Abstract
Integration host factor (IHF) is an Escherichia coli protein involved in (i) condensation of the bacterial nucleoid and (ii) regulation of a variety of cellular functions. In its regulatory role, IHF binds to a specific sequence to introduce a strong bend into the DNA; this provides a duplex architecture conducive to the assembly of site-specific nucleoprotein complexes. Alternatively, the protein can bind in a sequence-independent manner that weakly bends and wraps the duplex to promote nucleoid formation. IHF is also required for the development of several viruses, including bacteriophage lambda, where it promotes site-specific assembly of a genome packaging motor required for lytic development. Multiple IHF consensus sequences have been identified within the packaging initiation site (cos), and we here interrogate IHF–cos binding interactions using complementary electrophoretic mobility shift (EMS) and analytical ultracentrifugation (AUC) approaches. IHF recognizes a single consensus sequence within cos (I1) to afford a strongly bent nucleoprotein complex. In contrast, IHF binds weakly but with positive cooperativity to nonspecific DNA to afford an ensemble of complexes with increasing masses and levels of condensation. Global analysis of the EMS and AUC data provides constrained thermodynamic binding constants and nearest neighbor cooperativity factors for binding of IHF to I1 and to nonspecific DNA substrates. At elevated IHF concentrations, the nucleoprotein complexes undergo a transition from a condensed to an extended rodlike conformation; specific binding of IHF to I1 imparts a significant energy barrier to the transition. The results provide insight into how IHF can assemble specific regulatory complexes in the background of extensive nonspecific DNA condensation.
Escherichia coli integration host factor (IHF) is a basic, heterodimeric 22 kDa DNA binding protein that belongs to a class of histone-like proteins capable of bending and wrapping DNA into condensed structures.1−5 As such, one of its biological functions is condensation of the bacterial nucleoid along with other basic proteins such as DPS, HU, and H-NS.1,3,4 In this case, multiple IHF, HU, and H-NS proteins assemble onto DNA in a sequence-independent (nonspecific) manner to condense the duplex. IHF is unique, however, in that it also binds to specific DNA sequences with high affinity.5,6 This specific DNA binding activity is associated with the regulation of a number of cellular processes, including transcription,7 DNA replication,8 and site-specific recombination.6,9
In addition to these host functions, IHF is required for the development of several viruses, including bacteriophage lambda. Indeed, IHF was originally described as a host protein that is required for site-specific integration of the lambda genome into the E. coli genome during lysogeny.10,11 A number of IHF recognition sequences (H-elements) have been identified in the viral synapse site (attP), and specific binding of IHF to these elements is required to assemble the integrase complex.10 IHF also plays an important role in the lytic pathway of lambda development. A variety of studies have demonstrated that the host protein stimulates virus development in vivo(12,13) and genome packaging reactions in vitro.14−18
Terminase enzymes are responsible for viral genome packaging,19 and we have demonstrated that IHF promotes the assembly of a terminase motor complex at cos, the packaging initiation site in a lambda genome (Figure 1A).18,20 Our lab is interested in the thermodynamic features of packaging motor assembly; unfortunately, this represents a cooperative, multipartite interaction of four heterotrimeric terminase protomers and an indeterminate number of IHF proteins with multiple, putative recognition elements dispersed within the ∼270 bp cos sequence (Figure 1B).15,18,21,22 This presents an extremely complex system from which to dissect detailed mechanistic information. Therefore, as a first step toward biochemical characterization of these viral genome packaging complexes, we here characterize the most fundamental of these interactions, binding of IHF to the lambda cos sequence. The results provide insight into the general mechanism by which IHF can promote the assembly of specific regulatory complexes in the context of a vast excess of nonspecific nucleoid formation within the cell.
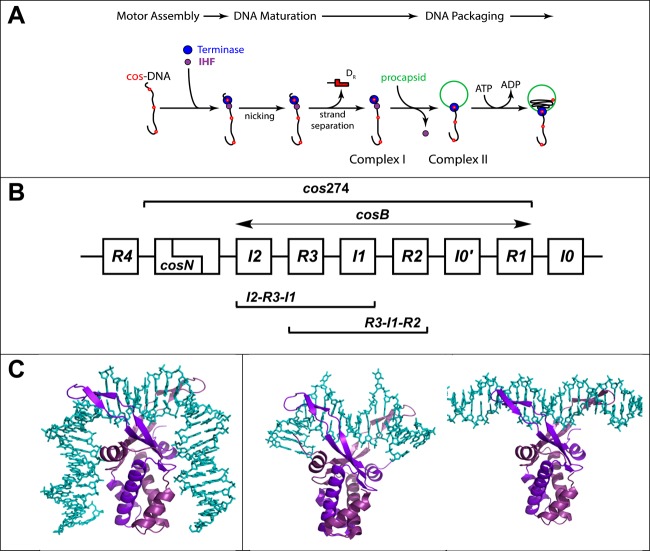
Figure 1.
(A) Assembly of a viral genome maturation and packaging complex at the cos site of the lambda genome. The terminase protomer is composed of one large gpA subunit tightly associated with two smaller gpNu1 subunits. Four protomers and an indeterminate number of IHF dimers cooperatively assemble at a cos sequence of a genome concatemer to engender the packaging motor complex; cos (red dots) represents the junction of two genomes in a concatemer and serves as the packaging initiation site. Terminase and IHF are depicted as blue and purple circles, respectively, for the sake of simplicity. The assembled motor nicks the duplex at cosN to yield the 12-base “sticky” end of the genome (complex I). This intermediate binds a procapsid to yield the functional packaging motor (complex II), which translocates viral DNA into the shell. (B) Detail of the cos region of the lambda genome. The sequence is multipartite consisting of cosN (nicking) and cosB (binding) subsites; cosB extends from I2 to R1 elements. The gpNu1 subunit specifically interacts with the three R elements, and several putative IHF consensus sequences have been identified (I0–I4). The model duplexes used in this study are indicated in the Figure: cos274 (274 bp), [R3-I1-R2] (75 bp), [I2-R3-I1] (75 bp), I1 (27 bp), and R3 (27 bp). (C) Structural models for IHF–DNA nucleoprotein complexes. The left panel shows the crystal structure of IHF bound in a specific complex with the H′ element of attP (PDB entry 1OWF) showing a duplex bend angle of >160°. The DNA binding site size in this complex is ∼34 bp. The middle panel shows the cocrystal structure of Anabaena HU protein bound in a nonspecific complex (PDB entry 1P71) depicting a “weak” (∼105°) bend in the duplex that is found in condensed, nucleoid DNA. The DNA binding site size in this complex is ∼20 bp. The right panel shows the structural model for IHF bound in a nonspecific, linear complex. The model was constructed using MacPymol by manually docking the crystal structure of IHF onto the minimal nonspecific R3 duplex. The DNA binding site size in this complex is ∼8 bp. In all structures, DNA is colored cyan and the α and β subunits of IHF are colored light and dark purple, respectively.
Experimental Procedures
DNA Substrates
DNA oligonucleotides (unmodified, 5′-end-labeled with IRDye 700, and 5′-end-labeled with 6-carboxyfluorescein) were obtained from Integrated DNA Technologies (Coralville, IA). Preparation of the duplex substrates used in this study is described in the Supporting Information. The molecular weights and extinction coefficients of the DNA substrates were calculated on the basis of their sequences and the molecular weights and extinction coefficients of any appended dyes.23 The concentration of the oligonucleotide strands and DNA duplexes was determined spectrally using their calculated extinction coefficients.
Purification of Integration Host Factor (IHF)
IHF was purified from HN880, a heat-inducible IHF-overproducing strain (a kind gift of H. Nash, National Institutes of Health, Bethesda, MD), as described previously.24 The concentration of IHF was determined spectrally (ε276 = 5800 M–1 cm–1), and the purified protein was analyzed by sedimentation velocity analytical ultracentrifugation; this affords an s(20,w) of 1.96 S (data not shown) and an experimental molecular weight of 22.2 kDa, which compares to the theoretical value based on the protein sequence (22.0 kDa). Importantly, there was no evidence of dissociation of the heterodimer or further self-association or aggregation of the protein in the concentration range utilized in this study.
