Abstract
Pif1 is a helicase involved in the maintenance of nuclear and mitochondrial genomes in eukaryotes. Here we report a new activity of Saccharomyces cerevisiae Pif1, annealing of complementary DNA strands. We identified preferred substrates for annealing as those that generate a duplex product with a single-stranded overhang relative to a blunt end duplex. Importantly, we show that Pif1 can anneal DNA in the presence of ATP and Mg2+. Pif1-mediated annealing also occurs in the presence of single-stranded DNA binding proteins. Additionally, we show that partial duplex substrates with 3′-single-stranded overhangs such as those generated during double-strand break repair can be annealed by Pif1.
Helicases are defined as a class of proteins that move directionally on nucleic acids (NA) separating the strands of duplex NA using the energy of NTP hydrolysis (reviewed in refs (1−4)). However, an increasing number of helicases have been shown to anneal or rewind complementary strands of oligonucleotides in the presence or absence of ATP.5−11 Annealing helicases are proposed to use their strand annealing activity for stabilization of stalled replication forks, double-strand break (DSB) repair, telomere metabolism, chromatin remodeling, and regulation of transcription (reviewed in ref (12)).
A number of helicases have been shown to exhibit strand annealing activity, including all five members of the human RecQ family (reviewed in ref (13)), Dna2,14 TWINKLE,9 and UvsW.15 HARP and AH2 are the only two DNA helicases that are known to have a requirement for ATP for strand annealing activity.8,16
Saccharomyces cerevisiae Pif1 is known to participate in both mitochondrial and nuclear DNA replication where its translocation on single-stranded DNA (ssDNA) is tightly coupled to ATP hydrolysis.17 Pif1 has been shown to dimerize upon DNA binding.18 However, the monomeric form translocates on ssDNA, unwinds double-stranded DNA (dsDNA), albeit with an efficiency lower than that of a dimer, and has been proposed to be involved in protein displacement.19 Pif1 is involved in several processes that are essential for DNA replication such as Okazaki fragment maturation and replication through G-quadruplex motifs and through replication fork barriers in rDNA (reviewed in ref (20)). Pif1 has been shown to interact with the single-stranded DNA binding protein Rim1, an important component of the yeast mitochondrial DNA replication complex.21 Pif1 is phosphorylated in response to DNA damage,22 which may direct it to stalled replication forks. Overexpression of Pif1 decreases the rate of cell growth and causes increased levels of formation of Mre11 and Rfa1 foci, indicative of DNA damage, in both telomeric and nontelomeric regions.23 Pif1 is involved in break-induced repair (BIR)24−26 as well as recombination-dependent telomere maintenance in the absence of telomerase.27
Strand annealing activity of annealing helicases has been implicated in DNA DSB repair mechanisms.12 There are three main pathways that repair DSBs: classical nonhomologous end joining (cNHEJ), alternative nonhomologous end joining (aNHEJ), which is also called microhomology-mediated end joining (MMEJ), and homologous recombination (HR).28 The cNHEJ pathway involves direct ligation of the broken ends. Both HR and aNHEJ/MMEJ require broken end resection that generates 3′-ssDNA tails, relying on annealing of complementary ssDNA to complete the repair process. Several helicases that are known to participate in DSB repair are also known to possess strand annealing activity.29−34
Some helicases implicated in telomere metabolism are known to possess strand annealing activity,14,35−37 including human Pif1.7,38 It is well established that Pif1 functions as a negative regulator of telomere length.39−42 The single-stranded telomeric overhang may require the strand annealing activity of an annealing helicase to form a more stable structure such as a T-loop.
Importantly, annealing activity exhibited by a helicase must be accounted for to interpret biochemical results. Annealing can compete with DNA unwinding to affect the outcome of DNA unwinding experiments in vitro.43 Therefore, understanding the biochemical mechanism for annealing is necessary for the interpretation of helicase unwinding activity.
Experimental Procedures
Oligonucleotides and Proteins
DNA oligonucleotides were purchased from Integrated DNA Technologies, purified using denaturing polyacrylamide gel electrophoresis,44 quantified by UV absorbance using calculated extinction coefficients,45 and radiolabeled as described previously.46 The nuclear isoform of yeast Pif1 and yeast Rim1 were purified as described previously.21 Yeast RPA was a generous gift from M. Wold. Dextran sulfate (molecular weight of 6500–10000 Da) from Sigma was dialyzed extensively against water to remove sodium.
DNA Unwinding Assay
All concentrations listed are after initiation of the DNA unwinding reaction. Unwinding experiments were performed at 25 °C in a buffer containing 25 mM HEPES (sodium salt) (pH 7.5), 50 mM NaCl, 10 mM MgCl2, 0.1 mM EDTA, 2 mM β-ME, and 0.1 mg/mL BSA. Reaction mixtures contained 2 nM DNA substrate and 5 mM ATP, and reactions were initiated upon addition of 200 nM Pif1 and 60 nM DNA trap complementary to the unlabeled strand. Reactions were quenched with 200 mM EDTA, 0.6% SDS, 0.1% bromophenol blue, 0.1% xylene cyanol, 6% glycerol, and 112 mM T70 (in nucleotides) to sequester proteins after the reaction. The substrate and ssDNA product were resolved on a 20% native polyacrylamide gel, detected using a PhosphorImager, and quantified using ImageQuant. The amount of product formed over time was plotted and fit to a single exponential.
Strand Annealing Assay
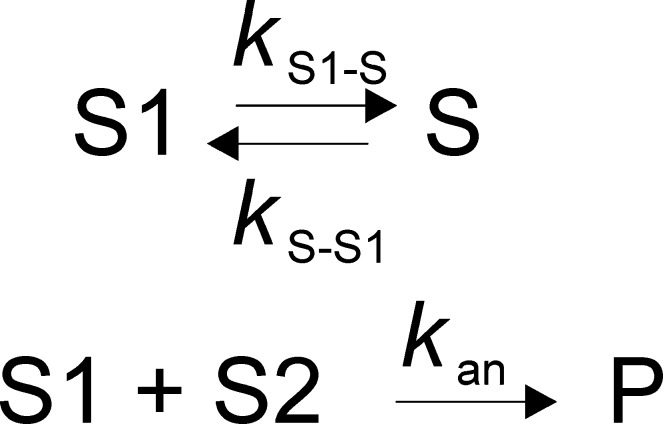
Pairs of complementary ssDNA substrates that were used for annealing assays are listed in Table 1. The annealing experiments were performed at 25 °C in a reaction buffer containing 25 mM HEPES (sodium salt) (pH 7.5), 50 mM NaCl, 0.1 mM EDTA, 2 mM β-ME, and 0.1 mg/mL BSA. All concentrations listed are final, after initiation of the reaction. The two complementary strands with one of them radiolabeled were mixed with 200 nM Pif1 (unless otherwise indicated in the figure legend) to initiate the reaction. For some experiments, ATP and MgCl2 were added with the Pif1. The ratio of radiolabeled ssDNA (2 nM) to unlabeled complementary ssDNA (2.6 nM) used for the assay was 1:1.3. Aliquots of the reaction were quenched at the indicated times with 200 mM EDTA, 1% SDS, 100 nM DNA trap, and 5 mg/mL dextran sulfate, which serves as a protein trap because of its polyanionic nature. Samples were immediately loaded on a native 20% polyacrylamide gel. The amounts of annealed DNA and ssDNA were quantified using a PhosphorImager and ImageQuant. Spontaneous annealing was determined by excluding the enzyme in the assay mixture. Data were fit to the simplest annealing mechanism that could account for the data (Schemes 1–3) using KinTek Explorer,47 where S1 and S2 are the two complementary strands of ssDNA and P is the duplex DNA product. In the absence of ATP, reactions with some substrates did not go to completion, suggesting an equilibrium between different substrate conformations as shown in Scheme 1. For reactions that included ATP, a simple annealing mechanism (Scheme 2) was used to fit the data. For reactions in which the product contained a 5′-ssDNA overhang and ATP was present, the reverse reaction (unwinding) was also included (Scheme 3). The rate constant kS1–S is associated with conversion of substrate (S1) to a conformation that is not capable of annealing, and kS–S1 is the rate constant for the conversion of S to S1. kS–S1 was not well-defined but does not affect the rate constant for annealing. The second-order rate constant for annealing is kan, and the rate constant for unwinding is kun.
