Abstract
MicroRNAs post-transcriptionally regulate the expression of approximately 60% of the mammalian genes, and have an important role in maintaining the differentiated state of somatic cells through the expression of unique tissue-specific microRNA sets. Likewise, the stemness of pluripotent cells is also sustained by embryonic stem cell-enriched microRNAs, which regulate genes involved in cell cycle, cell signaling and epigenetics, among others. Thus, microRNAs work as modulator molecules that ensure the appropriate expression profile of each cell type. Manipulation of microRNA expression might determine the cell fate. Indeed, microRNA-mediated reprogramming can change the differentiated status of somatic cells towards stemness or, conversely, microRNAs can also transform stem- into differentiated-cells both in vitro and in vivo. In this Review, we outline what is currently known in this field, focusing on the applications of microRNA in tissue engineering.
Keywords: Cell fate, ESC, iPSC, microRNA, stemness, tissue engineering.
INTRODUCTION
MicroRNAs (miRNAs) are small non-coding RNAs (ncRNAs) of approximately 22 nucleotides responsible for specific regulation of gene expression in a post-transcriptional manner, and, thereby, have an important role in several biological processes, such as development, cell proliferation, and apoptosis, among others [1-3]. Their genes correspond to 1-3% of all genes of the genome [4], and may be responsible for the regulation of approximately 60% of the coding genes [5]. The specificity of miRNAs is given by the seed region (nucleotides 2 to 8) that requires full complementarity to the mRNA-target [6-8]. The RNase III members, Drosha and Dicer, together with their specific partners, Dgcr-8 and TRBP, are crucial for miRNAs biogenesis, since they slice, respectively, primary and precursor miRNAs to yield the mature ones [9-12].
In vertebrates, approximately 70% of the miRNA genes are intragenic, usually being expressed in synergy with the host gene [13]. Forty percent of all miRNAs are organized in clusters and transcribed in a polycistronic fashion. Usually, these clustered miRNAs are members of a family, showing overlapping functions [13, 14]. Each cell type has a combination of either isolated or clustered expressing miRNAs, which regulate coding genes in a tissue-specific manner and, therefore, they are essential to the maintenance of cell identity and functional phenotype [15]. In this article, we review miRNAs involved in pluripotency maintenance, cell fate decision, differentiation-state safeguarding, focusing on how this knowledge has been used in tissue engineering.
MiRNAs IN STEM CELLS
Embryonic stem cells (ESCs) are derived from pre-implantation blastocysts, and have the ability of self-renewal and the latent capacity to differentiate in cells of all three embryonic germ layers, therefore being pluripotent [16, reviewed in 17]. Because of these remarkable characteristics, ESCs have been extensively used as model in developmental and therapeutic studies, including tissue regeneration, transplantation, and drug screenings [18-21]. To sustain the undifferentiated state, they have a unique gene expression profile [22-26] regulated by a highly expressed set of transcriptional factors, including Oct4, Nanog, and Sox2 that, in this scenario, have overlapping functions [27-29]. In addition to OCT4 and NANOG, the most commonly used markers of ESCs are the cell surface antigens (SSEA-3, SSEA-4, TRA-1-60, TRA-1-81) and tissue-nonspecific alkaline phosphatase [30]. Mouse pluripotent stem cells can also be derived from the epiblast of peri-implantation blastocysts, and are called EpiSC [31, 32]. These are epigenetically more similar to human ESCs (hESCs), being dependent on bFGF for self maintenance [33]. The human equivalent of mouse ESCs (mESCs), which is dependent on LIF, small molecule inhibitors of ERK1/ERK2 and GSK3b signaling, and express SSEA-1, has only recently been derived from human blastocysts and appear to be in an earlier epigenetic developmental state – therefore, they are called naïve hESCs, whereas the FGF-dependent hESCs are called primed hESCs [34, 35].
Epigenetic modifications are also essential for pluripotency maintenance. The early steps of embryonic development are marked by global DNA demethylation, and this “permissive” epigenetic state at pre-implantational phase is pivotal for the expression of the above mentioned transcription factors [36]. By the time of implantation, methylation of CpG dinucleotides is reestablished throughout the genome as result of the de novo activation of highly expressed DNA methyltransferases Dnmt3a and Dnmt3b, which triggers the differentiation process [37, 38]. When most of the DNA methylation marks have already been established, the expression of many genes belonging to the epigenetic machinery is decreased, concomitantly with the differentiation process [39, reviewed in 40].
More recently, the maintenance of pluripotency has also been associated to miRNA expression. Since the first discoveries of the regulatory effect of small ncRNAs lin-4 and let-7 in Caenorhabditis elegans, it is well established that miRNAs are involved in development [1, 41]. Indeed, disruption of Dicer gene in mice compromises the entire miRNA biogenesis and is lethal early in development [42]. Although mouse Dicer-null ESCs are viable, they have a slow proliferation rate and fail to differentiate [43, 44]. Mouse Dgcr8-deficient ESCs also exhibit delayed proliferation rates with prolonged G0 and G1 phases [45], and when induced to differentiation, show aberrant expression of specific differentiation markers, such as a delayed expression of primitive ectoderm marker (FgF5), a weak or absent expression of endoderm (Hnf4a and Afp), mesoderm (Brachyury, Bmp4, Gata) and ectoderm (Sox1) markers, and incomplete repression of pluripotency [43, 46]. Additionally, mouse Dicer-null ESCs showed significant hypomethylation of the genome, including the promoter of Oct4 gene (also known as Pou5f1 gene) [47, 48], which impairs differentiation.
Three miRNAs clusters hsa-miR-371-373 (ortholog of the mouse cluster mmu-miR-290-295), hsa/mmu-miR-302-367 and hsa/mmu-miR-17-92 are highly expressed in ESCs [49-54], therefore known as ESC-enriched miRNAs. They share similar seed sequences, suggesting an overlapping regulation of their targets [45, 55].
The hsa-miR-371-373 cluster transcribes four miRNAs [50], whereas its murine counterpart, mmu-miR-290-295 transcribes seven [49], and accounts for up to 70% of the total miRNAs expressed in mESCs [56]. At the stage of four to eight cells, mouse embryos already show miR-290-295 expression, which decreases after embryonic day 6.5 [57], and during in vitro differentiation of ESCs to embryoid bodies [52]. These miRNAs are involved in (a) the regulation of transition from G1 to S phase of the cell cycle through targeting of cell-cycle inhibitors [45]; (b) repression of mesoderm and primordial germ cell differentiation pathways [58]; (c) repression of epithelial-mesenchymal transition [59]. Transfection of miRNAs belonging to the miR-290-295 cluster restores many of the defects exhibited by mouse Dicer-null ESCs. This phenomenon was seen by two independent studies showing that miR-290-295 members inhibit the expression of the retinoblastoma-like 2 protein (Rbl2), which is a transcriptional repressor of DNA methyltransferases – Dnmt3a and Dnmt3b [47, 48]. Thus, the authors of these two studies [47, 48] proposed that this is the major mechanism by which cells regulate DNA de novo methylation during early development. Accordingly, introduction of miR-290, miR-302 and miR-17-92 mimics was able to re-establish the proliferation rates of Dgcr8-deficient ESCs [45]. However, miR-290-295 function is still controversial, since recently it was shown that, although Rbl2 mRNA levels are increased in mouse Dicer-null ESCs, its protein remains at low levels [60].
