Abstract
Cilia are microtubule-based cellular organelles that mediate signal transduction. Cilia are organized into several structurally and functionally distinct compartments: the basal body, the transition zone (TZ), and the cilia shaft. In vertebrates, the cystoprotein Inversin localizes to a portion of the cilia shaft adjacent to the TZ, a region termed the “Inversin compartment” (InvC). The mechanisms that establish and maintain the InvC are unknown. In the roundworm C. elegans, the cilia shafts of amphid channel and phasmid sensory cilia are subdivided into two regions defined by different microtubule ultrastructure: a proximal doublet-based region adjacent to the TZ, and a distal singlet-based region. It has been suggested that C. elegans cilia also possess an InvC, similarly to mammalian primary cilia. Here we explored the biogenesis, structure, and composition of the C. elegans ciliary doublet region and InvC. We show that the InvC is conserved and distinct from the doublet region. nphp-2 (the C. elegans Inversin homolog) and the doublet region genes arl-13, klp-11, and unc-119 are redundantly required for ciliogenesis. InvC and doublet region genes can be sorted into two modules—nphp-2+klp-11 and arl-13+unc-119—which are both antagonized by the hdac-6 deacetylase. The genes of this network modulate the sizes of the NPHP-2 InvC and ARL-13 doublet region. Glutamylation, a tubulin post-translational modification, is not required for ciliary targeting of InvC and doublet region components; rather, glutamylation is modulated by nphp-2, arl-13, and unc-119. The ciliary targeting and restricted localization of NPHP-2, ARL-13, and UNC-119 does not require TZ-, doublet region, and InvC-associated genes. NPHP-2 does require its calcium binding EF hand domain for targeting to the InvC. We conclude that the C. elegans InvC is distinct from the doublet region, and that components in these two regions interact to regulate ciliogenesis via cilia placement, ciliary microtubule ultrastructure, and protein localization.
Author Summary
Cilia are sensory organelles that are found on most types of human cells and play essential roles in diverse processes ranging from vision and olfaction to embryonic symmetry breaking and kidney development. Individual cilia are divided into multiple functionally and compositionally distinct compartments, including a proximal “Inversin” compartment, which is located near the base of cilia. We used the nematode C. elegans, a well-defined animal model of cilia biology, to characterize the genetics, components, and defining properties of the proximal cilium. The Inversin compartment is conserved in C. elegans, and is established independent of another proximal ciliary region, the microtubule doublet-based region. We showed how components of both the doublet region and the Inversin compartment genetically interact to regulate many pathways linked to core aspects of cilia biology, including ciliogenesis, cilia placement, cilia ultrastructure, microtubule stability, and the protein composition of ciliary compartments. In addition to expanding and clarifying our knowledge of basic cilia biology, these results also have direct implications for human health research because several of the genes and pathways explored in our work are linked to ciliopathies, a group of diseases caused by dysfunctional cilia.
Introduction
Cilia are cellular “antennae” that mediate the transduction of environmental signals into intracellular pathways. Cilia play an integral role in many cellular functions, including developmental signaling, symmetry breaking, cell-cell adhesion, cell-cycle control, stress response, and DNA damage response (e.g., [1]–[6]). The vast majority of cilia share a set of evolutionarily conserved features: cilia are supported by a microtubule-based backbone, the axoneme; are built by intraflagellar transport (IFT), a microtubule motor driven cargo transport system [7]; and can be divided into structurally and functionally distinct compartments. These compartments include the microtubule triplet basal body which roots the cilium to the cell, the microtubule doublet transition zone (TZ) which anchors the cilium to the membrane, and the microtubule doublet cilia shaft where IFT occurs. The basal body and TZ also act as selective filters for inbound and outbound ciliary cargo, functioning through physical occlusion and cargo-specific recognition mechanisms [7]–[9]. The cilia shaft has traditionally been treated as an undifferentiated whole [10], though recent evidence has shed light on subdivisions of the cilia shaft [11].
Inversin/Nephrocystin-2 specifically localizes to the Inversin compartment (InvC), a proximal portion of the cilia shaft adjacent to the TZ [12]. This region has been suggested to play a role in signal transduction and amplification [13]–[15], cilia placement, and ciliogenesis [16], [17]. Products of several genes—including INVS/NPHP2, NPHP3, NEK8/NPHP9, and ANKS6/NPHP16—localize to the InvC. Interactions between InvC genes and other cilia genes have only recently begun to be explored and have not been well generalized across animal and cell culture models [18]–[22].
The mechanisms that initially establish the InvC are currently unknown, though recruitment pathways for several InvC components are known. Work in vertebrate models has shed light on an InvC-specific physical interaction complex composed of Inversin, Nek8, Nphp3, and Anks6 [23]–[25]. Nek8, Nphp3, and Anks6 localize to the InvC in an Inversin-dependent manner, but Inversin itself localizes independently of the other proteins [23], [24]. Unc119b—a possible, though not proven, InvC component—may mediate an InvC-targeting pathway. In mammalian cells, Unc119b binds myristoylated cargo, including Nphp3, and shuttles it into the cilium. Once the Unc119b-Nphp3 complex has translocated into the cilium, the small GTPase Arl3 triggers Unc119b to release bound cargo [26]. InvC components other than Nphp3 are not known to be myristoylated and shuttled via Unc119b, suggesting additional InvC targeting pathways must exist. Whether Unc119b is required for the localization of Inversin and how Unc119b itself is targeted to the proximal cilium is unknown.
The nematode Caenorhabditis elegans is a well-studied model of cilia biology [27]. C. elegans possesses a ciliated nervous system [28]–[31] which is primarily used by the roundworm to detect internal and external cues and signals. Amphid channel cilia in the head and phasmid cilia in the tail are exposed to and sense the external environment through cuticular pores [32], [33]. Unlike most mammalian primary cilia, the cilia shafts of C. elegans amphid channel and phasmid cilia are divided into two regions: a proximal microtubule doublet-based region attached to the TZ, and a distal microtubule singlet-based region that extends from the doublet region [32], [33]. As both the doublet region of C. elegans cilia and the InvC of mammalian primary cilia lie at the proximal end of the cilium, directly adjacent to the TZ at the cilia base, and constitute only a portion of the length of the cilia shaft, previous work has viewed them as compositionally and functionally similar [13], [17], [34]. The relationship between the mammalian InvC and the C. elegans doublet region of cilia has not been well characterized; Here we present evidence that the InvC and the doublet region are distinct, but overlapping, ciliary regions.
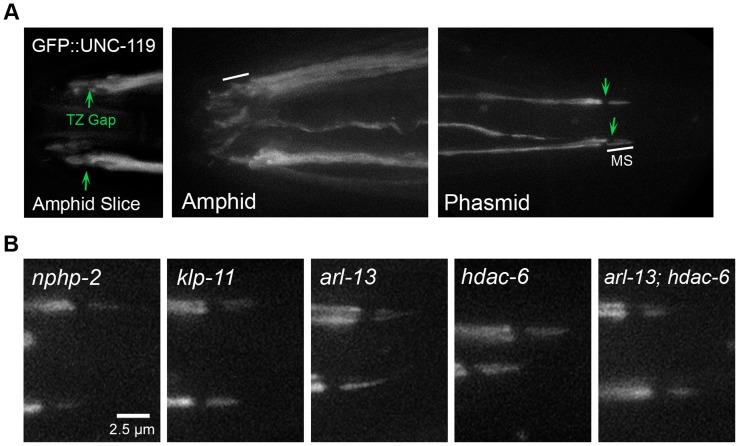
The C. elegans genome encodes orthologs for several of the mammalian InvC-associated proteins, including Inversin itself (NPHP-2), Unc119b (UNC-119), Arl3 (ARL-3), and possibly Nek8 (the uncharacterized paralogous pair NEKL-1 and NEKL-2), but likely not Nphp3 or Anks6. Of these, NPHP-2 and ARL-3 have previously been shown to be doublet region-localizing in C. elegans [17], [35]. C. elegans also possess several doublet region-enriched proteins, which are not InvC restricted in mammalian primary cilia; these include the kinesin-II IFT motor KLP-11 and the membrane-associated small GTPase ARL-13 [35]. The IFT motors Kinesin-II and OSM-3 work cooperatively to carry the IFT assemblies IFT-A and IFT-B and to build the doublet region—OSM-3 alone is sufficient to build the singlet region [36]. ARL-13 likely stabilizes the interaction between IFT-A and IFT-B particles and is required for ultrastructural integrity of the doublet region [35], [37]. arl-13 mutants exhibit multiple ciliary defects, some of which can be suppressed by deletion of histone deacetylase hdac-6, through an unknown mechanism [35]. Mammalian HDAC6 also antagonizes ciliogenesis in mammalian primary cilia: HDAC6 knockouts can suppress ciliogenesis defects arising from INVS/NPHP2 RNAi in MDCK cells [16].
In this work, we aimed to molecularly dissect the proximal cilium of C. elegans and to gain insight into the nature of the InvC and doublet region. We examined interactions between genes associated with the doublet region, determined the territories and localization dependencies of the protein products of these genes, and performed ultrastructural analysis of deletion mutants of these genes. We find that the InvC is conserved in C. elegans, is established early in development, and is distinct from the doublet region. nphp-2 interacts with doublet region genes to regulate cilia placement, microtubule ultrastructural patterning, tubulin glutamylation, and territory sizes of NPHP-2 and ARL-13. Finally, we show that nphp-2, arl-13, klp-11, and unc-119 fall into two parallel redundant genetic modules, and that interactions between the two modules are modulated by hdac-6 and arl-3. Together, the InvC and the doublet region function in concert to regulate many critical aspects of ciliogenesis and cilia biology.
Results
Genetic interactions between nphp-2 and arl-13 are modulated specifically by hdac-6
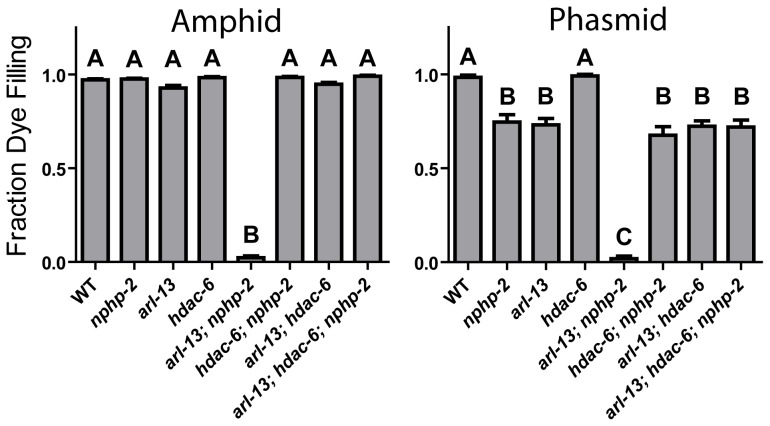
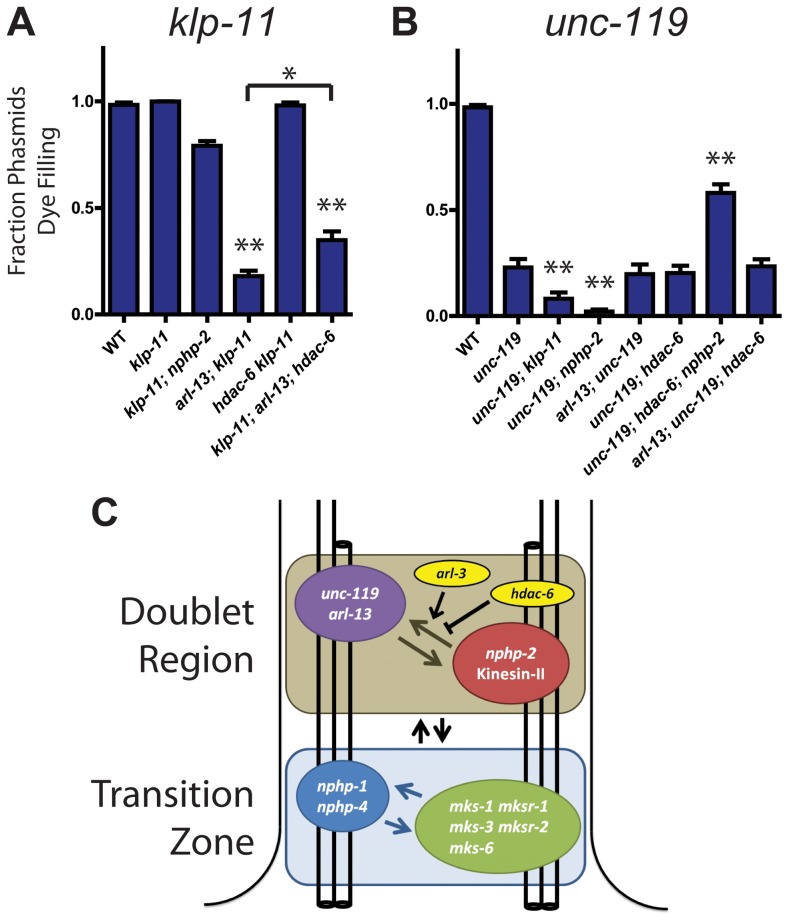
nphp-2 and arl-13 single mutants have statistically similar, moderate ciliogenic defects (Fig. 1). As hdac-6 and arl-3 modulate several arl-13 phenotypes [35], we sought to determine if nphp-2 and arl-13 genetically interact and if hdac-6 and arl-3 modulate nphp-2 phenotypes. We examined double, triple, and quadruple mutant combinations using “dye filling” of ciliated neurons as a gross indicator of ciliogenesis and cilia integrity [32]. Properly formed and placed cilia are environmentally exposed and take up fluorescent DiI dye, whereas stunted and misplaced cilia are not exposed and cannot take up DiI. Unlike the mild dye-filling defects (Dyf) of nphp-2 and arl-13 single mutants, arl-13; nphp-2 double mutants were severely synthetic dye-filling defective (SynDyf) in both the amphids and phasmids (Fig. 1). hdac-6 deletion did not suppress nphp-2 or arl-13 Dyf; this is contrary to previously published data indicating that hdac-6 can partially suppress the weak arl-13 single mutant Dyf, and may be due to a difference in dye filling or scoring method [35] (Materials and Methods). However, hdac-6 suppressed the arl-13; nphp-2 severe SynDyf phenotype to the mild Dyf severity of the single mutants in both amphids and phasmids (Fig. 1). hdac-6 may function by suppressing defects arising from one of the pathways or at a point where the two pathways converge. Combined, this data indicates that arl-13 and nphp-2 act in partially redundant parallel pathways antagonized by hdac-6.
