Nuclear receptor–binding, SET-domain containing 3 (NSD3) is the first protein methyltransferase essential for early neural crest development. NSD3 is required for neural crest gene expression but not for H3K36 dimethylation of most neural crest genes. NSD3-related methyltransferase activity independently regulates neural crest migration.
Abstract
Neural crest precursors express genes that cause them to become migratory, multipotent cells, distinguishing them from adjacent stationary neural progenitors in the neurepithelium. Histone methylation spatiotemporally regulates neural crest gene expression; however, the protein methyltransferases active in neural crest precursors are unknown. Moreover, the regulation of methylation during the dynamic process of neural crest migration is unclear. Here we show that the lysine methyltransferase NSD3 is abundantly and specifically expressed in premigratory and migratory neural crest cells. NSD3 expression commences before up-regulation of neural crest genes, and NSD3 is necessary for expression of the neural plate border gene Msx1, as well as the key neural crest transcription factors Sox10, Snail2, Sox9, and FoxD3, but not gene expression generally. Nevertheless, only Sox10 histone H3 lysine 36 dimethylation requires NSD3, revealing unexpected complexity in NSD3-dependent neural crest gene regulation. In addition, by temporally limiting expression of a dominant negative to migratory stages, we identify a novel, direct requirement for NSD3-related methyltransferase activity in neural crest migration. These results identify NSD3 as the first protein methyltransferase essential for neural crest gene expression during specification and show that NSD3-related methyltransferase activity independently regulates migration.
INTRODUCTION
The neural crest is a migratory cell population that forms a remarkable variety of cell types in vertebrate embryos. Initially part of the neurepithelium, neural crest cells are unique in relation to other neural precursors in their gene expression and developmental potential. Once motile, neural crest cells must coordinate complex migratory behaviors to reach their destinations, where they undergo controlled differentiation into diverse structures (Le Douarin and Kalcheim, 1999). A key question in neural crest development is: What regulates the restricted and sequential implementation of the neural crest genetic program to orchestrate this complex and varied sequence of events (Prasad et al., 2012)?
Histone methylation was recently implicated in the spatiotemporal regulation of gene expression in neural crest precursors. The delay between neural crest induction during gastrulation (Basch and Bronner-Fraser, 2006) and the initial expression of neural crest genes during neurulation (Khudyakov and Bronner-Fraser, 2009) correlates with repressive trimethylation (me3) of lysine 9 (K9) on histone H3 (H3K9me3) near the transcription start site of two key neural crest transcription factors, Sox10 and Snail2 (Strobl-Mazzulla et al., 2010). The demethylase KDM4A, which removes H3K9me3, allows Sox10 and Snail2 to be expressed (Strobl-Mazzulla et al., 2010). Meanwhile, neural crest enhancers are enriched for H3K4me3 and H3K4me1, the latter of which binds a CHD7/PBAF chromatin remodeling complex (Bajpai et al., 2010; Rada-Iglesias et al., 2012). Once expressed, Sox10 and Snail2 gene bodies are marked with H3 lysine 36 (K36) me3 or H3K36me3 and H3K9me3, respectively (Strobl-Mazzulla et al., 2010), both of which prevent spurious transcriptional initiation of actively transcribed genes (Carrozza et al., 2005; Keogh et al., 2005; Hahn et al., 2011). Nevertheless, to investigate the dynamics of and requirement for these neural crest–specific patterns of histone methylation and define the functional consequences of protein methylation during implementation of the neural crest developmental program, the protein methyltransferases that are necessary for neural crest gene expression must be identified.
It is becoming apparent that protein methylation also regulates migration. Metastatic cancer cells, which have morphological and molecular similarities to neural crest cells (Acloque et al., 2009; Theveneau and Mayor, 2012), overexpress lysine methyltransferases like NSD2, Ezh2, and G9a (Chen et al., 2010; Chase and Cross, 2011; Ezponda et al., 2013). These methyltransferases transcriptionally silence epithelial factors during the transition to a metastatic, mesenchymal state and, in the case of NSD2, promote migration (Cao et al., 2008; Chen et al., 2010; Min et al., 2010; Dong et al., 2012; Ren et al., 2012; Ezponda et al., 2013). Moreover, melanoma cell motility requires chromatin condensation (Gerlitz and Bustin, 2010). Crucially, this mechanism is not unique to metastasis, as protein methylation is also necessary for neural crest migration (Vermillion et al., 2014). To define methylation as a novel, general regulatory mechanism for motility and to characterize the regulation, function, and targets of methylation in migratory cells, it is essential to identify the methyltransferases that catalyze this methylation during neural crest migration.
Nuclear receptor–binding, SET (suppressor of variegation3-9, Enhancer of Zeste and Trithorax)–domain containing 3 (NSD3; also known as WHSC1L1) is a lysine methyltransferase that is up-regulated as a consequence of neural crest induction (Gammill and Bronner-Fraser, 2002; Adams et al., 2008). NSD3 is an oncogene that is amplified in breast, pancreatic, and lung cancers (Angrand et al., 2001; Tonon et al., 2005; Guffanti et al., 2009; Yang et al., 2010; Dutt et al., 2011). In addition, translocations involving NSD3 cause acute myeloid leukemia (Rosati et al., 2002; Morishita and di Luccio, 2011; Wagner and Carpenter, 2012). NSD3 is an H3K36 mono/dimethylase (Li et al., 2009; Kuo et al., 2011; Wagner and Carpenter, 2012) that can also affect H3K36 trimethylation in vivo (Rahman et al., 2011). H3K36 methylation is a mark of actively transcribed genes that is laid down on nucleosomes in the wake of RNA polymerase to repress spurious transcriptional initiation outside of the transcription start site (Bannister et al., 2005; Carrozza et al., 2005; Keogh et al., 2005; Pokholok et al., 2005; Carvalho et al., 2013). In complex with other chromatin modifiers, NSD3 is believed to affect transcriptional initiation and elongation through H3K36 methylation of gene bodies (Fang et al., 2010; Rahman et al., 2011); however, the mechanism for this is unclear. NSD-mediated H3K36 dimethylation (H3K36me2) can be transcription independent (Rechtsteiner et al., 2010), and high H3K36me2 does not always predict gene expression (Kuo et al., 2011), indicating that they are not necessarily related, and that our understanding of NSD function, gene expression, and H3K36 methylation are far from complete. NSD family members serve nonredundant functions due to their distinct histone-binding specificities (He et al., 2013); whereas knockouts of NSD1 (Rayasam et al., 2003) and NSD2 (Nimura et al., 2009) reveal crucial roles in development, the requirement for NSD3 in the embryo, and more specifically in the neural crest, has not been determined.
Here we report that NSD3 is essential for neural crest formation and migration. NSD3 knockdown leads to striking disruptions in neural crest gene expression that are not strictly correlated with changes in H3K36 dimethylation, raising intriguing possibilities regarding H3K36me2 function and NSD3 activity during neural crest specification. Furthermore, by taking the unique approach of temporally restricting expression of a dominant negative to migratory stages, we demonstrate a direct, separable role for NSD3-related methyltransferase activity during neural crest migration.
RESULTS
Premigratory and migratory neural crest cells express NSD3
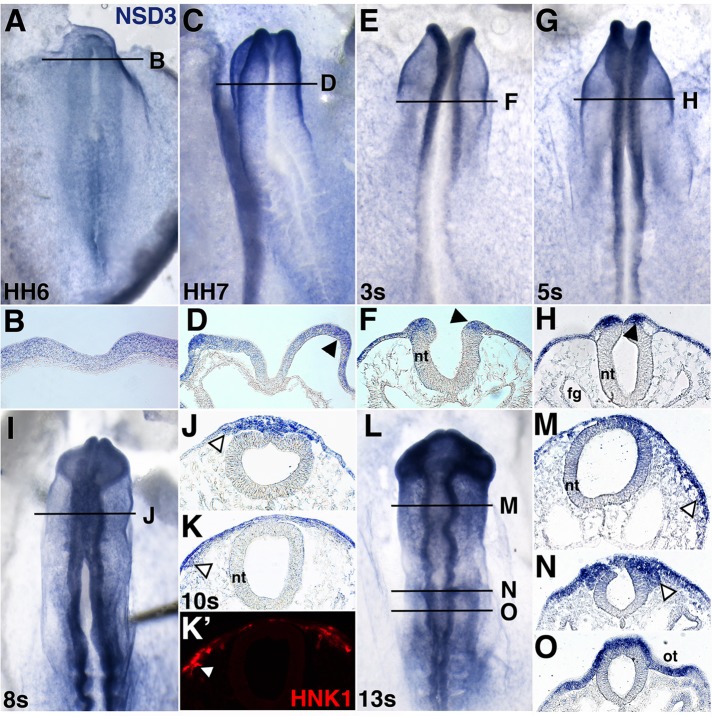
To identify the stages when NSD3 is relevant to neural crest development, we defined NSD3 spatiotemporal expression in chick embryos by in situ hybridization. NSD3 transcripts were undetectable until Hamburger and Hamilton (HH; Hamburger and Hamilton, 1951) stage 6, when NSD3 was up-regulated in the neural plate (Figure 1, A and B), preceding the onset of neural crest–specifier gene expression (Khudyakov and Bronner-Fraser, 2009; Betancur et al., 2010). By HH stage 7, NSD3 mRNA was most abundant at rostral neural plate borders (Figure 1, C and D), and within neural tissue, it was restricted to neural folds by 3 somites (HH stage 8–; Figure 1, E and F). At 5 somites (HH stage 8+), NSD3 was strongly expressed in the dorsal neural tube (Figure 1, G and H), which contains neural crest-specifier gene–expressing premigratory neural crest cells (Khudyakov and Bronner-Fraser, 2009). In embryos ranging from 8 (HH stage 9+) to 13 somites (HH stage 11), abundant NSD3 expression persisted in midbrain (Figure 1, I–M) and hindbrain (Figure 1, L and N) migratory neural crest cells that costained for the migratory neural crest cell marker HNK1 (Figure 1K′). Nonneural ectoderm and the otic placode also expressed NSD3. In contrast, related methyltransferases NSD1 and NSD2 were expressed at low, relatively uniform levels throughout early chicken embryos, with only slight enrichment of NSD2 in premigratory and NSD1 in migratory neural crest cells (Supplemental Figure S1). The comparatively abundant, restricted expression of NSD3 in neural crest precursors/cells throughout neural crest specification and migration suggests that NSD3 is the predominant neural crest NSD methyltransferase and could have neural crest–specific developmental functions.
FIGURE 1:
Neural crest cells express NSD3. NSD3 mRNA was visualized in HH stages 6 (A,B), 7 (C, D), 8– (3 somites [3s]; E, F) 8+ (5s; G, H), 9+ (8s; I, J), 10 (10s; K, K′), and 11 (13s; L–O) chick embryos by whole-mount in situ hybridization. Embryos were sectioned at the levels indicated in whole-mount views. NSD3 mRNA is first expressed throughout the rostral neural plate (A, B), but becomes restricted to premigratory (C–H; arrowheads in D, F, H) and migratory neural crest cells (open arrowheads) in the midbrain (I–M) and hindbrain (L, N, O). HNK1 immunostaining (K′) confirms the NSD3 is expressed (K) in 10-somite midbrain migratory neural crest cells. Nonneural ectoderm and otic placode also express NSD3. (A, C, E, G, I, L) Dorsal view, anterior to the top; (B, D, F, H, J, K, M–O) transverse sections, dorsal up. fg, foregut; nt, neural tube; ot, otic placode; s, somite.