The other highly expressed miRNA cluster in ESCs, miR-302-367, transcribes eight and five miRNAs, in human and mice, respectively [49, 50]. This cluster confers stemness proprieties to hESCs by controlling LEFTY1 and LEFTY2 expression, two inhibitors of TGFβ/ Nodal pathway that have an essential role in signaling early cell fate determination [61]. This cluster is also important to control cell cycle, since its inhibition leads to arrest of hESCs in G1 phase by targeting CYCLIN D1 [62].
Finally, miR-17-92 cluster comprises six miRNAs in human and mice, is overexpressed in mESC and hESC [50, 51], and has an important role in cell cycle regulation [63, 64]. Despite being associated to maintenance of ESC pluripotency, this cluster is also widely expressed in many cell types [65-67]. In humans, its overexpression may lead to several malignancies, since it is located at the genome region 13q31-q32 that is frequently found amplified in lymphomas and other cancer types [68, 69].
Interestingly, expression profiles for mESCs and EpiSCs reveal that they have differences in the expression of several miRNAs, including these important ESC-enriched miRNA clusters, miR-17-92, miR-290-295 and miR-302-267. The former two are more highly expressed in mESC, whereas the latter is in EpiSC [51]. Additionally, although barely expressed, members of Let-7 miRNA family, which are differentiation markers, are enriched in EpiSC in comparison to mESC, and that may reflect a degree of commitment to differentiation. Thus, miRNAs may have redundant and specific roles in regulation of pluripotency [51]. In humans, hsa-miR-302b expression is indicative of pluripotency in naïve and primed hESC, while expression of hsa-miR-371-373, the human ortholog of mmu-miR-290-295, is increased in naïve when compared to primed hESCs [33, 55].
Nevertheless, Oct4, Sox2 and Nanog regulate miR-290-295 and miR-302-367 gene clusters in mice and humans, reinforcing miRNAs role in pluripotency control and in the early steps of differentiation [56, 62, 70].
Adult stem cells, such as mesenchymal, bone marrow and hematopoietic stem cells are also capable of self-renewal and have the plasticity to differentiate into one or multiple cell types, functioning as a quiescent reservatory for tissue maintenance and repair throughout the life span [reviewed in 71]. As its embryonic counterparts, they have miRNAs that may participate in the maintenance of cell identity [72, 73], such as miR-489 that is highly expressed in mouse muscle stem cells (satellite cells), but is downregulated during cell activation [74].
MiRNAs IN INDUCED PLURIPOTENT STEM CELLS
Since the reversion of differentiated fibroblasts into pluripontent cells by the introduction of the defined reprogramming factors OCT4, SOX2, KLF4, and c-MYC (OSKM) in 2006, induced pluripotent stem cells (iPSCs) have been the center of many studies in cell therapy [75, 76]. IPSCs raise great interest in regenerative medicine because of their potential to overcome the issue of histocompatibility between cells and patient. Different methods have been used to deliver the reprogramming factors, including (a) integrative retrovirus vectors; (b) non-integrative vectors, such as adenovirus, Sendai virus, and plasmids; (c) DNA-free transfections, such as mRNAs and fusion proteins; and (d) excision after integration, such as piggyBac transposon and Retrovirus with loxP construction followed by Cre recombinase mRNA transfection [reviewed in 77]. Accordingly, new approaches to promote a better understanding of the mechanisms involved in maintenance of pluripotency, to achieve higher reprogramming efficiency, and to guarantee the safe use of iPSCs to therapy are of great importance. In this context, the introduction of ESC-enriched miRNAs has been used to improve cell reprogramming.
Transient transfection of miRNAs miR-291-3p, miR-294, and miR-295 enhances the efficiency of the reprogramming of mouse embryonic fibroblasts achieved by retroviral deliver of Oct4, Sox2 and Klf4. Among them, miR-294 showed the best results, increasing tenfold the efficiency rates [78]. Similarly, the expression of miRNAs from the miR-302-367 cluster also enhances retroviral reprogramming of human fibroblasts with OCT4, SOX2 and KLF4, either with or without c-MYC [79]. Conversely, inhibition of Let-7 family members, which are known to be robust maintainers of the differentiated state in mouse embryonic fibroblasts, enhanced in over fourfold the efficiency of reprogramming carried out by Oct4, Sox2 and Klf4 [80]. Other examples of miRNAs expression manipulation to improve the efficiency of reprogramming are the transfection of miR-93 and miR-106b [81], the knockout of miR-34 [82] and miR-199a-3p [83], among others [84-86].
Strikingly, the reprogramming of human skin cancer cells [87] and human fibroblasts [88] into a pluripotent state by the introduction of miR-302-367 cluster per se was reported. However, few studies have used this methodology recently, and reprogramming of somatic cells through transfection of members of the cluster miR-302-367 alone [89] or combined with miR-200c or miR-369 [90] yield no clones or resulted in a low efficient rate. Nevertheless, the use of miRNAs as OSKM adjuvant to produce iPSC might be a good strategy to improve the efficiency of somatic cells reprogramming [89]. As any other cell-therapy approach, miRNA use must be in consonance with the applications intended for iPSCs, since some differentiated cells retain more plasticity than others, and some cell types present a more robust epigenetic memory after reprogramming [reviewed in 91].
MiRNAs IN CELL FATE DECISION
Cell differentiation is a complex pathway that depends on both activation of lineage-specific genes and repression of pluripotency-related ones. However, a coordinated modulation of Oct4, Sox2 and Nanog expression in early steps of differentiation process contributes to specific germ layer induction of mESCs, despite combined expression of Oct4 and Sox2 suppresses germ layer differentiation [92]. Therefore, high Oct4 or Sox2 levels promote mesendodermal or neural ectoderm differentiation, respectively, while Nanog downregulation is decisive for lineage commitment [92]. Similar phenomenon was seen in a study using hESCs, whereas each factor is per se involved in a specific cell fate [93]. Likewise, miR-302-367 cluster that, as seen before, has an important role in the maintenance of pluripotent cells, is also expressed in the human endodermal lineage [94], providing evidence that this cluster has a role in organogenesis. Indeed, hsa-miR-302-367 cluster promoter is targeted by GATA6 transcription factor in early stages of lung epithelial development, promoting the proliferation of lung endoderm progenitor cells, proper apical-basal polarity and preventing its complete differentiation. Therefore, this cluster seems to be essential for the correct development of a single-layered lung epithelium [95]. Additionally, miR-17-92 that is also enriched in hESC [50], has an important role in the early stages of lung morphogenesis, regulating the proliferation-differentiation balance of lung epithelial progenitor cells [65]. As the cells commit to differentiation, Oct4, Sox2 and Nanog are downregulated, and consequently the clusters regulated by them, miR-290-295 and miR-302-367 in mice and human, respectively, are also silenced [56, 62]. Prior to being silenced, Oct4, Sox2 and Nanog also upregulate the expression of some miRNAs specifically associated with differentiation in mESCs, such as miR-9, miR124a, miR-155 and miR-708, which at least are in part responsible for proper cell fate determination [56]. Indeed, miRNAs are essential for ESCs specific-differentiation and maintenance of the differentiated status. Accordingly, miRNAs expression is frequently globally downregulated in tumors, which are less differentiated cells [15, 96].