Figure 1. The synthetic dye-filling defective phenotype of arl-13; nphp-2 mutants is modulated by hdac-6.
hdac-6 deletion suppresses arl-13; nphp-2 double mutant phenotypes. In phasmids, nphp-2 and arl-13 single mutants are mildly Dyf, which is not suppressed by hdac-6 deletion. In both amphids and phasmids, arl-13; nphp-2 is severely SynDyf and was suppressed by hdac-6. Data in both panels was analyzed with pairwise Mann-Whitney U-test between all groups, followed by the Holm-Bonferroni multiple comparison adjustment with a total alpha of 0.01. Groups from either panel sharing a capital letter are not significantly different, whereas groups from either panel with different capital letters do differ significantly.
arl-3 has also been implicated as a modulator of the arl-13 pathway [35]. We found that, unlike the interactions with hdac-6, interactions with arl-3 are cell-type specific (S1A Figure). Both arl-3 single mutants and arl-3; hdac-6 double mutants were nonDyf in both amphids and phasmids. In amphids, arl-13; arl-3 was mildly SynDyf, whereas arl-3; nphp-2 is nonDyf (cf. Fig. 1 and S1A Figure). hdac-6 deletion suppressed the arl-13; arl-3 phenotype to a severity similar to that of the arl-13 single mutants. In phasmids, both arl-13; arl-3 and arl-3; nphp-2 double mutants exhibited a moderate SynDyf phenotype. hdac-6 suppressed arl-13; arl-3 defects, but not arl-3; nphp-2 defects. Strikingly, in both amphids and phasmids, arl-3 deletion prevented the hdac-6 Dyf suppression in the arl-13; hdac-6; nphp-2 triple mutant described above. The SynDyf phenotype of arl-13; arl-3 is qualitatively different from the suppression of arl-13 Dyf defects by arl-3 found previously [35]. Similarly to the difference in hdac-6 suppression of arl-13 defects discussed above, this may be due to different scoring methods or assay conditions. We conclude that arl-3 functions parallel to both nphp-2 and arl-13 pathways, and likely lies in the same regulatory pathway as, but acts antagonistically to, hdac-6.
nphp-2 and arl-13 genetically interact to regulate amphid cilia ultrastructure
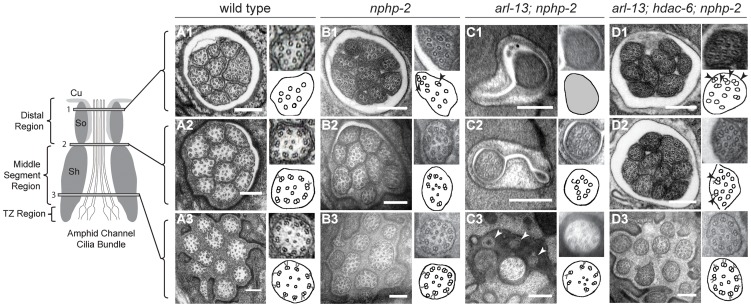
To gain a better understanding of the defects present in nphp-2 single and arl-13; nphp-2 double mutants, and to examine the effects of hdac-6 mediated suppression of the double mutant defects, we used serial-section transmission electron microscopy (TEM) to examine the ultrastructure of amphid channel cilia. Wild-type amphid cilia are divided into three segments based on microtubule ultrastructure: the TZ, the doublet region, and the singlet region (Fig. 2A). One of the two microtubules within a doublet, the A-tubule, extends to form the microtubule singlet seen in the distal cilium (Fig. 2A1); the second tubule of the doublet, the B-tubule, terminates at the distal end of the doublet region. The doublet microtubules of the TZ and doublet region are arranged in a circular pattern in close proximity to the ciliary membrane (Fig. 2A2, 2A3). Within a particular cilium, microtubule ultrastructural characteristics—the location in the lumen, membrane association, and singlet/doublet architecture—are similar across all nine outer microtubule doublets.
Figure 2. nphp-2 single, arl-13; nphp-2 double, and arl-13; hdac-6; nphp-2 triple mutants exhibit defects in ciliary ultrastructure.
Horizontal panels indicated by 1, 2, and 3 correspond to singlet region, doublet region-to-singlet region transition, and TZ levels, respectively, and are comparable across the genotypes. The insets show an enlarged view of the region within the white box, and are diagrammed in the accompanying cartoon. All scale bars are 250 nm. (A1–3) In wild-type amphids, all doublet B-tubules within a cilium have similar spans. (A1) Distal microtubule singlets are devoid of B-tubules. B-tubule containing microtubule doublets are present in (A2) the doublet region and (A3) the TZ. (B1–3) In nphp-2 animals, amphid channel cilia are shifted lengthwise with respect to each other (cf. Fig. 6A). Within a cilium, spans of microtubule doublet B-tubules are asynchronous; arrows in insets indicate these microtubules. TZ Y-links are disorganized (C1–3) In arl-13; nphp-2 animals, most amphid channel cilia are absent. (C1) The distal end is filled with electron dense material with unresolvable microtubules. (C2) A single, stub-like cilium is visible, consisting of only microtubule singlets. (C3) TZ microtubules are abnormal with some missing microtubule doublets. We also observe vesicle-like structures at this level that are indicated by arrows. (D1–3) In arl-13; hdac-6; nphp-2 animals, most amphid cilia are visible. Ectopic singlets and doublets are still present. Cilia are shifted posteriorly towards the tail. (D1–2) insets show asynchronous microtubules within cilia.
In nphp-2 single mutants, in a given amphid cross-section at a single level, we observe singlet regions of some cilia, doublet regions of other cilia, and TZs of the remaining cilia. This is consistent with amphid cilia that were shifted lengthwise with respect to each other, indicating a potential anchoring defect (Fig. 2B3). Additionally, within a given cilium, doublet and singlet microtubule spans were no longer aligned. In sections across the proximal axoneme, this appeared as singlets amongst the expected doublets, and in sections across the distal axoneme, this appeared as doublets interspersed between the expected singlets (Fig. 2B1–3). These defects indicate that nphp-2 is required both for microtubule patterning and for cilia anchoring. While the chemical fixation method utilized here does not preserve Y-link ultrastructure well, nphp-2 mutants exhibited significantly greater Y-link disorder than observed in wild-type animals. The TZ defects seen in nphp-2 animals may be related to both the lengthwise cilia shift and the TZ-placement defect previously reported in nphp-2 mutants [17]. This set of defects has not been reported in any other C. elegans cilia mutant, suggesting that nphp-2 functions in a novel capacity.
arl-13 single mutants also exhibit a range of ultrastructural defects [35], [37]. Doublets are observed in the central lumen of the cilium, which may originate from either displaced outer doublets or mispatterned inner singlets. In both C. elegans arl-13 and hennin/ARL13B mouse mutants, there is also an increased frequency of early B-tubule detachment from the A-tubule, indicative of microtubule stability or patterning defects [38]. Similar to nphp-2 mutants, ectopic microtubule singlets were sometimes visible in the doublet region of arl-13 worms [35], [37]. arl-13; nphp-2 double mutants exhibited extreme ultrastructural defects, likely causative of the severe Dyf phenotype (Fig. 2C1–3). Cilia were almost completely absent from the amphid channel pore. No other cilia were visible even at the distal dendritic level. In one instance, a single cilium was visible in TEM sections. At the TZ level, an incomplete set of doublets was visible within the single cilium (Fig. 2C3). Closer to the socket cell/sheath cell transition, a set of singlets was visible (Fig. 2C2). At the distal pore, we were unable to resolve any internal structure due to electron dense material filling the cilium (Fig. 2C1).
Remarkably, in arl-13; hdac-6; nphp-2 triple mutants almost all defects observed in the double mutant were suppressed (Fig. 2D1–3). The only defects remaining were misplaced TZs (Fig. 2D3), ectopic singlets in the doublet region (Fig. 2D2), and ectopic doublets in the singlet region (Fig. 2D1), similar to those present in the nphp-2 single mutant. We did not observe inner doublets as reported in arl-13 single mutants [35], [37]. These results are consistent with the observed hdac-6 suppression of arl-13; nphp-2 Dyf defects.
Because nphp-2 and arl-13 both exhibit ectopic microtubule singlets, nphp-2 mutants exhibit ectopic microtubule doublets, and nphp-2; arl-13 double mutants exhibit severe defects in ciliogenesis, we conclude that nphp-2 and arl-13 function together redundantly in regulation of microtubule patterning and ciliogenesis. Because ciliogenic defects are suppressed by hdac-6, but ectopic doublets and singlets are not, ciliogenesis and microtubule patterning may be independently regulated.
NPHP-2 and ARL-13 do not require TZ- and doublet region-associated genes for ciliary targeting
Localization of InvC and doublet region components can be broken down into three steps: first the protein is targeted to the cilia base, second, the protein is imported into the cilium, and third, the protein is restricted to a subdomain of the cilium [23]. The factors required for the initial establishment of the InvC and doublet region cilia targeting and localization restriction are unknown. The TZ functions as a regulator of ciliary protein import (Reviewed in [39]), and has been implicated in the import of InvC and doublet region components in mammalian cilia [40], [41]. As both nphp-2 and arl-13 genetically interact with TZ-associated genes [17], [37], we wanted to determine if NPHP-2 and ARL-13 ciliary targeting and import requires TZ components. In C. elegans, TZ genes are organized into two genetic and physical modules—the mks module and the nphp-1+nphp-4 module [8], [17], [25], [42]. We examined the localization of NPHP-2 and ARL-13 in mutants missing a component of each module (Fig. 3A,C).
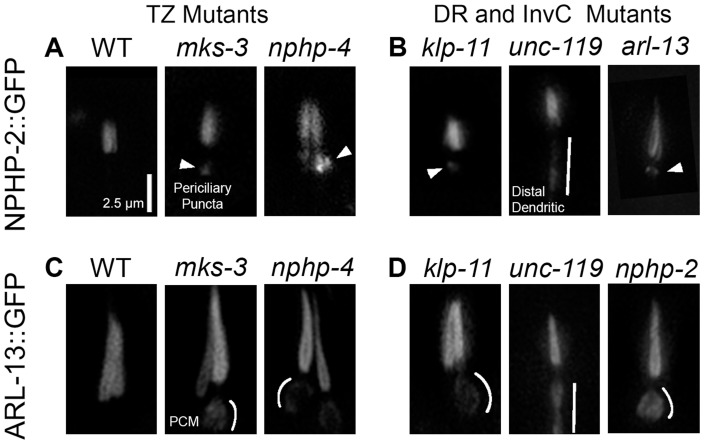
Figure 3. NPHP-2 and ARL-13 do not require TZ-, doublet region-, and InvC-associated genes for ciliary targeting.
(A) In wild type (WT), NPHP-2 is restricted to the proximal cilium. In both nphp-4 and mks-3 mutants NPHP-2::GFP was targeted to the cilium and restricted to the post-TZ proximal cilium. Several NPHP-2::GFP puncta were visible in the periciliary compartment. (B) In doublet region and InvC mutants, NPHP-2::GFP was targeted to the cilium and restricted to the post-TZ proximal cilium in klp-11, arl-13, and unc-119 mutants. arl-13 and klp-11 mutants exhibited periciliary NPHP-2::GFP puncta, and unc-119 mutants exhibited distal dendritic accumulation of NPHP-2::GFP. (C) In WT, ARL-13::GFP localizes to the proximal cilium. In both nphp-4 and mks-3 mutants, ARL-13::GFP was targeted to the cilium and restricted to the post-TZ proximal cilium. ARL-13::GFP also mislocalized to the periciliary membrane compartment in TZ mutants. (D) In nphp-2 and klp-11 mutants, ARL-13::GFP was targeted to the cilium and restricted to the post-TZ proximal cilium. In these mutants, ARL-13::GFP also mislocalized to the periciliary membrane compartment, and in unc-119 mutants, ARL-13::GFP mislocalized to the distal dendrite. Periciliary puncta – arrowheads, periciliary membrane – white arc, distal dendrite/periciliary accumulation – white bar. Periciliary membrane localization was judged by a visible enrichment of ARL-13::GFP on the edges of the periciliary compartment without a concomitant enrichment in the interior lumen of the periciliary region.
NPHP-2::GFP signal length is similar in wild-type and nphp-2 backgrounds (1.77±0.23 um vs 1.74±0.22 um, st. dev.) and NPHP-2::GFP rescues the SynDyf phenotype of the nphp-2 nphp-4 double mutant (Fig. 4B) [17]. These results indicate that this reporter reflects NPHP-2 functional and endogenous localization. In both nphp-4 and mks-3 single mutants, NPHP-2::GFP was properly targeted to and imported into the cilium. Mislocalized NPHP-2::GFP puncta in the periciliary region were sometimes visible (Fig. 3A). In mks-3; nphp-4 double mutants, there were severe ciliogenic and dendritic extension errors, as previously reported [17], [42]; in phasmid cilia that were visible and placed properly, NPHP-2::GFP localization appeared as in mks-3 and nphp-4 single mutants (S2A Figure). In a wild-type background, ARL-13::GFP localized exclusively to the doublet region in amphid channel and phasmid neurons (Fig. 3C). Like NPHP-2::GFP, ARL-13::GFP was targeted to and imported into the cilium properly in mks-3 and nphp-4 mutants. Similar to published reports, we also observed ARL-13::GFP mislocalization to the periciliary membrane, as judged by a fluorescent “fringe” surrounding the periciliary region where the membrane lies (Fig. 3C, enhanced contrast in S2B Figure), which has been suggested to be due to a failure of the TZ diffusion barrier [34]. In amphid cilia, like in phasmid cilia, both NPHP-2 and ARL-13 were targeted to the cilium and imported properly, and both infrequently exhibited mild mislocalization (S3 Figure).