Deleting the NSD3 methyltransferase domain creates a dominant negative
NSD3 is a SET-domain lysine methyltransferase (Angrand et al., 2001; Kim et al., 2006; Li et al., 2009). In other lysine methyltransferases, mutation or deletion of the SET domain results in dominant-negative activity (Roopra et al., 2004; Lee et al., 2005; Houston et al., 2008; Huang, 2008; Joshi et al., 2008; Tanaka et al., 2008; Fujiki et al., 2009). To create a dominant negative and investigate a role for NSD3 in neural crest development, we truncated NSD3 at the start of the SET domain (NSD3Δ1707, Figure 2A) and drove expression with the green fluorescent protein (GFP) bicistronic expression plasmid pMES (Swartz et al., 2001).
FIGURE 2:
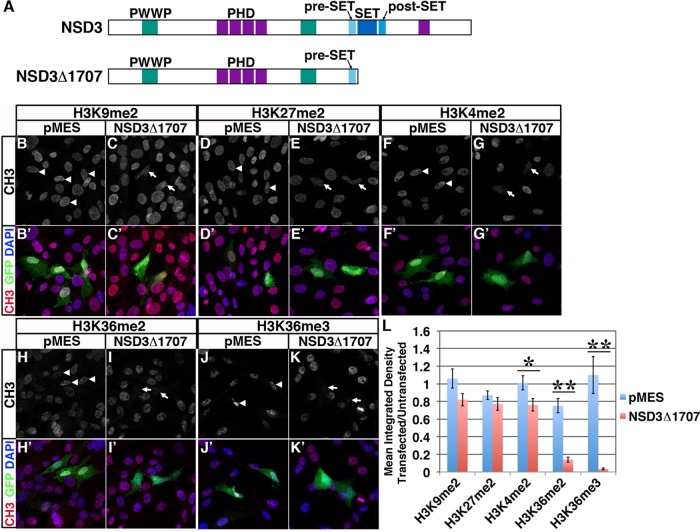
NSD3Δ1707 prevents H3K36me2 and me3. (A) Diagram of the protein domain organization of NSD3 (top) and NSD3Δ1707 (bottom). PHD, plant homeodomain fingers; PWWP, Pro-Trp-Trp-Pro domain; SET, suppressor of variegation3-9, Enhancer of Zeste and Trithorax domain. (B–K) Chick fibroblast DF-1 cells were transfected with the GFP (green) bicistronic expression plasmid pMES (B, D, F, H, J) or pMES driving NSD3Δ1707 (C, E, G, I, K). Twenty-four hours after transfection, cells were fixed and stained with anti-methyl (CH3; red) antibodies against dimethylated (me2) lysines H3K9me2 (B, C), H3K27me2 (D, E), H3K4me2 (F, G), or H3K36me2 (H, I) or trimethylated (me3) lysine H3K36me3 (J, K) and DAPI (blue). (L) Mean ± SEM of the ratios of the mean integrated density of fluorescence between cells transfected with pMES or NSD3Δ1707 and neighboring untransfected cells are shown for H3K9me2, H3K27me2, H3K4me2, H3K36me2, and H3K36me3. A ratio of 1 would indicate equal staining between transfected and untransfected cells. White arrowhead, pMES- transfected cell; white arrow, NSD3Δ1707-transfected cell. n ≥ 20 fields of cells/condition. Student's t test was used for statistical analysis; *significant, p < 0.05; **highly significant, p < 0.001.
We characterized SET domain–truncated NSD3Δ1707 by evaluating its effect on the known NSD3 target, histone H3. In nucleosomes, NSD3 dimethylates H3 lysine 36 (Li et al., 2009; Qiao et al., 2011). Thus, if NSD3Δ1707 specifically interferes with NSD3 and related methyltransferases, it should block H3K36me2 and, as a result, accumulation of H3K36me3 (Rahman et al., 2011; Wagner and Carpenter, 2012) but not affect methylation at other lysines, such as H3K9me2. As an additional test of NSD3Δ1707 activity, we also evaluated H3K4me2 and H3K27me2, as these marks have been attributed to NSD3 in less stringent assays with peptides and recombinant histones (Kim et al., 2006; Li et al., 2009). To this end, we compared global levels of H3K4me2, H3K27me2, H3K9me2, H3K36me2, and H3K36me3 in chick fibroblasts (which endogenously express NSD3; Supplemental Figure S2H) transfected with pMES or NSD3Δ1707. Because cell transfection is <100% efficient and because a Western blot would represent an average of the population, we visualized methyl marks in individual cells by immunofluorescence, expressing the intensity of fluorescence as a ratio of transfected cells to untransfected neighboring cells to control for physiological and experimental variations in signal strength. Whereas NSD3Δ1707 did not significantly alter levels of H3K9 or H3K27 dimethylation (Figure 2, C, E, and L), NSD3Δ1707 transfection did cause a slight but significant reduction in H3K4 dimethylation (Figure 2, G and L; p = 0.023). Most strikingly, H3K36me2 and me3 were dramatically reduced by NSD3Δ1707 transfection compared with pMES transfection (Figure 2, H–L, H3K36me2, p = 5.74 × 10−9; H3K36me3, p = 1.09 × 10−5), indicating that NSD3Δ1707 prevents H3K36 dimethylation and trimethylation in chick cells.
To address the possibility that NSD3Δ1707 elicits a methyltransferase-independent overexpression phenotype rather than a dominant-negative effect, we created a full-length NSD3 expression construct traced by bicistronic expression of mCherry (pMESmCh-NSD3; Roffers-Agarwal et al., 2012). We transfected chick cells with pMESmCh-NSD3 or pMES-mCherry and stained for H3K36me2 and me3 as well as H3K27me2 as a negative control. Of interest, we found that NSD3 overexpression did not alter levels of H3K36me2 (Supplemental Figure S2, D and G) but did deplete H3K36me3 compared with pMES-mCherry transfection (Supplemental Figure S2, E–G; p = 1.61 × 10−5). The reason NSD3 overexpression prevents H3K36me3 is unclear, but it could be the result of overexpressed NSD3 binding up limiting shared cofactors necessary for H3K36 trimethylase activity, excess NSD3 remaining bound to H3K36me2 and blocking conversion to H3K36me3, recruitment of an H3K36me3 demethylase, or other, unanticipated considerations. In any event, these reciprocal outcomes, with H3K36me2 maintained by NSD3 overexpression (Supplemental Figure S2) but blocked by NSD3Δ1707 (Figure 2), indicate that NSD3Δ1707 is a dominant negative that interferes with the activity of H3K36 dimethylases like NSD3 that are expressed in DF-1 cells (Supplemental Figure S2H).
Dominant-negative NSD3 disrupts Sox10 and Snail2 expression in premigratory neural crest cells
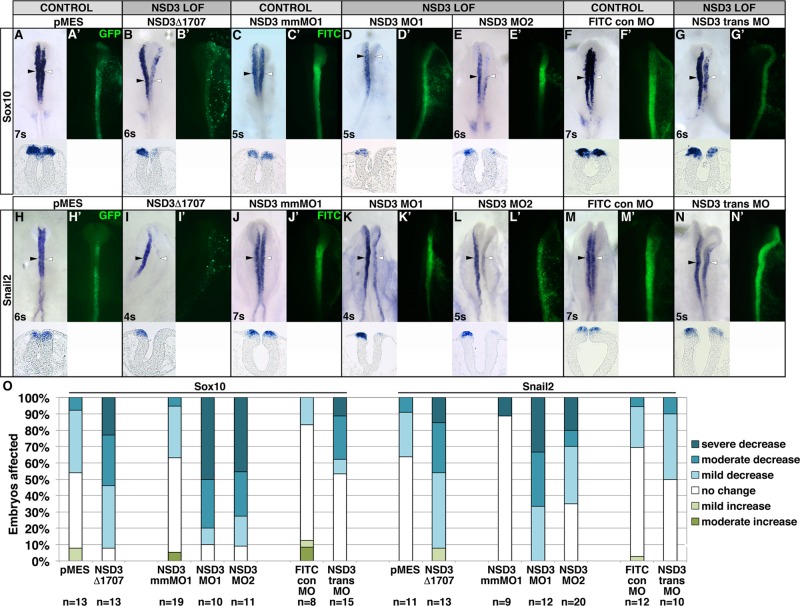
The genomic loci of neural crest transcription factors Sox10 and Snail2 exhibit H3K36 trimethylation in neural crest cells (Strobl-Mazzulla et al., 2010), consistent with H3K36me3 being a mark of active chromatin that represses spurious initiation of actively transcribed genes (reviewed in Wagner and Carpenter, 2012). To investigate the necessity of this methylation for Sox10 and Snail2 expression, we used NSD3Δ1707 to disrupt H3K36me2 and me3 during neural crest specification. We electroporated HH stage 4–6 embryos with NSD3Δ1707 or pMES, incubated to four to seven somites, and analyzed Sox10 and Snail2 expression by in situ hybridization. NSD3Δ1707 significantly reduced Sox10 and Snail2 expression in >90% of embryos (Sox10: Figure 3, B and O, 92.3% with reduced expression, p = 0.038; Snail2: Figure 3, I and O, 92.3% with reduced expression, p = 0.0042; see Supplemental Figure S3, A–C, for representative embryos in each scoring class), compared with a minority of embryos exhibiting mild defects after pMES electroporation (Sox10: Figure 3, A and O, 46.2% with reduced expression; Snail2: Figure 3, H and O, 36.4% with reduced expression). This outcome was not due to indirect effects on cell death or proliferation, as the frequency of cleaved caspase 3 and phospho-histone H3–immunopositive cells was not significantly changed in the targeted versus untargeted dorsal neural tube or in GFP-positive cells of embryos electroporated with NSD3Δ1707 or pMES (Supplemental Figure S4). The ability of SET domain–deleted NSD3 to interfere with neural crest specification suggests that the methyltransferase activity of NSD3 and its relatives is essential in the neural crest.
