomal proteins have been identified as phosphorylated (reviewed in Bussell et al., 2013). The four ABC transporters are particularly interesting since the peroxisomal CTS/PXA1/PED3 fatty acid importer is also an ABC transporter (reviewed in Hu et al., 2012). This is interesting because few peroxisomal metabolite transporters are known and proteomic identification of peroxisomal membrane proteins is difficult (Reumann, 2011).
Table 1.
Arabidopsis proteins that share the KRR> tripeptide with line CS84743 or the PSL> tripeptide of line CS84812
| Gene ID | Arabidopsis Proteins ending KRR> |
|---|---|
| AT1G34310 | ARF12: auxin response factor 12 |
| AT1G35520 | ARF15: auxin response factor 15 |
| AT3G25900 | HMT1: Homocysteine S-methyltransferase |
| AT5G40530 | S-adenosyl-L-met-dependent methyltransferase |
| AT1G15110 | PSS1: base-exchange-type phosphatidylserine synthase |
| AT5G67290 | FAD-dependent oxidoreductase |
| AT5G19670 | Exostosin-like |
| AT2G02880 | Mucin-related |
| AT1G03760 | Prefoldin chaperone subunit |
| AT5G13290 | CRN: pseudokinase |
| AT4G24400 | CIPK08: CBL-interacting protein kinase 8 |
| AT1G79200 | SCI1: cyclin dependent kinase inhibitor |
| AT1G13790 | FDM4: SGS3 homolog for DNA methylation |
| AT1G30460 | CPSF30: cleavage and polyadenylation specificity factor 30 |
| AT3G25440 | RNA-binding CRS1 / YhbY (CRM) domain protein |
| AT2G23340 | DEAR3: DREB and EAR motif protein 3 |
| AT5G49400 | Zinc knuckle (CCHC-type) family protein |
| AT4G16630 | DEA(D/H)-box RNA helicase 28 |
| AT2G42650 | Ribosomal protein L1p/L10e family |
| AT2G39350 | ABCG1:ABC transporter G-family |
| AT5G13580 | ABCG6: ABC transporter G-family |
| AT3G55090 | ABCG16: ABC transporter G-family |
| AT3G53510 | ABCG20: ABC transporter G-family |
| AT1G76730 | COG0212: unknown |
| AT2G33420 | unknown |
| AT3G01060 | unknown |
| AT1G04470 | unknown |
| Gene ID | Arabidopsis Proteins ending PSL> |
|---|---|
| AT1G77130 | GUX3/PGSIP2: glucuronyl transferase 3 |
| AT3G52790 | peptidoglycan-binding LysM domain-containing |
| AT1G51610 | MTPc4: metal transporter/cation efflux 4 |
| AT2G30390 | FC2: ferrochelatase 2 |
| AT2G22840 | GRF1: growth-regulating factor 1 |
| AT3G05640 | Serine/threonine phosphatase 2C family Group E |
| AT3G24550 | PERK1: proline-rich extensin-like receptor kinase 1 |
| AT3G20110 | CYP705A20: cytochrome P450 |
| AT4G32180 | PANK2: pantothenate kinase 2 |
| AT1G61000 | Nuf2 domain-containing protein |
| AT2G26920 | Ubiquitin-associated/translation elongation factor |
| AT5G12120 | Ubiquitin-associated/translation elongation factor |
| AT4G35270 | NLP2: NIN-like protein 2 |
| AT1G47640 | unknown |
Discussion
Based on our observations of Arabidopsis transgenic lines CS84743 and CS84812, we have determined that the C-terminal peptides IKRR> and KPSL> direct GFP to accumulate in peroxisomes. Unfortunately, these amino acid sequences were derived from short stretches of the non-coding regions of genes. Hence these data do not report the location of native Arabidopsis proteins. However, this work is similar to investigations that have used synthetic fusion proteins to test the flexibility of the PTS1 pathway for delivering proteins to peroxisomes (Mullen et al., 1997; Reumann et al., 2007; Lingner et al., 2011). These studies have been important in developing and testing algorithms for predicting PTS1s from amino acid sequence data. In several cases, tripeptides following the PTS1 consensus and predicted to be PTS1s by search algorithms have been incapable of targeting fluorescent proteins to peroxisomes (Lingner et al., 2011).