Electrophoretic Mobility Shift (EMS) Studies
Equilibrium binding experiments were performed in 20 mM Tris buffer (pH 8 at 4 °C) containing 55 mM NaCl, 1 mM EDTA, 7 mM β-me, 2 mM spermidine, and 10% (v/v) glycerol. The IRDye 700-labeled DNA substrate was included at a final concentration of 4 nM, and IHF was added as indicated in each individual experiment. The binding reaction mixtures were incubated at room temperature for 20 min and then loaded onto an 8% polyacrylamide gel (80:1 acrylamide/bisacrylamide mixture for the 274 bp substrates; 29:1 acrylamide/bisacrylamide mixture for the 75 and 27 bp substrates). The gels were run at 15 V/cm in 0.5× TBE at 4 °C for 1 h, and scanned using an Odyssey scanner (LI-COR Biosciences). The DNA present as unbound (free) and gel-retarded (bound) species was quantified using the ImageQuant software package (GE Healthcare Life Sciences), and the fraction of bound DNA (Fbound) was calculated according to
Analysis of the EMS Binding Data Using a Nonspecific Finite Lattice DNA Binding Model
Record and co-workers, building on the elegant work of McGee and von Hippel,25 have developed a model that is essentially a Scatchard formulation that takes into account the nonspecific, cooperative binding of a ligand (e.g., protein) to a one-dimensional lattice of finite length (e.g., DNA).26 The binding isotherm is described by
| 1a |
where [protein]free represents the concentration of free protein in the titration mixture, n is the nonspecific protein binding site size in base pairs, N is the duplex length in base pairs, υns represents the protein binding density (moles of protein bound per base pair in the duplex), Kns is the intrinsic equilibrium constant for protein binding to the nonspecific site, and (ff)n−1 is the probability of finding n – 1 free base pairs adjacent to any given unbound base pair in the duplex (i.e., an unoccupied protein binding site of size n).
where ω is the cooperativity factor and
To apply this model to EMS data, which yields the fraction of bound duplex (Fbound) at a given concentration of IHF, we need expressions that describe this experimentally observed value. The fraction of free duplex (Ffree) can be calculated according to the following equation27
| 1b |
and therefore
| 1c |
Equation 1a can be rearranged to generate an expression for (ff)N−1, and upon substitution into eq 1c and rearrangement, it can be shown that
| 1d |
where [protein]total and [DNA] represent the total molar concentrations of protein and DNA, respectively, added to the reaction mixture. Importantly, this formulation includes the nearest neighbor cooperativity parameter (ω, embedded within ff), which describes cooperative protein assembly on the duplex.
To constrain the analysis and provide well-resolved parameters, an ensemble of EMS data for cos274, [R3-I1-R2], and [I2-R3-I1] model duplexes (in triplicate, representative data shown in Figures 2A, 3C, and 3D, respectively) were globally fit to eq 1d by nonlinear least-squares (NLLS) analytical methods using Scientist (Micromath Scientific Software). The duplex length (N) was held as a local constant for each duplex. The IHF binding site size (n = 8)28 and duplex concentration ([DNA]) were held as global constants. Kns, υns, and ω were global variables that were allowed to float to their best values. The best fit of the ensemble of data is shown as solid lines in Figure 4A.
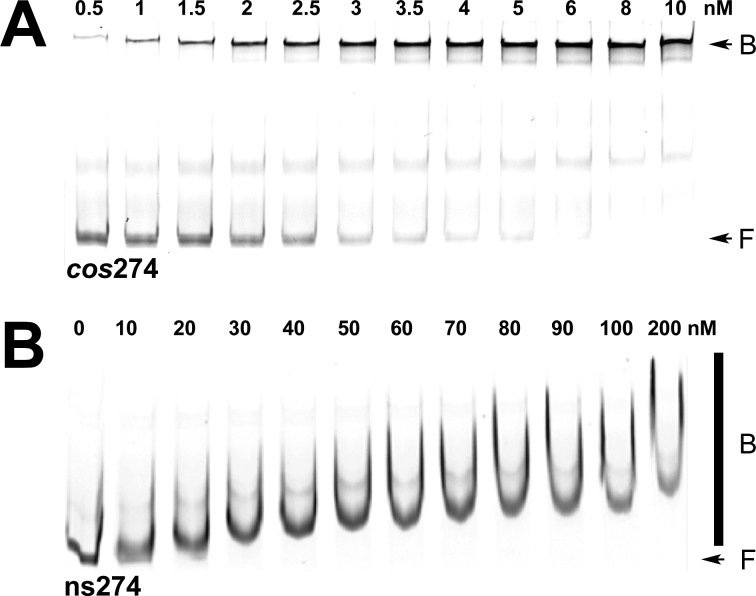
Figure 2.
Electrophoretic mobility shift (EMS) studies of binding of IHF to specific (cos274) and nonspecific (ns274) DNA substrates. (A) Representative polyacrylamide gel showing that IHF binds to the specific cos274 substrate to afford a distinct retarded complex. The positions of free (F) and bound (B) DNA complexes are indicated with arrows at the right of the gel image. The band in the middle of the gel represents a contaminant in the IRDye-labeled duplex (Supporting Information). It is unaffected in the titration study and was not considered in the calculation of Fbound. (B) Representative polyacrylamide gel showing that IHF binds to the nonspecific ns274 substrate to afford a concentration-dependent shift and smear on the gel. The positions of free (F) DNA and the bound (B) DNA complexes are indicated at the right of the gel image with an arrow and bar, respectively.
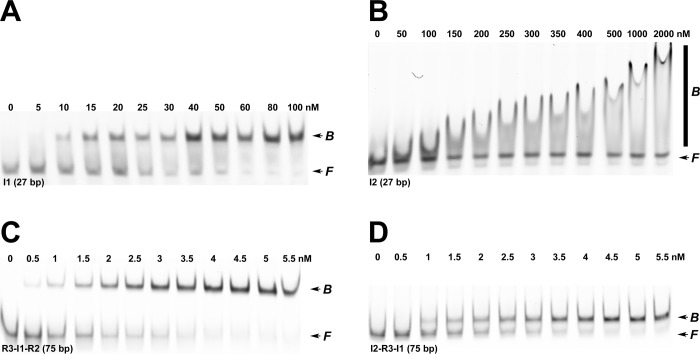
Figure 3.
EMS studies of binding of IHF to minimal duplex substrates. (A) Representative polyacrylamide gel showing that IHF binds to the minimal 27 bp I1-specific substrate to afford a distinct retarded complex. We note that upward “smearing” of the retarded band is observed at IHF concentrations of >100 nM (not shown). (B) Representative polyacrylamide gel showing that IHF binds to the minimal 27 bp I2 substrate to afford a smear on the gel. (C) Representative polyacrylamide gel showing that IHF binds to the 75 bp [R3-I1-R2] duplex substrate to afford a distinct retarded complex. (D) Representative polyacrylamide gel showing that IHF binds to the 75 bp [I2-R3-I1] duplex substrate to afford a distinct retarded complex. The positions of free and bound DNA complexes are indicated at the right of each gel image.
Figure 4.
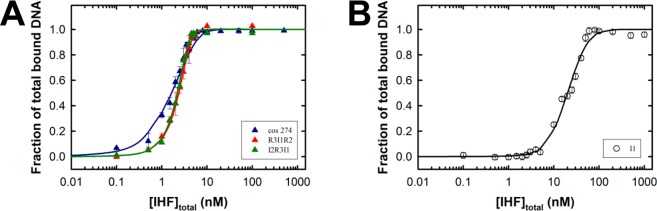
Quantitative analysis of EMS binding data. The EMS data (representative data presented in Figures 2 and 3) were converted to fraction bound DNA versus IHF concentration as described in Experimental Procedures. (A) Ensemble of EMS data for binding of IHF to the cos274 (blue), [R3-I1-R2] (red), and [I2-R3-I1] (green) duplexes. Each data point is the average of at least three separate experiments with the standard deviation indicated with error bars. The ensemble of data was simultaneously analyzed according to the nonspecific finite lattice DNA binding model as described in Experimental Procedures. The solid lines represent the best fits of the data, and the derived binding parameters are presented in Table 1. (B) EMS data for binding of IHF to the minimal I1-specific duplex. Each data point is the average of at least three separate experiments with the standard deviation indicated with error bars. The data were analyzed according to the competitive specific/nonspecific finite lattice DNA binding model as described in Experimental Procedures. The solid line represents the best fit of the data, and the derived binding parameters are presented in Table 1.
Analysis of the EMS Binding Data Using a Competitive Specific and Nonspecific Finite Lattice DNA Binding Model
Record and co-workers have further developed a “competitive binding” model that describes the interaction of protein with a short duplex that contains a single specific binding element.26 In this model, the protein can bind to its cognate element in a specific binding interaction, or it can bind in a distinct nonspecific binding mode, but not both simultaneously; i.e., the two binding modes compete with each other. Senear and co-workers have implemented this approach to interrogate EMS data,29 which is described by eq 1d, except that the total protein concentration is described by26
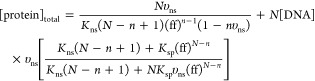 |
2 |
The first term describes the concentration of free IHF and the second term is the concentration of IHF bound to I1 in a specific or nonspecific manner. This modification was incorporated into eq 1d, and the EMS data for the I1 model duplex (in triplicate, representative data shown in Figure 3A) were fit to the model by NLLS analytical methods using Scientist (Micromath Scientific Software). The IHF binding site size (n = 8), duplex length (N = 27), and [DNA] were held constant while Kns, Ksp, υns, and ω were fitting variables that were allowed to float to their best values. The best fit of the data is shown as a solid line in Figure 4B
Sedimentation Velocity Analytical Ultracentrifugation (SV-AUC) Studies
Sedimentation velocity experiments were conducted with a Beckman Coulter XL-I ultracentrifuge fitted with an Aviv Biomedical fluorescence detection system (FDS) (λex = 488 nm; λem = 505–565 nm). Unless otherwise noted, each sample (400 μL) contained 4 nM 6-FAM-labeled DNA and the indicated concentration of IHF in 20 mM Tris buffer (pH 8) containing 55 mM NaCl, 1 mM EDTA, 7 mM β-ME, 2 mM spermidine, and 5% (v/v) glycerol. The samples were placed into preassembled 12 mm Epon charcoal-filled double-sector centerpieces along with a fluorescein reference cell. Fluorescence data were collected at 7 °C using a rotor speed of 42000 rpm and a spacing of 2 mm with five averages per position. Importantly, IHF protein added to the samples is “invisible” because only DNA contains the fluorescent label. The raw data were analyzed using the Sedfit data analysis package,30 using a continuous c(s) approach, and the weight-average sedimentation coefficients, ⟨s*⟩, were calculated by integration of the c(s) distribution.