FIGURE 3:
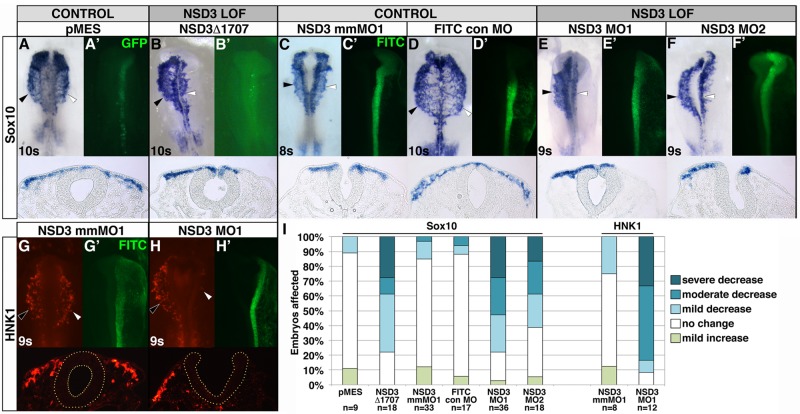
NSD3 is required for Sox10 and Snail2 expression in premigratory neural crest cells. (A–N) After unilateral electroporation at gastrula stages 4–5 with 3 μg/μl GFP bicistronic expression plasmid pMES (A, H), 3 μg/μl pMES driving NSD3Δ1707 (B, I), 0.75 mM 5– base pair mismatch MO (NSD3 mmMO1; C, J), 0.75 mM NSD3 MO1 (D, K), 0.92 mM NSD3 MO2 (E,L), 1.0 mM NSD3 translation blocking MO (NSD3 trans MO; F, M), or 1.0 mM FITC control MO (FITC con MO; G, N), embryos were incubated to four to seven somites, and Sox10 (A G) or Snail2 (H N) was visualized by in situ hybridization. (A′–N′) GFP- or FITC-tagged MO targeting. Dorsal view, anterior up. Corresponding cross sections are shown beneath dorsal views of embryos. White arrowhead, targeted side; black arrowhead, untargeted side; s, somite. (O) Embryos were categorized by the severity of decrease in Sox10 and Snail2 expression, comparing the targeted side of the embryo to the untargeted control side.
NSD3 is required for neural crest gene expression in premigratory neural crest cells
Because NSD3 is the predominant NSD-family methyltransferase in neural crest precursors (Figure 1 and Supplemental Figure S1), we used antisense morpholino oligonucleotides (MOs) to specifically knock down NSD3 in neural crest cells and test its requirement directly. We designed two fluorescein isothiocyanate (FITC)–tagged MOs to block either the NSD3 exon 3 splice acceptor (NSD3 MO1) or splice donor site (NSD3 MO2; Supplemental Figure S5A), resulting in the excision of exon 3 (Supplemental Figure S5B). PCR using cDNA synthesized from head folds of embryos electroporated with NSD3 MO1 showed the expected smaller NSD3 splice product, whereas electroporation with NSD3 MO2 caused a smear suggestive of nonsense-mediated decay (Supplemental Figure S5C). The NSD3 MO1–induced band was sequenced and verified as NSD3 exons 1 and 2 spliced to exon 4 with a resulting frame shift and premature stop codon. This transcript excludes all known functional domains in NSD3, such as the SET domain encoded by exons 18–21 (Angrand et al., 2001). Thus NSD3 MO1 and MO2 alter NSD3 splicing in the expected manner.
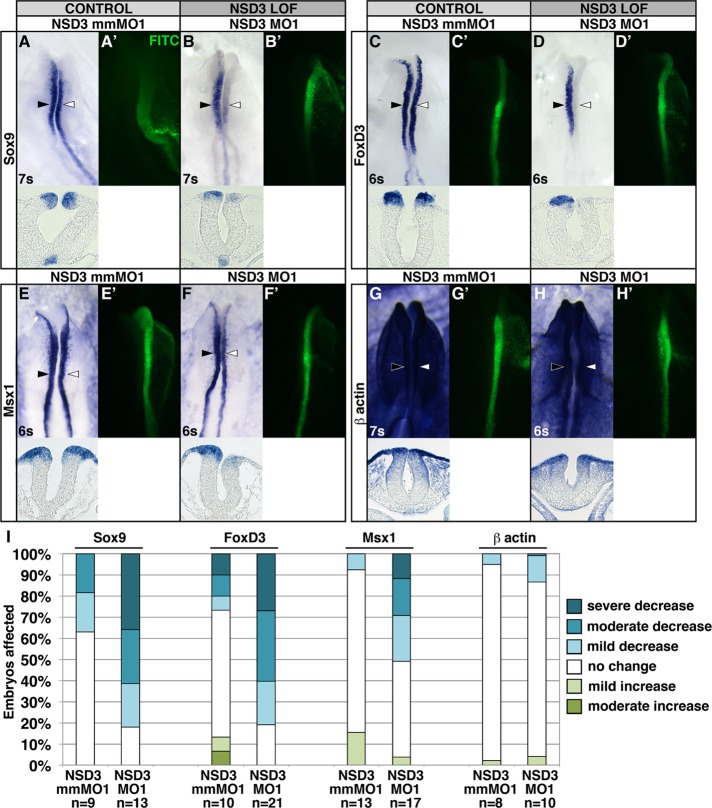
First, we evaluated the effect of NSD3 knockdown on neural crest gene expression during specification. Because 1.0 mM NSD3 MO1 exceeded the effective and specific dose range (Supplemental Figure S6A; Moulton and Yan, 2008), we unilaterally electroporated 0.75 mM NSD3 MO1 or a 5–base pair mismatch of NSD3 MO1 (NSD3 mmMO1), or 0.92 mM NSD3 MO2 into late gastrula–stage embryos (HH stages 4–5), and evaluated Sox10 and Snail2 expression by in situ hybridization. NSD3 MO1 and MO2 significantly impaired Sox10 expression on the targeted side of the embryo (Figure 3, D, E, and O; MO1, 90.0% with reduced expression, p = 3.0 × 10−4 compared with mmMO1; MO2, 90.9% with reduced expression, p = 6.4 × 10−4 compared with mmMO1), whereas mmMO1 did not alter Sox10 expression in the majority of embryos (Figure 3, C and O; mmMO1: 36.8% with reduced expression). Meanwhile Snail2 expression was reduced or lost in all embryos electroporated with NSD3 MO1 (Figure 3, K and O; 100% with reduced expression, p = 6.8 × 10−5) and most embryos electroporated with NSD3 MO2 (Figure 3, L and O; 65.0% with reduced expression, p = 0.048), compared with only 11.1% with NSD3 mmMO1 (Figure 3, J and O). This phenotype was not due to off-target effects on cell proliferation or death (Eisen and Smith, 2008), as the frequency of phospho-histone H3 and cleaved caspase 3 immunopositive cells was not significantly changed in the targeted versus untargeted dorsal neural tube of NSD3 MO1 or mmMO1 electroporated embryos (Supplemental Figure S7). Moreover, the splice-blocking phenotypes were not due to production of short, truncated NSD3 protein, because MO2 produced very little misspliced transcript (Supplemental Figure S5C) and because a NSD3 translation blocking MO (trans MO) also disrupted Sox10 expression (Figure 3, G and O; trans MO, 46.7% with reduced expression; FITC control MO, 16.7% with reduced expression; p = 0.0011) and showed a slight effect on Snail2 (Figure 3, N and O; trans MO, 50.0% with reduced expression; FITC control MO, 30.6% with reduced expression; p = 0.34). Although trans MO was less potent, it, like MO1 and MO2, affected Sox10 more strongly than Snail2. That three MOs phenocopy the dominant negative (Figure 4, B, I, and O) with different severities suggests that knockdown is specific and that inhibition of NSD3 can account for the dominant-negative phenotype.
FIGURE 4:
NSD3 is required for neural crest and neural plate border gene expression but not for gene expression generally. (A–H) After unilateral electroporation at gastrula stages 4–5 with 0.5 mM mmMO1 (A, C, E, G) or 0.5 mM MO1 (B, D, F, H), embryos were incubated to four to seven somites, and Sox9 (A, B), FoxD3 (C, D), Msx1 (E, F), or β-actin (G, H) was visualized by in situ hybridization. (A′–H′) FITC-tagged MO targeting. Dorsal view, anterior up. Corresponding cross sections are shown beneath dorsal views of embryos. White arrowhead, targeted side; black arrowhead, untargeted side; s, somite. (I) Embryos were categorized by the severity of decrease in expression, comparing the targeted side of the embryo to the untargeted control side. Low β-actin signal internally was caused by inefficient penetration of colorimetric substrates over the 15 min needed to stain the neural folds adequately.
To determine whether the requirement for NSD3 is Sox10 and Snail2 specific or affects neural crest specification generally, we repeated the experiment using MO1 and evaluated Sox9, FoxD3, Msx1, and β-actin expression. Like Sox10 and Snail2, NSD3 is required for expression of the neural plate border gene Msx1 (Figure 4, E, F, and I; MO1, 51.0% with reduced expression; mmMO1, 7.7% with reduced expression; p = 7.2 × 10−5) and neural crest transcription factors (Sox9: Figure 4, A, B, and I; MO1, 82.1% with reduced expression; mmMO, 37.0% with reduced expression; p = 7.7 × 10−5; FoxD3: Figure 4, C, D, and I; MO1, 81.0% with reduced expression; mmMO1: 26.7% with reduced expression; p = 1.5 × 10−5). Meanwhile, β-actin expression is NSD3 independent (Figure 4, G–I; MO1, 13.3% with reduced expression; mmMO1, 5.2% with reduced expression; p = 0.49). Thus NSD3 is essential for expression of key transcription factors in the neural crest gene regulatory network during neural crest specification (Betancur et al., 2010; Prasad et al., 2012) but not gene expression generally. Furthermore, this targeted gene knockdown shows that NSD3 has a neural crest–specific role for which NSD1 and 2 (Supplemental Figure S1) do not compensate, consistent with the nonoverlapping functions of NSD proteins due to their unique histone-binding specificities (He et al., 2013).
