Abstract
Background and Aim
The accuracy for predicting virological outcomes of peginterferon-α and ribavirin therapy in patients with chronic hepatitis C is limited to approximately 80%, even with IL28B genotyping. Our in vitro study revealed that the numbers of (TA) dinucleotide repeats [(TA)n] of rs72258881, which is located in the promoter region of IL28B gene, might regulate IL28B transcription. We aimed to evaluate the usefulness of these host factors for predicting virological outcomes of this therapy in response-guided clinical settings.
Methods
A nationwide, multi-center prospective study in Japan determined IL28B (rs8099917) genotype, (TA)n of rs72258881, and amino acid substitutions of hepatitis C virus and used these for multivariate analysis together with other parameters at pretreatment.
Results
After enrolling 215 patients with genotype 1 and high viral load from 23 hospitals between October 2009 and February 2011, intent-to-treat analysis identified 202 patients in whom the final virological outcomes could be determined. Non-virological response by non-TT genotype was predicted with 79.7% accuracy. When combined with the (TA)n, the incidences of virological response tended to be higher in the longer (TA)n group, regardless of rs8099917 genotype. Multivariate logistic regression analysis revealed that rs8099917 non-TT genotype (P < 0.001), shorter (TA)n (P = 0.011), mutation of amino acid 70 in the virus core region (P = 0.029), and lower levels of serum albumin (P = 0.036) were independently associated with non-virological response.
Conclusions
IL28B genotype and (TA)n of rs72258881 may independently affect virological outcomes of peginterferon-α and ribavirin as host factors, even in response-guided therapy.
Keywords: (TA) dinucleotide repeat, chronic hepatitis C, IL28B, response-guided therapy
Introduction
Globally as many as 150 millions of people are infected by hepatitis C virus (HCV) and every year approximately 350 000 patients die of HCV-related liver diseases, such as liver cirrhosis or hepatocellular carcinoma.1 The standard care for the treatment of chronic hepatitis C has been PEGylated interferon-α (PEG-IFN-α) and ribavirin (P/R) with sustained virological response (SVR) rates as high as 50% achieved, even in difficult-to-treat combination of HCV genotype 1 and high viral load.2 Recently introduced protease inhibitors, such as boceprevir3,4 or telaprevir,5,6 can improve the SVR rate as much as 70–80% in IFN-naïve patients. Furthermore, the era of interferon-free treatment with only-oral directly-acting antivirals has just arrived.7–9 However, elderly HCV-infected patients are often unable to either tolerate or afford these new treatment regimens. In such circumstances, it is very important to identify easy-to-treat patients prior to treatment. Factors that contribute to the success of drug-mediated eradication of HCV include host factors (such as age, gender, the extent of liver fibrosis, and insulin resistance10,11), viral factors (such as HCV genotype, viral load, and amino acid substitutions in core12 and NS5A13 regions), as well as drug-related factors (such as treatment regimens, adherence to these regimens, total drug doses, and the duration of treatment).14 The most epoch-making discoveries in this field were that single nucleotide polymorphisms (SNPs) in the IL28B gene (rs8099917, rs12979860) can predict virological response (VR) to P/R.15–19 However, all these findings were based primarily on retrospective studies with per protocol analysis. Furthermore, even if those factors are all concentrated, the accuracy with which therapeutic outcomes can be predicted still remains approximately 80%. We recently reported that the number of (TA) dinucleotide repeats [(TA)n] of rs72258881, which is located in the promoter region of IL28B gene, might regulate its transcription.20 Here we report our efforts to verify the role of (TA)n of rs72258881, by conducting a prospective multi-center cohort study with intent-to-treat analysis, in Japanese patients infected with HCV genotype 1 who were treated by response-guided therapy (RGT) with P/R.
Methods
Study Design
From October 2009 to February 2011, 233 patients with chronic hepatitis C were prospectively enrolled from nationwide 23 hospitals in Japan (Trial Registration: UMIN-CTR000002580); however, 18 patients were considered to be ineligible and excluded from this study because of violation of the following entry criteria: (1) infection with HCV serotype 121 or genotype 1 (1a or 1b)22 without co-infection with hepatitis B virus or human immunodeficiency virus; (2) pretreatment HCV RNA levels ≥ 5.0 log10 IU/mL, as determined using a quantitative real-time PCR method (COBAS AmpliPrep/COBAS TaqMan HCV test; Roche Molecular Systems, Pleasanton, CA, USA); (3) standard P/R therapy according to the American Association of the Study of the Liver Diseases (AASLD) guidelines.23 Consequently, 215 patients met the entry criteria and were treated with weekly administration of PEG-IFN-α2a (Chugai Pharmaceutical, Tokyo, Japan) and daily administration of ribavirin (Chugai Pharmaceutical), or with weekly administration of PEG-IFN-α2b (MSD Co., Tokyo, Japan) and daily administration of ribavirin (MSD Co.). Whereas the dose of PEG-IFN-α2a was 180 μg, regardless of the patient's body weight, doses of PEG-IFN-α2b were adjusted based on the patient's body weight: respective weekly doses of PEG-IFN-α2b for patients < 45 kg, ≥ 45 kg, and < 60 kg; ≥ 60 kg and < 75 kg; ≥ 75 kg and < 90 kg; and ≥ 90 kg were given 60 μg, 80 μg, 100 μg, 120 μg, and 150 μg of PEG-IFN-α2b. Respective daily doses of ribavirin for patients < 60 kg, ≥ 60 kg and < 80 kg, and ≥ 80 kg were given 600 mg, 800 mg, and 1000 mg. Dose modifications of PEG-IFN-α or ribavirin, relating to adverse events, were based on the manufacturers' recommendations.
The treatment duration was determined based on RGT according to guidelines of AASLD23 and the Japan Society of Hepatology (JSH).24 Patients in whom serum HCV RNA had disappeared within 12 weeks after starting therapy received a 48-week treatment regimen. Patients in whom serum HCV RNA was still detectable at 12 weeks, but not at 24 weeks after starting therapy received a 72-week extended treatment regimen.
