Alternative splicing of human NT5E generates CD73S, an endoplasmic reticulum–associated and dimerization-deficient glycoprotein that lacks enzymatic activity. CD73S functions as a negative regulator of canonical CD73 by promoting its proteasomal degradation, which may have significance in chronic liver disease and liver cancer.
Abstract
Ecto-5′-nucleotidase (CD73), encoded by NT5E, is the major enzymatic source of extracellular adenosine. CD73 controls numerous pathophysiological responses and is a potential disease target, but its regulation is poorly understood. We examined NT5E regulation by alternative splicing. Genomic database analysis of human transcripts led us to identify NT5E-2, a novel splice variant that was expressed at low abundance in normal human tissues but was significantly up-regulated in cirrhosis and hepatocellular carcinoma (HCC). NT5E-2 encodes a shorter CD73 isoform we named CD73S. The presence of CD73S protein, which lacks 50 amino acids, was detected in HCC using an isoform-specific antibody. A noncanonical mouse mRNA, similar to human CD73S, was observed, but the corresponding protein was undetectable. The two human isoforms exhibited functional differences, such that ectopic expression of canonical CD73 (CD73L) in human HepG2 cells was associated with decreased expression of the proliferation marker Ki67, whereas CD73S expression did not have an effect on Ki67 expression. CD73S was glycosylated, catalytically inactive, unable to dimerize, and complexed intracellularly with the endoplasmic reticulum chaperone calnexin. Furthermore, CD73S complexed with CD73L and promoted proteasome-dependent CD73L degradation. The findings reveal species-specific CD73 regulation, with potential significance to cancer, fibrosis, and other diseases characterized by changes in CD73 expression and function.
INTRODUCTION
Ecto-5′-nucleotidase (CD73) is a glycosyl-phosphatidylinositol–linked plasma membrane glycoprotein that is expressed on multiple cell types and in different tissues (Misumi et al., 1990; Resta et al., 1993; Knapp et al., 2012). CD73 catalyzes the conversion of extracellular AMP to adenosine (Resta et al., 1998; Zimmermann et al., 2012). Adenosine controls numerous physiological responses by activating one of four subtypes of G-protein–coupled receptors (Antonioli et al., 2013a; Chen et al., 2013). By virtue of its role as the major extracellular source of adenosine, CD73 has emerged as an important regulator of tissue homeostasis and pathophysiologic responses related to immunity, inflammation, pain, ischemia, tissue fibrosis, and cancer (Colgan et al., 2006; Zylka, 2011; Beavis et al., 2012; Antonioli et al., 2013b; Roberts et al., 2013). The only human disease linked to mutations in NT5E (the CD73-encoding gene) is calcification of joints and arteries (CALJA), an adult-onset condition characterized by joint pain (St Hilaire et al., 2011). The mechanism of how loss of CD73 function is related to the pathogenesis of this disease is not known, although the disease-associated CD73 isoforms appear to have trafficking defects (Fausther et al., 2014).
In addition to its direct genetic link to a human disease, there is evidence for the involvement of CD73 in cancer. For example, there is a positive correlation in triple-negative breast cancer between CD73 transcript levels and poor prognosis, as well as to resistance to chemotherapy (Loi et al., 2013). The findings in human breast cancer correlate with mouse models that have demonstrated limited tumor growth and metastasis upon ablation of CD73 function (Stagg et al., 2010, 2011; Wang et al., 2011; Allard et al., 2014). The apparent tumor-promoting roles of CD73 appear to be linked to its ability to generate adenosine, which acts as a potent suppressor of antitumor T-cell responses (Zhang, 2010; Beavis et al., 2012; Sitkovsky et al., 2014). However, nonenzymatic CD73 functions, which are poorly understood, may also be involved (Mikhailov et al., 2008; Terp et al., 2013).
Aside from its well-known roles in cancer and immunity, CD73 has recently emerged as a novel regulator of disease pathology in multiple liver disease models in mice. Specifically, CD73 promotes drug-induced hepatocellular injury and Mallory–Denk body inclusion formation (Snider et al., 2013), fibrosis (Peng et al., 2008; Fausther et al., 2012), and steatosis (Peng et al., 2009). In contrast to the animal models, dramatic decreases in NT5E mRNA are observed in human hepatitis C (HCV) and nonalcoholic fatty liver disease (NAFLD) livers (Snider et al., 2013), which suggests potential species-specific differences in CD73 regulation and/or function.
These observations, in combination with the fact that relatively little is known regarding CD73 regulation, prompted us to address the potential existence of alternative splice variants that may encode CD73 with different functions. Because alternative splicing is known to be a major driver for species-specific gene regulation (Barbosa-Morais et al., 2012; Merkin et al., 2012), understanding the major differences between CD73 in preclinical models and humans should help facilitate the translation of basic findings into the clinic. In the present study, we report on the identification of CD73S, a functionally distinct isoform of CD73 that is generated by alternative splicing of NT5E in the setting of human liver cirrhosis and hepatocellular carcinoma.
RESULTS
Human NT5E is regulated by alternative splicing in cirrhosis and hepatocellular carcinoma
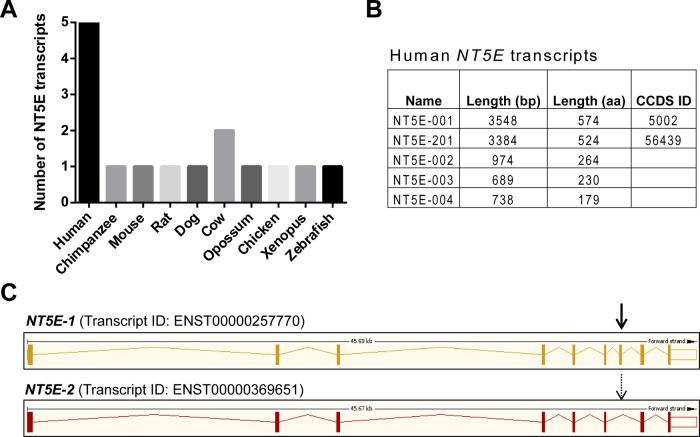
Using the Ensembl genome browser (Flicek et al., 2014), we compared the alternative splicing pattern of human NT5E to nine other vertebrate species. Whereas human NT5E encodes five transcripts, all other species, except for cow, are predicted to have one NT5E transcript (Figure 1A). Aside from NT5E-001, which encodes the full-length canonical CD73 protein, NT5E-201 (Figure 1B) is the only other human transcript annotated by the Collaborative Consensus Coding Sequence (CCDS) project (Harte et al., 2012) as being highly likely to encode a protein. Therefore we focused our subsequent analysis on comparing these two transcripts, and we refer to NT5E-001 and NT5E-201 as NT5E-1 and NT5E-2, respectively. The only difference between the two transcripts is the absence of exon 7 in NT5E-2 (Figure 1C).
FIGURE 1:
The human NT5E gene is regulated by alternative splicing. (A) Species comparison of the number of NT5E predicted transcripts using the Ensembl database. (B) Human NT5E has five splice variants, all predicted to be protein coding (bp, base pairs; aa, amino acids), but only two are annotated by CCDS. (C) The CCDS-validated transcripts NT5E-001 (NT5E-1) and NT5E-201 (NT5E-2), which differ with respect to the presence of exon 7 (arrow) in NT5E-1, are shown schematically.