Similarly, adult stem cells also have miRNAs involved in the commitment of their differentiation. For instance, expression of miR-590 and miR-199a in adult cardiomyocyte promotes re-entrance in cell cycle, resulting in cardiac repair in an ex-vivo mouse model [97].
One of the first miRNAs recognized by its role in differentiated tissues was let-7 [41]. With its orthologs organized in large families along the vertebrate genomes, Let-7 is up-regulated in differentiating and differentiated mouse cells [98-100]. Although mature Let-7 is poorly expressed in mESCs, its primary transcript is abundant [56]. The processing from pri-Let-7 to Let-7 mature duplex is inhibited by the RNA binding protein Lin28, which prevents differentiation and stabilizes mESC status [101].
Since then, many other miRNAs have been reported as having an important role in early steps of differentiation and maintenance of the differentiated status. Examples are miR-21 and miR-22 which were also reported as overexpressed in differentiated cells [49]. Indeed, Nanog and Sox2 are direct targets of miR-21 [102], and this miRNA may have an important role promoting adipocyte differentiation [103] as well as in bone formation, since it is overexpressed during the initial steps of osteogenic differentiation [104]. MiR-22, by its turn, has been reported as a maintainer of progenitor cells in murine mammary epithelium [99], promoter of osteogenic differentiation and inhibitor of adypogenic differentiation [105]. Moreover, a set of miRNAs was found to be up-regulated (miR-297, miR-96, miR-214, miR-125a, miR-424, miR-21, miR-29c, miR-7) or down-regulated (miR-376a) in mouse blastocysts when compared to the morula stage, indicating that they are involved in trophectoderm determination [106]. Furthermore, the different miRNA profiles characterizing the three germ layers in gastrulating embryo implicate the involvement of miRNAs in the differentiation of mesoderm, endoderm, and ectoderm [107-109]. In the Supplementary Table S1 (227KB, pdf) we show a comprehensive list of miRNAs involved in tissue regulation, organogenesis and development.
Once differentiation is established, each cell type will express its own set of miRNAs. Accordingly, tissues with the same ontogenetic origin have similar expression profiles, which are different from those of tissues originating from different embryonic layers [15]. Since miRNAs have been widely implicated in the control of stem cells fate, a better understanding of the relationships among miRNAs, transcription factors, signaling pathways, chromatin remodeling factors, and extracellular clues have a pivotal importance in developing new strategies to tissue engineering.
MiRNAs AS PROMISING TOOLS FOR TISSUE ENGINEERING
Tissue engineering (TE) is an interdisciplinary field that combines cells, engineered materials, and biomedical technology towards the development of bio-artificial tissue-like structures to restore, replace, maintain or improve the function of tissues or organs [110].
Currently, several tissues and organs are being engineered [111-114]. The obstacles in TE are to attain specific cell types, to develop appropriated scaffolds, and to promote the release of growth factors and other molecules from scaffolds in order to resemble organogenesis [115, 116]. The success of implant engineered tissues is also challenged by the difficult formation of blood vessel network, tissue innervation, and by inflammatory and immunological responses [reviewed in 117], making the transition from research stage to clinical trials limited to avascular or thin tissues, such as cartilage and skin [118, 119]. Given the role of miRNAs in many biological processes, including cell differentiation and maintenance of cell identity, modulation of these small molecules in combination with stem cells and/or bioartificial scaffolds has been providing encouraging results. Indeed, strategies for vascularization of bioartificial tissues, which are insofar mainly based on the delivery of angiogenic growth factors [120-122], have recently advanced with the use of miR-132 that indirectly induces Ras overexpression, enhancing neovascularization rate [123]. When this miRNA is encapsulated in biodegradable polymer nanoparticles, it improves vessel formation in human endothelial cells transplanted in immunodefficient mice. This approach allows the release of these small RNAs for weeks, longer than conventional lipid-based transfection [124]. Similarly, localized and sustained expression of miR-26a in vivo positively regulates osteogenesis-angiogenesis coupling, therefore, providing an enhanced efficiency in bone regeneration [125]. Zhang and colleagues have also reported that the inhibition of miR-29 in bioengineered vessels may increase the expression of its target gene, ELN, which has a major function in maintaining the integrity of the extracellular matrix of arteries [126].
Other studies also showed that TE could be potentially improved by the transfection of cells with miR-21, since it promoted high proliferation rates and high matrix synthesis of rat chondrocytes cultured in atelocollagen gel [127], and by implanting scaffold embedded with miR-29b in cutaneous injury, which was able to improve extracellular matrix remodeling of treated excisional wounds [116].
However, the most studied miRNAs in TE are related to directing cell fate. Examples of this are (a) the inhibition of miR-133, which enhances skeletal murine myoblast differentiation and the response to electrical stimulation in three-dimensional (3D) bioartificial muscle [128]; (b) the transfection of miR-206 in satellite cells, which increases their differentiation in a bioartificial muscle construct [129]; (c) the introduction of miR-148b mimic and miR-489 inhibitor, which improves osteogenesis from human mesenchymal stem cells and the expression of osteogenic markers, also in a 3D scaffold [130], (d) the usage of nano-bioglass ceramic particles (nBGC) that stimulates miR-30 expression in osteoblastic cells, inducing their differentiation [131], (e) the transfection of miR-31 inhibitor in bone marrow stromal cells, which increases osteogenic differentiation, bone mineral density, biocompatibility and regeneration rate [132]. Finally, miRNAs seem to have an important role in the development of engineered tissues with a refined architecture. For instance, articular cartilage is subdivided in specific zones that seem to be frequently lost in monolayer expanding cultures of chondrocytes. Superficial zone-specific miRNAs expression is also lost in this process, and the TGF-β-directed differentiation of chondrocytes in a 3D agarose culture was able to reestablish their expression. Therefore, manipulation of miRNA expression might be useful to the correct assembly of a complex engineered tissue [133]. A complete list containing all miRNAs used so far in TE is reported in Table 1. Thus, we expect that miRNAs will become an increasingly important tool for controlling cell fate for TE, and the prominent candidates to this purpose are listed in Supplementary Table S1 (227KB, pdf) .
Table 1.