VR was defined as achieving SVR or transient virological response (TVR); whereas SVR was defined as undetectable HCV RNA in serum 24 weeks after the cessation of treatment, TVR was defined as undetectable HCV RNA at the cessation of treatment with reappearance of HCV RNA in serum thereafter. Non-virological response (NVR), which was defined as detectable viremia throughout the 24 weeks of P/R therapy, was classified as one of two categories. The first of these, “null responder”, was defined as < 2 log-unit decline in the serum levels of HCV RNA from the pretreatment baseline value within the first 12 weeks and detectable viremia at 24 weeks after the start of P/R. The second, “partial responder”, was defined as ≥ 2 log-unit decline in the serum levels of HCV RNA from the pretreatment baseline value within the first 12 weeks and detectable viremia at 24 weeks after the start of P/R. Patients whose treatment was withdrawn due either to the presence of serum HCV RNA after 24 weeks of therapy or to viral breakthrough (VBT) were also included in this study for intent-to-treat analysis. VBT was included in NVR, and was subclassified into “null responder” or “partial responder”, according to the above criteria. Adherence to PEG-IFN-α and ribavirin up to 12 weeks after the start of P/R were calculated.
The study protocol (Fig. 1) complied with the Helsinki Declaration and was approved by the ethics committee of each participating institution. At the time of enrollment, written informed consent was obtained for the collection and storage of serum and peripheral blood.
Figure 1.
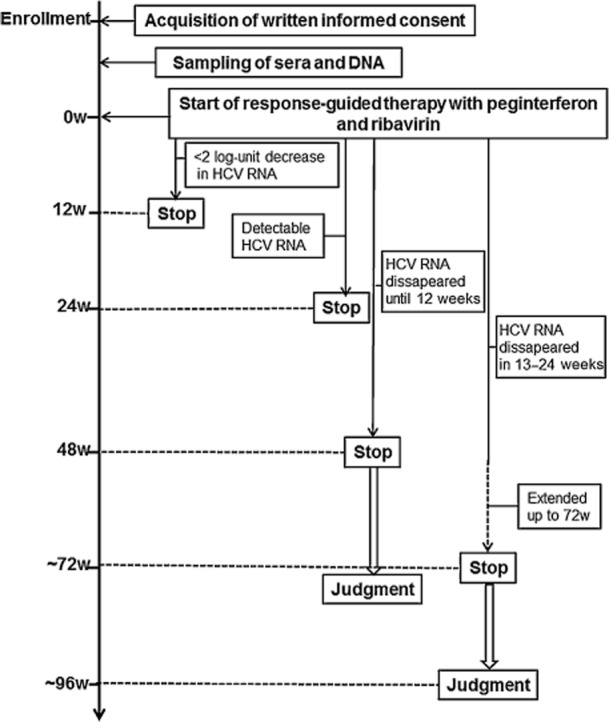
Study protocol for response-guided therapy. Durations of treatments were determined depending on the virological responses to peginterferon-α and ribavirin. If the serum hepatitis C virus (HCV) RNA became undetectable within 12 weeks (w), the treatment was stopped at 48 weeks. If the serum HCV RNA disappeared between 13 to 24 weeks after treatment, the duration was extended up to 72 weeks. Both cases were considered to meet the standard of care.
DNA Extraction
Genomic DNA was extracted from the buffy coat fraction of patients' whole blood using a GENOMIX kit (Talent SRL; Trieste, Italy).
IL28B genotyping
We previously reported that the rs8099917 polymorphism is a better predictor of the response of P/R to chronic hepatitis C in Japanese patients than any other SNPs reported near the IL28B gene.25 Therefore, the rs8099917 polymorphism was genotyped using the InvaderPlus assay (Third Wave Japan, Tokyo, Japan), which combines the polymerase chain reaction (PCR) and the invasive signal amplification reaction.26,27 In this prospective cohort study, in order to meet the requirements of RGT, both the doctors and the patients were blinded to the results of IL28B genotyping until final determination of virological outcomes.
Detection of amino acid substitutions in core and NS5A regions of HCV
Amino acids 70 and 91 of the HCV core region and the amino acid sequence of interferon-sensitivity determining region (ISDR: residues 2209–2248 of the NS5A region) were determined by direct sequencing, as previously reported.12,13
TA repeat genotyping
To determine the genotype of the TA repeat polymorphism, we used GeneScan analysis to detect the fragment size of the fluorescently labeled PCR amplicon. This method requires the use of nested PCR to prevent amplification of IL-29 region with high sequence similarity to regions within the IL-28A and IL-28B genes. The details of the nested PCR and GeneScan analysis may be found in the online version at the publisher's web-site as Supporting Information Table S1. The repeat number was validated by capillary sequencing as described previously.20
Evaluation of liver fibrosis by a simple noninvasive index (FIB-4)
The Fibrosis 4 (FIB-4) index that was used to evaluate liver fibrosis in each patient correlates well with hepatic fibrosis (as determined by liver biopsy) and requires only readily available clinical parameters for its determination.28
Statistical Analysis
Quantitative variables were expressed as the mean ± standard deviation, unless otherwise specified. Categorical variables were compared using Pearson's χ2-test or Fisher's exact test. Continuous variables were compared using the Mann–Whitney U-test. Multivariate and simultaneous logistic regression analysis was performed to determine predictive factors for NVR, by using the variables which were found to be P < 0.150 by univariate analysis. In addition, a decision tree modeled these pretreatment factors to predict NVR. All P values were two-tailed, and P < 0.05 was considered statistically significant. Data analyses were performed using IBM SPSS Statistics 20 (IBM, Armonk, NY, USA).
Results
Treatment profiles and virological outcomes
In this prospective study, 215 patients infected by HCV of serotype 1 or genotype 1 were eligible. The sub-genotypes of HCV were as follows: 1a (n = 2), 1b (n = 208), 1b + 2b (n = 1), and indeterminate (n = 4). By the end of November 2012, virological outcomes had been determined in 202 patients, except for 13 patients (6%) who were lost to follow-up. Whereas all of these patients were treated with P/R, 160 patients (74%) completed standard of care (SOC) treatment for at least 48 weeks, the remaining 55 patients had to withdraw from treatment owing to either serious adverse events (SAE) in 25 patients (12%), poor response in 24 patients (11%), or other unrelated causes in 6 patients (3%). The SAE or unrelated causes responsible for the termination of the treatment were described in the small inset of Figure 2.
