Key Points
Inhibiting LEDGF interaction with a novel fragment of MLL represents an attractive approach to develop new drugs for MLL leukemias.
Structural studies reveal a new pocket on the LEDGF IBD suitable for targeting by small-molecule inhibitors.
Abstract
Lens epithelium-derived growth factor (LEDGF) is a chromatin-associated protein implicated in leukemia and HIV type 1 infection. LEDGF associates with mixed-lineage leukemia (MLL) fusion proteins and menin and is required for leukemic transformation. To better understand the molecular mechanism underlying the LEDGF integrase-binding domain (IBD) interaction with MLL fusion proteins in leukemia, we determined the solution structure of the MLL-IBD complex. We found a novel MLL motif, integrase domain binding motif 2 (IBM2), which binds to a well-defined site on IBD. Point mutations within IBM2 abolished leukemogenic transformation by MLL-AF9, validating that this newly identified motif is essential for the oncogenic activity of MLL fusion proteins. Interestingly, the IBM2 binding site on IBD overlaps with the binding site for the HIV integrase (IN), and IN was capable of efficiently sequestering IBD from the menin-MLL complex. A short IBM2 peptide binds to IBD directly and inhibits both the IBD-MLL/menin and IBD-IN interactions. Our findings show that the same site on IBD is involved in binding to MLL and HIV-IN, revealing an attractive approach to simultaneously target LEDGF in leukemia and HIV.
Introduction
Lens epithelium–derived growth factor (LEDGF) is a chromatin-associated protein belonging to the hepatoma-derived growth factor family.1 It functions as a transcriptional coactivator activating stress-related genes2,3 and promotes growth and survival of lens epithelial cells, keratinocytes, and fibroblasts.4 Accumulating data involve LEDGF in acute myeloid leukemia (AML). Recurring chromosomal translocations of LEDGF with nucleoporin NUP98 gene have been identified in pediatric AML.5,6 Furthermore, the LEDGF/p75 isoform is consistently upregulated in a subset of AMLs resistant to chemotherapy.7 LEDGF is a critical cofactor of mixed-lineage leukemia (MLL) fusion proteins required for leukemic transformation.8 Importantly, disruption of the LEDGF interaction with MLL fusion proteins abolishes development of acute leukemia in vivo, suggesting that the LEDGF-MLL interaction represents a valuable target for development of novel antileukemic drugs.8
LEDGF plays an essential role in HIV type 1 (HIV-1) pathogenesis.9 LEDGF is a key host protein required for chromatin binding activity of HIV-1 integrase (IN).10 The catalytic domain of HIV-IN binds to the C-terminal integrase-binding domain (IBD) of LEDGF,11 and multiple studies validated this protein-protein interaction as necessary for HIV-1 replication.9,11,12 Targeting the IBD-IN interaction attracted strong attention as a valuable approach to develop novel antiretroviral drugs for HIV infection,13 and a number of studies reported small molecules and peptidomimetics binding to the HIV-IN and blocking its interaction with the LEDGF IBD. Such compounds interfere with HIV-IN activity and inhibit replication of HIV-1 in cell culture.14,15
Biochemical and structural studies revealed that the LEDGF IBD also interacts with menin and MLL.8,16 Therefore, the IBD plays a dual role as a cofactor of MLL fusion proteins in leukemia and as a cellular cofactor of viral IN in HIV infection. Consequently, protein-protein interactions of LEDGF IBD might constitute an attractive target for development of novel therapeutics. However, molecular details of IBD interaction with MLL and menin are not entirely understood. A recently reported crystal structure of the menin-MLL-LEDGF complex revealed that a helical fragment of MLL spanning residues 113 to 134 is involved in the interactions with LEDGF IBD.16 On the other hand, functional studies have shown that deletion of MLL residues 123 to 153 abolished MLL binding to LEDGF,8 revealing that a longer fragment might be required for formation of the MLL-LEDGF complex. In order to understand the molecular mechanism underlying the IBD interaction with MLL fusion proteins, we determined the solution structure of the MLL-IBD complex. Importantly, we identified a novel motif within MLL, which directly interacts with IBD and is critical for leukemogenic activity of MLL-AF9. Interestingly, we also found that the same site on IBD is involved in binding to this newly identified MLL fragment and to the HIV-IN. Our work provides an important structural basis to simultaneously target the protein-protein interactions of LEDGF in leukemia and HIV.
Methods
Molecular biology
LEDGF constructs for bacterial expression have been ordered from GenScript and cloned into pET32a. Full-length LEDGF complementary DNA was amplified using reverse transcription–polymerase chain reaction (RT-PCR) from messenger RNA extracted from HEK293 cells. The expression vector pCMV Flag-MLL-AF9 was prepared using pMSCV Flag-MLL-AF9 as template.17 Complementary DNA encoding IN and MLL110-160 constructs was synthesized by GeneArt Strings (Invitrogen). Detailed description of constructs is provided in the supplemental Methods (see the Blood Web site).
Protein expression and purification
IBD, MLL fragments, and the MLL-IBD fusions for in vitro studies were expressed in Escherichia coli as insoluble proteins. Inclusion bodies were solubilized in 6 M guanidine hydrochloride and refolded on-column bound to Ni-NTA resin (Qiagen). The thioredoxin-His6 tag has been cleaved by 3C protease, and final proteins were purified by ion exchange chromatography using SP Sepharose (GE Healthcare). IBD was concentrated and dialyzed into 50 mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid, pH 7.5, 50 mM NaCl, and 1 mM dithiothreitol. All other proteins were concentrated and dialyzed into 50 mM tris(hydroxymethyl)aminomethane-HCl, pH 7.5, 50 mM NaCl, and 1 mM β-mercaptoethanol. Full-length human menin was purified as described elsewhere.18 Proteins labeled with stable isotopes (13C, 15N) were expressed in M9 minimal media. HIV-IN was captured by HisTrap HP (GE Healthcare) followed by ion exchange chromatography on a Q Sepharose column (GE Healthcare).
Nuclear magnetic resonance (NMR) binding experiments
Mapping of IBD-MLL interaction was performed using 15N-labeled IBD and unlabeled MLL fragments. To determine binding affinity, 70 μM 15N-labeled IBD was titrated with MLL1-160 and the 2 mutants F129A and F148A,L149A (FLAA), and chemical shift perturbations were measured from heteronuclear single quantum coherence (HSQC) spectra. The Kd values were determined using a previously established procedure.19
The IBD2 peptide corresponding to the MLL 146-153 fragment was ordered from GenScript.