Analysis of the Sedimentation Velocity AUC Data
The weight-average sedimentation coefficient obtained above captures all of the species present during a titration experiment. Because fluorescence optics were used in our studies, only those complexes that contain the fluorescent DNA substrate contribute to the observed signal. We first consider a duplex of nonspecific sequence (e.g., the R3 duplex). In this case, ⟨s*⟩ reflects unbound DNA (Ffree) plus that in complex with protein (Fbound) and
| 3a |
where sfree* is the sedimentation coefficient of free (unbound) DNA and ⟨sbound-NS⟩ is the weight-average sedimentation coefficient for the ensemble of nonspecific complexes at a given concentration of IHF. Note that Fbound represents DNA with at least one IHF dimer bound in a nonspecific manner. The expressions of Ffree (eq 1b) and Fbound (eq 1d) were substituted into eq 3a to afford an expression that describes ⟨s*⟩ as a function of [IHF].
In the case of a duplex that contains a specific binding element (e.g., the I1 duplex), the weight-average sedimentation coefficient must consider not only nonspecific binding interactions as described above but also the contribution of the specific IHF–DNA complex to the experimentally observed ⟨s*⟩. In this case
| 3b |
where the right-hand term incorporates the contribution of the specific IHF–DNA complex into the experimentally observed ⟨s*⟩. During a titration experiment, Ffree is obtained from eq 1b and the fraction of DNA bound in a specific complex is described by26
| 3c |
and Fbound-NS can be calculated according to
These values were substituted into eq 3b to yield an expression that describes ⟨s*⟩ as a function of [IHF].
Analysis of the experimental data using the models presented above requires fitting of multiple parameters that if allowed to float in an unconstrained NLLS analysis would likely contribute to lower precision in the derived values. To constrain the analysis and provide better resolved values, the ensemble of AUC data for the R3 and I1 duplexes was analyzed globally, as follows. The R3 data were modeled to the finite nonspecific binding model (eq 3a), and the I1 data were simultaneously fit to the competition binding model (eq 3b). The nonspecific site size (n = 8), duplex length (N = 27), and experimentally determined sedimentation coefficient for free DNA (sfree* = 1.96) were held fixed as global constants; ⟨sbound-NS⟩ and sbound-SP* were local variables used in the finite nonspecific binding and competition binding equations, respectively, and υns, Kns, Ksp, and ω were global variables that were allowed to float to their best values by NLLS analytical methods using Scientist (Micromath Scientific Software). The best fits of the data are shown as solid lines in Figure 5C.
Figure 5.
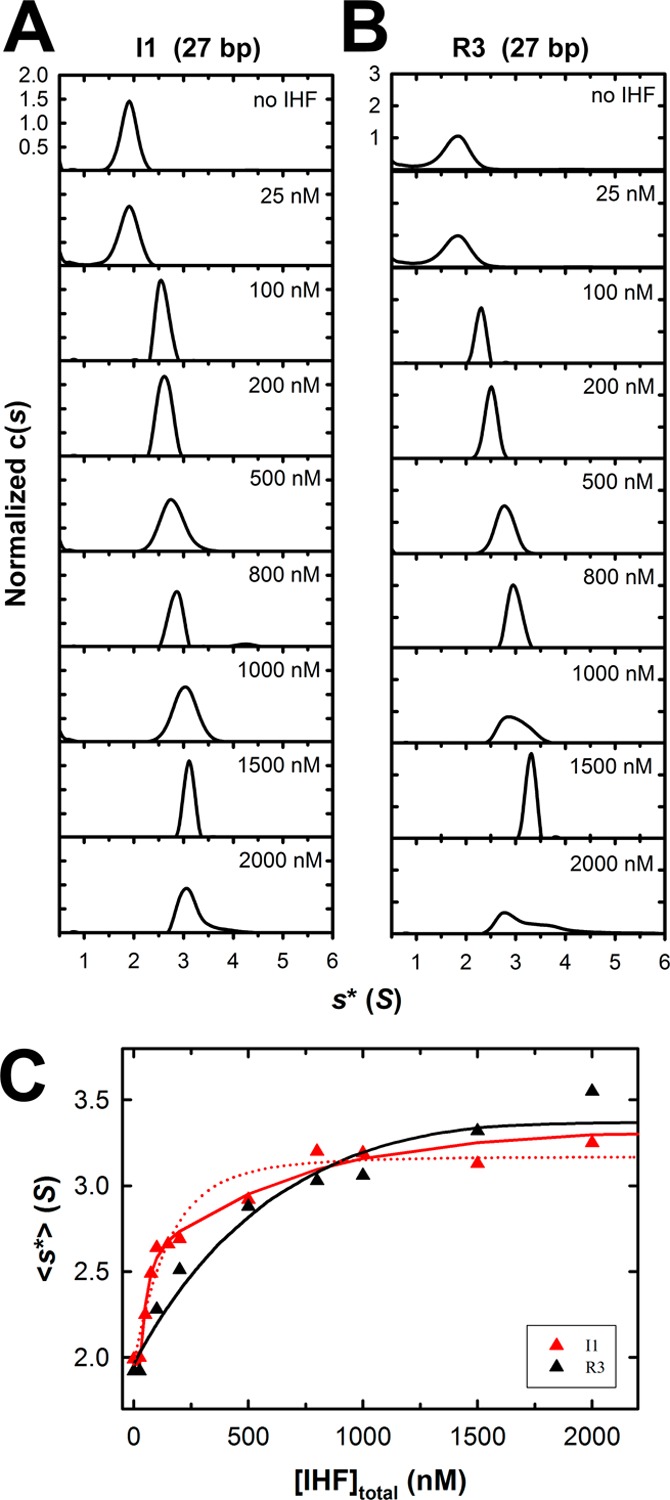
Interrogation of binding of IHF to minimal substrates using sedimentation velocity analytical ultracentrifugation (SV-AUC). Increasing concentrations of IHF were added to the minimal I1 (specific) and R3 (nonspecific) duplex substrates, and their sedimentation behavior was monitored by SV-AUC as described in Experimental Procedures. The c(s) distribution for each binding experiment was calculated using Sedfit. (A) Normalized c(s) profiles for the specific I1 duplex (27 bp). (B) Normalized c(s) profiles for the nonspecific R3 duplex (27 bp). (C) Weight-average sedimentation coefficients for each of the c(s) distributions shown in panels A (red triangles, I1) and B (black triangles, R3) were calculated using Sedfit and are plotted as a function of IHF concentration. The red dotted line represents the best fit of the I1 binding data to the nonspecific finite lattice DNA binding model, which does not adequately describe the data. The solid lines represent the best fit of simultaneous (global) analysis of the R3 (black) and I1 (red) binding data to (i) nonspecific finite lattice DNA binding and (ii) competitive specific/nonspecific finite lattice DNA models, respectively. The binding parameters derived from global analysis are presented in Table 1.
Results
IHF Binds to cos-DNA and Nonspecific Duplexes To Afford Distinctly Different Complexes
We previously examined binding of IHF to a 272 bp DNA substrate that models the full-length cos sequence found in concatemeric lambda DNA.20 Our prior study utilized radiolabeled duplex substrates under equilibrium binding conditions (10 pM DNA). To directly compare EMS data to the results of AUC studies described below, the EMS experiment was repeated to confirm that identical results are obtained with an IRDye-labeled cos-containing duplex [cos274 (Figure 1B)] present at an elevated concentration (4 nM). As anticipated, IHF binds to cos274 in a concentration-dependent manner to form a discrete, strongly retarded band in the gel (Figure 2A). Circular permutation studies have demonstrated that this pattern reflects binding of IHF to cosB to introduce a severe bend (120°) in the duplex.20 The quality of the retarded band is insensitive to IHF concentrations up to 1 μM; however, an upward “smearing” of the band is observed at greater concentrations, likely because of nonspecific binding at these elevated concentrations (see below).31 The EMS data were analyzed according to a phenomenological Hill model (eq S1 of the Supporting Information), which yields a Kapp of (1.2 ± 0.4) × 109 M–1 and a Hill coefficient n of 2 ± 1; these values are virtually identical with those obtained previously in our lab20 and with those obtained for binding of IHF to duplexes containing a high-affinity H′-element of attP.28,29,32 Importantly, our prior study utilized radiolabeled duplex substrates (no dye), and the correspondence of the results indicates that the IR tag used in the study presented here does not influence IHF binding.