Figure 2.
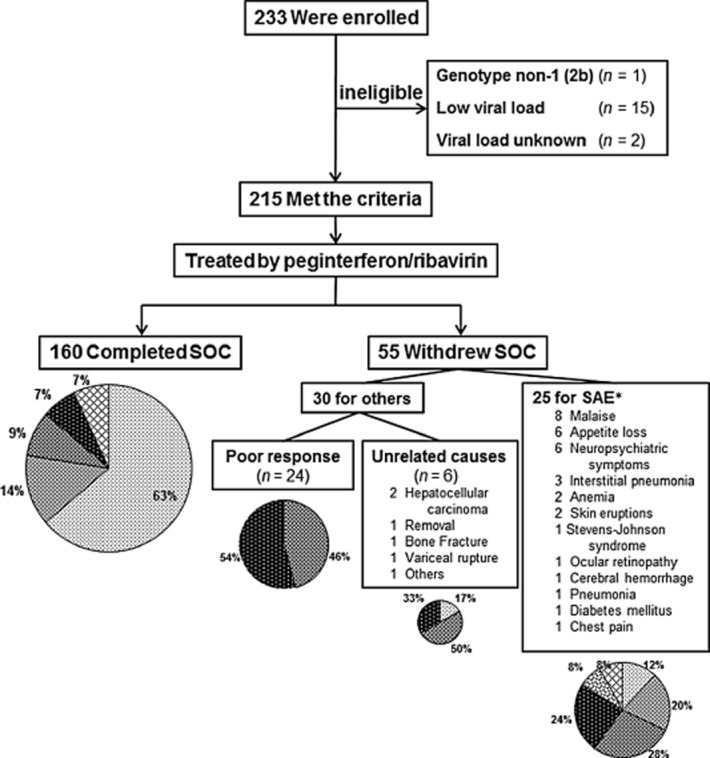
Enrollment and outcomes. Among 233 patients enrolled in this study, 18 patients were ineligible for the following reasons: genotype non-1 (2b) (n = 1); low viral load (n = 15); and unknown viral load (n = 2). Consequently, 215 patients met the entry criteria, and were treated with peginterferon and ribavirin. Among them, 160 patients completed standard of care (SOC). The remaining 55 patients were withdrawn from SOC, as detailed earlier. The virological outcomes with intent-to-treat analysis, as detailed in the Methods section, were shown as a pie chart for each group. *Serious adverse events were duplicated in some patients. RGT, response-guided therapy; SAE, serious adverse events; SVR, sustained virological response; TVR, transient virological response; NVR, non-virological response; R, responder; f/u, follow-up.  , SVR;
, SVR;  , TVR;
, TVR;  , Partial R (NVR);
, Partial R (NVR);  , Null R (NVR);
, Null R (NVR);  , undetemined (NVR);
, undetemined (NVR);  , Lost-to-f/u.
, Lost-to-f/u.
As shown in Figure 2, if the patients completed at least 48 weeks of P/R (SOC) under conditions that observed the requirements of RGT, the SVR rate was as high as 63%, and the incidences of VR (the sum of SVR and TVR) was 77%. In the patients where the treatment was terminated for SAE or unrelated causes, the respective incidences of VR were reduced to 32% or 17%, respectively. In particular, treatment of 24 patients had to be terminated owing to the poor response to P/R, as defined by the requirements of RGT. This resulted in 11 “partial responders” (46%) and 13 “null responders” (54%). In addition, 9 cases developed viral breakthrough, and were subclassified into “partial responder”: 6 of them completed SOC, while 3 stopped treatment because of the poor response to P/R (data not shown).
Patients' characteristics and IL28B genotypes
Whereas 154 individuals (71.6%) had the rs8099917 TT genotype (major-homo), 60 had the TG (hetero) genotype and 1 had the GG (minor-homo) genotype. The patients were classified into two groups, TT and non-TT (TG/GG), according to their rs8099917 genotypes, and their characteristics were compared. As shown in Table 1, lower levels of γ-GTP (P < 0.001), higher levels of total cholesterol (T.Chol; P = 0.013), higher levels of low-density lipoprotein cholesterol (LDL-C; P < 0.001), and lower levels of α-fetoprotein (AFP; P = 0.009) were significantly associated with TT genotype. The percentages of wild type of core 70 amino acid of patients with the TT genotype were significantly higher than those of patients with either TG or GG genotypes (71.7% vs 30.5%, P < 0.001).
Table 1.
Comparisons of host and viral factors between IL28B TT and TG/GG genotypes
| Factors | TT genotype (154) | TG/GG genotype (61) | P value |
|---|---|---|---|
| Age (years) | 58 ± 11 (154) | 58 ± 12 (61) | 0.679 |
| Gender (male/female) | 88/66 | 31/30 | 0.448 |
| Body weight (kg) | 60.2 ± 11.2 (149) | 58.7 ± 12.5 (60) | 0.318 |
| IFN naïve/experienced | 123/31 | 47/14 | 0.711 |
| PEG-IFN-α2a/-α2b | 23/131 | 11/50 | 0.679 |
| Albumin (g/dL) | 4.1 ± 0.5 (153) | 4.1 ± 0.4 (60) | 0.721 |
| AST (U/L) | 56 ± 37 (154) | 66 ± 49 (61) | 0.332 |
| ALT (U/L) | 70 ± 52 (154) | 77 ± 59 (61) | 0.422 |
| T.Bil (mg/dL) | 0.86 ± 0.33 (150) | 0.84 ± 0.37 (61) | 0.658 |
| ALP (U/L) | 268 ± 93 (154) | 254 ± 87 (61) | 0.509 |
| γ-GTP (U/L) | 46 ± 43 (154) | 78 ± 99 (61) | < 0.001 |
| T.Chol (mg/dL) | 172 ± 39 (151) | 160 ± 29 (58) | 0.013 |
| LDL-C (mg/dL) | 103 ± 28 (132) | 84 ± 27 (57) | < 0.001 |
| FBS (mg/dL) | 101 ± 21 (130) | 108 ± 29 (53) | 0.076 |
| IRI (μU/mL) | 10.9 ± 9.5 (66) | 15.5 ± 20.9 (26) | 0.541 |
| AFP (ng/mL) | 14.4 ± 67.1 (141) | 17.8 ± 26.2 (57) | 0.009 |
| HCV RNA (Log IU/mL) | 6.5 ± 0.5 (154) | 6.4 ± 0.6 (61) | 0.243 |
| WBC (/μL) | 4917 ± 1367 (154) | 4940 ± 1105 (61) | 0.525 |
| Hemoglobin (g/dL) | 13.8 ± 1.3 (154) | 13.7 ± 1.7 (61) | 0.633 |
| Platelets (x104/μL) | 16.5 ± 5.9 (154) | 16.9 ± 4.7 (61) | 0.347 |
| FIB-4 index | 2.82 ± 1.77 (154) | 2.80 ± 1.76 (61) | 0.994 |
| Core 70 amino acid (wild/mutant) | 109/43 | 18/41 | < 0.001 |
| Core 91 amino acid (wild/mutant) | 102/50 | 38/21 | 0.747 |
| ISDR mutation (n = 0–1/2-) | 126/25 | 50/7 | 0.523 |
Data are shown as mean ± standard deviation. Figures in parentheses are the numbers of data available in each factor. Significant P values are shown in bold.