Table 1. Oligonucleotide Sequences.
| product | type | substrate | sequence |
|---|---|---|---|
| 70T-30bp | 5′-overhang | 70T-30nt | 5′-(T)70CTGCTGCCATGTCACGCTGATGTCGCCTGT-3′ |
| 30nt CS | 5′-ACAGGCGACATCAGCGTGACATGGCAGCAG-3′ | ||
| 30bp | blunt end | 30nt | 5′-CTGCTGCCATGTCACGCTGATGTCGCCTGT-3′ |
| 30nt CS | 5′-ACAGGCGACATCAGCGTGACATGGCAGCAG-3′ | ||
| 20T-30bp | 5′-overhang | 20T-30nt | 5′-(T)20CTGCTGCCATGTCACGCTGATGTCGCCTGT-3′ |
| 30nt CS | 5′-ACAGGCGACATCAGCGTGACATGGCAGCAG-3′ | ||
| 20T-30bp-20T | fork | 20T-30nt | 5′-(T)20CTGCTGCCATGTCACGCTGATGTCGCCTGT-3′ |
| 30nt CS-20T | 5′-ACAGGCGACATCAGCGTGACATGGCAGCAG(T)20-3′ | ||
| 20T-20T-30bp | dual 5′-overhang | 20T-30nt | 5′-(T)20CTGCTGCCATGTCACGCTGATGTCGCCTGT-3′ |
| 20T-30nt CS | 5′-(T)20ACAGGCGACATCAGCGTGACATGGCAGCAG-3′ | ||
| 30bp-20T | 3′-overhang | 30nt | 5′-CTGCTGCCATGTCACGCTGATGTCGCCTGT-3′ |
| 30nt CS-20T | 5′-ACAGGCGACATCAGCGTGACATGGCAGCAG(T)20-3′ | ||
| 80bp blunt | blunt end | 80nt | 5′-GACGGGGAAAGCCGGCGAACGTGGCGAGAAAGGAAGGGAAGAAAGCGAAAGGAGCGGGCGCTAGGGCGCTGGCAAGTGTA-3′ |
| 80nt CS | 5′-ACACTTGCCAGCGCCCTAGCGCCCGCTCCTTTCGCTTTCTTCCCTTCCTTTCTCGCCACGTTCGCCGGCTTTCCCCGTC-3′ | ||
| 90bp blunt | blunt end | 30bp-30nt | 5′-GACCGTGGCGAGACAGGAAGTGAAGAGAGCGACAGGAGCGTGCCGTAGTGCGCTGGCAGC-3′a |
| 5′-GCTCTCTTCACTTCCTGTCTCGCCACGGTC-3′a | |||
| 30nt-30bp | 5′-GACCGTGGCGAGACAGGAAGTGAAGAGAGCGCTGCCAGCGCACTACGGCACGCTCCTGTC-3′a | ||
| 5′-GCTCTCTTCACTTCCTGTCTCGCCACGGTC-3′a | |||
| 90bp blunt-mut | 30bp-30nt | 5′-GACCGTGGCGAGACAGGAAGTGAAGAGAGCGACAGGAGCGTGCCGTAGTGCGCTGGCAGC-3′a | |
| 5′-GCTCTCTTCACTTCCTGTCTCGCCACGGTC-3′a | |||
| 30nt-30bp-mut | 5′-GACCGTGGCGAGACAGGAAGTGAAGAGAGCGCTGCCAGCGCACATGCGCACGCTCCTGTC-3′a | ||
| 5′-GCTCTCTTCACTTCCTGTCTCGCCACGGTC-3′a |
Underlined regions are duplex in the substrate.
Scheme 1. Annealing Mechanism in the Absence of ATP.
Scheme 3. Annealing Mechanism in the Presence of ATP for Reactions in Which the Product Contains a 5′-ssDNA Overhang.
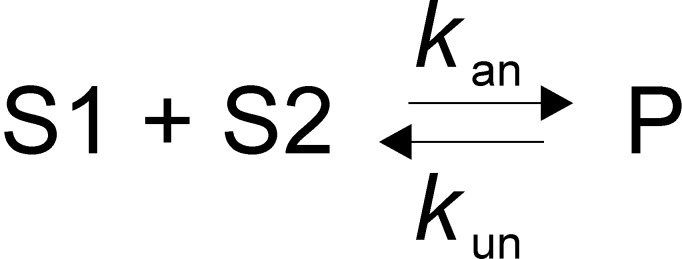
Scheme 2. Annealing Mechanism in the Presence of ATP for Reactions in Which the Annealed Product Is Not an Unwinding Substrate for Pif1.
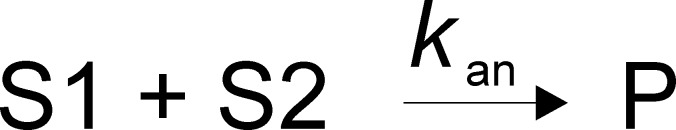
Förster Resonance Energy Transfer (FRET) Assay for Binding
5′-Cy3-labeled and 3′-Cy5-labeled, noncomplementary, eight-nucleotide oligonucleotides were incubated in buffer [25 mM HEPES (sodium salt) (pH 7.5), 50 mM NaCl, 0.1 mM EDTA, 2 mM β-ME, and 0.1 mg/mL BSA] at a final concentration of 100 nM each in a fluorometer maintained at 25 °C with a circulating water bath. Samples were excited at 550 ± 2 nm, and emission was monitored at 668 ± 4 nm. The oligonucleotide sequences were 5′-GTCACACT-Cy5-3′ and 5′-Cy3-AGCATCAG-3′. Pif1 was titrated into the solution containing one or both oligonucleotides. The increase in fluorescence corresponds to the increase in the level of FRET due to addition of Pif1. The change in fluorescence emission of samples titrated with Pif1 was corrected for sample dilution by subtracting the change in the fluorescence emission of oligonucleotide samples titrated with equal volumes of protein sample buffer.
Results
Removing the DNA Trap from an Unwinding Reaction Mixture Results in a Reduced Level of Product Formation
A likely role for Pif1 in promoting strand annealing activity was identified when an unwinding experiment showed a disparity in product formation with or without a DNA trap (Figure 1). Pif1-catalyzed unwinding experiments were performed in the presence or absence of a DNA trap that captures the unlabeled displaced strand, leaving the unwound radiolabeled strand as the ssDNA product. Less ssDNA was observed from Pif1-catalyzed unwinding of the 70T-30bp substrate when the DNA trap was not included (Figure 1b). In the absence of a DNA trap, the amount of ssDNA generated reached a plateau at ∼50%, suggesting that the unwound products were reannealed and an equilibrium had been reached between unwinding and reannealing activities, as has been shown previously for WRN and BLM helicases.48 These results suggest that Pif1 might promote strand annealing.
Figure 1.
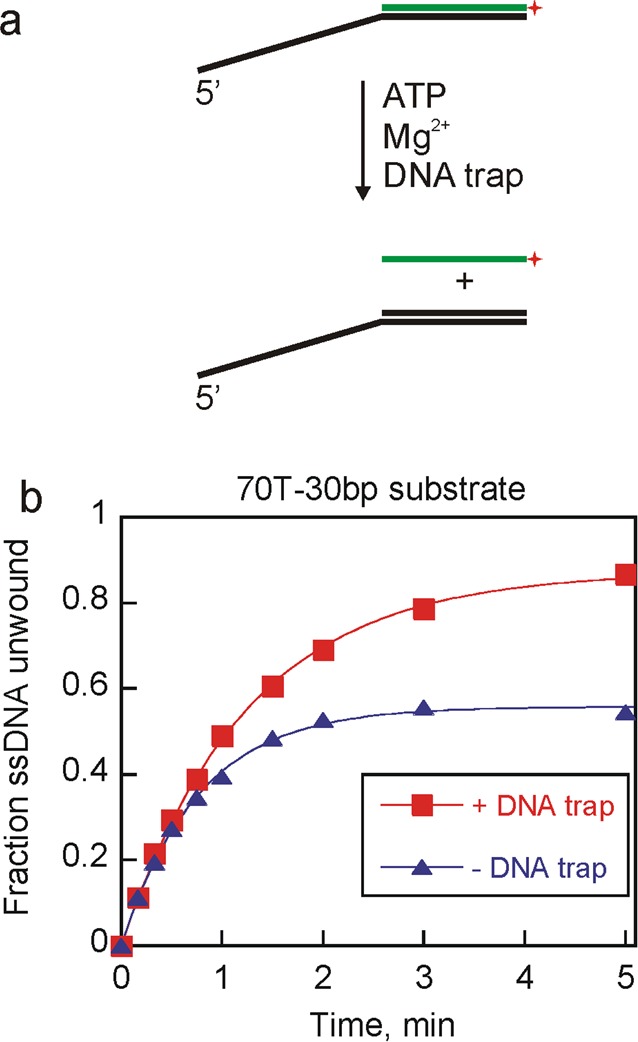
Pif1-catalyzed unwinding yielded differential product appearance in the presence or absence of DNA trap. (a) Duplex DNA substrate was mixed with Pif1 in the presence of ATP and MgCl2 to form ssDNA product. A DNA trap complementary to the unlabeled strand was included in some reactions. (b) Unwinding experiments were initiated by mixing 2 nM 70T-30bp substrate, ATP, and MgCl2 with 200 nM Pif1. Reactions were performed either in the absence (triangles) or in the presence (squares) of unlabeled 60 nM DNA trap. Fitting the data to the equation for a single exponential resulted in unwinding rate constants of 0.80 ± 0.02 and 1.3 ± 0.04 min–1 for unwinding in the presence and absence of DNA trap, respectively. The amplitudes of the product formation curves are 0.87 ± 0.01 and 0.56 ± 0.01 for unwinding in the presence and absence of a DNA trap, respectively.