MicroRNAs employed for tissue engineering approaches.
| MicroRNA | MicroRNA delivery | Result of microRNA manipulation | Ref. |
|---|---|---|---|
| miR-1 and miR-206 | Myogenic progenitor cells were transfected with each microRNA separately and cultured in a 3-D culture system. | Improvement of myogenic progenitor cells differentiation. |
[129] |
| miR-21 | Chondrocytes were cultured on an atelocollagen gel complexed with the miRNA. | Improvement of proliferation and matrix synthesis of the chondrocytes. | [127] |
| miR-26a | Bone marrow mesenchymal cells were cultured in an hydrogel system that releases a chemically modified miR-26a, which is a miRNA enhancer (agomiR-26a), and then this construction was implanted into calvarial bone defect in mice. |
Improvement of bone regeneration and modulation of angiogenesis- osteogenesis coupling. | [125] |
| miR-29a | Human vascular smooth muscle cells were seeded with polyglycolic acid scaffolds in the presence of miRNA-29a inhibitor. |
Improvement of elastin levels in bioengineered human vessels. |
[126] |
| miR-29b | Fibroblasts were cultured in collagen-based scaffolds doped with miR-29b and these were applied to rat wound model. | Improvement of the wound healing response through reduced wound contraction and reduction of collagen type I production after the injury. | [116] |
| miR-30c | Treatment of osteoblastic cells with nano-bioglass ceramic particules (nBGC) was able to induce miR-30c expression. | The indirect upregulation of this miRNA may lead to the osteoblastic differentiation. | [131] |
| miR-31 | Osteo-inductive bone marrow stromal stem cells transduced with anti-miR-31 lentiviral vectors were seeded on polyglycerol sebacate scaffolds and used to repair critical-sized calvarial defects in rats. |
Improvement of ostogenic differentiation, biocompatibility and regeneration rate in the repair of in vivo large bone defects. |
[132] |
| miR-132 | The microRNA was encapsulated in a targeted biodegradable polymer nanoparticle and delivered to endothelial cells before transplantation. |
Improvement of endothelial cells transplantation through vascularization enhancing. | [124] |
| miR-133 | Myoblasts were transfected with anti-miR-133 and cultured in a collagen/matrigel construct. | Improvement of myogenic differentiation and increased peak forces after electrical stimulation. | [128] |
| miR-221, miR-222, miR-140, miR-143, and miR-145 | In 3D agarose cultures, chondrocytes treated with TGF-β1 showed downregulation of miR-221 and miR-222 expression and inscreased expression of miR-140, miR-143 and miR-145. |
Alterations in microRNAs expression due to treatment of cells with TGF-β1, which is known to enhance chondrocytic differentiation, may represent a promising role in the tissue engineering of the articular cartilage superficial zone. |
[133] |
| miR-148b | Rat mesenchymal stem-cells were transfected with the microRNA lyophilized on a microporous titanium implant. | Improvement of osteogenic differentiation of the stem cells. |
[135] |
| miR-148b and miR-489 |
Human mesenchymal stem-cells were transfected with miR-148b mimic and miR-489 inhibitor and cultured in a 2-D surface or capsuled in a 3-D scaffold. |
Improvement of osteogenesis through the sensibilization of the cells to osteogenic signals. | [130] |
Finally, miRNAs might potentially be used to monitor the graft status, since in a mouse model of heart transplantation, allograft rejection seems to be associated with specific miRNA signatures. Moreover, miR-182 was found overexpressed in peripheral blood mononuclear cells and plasma in mice with allograft rejection [134].
CONCLUSION
MicroRNAs have an essential role in maintenance of cell pluripotent and differentiated states, as well as in cell fate decisions, working as modulators of cell identity. Accordingly, these small regulators might (a) assist the reprogramming of iPSC, an important source of cells for TE; (b) direct and maintain tissue-specific differentiation; (c) guarantee proper vascularization of engineered tissue. A better understanding of miRNAs involvement in tissue formation, regeneration and function will provide more efficient engineered tissues. Thus, on the whole, despite few studies have been performed so far, the results are very promising and warrant remarkable advances in the next future.
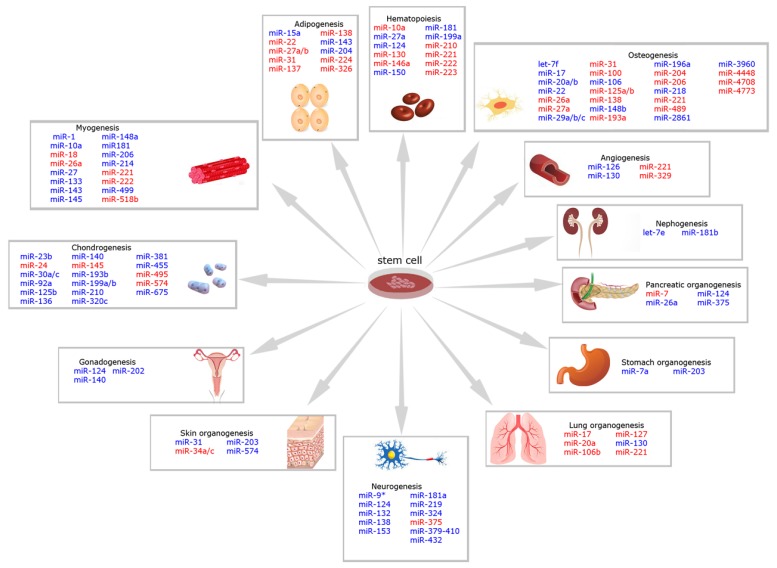
Fig. (1).
MiRNAs involved in cell fate during genesis and development of different tissues and organs in mammals. In blue, miRNAs that positively regulate differentiation; in red, miRNAs that negatively regulate differentiation. Images were obtained from www.shutterstock.com website. This figure graphically represents Supplementary Table S1. For further information about each miRNA, please see referred Table.
ACKNOWLEDGEMENTS
The authors thank Emerson Augusto da Silva for figure design. This work was supported by the Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) and the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) (Brazil). We apologize in advance to the authors whose important papers have not been cited here due to space constraints.
CONFLICT OF INTEREST
The authors confirm that this article content has no conflicts of interest.
SUPPLEMENTARY MATERIAL
Supplementary material is available on the publishers web site along with the published article.