Structure determination of MLL-IBD
NMR spectra were measured on Bruker Avance III 600 MHz NMR spectrometers at 25°C. Protein samples contained 0.34 mM 13C/15N-labeled MLL-IBD in 50 mM phosphate buffer, pH 6.5, and 50 mM NaCl supplemented with 5% D2O. Backbone assignment was completed based on triple-resonance experiments, and the structure has been calculated using Cyana 2.120 based on cross peaks derived from Nuclear Overhauser Effect Spectroscopy spectra and dihedral angle restraints from Talos+.21 The more detailed procedure is described in the supplemental Data. The coordinates for the MLL-IBD structure have been deposited in the Protein Data Bank (PDB) with the accession number 2MTN.
Isothermal titration calorimetry (ITC)
Menin-MLL1-160 complex was purified by size exclusion chromatography and titrated with IBD using a VP–Isothermal Titration Calorimeter. Menin-MLL1-160 complex was used at 20 μM concentration, 1000-fold above Kd for menin-MLL1-160 interaction.18 Further details are provided in supplemental Methods.
Coimmunoprecipitation (co-IP) of MLL-AF9 with menin and LEDGF
HEK293 cells were cotransfected by pCMV-Flag-MLL-AF9 or pCMV-Flag-MLL-AF9-FLAA with pMSCV-Myc-menin and pMSCV-LEDGF-HA plasmids using FuGENE 6 (Promega). Forty-eight hours after transfection, whole-cells lysates were immunoprecipitated with anti-FLAG M-2 Magnetic Beads (M8823) (Sigma-Aldrich) and analyzed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis and western blotting using anti-N-MLL antibody (05-764; Millipore), anti-β-actin antibody (A00702; GenScript), anti-menin antibody, and anti-HA tag antibody (2367S; Cell Signaling).
AlphaLISA assays
To assess the inhibition of IBD-menin-MLL interaction, we optimized AlphaLISA assay using 62.5 nM His-Trx-MLL160, 62.5 nM menin, and 62.5 nM Flag-IBD. Proteins were incubated for 1 hour in buffer containing 50 mM 4-morpholinepropanesulfonic acid (pH 7.25), 50 mM NaCl, 1 mM (tris[2-carboxyethyl]phosphine), 0.02% (weight to volume ratio) bovine serum albumin, and 0.025% (volume to volume ratio) Tween-20. Ni-chelate-coated donor beads and anti-Flag acceptor beads (10 μg/mL final concentration) were added and incubated for 1 hour. The luminescence signal was recorded using excitation at 680 nm and emissions at 615 nm using the PHERAstar microplate reader (BMG). Competition experiments with either integrase domain binding motif (IBM) 2 peptide or HIV-IN were performed with twofold serial dilutions. Data were analyzed using the Origin 7.0.
To assess the inhibition of HIV-IN interaction with IBD, we employed a very similar protocol as discussed previously using 62 nM His-HIV-IN and 1 μM Flag-IBD. Ni-chelate-coated donor beads and anti-Flag acceptor beads were used at 2.5 μg/mL concentration. Competition experiments were performed with IBM2 using threefold serial dilutions.
Virus production and bone marrow transformation assays
The pMSCV (for MLL-AF9 and the F129A and FLAA mutants) constructs were transfected using FuGENE 6 reagent (Roche) into Plat-E cells selected in puromycin (1 μg/mL) and blasticidin (10 μg/mL). Media containing the recombinant retrovirus were collected for transduction at 48 and 72 hours posttransfection. Bone marrow transformation assays were performed by transducing Lin-BM cells with retrovirus containing MLL-AF9 (or mutants) as described previously.17 To assess the growth, the transduced cells were cultured in Iscove modified Dulbecco medium supplemented with 15% fetal bovine serum, 1% penicillin/streptomycin, stem cell factor, and interleukin (IL) 3 and counted daily for 6 days. Colony formation was assessed by plating transformed cells in methylcellulose medium (STEMCELL Technologies), supplemented with IL-3, IL-6, stem cell factor, granulocyte macrophage–colony-stimulating factor, and 1 mg/mL G418. After 3 rounds of plating, colony-forming unit (with >50 cells) was scored under the microscope; colonies were stained using 0.1% iodonitrotetrazolium chloride and scanned. Cells harvested at the end of the experiment were cytospun and stained with the Hema 3 Stain Kit (Thermo Fisher Scientific). Images were acquired using a ×100 lens under an Olympus BX-51 microscope (Olympus).
Quantitative RT-PCR (qRT-PCR)
Reverse transcription and quantitative PCR were performed as previously described.17 TaqMan primer probes for HOXA9 and β-ACTIN were purchased from Applied Biosystems. Expression levels were analyzed using the comparative ΔΔCt method as described in ABI User Bulletin #2.
Chromatin immunoprecipitation (ChIP)
ChIP assays were performed as previously described using cells transfected with Hoxa9-luciferase (LUC) promoter, pMSCV-MLL-AF9, or the F129A or FLAA mutant plasmids.22 Antibodies against menin, LEDGF, AF9, and IgG were obtained from Bethyl Laboratories Inc. Quantitative PCR was performed on the precipitated DNAs with TaqMan primers and probes from Applied Biosystems. TaqMan probes for Hoxa9 were the same as used previously.22
Dual luciferase assay
HEK293 cells were cotransfected with pMSCV-MLL-AF9, pCMV-LEDGF, MSCV-menin, Renilla luciferase reporter (internal control), Hoxa9-LUC reporter, and increasing concentrations of pMIGR1-IN with FuGENE 6 according to the manufacturer’s instructions. Luciferase assays were performed using the Dual Luciferase Assay Kit (Promega) according to the manufacturer’s instructions. Emission was detected using a Monolight 3010 Luminometer (BD Biosciences).
Bone marrow cotransduction assay
Lin-c-Kit+ cells isolated from C57BL/6 mice were transducted with MSCV-neo-Flag-MLL-AF9. The following day, the cells were transduced with retroviruses packaged with MSCV-puro-IN or MSCV-puro-MLL110-160, MSCV-puro-MLL110-160 FLAA, MSCV-puro-MLL110-160 F129A. Cells were selected with G418 (1 mg/mL, Gibco) and puromycin (2 µg/mL, Sigma) for a week. Following the selection, cells were cultured for 2 weeks. Viable cells were counted using trypan blue staining. Following day 14, expression of Hoxa9 was analyzed using qRT-PCR, c-Kit level by fluorescence-activated cell sorter, and morphology of cells by Wright-Giemsa staining. In addition, 3 rounds of serial replating assay were performed. The more detailed procedure is described in the supplemental Data.