Pif1 Promotes Strand Annealing
Pif1’s ability to anneal complementary strands was initially investigated in the absence of ATP and Mg2+ to eliminate the effects of unwinding. A pair of complementary ssDNA substrates that would generate a partial duplex product were used to test Pif1’s ability to anneal (Figure 2a). The annealed products were separated from ssDNA on a native polyacrylamide gel (Figure 2b). Annealing of 70T-30nt with a 30nt complementary strand generates a 70T-30bp product (Table 1). Approximately 80% of the 70T-30bp product was formed in <2 min, whereas no product formation was observed in the absence of the enzyme during the time period of the reaction (Figure 2c), indicating Pif1 can promote annealing of complementary strands.
Figure 2.
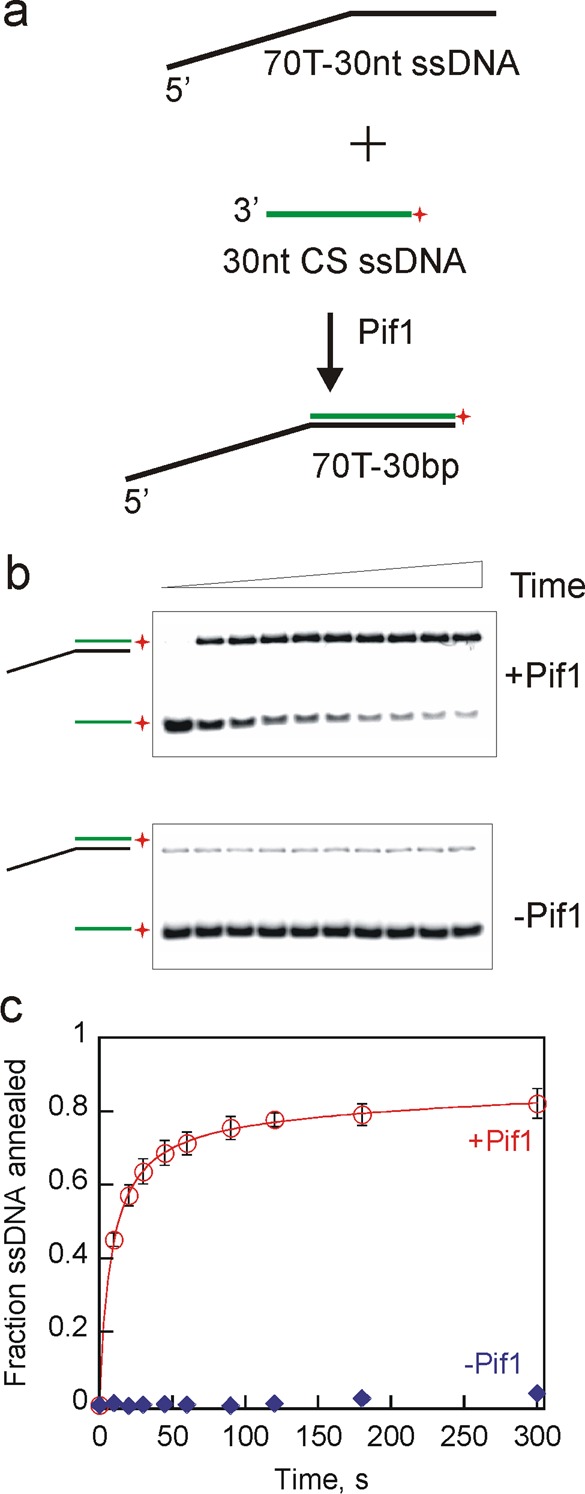
Pif1 exhibited robust strand annealing activity in the absence of ATP and MgCl2. (a) Schematic illustration for two partially complementary ssDNAs (70T-30nt and 30nt CS) annealing to generate a partial duplex DNA, 70T-30bp. Pif1 (200 nM) was incubated with 70T-30nt (2.6 nM) and 30nt CS (2 nM) for increasing times, and the reactions were stopped by mixing with a quench solution containing 200 mM EDTA, 1% SDS, 100 nM DNA trap, and 5 mg/mL dextran sulfate. (b) Representative gel images of annealed products formed in the presence of Pif1 (top) and in the absence of Pif1 (bottom) as a function of time. (c) Fraction of ssDNA annealed in the presence of Pif1 (circles). Spontaneous annealing was measured in the absence of Pif1 (diamonds). Error bars represent the standard deviation of three independent experiments.
Pif1 Can Bind Multiple Oligonucleotides Simultaneously
A FRET binding experiment was designed to investigate the ability of Pif1 to simultaneously bind multiple DNA strands (Figure 3a). Like other superfamily 1 helicases, Pif1 sequesters approximately eight nucleotides on the basis of a repeating pattern of nucleotide protection of approximately eight nucleotides in the presence of Pif1 in KMnO4 footprinting experiments (Figure S1 of the Supporting Information), so the ability of Pif1 to concurrently bind multiple oligonucleotides was investigated using Cy3- and Cy5-labeled noncomplementary 8-mers because these should bind to Pif1 with a 1:1 stoichiometry when considering only the helicase binding site. Upon titration of a mixture of the Cy3- and Cy5-labeled oligonucleotides with Pif1, FRET should be observed if multiple oligonucleotides bind to a single Pif1 molecule or to adjacent Pif1 molecules in a Pif1 oligomer (Figure 3a). Pif1 has been reported to dimerize in the presence of DNA.18 If each Pif1 monomer in the dimer binds a separate oligonucleotide, the FRET probes should be in the proximity. Alternatively, sites on the surface of Pif1 could interact with additional DNA molecules to juxtapose the fluorophores. We find that Pif1 is able to bind multiple oligonucleotides simultaneously, resulting in an increased FRET efficiency (Figure 3b), similar to that observed previously for hepatitis C virus NS349 and human Rad52.50 A similar increase in the FRET efficiency was observed when the concentration of DNA (2 nM) was the same as that used in the annealing experiments (Figure S2 of the Supporting Information). This activity provides a mechanism by which Pif1 can increase the local concentration of two DNA strands, which should facilitate annealing.
Figure 3.
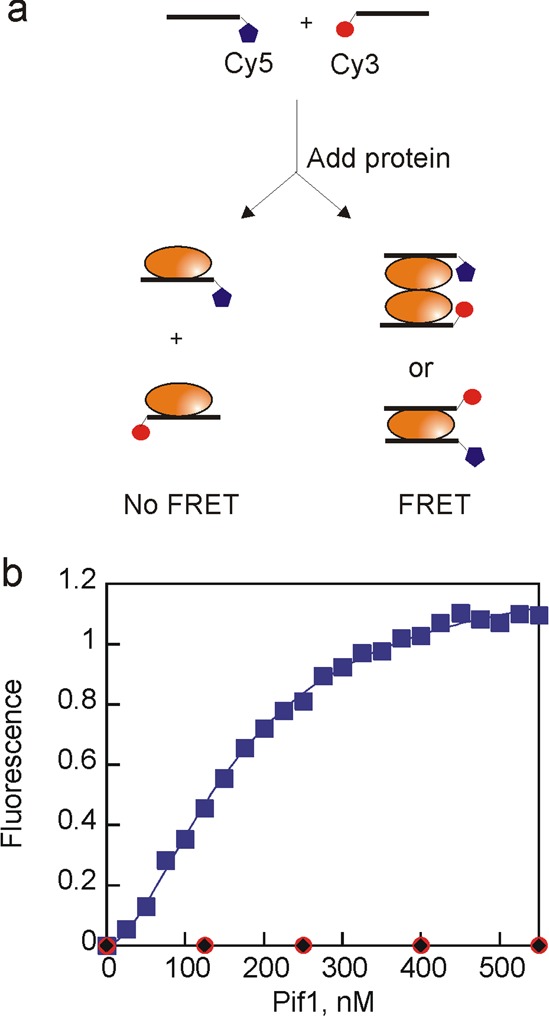
Pif1 can bind multiple strands of DNA. (a) Schematic illustration of fluorescence titration. Two noncomplementary 8-mer oligonucleotides (100 nM) were mixed and then titrated with Pif1. Fluorescence emission was measured at 668 nm with excitation at 550 nm. (b) Increase in Cy5 fluorescence emission as a function of Pif1 concentration (blue). Data were fit to the Hill equation to obtain the concentration of protein at which half of the maximal FRET change is observed, 200 nM Pif1, and the Hill coefficient of 2.1. Duplicate experiments produced similar results. No increase in fluorescence was observed in control reactions in which Pif1 was added to only the Cy3-labeled oligonucleotide (red) or only the Cy5-labeled oligonucleotide (black).