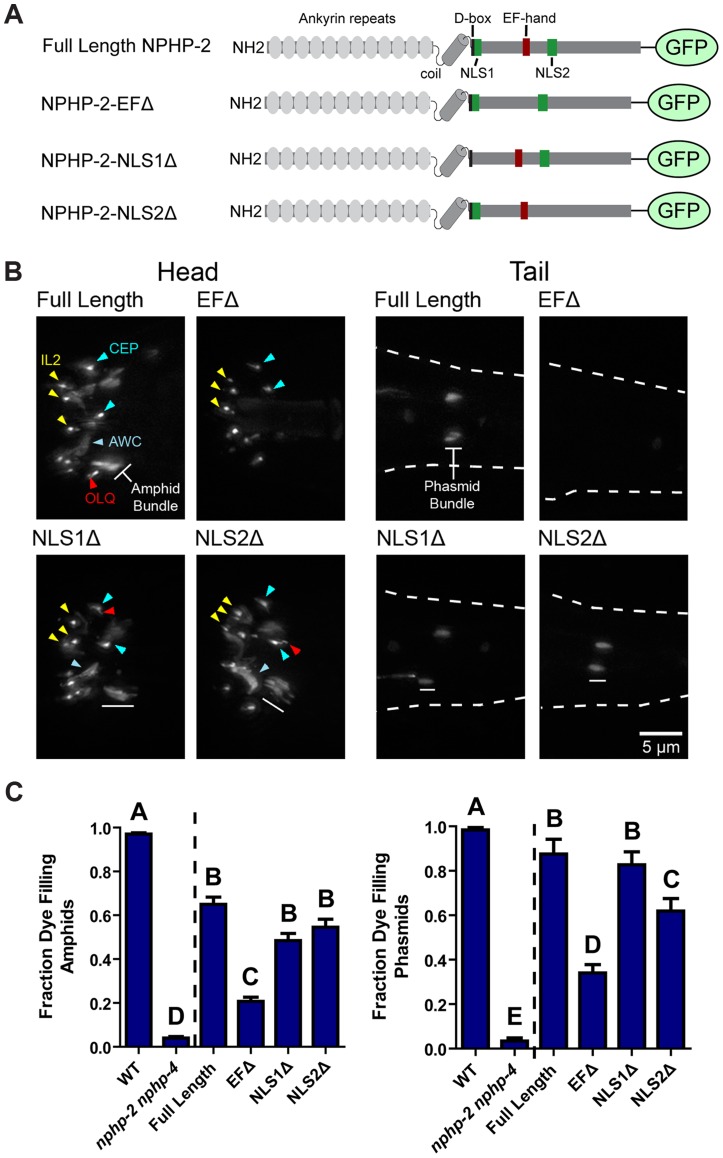
Figure 4. The EF hand is necessary for proper NPHP-2 localization and function.
(A) Diagram of the domains present in Inversin, NPHP-2, and each of the NPHP-2 domain deletion constructs. (B) Localization of GFP-tagged NPHP-2 domain deletion constructs. In the head, full length NPHP-2::GFP localizes to the InvC of amphid channel cilia, to the base of IL2 cilia, and to the base of either OLQ or CEP cilia. In the tail (outlined by dashes), NPHP-2::GFP localizes to the InvC of phasmid cilia. Localization of both NPHP-2-NLS1Δ::GFP and NPHP-2-NLS2Δ::GFP appeared roughly wild-type in all cell types, though both constructs appeared enriched in AWC wing cilia. NPHP-2-EFΔ::GFP is present in IL2 and CEP cilia but fails to localize to amphid channel and phasmid cilia. (C) NPHP-2-NLS1Δ::GFP, NPHP-2-NLS2Δ::GFP, and NPHP-2-EFΔ::GFP can rescue nphp-2 nphp-4 amphid and phasmid dye-filling defects, but NPHP-2-EFΔ::GFP rescue is significantly worse than full length NPHP-2::GFP in both amphid and phasmid neurons. Yellow arrowheads - IL2 cilia, red arrowheads - OLQ, blue arrowheads - CEP cilia, white bar - amphid/phasmid bundle. Letters indicate statistically distinct groups. Data was analyzed with pairwise Mann-Whitney U-test against both positive and negative controls, followed by the Holm-Bonferroni multiple comparison adjustment with a total alpha of 0.01. Non-transgene expressing siblings were used as negative controls in rescue experiments.
We investigated whether doublet region-associated genes were required for doublet region restriction of NPHP-2::GFP and ARL-13::GFP. hdac-6 had no obvious effect on the localization of NPHP-2::GFP (S2A Figure). NPHP-2::GFP was targeted and restricted to the proximal cilium in both klp-11 and arl-13 mutants. In both mutants, periciliary puncta similar to those seen in TZ-associated mutants were visible (Fig. 3B). unc-119 mutants also exhibited proper NPHP-2::GFP ciliary targeting. In unc-119 mutants, NPHP-2::GFP exhibited a unique distal dendrite mislocalization pattern, distinct from the periciliary puncta seen in other TZ and doublet region mutants (Fig. 3B).
In klp-11 and nphp-2 mutants, ARL-13::GFP was restricted to a proximal portion of the cilium as in wild type, but with a periciliary membrane mislocalization pattern similar to TZ mutants. ARL-13::GFP also did not require unc-119 for ciliary localization. In unc-119 mutants, ARL-13::GFP and NPHP-2::GFP localized along the distal dendrite in a similar manner (Fig. 3D).
In all TZ, doublet region, and InvC mutants examined, NPHP-2::GFP and ARL-13::GFP still localized to the cilium, suggesting that either unknown factors or redundant pathways are required for establishing ciliary territories. Periciliary mislocalization was observed for both reporters across all mutant backgrounds. This suggests that either a delicate, easily perturbed interaction network is required for NPHP-2 and ARL-13 ciliary import/export, or that the overexpressed reporter constructs are “leaking” out of the cilium in sensitized mutant backgrounds, or a combination of both possibilities.
NPHP-2 requires its EF-hand for proper localization and function
We next looked to determine which domains of NPHP-2 were required for InvC localization. In mammalian models, several domains in Inversin are required for ciliary targeting and InvC restriction: the ankyrin repeat region, IQ2 domain, and ninein-homologous region (Fig. 4A) [12], [43]. The IQ and ninein homology domains are not conserved in C. elegans NPHP-2; only the ankyrin repeat region, hydroxylation motif (S8 Figure), and two nuclear localization signals (NLSs) are conserved (Fig. 4A) [17]. NLS motifs are hypothesized to play a role in ciliary protein import, and are required for ciliary import of the IFT motor KIF17 [9]. NPHP-2 also contains a predicted calcium binding EF-hand which may function in the same calcium detection capacity as the IQ domain of Inversin [17].
We built NPHP-2::GFP constructs missing the EF-hand (NPHP-2-EFΔ::GFP, residues 520–533), NLS1 (NPHP-2-NLS1Δ::GFP, residues 441–446), or NLS2 (NPHP-2-NLS2Δ::GFP, residues 598–603) (Fig. 4A). Full-length NPHP-2::GFP localized to a short proximal region of amphid channel and phasmid cilia as well as IL, CEP, OLQ, amphid channel and phasmid cilia, and the AWC wing cilia (Fig. 4B). Native promoter driven NPHP-2::GFP was not visible in AWB cilia, as reported previously for AWB-specific promoter driven NPHP-2 [13]. NPHP-2-EFΔ::GFP was generally faint or absent in amphid channel, AWC, and phasmid cilia, indicating that the EF-hand is strictly required for normal localization of NPHP-2 in these cell types. NPHP-2-EFΔ::GFP properly localized in IL cilia, indicating that the fluorescent reporter was being synthesized and folded correctly. NPHP-2-EFΔ::GFP was additionally present in either CEP or OLQ cilia, though specific identification was difficult due to their close proximity. Deletion of either NLS1 or NLS2 did not perturb NPHP-2 localization (Fig. 4B).
We next tested whether these domain deletion constructs were functional by attempting to rescue the severe Dyf phenotype of nphp-2 nphp-4 mutants; nphp-4 single mutants are nonDyf, allowing for easy determination of rescue [17]. Rescue of the nphp-2 nphp-4 SynDyf phenotype by NPHP-2-NLS1Δ::GFP and NPHP-2-NLS2Δ::GFP was comparable to rescue by full length NPHP-2::GFP. NPHP-2-EFΔ::GFP only weakly rescued the SynDyf phenotype, indicating that the EF-hand is critical for both normal function and localization of NPHP-2 (Fig. 4C). These results indicate that the calcium-binding EF-hand plays a significant role in both localization and function of NPHP-2 in amphid channel and phasmid cilia, but is dispensable for localization in cilia of CEP, OLQ, and inner labial neurons.
UNC-119 is associated with the doublet region
Mammalian Unc119b localizes to the proximal cilium and is required for the targeting of several proteins to the InvC [26], and C. elegans unc-119 is required for singlet region biogenesis in amphid cilia [44], suggesting a compartment-specific role. We therefore examined the localization of posm-6::GFP::UNC-119. In phasmid cilia, GFP::UNC-119 localization was similar to NPHP-2::GFP and ARL-13::GFP: its localization was restricted only to a small proximal portion of the cilium and was excluded from the TZ. Similar to localization in phasmid cilia, GFP::UNC-119 likely localized to the doublet region and not the entire cilium of amphid cilia (Fig. 5A). GFP::UNC-119 was not motile in cilia.
Figure 5. UNC-119 localizes to the proximal cilium in phasmids and does not require DR and InvC genes to target the cilium.
(A) N-terminal GFP tagged UNC-119 localizes to dendrites and cilia of amphid channel and phasmid neurons. The TZ gap (arrow) can be discerned in the cilia of amphid channel neurons. In cilia of the CEP and OLQ neurons, GFP::UNC-119 fluorescence was faint and infrequently visible. In phasmid cilia, GFP::UNC-119 localizes to the doublet region, and is excluded from the TZ (arrow) and singlet region. (B) GFP::UNC-119 localization in doublet region- and InvC-associated mutants. GFP::UNC-119 localizes to the proximal cilium in all mutant backgrounds examined.
We also examined the dependence of UNC-119 localization on doublet region-associated genes. Specific amphid mislocalization was difficult to determine, and we therefore focused on phasmid cilia (S4 Figure). In phasmid cilia, GFP::UNC-119 did not require nphp-2, arl-13, hdac-6, or klp-11 for ciliary targeting or restriction to the proximal cilium (Fig. 5B).
IFT motors and unc-119 genetically interact with doublet region-associated genes
We examined the genetic interactions between the doublet region-associated genes nphp-2, arl-13, and their modulators hdac-6 and arl-3 with the two IFT motors, osm-3 and klp-11. In both amphids and phasmids, arl-13; klp-11 was SynDyf [37]. arl-13; klp-11 dye-filling defects were slightly suppressed by deletion of hdac-6 (Fig. 6A, S5A Figure). klp-11; arl-3 double mutants yielded a very mild SynDyf phenotype (S1B Figure). osm-3 single mutants are missing a singlet region and are completely Dyf, which precludes searching for synthetic interactors of osm-3. Instead, we assayed osm-3 double mutants for suppression of the severe dye-filling defect; in no mutant background examined was the defect suppressed (Fig. 6B, S5B Figure).
Figure 6. klp-11 and unc-119 genetically interact with arl-13 and nphp-2 in an hdac-6 dependent manner.
(A) klp-11 single mutants are not Dyf. klp-11 is SynDyf with arl-13, which is partially suppressed by deletion of hdac-6. (B) unc-119 single mutants are moderately Dyf. unc-119 is SynDyf with both klp-11 and nphp-2. unc-119; hdac-6; nphp-2 triple mutants exhibit suppression of the unc-119 Dyf phenotype. (C) Diagram of interactions between klp-11, arl-13, nphp-2, unc-119, and hdac-6, and between DR and TZ genes, based on SynDyf phenotypes presented in panels A and B and in Fig. 1 and S5 Figure. The T-bar indicates hdac-6 mediated suppression of SynDyf phenotypes. Data was analyzed with pairwise Mann-Whitney U-test between wild type, double mutants, and their respective single mutants, followed by the Holm-Bonferroni multiple comparison adjustment. *, significant versus single mutants at a total alpha of 0.05. **, significant versus single mutants at a total alpha of 0.01. nphp-2, arl-13, hdac-6, and arl-13; hdac-6 Dyf data is presented in Fig. 1.
We next examined genetic interactions between unc-119 and other doublet region-associated genes. unc-119 single mutants were severely Dyf, with the phasmid phenotype being less severe than the amphids (Fig. 6B, S5A Figure) [44]. In phasmids, both nphp-2 and klp-11 were SynDyf with unc-119. Surprisingly, we found that hdac-6 suppressed Dyf defects in the unc-119; nphp-2 double mutant to the level of the nphp-2 single mutant (Fig. 6B). arl-3 deletion was also able to significantly suppress the unc-119 Dyf phenotype in both amphids and phasmids (S1B Figure).
Genetic interactions between nphp-2, arl-13, klp-11, unc-119, hdac-6, and arl-3 are summarized in Fig. 6C. Doublet region-associated genes fall into two redundant pathways or modules: nphp-2+klp-11 and arl-13+unc-119. Deletion of two genes within a module does not result in an increase in Dyf severity over that present in single mutants, but deletion of any two genes from different modules yields a SynDyf phenotype. Interactions between these modules are regulated by hdac-6 and arl-3.
Double mutants with a deletion in a single TZ gene and a single doublet region gene are SynDyf. To determine whether hdac-6 mediated suppression of SynDyf defects extended to these cross-compartmental genetic interactions, we assayed for suppression of SynDyf defects in nphp-2 nphp-4 mutants by hdac-6 and arl-3. We found that neither hdac-6 nor arl-3 suppressed the severe nphp-2 nphp-4 SynDyf defect (S5D Figure). This suggests that hdac-6 functions specifically in doublet region pathways.
Axonemal glutamylation is downstream of the action of nphp-2, arl-13, unc-119, and hdac-6
Post-translational glutamylation predominantly occurs on the C-terminal tails of α- and β-tubulin of axonemal B-tubules [45]–[49], and regulates microtubule stability and IFT motor function [50]. Glutamylation is specifically associated with the doublet region, as B-tubules define and are only present in the doublet region. Additionally, in Chlamydomonas and Paramecium, TZ microtubules are not glutamylated [45], [51]. In C. elegans and vertebrates, mutants with defects in tubulin glutamylation or arl-13 exhibit B-tubule degeneration [38], [50], [52]. We therefore determined whether doublet region associated genes regulated tubulin glutamylation, or whether tubulin glutamylation specified the localization of doublet region proteins.