AFP, α-fetoprotein; ALP, alkaline phosphatase; ALT, alanine aminotransferase; AST, aspartate aminotransferase; FBS, fasting blood sugar; HCV, hepatitis C virus; IRI, immune-reactive insulin; ISDR, interferon-sensitivity determining region; LDL-C, low-density lipoprotein cholesterol; T.Bil, total bilirubin; T.Chol, total cholesterol; WBC, white blood cell; γ-GTP, γ-glutamyl transpeptidase.
Virological response, IL28B genotypes, and (TA)n of rs72258881
Intent-to-treat analysis of the entire cohort indicated SVR, TVR, and NVR rates of 49.3%, 12.6%, and 32.1%, respectively. As shown in Figure 3a, serum HCV RNA disappeared significantly earlier in patients with TT genotype than in those with either of the TG or GG genotype. Assessment of the usefulness of the rs8099917 non-TT genotype to predict NVR among 202 patients in whom the final virological outcome could be determined indicate a sensitivity of 63.8% (44/[44 + 25]); specificity of 88.0% (117/[117 + 16]); positive predictive value of 73.3% (44/[44 + 16]); negative predictive value of 82.4% (117/[25 + 117]), and an accuracy of 79.7% ([44 + 117]/202).
Figure 3.
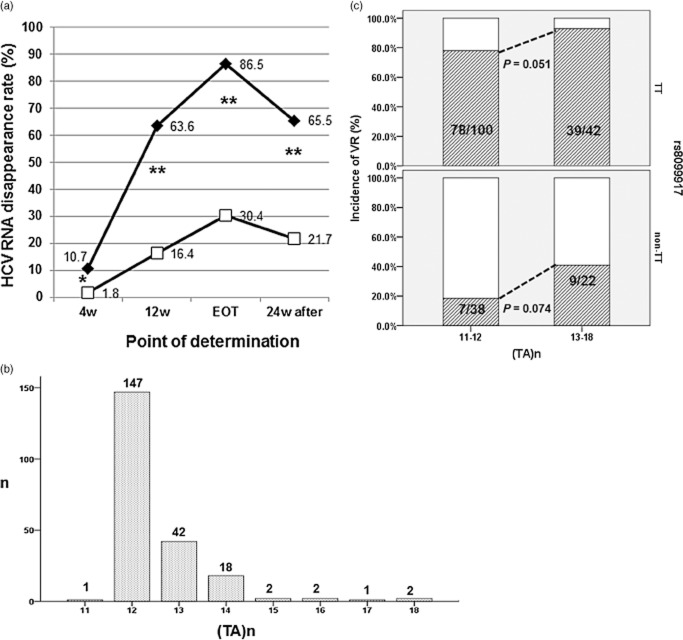
Genotype of rs8099917, (TA) dinucleotide repeat [(TA)n] of rs72258881 and virological response. (a) Hepatitis C virus (HCV) RNA disappearance rate. Serum HCV RNA disappeared significantly earlier in patients with TT genotype than in those with TG/GG genotypes. *P < 0.05, **P < 0.001. The abbreviation used was: EOT, end of treatment; HCV, hepatitis C virus; w, weeks.  , TT genotype;
, TT genotype;  , TG/GG genotype. (b) Distributions of (TA)n in this cohort. The most frequent (TA)n was 12 (n = 147: 68.4%). In 67 patients (31.2%), the numbers were more than 12. (c) The incidences of virological response (VR) in four groups stratified by rs8099917 genotype and (TA)n. The longer (TA)n might favor virological responses to PEGylated interferon-α and ribavirin, regardless of the IL28B genotype.
, TG/GG genotype. (b) Distributions of (TA)n in this cohort. The most frequent (TA)n was 12 (n = 147: 68.4%). In 67 patients (31.2%), the numbers were more than 12. (c) The incidences of virological response (VR) in four groups stratified by rs8099917 genotype and (TA)n. The longer (TA)n might favor virological responses to PEGylated interferon-α and ribavirin, regardless of the IL28B genotype.
As shown in Figure 3b, the (TA)n of rs72258881 varied from 11 to 18, with the most frequent numbers of repeats being 12 (n = 147; 68.4%). Given that more than 12 repeats were found in 67 patients (31.2%), the cohort was divided into 2 × 2 groups, according to rs8099917 genotype (TT or non-TT) and (TA)n (n = 11–12 or n = 13–18), and the incidences of VR were calculated as [SVR + TVR]/[SVR + TVR + NVR]. As shown in Figure 3c, the incidences of VR tended to be higher in the group with longer (TA)n than in the group with shorter (TA)n, regardless of the rs8099917 genotype. In particular, in patients with the non-TT genotype, the longer (TA)n might increase VR more than the twofold, relative to that for patients with a shorter (TA)n [compare 18.4% in (TA)11–12 vs 40.9% in (TA)13–18, P = 0.074].