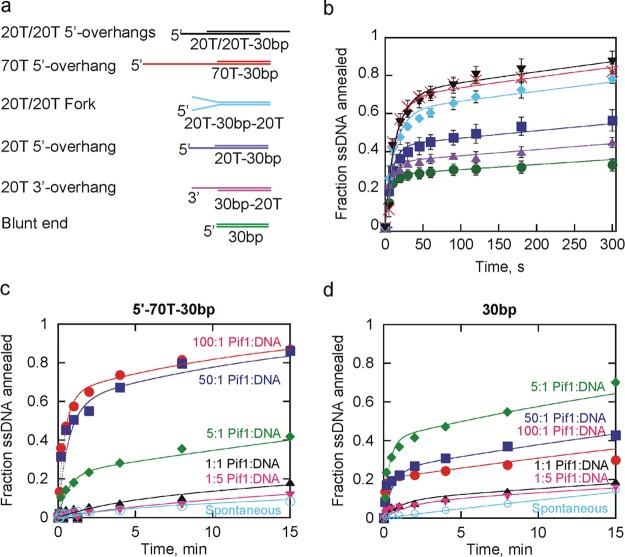
Substrate Preference for Annealing by Pif1
The effect of changes in the length and type of ssDNA overhangs, such as a short overhang versus a long overhang, a 3′-overhang versus a 5′-overhang, and one versus two overhangs, on Pif1-promoted strand annealing was investigated. Several substrates (Table 1) that would generate a 30bp duplex product (Figure 4a) with varying length overhangs were tested in the absence of ATP and Mg2+ to eliminate complicating effects of unwinding (Figure 4b). Data were fit to the annealing mechanism in Scheme 1. The constants obtained from the fit are listed in Table 2. The second-order rate constant for annealing is similar for each of the substrate pairs. What varies between the substrates is the fraction of substrate that is available for annealing, indicated by the rate constant for conversion of S1 to S, where S is substrate that is in a conformation that is not amenable to annealing. (A similar process could occur with S2. However, we included this only for one of the substrate strands to limit the number of variables in the fit and use the simplest mechanism possible to fit the data.) S could be some conformation in which the bases are not fully exposed because of short regions of base pairing within the substrate or the potential sequestration of bases by an excess of Pif1 bound to the substrate. A modest difference in the fraction of ssDNA annealed is observed for the products containing a 5′-overhang or 3′-overhang, with the 5′-overhang being the preferred product. In general, longer substrates are preferred (Figure 4b), but the length is not the only factor that influences annealing. Two different products, each containing two 20nt ssDNA overhangs, were compared. The forked duplex (cyan diamonds) has a ssDNA overhang on the 5′-end of one strand and the 3′-end of the other. The dual 5′-overhang product has 20nt ssDNA overhangs on each 5′-end (black triangles). The dual 5′-overhang product is formed preferentially over the forked product even though the total length of the substrates and overhangs is equivalent, again indicating that 5′-overhangs are preferred. The dual-5′-overhang product (two 20T overhangs) is annealed similarly to a product with a single 70T 5′-overhang even though the total length is reduced, indicating that the location of overhangs is important, in addition to total length. Although the ratio of Pif1 to DNA strands is the same for each of the substrate pairs utilized, the ratio of Pif1 to binding sites varies significantly between the substrate pairs due to the variation in substrate length. We note that the substrate preference reported at saturating enzyme concentrations may not be applicable when the enzyme concentration is not saturating because of the difference in the number of binding sites.
Figure 4.
Effect of varying ssDNA overhangs and Pif1 concentrations on annealing. (a) DNA substrates that can anneal to form a 30bp duplex are shown with the varying 5′- and 3′-overhangs indicated. The substrates are named according to the length and type (5′ or 3′) of ssDNA overhang and the length of duplex. They are shown in order from the most efficiently to least efficiently annealed. (b) Plot of the fraction of ssDNA annealed for the 30bp blunt end duplex (●), 20T 5′-overhang (■), 20T/20T fork (◆), 20T 3′-overhang (▲), 70T 5′-overhang (×), and 20T/20T dual 5′-overhangs (▼). Error bars represent the standard deviation of three independent experiments. Data were fit to Scheme 1, and the constants from the fits are listed in Table 2. (c) Annealing of the 70T 5′-overhang product measured at 200, 100, 10, 2, 0.4, and 0 nM Pif1, keeping the concentrations of the substrate strands constant at 2 nM radiolabeled 30nt CS and 2.6 nM 70T-30nt. Duplicate experiments produced similar results. Data were fit to Scheme 1 to obtain the second-order rate constant for annealing and the rate constant for conversion of S1 to S (1.9 × 107 M–1 s–1 and 0.022 s–1 for a 100:1 Pif1:DNA ratio, 1.9 × 107 M–1 s–1 and 0.037 s–1 for a 50:1 Pif1:DNA ratio, and 4.4 × 106 M–1 s–1 and 0.030 s–1 for a 5:1 Pif1:DNA ratio, respectively). Data obtained at 1:1 and 1:5 Pif1:DNA ratios and in the absence of Pif1 were not fit because of the small quantities of product formed. (d) Annealing of the 30bp blunt end duplex under the same conditions described for panel c. Duplicate experiments produced similar results. Data were fit to Scheme 1 to obtain the second-order rate constant for annealing and the rate constant for conversion of S1 to S (1.5 × 107 M–1 s–1 and 0.14 s–1 for a 100:1 Pif1:DNA ratio, 1.5 × 107 M–1 s–1 and 0.14 s–1 for a 50:1 Pif1:DNA ratio, and 1.1 × 107 M–1 s–1 and 0.078 s–1 for a 5:1 Pif1:DNA ratio, respectively). Data obtained at 1:1 and 1:5 Pif1:DNA ratios and in the absence of Pif1 were not fit because of the small quantities of product formed.
Table 2. Annealing Rate Constants.
| product | type | kan (M–1 s–1) | kS1–S (s–1) |
|---|---|---|---|
| 70T-30bp | 70T 5′-overhang | (2.3 ± 0.2) × 107 | 0.013 ± 0.003 |
| 20T-20T-30bp | dual 20T/20T 5′-overhang | (2.3 ± 0.2) × 107 | 0.020 ± 0.009 |
| 20T-30bp-20T | fork | (3.6 ± 1.1) × 107 | 0.037 ± 0.012 |
| 20T-30bp | 20T 5′-overhang | (2.1 ± 0.2) × 107 | 0.064 ± 0.013 |
| 30bp-20T | 20T 3′-overhang | (3.0 ± 0.1) × 107 | 0.13 ± 0.01 |
| 30bp | blunt end | (2.0 ± 0.3) × 107 | 0.13 ± 0.03 |
Effect of Enzyme Concentration on Annealing
Annealing experiments, thus far, included a large excess of Pif1 relative to DNA, similar to that present in the unwinding reaction mixtures shown in Figure 1 that initially indicated that annealing might be occurring. If Pif1 oligomerization is involved in annealing, then annealing activity should decrease if the Pif1:DNA ratio is decreased. Panels c and d of Figure 4 show the results of varying the quantity of Pif1 relative to the ssDNA substrates. The level of product formation (70T-30bp) increases dramatically when the Pif1 concentration is increased to a level 50–100-fold greater than the DNA concentration (Figure 4c). When the Pif1:DNA ratio is varied with the 30bp blunt end duplex (Figure 4d), the level of product formation is again low when Pif1 and the DNA are present at similar concentrations. However, more product was observed when only a small excess of Pif1 (5:1) was present than when a large excess of Pif1 was present (100:1). This suggests that when excess Pif1 is present with ssDNA substrates that can anneal to form a blunt end product, the excess enzyme may sequester the bases and actually slow annealing relative to a small excess of enzyme, although annealing by the presence of a large excess of Pif1 is still enhanced relative to spontaneous annealing. These results suggest that sequestration of the duplex-forming region by Pif1 can have two effects. Pif1 binding can prevent annealing due to sequestration of the DNA. This is illustrated by the results in Figure 4d that show that more blunt end duplex product is formed at a 5:1 Pif1:DNA ratio than at a 50:1 or 100:1 ratio. Second, binding of enzyme to the duplex-forming region can perpetuate annealing because of the enzyme’s ability to promote annealing. When the product has a ssDNA overhang, increasing the Pif1 concentration increases the level of annealing (Figure 4c), likely because of the increased number of binding sites on the substrates requiring a higher Pif1:DNA ratio to result in sufficient Pif1 binding in or near the complementary regions of the substrates. However, when the product is a blunt end duplex, that ratio is more complicated because all binding events sequester annealing sites (Figure 4d). Binding events on substrates that will anneal to form products with overhangs sequester regions that are not necessarily annealing sites, whereas all binding events on substrates that will anneal to form blunt end duplexes sequester annealing sites. Because of the difference in the number of binding sites for the enzyme on the two sets of substrate pairs, it is difficult to compare the rates and quantities of product formed at the various Pif1:DNA ratios because the quantity of Pif1 required to saturate the sets of substrate pairs is different.