In stark contrast, IHF binds ns274, a 274 bp duplex of nonspecific sequence, to yield a diffuse smear in the gel that becomes progressively retarded in a concentration-dependent manner (Figure 2B). This pattern is consistent with (i) weak binding interactions that result in dissociation of the nucleoprotein complex in the gel and/or (ii) binding of multiple IHF dimers to the duplex in a concentration-dependent manner. Both of these features are consistent with the role of IHF in nucleoid assembly, and we presume that this smeared pattern indicates weak, cooperative binding of IHF to nonspecific DNA. Whatever the case, the smears preclude quantitation of the binding data; however, visual inspection of the gel suggests that IHF binds to this nonspecific duplex with an apparent affinity 5–10-fold lower than that of the cos-containing duplex, based on the disappearance of the free DNA band.
IHF Binds to a Minimal I1 Consensus Sequence To Yield a Discrete, Bent Complex
Xin and Feiss identified several potential IHF binding elements within cos (Figure 1B), based on the H-element consensus sequence originally defined by Craig and Nash.10,33 The putative elements possess varying degrees of sequence homology to the well-characterized H-elements but differ from the “conventional” IHF recognition sequences with respect to their spacing and orientation within cos. DNase I and hydroxy radical footprinting studies have demonstrated that the I1 element is strongly protected by IHF, that the I2 element may be weakly protected, but that there is little to no protection at any of the other proposed elements.33−35 To dissect the binding interactions more fully and to derive thermodynamic binding parameters, we utilized minimal duplex substrates to define the affinity of IHF for the putative specific elements found within cos.
We first examined binding of IHF to I1, a 27 bp duplex comprising the I1 consensus sequence. As anticipated, IHF binds to I1 in a concentration-dependent manner to yield a discrete gel-retarded band in the EMS assay (Figure 3A). Analysis of the data yields a Kapp of (9.6 ± 0.5) × 107 M–1 and a Hill coefficient n of 2 ± 1. Importantly, the apparent affinity of IHF for this minimal substrate is an order of magnitude lower than that observed with the full-length cos substrate (Table S1 of the Supporting Information). This is discussed further below. We next examined binding of IHF to I2, a 27 bp duplex comprising the I2 element of cos (Figure 2B) that has been reported to weakly bind IHF.33 The EMS data presented in Figure 3B indicate that IHF binds to this putative element to afford a diffuse smear in the gel, a pattern virtually identical to that observed for binding of IHF to the ns274 duplex (Figure 2B) and to R3, a 27 bp “nonspecific” DNA duplex (Figure S2 of the Supporting Information). This pattern is typical of binding of IHF to duplexes of nonspecific sequence,20,29 and we interpret the data to indicate that the I2 sequence is not recognized as a specific binding element by IHF.
To further interrogate interactions of IHF with cos, we utilized [R3-I1-R2], a 76 bp duplex that contains a single, centrally located I1 element (Figure 1B). IHF binds to this duplex with high affinity, resulting in a distinct gel-retarded band (Figure 3C), a pattern that is consistent with the formation of a specific and strongly bent complex. Analysis of the binding data according to a Hill model yields a Kapp of (9.2 ± 0.4) × 108 M–1 and a Hill coefficient n of 3 ± 1. Virtually identical results were obtained with [I2-R3-I1], a 75 bp duplex that contains both I2 and I1 elements located at the ends of the duplex rather than the center (Figure 3D). Of note, the apparent affinity of IHF for these 76 bp model duplexes is similar to that observed for the full-length cos274 model substrate, all of which are an order of magnitude greater than that observed with the minimal I1 duplex (27 bp) (Table S1 of the Supporting Information). We further note that the complex formed with [R3-I1-R2] is more strongly retarded than that observed with the [I2-R3-I1] duplex. This precludes the possibility of cooperative binding of IHF to the I2 element in the presence of I1 in cis, which would result in a slower migrating complex, the opposite of what is observed. Within this context, an empirical relationship exists between duplex bending and nucleoprotein complex migration in a gel; the degree of retardation is greater when the bend is introduced in the center versus the end of a duplex of a given length (circular permutation analysis).36 Thus, we interpret the observed EMS patterns for the [R3-I1-R2] and [I2-R3-I1] duplexes to indicate that IHF binds with specificity only to the I1 element to introduce a solitary bend into both binding substrates.
Dissection of cos-Specific and Nonspecific IHF Binding Interactions Using EMS
The EMS studies indicate that I1 is the only element within cos that displays specific and high affinity for IHF. On the surface, this predicts that binding of IHF to its cognate I1 element should be described well by a simple Langmuir binding model and that the Hill coefficient should be 1; this is obviously not the case. This phenomenon has been previously described, and it has been attributed to nonspecific IHF binding interactions that are superimposed on the specific binding event.32 To directly test this hypothesis and to define the nonspecific binding parameters for IHF, we first adapted a nonspecific finite lattice DNA binding model as outlined by Record and co-workers.26 This analysis yields a nonspecific binding constant (Kns) and a nearest neighbor cooperativity factor (ω) that best describe the assembly of multiple proteins onto a long duplex of finite length, and of nonspecific sequence. Unfortunately, quantitation of binding of IHF to nonspecific DNA by EMS is not possible because of the diffuse nature of the retarded complexes (Figures 2B and 3B and Figure S2 of the Supporting Information). We therefore utilized EMS data for the [R3-I1-R2], [I2-R3-I1], and cos274 duplex substrates, which are composed of predominantly nonspecific DNA sequence. The ensemble of data was globally fit to the nonspecific finite lattice DNA binding model as described in Experimental Procedures (eq 1c), which yields an excellent fit to all of the data sets well (Figure 4A). The global analysis resolves a constrained nonspecific binding constant Kns of (7.0 ± 0.2) × 106 M–1 and the nearest neighbor cooperativity factor (ω = 37 ± 2). The positive value for ω is interpreted to indicate that binding of IHF to a duplex increases the probability of a second IHF dimer binding next to it by 37-fold, consistent with the greater than unity values for the Hill coefficient observed above.
We note that the resolved Kns obtained from this initial analysis is greater than published values (Kns ∼ 6.6 × 105 M–1).26,28,29 We hypothesized that this might reflect high-affinity specific binding of IHF to the I1 element, which is present in all of the duplexes used in the global analysis, and that this contributes to the apparent nonspecific binding affinity. To address this question and to further resolve the specific DNA binding constant (Ksp), we utilized a competitive specific/nonspecific finite lattice DNA binding model originally described by Record and co-workers26 and implemented for EMS data by Senear and co-workers.29 This model assumes that IHF can bind a short duplex containing a single I1 element in either (i) a specific (strongly bent) complex or (ii) a nonspecific (weakly bent/wrapped) complex, but not both simultaneously. The EMS data for the minimal I1 duplex were used in this analysis because its length is comparable to the site size required for specific IHF binding interactions (∼30 bp)29 and is only ∼3 times larger than the estimated nonspecific binding site size (8 bp).28 Analysis of the EMS data according to this model as described in Experimental Procedures (eqs 1c and 2) yields an excellent fit (Figure 4B) and returns a Kns of (2.9 ± 0.3) × 106 M–1 and an ω of 9.7 ± 2.6 (Table 1). These values are commensurate with previously published studies that examined binding of IHF to the H1′ element of attP using calorimetric approaches.28 In addition, the analysis provides a constraint on the upper limit of the IHF-specific DNA binding constant for the I1 element (Ksp ≤ 6.9 × 109 M–1). This is addressed further below.
Table 1. Analysis of the EMS and AUC Binding Data.
| EMS |
AUC | |||
|---|---|---|---|---|
| nonspecific finite lattice modela | competitive specific/nonspecific finite lattice modelb | competitive specific/nonspecific finite lattice modelc | hydrodynamic modelingd | |
| Kns | (7.0 ± 0.2) × 106 M–1 | (2.9 ± 0.3) × 106 M–1 | (1.7 ± 0.6) × 106 M–1 | |
| ω | 37 ± 2 | 9.7 ± 2.6 | 10 ± 4 | |
| Ksp | – | ≤6.9 × 109 M–1 | (2.0 ± 1.6) × 108 M–1 | |
| – | – | ssp* = 2.59 ± 0.12 S | ssp = 2.36 S | |
| – | – | ⟨sns*⟩ = 3.37 ± 0.04 S | sns = 3.52 S | |
EMS binding data for the cos274, [R3-I1-R2], and [I2-R3-I1] duplexes presented in Figure 4A were simultaneously analyzed (globally fit) according to the nonspecific finite lattice DNA binding model as outlined in Experimental Procedures. The best fit of the ensemble of data is presented as a solid lines in Figure 4A.