REFERENCES
- 1.Lee RC, Feinbaum RL, Ambros V. The C.elegans heterochronic gene lin-4 encodes small RNAs with antisense complementary to lin-14. Cell. 1993;75:843–54. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 2.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C.elegans. Cell. 1993;75:855–62. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 3.Pasquinelli AE, Reinhart BJ, Slack F , et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–9. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- 4.Zhao Y, Srivastava D. A developmental view of microRNA function. Trends Biochem Sci. 2007;32:189–97. doi: 10.1016/j.tibs.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 5.Friedman RC, Farh KK, Burge CB , et al. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lewis BP, Shih IH, Jones-Rhoades MW , et al. Prediction of mammalian microRNA targets. Cell. 2003;115:787–98. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 7.Doench JG, Sharp PA. Specificity of microRNA target selection in translational repression. Genes Dev. 2004;18:504–11. doi: 10.1101/gad.1184404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing. often flanked by adenosnes.indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 9.Zhang H, Kolb FA, Brondani V , et al. Human Dicer preferentially cleaves dsRNAs at their termini without a requirement for ATP. EMBO J. 2002;21:5875–85. doi: 10.1093/emboj/cdf582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee Y, Ahn C, Han J , et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–9. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 11.Han J, Lee Y, Yeom KH , et al. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–27. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chendrimada TP, Gregory RI, Kumaraswamy E , et al. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–4. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rodriguez A, Griffiths-Jones S, Ashurst JL , et al. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004;14:1902–10. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee Y, Kim M, Han J , et al. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–60. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu J, Getz G, Miska EA , et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–8. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 16.Thomson JA, Itskovitz-Eldor J, Shapiro SS , et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–7. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 17.Gokhale PJ, Andrews PW. Characterization of human pluripotent stem cells. Neuroreport. 2013;24:1031–4. doi: 10.1097/WNR.0000000000000040. [DOI] [PubMed] [Google Scholar]
- 18.Marolt D, Campos IM, Bhumiratana S , et al. Engineering bone tissue from human embryonic stem cells. Proc Natl Acad Sci U S A. 2012;109:8705–9. doi: 10.1073/pnas.1201830109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kerr CL, Letzen BS, Hill CM , et al. Efficient differentiation of human embryonic stem cells into oligodendrocyte progenitors for application in a rat contusion model of spinal cord injury. Int J Neurosci. 2010;120:305–13. doi: 10.3109/00207450903585290. [DOI] [PubMed] [Google Scholar]
- 20.Bosnjak ZJ, Yan Y, Canfield S , et al. Ketamine induces toxicity in human neurons differentiated from embryonic stem cells via mitochondrial apoptosis pathway. Curr Drug Saf. 2012;7:106–19. doi: 10.2174/157488612802715663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bahrami SB, Veiseh M, Dunn AA , et al. Temporal changes in Hox gene expression accompany endothelial cell differentiation of embryonic stem cells. Cell Adh Migr. 2011;5:133–41. doi: 10.4161/cam.5.2.14373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang Y, Wang H, Chang KH , et al. Transcriptome dynamics during human erythroid differentiation and development. Genomics. 2013;102:431–41. doi: 10.1016/j.ygeno.2013.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Assou S, Cerecedo D, Tondeur S , et al. A gene expression signature shared by human mature oocytes and embryonic stem cells. BMC Genomics. 2009;10 doi: 10.1186/1471-2164-10-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ivanova NB, Dimos JT, Schaniel C , et al. A stem cell molecular signature. Science. 2002;298:601–4. doi: 10.1126/science.1073823. [DOI] [PubMed] [Google Scholar]
- 25.Sato N, Sanjuan IM, Heke M , et al. Molecular signature of human embryonic stem cells and its comparison with the mouse. Dev Biol. 2003;260:404–13. doi: 10.1016/s0012-1606(03)00256-2. [DOI] [PubMed] [Google Scholar]
- 26.Azuara V, Perry P, Sauer S , et al. Chromatin signatures of pluripotent cell lines. Nat Cell Biol. 2006;8:532–8. doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- 27.Avilion AA, Nicolis SK, Pevny LH , et al. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev. 2003;17:126–40. doi: 10.1101/gad.224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chambers I, Colby D, Robertson M , et al. Functional expression cloning of Nanog. a pluripotency sustaining factor in embryonic stem cells. . Cell. 2003;113:643–55. doi: 10.1016/s0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- 29.Nichols J, Zevnik B, Anastassiadis K , et al. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998;95:379–91. doi: 10.1016/s0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- 30.Adewumi O, Aflatoonian B, Ahrlund-Richter L , et al. Characterization of human embryonic stem cell lines by the International Stem Cell Initiative. Nat Biotechnol. 2007;25:803–16. doi: 10.1038/nbt1318. [DOI] [PubMed] [Google Scholar]
- 31.Brons IG, Smithers LE, Trotter MW , et al. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature. 2007;448:191–5. doi: 10.1038/nature05950. [DOI] [PubMed] [Google Scholar]
- 32.Tesar PJ, Chenoweth JG, Brook FA , et al. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 2007;448:196–9. doi: 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- 33.Ware CB, Nelson AM, Mecham B , et al. Derivation of naive human embryonic stem cells. Proc Natl Acad Sci U S A. 2014;111:4484–9. doi: 10.1073/pnas.1319738111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gafni O, Weinberger L, Mansour AA , et al. Derivation of novel human ground state naive pluripotent stem cells. Nature. 2013;504:282–6. doi: 10.1038/nature12745. [DOI] [PubMed] [Google Scholar]
- 35.Chan YS, Goke J, Ng JH , et al. Induction of a human pluripotent state with distinct regulatory circuitry that resembles preimplantation epiblast. Cell Stem Cell. 2013;13:663–75. doi: 10.1016/j.stem.2013.11.015. [DOI] [PubMed] [Google Scholar]
- 36.Surani MA, Hayashi K, Hajkova P. Genetic and epigenetic regulators of pluripotency. Cell. 2007;128:747–62. doi: 10.1016/j.cell.2007.02.010. [DOI] [PubMed] [Google Scholar]
- 37.Monk M, Boubelik M, Lehnert S. Temporal and regional changes in DNA methylation in the embryonic. extraembryonic and germ cell lineages during mouse embryo development. Development. 1987;99:371–82. doi: 10.1242/dev.99.3.371. [DOI] [PubMed] [Google Scholar]
- 38.Kafri T, Ariel M, Brandeis M , et al. Developmental pattern of gene-specific DNA methylation in the mouse embryo and germ line. Genes Dev. 1992;6:705–14. doi: 10.1101/gad.6.5.705. [DOI] [PubMed] [Google Scholar]