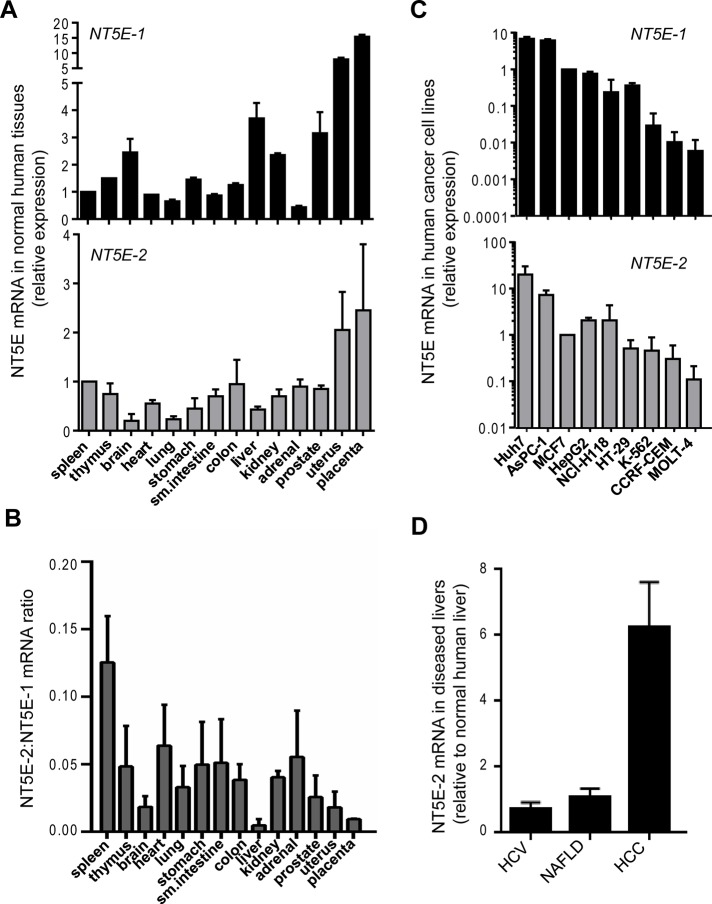
We found that the two transcripts had comparable relative distribution profiles across 14 normal human tissues (Figure 2A), but NT5E-2 was expressed at significantly lower levels than NT5E-1 (Figure 2B). We also evaluated the relative expression of these two isoforms in nine human cancer cell lines (Figure 2C) and found that the two hepatocellular carcinoma (HCC) cell lines Huh7 and HepG2 expressed among the highest levels of the two transcripts relative to the other tested cell lines (AsPC-1, MCF7, NCI-H118, HT-29, K-562, CCRF-CEM, and MOLT-4).Therefore we focused our subsequent analysis on NT5E expression in normal and diseased livers. Because alternative splicing of genes is known to be altered in disease states (David and Manley, 2010; Singh and Cooper, 2012), we evaluated the expression of NT5E-2 in chronic human liver diseases, including HCV, NAFLD, and HCC. Whereas expression of NT5E-2 did not differ significantly in HCV and NAFLD compared with normal livers, it was dramatically increased in HCC surgical specimens (Figure 2D), pointing to a disease-specific regulation. Furthermore, relative to normal human liver, the expression levels of the NT5E-2 transcript were increased by one to two orders of magnitude in the HCC cell lines (Figure 3A), whereas NT5E-1 expression was either unchanged (Huh7) or decreased (HepG2).
FIGURE 2:
Comparison of the mRNA levels of NT5E-1 and NT5E-2 in normal human tissues, cancer cell lines, and diseased human livers. (A) NT5E-1 (NM_002525) and NT5E-2 (NM_001204813) mRNA levels in different normal human tissues (fold change over spleen, which is set to 1). (B) Relative abundance of NT5E-2 to NT5E-1 in normal human tissues. (C) Relative abundance of NT5E-1 and NT5E-2 in different human cancer cell lines (MCF7 = 1). (D) Comparison of NT5E-2 mRNA in livers of patients with HCV (n = 4), NAFLD (n = 4), and HCC (n = 6) to normal human livers (n = 2).
FIGURE 3:
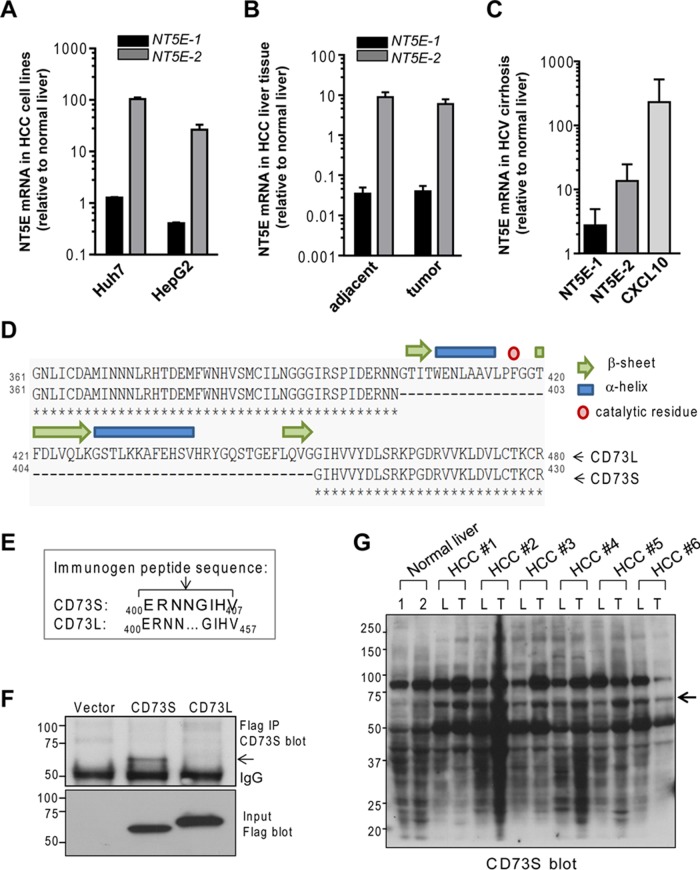
NT5E-2 is up-regulated in cirrhosis, and HCC and encodes a shorter CD73 protein (CD73S), which is functionally distinct from canonical CD73 (CD73L). (A–C) Relative expression of NT5E-1 and NT5E-2 mRNA in HCC cell lines (A), tumors and adjacent nontumor tissue from HCC surgical specimen obtained from six patients (B), and biopsies from patients with HCV-associated cirrhosis of the liver (C; clinical information on the human HCC and cirrhosis samples provided in Supplemental Table S2). CXCL10 is included as a positive control for HCV cirrhosis samples (Brownell and Polyak, 2013). (D) Protein sequence alignment of the C-termini of CD73L (NP_002517) and CD73S (NP_001191742). The 50 residues (404–453) missing in CD73S form three β-strands and two α-helices and include a catalytic residue (Phe-417). (E) Sequence of the synthetic peptide (ERNNGIHV) used to generate rabbit anti-CD73S antibodies. (F) Detection of total Flag-CD73S and Flag-CD73L protein (bottom) and validation of the CD73S antibody reactivity in Flag immunoprecipitates of transfected HEK293T cell lysates. CD73S and CD73L have predicted molecular weight of 58 and 63 kDa, respectively, but migrate at ∼67 and ∼72 kDa because of glycosylation and the Flag tag. (G) CD73S immunoblot of total tissue lysates from two normal human livers (1, 2) and six HCC paired tumors (T) and adjacent uninvolved liver (L) tissues (HCC 1–6; same as those used in B).