Factors associated with NVR
After excluding 13 patients who were lost to follow-up, we attempted to identify the variables that were associated with final virological outcome. As shown in Table 2, univariate analysis indicated that nine variables were significantly different or tended to be different between VR and NVR. NVR was associated with higher levels of serum AST, γ-GTP, and FIB-4, but with lower levels of T.Chol. Whereas the core 70-mino acid mutation was a viral factor related to NVR, the rs8099917 non-TT genotype and shorter (TA)n were host factors related to NVR. For nine variables for which univariate analysis indicated that the P value less than 0.15, multivariate logistic regression analysis identified four variables for the prediction of NVR: HCV core 70 amino acid mutation, rs8099917 non-TT genotype, shorter (TA)n, and the lower levels of serum albumin. Finally, a decision tree temporarily modeled 22 pretreatment factors (see the legend to Fig. 4) to predict NVR. For this purpose, 13 patients who were lost to follow-up were excluded from the analysis, in order to avoid their influence on final decision. As shown in Figure 4, if the rs8099917 genotype was primarily selected as the predictive factor (P < 0.001, χ2 = 57.647), then the shorter (TA)n attributed to the NVR in patients with the TT genotype (P = 0.023, χ2 = 5.166). In cases with the shorter (TA)n (n = 11 or 12), the presence of the core 70 amino acid mutation in HCV might significantly increase the incidences of NVR (P = 0.008, χ2 = 9.115). On the other hand, in the patients with non-TT genotype, higher viral load was the second most powerful determinant of NVR (P = 0.001, χ2 = 15.645).
Table 2.
Univariate and multivariate analyses of patients with chronic hepatitis C treated with pegylated interferon-α and ribavirin with respect to VR and NVR
| Variable | Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|---|
| VR (133) | NVR (69) | P value | B | P value | Odds ratio | 95% CI | |
| Gender (Male/Female) | 74/59 | 38/31 | 1.000 | ||||
| Age (years) | 58 ± 11 (133) | 59 ± 11 (69) | 0.973 | ||||
| Body weight (kg) | 59.9 ± 11.5 (130) | 59.7 ± 11.6 (68) | 0.992 | ||||
| Hx. of IFN treatment (naïve/experienced) | 108/25 | 52/17 | 0.363 | ||||
| PEG-IFN-α2a/-α2b | 18/115 | 12/57 | 0.533 | ||||
| Albumin (g/dL) | 4.2 ± 0.5 (133) | 4.0 ± 0.4 (67) | 0.148 | 0.118 | 0.036 | 1.125 | 1.008–1.256 |
| AST (U/L) | 52 ± 34 (133) | 70 ± 50 (69) | 0.022 | −0.001 | 0.919 | 0.999 | 0.983–1.016 |
| ALT (U/L) | 66 ± 47 (133) | 81 ± 64 (69) | 0.244 | ||||
| T.Bil (mg/dL) | 0.88 ± 0.32 (129) | 0.81 ± 0.35 (69) | 0.085 | 0.102 | 0.083 | 1.108 | 0.987–1.243 |
| ALP (U/L) | 258 ± 80 (133) | 273 ± 109 (69) | 0.378 | ||||
| γ-GTP (U/L) | 45 ± 43 (133) | 75 ± 95 (69) | < 0.001 | −0.002 | 0.508 | 0.998 | 0.991–1.005 |
| T.Chol (mg/dL) | 172 ± 39 (130) | 161 ± 32 (66) | 0.016 | 0.004 | 0.556 | 1.004 | 0.991–1.017 |
| HCV RNA (Log IU/mL) | 6.5 ± 0.6 (133) | 6.6 ± 0.5 (69) | 0.384 | ||||
| WBC (/μL) | 4982 ± 1248 (133) | 4784 ± 1271(69) | 0.275 | ||||
| Hemoglobin (g/dL) | 13.9 ± 1.2 (133) | 13.7 ± 1.8 (69) | 0.308 | ||||
| Platelets (× 104/μL) | 17.0 ± 5.9 (133) | 16.0 ± 5.0 (69) | 0.377 | ||||
| FIB-4 index | 2.7 ± 1.7 (133) | 3.1 ± 1.8 (69) | 0.035 | 0.055 | 0.755 | 1.056 | 0.749–1.490 |
| Core 70 amino acid (wild/mutant) | 93/37 | 23/45 | < 0.001 | −0.914 | 0.029 | 0.401 | 0.177–0.910 |
| Core 91 amino acid (wild/mutant) | 92/38 | 42/26 | 0.205 | ||||
| ISDR mutation (n = 0–1/2-) | 108/20 | 59/8 | 0.528 | ||||
| rs8099917 (TT/non-TT) | 117/16 | 25/44 | < 0.001 | −2.735 | < 0.001 | 0.065 | 0.025–0.171 |
| (TA)n (n = 11–12/13–18) | 85/48 | 53/16 | 0.079 | −1.226 | 0.011 | 0.294 | 0.114–0.757 |
| PEG-IFN adherence (%) | 95.5 ± 10.3 (132) | 89.7 ± 17.3 (64) | 0.093 | ||||
| Ribavirin adherence (%) | 94.8 ± 10.9 (133) | 90.3 ± 17.7 (64) | 0.232 | ||||
Data are shown as mean ± standard deviation. Figures in parentheses are the numbers of data available in each variable. Multivariate and simultaneous logistic regression analysis was performed to determine predictive factors for NVR, by using nine variables which were found to be P < 0.150 by univariate analysis (albumin, AST, T.Bil, γ-GTP, T.Chol, fib-4 index, Core 70 amino acid, rs8099917, (TA)n). In addition, PEG-IFN adherence and ribavirin adherence were excluded from this analysis, since these two variables could not be available at pretreatment. The corresponding references in categorical variables were as follows: wild (Core 70 amino acid); TT (rs8099917); n = 13–18 [(TA)n]. Significant P values are shown in bold. The calculated values for serum albumin and T.Bil by multivariate logistic regression analysis correspond to those per 0.1 of increase.