Effect of ATP and Mg2+ on Annealing by Pif1
Helicases hydrolyze ATP, so the strand annealing activity was examined under conditions in which strand separation can also occur. Annealing by Pif1 was tested in the presence or absence of ATP in the presence of MgCl2 using the 70T-30bp product (Figure 5). After the initial rapid phase, the level of product formation reached a plateau, indicating that unwinding and annealing were at equilibrium. This is similar to previous observations with Dda helicase in which the annealing product serves as a substrate for strand separation.43 Data collected in the presence of ATP and Pif1 were fit to Scheme 3 to obtain a second-order rate constant of (5.1 ± 0.3) × 106 M–1 s–1 for annealing and a rate constant of 0.021 ± 0.004 s–1 for unwinding. Data collected in the absence of ATP but in the presence of Pif1 were fit to Scheme 1, and a second-order rate constant of (1.3 ± 0.2) × 107 M–1 s–1 for annealing was obtained. In the absence of Pif1, data were fit to Scheme 2, and the second-order rate constants for annealing were (1.7 ± 0.9) × 105 and (5.8 ± 0.3) × 105 M–1 s–1 in the presence and absence of ATP, respectively. The reduced spontaneous annealing rate in the presence of ATP is likely due to sequestration of Mg2+ by ATP, thereby weakening the ability of Mg2+ to stabilize the duplex. Notably, Pif1 enhances the spontaneous annealing rate 20-fold in the absence of ATP and 30-fold in the presence of ATP.
Figure 5.
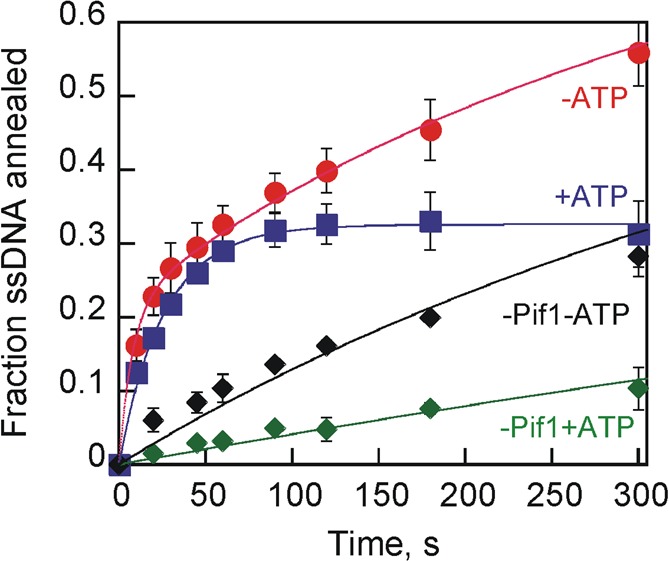
Effect of ATP and Mg2+ on Pif1 strand annealing activity. Shown are the kinetic plots observed for Pif1 (200 nM) catalyzed annealing of 70T-30nt (2 nM) and 30nt CS (2.6 nM) to generate 70T-30bp in the presence (blue) or absence (red) of 5 mM ATP. Control reaction mixtures in the presence (green) or absence (black) of ATP lacked Pif1. All reaction mixtures contained 10 mM MgCl2. Data collected in the presence of ATP and Pif1 were fit to Scheme 3 to obtain a second-order rate constant of (5.1 ± 0.3) × 106 M–1 s–1 for annealing and a rate constant of 0.021 ± 0.004 s–1 for unwinding. Data collected in the absence of ATP but in the presence of Pif1 were fit to Scheme 1, and a second-order rate constant of (1.3 ± 0.2) × 107 M–1 s–1 for annealing and a rate constant of 0.094 ± 0.004 s–1 for conversion between S1 and S were obtained. In the absence of Pif1, data were fit to Scheme 2, and the second-order rate constants for annealing were (1.7 ± 0.9) × 105 and (5.8 ± 0.3) × 105 M–1 s–1 in the presence and absence of ATP, respectively.
Effect of Duplex Length on Annealing by Pif1
Pif1-promoted strand annealing experiments in the presence of ATP and MgCl2 using 70T-30bp (Figure 5) product showed that the unwinding and annealing activities of Pif1 reached equilibrium. It is well-established that Pif1 has relatively low processivity for unwinding,17,19 suggesting that an increase in duplex length should shift the equilibrium toward annealing. In addition, substrates that would generate an annealed product that is a blunt end duplex or has a 3′-ssDNA overhang should produce more product in an annealing reaction because the product should not be unwound by the 5′-to-3′ helicase activity of Pif1. To test this, two complementary ssDNAs that can generate an 80bp blunt DNA product were used (Figure 6a). Pif1 strand annealing activity in the presence of ATP and MgCl2 led to nearly complete annealing of the substrates (Figure 6b) relative to annealing of substrates that produce a product that could be readily unwound (Figure 5). The rate constant for annealing based on a fit of the data to a simple annealing mechanism (Scheme 2) is 3.0 × 106 M–1 s–1. When ATP was absent, the second-order rate constant for annealing was 2.9 × 107 M–1 s–1. Faster annealing in the absence of ATP suggests that even though the final product of the annealing reaction is not a substrate for Pif1 unwinding, a partially annealed intermediate could be a substrate for unwinding by Pif1. Spontaneous strand annealing was observed for formation of the 80bp blunt DNA product, with a rate constant of 3.0 × 105 M–1 s–1. This is similar to previous DNA hybridization rate measurements of 2.0 × 105 to 1.2 × 106 M–1 s–1, depending on the sequence.51 However, annealing by Pif1 was 10-fold faster than spontaneous annealing.
Figure 6.
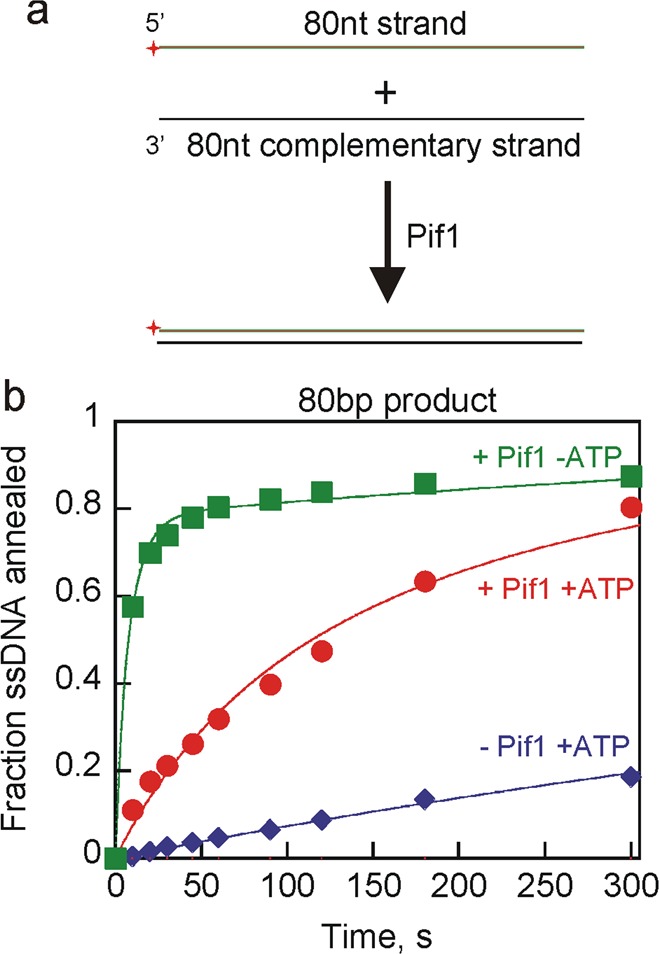
Pif1-catalyzed strand annealing using longer DNA substrates. (a) Schematic illustration for strand annealing experiments using 80nt strand and 80nt CS to generate an 80bp blunt end dsDNA product. (b) Results for Pif1 (200 nM) annealing of the 80nt strand (2.6 nM) with radiolabeled 80nt CS (2 nM) to generate an 80bp blunt DNA product in the presence of ATP and MgCl2 (red circles). Annealing by Pif1 in the absence of ATP but in the presence of MgCl2 is shown as green squares. Spontaneous annealing in the absence of enzyme in the presence of ATP and MgCl2 was also measured (blue diamonds). Data were fit to a simple annealing mechanism (Scheme 2) to obtain second-order rate constants for annealing of 3.0 × 106 M–1 s–1 in the presence of Pif1 and ATP, 2.9 × 107 M–1 s–1 in the presence of Pif1 but in the absence of ATP, and 3.0 × 105 M–1 s–1 in the absence of Pif1.