Given these findings, we compared the regulation of NT5E-1/NT5E-2 in HCC tumors and adjacent nontumor liver tissues using an independent set of surgical specimens from those shown in Figure 2D. Again, we noted a significant (sixfold to eightfold) increase in NT5E-2 in HCC tumors, which was similar to the adjacent uninvolved livers (Figure 3B). In contrast, NT5E-1 mRNA was dramatically decreased (by >90%) in HCC tumors and adjacent livers relative to normal livers (Figure 3B). Because up-regulation of NT5E-2 was also observed in the adjacent nontumor tissue, which is frequently cirrhotic, we analyzed NT5E expression in liver biopsies from patients with confirmed HCV-related cirrhosis (in the absence of HCC) and found that NT5E-2 was elevated in these livers (Figure 3C) to an extent similar to what is seen in HCC (Figure 3B), thereby indicating that NT5E-2 up-regulation occurs before HCC development in the context of cirrhosis.
Alternative splicing of NT5E generates CD73S, a functionally distinct CD73 protein
The NT5E-2 transcript is predicted to encode a shorter protein, which we termed CD73S (S, short; Figure 3D). NT5E-1 encodes canonical CD73, which we refer to as CD73L (L, long). Human CD73S lacks amino acids 404–453, encoded by the missing exon 7 and located in the C-terminus. The C-terminus contains the substrate-binding domain and the interface important for forming the functional CD73 homodimer expressed at the cell surface (Knapp et al., 2012). Based on this, CD73S is predicted to have compromised activity as an AMPase because it lacks a critical catalytic residue (Phe-417). Furthermore, the missing secondary structure (three β-strands and two α-helices; Figure 3D) suggests that this protein would have different folding and dimerization properties.
To assess the presence of CD73S in HCC, we generated a rabbit antibody directed to the peptide ERNNGIHV representing the unique junction present in CD73S but not CD73L (Figure 3E). DNA encoding Flag epitope-tagged versions of CD73L and CD73S or control vector was transfected into HEK293T cells, and the presence of both proteins was assessed by a Flag immunoblot (Figure 3F). This validated that NT5E-2 encodes the shorter (∼67 kDa) CD73S compared with CD73L (∼72 kDa) protein. Furthermore, the rabbit anti-CD73S antibody selectively recognized CD73S in the overexpression system (Figure 3F, top) and detected an ∼67-kDa protein in HCC but not normal livers (Figure 3G, arrow), although the antibody also exhibited nonspecific binding to other proteins in normal and HCC tissue lysates. Blotting with preimmune rabbit serum did not reveal differences between normal and HCC livers (Supplemental Figure S1). In agreement with the mRNA results (Figure 3B), both the HCC tumors and adjacent livers expressed the 67-kDa protein (Figure 3G), which we predict is CD73S.
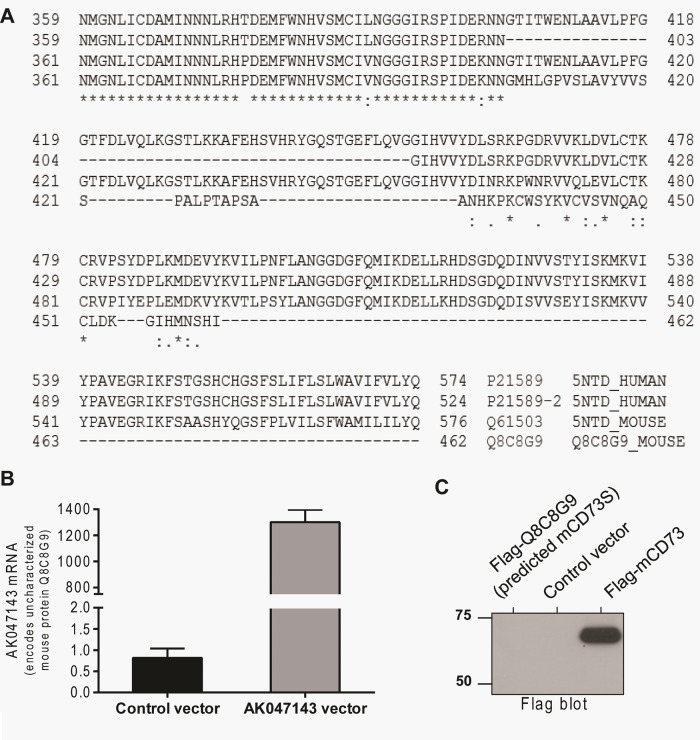
We also assessed the expression of a putative mouse protein (Uniprot ID Q8C8G9) that shares some sequence similarity to human CD73S (Figure 4A). Despite robust mRNA detection upon overexpression of the corresponding cDNA in primary mouse hepatocytes, we did not detect any protein (Figure 4, B and C). Using a different cell host (BHK-21 cells), addition of a proteasome inhibitor and analysis of the cell culture medium also did not reveal protein expression in the cells or the culture medium (unpublished data), suggesting that CD73S is a human-specific variant.
FIGURE 4:
Lack of evidence for mouse CD73S protein expression. (A) Sequence alignment of the C-termini of human CD73L (Uniprot ID P21589), human CD73S (P21589-2), mouse CD73 (Q61503), and a putative mouse protein (Q8C8G9), which shares sequence similarity to the human CD73S variant. (B) Overexpression of AK047143, which encodes the putative protein Q8C8G9, results in robust mRNA expression in primary mouse hepatocytes. (C) Flag immunoblot of lysates from transfected mouse hepatocytes demonstrating lack of Q8C8G9 protein expression (left lane).