EMS data for the minimal I1 duplex presented in Figure 4B were fit to the competitive specific/nonspecific finite lattice DNA binding model as outlined in Experimental Procedures. The best fit is presented as a solid line in Figure 4B.
SV-AUC binding data for the I1 and R3 minimal duplexes presented in Figure 5 were simultaneously analyzed (globally fit) according to the competitive specific/nonspecific finite lattice DNA binding model and the nonspecific finite lattice DNA binding model, respectively, as oulined in Experimental Procedures. The best fit of the ensemble of data is presented as solid lines in Figure 5C.
Hydropro was used to determine the theoretical sedimentation coefficients for the specific IHF-I1 complex (ssp based on high-resolution structural data43) and the nonspecific IHF–R3 complex [sns based on a structural model of three IHF dimers bound to the 27 bp duplex (Supporting Information)].
In summary, the EMS data indicate that (i) IHF binds weakly and with modest cooperativity to duplexes of nonspecific sequence resulting in diffuse smears on the gel regardless of duplex length, (ii) the 27 bp I1 sequence is the sole element within cos that is recognized as a specific IHF binding element, (iii) I1 is necessary and sufficient to afford a discrete, ostensibly strongly bent complex in the gel, regardless of duplex length, and (iv) the apparent affinity of IHF for the I1 element is strongly influenced by duplex length resulting from superimposed cooperative, nonspecific DNA binding interactions.
Dissection of cos-Specific and Nonspecific Binding Interactions Using Analytical Ultracentrifugation
The EMS studies presented above demonstrate that while IHF binds to the minimal I1 duplex to yield a distinct (specific) retarded band, nonspecific binding interactions are also observed in a similar concentration range (compare panels A and B of Figure 3). This feature precludes an accurate resolution of the specific I1 binding constant (Ksp) using the EMS data because the specific binding event is likely polluted with nonspecific binding interactions.26 We therefore turned to sedimentation velocity analytical ultracentrifugation (SV-AUC) to interrogate the thermodynamic features describing binding of IHF to the minimal 27 bp cos-specific (I1) and nonspecific (R3) duplexes.
The AUC studies utilized binding conditions identical to those used for the EMS experiments, except that the duplexes were labeled with 6-carboxyfluorescein (6-FAM). Sedimentation velocity data were collected using fluorescence optics to selectively monitor sedimentation of the DNA. The SV-AUC data were analyzed using Sedfit, which yields experimental sedimentation coefficients of 1.96 S for both the I1 and R3 duplexes (Figure 5A,B). Incremental addition of IHF to either duplex results in a progressive, concentration-dependent increase in s*, consistent with the formation of IHF·DNA nucleoprotein complexes of increasing mass. Importantly, binding of IHF to the I1 duplex is observed at concentrations well beyond that required to fully saturate the specific binding interaction observed in the EMS study [<50 nM (Figure 4B)]. This observation confirms that while EMS exposes the initial specific binding interaction, it does not unmask additional lower-affinity nonspecific binding to the duplex. In contrast, AUC additionally captures the nonspecific binding interactions, and the two experimental approachs are strongly complementary.
The individual c(s) distributions displayed in Figure 5 were integrated using Sedfit to obtain a weight-average sedimentation coefficient, ⟨s*⟩, which reflects the ensemble distribution of nucleoprotein complexes in solution. A plot of ⟨s*⟩ as a function of IHF concentration is shown in Figure 5C. Close inspection of the data reveals that while IHF binds to the nonspecific duplex (R3, black) in a monotonic manner, the binding curve for the specific duplex (I1, red) appears to be biphasic. This suggests that an additional, high-affinity binding interaction is superimposed on nonspecific DNA binding. We hypothesized that this high-affinity transition represents specific binding of IHF to the I1 element, which is not present in the R3 duplex; the second lower-affinity transition reflects nonspecific DNA interactions available to both duplexes. To directly test this hypothesis, we adapted the binding models developed by Record and co-workers26 to analyze the SV-AUC data as described in Experimental Procedures. To constrain the resolved parameters, we performed a global fit of the ensemble of AUC data as follows. First, the binding data were fit to the nonspecific finite lattice DNA binding model (eq 3a). While this analysis provides an excellent fit to the R3 binding data, the I1 binding isotherm is poorly described by this model (red dashed line, Figure 5C) and affords unrealistic binding parameters (not shown). We therefore analyzed the data using a more sophisticated approach; the R3 (nonspecific) binding data were fit to the nonspecific finite lattice DNA binding model (eq 3a), while the I1 (cos-specific) binding data were simultaneously fit to the competitive specific/nonspecific finite lattice DNA binding model (eq 3b). Importantly, the common parameters Kns and ω were allowed to float as global parameters. This approach results in an excellent fit to both data sets (Figure 5C); the monotonic interaction of IHF with nonspecific DNA is described well by the nonspecific finite lattice DNA binding model (black line), while the competitive specific/nonspecific finite lattice DNA binding model captures the biphasic transition of specific plus superimposed nonspecific binding events (solid red line). This analysis returns a nonspecific binding constant Kns of (1.7 ± 0.6) × 106 M–1 and a nearest neighbor cooperativity ω of 10 ± 4, which are in a good agreement with the EMS results (Table 1). Furthermore, because of the constraints imposed by global analysis of the data, the specific binding constant is also resolved with reasonable precision [Ksp = (2.0 ± 1.6) × 108 M–1]. This value is commensurate with those observed for binding of IHF to the high-affinity H′ element of attP despite the fact that the I1 element diverges from the cannonical high-affinity sequence.29,33 Moreover, the specificity parameter for binding of IHF to the I1 element (Ksp/Kns = 117) is harmonious with that observed for binding of IHF to H′ derived from calorimetry approaches.28 Finally, the analysis resolves the weight-average sedimentation coefficient for the nonspecific IHF·DNA complexes (⟨s*ns⟩ = 3.37 ± 0.04 S) and the sedimentation coefficient for the specific IHF·DNA binary complex (ssp = 2.59 ± 0.12 S). These experimentally resolved values are in good agreement with hydrodynamic modeling of both the specific complex (ssp = 2.36 S, based on the IHF·DNA crystal structure) and nonspecific complex [sns = 3.52 S, based on a structural model of IHF bound to the R3 duplex (Table 1)].
Multiple IHF Dimers Assemble on Duplex DNA To Afford Higher-Order Nucleoprotein Complexes
We next utilized SV-AUC to examine binding of IHF to the full-length cos274 and ns274 model duplexes. The SV-AUC data were analyzed using Sedfit, and the c(s) distributions are shown in Figure 6. Analysis of the data in the absence of IHF yields experimental sedimentation coefficients of 4.1 for both the cos274 and ns274 duplexes. Incremental addition of IHF to the specific cos274 duplex results in a progressive, concentration-dependent increase in the experimental sedimentation coefficient to a maximal value of ∼10 S (Figure 6A). Given the magnitude of the sedimentation coefficient and considering the high concentration of IHF required to reach saturation, it is likely that this complex reflects not only specific binding of IHF to the I1 element but also the assembly of multiple IHF dimers in a nonspecific manner. As noted above, these subsequent binding events are not detected in the EMS studies except at IHF concentrations of >1 μM.
Figure 6.