Results
Two separate MLL motifs are involved in the interaction with LEDGF IBD
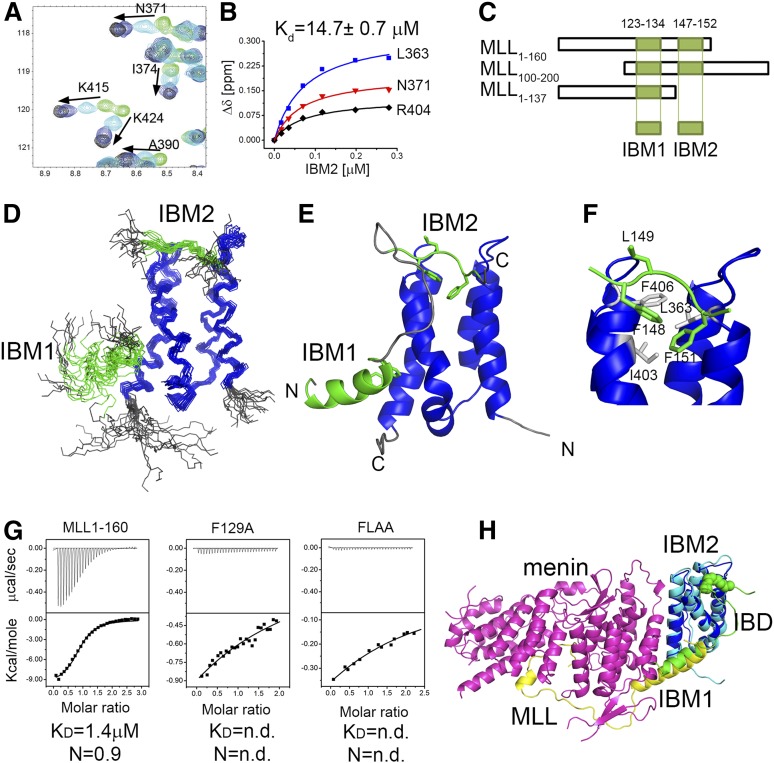
To accurately map the MLL fragment necessary for the interaction with LEDGF, we tested binding of various MLL fragments to the IBD of LEDGF using NMR. First, we found that MLL1-160 binds to the IBD in the absence of menin with moderate affinity (Kd = 14.7 ± 0.7 µM) (Figure 1A-B). Further mapping revealed that the MLL fragment comprising residues 100 to 160 is required for binding to the IBD (Figure 1C and supplemental Figure 1). Interestingly, a recently published crystal structure of menin-MLL-LEDGF complex lacks any contacts involving MLL upstream of residue 135.16 To understand the role of MLL residues in binding to the IBD, we determined the solution structure of MLL-IBD complex using NMR spectroscopy. To overcome a modest affinity, we designed a covalent complex by fusion of MLL to the IBD, which is a frequently adopted approach for structure determination of protein-protein complexes by NMR.23,24
Figure 1.
Characterization of MLL-IBD interaction and the solution structure of MLL-IBD complex. (A) HSQC spectrum for 70 μM IBD (green) titrated with 35 μM (cyan), 120 μM (blue), and 280 μM MLL160 (black). (B) Determination of IBD-MLL160 binding affinity from NMR titrations. Representative titration curves for 3 residues are shown, and the average Kd is reported. (C) Schematics of MLL constructs used for mapping the IBM1 and IBM2 binding motifs. (D) The 20 lowest energy structures of MLL-IBD. Well-ordered MLL fragments corresponding to residues 124 to 133 (IBM1) and 147 to 152 (IBM2) are shown in green, and IBD (residues 349-428) is blue. (E) Representative structure of MLL-IBD; coloring is the same as in panel D. (F) Details of IBM2-IBD interaction. Key residues involved in the contacts are green (IBM2) and gray (IBD). (G) Characterization of binding affinity using ITC. Equimolar complexes of menin with MLL160, MLL160 F129A, and MLL160 FLAA were titrated with IBD. Experiments were performed twice, and Kd and stoichiometry (N) are shown for representative experiments. (H) Model of menin-MLL-LEDGF complex based on solution MLL-IBD structure and previously published crystal structure (PDB code 3U88).
The MLL-IBD fusion construct optimized for structural studies included a 110 to 160 fragment of MLL fused to a 337 to 442 fragment of LEDGF encompassing the IBD. Comparison of the 1H-15N HSQC spectra of the IBD saturated with MLL1-160 with the spectra of the MLL110-160-IBD fusion clearly shows that linking of the 2 protein fragments did not affect the structure of the complex (supplemental Figure 2). We then determined the structure of the MLL-IBD complex in solution using a total of 1255 distance restraints, including 86 inter–MLL-IBD distances (supplemental Table 1). The overall structure of the complex is well defined with 2.0 Å backbone root mean square deviation for ordered IBD and MLL fragments in the 20 lowest energy conformers. Based on the structure, we found that the IBD interacts with MLL110-160 via 2 distinct fragments, which we labeled, respectively, IBM1 (residues 123-134) and IBM2 (residues 147-152) (Figure 1C-E). Although the IBM1 has been reported in the crystal structure,16 the IBM2 has not been observed before and represents a novel motif within MLL.
The IBM1 and IBM2 motifs form independent contacts with the IBD and are separated by a flexible linker that is 12 amino acids long. The IBM1 is an α-helical motif, which packs onto the site formed by 2 C-terminal helices of the IBD (Figure 1D-E). Interestingly, this helix is much longer in the ternary menin-MLL-IBD complex and contacts both menin and the IBD (Figure 1H). A second motif, IBM2, is short and interacts with the IBD in an extended conformation. The key contacts involve packing of MLL IBM2 side chains of F148 and F151 into a concave hydrophobic face of the IBD lined by L363, I403, and F406 (Figure 1F).
Both IBM1 and IBM2 are necessary for high-affinity association of LEDGF with menin-MLL complex
It has been previously reported that menin is necessary to tether MLL to LEDGF.8 Here, we provide evidence that MLL directly binds to the IBD in the absence of menin, albeit with moderate affinity, Kd = 14.7 µM (Figure 1A-B). To assess how menin affects the MLL-IBD interaction, we employed ITC and quantified binding of the IBD to the menin-MLL1-160 complex. As compared with the binary interaction, IBD binds to the menin-MLL complex with ∼10-fold increased affinity (Kd = 1.4 µM, Figure 1G).