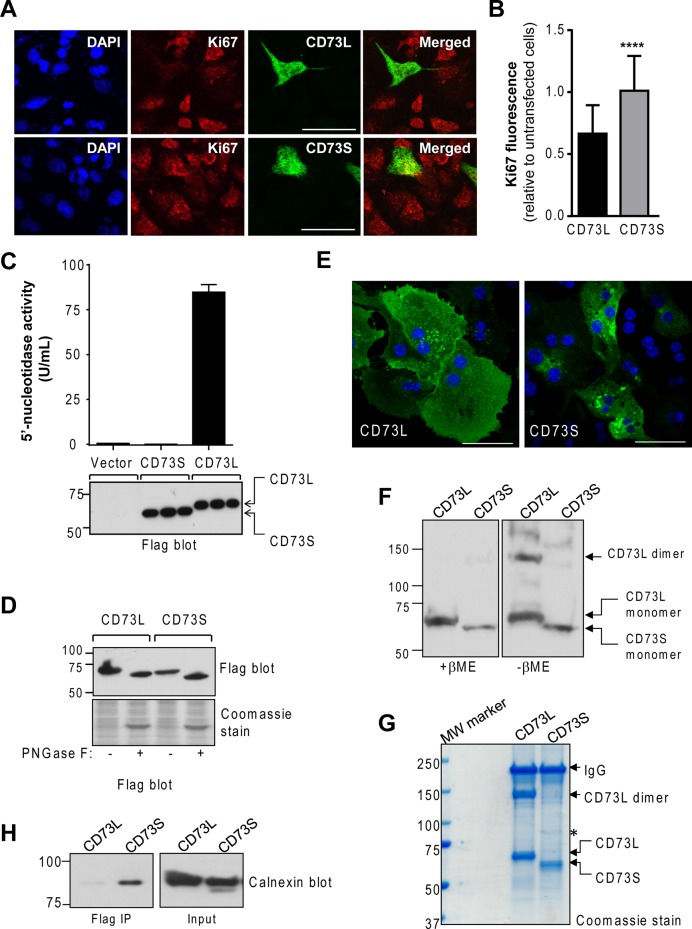
To determine whether CD73S and CD73L exhibit functional differences, we transfected each isoform into HepG2 cells and monitored the expression of the cell proliferation marker Ki67. The cells expressing CD73L had a significant decrease in Ki67 staining, whereas CD73S expression did not significantly alter Ki67 expression relative to neighboring untransfected cells (Figure 5, A and B). Therefore CD73L and CD73S exhibit functional differences with respect to their ability to modulate the proliferation potential of cultured human HCC cells.
FIGURE 5:
Molecular and functional characteristics of CD73S. (A) Coimmunofluorescence analysis of the proliferation marker Ki67 (red) and Flag-CD73S/L (green) in transfected HepG2 cells. Blue, 4′,6-diamidino-2-phenylindole (DAPI); bar, 20 μm. (B) Quantification of Ki67 immunofluorescence staining in CD73-expressing cells relative to neighbor untransfected cells (30 CD73+ cells and 150 CD73− untransfected cells were counted per condition). ****p < 0.0001, unpaired t test. (C) Measurement of 5′-nucleotidase activity in transfected HEK293T cell lysates. Samples expressing equal protein levels (bottom) were analyzed in triplicate. (D) Treatment of CD73-transfected HEK293T cell lysates with peptide-N-glycosidase F results in similar deglycosylation of both isoforms. (E) Immunofluorescence-based localization of Flag-CD73L or Flag-CD73S in primary mouse hepatocytes. Blue, DAPI; bar, 20 μm. (F) Flag immunoblot of Flag-CD73L– and -CD73S–expressing HEK293T total cell lysates analyzed under reducing (+β-mercaptoethanol, βME) or nonreducing (–βME) conditions, showing that CD73S does not dimerize. (G) Coomassie-stained gel of Flag immunoprecipitates of the nonreduced samples shown in F. Asterisk denotes a unique protein present in CD73S immunoprecipitates, identified by mass spectrometry as calnexin. (H) Biochemical validation that CD73S coimmunoprecipitates with calnexin.
CD73S is a dimerization-deficient, endoplasmic reticulum–associated glycoprotein that lacks 5′-nucleotidase activity
To understand the reason behind the functional differences between CD73S and CD73L in the HepG2 cells, we analyzed the molecular characteristics of CD73S. As predicted from the protein sequence, we found that CD73S completely lacks 5′-nucleotidase activity (Figure 5C). The glycosylation sites on CD73, Asn-311/333/403; Chen et al., 2009), are present in both isoforms and, consistent with this, treatment with peptide-N-glycosidase F resulted in deglycosylation of both isoforms (Figure 5D). Of note, upon expression in primary mouse hepatocytes, human CD73L exhibited plasma membrane and intracellular distribution, whereas human CD73S was primarily intracellular (Figure 5E). Another major difference between CD73L and CD73S was the lack of disulfide-linked homodimer formation in the latter, as determined by immunoblot and Coomassie stain of Flag-CD73–transfected cell lysates and Flag immunoprecipitates under reducing and nonreducing conditions (Figure 5, F and G). The Coomassie stain further revealed the presence of an ∼90-kDa protein that coimmunoprecipitated with CD73S but not CD73L (Figure 5G, asterisk). This protein was identified by mass spectrometry (not shown) as the endoplasmic reticulum (ER) chaperone calnexin, which was validated biochemically (Figure 5H). Coimmunofluorescence staining of CD73S and another ER marker protein, PDI, also revealed colocalization (Supplemental Figure S2A). These data demonstrate that CD73S has molecular and enzymatic properties that are significantly different from those of CD73L.
CD73S negatively regulates CD73L activity and protein expression in a proteasome-dependent manner
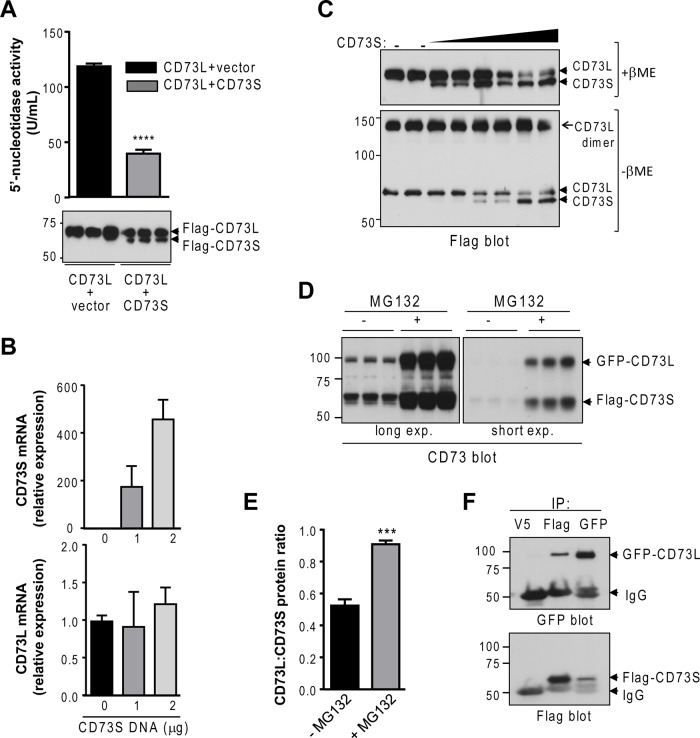
To assess whether there is cross-regulation between the two human CD73 isoforms, we transfected equal amounts of DNA encoding CD73L in combination with either control vector or CD73S and found that the presence of CD73S caused >50% decrease in the 5′-nucleotidase activity of CD73L, which correlated with decreased CD73L protein expression (Figure 6A). This effect was specific to CD73S, since control overexpression of excess Flag-GAPDH did not result in the same decrease in CD73L protein levels (Supplemental Figure S2B), and the effect was observed regardless of the epitope tag, since GFP-CD73L was similarly down-regulated in the presence of CD73S (Supplemental Figure S2C). Furthermore, the inhibitory effect of CD73S was not limited to the ectonucleotidase CD73L, since CD73S also down-regulated the expression of the cytosolic 5′-nucleotidase IA in a co-overexpression system (Supplemental Figure S2D).