The PSL> tripeptide found in line CS84812 falls within the consensus of a minor PTS1 and has an enhancing K residue at the −4 position, so it is not surprising to see it functioning as a PTS1. In contrast, the KRR> tripeptide found in line CS84743 is well outside even the minor PTS1 consensus. This raises the question of whether IKRR> directs the fusion protein to utilize the PTS1 pathway or some other peroxisomal import pathway. IKRR> is probably not functioning as a PTS2 or mPTS since the sequence is not N-terminal, nor is the fusion protein in the peroxisomal membranes. One possibility is that the Ala10-EF-IKRR sequence allows the fusion protein to interact with another protein destined for peroxisomes by a protein-protein “piggyback” mechanism. Another question raised by our results is why these tripeptide sequences have never been found on peroxisomal proteins, even by proteomic studies. It is possible that the Arabidopsis proteins that contain these tripeptide sequences accumulate too low concentrations under native conditions, levels too low to be detected by proteomic analysis (Reumann, 2011). Furthermore, proteomics studies have only been conducted on peroxisomes purified from a limited number of organs under a small number of environmental/developmental conditions (Reumann, 2011). It is possible that the Arabidopsis proteins that utilize KRR> and PSL> are expressed in limited regions of the plant or under limited circumstances.
The discovery of these two new PTS1 sequences may alter our understanding of peroxisome function beyond the requirements for protein import. We have identified forty-one Arabidopsis proteins that share the PSL> or KRR> tripeptides and may therefore be peroxisomal. We do not expect all of these proteins to be peroxisomal since even major PTS1s do not always target proteins to peroxisomes. Two major factors can override the presence of a PTS1: the presence of an N-terminal sequence for nuclear, mitochondrial, plastid, or ER targeting; and the burying of the PTS1 within the protein, where it is inaccessible by the import machinery (Neuberger et al., 2003; Wolf et al., 2010). In addition, minor PTS1s often require upstream enhancing sequences to promote successful targeting (Lingner et al., 2011). Whether any of the proteins identified in Table 1 are truly peroxisome-targeted could be determined experimentally by full length fusions to GFP. Alternatively, an informatics approach could find their homologs in other species and determine whether they also have PTS1-like sequences as has been shown for many confirmed peroxisome proteins (Reumann et al., 2012). Regardless, it seems appropriate to add these two targeting signals to the training sets used for prediction algorithms so that future bioinformatic searches for peroxisomal proteins will be more complete.
Supplementary Material
Fluorescence from Peroxisome-mCherry marker is not visible in the GFP channel. Mature leaves of wild-type Arabidopsis were infiltrated with Agrobacterium carrying a construct for peroxisome-targeted mCherry fluorescent protein. Epifluorescence images using GFP filter set (A) show only autofluorescence of chlorophyll in chloroplasts. Image of the same region using the mCherry filter set (B) shows peroxisomes labeled with mCherry fluorescent protein. Merged image (C) shows combined signals. Scale bar = 5μm.
Acknowledgments
This project was supported by Award Number SC2GM098160 from the National Institute of General Medical Sciences. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of General Medical Sciences or the National Institutes of Health. Special thanks to Paula M. Hauck for editing the manuscript.