In order to test whether both IBM1 and IBM2 motifs in MLL are required for high-affinity binding of IBD to the menin-MLL complex, we used mutagenesis. We introduced the mutation of F129A in IBM1, which has been shown previously to abolish MLL-ENL binding to LEDGF.8 To impair interaction of IBM2 with IBD, we introduced single F148A and double F148A and L149A (FLAA) mutations (Figure 1F). We assessed how these mutations affect binary interactions between MLL1-160 and the IBD, and using NMR, we found that F129A has a rather weak effect (supplemental Figure 3), F148A decreased the binding ∼10-fold, and the FLAA mutation almost completely abolished this interaction (supplemental Figure 3). Subsequently, we employed ITC to test the impact of mutations within IBM1 and IBM2 on formation of the ternary complex with menin and IBD. Both F129A and FLAA mutations result in a very severe decrease in binding affinity of the IBD to the menin-MLL1-160 complex (Figure 1G). Altogether, these data demonstrate that the intact fragment of MLL containing both IBM1 and IBM2 is required to maintain high-affinity menin-MLL-IBD complex.
IBM2 is required for leukemogenic activity of MLL-AF9
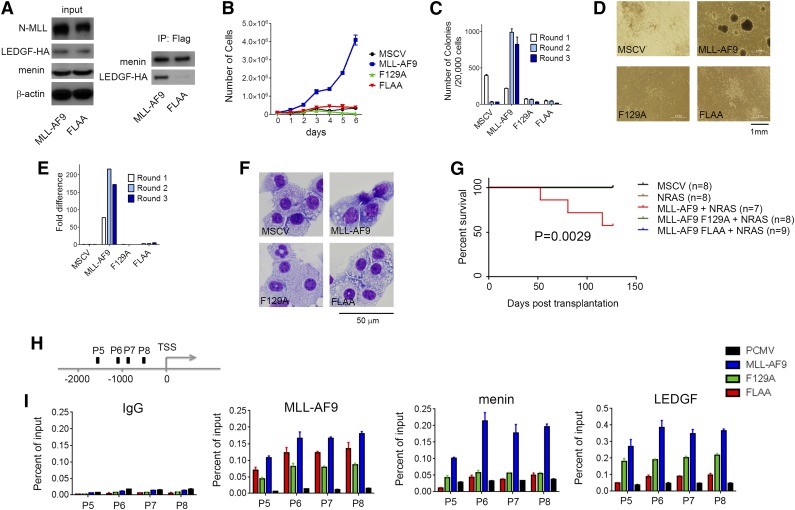
Interaction of LEDGF with the menin-MLL-fusion protein complex is essential for oncogenic activity of MLL fusion proteins.8 The F129A mutation has previously been shown to abolish binding of LEDGF to the menin-MLL-ENL complex as assessed by co-IP,8 which is consistent with our binding data (Figure 1G). To assess the role of the second motif, IBM2, we introduced FLAA mutation into MLL-AF9. The co-IP clearly shows that MLL-AF9 interacts simultaneously with menin and LEDGF (Figure 2A). On the contrary, the MLL-AF9 FLAA mutant interacts only with menin but not with LEDGF (Figure 2A). These results demonstrate that binding of LEDGF to the menin-MLL-AF9 requires intact IBM2, recapitulating the in vitro binding data.
Figure 2.
Intact IBM2 is necessary for leukemogenic transformation by MLL-AF9. (A) Co-IP demonstrates that mutation FLAA in MLL-AF9 abolished the interaction with LEDGF but not with menin. HEK293 cells were transduced with Myc-menin, LEDGF-HA, and Flag-MLL-AF9 or Flag-MLL-AF9 FLAA and coimmunoprecipitated using anti-Flag antibody. (B) Growth curves for bone marrow cells (BMCs) transformed with MSCV vector, MLL-AF9, and the F129A and FLAA mutants. (C) Activity of MLL-AF9 and the mutants in colony formation assay in transformed BMCs. Colony counts after 3 rounds of replating (C) and representative colonies (D) are shown. (E) qRT-PCR in BMCs transformed with MLL-AF9 and the 2 mutants for 3 replating rounds showing expression of Hoxa9. Expression of Hoxa9 has been normalized to β-actin and is referenced to the MSCV vector. (F) Wright-Giemsa-stained cytospins demonstrating differentiation of BMCs transformed with MSCV, MLL-AF9, and the 2 mutants. (G) Survival of mice transplanted with MSCV-MLL-AF9 and F129 and FLAA mutants. NRAS has been included to accelerate development of leukemia; n represents number of animals in each group. (H) Schematic diagram showing the endogenous Hoxa9 locus and probes used in the ChIP assay. TSS, transcription start site. (I) ChIP assays performed in HEK293 cells transduced with MLL-AF9 and mutants F129A and FLAA to determine the recruitment of MLL-AF9, menin, and LEDGF to the Hoxa9 locus with 4 different probes. Antibodies against AF9, menin, and LEDGF were used.
To test whether both the IBM1 and IBM2 motifs are required for the oncogenic activity of an MLL fusion protein, we transduced murine BMCs with MLL-AF9 and the 2 mutants, F129A and FLAA. We found that only MLL-AF9 was capable of efficiently transforming BMCs and sustained proliferation (Figure 2B), whereas the BMCs transformed with either of the 2 mutants had severe defects in proliferation (Figure 2B). The BMCs transformed with MLL-AF9 form colonies in methylcellulose media and can be serially replated (Figure 2C). However, both F129A and FLAA mutations almost completely abolished colony-forming unit potential (Figure 2C-D). The oncogenic activity of MLL fusion proteins is also associated with high expression of Hoxa9, which is a downstream target of MLL-AF9.25 To test whether disruption of the LEDGF interaction with menin-MLL-AF9 affects Hoxa9 expression, we performed qRT-PCR. The BMCs expressing F129A and FLAA mutants show very low levels of Hoxa9 as compared with MLL-AF9–transformed BMCs (Figure 2E). Transformation with MLL-AF9 leads to a block in cellular differentiation of hematopoietic cells. Comparison of the morphology of BMCs transformed with MLL-AF9 and the 2 mutants revealed a more differentiated phenotype for cells harboring F129A and FLAA mutants compared with MLL-AF9 (Figure 2F), as assessed by increased cell size, higher cytoplasm to nuclear ratio, and highly vacuolated cytoplasm. Furthermore, we tested whether F129A and FLAA mutations would impair development of leukemia in vivo. Although nearly half of animals transplanted with MLL-AF9 developed leukemia, we did not observe any disease in mice transplanted with MLL-AF9 bearing either F129 or FLAA mutations (Figure 2G; supplemental Figure 4). Altogether, these results demonstrate that mutations within the IBM1 and IBM2 that abolish the interaction of MLL with LEDGF result in loss of transforming properties by MLL-AF9.