FIGURE 6:
CD73S negatively regulates CD73L expression in a proteasome-dependent manner. (A) Measurement of 5′-nucleotidase activity in lysates from HEK293T cells transfected with equal amount of Flag-CD73L cDNA together with empty vector or Flag-CD73S. ****p < 0.0001, unpaired t test. (B) Measurement of CD73L (endogenous) and CD73S (transfected) mRNA in Huh-7 cells, showing that CD73S does not affect CD73L mRNA. (C) Determination of the effect of increasing levels of CD73S on CD73L dimer formation, which is detected under nonreducing conditions (–βME). (D) CD73 immunoblot of CD73-transfected HEK-293T lysates (±MG132). The same membrane under two exposure times from a triplicate experiment is shown. (E) Quantification of blot from D, showing that MG132 restores CD73L protein levels. ***p < 0.001, unpaired t test. (F) Reciprocal coimmunoprecipitation of CD73S/L-transfected HEK293T lysates, demonstrating that CD73S complexes with CD73L. Anti-V5 antibody was used as a negative control.
The inhibitory effect of CD73S on CD73L expression did not involve decreased mRNA, since the levels of endogenous CD73L mRNA in the presence of CD73S overexpression were unchanged relative to control levels (Figure 6B); similar results were obtained when both isoforms were co-overexpressed (unpublished data). In addition, the presence of increasing levels of CD73S protein did not significantly affect the expression of the CD73L dimer (Figure 6C), which is typically found at the cell surface, suggesting that CD73S inhibits CD73L protein expression most likely during processing in the ER. To that end, addition of MG132 restored CD73L protein in the presence of CD73S (Figure 6, D and E), indicating that CD73S promotes proteasome-dependent degradation of CD73L. Reciprocal coimmunoprecipitation of the two isoforms revealed that a fraction of CD73L is associated in a complex with CD73S (Figure 6F), indicating that CD73L degradation may result from an interaction between the two CD73 isoforms. Collectively these data demonstrate that CD73S functions as a negative regulator of CD73L protein expression, and possibly cytoplasmic nucleotidases as well.
DISCUSSION
Alternative splicing as a mechanism for species-specific CD73 regulation
We demonstrate that alternative splicing is a novel mode of human CD73 regulation. Alternative splicing of pre-mRNA is a mechanism by which multiple mRNA and protein isoforms are generated from a limited set of genes, which promotes functional complexity of organisms (Matera and Wang, 2014). Important protein properties, including intracellular localization, posttranslational modifications, turnover, enzymatic activity, and the shaping of signaling networks, are dictated by alternative splicing; it is estimated that >90% of multiexon human genes are subject to such regulation (Barbosa-Morais et al., 2012; Merkin et al., 2012; Singh and Cooper, 2012). Cross-species comparisons using RNA-Seq revealed that, unlike tissue-specific gene expression, which is largely conserved, alternative splicing differs significantly between species (Barbosa-Morais et al., 2012). Therefore alternative splicing of human NT5E may contribute to some of the previously reported differences between human and mouse CD73. For example, CD73 is abundantly expressed on afferent lymphatic vessels in humans but not in mice (Algars et al., 2011), whereas the opposite pattern has been observed in regulatory T-cells (Toth et al., 2013). Given the important biological functions of CD73 and its potential as a disease target, better understanding of species-specific CD73 regulation could facilitate translation of preclinical findings to the clinic.
Alternative splicing as a mechanism for CD73 regulation in human diseases
Our findings show that NT5E-2 mRNA and its protein product, CD73S, are not abundant in normal human tissues but are induced in HCV-associated cirrhosis and HCC. Pre-mRNA splicing factors and events are known to be deregulated in human cancers (David and Manley, 2010; Singh and Cooper, 2012; De Craene and Berx, 2013), including HCC (Chan et al., 2014; Qi et al., 2014). For example, increased activities of the splicing factors CUGBP1, hnRNPH, hnRNPA1, hnRNPA2B1, and SF2/ASF are believed to contribute to a switch in insulin receptor (IR) isoform expression in HCC (Chettouh et al., 2013). IR-B, which mediates the metabolic effects of insulin, is the predominant isoform in normal adult liver. However, during hepatocyte neoplastic transformation, IR expression switches to the IR-A isoform, which promotes cell proliferation (Chettouh et al., 2013). Another example is the expression of a metastasis-promoting, N-terminally truncated isoform of carboxypeptidase E (CPE), which is generated by alternative splicing of the CPE gene in HCC and other cancers (Lee et al., 2011). Nothing is known regarding the involvement of CD73 in progression to cirrhosis and HCC development. Studies in mouse models demonstrated a promoting role of CD73 in hepatocellular injury (Snider et al., 2013), fibrosis (Peng et al., 2008; Fausther et al., 2012), and steatosis (Peng et al., 2009), with the last two likely involving adenosine receptor signaling. However, NT5E-1 is dramatically decreased in HCV and NAFLD (Snider et al., 2013) as well as HCC (this study). In light of the functional differences between CD73S and CD73L, any association between NT5E expression and human disease outcomes should be carefully interpreted if the data were generated using microarray platforms (e.g., Affymetrix) that do not distinguish between NT5E-1 and NT5E-2 expression.
Regulation of CD73L protein levels via CD73S-CD73L complex formation
The association of CD73 with the ER chaperone calnexin, combined with its intracellular localization, suggests that CD73S may be recognized by the ER quality control system as a misfolded glycoprotein (Vembar and Brodsky, 2008). Furthermore, the ability of CD73S to complex with CD73L and promote CD73L degradation in a proteasome-dependent manner provides a novel mechanism for regulation of CD73 protein levels. It remains to be determined whether NT5E-2 mRNA and CD73S protein induction is associated with CD73 decreases observed in other cancers, such as malignant melanoma (Wang et al., 2012), in which methylation-dependent transcriptional silencing of NT5E appears to be involved. However, given the growing appreciation for epigenetic mechanisms in alternative splicing regulation (Luco et al., 2011), these two pathways may converge to regulate CD73 expression in human cancers, as well as in other diseases.
CD73S as a potential negative regulator of proapoptotic and antiproliferative functions of adenosine in HCC
The inverse association between CD73L expression and Ki67 positivity in HepG2 cells suggests that CD73L may function to limit hepatoma cell proliferation. This may involve adenosine because of the known proapoptotic and antiproliferative functions of adenosine or adenosine A3 receptor agonists in these cells (Hermes et al., 2007; Cohen et al., 2011; Tamura et al., 2012). Given that CD73S promotes CD73L down-regulation, one potential function of CD73S in HCC may be to limit the antiproliferative and proapoptotic function of CD73L-generated adenosine (Figure 7). The effects of adenosine in cancer are complex and can result in different outcomes, depending upon which receptor is activated (Antonioli et al., 2013a). In general, activation of the adenosine receptors A1, A2A, and A2B promotes cancer cell proliferation, whereas activation of the A3 receptor blocks it (Antonioli et al., 2013a). Furthermore, activation of A3 receptor promotes apoptosis of cancer cells, and the adenosine A3 receptor agonist CF102 has antitumor effects in the liver (Cohen et al., 2011). Additional studies will be necessary to assess whether NT5E-2 mRNA and CD73S protein levels correlate with liver disease progression and the ultimate development of HCC.