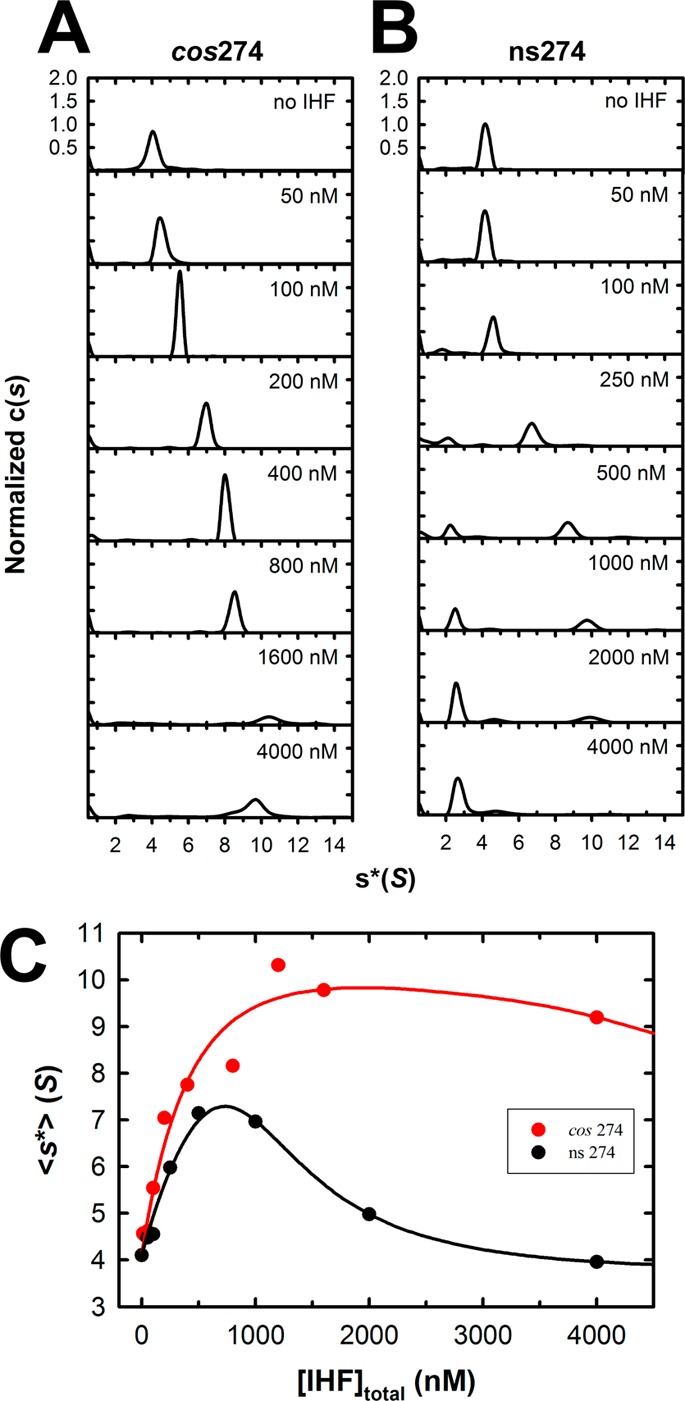
Interrogation of binding of IHF to full-length cos274 and ns274 duplex substrates using sedimentation velocity analytical ultracentrifugation (SV-AUC). Increasing concentrations of IHF were added to each duplex, and their sedimentation behavior was monitored by SV-AUC as described in Experimental Procedures. The c(s) distribution for each binding experiment were calculated using Sedfit. (A) Normalized c(s) profiles for the specific cos274 duplex. (B) Normalized c(s) profiles for the nonspecific ns274 duplex. (C) Weight-average sedimentation coefficients for each of the c(s) distributions shown in panel A (red circles, cos274) and panel B (black circles, ns274) were calculated using Sedfit and are plotted as a function of IHF concentration. The solid lines represent the best fits of simultaneous (global) analysis of the ensemble of binding data to the DNA unbending model (Scheme 1) as described in Supporting Information. The binding parameters derived from global analysis are presented in Table S2 of the Supporting Information.
A similar binding pattern is initially observed with the nonspecific ns274 substrate, and the evolution of an ∼10 S complex is apparent; however, this undergoes a transition to a distinct, very slowly sedimenting ∼2.5 S species that appears at IHF concentrations of ≥100 nM (Figure 6B). The appearance of two distinct peaks in the c(s) distribution suggests that interconversion between the two disparate conformations is slow relative to the time scale of the AUC experiment (hours).
The individual c(s) distributions were integrated using Sedfit to obtain a weight-average sedimentation coefficient, and a plot of ⟨s*⟩ as a function of IHF concentration is shown in Figure 6C. As expected, binding of IHF to ns274 (black circles) results in an initial increase in ⟨s*⟩ followed by a dramatic decrease, which reflects the concentration-dependent evolution of the 2.5 S species. A similar though less striking trend in ⟨s*⟩ is observed with the specific cos274 substrate (red circles). This phenomenon is discussed below.
Discussion
IHF plays an important role in a variety of biological processes, including condensation of the bacterial nucleoid and regulation of cellular processes such as DNA replication and transcription.6 In the former case, this involves binding to DNA in a nonspecific manner, resulting in a condensed and compacted nucleoprotein structure. In contrast, the regulatory duties of IHF require specific recognition of a high-affinity cognate binding site and introduction of a strong bend into the duplex; this provides an architecture conducive to the cooperative assembly of additional proteins at that site to engender functional, higher-order nucleoprotein complexes.5,6 The manner by which IHF fulfills these two disparate roles is of significant biological interest.
IHF also plays an important role in the development of several viruses for which lambda serves as a prototype. In this case, IHF binds specifically to the cos region of the lambda genome and cooperatively assembles the terminase packaging motor complex. Multiple putative I-elements have been identified within cos, but the data presented here demonstrate that IHF binds with high affinity to only I1. Importantly, IHF binds to I1-containing substrates to afford a discrete band on the EMS gel regardless of position within the duplex or duplex length. The intrinsic affinity for the minimal 27 bp I1 element is an order of magnitude lower than that observed with longer substrates, however, which is the result of superimposed cooperative binding of IHF in a nonspecific manner. EMS is able to discern the initial high-affinity, specific complex formed between IHF and I1-containing duplexes but does not readily resolve subsequent nonspecific binding events. In contrast, SV-AUC readily reveals these higher-order nucleoprotein complexes in a concentration-dependent manner. Thus, EMS and SV-AUC techniques provide a complementary view of these complex interactions, which has allowed a thermodynamic dissection of these two IHF binding modes using model cos substrates.
Model for a Nucleoid to Linear Rod Transition
While the original intent of this work was to characterize binding of IHF to cos as a foundation for the thermodynamic dissection of cooperative packaging motor assembly, the results have much broader implications. The EMS data indicate that IHF binds to nonspecific duplexes, yielding an increasingly smeared pattern that reflects the assembly of nucleoprotein complexes of increasing mass with increasing protein concentration. Consistently, the SV-AUC data show an analogous increase in ⟨s*⟩; however, the data for binding of IHF to the ns275 duplex are remarkable. The initial increase in ⟨s*⟩ is followed by a transition to a smaller 2.5 S species despite the fact that the mass of the complex continues to increase, as evidenced by the EMS data. The fact that the ⟨s*⟩ of the complex is smaller than that of free DNA suggests the former is fairly rigid in solution as compared to the wormlike nature of the unbound duplex.44 How can this be rationalized?
The sedimentation behavior of a particle in solution depends on not only its mass but also its shape
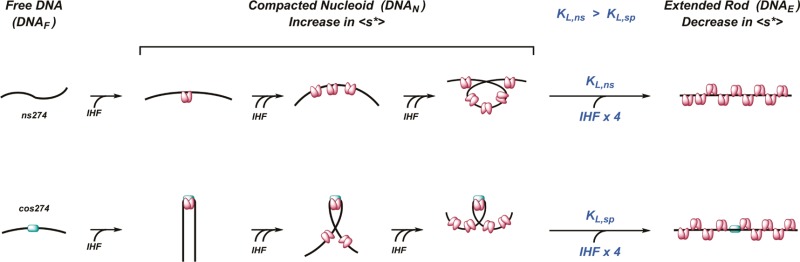
where f is the frictional coefficient of the complex. Compact particles of a given mass have a minimal frictional coefficient, whereas this value increases as the shape of the particle becomes more asymmetric. Thus, the most parsimonious interpretation of the SV-AUC data is that the 2.5 S species, while large in mass, is also significantly extended in shape. We suggest that the observed SV-AUC behavior reflects the histone-like properties of IHF. Initially, the protein binds to nonspecific DNA with modest cooperativity to afford condensed nucleoprotein complexes; this serves to compact the DNA in a sequence-independent fashion (see Scheme 1).5,6,37 The combined effect of increased mass and duplex compaction (small f) leads to an increase in ⟨s*⟩ up to a maximal value where the condensed nucleoid state is saturated with protein. The occluded site size in the condensed state is greater than that found in a linear complex (see Figure 1C).5,28 Thus, the duplex must undergo a transition to an extended rod conformation to accommodate a full complement of protein, a transition that is driven by the binding of additional IHF dimers. This transition significantly increases the frictional coefficient of the complex (large f), resulting in a concomitant decrease in ⟨s*⟩.
Scheme 1. Model for IHF Nucleoprotein Complexes.
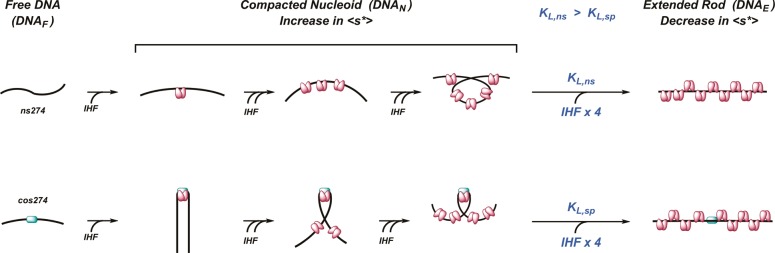
Details are provided in the text.
The model proposed above is consistent with electron microscopy studies that show IHF initially binds to DNA to afford compact nucleoids, but that these undergo a transition to rigid, extended rodlike structures with increasing concentrations of the protein.4 This feature is shared with other histone-like proteins, including HU38 and H-NS,39 and these extended nucleoprotein conformations have been proposed to play an important role in DNA organization within the nucleoid. Whatever the case, the solution-based AUC studies presented here nicely complement the structural EM studies, both of which indicate that elongated, nonspecific nucleoprotein complexes are formed at high IHF binding densities.