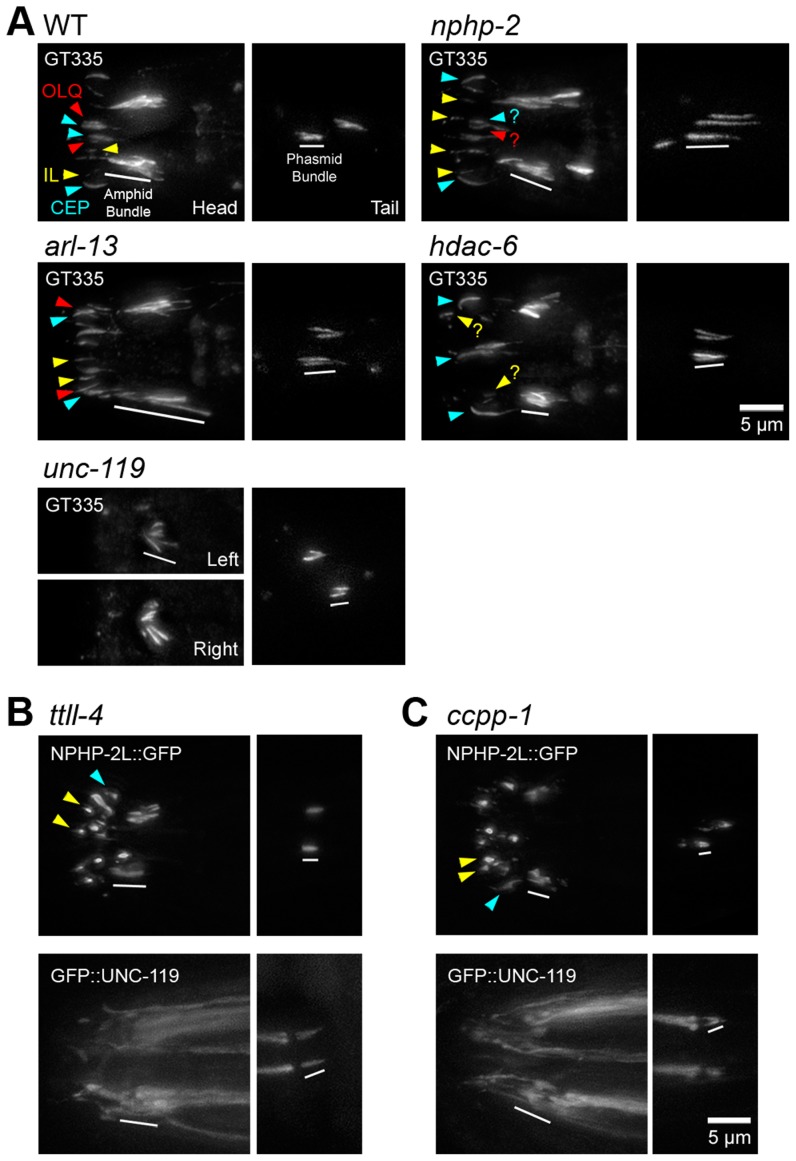
In wild-type animals, the anti-glutamylated tubulin antibody GT335 labeled the doublet region of amphid channel and phasmid cilia (Fig. 7A) [52]. nphp-2 mutants exhibited characteristic cilia displacement in the amphids, but no qualitative changes in head cilia glutamylation. The glutamylation signal in nphp-2 phasmid cilia ranged from wild-type-like to extremely elongated, which is consistent with the TEM observation of B-tubules extending into the distal axoneme (Fig. 2B). arl-13 mutants exhibited elongated staining in amphid channel cilia. Amphid staining in unc-119 mutants was extremely shortened and cilia were angled inwards. hdac-6 mutants appeared to have shortened GT335 staining of amphid channel cilia. We also observed significant differences in the length of the phasmid GT335 ciliary signal. Both arl-13 (3.95±0.25 µm) and nphp-2 (4.30±0.32 µm) mutants had phasmid staining significantly longer than in wild type (2.79±0.07 µm), while unc-119 (2.40±0.05 µm) mutants had staining significantly shorter (S7E Figure). The length of GT335 staining in amphid cilia was not quantified due to the difficulty of unbiased measurement of a single cilium within the amphid bundle.
Figure 7. nphp-2, arl-13, and hdac-6 regulate glutamylation in head and tail cilia.
(A) Worms stained with GT335 anti-glutamylated tubulin antibody. In WT background, GT335 labels amphid and phasmid doublet region microtubules, and CEP doublet region and singlet region microtubules consistently, and OLQ cilia inconsistently. CEP distal microtubule singlets were also labelled by GT335. Inner labial cilia were also glutamylated, but specific IL1 and IL2 identification was not possible. nphp-2 mutants showed characteristic posterior shifted cilia, and glutamylation in the head looked similar to WT, with infrequent weak inner labial cilia staining. Phasmids exhibited an elongated glutamylation signal. arl-13 mutants exhibited elongated staining of amphid bundle, CEP, and OLQ cilia. Amphid staining in unc-119 mutants was extremely shortened and cilia were angled inwards. unc-119 mutants exhibited almost no staining of CEP, IL, and OLQ cilia. hdac-6 mutants displayed shorter amphid bundle staining, and reduced IL and OLQ staining. (B) In ttll-4 mutants, NPHP-2::GFP and GFP::UNC-119 localize similarly to WT. (C) In ccpp-1 mutants, both NPHP-2::GFP and GFP::UNC-119 localize similarly to in WT, and GFP::UNC-119 is present in the TZ and accumulates at the distal dendrite. Yellow arrowheads – IL2 cilia, red arrowheads - OLQ cilia, blue arrowheads – CEP cilia, white bar – amphid/phasmid bundle.
To determine if doublet region-associated protein localization was dependent on tubulin glutamylation status, we examined the localization of NPHP-2::GFP and GFP::UNC-119 in ccpp-1 and ttll-4 mutants. Mutants of ccpp-1, which encodes a tubulin deglutamylase, display degenerating amphid channel and phasmid cilia with a concomitant dye-filling defect; this is suppressed by deletion of the opposing glutamylase, encoded by the tubulin tyrosine ligase-like gene ttll-4 [50]. In each mutant, NPHP-2::GFP and GFP::UNC-119 reporters were targeted to cilia and were restricted to the proximal cilium similarly to wild-type (Fig. 7B,C). This was surprising in the case of ccpp-1 mutants, as cilia degenerate as the worm ages (Fig. 7B,C). These reporters were still doublet region-associated in earlier larval stages of ccpp-1 mutants when ciliary degeneration was not as severe (S9C-D Figure). Combined, these results indicate that nphp-2, arl-13, unc-119, and hdac-6 lie upstream in regulation of tubulin glutamylation pathways, and the localization patterns of their protein products are not defined by tubulin glutamylation.
Doublet region protein territories are genetically regulated
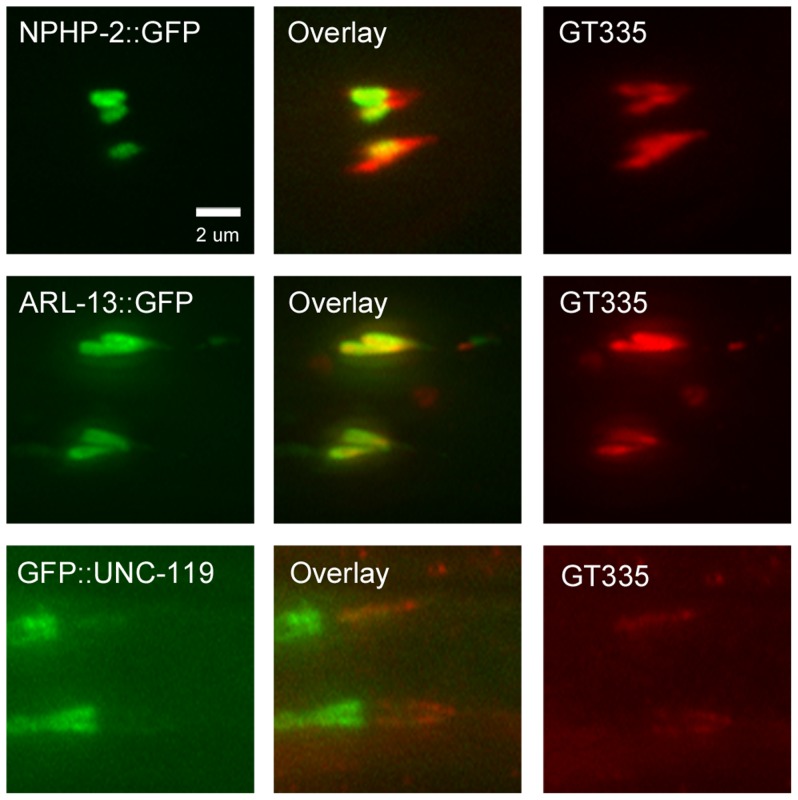
To allow for a more direct comparison of subciliary localization, we stained transgenic NPHP-2::GFP, ARL-13::GFP, and GFP::UNC-119 strains with GT335 (Fig. 8). NPHP-2::GFP did not fully overlap with GT335, only colabelling the proximal portion of the doublet region of amphid and phasmid cilia. However, ARL-13::GFP colabelled with a greater portion of the GT335 doublet region signal in amphid cilia than did NPHP-2::GFP, and completely colabelled with GT335 in phasmid cilia (Fig. 8, S6A Figure); this suggests that ARL-13 is associated with the microtubule doublets that define the doublet region. GFP::UNC-119 colabelled with either a significant fraction of the length of the GT335 signal. Additionally, GFP::UNC-119 did not extend beyond the GT335 labelled doublet region, indicating that GFP::UNC-119 is excluded from the TZ, which is not labelled by GT335.
Figure 8. NPHP-2::GFP, ARL-13::GFP, and GFP::UNC-119 colabel with GT335 staining.
In amphid channel and phasmid cilia, NPHP-2::GFP, ARL-13::GFP, and GFP::UNC-119 signals overlap with GT335 staining. ARL-13::GFP and GT335 staining overlap completely. NPHP-2::GFP does not overlap completely with GT335. TZ staining by GT335 is frequently visible, as in the NPHP-2::GFP overlay. Though the signal of GFP::UNC-119 is dim due to the staining procedure, GFP::UNC-119 appears not to overlap completely with GT335. Data is quantified in S6A Figure.
We also examined the territory length of each of the fluorescent reporters as a fraction of the total cilia length. Transgenic animals were incubated with DiI to label the length of the cilium. NPHP-2::GFP marked a significantly shorter fraction of the length of the cilium than did ARL-13::GFP or GFP::UNC-119 (S6C Figure).
To understand how the localization of doublet region-associated proteins is genetically regulated, we measured the length of the cilium marked by NPHP-2::GFP, ARL-13::GFP, GFP::UNC-119, and KAP-1::GFP—a component of Kinesin-II—in different mutant backgrounds (S7 Figure). ARL-13::GFP and NPHP-2::GFP have interdependent localizations: in arl-13 mutants, the NPHP-2::GFP territory was extended along the cilium, and in nphp-2 mutants, the ARL-13::GFP territory was extended. unc-119 mutants exhibited shortened ARL-13::GFP and NPHP-2::GFP territories (S7A-B Figure). Additionally, klp-11 mutants had a shorter NPHP-2::GFP, but not ARL-13::GFP, localization signal (S7A Figure). No significant differences were found in the territory lengths of GFP::UNC-119 and KAP-1::GFP in any of the mutant backgrounds (S7C-D Figure). The length of the tubulin glutamylation signal is also genetically controlled; the GT335 signal is shortened in unc-119 mutants, and elongated in nphp-2 and arl-13 mutants (S7E Figure).
In sum, the NPHP-2::GFP territory size is shorter than the territories of doublet region components ARL-13 and Kinesin-II, marks a shorter length of the cilium than ARL-13 and UNC-119, is shorter than the doublet region-linked glutamylated tubulin signal, and colabels only a portion of both amphid channel and phasmid GT335 staining. We conclude that NPHP-2 marks a region of the cilium distinct from the doublet region, and propose that this region is analogous to the InvC of mammalian cilia.
Discussion
In this study, we present evidence that (1) C. elegans possesses a conserved InvC compartment, (2) interactions between nphp-2 and arl-13 regulate microtubule ultrastructural patterning, (3) InvC and doublet region sizes are distinct and genetically regulated, (4) hdac-6 and arl-3 modulate interactions between nphp-2, arl-13, klp-11, and unc-119, (5) and that microtubule glutamylation is downstream of the action of InvC and doublet region genes. Additionally, we found that genes associated with the proximal cilium (TZ, InvC, and doublet region) can be grouped into parallel genetic modules, which interact to drive ciliary anchoring and proper ciliogenesis. Finally, we addressed several possible mechanisms for the ciliary targeting and InvC restriction of NPHP-2.
Limitations of overexpression reporter constructs
In the last twenty years, fluorescent tagging of proteins has allowed for unparalleled insight into in vivo localization and transport mechanisms. However, the primary method of introducing transgenes into C. elegans, microinjection, yields extrachromosomal arrays containing many copies of the reporter construct [53]. Many aspects of ciliogenesis are tightly regulated, and may require stoichiometric quantities of components for proper function. We previously showed that overexpression of proteins may lead to defects in ciliogenesis and IFT, resulting in the SynDyf phenotype [17], [54].
We sought to minimize these effects on our conclusions by avoiding direct comparisons between multiple reporters, and using antibody colabeling when comparisons were required. In wild-type and mutant backgrounds, we tested all constructs for dominant defects in ciliogenesis (S1 Table). In the respective mutant background, we routinely test for rescue of mutant phenotype, which indicates that the reporter is functional (Fig. 4A) [17], [29], [50], [54]–[57]. For example, NPHP-2::GFP localization is similar in wild-type and nphp-2 mutant backgrounds, and rescues the nphp-2 nphp-4 SynDyf phenotype (Fig. 4B) [17]. Therefore we can conclude that NPHP-2::GFP provides and accurate and functional reflection of the endogenous localization pattern.
The use of ARL-13::GFP is well established in the literature, with five papers examining ARL-13::GFP localization in amphid and phasmid cilia [34], [35], [37], [58], [59], and a sixth paper examining ARL-13::GFP localization in AFD cilia [60]. In two papers, the ARL-13::GFP construct is the same as was used in our work [35], [59]. Moreover, this ARL-13::GFP reporter is functional [35] and displays a similar localization in wild-type and arl-13 backgrounds (Jinghua Hu, personal communication). In three papers, ARL-13::GFP reporters are used to determine subciliary localization of ARL-13::GFP [34], [59], [60]. Additionally, the ARL-13::GFP transgenic lines used in the papers by the Hu and Blacque labs were built independently and exhibit similar localizations.
In the future, genomic engineering (e.g., CRISPR) will allow for easy single-copy fluorescent tagging of proteins and in vivo analysis, addressing many of these concerns [61]. These techniques are not a panacea though, as several genes, including nphp-2, may be expressed at too low a level for single-copy tagged constructs to be visible without advanced microscopic techniques (personal communication, Knudra Transgenics).