FIGURE 7:
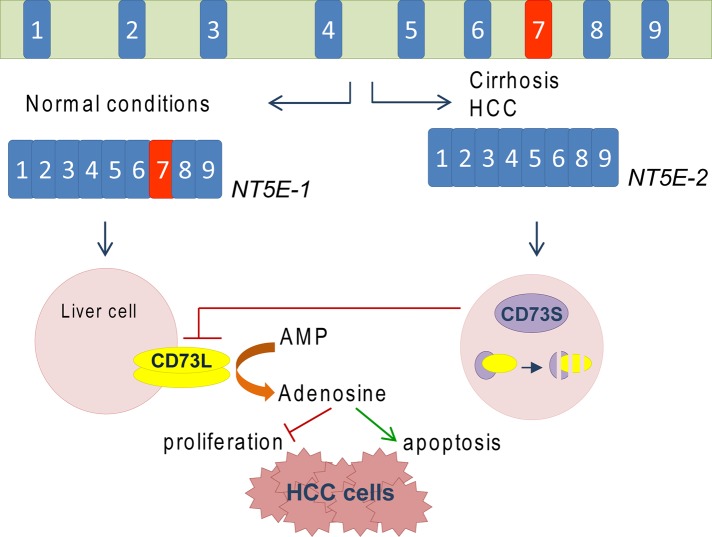
Proposed model for NT5E and CD73 regulation in the context of liver cirrhosis and HCC. Pre-mRNA splicing of NT5E under normal conditions results in expression of the nine-exon NT5E-1 transcript, which encodes canonical CD73 (CD73L). In the context of cirrhosis and HCC, there is significant up-regulation of the alternative splice variant NT5E-2, which encodes CD73S. CD73S is an ER-associated glycoprotein that does not dimerize and lacks 5′-nucleotidase activity. CD73S complexes with CD73L and promotes CD73L degradation via the proteasome. A potential function of CD73S may be to limit the antiproliferative and proapoptotic functions of CD73L-generated adenosine. The cell type(s) affected by CD73S induction within the diseased liver remain to be defined.
MATERIALS AND METHODS
Antibodies and reagents
We used mouse anti-Flag M2 clone (Sigma-Aldrich, St. Louis, MO), rabbit anti-turboGFP (Origene, Rockville, MD), mouse anti-V5 (Invitrogen, Carlsbad, CA), rabbit anti-CD73 (Novus, Littleton, CO), and rabbit anti-calnexin, -PDI, and -Ki67 (Cell Signaling, Danvers MA). Rabbit anti-CD73S antibody was generated by Abbiotec (San Diego, CA) using a 90-d immunization protocol with the peptide C-ERNNGIHV conjugated to KLH, followed by immunoglobulin G purification using affinity chromatography. cDNAs encoding human myc-DDK (Flag)-CD73L, -cN-IA, and turboGFP-tagged CD73L in pCMV6-Entry vector were purchased from Origene. cDNAs encoding myc-DDK tagged CD73S and mouse AK047143 in pCMV6-Entry vector was synthesized by Blue Heron Biotechnology, Bothell, WA.
Quantitative PCR
RNA extraction, quantitative PCR (qPCR), and data analysis were performed as described (Snider et al., 2011) using sequence-specific primers (Supplemental Table S1).
Human RNA and tissues
Normal human tissues RNA panels were obtained from Clontech (Mountain View, CA). Diseased human livers were from biopsies (HCV, NAFLD) or surgical specimens (HCC) that were collected at the University of Michigan under an approved human subjects protocol and are described in Supplemental Table S2. None of the HCC patients received pharmacologic therapy before the surgery to remove the tumor(s). The normal control livers that were used for immunoblotting were obtained from the National Disease Research Interchange, and the control livers that were used for qPCR analysis were from biopsies and previously described (Snider et al., 2013).
Cell culture, transfections, and immunofluorescence analysis
The human HCC cell line Huh7 was obtained from the Japanese Collection of Research Bioresources (JCRB), and all other cell lines were obtained from the American Type Culture Collection (ATCC; Manassas, VA) and cultured according to ATCC recommendations. Embryonic kidney (HEK293T) and HCC (HepG2) human cells were cultured in DMEM and EMEM, respectively, supplemented with 10% fetal bovine serum. The cell lines were transfected with CD73S and/or CD73L plasmid DNA using Lipofectamine LTX (Invitrogen). Primary mouse hepatocytes were isolated and transfected for 24 h as described (Snider et al., 2011). MG132 (10 μg/ml) was added for 12–14 h after the transfection. For the immunofluorescence analyses, cells were fixed, stained, and imaged as described (Snider et al., 2011).
CD73 enzyme activity, immunoprecipitation, and immunoblotting
CD73 enzymatic activity was assessed biochemically by its ability to catalyze the enzymatic hydrolysis of 5′-inosine monophosphate (5′-IMP) to form inosine as described (Snider et al., 2013). Immunoprecipitation of control and CD73-expressing cell lysates was performed using the designated antibodies conjugated to Protein G-Dynabeads (Invitrogen, Carlsbad, CA) for 3 h at 4ºC. Immunoblotting was performed as described (Snider et al., 2013).
Data analysis
Statistical analysis was done using Prism 6 (GraphPad Software). Photoshop (CS2; Adobe) was used for densitometry and quantification of the Ki67 fluorescence (5000-pixel areas were measured for each cell).
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grants R01 DK52951 (M.B.O.), K01 DK093776 (N.T.S.), and K08 CA151414 (T.H.W.). The work also used Core Services and pilot grant funding supported by Grants DK089503 and DK034933 to the University of Michigan. The authors thank Anna Lok for providing the human liver cirrhosis biopsy specimens used in this study.
Abbreviations used:
- CD73L
long protein isoform of CD73
- CD73S
short protein isoform of CD73
- HCC
hepatocellular carcinoma
- HCV
hepatitis C virus
- NAFLD
nonalcoholic fatty liver disease.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E14-06-1167) on October 8, 2014.