A Specific IHF Complex Inhibits the Nucleoid–Rod Transition
Close inspection of binding of IHF to the specific cos274 duplex reveals a similar trend in the AUC, an initial increase in ⟨s*⟩ followed by a decrease as a function of IHF concentration; however, the decrease is quite modest compared to that of the nonspecific duplex (see Figure 6C). We interpret these observations as follows; at low concentrations, IHF binds to the I1 element in cos274 to yield a specific, strongly bent complex (Scheme 1). This high-affinity binding event is not discerned during AUC because the mass change induced by binding the 20 kDa protein to the 169 kDa duplex is small.a This is followed by weak, nonspecific cooperative assembly of IHF on the duplex resulting in a concentration-dependent increase in ⟨s*⟩, as is observed with the ns274 duplex. At sufficiently elevated IHF concentrations (>1 μM), the observed ⟨s*⟩ declines as a result of a conformational change to the extended rod conformation. Importantly, the transition is strongly attenuated with the cos274 substrate as compared to the nonspecific ns274 duplex. This observation suggests that “unbending” of a duplex with a specifically bound IHF dimer is energetically more demanding than “unwrapping” of the compacted, nonspecific nucleoid complexes. In other words, the presence of a high-affinity, specifically bound IHF dimer presents a significant energy barrier to the transition. This interpretation is consistent with the EMS studies that show the specific bent IHF·cos-DNA complex remains unperturbed in the gel in the background of nonspecific binding up to 1 μM IHF while continuous smears are observed with nonspecific duplex substrates.
Energetic Analysis of the Rigid Rod Transition
A simple model that captures the hypotheses described above and describes the ensemble of nucleoprotein complexes present in solution during an IHF titration experiment is presented in Scheme 1. Initially, IHF binds weakly but cooperatively to nonspecific DNA to yield an ensemble of complexes that contain multiple copies of IHF (⟨DNAN⟩). At sufficiently high concentrations, the nucleoid is “unbent” to accommodate a full complement of IHF dimers, as described above. This equilibrium transition is described by an equilibrium constant (KL) and is driven by the binding of “m” IHF dimers to the saturated nucleoid (essentially a Hill binding model). We derive a mathematical expression for this “unbending” model, and the ensemble of experimental AUC data for binding of IHF to ns274 and cos274 DNA was globally fit to this model as described in the Supporting Information. While admittedly simplistic, the model describes the data remarkably well (see Figure 6C). The analysis predicts that (i) the nucleoid to rod transition is a cooperative process driven by the binding of at least four IHF dimers to the condensed nucleoid (“Hill” coefficient, m = 4) and (ii) “unbending” of a duplex with a specifically bound IHF dimer is energetically more demanding than “unwrapping” of the compacted, nonspecific nucleoid complexes. In other words, the presence of a specific IHF complex imparts a significant energy barrier to the extended rod transition [∼13-fold; ΔΔG ∼ 1.4 kcal/mol (Table S2 of the Supporting Information)]. The implications of this observation are discussed further below.
In summary, complementary EMS and AUC experiments combined with global analysis and mathematical modeling of the ensemble of data have allowed a constrained thermodynamic dissection of specific and nonspecific binding interactions of IHF with model DNA duplexes. The complementary techniques provide a powerful approach that can be applied to protein–DNA binding interactions in a general sense.
Biological Implications
Consistent with the data presented here, all published studies indicate that the affinity of IHF for its cognate element is in the low nanomolar range.5,20 In contrast, the concentration of IHF in an E. coli cell has been estimated to be ∼10 μM and up to 100 μM in exponentially growing and stationary phase cells, respectively.2,5,16,40 On the surface, this predicts that all of the specific DNA binding sites in a cell should be occupied by IHF at all times, a situation that belies its role as a regulator of nucleoprotein complex assembly; however, the relative concentration of nonspecific DNA provided by the E. coli genome must drive the bulk of intracellular IHF into nonspecific nucleoid complexes. Indeed, this has been observed experimentally,41 and the concentration of “free” IHF in the cell is estimated to be in the low nanomolar range.40,42 Record and co-workers have suggested that specific IHF·DNA complexes are more likely to form when the IHF concentration is limiting.28 The data presented here further suggest that specific, strongly bent regulatory complexes, once formed, are relatively stable in the background of excess nonspecific binding interactions. Furthermore, one must consider that IHF does not act in isolation, and its role is to provide a specific duplex architecture conducive to the assembly of additional regulatory proteins at that site. These cooperative interactions provide additional binding energy that serves to further stabilize the specific, higher-order nucleoprotein complexes. The corollary to this conclusion is that a specifically bound IHF dimer may impose an impediment to nucleosome assembly at that site, which would further promote the assembly of specific regulatory complexes in the cell. In summary, these features may allow the specific regulatory functions of IHF to be expressed in the background of a vast excess of nonspecific nucleoid condensation. This provides an ideal mechanism whereby IHF can fulfill its multiple and diverse roles within the cell: nonspecific nucleoid formation versus specific regulatory complex assembly.
The capacity of IHF to assemble specific nucleoprotein complexes in the context of a vast excess of nonspecific DNA similarly plays a critical role during viral infection. While the cos sequence represents less than 0.2% of the lambda genome length, IHF promotes site-specific assembly of the packaging complexes at that site. We previously demonstrated that IHF and the small subunit of lambda terminase cooperatively bind and bend cos-containing DNA duplexes.20 The functional motor complex is composed of eight copies of this small terminase subunit plus four copies of the large, catalytic subunit,15,18,21,22 and preliminary data indicate that IHF and the terminase subunits cooperatively assemble at cos (manuscript in preparation). The study presented here sets the stage for detailed biochemical interrogation of this complicated assembly process, and ongoing studies in our lab are directed at dissecting the thermodynamic parameters describing these essential steps in virus development.
Acknowledgments
Analytical ultracentrifugation studies were conducted at the University of Washington Center for Intracellular Delivery of Biologics. We thank Dr. John Sumida for guidance on the AUC fluorescence detection system and for helpful discussions. We are further indebted to Dr. Karl Maluf for helpful discussions and critical review of the manuscript.
Glossary
Abbreviations
- EMS
electrophoretic mobility shift
- AUC
analytical ultracentrifugation
- SE-AUC
sedimentation equilibrium analytical ultracentrifugation
- SV-AUC
sedimentation velocity analytical ultracentrifugation
- cos
cohesive end site of the lambda genome
- H-element
specific IHF recognition elements found within the synapse sites of the lambda and E. coli genomes
- I-element
specific IHF recognition elements found within the cos sequence of the lambda genome
- PDB
Protein Data Bank.
Supporting Information Available
Protocol for duplex substrate preparation, mathematical models for the Hill equation and for a nucleoid to linear rod transition, analysis of the duplex unbending data, hydrodynamic modeling of the specific and nonspecific IHF nucleoprotein complexes, and EMS data for binding of IHF to ns247 duplex. This material is available free of charge via the Internet at http://pubs.acs.org.
Author Contributions
S.J.S. and T.-C.Y. contributed equally to this work.