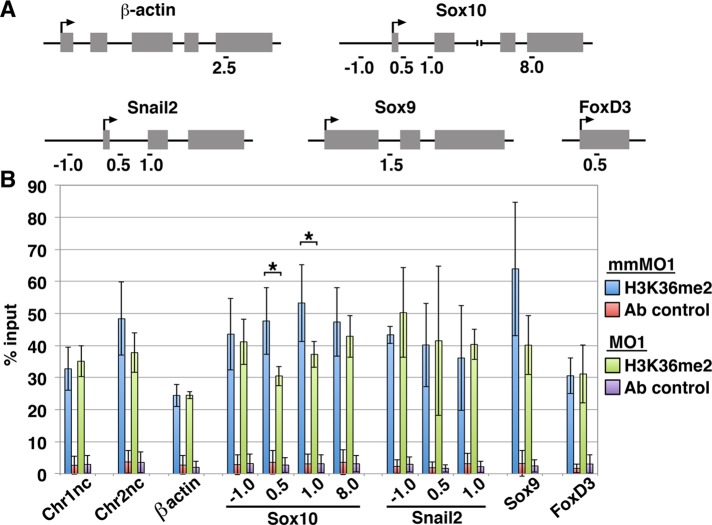
NSD3 specifically methylates the Sox10 gene
NSD3 binds target gene promoters and coding regions and is believed to affect transcriptional initiation and elongation because gene silencing in NSD3-deficient cells is associated with loss of gene body H3K36 methylation (Fang et al., 2010; Rahman et al., 2011; Wagner and Carpenter, 2012). Thus NSD3 could affect neural crest gene expression (Figures 3 and 4) by H3K36 dimethylating these genes. To investigate this hypothesis, we bilaterally electroporated embryos with NSD3 MO1 or mmMO1 and dissected cranial neural tubes at four to eight somites, when neural crest gene expression was NSD3 dependent (Figures 3 and 4). Because chromatin immunoprecipitation (ChIP)–rated NSD3 antibodies are not available, we then performed ChIP using a validated H3K36me2 antibody (Egelhofer et al., 2011). Because H3K36me2 is highest immediately upstream and downstream of transcription start sites and also marks intergenic regions and constitutive heterochromatin (Bell et al., 2007; Rechsteiner et al., 2010; Chantalat et al., 2011; Kuo et al., 2011), we evaluated the effects of NSD3 knockdown on H3K36me2 occupancy at several locations. Compared to mmMO1-electroporated tissue, NSD3 knockdown did not affect H3K36me2 occupancy at open reading frame–free, intergenic regions of chromosome 1 (Figure 5B; Chr1nc, control for Sox10, p = 0.32) and chromosome 2 (Chr2nc, control for Snail 2, p = 0.12). Similarly, H3K36me2 at the β-actin gene was NSD3 independent (Figure 5B; p = 0.49), indicating that NSD3 knockdown does not affect gene body H3K36 dimethylation generally. In contrast, NSD3 knockdown reduced H3K36me2 occupancy at the Sox10 gene 0.5 and 1.0 kb from the transcription start site (Figure 5, A and B; p = 0.026 and 0.047, respectively). On the other hand, NSD3 knockdown did not diminish H3K36me2 occupancy in the Sox10 promoter or 3′ end, at any location in Snail2, or in the Sox9 or FoxD3 gene bodies (Figure 5, A and B; Sox10, p = 0.38 at −1.0 kb, p = 0.29 at 8.0 kb; Snail2: p = 0.22 at −1.0 kb, p = 0.47 at 0.5 kb, p = 0.35 at 1.0 kb; Sox9: 0.072 at 1.0 kb; FoxD3: p = 0.47 at 0.5 kb). Therefore NSD3 is required for H3K36 dimethylation of Sox10 but not other neural crest transcription factors, despite being required for the expression of all four genes (Figures 3 and 4). This disconnect indicates that the regulation of neural crest specification by NSD3 is more complex than direct H3K36 dimethylation of neural crest genes.
FIGURE 5:
NSD3 dimethylates H3K36 at the Sox10 locus. (A) Primer locations in β-actin, Sox10, Snail2, Sox9, and FoxD3 genes used for quantitative PCR after chromatin immunoprecipitation, named by their distance in kilobases from the transcription start site (arrow). (B) Average percentage input (mean ± SD) recovered in three independent chromatin immunoprecipitation experiments performed with 30 pooled NSD3 mmMO1- or MO1-electroporated neural tubes, assaying H3K36me2 occupancy at 12 genomic loci (Chr1nc [chromosome 1 negative control], Chr2nc [chromosome 2 negative control], β-actin, four regions within Sox10, three regions within Snail2, Sox9, and FoxD3). Input recovered is in the normal range for a methylated histone (Cell Signaling Technology, www.cellsignal.com/support/faq_chip.html#a11) and consistent with levels of H3K36me2 occupancy in other systems (e.g., Blackledge et al., 2010; Asangani et al., 2013; fold enrichment in Figure 5 ranges from 5 to 300). Normal rabbit IgG was an immunoprecipitation control (Ab control). Student's t test was used for statistical analysis; *significant; p < 0.05.
Sustained NSD3 loss of function prevents neural crest migration
Because NSD3 expression persists in migratory neural crest cells (Figure 1), we reasoned that NSD3 might also be required for neural crest migration. First, we examined the effects of constitutive NSD3 loss of function. We electroporated NSD3Δ1707, pMES, NSD3 MO1, MO2, mmMO1, or FITC control MO into HH stage 4–6 embryos, incubated until 8–11 somites, and identified migrating neural crest cells by Sox10 in situ hybridization. NSD3Δ1707-targeted neural crest cells remained in the neural tube or migrated a shorter distance than those on the untargeted side in 77.8% of embryos, compared with only 11.1% with pMES (Figure 6, A, B, and I; p = 0.011). Similarly, Sox10-positive neural crest cell migration distance was reduced in 77.8% of embryos electroporated with NSD3 MO1, with more than one-half of the embryos moderately or severely decreased (Figure 6, E and I; p = 5.3 × 10−7 compared with mmMO1). In contrast, only 15.2% of NSD3 mmMO1-electroporated embryos were affected (Figure 6, C and I). Similarly, neural crest migration was curtailed on the targeted side of 61.1% of NSD3 MO2-electroporated embryos, compared with 11.8% of embryos electroporated with a standard FITC control MO (Figure 6, D, F, and I; p = 0.031). Because NSD3 knockdown interferes with Sox10 expression (Figure 3) and NSD3-dependent neural crest migration defects could reflect impaired Sox10 expression rather than faulty migration, we also evaluated HNK1 immunofluorescence, a marker expressed once neural crest cells are migratory. HNK1 immunoreactivity confirmed that NSD3 knockdown was incompatible with migration: 91.7% of NSD3 MO1–electroporated embryos exhibited decreased migration on the targeted side, with >80% severely or moderately affected, compared with 25.0% mildly affected NSD3 mmMO1 embryos (Figure 6, G–I, p = 0.0023). Of importance, both NSD3 MO1 and MO2 produce the same phenotype (compare Figure 6, E and F; also see Figure 6I) in a dose-dependent manner (Supplemental Figure S6, B and C), indicating specificity (Eisen and Smith, 2008; Moulton and Yan, 2008). Neural crest migration defects achieved by two different loss-of-function techniques—morpholino and dominant negative—indicate that NSD3 is necessary for neural crest migration.
FIGURE 6:
NSD3 is required for neural crest migration. (A–H) Embryos were unilaterally electroporated at gastrula stages 4–6 with 6-8.6 μg/μl GFP bicistronic expression plasmid pMES (A), 6–8.6 μg/μl pMES driving NSD3Δ1707 (B), 0.75 mM 5–base pair mismatch MO (NSD3 mmMO1; C, G), 0.80 mM FITC standard control MO (D), 0.75 mM NSD3 MO1 (E, H), or 0.92 mM NSD3 MO2 (F). After incubation to 8–11 somites, neural crest cells were visualized by in situ hybridization for Sox10 (A–F) or immunofluorescence for HNK1 (G, H). (A′–H′) GFP- or FITC-tagged MO targeting. Dorsal view, anterior up. Corresponding cross sections are shown beneath dorsal views. Yellow dots outline the neural tube in G and H. White arrowhead, targeted side; black arrowhead, untargeted side; s, somite. (I) Embryos were categorized by the distance targeted neural crest cells migrated compared with the untargeted control side.
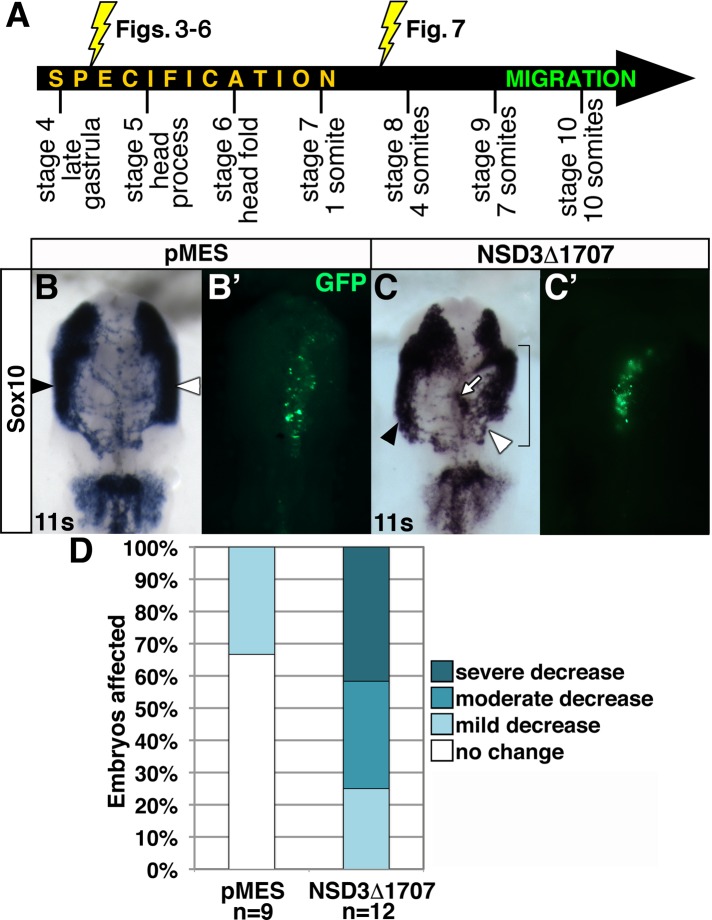
NSD3-related methyltransferase activity is required during neural crest migration independent of its role during specification
Sustained NSD3 knockdown shows that NSD3 is required for neural crest migration (Figure 6) but does not resolve whether NSD3 acts in migratory neural crest cells or is indirectly required due to its essential role during neural crest specification (Figures 3 and 4). Strikingly, neural crest migration was defective in embryos with either reduced (Figure 6E) or normal levels (Figure 6F) of Sox10 expression, suggesting that NSD3 regulates migration independent of its role during specification (Figures 3 and 4). To test this possibility directly, we set out to block NSD3 function in migratory neural crest cells without affecting their specification. However, since NSD3 expression begins at stage 6 (Figure 1), NSD3 MO electroporation after this stage will not elicit an effect until accumulated NSD3 protein and its relatively stable target methylation turn over (Barth and Imhof, 2010). Instead, we used NSD3Δ1707. Although this reagent is not necessarily specific to NSD3, it will disrupt the activity of existing NSD3-related methyltransferases upon its translation and after methyl mark turnover. Because NSD proteins are functionally nonredundant (He et al., 2013) and NSD3 has neural crest–specific functions for which NSD1 and 2 do not compensate (Figures 3–5), and because NSD3 is the predominant NSD methyltransferase in the neural crest, particularly at migratory stages (Figure 1 and Supplemental Figure S1), NSD3Δ1707 will reveal NSD methyltransferase–dependent migration defects relevant to NSD3.
We electroporated NSD3Δ1707 or pMES into one- to four-somite cranial neural folds (HH stages 7–8; Figure 7A). After incubating electroporated embryos to migratory stages, we visualized migratory neural crest cells using Sox10 in situ hybridization. Delays in neural crest migration were apparent in 100% of NSD3Δ1707-electroporated embryos (Figure 7, C and D). Of these, 75% exhibited moderate to severe defects, compared with mild disruptions in only 33.3% of pMES electroporated embryos (Figure 7, B and D, p = 3.8 × 10−4). NSD3Δ1707-expressing neural crest cells emigrated from the neural tube but appeared to halt en route (Figure 7C), suggesting that dominant-negative activity commenced in actively migrating cells. This effect was sometimes apparent throughout the population, so that the extent of migration was reduced compared with the untargeted side of the embryo (Figure 7C, white arrowhead), as seen after gastrula-stage MO electroporation (Figure 6, B, E, and F). Alternatively, groups of cells trailed the rest of the normally migrating population or did not migrate away from the midline (Figure 7C, white arrow). These data indicate that methylation by NSD3 or a related methyltransferase directly regulates neural crest migration.