Mutations in IBMs disrupt cooperative assembly of menin-MLL-AF9-LEDGF complex on Hoxa9
To test whether disruption of the MLL-LEDGF interaction impairs the recruitment of MLL-AF9 to Hoxa9 loci, we performed ChIP experiments. We found that the presence of F129A and FLAA mutations in MLL-AF9 significantly decreased the occupancy of MLL-AF9 at 4 distinct sites on the Hoxa9 promoter region (Figure 2H-I). Importantly, we also observed strong loss of menin binding and a very significant decrease in the level of LEDGF at Hoxa9 for both MLL-AF9 mutants (Figure 2H-I). These findings strongly suggest that the ternary complex of menin, MLL-AF9, and LEDGF cooperatively assembles on Hoxa9.
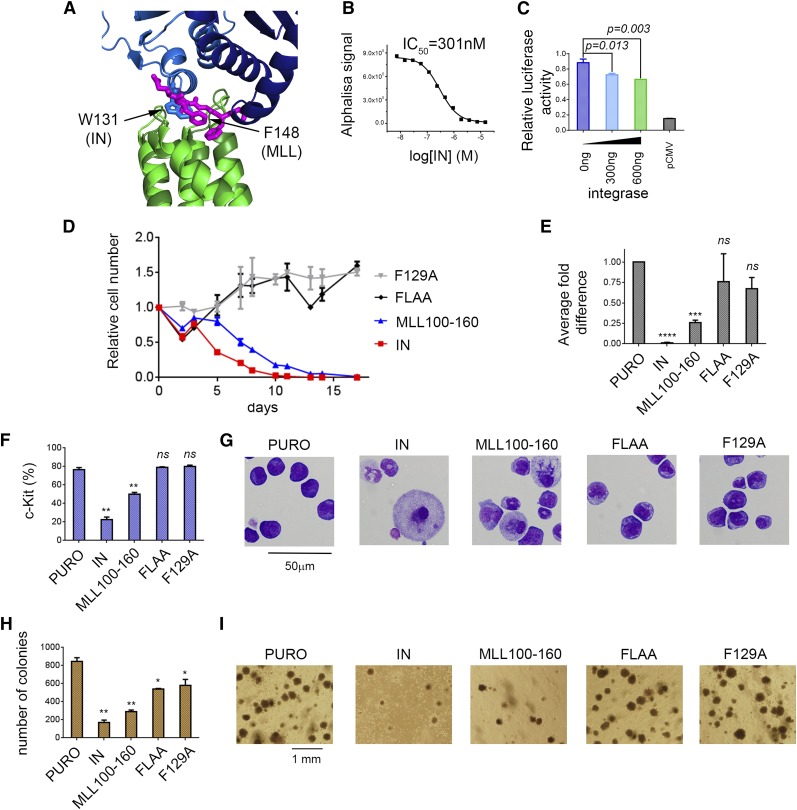
HIV-IN sequesters IBD from menin-MLL-LEDGF complex
LEDGF plays an intriguing dual role being a component of the menin-MLL and menin-MLL fusion complexes as well as a binding target of HIV IN required for the integration of the HIV genome into host chromatin.8,9 Both activities of LEDGF require the IBD, which is involved in the protein-protein interactions with menin-MLL and IN. Interestingly, the analysis of the crystal structure of the IN-IBD complex26 and our structure of MLL-IBD revealed that the IN binding site on IBD overlaps with the binding site of the IBM2 MLL motif (Figure 3A). For example, the W131 from IN occupies the same position as F148 from IBM2 (Figure 3A). Based on the structural analysis, we hypothesized that IN should compete with MLL for binding to the IBD. Because the IN interacts with IBD with 2 orders of magnitude higher affinity (Kd∼10 nM)27 than IBD binds to menin-MLL, we anticipated that IN should be capable of sequestering IBD from the ternary complex with menin-MLL. To test this, we developed an AlphaLISA assay (see supplemental Methods), and, as predicted, IN proved to be a very potent competitor, disrupting the IBD-menin-MLL interaction with 50% inhibition concentration (IC50) = 301 nM (Figure 3B).
Figure 3.
The same site on IBD interacts with IBM2 and HIV-IN. (A) Superposition of MLL-IBD structure (MLL IBM2 is shown in magenta, and IBD is green) onto IBD-IN complex (IN dimer is blue; based on PDB structure 2B4J). W131 from the IN binds to the same site on IBD as F148 from IBM2 motif. (B) IN disrupts menin-MLL-IBD complex in AlphaLISA assay by sequestering IBD from menin-MLL. (C) Dual luciferase assay performed in HEK293 cells with Hoxa9-LUC reporter. Cells were cotransfected with menin, MLL-AF9, LEDGF, and increasing amounts of IN. (D-I) Expression of IN and MLL110-160 impairs transformation of BMCs with MLL-AF9. BMCs were cotransduced with MSCV-neo-MLL-AF9 and MSCV-puro containing IN, MLL110-160, and MLL110-160 F129A or MLL110-160 FLAA. Expression of IN and MLL fragments has been validated using qRT-PCR (supplemental Figure 5). (D) Growth curves normalized to MSCV-puro. (E) Expression of Hoxa9 in transformed cells (day 14). (F) Expression of c-Kit (day 14). (G) Wright-Giemsa-stained cytospins (day 14). (H) Colony counts after 3 rounds of replating. (I) Representative colonies in round 3.
To further assess whether IN disrupts the LEDGF-menin-MLL-AF9 interaction in cells, we employed the luciferase reporter assay. We expected that overexpression of IN would reduce transactivation of the Hoxa9 promoter induced by LEDGF-menin-MLL-AF9. Indeed, we observed a dose-dependent reduction of Hoxa9 transactivation activity induced by HIV-IN expression (Figure 3C). Therefore, IN sequesters LEDGF from the menin-MLL fusion complex and reduces transactivation activity of the MLL-AF9 fusion protein. Intriguingly, these results suggest that overexpression of HIV-IN may reverse the leukemogenic activity of MLL fusion proteins in leukemia.