- 39.Kantor B, Makedonski K, Shemer R , et al. Expression and localization of components of the histone deacetylases multiprotein repressory complexes in the mouse preimplantation embryo. Gene Expr Patterns. 2003;3:697–702. doi: 10.1016/j.modgep.2003.07.003. [DOI] [PubMed] [Google Scholar]
- 40.Dean W, Lucifero D, Santos F. DNA methylation in mammalian development and disease. Birth Defects Res C Embryo Today. 2005;75:98–111. doi: 10.1002/bdrc.20037. [DOI] [PubMed] [Google Scholar]
- 41.Reinhart BJ, Slack FJ, Basson M , et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–6. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 42.Bernstein E, Kim SY, Carmell MA , et al. Dicer is essential for mouse development. Nat Genet. 2003;35:215–7. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- 43.Kanellopoulou C, Muljo SA, Kung AL , et al. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 2005;19:489–501. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Murchison EP, Partridge JF, Tam OH , et al. Characterization of Dicer-deficient murine embryonic stem cells. Proc Natl Acad Sci U S A. 2005;102:12135–40. doi: 10.1073/pnas.0505479102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Y, Baskerville S, Shenoy A , et al. Embryonic stem cell-specific microRNAs regulate the G1-S transition and promote rapid proliferation. Nat Genet. 2008;40:1478–83. doi: 10.1038/ng.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang Y, Medvid R, Melton C , et al. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat Genet. 2007;39:380–5. doi: 10.1038/ng1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Benetti R, Gonzalo S, Jaco I , et al. A mammalian microRNA cluster controls DNA methylation and telomere recombination via Rbl2-dependent regulation of DNA methyltransferases. Nat Struct Mol Biol. 2008;15:268–79. doi: 10.1038/nsmb.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sinkkonen L, Hugenschmidt T, Berninger P , et al. MicroRNAs control de novo DNA methylation through regulation of transcriptional repressors in mouse embryonic stem cells. Nat Struct Mol Biol. 2008;15:259–67. doi: 10.1038/nsmb.1391. [DOI] [PubMed] [Google Scholar]
- 49.Houbaviy HB, Murray MF, Sharp PA. Embryonic stem cell-specific microRNAs. Dev Cell. 2003;5:351–8. doi: 10.1016/s1534-5807(03)00227-2. [DOI] [PubMed] [Google Scholar]
- 50.Suh MR, Lee Y, Kim JY , et al. Human embryonic stem cells express a unique set of microRNAs. Dev Biol. 2004;270:488–98. doi: 10.1016/j.ydbio.2004.02.019. [DOI] [PubMed] [Google Scholar]
- 51.Jouneau A, Ciaudo C, Sismeiro O , et al. Naive and primed murine pluripotent stem cells have distinct miRNA expression profiles. RNA. 2012;18:253–64. doi: 10.1261/rna.028878.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen C, Ridzon D, Lee CT , et al. Defining embryonic stem cell identity using differentiation-related microRNAs and their potential targets. Mamm Genome. 2007;18:316–27. doi: 10.1007/s00335-007-9032-6. [DOI] [PubMed] [Google Scholar]
- 53.Lakshmipathy U, Love B, Goff LA , et al. MicroRNA expression pattern of undifferentiated and differentiated human embryonic stem cells. Stem Cells Dev. 2007;16:1003–16. doi: 10.1089/scd.2007.0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Barad O, Meiri E, Avniel A , et al. MicroRNA expression detected by oligonucleotide microarrays: system establishment and expression profiling in human tissues. Genome Res. 2004;14:2486–94. doi: 10.1101/gr.2845604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stadler B, Ivanovska I, Mehta K , et al. Characterization of microRNAs involved in embryonic stem cell states. Stem Cells Dev. 2010;19:935–50. doi: 10.1089/scd.2009.0426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Marson A, Levine SS, Cole MF , et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134:521–33. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Medeiros LA, Dennis LM, Gill ME , et al. Mir-290-295 deficiency in mice results in partially penetrant embryonic lethality and germ cell defects. Proc Natl Acad Sci USA. 2011;108:14163–8. doi: 10.1073/pnas.1111241108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zovoilis A, Smorag L, Pantazi A , et al. Members of the miR-290 cluster modulate in vitro differentiation of mouse embryonic stem cells. Differentiation. 2009;78:69–78. doi: 10.1016/j.diff.2009.06.003. [DOI] [PubMed] [Google Scholar]
- 59.Lüningschrör P, Stocker B, Kaltschmidt B , et al. miR-290 cluster modulates pluripotency by repressing canonical NF-kappaB signaling. Stem Cells. 2012;30:655–64. doi: 10.1002/stem.1033. [DOI] [PubMed] [Google Scholar]
- 60.Ip J, Canham P, Choo KH , et al. Normal DNA methylation dynamics in DICER1-deficient mouse embryonic stem cells. PLoS Genet. 2012;8:e1002919. doi: 10.1371/journal.pgen.1002919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Barroso-delJesus A, Lucena-Aguilar G, Sanchez L , et al. The Nodal inhibitor Lefty is negatively modulated by the microRNA miR-302 in human embryonic stem cells. FASEB J. 2011;25:1497–508. doi: 10.1096/fj.10-172221. [DOI] [PubMed] [Google Scholar]
- 62.Card DA, Hebbar PB, Li L , et al. Oct4/Sox2-regulated miR-302 targets cyclin D1 in human embryonic stem cells. Mol Cell Biol. 2008;28:6426–38. doi: 10.1128/MCB.00359-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.O'Donnell KA, Wentzel EA, Zeller KI , et al. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–43. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 64.Woods K, Thomson JM, Hammond SM. Direct regulation of an oncogenic micro-RNA cluster by E2F transcription factors. J Biol Chem. 2007;282:2130–4. doi: 10.1074/jbc.C600252200. [DOI] [PubMed] [Google Scholar]
- 65.Lu Y, Thomson JM, Wong HY , et al. Transgenic over-expression of the microRNA miR-17-92 cluster promotes proliferation and inhibits differentiation of lung epithelial progenitor cells. Dev Biol. 2007;310:442–53. doi: 10.1016/j.ydbio.2007.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ventura A, Young AG, Winslow MM , et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132:875–86. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xiao C, Srinivasan L, Calado DP , et al. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat Immunol. 2008;9:405–14. doi: 10.1038/ni1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ota A, Tagawa H, Karnan S , et al. Identification and characterization of a novel gene. C13of25.as a target for 13q31-q32 amplification in malignant lymphoma. Cancer Res . 2004;64:3087–95. doi: 10.1158/0008-5472.can-03-3773. [DOI] [PubMed] [Google Scholar]
- 69.Tagawa H, Seto M. A microRNA cluster as a target of genomic amplification in malignant lymphoma. Leukemia. 2005;19:2013–6. doi: 10.1038/sj.leu.2403942. [DOI] [PubMed] [Google Scholar]
- 70.Barroso-delJesus A, Romero-Lopez C, Lucena-Aguilar G , et al. Embryonic stem cell-specific miR302-367 cluster: human gene structure and functional characterization of its core promoter. Mol Cell Biol. 2008;28:6609–19. doi: 10.1128/MCB.00398-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Snippert HJ, Clevers H. Tracking adult stem cells. EMBO Rep. 2011;12:113–22. doi: 10.1038/embor.2010.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schoolmeesters A, Eklund T, Leake D , et al. Functional profiling reveals critical role for miRNA in differentiation of human mesenchymal stem cells. PLoS One. 2009;4:e5605. doi: 10.1371/journal.pone.0005605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kim YJ, Bae SW, Yu SS , et al. miR-196a regulates proliferation and osteogenic differentiation in mesenchymal stem cells derived from human adipose tissue. J Bone Miner Res. 2009;24:816–25. doi: 10.1359/jbmr.081230. [DOI] [PubMed] [Google Scholar]