FIGURE 7:
NSD3 or a related methyltransferase regulates neural crest migration independent of specification. (A) Whereas electroporation at stage 4+ knocks down NSD3 or interferes with NSD3-related activity throughout early neural crest development, electroporation at stages 7–8 limits NSD3 loss of function to migratory stages. (B, C) Embryos were unilaterally electroporated at one to four somites with 6–8 μg/μl GFP bicistronic expression plasmid pMES (B) or pMES driving NSD3Δ1707 (C). After incubation to migration stages, neural crest cells were visualized by Sox10 in situ hybridization. Bracket, targeted cells; white arrow, neural crest cells that have not migrated. (B′, C′) GFP construct targeting. Dorsal view, anterior up. White arrowhead, targeted side; black arrowhead, untargeted side; s, somite. (D) Embryos were categorized by the distance MO-targeted neural crest cells migrated compared with the untargeted control side. All embryos electroporated with NSD3Δ1707 had impaired neural crest migration.
DISCUSSION
Although histone methylation has been implicated in the spatiotemporal regulation of neural crest gene expression during specification, the relevant methyltransferases were unknown. In addition, a requirement for a methylation regulatory factor during neural crest migration independent of its role during specification was untested. We show that premigratory and migratory neural crest cells abundantly express the lysine methyltransferase NSD3 and that NSD3 is necessary for neural crest gene expression during neural crest specification. NSD3 is specifically required for H3K36 dimethylation of Sox10, but not β-actin, intergenic H3K36me2, or, curiously, Snail2, Sox9, or FoxD3. Moreover, NSD3-related methyltransferase activity is essential for migration independent of its role during specification. Together, these findings identify NSD3 as the first protein methyltransferase necessary for neural crest specification, show that NSD3 exhibits unexpected complexity in its regulation of neural crest genes, and reveal a novel, crucial role for NSD3-related methyltransferase activity in neural crest migration.
NSD3 regulates neural crest specification
NSD3 knockdown efficiently blocked expression of key neural crest transcription factors Sox10, Snail2, Sox9, and FoxD3, as well as the neural plate border marker Msx1 (Figures 3 and 4). Although rescuing the phenotype with exogenous NSD3 would be the most stringent way to prove that this effect is a specific consequence of NSD3 knockdown, NSD3 overexpression also disrupts neural crest development (unpublished data), likely because it prevents H3K36me3 (Supplemental Figure S2, F and G). Thus we were unable to identify an NSD3 dose that rescued normal development, which is common for genes for which the timing and exact level of expression are critical (Eisen and Smith, 2008). Alternate indications of specificity (Moulton and Yan, 2008) are that two different splice-blocking MOs targeting a single internal exon elicited the same phenotype (Supplemental Figure S5 and Figure 3) and that a translation-blocking MO gave a hypomorphic version of the phenotype. Moreover, NSD3 knockdown specifically affected neural plate border/neural crest gene expression (Figures 3 and 4), as well as H3K36me2 occupancy on the Sox10 transcribed region (Figure 5), and gave a phenotype that was dose dependent (Supplemental Figure S6, B and C), independent of effects on cell death and proliferation (Supplemental Figure S7), not observed with control or 5–base pair mismatch MO, and phenocopied by an alternative loss-of-function approach (NSD3Δ1707; Figure 3). These data argue that the phenotype is specifically due to NSD3 knockdown (Eisen and Smith, 2008; Moulton and Yan, 2008) and indicate that NSD3 is essential for neural crest development.
Whereas neural crest enhancer and promoter methylation (Strobl-Mazzulla et al., 2010; Rada-Iglesias et al., 2012) and chromatin modifiers (Bajpai et al., 2010) have been implicated in the regulation of neural crest gene expression during specification, NSD3 is the first protein methyltransferase to be required for this process. It is intriguing that NSD3 is up-regulated in neural crest precursors just before neurulation (Figure 1) and the stage at which neural crest “specifier” transcription factor expression commences (Khudyakov and Bronner-Fraser, 2009). This suggests that the spatiotemporal regulation of NSD3 expression in the neural plate border and neural folds (Figure 1) is another factor, along with KDM4A expression and loss of H3K9me3 demethylation (Strobl-Mazzulla et al., 2010), that determines the timing and location of neural crest gene transcription.
How does NSD3 regulate neural crest gene expression? One possibility is that NSD3-mediated H3K36me2 is necessary for transcriptional initiation and elongation (Fang et al., 2010; Rahman et al., 2011; Wagner and Carpenter, 2012) or creates a chromatin state that promotes transcription (Kuo et al., 2011). Concomitant loss of Sox10 expression (Figure 3, B, D, E, G, and O) and reduction of H3K36me2 (Figure 5B) on the Sox10 gene body in NSD3-deficient embryos supports this mechanism and validates our assay. However, NSD3 knockdown did not alter Snail2, Sox9, or FoxD3 H3K36me2 occupancy (Figure 5B), even as the same MO reduced or abolished embryonic expression of these genes (Figures 3 and 4). Therefore NSD3 may regulate neural crest gene expression directly (e.g., Sox10) and indirectly by H3K36 dimethylating gene bodies of transcription factors required for Snail2, Sox9, and FoxD3 expression. Alternatively, given that NSD3 is required for expression of these genes (Figures 3 and 4), although their H3K36me2 occupancy is for the most part NSD3 independent (Figure 5), it is also possible that NSD3 affects neural crest gene expression by a mechanism other than histone methylation. NSD proteins bind promoters (Lucio-Eterovic et al., 2010; Kuo et al., 2011; Rahman et al., 2011) and contain a domain believed to mediate nonhistone protein interactions unique to each NSD family member (He et al., 2013). Thus another possibility is that NSD3 binds and methylates nonhistone proteins such as transcription factors at neural crest promoters to regulate neural crest gene expression. In support of this idea, NSD1 methylates the p65 subunit of NF-κB, leading to transcriptional activation (Lu et al., 2010). It is also possible that chromatin context has an effect; for example, H3K9me3 and H3K36me3 occupancies are equivalent over the Snail2 gene body, whereas H3K36me3 predominates over H3K9me3 in Sox10 (Strobl-Mazzulla et al., 2010). Experiments are underway to test these mechanisms.
Our results also inform understanding of H3K36 methylation generally. It is interesting that NSD3 knockdown only affected H3K36me2 occupancy at the 5′ end of the Sox10 gene, where it was never completely abolished (Figure 5B). This indicates that methyltransferases in addition to NSD3 regulate H3K36me2 in neural crest cells and that this histone mark likely serves multiple functions. Residual H3K36 methylation is similarly observed in NSD3-deficient human cells (Rahman et al., 2011) and throughout the genome in NSD mutant Caenorhabditis elegans (Rechtsteiner et al., 2010). Thus, in our system, it will be important to determine whether persistent, NSD3-independent H3K36me2 is neural crest specific or whether there is a pattern of basal, generic H3K36me2 genome-wide. It is striking that residual H3K36me2 levels (Figure 5B, Sox10, 0.5 and 1.0 kb, green bars) were remarkably consistent (i.e., small SD) and tightly regulated. This contrasts with generally highly variable H3K36me2 occupancy of neural crest genes in control embryos (Figure 5B, blue bars) and at most NSD3-independent sites in MO1 embryos. Neural crest gene H3K36me2 variability could reflect normal epigenetic variation or could indicate that H3K36me2 status is particularly volatile on neural crest genes between four and eight somites, when samples were pooled for analysis.
Neural crest migration requires methylation
By temporally restricting expression of NSD3Δ1707, we were able to bypass the requirement for NSD3 during neural crest specification and reveal that NSD3-related methyltransferase activity is also necessary in migratory neural crest cells. Although a DNA construct can produce active protein ∼4 h after electroporation into chick embryos (Sauka-Spengler and Barembaum, 2008), methyl marks must also turn over after accumulation of NSD3Δ1707 for a phenotype to be apparent. H3K36me2 has an ∼0.5-d half-life in human cell culture (Barth and Imhof, 2010), and thus we expected a delay between NSD3Δ1707 electroporation and the onset of a phenotype. Moreover, we anticipated that the effect would be stochastic if methyl mark turnover were limiting. Indeed, NSD3Δ1707-electroporated neural crest cells appeared to be arrested or defective as they migrated at a range of distances from the neural tube at 10 h postelectroporation (Figure 7). Although NSD3Δ1707 is not necessarily specific for NSD3, as NSD3 is the predominant NSD methyltransferase in the neural crest, particularly at migratory stages (Figure 1 and Supplemental Figure S1) and NSD3 binds different chromatin motifs than NSD1 and 2 (He et al., 2013) and exhibits neural crest–specific functions (Figures 3–5), this result is consistent with a requirement for NSD3 during neural crest migration. In support of this conclusion, NSD3 MO–electroporated neural crest cells that expressed normal levels of Sox10 (i.e., were normally specified) failed to migrate or exhibited impaired migration (Figure 6F), suggesting that NSD3 regulates migration independent of specification. Of importance, the ability of NSD3 lacking the SET domain to interfere with neural crest migration (Figures 6 and 7) suggests that methyltransferase activity, and not another domain or function, is crucial for this process. Although constitutive knockdown of other chromatin regulatory factors impairs migration (Bajpai et al., 2010; Strobl-Mazzulla et al., 2010), this is the first time the requirement for a methylation-associated factor during neural crest migration has been separated from its role during specification.
How NSD3 might act during neural crest migration is unclear. Although H3K36me2 ChIP of migratory neural crest cells would address the possibility that histone methylation is defective, NSD3-deficient cells fail to migrate (Figures 6 and 7), precluding the use of migratory neural crest culture–based ChIP (Rada-Iglesias et al., 2012). Alternatively, MO-electroporated embryonic heads could be used (Strobl-Mazzulla et al., 2010); however, other cell types would predominate and complicate analysis of the migratory neural crest epigenetic signature. It is also less obvious which targets to analyze when assaying migration. Neural crest specification is by nature transcriptional, involving a network of core transcription factors, the expression and output of which will be affected by histone modifications (Betancur et al., 2010; Prasad et al., 2012). However, migration is a dynamic process subject to extensive posttranslational regulation (Rottner and Stradal, 2011; Boulter et al., 2012; Schaefer et al., 2012), including methylation of nonhistone proteins (Vermillion et al., 2014). Although NSD3 might epigenetically regulate migration-essential gene expression or could enable chromatin condensation that is required for motility (Gerlitz and Bustin, 2010), it is also possible that NSD3, like NSD1 (Lu et al., 2010), methylates nonhistone proteins to modulate their activity during migration (Vermillion et al., 2014). Testing this hypothesis is an important priority.
MATERIALS AND METHODS
Chicken embryo culture
Fertile chicken embryos were obtained from local sources, incubated at 37–38°C in a humidified incubator (G.Q.F. Manufacturing, Savannah, GA), and staged according to Hamburger and Hamilton (1951) or by counting somite pairs.