REFERENCES
- Algars A, Karikoski M, Yegutkin GG, Stoitzner P, Niemela J, Salmi M, Jalkanen S. Different role of CD73 in leukocyte trafficking via blood and lymph vessels. Blood. 2011;117:4387–4393. doi: 10.1182/blood-2010-11-321646. [DOI] [PubMed] [Google Scholar]
- Allard B, Turcotte M, Spring K, Pommey S, Royal I, Stagg J. Anti-CD73 therapy impairs tumor angiogenesis. Int J Cancer. 2014;134:1466–1473. doi: 10.1002/ijc.28456. [DOI] [PubMed] [Google Scholar]
- Antonioli L, Blandizzi C, Pacher P, Hasko G. Immunity, inflammation and cancer: a leading role for adenosine. Nat Rev Cancer. 2013a;13:842–857. doi: 10.1038/nrc3613. [DOI] [PubMed] [Google Scholar]
- Antonioli L, Pacher P, Vizi ES, Hasko G. CD39 and CD73 in immunity and inflammation. Trends Mol Med. 2013b;19:355–367. doi: 10.1016/j.molmed.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa-Morais NL, Irimia M, Pan Q, Xiong HY, Gueroussov S, Lee LJ, Slobodeniuc V, Kutter C, Watt S, Colak R, et al. The evolutionary landscape of alternative splicing in vertebrate species. Science. 2012;338:1587–1593. doi: 10.1126/science.1230612. [DOI] [PubMed] [Google Scholar]
- Beavis PA, Stagg J, Darcy PK, Smyth MJ. CD73: a potent suppressor of antitumor immune responses. Trends Immunol. 2012;33:231–237. doi: 10.1016/j.it.2012.02.009. [DOI] [PubMed] [Google Scholar]
- Brownell J, Polyak SJ. Molecular pathways: hepatitis C virus, CXCL10, and the inflammatory road to liver cancer. Clin Cancer Res. 2013;19:1347–1352. doi: 10.1158/1078-0432.CCR-12-0928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan TH, Lin CH, Qi L, Fei J, Li Y, Yong KJ, Liu M, Song Y, Chow RK, Ng VH, et al. A disrupted RNA editing balance mediated by ADARs (Adenosine DeAminases that act on RNA) in human hepatocellular carcinoma. Gut 63, 832–843. 2014 doi: 10.1136/gutjnl-2012-304037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JF, Eltzschig HK, Fredholm BB. Adenosine receptors as drug targets—what are the challenges. Nat Rev Drug Discov. 2013;12:265–286. doi: 10.1038/nrd3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Jiang X, Sun D, Han G, Wang F, Ye M, Wang L, Zou H. Glycoproteomics analysis of human liver tissue by combination of multiple enzyme digestion and hydrazide chemistry. J Proteome Res. 2009;8:651–661. doi: 10.1021/pr8008012. [DOI] [PubMed] [Google Scholar]
- Chettouh H, Fartoux L, Aoudjehane L, Wendum D, Claperon A, Chretien Y, Rey C, Scatton O, Soubrane O, Conti F, et al. Mitogenic insulin receptor-A is overexpressed in human hepatocellular carcinoma due to EGFR-mediated dysregulation of RNA splicing factors. Cancer Res. 2013;73:3974–3986. doi: 10.1158/0008-5472.CAN-12-3824. [DOI] [PubMed] [Google Scholar]
- Cohen S, Stemmer SM, Zozulya G, Ochaion A, Patoka R, Barer F, Bar-Yehuda S, Rath-Wolfson L, Jacobson KA, Fishman P. CF102 an A3 adenosine receptor agonist mediates anti-tumor and anti-inflammatory effects in the liver. J Cell Physiol. 2011;226:2438–2447. doi: 10.1002/jcp.22593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colgan SP, Eltzschig HK, Eckle T, Thompson LF. Physiological roles for ecto-5’-nucleotidase (CD73) Purinergic Signal. 2006;2:351–360. doi: 10.1007/s11302-005-5302-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David CJ, Manley JL. Alternative pre-mRNA splicing regulation in cancer: pathways and programs unhinged. Genes Dev. 2010;24:2343–2364. doi: 10.1101/gad.1973010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Craene B, Berx G. Regulatory networks defining EMT during cancer initiation and progression. Nat Rev Cancer. 2013;13:97–110. doi: 10.1038/nrc3447. [DOI] [PubMed] [Google Scholar]
- Fausther M, Lavoie EG, Goree JR, Baldini G, Dranoff JA. NT5E mutations that cause human disease are associated with intracellular mistrafficking of NT5E protein. PLoS One. 2014;9:e98568. doi: 10.1371/journal.pone.0098568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fausther M, Sheung N, Saiman Y, Bansal MB, Dranoff JA. Activated hepatic stellate cells upregulate transcription of ecto-5’-nucleotidase/CD73 via specific SP1 and SMAD promoter elements. Am J Physiol Gastrointest Liver Physiol. 2012;303:G904–G914. doi: 10.1152/ajpgi.00015.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flicek P, Amode MR, Barrell D, Beal K, Billis K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fitzgerald S, et al. Ensembl 2014. Nucleic Acids Res. 2014;42:D749–D755. doi: 10.1093/nar/gkt1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harte RA, Farrell CM, Loveland JE, Suner MM, Wilming L, Aken B, Barrell D, Frankish A, Wallin C, Searle S, et al. Tracking and coordinating an international curation effort for the CCDS Project. Database (Oxford) 2012. 2012:bas008. doi: 10.1093/database/bas008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermes M, Osswald H, Kloor D. Role of S-adenosylhomocysteine hydrolase in adenosine-induced apoptosis in HepG2 cells. Exp Cell Res. 2007;313:264–283. doi: 10.1016/j.yexcr.2006.10.003. [DOI] [PubMed] [Google Scholar]
- Knapp K, Zebisch M, Pippel J, El-Tayeb A, Muller CE, Strater N. Crystal structure of the human ecto-5’-nucleotidase (CD73): insights into the regulation of purinergic signaling. Structure. 2012;20:2161–2173. doi: 10.1016/j.str.2012.10.001. [DOI] [PubMed] [Google Scholar]
- Lee TK, Murthy SR, Cawley NX, Dhanvantari S, Hewitt SM, Lou H, Lau T, Ma S, Huynh T, Wesley RA, et al. An N-terminal truncated carboxypeptidase E splice isoform induces tumor growth and is a biomarker for predicting future metastasis in human cancers. J Clin Invest. 2011;121:880–892. doi: 10.1172/JCI40433. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Loi S, Pommey S, Haibe-Kains B, Beavis PA, Darcy PK, Smyth MJ, Stagg J. CD73 promotes anthracycline resistance and poor prognosis in triple negative breast cancer. Proc Natl Acad Sci USA. 2013;110:11091–11096. doi: 10.1073/pnas.1222251110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luco RF, Allo M, Schor IE, Kornblihtt AR, Misteli T. Epigenetics in alternative pre-mRNA splicing. Cell. 2011;144:16–26. doi: 10.1016/j.cell.2010.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matera AG, Wang Z. A day in the life of the spliceosome. Nat Rev Mol Cell Biol. 2014;15:108–121. doi: 10.1038/nrm3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkin J, Russell C, Chen P, Burge CB. Evolutionary dynamics of gene and isoform regulation in mammalian tissues. Science. 2012;338:1593–1599. doi: 10.1126/science.1228186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikhailov A, Sokolovskaya A, Yegutkin GG, Amdahl H, West A, Yagita H, Lahesmaa R, Thompson LF, Jalkanen S, Blokhin D, et al. CD73 participates in cellular multiresistance program and protects against TRAIL-induced apoptosis. J Immunol. 2008;181:464–475. doi: 10.4049/jimmunol.181.1.464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misumi Y, Ogata S, Ohkubo K, Hirose S, Ikehara Y. Primary structure of human placental 5’-nucleotidase and identification of the glycolipid anchor in the mature form. Eur J Biochem/FEBS. 1990;191:563–569. doi: 10.1111/j.1432-1033.1990.tb19158.x. [DOI] [PubMed] [Google Scholar]