Abbreviations
- PTS1
type 1 peroxisomal targeting signal
- GFP
green fluorescent protein
- UTR
untranslated region
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arai Y, Hayashi M, Nishimura M. Proteomic analysis of highly purified peroxisomes from etiolated soybean cotyledons. Plant Cell Physiol. 2008;49:526–39. doi: 10.1093/pcp/pcn027. [DOI] [PubMed] [Google Scholar]
- Babujee L, Wurtz V, Ma C, Lueder F, Soni P, van Dorsselaer A, et al. The proteome map of spinach leaf peroxisomes indicates partial compartmentalization of phylloquinone (vitamin K1) biosynthesis in plant peroxisomes. J Exp Bot. 2010;61:1441–53. doi: 10.1093/jxb/erq014. [DOI] [PubMed] [Google Scholar]
- Bussell JD, Behrens C, Ecke W, Eubel H. Arabidopsis peroxisome proteomics. Front Plant Sci. 2013;4:101. doi: 10.3389/fpls.2013.00101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler SR, Ehrhardt DW, Griffitts JS, Somerville CR. Random GFP::cDNA fusions enable visualization of subcellular structures in cells of Arabidopsis at a high frequency. Proc Natl Acad Sci U S A. 2000;97:3718–23. doi: 10.1073/pnas.97.7.3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer JM, McNew JA, Goodman JM. The sorting sequence of the peroxisomal integral membrane protein PMP47 is contained within a short hydrophilic loop. J Cell Biol. 1996;133:269–80. doi: 10.1083/jcb.133.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eubel H, Meyer EH, Taylor NL, Bussell JD, O’Toole N, Heazlewood JL, et al. Novel proteins, putative membrane transporters, and an integrated metabolic network are revealed by quantitative proteomic analysis of Arabidopsis cell culture peroxisomes. Plant Physiol. 2008;148:1809–29. doi: 10.1104/pp.108.129999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukao Y, Hayashi M, Hara-Nishimura I, Nishimura M. Novel glyoxysomal protein kinase, GPK1, identified by proteomic analysis of glyoxysomes in etiolated cotyledons of Arabidopsis thaliana. Plant Cell Physiol. 2003;44:1002–12. doi: 10.1093/pcp/pcg145. [DOI] [PubMed] [Google Scholar]
- Fukao Y, Hayashi M, Nishimura M. Proteomic analysis of leaf peroxisomal proteins in greening cotyledons of Arabidopsis thaliana. Plant Cell Physiol. 2002;43:689–96. doi: 10.1093/pcp/pcf101. [DOI] [PubMed] [Google Scholar]
- Gamborg OL, Miller RA, Ojima K. Nutrient requirements of suspension cultures of soybean root cells. Exp Cell Res. 1968;50:151–8. doi: 10.1016/0014-4827(68)90403-5. [DOI] [PubMed] [Google Scholar]
- Hu J, Baker A, Bartel B, Linka N, Mullen RT, Reumann S, et al. Plant peroxisomes: Biogenesis and function. Plant Cell. 2012;24:2279–303. doi: 10.1105/tpc.112.096586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamigaki A, Mano S, Terauchi K, Nishi Y, Tachibe-Kinoshita Y, Nito K, et al. Identification of peroxisomal targeting signal of pumpkin catalase and the binding analysis with PTS1 receptor. Plant J. 2003;33:161–75. doi: 10.1046/j.0960-7412.2003.001605.x. [DOI] [PubMed] [Google Scholar]
- Kato A, Takeda-Yoshikawa Y, Hayashi M, Kondo M, Hara-Nishimura I, Nishimura M. Glyoxysomal malate dehydrogenase in pumpkin: Cloning of a cDNA and functional analysis of its presequence. Plant Cell Physiol. 1998;39:186–95. doi: 10.1093/oxfordjournals.pcp.a029356. [DOI] [PubMed] [Google Scholar]