ALP, alkaline phosphatase; ALT, alanine aminotransferase; AST, aspartate aminotransferase; CI, confidence interval; HCV, hepatitis C virus; Hx., history; ISDR, interferon-sensitivity determining region; NVR, non-virological response; PEG-IFN, pegylated interferon; T.Bil, total bilirubin; T.Chol, total cholesterol; VR, virological response; WBC, white blood cell; γGTP, γ-glutamyl transpeptidase.
Figure 4.
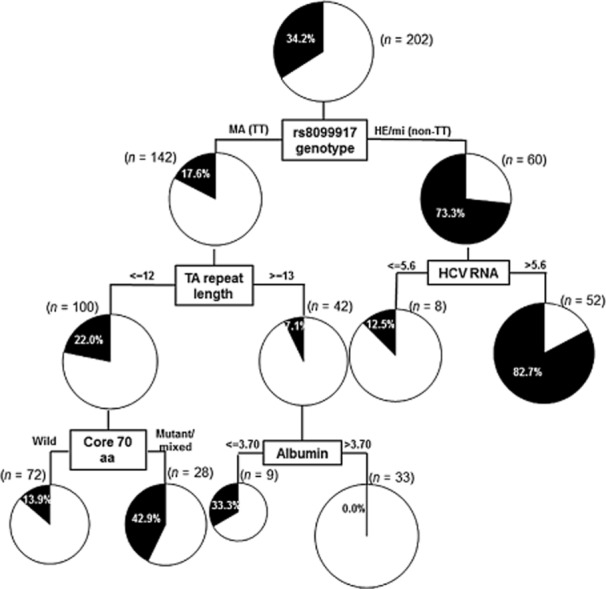
Decision tree analysis for the virological outcomes. Boxes indicate the factors used for splitting and the cut-off value for the split. Pie charts indicate the rate of non-virological response for each group of patients after splitting. A total of 202 patients were included in this analysis, after excluding 13 patients who were lost to follow-up, in order to avoid the influence on final decision. Among 22 pretreatment factors (gender, prior history of interferon, pegylated interferon regimen, age, body weight; serum albumin, aspartate aminotransferase, alanine aminotransferase, total bilirubin, alkaline phosphatase, γ-glutamyl transpeptidase, total cholesterol; white blood cell, hemoglobin, platelets; FIB-4; serum levels of hepatitis C virus (HCV) RNA (reverse transcription–polymerase chain reaction), core 70/91 amino acid mutation, interferon-sensitivity determining region mutation; rs8099917 genotype, TA repeat length) tested for their abilities to predict non-virological responses, determinations of (TA)n of rs72258881 and/or the HCV core 70 amino acid substitution were useful, especially for patients with the TT genotype. In the patients with a non-TT genotype, HCV viral load was the second most important determinant of virological response. MA, major-homo; HE, hetero; mi, minor-homo; aa, amino acid. The units used to measure levels of albumin and HCV RNA were g/dL and Log IU/mL, respectively.
Discussion
This is the first prospective evaluation of the usefulness of the rs8099917 SNP of the IL28B gene to predict virological outcome of RGT with P/R in the patients with chronic hepatitis C. Given that RGT has been accepted by AASLD,2,23 European Association for the Study of the Liver (EASL),29 and JSH24 as the standard interferon-based treatment, it is often challenging to perform conventional per-protocol analysis in real clinical settings. With intent-to-treat analysis, the rates of SVR, TVR, and NVR were 49.3%, 12.6%, and 32.1%, respectively. The SVR rate in our cohort was as much as 10% higher than that in a report based on non-RGT with intent-to-treat analysis.30 This might substantiate the value of RGT for the treatment of chronic hepatitis C. Differences between the TT genotype and non-TT genotype of rs8099917 in several background features are of interest. For instance, serum levels of γ-GTP and AFP were higher, while those of T.Chol and LDL-C were lower in patients with the non-TT genotype, compared with those with the TT genotype. Especially for the association of γ-GTP and LDL-C levels with IL28B genotypes, quite similar results were recently reported for HALT-C study31 and our retrospective study,32 respectively. However, the precise mechanisms by which IL28B genotypes affect levels of γ-GTP and LDL-C have yet to be elucidated. The current study confirmed that the core 70 amino acid mutation is more frequently associated with the non-TT genotype than with TT genotype.33 The rs8099917 genotype could clearly differentiate between the effects of P/R on the disappearance of serum HCV RNA, which ultimately leads to SVR rate at 65.5% and 21.7%, in the TT and non-TT genotypes, respectively. However, given the less than 80% accuracy in predicting NVR in non-TT genotypes, there is likely a fair level of discrepancy between predicted and actual virological outcome. We have recently reported that the (TA) dinucleotide repeat of rs72258881, which lies in the promoter region of IL28B gene, can affect the transcriptional activity in a (TA)n length-dependent manner in vitro.20 The current study has indicated that in clinical settings, increases in (TA)n tended to increase the incidences of VR in patients regardless of rs8099917 genotypes. Although the current study did not directly assess the expression of IL28B gene at the mRNA or protein levels, further investigation of mechanistic basis of the length-dependent effects of (TA)n on VR might help to elucidate how the responsiveness of patients to P/R is regulated by their levels of IL28B expression.
Univariate analysis indicated that NVR was associated with significantly higher levels of serum AST, γ-GTP, and FIB-4, together with lower levels of T.Chol, than for VR. These findings are consistent with those of our previous retrospective study that involved per-protocol analysis.32 Regarding the viral factor, the core 70 amino acid mutation was significantly correlated with NVR, as reported previously.12 Multivariate logistic regression analysis revealed that rs8099917 non-TT genotype, shorter (TA)n, core 70 amino acid mutation, and the lower levels of serum albumin were independently associated with NVR, in this prospective cohort with RGT. A decision tree analysis might demonstrate more clearly the clinical implications to measure simultaneously these host and viral factors at pretreatment.