The nature of the doublet region
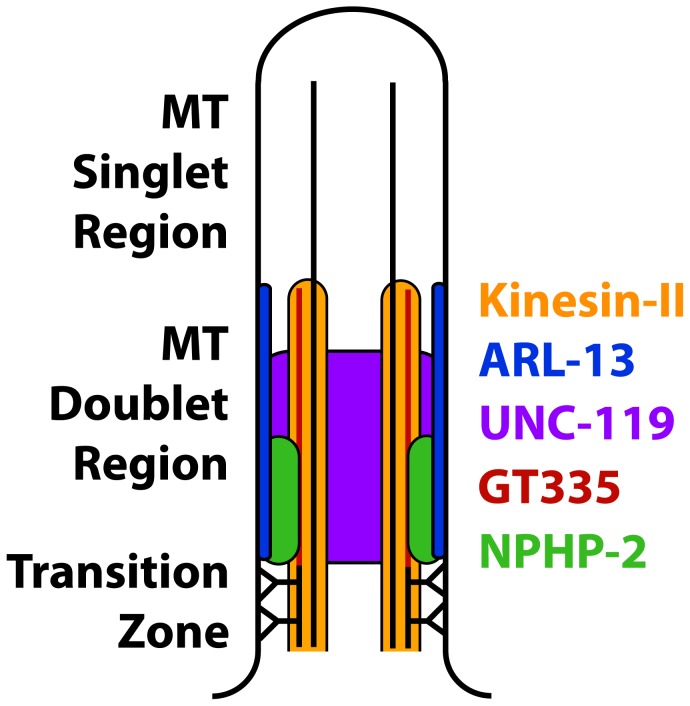
The C. elegans doublet region and the mammalian proximal InvC have been considered analogous [11], [13], [17], [34], [37]. In C. elegans, a number of factors have been associated with the doublet region, including NPHP-2, ARL-13, UNC-119, ARL-3, HDAC-6, the Kinesin-II components KAP-1/KLP-11/KLP-20 [35]–[37], and glutamylated tubulin [50]. However, mammalian orthologs of many C. elegans doublet region proteins localize along the entire cilium [26], [62]–[71]; this casts doubt on the equivalence between the C. elegans doublet region and the mammalian InvC. It is likely that the C. elegans ciliary doublet region is analogous to the entire mammalian ciliary doublet-based cilia shaft, and that the mammalian InvC is analogous to a C. elegans InvC (modelled in Fig. 9).
Figure 9. Model of the composition of the proximal cilium.
ARL-13 is depicted as membrane associated based on published characterizations [37]. NPHP-2 is depicted as membrane associated based on the membrane association of Inversin in mammalian primary cilia, and because in C. elegans NPHP-2 reporters appear membrane associated by casual observation [12], [17]. Kinesin-II is microtubule associated, and UNC-119 is depicted nonspecifically because of the diffuse localization of GFP::UNC-119. Poly-glutamylated tubulin is depicted as a modification of the B-tubule, as reported [45].
NPHP-2 localization requires an EF-hand
Calcium signaling plays a crucial role in signal transduction and ciliary function [72], [73]; cilia have high intraciliary calcium concentrations, and many TZ proteins possess calcium binding domains [74]. The NPHP-2 vertebrate homolog, Inversin, has two identified calmodulin-binding IQ domains [75], [76], one of which is required for proper localization [12]. Calmodulin detects intracellular calcium concentrations through a calcium-binding EF-hand. Though C. elegans NPHP-2 does not encode a predicted IQ domain, it does possess an EF-hand. This EF-hand is required for the localization and function of NPHP-2 in amphid and phasmid cilia, similarly to the IQ2 domain of Inversin. A significant difference exists between the EF-hand of NPHP-2 and the IQ2 domain of Inversin: deletion of the EF-hand of NPHP-2 results in a complete lack of ciliary localization, whereas deletion of the IQ2 domain of Inversin results in a mislocalization of Inversin along the entire cilium. In both systems, Inversin/NPHP-2 no longer localizes to the InvC. This suggests that Ca2+ detection/binding by and the subsequent hypothetical modulation of the activity of Inversin/NPHP-2 is a critical, conserved feature of the protein. Two possibilities arise for the function of these domains: Ca2+ specifies the localization of NPHP-2, modulates the activity of the protein, or both.
UNC-119 is a proximal ciliary protein
We found that UNC-119 localizes to the proximal cilium and is excluded from the distal region. GFP::UNC-119 and GT335 colabel, indicating that GFP::UNC-119 is excluded from the TZ, as TZ microtubules are not glutamylated (See Fig. 8, in which GT335 colabels with TZ-excluded ARL-13::GFP and NPHP-2::GFP). In C. elegans, UNC-119 labels a shorter portion of the cilium than ARL-13 or GT335, markers associated with the doublet region. Additionally, mammalian Unc119b physically interacts with the InvC component Nphp3 and is proximally restricted in cilia of RPE cells, suggesting that Unc119b is associated with the InvC [26]. However, GFP::UNC-119 marked a larger portion of the cilium than did NPHP-2::GFP.
The C. elegans genome encodes homologs of many of many Unc119b shuttle proteins, including Unc119b, Arl3, and RP2, and two myristoylated ciliary proteins which require unc-119 for ciliary localization [77]. arl-3 genetically interacts with unc-119 and nphp-2, suggesting that in C. elegans the components of the shuttle are in place. These shuttle components do not appear to be required for the localization of NPHP-2, as NPHP-2 is imported into the cilium in unc-119 and arl-3 mutants. In unc-119 mutants, NPHP-2 exhibited a unique distal dendritic localization pattern that cannot be attributable to TZ leakage; it is unknown whether this population represents NPHP-2 that has not been properly imported into the cilium or is mistargeted NPHP-2.
The function of hdac-6
In both mammalian systems and C. elegans, Hdac6/hdac-6 functions as an antagonist of ciliogenesis and cilia stability [16], [62]. In mammalian primary cilia, Hdac6 functions as an α-tubulin K40 deacetylase regulates microtubule function and stability [62], [78]–[81]. The C. elegans genome encodes a single α-tubulin with the acetylatable residue K40, MEC-12. However, there is no direct evidence for mec-12 expression in amphid and phasmid neurons, and the anti-α-tubulin-K40 antibody 6-11b-1 does not label amphid and phasmid cilia in wild-type animals or hdac-6 mutants (S10 Figure) [82]. Alternative tubulin acetylation sites may exist, including on β-tubulin [83]; hdac-6 could deacetylate these secondary sites. NPHP-2 contains a predicted N-terminal acetylation site (at 2S) which may be deacetylated by HDAC-6; this may modulate binding between NPHP-2 and its targets [84]. Additionally, HDAC-6 may have other unidentified ciliary targets [35]. Determining the mechanism by which HDAC-6 acts as a genetic modifier of InvC and doublet region gene defects is an important future direction.
Doublet region and InvC components modulate tubulin post-translational modification
Tubulin glutamylation is associated with the proximal portions of microtubule B-tubules in C. elegans cilia, mouse spermatozoa flagella, and Chlamydomonas flagella [47], [50], [85]. In C. elegans, microtubule glutamylation is linked to microtubule ultrastructure, stability, and maintenance [50], [86]. Multiple doublet region genes regulate microtubule glutamylation, and we observed a correlation between ectopic glutamylation and ectopic microtubule doublets. Additionally, ciliary targeting of doublet region proteins is not dependent on glutamylation status. Therefore, nphp-2, arl-13, hdac-6, and unc-119 function upstream of microtubule glutamylation, which may enable them to exert an influence on microtubule patterning, IFT, ciliogenesis, and, in the case of unc-119, singlet region biogenesis. These pathways may be conserved: in Arl13b/hennin mutant mice, ciliary microtubule glutamylation intensity is reduced, and microtubule B-tubules have ultrastructural defects [38], [64].
Doublet region- and InvC-associated genes form genetic modules
In C. elegans, TZ-associated genes can be grouped into two distinct, partially redundant genetic and physical modules [8], [17], [42]. Doublet region- and InvC-associated genes may be grouped in a similar manner into a nphp-2+klp-11 module and an arl-13+unc-119 module. hdac-6 appears to function outside the two modules, negatively regulating both: deletion of hdac-6 in SynDyf double mutants suppresses the SynDyf phenotype. In mammalian primary cilia, Hdac6 also plays an antagonistic role, destabilizing cilia through deacetylation of tubulin, a pathway suppressed by Inversin [16]. arl-3 may function outside of the two modules in a cell-type specific manner; in phasmids it genetically interacts with components from both modules, but in amphids arl-3 only genetically interacts with only the arl-13+unc-119 module and not the nphp-2+klp-11 module. In amphid cilia, nphp-2 also does not interact with the TZ SynDyf network.
Curiously, a further two genetic module organization exists between TZ genes and the InvC/doublet region genes. nphp-2 nphp-4, mks-3; nphp-2, and arl-13; nphp-4 double mutants exhibit severe ciliogenic defects [17]. Deletion of one TZ gene from either TZ module and one doublet region gene from either doublet region module yields a SynDyf phenotype, though not all combinations have been tested. The TZ SynDyf network and the doublet region SynDyf network can be thought of as two genetically interacting “super-modules”, each consisting of two to three sub-modules described in this and previous work [8], [17], [25], [42].
Origin of the Inversin Compartment
The localization requirements for multiple InvC localizing components have been previously determined, but how the InvC is initially established is not known.
In C. elegans, the InvC is likely established early in cilia development, as NPHP-2 is proximally restricted in phasmid cilia as early as the first larval L1 stage immediately following hatching (S9A Figure), and is not motile (S9B Figure), unlike the larval stage-dependent dynamic localization of ARL-13 [34]. We have eliminated several mechanisms for the establishment of the InvC. Ciliary ultrastructure does not seem to play a role, as in both mammals and C. elegans, the localization of Inversin/NPHP-2 is associated with only a sub-portion of the doublet region where there are no identifiable ultrastructural features [12]. Tubulin post-translational modifications also do not appear to specify the InvC, as nphp-2 (and genetically interacting doublet region components) lies upstream of glutamylation pathways. The TZ does not appear to play a major role in specifying the InvC. NPHP-2 still localizes and is restricted to the proximal cilium in TZ single and double mutants. IFT is another candidate mechanism, but we found that although IFT components genetically interact with InvC and doublet region associated genes, Kinesin-II is not required for NPHP-2 localization. In mammalian cilia, the Unc119b shuttle is required for the ciliary import of Nphp3; this activity is upstream of the action of Inversin in Nphp3 localization. In C. elegans phasmid cilia, NPHP-2 does not require either unc-119 or its effector arl-3 for ciliary import or InvC restriction (S2A Figure).
Several mechanisms for establishing the InvC remain. First, calcium may play a role. Both Inversin and NPHP-2 require a calcium binding domain for InvC localization [12]; the origin and nature of the calcium signal these domains detect is unknown. Second, the InvC may also be initially established by a diffusion of factors from the cilia base. A third possibility is that cilia membrane composition may help define the InvC. The cilium has a distinct membrane composition from the plasma membrane, and the different ciliary subregions may also have differential membrane composition.
Final summary
We propose that the logic underlying the establishment of the NPHP-2/Inversin compartment may be similar in C. elegans and mammals, in a manner independent of microtubule ultrastructure. We have shown that doublet region- and InvC-associated genes interact to guide ciliogenesis, cilia placement, cilia ultrastructure, protein composition, and tubulin post-translational modification. The next challenge is to determine what initially patterns the doublet region and InvC, and to understand the function of these cilia regions.
Materials and Methods
General molecular biology
Standard protocols were followed for all molecular biological procedures. PCR amplification using Taq polymerase (New England BioLabs, Ipswich, MA, USA) was used for genotyping deletion alleles, and was followed by restriction digest for SNP diagnosis. PCR amplification for construction of transgenic constructs was performed with Phusion High fidelity DNA polymerase (New England BioLabs), templated off C. elegans genomic DNA. Sequencing was performed offsite (GeneWiz, South Plainfield, NJ, USA). PCR primer and construct sequences are available upon request.
Bioinformatics and computer tools
Protein BLAST was used to find sequence orthologs [87]. All protein sequence information other than that of C. elegans was provided by NCBI, and all C. elegans nucleotide and protein sequences were provided by WormBase (Releases WS229 and WS234). Structural motif and domain predictions were generated by MotifScan [88]. Acetylation motifs were identified using NetAcet 1.0 [84]. Coiled-coil regions were identified with COILS [89]. ApE 2.0.36 was used for sequence manipulation, annotation, and restriction site identification.
Strains and maintenance
All strains were cultured at room temperature, unless otherwise noted, under standard conditions [90]. Transgenic strains using pha-1 selection were grown at 25°C, and pha-1(e2123) mutants were grown at 15°C. Deletion alleles were outcrossed to him-5 at least four times. Strains used in this study are listed in S2A Table. Alleles used were as follows: nphp-2(gk653), nphp-4(tm925), mks-3(tm2547), arl-13(gk513), unc-119(ed3), hdac-6(ok3203), arl-3(tm1703), him-5(e1490), osm-3(p802), klp-11(tm324), ccpp-1(ok1821), and ttll-4(tm3310). Primers used for diagnosis are listed in S2B Table. All transgenic strains used in Fig. 1–9 were tested using dye-filling for dominant negative defects in ciliogenesis. We observed no adverse effects in NPHP-2::GFP and GFP::UNC-119 transgenic strains but did find dominant negative defects in ARL-13::GFP transgenic strains (S1 Table).
The full length isoform of NPHP-2 was used in all NPHP-2 reporter constructs. The full length isoform, NPHP-2L, differs from the shorter isoform, NPHP-2S, in that the shorter isoform is missing 22 non-conserved amino acids of unknown function. Both isoforms have similar subciliary localization [17].
Electron microscopy
nphp-2 and arl-13; hdac-6; nphp-2 young adult animals were fixed using 3.5% glutaraldehyde +1% PFA in 0.1M HEPES and then in 1% OsO4+1.25% K4Fe(CN)4 in 0.1M HEPES. Samples were infiltrated and embedded in Embed-812 plastic resin. arl-13; nphp-2 young adult animals were fixed using high-pressure freeze fixation and freeze substitution in 2% OsO4+2% water in acetone as the primary fixative [91]. Samples were slowly freeze substituted in an RMC freeze substitution device, before infiltration with Embed-812 plastic resin. Images for wild-type animals fixed by a comparable immersion fixation method (cf. [32]) are now curated by the Hall lab at Einstein courtesy of E. Hedgecock. These wild type images are also available online at www.wormimage.org.
For TEM, serial sections (70 nm thickness) of fixed animals were collected on copper slot grids coated with formvar and evaporated carbon and stained with 4% uranyl acetate in 70% methanol, followed by washing and incubating with aqueous lead citrate. Images were captured on a Philips CM10 transmission electron microscope at 80 kV with a Morada 11 megapixel TEM CCD camera driven by iTEM software (Olympus Soft Imaging Solutions).
For each strain, we imaged three individuals that were fixed chemically. Additionally, we were concerned that the severe defects seen in the arl-13; nphp-2 double mutant were partially due to the harsh chemical fixation method. We fixed a fourth double mutant using high pressure freeze (HPF) fixation, which introduced fewer artifacts to confirm the validity of the chemical fixation data. We used the same strain in the construction of the double arl-13; nphp-2 and triple arl-13; hdac-6; nphp-2 mutants as was used in the previously published EM of the arl-13 single mutant.