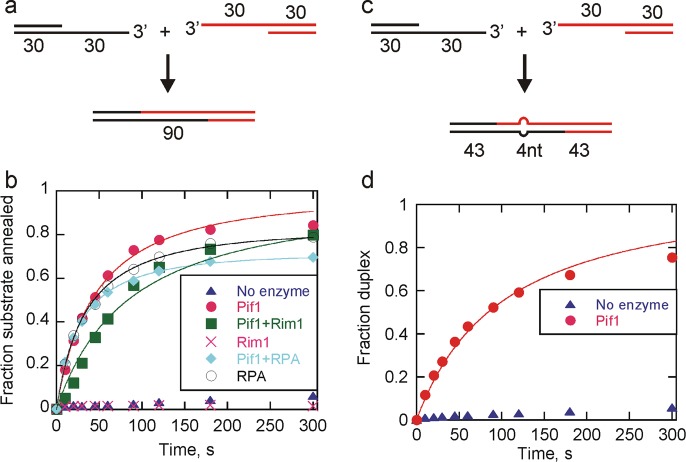
Pif1 Promotes Annealing of Partial Duplexes with 3′-Overhangs
Repair of double-strand breaks in cells can occur by various pathways, most of which involve resection by a nuclease, resulting in complementary 3′-overhangs. Two partial duplex substrates (each containing 30bp and a 30nt 3′-ssDNA overhang) (Figure 7a) were used to investigate the ability of Pif1 to catalyze annealing of such substrates. These substrates are similar to those that could occur during double-strand break repair by the aNHEJ or MMEJ pathway, although sequences involved in aNHEJ often contain regions of noncomplementarity within the region to be annealed. In the absence of enzyme, very little annealing was observed; however, in the presence of Pif1, annealing was much more rapid (Figure 7b). During double-strand break repair, after end resection, the resulting ssDNA is thought to be coated by single-stranded DNA binding proteins. Because Pif1 is involved in genome maintenance in both the yeast nucleus and mitochondria, the ability of Pif1 to anneal in the presence of yeast RPA, the nuclear single-stranded DNA binding protein, and Rim1, the yeast mitochondrial single-stranded DNA binding protein, was investigated (Figure 7b). Annealing was similar in the presence and absence of single-stranded DNA binding proteins with rate constants of 9.2 × 106, 1.5 × 107, and 4.8 × 106 M–1 s–1 for annealing by Pif1 alone, Pif1 and RPA, and Pif1 in the presence of Rim1, respectively. No annealing was observed by Rim1, but RPA alone annealed like Pif1 alone with a rate constant of 1.2 × 107 M–1 s–1. Spontaneous annealing was negligible within this time frame. The ability of Pif1 to anneal substrates with 3′-overhangs in the presence of single-stranded DNA binding proteins suggests that Pif1 annealing activity could function in processes such as double-strand break repair in addition to its known role in the regulation of telomerase at DSBs39−42 and its recent description in BIR.24−26
Figure 7.
Pif1-catalyzed annealing of partial duplex substrates with 3′-overhangs. (a) Schematic illustration for strand annealing experiments using two substrates with 30bp duplexes and complementary 30nt overhangs to generate a 90bp blunt end dsDNA product. (b) Shown are results for Pif1 (200 nM) annealing of 30nt-30bp (2.6 nM) and radiolabeled 30bp-30nt (2 nM) to generate a 90bp blunt end DNA product in the presence of ATP and MgCl2 (circles). Pif1 annealing in the presence of 200 nM Rim1 (squares) and 200 nM RPA (diamonds) was also measured. Annealing activity of RPA alone is shown as empty circles, Rim1 alone is shown as X. Spontaneous annealing in the absence of enzyme was also measured (triangles). Data were fit to a simple annealing mechanism (Scheme 2) to obtain second-order rate constants for annealing by Pif1 (9.2 × 106 M–1 s–1), Pif1 and RPA (1.5 × 107 M–1 s–1), Pif1 and Rim1 (4.8 × 106 M–1 s–1), and RPA (1.2 × 107 M–1 s–1). (c) Schematic illustration for strand annealing experiments using two substrates with 30bp duplexes and 30nt overhangs containing 4nt of mismatch in the middle of otherwise complementary sequences to generate a 90bp blunt end dsDNA product containing a 4nt bubble. (d) Results for Pif1 (200 nM)-catalyzed annealing of 30nt-30bp-mut (2.6 nM) and radiolabeled 30bp-30nt (2 nM) to generate a 90bp blunt end DNA product containing a 4nt bubble in the presence of ATP and MgCl2 (circles). Spontaneous annealing in the absence of enzyme was also measured (triangles). Data were fit to a simple annealing mechanism (Scheme 2) to obtain a second-order rate constant for annealing by Pif1 (4.1 × 106 M–1 s–1).
Human RPA has been shown to both inhibit and stimulate spontaneous strand annealing depending on the substrate type.52 Yeast RPA also appears to stimulate annealing of some substrates (Figure 7b), but with other substrates and conditions, annealing is inhibited by RPA.53,54 RPA has been previously shown to have varying effects on annealing depending on the specific substrate. Annealing by Rad52 is inhibited by yeast RPA on unstructured DNA substrates, but RPA stimulated Rad52 annealing of denatured plasmid DNA, suggesting that RPA may eliminate secondary structures in DNA to enhance annealing by Rad52.53 The differential effect of RPA on the annealing activity of different substrates may be related to the ability of RPA to remove secondary structure. It appears that yeast RPA, like human RPA, can either inhibit or stimulate annealing of complementary DNA, depending on the specific sequence and conditions.
Repair by the aNHEJ pathway often involves annealing of two partially complementary ssDNAs. Therefore, annealing of 3′-overhang partial duplex substrates with a 4nt mismatch in the middle of the otherwise homologous overhangs (Figure 7c) was investigated (Figure 7d). These substrates may approximate those that are annealed during repair by the aNHEJ pathway, although the length of the complementary regions and the degree of mismatch could vary from those used here. Annealing of substrates with a mismatch in the complementary overhang by Pif1 occurred at a rate of 4.1 × 106 M–1 s–1, which is similar to the rate constant for annealing fully complementary sequences of similar length (Figure 7b), suggesting a possible role for Pif1 in repair of sequences with partial homology.
Discussion
Pif1 showed robust annealing of 30nt DNAs in the absence of ATP and MgCl2 (Figures 2 and 4). There are many factors that influence the quantity of product formed in an annealing reaction. First, the position of the ssDNA tail on the product affects annealing with overhangs on the 5′-end preferred (Figure 4b). Second, the ratio of Pif1 to binding sites on the DNA influences annealing as seen by the inhibition of annealing of the 30bp duplex at very high Pif1:DNA ratios (Figure 4d). The rate and amplitude of product formed vary depending on the ratio of enzyme to binding sites. Finally, the inherent ability of the enzyme to melt the dsDNA product influences the quantity of annealed product formed (Figure 5). In the presence of ATP and MgCl2 (Figure 5), the reaction reached an equilibrium between unwinding and annealing. Pif1’s strand annealing activity in the presence of ATP and MgCl2 was more efficient with longer duplex substrates (Figure 6). This is likely because of the decreased level of unwinding due to the nonprocessive nature of Pif117,55 resulting in the equilibrium between unwinding and annealing being shifted further toward annealing. These results indicate that the antagonizing functions of Pif1 could be regulated by the type of substrate it processes.
Human Pif1 has been shown to have annealing activity.7,38 It is possible that many Pif1 family members might possess strand annealing functions in a manner similar to that of the RecQ family helicases (reviewed in ref (13)). Members of the Pif1 and RecQ families are known to participate in some of the same biological processes, such as telomere regulation, Okazaki fragment maturation, homologous recombination, G-quadruplex processing, and DNA repair.34,56 It is possible that coordinated unwinding and annealing activities of these two helicase families are required to achieve some of their biological functions.
The mechanism of strand annealing by helicases and its biological consequences remain largely unknown. An important question is how two competing activities of the same helicase are regulated. A simple view is that an annealing helicase has to perform either unwinding or annealing depending on the specific biological function and the type of substrate encountered; it could be regulated by the microenvironment at the site of action. Alternatively, a coordinated action of unwinding and annealing activities may be required to achieve the necessary biological function. Several possible mechanisms have been proposed to regulate helicase annealing activity, such as the presence of an annealing domain, protein oligomerization, ATP hydrolysis as a switch between unwinding and annealing, interactions of helicases with other proteins, and post-translational modification of the helicase.12 Pif1 is known to be phosphorylated in response to DNA damage,22 and the effect of this modification on the equilibrium between unwinding and annealing is unknown.
Because Pif1 binds to ssDNA, there are two types of substrates on which Pif1 might act: either 5′-ssDNA tails or 3′-ssDNA tails. Pif1 is a 5′-to-3′ helicase, so its helicase activity and annealing activity are expected to be in competition on substrates with 5′-ssDNA tails. On substrates with 3′-ssDNA tails, translocation could displace bound proteins57,58 and facilitate annealing of the resulting ssDNA. Interestingly, most mechanisms of repair of DNA DSBs occur through generation of 3′-ssDNA tails by a 5′-to-3′ resection mechanism (reviewed in ref (28)). Pif1 is known to participate in DSB repair22,39 and has a role in BIR.24−26 Annealing of complementary 3′-ssDNA overhangs must occur in the final steps of DSB repair, and annealing of 3′-ssDNA overhangs was enhanced in the presence of Pif1, even in the presence of single-stranded DNA binding proteins (Figure 7b) or mismatches in the DNA sequence (Figure 7d).
In general, helicases are thought to unwind DNA. However, the discovery of annealing helicases5,10 and UvsW rewinding59 indicates duplex formation can also be catalyzed by helicases. This rewinding activity has been proposed to play a role in the stabilization of stalled replication forks, DNA repair, transcription, telomere metabolism, and chromatin remodeling.12
In BIR, Pif1 has been proposed to be located both with the polymerase at the leading edge of the migrating D-loop and at the trailing edge of the D-loop.24−26 The trailing Pif1 has been suggested to relieve topological stress and displace the newly extended strand. Pif1’s ability to anneal complementary strands provides another possible role for Pif1 in this process. At the trailing edge of the migrating D-loop, the newly synthesized strand must be displaced and the template strands must reanneal. Pif1 can catalyze both of these processes.