- Lee MS, Mullen RT, Trelease RN. Oilseed isocitrate lyases lacking their essential type 1 peroxisomal targeting signal are piggybacked to glyoxysomes. Plant Cell. 1997;9:185–97. doi: 10.1105/tpc.9.2.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingner T, Kataya AR, Antonicelli GE, Benichou A, Nilssen K, Chen XY, et al. Identification of novel plant peroxisomal targeting signals by a combination of machine learning methods and in vivo subcellular targeting analyses. Plant Cell. 2011;23:1556–72. doi: 10.1105/tpc.111.084095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur J, Mathur N, Hulskamp M. Simultaneous visualization of peroxisomes and cytoskeletal elements reveals actin and not microtubule-based peroxisome motility in plants. Plant Physiol. 2002;128:1031–45. doi: 10.1104/pp.011018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen RT, Lee MS, Flynn CR, Trelease RN. Diverse amino acid residues function within the type 1 peroxisomal targeting signal. Implications for the role of accessory residues upstream of the type 1 peroxisomal targeting signal. Plant Physiol. 1997;115:881–9. doi: 10.1104/pp.115.3.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen RT, Trelease RN. The sorting signals for peroxisomal membrane-bound ascorbate peroxidase are within its C-terminal tail. J Biol Chem. 2000;275:16337–44. doi: 10.1074/jbc.M001266200. [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiologia Plantarum. 1962;15:473–97. [Google Scholar]
- Nelson BK, Cai X, Nebenfuhr A. A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 2007;51:1126–36. doi: 10.1111/j.1365-313X.2007.03212.x. [DOI] [PubMed] [Google Scholar]
- Neuberger G, Maurer-Stroh S, Eisenhaber B, Hartig A, Eisenhaber F. Prediction of peroxisomal targeting signal 1 containing proteins from amino acid sequence. J Mol Biol. 2003;328:581–92. doi: 10.1016/s0022-2836(03)00319-x. [DOI] [PubMed] [Google Scholar]
- Reumann S. Specification of the peroxisome targeting signals type 1 and type 2 of plant peroxisomes by bioinformatics analyses. Plant Physiol. 2004;135:783–800. doi: 10.1104/pp.103.035584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reumann S. Toward a definition of the complete proteome of plant peroxisomes: Where experimental proteomics must be complemented by bioinformatics. Proteomics. 2011;11:1764–79. doi: 10.1002/pmic.201000681. [DOI] [PubMed] [Google Scholar]
- Reumann S, Babujee L, Ma C, Wienkoop S, Siemsen T, Antonicelli GE, et al. Proteome analysis of Arabidopsis leaf peroxisomes reveals novel targeting peptides, metabolic pathways, and defense mechanisms. Plant Cell. 2007;19:3170–93. doi: 10.1105/tpc.107.050989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reumann S, Buchwald D, Lingner T. PredPlantPTS1: A web server for the prediction of plant peroxisomal proteins. Front Plant Sci. 2012;3:194. doi: 10.3389/fpls.2012.00194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reumann S, Quan S, Aung K, Yang P, Manandhar-Shrestha K, Holbrook D, et al. In-depth proteome analysis of Arabidopsis leaf peroxisomes combined with in vivo subcellular targeting verification indicates novel metabolic and regulatory functions of peroxisomes. Plant Physiol. 2009;150:125–43. doi: 10.1104/pp.109.137703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluter A, Fourcade S, Ripp R, Mandel JL, Poch O, Pujol A. The evolutionary origin of peroxisomes: An ER-peroxisome connection. Mol Biol Evol. 2006;23:838–45. doi: 10.1093/molbev/msj103. [DOI] [PubMed] [Google Scholar]
- Wolf J, Schliebs W, Erdmann R. Peroxisomes as dynamic organelles: Peroxisomal matrix protein import. FEBS J. 2010;277:3268–78. doi: 10.1111/j.1742-4658.2010.07739.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fluorescence from Peroxisome-mCherry marker is not visible in the GFP channel. Mature leaves of wild-type Arabidopsis were infiltrated with Agrobacterium carrying a construct for peroxisome-targeted mCherry fluorescent protein. Epifluorescence images using GFP filter set (A) show only autofluorescence of chlorophyll in chloroplasts. Image of the same region using the mCherry filter set (B) shows peroxisomes labeled with mCherry fluorescent protein. Merged image (C) shows combined signals. Scale bar = 5μm.