Imaging
Animals were mounted on 5% Noble agar pads on standard microscope slides, and immobilized with a five minute incubation in 10 mM sodium azide. Worms were imaged using a Zeiss Plan-AXIOCHROMA 100X 1.4NA oil objective on a Zeiss Axio Imager.D1M (Zeiss, Oberkochen, Germany) with a Retiga-SRV Fast 1394 digital camera (Q-Imaging, Surrey, BC, Canada). Exposure time for antibodies was 100 ms, and exposure time for GFP fluorophores was 250 ms. Images were captured and manipulated using Metamorph software (Version 7.6.1.0, MDS Analytical technologies, Sunnyvale, CA, USA). Image stacks were 3D deconvolved using Auto Deblur software (Version 1.4.1, Media Cybernetics, Bethesda, MD, USA). Figures and diagrams were created with Adobe Photoshop CS3 (Version 10.0, Adobe Systems, San Jose, CA, USA) and Adobe Illustrator (Version 13.0.0, Adobe Systems). Image brightness and contrast were modified uniformly across an image, but gamma was not adjusted from 1.00. Brightness manipulations are similar, but not identical, across panels and figures. Significant variations in absolute intensity are noted where appropriate. For all strains, unless noted, worms were picked at L4 stage 24 hours before imaging.
Statistical analysis
All statistical analysis was performed with a combination of GraphPad Prism (Version 5.01, GraphPad Software, La Jolla, CA, USA) and Microsoft Excel (Version 14.0.7106, 32-Bit, Microsoft Corporation, Seattle, WA, USA). Sample size (n) for all figures is listed in S3 Table. Minimum p value for significance was set at 0.01 for all analyses unless otherwise specified. All parametric and continuous data types were analyzed using unpaired t-tests with Welch's correction to avoid assumption of equal variance. When multiple t-tests were performed on related data sets presented together, the Holm-Bonferroni multiple comparison adjustment was used to ensure the total alpha for the analysis did not exceed 0.01. All nonparametric and discontinuous data types were analyzed using Mann-Whitney U-test. Similar to the analysis of continuous data types, the Holm-Bonferroni multiple comparison adjustment was employed to ensure total alpha for all related comparisons did not exceed 0.01. Specific pairwise comparisons made are described in figure legends. Letters on graphs indicate statistically distinct groups, e.g., all groups marked ‘A’ are significantly different from all groups marked ‘B’.
Dye-filling assays
Staged young adult hermaphrodites were washed of plates with M9, and then rinsed three further times in M9, using gentle centrifugation to pellet the worms between rinses. Worms were then incubated in 40 µg/mL DiI (2.5 mg/mL dimethyl formamide stock, diluted 1∶1000 in M9) (Invitrogen) for 30 minutes in the dark. Worms were then rinsed three times in M9 as before, and were then placed on a seeded plate for a further 30-60 minutes to recover and flush dye from the digestive tract. Animals were anesthetized with 10 mM sodium azide and then immediately scored on a compound microscope (see Imaging section) using for dye-filling by manual counting of filled cell bodies. Cell body counts within the amphid or phasmid organs were averaged together to yield the average number of cells filling per organ per worm, and subjected to statistical analysis. Ectopically dye-filled neurons (e.g., IL2s) were not included in the total count.
Antibody staining
Antibody staining was performed under the standard Ruvkun-Finney protocol (Anatomical Methods, [92]) using GT335, an antibody against branch point single and polyglutamylated tubulin. Both primary (GT335, mouse, 1∶100, Enzo Life Sciences) and secondary antibody (Alexa Flour 568 goat anti-mouse, 1∶2000 dilution, Life Technologies) washes were performed at 4°C overnight. Young adult hermaphrodites were selected for imaging using the number of eggs in the animal—between one and ten—as a proxy for age.
Supplemental Information
arl-3 genetically interacts with InvC and doublet region associated genes. Dye filling was used to test for synthetic interactions between InvC and doublet region genes. (A) arl-3 interacts with nphp-2 and arl-13 in a sensillum-specific manner. arl-3 single mutants and arl-3; hdac-6 double mutants are nonDyf in amphids and phasmids. In both amphids and phasmids, arl-13; arl-3 double mutants are mildly SynDyf, which is suppressed by hdac-6 deletion. arl-3 deletion did not suppress arl-13; nphp-2 defects. hdac-6 mediated suppression of arl-13; nphp-2 defects requires arl-3. In phasmids, but not amphids, nphp-2 genetically interacted with arl-3, but this was not suppressed by hdac-6 deletion. (B) arl-3 genetically interacts with unc-119 and klp-11 in amphids and phasmids. arl-3 deletion moderately suppressed unc-119 single mutant Dyf. klp-11; arl-3 was slightly SynDyf. Data in panel A was statistically analyzed in conjunction with data from Fig. 1 because of the overlap in genotypes examined. Both panel A and Fig. 1 were analyzed with pairwise Mann-Whitney U-test between all groups, followed by the Holm-Bonferroni multiple comparison adjustment with a total alpha of 0.01. Genotypes from either panel A of this figure or Fig. 1 sharing a capital letter are not significantly different, whereas groups from either panel with different capital letters do differ significantly. Data panel B was analyzed using pairwise Mann-Whitney U-test between double mutants, their respective single mutants, and wild type followed by the Holm-Bonferroni multiple comparison adjustment with a total alpha of 0.01. **, double mutant phenotype is significantly different from both respective single mutants.
(TIF)
Localization requirements of NPHP-2::GFP and ARL-13::GFP in phasmid cilia. (A) NPHP-2::GFP does not require mks-3; nphp-4, hdac-6, or arl-13; hdac-6 for ciliary targeting or restriction to the proximal cilium. (B) Contrast enhanced version of Fig. 3C-D. ARL-13::GFP mislocalizes to the periciliary compartment in TZ and doublet region mutants.
(TIF)
NPHP-2 and ARL-13 do not require TZ-, doublet region-, and InvC-associated genes for ciliary targeting in amphids. (A) Localization of NPHP-2::GFP is subtly disrupted in mks-3, nphp-4 and mks-3; nphp-4 mutants. Periciliary puncta in amphid channel cilia and altered staining in IL2, CEP, and OLQ cilia are visible. (B) The NPHP-2::GFP amphid bundle is shortened in klp-11 and unc-119 mutants, and elongated in arl-13 mutants. Periciliary puncta are visible in klp-11 and arl-13 mutants. hdac-6 deletion does not suppress the arl-13 phenotype. (C) ARL-13::GFP localizes primarily to the doublet region of amphid channel cilia, and to a nonspecific proximal region of IL2 cilia. In mks-3 and nphp-4 mutants, ARL-13::GFP localization in amphid channel cilia looks grossly wild-type. CEP and OLQ cilia show increased ARL-13::GFP localization. (D) unc-119 mutants exhibit distal dendritic accumulation of ARL-13::GFP. klp-11 and nphp-2 mutants exhibit increased CEP and OLQ ARL-13::GFP localization. Arrowheads indicate periciliary puncta. Bar indicates distal dendritic localization.
(TIF)
Amphid UNC-119 localization in InvC and doublet region mutants. GFP::UNC-119 appears similar in wild-type (Fig. 5A), nphp-2, and hdac-6 amphid channel cilia. klp-11 and arl-13 exhibit an accumulation of GFP::UNC-119 in amphid channel, OLQ, CEP, and inner labial cilia. hdac-6 deletion does not suppress GFP::UNC-119 mislocalization in arl-13 mutants. Green arrow – TZ gap, yellow arrowheads – IL2 cilia, red arrowheads - OLQ cilia, blue arrowheads – CEP cilia, white bar – amphid bundle.
(TIF)
Amphid dye-filling of IFT and unc-119 mutants. (A) klp-11 single mutants are not Dyf. klp-11 is SynDyf with arl-13, which is suppressed by deletion of hdac-6. (B) unc-119 single mutants are severely Dyf. hdac-6; nphp-2 suppresses the unc-119 Dyf phenotype to a small degree. (C) osm-3 single mutants are both severely amphid and phasmid Dyf. In no double mutant was this suppressed. (D) hdac-6; nphp-2 nphp-4 and arl-3; nphp-2 nphp-4 worms were assayed for suppression of SynDyf defects. Neither strain exhibited significant suppression as compared to the nphp-2 nphp-4 double mutant. Data was analyzed with pairwise Mann-Whitney U-test between wild type, double mutants, triple mutants and their respective single mutants, followed by the Holm-Bonferroni multiple comparison adjustment. **, significant versus single mutants at a total alpha of 0.01.
(TIF)
NPHP-2::GFP marks a significantly smaller region of the cilium than ARL-13::GFP and GFP::UNC-119. (A) Worms expressing NPHP-2::GFP, ARL-13::GFP, and GFP::UNC-119 reporters were stained with GT335. The ratio of the length of each reporter localization pattern to the length of the entire cilium, minus the TZ was computed per-cilium. NPHP-2::GFP marks a significantly shorter proportion of the cilium than either ARL-13::GFP or GFP::UNC-119. (B) Absolute lengths of data presented in panel A. (C) Worms expressing NPHP-2::GFP, ARL-13::GFP, and GFP::UNC-119 reporters were incubated with DiI to label cilia. The ratio of the length of each reporter localization pattern to the length of the entire cilium, minus the TZ was computed per-cilium. NPHP-2::GFP marks a significantly shorter proportion of the cilium than either ARL-13::GFP or GFP::UNC-119. (D) Absolute lengths of data presented in panel C. Phasmid cilia lengths in transgenic strains as measured using DiI staining, and reporter localization size. Data in each panel was analyzed with pairwise t-tests with Welch's Correction, followed by the Holm-Bonferroni multiple comparison adjustment for a total alpha of 0.01.
(TIF)
Doublet region- and InvC-associated genes regulate the localization patterns of doublet region and InvC components. Each data point represents the averaged lengths of the GFP signal or immunofluorescence in all visible and distinct phasmid cilia within a single animal. (A) NPHP-2::GFP localization length is significantly decreased in klp-11 and unc-119 mutants, and significantly increased in arl-13 mutants. hdac-6 deletion partially suppresses the arl-13 phenotype. (B) ARL-13::GFP localization length is increased in nphp-2 and decreased in unc-119 mutants. (C) GFP::UNC-119 localization length does not change significantly in any strain. (D) KAP-1 localization length measurements were more variable that the other reporters due to a faint KAP-1::GFP signal, but were not significantly different from wild type in any strain. Localization length was significantly altered in nphp-2 mutants. (E) nphp-2 and arl-13 had significantly longer GT335 signals than in wild type. nphp-2 mutants have increased variability in GT335 signal lengths compared to WT. (F) Diagram representing genetic control of doublet region protein localization in phasmid cilia. An arrow between a gene and a protein/GT335 indicates that the gene is required for the protein/GT335 to label its entire territory. A T-bar between a gene and a protein/GT335 indicates that the gene restricts the size of the protein territory/GT335 signal length. Data was analyzed with an unpaired t-test with Welch's Correction against wild type, followed by the Holm-Bonferroni multiple comparison adjustment for all comparisons in a given panel. **p<0.01.
(TIF)
NPHP-2 contains a strongly conserved hydroxylation motif and a predicted acetylation site. (A) Hydroxylation motif is highlighted in green. Hydroxylated asparagine is highlighted in blue. ANKS6 and INVS alignment are from [24]. (B) NPHP-2 domain model. NPHP-2 contains a predicted acetylation site at 2S, and two predicted SUMOylation sites. Green letters indicate modified residues. Acetylation and SUMOylation motifs do not appear to be conserved.
(TIF)
(A) NPHP-2::GFP localizes to the proximal portion of the cilium in L1 animals. (B) Kymograph of NPHP-2::GFP in phasmid cilia. No movement of NPHP-2::GFP was detectable. Total duration of recording was 40 seconds. (C) NPHP-2::GFP localizes to amphid and phasmid cilia in L1 and L4 stage ccpp-1 mutants. (D) GFP::UNC-119 localizes to amphid and phasmid cilia in L1 and L4 stage ccpp-1 mutants. White bars indicate ciliary GFP::UNC-119 localization. White arrow indicates abnormal cilia base accumulation of GFP::UNC-119.
(TIF)
The anti-acetylated tubulin antibody 6-11b-1 does not label amphid channel and phasmid cilia in either WT animals or hdac-6 mutants. 6-11b-1 antibody labels only dendrites of the touch neurons. hdac-6 deletion does not increase the amount of acetylation detected in touch neurons. Non-specific labelling is seen in the buccal cavity, and not in amphid cilia, as determined by analysis of 3D image stacks.
(TIF)
Transgenic strains used in this study have mild to no ciliogenic defects. Worms from each transgenic strain imaged or otherwise quantified in Fig. 1-9—other than strains with a Dyf or SynDyf background—were assayed for dye-filling defects to determine if there were any gross defects in ciliogenesis or cilia placement. Scores reported are average fraction of cell bodies taking up dye, plus/minus std. err. Yellow cells mark possible dominant negative defects. Transgenic strains expressing NPHP-2::GFP and GFP::UNC-119 did not exhibit defects, but strains expressing ARL-13::GFP all exhibited mild defects.
(XLSX)
List of strains and PCR deletion diagnosis primers used in this work. Strains are organized by the figures in which they appear.
(XLS)
Experimental sample sizes for figures with statistical analysis.