Expression of IN and MLL110-160 impairs leukemogenic transformation
To validate whether expression of HIV-IN or an MLL fragment impairs oncogenic transformation by MLL-AF9, we cotransduced BMCs with MLL-AF9 and IN or MLL110-160. Expression of IN resulted in a significant growth inhibition of MLL-AF9–transformed cells, reduction in Hoxa9 expression, induction of differentiation as evident from cell morphology changes, and a decrease in c-Kit expression quantified by flow cytometry (Figure 3D-I; supplemental Figure 5). Expression of IN also resulted in strong reduction in colony formation of MLL-AF9–transformed BMCs. These data clearly demonstrate that IN impairs the MLL-AF9 fusion protein–mediated oncogenic transformation. We also tested whether expression of a short fragment of MLL encompassing the IBD binding motifs (residues 110-160) affects the activity of MLL-AF9. Indeed, the MLL110-160 fragment significantly reduced Hoxa9 expression, enhanced differentiation, and reduced colony formation of MLL-AF9–transformed cells suggesting a dominant negative function (Figure 3D-I; supplemental Figure 5). On the contrary, neither the F129A nor the FLAA mutant inhibited transformation mediated by MLL-AF9 (Figure 3D-I; supplemental Figure 5), implying that the dominant negative effect of MLL110-160 is specific.
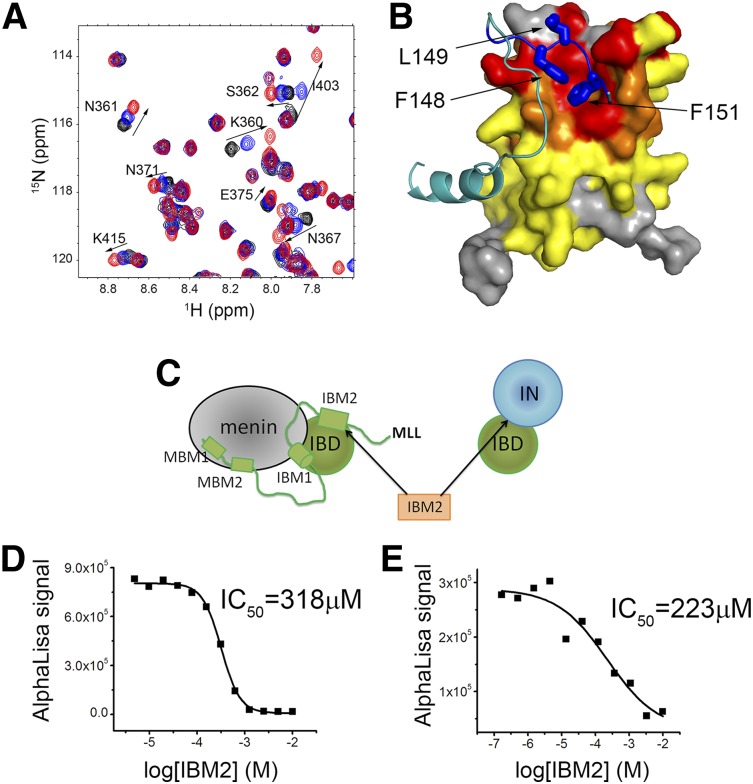
IBM2 peptide disrupts menin-MLL-IBD and IN-IBD complexes
Targeting the interaction of LEDGF with the menin-MLL may constitute an attractive approach to reverse the oncogenic activity of MLL fusion proteins in leukemia.8 Our structural studies revealed that IBM2 of MLL interacts with a concave hydrophobic site on IBD, which may represent a “druggable” site to develop small-molecule or peptidomimetic inhibitors. To validate this hypothesis, we assessed whether a short peptide corresponding to IBM2 can bind to the IBD. Indeed, an 8-amino-acid-long peptide derived from IBM2 directly binds to IBD and induces chemical shift perturbations on NMR spectra of IBD consistent with binding to the IBM2 site (Figure 4A-B). Based on the NMR titration experiment, we estimated that IBM2-derived peptide binds to IBD with ∼10-fold reduced affinity as compared with MLL1-160.
Figure 4.
IBM2 peptide binds to IBD and disrupts menin-MLL-IBD and IN-IBD interactions. (A) HSQC spectrum of 70 µM IBD (black) titrated with 280 μM (blue) and 1 mM (red) IBM2 peptide. (B) Mapping of chemical shift perturbations onto the structure of MLL-IBD complex upon binding of IBM2 peptide. Color coding represents magnitude of chemical shift perturbations: Δσ < 0.025 ppm (yellow), 0.025 < Δσ < 0.08 ppm (orange), and Δσ > 0.08 ppm (red). The MLL fragment is shown in cyan, and the key residues in IBM2 motif are blue. (C) Schematics rationalizing the use of IBM2 peptide as competitor of menin-MLL-IBD and IN-IBD. The IC50 values for competition experiments using IBM2 with menin-MLL-IBD and IN-IBD are shown, respectively, in panels D and E.
As shown previously, the IBM2 site on IBD represents a binding site for both MLL and IN. Therefore, IBM2-derived peptide should disrupt the IBD-menin-MLL and IBD-IN interactions (Figure 4C). Indeed, using the AlphaLISA assays, we found that this peptide inhibits these 2 complexes with moderate affinity (IC50 = 318 μM and IC50 = 223 μM, respectively) (Figure 4D-E). Although modest in vitro activity of the IBM2-derived peptide is insufficient to demonstrate cellular activity, it provides an important proof of concept that a short peptide can directly bind to the IBD and inhibit its interaction with both MLL-menin and IN. This finding paves the way toward development of more potent peptidomimetics or small-molecule inhibitors of the protein-protein interactions between IBD and MLL as a future potential therapeutic method for both MLL leukemia and HIV infection.
Discussion
In this work, we determined the structure of the MLL-IBD complex and found a new motif within MLL, labeled here as IBM2, that is involved in binding to the IBD of LEDGF. The newly identified IBM2 motif is necessary for the formation of a high-affinity complex between LEDGF and menin-MLL-AF9. The mutation of IBM2 within MLL-AF9 disrupts its interaction with LEDGF and abolishes the leukemogenic activity of MLL-AF9. Interestingly, the IBM2 binding site (but not IBM1) on the IBD overlaps with the binding site for HIV-IN. We have further demonstrated that binding of IBD to IN and menin-MLL is mutually exclusive. A short peptide corresponding to the MLL IBM2 fragment directly binds to the IBD and disrupts both the IBD-menin-MLL and IBD-IN complexes (Figure 4). Altogether, these experiments validate the IBM2 site on IBD as an attractive site to develop therapeutic agents for MLL leukemias as well as to block the HIV-IN activity critical to virus replication.