A biologically relevant activity of Pif1 is the unfolding of quadruplex DNA structures that form due to folding of G-rich sequences. In the absence of Pif1, such structures cause stalling of DNA replication, leading to increased genomic instability.61 G-Rich sequences can spontaneously fold into quadruplex structures; therefore, a complementary strand must anneal to the G-rich sequence prior to refolding to form duplex DNA. The ability of Pif1 to anneal two strands may allow the enzyme to melt quadruplex DNA, followed by annealing of the complementary strand prior to refolding of the quadruplex structure. In the absence of annealing, the quadruplex structure can simply refold after Pif1-catalyzed unfolding.62 Therefore, annealing activity may be beneficial to Pif1’s ability to unfold quadruplex structures and reduce genomic instability. Pif1 has been suggested to be involved in alternative lengthening of telomeres (ALT),27 a recombination-based pathway that maintains telomeres in a telomerase-independent manner in 10–15% of cancer cells.60 The role for Pif1 during ALT might be related to its ability to unfold quadruplex DNA that can form from telomeric sequences, followed by rapid annealing of the G-rich telomeric strand to the complementary C-rich strand.
Annealing of complementary single strands is a common observation for DNA binding proteins that must be interpreted with caution. DNA in single-stranded form can exhibit greatly varying degrees of intramolecular interactions that can reduce the rate of intermolecular base pairing. Different sequences have different rates of spontaneous annealing.51 Therefore, it is expected that a large variation of annealing rates exists. Hence, the ability of a protein that binds to single-stranded DNA to catalyze annealing, versus melting, will depend on the sequence and the strength of the protein–DNA interaction. Furthermore, proteins that have multiple binding sites on their surface are likely to exhibit complex behavior when encountering complementary single-stranded DNA. An example of such complex behavior is exhibited by the RPA protein. Both human52 and yeast53 RPA can stimulate or inhibit annealing in vitro depending on the type of substrate and conditions. It is feasible that Pif1 and other positively charged enzymes simply act as chemical attractants of negatively charged DNA. However, even if the enhancement in annealing of ssDNA strands by Pif1 is simply a result of Pif1 binding and bringing multiple strands into the proximity of each other, this could still be relevant within a cell.
Aside from its biological implications, an understanding of the strand annealing activity of Pif1 is essential for the appropriate design of biochemical experiments. As shown in Figure 1, if unwinding experiments are performed in the absence of a DNA trap to capture the unlabeled displaced strand, the amplitude of the product formation curve is reduced. This reduction in amplitude could be interpreted as inefficient unwinding if annealing is not considered. Design and interpretation of experiments to measure DNA unwinding by Pif1 should take into account this enzyme’s ability to facilitate annealing of complementary strands.
Acknowledgments
We thank Mark S. Wold for providing RPA protein.
Glossary
Abbreviations
- NA
nucleic acids
- DSB
double-strand break
- ssDNA
single-stranded DNA
- dsDNA
double-stranded DNA
- BIR
break-induced repair
- cNHEJ
classical nonhomologous end joining
- aNHEJ
alternative nonhomologous end joining
- MMEJ
microhomology-mediated end joining
- HR
homologous recombination
- β-ME
β-mercaptoethanol
- BSA
bovine serum albumin
- SDS
sodium dodecyl sulfate
- ALT
alternative lengthening of telomeres.
Supporting Information Available
Two supplemental figures (Figures S1 and S2). This material is available free of charge via the Internet at http://pubs.acs.org.
Author Present Address
† R.R.-B.: Department of Pathology and Immunology, Washington University School of Medicine, 660 S. Euclid Ave., St. Louis, MO 63110.
Author Contributions
R.R.-B. and A.K.B. contributed equally to this work.
This work was supported by National Institutes of Health Grant R01 GM098922 to K.D.R. and by the Arkansas Biosciences Institute, the major research component of the Arkansas Tobacco Settlement Proceeds Act of 2000.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Byrd A. K.; Raney K. D. (2012) Superfamily 2 helicases. Front. Biosci. 17, 2070–2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raney K. D.; Byrd A. K.; Aarattuthodiyil S. (2013) Structure and Mechanisms of SF1 DNA Helicases. Adv. Exp. Med. Biol. 767, 17–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilhooly N. S.; Gwynn E. J.; Dillingham M. S. (2013) Superfamily 1 helicases. Front. Biosci. , Scholar Ed. 5, 206–216. [DOI] [PubMed] [Google Scholar]
- Brosh R. M. Jr. (2013) DNA helicases involved in DNA repair and their roles in cancer. Nat. Rev. Cancer 13, 542–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdez B. C.; Henning D.; Perumal K.; Busch H. (1997) RNA-unwinding and RNA-folding activities of RNA helicase II/Gu: Two activities in separate domains of the same protein. Eur. J. Biochem. 250, 800–807. [DOI] [PubMed] [Google Scholar]
- Machwe A.; Xiao L.; Groden J.; Matson S. W.; Orren D. K. (2005) RecQ family members combine strand pairing and unwinding activities to catalyze strand exchange. J. Biol. Chem. 280, 23397–23407. [DOI] [PubMed] [Google Scholar]
- Gu Y.; Masuda Y.; Kamiya K. (2008) Biochemical analysis of human PIF1 helicase and functions of its N-terminal domain. Nucleic Acids Res. 36, 6295–6308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusufzai T.; Kadonaga J. T. (2008) HARP is an ATP-driven annealing helicase. Science 322, 748–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen D.; Nandakumar D.; Tang G. Q.; Patel S. S. (2012) Human mitochondrial DNA helicase TWINKLE is both an unwinding and annealing helicase. J. Biol. Chem. 287, 14545–14556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia P. L.; Liu Y.; Jiricny J.; West S. C.; Janscak P. (2004) Human RECQ5β, a protein with DNA helicase and strand-annealing activities in a single polypeptide. EMBO J. 23, 2882–2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaue D.; Kobbe D.; Kemmerich F.; Kozikowska A.; Puchta H.; Seidel R. (2013) Fork sensing and strand switching control antagonistic activities of RecQ helicases. Nat. Commun. 4, 2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y. (2012) Unwinding and rewinding: Double faces of helicase?. J. Nucleic Acids 2012, 140601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vindigni A.; Hickson I. D. (2009) RecQ helicases: Multiple structures for multiple functions?. HFSP J. 3, 153–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda-Sasa T.; Polaczek P.; Campbell J. L. (2006) Single strand annealing and ATP-independent strand exchange activities of yeast and human DNA2: Possible role in Okazaki fragment maturation. J. Biol. Chem. 281, 38555–38564. [DOI] [PubMed] [Google Scholar]
- Nelson S. W.; Benkovic S. J. (2007) The T4 phage UvsW protein contains both DNA unwinding and strand annealing activities. J. Biol. Chem. 282, 407–416. [DOI] [PubMed] [Google Scholar]
- Yusufzai T.; Kadonaga J. T. (2010) Annealing helicase 2 (AH2), a DNA-rewinding motor with an HNH motif. Proc. Natl. Acad. Sci. U.S.A. 107, 20970–20973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanagoudr-Bhojappa R.; Chib S.; Byrd A. K.; Aarattuthodiyil S.; Pandey M.; Patel S. S.; Raney K. D. (2013) Yeast Pif1 helicase exhibits a one-base-pair stepping mechanism for unwinding duplex DNA. J. Biol. Chem. 288, 16185–16195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barranco-Medina S.; Galletto R. (2010) DNA binding induces dimerization of Saccharomyces cerevisiae Pif1. Biochemistry 49, 8445–8454. [DOI] [PubMed] [Google Scholar]
- Galletto R.; Tomko E. J. (2013) Translocation of Saccharomyces cerevisiae Pif1 helicase monomers on single-stranded DNA. Nucleic Acids Res. 41, 4613–4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung W. H. (2014) To Peep into Pif1 Helicase: Multifaceted All the Way from Genome Stability to Repair-Associated DNA Synthesis. J. Microbiol. 52, 89–98. [DOI] [PubMed] [Google Scholar]