(XLSX)
Acknowledgments
We thank L. Gunther and G. Perumal for help with high pressure freezing procedures. We thank E. Hedgecock for contribution of archival images to the Hall lab collection. We thank members of the Barr lab, including Robert O'Hagan, for critical reading of the manuscript and assistance with reagents and procedures. We also thank Jinghua Hu, Oliver Blacque, and the C. elegans Genetics Center for strains and reagents, the Japanese National BioResource Project and the C. elegans Gene Knockout Consortium for deletion mutants, Wormbase for sequence information. We are also grateful to the Rutgers C. elegans community, because you can't start a fire without a spark.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was funded by the National Institutes of Health OD010943 (DHH) and DK074746 (MMB). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Ke YN, Yang WX (2014) Primary cilium: An elaborate structure that blocks cell division? Gene 547(2): 175–185. [DOI] [PubMed] [Google Scholar]
- 2. Fry AM, Leaper MJ, Bayliss R (2014) The primary cilium: Guardian of organ development and homeostasis. Organogenesis 10(1): 62–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Goggolidou P (2014) Wnt and planar cell polarity signaling in cystic renal disease. Organogenesis 10(1): 86–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yoshiba S, Hamada H (2014) Roles of cilia, fluid flow, and Ca2+ signaling in breaking of left-right symmetry. Trends Genet 30(1): 10–17. [DOI] [PubMed] [Google Scholar]
- 5. Yuan S, Sun Z (2013) Expanding horizons: Ciliary proteins reach beyond cilia. Annu Rev Genet 47: 353–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jackson PK (2013) Nek8 couples renal ciliopathies to DNA damage and checkpoint control. Mol Cell 51(4): 407–408. [DOI] [PubMed] [Google Scholar]
- 7. Rosenbaum JL, Witman GB (2002) Intraflagellar transport. Nat Rev Mol Cell Biol. 3(11): 813–825. [DOI] [PubMed] [Google Scholar]
- 8. Williams CL, Li C, Kida K, Inglis PN, Mohan S, et al. (2011) MKS and NPHP modules cooperate to establish basal body/transition zone membrane associations and ciliary gate function during ciliogenesis. J Cell Biol 192(6): 1023–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dishinger JF, Kee HL, Jenkins PM, Fan S, Hurd TW, et al. (2010) Ciliary entry of the kinesin-2 motor KIF17 is regulated by importin-beta2 and RanGTP. Nat Cell Biol 12(7): 703–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Satir P, Pedersen LB, Christensen ST (2010) The primary cilium at a glance. J Cell Sci 123(Pt 4): 499–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blacque OE, Sanders AA. (2014) Compartments within a compartment: What can tell us about ciliary subdomain composition, biogenesis, function, and disease. Organogenesis 10(1). [DOI] [PMC free article] [PubMed]
- 12. Shiba D, Yamaoka Y, Hagiwara H, Takamatsu T, Hamada H, et al. (2009) Localization of Inv in a distinctive intraciliary compartment requires the C-terminal ninein-homolog-containing region. J Cell Sci 122(Pt 1): 44–54. [DOI] [PubMed] [Google Scholar]
- 13. Wojtyniak M, Brear AG, O'Halloran DM, Sengupta P (2013) Cell- and subunit-specific mechanisms of CNG channel ciliary trafficking and localization in C. elegans . J Cell Sci 126(Pt 19): 4381–4395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gong Z, Son W, Chung YD, Kim J, Shin DW, et al. (2004) Two interdependent TRPV channel subunits, inactive and nanchung, mediate hearing in drosophila. J Neurosci 24(41): 9059–9066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. French DA, Badamdorj D, Kleene SJ (2010) Spatial distribution of calcium-gated chloride channels in olfactory cilia. PLoS One 5(12): e15676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mergen M, Engel C, Muller B, Follo M, Schafer T, et al. (2013) The nephronophthisis gene product NPHP2/Inversin interacts with aurora A and interferes with HDAC6-mediated cilia disassembly. Nephrol Dial Transplant 28(11): 2744–2753. [DOI] [PubMed] [Google Scholar]
- 17. Warburton-Pitt SR, Jauregui AR, Li C, Wang J, Leroux MR, et al. (2012) Ciliogenesis in Caenorhabditis elegans requires genetic interactions between ciliary middle segment localized NPHP-2 (Inversin) and transition zone-associated proteins. J Cell Sci 125(Pt 11): 2592–2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Otto EA, Schermer B, Obara T, O'Toole JF, Hiller KS, et al. (2003) Mutations in INVS encoding Inversin cause nephronophthisis type 2, linking renal cystic disease to the function of primary cilia and left-right axis determination. Nat Genet 34(4): 413–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Olbrich H, Fliegauf M, Hoefele J, Kispert A, Otto E, et al. (2003) Mutations in a novel gene, NPHP3, cause adolescent nephronophthisis, tapeto-retinal degeneration and hepatic fibrosis. Nat Genet. [DOI] [PubMed]
- 20. Bergmann C, Fliegauf M, Bruchle NO, Frank V, Olbrich H, et al. (2008) Loss of Nephrocystin-3 function can cause embryonic lethality, Meckel-Gruber-like syndrome, situs inversus, and renal-hepatic-pancreatic dysplasia. Am J Hum Genet 82(4): 959–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Otto EA, Trapp ML, Schultheiss UT, Helou J, Quarmby LM, et al. (2008) NEK8 mutations affect ciliary and centrosomal localization and may cause nephronophthisis. J Am Soc Nephrol 19(3): 587–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cantagrel V, Silhavy JL, Bielas SL, Swistun D, Marsh SE, et al. (2008) Mutations in the cilia gene ARL13B lead to the classical form of Joubert syndrome. Am J Hum Genet 83(2): 170–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shiba D, Manning DK, Koga H, Beier DR, Yokoyama T (2010) Inv acts as a molecular anchor for Nphp3 and Nek8 in the proximal segment of primary cilia. Cytoskeleton (Hoboken) 67(2): 112–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hoff S, Halbritter J, Epting D, Frank V, Nguyen TM, et al. (2013) ANKS6 is a central component of a nephronophthisis module linking NEK8 to INVS and NPHP3. Nat Genet 45(8): 951–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sang L, Miller JJ, Corbit KC, Giles RH, Brauer MJ, et al. (2011) Mapping the NPHP-JBTS-MKS protein network reveals ciliopathy disease genes and pathways. Cell 145(4): 513–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wright KJ, Baye LM, Olivier-Mason A, Mukhopadhyay S, Sang L, et al. (2011) An ARL3-UNC119-RP2 GTPase cycle targets myristoylated NPHP3 to the primary cilium. Genes Dev 25(22): 2347–2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Inglis PN, Ou G, Leroux MR, Scholey JM. (2007) The sensory cilia of Caenorhabditis elegans. WormBook: 1–22. [DOI] [PMC free article] [PubMed]
- 28. Barr MM, Sternberg PW (1999) A polycystic kidney-disease gene homologue required for male mating behaviour in C. elegans . Nature 401(6751): 386–389. [DOI] [PubMed] [Google Scholar]
- 29. Morsci NS, Barr MM (2011) Kinesin-3 KLP-6 regulates intraflagellar transport in male-specific cilia of Caenorhabditis elegans . Curr Biol 21(14): 1239–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang J, Schwartz HT, Barr MM (2010) Functional specialization of sensory cilia by an RFX transcription factor isoform. Genetics 186(4): 1295–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bae YK, Kim E, L'Hernault S W, Barr MM (2009) The CIL-1 PI 5-phosphatase localizes TRP polycystins to cilia and activates sperm in C. elegans . Curr Biol 19(19): 1599–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Perkins LA, Hedgecock EM, Thomson JN, Culotti JG (1986) Mutant sensory cilia in the nematode Caenorhabditis elegans . Dev Biol 117(2): 456–487. [DOI] [PubMed] [Google Scholar]
- 33. Doroquez DB, Berciu C, Anderson JR, Sengupta P, Nicastro D (2014) A high-resolution morphological and ultrastructural map of anterior sensory cilia and glia in Caenorhabditis elegans . Elife 3: e01948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cevik S, Sanders AA, Van Wijk E, Boldt K, Clarke L, et al. (2013) Active transport and diffusion barriers restrict joubert syndrome-associated ARL13B/ARL-13 to an Inv-like ciliary membrane subdomain. PLoS Genet 9(12): e1003977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Li Y, Wei Q, Zhang Y, Ling K, Hu J (2010) The small GTPases ARL-13 and ARL-3 coordinate intraflagellar transport and ciliogenesis. J Cell Biol 189(6): 1039–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Snow JJ, Ou G, Gunnarson AL, Walker MR, Zhou HM, et al. (2004) Two anterograde intraflagellar transport motors cooperate to build sensory cilia on C. elegans neurons. Nat Cell Biol 6(11): 1109–13. [DOI] [PubMed] [Google Scholar]
- 37. Cevik S, Hori Y, Kaplan OI, Kida K, Toivenon T, et al. (2010) Joubert syndrome Arl13b functions at ciliary membranes and stabilizes protein transport in Caenorhabditis elegans . J Cell Biol 188(6): 953–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Caspary T, Larkins CE, Anderson KV (2007) The graded response to sonic hedgehog depends on cilia architecture. Dev Cell 12(5): 767–778. [DOI] [PubMed] [Google Scholar]
- 39. Reiter JF, Blacque OE, Leroux MR (2012) The base of the cilium: Roles for transition fibres and the transition zone in ciliary formation, maintenance and compartmentalization. EMBO Rep 13(7): 608–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nakata K, Shiba D, Kobayashi D, Yokoyama T (2012) Targeting of Nphp3 to the primary cilia is controlled by an N-terminal myristoylation site and coiled-coil domains. Cytoskeleton (Hoboken) 69(4): 221–234. [DOI] [PubMed] [Google Scholar]
- 41. Humbert MC, Weihbrecht K, Searby CC, Li Y, Pope RM, et al. (2012) ARL13B, PDE6D, and CEP164 form a functional network for INPP5E ciliary targeting. Proc Natl Acad Sci U S A 109(48): 19691–19696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Williams CL, Masyukova SV, Yoder BK (2010) Normal ciliogenesis requires synergy between the cystic kidney disease genes MKS-3 and NPHP-4. J Am Soc Nephrol 21(5): 782–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Watanabe D, Saijoh Y, Nonaka S, Sasaki G, Ikawa Y, et al. (2003) The left-right determinant Inversin is a component of node monocilia and other 9+0 cilia. Development 130(9): 1725–1734. [DOI] [PubMed] [Google Scholar]
- 44. Ou G, Koga M, Blacque OE, Murayama T, Ohshima Y, et al. (2007) Sensory ciliogenesis in Caenorhabditis elegans: Assignment of IFT components into distinct modules based on transport and phenotypic profiles. Mol Biol Cell 18(5): 1554–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lechtreck KF, Geimer S (2000) Distribution of polyglutamylated tubulin in the flagellar apparatus of green flagellates. Cell Motil Cytoskeleton 47(3): 219–35. [DOI] [PubMed] [Google Scholar]
- 46. Suryavanshi S, Edde B, Fox LA, Guerrero S, Hard R, et al. (2010) Tubulin glutamylation regulates ciliary motility by altering inner dynein arm activity. Curr Biol 20(5): 435–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Fouquet JP, Prigent Y, Kann ML (1996) Comparative immunogold analysis of tubulin isoforms in the mouse sperm flagellum: Unique distribution of glutamylated tubulin. Mol Reprod Dev 43(3): 358–365. [DOI] [PubMed] [Google Scholar]
- 48. Edde B, Rossier J, Le Caer JP, Desbruyeres E, Gros F, et al. (1990) Posttranslational glutamylation of alpha-tubulin. Science 247(4938): 83–5. [DOI] [PubMed] [Google Scholar]
- 49. Sharma N, Bryant J, Wloga D, Donaldson R, Davis RC, et al. (2007) Katanin regulates dynamics of microtubules and biogenesis of motile cilia. J Cell Biol 178(6): 1065–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. O'Hagan R, Piasecki BP, Silva M, Phirke P, Nguyen KC, et al. (2011) The tubulin deglutamylase CCPP-1 regulates the function and stability of sensory cilia in C. elegans . Curr Biol 21(20): 1685–1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kann ML, Soues S, Levilliers N, Fouquet JP (2003) Glutamylated tubulin: Diversity of expression and distribution of isoforms. Cell Motil Cytoskeleton 55(1): 14–25. [DOI] [PubMed] [Google Scholar]
- 52. Pathak N, Obara T, Mangos S, Liu Y, Drummond IA (2007) The zebrafish fleer gene encodes an essential regulator of cilia tubulin polyglutamylation. Mol Biol Cell 18(11): 4353–4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Berkowitz LA, Knight AL, Caldwell GA, Caldwell KA (2008) Generation of stable transgenic C. elegans using microinjection. J Vis Exp (18) pii: 833. doi(18): 10.3791/833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Jauregui AR, Nguyen KC, Hall DH, Barr MM (2008) The Caenorhabditis elegans nephrocystins act as global modifiers of cilium structure. J Cell Biol 180(5): 973–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Barr MM, DeModena J, Braun D, Nguyen CQ, Hall DH, et al. (2001) The Caenorhabditis elegans autosomal dominant polycystic kidney disease gene homologs lov-1 and pkd-2 act in the same pathway. Curr Biol 11(17): 1341–1346. [DOI] [PubMed] [Google Scholar]
- 56. Peden EM, Barr MM (2005) The KLP-6 kinesin is required for male mating behaviors and polycystin localization in Caenorhabditis elegans . Current Biology 15(5): 394–404. [DOI] [PubMed] [Google Scholar]
- 57. Bae YK, Kim E, L'hernault SW, Barr MM (2009) The CIL-1 PI 5-phosphatase localizes TRP polycystins to cilia and activates sperm in C. elegans . Curr Biol 19(19): 1599–1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Blacque OE, Perens EA, Boroevich KA, Inglis PN, Li C, et al. (2005) Functional genomics of the cilium, a sensory organelle. Curr Biol 15(10): 935–41. [DOI] [PubMed] [Google Scholar]
- 59. Li Y, Zhang Q, Wei Q, Zhang Y, Ling K, et al. (2012) SUMOylation of the small GTPase ARL-13 promotes ciliary targeting of sensory receptors. J Cell Biol 199(4): 589–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nguyen PA, Liou W, Hall DH, Leroux MR. (2014) Ciliopathy proteins establish a bipartite signaling compartment in a C. elegans thermosensory neuron. J Cell Sci. [DOI] [PMC free article] [PubMed]
- 61. Frokjaer-Jensen C (2013) Exciting prospects for precise engineering of Caenorhabditis elegans genomes with CRISPR/Cas9 Genetics. 195(3): 635–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Pugacheva EN, Jablonski SA, Hartman TR, Henske EP, Golemis EA (2007) HEF1-dependent Aurora A activation induces disassembly of the primary cilium. Cell 129(7): 1351–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Hori Y, Kobayashi T, Kikko Y, Kontani K, Katada T (2008) Domain architecture of the atypical arf-family GTPase Arl13b involved in cilia formation. Biochem Biophys Res Commun 373(1): 119–124. [DOI] [PubMed] [Google Scholar]