- 74.Cheung TH, Quach NL, Charville GW , et al. Maintenance of muscle stem-cell quiescence by microRNA-489. Nature. 2012;482:524–8. doi: 10.1038/nature10834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–76. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 76.Takahashi K, Tanabe K, Ohnuki M , et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–72. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 77.Zhou YY, Zeng F. Integration-free methods for generating induced pluripotent stem cells. Genomics Proteomics Bioinformatics. 2013;11:284–7. doi: 10.1016/j.gpb.2013.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Judson RL, Babiarz JE, Venere M , et al. Embryonic stem cell-specific microRNAs promote induced pluripotency. Nat Biotechnol. 2009;27:459–61. doi: 10.1038/nbt.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Subramanyam D, Lamouille S, Judson RL , et al. Multiple targets of miR-302 and miR-372 promote reprogramming of human fibroblasts to induced pluripotent stem cells. Nat Biotechnol. 2011;29:443–8. doi: 10.1038/nbt.1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Melton C, Judson RL, Blelloch R. Opposing microRNA families regulate self-renewal in mouse embryonic stem cells. Nature. 2010;463:621–6. doi: 10.1038/nature08725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li Z, Yang CS, Nakashima K , et al. Small RNA-mediated regulation of iPS cell generation. EMBO J. 2011;30:823–34. doi: 10.1038/emboj.2011.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Choi YJ, Lin CP, Ho JJ , et al. miR-34 miRNAs provide a barrier for somatic cell reprogramming. Nat Cell Biol. 2011;13:1353–60. doi: 10.1038/ncb2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang J, He Q, Han C , et al. p53-facilitated miR-199a-3p regulates somatic cell reprogramming. Stem Cells. 2012;30:1405–13. doi: 10.1002/stem.1121. [DOI] [PubMed] [Google Scholar]
- 84.Bao X, Zhu X, Liao B , et al. MicroRNAs in somatic cell reprogramming. Curr Opin Cell Biol. 2013;25:208–14. doi: 10.1016/j.ceb.2012.12.004. [DOI] [PubMed] [Google Scholar]
- 85.Moradi S, Asgari S, Baharvand H. Harmonies played by microRNAs in cell fate reprogramming. Stem Cells. 2014;32:3–15. doi: 10.1002/stem.1576. [DOI] [PubMed] [Google Scholar]
- 86.Yang CS, Rana TM. Learning the molecular mechanisms of the reprogramming factors: let's start from microRNAs. Mol Biosyst. 2013;9:10–7. doi: 10.1039/c2mb25088h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lin SL, Chang DC, Chang-Lin S , et al. Mir-302 reprograms human skin cancer cells into a pluripotent ES-cell-like state. RNA. 2008;14:2115–24. doi: 10.1261/rna.1162708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Anokye-Danso F, Trivedi CM, Juhr D , et al. Highly efficient miRNA-mediated reprogramming of mouse and human somatic cells to pluripotency. Cell Stem Cell. 2011;8:376–88. doi: 10.1016/j.stem.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hu S, Wilson KD, Ghosh Z , et al. MicroRNA-302 increases reprogramming efficiency via repression of NR2F2. Stem Cells. 2013;31:259–68. doi: 10.1002/stem.1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Miyoshi N, Ishii H, Nagano H , et al. Reprogramming of mouse and human cells to pluripotency using mature microRNAs. Cell Stem Cell. 2011;8:633–8. doi: 10.1016/j.stem.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 91.González F, Boue S, Izpisua Belmonte JC. Methods for making induced pluripotent stem cells: reprogramming à la carte. Nat Rev Genet. 2011;12:231–42. doi: 10.1038/nrg2937. [DOI] [PubMed] [Google Scholar]
- 92.Thomson M, Liu SJ, Zou LN , et al. Pluripotency factors in embryonic stem cells regulate differentiation into germ layers. Cell. 2011;145:875–89. doi: 10.1016/j.cell.2011.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wang Z, Oron E, Nelson B , et al. Distinct lineage specification roles for NANOG.CT4.and SOX2 in human embryonic stem cells. Cell Stem Cell . 2012; 10:440–54. doi: 10.1016/j.stem.2012.02.016. [DOI] [PubMed] [Google Scholar]
- 94.Hinton A, Afrikanova I, Wilson M , et al. A distinct microRNA signature for definitive endoderm derived from human embryonic stem cells. Stem Cells Dev. 2010;19:797–807. doi: 10.1089/scd.2009.0224. [DOI] [PubMed] [Google Scholar]
- 95.Tian Y, Zhang Y, Hurd L , et al. Regulation of lung endoderm progenitor cell behavior by miR302/367. Development. 2011;138:1235–45. doi: 10.1242/dev.061762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Baskerville S, Bartel DP. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA. 2005;11:241–7. doi: 10.1261/rna.7240905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Eulalio A, Mano M, Dal Ferro M , et al. Functional screening identifies miRNAs inducing cardiac regeneration. Nature. 2012;492:376–81. doi: 10.1038/nature11739. [DOI] [PubMed] [Google Scholar]
- 98.Wulczyn FG, Smirnova L, Rybak A , et al. Post-transcriptional regulation of the let-7 microRNA during neural cell specification. FASEB J. 2007;21:415–26. doi: 10.1096/fj.06-6130com. [DOI] [PubMed] [Google Scholar]
- 99.Ibarra I, Erlich Y, Muthuswamy SK , et al. A role for microRNAs in maintenance of mouse mammary epithelial progenitor cells. Genes Dev. 2007;21:3238–43. doi: 10.1101/gad.1616307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Viñas JL, Ventayol M, Brune B , et al. miRNA let-7e modulates the Wnt pathway and early nephrogenic markers in mouse embryonic stem cell differentiation. PLoS One. 2013;8:e60937. doi: 10.1371/journal.pone.0060937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Boyer LA, Lee TI, Cole MF , et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–56. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kang M, Yan LM, Zhang WY , et al. Role of microRNA-21 in regulating 3T3-L1 adipocyte differentiation and adiponectin expression. Mol Biol Rep. 2013;40:5027–34. doi: 10.1007/s11033-013-2603-6. [DOI] [PubMed] [Google Scholar]
- 104.Eguchi T, Watanabe K, Hara ES , et al. OstemiR: a novel panel of microRNA biomarkers in osteoblastic and osteocytic differentiation from mesencymal stem cells. PLoS One. 2013;8:e58796. doi: 10.1371/journal.pone.0058796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Huang S, Wang S, Bian C , et al. Upregulation of miR-22 promotes osteogenic differentiation and inhibits adipogenic differentiation of human adipose tissue-derived mesenchymal stem cells by repressing HDAC6 protein expression. Stem Cells Dev. 2012;21:2531–40. doi: 10.1089/scd.2012.0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Viswanathan SR, Mermel CH, Lu J , et al. microRNA expression during trophectoderm specification. PLoS One. 2009;4:e6143. doi: 10.1371/journal.pone.0006143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Berardi E, Pues M, Thorrez L , et al. MiRNAs in ESC differentiation. Am J Physiol Heart Circ Physiol. 2012;303:H931–H9. doi: 10.1152/ajpheart.00338.2012. [DOI] [PubMed] [Google Scholar]