Morpholinos and DNA constructs
Morpholino antisense oligonucleotides (MOs) 3′ modified with fluorescein (FITC) were obtained from GeneTools (Philomath, OR) with the following sequences: NSD3 splice acceptor MO (NSD3 MO1), TGCACCTGGAGAGAACAACACAAGC; 5–base pair mismatch control NSD3 splice acceptor MO (NSD3 mmMO1), TGCACGTCGAGACAACAACAGAACC (mismatches underlined); NSD3 splice donor MO (NSD3 MO2), TGGTGACCTTCCTCCCTTACCTCTT; and NSD3 translation-blocking MO (NSD3 trans MO), AACCAGCAGCATCCCCAAGGTCCAT. The full-length NSD3 overexpression construct was PCR amplified from chick 6- to 10-somite cDNA and cloned into pMES-mCherry (Roffers-Agarwal et al., 2012). NSD3Δ1707 was created by PCR amplifying NSD3 coding sequences up to amino acid 1706 and cloning into pMES (Swartz et al., 2001).
Embryo electroporation
MOs or DNA constructs were unilaterally electroporated ex ovo at HH stages 4–5 or in ovo at one to four somites as previously described (Gammill and Krull, 2011). All MOs were coelectroporated with 300 ng/μl pCS2-MT construct (Turner and Weintraub, 1994) as carrier to increase the efficiency of delivery. After electroporation, embryos were incubated to appropriate stages, fixed for 1 h at room temperature with 4% paraformaldehyde, rinsed with phosphate-buffered saline (PBS) plus 0.1% Tween, and imaged with a Zeiss (Pleasanton, CA) Discovery V8 stereoscope with an AxioCam MRc5 camera and Zeiss AxioVision software to record targeting fluorescence.
In situ hybridization
In situ hybridization was performed as described previously (Wilkinson, 1992) with probes against NSD3 (Adams et al., 2008), Sox10 (Cheng et al., 2000), Snail2 (Gammill and Bronner-Fraser, 2002), FoxD3 (Kos et al., 2001), Sox9 (Kordes et al., 2005), and Msx1 (clone ChEST900p21; Boardman et al., 2002). One kilobase of chick NSD1 and NSD2 sequence excluding conserved domains were PCR amplified from 4- to 12-somite chick cDNA and subcloned into pBluescript for probe synthesis. Chick β-actin sequence (GenBank accession number L08165) between 201 and 1350 nucleotides was PCR amplified and cloned into pCRII-TOPO. Whole-mount embryos were visualized with a Zeiss Discovery V8 stereoscope, and images were acquired with a Zeiss AxioCam MRc5.
Evaluation of phenotypes
After in situ hybridization, embryos were scored, comparing the targeted to the untargeted side in the cranial region with the highest efficiency of transfection (FITC or GFP fluorescence). In embryos electroporated at stages 4–5 (Figures 3, 4, and 6), the severity of the change was categorized as severe decrease (absence or major reduction in premigratory gene expression or failure to migrate), moderate decrease (approximately half the intensity of premigratory gene expression, or about half the distance migrated), or mild decrease (premigratory gene expression minimally reduced or a slight reduction in the distance migrated; see Supplemental Figure S2). Whole-mount embryos were scored, with the exception of β-actin–stained embryos, for which four sections per embryo were scored. In embryos electroporated at one to four somites (Figure 7), neural crest cells could be affected while in transit, and both leading and trailing migratory neural crest cells were scored. In these embryos, a severe decrease indicated that most migrating cells (leading edge or trailing) were delayed; a moderate decrease indicated approximately half the cells were behind in their migration; and a mild decrease indicated a minority of cells exhibited defective migration, evident either as a small difference in migration distance or as a group of lagging cells. The statistical significance of the effects was calculated by Fisher's exact test in R (R Development Core Team, 2011).
Histology
Embryos were infiltrated with 5 and 15% sucrose, mounted in 7.5% gelatin in 15% sucrose, frozen with liquid nitrogen, and stored at −80°C. Embryos were sectioned at 10–20 μm with a Leica (Buffalo Grove, IL) CM1900 cryostat.
Cell culture
DF-1 cells were cultured in DMEM supplemented with 2 mM l-glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin (Life Technologies, Grand Island, NY), 10% fetal bovine serum (VWR International, Radnor, PA), and 5 μg/ml Plasmocin (InvivoGen, San Diego, CA). For transfection, cells were plated on coverslips, and pMES, pMES-NSD3Δ1707, pMES-mCherry, or pMESmCh-NSD3 was delivered with Lipofectamine 2000 according to the manufacturer's instructions (Life Technologies). After 24 h, cells were fixed with 4% paraformaldehyde for 15 min at room temperature and rinsed with PBS plus 0.1% Triton X-100 before immunofluorescence analysis. Images were acquired with Zeiss LSM710 laser scanning confocal microscope and analyzed with ImageJ software (National Institutes of Health, Bethesda, MD). Images were thresholded, and nuclei were selected as regions of interest only if they were nonoverlapping, within the correct focal plane, and not actively dividing. For each transfected nucleus that was analyzed, four to six adjacent, untransfected nuclei were analyzed to account for variability of staining within each field. At least 20 fields of cells were imaged for each condition (5 fields imaged/coverslip, with transfections performed in duplicate on two separate occasions). Statistical analysis was performed in Excel (Microsoft, Redmond, WA).
Antibodies and immunofluorescence
Sections and cells were blocked with 5% fetal bovine serum in PBS plus 0.1% Triton X-100 and incubated with 1:5 anti-HNK1 (American Type Culture Collection, Manassas, VA), 1:200 anti–cleaved caspase 3 (Cell Signaling Technology, Danvers, MA), 1:250 anti–phospho-histone H3 (EMD Millipore, Billerica, MA), 1:100 anti–dimethyl H3K36 (Cell Signaling Technology), 1:25–1:50 anti–trimethyl H3K36 (Cell Signaling Technology), 1:100 anti–dimethyl H3K9 (EMD Millipore), 1:100 anti–dimethyl H3K27 (EMD Millipore), or 1:250 anti–dimethyl H3K4 (Epigentek, Farmingdale, NY), followed by the appropriate secondary antibody (1:250; Rhodamine Red-X anti–immunoglobulin M (IgM) or Rhodamine Red-X anti-rabbit; Jackson ImmunoResearch, West Grove, PA). Sections and cells were mounted with PermaFluor (Thermo Fisher Scientific, Waltham, MA) containing 1 μg/ml 4′,6-diamidino-2-phenylindole (DAPI), and images were acquired using a Zeiss LSM710 laser scanning confocal microscope.
Chromatin immunoprecipitation
After electroporation at HH stages 4–5, neural tubes well targeted with 0.5 mM NSD3 MO1 or mmMO1 were dissected from embryos with four to eight somites, snap frozen, and stored at −80°C. Three independent experiments with 30 neural tubes each for MO1 and mmMO1 were performed. Tubes were homogenized in nuclei extraction buffer (0.5% NP-40, 0.25% Triton X-100, 10 mM Tris-HCl, pH 7.4, 3 mM CaCl2, 0.25 M sucrose, 1 mM dithiothreitol, 0.2 mM phenylmethylsulfonyl fluoride, and EDTA-free protease inhibitors [Complete; Roche, Indianapolis, IN]) and cross-linked with 1% formaldehyde for 10 min at room temperature, followed by 5 min of 125 mM glycine. After centrifugation, the pellet was washed three times with cold PBS with protease inhibitors, snap frozen, and stored at −80°C. After resuspension in SDS lysis buffer (1% SDS, 50 mM Tris, pH 8.0, 10 mM EDTA, and EDTA-free protease inhibitors), chromatin was sheared using a Bioruptor Standard sonicator (Diagenode, Denville, NJ), for 15 min (45 s on, 15 s off). After centrifugation, supernatant was diluted 1:10 with ChIP dilution buffer (16.7 mM Tris-Cl, pH 8.0, 1.1% Triton X-100, 167 mM NaCl, and EDTA-free protease inhibitors). Equal volumes were combined with 5 μg of H3K36me2 antibody (Abcam, Cambridge, MA) or normal rabbit IgG antibody (Cell Signaling Technology) and incubated at 4°C overnight. Protein A Dynabeads (Life Technologies) were blocked with 0.5% bovine serum albumin overnight at 4°C, and then chromatin, antibodies, and beads were incubated at 4°C for 2 h. Bead complexes were washed extensively with RIPA buffer (50 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid–KOH, pH 8, 500 mM LiCl, 1 mM EDTA, 1% NP-40, 0.7% Na-deoxycholate, 1× protease inhibitors), followed by TE/NaCl (10 mM Tris-HCl, 1 mM EDTA pH 8, 50 mM NaCl), magnetically isolated, and eluted in elution buffer (50 mM Tris-HCl, pH 8.0, 10 mM EDTA, 1% SDS) by incubating at 65°C for 20 min with vortexing every 2 min. After centrifugation, 200 mM NaCl was added to supernatant, and cross-linking was reversed by incubating at 60°C overnight. DNA was isolated with the Qiaquick PCR purification kit (Qiagen, Velencia, CA) and quantitatively amplified in duplicate using SYBR Green QPCR reagent (Stratagene, La Jolla, CA) on an MX3000P thermocycler (Stratagene). All primers were previously described (Strobl-Mazzulla et al., 2010), except as follows: Snail2 0.5 kb, forward primer, CTTATTCGGGAAGCCAAAAA, and reverse primer, GGCTTTGCTTCAGGTTTCAG; Sox9 1.5 kb, forward primer, GGAGGGGAATCCTAAATCCA, and reverse primer, TGACCAAACGAGAGGTGTGA; and FoxD3 0.5 kb, forward primer, CAACCGCTTCCCCTACTACC, and reverse primer, CGAACATGTCCTCAGACTGC). The β-actin primers, previously indicated as 0.5 kb downstream from the transcription start site (Strobl-Mazzulla et al., 2010), are actually 2.5 kb downstream and were thus renamed. We used 0.1% of the immunoprecipitated sample as the input. Statistical analysis was performed in Excel (Microsoft).
Supplementary Material
Acknowledgments
We thank Sumit Agarwal for assistance with subcloning and Micah Gearhart, Mark Murphy, and Corinne Fairchild for help with chromatin immunoprecipitation. We are grateful to Ryan Shanley of the Biostatistics and Bioinformatics shared resource of the University of Minnesota Masonic Cancer Center and Wuming Gong for help with statistical analyses, as well as for the resources of the Minnesota Supercomputing Institute. We give many thanks to Yi-Chuan Cheng for the Sox10 plasmid, Katie Vermillion and Julaine Roffers-Agarwal for help with blind scoring, Julaine Roffers-Agarwal and York Marahrens for comments on the manuscript, and Yasuhiko Kawakami for feedback over the course of this project. This work was supported by the National Science Foundation IOS-1052102 (L.S.G.), Basil O’Connor Starter Scholar Research Grant Award 5-FY09-39 from the March of Dimes (L.S.G.), National Institutes of Health F32 DE021651 (B.J.F.), and a University of Minnesota Developmental Biology Center Training Grant (B.J.F.).