Targeting LEDGF-menin-MLL interaction represents an attractive approach for development of novel therapeutics for MLL leukemias.8,28 We have recently developed small molecules that bind to menin and inhibit the menin-MLL interaction and demonstrate potent and selective activity in MLL leukemia cells.29,30 Such an approach, however, may have potentially undesired consequences as menin functions as a tumor suppressor in endocrine tissues.31 Therefore, inhibitors of the activity of MLL fusions that would not affect menin function are highly desired. In this work, we found a novel MLL motif, IBM2, which interacts with IBD and is as essential for leukemogenic activity of MLL-AF9. We also demonstrated that a short peptide derived from MLL and corresponding to IBM2 motif is sufficient to bind to IBD and disrupt the IBD-menin-MLL complex. Analysis of the MLL-IBD structure shows that IBM2 binds in an extended conformation and the side chains of F148 and F151 form contacts with the concave hydrophobic site on the IBD, strongly indicating that it represents a “druggable” site. Therefore, it is likely that more potent ligands, such as small molecules or peptidomimetics, can be developed as a potential therapeutic approach for MLL leukemias. The structure of the MLL-IBD complex reported here will significantly facilitate development of such inhibitors.
Interestingly, the structure of MLL-IBD revealed that the IBD utilizes the same site to bind MLL as well as IN. This finding has 2 key implications. First, because of high-affinity binding, IN can sequester LEDGF from the menin-MLL complex. We demonstrated that expression of the IN significantly reduces transformation activity of MLL-AF9 (Figure 3) demonstrating that HIV-IN acts as a cellular inhibitor suppressing activity of MLL fusion proteins. Second, potent small-molecule or peptidomimetic inhibitors that bind to the IBM2 site will disrupt the interaction of LEDGF with IN and therefore should inhibit the replication of HIV. Development of IN-LEDGF interaction inhibitors has been recognized as an attractive target for new antiviral drugs, and multiple studies have identified small-molecule inhibitors, all of which bind to IN.13 However, ligands binding to the LEDGF IBD are more desired to avoid resistance resulting from mutation of the viral IN.32 Recently, peptides derived from LEDGF have been reported to disrupt MLL-LEDGF interaction and delay development of leukemia in an animal model of MLL-AF9.28 However, no potent small-molecule inhibitors directly binding to the IBD have been identified to date, and the structure of MLL-IBD reported here may significantly facilitate such efforts.
In summary, LEDGF plays a dual role as an oncogenic cofactor of MLL fusion proteins and also as a binding partner of HIV-IN required for viral infection. The finding that the same site on IBD is involved in binding of MLL and IN paves the way toward development of novel therapeutic agents with dual applications for both MLL leukemias and HIV.
Acknowledgments
This work was supported by research scholar grants from the American Cancer Society (RSG-11-082-01-DMC) (T.C.) (RSG-13-130-01-CDD) (J.G.) and grants from the National Institutes of Health, National Cancer Institute (R01CA160467) (J.G.) (R01CA151425) (J.L.H.).
Footnotes
The data reported in this article have been deposited in the Protein Data Bank database (accession number 2MTN).
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: M.J.M. purified proteins; M.J.M. and T.C. performed NMR experiments and determined the structure; T.P., S.H., and A.G.M. performed cellular assays; J.P. and A.Y. performed biochemical experiments; H.M. performed the in vivo experiment; J.L.H., A.G.M., J.G., and T.C. planned the experiments and wrote the manuscript with input from all authors.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Tomasz Cierpicki, University of Michigan, Department of Pathology, 1150 West Medical Center Dr, MSRB1, Room 4510, Ann Arbor, MI 48109; e-mail: tomaszc@umich.edu.
References
- 1.Dietz F, Franken S, Yoshida K, Nakamura H, Kappler J, Gieselmann V. The family of hepatoma-derived growth factor proteins: characterization of a new member HRP-4 and classification of its subfamilies. Biochem J. 2002;366(pt 2):491–500. doi: 10.1042/BJ20011811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ge H, Si Y, Roeder RG. Isolation of cDNAs encoding novel transcription coactivators p52 and p75 reveals an alternate regulatory mechanism of transcriptional activation. EMBO J. 1998;17(22):6723–6729. doi: 10.1093/emboj/17.22.6723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Singh DP, Fatma N, Kimura A, Chylack LT, Jr, Shinohara T. LEDGF binds to heat shock and stress-related element to activate the expression of stress-related genes. Biochem Biophys Res Commun. 2001;283(4):943–955. doi: 10.1006/bbrc.2001.4887. [DOI] [PubMed] [Google Scholar]
- 4.Singh DP, Kimura A, Chylack LT, Jr, Shinohara T. Lens epithelium-derived growth factor (LEDGF/p75) and p52 are derived from a single gene by alternative splicing. Gene. 2000;242(1-2):265–273. doi: 10.1016/s0378-1119(99)00506-5. [DOI] [PubMed] [Google Scholar]
- 5.Hussey DJ, Moore S, Nicola M, Dobrovic A. Fusion of the NUP98 gene with the LEDGF/p52 gene defines a recurrent acute myeloid leukemia translocation. BMC Genet. 2001;2:20. doi: 10.1186/1471-2156-2-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morerio C, Acquila M, Rosanda C, et al. t(9;11)(p22;p15) with NUP98-LEDGF fusion gene in pediatric acute myeloid leukemia. Leuk Res. 2005;29(4):467–470. doi: 10.1016/j.leukres.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 7.Huang TS, Myklebust LM, Kjarland E, et al. LEDGF/p75 has increased expression in blasts from chemotherapy-resistant human acute myelogenic leukemia patients and protects leukemia cells from apoptosis in vitro. Mol Cancer. 2007;6:31. doi: 10.1186/1476-4598-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yokoyama A, Cleary ML. Menin critically links MLL proteins with LEDGF on cancer-associated target genes. Cancer Cell. 2008;14(1):36–46. doi: 10.1016/j.ccr.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Llano M, Saenz DT, Meehan A, et al. An essential role for LEDGF/p75 in HIV integration. Science. 2006;314(5798):461–464. doi: 10.1126/science.1132319. [DOI] [PubMed] [Google Scholar]