- 64. Larkins CE, Aviles GD, East MP, Kahn RA, Caspary T (2011) Arl13b regulates ciliogenesis and the dynamic localization of Shh signaling proteins. Mol Biol Cell 22(23): 4694–4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Satish Tammana TV, Tammana D, Diener DR, Rosenbaum J (2013) Centrosomal protein CEP104 (Chlamydomonas FAP256) moves to the ciliary tip during ciliary assembly. J Cell Sci 126(Pt 21): 5018–5029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Lee JE, Silhavy JL, Zaki MS, Schroth J, Bielas SL, et al. (2012) CEP41 is mutated in Joubert syndrome and is required for tubulin glutamylation at the cilium. Nat Genet 44(2): 193–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Zhou C, Cunningham L, Marcus AI, Li Y, Kahn RA (2006) Arl2 and Arl3 regulate different microtubule-dependent processes. Mol Biol Cell 17(5): 2476–2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Wu Y, Dai XQ, Li Q, Chen CX, Mai W, et al. (2006) Kinesin-2 mediates physical and functional interactions between Polycystin-2 and Fibrocystin. Hum Mol Genet 15(22): 3280–92. [DOI] [PubMed] [Google Scholar]
- 69. Follit JA, Xu F, Keady BT, Pazour GJ (2009) Characterization of mouse IFT complex B. Cell Motil Cytoskeleton 66(8): 457–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Fan S, Hurd TW, Liu CJ, Straight SW, Weimbs T, et al. (2004) Polarity proteins control ciliogenesis via kinesin motor interactions. Curr Biol 14(16): 1451–1461. [DOI] [PubMed] [Google Scholar]
- 71. Sanchez de Diego A, Alonso Guerrero A, Martinez-A C, van Wely KH (2014) Dido3-dependent HDAC6 targeting controls cilium size. Nat Commun 5: 3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. DiPetrillo CG, Smith EF (2013) Methods for analysis of calcium/calmodulin signaling in cilia and flagella. Methods Enzymol 524: 37–57. [DOI] [PubMed] [Google Scholar]
- 73. Jin X, Mohieldin AM, Muntean BS, Green JA, Shah JV, et al. (2014) Cilioplasm is a cellular compartment for calcium signaling in response to mechanical and chemical stimuli. Cell Mol Life Sci 71(11): 2165–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Collingridge P, Brownlee C, Wheeler GL (2013) Compartmentalized calcium signaling in cilia regulates intraflagellar transport. Curr Biol 23(22): 2311–2318. [DOI] [PubMed] [Google Scholar]
- 75. Morgan D, Goodship J, Essner JJ, Vogan KJ, Turnpenny L, et al. (2002) The left-right determinant Inversin has highly conserved ankyrin repeat and IQ domains and interacts with calmodulin. Hum Genet 110(4): 377–384. [DOI] [PubMed] [Google Scholar]
- 76. Yasuhiko Y, Imai F, Ookubo K, Takakuwa Y, Shiokawa K, et al. (2001) Calmodulin binds to Inv protein: Implication for the regulation of Inv function. Dev Growth Differ 43(6): 671–681. [DOI] [PubMed] [Google Scholar]
- 77. Zhang H, Constantine R, Vorobiev S, Chen Y, Seetharaman J, et al. (2011) UNC119 is required for G protein trafficking in sensory neurons. Nat Neurosci 14(7): 874–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Hubbert C, Guardiola A, Shao R, Kawaguchi Y, Ito A, et al. (2002) HDAC6 is a microtubule-associated deacetylase. Nature 417(6887): 455–458. [DOI] [PubMed] [Google Scholar]
- 79. Zhang Y, Li N, Caron C, Matthias G, Hess D, et al. (2003) HDAC-6 interacts with and deacetylates tubulin and microtubules in vivo. EMBO J 22(5): 1168–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Matsuyama A, Shimazu T, Sumida Y, Saito A, Yoshimatsu Y, et al. (2002) In vivo destabilization of dynamic microtubules by HDAC6-mediated deacetylation. EMBO J 21(24): 6820–6831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Verhey KJ, Gaertig J (2007) The tubulin code. Cell Cycle 6(17): 2152–60. [DOI] [PubMed] [Google Scholar]
- 82. Fukushige T, Siddiqui ZK, Chou M, Culotti JG, Gogonea CB, et al. (1999) MEC-12, an alpha-tubulin required for touch sensitivity in C. elegans . J Cell Sci 112(Pt 3): 395–403. [DOI] [PubMed] [Google Scholar]
- 83. Chu CW, Hou F, Zhang J, Phu L, Loktev AV, et al. (2011) A novel acetylation of beta-tubulin by san modulates microtubule polymerization via down-regulating tubulin incorporation. Mol Biol Cell 22(4): 448–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Kiemer L, Bendtsen JD, Blom N (2005) NetAcet: Prediction of N-terminal acetylation sites. Bioinformatics 21(7): 1269–1270. [DOI] [PubMed] [Google Scholar]
- 85. Kubo T, Yanagisawa HA, Yagi T, Hirono M, Kamiya R (2010) Tubulin polyglutamylation regulates axonemal motility by modulating activities of inner-arm dyneins. Curr Biol 20(5): 441–5. [DOI] [PubMed] [Google Scholar]
- 86. Kimura Y, Kurabe N, Ikegami K, Tsutsumi K, Konishi Y, et al. (2010) Identification of tubulin deglutamylase among Caenorhabditis elegans and mammalian cytosolic carboxypeptidases (CCPs). J Biol Chem 285(30): 22936–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, et al. (1997) Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res 25(17): 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Hau J, Muller M, Pagni M (2007) HitKeeper, a generic software package for hit list management. Source Code Biol Med 2: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Lupas A (1997) Predicting coiled-coil regions in proteins. Curr Opin Struct Biol 7(3): 388–393. [DOI] [PubMed] [Google Scholar]
- 90. Brenner S (1974) The genetics of Caenorhabditis elegans . Genetics 77(1): 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Weimer RM (2006) Preservation of C. elegans tissue via high-pressure freezing and freeze-substitution for ultrastructural analysis and immunocytochemistry. Methods Mol Biol 351: 203–221. [DOI] [PubMed] [Google Scholar]
- 92.Altun ZF, Herndon LA, Crocker C, Lints R, Hall DH. (2013) WormAtlas. 2013.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
arl-3 genetically interacts with InvC and doublet region associated genes. Dye filling was used to test for synthetic interactions between InvC and doublet region genes. (A) arl-3 interacts with nphp-2 and arl-13 in a sensillum-specific manner. arl-3 single mutants and arl-3; hdac-6 double mutants are nonDyf in amphids and phasmids. In both amphids and phasmids, arl-13; arl-3 double mutants are mildly SynDyf, which is suppressed by hdac-6 deletion. arl-3 deletion did not suppress arl-13; nphp-2 defects. hdac-6 mediated suppression of arl-13; nphp-2 defects requires arl-3. In phasmids, but not amphids, nphp-2 genetically interacted with arl-3, but this was not suppressed by hdac-6 deletion. (B) arl-3 genetically interacts with unc-119 and klp-11 in amphids and phasmids. arl-3 deletion moderately suppressed unc-119 single mutant Dyf. klp-11; arl-3 was slightly SynDyf. Data in panel A was statistically analyzed in conjunction with data from Fig. 1 because of the overlap in genotypes examined. Both panel A and Fig. 1 were analyzed with pairwise Mann-Whitney U-test between all groups, followed by the Holm-Bonferroni multiple comparison adjustment with a total alpha of 0.01. Genotypes from either panel A of this figure or Fig. 1 sharing a capital letter are not significantly different, whereas groups from either panel with different capital letters do differ significantly. Data panel B was analyzed using pairwise Mann-Whitney U-test between double mutants, their respective single mutants, and wild type followed by the Holm-Bonferroni multiple comparison adjustment with a total alpha of 0.01. **, double mutant phenotype is significantly different from both respective single mutants.
(TIF)
Localization requirements of NPHP-2::GFP and ARL-13::GFP in phasmid cilia. (A) NPHP-2::GFP does not require mks-3; nphp-4, hdac-6, or arl-13; hdac-6 for ciliary targeting or restriction to the proximal cilium. (B) Contrast enhanced version of Fig. 3C-D. ARL-13::GFP mislocalizes to the periciliary compartment in TZ and doublet region mutants.
(TIF)
NPHP-2 and ARL-13 do not require TZ-, doublet region-, and InvC-associated genes for ciliary targeting in amphids. (A) Localization of NPHP-2::GFP is subtly disrupted in mks-3, nphp-4 and mks-3; nphp-4 mutants. Periciliary puncta in amphid channel cilia and altered staining in IL2, CEP, and OLQ cilia are visible. (B) The NPHP-2::GFP amphid bundle is shortened in klp-11 and unc-119 mutants, and elongated in arl-13 mutants. Periciliary puncta are visible in klp-11 and arl-13 mutants. hdac-6 deletion does not suppress the arl-13 phenotype. (C) ARL-13::GFP localizes primarily to the doublet region of amphid channel cilia, and to a nonspecific proximal region of IL2 cilia. In mks-3 and nphp-4 mutants, ARL-13::GFP localization in amphid channel cilia looks grossly wild-type. CEP and OLQ cilia show increased ARL-13::GFP localization. (D) unc-119 mutants exhibit distal dendritic accumulation of ARL-13::GFP. klp-11 and nphp-2 mutants exhibit increased CEP and OLQ ARL-13::GFP localization. Arrowheads indicate periciliary puncta. Bar indicates distal dendritic localization.
(TIF)
Amphid UNC-119 localization in InvC and doublet region mutants. GFP::UNC-119 appears similar in wild-type (Fig. 5A), nphp-2, and hdac-6 amphid channel cilia. klp-11 and arl-13 exhibit an accumulation of GFP::UNC-119 in amphid channel, OLQ, CEP, and inner labial cilia. hdac-6 deletion does not suppress GFP::UNC-119 mislocalization in arl-13 mutants. Green arrow – TZ gap, yellow arrowheads – IL2 cilia, red arrowheads - OLQ cilia, blue arrowheads – CEP cilia, white bar – amphid bundle.
(TIF)
Amphid dye-filling of IFT and unc-119 mutants. (A) klp-11 single mutants are not Dyf. klp-11 is SynDyf with arl-13, which is suppressed by deletion of hdac-6. (B) unc-119 single mutants are severely Dyf. hdac-6; nphp-2 suppresses the unc-119 Dyf phenotype to a small degree. (C) osm-3 single mutants are both severely amphid and phasmid Dyf. In no double mutant was this suppressed. (D) hdac-6; nphp-2 nphp-4 and arl-3; nphp-2 nphp-4 worms were assayed for suppression of SynDyf defects. Neither strain exhibited significant suppression as compared to the nphp-2 nphp-4 double mutant. Data was analyzed with pairwise Mann-Whitney U-test between wild type, double mutants, triple mutants and their respective single mutants, followed by the Holm-Bonferroni multiple comparison adjustment. **, significant versus single mutants at a total alpha of 0.01.
(TIF)
NPHP-2::GFP marks a significantly smaller region of the cilium than ARL-13::GFP and GFP::UNC-119. (A) Worms expressing NPHP-2::GFP, ARL-13::GFP, and GFP::UNC-119 reporters were stained with GT335. The ratio of the length of each reporter localization pattern to the length of the entire cilium, minus the TZ was computed per-cilium. NPHP-2::GFP marks a significantly shorter proportion of the cilium than either ARL-13::GFP or GFP::UNC-119. (B) Absolute lengths of data presented in panel A. (C) Worms expressing NPHP-2::GFP, ARL-13::GFP, and GFP::UNC-119 reporters were incubated with DiI to label cilia. The ratio of the length of each reporter localization pattern to the length of the entire cilium, minus the TZ was computed per-cilium. NPHP-2::GFP marks a significantly shorter proportion of the cilium than either ARL-13::GFP or GFP::UNC-119. (D) Absolute lengths of data presented in panel C. Phasmid cilia lengths in transgenic strains as measured using DiI staining, and reporter localization size. Data in each panel was analyzed with pairwise t-tests with Welch's Correction, followed by the Holm-Bonferroni multiple comparison adjustment for a total alpha of 0.01.
(TIF)
Doublet region- and InvC-associated genes regulate the localization patterns of doublet region and InvC components. Each data point represents the averaged lengths of the GFP signal or immunofluorescence in all visible and distinct phasmid cilia within a single animal. (A) NPHP-2::GFP localization length is significantly decreased in klp-11 and unc-119 mutants, and significantly increased in arl-13 mutants. hdac-6 deletion partially suppresses the arl-13 phenotype. (B) ARL-13::GFP localization length is increased in nphp-2 and decreased in unc-119 mutants. (C) GFP::UNC-119 localization length does not change significantly in any strain. (D) KAP-1 localization length measurements were more variable that the other reporters due to a faint KAP-1::GFP signal, but were not significantly different from wild type in any strain. Localization length was significantly altered in nphp-2 mutants. (E) nphp-2 and arl-13 had significantly longer GT335 signals than in wild type. nphp-2 mutants have increased variability in GT335 signal lengths compared to WT. (F) Diagram representing genetic control of doublet region protein localization in phasmid cilia. An arrow between a gene and a protein/GT335 indicates that the gene is required for the protein/GT335 to label its entire territory. A T-bar between a gene and a protein/GT335 indicates that the gene restricts the size of the protein territory/GT335 signal length. Data was analyzed with an unpaired t-test with Welch's Correction against wild type, followed by the Holm-Bonferroni multiple comparison adjustment for all comparisons in a given panel. **p<0.01.
(TIF)
NPHP-2 contains a strongly conserved hydroxylation motif and a predicted acetylation site. (A) Hydroxylation motif is highlighted in green. Hydroxylated asparagine is highlighted in blue. ANKS6 and INVS alignment are from [24]. (B) NPHP-2 domain model. NPHP-2 contains a predicted acetylation site at 2S, and two predicted SUMOylation sites. Green letters indicate modified residues. Acetylation and SUMOylation motifs do not appear to be conserved.
(TIF)
(A) NPHP-2::GFP localizes to the proximal portion of the cilium in L1 animals. (B) Kymograph of NPHP-2::GFP in phasmid cilia. No movement of NPHP-2::GFP was detectable. Total duration of recording was 40 seconds. (C) NPHP-2::GFP localizes to amphid and phasmid cilia in L1 and L4 stage ccpp-1 mutants. (D) GFP::UNC-119 localizes to amphid and phasmid cilia in L1 and L4 stage ccpp-1 mutants. White bars indicate ciliary GFP::UNC-119 localization. White arrow indicates abnormal cilia base accumulation of GFP::UNC-119.
(TIF)
The anti-acetylated tubulin antibody 6-11b-1 does not label amphid channel and phasmid cilia in either WT animals or hdac-6 mutants. 6-11b-1 antibody labels only dendrites of the touch neurons. hdac-6 deletion does not increase the amount of acetylation detected in touch neurons. Non-specific labelling is seen in the buccal cavity, and not in amphid cilia, as determined by analysis of 3D image stacks.
(TIF)
Transgenic strains used in this study have mild to no ciliogenic defects. Worms from each transgenic strain imaged or otherwise quantified in Fig. 1-9—other than strains with a Dyf or SynDyf background—were assayed for dye-filling defects to determine if there were any gross defects in ciliogenesis or cilia placement. Scores reported are average fraction of cell bodies taking up dye, plus/minus std. err. Yellow cells mark possible dominant negative defects. Transgenic strains expressing NPHP-2::GFP and GFP::UNC-119 did not exhibit defects, but strains expressing ARL-13::GFP all exhibited mild defects.
(XLSX)
List of strains and PCR deletion diagnosis primers used in this work. Strains are organized by the figures in which they appear.
(XLS)
Experimental sample sizes for figures with statistical analysis.
(XLSX)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.