Abbreviations used:
- ChIP
chromatin immunoprecipitation
- H3
histone H3
- HH
Hamburger and Hamilton
- K36
lysine 36
- me2
dimethylation
- me3
trimethylation
- MO
morpholino oligonucleotide
- SET
suppressor of variegation3-9, Enhancer of Zeste and Trithorax.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-12-0744) on October 15, 2014.
REFERENCES
- Acloque H, Adams MS, Fishwick K, Bronner-Fraser M, Nieto MA. Epithelial-mesenchymal transitions: the importance of changing cell state in development and disease. J Clin Invest. 2009;119:1438–1449. doi: 10.1172/JCI38019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams MS, Gammill LS, Bronner-Fraser M. Discovery of transcription factors and other candidate regulators of neural crest development. Dev Dyn. 2008;237:1021–1033. doi: 10.1002/dvdy.21513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angrand PO, Apiou F, Stewart AF, Dutrillaux B, Losson R, Chambon P. NSD3, a new SET domain-containing gene, maps to 8p12 and is amplified in human breast cancer cell lines. Genomics. 2001;74:79–88. doi: 10.1006/geno.2001.6524. [DOI] [PubMed] [Google Scholar]
- Asangani IA, Ateeq B, Cao Q, Dodson L, Pandhi M, Kunju LP, Mehra R, Lonigro RJ, Siddiqui J, Palanisamy N, et al. Characterization of the EZH2-MMSET histone methyltransferase regulatory axis in cancer. Mol Cell. 2013;49:80–93. doi: 10.1016/j.molcel.2012.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajpai R, Chen DA, Rada-Iglesias A, Zhang J, Xiong Y, Helms J, Chang CP, Zhao Y, Swigut T, Wysocka J. CHD7 cooperates with PBAF to control multipotent neural crest formation. Nature. 2010;463:958–962. doi: 10.1038/nature08733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister AJ, Schneider R, Myers FA, Thorne AW, Crane-Robinson C, Kouzarides T. Spatial distribution of di- and tri-methyl lysine 36 of histone H3 at active genes. J Biol Chem. 2005;280:17732–17736. doi: 10.1074/jbc.M500796200. [DOI] [PubMed] [Google Scholar]
- Barth TK, Imhof A. Fast signals and slow marks: the dynamics of histone modifications. Trends Biochem Sci. 2010;35:618–626. doi: 10.1016/j.tibs.2010.05.006. [DOI] [PubMed] [Google Scholar]
- Basch ML, Bronner-Fraser M. Neural crest inducing signals. Adv Exp Med Biol. 2006;589:24–31. doi: 10.1007/978-0-387-46954-6_2. [DOI] [PubMed] [Google Scholar]
- Bell O, Wirbelauer C, Hild M, Scharf AN, Schwaiger M, MacAlpine DM, Zilbermann F, van Leeuwen F, Bell SP, Imhof A, et al. Localized H3K36 methylation states define histone H4K16 acetylation during transcriptional elongation in Drosophila. EMBO J. 2007;26:4974–4984. doi: 10.1038/sj.emboj.7601926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betancur P, Bronner-Fraser M, Sauka-Spengler T. Assembling neural crest regulatory circuits into a gene regulatory network. Annu Rev Cell Dev Biol. 2010;26:581–603. doi: 10.1146/annurev.cellbio.042308.113245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackledge NP, Zhou JC, Tolstorukov MY, Farcas AM, Park PJ, Klose RJ. CpG islands recruit a histone H3 lysine 36 demethylase. Mol Cell. 2010;38:179–190. doi: 10.1016/j.molcel.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boardman PE, Sanz-Exquerro J, Overton IM, Burt DW, Bosch E, Fong WT, Tickle C, Brown WRA, Wilson SA, Hubbard SJ. A comprehensive collection of chicken cDNAs. Curr Biol. 2002;12:1965–1969. doi: 10.1016/s0960-9822(02)01296-4. [DOI] [PubMed] [Google Scholar]
- Boulter E, Estrach S, Garcia-Mata R, Feral CC. Off the beaten paths: alternative and crosstalk regulation of Rho GTPases. FASEB J. 2012;26:469–479. doi: 10.1096/fj.11-192252. [DOI] [PubMed] [Google Scholar]
- Cao Q, Yu J, Dhanasekaran SM, Kim JH, Mani RS, Tomlins SA, Mehra R, Laxman B, Cao X, Kleer CG, et al. Repression of E-cadherin by the polycomb group protein EZH2 in cancer. Oncogene. 2008;27:7274–7284. doi: 10.1038/onc.2008.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, Shia WJ, Anderson S, Yates J, Washburn MP, Workman JL. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005;123:581–592. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- Carvalho S, Raposo AC, Martins FB, Grosso AR, Sridhara SC, Rino J, Carmo-Fonseca M, de Almeida SF. Histone methyltransferase SETD2 coordinates FACT recruitment with nucleosome dynamics during transcription. Nucleic Acids Res. 2013;41:2881–2893. doi: 10.1093/nar/gks1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chantalat S, Depaux A, Hery P, Barral S, Thuret JY, Dimitrov S, Gerard M. Histone H3 trimethylation at lysine 36 is associated with constitutive and facultative heterochromatin. Genome Res. 2011;21:1426–1437. doi: 10.1101/gr.118091.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase A, Cross NC. Aberrations of EZH2 in cancer. Clin Cancer Res. 2011;17:2613–2618. doi: 10.1158/1078-0432.CCR-10-2156. [DOI] [PubMed] [Google Scholar]
- Chen MW, Hua KT, Kao HJ, Chi CC, Wei LH, Johansson G, Shiah SG, Chen PS, Jeng YM, Cheng TY, et al. H3K9 histone methyltransferase G9a promotes lung cancer invasion and metastasis by silencing the cell adhesion molecule Ep-CAM. Cancer Res. 2010;70:7830–7840. doi: 10.1158/0008-5472.CAN-10-0833. [DOI] [PubMed] [Google Scholar]
- Cheng Y, Cheung M, Abu-Elmagd MM, Orme A, Scotting PJ. Chick sox10, a transcription factor expressed in both early neural crest cells and central nervous system. Brain Res Dev Brain Res. 2000;121:233–241. doi: 10.1016/s0165-3806(00)00049-3. [DOI] [PubMed] [Google Scholar]
- Dong C, Wu Y, Yao J, Wang Y, Yu Y, Rychahou PG, Evers BM, Zhou BP. G9a interacts with Snail and is critical for Snail-mediated E-cadherin repression in human breast cancer. J Clin Invest. 2012;122:1469–1486. doi: 10.1172/JCI57349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutt A, Ramos AH, Hammerman PS, Mermel C, Cho J, Sharifnia T, Chande A, Tanaka KE, Stransky N, Greulich H, et al. Inhibitor-sensitive FGFR1 amplification in human non-small cell lung cancer. PLoS One. 2011;6:e20351. doi: 10.1371/journal.pone.0020351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egelhofer TA, Minoda A, Klugman S, Lee K, Kolasinska-Zwierz P, Aleksevendo AA, Cheung MS, Day DS, Gadel S, Gorchakov AA, et al. An assessment of histone-modification antibody quality. Nat Struct Mol Biol. 2011;18:91–93. doi: 10.1038/nsmb.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen JS, Smith JC. Controlling morpholino experiments: don’t stop making antisense. Development. 2008;135:1735–1743. doi: 10.1242/dev.001115. [DOI] [PubMed] [Google Scholar]
- Ezponda T, Popovic R, Shah MY, Martinez-Garcia E, Zheng Y, Min DJ, Will C, Neri A, Kelleher NL, Yu J, Licht JD. The histone methyltransferase MMSET/WHSC1 activates TWIST1 to promote an epithelial-mesenchymal transition and invasive properties of prostate cancer. Oncogene 32, 2882–2890. 2013 doi: 10.1038/onc.2012.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang R, Barbera AJ, Xu Y, Rutenberg M, Leonor T, Bi Q, Lan F, Mei P, Yuan GC, Lian C, et al. Human LSD2/KDM1b/AOF1 regulates gene transcription by modulating intragenic H3K4me2 methylation. Mol Cell. 2010;39:222–233. doi: 10.1016/j.molcel.2010.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiki R, Chikanishi T, Hashiba W, Ito H, Takada I, Roeder RG, Kitagawa H, Kato S. GlcNAcylation of a histone methyltransferase in retinoic-acid-induced granulopoiesis. Nature. 2009;459:455–459. doi: 10.1038/nature07954. [DOI] [PubMed] [Google Scholar]
- Gammill LS, Bronner-Fraser M. Genomic analysis of neural crest induction. Development. 2002;129:5731–5741. doi: 10.1242/dev.00175. [DOI] [PubMed] [Google Scholar]
- Gammill LS, Krull CE. Embryological and genetic manipulation of chick development. Methods Mol Biol. 2011;770:119–137. doi: 10.1007/978-1-61779-210-6_5. [DOI] [PubMed] [Google Scholar]
- Gerlitz G, Bustin M. Efficient cell migration requires global chromatin condensation. J Cell Sci. 2010;123:2207–2217. doi: 10.1242/jcs.058271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guffanti A, Iacono M, Pelucchi P, Kim N, Solda G, Croft LJ, Taft RJ, Rizzi E, Askarian-Amiri M, Bonnal RJ, et al. A transcriptional sketch of a primary human breast cancer by 454 deep sequencing. BMC Genomics. 2009;10:163. doi: 10.1186/1471-2164-10-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MA, Wu X, Li AX, Hahn T, Pfeifer GP. Relationship between gene body DNA methylation and intragenic H3K9me3 and H3K36me3 chromatin marks. PLoS One. 2011;6:e18844. doi: 10.1371/journal.pone.0018844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamburger V, Hamilton HL. A series of normal stages in the development of the chick embryo. J Morphol. 1951;88:49–92. [PubMed] [Google Scholar]
- He C, Li F, Zhang J, Wu J, Shi Y. The methyltransferase NSD3 has chromatin-binding motifs, PHD5-C5HCH, that are distinct from other NSD (nuclear receptor SET domain) family members in their histone H3 recognition. J Biol Chem. 2013;288:4692–4703. doi: 10.1074/jbc.M112.426148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houston SI, McManus KJ, Adams MM, Sims JK, Carpenter PB, Hendzel MJ, Rice JC. Catalytic function of the PR-Set7 histone H4 lysine 20 monomethyltransferase is essential for mitotic entry and genomic stability. J Biol Chem. 2008;283:19478–19488. doi: 10.1074/jbc.M710579200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S. Histone methylation and the initiation of cancer. In: Tollefsbol T, editor. Cancer Epigenetics. Boca Raton, FL: CRC Press; 2008. pp. 109–150. [Google Scholar]
- Joshi P, Carrington EA, Wang L, Ketel CS, Miller EL, Jones RS, Simon JA. Dominant alleles identify SET domain residues required for histone methyltransferase of Polycomb repressive complex 2. J Biol Chem. 2008;283:27757–27766. doi: 10.1074/jbc.M804442200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keogh MC, Kurdistani SK, Morris SA, Ahn SH, Podolny V, Collins SR, Schuldiner M, Chin K, Punna T, Thompson NJ, et al. Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell. 2005;123:593–605. doi: 10.1016/j.cell.2005.10.025. [DOI] [PubMed] [Google Scholar]