There are several limitations to this study. First, the prospective study design prompted us to evaluate the virological response by performing intent-to-treat analysis, rather than strict per-protocol analysis. Especially, in patients for whom treatment needed to be terminated prematurely owing to SAE or other unrelated causes, virological outcomes were worse than expected. Nonetheless, the results of this study appear to reflect the real clinical settings used for the treatment of chronic hepatitis C. Another limitation was that the cohort contained fewer patients with non-TT genotype of rs8099917 than those with TT genotype. This might explain the unexpected observation that decision tree analysis did not select (TA)n as a predictive factor in the group with non-TT genotype. External validation is needed to establish whether the results of our prospective study could apply more generally. However, a requirement for such studies is that the indications of current and future direct-acting antiviral agents for chronic hepatitis C should be further clarified in clinical settings.
In conclusion, the IL28B genotype and (TA)n of rs72258881 may independently affect the virological outcomes of RGT with P/R for chronic hepatitis C. At a minimum, when considering P/R-based regimens for chronic hepatitis C, pretreatment determinations of both genotypes as well as the core 70 amino acid mutation of HCV are promising cost-effective tools to predict VR.
Acknowledgments
The authors thank Dr. Yasuyuki Kohjima (NHO Kyushu Medical Center), Dr. Kyoko Fukuhara and Dr. Ken-ichi Ikejima (Juntendo University), Dr. Kunio Nakane (Akita City Hospital), Dr. Hirohito Yoneyama (Kagawa Medical University School of Medicine), Dr. Shinya Maekawa (Yamanashi University School of Medicine), Dr. Haruhisa Nakao (Aichi Medical University School of Medicine), Dr. Hiroshi Mitsui and Dr. Naoaki Hashimoto (Tokyo Teishin Hospital), Dr. Takeya Tsutsumi and Dr. Hiroshi Yotsuyanagi (The University of Tokyo), Dr. Hideaki Miura and Dr. Haruki Yamada (Social Insurance Chuo General Hospital), Dr. Keiichi Hirata and Dr. Shigeki Hayashi (NHO Disaster Medical Center), Dr. Koichi Takaguchi (Kagawa Prefectural Central Hospital), Dr. Shogo Ohkoshi and Dr. Yutaka Aoyagi (Niigata University School of Medicine), Dr. Akihisa Miyazaki (Juntendo University, Nerima Hospital), Dr. Hiroshi Adachi (Tonami General Hospital), Dr. Tsutomu Ogawa (Takamatsu Red Cross Hospital), Dr. Masaaki Korenaga, Dr. Keisuke Hino (Kawasaki Medical School Hospital), Dr. Hideyuki Nomura (SHIN-KOKURA Hospital), Dr. Mikio Yanase (National Center for Global Health and Medicine Hospital) for assistance with the collection of clinical samples.
Supporting Information
Additional Supporting Information may be found in the online version of this article at the publisher's web-site
Methods of TA repeat genotyping.
References
- 1.World Health Organization. 2014. WHO Fact sheet N°164, Updated April. Hepatitis C. Cited 19 June 2014. Available from URL: http://www.who.int/mediacentre/factsheets/fs164/en/index.html.
- 2.Ghany MG, Nelson DR, Strader DB, Thomas DL, Seeff LB American Association for Study of Liver Diseases. An update on treatment of genotype 1 chronic hepatitis C virus infection: 2011 Practice guideline by the American Association for the Study of Liver Diseases. Hepatology. 2011;54:1433–1443. doi: 10.1002/hep.24641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Poordad F, McCone J, Jr, Bacon BR, et al. Boceprevir for untreated chronic HCV genotype 1 infection. N. Engl. J. Med. 2011;364:1195–1206. doi: 10.1056/NEJMoa1010494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bacon BR, Gordon SC, Lawitz E, et al. Boceprevir for previously treated chronic HCV genotype 1 infection. N. Engl. J. Med. 2011;364:1207–1217. doi: 10.1056/NEJMoa1009482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jacobson IM, McHutchison JG, Dusheiko G, et al. Telaprevir for previously untreated chronic hepatitis C virus infection. N. Engl. J. Med. 2011;364:2405–2416. doi: 10.1056/NEJMoa1012912. [DOI] [PubMed] [Google Scholar]
- 6.McHutchison JG, Manns MP, Muir AJ, et al. Telaprevir for previously treated chronic HCV infection. N. Engl. J. Med. 2010;362:1292–1303. doi: 10.1056/NEJMoa0908014. [DOI] [PubMed] [Google Scholar]
- 7.Chayama K, Takahashi S, Toyota J, et al. Dual therapy with the nonstructural protein 5A inhibitor, daclatasvir, and the nonstructural protein 3 protease inhibitor, asunaprevir, in hepatitis C virus genotype 1b-infected null responders. Hepatology. 55:742–748. doi: 10.1002/hep.24724. [DOI] [PubMed] [Google Scholar]
- Gane EJ, Stedman CA, Hyland RH, et al. Nucleotide polymerase inhibitor sofosbuvir plus ribavirin for hepatitis C. N. Engl. J. Med. 2013;368:34–44. doi: 10.1056/NEJMoa1208953. [DOI] [PubMed] [Google Scholar]
- 9.Poordad F, Lawitz E, Kowdley KV, et al. Exploratory study of oral combination antiviral therapy for hepatitis C. N. Engl. J. Med. 2013;368:45–53. doi: 10.1056/NEJMoa1208809. [DOI] [PubMed] [Google Scholar]
- 10.Guedj H, Guedj J, Negro F, et al. The impact of fibrosis and steatosis on early viral kinetics in HCV genotype 1-infected patients treated with Peg-IFN-alfa-2a and ribavirin. J. Viral Hepat. 2012;19:488–496. doi: 10.1111/j.1365-2893.2011.01569.x. [DOI] [PubMed] [Google Scholar]
- 11.Harrison SA, Hamzeh FM, Han J, Pandya PK, Sheikh MY, Vierling JM. Chronic hepatitis C genotype 1 patients with insulin resistance treated with pioglitazone and peginterferon alpha-2a plus ribavirin. Hepatology. 2012;56:464–473. doi: 10.1002/hep.25661. [DOI] [PubMed] [Google Scholar]