- Ramanagoudr-Bhojappa R.; Blair L. P.; Tackett A. J.; Raney K. D. (2013) Physical and functional interaction between yeast Pif1 helicase and Rim1 single-stranded DNA binding protein. Nucleic Acids Res. 41, 1029–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makovets S.; Blackburn E. H. (2009) DNA damage signalling prevents deleterious telomere addition at DNA breaks. Nat. Cell Biol. 11, 1383–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang M.; Luke B.; Kraft C.; Li Z.; Peter M.; Lingner J.; Rothstein R. (2009) Telomerase is essential to alleviate pif1-induced replication stress at telomeres. Genetics 183, 779–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung W. H.; Zhu Z.; Papusha A.; Malkova A.; Ira G. (2010) Defective resection at DNA double-strand breaks leads to de novo telomere formation and enhances gene targeting. PLoS Genet. 6, e1000948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson M. A.; Kwon Y.; Xu Y.; Chung W. H.; Chi P.; Niu H.; Mayle R.; Chen X.; Malkova A.; Sung P.; Ira G. (2013) Pif1 helicase and Poldelta promote recombination-coupled DNA synthesis via bubble migration. Nature 502, 393–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini N.; Ramakrishnan S.; Elango R.; Ayyar S.; Zhang Y.; Deem A.; Ira G.; Haber J. E.; Lobachev K. S.; Malkova A. (2013) Migrating bubble during break-induced replication drives conservative DNA synthesis. Nature 502, 389–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewar J. M.; Lydall D. (2010) Pif1- and Exo1-dependent nucleases coordinate checkpoint activation following telomere uncapping. EMBO J. 29, 4020–4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deriano L.; Roth D. B. (2013) Modernizing the nonhomologous end-joining repertoire: Alternative and classical NHEJ share the stage. Annu. Rev. Genet. 47, 433–455. [DOI] [PubMed] [Google Scholar]
- Stevnsner T.; Muftuoglu M.; Aamann M. D.; Bohr V. A. (2008) The role of Cockayne Syndrome group B (CSB) protein in base excision repair and aging. Mech. Ageing Dev. 129, 441–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postow L.; Woo E. M.; Chait B. T.; Funabiki H. (2009) Identification of SMARCAL1 as a component of the DNA damage response. J. Biol. Chem. 284, 35951–35961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusufzai T.; Kong X.; Yokomori K.; Kadonaga J. T. (2009) The annealing helicase HARP is recruited to DNA repair sites via an interaction with RPA. Genes Dev. 23, 2400–2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budd M. E.; Campbell J. L. (2009) Interplay of Mre11 nuclease with Dna2 plus Sgs1 in Rad51-dependent recombinational repair. PLoS One 4, e4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein K. A.; Gangloff S.; Rothstein R. (2010) The RecQ DNA helicases in DNA repair. Annu. Rev. Genet. 44, 393–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochman M. L.; Sabouri; Zakian V. A. (2010) Unwinding the functions of the Pif1family helicases. DNA Repair 9, 237–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opresko P. L.; Otterlei M.; Graakjaer J.; Bruheim P.; Dawut L.; Kolvraa S.; May A.; Seidman M. M.; Bohr V. A. (2004) The Werner syndrome helicase and exonuclease cooperate to resolve telomeric D loops in a manner regulated by TRF1 and TRF2. Mol. Cell 14, 763–774. [DOI] [PubMed] [Google Scholar]
- Opresko P. L.; Mason P. A.; Podell E. R.; Lei M.; Hickson I. D.; Cech T. R.; Bohr V. A. (2005) POT1 stimulates RecQ helicases WRN and BLM to unwind telomeric DNA substrates. J. Biol. Chem. 280, 32069–32080. [DOI] [PubMed] [Google Scholar]
- Ghosh A. K.; Rossi M. L.; Singh D. K.; Dunn C.; Ramamoorthy M.; Croteau D. L.; Liu Y.; Bohr V. A. (2012) RECQL4, the protein mutated in Rothmund-Thomson syndrome, functions in telomere maintenance. J. Biol. Chem. 287, 196–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George T.; Wen Q.; Griffiths R.; Ganesh A.; Meuth M.; Sanders C. M. (2009) Human Pif1 helicase unwinds synthetic DNA structures resembling stalled DNA replication forks. Nucleic Acids Res. 37, 6491–6502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz V. P.; Zakian V. A. (1994) The Saccharomyces PIF1 DNA helicase inhibits telomere elongation and de novo telomere formation. Cell 76, 145–155. [DOI] [PubMed] [Google Scholar]
- Zhou J.; Monson E. K.; Teng S. C.; Schulz V. P.; Zakian V. A. (2000) Pif1p helicase, a catalytic inhibitor of telomerase in yeast. Science 289, 771–774. [DOI] [PubMed] [Google Scholar]
- Boule J. B.; Vega L. R.; Zakian V. A. (2005) The yeast Pif1p helicase removes telomerase from telomeric DNA. Nature 438, 57–61. [DOI] [PubMed] [Google Scholar]
- Boule J. B.; Zakian V. A. (2010) Characterization of the helicase activity and anti-telomerase properties of yeast Pif1p in vitro. Methods Mol. Biol. 587, 359–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raney K. D.; Benkovic S. J. (1995) Bacteriophage T4 Dda helicase translocates in a unidirectional fashion on single-stranded DNA. J. Biol. Chem. 270, 22236–22242. [DOI] [PubMed] [Google Scholar]
- Morris P. D.; Tackett A. J.; Babb K.; Nanduri B.; Chick C.; Scott J.; Raney K. D. (2001) Evidence for a functional monomeric form of the bacteriophage T4 Dda helicase. Dda does not form stable oligomeric structures. J. Biol. Chem. 276, 19691–19698. [DOI] [PubMed] [Google Scholar]
- Carroll S. S.; Benseler F.; Olsen D. B. (1996) Preparation and use of synthetic oligoribonucleotides as tools for study of viral polymerases. Methods Enzymol. 275, 365–382. [DOI] [PubMed] [Google Scholar]
- Byrd A. K.; Raney K. D. (2004) Protein displacement by an assembly of helicase molecules aligned along single-stranded DNA. Nat. Struct. Mol. Biol. 11, 531–538. [DOI] [PubMed] [Google Scholar]
- Johnson K. A.; Simpson Z. B.; Blom T. (2009) Global Kinetic Explorer: A new computer program for dynamic simulation and fitting of kinetic data. Anal. Biochem. 387, 20–29. [DOI] [PubMed] [Google Scholar]
- Machwe A.; Lozada E. M.; Xiao L.; Orren D. K. (2006) Competition between the DNA unwinding and strand pairing activities of the Werner and Bloom syndrome proteins. BMC Mol. Biol. 7, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings T. A.; Mackintosh S. G.; Harrison M. K.; Sikora D.; Sikora B.; Dave B.; Tackett A. J.; Cameron C. E.; Raney K. D. (2009) NS3 helicase from the hepatitis C virus can function as a monomer or oligomer depending on enzyme and substrate concentrations. J. Biol. Chem. 284, 4806–4814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimme J. M.; Honda M.; Wright R.; Okuno Y.; Rothenberg E.; Mazin A. V.; Ha T.; Spies M. (2010) Human Rad52 binds and wraps single-stranded DNA and mediates annealing via two hRad52-ssDNA complexes. Nucleic Acids Res. 38, 2917–2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y.; Wolf L. K.; Georgiadis R. M. (2006) Secondary structure effects on DNA hybridization kinetics: A solution versus surface comparison. Nucleic Acids Res. 34, 3370–3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartos J. D.; Willmott L. J.; Binz S. K.; Wold M. S.; Bambara R. A. (2008) Catalysis of strand annealing by replication protein A derives from its strand melting properties. J. Biol. Chem. 283, 21758–21768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama T.; New J. H.; Kowalczykowski S. C. (1998) DNA annealing by RAD52 protein is stimulated by specific interaction with the complex of replication protein A and single-stranded DNA. Proc. Natl. Acad. Sci. U.S.A. 95, 6049–6054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng S. K.; Gibb B.; de Almeida M. J.; Greene E. C.; Symington L. S. (2014) RPA antagonizes microhomology-mediated repair of DNA double-strand breaks. Nat. Struct. Mol. Biol. 21, 405–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahaye A.; Leterme S.; Foury F. (1993) PIF1 DNA helicase from Saccharomyces cerevisiae. Biochemical characterization of the enzyme. J. Biol. Chem. 268, 26155–26161. [PubMed] [Google Scholar]
- Singh D. K.; Ghosh A. K.; Croteau D. L.; Bohr V. A. (2012) RecQ helicases in DNA double strand break repair and telomere maintenance. Mutat. Res. 736, 15–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedinger P.; Hochstrasser M.; Victor Jongeneel C.; Alberts B. M. (1983) Properties of the T4 bacteriophage DNA replication apparatus: The T4 dda DNA helicase is required to pass a bound RNA polymerase molecule. Cell 34, 115–123. [DOI] [PubMed] [Google Scholar]
- Byrd A. K.; Raney K. D. (2006) Displacement of a DNA binding protein by Dda helicase. Nucleic Acids Res. 34, 3020–3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manosas M.; Perumal S. K.; Bianco P.; Ritort F.; Benkovic S. J.; Croquette V. (2013) RecG and UvsW catalyse robust DNA rewinding critical for stalled DNA replication fork rescue. Nat. Commun. 4, 2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesare A. J.; Reddel R. R. (2010) Alternative lengthening of telomeres: Models, mechanisms and implications. Nat. Rev. Genet. 11, 319–330. [DOI] [PubMed] [Google Scholar]
- Paeschke K.; Bochman M. L.; Garcia P. D.; Cejka P.; Friedman K. L.; Kowalczykowski S. C.; Zakian V. A. (2013) Pif1 family helicases suppress genome instability at G-quadruplex motifs. Nature 497, 458–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R.; Zhang J.; Bochman M. L.; Zakian V. A.; Ha T. (2014) Periodic DNA patrolling underlies diverse functions of Pif1 on R-loops and G-rich DNA. eLife 3, e02190. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.