- Khudyakov J, Bronner-Fraser M. Comprehensive spatiotemporal analysis of early chick neural crest network genes. Dev Dyn. 2009;238:716–723. doi: 10.1002/dvdy.21881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SM, Kee HJ, Eom GH, Choe NW, Kim JY, Kim YS, Kim SK, Kook H, Seo SB. Characterization of a novel WHSC1-associated SET domain protein with H3K4 and H3K27 methyltransferase activity. Biochem Biophys Res Commun. 2006;345:318–323. doi: 10.1016/j.bbrc.2006.04.095. [DOI] [PubMed] [Google Scholar]
- Kordes U, Cheng Y-C, Scotting PJ. Sox group E gene expression distinguishes different types and maturational stages of glial cells in developing chick and mouse. Brain Res Dev Brain Res. 2005;157:209–213. doi: 10.1016/j.devbrainres.2005.03.009. [DOI] [PubMed] [Google Scholar]
- Kos R, Reedy MV, Johnson RL, Erickson CA. The winged-helix transcription factor FoxD3 is important for establishing the neural crest lineage and repressing melanogenesis in avian embryos. Development. 2001;128:1467–1479. doi: 10.1242/dev.128.8.1467. [DOI] [PubMed] [Google Scholar]
- Kuo AJ, Cheung P, Chen K, Zee BM, Kioi M, Lauring J, Xi Y, Park BH, Shi X, Garcia BA, et al. NSD2 links dimethylation of histone H3 at lysine 36 to oncogenic programming. Mol Cell. 2011;44:609–620. doi: 10.1016/j.molcel.2011.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Douarin N, Kalcheim C. The Neural Crest. Cambridge, UK: Cambridge University Press; 1999. [Google Scholar]
- Lee SH, Oshige M, Durant ST, Rasila KK, Williamson EA, Ramsey H, Kwan L, Nickoloff JA, Hromas R. The SET domain protein Metnase mediates foreign DNA integration and links integration to nonhomologous end-joining repair. Proc Natl Acad Sci USA. 2005;102:18075–18080. doi: 10.1073/pnas.0503676102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Trojer P, Xu CF, Cheung P, Kuo A, Drury WJ, 3rd, Qiao Q, Neubert TA, Xu RM, Gozani O, Reinberg D. The target of the NSD family of histone lysine methyltransferases depends on the nature of the substrate. J Biol Chem. 2009;284:34283–34295. doi: 10.1074/jbc.M109.034462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu T, Jackson MW, Wang B, Yang M, Chance MR, Miyagi M, Gudkov AV, Stark GR. Regulation of NF-kappaB by NSD1/FBXL11-dependent reversible lysine methylation of p65. Proc Natl Acad Sci USA. 2010;107:46–51. doi: 10.1073/pnas.0912493107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucio-Eterovic AK, Singh MM, Gardner JE, Veerappan CS, Rice JC, Carpenter PB. Role for the nuclear receptor-binding SET domain protein 1 (NSD1) methyltransferase in coordinating lysine 36 methylation at histone 3 with RNA polymerase II function. Proc Natl Acad Sci USA. 2010;107:16952–16957. doi: 10.1073/pnas.1002653107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min J, Zaslavsky A, Fedele G, McLaughlin SK, Reczek EE, De Raedt T, Guney I, Strochlic DE, Macconaill LE, et al. An oncogene-tumor suppressor cascade drives metastatic prostate cancer by coordinately activating Ras and nuclear factor-kappaB. Nat Med. 2010;16:286–294. doi: 10.1038/nm.2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morishita M, di Luccio E. Cancers and the NSD family of histone lysine methyltransferases. Biochim Biophys Acta. 2011;1816:158–163. doi: 10.1016/j.bbcan.2011.05.004. [DOI] [PubMed] [Google Scholar]
- Moulton JD, Yan Y-L. Using morpholinos to control gene expression. Curr Protoc Mol Biol. 2008 doi: 10.1002/0471142727.mb2608s83. 83, 26.28.21–26.28.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimura K, Ura K, Shiratori H, Ikawa M, Okabe M, Schwartz RJ, Kaneda Y. A histone H3 lysine 36 trimethyltransferase links Nkx2-5 to Wolf-Hirschhorn syndrome. Nature. 2009;460:287–291. doi: 10.1038/nature08086. [DOI] [PubMed] [Google Scholar]
- Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, Lee TI, Bell GW, Walker K, Rolfe PA, Herbolsheimer E, et al. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122:517–527. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- Prasad MS, Sauka-Spengler T, LaBonne C. Induction of the neural crest state: control of stem cell attributes by gene regulatory, post-transcriptional and epigenetic interactions. Dev Biol. 2012;366:10–21. doi: 10.1016/j.ydbio.2012.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao Q, Li Y, Chen Z, Wang M, Reinberg D, Xu RM. The structure of NSD1 reveals an autoregulatory mechanism underlying histone H3K36 methylation. J Biol Chem. 2011;286:8361–8368. doi: 10.1074/jbc.M110.204115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Prescott S, Brugmann SA, Swigut T, Wysocka J. Epigenomic annotation of enhancers predicts transcriptional regulators of human neural crest. Cell Stem Cell. 2012;11:633–648. doi: 10.1016/j.stem.2012.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman S, Sowa ME, Ottinger M, Smith JA, Shi Y, Harper JW, Howley PM. The Brd4 extraterminal domain confers transcription activation independent of pTEFb by recruiting multiple proteins, including NSD3. Mol Cell Biol. 2011;31:2641–2652. doi: 10.1128/MCB.01341-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayasam GV, Wendling O, Angrand PO, Mark M, Niederreither K, Song L, Lerouge T, Hager GL, Chambon P, Losson R. NSD1 is essential for early post-implantation development and has a catalytically active SET domain. EMBO J. 2003;22:3153–3163. doi: 10.1093/emboj/cdg288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. R Foundation for Statistical Computing. Vienna, Austria: 2011. R: A Language and Environment for Statistical Computing. www.R-project.org/ [Google Scholar]
- Rechtsteiner A, Ercan S, Takasaki T, Phippen TM, Egelhofer TA, Wang W, Kimura H, Lieb JD, Strome S. The histone H3K36 methyltransferase MES-4 acts epigenetically to transmit the memory of germline gene expression to progeny. PLoS Genet. 2010;6:e1001091. doi: 10.1371/journal.pgen.1001091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren G, Baritaki S, Marathe H, Feng J, Park S, Beach S, Bazeley PS, Beshir AB, Fenteany G, Mehra R, et al. Polycomb protein EZH2 regulates tumor invasion via the transcriptional repression of the metastasis suppressor RKIP in breast and prostate cancer. Cancer Res. 2012;72:3091–3104. doi: 10.1158/0008-5472.CAN-11-3546. [DOI] [PubMed] [Google Scholar]
- Roffers-Agarwal J, Hutt KJ, Gammill LS. Paladin is an antiphosphatase that regulates neural crest cell formation and migration. Dev Biol. 2012;371:180–190. doi: 10.1016/j.ydbio.2012.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roopra A, Qazi R, Schoenike B, Daley TJ, Morrison JF. Localized domains of G9a-mediated histone methylation are required for silencing of neuronal genes. Mol Cell. 2004;14:727–738. doi: 10.1016/j.molcel.2004.05.026. [DOI] [PubMed] [Google Scholar]
- Rosati R, La Starza R, Veronese A, Aventin A, Schwienbacher C, Vallespi T, Negrini M, Martelli MF, Mecucci C. NUP98 is fused to the NSD3 gene in acute myeloid leukemia associated with t(8;11)(p11.2;p15) Blood. 2002;99:3857–3860. doi: 10.1182/blood.v99.10.3857. [DOI] [PubMed] [Google Scholar]
- Rottner K, Stradal TE. Actin dynamics and turnover in cell motility. Curr Opin Cell Biol. 2011;23:569–578. doi: 10.1016/j.ceb.2011.07.003. [DOI] [PubMed] [Google Scholar]
- Sauka-Spengler T, Barembaum M. Gain- and loss-of-function approaches in the chick embryo. Methods Cell Biol. 2008;87:237–256. doi: 10.1016/S0091-679X(08)00212-4. [DOI] [PubMed] [Google Scholar]
- Schaefer A, Nethe M, Hordijk PL. Ubiquitin links to cytoskeletal dynamics, cell adhesion and migration. Biochem J. 2012;442:13–25. doi: 10.1042/BJ20111815. [DOI] [PubMed] [Google Scholar]
- Strobl-Mazzulla PH, Sauka-Spengler T, Bronner-Fraser M. Histone demethylase JmjD2A regulates neural crest specification. Dev Cell. 2010;19:460–468. doi: 10.1016/j.devcel.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swartz ME, Eberhart J, Pasquale EB, Krull CE. EphA4/ephrin-A5 interactions in muscle precursor cell migration in the avian forelimb. Development. 2001;128:4669–4680. doi: 10.1242/dev.128.23.4669. [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Nakayama Y, Taniguchi M, Kioussis D. Regulation of early T cell development by the PHD finger of histone lysine methyltransferase ASH1. Biochem Biophys Res Commun. 2008;365:589–594. doi: 10.1016/j.bbrc.2007.10.159. [DOI] [PubMed] [Google Scholar]
- Theveneau E, Mayor R. Neural crest delamination and migration: from epithelium-to-mesenchyme transition to collective cell migration. Dev Biol. 2012;366:34–54. doi: 10.1016/j.ydbio.2011.12.041. [DOI] [PubMed] [Google Scholar]
- Tonon G, Wong KK, Maulik G, Brennan C, Feng B, Zhang Y, Khatry DB, Protopopov A, You MJ, Aguirre AJ, et al. High-resolution genomic profiles of human lung cancer. Proc Natl Acad Sci USA. 2005;102:9625–9630. doi: 10.1073/pnas.0504126102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner DL, Weintraub H. Expression of achaete-scute homolog 3 in Xenopus embryos converts ectodermal cells to a neural fate. Genes Dev. 1994;8:1434–1447. doi: 10.1101/gad.8.12.1434. [DOI] [PubMed] [Google Scholar]
- Vermillion KL, Lidberg KA, Gammill LS. Cytoplasmic protein methylation is essential for neural crest migration. J Cell Biol. 2014;204:95–109. doi: 10.1083/jcb.201306071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner EJ, Carpenter PB. Understanding the language of Lys36 methylation at histone H3. Nat Rev Mol Cell Biol. 2012;13:115–126. doi: 10.1038/nrm3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson D. Situ Hybridization: A Practical Approach. Oxford, UK: Oxford University Press; 1992. Whole mount in situ hybridization of vertebrate embryos; pp. 75–83. [Google Scholar]
- Yang ZQ, Liu G, Bollig-Fischer A, Giroux CN, Ethier SP. Transforming properties of 8p11-12 amplified genes in human breast cancer. Cancer Res. 2010;70:8487–8497. doi: 10.1158/0008-5472.CAN-10-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.