- 10.Maertens G, Cherepanov P, Pluymers W, et al. LEDGF/p75 is essential for nuclear and chromosomal targeting of HIV-1 integrase in human cells. J Biol Chem. 2003;278(35):33528–33539. doi: 10.1074/jbc.M303594200. [DOI] [PubMed] [Google Scholar]
- 11.Cherepanov P, Devroe E, Silver PA, Engelman A. Identification of an evolutionarily conserved domain in human lens epithelium-derived growth factor/transcriptional co-activator p75 (LEDGF/p75) that binds HIV-1 integrase. J Biol Chem. 2004;279(47):48883–48892. doi: 10.1074/jbc.M406307200. [DOI] [PubMed] [Google Scholar]
- 12.De Rijck J, Vandekerckhove L, Gijsbers R, et al. Overexpression of the lens epithelium-derived growth factor/p75 integrase binding domain inhibits human immunodeficiency virus replication. J Virol. 2006;80(23):11498–11509. doi: 10.1128/JVI.00801-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Christ F, Debyser Z. The LEDGF/p75 integrase interaction, a novel target for anti-HIV therapy. Virology. 2013;435(1):102–109. doi: 10.1016/j.virol.2012.09.033. [DOI] [PubMed] [Google Scholar]
- 14.Christ F, Voet A, Marchand A, et al. Rational design of small-molecule inhibitors of the LEDGF/p75-integrase interaction and HIV replication. Nat Chem Biol. 2010;6(6):442–448. doi: 10.1038/nchembio.370. [DOI] [PubMed] [Google Scholar]
- 15.Christ F, Shaw S, Demeulemeester J, et al. Small-molecule inhibitors of the LEDGF/p75 binding site of integrase block HIV replication and modulate integrase multimerization. Antimicrob Agents Chemother. 2012;56(8):4365–4374. doi: 10.1128/AAC.00717-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang J, Gurung B, Wan B, et al. The same pocket in menin binds both MLL and JUND but has opposite effects on transcription. Nature. 2012;482(7386):542–546. doi: 10.1038/nature10806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muntean AG, Tan J, Sitwala K, et al. The PAF complex synergizes with MLL fusion proteins at HOX loci to promote leukemogenesis. Cancer Cell. 2010;17(6):609–621. doi: 10.1016/j.ccr.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grembecka J, Belcher AM, Hartley T, Cierpicki T. Molecular basis of the mixed lineage leukemia-menin interaction: implications for targeting mixed lineage leukemias. J Biol Chem. 2010;285(52):40690–40698. doi: 10.1074/jbc.M110.172783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tugarinov V, Kay LE. Quantitative NMR studies of high molecular weight proteins: application to domain orientation and ligand binding in the 723 residue enzyme malate synthase G. J Mol Biol. 2003;327(5):1121–1133. doi: 10.1016/s0022-2836(03)00238-9. [DOI] [PubMed] [Google Scholar]
- 20.Güntert P. Automated NMR structure calculation with CYANA. Methods Mol Biol. 2004;278:353–378. doi: 10.1385/1-59259-809-9:353. [DOI] [PubMed] [Google Scholar]
- 21.Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J Biomol NMR. 2009;44(4):213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang J, Muntean AG, Hess JL. ECSASB2 mediates MLL degradation during hematopoietic differentiation. Blood. 2012;119(5):1151–1161. doi: 10.1182/blood-2011-06-362079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu Y, Chen W, Gaudet J, et al. Structural basis for recognition of SMRT/N-CoR by the MYND domain and its contribution to AML1/ETO’s activity. Cancer Cell. 2007;11(6):483–497. doi: 10.1016/j.ccr.2007.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leach BI, Kuntimaddi A, Schmidt CR, Cierpicki T, Johnson SA, Bushweller JH. Leukemia fusion target AF9 is an intrinsically disordered transcriptional regulator that recruits multiple partners via coupled folding and binding. Structure. 2013;21(1):176–183. doi: 10.1016/j.str.2012.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yokoyama A, Somervaille TC, Smith KS, Rozenblatt-Rosen O, Meyerson M, Cleary ML. The menin tumor suppressor protein is an essential oncogenic cofactor for MLL-associated leukemogenesis. Cell. 2005;123(2):207–218. doi: 10.1016/j.cell.2005.09.025. [DOI] [PubMed] [Google Scholar]
- 26.Cherepanov P, Ambrosio AL, Rahman S, Ellenberger T, Engelman A. Structural basis for the recognition between HIV-1 integrase and transcriptional coactivator p75. Proc Natl Acad Sci USA. 2005;102(48):17308–17313. doi: 10.1073/pnas.0506924102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vandegraaff N, Devroe E, Turlure F, Silver PA, Engelman A. Biochemical and genetic analyses of integrase-interacting proteins lens epithelium-derived growth factor (LEDGF)/p75 and hepatoma-derived growth factor related protein 2 (HRP2) in preintegration complex function and HIV-1 replication. Virology. 2006;346(2):415–426. doi: 10.1016/j.virol.2005.11.022. [DOI] [PubMed] [Google Scholar]
- 28.Méreau H, De Rijck J, Cermáková K, et al. Impairing MLL-fusion gene-mediated transformation by dissecting critical interactions with the lens epithelium-derived growth factor (LEDGF/p75). Leukemia. 2013;27(6):1245–1253. doi: 10.1038/leu.2013.10. [DOI] [PubMed] [Google Scholar]
- 29.Grembecka J, He S, Shi A, et al. Menin-MLL inhibitors reverse oncogenic activity of MLL fusion proteins in leukemia. Nat Chem Biol. 2012;8(3):277–284. doi: 10.1038/nchembio.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shi A, Murai MJ, He S, et al. Structural insights into inhibition of the bivalent menin-MLL interaction by small molecules in leukemia. Blood. 2012;120(23):4461–4469. doi: 10.1182/blood-2012-05-429274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marx SJ. Molecular genetics of multiple endocrine neoplasia types 1 and 2. Nat Rev Cancer. 2005;5(5):367–375. doi: 10.1038/nrc1610. [DOI] [PubMed] [Google Scholar]
- 32.Hombrouck A, De Rijck J, Hendrix J, et al. Virus evolution reveals an exclusive role for LEDGF/p75 in chromosomal tethering of HIV. PLoS Pathog. 2007;3(3):e47. doi: 10.1371/journal.ppat.0030047. [DOI] [PMC free article] [PubMed] [Google Scholar]