- 12.Akuta N, Suzuki F, Sezaki H, et al. Association of amino acid substitution pattern in core protein of hepatitis C virus genotype 1b high viral load and non-virological response to interferon-ribavirin combination therapy. Intervirology. 2005;48:372–380. doi: 10.1159/000086064. [DOI] [PubMed] [Google Scholar]
- 13.Enomoto N, Sakuma I, Asahina Y, et al. Mutations in the nonstructural protein 5A gene and response to interferon in patients with chronic hepatitis C virus 1b infection. N. Engl. J. Med. 1996;334:77–81. doi: 10.1056/NEJM199601113340203. [DOI] [PubMed] [Google Scholar]
- 14.Manns MP. Adherence to combination therapy: influence on sustained virologic response and economic impact. Gastroenterol. Clin. North Am. 2004;33:11–24. doi: 10.1016/j.gtc.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 15.Tanaka Y, Nishida N, Sugiyama M, et al. Genome-wide association of IL28B with response to pegylated interferon-alpha and ribavirin therapy for chronic hepatitis C. Nat. Genet. 2009;41:1105–1109. doi: 10.1038/ng.449. [DOI] [PubMed] [Google Scholar]
- 16.Suppiah V, Moldovan M, Ahlenstiel G, et al. IL28B is associated with response to chronic hepatitis C interferon-alpha and ribavirin therapy. Nat. Genet. 2009;41:1100–1104. doi: 10.1038/ng.447. [DOI] [PubMed] [Google Scholar]
- 17.Thomas DL, Thio CL, Martin MP, et al. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature. 2009;461:798–801. doi: 10.1038/nature08463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ge D, Fellay J, Thompson AJ, et al. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 2009;461:399–401. doi: 10.1038/nature08309. [DOI] [PubMed] [Google Scholar]
- 19.Rauch A, Kutalik Z, Descombes P, et al. Genetic variation in IL28B is associated with chronic hepatitis C and treatment failure: a genome-wide association study. Gastroenterology. 2010;138:1338–1345. doi: 10.1053/j.gastro.2009.12.056. [DOI] [PubMed] [Google Scholar]
- 20.Sugiyama M, Tanaka Y, Wakita T, Nakanishi M, Mizokami M. Genetic variation of the IL-28B promoter affecting gene expression. PLoS ONE. 2011;6:e26620. doi: 10.1371/journal.pone.0026620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsukiyama-Kohara K, Yamaguchi K, Maki N, et al. Antigenicities of group I and II hepatitis C virus polypeptides—molecular basis of diagnosis. Virology. 1993;192:430–437. doi: 10.1006/viro.1993.1058. [DOI] [PubMed] [Google Scholar]
- 22.Ohno T, Mizokami M, Wu RR, et al. New hepatitis C virus (HCV) genotyping system that allows for identification of HCV genotypes 1a, 1b, 2a, 2b, 3a, 3b, 4, 5a, and 6a. J. Clin. Microbiol. 1997;35:201–207. doi: 10.1128/jcm.35.1.201-207.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ghany MG, Strader DB, Thomas DL, Seeff LB American Association for the Study of Liver Diseases. Diagnosis, management, and treatment of hepatitis C: an update. Hepatology. 2009;49:1335–1374. doi: 10.1002/hep.22759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Izumi N, Nishiguchi S, Hino K, et al. Management of hepatitis C; Report of the Consensus Meeting at the 45th Annual Meeting of the Japan Society of Hepatology (2009) Hepatol. Res. 2010;40:347–368. doi: 10.1111/j.1872-034X.2010.00642.x. [DOI] [PubMed] [Google Scholar]
- 25.Ito K, Higami K, Masaki N, et al. The rs8099917 polymorphism, determined by a suitable genotyping method, is a better predictor for response to pegylated interferon-alpha/ribavirin therapy in Japanese patients than other SNPs associated with IL28B. J. Clin. Microbiol. 2011;49:1853–1860. doi: 10.1128/JCM.02139-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lyamichev VI, Kaiser MW, Lyamicheva NE, et al. Experimental and theoretical analysis of the invasive signal amplification reaction. Biochemistry. 2000;39:9523–9532. doi: 10.1021/bi0007829. [DOI] [PubMed] [Google Scholar]
- 27.Hall JG, Eis PS, Law SM, et al. Sensitive detection of DNA polymorphisms by the serial invasive signal amplification reaction. Proc. Natl. Acad. Sci. U.S.A. 2000;97:8272–8277. doi: 10.1073/pnas.140225597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sterling RK, Lissen E, Clumeck N, et al. Development of a simple noninvasive index to predict significant fibrosis in patients with HIV/HCV coinfection. Hepatology. 2006;43:1317–1325. doi: 10.1002/hep.21178. [DOI] [PubMed] [Google Scholar]
- 29.European Association for the Study of the Liver. EASL Clinical Practice Guidelines: management of hepatitis C virus infection. J. Hepatol. 2011;55:245–264. doi: 10.1016/j.jhep.2011.02.023. [DOI] [PubMed] [Google Scholar]
- 30.McHutchison JG, Lawitz EJ, Shiffman ML, et al. Peginterferon alfa-2b or alfa-2a with ribavirin for treatment of hepatitis C infection. N. Engl. J. Med. 2009;361:580–593. doi: 10.1056/NEJMoa0808010. [DOI] [PubMed] [Google Scholar]
- 31.Everhart JE, Wright EC. Association of γ-glutamyltransferase (GGT) activity with treatment and clinical outcomes in chronic hepatitis C (HCV) Hepatology. 2013;57:1725–1733. doi: 10.1002/hep.26203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saito H, Ito K, Sugiyama M, et al. Factors responsible for the discrepancy between IL28B polymorphism prediction and the viral response to peginterferon plus ribavirin therapy in Japanese chronic hepatitis C patients. Hepatol. Res. 2012;42:958–965. doi: 10.1111/j.1872-034X.2012.01013.x. [DOI] [PubMed] [Google Scholar]
- 33.Akuta N, Suzuki F, Hirakawa M, et al. Amino acid substitution in hepatitis C virus core region and genetic variation near the interleukin 28B gene predict viral response to telaprevir with peginterferon and ribavirin. Hepatology. 2010;52:421–429. doi: 10.1002/hep.23690. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Methods of TA repeat genotyping.