- 108.Parsons XH. MicroRNA profiling reveals distinct mechanisms governing cardiac and neural lineage-specification of pluripotent human embryonic stem cells. J Stem Cell Res Ther. 2012;2 doi: 10.4172/2157-7633.1000124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Tzur G, Levy A, Meiri E , et al. MicroRNA expression patterns and function in endodermal differentiation of human embryonic stem cells. PLoS One. 2008;3:e3726. doi: 10.1371/journal.pone.0003726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Langer R, Vacanti JP. Tissue engineering. Science. 1993;260:920–6. doi: 10.1126/science.8493529. [DOI] [PubMed] [Google Scholar]
- 111.Scuderi N, Onesti MG, Bistoni G , et al. The clinical application of autologous bioengineered skin based on a hyaluronic acid scaffold. Biomaterials. 2008;29:1620–9. doi: 10.1016/j.biomaterials.2007.12.024. [DOI] [PubMed] [Google Scholar]
- 112.Hollander AP, Dickinson SC, Sims TJ , et al. Maturation of tissue engineered cartilage implanted in injured and osteoarthritic human knees. Tissue Eng. 2006;12:1787–98. doi: 10.1089/ten.2006.12.1787. [DOI] [PubMed] [Google Scholar]
- 113.Petersen TH, Calle EA, Zhao L , et al. Tissue-engineered lungs for in vivo implantation. Science. 2010;329:538–41. doi: 10.1126/science.1189345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Macchiarini P, Jungebluth P, Go T , et al. Clinical transplantation of a tissue-engineered airway. Lancet. 2008;372:2023–30. doi: 10.1016/S0140-6736(08)61598-6. [DOI] [PubMed] [Google Scholar]
- 115.Wang X, Wenk E, Zhang X , et al. Growth factor gradients via microsphere delivery in biopolymer scaffolds for osteochondral tissue engineering. J Control Release. 2009;134:81–90. doi: 10.1016/j.jconrel.2008.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Monaghan M, Browne S, Schenke-Layland K , et al. A collagen-based scaffold delivering exogenous microRNA-29b to modulate extracellular matrix remodeling. Mol Ther. 2014;22:786–96. doi: 10.1038/mt.2013.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Berthiaume F, Maguire TJ, Yarmush ML. Tissue engineering and regenerative medicine: history. progess.and challenges. . Annu Rev Chem Biomol Eng. 2011; 2:403–30. doi: 10.1146/annurev-chembioeng-061010-114257. [DOI] [PubMed] [Google Scholar]
- 118.Mertsching H, Schanz J, Steger V , et al. Generation and transplantation of an autologous vascularized bioartificial human tissue. Transplantation. 2009;88:203–10. doi: 10.1097/TP.0b013e3181ac15e1. [DOI] [PubMed] [Google Scholar]
- 119.Lysaght MJ, Jaklenec A, Deweerd E. Great expectations: private sector activity in tissue engineering. regenerative mediine.and stem cell therapeutics. Tissue Eng Part A . 2008; 14:305–15. doi: 10.1089/tea.2007.0267. [DOI] [PubMed] [Google Scholar]
- 120.des Rieux A, Ucakar B, Mupendwa BP , et al. 3D systems delivering VEGF to promote angiogenesis for tissue engineering. J Control Release. 2011;150:272–8. doi: 10.1016/j.jconrel.2010.11.028. [DOI] [PubMed] [Google Scholar]
- 121.Ekaputra AK, Prestwich GD, Cool SM , et al. The three-dimensional vascularization of growth factor-releasing hybrid scaffold of poly (epsilon-caprolactone)/collagen fibers and hyaluronic acid hydrogel. Biomaterials. 2011;32:8108–17. doi: 10.1016/j.biomaterials.2011.07.022. [DOI] [PubMed] [Google Scholar]
- 122.Saif J, Schwarz TM, Chau DY , et al. Combination of injectable multiple growth factor-releasing scaffolds and cell therapy as an advanced modality to enhance tissue neovascularization. Arterioscler Thromb Vasc Biol. 2010;30:1897–904. doi: 10.1161/ATVBAHA.110.207928. [DOI] [PubMed] [Google Scholar]
- 123.Katare R, Riu F, Mitchell K , et al. Transplantation of human pericyte progenitor cells improves the repair of infarcted heart through activation of an angiogenic program involving microRNA-132. Circ Res. 2011;109:894–906. doi: 10.1161/CIRCRESAHA.111.251546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Devalliere J, Chang WG, Andrejecsk JW , et al. Sustained delivery of proangiogenic microRNA-132 by nanoparticle transfection improves endothelial cell transplantation. FASEB J. 2014;28:908–22. doi: 10.1096/fj.13-238527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Li Y, Fan L, Liu S , et al. The promotion of bone regeneration through positive regulation of angiogenic-osteogenic coupling using microRNA-26a. Biomaterials. 2013;34:5048–58. doi: 10.1016/j.biomaterials.2013.03.052. [DOI] [PubMed] [Google Scholar]
- 126.Zhang P, Huang A, Ferruzzi J , et al. Inhibition of microRNA-29 enhances elastin levels in cells haploinsufficient for elastin and in bioengineered vessels – brief report. Arterioscler Thromb Vasc Biol. 2012;32:756–9. doi: 10.1161/ATVBAHA.111.238113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kongcharoensombat W, Nakasa T, Ishikawa M , et al. The effect of microRNA-21 on proliferation and matrix synthesis of chondrocytes embedded in atelocollagen gel. Knee Surg Sports Traumatol Arthrosc. 2010;18:1679–84. doi: 10.1007/s00167-010-1111-7. [DOI] [PubMed] [Google Scholar]
- 128.Rhim C, Cheng CS, Kraus WE , et al. Effect of microRNA modulation on bioartificial muscle function. Tissue Eng Part A. 2010;16:3589–97. doi: 10.1089/ten.tea.2009.0601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Koning M, Werker PM, van der Schaft DW , et al. MicroRNA-1 and microRNA-206 improve differentiation potential of human satellite cells: a novel approach for tissue engineering of skeletal muscle. Tissue Eng Part A. 2012;18:889–98. doi: 10.1089/ten.TEA.2011.0191. [DOI] [PubMed] [Google Scholar]
- 130.Mariner PD, Johannesen E, Anseth KS. Manipulation of miRNA activity accelerates osteogenic differentiation of hMSCs in engineered 3D scaffolds. J Tissue Eng Regen Med. 2012;6:314–24. doi: 10.1002/term.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Moorthi A, Vimalraj S, Avani C , et al. Expression of microRNA-30c and its target genes in human osteoblastic cells by nano-bioglass ceramic-treatment. Int J Biol Macromol. 2013;56:181–5. doi: 10.1016/j.ijbiomac.2013.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Deng Y, Bi X, Zhou H , et al. Repair of critical-sized bone defects with anti-miR-31-expressing bone marrow stromal stem cells and poly(glycerol sebacate) scaffolds. Eur Cell Mater. 2014;27:13–25. doi: 10.22203/ecm.v027a02. [DOI] [PubMed] [Google Scholar]
- 133.Hong E, Reddi AH. Dedifferentiation and redifferentiation of articular chondrocytes from surface and middle zones: changes in microRNAs-221/-222.140.and -143/145 expression. Tissue Eng Part A . 2013; 19:1015–22. doi: 10.1089/ten.TEA.2012.0055. [DOI] [PubMed] [Google Scholar]
- 134.Wei L, Wang M, Qu X , et al. Differential expression of microRNAs during allograft rejection. Am J Transplant. 2012;12:1113–23. doi: 10.1111/j.1600-6143.2011.03958.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Song W, Wu K, Yan J , et al. MiR-148b laden titanium implant promoting osteogenic differentiation of rat bone marrow mesenchymal stem cells. RSC Adv. 2013;3:11292–300. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material is available on the publishers web site along with the published article.