- Peng Z, Borea PA, Varani K, Wilder T, Yee H, Chiriboga L, Blackburn MR, Azzena G, Resta G, Cronstein BN. Adenosine signaling contributes to ethanol-induced fatty liver in mice. J Clin Invest. 2009;119:582–594. doi: 10.1172/JCI37409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Z, Fernandez P, Wilder T, Yee H, Chiriboga L, Chan ES, Cronstein BN. Ecto-5’-nucleotidase (CD73)-mediated extracellular adenosine production plays a critical role in hepatic fibrosis. FASEB J. 2008;22:2263–2272. doi: 10.1096/fj.07-100685. [DOI] [PubMed] [Google Scholar]
- Qi L, Chan TH, Tenen DG, Chen L. RNA editome imbalance in hepatocellular carcinoma. Cancer Res. 2014;74:1301–1306. doi: 10.1158/0008-5472.CAN-13-3485. [DOI] [PubMed] [Google Scholar]
- Resta R, Hooker SW, Hansen KR, Laurent AB, Park JL, Blackburn MR, Knudsen TB, Thompson LF. Murine ecto-5’-nucleotidase (CD73): cDNA cloning and tissue distribution. Gene. 1993;133:171–177. doi: 10.1016/0378-1119(93)90635-g. [DOI] [PubMed] [Google Scholar]
- Resta R, Yamashita Y, Thompson LF. Ecto-enzyme and signaling functions of lymphocyte CD73. Immunol Rev. 1998;161:95–109. doi: 10.1111/j.1600-065x.1998.tb01574.x. [DOI] [PubMed] [Google Scholar]
- Roberts V, Lu B, Rajakumar S, Cowan PJ, Dwyer KM. The CD39-adenosinergic axis in the pathogenesis of renal ischemia-reperfusion injury. Purinergic Signal. 2013;9:135–143. doi: 10.1007/s11302-012-9342-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh RK, Cooper TA. Pre-mRNA splicing in disease and therapeutics. Trends Mol Med. 2012;18:472–482. doi: 10.1016/j.molmed.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitkovsky MV, Hatfield S, Abbott R, Belikoff B, Lukashev D, Ohta A. Hostile, hypoxia-A2-adenosinergic tumor biology as the next barrier to overcome for tumor immunologists. Cancer Immunol Res. 2014;2:598–605. doi: 10.1158/2326-6066.CIR-14-0075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snider NT, Griggs NW, Singla A, Moons DS, Weerasinghe SV, Lok AS, Ruan C, Burant CF, Conjeevaram HS, Omary MB. CD73 (ecto-5’-nucleotidase) hepatocyte levels differ across mouse strains and contribute to mallory-denk body formation. Hepatology. 2013;58:1790–1800. doi: 10.1002/hep.26525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snider NT, Weerasinghe SV, Singla A, Leonard JM, Hanada S, Andrews PC, Lok AS, Omary MB. Energy determinants GAPDH and NDPK act as genetic modifiers for hepatocyte inclusion formation. J Cell Biol. 2011;195:217–229. doi: 10.1083/jcb.201102142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stagg J, Divisekera U, Duret H, Sparwasser T, Teng MW, Darcy PK, Smyth MJ. CD73-deficient mice have increased antitumor immunity and are resistant to experimental metastasis. Cancer Res. 2011;71:2892–2900. doi: 10.1158/0008-5472.CAN-10-4246. [DOI] [PubMed] [Google Scholar]
- Stagg J, Divisekera U, McLaughlin N, Sharkey J, Pommey S, Denoyer D, Dwyer KM, Smyth MJ. Anti-CD73 antibody therapy inhibits breast tumor growth and metastasis. Proc Natl Acad Sci USA. 2010;107:1547–1552. doi: 10.1073/pnas.0908801107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Hilaire C, Ziegler SG, Markello TC, Brusco A, Groden C, Gill F, Carlson-Donohoe H, Lederman RJ, Chen MY, Yang D, et al. NT5E mutations and arterial calcifications. N Engl J Med. 2011;364:432–442. doi: 10.1056/NEJMoa0912923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Kanno T, Fujita Y, Gotoh A, Nakano T, Nishizaki T. A(2a) adenosine receptor mediates HepG2 cell apoptosis by downregulating Bcl-X(L) expression and upregulating Bid expression. J Cell Biochem. 2012;113:1766–1775. doi: 10.1002/jcb.24048. [DOI] [PubMed] [Google Scholar]
- Terp MG, Olesen KA, Arnspang EC, Lund RR, Lagerholm BC, Ditzel HJ, Leth-Larsen R. Anti-human CD73 monoclonal antibody inhibits metastasis formation in human breast cancer by inducing clustering and internalization of CD73 expressed on the surface of cancer cells. J Immunol. 2013;191:4165–4173. doi: 10.4049/jimmunol.1301274. [DOI] [PubMed] [Google Scholar]
- Toth I, Le AQ, Hartjen P, Thomssen A, Matzat V, Lehmann C, Scheurich C, Beisel C, Busch P, Degen O, et al. Decreased frequency of CD73+CD8+ T cells of HIV-infected patients correlates with immune activation and T cell exhaustion. J Leukocyte Biol. 2013;94:551–561. doi: 10.1189/jlb.0113018. [DOI] [PubMed] [Google Scholar]
- Vembar SS, Brodsky JL. One step at a time: endoplasmic reticulum-associated degradation. Nat Rev Mol Cell Biol. 2008;9:944–957. doi: 10.1038/nrm2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Fan J, Thompson LF, Zhang Y, Shin T, Curiel TJ, Zhang B. CD73 has distinct roles in nonhematopoietic and hematopoietic cells to promote tumor growth in mice. J Clin Invest. 2011;121:2371–2382. doi: 10.1172/JCI45559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Lee S, Nigro CL, Lattanzio L, Merlano M, Monteverde M, Matin R, Purdie K, Mladkova N, Bergamaschi D, et al. NT5E (CD73) is epigenetically regulated in malignant melanoma and associated with metastatic site specificity. Br J Cancer. 2012;106:1446–1452. doi: 10.1038/bjc.2012.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B. CD73: a novel target for cancer immunotherapy. Cancer Res. 2010;70:6407–6411. doi: 10.1158/0008-5472.CAN-10-1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann H, Zebisch M, Strater N. Cellular function and molecular structure of ecto-nucleotidases. Purinergic Signal. 2012;8:437–502. doi: 10.1007/s11302-012-9309-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zylka MJ. Pain-relieving prospects for adenosine receptors and ectonucleotidases. Trends Mol Med. 2011;17:188–196. doi: 10.1016/j.molmed.2010.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.