This work was supported by National Institutes of Health Grant 5R01GM088186 and Washington State Life Sciences Discovery Fund Grant 2496490.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Footnotes
Supplementary Material
References
- Azam T. A.; Ishihama A. (1999) Twelve Species of the Nucleoid-associated Protein from Escherichia coli: Sequence Recognition Specificity and DNA Binding Affinity. J. Biol. Chem. 274, 33105–33113. [DOI] [PubMed] [Google Scholar]
- Azam T. A.; Iwata A.; Nishimura A.; Udea S.; Ishihama A. (1999) Growth Phase-Dependent Variation in Protein Composition of the Escherichia coli Nucleoid. J. Bacteriol. 181, 6361–6370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drlica K.; Rouviere-Yaniv J. (1987) Histonelike Proteins of Bacteria. Microbiol. Rev. 51, 301–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar T.; Petrov A. S.; Vitko J. R.; Santai C. T.; Harvey S. C.; Mukerji I.; Hud N. V. (2009) Integration Host Factor (IHF) Dictates the Structure of Polyamine-DNA Condensates: Implications for the Role of IHF in the Compaction of Bacterial Chromatin. Biochemistry 48, 667–675. [DOI] [PubMed] [Google Scholar]
- Swinger K. K.; Rice P. A. (2004) IHF and HU: Flexible Architects of Bent DNA. Curr. Opin. Struct. Biol. 14, 28–35. [DOI] [PubMed] [Google Scholar]
- Friedman D. I. (1988) Integration host factor: A protein for all reasons. Cell 55, 545–554. [DOI] [PubMed] [Google Scholar]
- Arfin S. M.; Long A. D.; Ito E. T.; Tolleri L.; Riehle M. M.; Paegle E. S.; Hatfield G. W. (2000) Global Gene Expression Profiling in Escherichia coli K12: The Effects of Integration Host Factor. J. Biol. Chem. 275, 29672–29684. [DOI] [PubMed] [Google Scholar]
- Hwang D. S.; Kornberg A. (1992) Origin of the Replication Origin of Escherichia coli by DnaA Protein with Protein HU or IHF. J. Biol. Chem. 267, 23083–23086. [PubMed] [Google Scholar]
- Goodman S. D.; Nicholson S. C.; Nash H. A. (1992) Deformation of DNA during site-specific recombination of bacteriophage lambda: Replacement of IHF protein by HU protein or sequence-directed bends. Proc. Natl. Acad. Sci. U.S.A. 89, 11910–11914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig N. L.; Nash H. A. (1984) E. coli integration host factor binds to specific sites in DNA. Cell 39, 707–716. [DOI] [PubMed] [Google Scholar]
- Nash H. A.; Robertson C. A.; Flamm E.; Weisberg R. A.; Miller H. I. (1987) Overproduction of Escherichia coli integration host factor, a protein with nonidentical subunits. J. Bacteriol. 169, 4124–4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendelson I.; Gottesman M.; Oppenheim A. B. (1991) HU and Integration Host Factor Function as Auxiliary Proteins in Cleavage of Phage Lambda Cohesive Ends by Terminase. J. Bacteriol. 173, 1670–1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin W.; Cai Z. H.; Feiss M. (1993) Function of IHF in lambda DNA packaging. II. Effects of mutations altering the IHF binding site and the intrinsic bend in cosB on lambda development. J. Mol. Biol. 230, 505–515. [DOI] [PubMed] [Google Scholar]
- Yang Q.; Catalano C. E. (2003) Biochemical characterization of bacteriophage lambda genome packaging in vitro. Virology 305, 276–287. [DOI] [PubMed] [Google Scholar]
- Maluf N. K.; Yang Q.; Catalano C. E. (2005) Self-association properties of the bacteriophage lambda terminase holoenzyme: Implications for the DNA packaging motor. J. Mol. Biol. 347, 523–542. [DOI] [PubMed] [Google Scholar]
- Gaussier H.; Yang Q.; Catalano C. E. (2006) Building a virus from scratch: Assembly of an infectious virus using purified components in a rigorously defined biochemical assay system. J. Mol. Biol. 357, 1154–1166. [DOI] [PubMed] [Google Scholar]
- Yang Q.; Catalano C. E.; Maluf N. K. (2009) Kinetic Analysis of the Genome Packaging Reaction in Bacteriophage Lambda. Biochemistry 48, 10705–10715. [DOI] [PubMed] [Google Scholar]
- Andrews B. T.; Catalano C. E. (2012) The Enzymology of a Viral Genome Packaging Motor is Influenced by the Assembly State of the Motor Subunits. Biochemistry 51, 9342–9353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalano C. E. (2005) Viral Genome Packaging Machines: An Overview. In Viral Genome Packaging Machines: Genetics, Structure, and Mechanism (Catalano C. E., Ed.) pp 1–4, Kluwer Academic/Plenum Publishers, New York. [Google Scholar]
- Ortega M. E.; Catalano C. E. (2006) Bacteriophage lambda gpNu1 and Escherichia coli IHF proteins cooperatively bind and bend viral DNA: Implications for the assembly of a genome-packaging motor. Biochemistry 45, 5180–5189. [DOI] [PubMed] [Google Scholar]
- Maluf N. K.; Gaussier H.; Bogner E.; Feiss M.; Catalano C. E. (2006) Assembly of bacteriophage lambda terminase into a viral DNA maturation and packaging machine. Biochemistry 45, 15259–15268. [DOI] [PubMed] [Google Scholar]
- Andrews B. T.; Catalano C. E. (2013) Strong Subunit Coordination Drives a Powerful Viral DNA Packaging Motor. Proc. Natl. Acad. Sci. U.S.A. 110, 5909–5914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stothard P. (2000) The Sequence Manipulation Suite: JavaScript programs for analyzing and formatting protein and DNA sequences. Biotechniques 28, 1102–1104. [DOI] [PubMed] [Google Scholar]
- Chang J. R.; Andrews B. T.; Catalano C. E. (2012) Energy Independent Helicase Activity of a Viral Genome Packaging. Biochemistry 51, 391–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGhee J. D.; von Hippel P. H. (1974) Theoretical Aspects of DNA-Protein Interactions: Co-operative and Non-co-operative Binding of Large Ligands to a One-dimensional Homogeneous Lattice. J. Mol. Biol. 86, 469–489. [DOI] [PubMed] [Google Scholar]
- Tsodikov O. V.; Holbrook J. A.; Shkel I. A.; Record M. T. Jr. (2001) Analytic Binding Isotherms Describing Competitive Interactions of a Protein Ligand with Specific and Nonspecific Sites on the Same DNA Oligomer. Biophys. J. 81, 1960–1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T.-C.; Maluf N. K. (2014) Characterization of the non-specific DNA binding properties of the adenoviral IVa2 protein. Biophys. Chem. 193–194, 1–8. [DOI] [PubMed] [Google Scholar]
- Holbrook J. A.; Tsodikov O. V.; Saecker R. M.; Record M. T. Jr. (2001) Specific and non-specific interactions of integration host factor with DNA: Thermodynamic evidence for disruption of multiple IHF surface salt-bridges coupled to DNA binding. J. Mol. Biol. 310, 379–401. [DOI] [PubMed] [Google Scholar]
- Aeling K. A.; Opel M. L.; Steffen N. R.; Tretyachenko-Ladokhina V.; Hatfield G. W.; Lathrop R. H.; Senear D. F. (2006) Indirect recognition in sequence-specific DNA binding by Escherichia coli integration host factor: The role of DNA deformation energy. J. Biol. Chem. 281, 39236–39248. [DOI] [PubMed] [Google Scholar]
- Schuck P. (2000) Size-Distribution Analysis of Macromolecules by Sedimentation Velocity Ultracentrifugation and Lamm Equation Modeling. Biophys. J. 78, 1606–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanyal S. J. (2013) Cooperative Assembly of Terminase and Integration Host Factor at the Packaging Initiation Site of Bacteriophage Lambda. In Department of Medicinal Chemistry, p 167, University of Washington, Seattle. [Google Scholar]
- Dhavan G. M.; Crothers D. M.; Chance M. R.; Brenowitz M. (2002) Concerted Binding and Bending of DNA by Escherichia coli Integration Host Factor. J. Mol. Biol. 315, 1027–1037. [DOI] [PubMed] [Google Scholar]
- Xin W. N.; Feiss M. (1988) The interaction of Escherichia coli integration host factor with the cohesive end sites of phages lambda and 21. Nucleic Acids Res. 16, 2015–2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin W.; Feiss M. (1993) Function of IHF in lambda DNA packaging. I. Identification of the strong binding site for integration host factor and the locus for intrinsic bending in cosB. J. Mol. Biol. 230, 492–504. [DOI] [PubMed] [Google Scholar]
- Morse B. K.; Michalczyk R.; Kosturko L. D. (1994) Multiple molecules of integration host factor (IHF) at a single DNA binding site, the bacteriophage λ cos I1 site. Biochimie 76, 1005–1017. [DOI] [PubMed] [Google Scholar]
- Thompson J. F.; Landy A. (1988) Empirical Estimation of Protein-Induced DNA Bending Angles: Aplications to Lambda Site-specific Recombination Complexes. Nucleic Acids Res. 16, 9687–9705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khrapunov S.; Brenowitz M.; Rice P. A.; Catalano C. E. (2006) Binding then bending: A mechanism for wrapping DNA. Proc. Natl. Acad. Sci. U.S.A. 103, 19217–19218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh J.; Saecker R. M.; Record M. T. Jr. (2008) DNA Binding Mode Transitions of Escherichia coli HUαβ: Evidence for Formation of a Bent DNA–Protein Complex on Intact, Linear Duplex DNA. J. Mol. Biol. 383, 324–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.; Chen H.; Kenney L. J.; Yan J. (2010) A divalent switch drives H-NS/DNA-binding conformations between stiffening and bridging modes. Genes Dev. 24, 339–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murtin C.; Engelhorn M.; Geiselmann J.; Frédéric B. (1989) A Quantitative UV Laser Footprinting Analysis of the Interaction of IHF with Specific Binding Sites: Re-evaluation of the Effective Concentration of IHF in the Cell. J. Mol. Biol. 284, 949–961. [DOI] [PubMed] [Google Scholar]
- Azam T. A.; Hiraga S.; Ishihama A. (2000) Two types of localization of the DNA-binding proteins within the Escherichia coli nucleoid. Genes Cells 5, 613–626. [DOI] [PubMed] [Google Scholar]
- Yang S. W.; Nash H. A. (1995) Comparison of protein binding to DNA in vivo and in vitro: Defining an effective intracellular target. EMBO J. 14, 6292–6300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice P. A.; Yang S.-W.; Mizuuchi K.; Nash H. A. (1996) Crystal Structure of an IHF-DNA Complex: A Protein-Induced DNA U-Turn. Cell 87, 1295–1306. [DOI] [PubMed] [Google Scholar]
- Koenderink G. H.; Planken K. L.; Roozendaal R.; Philipse A. P. (2005) Monodisperse DNA restriction fragments: II. Sedimentation velocity and equilibrium experiments. J. Colloid Interface Sci. 291, 126–134. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.